Abstract
Small noncoding RNAs are central regulators of genome activity and stability. Their regulatory function typically involves sequence similarity with their target sites, but understanding the criteria by which they specifically recognize and regulate their targets across the genome remains a major challenge in the field, especially in the face of the diversity of silencing pathways involved. The dominance hierarchy among self-incompatibility alleles in Brassicaceae is controlled by interactions between a highly diversified set of small noncoding RNAs produced by dominant S-alleles and their corresponding target sites on recessive S-alleles. By controlled crosses, we created numerous heterozygous combinations of S-alleles in Arabidopsis halleri and developed an real-time quantitative PCR assay to compare allele-specific transcript levels for the pollen determinant of self-incompatibility (SCR). This provides the unique opportunity to evaluate the precise base-pairing requirements for effective transcriptional regulation of this target gene. We found strong transcriptional silencing of recessive SCR alleles in all heterozygote combinations examined. A simple threshold model of base pairing for the small RNA–target interaction captures most of the variation in SCR transcript levels. For a subset of S-alleles, we also measured allele-specific transcript levels of the determinant of pistil specificity (SRK), and found sharply distinct expression dynamics throughout flower development between SCR and SRK. In contrast to SCR, both SRK alleles were expressed at similar levels in the heterozygote genotypes examined, suggesting no transcriptional control of dominance for this gene. We discuss the implications for the evolutionary processes associated with the origin and maintenance of the dominance hierarchy among self-incompatibility alleles.
Keywords: dominance/recessivity, sporophytic self-incompatibility, RT-qPCR, allele-specific expression assay, Arabidopsis halleri
SMALL noncoding RNAs (sRNAs) are short RNA molecules (20–25 nt) with a range of regulatory functions (Vazquez et al. 2010; Aalto and Pasquinelli 2012). The best-known members of this class of molecules are microRNAs (miRNAs), which are typically involved in post-transcriptional gene silencing and regulate the activity of their target gene in trans by either messenger RNA (mRNA) cleavage (quickly followed by degradation) or by blocking translation (Li et al. 2014). In some cases, the action of miRNAs leads to the production of secondary phased short interfering RNAs (pha-siRNAs) by their target coding or noncoding sequence, which in turn can regulate other downstream targets (Fei et al. 2013). Another major set of sRNAs is heterochromatic short interfering RNAs (hc-siRNAs), which mediate transcriptional silencing of repeat sequences in the genome through epigenetic modification by the RNA-dependent DNA methylation pathway (Matzke et al. 2009).
Both miRNAs and siRNAs guide their effector molecules (members of the ARGONAUTE gene family: AGO1 and AGO4, respectively) to their target sites by sequence similarity through base pairing. For plant miRNAs, all functionally verified interactions appear to involve very high sequence similarity with the target sequence (Wang et al. 2015). However, high base-pairing complementarity is not the sole determinant of target specificity, and the positions of mismatches in the miRNA:target duplex are also important. Indeed, expression assays showed that while individual mismatches typically have limited functional consequences, they can also entirely inactivate the interaction when present at specific positions, for example, the 10th and 11th nucleotide, corresponding to the site of cleavage (Jones-Rhoades et al. 2006). Furthermore, the position of mismatches along the miRNA:target duplex also seems to be crucial, with a greater tolerance in the 3′ than the 5′ region of the miRNA (up to four mismatches generally have limited functional consequences in the 3′ region, while only two mismatches in the 5′ region seem sufficient to abolish the target recognition capability; Mallory et al. 2004; Parizotto et al. 2004; Schwab et al. 2005; Liu et al. 2014). These observations have led to the formulation of general “rules” for miRNA targeting (Axtell and Meyers 2018), but at the same time they have also revealed a large number of exceptions. As a result, in silico prediction of miRNA target sites currently remains difficult (Ding et al. 2012; Axtell and Meyers 2018). For other types of sRNAs (pha-siRNAs and hc-siRNAs), even less is known about the base-pairing requirements for targeting, mostly because of the absence of experimentally confirmed examples of discrete, single-siRNA target sites either in cis or in trans (Wang et al. 2015).
In this context, the recent discovery by Tarutani et al. (2010), Durand et al. (2014), and Yasuda et al. (2016) of a highly diversified set of sRNAs at the gene cluster controlling self-incompatibility (SI) in Brassicaceae provides an experimentally tractable model to evaluate the base-pairing requirements for silencing by a set of sRNAs regulating expression of a single gene. Sporophytic SI is a genetic system that evolved in several hermaphroditic plant lineages to enforce outcrossing by preventing self-fertilization, hence avoiding inbreeding depression (De Nettancourt 2001). In the Brassicaceae family, SI is controlled by a single genomic region called the “S-locus,” which contains two tightly linked genes, SCR and SRK, that encode the pollen S-locus cysteine-rich and the stigma S-locus receptor kinase recognition proteins, respectively. This system involves a polymorphism in which multiple deeply diverged alleles are maintained, and a large number of S-alleles is typically found in natural populations of self-incompatible species (Castric and Vekemans 2004). With such a large diversity of S-alleles, most individual plants are heterozygotes at the S-locus. Yet in many cases, only one of the two S-alleles in a heterozygous genotype is expressed at the phenotypic level in either pollen or pistil, as can be revealed by controlled pollination assays on pollen or pistil tester lines (e.g., Llaurens et al. 2008; Durand et al. 2014). Which S-allele is expressed is determined by the alleles’ relative position in a dominance hierarchy. In the genus Brassica, pollen dominance phenotypes are controlled by transcriptional silencing of recessive alleles by dominant ones (Schopfer et al. 1999; Kakizaki et al. 2003). Silencing is caused by 24 nt-long trans-acting sRNAs produced by dominant S-alleles and capable of targeting a DNA sequence in the promoter sequence of the SCR gene of recessive S-alleles, provoking DNA methylation (Shiba et al. 2006). Details of how these sRNAs achieve their silencing function remain incompletely understood (Finnegan et al. 2011), but it is clear that their biogenesis is similar to that of miRNAs (i.e., they are produced by a short hairpin structure), while their mode of action is reminiscent of that of siRNAs (i.e., the transcriptional gene silencing functions through recruitment of the methylation machinery). Strikingly, the full dominance hierarchy in the Brassica genus seems to be controlled by just two sRNAs called Smi and Smi2 (Tarutani et al. 2010; Yasuda et al. 2016). Smi and Smi2 target distinct DNA sequences, both located in the promoter region of SCR, and both seem to involve DNA methylation and 24-nt active RNA molecules.
However, the dominance hierarchy in Brassica is peculiar in that only two ancestral allelic lineages segregate in that genus [(the class I and class II alleles, see, e.g., Leducq et al. (2014)], whereas other self-incompatible species in Brassicaceae typically retain dozens of highly divergent ancestral allelic lineages (Castric and Vekemans, 2004; Genete et al. 2020). A recent study showed that in Arabidopsis halleri, a Brassicaceae species with multiple allelic lineages at the S-locus, the dominance hierarchy among S-alleles in pollen is controlled by as many as eight different sRNA precursor families and their target sites, whose interactions collectively determine the position of the alleles along the hierarchy (Durand et al. 2014). In that genus, much less is known about the mechanisms by which the predicted sRNA–target interactions translate into the dominance phenotypes. First, the expression dynamics of the SCR gene across flower development stages are poorly known. Kusaba et al. (2002) measured expression of SCR alleles in A. lyrata, but focused on only two S-alleles [SCRa and SCRb, also known as AlSCR13 and AlSCR20, respectively, in Mable et al. (2003)] and found striking differences in their expression dynamics in anthers. Hence, the developmental stage at which the transcriptional control of dominance in pollen should be tested is not precisely known. Second, while these studies confirmed monoallelic expression, consistent with the observed dominance relationship between the two alleles (SCRb > SCRa; Kusaba et al. 2002), they measured only a single heterozygous combination among the myriad possible combinations of S-alleles (at least 43 S-alleles; Genete et al. 2020). Hence, complete experimental validation of the transcriptional control of dominance among S-alleles in the Arabidopsis genus is still lacking. Third, Durand et al. (2014) noted that sRNA–target interaction predictions occasionally did not agree with the observed dominance phenotype. In particular, they identified pairs of S-alleles where no sRNA produced by the dominant allele was expected to target the SCR gene of the recessive one, yet a dominance phenotype was observed in controlled crosses (e.g., Ah04 > Ah03), suggesting the possibility that mechanisms other than transcriptional control may be acting. Conversely, in other rare cases, sRNAs produced by a recessive S-allele were predicted to target the SCR gene of a more dominant allele, suggesting exceptions to the set of base-pairing rules used to predict target sites. Fourth, the target sites for the two sRNAs in Brassica were both located in the promoter sequence (Tarutani et al. 2010; Yasuda et al. 2016), and can thus reasonably be expected to prevent transcriptional initiation through local modification of the chromatin structure associated with DNA methylation. However, besides some mapping in the promoter, many of the predicted sRNA target sites in A. halleri instead mapped to the SCR intron or the intron–exon boundary (Durand et al. 2014), which suggests that distinct silencing pathways might be acting (Cuerda-Gil and Slotkin 2016). Thus, it remains to be determined whether transcriptional control is also valid when the targets are located elsewhere in the SCR gene. Finally, the dominance hierarchy at the female determinant SRK differs from that at SCR, codominance being more frequent than in the pollen both in Brassica (Hatakeyama et al. 2001) and in A. halleri (Llaurens et al. 2008). However, at this stage it remains unclear whether dominance in pistils is associated with SRK expression differences since the number of allelic pairs tested has remained limited, both in Brassica and Arabidopsis (Suzuki et al. 1999; Kusaba et al. 2002).
Here, we take advantage of the fact that pollen dominance interactions in Arabidopsis SI are controlled by the diversity of sRNAs and of their target sites to determine the base-pairing requirements for successful sRNA-mediated transcriptional silencing of recessive SCR alleles. We first used controlled crosses to obtain a collection of 88 A. halleri plants where nine S-alleles were placed in various homozygous and heterozygote combinations, for which pairwise dominance interactions had been phenotypically determined. We then developed and validated a quantitative PCR (qPCR) protocol for allele-specific expression of the nine SCR and a subset of five SRK alleles in A. halleri. This enabled us to analyze the expression dynamics of each of these alleles in four flower developmental stages and test the transcriptional control of dominance for both genes in many genotypic combinations. We quantified the strength of silencing of recessive SCR alleles and propose a quantitative threshold model for how sequence identity between the small noncoding RNAs and their target sites results in silencing.
Overall, our results advance our understanding of the SI system in the Brassicaceae family. They also offer a detailed study of the way dominance/recessivity interactions can arise in a system where they are controlled in an unusual way. Third, they provide insight into the mechanism involved, which involves sRNA species. The increased understanding of how this system works also sheds some light on the evolutionary origin and maintenance of the S-locus dominance hierarchy in Brassicaceae.
Materials and Methods
Plant material
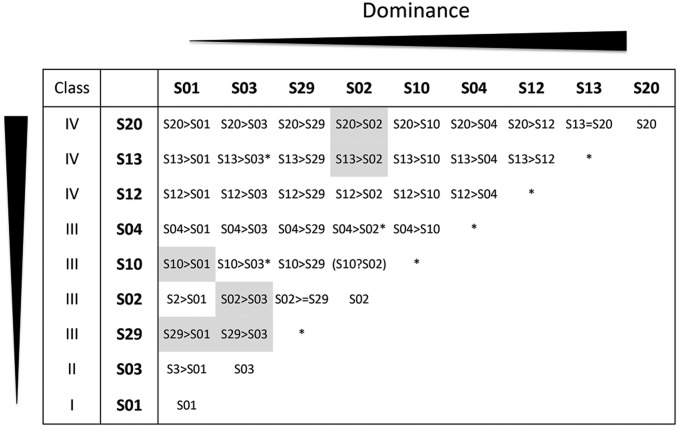
We used controlled crosses to create a collection of 88 A. halleri plants containing nine different S-alleles (S01, S02, S03, S04, S10, S12, S13, S20, and S29) in a total of 37 of all 45 possible homozygous and heterozygous combinations (Figure 1). Some S-locus genotypes were obtained independently by controlled crosses of different parental plants and were considered below as “biological replicates” (with different genetic backgrounds, averaging n = 2.05 biological replicates per S-locus genotype; Supplemental Material, Table S1 and Table S2). Three plants were cloned by taking cuttings and considered as “clone replicates” (identical genetic background, Table S1) that we used to evaluate the expression variance associated with different genetic backgrounds.
Figure 1.
Pairwise dominance interactions in pollen between the nine A. halleri S-alleles included in this study. Gray-shaded cells indicate pairwise dominance interactions inferred from phylogenetic classes rather than directly determined phenotypically. Star symbols indicate genotypes that were not available for the transcriptional analysis. Genotype S10S02 is shown in parentheses with a question mark to indicate that this dominance relationship is currently not known experimentally, and cannot be determined from the phylogeny because these two alleles belong to the same phylogenetic class (shown on the left side of the figure). The possibly unusual dominance interaction between S02 and S29 is indicated by the symbol “>=” (see text for details). Details of the crosses and references for the raw phenotypic data used in this figure are reported in Table S3.
Each plant was genotyped at the S-locus using the PCR-based protocol described in Llaurens et al. (2008). Dominance interactions between S-allele pairs in heterozygotes were either taken from Llaurens et al. (2008), Durand et al. (2014), or Leducq et al. (2014), or newly determined by controlled pollination assays following the protocol of Durand et al. (2014). In a few instances where the relative dominance of the two alleles had not been determined, these were inferred from the phylogeny of SRK alleles, which is largely consistent with the dominance hierarchy (Durand et al. 2014) and was used as a basis to define four classes of S-alleles (Prigoda et al. 2005). The pairwise dominance interactions between these alleles as determined by pollen and pistil compatibility phenotypes of heterozygous plants are reported in Figure 1 and Table S3.
RNA extraction and reverse transcription
On each plant, we collected flower buds at four developmental stages: (1) five highly immature inflorescence extremities [> 2.5 days before opening, buds smaller than 0.5 mm, and stages 1–10 in A. thaliana according to Smyth et al. (1990)]; (2) 10 immature buds (2.5 days before opening, between 0.5 and 1 mm, approximately stage 11); (3) 10 mature buds (1 day before opening, longer than 1 mm, and approximately stage 12); and (4) 10 open flowers (approximately stages 13–15). These stages were characterized by establishing the size distribution within each stage and measuring the time to flower opening based on 10 buds for each stage overall. Samples collected were flash-frozen in liquid nitrogen, then stored at −80° before RNA extraction. Tissues were finely ground with a FastPrep-24 5G Benchtop Homogenizer (model number 6004-500; MP Biomedicals) equipped with a Coolprep 24 × 2 ml adapter (6002-528) and FastPrep Lysis Beads & Matrix tube D. Total RNAs were extracted with the Arcturus “Picopure RNA isolation” kit from Life Science (PN: KIT0204) according to the manufacturer’s protocol, including a step of incubation with DNAse to remove genomic DNA (gDNA) contamination. We normalized samples by using 1 mg of total RNA to perform reverse transcription using the RevertAid Fermentas enzyme following the manufacturer’s instructions.
Primer design
A major challenge for studying the expression of multiple S-alleles is their very sequence divergence (Goubet et al. 2012), precluding the possibility of designing qPCR primers that amplify all alleles of either the SRK or the SCR locus. Hence, we designed qPCR primers specifically targeted to each SCR and SRK allele, and for each heterozygous genotype we independently measured expression of both alleles of each gene. Primers were designed based on genomic sequences from BAC clones (Goubet et al. 2012; Durand et al. 2014; Novikova et al. 2016), with lengths of ∼20 nucleotides, GC content ∼50%, and a target amplicon size around 150 nucleotides (Figure S1). The coding sequence of SCR is composed of two small exons separated by one large intron, and that of SRK is composed of seven exons. Whenever possible, we placed primers on either side of an intron to identify and discard amplification from residual gDNA. However, this was not always possible, because the coding sequence of the SCR gene is short; for SCR01 and SCR20, both primers were within the same exon. For SRK alleles, the primers were again designed on either side of the first intron or spanning the first and second introns (Figure S2). Given the effort of optimizing new qPCR primers, and because no differences in transcript levels were previously observed between dominant and recessive SRK alleles (Suzuki et al. 1999; Kusaba et al. 2002), we decided to place most effort on SCR and optimized qPCR primers for all nine SCR alleles, but focused on five SRK alleles only. To obtain relative expression levels across samples, we used actin 8 (At1g49240) as a housekeeping gene for standardization after verifying that the A. thaliana and A. halleri sequences are identical at the primer positions (An et al. 1996). Primer sequences are reported in Table S4.
Quantitative real-time PCR
On each complementary DNA (cDNA) sample, at least three qPCR reactions (referred to below as “technical” replicates) were performed for actin 8 and for each of the S-alleles contained in the genotype (one S-allele for homozygotes and two S-alleles for heterozygotes). The runs were made on a LightCycler480 (Roche) with iTaq Universal SYBR Green Supermix (ref 172-5121; Bio-Rad, Hercules, CA). Amplified cDNA was quantified by the number of cycles at which the fluorescence signal was greater than the default threshold during the logarithmic phase of amplification using the LightCycler 480 software release 1.5.0 SP3. The CtSCR and CtSRK values of each technical replicate were normalized relative to the average Ctactin measure across the three replicates. The relative transcript levels are shown after normalization with actin amplification through the comparative 2−ΔCt method (Livak and Schmittgen 2001).
Validation of qPCR primers at the dilution limits
Given the very large nucleotide divergence between alleles of either SCR or SRK, cross-amplification is unlikely. However, to formally exclude this, we first performed cross-amplification experiments by using each pair of SCR primers on a set of cDNA samples from individual plants that did not contain that target SCR allele, but instead contained two other SCR alleles in various heterozygous genotypic combinations (n = 7 individuals on average). To evaluate our ability to measure expression of SCR alleles in biological situations where they are expected to be transcriptionally silenced, we then used a series of dilutions to explore the loss of linearity of the relationship between Ct and the dilution (with six to eight replicates per level). Then, we examined the shape of the melting curves to determine whether our measures at limiting dilutions reflected proper PCR amplification or the formation of primer dimers. Finally, we used water in place of cDNA to evaluate the formation of primer dimers in the complete absence of the target template DNA.
Expression dynamics and the effect of dominance
We used generalized linear mixed models (lme4 package in R; Bates et al. 2014) to decompose Ct values normalized by the actin 8 control (as the dependent variable) into the effects of five explanatory variables. Two of them were treated as fixed effects: developmental stage (four categories) and relative dominance of the allele studied in the genotype (three categories: recessive, dominant, and homozygous). Because expression of the different SCR (and SRK) alleles was quantified by different primer pairs with inevitably different amplification efficiencies, Ct values cannot be directly compared across alleles and accordingly we included the identity of SCR or SRK alleles as random effects. Biological and clone replicates were also treated as random effects, with clones nested within biological replicates [lmer(log(Ct_SCR.actine) ∼ stage*dominance_phenotype + (1|allele_measured:stage) + (1|Biological_replicate/Clone_replicate), Table S5]. We visually examined the normality of the residuals of the model under different distributions of 2−ΔCt, including Gaussian, γ, and Gaussian with logarithmic transformations. We tested whether the different S-alleles have different expression profiles across developmental stages, as suggested by Kusaba et al. (2002) for SCR in A. lyrata, using ANOVA to compare nested models in which a random effect for the interaction between the “allele-measured” and “stage” effects was either absent (model 1) or introduced (model 2; Table S5B), in addition to the fixed effect of stage. The existence of this interaction was tested separately for SCR and SRK.
Target features and silencing effect
Expression of SCR in heterozygous genotypes in A. halleri is controlled by sRNA-based regulatory machinery (Durand et al. 2014). We sought to determine how SCR transcript levels are affected by specific features of the sRNA–target interactions between S-alleles. We retrieved sRNA from our previous studies’ sequencing data from individuals carrying eight of the nine S-alleles considered [S01, S03, S04, S10, S12, S13, and S20 from Durand et al. (2014) and S02 from Novikova et al. (2016)]. No sRNA sequencing data were available for S29. We used these sRNA sequences to determine the complete set of sRNA molecules uniquely produced by the annotated sRNA precursors of each of these eight S-alleles. To do this, we mapped the sRNA reads to the sRNA precursor sequences carried by the respective S-alleles, excluding those that mapped to other locations in the closely related A. lyrata genome (Durand et al. 2014). For each sRNA produced by a given S-allele, we then predicted putative target sites in the SCR gene of all other S-alleles, including 2 kb of genomic nucleotide sequence both upstream and downstream of SCR, using the dedicated alignment algorithm and scoring matrix described in Durand et al. (2014). Briefly, this algorithm quantifies alignment quality by a scoring system based on adding positive or negative values for matching nts (+1), and mismatches and gaps (−1), taking into account the noncanonical G:U interaction (−0.5). For each pair of alleles considered, only the sRNA–target combination with the highest score was selected for further analysis (Table S6). The analysis was performed regardless of the dominance relationship (i.e., we predicted putative target sites of sRNAs produced by dominant S-alleles onto recessive S-alleles, and reciprocally from recessive S-alleles onto dominant S-alleles). Because the mechanisms by which silencing is achieved remained unclear at this stage, we did not filter these sRNAs further in terms of length or identity of the 5′ nucleotide, in line with Durand et al. (2014). When the target with the highest score involved an sRNA with a noncanonical size (anything but 21 or 24 nt), we also reported the best target score among the set of 21- and 24-nt sRNA molecules produced by the same S-allele (Table S6). We used Akaike Information Criteria (AIC) to compare how well different base-pairing scores for target site identification predicted the level of SCR expression (and hence the silencing phenomenon), varying the threshold from 14 to 22. Lower values of AIC are associated with better fit of the model. We then added a new fixed effect in our basal model to test whether targets at different positions along the SCR gene (five categories: 5′ portion, exons, intron, overlapping the exon–intron boundary, or 3′ portion of the gene) are associated with different silencing strengths. For this analysis, we included only targets above the threshold identified (score ≥ 18, see Results).
Effective silencing of recessive SCR alleles in Brassica rapa depends upon combinations of individual sequence mismatches between the Smi and Smi2 sRNAs and their target sites in the class II alleles (Yasuda et al. 2016), but interactions in that study were predicted based on raw counts of nucleotide mismatches, and were thus not directly comparable to our results. To determine whether the base-pairing requirements for silencing are similar, we reanalyzed these interactions using our scoring system to compare the sRNA–target alignment scores between Brassica and Arabidopsis (Tarutani et al. 2010; Yasuda et al. 2016).
Finally, we used the phylogeny in Durand et al. (2014) to classify sRNA–target interactions into “recent” (mir867 and mirS4) and “ancient” (mirS1, mirS2 mirS3, mirS5, mir1887, and mir4239). Based on this classification, we used a linear regression to compare the alignment score for recent and ancient sRNAs, and tested the hypothesis that interactions with base-pairing scores above the threshold at which silencing was complete correspond to recently emerged interactions that have not yet accumulated mismatches.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.c.4877223.v3.
Results
Validation of the qPCR protocol and the allele-specific primers
The specificity test confirmed the absence of cross-amplification between alleles, as the Ct measures for water control and cross-amplification were comparably high (around Ct = 34), and both were higher than the positive controls (median Ct = 22; Figure S3). Overall, serial dilutions of the template cDNA confirmed the linearity of the Ct measure within the range of values observed for a given allele across the different conditions examined (Figure S4A). Because we aimed to study silencing, we then explored how signal was lost at the dilution limits. As expected, linearity started to be lost at very low cDNA concentrations (in particular for alleles SCR01, SCR02, SCR04, SCR13, and SCR20; Figure S4A), and examination of melting curves under these conditions indicated the formation of primer dimers rather than the expected transcripts. Hence, we note that comparing levels of expression for a given allele between different recessive contexts (e.g., when silenced by different sRNAs) will be challenging, especially for the abovementioned alleles. The dilution series was also linear for most SRK alleles (Figure S4B), except for SRK12 (data not shown), which was excluded from further analyses.
SCR and SRK expression dynamics across flower development stages
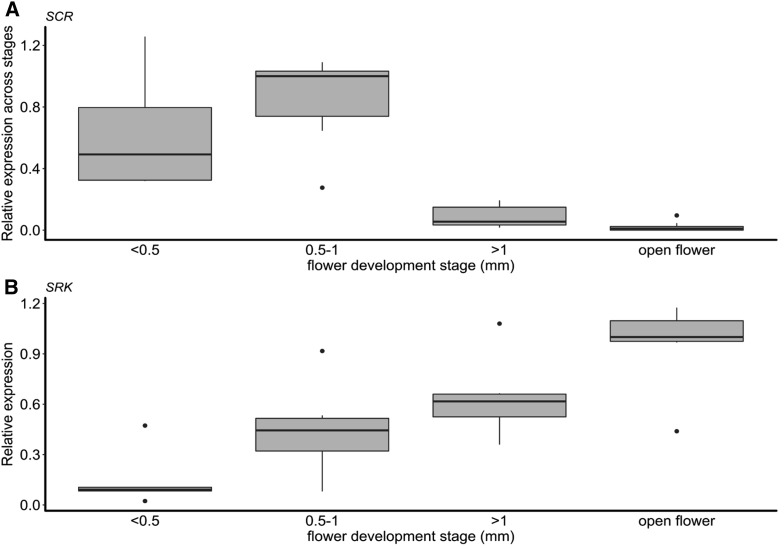
In total, we performed 344 RNA extractions and RT-PCRs from the 37 different S-locus genotypes sampled at four developmental stages. For SCR, we measured 1838 CtSCR/Ctactin expression ratios (i.e., an average of 26.9 expression measures per S-allele in each diploid genotype; Table S1). For SRK, we measured 480 CtSRK/Ctactin ratios (i.e., an average of 11.1 expression measures per S-allele in each diploid genotype; Table S2). Distribution of the residuals of the generalized mixed linear model was closest to normality after logarithmic transformation of the ratios (Figure S5). As expected, measured expression levels for a given S-locus genotype were more repeatable across clones than across biological replicates (deviance estimates of 0.40 and 1.08, respectively; Table S5A). The deviance associated with the allele’s expression dynamics was higher (deviance = 4.56), although we note that the technical error was also important (deviance = 6.08; Table S5A). We first examined the expression dynamics of the different SCR alleles across developmental stages. Because recessive SCR alleles were consistently silenced (see below), for this analysis we focused only on genotypes in which each focal allele was known to be phenotypically dominant (Figure 2A). Overall, we observed strong differences between stages (F-value: 10.76, P-value: 5.7e−5; Table S5C), with high expression of SCR in buds at early developmental stages (< 0.5–1 mm) and low expression in late buds right before opening and in open flowers. This pattern is consistent with degeneration of the anther tapetum in later stages, as SCR is expected to be expressed in this cellular layer. The expression dynamics of SRK differed sharply from those of SCR, with monotonically increasing expression during flower development (lowest in immature buds < 0.5 mm and highest in open flowers; see Figure 2B; F-value: 4.411, P-value: 0.007; Table S5H). We detected differences in the expression dynamics between SCR alleles (χ2: 308.19, P-value < 2.2e−16; Table S5B) in line with Kusaba et al. (2002), and also for SRK (χ2: 6.9103, P-value 0.00857; Table S5G).
Figure 2.
Expression dynamics of (A) SCR and (B) SRK during flower development, from early buds (< 0.5 mm) to open flowers. For SCR, only genotypes in which a given allele was either dominant or codominant were included (recessive SCR alleles were strongly silenced at all stages and were therefore not informative here). All genotypes are shown for SRK. For each allele, 2−ΔCt values were normalized relative to the developmental stage with the highest expression. For each stage, the thick horizontal line represents the median, and the box represents the first and third quartiles. The upper whisker extends from the hinge to the largest value no further than 1.5 x interquartile range from the hinge (or distance between the first and third quartiles). The lower whisker extends from the hinge to the smallest value at most 1.5 x interquartile range of the hinge and the black dots represents outlier values.
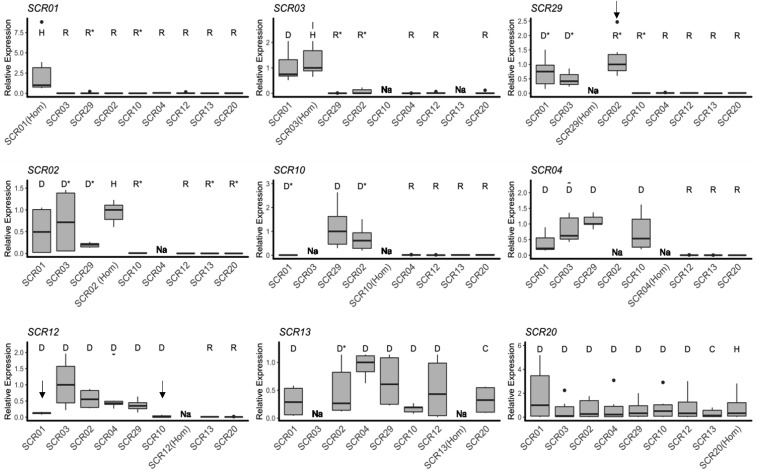
Transcriptional control
Based on this clarified dynamics of transcripts abundance, we reduced noise by focusing on the <0.5 and 1 mm stages (the most informative developmental stage) and averaged 2−ΔCt values across these two stages to compare the expression of a given focal SCR allele between genotypic contexts where it was either dominant or recessive relative to the other allele present. Of the 54 pairs where the dominance phenotype had been established by controlled crosses and qPCR results were available for both SCR alleles (Table S3), 51 (94.4%) are associated with strongly asymmetrical transcript levels, with higher expression of the dominant SCR allele (Figure 3). Our results are thus largely consistent with the hypothesis of transcriptional control of the dominance hierarchy in pollen genotypic combinations. SCR transcripts of the most recessive allele S01 were detected only in the S01/S01 homozygote, and not in any other genotypic combination. Going up the dominance hierarchy, SCR expression was detected in an increasing number of heterozygous combinations, in agreement with the phenotypic dominance (Figure 3). The two most dominant alleles, SCR13 and SCR20, were expressed in all heterozygous contexts, including when heterozygous with one another (S13/S20), also as expected given their observed codominance (Durand et al. 2014). Only three exceptions were found (indicated by arrows on Figure 3). We observed low expression for both SCR01 and SCR12 in S01/S12 heterozygotes and for both SCR10 and SCR12 in the S10/S12 genotype, which is not consistent with the documented phenotypic dominance of these alleles in pollen (S12 > S01 and S12 > S10; see Figure 1 and Table S3). We also detected expression of both SCR02 and SCR29 in the heterozygous S02/S29 genotype, which might explain the unusual phenotypic data indicating robust rejection of pollen from this heterozygous genotype by the [S02] tester line, but only partial compatibility with the [S29] tester line (Table S3). The dominance interaction between these two alleles may therefore be partial, at both the transcriptional and phenotypic levels. Interestingly, these two alleles belong to a phylogenetic class of S-alleles (class III in Prigoda et al. 2005), which in A. lyrata tend to show inconsistent (or leaky) SI responses (Kusaba et al. 2001).
Figure 3.
Expression of individual SCR alleles in different genotypic contexts. Pollen dominance statuses of the S-allele whose expression is measured relative to the other allele in the genotype as determined by controlled crosses are represented by different letters (D: dominant; C: codominant; R: recessive; U: unknown; and H: Homozygote; Table S3). In a few instances, relative dominance statuses of the two alleles had not been resolved phenotypically and were inferred from the phylogeny (marked by asterisks). Thick horizontal bars represent the median of 2−ΔCt values, first and third quartiles are indicated by the upper and lower limits of the boxes. The upper whisker extends from the hinge to the largest value no further than 1.5 x interquartile range from the hinge (or distance between the first and third quartiles). The lower whisker extends from the hinge to the smallest value at most 1.5 x interquartile range of the hinge and the black dots represents outlier values. We normalized values relative to the highest median across heterozygous combinations within each panel. Alleles are ordered from left to right and from top to bottom according to their position along the dominance hierarchy, with SCR01 the most recessive and SCR13 and SCR20 the most dominant alleles. Under a model of transcriptional control of dominance, high expression is expected when a given allele is either dominant or codominant, and low expression when it is recessive. Exceptions to this model are marked by black vertical arrows and discussed in the text. “Na” marks genotypes that were not available.
Overall, with these three exceptions, we observed striking differences in transcript levels of the same SCR allele depending on the relative phenotypic dominance status in different genotypes (F-value = 19.538; P-value < 2.2e−16; Table S5C), suggesting complete silencing of recessive SCR alleles. Specifically, we observed an average 145-fold decrease in transcript abundance in genotypes where a given focal allele was phenotypically recessive as compared to genotypes in which the same allele was dominant. We note that the silencing was so strong that the Ct values associated with recessive SCR transcripts were comparable with those of the negative controls (Figure S3) and close to the detection limits of our method, such that the magnitude of the calculated fold-change value is probably underestimated. In contrast, we found no significant effect of dominance in pistils on SRK expression (F-value: 6.8884; P-value: 0.068244; Figure S6 and Table S5H), confirming the absence of transcriptional control of dominance for SRK.
Target features and silencing effect
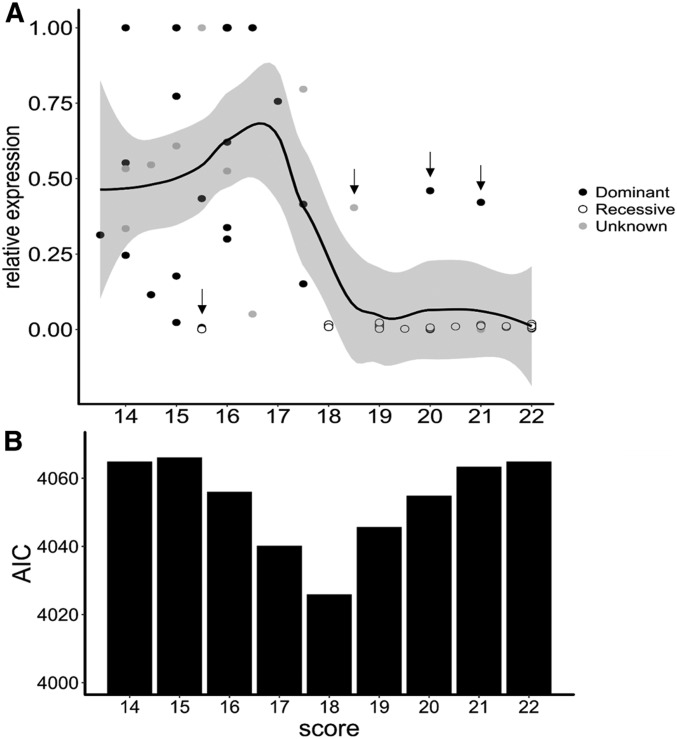
Levels of SCR expression of any given focal allele varied sharply with the alignment score of the best target available for the repertoire of canonical sRNAs produced by the other allele present in the genotype (Figure 4A). Specifically, we observed high average levels of SCR transcripts when the score of their best predicted target was low, but consistently low levels of SCR transcripts when the score of the best target was high (Figure 4A and Table S5D). Strikingly, the transition between high expression and low expression was abrupt (around an alignment score of 18, Figure 4B), suggesting a sharp threshold effect rather than a quantitative model for transcriptional silencing.
Figure 4.
Base-pairing requirements for the transcriptional control of SCR alleles by small RNAs (sRNAs) suggest a threshold model. (A) Relative expression of SCR alleles as a function of the alignment score of the “best” interaction between the focal allele (including 2 kb of sequence upstream and downstream of SCR) and the population of sRNAs produced by sRNA precursors of the other allele in the genotype. For each allele, expression was normalized relative to the genotype in which the 2−ΔCt value was highest. Dots are colored according to the dominance status of the focal SCR allele in each genotypic context (black: dominant; white: recessive; and gray: undetermined). The black line corresponds to a local regression obtained by a smooth function (loess function, span = 0.5) in the ggplot2 package (Wickham 2009) and the gray area covers the 95% C.I. Vertical arrows point to observations that do not fit the threshold model of transcriptional control and are represented individually on Figure 5. (B) Bar plot of the Akaike Information Criteria (AIC) quantifying the fit of the generalized linear model for different target alignment scores used to define functional targets. Lower AIC values indicate a better fit, indicating that a threshold score of 18 to define functional sRNA–target interactions provides the best explanatory power of the variation in SCR transcript levels in heterozygotes.
In two cases, the SCR gene of a dominant allele (S20) was predicted to be targeted by an sRNA with a score above the threshold but still had high relative expression (in agreement with the dominant phenotype established by controlled crosses), confirming the absence of silencing (target of Ah04mir4239 on SCR20, score = 20, and target of Ah10mir4239 on SCR20, score = 21; Figure 5A). Hence, these two targeting predictions do not seem to result in functional interactions and may correspond to false positives. Examining these two exceptions in detail did not reveal mismatches at the 10–11th nucleotide position, suggesting that mismatches at other positions have rendered these sRNA–target interactions inactive (Figure 5A). We note that the target of Ah10mir4239 with the highest score is predicted for an sRNA of noncanonical size (25 nt), but that this precursor also produces a canonical 24-nt sRNA (sRNA precursors commonly produce a variety of different molecules called “isomirs”) with a score above the threshold (score = 20, Table S6). These two sRNAs (Ah04mir4239 and Ah10mir4239) have a 5′ nucleotide different from the expected “A” for 24-nt sRNAs, possibly suggesting that loading into an improper AGO protein may have rendered these predicted interactions inactive. Another exception concerns the observed low score (15.5) for the best match between an sRNA from the dominant allele Ah04mirS4 and its best putative target at the recessive SCR03 (Figure 5B). Whether SCR04 silences SCR03 through this unusual target or through another elusive mechanism remains to be discovered.
Figure 5.
Predicted sRNA–target interactions that do not fit with the documented dominance phenotype or the measured expression. For each alignment, the sequence on top is the small RNA (sRNA) and the bottom sequence is the best predicted target site on the SCR gene sequence (including 2 kb of sequence upstream and downstream of SCR). (A) sRNA targets with a score > 18, while the S-allele producing the sRNA is phenotypically recessive over the S-allele containing the SCR sequence. (B) sRNA target with a score < 18, while the S-allele producing the sRNA (S04) is phenotypically dominant over the S-allele containing the SCR sequence and transcript levels of the SCR03 allele are accordingly very low. This is the best target we could identify on SCR03 for sRNAs produced by S04.
In spite of the generally very low expression of all recessive alleles, we found marginal evidence that the strength of silencing experienced by a given SCR allele varies across genotypic combinations for a given allele (F-value = 2.221, P-value = 0.0756; Table S5I). However, there was no evidence that the position of the target site in the measured allele (promoter, intron, intron–exon boundary, or upstream vs. downstream) could explain this variation (F-value = 1.7061, P-value = 0.1928; Table S5E). We also found no effect of the inferred age of the miRNA on the mean alignment score (mean = 20.41 and 20.22 for recent or ancient miRNAs, respectively; F-value: 0.0362; P-value = 0.8504; Table S5J). Finally, we compared the alignment scores observed here in Arabidopsis with those in Brassica for Smi and Smi2 on their SCR target sequences. A clear threshold was also observed, but in Brassica the alignment score threshold distinguishing dominant from recessive interactions was 16.5 instead of 18 (Table S6), suggesting distinct base-pairing requirements for effective silencing in these two systems.
Discussion
Here, we build upon the interallelic regulatory network controlling dominance interactions revealed in Durand et al. (2014) to determine the base-pairing requirement for sRNA silencing in the Arabidopsis SI system. We first clarified several aspects of the expression pattern of the SI genes in A. halleri and confirmed that the dominance interactions in Arabidopsis involve transcriptional regulation of the SCR gene.
Expression profile of pollen and pistil SI alleles in A. halleri
Earlier accounts had suggested that alleles of the allelic series may differ from one another in their expression dynamics over developmental stages (Kusaba et al. 2002). In line with Suzuki et al. (1999), Schopfer et al. (1999), Takayama et al. (2000), Shiba et al. (2002), and Kakizaki et al. (2003), we found maximal expression of SCR in early buds but low or no expression at the open-flower stage. This expression pattern is consistent with in situ hybridization experiments showing that SCR transcripts are localized in the tapetum, a specialized layer of cells involved in pollen grain coating (Iwano et al. 2003), which undergoes apoptosis and is quickly degraded as the pollen grains develop inside the anther (Murphy and Ross 1998; Takayama et al. 2000). We confirmed differences in the temporal dynamics of expression among alleles, as suggested by Kusaba et al. (2002) in A. lyrata, possibly as the result of sequence divergence of the promotor sequences of the different SCR alleles. Finally, we confirmed that, unlike SCR, transcript levels of SRK increased steadily during flower development and were very low in early buds, consistent with the observation that SI in Brassicaceae can be experimentally overcome to obtain selfed progenies by “bud pollination” (Pearson 1929).
Generality of the transcriptional control of dominance in Arabidopsis SI
We then confirmed the generality of the transcriptional control of dominance for SCR, with as many as 96.3% of the documented dominance interactions associated with monoallelic expression of the dominant SCR allele and complete silencing of the recessive SCR allele in heterozygote genotypes. Even in the single heterozygous genotype where, in our previous study (Durand et al. 2014), no sRNA produced by the phenotypically dominant allele was predicted to target the sequence of the phenotypically recessive SCR allele (e.g., S04 > S03), transcripts from the recessive SCR03 allele were undetectable. This suggests either that some functional sRNAs or targets have remained undetected by previous sequencing and/or by our in silico prediction procedures, or that mechanisms other than sRNAs may cause transcriptional silencing for some S-allele combinations. Regardless of the underlying cause, the generality in the transcriptional control of dominance suggests that simply comparing transcript levels between the two alleles in a heterozygote could be used as a first approximation to determine their relative dominance levels. In contrast, we confirmed the absence of transcriptional control for SRK, where both alleles were consistently expressed at similar levels in the heterozygous genotypes examined, irrespective of the (pistil) dominance phenotype. For SRK, other dominance mechanisms must therefore be acting, which are yet to be discovered (e.g., Naithani et al. 2007).
Variation in the strength of silencing of sRNA targets along the SCR gene
An important feature of the silencing phenomenon in pollen is that the decrease of transcript levels for recessive SCR alleles was very strong in heterozygous genotypes, with transcript levels of recessive alleles below the limits of detection in most cases. This is in line with the strong transcriptional silencing by heterochromatic siRNAs [typically very strong for transposable element sequences, see Marí-Ordóñez et al. (2013)], while post-transcriptional gene silencing by miRNAs can be more quantitative (Liu et al. 2014). As a result of this strong decrease of transcript levels, the strength of silencing appears independent of whether the sRNA target in the SCR gene is in the promoter vs. the intron, although we note our low power to distinguish among transcript levels of recessive alleles, which were all extremely low. It remains to be discovered whether different locations of the sRNA targets (Durand et al. 2014) imply different transcriptional silencing mechanisms.
A simple threshold model for sRNA-based silencing
Based on the many allelic combinations where we could compare the predicted target sites with the level of transcriptional silencing, we find that a simple threshold model for base pairing between sRNAs and their target sites captures most of the variation in SCR expression in heterozygotes. This result provides direct experimental validation of the ad hoc criteria used in Durand et al. (2014). However, our results also indicate that this quantitative threshold does not fully capture the complexity of targeting interactions. Indeed, in three of the 54 cases tested, this simple threshold model would wrongly predict targeting of a dominant SCR allele by an sRNA from a more recessive allele, as the dominant SCR allele was actually expressed at normal levels, with no sign of silencing in these heterozygotes (Figure 5A). The targeting interaction may be abolished by defects in the sRNA itself (e.g., for Ah04mir4239 the 5′ nucleotide is a G, while the majority of functional 24-nt sRNA molecules end with a 5′A, which may interfere with loading in the appropriate AGO protein). Alternatively, the targeting interactions may be abolished by the position of the mismatches (at positions 14 and 22 of the Ah10mir4239, and at positions 13 and 21 of the Ah04mir4239, both on SCR20). Similarly, a single mismatch at position 10 in the Smi interaction in Brassica (Tarutani et al. 2010) and in other miRNA–target interactions (Franco-Zorrilla et al. 2007) resulted in loss of the interaction function (Table S6). Interestingly, quantitative differences may exist between Arabidopsis and Brassica, as the experimentally validated targets in Brassica (Tarutani et al. 2010; Yasuda et al. 2016) correspond to a lower base-pairing threshold than our estimate in Arabidopsis. For Brassica, both class I and class II alleles have Smi sequences, but a mismatch at the 10th position was proposed to explain why the class II Smi is not functional. Here, we show that this mismatch drives the alignment score below the 16.5 threshold and could explain the loss-of-function, regardless of its position. Overall, although these sRNAs achieve their function in a way that may differ sharply from classical miRNAs (DNA methylation vs. mRNA cleavage), our results suggest that the sRNA–target complementarity rules for silencing are qualitatively consistent in the two cases (Liu et al. 2014). Better understanding of the molecular pathway through which these sRNAs epigenetically silence their target gene (SCR) will now be key to determining whether this threshold model can be generalized to more classical siRNAs found across the genome, as evidence is still missing for such classes of sRNAs.
Implications for the evolution of the dominance hierarchy
The existence of a threshold model has important implications for how the dominance hierarchy can evolve. Our model suggests that a single SNP can be sufficient to turn a codominant interaction into dominance (and vice versa), making this a relatively trivial molecular event. Yasuda et al. (2016) observed this change in B. rapa, where single-SNP differences at the sRNA Smi2 changed the interactions with its SCR target sequence and resulted in a linear dominance hierarchy among the species’ four class II S-alleles. Strikingly, we observed some cases of base pairing at sRNA–target interactions with very high alignment scores (up to 22, i.e., above the threshold at which transcriptional silencing was already complete, which occurred at a score of 18). Under our simple threshold model, such interactions are not expected since acquiring a more perfect target is not expected to give a further fitness gain. These interactions might simply reflect the recent emergence of these silencing interactions. Indeed, one model for the emergence of new miRNAs in plant genomes involves a partial duplication of the target gene, which entails perfect complementarity at the time of origin with degradation over time by the accumulation of mutations (Allen et al. 2004). Under this scenario, the higher-than-expected levels of sRNA–target complementarity could reflect the recent origin of these sRNAs. However, we found no evidence of a difference in alignment scores for young vs. old sRNA precursors. A second possibility is that selection for developmental robustness acts to ensure monoallelic expression of SCR (especially during stress events; Boukhibar and Barkoulas 2016), since biallelic expression in pollen results in increased rejection by pistils and thus greatly reduces a plant’s reproductive fitness (Llaurens et al. 2009). Indeed, we observed strong variation in overall SCR expression when the sRNA target score of the companion allele was below the threshold in the benign greenhouse conditions under which we grew our plants, and it is possible that the epigenetic machinery may be weaker under stress conditions, and require stronger base pairing to achieve proper silencing. Finally, a third possibility is that sRNA–target complementarity above the threshold reflects the pleiotropic constraint of having a given sRNA from a dominant allele control silencing of the complete set of target sequences from the multiple recessive alleles segregating, and reciprocally of having a given SCR target in a recessive allele maintaining a molecular match with a given sRNA distributed among a variety of dominant alleles. Comparing the complementarity scores of sRNA–target interactions among sRNAs or targets that contribute to high vs. low numbers of dominance/recessive interactions will now require a more complete description of the sRNA–target regulatory network among the larger set of S-alleles segregating in natural populations.
Acknowledgments
We thank Sylvain Billiard and Isabelle de Cauwer for statistical advice and discussions, Romuald Rouger and Anne Duputié for help with producing figures, and Alexis Sarazin and three anonymous reviewers for comments on the manuscript. This work was funded by the European Research Council (NOVEL project, grant number 648321). N.B. was supported by a doctoral grant from the president of the Université de Lille-Sciences et Technologies and the French Ministry of Research. The authors also thank the Région Hauts-de-France, the Ministère de l’Enseignement Supérieur et de la Recherche (CPER Climibio), and the European Fund for Regional Economic Development for their financial support.
Author contributions: N.B., S.S., S.B., and A.-C.H. performed the molecular biology experiments. C.P. and E.S. obtained and took care of the plants. S.S., I.F.-L., and X.V. provided advice on the experimental strategy and interpretations. N.B. performed the statistical analyses. V.C. supervised the work. N.B. and V.C. wrote the manuscript.
Footnotes
Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.c.4877223.v3.
Communicating editor: J. Birchler
Literature Cited
- Aalto A. P., and Pasquinelli A. E., 2012. Small non-coding RNAs mount a silent revolution in gene expression. Curr. Opin. Cell Biol. 24: 333–340. 10.1016/j.ceb.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A. M., Sung G. H., Spatafora J. W. et al. , 2004. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290. 10.1038/ng1478 [DOI] [PubMed] [Google Scholar]
- An Y. Q., McDowell J. M., Huang S., McKinney E. C., Chambliss S., et al. , 1996. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10: 107–121. 10.1046/j.1365-313x.1996.10010107.x [DOI] [PubMed] [Google Scholar]
- Axtell M. J., and Meyers B. C., 2018. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 30: 272–284. 10.1105/tpc.17.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., and Walker S., 2014. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. [Google Scholar]
- Boukhibar L. M., and Barkoulas M., 2016. The developmental genetics of biological robustness. Ann. Bot. (Lond.) 117: 699–707. 10.1093/aob/mcv128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V., and Vekemans X., 2004. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Mol. Ecol. 13: 2873–2889. 10.1111/j.1365-294X.2004.02267.x [DOI] [PubMed] [Google Scholar]
- Castric V., Bechsgaard J., Schierup M. H., and Vekemans X., 2008. Repeated adaptive introgression at a gene under multiallelic balancing selection. PLoS Genet. 4: e1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D., and Slotkin R. K., 2016. Non-canonical RNA-directed DNA methylation. Nat. Plants 2: 16163 (erratum: Nat. Plants 3: 16211). 10.1038/nplants.2016.163 [DOI] [PubMed] [Google Scholar]
- De Nettancourt D., 2001. Incompatibility and Incongruity in Wild and Cultivated Plants. Springer-Verlag, New York: 10.1007/978-3-662-04502-2 [DOI] [Google Scholar]
- Ding J., Zhou S., and Guan J., 2012. Finding MicroRNA targets in plants: current status and perspectives. Genomics Proteomics Bioinformatics 10: 264–275. 10.1016/j.gpb.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E., Méheust R., Soucaze M., Goubet P. M., Gallina S. et al. , 2014. Dominance hierarchy arising from the evolution of a complex small RNA regulatory network. Science 346: 1200–1205. 10.1126/science.1259442 [DOI] [PubMed] [Google Scholar]
- Fei Q., Xia R., and Meyers B. C., 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. 10.1105/tpc.113.114652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E. J., Liang D., and Wang M., 2011. Self-incompatibility: Smi silences through a novel sRNA pathway. Trends Plant Sci. 16: 238–241. 10.1016/j.tplants.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I. et al. , 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Genete M., Castric V., and Vekemans X., 2020. Genotyping and de novo discovery of allelic variants at the Brassicaceae self-incompatibility locus from short-read sequencing data. Mol. Biol. Evol. 37: 1193–1201. 10.1093/molbev/msz258 [DOI] [PubMed] [Google Scholar]
- Goubet P. M., Bergès H., Bellec A., Prat E., Helmstetter N. et al. , 2012. Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet. 8: e1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K., Takasaki T., Suzuki G., Nishio T., Watanabe M. et al. , 2001. The S Receptor Kinase gene determines dominance relationships in stigma expression of self-incompatibility in Brassica. Plant J. 26: 69–76. 10.1046/j.1365-313x.2001.01009.x [DOI] [PubMed] [Google Scholar]
- Iwano M., Shiba H., Funato M., Shimosato H., Takayama S. et al. , 2003. Immunohistochemical studies on translocation of pollen S-haplotype determinant in self-incompatibility of Brassica rapa. Plant Cell Physiol. 44: 428–436. 10.1093/pcp/pcg056 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P., and Bartel B., 2006. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53. 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- Kakizaki T., Takada Y., Ito A., Suzuki G., Shiba H. et al. , 2003. Linear dominance relationship among four class-II S haplotypes in pollen is determined by the expression of SP11 in Brassica self-incompatibility. Plant Cell Physiol. 44: 70–75. 10.1093/pcp/pcg009 [DOI] [PubMed] [Google Scholar]
- Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J. B. et al. , 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643. 10.1105/tpc.13.3.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Tung C.-W., Nasrallah M. E., and Nasrallah J. B., 2002. Monoallelic expression and dominance interactions in anthers of self-incompatible Arabidopsis lyrata. Plant Physiol. 128: 17–20. 10.1104/pp.010790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leducq J.-B., Gosset C. C., Gries R., Calin K., Schmitt É., et al. 2014. Self-incompatibility in Brassicaceae: identification and characterization of SRK-like sequences linked to the S-locus in the tribe Biscutelleae. G3 (Bethesda) 4: 983–992. 10.1534/g3.114.010843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Reichel M., Li Y., and Millar A. A., 2014. The functional scope of plant microRNA-mediated silencing. Trends Plant Sci. 19: 750–756. 10.1016/j.tplants.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Liu Q., Wang F., and Axtell M. J., 2014. Analysis of complementarity requirements for plant MicroRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell 26: 741–753. 10.1105/tpc.113.120972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Llaurens V., Billiard S., Leducq J.-B., Castric V., Klein E. K. et al. , 2008. Does frequency-dependent selection with complex dominance interactions accurately predict allelic frequencies at the self-incompatibility locus in Arabidopsis halleri? Evolution 62: 2545–2557. 10.1111/j.1558-5646.2008.00469.x [DOI] [PubMed] [Google Scholar]
- Llaurens V., Billiard S., Castric V., Vekemans X., 2009. Evolution of dominance in sporophytic self-incompatibility systems: I. genetic load and coevolution of levels of dominance in pollen and pistil Evolution, 63: 2427–2437. 10.1111/j.1558-5646.2009.00709.x [DOI] [PubMed] [Google Scholar]
- Mable B. K., Schierup M. H., and Charlesworth D., 2003. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90: 422–431. 10.1038/sj.hdy.6800261 [DOI] [PubMed] [Google Scholar]
- Mallory A. C., Reinhart B. J., Jones-Rhoades M. W., Tang G., Zamore P. D. et al. , 2004. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5’ region. EMBO J. 23: 3356–3364. 10.1038/sj.emboj.7600340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V. et al. , 2013. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. 10.1038/ng.2703 [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., and Matzke A. J., 2009. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376. 10.1016/j.ceb.2009.01.025 [DOI] [PubMed] [Google Scholar]
- Murphy D. J., and Ross J. H., 1998. Biosynthesis, targeting and processing of oleosin-like proteins, which are major pollen coat components in Brassica napus. Plant J. 13: 1–16. [PubMed] [Google Scholar]
- Naithani S., Chookajorn T., Ripoll D. R., and Nasrallah J. B., 2007. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 104: 12211–12216. 10.1073/pnas.0705186104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova P. Y., Hohmann N., Nizhynska V., Tsuchimatsu T., Ali J. et al. , 2016. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat. Genet. 48: 1077–1082. 10.1038/ng.3617 [DOI] [PubMed] [Google Scholar]
- Parizotto E. A., Parizotto E. A., Dunoyer P., Dunoyer P., Rahm N. et al. , 2004. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18: 2237–2242. [corrigenda: Genes Dev. 30: 1251–1252 (2016)]. 10.1101/gad.307804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson O. H., 1929. Observations on the type of sterility in Brassica oleacea var. capitata. Proc. Am. Soc. Hortic. Sci. 26: 34–38. [Google Scholar]
- Prigoda N., Nassuth A., Mable B. K, 2005. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22: 1609–1620. 10.1093/molbev/msi153 [DOI] [PubMed] [Google Scholar]
- Schopfer C. R., Nasrallah M. E., and Nasrallah J. B., 1999. The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700. 10.1126/science.286.5445.1697 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M. et al. , 2005. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527. 10.1016/j.devcel.2005.01.018 [DOI] [PubMed] [Google Scholar]
- Shiba H., Iwano M., Entani T., Ishimoto K., Shimosato H. et al. , 2002. The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14: 491–504. 10.1105/tpc.010378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba H., Kakizaki T., Iwano M., Tarutani Y., Watanabe M. et al. , 2006. Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet. 38: 297–299. 10.1038/ng1734 [DOI] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., and Meyerowitz E. M., 1990. Early flower development in Arabidopsis. Plant Cell 2: 755–767. 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G., Kai N., Hirose T., Fukui K., Nishio T. et al. , 1999. Genomic organization of the S locus : identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shiba H., Iwano M., Shimosato H., Che F. S. et al. , 2000. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97: 1920–1925. 10.1073/pnas.040556397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutani Y., Shiba H., Iwano M., Kakizaki T., Suzuki G. et al. , 2010. Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466: 983–986. 10.1038/nature09308 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Legrand S., and Windels D., 2010. The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 15: 337–345. 10.1016/j.tplants.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Wang F., Polydore S., and Axtell M. J., 2015. More than meets the eye? Factors that affect target selection by plant miRNAs and heterochromatic siRNAs. Curr. Opin. Plant Biol. 27: 118–124. 10.1016/j.pbi.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2009. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Yasuda S., Wada Y., Kakizaki T., Tarutani Y., Miura-uno E. et al. , 2016. A complex dominance hierarchy is controlled by polymorphism of small RNAs and their targets. Nat. Plants 6: 16206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.c.4877223.v3.