Abstract
TRP channels function in many types of sensory receptor cells. Despite extensive analyses, an open question is whether there exists a family of auxiliary subunits, which could influence localization, trafficking, and function of TRP channels. Here, using Drosophila melanogaster, we reveal a previously unknown TRP interacting protein, INAF-C, which is expressed exclusively in the ultraviolet-sensing R7 photoreceptor cells. INAF-C is encoded by an unusual locus comprised of four distinct coding regions, which give rise to four unique single-transmembrane-containing proteins. With the exception of INAF-B, roles for the other INAF proteins were unknown. We found that both INAF-B and INAF-C are required for TRP stability and localization in R7 cells. Conversely, loss of just INAF-B greatly reduced TRP from other types of photoreceptor cells, but not R7. The requirements for TRP and INAF are reciprocal, since loss of TRP decreased the concentrations of both INAF-B and INAF-C. INAF-A, which is not normally expressed in photoreceptor cells, can functionally substitute for INAF-B, indicating that it is a third TRP auxiliary protein. Reminiscent of the structural requirements between Kv channels and KCNE auxiliary subunits, the codependencies of TRP and INAF depended on several transmembrane domains (TMDs) in TRP, and the TMD and the C-terminus of INAF-B. Our studies support a model in which the inaF locus encodes a family of at least three TRP auxiliary subunits.
Keywords: Drosophila melanogaster, TRP channel, inaF, β subunit, photoreceptor cells, phototransduction
DROSOPHILA TRP is the founding member of a large family of evolutionarily conserved cation channels (Montell and Rubin 1989; Hardie and Minke 1992), which have many critical roles, including broad roles in sensory reception (Venkatachalam and Montell 2007). TRP channels enable sensory cells to detect stimuli ranging from light to tastants, auditory cues, thermal stimuli, and others (Venkatachalam and Montell 2007). In fly photoreceptor cells, TRP, and a highly related protein, TRPL, culminate a signaling cascade that is initiated by light activation of rhodopsins, and engages a trimeric G-protein (Gq), thereby stimulating a phospholipase C (PLC) (Montell and Rubin 1989; Hardie and Minke 1992; Phillips et al. 1992; Niemeyer et al. 1996; Montell 2012).
Signaling cascades similar to the one used in fly photoreceptor cells are employed in multiple sensory cells in mammals. A virtually identical phototransduction cascade functions in mammalian intrinsically photosensitive retinal ganglion cells (Berson et al. 2002), which depend on related TRPC channels (Xue et al. 2011). Sweet, bitter, and umami tastes in mammalian taste receptor cells function through a cascade that is initiated by G-protein coupled receptors and engagement of Gq, PLC, and two TRPM channels (Liman et al. 2014; Dutta Banik et al. 2018). In many mammals, the detection of pheromones through the vomeronasal organ is mediated by a TRPC-dependent cascade (Leypold et al. 2002; Stowers et al. 2002; Lucas et al. 2003). TRP channels that are activated through GPCR, Gq, and PLC signaling are also used in many other cells and organs, including the brain (Nilius and Szallasi 2014).
High-resolution structures of mammalian TRP channels reveal that they consist of four pore-forming subunits, each with six transmembrane domains (TMDs) (Cao et al. 2013; Vangeel and Voets 2019; Pumroy et al. 2020; Wang et al. 2020). The structures of TRP channels are reminiscent of voltage-gated K+ channels (Kv) of the Shaker family, which consist of four α subunits with six TMDs each (Long et al. 2005). These Kv α subunits associate with small β subunits, which include one TMD (Abbott 2016a). Auxiliary subunits have a diversity of effects on the channels, including trafficking, subunit assembly, and channel stability and impact the biophysical properties of the channels (Li et al. 2006; Abbott 2016a). Interactions between the α and β subunits occur between multiple TMDs of the α subunit, and the TMD and C-terminus of the β subunit (Abbott 2016a).
Despite extensive studies on TRP channels from organisms ranging from worms to mammals, it is unclear whether there exists a family of TRP auxiliary subunits. Based on the Kv channels, excellent candidates include proteins with one TMD, and which interact with the TMDs of TRP channels through the TMD and C-termini of the candidate auxiliary subunits (Abbott 2016a). TRP auxiliary subunits should also be coexpressed with TRP in the microvillar portion of the photoreceptor cells, the rhabdomeres, where phototransduction takes place.
In this work, we identified Drosophila INAF-C—a previously unknown TRP interacting protein, with one predicted TMD. The compound eye includes ∼800 ommatidia, each with eight photoreceptor cells (Montell 2012). Remarkably, INAF-C and TRP associate in just the ultraviolet-sensing R7 cells. Flies encode four distinct INAF proteins, one of which, INAF-B, has previously been shown to impact on the concentration of TRP (Li et al. 1999; Cheng and Nash 2007). We demonstrate that INAF-B and INAF-C are both required for TRP localization and stability in R7 cells, while loss of INAF-B only causes a dramatic reduction in TRP in the other photoreceptor cells. The dependence is reciprocal, since loss of TRP causes instability of INAF-B and INAF-C. The mutual requirements for TRP and INAF depend on domains reminiscent of the domains required for interactions between α and β subunits of Kv channels. We propose that INAF proteins comprise a set of TRP auxiliary proteins. Related INAF proteins are encoded in mammals, suggesting that INAF represents a family of evolutionarily conserved TRP auxiliary proteins.
Materials and Methods
Fly stocks
inaFP106x was obtained from William Pak (Li et al. 1999), otduvi was from Claude Desplan (Tahayato et al. 2003), and the ER-150 transgenic flies were from Roger Hardie (Liu et al. 2020). We obtained the following stocks from the Bloomington Stock Center: trpMB03672 (trpMB; stock 23636) and trplMB10553 (trplMB; stock 29134). trp343 (Montell and Rubin 1989), trpl302 (Niemeyer et al. 1996), rh1I17 (O’Tousa et al. 1985), norpAP24 (Bloomquist et al. 1988), inaD1 (Tsunoda et al. 1997), and boss1 (Reinke and Zipursky 1988) were described previously.
Purification of SBP::TRP and identification of TRP-interacting proteins by mass spectrometry
Transgenic flies that express SBP::TRP in all photoreceptor cells (P[SBP::trp]) were generated previously (Chen et al. 2015). We introduced this transgene in a trp343 background, and affinity purified SBP::TRP, according to methods we used to purify XPORT-B::SBP (Chen et al. 2015) with minor modifications. Briefly, we homogenized heads from 10 g w1118 and P[SBP::trp];trp343 flies in extraction buffer [100 mM Tris∙HCl pH 8, 150 mM NaCl, 1 mM EDTA, cOmplete protease inhibitor (Catalog# 11697498001; Roche)]. After pelleting the membranes, we suspended the pellets in extraction buffer containing 0.2% Triton X-100 at 4° for 2 hr. For affinity purification, we equilibrated and washed columns of Strep-Tactin Superflow plus beads (Cat no./ID 30004; Qiagen) with extraction buffer containing 0.2% Triton X-100, and eluted with extraction buffer containing 0.2% Triton X-100 plus 2.5 mM desthiobiotin. The second elutant fraction was concentrated using an Amicon Ultracel-3K centrifuge filter. The mass spectrometry to identify peptides released by trypsin digestion of TRP-interacting proteins was performed at the Johns Hopkins Mass Spectrometry and Proteomics Facility (https://msf.johnshopkins.edu).
Homologous recombination to generate new inaF alleles
We created the inaFΔA, inaFΔB, inaFΔC, inaFΔBC, and inaFΔD alleles by ends-out homologous recombination (Gong and Golic 2003). The mutations deleted the exons illustrated in Figure 1F. The nucleotides specifying the coding regions of each isoform are provided, following by nucleotides that were deleted to generate the indicated alleles: inaFΔA (A coding region 1–270; deleted −9 to 336), inaFΔB (B coding region 1–246; deleted −550 to 345), inaFΔC (C coding region 1–423; deleted −313 to 965), inaFΔBC (B coding region 1–246; deleted −550 to 1981), inaFΔD (coding region of the first inaF-D exon: 1–226; deleted −230 to 277), and inaFΔABD (A coding region 1–270; deleted −1179 to 1253, which also deletes 1 to 21 of the B coding region, and combines the same deletion as in inaFΔD). The plasmids for making donor lines to create the deletions by ends-out homologous recombination were made by inserting PCR amplified genomic DNA fragments flanking the target deletions into pW35 vector. Two loxP sites were introduced during cloning so that the white marker gene could be subsequently floxed out. Knockout flies were verified by PCR, and the white marker was excised by genetically introducing Cre recombinase (1092; Bloomington Stock), leaving a 34-bp loxP site (ATAACTTCGTATAATGTATGCTATACGAAGTTAT). The deletions in inaFΔA, inaFΔB, inaFΔC, inaFΔBC, and inaFΔD were 0.34, 0.90, 1.28, 2.53, and 0.51 kb, respectively. The two deletions in inaFΔABD were 2.43 and 0.51 kb, respectively. All of the deletions were confirmed by sequencing PCR-amplified genomic DNA from the mutant alleles.
Figure 1.
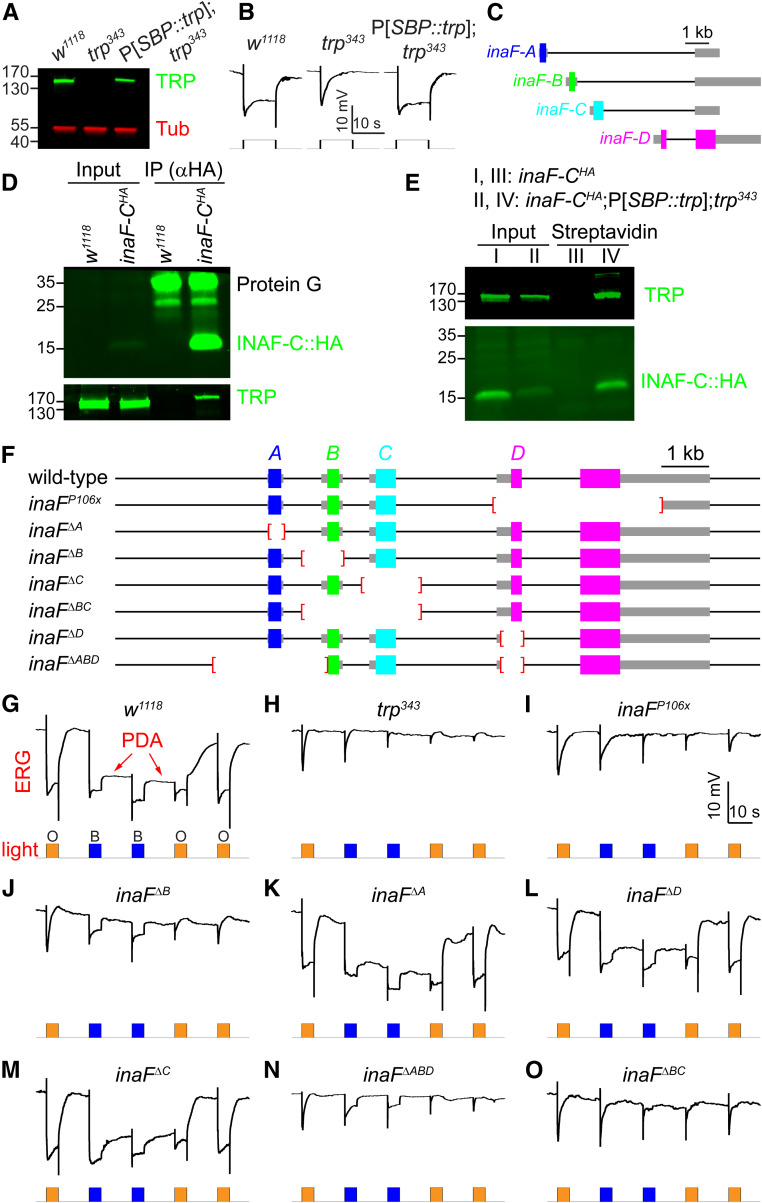
INAF-C associates with TRP, and contributes to the light response. (A) Expression level of TRP in P[SBP::trp] transgenic flies. Western blot showing the expression levels of TRP in head extracts prepared from w1118 and P[SBP::trp];trp343 flies. The blot was probed with rabbit anti-TRP and mouse anti-tubulin (Tub). Protein size markers (kDa) are indicated. (B) Representative ERG responses to orange light using the indicated flies; n ≥ 5 for each genotype. Time and mV scale bars are presented. (C) The four inaF mRNAs encode distinct proteins: INAF-A, INAF-B, INAF-C, and INAF-D. The colored rectangles indicate the protein-coding regions, the thick gray lines represent untranslated regions, and the thin black lines indicate introns. A DNA scale bar is provided. (D) Co-IPs to assess whether TRP associates with INAF-C::HA. Anti-HA (mouse) was used to perform IPs using head extracts prepared from w1118 and inaF-CHA, which harbors an HA tag knocked into the C terminus of INAF-C. The IPs were fractionated by SDS-PAGE, and Western blots probed with rabbit anti-HA (top; recognizes INAF-C::HA) or rabbit anti-TRP (bottom) primary antibodies and LI-COR anti-rabbit IRDye 800CW secondary antibodies. The positions of protein size markers (kDa) are shown to the left. The rabbit anti-HA also reacted with Protein G, which was released from Protein G beads upon elution. (E) Pulldown assays to determine whether INAF-C associates with TRP. Head extracts were prepared from inaF-CHA and inaF-CHA;P[SBP::trp];trp343 flies, the latter of which expresses the SBP::trp transgene in a trp343 null background. Head extracts were incubated with streptavidin beads, and the eluted proteins (streptavidin lanes) were fractionated by SDS-PAGE. The Western blots were probed with rabbit anti-TRP (top) and rabbit anti-HA (bottom) to identify TRP and INAF-C::HA, respectively. The input represents 4% of the extracts offered in the assays (D and E), and protein size markers (kDa) are shown to the left. (F) Wild-type inaF and inaF mutant alleles. Deletions are indicated by red brackets. (G–O) Representative ERG responses from the indicated genotypes (n ≥ 5 for each). A prolonged depolarizing afterpotential (PDA) is indicated in (G). Orange (O) and blue light (B) stimuli (5 sec/pulse) are indicated. Time and mV scales are indicated.
Generation of inaFΔABD
To create inaFΔABD flies, we used CRISPR/Cas9 and nonhomologous end joining (NHEJ) to introduce a mutation affecting inaF-A and inaF-B in an inaFΔD background. To do so, we first generated transgenic flies harboring an inaF-B gRNA targeting a sequence near the translation initiation site of inaF-B using the pCFD3 vector (Port et al. 2014): 5′ GAGCGGACCGTCGGCACTGA TGG 3′ (PAM sequence is underlined), and the inaF-B gRNA transgene was integrated into the attp40 site by PhiC31 integrase-mediated transgenesis (BestGene). We combined the inaF-B gRNA transgene with a genetically encoded Cas9 (vas-Cas9 VK00027; http://flybase.org/reports/FBti0154822.html), and crossed the two components into the inaFΔD background.
We screened 11 individual lines by performing electroretinograms (ERGs). Four lines exhibited a transient response to orange light, and a sustained response to bright blue light. The other seven lines showed normal responses to orange light and normal prolonged depolarizing afterpotential (PDA) upon exposure to bright blue light. We genotyped two lines with altered ERGs. One had a small indel (8 bp deletion and 3 bp insertion) and the other had a 2432 bp deletion. We named this latter allele inaFΔABD (Figure 1F) because its deletion completely removed inaF-A (A coding region 1–270; deleted −1179 to 1253) and the 5′ end of inaF-B (B coding region 1–246; deleted up to +21).
Generating inaF-AHA, inaF-BHA, inaF-CHA and inaF-DHA-P2A-QF knock-in flies
To insert the sequences encoding a hemagglutinin (HA) tag (YPYDVPDYA) or an HA-P2A-QF cassette (encoding an HA tag, P2A peptide, and QF2) in frame, immediately 5′ to the stop codon of the target gene, we used the pHD-ScarlesDsRed vector [Drosophila Genomics Resource Center (DGRC) #1364) as the donor plasmid to perform CRISPR/Cas9-mediated scarless genomic editing (http://flycrispr.molbio.wisc.edu/scarless). After verifying the intended homology-directed repair events, the 3×P3-DsRed marker gene was removed by genetically introducing a PBac transposase. After removing the 3×P3-DsRed, the knock-in was confirmed by sequencing PCR-amplified genomic DNA from the respective knock-in flies.
Generating transgenic flies expressing QF under control of the inaF-C promoter
To express QF under control of the inaF-C transcriptional control region, we prepared genomic DNA from w1118 flies, and PCR amplified a 0.8 kb genomic fragment from the region flanking the 5′ end of inaF-C using the following primers: 5′TTATGCTAGCGGATCCGATCGGATGGCTATCATTTAGTTAGCC3′ and 5′CGGCATGTTGGAATTCTACTGCGGATATGTACTTTTCTGGTCG3′). We inserted the inaF-C 5′ flanking genomic DNA into the pattB-QF-hsp70 vector (Potter et al. 2010), introduced the transgenes at the ZH-86Fb site (Bischof et al. 2007) by PhiC31 integrase-mediated transgenesis (BestGene), and removed the white marker gene with a genetically encoded Cre recombinase (1501; Bloomington Drosophila Stock Center) to create the wild-type transgene inaF-C•QF. The CC→TA mutation and the 6-nt deletion (Δ6) in the RCSI-like element of inaF-C (Figure 3C) were introduced by PCR-mediated mutagenesis. Transgenic flies expressing QF under control of the 0.8-kb inaF-C promoter with the CC→TA or Δ6 mutations in the RCSI (inaF-CCC→TA•QF and inaF-CΔ6•QF, respectively) were generated as described above for producing wild-type inaF-C•QF. The transgenic lines were combined with a 10×QUAS-6×GFP inserted into the attp2 site (Shearin et al. 2014). Prior to conducting these genetic crosses, the white marker in the 10×QUAS-6×GFP transgene was mutated by CRISPR/Cas9 (Liu et al. 2020) so that the animals were white eyed (P[w-,10×QUAS-6×GFP]). Flies that were trans-heterozygous for one of the three inaF-C•QF transgenes and the P[w-,10×QUAS-6×GFP] transgene were costained with anti-GFP and anti-Rh1.
Figure 3.
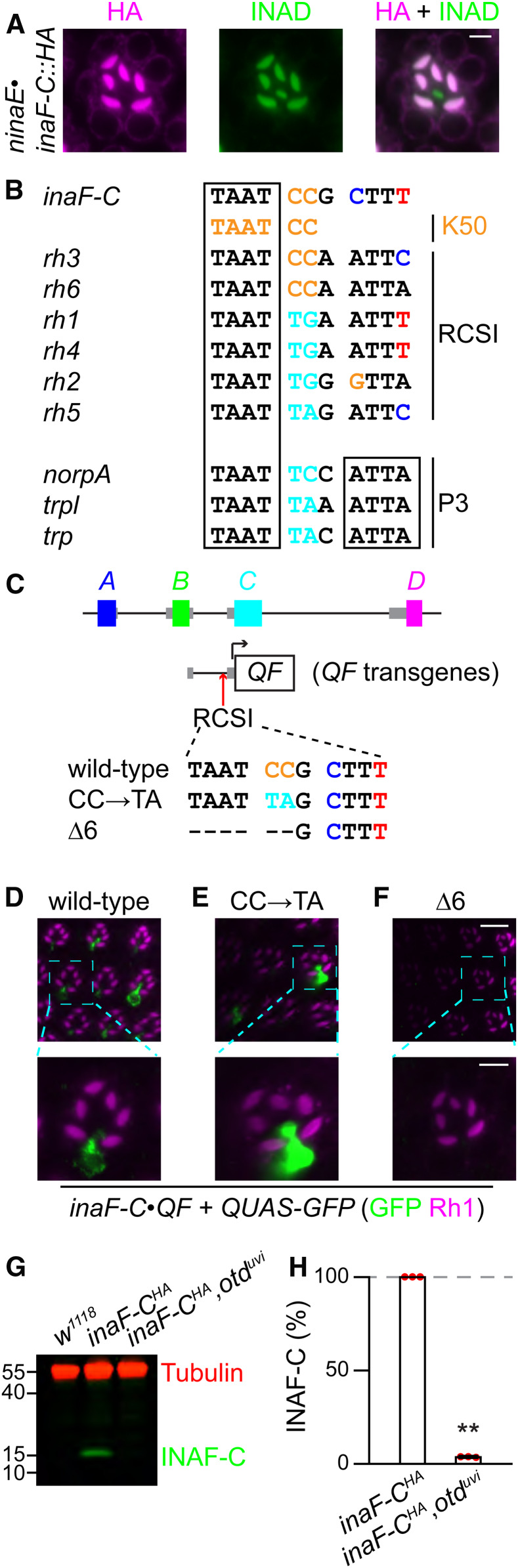
Expression of inaF-C in R7 cells depends on the rhodopsin core sequence I (RCSI). (A) An ommatidium (R7 layer) from flies expressing inaF-C::HA under control of the ninaE promoter (ninaE•inaF-C::HA) were costained with mouse anti-HA (magenta) and rabbit anti-INAD (green); bar, 3 μm. (B) Comparison between the RCSI elements in the promoter of rhodopsins, the palindromic P3 motif in several broadly expressed photoreceptor genes (norpA, trpl, and trp), and the K50 motif in the promoters of inaF-C, rh3, and rh6. (C) Mutations in the inaF-C RCSI sequence introduced into an 0.8 kb region of the inaF-C promoter (−770 to −1, relative to the translation start codon). The three inaF-C promoter versions were fused directed to the coding region of QF and used to generate transgenic flies. (D–F) The number of R7 cells expressing INAF-C is reduced or eliminated by mutations in the inaF-C RCSI sequence. QF expressed under control of either the wild-type or mutated inaF-C promoter sequences (inaF-C•QF) was used to drive QUAS-GFP. Shown are optical sections of compound eyes from the indicated flies stained with anti-GFP (Green) and anti-Rh1 (magenta). The scale bars in the upper and lower panels indicate 10 and 4 μm, respectively. (D) inaF-Cwild-type•QF. (E) inaF-CCC→TA•QF. (F) inaF-CΔ6•QF. (G) Extracts from 0.5 head equivalents from the indicated flies were fractionated by SDS-PAGE, and Western blots were probed with mouse anti-tubulin (loading control) and rabbit anti-HA (recognizes INAF-C::HA). Protein size markers are indicated (kDa). (H) Quantification of INAF-C levels obtained from Western blot analyses represented in (G).
Creating transgenic flies expressing TRP-TRPL chimeras
We previously generated flies expressing TRP-TRPL chimeras I–V (Chen et al. 2015). To produce transgenic flies expressing TRP-TRPL chimera VI–IX, each with an N-terminal His-SBP tag and expressed under the transcriptional control of ninaE, we first created the pattB[ninaE•His-SBP] vector. To do so, we subcloned into the pattB vector (Bischof et al. 2007) a 2.96 kb DNA fragment (nucleotide: −2963 to −1) 5′ of the ninaE translational start codon together with the sequences coding for the His-SBP tag, and a 0.70-kb DNA fragment (nucleotide: 1587 to 2288) 3′ of the ninaE stop codon (nucleotide: 1484–1486). We then subcloned DNA sequences encoding the following TRP/TRPL chimeras so that they were in frame with the N-terminal His-SBP tag: VI, TRP (1–328)-TRPL (336–423)-TRP (417–1275); VII, TRP (1–416)-TRPL (424–463)-TRP (457–1275); VIII, TRP (1–456)-TRPL (464–555)-TRP (549–1275); IX, TRP (1–548)-TRPL (556–671)-TRP (665–1275). After verifying the constructs by DNA sequencing, the VI–IX transgenes were integrated into the ZH-86Fb attp docking site (BestGene) and crossed into trpl302;trpMB and inaFP106x;trpl302;trpMB mutant backgrounds. The transgenes encoding the I–V chimeras, which were previously established in a trplMB;trpMB background (both trplMB and trpl302 are null alleles) (Chen et al. 2015), were crossed into an inaFP106x background so that each Chimera I–V was expressed in an inaFP106x;trplMB;trpMB background. The concentrations of the TRP/TRPL chimeras were then compared in the double and triple mutant backgrounds (without and with the inaFP106x mutation) to determine the effects of the INAF proteins on levels of these chimeras.
Transgenic flies expressing inaF under control of the ninaE promoter
To generate ninaE•inaF-A/B/C/D and ninaE•inaF-C::HA transgenic flies, the coding regions of inaF-A, inaF-B, inaF-C, inaF-D, and inaF-C::HA were expressed under control of the ninaE promoter by subcloning the inaF sequences into a P element vector with ninaE promoter so that the coding regions were 3′ to the ninaE transcriptional control region. The transgenic flies were generated by P-element-mediated transformation (BestGene) in a w1118 background. The w+ marker served to identify the transgenic flies. We created a white-eyed version of ninaE•inaF-C::HA by mutating the white marker using the CRISPR/Cas9-based “white eraser” (Liu et al. 2020).
To generate the following inaF constructs, we first subcloned a 2.96 kb ninaE promoter region (nucleotides −2963 to −1, which were 5′ to the translational start codon of ninaE) together with 0.70 kb DNA of ninaE 3′ fragment (nucleotide: 1587 to 2288) into the pattB vector (Bischof et al. 2007) to create the pattB[ninaE] vector. We then subcloned sequences encoding inaF-B::HA and inaF-D::HA into pattB[ninaE]. We also subcloned the DNA sequences encoding the following INAF-B/INAF-D chimeras fused to HA tags at the C termini (amino acid indicated in parentheses) into pattB[ninaE]: (1) BD1::HA consists of INAF-B (1–64)-INAF-D (62–353)-HA tag, (2) BD2::HA consists of INAF-D (1–61)-INAF-B (65–81)-HA tag, and (3) BD3::HA consists of INAF-D (1–38)-INAF-B (42–81)-HA tag. The transgenic flies (generated by BestGene) contained insertions introduced into the ZH-51C attp docking site (Bischof et al. 2007). The white marker used to identify the transgenes was removed by genetically introducing a transgenic copy of Cre recombinase.
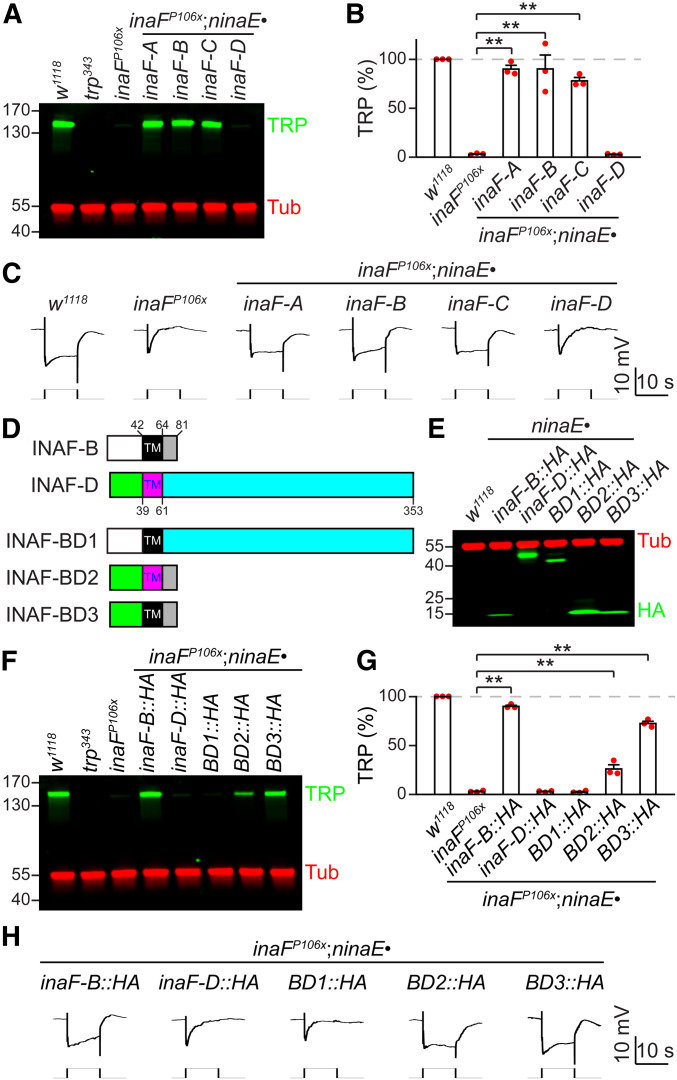
All transgenes were analyzed in flies that were homozygous for the insertions, except in Figure 7, F–H in which the male inaFP106x flies were tested with only one copy of the transgenes inserted into the ZH-51C attp site on the second chromosome.
Figure 7.
Mapping domains in INAF-B critical for TRP expression. (A–C) Testing whether INAF-A, C, or D can substitute for INAF-B in R1-R6 cells. Each inaF coding region was expressed under the control of the ninaE promoter in an inaFP106xmutant background (inaFP106x;ninaE•inaF-A, B, C or D). (A) Head extracts were prepared from the indicated flies and a Western blot was probed with rabbit anti-TRP and mouse anti-tubulin (Tub, loading control). Protein size markers (kDa) are shown. (B) Quantification of the relative TRP level in the indicated flies, based on Western blot analyses represented in (A); n = 3. (C) ERG responses to orange light in the indicated flies; n ≥ 5 for each genotype. (D) Cartoons illustrating INAF-B/INAF-D chimeras. TM indicates the distinct single transmembrane segments in INAF-B and INAF-D. The numbers indicate amino acid residues. The white and green rectangles represent the N-termini of INAF-B and INAF-D. The gray and turquoise rectangles stand for the C-termini of INAF-B and INAF-D. (E) Western blot showing expression of INAF-B/INAF-D (BD) chimeras. Head extracts were prepared from flies expressing the indicated transgenes under control of the ninaE promoter, and fractionated by SDS-PAGE. The blot was probed with rabbit anti-HA (recognizes INAF-B::HA, INAF-D::HA and INAF-B/INAF-D: BD1, BD2 and BD3) and mouse anti-Tub. The positions of protein size markers (kDa) are indicated. (F–H) Determining the domain(s) in INAF-B essential for its function. (F) Head extracts were prepared from inaFP106x flies expressing the indicated inaF chimeric transgenes under control of the ninaE promoter, and fractionated by SDS-PAGE. A Western blot was probed with rabbit anti-TRP and mouse anti- Tub. The positions of protein size markers (kDa) are indicated. (G) Quantification of Western blot data represented in (F); n = 3. Error bars represent SEMs; **P < 0.01 (one-way ANOVA with Holm–Sidak post hoc analyses). (H) ERG responses to orange light in the indicated flies; n ≥ 5 for each genotype.
Sources of antibodies
Mouse anti-Rh3 (2B1) and mouse anti-Rh4 (11E6) (Chou et al. 1999) were provided by Steve Britt (University of Texas at Austin). Rabbit polyclonal anti-TRPL (Niemeyer et al. 1996) was provided by Charles Zuker (Columbia University). Rabbit polyclonal anti-Rh1 (Satoh et al. 2005) was provided by Donald Ready (Purdue University). We purchased the following antibodies from the companies indicated: rabbit anti-HA (715500; Invitrogen), mouse anti-HA (H3663; Sigma), chicken anti-GFP (A10262; Invitrogen), rabbit anti-Actin (ab1801; Abcam), mouse anti-SBP tag (sc-101595; Santa Cruz Biotechnology), mouse anti-Rh1 (4C5; DSHB) and mouse anti-Tubulin (12G10; DSHB). We previously described anti-NORPA (Wang et al. 2005), anti-INAD (Wes et al. 1999), and anti-TRP (Chevesich et al. 1997). Goat anti-mouse IRDye 680LT (LI-COR 926–68020) and Donkey anti-rabbit IRDye 800CW (LI-COR 926–32213) were used as the secondary antibodies for Western-blots, after using mouse and rabbit primary antibodies, respectively. To perform the whole-mount immunostaining, we used the following secondary antibodies (Invitrogen) in combination with the primary antibodies indicated in parentheses: Alexa Fluor 488 A11001 (mouse anti-HA), Alexa Fluor 488 A11008 (rabbit anti-HA and rabbit anti-TRP), Alexa Fluor 488 A11034 (rabbit anti-TRP), Alexa Fluor 488 A11039 (chicken anti-GFP), Alexa Fluor 568 A11004 (mouse anti-HA, mouse anti-Rh1, mouse anti-Rh3 and mouse anti-Rh4), Alexa Fluor 568 A11036 (rabbit anti-TRP), and Alexa Fluor 633 A21070 (rabbit anti-TRP and rabbit anti-INAD).
Immunoprecipitation and pulldown assays
Immunoprecipitations using anti-HA dynabeads:
To determine whether TRP co-immunoprecipitates with INAF-C, we used knock-in flies expressing INAF-C::HA to perform co-immunoprecipitations (co-IPs) with anti-HA. We homogenized 240 fly heads from w1118 (negative control) or inaF-CHA in 1440 μl extraction buffer [100 mM Tris∙HCl pH 8, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, cOmplete protease inhibitor (Catalog# 11697498001; Roche)]. After rotation at 4° overnight, the homogenates were centrifuged three times at 12,000 × g for 10 min, and the supernatants were retained.
To attach anti-HA to Dynabeads Protein G (Catalog# 10003D; Invitrogen), we first transferred 100 μl (∼3 mg) of the beads for each immunoprecipitation (IP) in individual tubes. The Dynabeads were pelleted using a DynaMag-2 magnet (Catalog# 12321D; Invitrogen) to remove the supernatants, and the beads were resuspended in 400 μl extraction buffer. The Dynabeads were then pelleted again on a DynaMag-2 to remove the supernatants, and 400 μl extraction buffer together with 10 μl anti-HA (Catalog# 1H3663, ∼10 μg; Sigma) were added to resuspend the beads. The tubes were rotated at room temperature for 10 min, and the beads were pelleted using a DynaMag-2 to remove the supernatants. The beads were then resuspended in 400 μl extraction buffer, pelleted using a DynaMag-2, and the supernatants were removed.
To perform the IPs, we added 1.2 ml of fly head extracts (equivalent to 200 heads) to the pelleted anti-HA-Dynabeads, and rotated the tubes at room temperature for 30 min. The anti-HA-Dynabeads were pelleted using a DynaMag-2 to remove the supernatants. After three rounds of washes with 400 μl extraction buffer, the beads were resuspended in 200 μl extraction buffer, transferred to a new tube, and the beads were pelleted using the DynaMag-2. The supernatants were removed, and 60 μl 2× Laemmli sample buffer was added to each tube to resuspend the beads. After heating at 70° for 10 min to release the interacting proteins from the beads, the beads were pelleted using the DynaMag-2. The supernatants were retained and used to perform Western blots, and the signals were detected using a LI-COR Odyssey system.
Streptavidin pulldown assays:
To determine whether INAF-C associates with TRP, we used TRP tagged with the streptavidin-binding peptide (SBP::TRP), so that we could pulldown the complex using Dynabeads MyOne Streptavidin T1 (Catalog# 65601; Invitrogen). We prepared head extracts as described above from inaF-CHA (negative control) and inaF-CHA;P[SBP::trp];trp343 flies.
To prepare the Dynabeads MyOne Streptavidin T1 (streptavidin beads) for the assays, we transferred 200 μl of the beads (2 mg) to each tube, added 1 ml of extraction buffer and 0.1% γ-globulins (Catalog#G5009; Sigma), pelleted the streptavidin beads using the DynaMag-2 for 2 min, and removed the supernatants. The beads were washed three times with 200 μl of extraction buffer and 0.1% γ-globulins.
To perform the pulldowns, the streptavidin beads were resuspended in 1.2 ml head extracts (equivalent to 200 heads), and the tubes were rotated at room temperature for 30 min. The beads were pelleted for 3 min using the DynaMag-2, the supernatants were removed, and the beads were washed four times with 400 μl extraction buffer and 0.1% γ-globulins. The beads were resuspended in 200 μl of extraction buffer, transferred to a new tube, pelleted for 3 min using the DynaMag-2, and the supernatants were removed. The beads were resuspended in 60 μl 2× Laemmli sample buffer, heated at 70° for 10 min, and pelleted using a DynaMag-2 for 3 min. The supernatants containing the released streptavidin pulldown complex were retained for Western blots.
To perform the Western blots, we first performed SDS-PAGE by loading 12 μl of head extracts (equivalent to two heads) as the “input,” and loaded 15 μl of the IP products or the streptavidin pulldown products (equivalent to ∼50 heads). Thus, the input represented ∼4% of the samples used in the assays. Rabbit anti-TRP polyclonal antibodies and rabbit anti-HA (Catalog# 715500; Invitrogen) were used for blotting TRP and HA respectively. The IP products common to the w1118 and inaF-CHA samples (between 25 and 35 kDa; Figure 1D) appeared to correspond to the protein G because we detected similar bands even when we eluted blank Dynabeads Protein G (Catalog# 10003D; Invitrogen) with 2× Laemmli sample buffer.
Western blots
The Western blots to assay protein levels directly from head extracts (not from IPs or pulldowns), were performed essentially as described previously (Chen et al. 2015). Briefly, protein extracts equivalent to 0.5 head from 1-day-old flies of indicated genotypes were fractionated by SDS-PAGE, Western blots were probed with the indicated primary antibodies and secondary antibodies, and the results were visualized and quantified using a LI-COR Odyssey or Odyssey CLx system. We quantified protein levels in mutant and transgenic flies relative to w1118 or the indicated controls. We normalized the levels of the indicated proteins by probing the same blots with either anti-tubulin or anti-actin. For Rh1 (Figure 5, G and H), we normalized the level of Rh1 to a ∼60 kDa nonspecific protein recognized by rabbit anti-Rh1. Quantification was based on three independent replicates of the Western blots.
Figure 5.
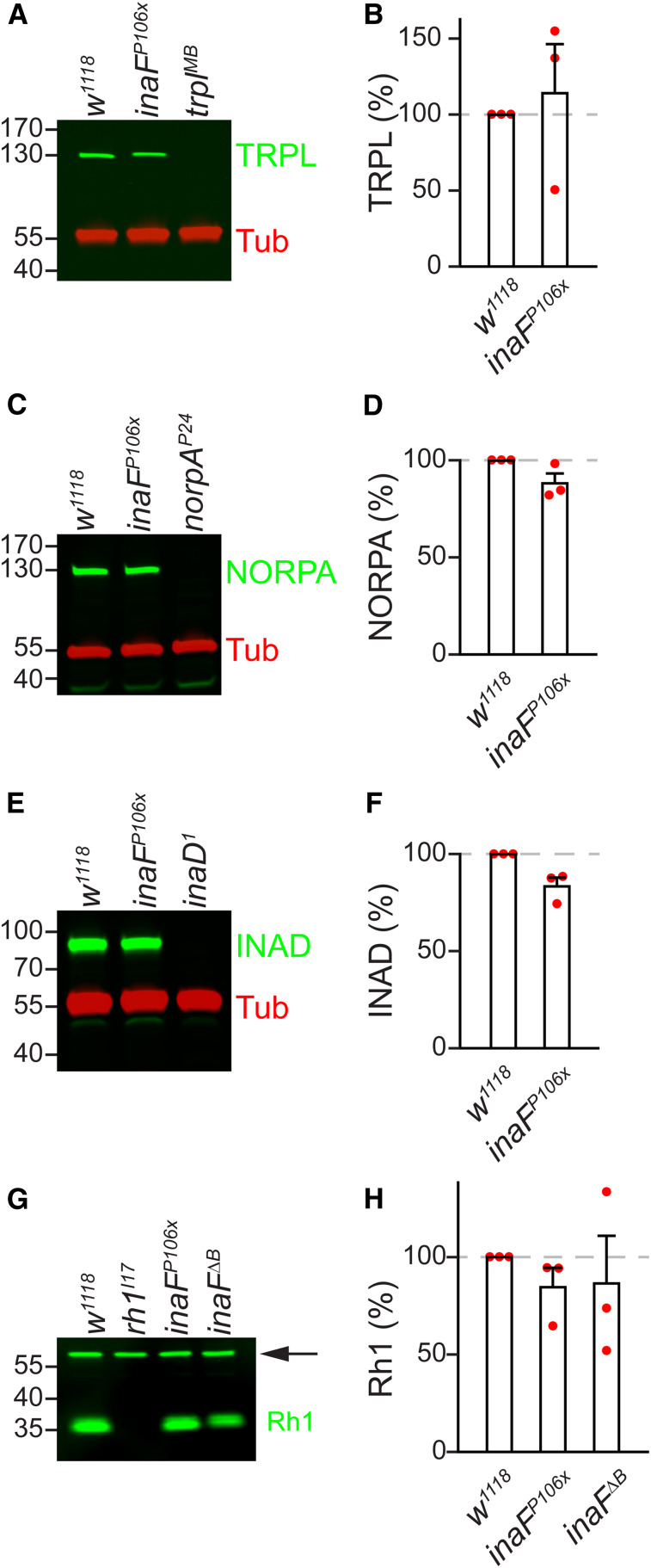
TRPL, NORPA, INAD, and Rh1 protein levels in inaFP106x. (A, C, E, and G) Head extracts were prepared from the indicated flies, fractionated by SDS-PAGE, and Western blots were probed with: (A) anti-TRPL, (C) anti-NORPA, (E) anti-INAD, and (G) anti-Rh1. Anti-Tubulin (Tub) staining provided a loading control in (A), (C), and (E), and a ∼60 kDa nonspecific band (indicated by the arrow) provided a loading control in (G). Protein size markers are indicated (kDa). (B, D, F, and H) Quantification showing relative expression levels of the proteins detected in the Western blots: (B) TRPL, (D) NORPA, (F) INAD, and (H) Rh1; n = 3. (B, D, and F) Unpaired Student’s t-tests; (H) one-way ANOVA.
Whole-mount immunostaining
Immunostainings of whole-mounts of compound eyes were performed as described (Chen et al. 2015), except for the employment of 1 μm optical sections in the current work. Briefly, after fixing the tissue for 2 hr on ice in PBS and 4% paraformaldehyde, the eyes were dissected in PBS and 0.1% Triton X-100, and incubated sequentially with the indicated primary and secondary antibodies. To-Pro3 (T3605) was used as the nuclear counterstain in the indicated experiments.
ERG recordings
ERG recordings using orange light were performed as described (Wes et al. 1999). Briefly, flies 1-day posteclosion were exposed to 10-sec pulses of bright orange light (∼30 mW/cm2) at a frequency of two pulses per minute. To assay PDAs, we exposed 1-day-old flies to 5-sec light interspersed by ∼12.6 sec between pulses. The sequence of light exposure was one orange (∼30 mW/cm2), two blue (∼1–2 mW/cm2) and two orange (∼30 mW/cm2). We used a Schott OG590 filter (590 nm long pass) for obtaining orange light, and a Schott BG28 filter (330–590 nm bandpass) for blue light, which was generated from a Newport Oriel Apex illuminator. >5 flies were analyzed per genotype.
Experimental design and statistical analyses
The bar plots represent the means ± SEMs. Individual data points are indicated in red. We used unpaired Student’s t-tests for two-sample comparisons, and one-way ANOVA with Holm–Sidak post hoc analyses for multiple comparisons. Statistically significant differences are indicated by asterisks (*P < 0.05, **P < 0.01).
Data availability
Strains, plasmids, and DNA sequences are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. The Supplemental Materials include three figures and two tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12340562.
Results
INAF-C is a TRP-interacting protein
To identify TRP-interacting proteins, we isolated TRP from fly photoreceptor cells and performed mass spectrometry. To conduct this analysis, we used flies expressing TRP fused at the N-terminus to a streptavidin-binding peptide tag (SBP::TRP) in all photoreceptor cells in the compound eye (Chen et al. 2015). We introduced the SBP::trp transgene (P[SBP::trp]) in a trp343 null mutant background and found that the fusion protein was expressed at 38 ± 6% of the wild-type TRP level (Figure 1A and Supplemental Material, Figure S1A). The SBP::TRP fusion protein was functional since it rescued the transient light response displayed by the trp343 null mutant (Figure 1B and Figure S1B). We prepared head extracts from P[SBP::trp];trp343 heads, purified SBP::TRP and its interacting proteins on streptavidin resin, and analyzed the peptides in the complex by mass spectrometry. As a negative control, we performed the identical procedure in parallel using extracts from control (w1118) flies, which do not express SBP::TRP. We conducted the analyses in duplicate using independent samples.
The mass spectrometry analyses were effective since we found peptides corresponding to proteins that are known to associate with TRP. The most prominent was the PDZ-containing scaffold protein INAD (Table S1), which binds to TRP and is required for retention of TRP in the rhabdomeres (Huber et al. 1996; Shieh and Zhu 1996; Chevesich et al. 1997; Li and Montell 2000). We also identified the phospholipase C (NORPA) and protein kinase C (INAC), both of which function in phototransduction and bind directly to INAD, and therefore interact indirectly with TRP (Huber et al. 1996; Chevesich et al. 1997; Tsunoda et al. 1997). We found multiple INAF-B positive peptides (Table S1), which is a protein that co-immunoprecipitates with TRP (Cheng and Nash 2007). However, the INAF-B peptides arose in only one of the two mass spectrometry analyses (Table S1), possibly because INAF-B is only 9 kDa, which is just above the exclusion limit of the filters used to concentrate the samples prior to performing the mass spectrometry.
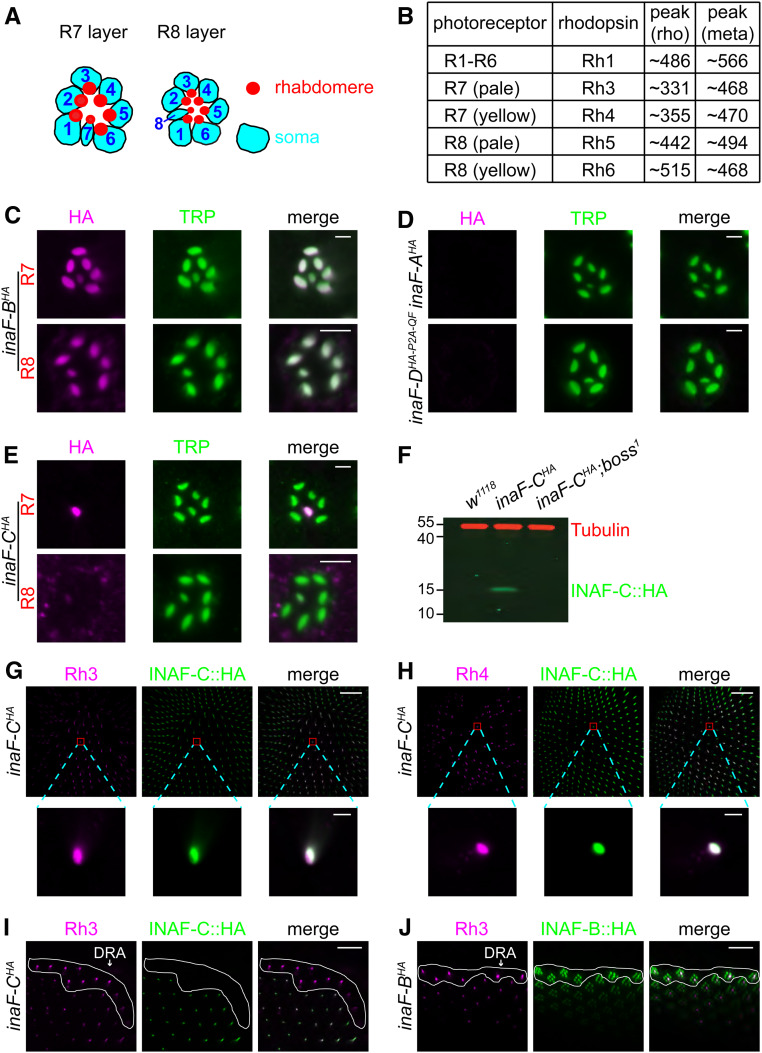
We found that INAF-C, a protein that was not previously known to associate with TRP, emerged in both mass spectrometry analyses (Table S1). INAF-C is one of four proteins encoded by the inaF locus through alternative 5′ exons (Figure 1C; INAF-A, INAF-B, INAF-C, and INAF-D) (Cheng and Nash 2007). Each INAF protein (size in amino acids: INAF-A, 89; INAF-B, 81; INAF-C, 140; and INAF-D, 353) includes a single predicted TMD (Cheng and Nash 2007). The amino acid sequence of each INAF protein is different from the others since they have unique coding regions (Figure 1C). INAF-A, INAF-B, and INAF-C isoforms share only modest homology (identities between: A and B, 36%; B and C, 24%; A and C, 18%), and there is virtually no primary amino acid sequence homology between these isoforms and INAF-D (identities between: A and D, 5.6%; B and D, 7.6%; C and D, 4.8%).
To provide an additional test as to whether INAF-C and TRP interact in vivo, we performed co-IPs. To conduct the co-IPs, we used gene editing to tag the C-terminus of the endogenous INAF-C protein with an in-frame HA epitope tag (INAF-C::HA). We pulled down INAF-C::HA with anti-HA, and found that TRP co-immunoprecipitated from inaF-CHA head extracts but not from w1118 control heads (Figure 1D). We then performed the reverse co-IP. We used streptavidin resin to pull down SBP::TRP from head extracts prepared from flies expressing both SBP::TRP and INAF-C::HA (inaF-CHA;P[SBP::trp];trp343), and found that INAF-C co-immunoprecipitated (Figure 1E). These findings demonstrate that INAF-C and TRP form a complex in vivo.
Elimination of INAF-B and INAF-C causes an ERG phenotype similar to trp
To explore whether knockout of inaF-C affects the photoresponse, we deleted the exon that uniquely encodes INAF-C (Figure 1F; inaFΔC). We also generated deletions that specifically disrupt production of INAF-A, INAF-B, or INAF-D (Figure 1F; inaFΔA, inaFΔB, and inaFΔD). We then performed ERG recordings, which measure the summed retinal responses to light. In control flies (w1118), orange light (580 nm) induces a response, which quickly terminates upon cessation of the stimulus (Figure 1G and Table S2). Bright blue light (480 nm) causes a similar initial light response. Upon cessation of blue light, there is a sustained response in the dark (PDA; Figure 1G and Table S2), because rhodopsin 1 (Rh1) remains active (Wang and Montell 2007; Pak et al. 2012). Rh1 is the major rhodopsin in the eye, which is expressed in six out of eight photoreceptor cells in each of ommatidium of the compound eye (R1–R6; Figure 2, A and B). Exposure to orange light is required to turn off Rh1 following a blue light stimulus, and thereby terminate the PDA. Blue light (480 nm) does not induce a PDA in the R7 and R8 photoreceptor cells, because the absorption maxima of the light-activated rhodopsins in these cells (Rh3–Rh6) allow them to be turned off with cessation of blue light (Figure 2B). Consequently, there is a partial decline in the ERG response after termination of the blue light (Figure 1G and Table S2).
Figure 2.
INAF-C is specifically expressed in R7 cells, while INAF-B is expressed in all photoreceptor cells. (A) Cartoons of photoreceptor cells. The R7 and R8 cells occupy the distal and proximal regions of the ommatidia, while the R1–R6 cells span both regions. The soma and rhabdomeres are indicated. (B) Summary of expression patterns of rhodopsins in most ommatidia in the compound eye. The table lists the photoreceptor cell expression patterns of the rhodopsins, and the absorption peaks (in nm) for each dark-adapted rhodopsin (rho) and light-activated metarhodopsin (meta). The two major types of ommatidia, pale and yellow, express Rh3/Rh5 and Rh4/Rh6, respectively. (C–E) Spatial distribution of the four INAF proteins in photoreceptor cells using flies with HA tags knocked into the C-termini of the endogenous INAF proteins. Shown are single-ommatidia stained with mouse anti-HA (magenta) and rabbit anti-TRP (green). In (C and E), the top panels show staining of the R7 layer, while the bottom panels show staining of the R8 layer. In (D), the panels show staining of the R7 layer of inaF-AHA and inaF-DHA-P2A-QF; bar, 3 μm. (F) Western blot of extracts from the indicated flies probed with mouse anti-Tubulin and rabbit anti-HA antibodies. The boss1 mutation prevents development of R7 cells. The positions of protein size markers (kDa) are indicated. (G and H) Multiple-ommatidia (R7 layer) from inaF-CHA flies, were costained with rabbit anti-HA (green; recognizes INAF-C::HA), and either mouse anti-Rh3 (magenta; G) or mouse anti-Rh4 (magenta; H). Higher magnification images of single-ommatidia are shown below. The scale bars in the upper and lower panels indicate 40 and 3 μm, respectively. (I and J) Multiple-ommatidia (R7 layer) from around the dorsal rim area (DRA, encircled by the white-lines) from inaF-CHA flies (I) and inaF-BHA flies (J) were costained with mouse anti-Rh3 (magenta) and rabbit anti-HA (green); bar, 20 μm.
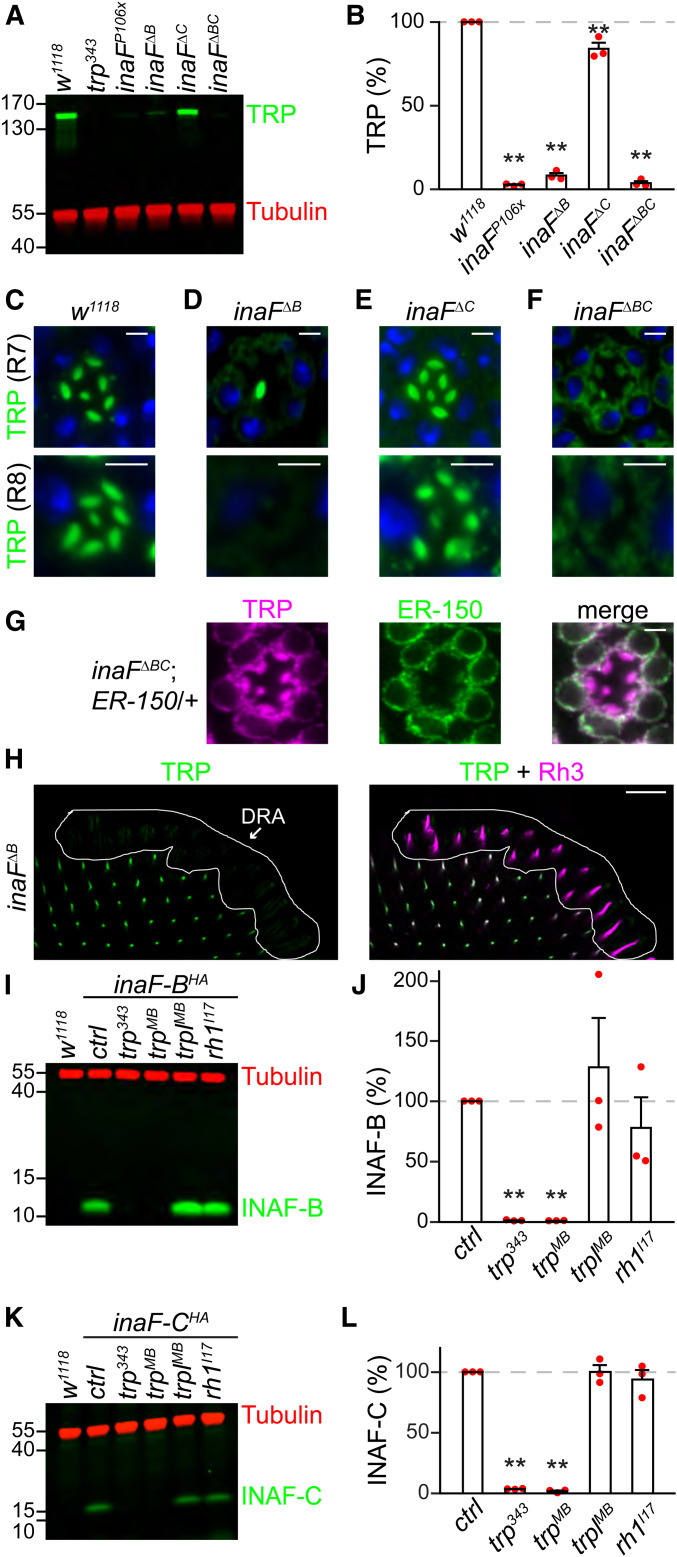
We used the PDA paradigm to characterize the impairments in the light responses in each inaF mutant allele. The trp343 null mutation causes a transient response to orange or blue light (Figure 1H) (Cosens and Manning 1969). Similarly, the inaFP106x mutation, which prevents splicing of all four inaF transcripts, results in a comparable transient light response (Li et al. 1999; Cheng and Nash 2007) (Figure 1, H and I and Table S2). Loss of inaF-B (Figure 1F; inaFΔB) also causes a transient response to orange light (Figure 1J and Table S2). However, in contrast to a previous study reporting that disruption of inaF-B alone is fully responsible for the inaFP106x phenotype (Cheng and Nash 2007), we found that the inaFΔB phenotype was not as strong as inaFP106x (Figure 1, I and J and Table S2). This was evident upon exposure to blue light, since inaFΔB flies exhibited a maintained rather than a transient response to blue light (Figure 1J and Table S2).
The demonstration that loss of INAF-B did not cause a phenotype as severe as inaFP106x indicated that at least one INAF isoform in addition to INAF-B contributes to the photoresponse. INAF-C was the best candidate since it associates with TRP, based on the mass spectrometry data and co-IP experiments. Therefore, we performed ERGs on inaFΔC flies. To complete the analyses of each INAF isoform, we included the inaFΔA and inaFΔD mutants. The inaFΔA and inaFΔD ERGs were indistinguishable from control flies (Figure 1, G, K, and L and Table S2), consistent with the observations that no INAF-A or INAF-D peptides were uncovered in the mass spectrometry analysis of the TRP complex. Surprisingly, elimination of INAF-C (inaFΔC) also had no impact on the ERG (Figure 1M and Table S2). To reconcile the difference between the inaFΔB and inaFP106x alleles, we generated flies that were missing inaF-B in combination with inaF-C (inaFΔBC; Figure 1F). We also created another inaF allele with deletions that prevent expression of inaF-A, inaF-B, and inaF-D, leaving inaF-C intact (inaFΔABD; Figure 1F). The inaFΔABD flies exhibited an ERG phenotype similar to the inaFΔB mutants (Figure 1, J and N and Table S2), indicating that neither INAF-A nor INAF-D contributes to the remaining photoresponse.
Of significance here, loss of both inaF-B and inaF-C caused a more pronounced ERG deficit than elimination of inaF-B alone. In particular, unlike inaFΔB flies, which display a sustained response to blue light, inaFΔBC flies show a transient response (Figure 1, J and O and Table S2) indistinguishable from the inaFP106X null allele. Thus, INAF-C contributes to the photoresponse.
R7-specific expression of INAF-C through an RCSI-like transcriptional motif
To provide an explanation for the stronger ERG phenotype resulting from elimination of both INAF-B and INAF-C, relative to removing INAF-B only, we examined the cellular distribution of the INAF proteins. Proteins required for phototransduction, such as TRP, are localized to the microvillar portion of photoreceptor cells—the rhabdomeres (Figure 2A). To reveal the potential expression patterns of each INAF protein, we used CRISPR/Cas9 to edit the inaF locus, thereby creating a complete set of four INAF isoforms with an endogenous C-terminal HA tag: INAF-A::HA, INAF-B::HA, INAF-C::HA (described above), and INAF-D::HA (inaF-AHA, inaF-BHA, inaF-CHA, and inaF-DHA-P2A-QF). INAF-B has been reported to colocalize with TRP in the distal region of the eye (Cheng and Nash 2007). We found that INAF-B and TRP co-colocalize in the rhabdomeres of all photoreceptor cells, including R8, in the proximal region of the eye (Figure 2C). Neither INAF-A nor INAF-D was detected in photoreceptor cells (Figure 2D), consistent with the lack of any INAF-A or INAF-D peptides interacting with SBP::TRP in the mass spectrometry analyses.
Unexpectedly, we found that INAF-C was restricted to the rhabdomeres of only one type of photoreceptor cell: R7 cells (Figure 2E). Because INAF-C was limited to R7 cells, it was undetectable in a mutant, boss1, missing R7 cells (Figure 2F). We performed double labeling with anti-HA and antibodies to rhodopsin 3 (Rh3) and rhodopsin 4 (Rh4), which are expressed in nonoverlapping subsets of R7 cells (Montell et al. 1987; Zuker et al. 1987). We found that every INAF-C-positive cell expressed either Rh3 or Rh4 (Figure 2, G and H). However, INAF-C was not expressed in R7 cells in the dorsal rim area (DRA), which expresses Rh3 but not Rh4 (Figure 2I) (Wernet et al. 2003). Rather, we found that INAF-B only was expressed in R7 cells in the DRA (Figure 2J).
The R7-specific expression of INAF-C raises the question as to the mechanism underlying this restricted expression pattern. To test whether there may be a block to stable production of INAF-C in R1–R6 cells at either the translational or post-translational level, we created transgenic flies expressing the inaF-C::HA coding sequence under the direct control of the ninaE (rh1) promoter (ninaE•inaF-C::HA). We then performed double-labeling with anti-HA and anti-INAD, which labels the rhabdomeres of all photoreceptor cells (Figure 3A) (Chevesich et al. 1997). We found that INAF-C::HA overlapped with INAD in the outer photoreceptor cells (R1-R6), but not in R7 cells (Figure 3A). The stable ectopic expression of INAF-C::HA in R1-R6 cells argues against a translational or post-translational mechanism preventing synthesis or stability of INAF-C in R1-R6 cells.
To explore a transcriptional mechanism for directing expression of inaF-C in R7 cells, we examined the DNA sequence flanking the 5′ end of the coding exon of inaF-C, and identified an 11-bp motif (TAATCCGCTTT) starting 81 nucleotides 5′ to the predicted transcriptional start site of inaF-C. This sequence, which begins with a 4 bp palindrome, is 100% conserved in the 5′ flanking region in inaF-C genes across multiple Drosophila species (Figure S2A). Furthermore, it resembles the Rhodopsin Core Sequence I (RCSI) in the promoters of fly rhodopsins (Figure 3B) (Fortini and Rubin 1990; Wilson et al. 1995; Papatsenko et al. 2001; Rister et al. 2015). However, the sequence in inaF-C does not include a perfect match to the 4 bp palindrome at the 3′ end of a related sequence in multiple broadly expressed phototransduction genes, such as trp (Figure 3B; P3 sequence: TAATYNRATTA; Y = C or T, R = A or G) (Fortini and Rubin 1990; Wilson et al. 1995; Papatsenko et al. 2001; Rister et al. 2015).
To test whether the RCSI-like motif was essential for inaF-C expression in R7 cells, we used the QF/QUAS binary expression system (Potter et al. 2010) to express GFP under the control of a 0.8 kb inaF-C 5′ flanking sequence encompassing the RCSI (inaF-C•QF + QUAS-GFP; Figure 3C). We found that GFP was expressed in R7 cells and not in R1–R6 or R8 cells (Figure S2B), demonstrating that the 0.8 kb promoter region recapitulated R7 expression of inaF-C (Figure 3D). The signals were distributed throughout the R7 cells since GFP is not restricted to the rhabdomeres. To determine if the RCSI-like sequence contributes to R7 expression, we mutated the two bases flanking the 3′ end of the invariant TAAT [Figure 3C; CC→TA] and found that this reduced expression to a small subset of R7 cells (9.7 ± 2.3%, n = 5; Figure 3E). When we introduced a 6-bp deletion within the RCSI-like motif that includes the invariant TAAT (Figure 3C; Δ6), GFP expression was eliminated in R7 cells (n = 8; Figure 3F). Therefore, inaF-C expression in R7 photoreceptor cells depends on the RCSI-like motif.
The RCSI-like motif in inaF-C begins with a “K50 motif” (TAATCC sequence, Figure 3B), which is the binding site for the Orthodentical (Otd) transcription factor (Rister et al. 2015). This sequence is also present in the in rh3 and rh6 RCSI sequence (Figure 3B). Because Otd regulates expression of several rhodopsin genes, including rh3 and rh6 (Tahayato et al. 2003), we examined whether it also regulated expression of INAF-C. We found that INAF-C was dramatically reduced in an otduvi mutant background (Figure 3, G and H).
TRP expression in R7 cells depends on both INAF-B and INAF-C
The observations that INAF-C is expressed exclusively in the rhabdomeres and interacts with TRP indicate that it is a subunit of the TRP complex. INAF-B and INAF-C include one TMD each, which is reminiscent of auxiliary subunits (β subunits) that interact with voltage-gated channels, such as KCNE proteins, which assemble with Kv channels (Kanda and Abbott 2012; Abbott 2016a,b). In addition, some β subunits are required for stability of the pore-forming subunits of Kv channels (Richards et al. 2004; Buraei and Yang 2010; Abbott 2012; Kanda and Abbott 2012; Sun et al. 2012). Similarly, one or more INAF proteins impact on TRP expression since TRP was greatly reduced in the inaFP106x mutant (Figure 4, A and B) (Li et al. 1999). In the inaFΔB mutant, TRP was also dramatically reduced, but not to the same extent as in inaFP106x flies (Figure 4, A and B). The requirement for INAF for expression of TRP did not extend to TRPL, a highly related channel required for phototransduction, since TRPL levels were unchanged in inaFP106x flies (Figure 5, A and B). Furthermore, INAF proteins were dispensable for expression of NORPA, INAD and Rh1, because the levels of these signaling proteins were unaffected in inaFP106x (Figure 5, C–H).
Figure 4.
INAF-B/INAF-C and TRP are mutually required for expression in photoreceptor cells. (A, I, and K) Head extracts were prepared from the indicated flies, 0.5 head equivalents/lane were fractionated by SDS-PAGE, and Western blots were probed with mouse anti-tubulin (loading control) and the following antibodies: (A) rabbit anti-TRP, (I) rabbit anti-HA (recognizes INAF-B::HA), and (K) rabbit anti-HA (recognizes INAF-C::HA). The anti-HA signals reflected the endogenous levels of INAF-B and INAF-C since the extracts were prepared from flies with HA-tags knocked into the endogenous inaF-B and inaF-C genes. Protein size markers are indicated (kDa). (B, J, and L) Relative levels of the following proteins in heads of the indicated flies, based on Western blot analyses shown to the left: (B) TRP, (J) INAF-B, and (L) INAF-C; n = 3. Error bars represent SEMs; **P < 0.01 (one-way ANOVA with Holm–Sidak post hoc analyses). (C–F) Optical sections from single-ommatidia from the compound eyes of w1118 flies and the indicated inaF mutants, stained with rabbit anti-TRP (green) and a nuclear counterstain, To-PRO3 (blue). Top: R7 layer; bottom: R8 layer; bar, 3 μm. (G) Optical sections from single-ommatidia (R7 layer) from the compound eyes of inaFΔBC;ER-150/+ flies, stained with rabbit anti-TRP (magenta) and chicken anti-GFP (green, recognizes ER-150). ER-150 is an ER-localized GCaMP, which was expressed under control of the ninaE promoter (Liu et al. 2020); bar, 3 μm. (H) Multiple ommatidia (R7 layer) from inaFΔB flies around the DRA, were costained with rabbit anti-TRP (green) and mouse anti-Rh3 (magenta). The left panel shows staining with anti-TRP, while the right panel shows anti-TRP and anti-Rh3 staining; bar, 20 μm.
To address the cellular basis for the low levels of TRP in the inaFΔB mutant, we stained compound eyes with anti-TRP. In control flies (w1118), TRP is expressed in the rhabdomeres of every photoreceptor cell (Figure 4C) (Montell and Rubin 1989; Chevesich et al. 1997). In contrast, in inaFΔB flies, TRP was dramatically reduced in R1–R6 and R8 cells, but remained at high levels in R7 cells (Figure 4D). Loss of INAF-C in the inaFΔC mutant mildly, but significantly, reduced the level of TRP (Figure 4, A and B), which remained in the rhabdomeres of all photoreceptor cells (Figure 4E). However, in inaFΔBC, the concentration of TRP was greatly reduced (Figure 4, A and B). To localize the residual TRP in the photoreceptor cells, we performed immunostaining and used a high gain to detect the signals. We found that a large proportion of TRP in inaFΔBC photoreceptor cells was perinuclear, suggesting localization to the ER (Figure 4F). In support of this proposal, the extra-rhabdomeral TRP largely colocalized with an ER marker (ER-150) (Liu et al. 2020), which is expressed in R1–R6 cells (Figure 4G). These results demonstrate that both INAF-B and INAF-C are required for high levels of expression of TRP in the rhabdomeres of R7 cells, consistent with the expression of both INAF-B and INAF-C in R7 cells (Figure 2, C and E). A different situation occurs in the R7 cells in the DRA, which express INAF-B but not INAF-C (Figure 2, I and J). In contrast to the rest of the eye, TRP was dramatically reduced in the R7 cells in the DRA of inaFΔB flies (Figure 4H).
To determine whether expression of INAF-B and INAF-C reciprocally depend on TRP, we performed Western blots. We found that INAF-B and INAF-C were undetectable in each of two trp null mutants, but were expressed normally in trplMB and ninaEI17 (rh1) null mutant flies (Figure 4, I–L). These data demonstrate that there is a mutual requirement between TRP and INAF-B/INAF-C for protein expression.
Transmembrane segments in TRP confer dependence on INAF
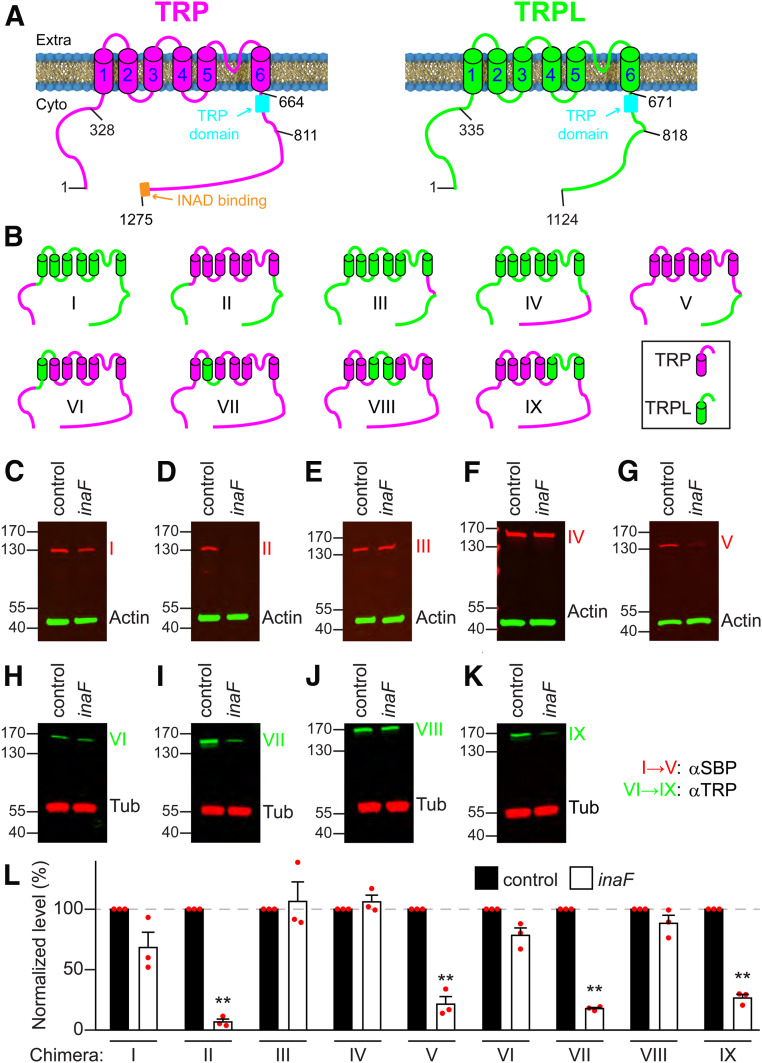
A large body of studies demonstrate that the β subunits interact with several TMDs in Kv, and do so primarily through their single TMD and cytoplasmic C termini (Abbott 2016a). However, it is not feasible to functionally express TRP in tissue culture cells for performing biochemistry, since TRP does not traffic out of the ER and is degraded. Therefore, to determine the domains in TRP that are required for conferring INAF-dependence in the native photoreceptor cells, we took advantage of the findings that TRPL expression is not dependent on any INAF protein (Figure 5, A and B). TRP and TRPL are comprised of 1275 and 1124 amino acids, respectively (Figure 6A), and share 39% overall identity (Montell and Rubin 1989; Phillips et al. 1992). Most of the identity spans the N-terminal 825 residues, which includes the N-terminal domain with ankyrin repeats, the six transmembrane segments, the TRP domain, and a small adjacent segment. The C-terminal ∼450 residue region of TRP is essentially unrelated to the C-terminal domain of TRPL.
Figure 6.
Mapping regions in TRP required for interaction with INAF-B. (A) Cartoons depicting the membrane topologies of TRP and TRPL. The barrels (1–6) indicate the S1–S6 transmembrane segments. The N- and C-termini are on the cytoplasmic (Cyto) side of the rhabdomeral membrane. The TRP domain, and the INAD binding site are indicated. 1275 and 1124 indicate the C-terminal residues of TRP and TRPL. The other numbers indicate some of the sites targeted for generating the TRP-TRPL chimeras. (B) Cartoons indicating TRP-TRPL chimera (I–IX). The regions from TRP and TRPL are represented in magenta and green, respectively. (C–K) Western blots showing the expression levels of each chimera (I–IX) in head extracts from inaFP106x;trpl;trp flies (inaF), or in trpl;trp flies, which provided an inaF+ background (control). Chimeras I–V and VI–IX were in trplMB;trpMB and trpl302;trpMB backgrounds, respectively. The transgenes encoding each chimera were expressed under control of the ninaE promoter. Anti-SBP rather than anti-TRP was used to monitor the levels of chimeras I–V, since these proteins were missing the epitope used to generate anti-TRP. Anti-actin served as the loading control. Chimeras VI–IX were detected with anti-TRP, and anti-tubulin (Tub) provided the control for normalization. The positions of protein size markers (kDa) are shown. (L) Quantification of the relative levels of the TRP-TRPL chimeras in inaFP106x;trpl;trp heads; n = 3. Error bars represent SEMs. **P < 0.01 Unpaired Student’s t-tests.
We tested a series of TRP/TRPL chimeras to identify a region of TRP that might transform TRPL into a channel that depended on INAF-B for protein expression. To do so, we used the ninaE promoter to drive expression of the chimeras, and compared their expression levels in control and inaFP106x flies. We found that swapping the conserved N-terminal domain or the conserved TRP domain from TRP into TRPL did not induce a requirement for INAF-B (chimeras I and III; Figure 6, B, C, E, and L). The highly divergent C-terminal domain from TRP also did not switch TRPL into a channel that needed INAF-B for protein expression (chimera IV; Figure 6, B, F, and L). Consistent with this finding, when we replaced the C-terminus of TRP with the C-terminus of TRPL, the chimeric protein did not lose its requirement for INAF-B for expression (chimera V; Figure 6, B, G, and L).
The preceding data suggest that the transmembrane domains in TRP are responsible for the dependence of TRP on INAF-B. To address this possibility, we tested a chimera comprised of the six TRP transmembrane segments (S1–S6) fused to the N- and C-termini of TRPL (chimera II; Figure 6B). The concentration of chimera II was greatly diminished in inaFP106x flies (Figure 6, D and L), indicating that the TRP transmembrane segments define the dependence on INAF-B. To narrow down which transmembrane segments (TMDs) confer dependence on INAF-B for expression, we created chimeras in which we replaced either one or two TMDs of TRP with the counterparts from TRPL (Figure 6B; chimeras VI–IX, TRPL TMDs in green). We then determined whether any of these chimeric proteins became resistant to the absence of INAF-B. Remarkably, expression of chimera VI—a protein comprised almost entirely of TRP sequences (1187 out of 1275 amino acids, ∼93% TRP), except for 88 TRPL residues that included S1—was not dependent on INAF-B (Figure 6, H and L). Chimera VIII, which consists primarily of TRP except for the region spanning S3 and S4, was also not dependent on INAF-B (Figure 6, J and L). In contrast, substitutions of S2 (Figure 6B; chimera VII), or S5, S6, and the intervening pore loop (Figure 6B; chimera IX), had no or only minimal effects, because these two chimeras greatly increase their expression in the presence of INAF-B (Figure 6, I, K, and L). Thus, S1 and S3-S4 TMDs are critical for conferring a requirement for INAF-B for TRP expression.
To explore whether any of the other three INAF proteins could functionally substitute for INAF-B, we ectopically expressed inaF-A, inaF-C, or inaF-D in R1-R6 cells under the control of ninaE promoter in an inaFP106x background. Either inaF-A or inaF-C, but not inaF-D restored TRP protein expression (Figure 7, A and B) and normal ERG responses (Figure 7C and Figure S3A). To test whether the inability of INAF-D to substitute for INAF-B was due to an obstacle in producing INAF-D in R1-R6 cells, we expressed the transgene encoding an HA tagged version of INAF-D under control of the ninaE promoter (ninaE•inaF-D::HA). INAF-D::HA was expressed, demonstrating that INAF-D could be produced ectopically in R1–R6 cells (Figure 7E).
To identify the region in INAF-B that is critical for promoting TRP expression, we created flies expressing HA epitope-tagged INAF-B/INAF-D chimeras (Figure 7D) in R1–R6 cells. All three chimeras (INAF-BD1, INAF-BD2, and INAF-BD3) were produced at levels comparable to, or higher than, INAF-B::HA (Figure 7E). When we replaced the C-terminal 17 residues of INAF-B (residues 65–81) with the much larger C-terminus of INAF-D (residues, 62–353), the fusion protein lost the ability to support TRP expression (INAF-BD1; Figure 7, F–H and Figure S3B). Conversely, a fusion protein consisting of the N-terminus of INAF-D fused to the TMD and C-terminus of INAF-B largely substituted for INAF-B (INAF-BD3; Figure 7, F–H and Figure S3B). Even a fusion protein consisting of the N-terminal region and the TMD from INAF-D fused to just the 17 C-terminal residues of INAF-B (residues 65–81) partially substituted for INAF-B (INAF-BD2; Figure 7, F–H).
Discussion
We propose that INAF comprises a family of proteins that serve as auxiliary subunits for TRP channels. Auxiliary subunits for other channels, such as the β subunit for Kv channels, associate, and colocalize, with the channel-forming α subunit and promote trafficking and protein stability, and modulate the biophysical properties of the channels (Abbott 2016a). INAF-B and INAF-C function as auxiliary subunits that contribute to the folding and assembly of TRP, which is necessary for exiting the ER. Consistent with this proposal, the residual TRP in inaFΔBC mutants is largely retained in the perinuclear zone—a region containing the ER.
Multiple parallels with β subunits support the model that INAF proteins are a set of TRP auxiliary subunits. Similar to β subunits, all four INAF proteins consist of one TMD. The interactions between α and β subunits of Kv channels depend on several TMDs in the α subunit, and the TMD and C-terminal domain of the β subunit (Abbott 2016a). We show that the codependency of TRP and INAF-B for expression and localization depend on multiple TMDs in TRP, and the TMD and C-terminus of INAF-B. However, it was not possible to conduct biophysical studies in tissue culture cells, since TRP is retained in the ER, even when we coexpressed TRP in insect S2 cells with INAF-B and two ER proteins that promote the transporting of TRP through the secretory pathway (XPORT-A and XPORT-B) (Rosenbaum et al. 2011; Chen et al. 2015).
While it remains to be determined whether INAF-B and INAF-C alter the activation kinetics, mean open time, deactivation kinetics, or some other aspect of the TRP channel, it is intriguing to speculate that the combination of INAF-B and INAF-C regulate TRP differently from INAF-B alone. If so, then the TRP-dependent conductance in R1-6 cells, which express INAF-B only, may be distinct from the TRP current in the ultraviolet-sensing R7 cells, which express INAF-B and INAF-C. In support of the idea that INAF-B and INAF-C are doing more than impacting TRP expression, elimination of just INAF-B or INAF-C has only minimal effects on the levels or localization of TRP in R7 cells.
We suggest that the regulation of TRP channels by the Drosophila INAF proteins is not limited to TRP in photoreceptor cells. Rather, INAF proteins may function more broadly in regulating TRP family members in other neurons. Consistent with this proposal, we found that INAF-A rescues loss of INAF-B in R1-6 cells, even though INAF-A is only 36% identical to INAF-B, and is not normally expressed in any photoreceptor cell. Thus, INAF-A is a third INAF family member that is capable of functioning as a TRP auxiliary protein. It seems plausible that INAF-A normally regulates TRP family members in extraretinal cells.
INAF-D is the INAF isoform most different from INAF-A, INAF-B, and INAF-C, since it has a much longer cytoplasmic C-terminal end, and there is virtually no primary amino acid sequence homology between INAF-D and the other three INAF proteins. Like INAF-A, the INAF-D protein is not expressed in photoreceptor cells. However, in contrast to INAF-A, it did not substitute for INAF-B, even though we were able to express it in R1–6 cells. Thus, INAF-D may be an auxiliary protein for a TRP channel that is relatively distantly related to the classical TRP in photoreceptor cells.
While we propose that INAF is a family of TRP auxiliary proteins, not all TRP channels depend on INAF. While loss of INAF-B/C causes a profound reduction in TRP levels, it has no impact on TRPL. This may reflect a notable difference in the spatial distribution of TRP and TRPL. While TRP is statically located in the rhabdomeres of wild-type flies, regardless of light conditions, TRPL undergoes dynamic light-dependent translocation from the rhabdomeres to the cell bodies in response to light (Bähner et al. 2002; Cronin et al. 2006). Thus, if INAF-B and INAF-C were constitutively bound to TRPL, as is the case with TRP, it might preclude light-dependent movement.
Future studies aimed at examining the expression of INAF-A and INAF-D offer to reveal roles of these proteins in regulating 1 or more of the 13 Drosophila TRP channels in extraretinal cells. If INAF-A and INAF-D interact with TRP proteins that are amenable to functional expression in vitro, it would provide the opportunity to determine whether INAF proteins regulate TRP channel currents, in addition to impacting on localization and stability of TRP, as is the case for INAF-B and INAF-C in photoreceptor cells. Finally, two INAF-related proteins (INAFM1 and INAFM2) are encoded in the mouse and human genomes (Cheng and Nash 2007). Whether these INAF-like proteins function as auxiliary proteins for mammalian TRPs would be exciting to address.
Acknowledgments
This work was supported by a grant to C.M. from the National Eye Institute (EY010852). We thank the Bloomington Drosophila Stock Center [National Institutes of Health (NIH) grant P40OD018537] for many of the stocks used in this study. We thank W. Pak, C. Desplan, and R. Hardie for fly stocks, S. Britt, C. Zuker, and D. Ready for antibodies, and the Drosophila Genomics Resource Center (NIH grant 2P40OD010949) for the pHD-ScarlesDsRed vector. We thank the Johns Hopkins Mass Spectrometry and Proteomics Facility for performing the mass spectrometry analyses, J. Luo for technical help with CRISPR/Cas9, and the flyCRISPR (http://flycrispr.molbio.wisc.edu) for bioinformatics.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12340562.
Communicating editor: Kate O’Connor-Giles
Literature Cited
- Abbott G. W., 2012. KCNE2 and the K (+) channel: the tail wagging the dog. Channels 6: 1–10. 10.4161/chan.19126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott G. W., 2016a KCNE1 and KCNE3: the yin and yang of voltage-gated K+ channel regulation. Gene 576: 1–13. 10.1016/j.gene.2015.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott G. W., 2016b KCNE4 and KCNE5: K+ channel regulation and cardiac arrhythmogenesis. Gene 593: 249–260. 10.1016/j.gene.2016.07.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähner M., Frechter S., Da Silva N., Minke B., Paulsen R. et al. , 2002. Light-regulated subcellular translocation of Drosophila TRPL channels induces long-term adaptation and modifies the light-induced current. Neuron 34: 83–93. 10.1016/S0896-6273(02)00630-X [DOI] [PubMed] [Google Scholar]
- Berson D. M., Dunn F. A., and Takao M., 2002. Phototransduction by retinal ganglion cells that set the circadian clock. Science 295: 1070–1073. 10.1126/science.1067262 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., and Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist B. T., Shortridge R. D., Schneuwly S., Perdew M., Montell C. et al. , 1988. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell 54: 723–733. 10.1016/S0092-8674(88)80017-5 [DOI] [PubMed] [Google Scholar]
- Buraei Z., and Yang J., 2010. The β subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90: 1461–1506. 10.1152/physrev.00057.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E., Liao M., Cheng Y., and Julius D., 2013. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504: 113–118. 10.1038/nature12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Chen H. C., and Montell C., 2015. TRP and rhodopsin transport depends on dual XPORT ER chaperones encoded by an operon. Cell Rep. 13: 573–584. 10.1016/j.celrep.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., and Nash H. A., 2007. Drosophila TRP channels require a protein with a distinctive motif encoded by the inaF locus. Proc. Natl. Acad. Sci. USA 104: 17730–17734. 10.1073/pnas.0708368104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevesich J., Kreuz A. J., and Montell C., 1997. Requirement for the PDZ domain protein, INAD, for localization of the TRP store-operated channel to a signaling complex. Neuron 18: 95–105. 10.1016/S0896-6273(01)80049-0 [DOI] [PubMed] [Google Scholar]
- Chou W. H., Huber A., Bentrop J., Schulz S., Schwab K. et al. , 1999. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development 126: 607–616. [DOI] [PubMed] [Google Scholar]
- Cosens D. J., and Manning A., 1969. Abnormal electroretinogram from a Drosophila mutant. Nature 224: 285–287. 10.1038/224285a0 [DOI] [PubMed] [Google Scholar]
- Cronin M. A., Lieu M. H., and Tsunoda S., 2006. Two stages of light-dependent TRPL-channel translocation in Drosophila photoreceptors. J. Cell Sci. 119: 2935–2944. 10.1242/jcs.03049 [DOI] [PubMed] [Google Scholar]
- Dutta Banik D., Martin L. E., Freichel M., Torregrossa A. M., and Medler K. F., 2018. TRPM4 and TRPM5 are both required for normal signaling in taste receptor cells. Proc. Natl. Acad. Sci. USA 115: E772–E781. 10.1073/pnas.1718802115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M. E., and Rubin G. M., 1990. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 4: 444–463. 10.1101/gad.4.3.444 [DOI] [PubMed] [Google Scholar]
- Gong W. J., and Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561. 10.1073/pnas.0535280100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie R. C., and Minke B., 1992. The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron 8: 643–651. 10.1016/0896-6273(92)90086-S [DOI] [PubMed] [Google Scholar]
- Huber A., Sander P., Gobert A., Bähner M., Hermann R. et al. , 1996. The transient receptor potential protein (Trp), a putative store-operated Ca2+ channel essential for phosphoinositide-mediated photoreception, forms a signaling complex with NorpA, InaC and InaD. EMBO J. 15: 7036–7045. 10.1002/j.1460-2075.1996.tb01095.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda V. A., and Abbott G. W., 2012. KCNE regulation of K+ channel trafficking - a Sisyphean task? Front. Physiol. 3: 231 10.3389/fphys.2012.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leypold B. G., Yu C. R., Leinders-Zufall T., Kim M. M., Zufall F. et al. , 2002. Altered sexual and social behaviors in trp2 mutant mice. Proc. Natl. Acad. Sci. USA 99: 6376–6381. 10.1073/pnas.082127599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Geng C., Leung H. T., Hong Y. S., Strong L. L. et al. , 1999. INAF, a protein required for transient receptor potential Ca2+ channel function. Proc. Natl. Acad. Sci. USA 96: 13474–13479. 10.1073/pnas.96.23.13474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. S., and Montell C., 2000. TRP and the PDZ protein, INAD, form the core complex required for retention of the signalplex in Drosophila photoreceptor cells. J. Cell Biol. 150: 1411–1422. 10.1083/jcb.150.6.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Um S. Y., and McDonald T. V., 2006. Voltage-gated potassium channels: regulation by accessory subunits. Neuroscientist 12: 199–210. 10.1177/1073858406287717 [DOI] [PubMed] [Google Scholar]
- Liman E. R., Zhang Y. V., and Montell C., 2014. Peripheral coding of taste. Neuron 81: 984–1000. 10.1016/j.neuron.2014.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. H., Chen Z., Oliva M. K., Luo J., Collier S. et al. , 2020. Rapid release of Ca2+ from endoplasmic reticulum mediated by Na+/Ca2+ exchange. J. Neurosci. 40: 3152–3164. 10.1523/JNEUROSCI.2675-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. B., Campbell E. B., and Mackinnon R., 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309: 897–903. 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Lucas P., Ukhanov K., Leinders-Zufall T., and Zufall F., 2003. A diacylglycerol-gated cation channel in vomeronasal neuron dendrites is impaired in TRPC2 mutant mice: mechanism of pheromone transduction. Neuron 40: 551–561. 10.1016/S0896-6273(03)00675-5 [DOI] [PubMed] [Google Scholar]
- Montell C., 2012. Drosophila visual transduction. Trends Neurosci. 35: 356–363. 10.1016/j.tins.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., and Rubin G. M., 1989. Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron 2: 1313–1323. 10.1016/0896-6273(89)90069-X [DOI] [PubMed] [Google Scholar]
- Montell C., Jones K., Zuker C., and Rubin G., 1987. A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J. Neurosci. 7: 1558–1566. 10.1523/JNEUROSCI.07-05-01558.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer B. A., Suzuki E., Scott K., Jalink K., and Zuker C. S., 1996. The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell 85: 651–659. 10.1016/S0092-8674(00)81232-5 [DOI] [PubMed] [Google Scholar]
- Nilius B., and Szallasi A., 2014. Transient receptor potential channels as drug targets: from the science of basic research to the art of medicine. Pharmacol. Rev. 66: 676–814. 10.1124/pr.113.008268 [DOI] [PubMed] [Google Scholar]
- O’Tousa J. E., Baehr W., Martin R. L., Hirsh J., Pak W. L. et al. , 1985. The Drosophila ninaE gene encodes an opsin. Cell 40: 839–850. 10.1016/0092-8674(85)90343-5 [DOI] [PubMed] [Google Scholar]
- Pak W. L., Shino S., and Leung H. T., 2012. PDA (Prolonged Depolarizing Afterpotential)-defective mutants: the story of nina’s and ina’s-pinta and santa maria, too. J. Neurogenet. 26: 216–237. 10.3109/01677063.2011.642430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D., Nazina A., and Desplan C., 2001. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech. Dev. 101: 143–153. 10.1016/S0925-4773(00)00581-5 [DOI] [PubMed] [Google Scholar]
- Phillips A. M., Bull A., and Kelly L. E., 1992. Identification of a Drosophila gene encoding a calmodulin-binding protein with homology to the trp phototransduction gene. Neuron 8: 631–642. 10.1016/0896-6273(92)90085-R [DOI] [PubMed] [Google Scholar]
- Port F., Chen H. M., Lee T., and Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Tasic B., Russler E. V., Liang L., and Luo L., 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141: 536–548. 10.1016/j.cell.2010.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy R. A., Fluck E. C. 3rd, Ahmed T., and Moiseenkova-Bell V. Y., 2020. Structural insights into the gating mechanisms of TRPV channels. Cell Calcium 87: 102168 10.1016/j.ceca.2020.102168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke R., and Zipursky S. L., 1988. Cell-cell interaction in the Drosophila retina: the bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell 55: 321–330. 10.1016/0092-8674(88)90055-4 [DOI] [PubMed] [Google Scholar]
- Richards M. W., Butcher A. J., and Dolphin A. C., 2004. Ca2+ channel β-subunits: structural insights AID our understanding. Trends Pharmacol. Sci. 25: 626–632. 10.1016/j.tips.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Rister J., Razzaq A., Boodram P., Desai N., Tsanis C. et al. , 2015. Single-base pair differences in a shared motif determine differential Rhodopsin expression. Science 350: 1258–1261. 10.1126/science.aab3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum E. E., Brehm K. S., Vasiljevic E., Liu C. H., Hardie R. C. et al. , 2011. XPORT-dependent transport of TRP and rhodopsin. Neuron 72: 602–615. 10.1016/j.neuron.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A. K., O’Tousa J. E., Ozaki K., and Ready D. F., 2005. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132: 1487–1497. 10.1242/dev.01704 [DOI] [PubMed] [Google Scholar]
- Shearin H. K., Macdonald I. S., Spector L. P., and Stowers R. S., 2014. Hexameric GFP and mCherry reporters for the Drosophila GAL4, Q, and LexA transcription systems. Genetics 196: 951–960. 10.1534/genetics.113.161141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh B.-H., and Zhu M.-Y., 1996. Regulation of the TRP Ca2+ channel by INAD in Drosophila photoreceptors. Neuron 16: 991–998. 10.1016/S0896-6273(00)80122-1 [DOI] [PubMed] [Google Scholar]
- Stowers L., Holy T. E., Meister M., Dulac C., and Koentges G., 2002. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science 295: 1493–1500. 10.1126/science.1069259 [DOI] [PubMed] [Google Scholar]
- Sun X., Zaydman M. A., and Cui J., 2012. Regulation of voltage-activated K+ channel Gating by transmembrane β subunits. Front. Pharmacol. 3: 63 10.3389/fphar.2012.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahayato A., Sonneville R., Pichaud F., Wernet M. F., Papatsenko D. et al. , 2003. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev. Cell 5: 391–402. 10.1016/S1534-5807(03)00239-9 [DOI] [PubMed] [Google Scholar]
- Tsunoda S., Sierralta J., Sun Y., Bodner R., Suzuki E. et al. , 1997. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature 388: 243–249. 10.1038/40805 [DOI] [PubMed] [Google Scholar]
- Vangeel L., and Voets T., 2019. Transient receptor potential channels and calcium signaling. Cold Spring Harb. Perspect. Biol. 11: a035048 10.1101/cshperspect.a035048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K., and Montell C., 2007. TRP channels. Annu. Rev. Biochem. 76: 387–417. 10.1146/annurev.biochem.75.103004.142819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Cheng X., Tian J., Xiao Y., Tian T. et al. , 2020. TRPC channels: structure, function, regulation and recent advances in small molecular probes. Pharmacol. Ther. 209: 107497 10.1016/j.pharmthera.2020.107497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., and Montell C., 2007. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 454: 821–847. 10.1007/s00424-007-0251-1 [DOI] [PubMed] [Google Scholar]
- Wang T., Xu H., Oberwinkler J., Gu Y., Hardie R. C. et al. , 2005. Light activation, adaptation, and cell survival functions of the Na+/Ca2+ exchanger CalX. Neuron 45: 367–378. 10.1016/j.neuron.2004.12.046 [DOI] [PubMed] [Google Scholar]
- Wernet M. F., Labhart T., Baumann F., Mazzoni E. O., Pichaud F. et al. , 2003. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115: 267–279. 10.1016/S0092-8674(03)00848-1 [DOI] [PubMed] [Google Scholar]
- Wes P. D., Xu X.-Z. S., Li H.-S., Chien F., Doberstein S. K. et al. , 1999. Termination of phototransduction requires binding of the NINAC myosin III and the PDZ protein INAD. Nat. Neurosci. 2: 447–453. 10.1038/8116 [DOI] [PubMed] [Google Scholar]
- Wilson D. S., Guenther B., Desplan C., and Kuriyan J., 1995. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell 82: 709–719. 10.1016/0092-8674(95)90468-9 [DOI] [PubMed] [Google Scholar]
- Xue T., Do M. T., Riccio A., Jiang Z., Hsieh J. et al. , 2011. Melanopsin signalling in mammalian iris and retina. Nature 479: 67–73. 10.1038/nature10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker C. S., Montell C., Jones K., Laverty T., and Rubin G. M., 1987. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: homologies with other signal-transducing molecules. J. Neurosci. 7: 1550–1557. 10.1523/JNEUROSCI.07-05-01550.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains, plasmids, and DNA sequences are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. The Supplemental Materials include three figures and two tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12340562.