Highlights
-
•
Implementation of SARS-CoV-2 genome testing by numerous laboratories in Austria.
-
•
Broad range of combinations of nucleic acid extraction and RT-PCR methods.
-
•
Variable analytical performance of different protocols.
-
•
Good performance of some analytical setups.
-
•
Potential for optimisation of pre-RT-PCR dilution steps.
Keywords: SARS-CoV-2, COVID-19 pandemic, Molecular test, Standardisation, Evaluation, External quality assessment
Abstract
Background
Broad and decentralised testing of SARS-CoV-2 RNA genomes is a WHO-recommended strategy to contain the SARS-CoV-2 pandemic by identifying infected cases in order to minimize onward transmission. With the need to increase the test capacities in Austria, nation-wide numerous laboratories rapidly implemented assays for molecular detection of SARS-CoV-2 based on real-time RT-PCR assays. The objective of this study was to monitor reliability of the laboratory results for SARS-CoV-2 RNA detection through an external quality assessment (EQA) scheme.
Methods
For this, the Center for Virology, Medical University of Vienna was tasked by the Federal Ministry of Social Affairs, Health, Care and Consumer Protection to perform the first Austrian EQA on SARS-CoV-2 which was organised in cooperation with the Austrian Association for Quality Assurance and Standardization of Medical and Diagnostic Tests (ÖQUASTA). Data were analysed on the basis of qualitative outcome of testing in relation to the nucleic acid (NA) extraction and detection methods used.
Results and Conclusion
A total of 52 laboratories participated, contributing results from 67 test panels comprising 42 distinct combinations of NA extraction and PCR reagents. By testing 3 positive (CT values: S1, 28.4; S2, 33.6; S3, 38.5) and 1 negative sample, no false-positive results were obtained by any of the laboratories. Otherwise, 40/67 tests (60 %) detected all positive samples correctly as positive, but 25/67 tests (37 %) did not detect the weakest positive sample (S3), and 3 % reported S2 and S3 as false-negative. Improvement in test sensitivity by focusing on NA extraction and/or PCR-based detection is recommended.
1. Introduction
Identification and rapid global spread of the novel SARS-CoV-2 coronavirus with first cases of the disease COVID-19 being detected in Europe in January 2020 [1] necessitated the development and deployment of diagnostic tests for the specific detection of SARS-CoV-2 genome sequences in human samples.
Following the online release of the first SARS-CoV-2 viral genome sequence on January 10 [2] an initial protocol for SARS-CoV-2 specific real-time reverse transcription (RT) PCR was presented under the leadership of the Institute of Virology at the Charité, Universitätsmedizin (Berlin, Germany) in cooperation with Erasmus Medical Center Rotterdam (The Netherlands) and Public Health England, London (UK) [3]. Reagents were made available through TIB Molbiol Berlin (Germany). Numerous other companies followed with similar PCR-based approaches and the World Health Organization (WHO) as well as the European Centre for Disease Prevention and Control (ECDC) provided a list of available laboratory tests for the molecular detection of SARS-CoV-2. In addition, online guidance to allow promoting the implementation of diagnostic protocols in laboratories other than expert laboratories was provided.
The initial WHO strategy recommended containment of SARS-CoV-2 by rapidly identifying cases and by tracing contact cases in order to minimize onward transmission [4]. This required broad and decentralised screening and the rapid increase in test capacities. Hence, in Austria many laboratories nation-wide began introducing SARS-CoV-2 molecular detection assays.
Reliable laboratory results are essential for rapid and accurate decision making in patient care [5]. In addition to internal quality control, participation in an External Quality Assessment (EQA) is an important tool that allows individual laboratories to have the reliability of their results monitored by an independent third party in comparison with other participating diagnostic laboratories [6].
Therefore, the Center for Virology, Medical University of Vienna was tasked by the Austrian Federal Ministry of Social Affairs, Health, Care and Consumer Protection to organise the first Austrian EQA scheme for qualitative molecular detection of SARS-CoV-2 RNA in May 2020. This EQA was performed in cooperation with the Austrian Association for Quality Assurance and Standardization of Medical and Diagnostic Tests (ÖQUASTA).
The objectives of this first EQA scheme were to assess the capability of individual laboratories to perform sensitive and reliable SARS-CoV-2 molecular diagnostics and to provide insights into the limits of test sensitivity in order to improve SARS-CoV-2 RNA detection capabilities.
2. Materials and methods
2.1. Sample preparation
The EQA test panel consisted of four samples (900 μL each), three positive and one negative, that were distributed by the Center for Virology to participant laboratories. Positive samples were three individual SARS-CoV-2 positive oro-nasopharyngeal swab samples 100-fold diluted with sterile sodium chloride solution (0.9 % NaCl). The negative sample consisted of the diluent, a 0.9 % NaCl solution. In order to guarantee stability of SARS-CoV-2 genomic RNA, all samples were stored at −20 °C and one panel was tested before shipment as follows: after thawing, samples were kept at ambient temperature for 2 days (anticipated maximum shipping time) before further storage at 4 °C for 4 days (maximal expected storage time before testing). Thereafter, samples were quantified with our in-house SARS-CoV-2 molecular detection assay. For this, nucleic acid (NA) was extracted from 200 μL input sample and eluted in 50 μL of elution buffer using the NucliSENS easyMAG platform (bioMérieux, France). For real-time RT-PCR, each 25 μL reaction mixture contained 12.5 μL of 2x reaction buffer, 0.4 μL of a 50 mM magnesium sulphate solution, 1 μL of SuperScript III RT/Platinum Taq Mix (SuperScript III One-Step RT-PCR System, Invitrogen, Germany), 0.5 μL each of LightMix® Modular SARS-CoV (COVID19) E-gene and LightMix® Modular EAV RNA Extraction Control (TIB Molbiol, Berlin, Germany) and 10 μL of eluted NA as template. Thermal cycling was performed using a Light Cycler 480 II (Roche, Vienna, Austria): reverse transcription at 55 °C for 20 min; denaturation at 94 °C for 3 min; 45 amplification cycles of 94 °C for 15 s and 58 °C for 30 s (acquisition steps); ending with a final incubation at 40 °C for 30 s.
The final cycle threshold (CT) values of the three positive samples were: 28.4 (S1), 33.6 (S2), 38.5 (S3) for the E-gene of the SARS-CoV-2 genome. The sample S4 was negative as expected.
2.2. Organisation of the EQA scheme and shipment of samples
A questionnaire for participation was sent out at the beginning of May to 82 laboratories in Austria known to have implemented molecular tests for detection of SARS-CoV-2 RNA genomes. Therein, laboratories were informed about the EQA scheme and in terms of participation the laboratories were asked to provide information about the implemented test systems including NA extraction platform and reagents for real-time RT-PCR. Samples were dispatched on May 18 by overnight-express delivery at ambient temperature conditions and participants were instructed to either immediately test or alternatively store the material at 4 °C until analysis and to perform testing in the same way as routine samples. Individual laboratories which intended to participate with more than one molecular test system received independently shipped test panels for each test system.
2.3. Data acquisition
Raw data and qualitative interpretation (positive, negative) were submitted to the EQA provider with a deadline of May 26 via a web portal, email, fax or paper. Individual results were scored as either correct or incorrect and summarised in a general report in addition to individual reports. Optionally, the participants had the possibility to report the target region(s) of the respective real-time RT-PCR assays and obtain CT or Cq values reported by all laboratories.
3. Results
3.1. Laboratory characteristics
Fifty-two laboratories participated with a total of 67 test panels. Forty-two laboratories participated with one and 10 laboratories with more than one test panel. In particular, 6/10 laboratories used two, three labs used three, and one lab used four distinct test systems. All laboratories responded in time and their results were submitted along with the raw data.
3.2. Overall SARS-CoV-2 molecular test performance
The negative sample S4 tested negative in all 67 individual panels (100 %). Otherwise, 40/67 test systems (59.7 %) detected all three positive samples as positive, 25/67 tests (37.3 %) found S3 (CT value of E-gene, 38.5) as negative, and 2/67 (3 %), found the S2 (CT value of E-gene, 33.6) and S3 as negative (Table 1 ).
Table 1.
Summary of EQA outcomes of participating laboratories.
| Total | all samples correct | S3 false-negative | S2 and S3 false-negative | |
|---|---|---|---|---|
| Test panels | 67 | 40 | 25 | 2 |
| Laboratories | 52 | 34* | 17 | 1 |
at least 1 molecular test system detected all samples correctly, when more than one test system was used in one and the same laboratory.
Analysis of data at the laboratory level showed that 34 labs (65.4 %) detected all samples correctly with at least one test system. S3 was incorrectly reported negative by 17/52 labs (32.7 %), and 1/52 labs (1.9 %) reported false-negative for samples S2 and S3.
Of the ten labs that used more than one test system, six labs reported identical (consistent) results for all systems and four labs reported contradictory results for the different test (Table 2 ).
Table 2.
Summary of EQA outcomes according to the number of different molecular test systems used.
| Number of laboratories |
|||||
|---|---|---|---|---|---|
| # of test panels per lab | Total | all samples correct | S3 false-negative | S2 and S3 false-negative | all samples correct with at least 1 test system |
| 1 | 42 | 27 | 15 | ||
| 2 | 6 | 3 | 1 | 2 | |
| 3 | 3 | 2 | 1 | ||
| 4 | 1 | 1 | |||
3.3. SARS-CoV-2 molecular test performance according to nucleic acid extraction platforms and real-time RT-PCR reagents
A total of 27 different NA extraction platforms were used including 6 fully- or semi-automated platforms with a combined NA extraction and amplification workflow (GeneXpert, Cobas 6800, BD Max, NeuMoDX, GenomEra, and AIGS), 15 semi-automatic NA extraction or pipetting platforms, and at least 5 distinct column-based NA isolation kits either manually or semi-automatically applied. The data are shown in Table 3 .
Table 3.
EQA outcomes per nucleic acid extraction methods used.
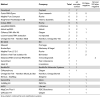 |
Grey-shaded: combined NA and amplification systems.
The most frequently used systems were GeneXpert (Cepheid), Quick-RNA/Zymo (Zymo), MagNa Pure compact (Roche), KingFisher/BioSprint 96 (Thermo Scientific), and Cobas 6800. Notably, two platforms, GeneXpert and Cobas 6800, were used by 12 distinct centers and none of these reported false-negative results (Table 3).
In terms of real-time RT-PCR reagents, 22 distinct assays were used (Table 4 ). The most frequently used reagent kits were from RealStar (Altona), ViroReal (Ingenetix), Xpert Xpress (Cepheid) and LightMix Modular (TIB Molbiol).
Table 4.
EQA outcomes per PCR reagents used.
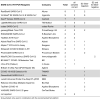 |
Grey-shaded: only used in combination with the recommended NA platform.
For the platforms GeneXpert, Cobas 6800, GenomEra, and AIGS the PCR reagents were used as recommended by the company. The remaining 18 distinct PCR assays were independently combined with 22 types of NA extraction, resulting in an overall of at least 38 distinct test systems. The outcomes are shown in Table 5 .
Table 5.
EQA test outcomes per NA method and detection assay.
| RealStar® SARS-CoV-2 | ViroReal® Kit SARS-CoV-2 & SARS-CoV | LightMix®Modular SARS-CoV | genesig®Real-Time PCR | RIDA®GENE SARS-CoV-2 | TaqMan 2019-nCoV | Abbott RealTime SARS-CoV-2 | Allplex™ 2019-nCoV Assay | EliGene® COVID19 BASIC A RT | PhoenixDx® SARS-CoV-2 | MutaPLEX® Coronavirus (SARS-CoV-2) | BD MAX™ COVID-19 Molecular Test | nCov-IP4 (Pasteur) | Brilliant III Ultra-Fast QPCR Master Mix | Luna® Universal Probe One-Step RT-qPCR | Reliance One-Step Multiplex Supermix | TaqPath COVID-19 | VIASURE SARS-CoV-2 S gene | not reported | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Quick-RNA/Zymo | 1,2,2,2 | 1 | 2,2 | ||||||||||||||||
| MagNa Pure Compact | 1 | 2 | 1,1 | 1 | 1 | ||||||||||||||
| KingFisher Flex/BioSprint 96 | 1,1,2 | 2 | 1,1 | ||||||||||||||||
| easyMAG/EMAG | 1 | 1 | 1 | ||||||||||||||||
| Abbott sp2000 | 1 | 1,2 | |||||||||||||||||
| QIAamp DNA Mini Kit | 1,1 | 2 | |||||||||||||||||
| column-based RNA extraction* | 1 | 1 | |||||||||||||||||
| Omega Bio-Tek - Hamilton NGS | 2 | 1 | |||||||||||||||||
| BD MAX | 1 | 2 | |||||||||||||||||
| Maxwell | 2 | 1 | |||||||||||||||||
| extraction Kit - Machery Nagel | 1,2 | ||||||||||||||||||
| MagNa Pure LC 2.0 | 2 | 2 | |||||||||||||||||
| EliGene Viral Fast Isolation Kit | 2 | 2 | |||||||||||||||||
| GenoXtract | 1 | ||||||||||||||||||
| Ideal 32 | 1 | ||||||||||||||||||
| NeuMoDX | 1 | ||||||||||||||||||
| QIAsymphony | 1 | ||||||||||||||||||
| Omega Bio-Tek - Hamilton MicroLab | 2 | ||||||||||||||||||
| Nimbus - StarMag | 2 | ||||||||||||||||||
| IndiMag 48 | 2 | ||||||||||||||||||
| MagCore Plus II | 2 | ||||||||||||||||||
| QIACube HT | 2 | ||||||||||||||||||
| MutaCLEAN® Universal RNA/DNA | 3,3 |
1: all samples correct; 2: S3 false-negative; 3: S2 and S3 false-negative.* not specified.
3.4. Quantitative SARS-CoV-2 molecular test performance
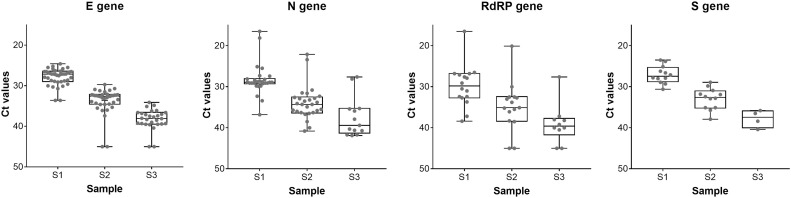
Finally, the participating laboratories had the possibility to report the target region(s) along with the achieved CT or Cq values of the respective PCR assays. The reported target regions were within the E-gene, N-gene, RdRP-gene, and S-gene, listed in order from most used to least. The best test performances (% correctly detected samples) were reported for assays targeting the E-gene as given in Table 6 and Fig. 1 .
Table 6.
Average CT values for the 4 most frequently reported gene targets.
| E (n = 40) |
N (n = 27) |
RdRP (n = 16) |
S (n = 12) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | 95 % C.I. | Positivity rate (%) | Mean | 95 % C.I. | Positivity rate (%) | Mean | 95 % C.I. | Positivity rate (%) | Mean | 95 % C.I. | Positivity rate (%) | |
| S1 | 27.8 | 27.2 - 28.4 | 100 | 28.3 | 26.8 - 29.8 | 100 | 29.8 | 27.1 - 32.5 | 100 | 27.1 | 25.7 - 28.4 | 100 |
| S2 | 33.6 | 32.6 - 34.6 | 95 | 33.9 | 32.3 - 35.5 | 100 | 34.9 | 31.6 - 38.2 | 81 | 32.9 | 31.3 - 34.6 | 100 |
| S3 | 38.3 | 37.3 - 39.2 | 63 | 37.4 | 34.5 - 40.4 | 48 | 39.1 | 35.6 - 42.5 | 50 | 37.8 | 34.5 - 41.1 | 33 |
Fig. 1.
Boxplot of CT values for each sample and the indicated target genes. Boxplots represent quartiles, showing 50 % IQR and median within the box.
4. Discussion
Of the 52 laboratories that participated in the EQA for molecular SARS-CoV-2 genome testing, 65 % achieved the maximum score possible, meaning that all 4 samples were reported correctly. Although none of the participating laboratories reported a false-positive result, 35 % of the laboratories did not detect the SARS-CoV-2 genome in sample S3 which had the lowest concentration (CT 38.5) of the 3 positive EQA test samples. A decrease in SARS-CoV-2 RNA in sample S3 during shipment is considered unlikely, as the stability of the viral RNA in the test panels was tested before dispatch under shipment conditions. In addition, shipment day and delivery time of the test panels was identical for all participants and no significant difference in the time of analysis was observed between the group which correctly analysed S3 as positive and that which tested S3 false-negative (p = 0.56; unpaired t-test). The observation that all except one laboratory detected sample S2 correctly, which had the second lowest concentration (CT 33.6), suggests that the sensitivity limit of the respective test systems may conform to viral loads between S2 and S3. To further clarify this point, a positive sample with a viral load between S2 and S3 will be included in future quality assessment rounds for SARS-CoV-2 genome testing.
Since numerous different real-time RT-PCR assays combined with a variety of different methods for NA extraction were used, no clear evidence for a substantial advantage of one system over another can be observed. Clearly, the best test performance was achieved for the fully automated test systems GeneXpert and Cobas 6800, which all detected the low positive sample S3. The high sensitivity of these two test systems is likely due to the high amount of input material for NA extraction combined with a high volume of extracted NA used for the SARS-CoV-2 genome detection. We obtained volumes for the NA extraction and real-time RT-PCR steps for other test systems and did not find a statistically significant difference between the group that identified sample S3 as positive and the group which failed to report a positive result (1: S3 correct, n = 20; 2: S3 false-negative, n = 16; p = 0.69; unpaired t-test). Nevertheless, high dilution in the NA extraction step and use of small aliquots of the extract in the RT-PCR step are not recommended, as they would lead to negative results in samples with low viral load.
This manuscript provides important information on the variety of NA extraction and RNA test systems currently used for routine diagnostics of acute SARS-CoV-2 infections as well as for large scale screening of asymptomatic persons. In particular, the viral loads of the three positive samples chosen for this first national EQA scheme intentionally included a sample with a low viral load (S3: CT 38.5) which was found in our patient cohort in about 1 % of the initially tested samples leading to COVID-19 diagnosis and in up to 25 % of consecutive samples of recovering patients at a later stage of infection.
Overall, most laboratories achieved reasonable test sensitivity, which is an important prerequisite for early detection of viral spread. Nevertheless, test sensitivity can be improved by a modification of the workflow, for example by modifying volumes in the NA extraction method and/or the volume used in the RT-PCR step. An increase in analytical sensitivity may also be critical when considering pooling [7].
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
I. Görzer: Conceptualization, Writing - original draft, Visualization. Ch. Buchta: Project administration, Formal analysis, Writing - review & editing. P. Chiba: Project administration, Formal analysis, Writing - review & editing. B. Benka: Resources, Project administration. J.V. Camp: Visualization, Writing - review & editing. E. Puchhammer-Stöckl: Supervision, Writing - review & editing. S.W. Aberle: Resources, Conceptualization, Supervision, Writing - review & editing.
Declaration of Competing Interest
The Authors have no conflicts of interest to declare.
Acknowledgements
We gratefully thank Tanja Amon, Andreas Rohorzka, and Ursula Sinzinger for excellent technical assistance.
Contributor Information
I. Görzer, Email: irene.goerzer@meduniwien.ac.at.
S.W. Aberle, Email: stephan.aberle@meduniwien.ac.at.
References
- 1.Spiteri G. First cases of coronavirus disease 2019 (COVID-19) in the WHO European Region, 24 January to 21 February 2020. Euro Surveill. 2020;25(9) doi: 10.2807/1560-7917.ES.2020.25.9.2000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.https://virological.org/t/novel-2019-coronavirus-genome/319.
- 3.Corman V.M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.https://www.who.int/publications/i/item/critical-preparedness-readiness-and-response-actions-for-covid-19.
- 5.Hallworth M.J. That’ 70%’ claim again. Ann. Clin. Biochem. 2018;55(4):517–518. doi: 10.1177/0004563218773778. [DOI] [PubMed] [Google Scholar]
- 6.Sciacovelli L. External quality assessment: an effective tool for clinical governance in laboratory medicine. Clin. Chem. Lab. Med. 2006;44(6):740–749. doi: 10.1515/CCLM.2006.133. [DOI] [PubMed] [Google Scholar]
- 7.Majid M., Omer S.O., Khwaja A.I. Optimising SARS-CoV-2 pooled testing for low-resource settings. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]