Abstract
In this Primer, Vance and Lee discuss two classes of membrane fusion-catalyzing proteins — i.e. fusogens — that are conserved between viruses and eukaryotes.
Main Text
Since their discovery, viruses have been associated with disease. Viruses must hijack a cell’s replication machinery to propagate, leading to a myriad of woes for the unfortunate organism chosen as a host. However, virus–human interactions are not always negative. Though the hundreds of viral species that thrive as human pathogens draw the lion’s share of our attention, growing evidence in the field of membrane fusion suggests that eukaryote–virus relationships are not always defined by conflict. This Primer will focus on two classes of fusogens (i.e. proteins that catalyze membrane fusion) that are shared between viruses and eukaryotes. The conservation of these fusogens in remarkably different organisms denotes a relationship between eukaryotes and viruses that is both ancient in origin and current in its impact.
Membrane fusion in viruses
The fusion of two membrane bilayers is critical to eukaryotic life, occurring in gamete fusion, muscle differentiation, ocular lens formation, neurotransmitter release, and placenta formation. Ironically, it is through a similar process that enveloped viruses gain access to eukaryotic cells, merging membranes to release viral genetic material within the cytoplasm. In all cases, membrane fusion is exceptionally unfavorable without the assistance of specialized proteins to overcome the repulsive forces between membranes. While few of these proteins have been discovered in eukaryotes, there are many known viral fusion proteins, also called viral glycoproteins or fusogens, that facilitate cell entry.
Viruses have evolved at least four classes of viral fusion protein with remarkable differences in tertiary structure and multimerization. Viral fusion proteins from class I form coiled-coil trimers (e.g. influenza, coronavirus, HIV, Ebola); class II proteins transition from dimers to trimers during fusion, producing an elongated ectodomain heavily composed of β sheets that settles into a hairpin trimer after fusion (e.g. Dengue fever virus, West Nile virus, Zika virus, tick-borne encephalitis virus); and class III proteins combine elements from the former two classes, taking on a post-fusion conformation that contains both a coiled-coil trimerization region similar to class I, and an elongated trimer of hairpins as in class II (e.g. vesicular stomatitis virus, herpes simplex virus 1, rabies virus).
Class I, II and III fusion proteins are produced by enveloped viruses to facilitate virus–host membrane fusion. In contrast, class IV fusion proteins — also called fusion-associated small transmembrane (FAST) proteins — are cell–cell fusogens made by reoviruses to merge multiple host cells into a syncytium, i.e. a multinucleate cell. This strategy helps the reovirus spread locally between cells and induces apoptotic pathways to promote eventual lysis and release of viral progeny. FAST proteins are much smaller than all other viral fusogens, producing ectodomains of around 20–40 amino acids that are structurally diverse between reovirus species, including polyproline helices and cysteine loops.
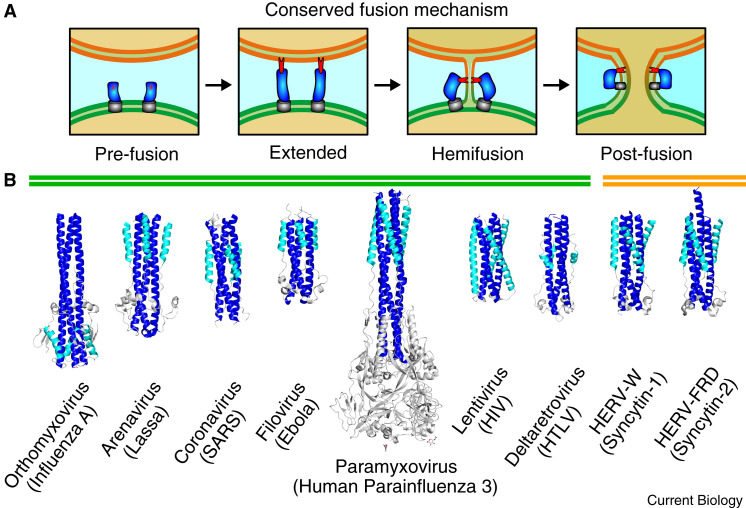
A cursory glance at the different classes of viral fusogen and their diverse structures would seem to indicate similarly diverse modes of fusion. While that is true for class IV viral glycoproteins, the enveloped viral fusogens (i.e. classes I, II and III) remarkably catalyze membrane fusion through a conserved mechanism (Figure 1 A). Briefly, viral fusion proteins are presented on the surface of the virion in a metastable state with their hydrophobic ‘fusion peptide’ (class I) or ‘fusion loop’ (all class II and III, and certain class I) safely buried in the fusogen. An external trigger, which can be receptor binding and/or a change in pH, initiates a conformational change to release the fusion peptide/loop so that it can embed in the host membrane. The extended intermediate forms a physical tether between the virus and host. The collapse of the short-lived extended intermediate forms a hemifusion state, where the two outer lipid layers fuse. Finally, the inner lipid leaflets merge and the fusion pore expands, as the fusogen forms the stable post-fusion conformation. Without these proteins and the large conformational changes they undergo, enveloped viruses would be unable to infect their hosts, making fusogens an obvious target for antiviral therapeutics.
Figure 1.
Conserved mechanism for membrane fusion and the class I fusogen superfamily
(A) Viral fusion proteins (blue) are displayed in a pre-fusion state, with their carboxy-terminal transmembrane region (grey) attached to the virion membrane (green), and their hydrophobic fusion peptide or fusion loop (red) safely buried. The protein then goes through a series of conformational changes, extending to embed its fusion peptide/loop in the host cell membrane (orange) before bringing the carboxy-terminal transmembrane region and fusion peptide/loop together in a post-fusion state. (B) Structures of the viral class I fusogens from major human pathogens are presented, including influenza A (PDB: 1HTM), Lassa virus (PDB: 5OMI), SARS-CoV (PDB: 1WNC), Ebola (PDB: 2EBO), human parainfluenza virus-3 (PDB: 1ZTM), HIV-1 (PDB: 3WFV), human T-lymphotropic virus-1 (PDB: 1MG1), as well as the two solved structures for human syncytin-1 (PDB: 6RX1) and syncytin-2 (PDB: 6RX3). The amino-terminal helix (heptad repeat-1) is coloured blue, the carboxy-terminal helix (heptad repeat-2) is coloured cyan, and the connecting chain reversal region (if present) is coloured gray. The structures shown are in their post-fusion trimeric states.
However, a beneficial aspect of eukaryote–virus interactions has begun to surface with recent studies into eukaryotic membrane fusion proteins and their striking similarity to their viral counterparts. While not entirely understood, a long-standing and ongoing link between viral and eukaryotic fusion proteins is now accepted, with researchers even grouping structurally similar fusogens from viruses and eukaryotes under shared superfamilies. To understand how this new chapter in viral identity has occurred — and to better define these superfamilies — it is important to start at the beginning: birth.
Class I fusogen superfamily: the discovery of syncytins
The class I viral fusion proteins are at the forefront of some of the deadliest viral infections of the modern era, including the influenza viruses, coronaviruses, Ebola virus, Lassa virus and HIV. Yet, their ubiquitous presence in host endogenous retroviruses (ERVs) means that they are also a part of the most intimate examples of virus–host coexistence. ERVs are the result of former retroviral infections of germline cells that attain a passive longevity via integration into a host’s genome. As such, they no longer depend on horizontal spread between hosts for their continued existence, instead being passed vertically through generations. Most ERVs have numerous copies throughout the genome, with ERV elements making up approximately 8% of the human genome. In comparison, only ∼1% of the total human genome is made up of protein-encoding genes.
Initially, ERV sequences retain their proviral structure with gag, pol, and env genes. As part of the endogenization process, most ERV genes undergo deleterious mutations over time, thereby nullifying any potential function their translated protein might have. However, this is not always the case. Some ERV transcripts are actively translated by their mammalian hosts, producing ‘domesticated’ viral proteins. For instance, the Gag-derived Fv1 protein promotes antiviral activity against exogenous viruses in mice, whilst another Gag-like protein (Arc) has been implicated in proper neuronal communication in animals.
Perhaps the most intriguing example of human ERV (HERV) protein functionalization came when two groups discovered that an env gene from HERV-W produced a still-functional class I fusion protein capable of merging multiple cells into a syncytium. Such syncytia are found at the placental interface between the embryo and the mother; as such, the preferential expression of HERV-W in embryo cells destined for the placental interface made this discovery all the more intriguing. This virally derived fusion protein was dubbed a syncytin, later termed syncytin-1 after the discovery of a second such protein from HERV-FRD, aptly named syncytin-2. Knockouts of similar proteins in mice revealed that syncytins are vital for the formation of the placenta, confirming that one of the fundamental aspects of mammalian birth relies on a formerly viral protein.
The fusion domains for syncytin-1 and 2 have been structurally determined (Figure 1B), showing the expected six-helix bundles (three protomers, two helices each) that define the post-fusion state of class I viral fusion proteins. Given their structural and evolutionary connections, syncytins and class I viral fusogens can be grouped together into the class I fusogen superfamily.
Class I fusogen superfamily: are viruses the founders of placental birth?
The importance of the syncytin discovery became apparent as syncytins from a wide range of organisms outside of primates were discovered. While there are many partially or fully conserved ERV fusogens throughout the tree of life, the requirements for a true syncytin have been defined as: specific expression in the placenta; definitive proof of cell–cell fusion ability; and conservation for at least 10 million years of evolution. Syncytins that have met these requirements have been found in all species in which they have been sought, including mice, rabbits, ruminants, carnivores, the shrew-like tenrecs, marsupials, and even a placental lizard.
The widespread nature of syncytins would suggest a viral insertion into an early ancestor of placenta-possessing animals that was then conserved throughout different speciation events due to its lynchpin role in birth. Surprisingly, analysis of the distinct syncytin sequences revealed that the different species attained these fusogens from separate viral insertions, contradicting a conserved ancestral insertion. The prevailing theory that reconciles the necessity of syncytins with their multiple insertions presumes that an early syncytin domestication did occur in placental ancestors; this insertion is thought to be responsible for the switch from egg-bearing to live-bearing reproduction strategies. However, superior fusion proteins were likely picked up from additional insertions over time and were subsequently conserved at the expense of the ancient syncytins, akin to a baton pass between successive syncytins. Indeed, examples of such obsolete syncytins undergoing evolutionary decay have been observed, including the non-fusogenic ERV3 in higher primates, and the pan-Mars-Env2 in marsupials.
Strong (albeit circumstantial) evidence suggests that the different syncytin insertions are responsible for the interspecies differences in placenta structure, specifically in the extent of contact between embryo cells and maternal blood. For example, the rise of the specific ruminant placenta that features the unique fusion of embryo cells with maternal cells correlates well with the capturing of the specific ruminant syncytin. Additionally, the placental structure found in hyenas shows a much higher level of embryo–mother contact relative to all other studied carnivores, possibly because of an additional, hyena-specific syncytin-like protein (Hyena-Env2). How syncytins impact placenta structure and which aspects of their molecular structures are responsible for the differences remain a mystery, due in part to the lack of structure–function data on non-human syncytins.
Class II fusogen superfamily: discovery of EFF-1 and HAP2
The paradigm-shifting discovery of syncytins has brought about many questions, not least of which is: are other eukaryotic fusion proteins derived from viruses? Sadly, the fusogens responsible for many non-placental processes remain a mystery in vertebrates (as does their relationship, if any, to viral fusion proteins). To extend our understanding of eukaryotic fusogens, researchers turned to ‘simpler’ eukaryotes, including green algae, plants, and nematodes.
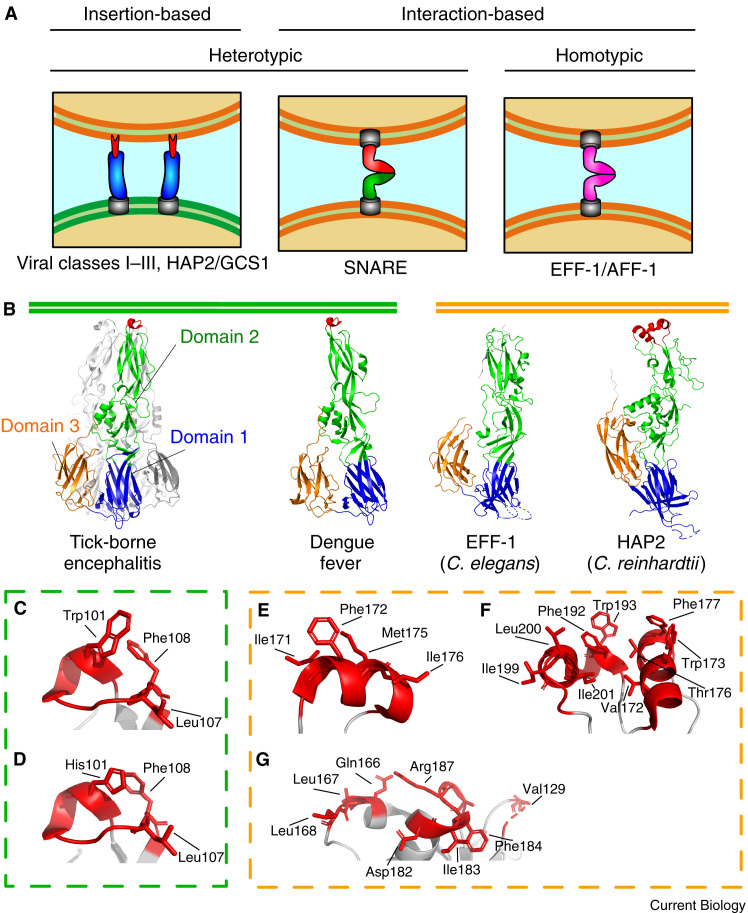
For decades, two proteins from Caenorhabditis elegans have been known to be crucial in the myriad of fusion activities that form nematode organs — the paralogous proteins epithelial fusion failure 1 (EFF-1) and anchor cell fusion failure 1 (AFF-1). In vitro experiments demonstrated that EFF-1 and AFF-1 — when transfected into naturally non-fusogenic cells — were able to induce the formation of syncytia, confirming them as definitive fusogens. Interestingly, this was only true when the proteins were expressed on the surfaces of each of the fusing cells, implying a fusion strategy that requires interactions between the same proteins expressed on opposite cells (homotypic). Said strategy is more like SNARE-mediated endosomal fusion than viral fusion (Figure 2 A).
Figure 2.
Alterations to the membrane fusion strategy, and the class II fusogen superfamily
(A) Snapshot representations of the viral membrane fusion strategy directly before the hemifusion intermediate, and two altered versions from eukaryotes. The strategies are separated into insertion-based (a fusion protein inserts into the opposing membrane) vs. interaction-based (proteins from both cells form complexes that bring the membranes together), and heterotypic (the cells are presenting different fusion proteins, or only one cell has a fusion protein) vs. homotypic (the cells present the same fusion protein to each other for fusion). (B) The tick-borne encephalitis virus post-fusion trimer (PDB: 1URZ), with a single protomer coloured according to the different domains. Domain 1 is a split Ig-like β sandwich (blue), domain 2 is an elongated ectodomain (green), and domain 3 is another β sandwich (orange). The fusion loop, if present, is coloured red. Similarly coloured representations of protomers from dengue virus type 1 (PDB: 4GT0), C. elegans EFF-1 (PDB: 4OJC), and CrHAP2 (PDB: 6DBS) are also shown. (C–G) Magnified images of the fusion loops for (C) tick-borne encephalitis, (D) dengue fever, (E) AtHAP2, (F) CrHAP2, and (G) TcHAP2 (PDB: 5OW4) are displayed in red. Residues capable of contributing to the hydrophobic surface are labeled.
Around the same time, a strong lead in the intensive search for the protein responsible for gamete fusion came from studies in both Arabidopsis thaliana and Lilium longiflorum, where genes encoding homologous transmembrane proteins — named hapless2 (HAP2) and generative cell specific 1 (GCS1), respectively — were found to be crucial for the production of fertile gametes. Similar proteins have been found throughout eukaryotes, with examples from the green alga Chlamyodomonas reinhardtii and the Plasmodium parasites localizing to male-equivalent gametes for some unknown role in fertilization. Even ciliate species that do not conform to the male–female paradigm possess HAP2 in some vital role. A lack of significant sequence identity to viral fusogens (and EFF-1/AFF-1) seemed to negate any obvious connection.
Therefore, it was quite the surprise when the structures of both EFF-1 from C. elegans and HAP2 from C. reinhardtii (CrHAP2) were determined, revealing striking resemblances to class II viral fusion proteins (Figure 2B). Despite the poor sequence similarity, the conserved structures show that EFF-1, HAP2, and the class II viral fusion proteins belong to another protein superfamily, dubbed the class II fusogens (also termed fusexins by Valansi et al. (2017)).
While all members of this superfamily are involved in membrane fusion, functional differences have been noted. Class II viral fusion proteins are proposed to undergo a change in multimerization that facilitates fusion, transitioning from a pre-fusion dimer to a post-fusion trimer. The pre-fusion multimerization state for HAP2 is not known, but EFF-1 has been shown to use a monomer-to-trimer transition instead, likely deviating to accommodate its homotypic strategy. EFF-1 also lacks a fusion loop, once again in line with a homotypic strategy that does not require insertion into the opposing membrane (Figure 2A).
In contrast, the structures of HAP2 from C. reinhardtii (CrHAP2), A. thaliana (AtHAP2) and Trypanosoma cruzi (TcHAP2) all possess fusion loops, presenting a series of hydrophobic residues outwards for membrane insertion. All these fusion loops show extensive variations, both between each other and in comparison to viral examples. Both dengue virus and tick-borne encephalitis virus fusion loops have a single loop containing a small helix that presents two aromatic residues (Trp101 or His101, and Phe108) and an aliphatic residue (Leu107) (Figure 2C,D). The AtHAP2 fusion loop also contains a single α helix, although it is longer and presents a different group of aromatic and aliphatic residues (Figure 2E). CrHAP2’s fusion loop is much more extensive, containing two α helices on either side of a smaller 310 helix; the multiple helices allow for many more hydrophobic residues to be presented to the membrane (Figure 2F). Contrastingly, the tip of the ectodomain from T. cruzi takes on a shallow, flat surface that — along with more traditional hydrophobic residues — incorporates larger charged/polar residues (i.e. arginine and glutamine) by stretching the residues horizontal to the membrane, thereby presenting the aliphatic portions of their side chains (Figure 2G). It has been theorized that the species-specific lipid complement in gamete membranes could be the driving evolutionary pressure behind the variation in these fusion loops, making the different structures a form of gametic isolation that promotes speciation.
Class II fusogen superfamily: the distribution of known eukaryotic cell–cell fusion proteins
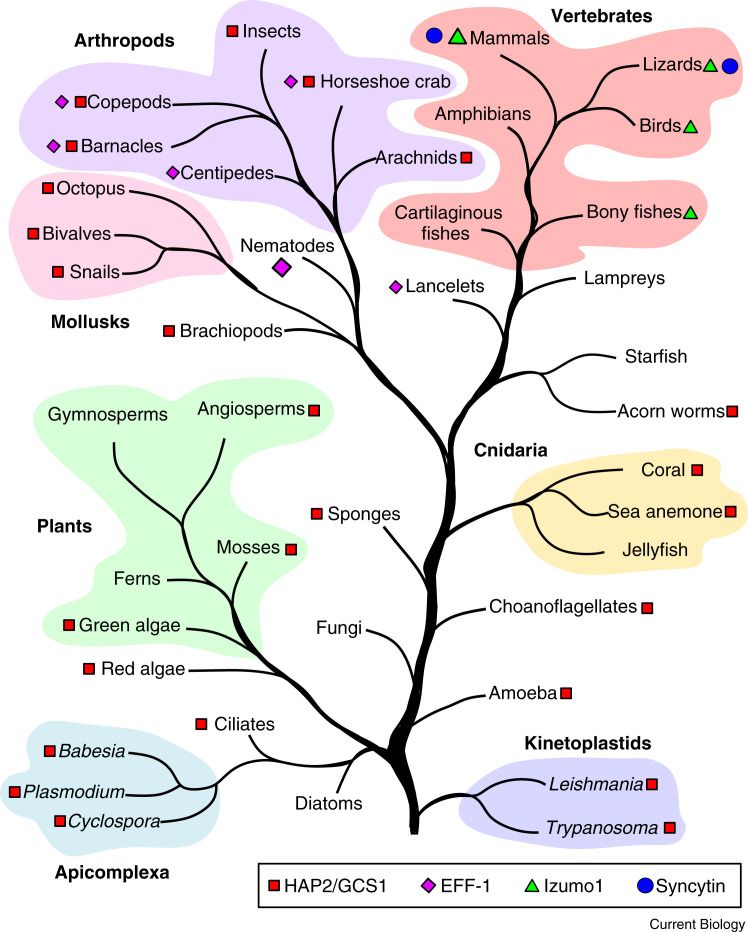
Analysis of sequence similarity was not effective at predicting links between viral and eukaryotic fusion proteins. However, sequence alignment searches can provide insight into the spread of these now-known fusion proteins in eukaryotes. For instance, EFF-1-like sequences are mostly localized to nematodes, with a few examples found in various arthropods (Figure 3 ). Lancelets also have an EFF-1-like protein, the fusion activity for which has been verified. The presence of EFF-1 in members of the superphyla Protostomia (includes arthropods, molluscs, and nematodes) and Deuterostomia (includes vertebrates, hemichordates and lancelets) means that the fusogen likely emerged before the divergence of these massive groups (between 550 and 900 million years ago). Whether this emergence was the result of a viral insertion or a divergence from a duplicated HAP2 gene remains a mystery.
Figure 3.
Distribution of fusion-related proteins throughout the tree of life
A graphical representation of the tree of life, simplified to include organisms predicted to harbour a fusion-related protein. A PSI-BLASTp search of the NCBI database was performed, using HAP2 (red square) from C. reinhardtii, A. thaliana, Centruroides sculpturatus, Nematostella vectensis and Trypanosoma cruzi, EFF-1 (purple diamond) from C. elegans and Branchiostoma floridae, and human Izumo1 (green triangle) as queries. Several major groupings are colour coded. Distances between branches are not drawn to a timescale.
In keeping with HAP2’s role in gamete fusion, sequences are found throughout eukaryotes, including: single-celled ciliates, kinetoplastids and apicomplexan parasites; a large variety of plants (algae, mosses, liverworts, and angiosperms); sea anemone and coral; cephalopods and snails of the Mollusca phylum; as well as many types of arthropods (insects, arachnids, and crustaceans) (Figure 3). The presence of HAP2 throughout eukaryotes implies an early emergence, perhaps sparking the adoption of sex as a reproductive strategy. Interestingly, no evidence of HAP2 orthologs has been found in fungi, diatoms, gymnosperm plants, or vertebrates — all groups that can participate in sexual reproduction. While certain absences may be explained by poor representation of sequenced genomes in the NCBI database, the complete absence in highly studied groups, like mammals, is especially peculiar. Indeed, the closest ancestor to humans to possess a HAP2 gene is the acorn worm — hardly an intimate relation (Figure 3). Though frustrating for researchers in the field, the absence of HAP2 in humans may make the HAP2 of human pathogens like Plasmodium falciparum, Cyclospora cayetanensis, T. cruzi, and Leishmania panamensis an intriguing target for the development of anti-parasitic inhibitors.
So, the fusogen behind human or mammalian gamete fusion remains a mystery. It appears likely that an early vertebrate ancestor picked up a new fusogen that replaced HAP2 (similar to the swapping of syncytins) and developed an undiscovered strategy for gamete fusion. Previously, at least three membrane-bound proteins — sperm Izumo1, egg Juno, and egg CD9 — were shown to be essential in gamete recognition and/or the fusion process. Izumo1 and Juno were thought to be the bona fide sperm fusogen and egg receptor, respectively, as these proteins are conserved in all mammals and constitutive knockout of either Izumo1 or Juno results in healthy but infertile mice. Although their interaction is essential for fertilization, ectopic expression of either Izumo1 or Juno in HEK293 cells was not sufficient to produce multinucleated cells. Along with the structural data distinguishing both proteins from any known fusogen structure, it has become apparent that neither is a true fusion protein. Interestingly, Izumo1 and HAP2 appear to be mutually exclusive, the former being widespread within and limited to higher-order vertebrates (Figure 3). Perhaps the conservation of Izumo1 and its receptor Juno is indicative of a conserved sperm–egg fusion strategy — complete with an as-yet undiscovered fusogen — throughout the higher vertebrates, one that is missing from HAP2-containing eukaryotes, and the cartilaginous fishes and lampreys in between.
Perspectives
The known eukaryotic fusion proteins can be split into two families, along with their viral counterparts. While the class I fusogens include both viral and retrovirally-derived proteins, the origin of the various members of the class II fusogen superfamily is less clear. It is only through structural studies that the links that connect the class II members were revealed. Indeed, this is an example of the value structural biology has brought to the field of membrane fusion.
The fusogen superfamilies — shared by viruses and eukaryotes alike — indicate an extensive, long-running coexistence between disparate organisms. In a millennium where the effects of the microbiome (the collection of microorganisms that reside on or within the human body) on human health have become common knowledge, the viral component of the microbiome has gone relatively unacknowledged. Continued work in the field of cell–cell fusion has shown how deeply our development as a species has relied on these viruses. Whether more virally-linked fusogens will be found remains an open question, one that will reveal the true extent of this relationship — a relationship where pathogenesis is not a prerequisite and symbiosis is a possibility.
Further Reading
- Avinoam O., Fridman K., Valansi C., Abutbul I., Zeev-Ben-Mordehai T., Maurer U.E., Sapir A., Danino D., Grünewald K., White J.M. Conserved eukaryotic fusogens can fuse viral envelopes to cells. Science. 2011;332:589–592. doi: 10.1126/science.1202333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero E., Fedry J., Legrand P., Krey T., Rey F.A. Species-specific functional regions of the green alga gamete fusion protein HAP2 revealed by structural studies. Structure. 2019;27:113–124.e4. doi: 10.1016/j.str.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E., Doe B., Goulding D., Wright G.J. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A., Vernochet C., Bawa O., Harper F., Pierron G., Opolon P., Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry J., Liu Y., Péhau-Arnaudet G., Pei J., Li W., Tortorici M.A., Traincard F., Meola A., Bricogne G., Grishin N.V. The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell. 2017;168:904–915.e10. doi: 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Dong X., Pinello J., Zhang J., Lu C., Iacob R.E., Engen J.R., Snell W.J., Springer T.A. Fusion surface structure, function, and dynamics of gamete fusogen HAP2. eLife. 2018;7 doi: 10.7554/eLife.39772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk M., Cornelis G., Vernochet C., Heidmann O., Dupressoir A., Conley A., Glickman S., Heidmann T. Capture of a hyena-specific retroviral envelope gene with placental expression associated in evolution with the unique emergence among carnivorans of hemochorial placentation in Hyaenidae. J. Virol. 2019;93:e01811–18. doi: 10.1128/JVI.01811-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N., Ikawa M., Isotani A., Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- Lavialle C., Cornelis G., Dupressoir A., Esnault C., Heidmann O., Vernochet C., Heidmann T. Paleovirology of “syncytins”, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.-Y., Edouard P., Howes S. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Pérez-Vargas J., Krey T., Valansi C., Avinoam O., Haouz A., Jamin M., Raveh-Barak H., Podbilewicz B., Rey F.A. Structural basis of eukaryotic cell–cell fusion. Cell. 2014;157:407–419. doi: 10.1016/j.cell.2014.02.020. [DOI] [PubMed] [Google Scholar]
- Pinello J.F., Lai A.L., Millet J.K., Cassidy-Hanley D., Freed J.H., Clark T.G. Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr. Biol. 2017;27:651–660. doi: 10.1016/j.cub.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz B. Virus and cell fusion mechanisms. Annu. Rev. Cell Dev. Biol. 2014;30:111–139. doi: 10.1146/annurev-cellbio-101512-122422. [DOI] [PubMed] [Google Scholar]
- Renard M., Varela P.F., Letzelter C., Duquerroy S., Rey F.A., Heidmann T. Crystal structure of a pivotal domain of human syncytin-2, a 40 million years old endogenous retrovirus fusogenic envelope gene captured by primates. J. Mol. Biol. 2005;352:1029–1034. doi: 10.1016/j.jmb.2005.07.058. [DOI] [PubMed] [Google Scholar]
- Valansi C., Moi D., Leikina E., Matveev E., Graña M., Chernomordik L.V., Romero H., Aguilar P.S., Podbilewicz B. Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell Biol. 2017;216:571–581. doi: 10.1083/jcb.201610093. [DOI] [PMC free article] [PubMed] [Google Scholar]