Introduction
Cerebral Small Vessel Disease (SVD) refers to the sum of all pathological processes that affect small vessels of the brain, including small arteries, arterioles, capillaries, and small veins1, as well as their interactions with perivascular structures. As such, SVD is the underlying pathology responsible for manifestations that are both common and diverse. Stroke, the second-leading cause of death and acquired disability worldwide, can be attributed to SVD in up to 20% of ischemic strokes and the vast majority of hemorrhagic strokes. SVD is also the most frequent pathology underlying vascular cognitive impairment and dementia as well as age-related depression and gait deterioration2. Finally, SVD associates with observable neuroimaging features such as cerebral white matter lesions, lacunes of presumed vascular origin, cerebral microbleeds (CMBs), dilated perivascular spaces, and total cerebral atrophy1,3. The prognostic relevance of some of these neuroimaging biomarkers is well established, being associated not only with higher risk of stroke and dementia4 but also with a decline in the ability to carry out daily activities1.
Genetic investigation of both rare familial SVD syndromes as well as complex sporadic late-life disease is a promising approach to understanding and discovering novel treatments for SVD. Cerebrovascular disease of all causes is a highly heritable trait. A family history of stroke is associated with a 3-fold increase in risk5, and two main manifestations of SVD, SVD-stroke and white matter lesions, are 16% and 50% heritable, respectively6,7. Further, monogenic disorders constitute up to 5% of all strokes overall8, and CADASIL, the most common familial stroke disorder, has SVD as a core manifestation.
We aim to provide the reader with an overview of the genes and corresponding biological pathways that have been found to be implicated in rare and common forms of SVD. We have elected to categorize the literature by gene or genetic locus rather than symptoms, signs, or syndromes to highlight their contributions to new insights about the pathogenic basis of SVD. We have grouped genetic associations with SVD into two broad mechanistic categories: arteriolosclerosis and amyloidosis.
Loci discovered for monogenic SVD: arteriolosclerosis-related
NOTCH3 (19p13):
Among monogenic SVD, Cerebral Autosomal Dominant Arteriopathy with Subcortical Ischemic Strokes and Leukoencephalopathy (CADASIL) is the most common, with a prevalence of approximately 5 per 100,000 in Northern England9. The clinical presentation is highly variable with a syndromic constellation that can include migraine, psychiatric disorders, cerebrovascular events, and progressive dementia9. Stroke and transient ischemic attack are the most frequently reported symptoms, present in 85% of symptomatic individuals with an average age of onset of 46 years. Although ischemic manifestations (usually presenting as lacunar syndromes) are the most common, cases of subcortical hemorrhage and cerebral microbleeds have also been described10. MRI changes are considered hallmarks of the disease, present in all individuals that have received the diagnosis. These consist of MRI-based white matter hyperintensities (WMH) with a symmetric and diffuse pattern involving periventricular areas and centrum semiovale. Involvement of the external capsule and anterior temporal lobes increases the suspect for the disease9. CADASIL results from mutations in the NOTCH3 gene (19p13), which encodes a transmembrane receptor expressed on vascular smooth muscle cells and pericytes11,12. The relation between NOTCH3 mutations and vascular smooth muscle cell pathology is confirmed by the derangement and accumulation of granular osmophilic material in the microvasculature. The typical CADASIL causing mutation impacts the number of cysteine residues, leading to misfolding of the receptor. Up to 65% of mutations are clustered in exon 4 of the gene13, but they can extend throughout exons 2–24. Sequencing of these regions is the appropriate method to confirm the diagnosis in patients with suggestive clinical presentation. Mutations not involving a cysteine residue or leading to loss-of-function have uncertain causative pathogenicity and pathological evaluation of skin biopsy is warranted in these cases13. Recent efforts from the Exome Aggregation Consortium (ExAC) have also shown that “classical” CADASIL mutations are more frequent than what expected from disease prevalence, suggesting the possibility that most mutations may lead to a milder phenotype, clinically concealed14. However, a clear genotype-phenotype correlation has not been found so far.
Despite these well-characterized associations in monogenic disease, evidence from GWAS supporting the role of NOTCH3 in sporadic SVD are lacking, with no recognized association between common NOTCH3 variants and SVD manifestations15. Notably, a single GWAS study on WMH in healthy subjects did find associations between common variants on EFEMP1 and higher WMH load16. The gene seems more involved in cell survival through Notch signaling, which instead seems to be spared in CADASIL-causing mutations.
HTRA1 (10q26):
Following the characterization of CADASIL, an autosomal recessive cerebral arteriopathy with subcortical infarcts and leukoencephalopathy (CARASIL) has also been described. As the name implies, CARASIL bears some syndromic similarities with CADASIL but is transmitted following an autosomal recessive pattern. Clinical description of disease is based on a few cases, mostly of Japanese origin17. Compared to CADASIL, cognitive decline begins even earlier, and migraine is not common. In addition, gait disturbance (usually slowly progressive spasticity in the lower extremities), low back pain (spondylosis deformans) and alopecia are the most characteristic features. HTRA1 is the gene mutated in the disease and its product is a serine protease involved in repression of transforming growth factor‐β (TGF‐β) signaling18.
A recent study found that common variants in the gene were associated with increased risk for SVD-stroke stroke19. Although these findings may suggest the involvement of the HTRA1 also in the pathogenesis of sporadic SVD, the associations did not reach threshold for genome-wide significance.
Together, these studies have contributed to the “matrisome” hypothesis of the pathogenesis of SVD20. The matrisome is comprised of proteins involved in processes that maintain tissue homeostasis. It has been proposed that agglomerations of defective NOTCH3 ectodomains lead to rearrangements in key proteins of the matrisome (such as tissue inhibitor of metalloproteinase 3 (TIMP 3) and vitronectin), and that HTRA1 mutations impair TGF-β1 signaling with an overall effect on pericyte proliferation and protein accumulation in vessel wall, metalloproteinase activity, blood brain barrier permeability and dysfunctional cerebral blood flow20,21.
For completeness, we cited here another genetically determined form of microangiopathy with ischemic and hemorrhagic strokes, cognitive deterioration, and leukoencephalopathy on MRI scanning: cathepsin A-related arteriopathy with strokes and leukoencephalopathy (CARASAL)22.
α-GAL (Xq22):
Anderson-Fabry (a.k.a. Fabry) Disease is caused by insufficient activity of the enzyme alpha-galactosidase A (α-GAL) with subsequent accumulation of glycosphingolipids (mainly globotriaosylceramid and galactosylceramide) in multiple tissues. Given the pleiotropic manifestations of the disease, prevalence varies in specific at-risk populations compared to the general public. For example, in patients presenting with cryptogenic ischemic stroke and aged 18 to 55 years, 0.5% were found to carry a α-GAL mutation23 .
The classical presentation involves childhood neuropathy (including burning pain in the extremities and hypohydrosis), gastrointestinal symptoms (most commonly nausea, abdominal pain, and postprandial diarrhea), corneal opacities, hearing loss, and angiokeratoma24. Stroke may result from either cardioembolism or direct vessel involvement. Deposits of sphingolipids in vascular endothelia and smooth muscle cause vessel stenosis, occlusion, and dilation with a demonstrated change in regional cerebral perfusion24. Up to 14% of known disease-causing carriers present with a late-onset phenotype. With advancing age and progression of glycosphingolipid accumulation, renal failure, heart disease and cerebrovascular disease manifest more prominently24. Patients with Fabry disease have a 5.5–12.2-fold increased risk of stroke compared to the general population and have a mean age at first stroke of approximately 40 years. Most patients suffer from ischemic strokes, both of small and large vessel subtypes, but intracerebral hemorrhage (ICH) has also been described25. Finally, a high burden of white matter lesions is frequently seen on MRI scans of patient’s with Fabry disease, far in excess of age-matched peers26.
Fabry disease is inherited in an X-linked pattern. Symptoms present earlier and are more severe in men, but up to 70% of women who have one altered copy of α-GAL also experience many of the classic features of the disorder. Several mutations (mostly missense or nonsense mutations) involving in the α-GAL gene have been reported, and while exon 7 contains ~22% of them, no clear enriched region for disease-causing mutations has been described27. Enzymatic analysis performed by using plasma or leukocytes may show a deficiency of α-GAL in males and may be normal in female (heterozygote) patients. Confirmation by molecular genetic testing remains the gold-standard.
COL4A1 and COL4A2 (13q34):
Type IV collagen is a component of basement membranes in blood vessels in virtually all tissues throughout the body. It provides the substrate for basement membrane to interact with nearby cells, and plays a role in cell migration, proliferation, differentiation and angiogenesis. There are six human genes associated with collagen IV (COL4A1-A6). The collagen IV expressed in vascular basement membrane is a heterotrimer made from the combination of the transcribed products of COL4A1 and COL4A2. Mutations in these genes are associated with small vessel disease in various organs, and penetrance is estimated to be nearly 100%28.
Manifestations of COL4A1 mutations range from rare classical syndromes with multiple highly-specific features to more common-appearing end organ events such as ICH. In general, clinical manifestations of COL4A1 mutations occur in four domains: ocular, renal, muscle, and cerebral manifestations.
Cerebral SVD is described in more than half of individuals with COL4A1 mutations. Essentially all established neuroradiologic markers of SVD have been reported: WMH affecting periventricular, subcortical and pontine regions usually bilateral and symmetrical with sparing of the temporal and occipital lobes and arcuate fibers; dilated perivascular spaces; lacunar infarcts; and microbleeds in both deep and lobar locations29. WMH, present in up to 63.5% of patients in a study with a mean age at MRI of 30 years, are the most common manifestation30. The second most common is stroke, specifically ICH but also ischemic. Both mouse experiments and clinical observation have linked COL4A1 mutations to trauma-related ICH, suggesting that triggering events (such as birth or later head trauma) result in recurrent intracerebral hemorrhages (ICH) in both young and old subjects with mutations29-31.
One additional cerebral SVD-specific COL4A1 syndrome, Pontine autosomal dominant microangiopathy and leukoencephalopathy (PADMAL), results from a slightly different pathogenic mechanism32. The causative mutation in the 3’ untranslated region (3’UTR) of COL4A1 alters a microRNA binding site and results in an over-expression of the gene, rather than a defect in protein folding. Patients with PADMAL present with dysarthria, ataxia, and stroke, as well as mood disturbance and dementia. As underlined by the name, the hallmark of the disease is involvement of the pons in all those affected. The definite diagnosis of a COL4A1-related disorder is made in a proband with suggestive findings and identification of a heterozygous pathogenic variant via sequence analysis of the gene.
More recently, large-scale genetic association studies have lent support to the hypothesis that variation in collagen type IV genes extends beyond rare Mendelian diseases. Two studies have identified common variants in COL4A2 in association with lacunar ischemic stroke and deep ICH20-33. Further, a meta-analysis of combined healthy and stroke participants identified a common variant in COL4A2 in association with WMH burden7.
Most mutations associated with syndromic presentations impact α-helix domains, which are necessary for trimerization into collagen IV. Failure of this process leads to intra- and extra-cellular accumulation of defective collagen with consequent structural instability of basement membranes28-34, loss of vessel wall integrity, and increased BBB permeability35. ER stress and activation of the unfolded protein response due to the high levels of misfolded collagen have been uncovered as additional cytotoxic ramifications that likely contribute to disease28. In contrast, variants discovered in association with sporadic SVD have thus far been intronic in nature, and the mechanisms through which they contribute to disease remain unclear20,33.
FOXC1 and FOXF2 (6p25):
Mutations in the Forkhead Box C1(FOXC1) gene lead to a clinical picture similar to COL4A1 mutations with ischemic strokes and cerebral SVD with ocular involvement. The latter is represented by anterior segment abnormalities, which can coexist with other dysmorphisms in the Axenfeld-Rieger Syndrome (ARS). Classical neuroimaging features of SVD are present with WMH and lacunar infarcts of very early onset. Patients with missense and nonsense mutations of FOXC1 were found to have all the neuroimaging spectrum of SVD. FOXC1 is highly expressed in brain pericytes that are critical components of the BBB, and expression of the gene is crucial for endothelial cell proliferation and blood vessel stability36.
Additional evidence of the role of this gene in SVD comes from GWAS data examining FOXC1 in 9,361 stroke-free subjects36. In this study, three single nucleotide polymorphism (SNPs), all of which influence FOXC1 transcript levels, were found to be associated with WMH burden. FOXF2, a paralogue of FOXC1, has also been identified in a GWAS of small-vessel stroke risk37. The pathogenic pathway that links FOXC1/FOXF2 genes to SVD may also include the PITX2 (pituitary homeobox 2) gene, a developmental transcription factor expressed in the neural crest36. PITX2 physically interacts with FOXC1, and variants in this gene are associated with increased risk for WMH. PITX2 is well known for its strong associations with atrial fibrillation and cardioembolic stroke risk36, and this linked pathway with FOXC1 could implicate a shared mechanism between these seemingly disparate etiopathology stroke mechanisms.
TREX1(3p21):
Retinal vasculopathy with cerebral leukodystrophy-like syndromes (RVCL) is a term that encompasses at least three previously described syndromes: cerebro-retinal vasculopathy (CRV), hereditary vascular retinopathy (HRV) and hereditary endotheliopathy with retinopathy, nephropathy, and stroke (HERNS). These syndromes share involvement of the retinal vasculature together with variable SVD features and nonuniform organ involvement with autoimmune features including systemic lupus38. Patients from these groups have all exhibited C-terminal frame-shift mutations in TREX1 (3p21), which encodes for a DNA exonuclease. Mutations in TREX1 lead to impairment in the apoptosis and interferon pathways39. RVCL patients exhibit the full spectrum of cerebral SVD with thickened vascular basement membrane and fibrinoid vascular necrosis. Neuroimaging demonstrates WMH, usually with punctate features, as well as ring-enhancing lesions that relapse and remit with time, and white matter calcifications. Neurological symptoms are less specific and include focal neurological deficits, migraine, cognitive impairment, psychiatric disturbances, and seizures38.
Loci discovered for monogenic SVD: amyloid-related
The genes highlighted so far contribute to biological pathways that lead to the pathological arteriosclerotic changes of SVD. The other predominant pathological variant of SVD is cerebral amyloid angiopathy (CAA). This term summarizes the features characterized by deposition of amyloid fibrils in cortical and leptomeningeal arterioles and arteries. Both Aβ and non-Aβ cerebral amyloid angiopathy are found in ICH1.
APP (21q21):
Mutations in the amyloid precursor protein gene (APP) gene lead to the Hereditary Cerebral Hemorrhage With Amyloidosis (HCHWA) syndrome40. Impairment in gene function results in classical CAA pathology, with deposition of misfolded amyloid fibrils in the walls of cerebral arterioles. The clinical phenotype is characterized by development of lobar ICH, usually between ages 45 and 65, with neuroimaging variably revealing WMH, superficial siderosis, and multifocal lesions of both hemorrhagic and ischemic nature.
No variants in APP were found in a small sample of sporadic CAA-related ICH41 and published lobar ICH GWAS have not identified genome-wide associations for common variants at APP42.
Loci discovered for sporadic SVD
Cerebral SVD is influenced by genetic and non-genetic exposures. Early epidemiological observations between family history and risk of stroke43 have been reinforced by large-scale genetic studies that have identified novel genes and potential mechanisms involved in SVD pathogenesis. Here we review the additional loci highlighted by the latest GWAS on three clinical manifestations and one neuroimaging features that are commonly used as markers of SVD, including SVD ischemic stroke, ICH, vascular dementia and WMH. GWAS loci do not automatically map onto specific genes, and so additional research will be needed to refine signals and identify which specific gene or genes at each location are responsible for the statistical association between variants and traits (Table 1). As such, we have categorized these associations by chromosome and genetic location rather than by nearby genes.
Table 1:
Summary of established loci associated with SVD manifestations and their source of discovery
| Genes and selected reference | Evidence from Mendelian and/or sporadic SVD manifestations |
SVD manifestations | OR (95% CI) for the risk allele of the top SNP derived from GWAS |
|---|---|---|---|
| NOTCH3(19p13) 9 | Mendelian disease | WMH, ischemic stroke, ICH | |
| HTRA1(10q26) 20 | both | WMH | |
| Ischemic stroke | 1.23(1.10–1.37) | ||
| α-GAL (Xq22) 24 | Mendelian disease | Ischemic stroke | |
| COL4A1/ COL4A2 (13q34) 20 | both | ICH | 1.21(1.07–1.37) |
| Ischemic stroke | 1.17(1.11–1.24) | ||
| WMH | 1.09(1.04–1.15) | ||
| FOXC/FOXF2 (6p25) 36 | both | Ischemic stroke | 1.08(1.02-1.14) |
| WMH | |||
| TREX1 (3p21) 39 | Mendelian disease | Ischemic stroke, WMH | |
| APP (21q21) 40 | Mendelian disease | ICH | |
| APOE ε (19q13) 44 | both | ICH | 1.51(1.23-1.85) |
| WMH | 1.63(1.004-2.64) | ||
| ZCCHC14 (16q24) 45 | sporadic disease | Stroke | 1.16(1.10-1.22) |
| WMH | 1.10(1.05-1.16) | ||
| WDR12/ICA1L (2q33) 6 | sporadic disease | Stroke | |
| WMH | 1.10(1.03–1.18) | ||
| PMF (11q22) 7,42 | sporadic disease | ICH | 1.44(1.27–1.64) |
| WMH | 1.06(1.01–1.12) |
Loci implicated in arteriolosclerosis-related SVD
SVD Ischemic stroke:
The latest and largest GWAS of stroke has arisen from the MEGASTROKE collaboration, leveraging 67,162 stroke cases and 454,450 controls recruited from sites worldwide6. This study has reinforced the idea that most associations between variants and stroke risk are restricted to a specific etiological stroke subtype. In analyses restricted to SVD-stroke following standard subtyping criteria46, MEGASTROKE reported associations with two loci at 16q24 and 2q33.
16q24:
This locus was previously identified as associated with SVD stroke and other cerebral SVD‐related phenotypes such as WMH45. Gene-expression studies suggest that the lead variant driving this association impacts the expression levels of the nearby ZCCHC14 gene in vascular cells, possibly through an effect on methylation. ZCCHC14 encodes a zinc-finger protein with roles in in nucleic acid binding and regulation of DNA transcription45. Its precise role in SVD remains unknown.
2q33:
This region is known to be highly pleiotropic and has been found to be associated with SVD-stroke as well as WMH burden (see below)6,7. Two of the genes implicated in driving this signal are WDR12 and ICA1L. WDR12 encodes the WD repeat domain 12 gene, a key component of the ribosome complex. Although the gene was first highlighted in GWAS of coronary artery disease47, a recent meta-analysis failed to link variants in the gene with carotid intima-media thickness48. Leading SVD-associated SNPs in WDR12 may also exert their biological effect through the ICA1L (Islet Cell Autoantigen 1 Like) gene, as they appear to mediate expression of this gene in frontal cortex49.
14q22:
This locus was identified through MEGASTROKE analyses of variants with low allele frequency (<5%), which revealed an excess of such variants at 14q22 in participants with large-artery as well as SVD stroke50. The GCH1 gene (GTP cyclohydrolase 1) maps onto this locus but does not reach genome-wide significance in the standard GWAS. Nevertheless, this finding does raise interest in a novel pathway involved in tetrahydrobiopterin synthesis, which may lead to stroke via alterations in the nitric oxide signaling that requires this cofactor.
12q24:
This locus was initially found to be associated with all ischemic stroke but not with any specific subtype37. A more recent effort has since shown that the effect of variants at 12q24 is strongest for SVD strokes37. Which gene among the several encoded at this locus is driving the observed association is uncertain, but fine-mapping analyses and testing of variants with predicted protein-damaging results have raised evidence that the SH2B3 gene may be the culprit6. SH2B3 is involved in a range of signaling activities for growth factor and cytokine receptors and has previously been associated with coronary artery disease, endothelial cell migration, and atherosclerosis6. These findings align with established evidence linking shared genetic influence between large artery atherosclerosis and SVD strokes1, and support a shared biological mechanism for atherosclerotic and arteriolosclerotic processes.
SVD-stroke is affected by higher disease heterogeneity compared to the other etiological stroke subtypes. This has led to an overdiagnosis and consequently a deflated heritability of SVD-stroke51. A better understanding of the genetic architecture of this complex disease can be achieved with a more detailed phenotyping of cases, for instance with neuroimaging51.
White matter hyperintensity
White matter hyperintensities are commonly defined as bilateral, typically symmetrical MRI signal abnormality on T2-weighted sequences3. Given the repeatedly demonstrated overlap between WMH and other more overt manifestations of cerebral SVD, they are considered a reliable biomarker4. GWAS of this trait have been performed within stroke as well as stroke-free populations, amid concern that WMH appearing in the presence of stroke may be pathogenically distinct from that which occurs in the aging non-stroke population.
17q25 locus:
The 17q25 locus was identified in association with WMH burden in stroke-free populations52. Among these, ACOX1 (acyl-coenzyme A oxidase 1) and UNC13D (Unc-13 Homolog D) are also involved in adrenoleukodystrophy, a rare genetic disease characterized by the breakdown or loss of myelin. This observation reinforces the importance of these genes in the biology of cerebral white matter. Additionally, ACOX1 is a key enzyme of the fatty acid beta-oxidation pathway. Involvement of the phosphorylation cascade in the pathogenesis of WMH is further supported by previously observed genetic associations between WMH burden and variants within the mitochondrial genome53. TRIM65 and TRIM47 (tripartite motif-containing genes) and WBP2 (WW domain binding protein 2 gene) are also mapped on this region and have been confirmed in a separate study17. Proteins encoded by these genes are involved in a broad range of biological processes including apoptosis and transcription regulation.
6q25:
This locus was discovered in a recent GWAS that included both stroke-affected and stroke-free individuals, for a total of 11,226 cases54. PLEKHG1 maps on this locus and is part of a family of Rho guanine nucleotide exchange factors. The protein encoded by PLEKHG1 has an intriguing role in the reorientation of cells in the vascular endothelium, suggesting that disruption responses to mechanical stress in endothelial cells may be involved in the formation of white matter lesions. The leading SNP associated with WMH at 6q25 has also been found increase SVD stroke risk by almost 10%54.
2p16:
This region was first identified in a large GWAS of WMH17, and subsequently independently validated54. 2p16 contains the EFEMP1 gene, which encodes for an extracellular matrix glycoprotein that has effects on vessel development and matrix metalloproteinase activity. These findings further support the role of the matrisome (see above) in the pathogenesis of SVD. EFEMP1 is also uniquely upregulated in gliomas and exerts at least some of its function through Notch3 signaling, highlighting once again the shared pathologic processes in sporadic and familial SVD.
5q14:
Common variants at 5q14 have been found in association with decreased white matter structural integrity as quantified via advanced neuroimaging techniques2. Among the several genes that map onto this locus, the leading target of interest has been VCAN (Versican), which encodes for the homonym extracellular matrix proteoglycan. Involvement of VCAN bolsters the hypothesis of the matrisome’s role in SVD pathogenesis, while simultaneously illustrating the utility of innovative biomarkers in GWAS of cerebral SVD.
Intracerebral hemorrhage
Non-traumatic ICH can be broadly divided into two categories, depending on the region of the bleeding: lobar (cerebral cortex with or without involvement of subcortical white matter) and non-lobar (basal ganglia, thalamus, cerebellum and brainstem).
1q22:
GWAS for non-lobar ICH, classically considered to be hypertensive in nature and related to arteriolosclerotic risk factors rather than amyloid-related processes, have identified novel associations at 1q2242. Two genes map on this region. The first, PMF1, encodes Polyamine Modulated Factor 1 which is a key coactivator of DNA-transcription. The second, SLC25A44, encodes for Solute Carrier Family 25 Member 44, which is part of a family of mitochondrial carrier proteins. Whether one or both of these genes are involved in the pathogenesis of ICH, and the mechanism through which these genes may drive the development of SVD remains unclear. The locus has apparent pleiotropic associations with both WMH as well as all-cause ischemic stroke16,37.
Additionally, recapitulating the epidemiologic association between non-lobar ICH and hypertension, a polygenic risk score for hypertension severity has also been shown to predict risk of non-lobar ICH55. While this finding provides evidence for a direct causal link between hypertension and non-lobar ICH, it does not in itself provide novel insights into unappreciated disease mechanisms.
Vascular dementia
Though SVD is implicated in up to 45% of dementia2, only one vascular dementia GWAS has been conducted to date. Using data from the population-based Rotterdam Study, a locus located on the X chromosome, close to the nuclear receptor for androgen (AR) gene (Xq12), was found to be associated with incident vascular dementia2. This locus has not yet been replicated in other studies.
Beyond incident vascular dementia risk, a polygenic risk score developed in association with SVD stroke has been found to correlate with general cognitive abilities in adulthood, suggesting a shared genetic contribution to SVD and adult cognition56. Furthermore, Alzheimer’s Disease (AD) and SVD stroke have been found to exhibit shared genetic contributions57, perhaps reflecting a co-contribution of genetic risk factors to commonly-encountered late-onset AD patients who share risk factors for both traits. Although a GWAS of neuropathological features showed that loci conferring risk for clinically-defined AD did not impact vascular brain injury phenotypes58, meta-analysis of AD and SVD stroke have identified an association at 17q25 near ATP5H (ATP Synthase Peripheral Stalk Subunit D). This gene codes for the mitochondrial adenosine triphosphate synthase that catalyzes mitochondrial ATP synthesis. These associations are supportive of the lipid transport pathway as likely mechanism shared among these traits57.
Loci implicated in amyloid-related SVD
Intracerebral hemorrhage:
APOE ε (19q13):
APOE ε2 and ε4 alleles represent the most powerful genetic risk factor for CAA-related ICH, and its effect size remains among the largest for any common allele on a sporadic human trait. The ε4 allele is also a potent genetic risk factor for Alzheimer’s Disease (AD). APOE epsilon alleles are a haplotype of two common variants, rs7412 and rs429358, which taken together define the epsilon allele status. Until recently, genome-wide genotyping arrays and imputation schema did not allow for the accurate capture of these two variants, and so published GWAS of CAA-related ICH have not demonstrated a signal in the APOE region42. Rather, dedicated studies of APOE have relied on direct genotyping of the APOE epsilon allele-defining variants and have demonstrated a strong and well-replicated signal44,59. The differential mechanisms predisposing to parenchymal (AD) vs. vascular (CAA) localization of amyloid protein remains an active area of investigation. Beyond CAA, preliminary evidence suggests that APOE is also involved in arteriolosclerosis, providing a potential bridge between the pathophysiologic mechanisms described herein. In a meta-analysis combining general and high-risk populations, carriers of APOE ε2 and/or ε4 were found to have increased WMH load, consistent with a role in arteriolosclerosis in addition to established amyloid-related processes60. Given the limited sensitivity and specificity, tests for APOE alleles are not indicated as screening or diagnostic procedures.
Mechanisms in Sporadic Small Vessel Disease
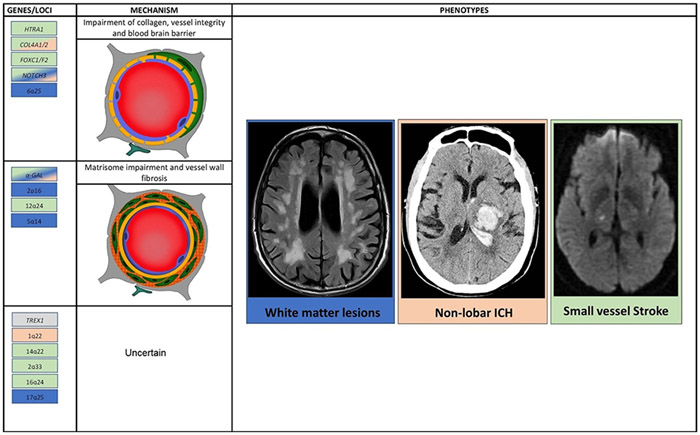
Dedicated post-GWAS studies are needed to functionally characterize the variants outline above in order to reliably identify the genes through which they exert their effects and link those effects to endophenotypes and biomarkers. Studies on hereditary forms of SVD and emerging associations in sporadic disease highlight possible pathological thematic pathways for non-amyloidogenic SVD (Figure 1). Orthogonal research approaches in model systems have converged on similar themes, and future research will increasingly rely on multidisciplinary approaches to functionally characterize genetic mechanisms of disease.
Figure 1:
Overview of pathological mechanisms that may be involved in cerebral small vessel disease (CSVD). Collagen dysfunction, distruption of vessel integrity, dissarangement of matrisome and increased permeabiliyt of blood brain barriers have been linked to genes and to macrosopic manifestations of CSVD, both in genetic epidemiology and studies on familial forms of the disease. The background color of genes and loci enlisted corresponds to at least one of the macroscopic manifestations of CSVD.
Collagen dysfunction:
Genetic mutations that alter the conformation of the chain of collagen lead to impaired deposition of collagen in vessel walls. This, in turn, results in weakening of basement membranes, loss of vessel wall integrity and increased BBB permeability35. The discovery that common variants in COL4A1/2 are associated with sporadic manifestations SVD20, reinforces the hypothesis of this pathway in SVD.
Vessel integrity and the BBB:
Studies on FOXC1 have suggested that impairment in BBB development and integrity may lead to the SVD spectrum. The gene is expressed in brain pericytes that are critical components of the BBB and is crucial for endothelial cell proliferation and blood vessel stability36. Mutations in the gene lead to the syndromic manifestations with SVD pathology and there are SNPs in the gene that are risk factor for SVD stroke and WMH36. These results suggest that an impairment of the BBB may be a key component of the SVD pathogenesis.
Matrisome:
Thanks to genetic characterization of NOTCH3, the matrisome’s role in disease has arisen to prominence, involving agglomerations of defective NOTCH3 ectodomain leading to derangements in tissue inhibitor of metalloproteinase 3 (TIMP 3) and vitronectin activity, with protein accumulation in vessel wall and dysfunctional cerebral blood flow22. Another protein that was found to aggregate with NOTCH3 is latent TGF-beta binding protein 1 (LTBP-1). This protein and its role in the pathway towards the vessel wall fibrosis is also suggested from the studies on CARASIL that showed that HTRA1 mutations impair transforming growth factor beta 1 (TGF-β1) signaling with effects on pericyte proliferation, metalloproteinase, blood brain barrier permeability21. Evidence from GWAS that support the role of NOTCH3 in SVD manifestation is debatable with few underpowered studies that failed to find an association between common variants and SVD manifestations15.
Future Directions
Clarifying the genetic basis for SVD carries the potential for new treatments for cerebrovascular disease, which, given the high prevalence of SVD in the population, could have an enormous impact on stroke prevention and healthy aging. Because cerebral small vessel tissue is largely inaccessible for direct study in living humans, the human genetic approach is a crucial way to derive information on the processes that lead to disease and its clinical ramifications.
Incorporating the results of these genetic studies into an improved working model of SVD pathogenesis and progression requires a series of steps from initial statistical associations to testing in functional model systems. In the near term, genetic associations may also impact risk stratification or even patient selection for clinical trials through application of polygenic risk scores (PRS) or testing of specific high-impact variants such as APOE ε4. In PRS, the small effects of many genetic variants across the genome are summed into an individualized score that reflects the cumulative genetic risk of a disease or trait. PRS have been shown to provide orthogonal information to identify individuals at highest risk for sporadic myocardial infarction61 at effect sizes that rival those for monogenic diseases. A similar polygenic risk approach may ultimately prove effective in modeling SVD, identifying subsets of individuals at highest risk for SVD stroke or ICH.
The inclusion of diverse ancestral backgrounds will be crucial to better understand the genetic architecture of complex diseases in general, to identify variants that may exert effects within specific racial or ethnic populations, and to improve our ability to map variants to genes through trans-ethnic fine mapping, which leverages differences in genetic linkage between diverse populations to refine the search for causal variants. Diverse ancestries are also critical to developing clinically useful genetic tests whose results are interpretable for all patients rather than only specific well-represented populations.
Human genetics has identified key genes and pathways involved in the pathogenesis and progression of cerebral SVD. While important diagnostically in some cases, studies examining the genetic foundations of disease can also lead to the identification of novel therapeutic targets, predict potential adverse effects to specific treatments, and stratify individualized risk. As proof of concept potentially valid for other monogenic disorders, the discovery of the Notch3 signaling pathways in CADASIL has led to novel promising therapeutic strategies aimed to silence the mutated Notch3, such as short hairpin RNA use and cysteine-corrective NOTCH3 exon skipping62.
Continued effort in this field holds great promise for improving the care of cerebral SVD in all its forms for the ultimate benefit of patients at risk for, or affected by, the disease.
Acknowledgments
Sources of funding
This study was supported by the following awards from the NINDS: K23NS086873, R01NS103924, 1R01NS093870. SM is supported by the American Heart Association/American Stroke Association fellowship (18POST34080063).
Footnotes
Disclosures
Dr. Rosand reports grants from OneMind; personal fees from Boehringer Ingelheim, Pfizer, New Beta Innovation, outside the submitted work. Dr. Anderson receives sponsored research support from the American Heart Association (Atrial Fibrillation Strategically Focused Research Network), and Bayer AG, and has consulted for ApoPharma, Inc.
References
- 1.Pantoni L Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol. 2010;9:689–701. [DOI] [PubMed] [Google Scholar]
- 2.Markus HS, Schmidt R. Genetics of Vascular Cognitive Impairment. Stroke; a journal of cerebral circulation. 2019;50:735–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. The Lancet Neurology. 2013;12:822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debette S, Markus HS. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ. 2010;341:c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seshadri S, Beiser A, Pikula A, Himali JJ, Kelly-Hayes M, Debette S, et al. Parental occurrence of stroke and risk of stroke in their children: the Framingham study. Circulation. 2010;121:1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature Genetics. 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traylor M, Zhang CR, Adib-Samii P, Devan WJ, Parsons OE, Lanfranconi S, et al. Genome-wide meta-analysis of cerebral white matter hyperintensities in patients with stroke. Neurology. 2016;86:146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto Y, Craggs L, Baumann M, Kalimo H, Kalaria RN. Review: molecular genetics and pathology of hereditary small vessel diseases of the brain. Neuropathol. Appl. Neurobiol 2011;37:94–113. [DOI] [PubMed] [Google Scholar]
- 9.Di Donato I, Bianchi S, De Stefano N, Dichgans M, Dotti MT, Duering M, et al. Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy (CADASIL) as a model of small vessel disease: update on clinical, diagnostic, and management aspects. BMC Med. 2017;15:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesnik Oberstein SA, van den Boom R, van Buchem MA, van Houwelingen HC, Bakker E, Vollebregt E, et al. Cerebral microbleeds in CADASIL. Neurology. 2001;57:1066–1070. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Zhang X, Leathers R, Makino A, Huang C, Parsa P, et al. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat. Med 2009;15:1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, et al. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J. Clin. Invest 2000;105:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutten JW, Haan J, Terwindt GM, van Duinen SG, Boon EM, Lesnik Oberstein SA. Interpretation of NOTCH3 mutations in the diagnosis of CADASIL. Expert Review of Molecular Diagnostics. 2014;14:593–603. [DOI] [PubMed] [Google Scholar]
- 14.Rutten JW, Dauwerse HG, Gravesteijn G, van Belzen MJ, van der Grond J, Polke JM, et al. Archetypal NOTCH3 mutations frequent in public exome: implications for CADASIL. Ann Clin Transl Neurol. 2016;3:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutten-Jacobs LCA, Traylor M, Adib-Samii P, Thijs V, Sudlow C, Rothwell PM, et al. Common NOTCH3 Variants and Cerebral Small-Vessel Disease. Stroke. 2015;46:1482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaaren BFJ, Debette S, Bis JC, Smith JA, Ikram MK, Adams HH, et al. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet. 2015;8:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tikka S, Baumann M, Siitonen M, Pasanen P, Pöyhönen M, Myllykangas L, et al. CADASIL and CARASIL. Brain Pathol. 2014;24:525–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiga A, Nozaki H, Yokoseki A, Nihonmatsu M, Kawata H, Kato T, et al. Cerebral small-vessel disease protein HTRA1 controls the amount of TGF-β1 via cleavage of proTGF-β1. Hum. Mol. Genet 2011;20:1800–1810. [DOI] [PubMed] [Google Scholar]
- 19.Rannikmäe K, Sivakumaran V, Millar H, Malik R, Anderson CD, Chong M, et al. COL4A2 is associated with lacunar ischemic stroke and deep ICH: Meta-analyses among 21,500 cases and 40,600 controls. Neurology. 2017;89:1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joutel A, Haddad I, Ratelade J, Nelson MT. Perturbations of the cerebrovascular matrisome: A convergent mechanism in small vessel disease of the brain? J Cereb Blood Flow Metab. 2016;36:143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baron-Menguy C, Domenga-Denier V, Ghezali L, Faraci FM, Joutel A. Increased Notch3 Activity Mediates Pathological Changes in Structure of Cerebral Arteries. Hypertension. 2017;69:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bugiani M, Kevelam SH, Bakels HS, Waisfisz Q, Ceuterick-de Groote C, Niessen HWM, et al. Cathepsin A-related arteriopathy with strokes and leukoencephalopathy (CARASAL). Neurology. 2016;87:1777–1786. [DOI] [PubMed] [Google Scholar]
- 23.Rolfs A, Fazekas F, Grittner U, Dichgans M, Martus P, Holzhausen M, et al. Acute cerebrovascular disease in the young: the Stroke in Young Fabry Patients study. Stroke. 2013;44:340–349. [DOI] [PubMed] [Google Scholar]
- 24.Schiffmann R Fabry disease [Internet] In: Handbook of Clinical Neurology. Elsevier; 2015. [cited 2019 May 8]. p. 231–248.Available from: https://linkinghub.elsevier.com/retrieve/pii/B9780444627025000172 [DOI] [PubMed] [Google Scholar]
- 25.Sims K, Politei J, Banikazemi M, Lee P. Stroke in Fabry disease frequently occurs before diagnosis and in the absence of other clinical events: natural history data from the Fabry Registry. Stroke. 2009;40:788–794. [DOI] [PubMed] [Google Scholar]
- 26.Rost NS, Cloonan L, Kanakis AS, Fitzpatrick KM, Azzariti DR, Clarke V, et al. Determinants of white matter hyperintensity burden in patients with Fabry disease. Neurology. 2016;86:1880–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta A, Beck M, Sunder-Plassmann G, editors. Fabry Disease: Perspectives from 5 Years of FOS [Internet]. Oxford: Oxford PharmaGenesis; 2006. [cited 2019 May 8]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK11586/ [PubMed] [Google Scholar]
- 28.Germain DP, Elliott PM, Falissard B, Fomin VV, Hilz MJ, Jovanovic A, et al. The effect of enzyme replacement therapy on clinical outcomes in male patients with Fabry disease: A systematic literature review by a European panel of experts. Mol Genet Metab Rep. 2019;19:100454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vahedi K, Alamowitch S. Clinical spectrum of type IV collagen (COL4A1) mutations: a novel genetic multisystem disease: Current Opinion in Neurology. 2011;24:63–68. [DOI] [PubMed] [Google Scholar]
- 30.Silvia L, MH S. COL4A1 Mutations as a Monogenic Cause of Cerebral Small Vessel Disease. Stroke. 2010;41:e513–e518. [DOI] [PubMed] [Google Scholar]
- 31.Meuwissen MEC, Halley DJJ, Smit LS, Lequin MH, Cobben JM, de Coo R, et al. The expanding phenotype of COL4A1 and COL4A2 mutations: clinical data on 13 newly identified families and a review of the literature. Genet. Med 2015;17:843–853. [DOI] [PubMed] [Google Scholar]
- 32.Verdura E, Hervé D, Bergametti F, Jacquet C, Morvan T, Prieto-Morin C, et al. Disruption of a miR-29 binding site leading to COL4A1 upregulation causes pontine autosomal dominant microangiopathy with leukoencephalopathy. Ann. Neurol 2016;80:741–753. [DOI] [PubMed] [Google Scholar]
- 33.Malik R, Rannikmäe K, Traylor M, Georgakis MK, Sargurupremraj M, Markus HS, et al. Genome-wide meta-analysis identifies 3 novel loci associated with stroke. Annals of Neurology. 2018;84:934–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo DS, Labelle-Dumais C, Gould DB. COL4A1 and COL4A2 mutations and disease: insights into pathogenic mechanisms and potential therapeutic targets. Hum. Mol. Genet 2012;21:R97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang CE, Wong SM, Van De Haar HJ, Staals J, Jansen JFA, Jeukens CRLPN, et al. Blood-brain barrier leakage is more widespread in patients with cerebral small vessel disease. Neurology. 2017;88:426–432. [DOI] [PubMed] [Google Scholar]
- 36.French CR, Seshadri S, Destefano AL, Fornage M, Arnold CR, Gage PJ, et al. Mutation of FOXC1 and PITX2 induces cerebral small-vessel disease. J. Clin. Invest 2014;124:4877–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chauhan G, Arnold CR, Chu AY, Fornage M, Reyahi A, Bis JC, et al. Identification of additional risk loci for stroke and small vessel disease: a meta-analysis of genome-wide association studies. The Lancet Neurology. 2016;15:695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stam AH, Kothari PH, Shaikh A, Gschwendter A, Jen JC, Hodgkinson S, et al. Retinal vasculopathy with cerebral leukoencephalopathy and systemic manifestations. Brain. 2016;139:2909–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice GI, Rodero MP, Crow YJ. Human disease phenotypes associated with mutations in TREX1. J. Clin. Immunol 2015;35:235–243. [DOI] [PubMed] [Google Scholar]
- 40.Kamp JA, Moursel LG, Haan J, Terwindt GM, Lesnik Oberstein SAMJ, Van Duinen SG, et al. Amyloid β in hereditary cerebral hemorrhage with amyloidosis-Dutch type. Reviews in the Neurosciences. 2014;25:641–51. [DOI] [PubMed] [Google Scholar]
- 41.Biffi A, Plourde A, Shen Y, Onofrio R, Smith EE, Frosch M, et al. Screening for familial APP mutations in sporadic cerebral amyloid angiopathy. PLoS ONE. 2010;5:e13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. American Journal of Human Genetics. 2014;94:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jerrard-Dunne P, Cloud G, Hassan A, Markus HS. Evaluating the genetic component of ischemic stroke subtypes: A family history study. Stroke. 2003;34:1364–9. [DOI] [PubMed] [Google Scholar]
- 44.Marini S, Crawford K, Morotti A, Lee MJ, Pezzini A, Moomaw CJ, et al. Association of Apolipoprotein E With Intracerebral Hemorrhage Risk by Race/Ethnicity: A Meta-analysis. JAMA Neurol. 2019;76:480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traylor M, Malik R, Nalls MA, Cotlarciuc I, Radmanesh F, Thorleifsson G, et al. Genetic variation at 16q24.2 is associated with small vessel stroke. Annals of neurology. 2017;81:383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams HP, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of Subtype of Acute Ischemic Stroke. Stroke. 1993;24:35–41. [DOI] [PubMed] [Google Scholar]
- 47.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature Genetics. 2009;41:334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natarajan P, Bis JC, Bielak LF, Cox AJ, Dörr M, Feitosa MF, et al. Multiethnic Exome-Wide Association Study of Subclinical Atherosclerosis. Circulation: Cardiovascular Genetics. 2016;9:511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jian X, Satizabal CL, Smith AV, Wittfeld K, Bis JC, Smith JA, et al. Exome Chip Analysis Identifies Low-Frequency and Rare Variants in MRPL38 for White Matter Hyperintensities on Brain Magnetic Resonance Imaging. Stroke. 2018;49:1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malik R, Traylor M, Pulit SL, Bevan S, Hopewell JC, Holliday EG, et al. Low-frequency and common genetic variation in ischemic stroke: The METASTROKE collaboration. Neurology. 2016;86:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traylor M, Bevan S, Baron J-C, Hassan A, Lewis CM, Markus HS. Genetic Architecture of Lacunar Stroke. Stroke. 2015;46:2407–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fornage M, Debette S, Bis JC, Schmidt H, Ikram MA, Dufouil C, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Annals of neurology. 2011;69:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson CD, Biffi A, Rahman R, Ross OA, Jagiella JM, Kissela B, et al. Common mitochondrial sequence variants in ischemic stroke. Annals of Neurology. 2011;69:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traylor M, Tozer DJ, Croall ID, Lisiecka Ford DM, Olorunda AO, Boncoraglio G, et al. Genetic variation in PLEKHG1 is associated with white matter hyperintensities (n = 11,226). Neurology. 2019;92:e749–e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falcone GJ, Biffi A, Devan WJ, Jagiella JM, Schmidt H, Kissela B, et al. Burden of risk alleles for hypertension increases risk of intracerebral hemorrhage. Stroke. 2012;43:2877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris SE, Malik R, Marioni R, Campbell A, Seshadri S, Worrall BB, et al. Polygenic risk of ischemic stroke is associated with cognitive ability. Neurology. 2016;86:611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Traylor M, Adib-Samii P, Harold D, Initiative ADN, International Stroke Genetics Consortium (ISGC) UKYLSDNA resource, Dichgans M, et al. Shared genetic contribution to Ischaemic Stroke and Alzheimer’s Disease. Annals of neurology. 2016;79:739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS genetics. 2014;10:e1004606–e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H, et al. Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann. Neurol 2010;68:934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schilling S, DeStefano AL, Sachdev PS, Choi SH, Mather KA, DeCarli CD, et al. APOE genotype and MRI markers of cerebrovascular disease: systematic review and meta-analysis. Neurology. 2013;81:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khera A, Chaffin M, Zekavat S, Collins R, Roselli C, P N, et al. Whole Genome Sequencing to Characterize Monogenic and Polygenic Contributions in Patients Hospitalized with Early-Onset Myocardial Infarction. Circulation. 2019;139:1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bersano A, Bedini G, Oskam J, Mariotti C, Taroni F, Baratta S, et al. CADASIL: Treatment and Management Options. Curr Treat Options Neurol. 2017;19:31. [DOI] [PubMed] [Google Scholar]