Abstract
While DNA serves as the blueprint of life, the distinct functions of each cell are determined by the dynamic expression of genes from the static genome. The amount and specific sequences of RNAs expressed in a given cell involves a number of regulated processes including RNA synthesis (transcription), processing, splicing, modification, polyadenylation, stability, translation, and degradation. As errors during mRNA production can create gene products that are deleterious to the organism, quality control mechanisms exist to survey and remove errors in mRNA expression and processing. Here, we will provide an overview of mRNA processing and quality control mechanisms that occur in Caenorhabditis elegans, with a focus on those that occur on protein-coding genes after transcription initiation. In addition, we will describe the genetic and technical approaches that have allowed studies in C. elegans to reveal important mechanistic insight into these processes.
Keywords: Caenorhabditis elegans, splicing, RNA editing, RNA modification, polyadenylation, quality control, WormBook
MATURE mRNAs are born from a series of co- and post-transcriptional processing events that act on a precursor mRNA (pre-mRNA) transcribed by RNA polymerase II (Pol II) (Figure 1). The majority of known eukaryotic mRNA processing events occur in Caenorhabditis elegans, including capping of the mRNA at the 5′ end soon after nascent RNA synthesis, splicing, modification, and polyadenylation. Additionally, many nascent mRNA molecules in C. elegans undergo RNA processing events not observed in other common metazoan model organisms. In particular, 84% of C. elegans genes are trans-spliced (Tourasse et al. 2017), a process where a capped 22 nucleotide (nt) RNA (referred to as a splice leader) is covalently attached to the 5′ end of the pre-mRNA [reviewed in Blumenthal (2012) and discussed in detail later in this chapter]. The presence of a trans-splicing pathway in C. elegans has permitted the evolution of operons throughout the nematode genome (Morton and Blumenthal 2011b). While eukaryotes typically transcribe genes in a monocistronic fashion (i.e., one gene per one mRNA), the C. elegans genome has ∼15% of protein-coding genes arranged in operons (Zorio et al. 1994; Blumenthal et al. 2002). These clusters of genes are transcribed from a single promoter as a polycistronic pre-mRNA which are cotranscriptionally processed into multiple mature mRNAs.
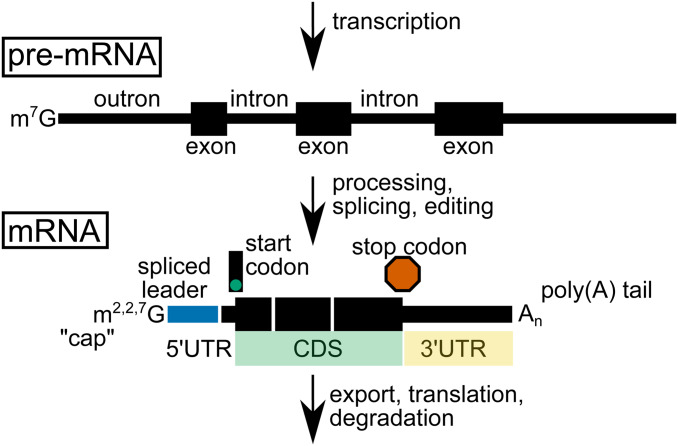
Figure 1.
Commonly used terms in the study of mRNA biogenesis and regulation. An mRNA begins its life with transcription and initially exists as a premature mRNA (pre-mRNA). The pre-mRNA includes more sequence than the mature mRNA will contain. The pre-mRNA is processed, spliced, and edited to give rise to the mature mRNA. Several commonly described features of the mature mRNA are indicated, including the trimethylguanosine “cap,” spliced leader (blue, which is appended after excision of the outron), 5′ UTR (the portion of the mRNA upstream of the start codon), CDS (green, coding DNA sequencing also called open reading frame (ORF), 3′ UTR (yellow, the region of mRNA downstream of the stop codon) and untemplated poly(A) tail. For simplicity, the pre-mRNA and mRNA are shown as discrete entities, though there is evidence that mRNA maturation occurs cotranscriptionally. The mature mRNA is exported from the nucleus, possibly translated, and eventually degraded.
The processing events of both monocistronic and polycistronic mRNAs are affected by the interaction of RNA binding proteins (RBPs) and other regulatory factors with sequence and structural elements located in noncoding regions of the mRNA, including the intron and untranslated regions (UTRs). In the Wormbase database, the gene sequence information contains the entire transcribed region, while the transcript sequence information contains the exonic information as well as the 5′ and 3′ UTRs to model the mature mRNA (Spieth et al. 2014). While these annotations are sufficient for many uses, the accuracy of transcript boundaries and structures is often improved by consulting the primary data. Several recent studies have annotated specific transcript features including 5′ ends (Saito et al. 2013), spliced leaders (Allen et al. 2011), spliced mRNA isoforms (Tourasse et al. 2017), 3′ ends (Mangone et al. 2010; Jan et al. 2011), and poly(A) tails (Lima et al. 2017).
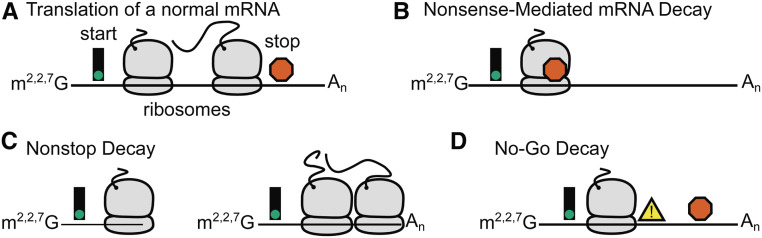
As errors in RNA processing events and transcription can lead to misexpression of proteins and/or aberrant protein function, cells employ a number of mRNA quality control pathways (Isken and Maquat 2007). The cellular machinery for mRNA processing and quality control is conserved in C. elegans. However, the nonessential nature of some of this machinery is unique to C. elegans, and provides a vantage point for determining important mechanistic insights, as well as an exploration of genetic interactions between individual gene regulatory processes and cellular pathways.
In addition to genetic screens, technical advances in exploring tissue- and cell-specific gene regulation have allowed these processes to be carefully dissected in C. elegans. For example, the combination of promoters that provide tissue- and/or cell-specific transcription, the compact genome organization, and the transparent nature of C. elegans allows for spatiotemporal analysis of the effects of RNA processing using fluorescent reporters in living animals (reviewed extensively in Gracida et al. 2016; Wani and Kuroyanagi 2017). Multichromatic reporters that are translated into different fluorescent proteins based on splicing of upstream exons have been particularly insightful in determining alternative splicing events that occur for specific transcripts in individual C. elegans neurons (Kuroyanagi et al. 2006, 2010; Norris et al. 2014).
Transcriptome-wide approaches to study effects of RNA processing and modification on gene expression in specific cells and/or tissues have also been extensively used in C. elegans. One approach uses animals with tissue/cell-type specific expression of an epitope-tagged poly(A) binding protein (PABP), which binds to the poly(A) tail of mRNAs (Roy et al. 2002). Immunoprecipitation of the tagged PABP from these animals followed by high-throughput sequencing has been used in C. elegans to identify alternative polyadenylation events that occur in transcripts expressed in intestines, muscle, neurons, seam cells, and hypodermal tissue (Blazie et al. 2015, 2017). A second global approach to identify transcripts present in specific cell- or tissue-types uses chemo-mechanical dissociation of animals expressing a fluorescent protein in the cell/tissue type of interest and fluorescence-assisted cell sorting to isolate those cells (Spencer et al. 2011, 2014). High-throughput sequencing of RNA has revealed splicing events that occur in a number of tissues in both larval and adult animals (Kaletsky et al. 2016, 2018) as well as RNA modification events that occur in neurons of larval animals (Deffit et al. 2017).
Together with behavioral and other genetic analyses, these techniques are providing a mechanistic understanding of how cell/tissue-specific splicing, modification, and polyadenylation contribute to organismal physiology. In this chapter, we will provide an introduction to each of these processes as well as the mRNA quality control pathways that help mitigate errors in mRNA processing.
RNA Editing and Modification
The central dogma suggests that mRNAs are simply faithful copies of the genome that serve as molecular instructions for protein production. However, there are a number of cellular processes that act on newly synthesized mRNA to alter genetic information. In addition to removal of large intronic sequences from nascent mRNA, individual nucleosides in RNA can be inserted, deleted, undergo base conversions, or be chemically modified. The processes of base insertion, deletion, and conversion were originally referred to as “RNA editing” to describe that the information in the RNA molecule is edited/changed from the genomic sequence (Gott and Emeson 2000). RNA editing can occur to varying extents both during development and in a cell/tissue-specific manner. This process is an important mechanism to regulate gene expression and function from a constant genome. Furthermore, as these events insert, delete, or alter base-pairing of nucleotides, RNA editing can effectively rewire genomic information to generate molecular and phenotypic diversity.
Chemical modification of RNA nucleosides occurs in all organisms (Frye et al. 2018). Over 100 chemically distinct and naturally occurring RNA modifications have been known for several decades. Many of these were initially identified in abundant RNA species such as ribosomal RNA (rRNA) and transfer RNA (tRNA), likely due to the abundance of these RNAs in cells. These modifications serve important functions in such RNAs (reviewed in (Sarin and Leidel 2014; Roundtree et al. 2017)), but here our focus will be on modifications in mRNAs. Due to advances in enrichment techniques and high-throughput sequencing, the identification of RNA modifications within mRNA have expanded greatly in recent years (reviewed in Helm and Motorin 2017; Peer et al. 2017). Thus far, the majority of RNA modifications identified in mRNA involve methylation of a specific nucleoside, such as N6-methyladenosine (m6A) and 5-methylcytosine (m5C) (Bohnsack et al. 2019; Shi et al. 2019). The biological consequences of these modifications are just beginning to be identified. Some modifications alter base-pairing of mRNA and directly impact gene expression, and some modifications have reader proteins that recognize the chemically altered nucleoside, and relay this information for gene regulatory and cell fate changes (Roundtree et al. 2017; Covelo-Molares et al. 2018; Casella et al. 2019).
While the identification of editing events and modifications present in the transcriptomes of human cells has expanded exponentially in recent years, the in vivo RNA modification landscape of animals is largely unknown. In this chapter, we will focus on the most well-established nucleoside changes that occur in C. elegans mRNA, which are adenosine (A)-to-inosine(I) RNA editing events. The machinery that catalyzes deamination of adenosine, as well as the biological and gene regulatory consequences of A-to-I editing, will be discussed (Table 1). In addition, we will describe a handful of reports of other RNA modifications in C. elegans and highlight future areas for expansion.
Table 1. Brief summary of RNA editing and modification factors in C. elegans.
| C. elegans | Human Ortholog | Domains | Description |
|---|---|---|---|
| Adenosine to inosine RNA editing | |||
| adr-1 | ADAD1 | dsRBD, adenosine deaminase | ADR-1 physically interacts with ADR-2 and promotes editing of certain transcripts, ADR-1 can also inhibit editing by an unknown mechanism |
| adr-2 | ADAR2 | dsRBD, adenosine deaminase | ADR-2 is the sole adenosine deaminase acting on mRNA in C. elegans |
| adbp-1 | None identified | ADBP-1 is important for nuclear localization of ADR-2 and RNA editing | |
| Possible mRNA editing and mRNA modification enzymes in C. elegans | |||
| cdd-1 | CDA | Cytidine deaminase | Loss of cdd-1 affects uridine metabolism, may also be involved in C-U editing of mRNA |
| cdd-2 | CDA | Cytidine deaminase | Loss of cdd-2 affects uridine metabolism, may also be involved in C-U editing of mRNA |
| nsun-2 | NSUN2 | RNA C5-methyltransferase, SAM- dependent methyltransferase | Based on homology, NSUN-2 is predicted to have cytosine methyltransferase activity on tRNA and mRNA |
| B0024.11 | PUS7 | Pseudouridine synthase | Based on homology, B0024.11 is predicted to have pseudouridylation activity on RNA |
| — | METTL3 | Adenine-N6-methyltransferase | No known homolog |
| — | METTL14 | Adenine-N6-methyltransferase | No known homolog |
| mett-10 | METTL16 | Adenine-N6-methyltransferase | Based on homology, METT-10 is predicted to have N6-methyltransferase activity on rRNA |
This table describes factors that are currently known to act in adenosine-to-inosine RNA editing in C. elegans. In addition, this table includes factors that are predicted to function in different types of RNA modification of mRNA.
Adenosine-to-inosine RNA editing
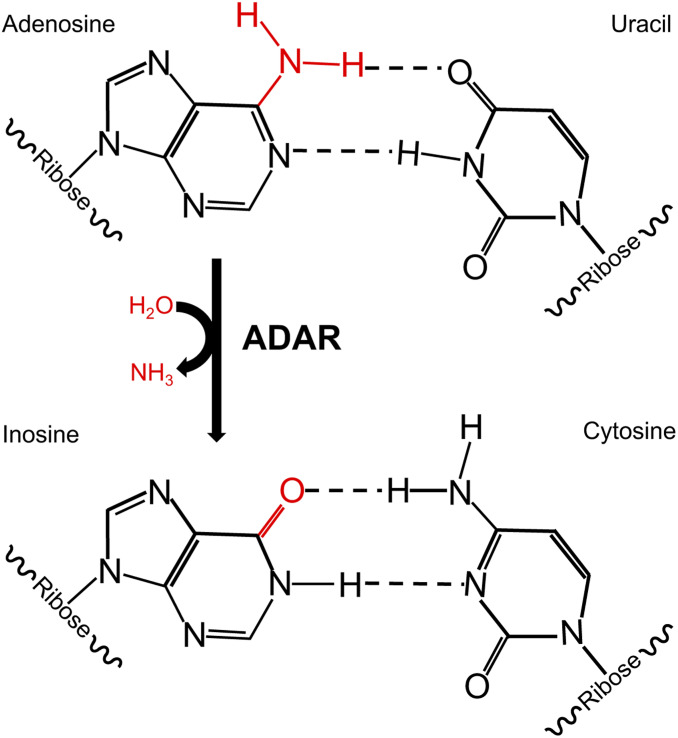
The adenosine deaminases that act on the RNA (ADAR) family of enzymes catalyze A-to-I RNA editing in all animals (Figure 2). These enzymes were first uncovered during studies of antisense RNA techniques in the developing Xenopus laevis embryo (Bass and Weintraub 1987; Rebagliati and Melton 1987). When double-stranded RNA (dsRNA) was injected into developing Xenopus embryos, the dsRNA appeared to unwind. It was later revealed that this “unwinding activity” was due to adenosine deamination to inosine (Bass and Weintraub 1988). As adenosine and inosine have different base-pairing capabilities (Figure 2), the conversion of adenosine to inosine results in mismatches in perfectly base-paired dsRNA, and the presence of multiple A-to-I editing events leads to decreased stability of dsRNA and can affect the cellular function of dsRNA. In C. elegans, editing can prevent entry of dsRNA into the RNA interference (RNAi) pathway, thus inhibiting silencing of gene expression (Knight and Bass 2002). In addition, as dsRNA signals the presence of foreign invaders to the vertebrate immune system, ADARs in vertebrates and C. elegans have been proposed to function in immunity as markers of endogenous (“self”) dsRNA (Eisenberg and Levanon 2018; Reich et al. 2018). These data, and the role of ADARs in regulating gene expression in both development and tissue-specific manners, will be described below.
Figure 2.
The impact of ADARs on RNA. ADARs use water (H2O) to catalyze the removal of an amine group (red) from adenosine, resulting in inosine. While adenosine in RNA base-pairs with uracil, inosine base-pairs with cytosine.
The C. elegans A-to-I editing machinery
The C. elegans adr-2 gene was identified through homology soon after the first mammalian A-to-I editing enzyme was cloned (Kim et al. 1994). Similar to all ADARs (Savva et al. 2012), C. elegans ADR-2 contains an N-terminal dsRNA binding domain (dsRBD) and a C-terminal deaminase domain (Hough et al. 1999). Interestingly, a recent biochemical study suggests that ADR-2 has an ∼100-fold weaker in vitro affinity for dsRNA compared to all other ADARs characterized to date (Rajendren et al. 2018). Despite this reduced affinity for dsRNA, it is well established that ADR-2 is responsible for all A-to-I editing events in C. elegans. The initial characterization of an adr-2 genetic mutant (gv42) revealed loss of in vitro dsRNA editing as well as complete loss of editing for a handful of endogenous mRNAs (Tonkin et al. 2002), a finding that was corroborated by more recent high-throughput sequencing studies using another adr-2 deletion [adr-2(ok735)] (Washburn et al. 2014). Together, these studies support the idea that ADR-2 is the only A-to-I mRNA editing enzyme in C. elegans.
Although ADR-2 is the only enzyme that catalyzes adenosine deamination in C. elegans, ADR-2 activity is regulated by ADR-1 (Tonkin et al. 2002; Washburn et al. 2014; Rajendren et al. 2018). As suggested by the name, C. elegans adr-1 has sequence similarity with the ADAR family. ADR-1 contains two N-terminal dsRBDs and a C-terminal domain that is homologous with the ADAR deaminase domain, though the C-terminal domain lacks the essential amino acids required for catalyzing adenosine deamination (Tonkin et al. 2002). Thus, ADR-1 is an editing-deficient member of the ADAR family. It is important to note that the numbering of the C. elegans ADR proteins does not relate directly to the mammalian ADAR proteins. In addition, both ADR-1 and ADR-2 are referred to as ADARs due to the conserved domain structure, not necessarily the ability to deaminate dsRNA.
The initial characterization of animals lacking adr-1 [adr-1(gv6)] revealed decreased editing of dsRNA in vitro (Tonkin et al. 2002). High-throughput sequencing approaches of a second adr-1 allele [adr-1(tm668)] and biochemical studies have indicated that ADR-1 binds to ADR-2 directly and ADR-2 target mRNAs to promote A-to-I editing at many sites in the transcriptome (Washburn et al. 2014; Rajendren et al. 2018). In addition, RNA-immunoprecipitation studies of ADR-2 from wild-type animals and those lacking adr-1 suggest that the ability of ADR-2 to interact with most mRNAs requires ADR-1 (Rajendren et al. 2018). Together, these data suggest a model in which ADR-1 binds to both ADR-2 and cellular mRNAs to deliver ADR-2 to specific adenosines to promote editing (Figure 3). However, it remains to be shown whether the ADR-1/ADR-2 complex exists on mRNAs in vivo, and, more specifically, whether the binding of ADR-2 binding to dsRNA is compatible with simultaneous binding to ADR-1. Future studies need to focus on identifying binding sites of ADR-1 and ADR-2 on cellular mRNAs and correlating that information with the extent of editing of specific adenosines in those mRNAs.
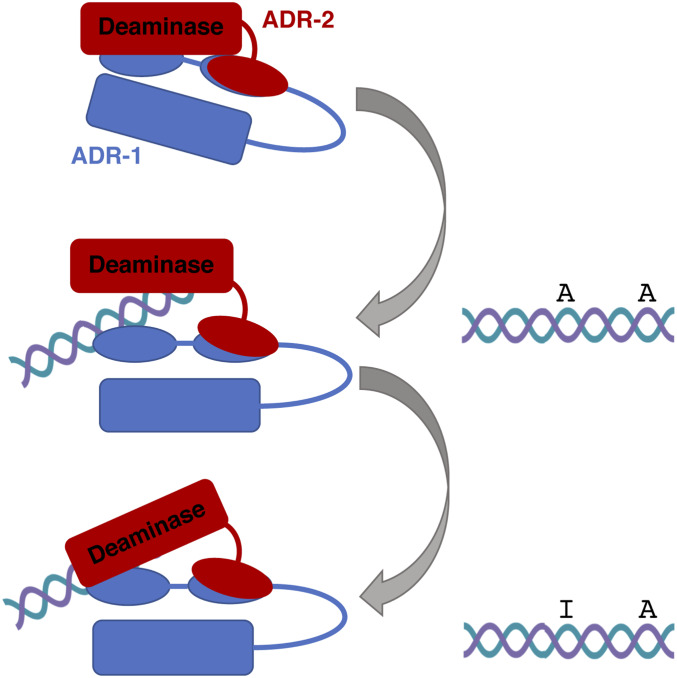
Figure 3.
Substrate recognition by the C. elegans ADARs. C. elegans ADR-1 (blue) contains two dsRNA binding domains (ovals) and a deaminase domain (red), but lacks critical amino acids to perform deamination. ADR-1 interacts physically with both ADR-2 (red) and target mRNAs (teal/purple dsRNA) to promote editing by ADR-2 at specific sites.
It is well-established that ADR-1 and ADR-2 function together for efficient editing of many mRNAs; however, ADR-1 is also known to inhibit ADR-2 activity at specific sites (Tonkin et al. 2002; Washburn et al. 2014). Interestingly, in the nervous system, where ADR-1 and ADR-2 function together to efficiently edit and regulate expression of important neural mRNAs (Deffit et al. 2017), ADR-1 has also been shown to inhibit editing of specific reporter mRNAs (Washburn and Hundley 2016). The ability of ADARs, and most RBPs, to use different modes of substrate recognition at different points in development, and in different tissues, is underexplored. Biochemical studies of RBPs indicate that in vitro recognition of RNA targets can be altered when multiple proteins are in a complex (Campbell et al. 2012); however, similar in vivo studies are lacking. Due to the technical advances to examine tissue-specific gene regulation and transgenic systems to express RBPs in specific cell and tissue types [described above and reviewed in Nance and Frokjaer-Jensen (2019)], studies in C. elegans will likely lead the field in addressing tissue- and developmental-specific functions of ADARs and RBPs in general.
The expression pattern of ADR-1 has been determined primarily by translational reporter fusions. Transgenic expression of an ADR-1::GFP fusion containing the adr-1 promoter and an N-terminal portion of ADR-1 fused to GFP is expressed in most cells of the nervous system throughout development (Tonkin et al. 2002). In addition, ADR-1 is expressed in the developing vulva of L4 animals, and loss of adr-1 leads to a protruding vulva phenotype in a small portion of animals (<10%) (Tonkin et al. 2002; Ganem et al. 2019). Historically, expression studies with the adr-2 promoter were lacking due to the inability to generate adr-2 transgenes, presumably because adr-2 exists in a six-gene operon (Hough et al. 1999; Tonkin et al. 2002), but, with the recent expansion in modern genome engineering technologies [i.e., CRISPR, reviewed in Dickinson and Goldstein (2016)], the technical hurdles to ADR-2 expression studies will likely be overcome. In a recent global analysis of single-cell transcriptome profiling of L2 animals, expression of adr-1 and adr-2 mRNA was reported to be expressed in neurons, muscle, hypodermis, intestine, and the germline (Cao et al. 2017), suggesting ADR-1 may be expressed more broadly than observed with the ADR-1::GFP fusion.
The role of editing and ADARs in many of these tissues is currently unknown. However, as animals individually lacking adr-1 or adr-2 and an animal lacking both genes are viable (Tonkin et al. 2002), C. elegans will be an important contributor to determining how tissue-specific gene regulation contributes to organismal function and development. In this regard, C. elegans adr mutants exhibit defects in chemotaxis (Tonkin et al. 2002), and recent high-throughput sequencing of isolated neural cells, combined with functional assays of genetic mutants, led to an identification of an edited mRNA (clec-41) that was critical for this behavioral defect (Deffit et al. 2017). It is important to note that in these same genetic mutants, namely adr-1(–) animals, altered editing and gene expression of clec-41 mRNA was not observed in RNA isolated from whole larval animals. Similar tissue-specific approaches may also prove informative for understanding the role of ADR-1 and ADR-2 in lifespan (Sebastiani et al. 2009; Ganem et al. 2019).
RNA editing in space and time
High-throughput sequencing studies of the C. elegans transcriptome have revealed tens of thousands of editing sites in protein-coding genes and long-noncoding RNAs (Wu et al. 2011; Washburn et al. 2014; Whipple et al. 2015; Zhao et al. 2015; Deffit et al. 2017; Goldstein et al. 2017; Reich et al. 2018; Ganem et al. 2019). These studies have used a number of different technical and computational approaches, including enriching for dsRNA (Reich and Bass 2019). All of these studies take advantage of the nonessential nature of A-to-I editing in C. elegans. Parallel analyses of sequencing data from the adr-2(–) animals facilitates accurate identification of A-to-I editing events from technical errors in sequencing data, including errors introduced by PCR, sequencing, and/or strain differences (Bass et al. 2012; Reich and Bass 2019). Two general trends that have emerged from these high-throughput studies are that editing levels are developmentally regulated (Zhao et al. 2015; Goldstein et al. 2017; Reich et al. 2018; Ganem et al. 2019) and that most editing events occur in noncoding regions of the transcriptome (Washburn et al. 2014; Whipple et al. 2015; Deffit et al. 2017).
High-throughput sequencing studies have identified most editing events in RNA isolated from animals early in development (Zhao et al. 2015; Reich et al. 2018), which is also when expression of adr-1 and adr-2 mRNA levels peak (Hundley et al. 2008). There are some reported differences in the exact “peak” of RNA editing (embryos vs. L1s), which may relate to secondary effects of starvation-induced transcript changes and/or differences in recovery time after L1 arrest. Interestingly, it has been shown that some mRNAs are expressed throughout development, but edited only at a specific stage. An example is egl-2, which is edited in embryos, but not L4 animals (Goldstein et al. 2017). In addition, it was recently shown that the ability of ADR-1 to promote ADR-2 editing of mRNAs is developmentally regulated, with ADR-1 regulatory function contributing to higher editing of specific mRNAs in L4 animals compared to embryos (Ganem et al. 2019). The consequences of ADARs and RNA editing for gene expression at specific developmental stages is beginning to emerge (Zhao et al. 2015; Reich et al. 2018; Ganem et al. 2019). However, as tissue-specific effects may be masked when examining RNA isolated from whole animals of different developmental stages, it will be important to combine the tools of tissue-specific studies with development to obtain a clear picture of the function of RNA editing on development.
The impact of A-to-I RNA editing on gene expression is not relayed by specific “reader” proteins, but rather is dictated by the location of the editing event within an RNA species. As the base-pairing properties of inosine mimic those of guanosine (Figure 2), A-to-I editing events alter the structure and sequence-specific interactions of the edited RNA. Editing events in open reading frames (ORFs) of mRNAs can alter base-pairing with tRNAs, thus changing the genetic code (referred to as recoding events), and, ultimately, the amino acid sequence of the protein. Recoding events play critical roles in proteomic diversity of ion channels and receptors in mammals, flies, and cephalopods (Tariq and Jantsch 2012; Rosenthal 2015; Keegan et al. 2017). However, only a handful of editing sites have been identified within the coding regions of C. elegans mRNAs [e.g., eight validated recoding events identified in one study (Zhao et al. 2015)], and there is currently no evidence that C. elegans recoding sites have functional consequences. The lack of identification of C. elegans recoding sites does not appear to be due to tissue-specific effects, as recent transcriptome-wide sequencing of mRNAs from isolated neural cells did not detect novel recoding editing sites, suggesting that the major role of A-to-I editing in the C. elegans nervous system is not to generate proteome diversity (Deffit et al. 2017).
A number of recent high-throughput sequencing studies have revealed that a majority of A-to-I editing events in the C. elegans transcriptome occur within introns (Zhao et al. 2015; Deffit et al. 2017; Reich et al. 2018). As ADR-2 is present in the nucleus (Ohta et al. 2008), and the important sequences for splicing (i.e., the branch point adenosine, splice donor, splice acceptor) all contain adenosines or guanosines, A-to-I editing within intronic regions has the potential to both create and disrupt splicing, though, to date, no studies have addressed the impact of RNA editing on splicing in C. elegans. With the many elegant technical tools available to study splicing, such as fluorescent splicing reporters for tissue-specific analysis, as well as the adr mutants, this is an emerging and important area for investigation.
Additionally, it has been shown recently that circular RNA species can be formed from back-splicing of intronic sequences (Wilusz 2018). High-throughput sequencing studies identified at least 1166 circular RNAs (circRNAs) in C. elegans, some of which accumulate during aging (Cortés-López et al. 2018). Studies from human cells indicate that A-to-I editing and ADARs influence circular RNA production (Ivanov et al. 2015). It was recently reported that regions of the C. elegans genome that are enriched for editing (identified by immunoprecipitation and high-throughput sequencing) significantly overlap with circular RNAs (Reich et al. 2018). However, the functional consequences of RNA editing and ADARs on C. elegans circRNAs, as well as the biological impact of circRNAs on C. elegans development and organismal function, are unknown.
As the 3′ UTRs of cellular mRNAs contain elements for post-transcriptional gene regulation, and thousands of editing sites in C. elegans occur in 3′ UTRs, these editing events have the potential to alter mRNA stability, localization, and translation (Hundley and Bass 2010). Global expression analyses have revealed small, but reproducible, decreases in mRNA expression of genes with edited 3′ UTRs in adr mutant embryos compared with wild-type embryos (Goldstein et al. 2017; Reich et al. 2018). These gene regulatory effects were not observed in later stage animals, suggesting developmental-specific roles for editing in gene regulation. However, it is also possible that A-to-I editing in 3′ UTRs regulates gene expression in specific tissues, and is masked by analysis of RNA isolated from whole animals, as described above. Moreover, in large part, functional studies of the role of individual editing events in noncoding regions serving to regulate C. elegans gene expression are lacking. With recent advances in the use of CRISPR to modify specific nucleotides of the C. elegans genome (Dickinson and Goldstein 2016), as well as techniques to analyze tissue-specific gene expression, it is now possible to start directly assessing the consequences of specific 3′ UTR editing events on gene expression.
ADARs regulate the levels and fates of endogenous dsRNA
In addition to the role of individual editing events in regulating gene expression, the ability of ADARs to bind and modify dsRNA impacts dsRNA recognition by other dsRBPs. The helical structure of dsRNA is A-form, and, thus, has a narrow major groove that prevents sequence-specific contacts with proteins. Therefore, dsRBPs recognize primarily the shape of dsRNA, and all characterized dsRBPs bind to dsRNA of any sequence (Bass 2006). The lack of specific binding by dsRBPs can result in an intersection of dsRNA-mediated pathways in vivo. For ADARs, this intersection has been well documented to impact the production of small RNAs and silencing of cellular RNAs, both of which are fields where C. elegans research has been at the forefront (Youngman and Claycomb 2014).
The biogenesis of most classes of small RNAs requires the action of nucleases that act on longer dsRNA precursors (Fischer 2010; Billi et al. 2014). In some instances, ADARs can bind and edit these dsRNA precursors, which can alter small RNA sequence and/or production (Nishikura 2016). The first evidence of this antagonistic function came from studies of transgene expression in C. elegans adr mutants (Knight and Bass 2002). Highly repetitive extrachromosomal arrays can give rise to dsRNA through overlapping sense and antisense transcription. Editing of these dsRNA molecules prevents recognition by Dicer, thus decreasing small interfering RNA (siRNA) production and allowing expression of the transgene. Repetitive transgenes expressed in a number of tissues, including muscle, intestine, and hypodermis, can be acted upon by ADR-2 to prevent silencing (Knight and Bass 2002; Ohta et al. 2008). Loss of nuclear localization of ADR-2 increases somatic transgene silencing (Ohta et al. 2008), while loss of RNA interference (RNAi) factors leads to reduced silencing (Knight and Bass 2002).
The ability of ADARs to protect cellular transcripts from small RNA processing is not limited to transgenes. Loss of ADARs affects the levels of mature microRNAs (miRNAs) (Warf et al. 2012). Interestingly, these effects are more prominent upon loss of adr-1 than adr-2, suggesting that ADAR binding to miRNA precursors antagonizes processing more than editing. The increases in mature miRNAs and consequent changes in the mRNA levels of miRNA target genes in the absence of ADARs has also been observed in mouse embryos (Vesely et al. 2012), suggesting a conserved role for ADARs in antagonizing small RNA processing.
Recently, a striking phenotype of frequent adult bursting was observed when C. elegans adr mutants were in an Enhanced RNAi (Eri) background (Reich et al. 2018). This synthetic genetic interaction was alleviated when core RNAi factors were absent. The rescue of adr phenotypes by loss of RNAi factors is consistent with previous studies that indicated loss of RNAi factors could restore the chemotaxis and lifespan defects of C. elegans adr mutants (Tonkin and Bass 2003; Sebastiani et al. 2009). Mechanistically, these data suggest that ADARs protect endogenous dsRNA from destruction by siRNA silencing pathways, and that aberrant destruction of endogenous dsRNA has developmental and neurological consequences (Pasquinelli 2018). Consistent with this model, loss of another C. elegans RNA binding protein, TDP-1, leads to increased dsRNA production and chemotaxis defects, the latter of which can be rescued by loss of RNAi factors (Saldi et al. 2014). Furthermore, recent high-throughput sequencing of small RNAs identified a class of 23 nt siRNAs that are homologous to the double-stranded regions of C. elegans edited mRNAs, and loss of adrs resulted in downregulation of these mRNAs (Reich et al. 2018). A previous study also identified small RNAs (22, 23, and 24 nt siRNAs) regulated by adr expression (Wu et al. 2011). However, in large part, the two studies do not overlap in identification of genomic loci, suggesting that ADARs may participate in regulating multiple, distinct small RNA generating pathways. These studies in C. elegans showcase the role of ADARs in counteracting immune responses to dsRNA, and are also consistent with the emerging view in mammals of ADARs in preventing aberrant recognition of endogenous dsRNA by the innate immune system (Samuel 2019). Together, these data suggest that the conserved, and perhaps primary, function of A-to-I editing is to protect self dsRNA.
Are other modifications present in C. elegans mRNAs?
The identification of mammalian ADARs coincided with sequencing of the C. elegans genome and stimulated the study of A-to-I editing in C. elegans. The other major class of deaminases in mammals, cytidine deaminases, have a handful of RNA targets, such as apolipoprotein B mRNA, but the majority of C-U edits occur at the DNA level (Salter et al. 2016). There has been one report of a C-U edited mRNA in C. elegans (Wang et al. 2004). The serendipitous discovery of C-U editing of the gld-2 mRNA occurred when researchers noticed discrepancy between cDNA sequences and genomic sequences. Further analysis indicated that C-U editing of gld-2 mRNA occurred in the germline, but not the soma (Wang et al. 2004). The biological function of the cytidine deamination of gld-2 mRNA is unknown. Interestingly, it was shown recently that two C. elegans cytidine deaminases, cdd-1 and cdd-2, are important for germline proliferation (Chi et al. 2016). This effect is likely due to changes in the cellular pyrimidine pool and the action of these enzymes on free nucleotides (Table 1). However, a direct role for these two enzymes and the seven other predicted cytidine deaminases in C. elegans (Wang et al. 2004) in modifying gld-2 mRNA has not been examined.
In contrast to the well-established editing enzymes that act on mRNA, the enzymes that deposit many other mRNA modifications have only recently been identified in mammals, flies, and yeast (Frye et al. 2018). Database searches of enzymes that catalyze some modifications (e.g., pseudouridylation and 5-methylcytosine) reveal potential homologous C. elegans genes (Dezi et al. 2016) (Table 1). Interestingly, for some modifications present in human mRNAs, such as m6A, the major modification enzymes are absent from the C. elegans genome, while homologs of enzymes that play a more minor role in humans are present in C. elegans, but not characterized to date (Table 1). Some RNA modifications, such as 5-hydroxymethylcytidine (hm5C), have been detected in C. elegans RNA, but the enzyme that performs this modification in humans, TET, does not have an easily identifiable homolog in C. elegans (Huber et al. 2015). Metabolic labeling experiments coupled to mass spectrometry have also identified several RNA modifications that occur in C. elegans (van Delft et al. 2017). These modifications responded dynamically to cellular and environmental stress, and were identified in both large (>200 nt) and small (<200 nt) RNAs. A functional consequence on gene expression in starved animals was identified for one of these modifications, 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), which occurs in the anticodon of select tRNAs. As tRNA and rRNA are much more abundant than mRNA, information about mRNA modification will be more difficult to obtain from global metabolic studies. Future studies using enrichment techniques for specific modifications (Helm and Motorin 2017), as well as genetic mutants, will be important to understand the C. elegans mRNA modification landscape. In addition, with the emerging use of technologies that allow direct sequencing of RNA (Kono and Arakawa 2019), detection of modifications in C. elegans mRNAs will increase over the coming years.
Splicing
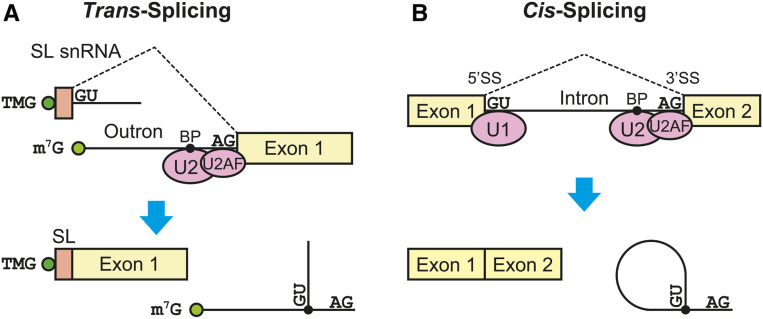
RNA splicing is a process that removes introns from a primary transcript and ligates exons. Eukaryotic pre-mRNA splicing entails two sequential transesterification reactions (branching and exon ligation) and is catalyzed by a large ribonucleoprotein complex termed the spliceosome. The spliceosome comprises five uridine-rich small nuclear RNAs (U snRNAs) and nearly 200 proteins that are highly conserved from yeast to humans (Shi 2017). The spliceosome assembles on pre-mRNAs and undergoes numerous conformational changes mediated by trans-acting proteins such as adenosine triphosphatases (ATPases)/RNA helicases. Near-atomic resolution structures of Saccharomyces cerevisiae and human spliceosomes at different stages of assembly, catalysis, and disassembly have been solved by cryoelectron microscopy (cryo-EM) in the last couple of years, and provide considerable mechanistic insight into how the spliceosome achieves the two transesterification reactions (Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016, 2017, 2018; Yan et al. 2016, 2017; Bai et al. 2017, 2018; Bertram et al. 2017a,b; Fica et al. 2017, 2019; Liu et al. 2017; Plaschka et al. 2017; Wilkinson et al. 2017; Zhang et al. 2017, 2018, 2019; Haselbach et al. 2018; Zhan et al. 2018a,b). In C. elegans, there are two categories of spliceosomal splicing: trans-splicing and cis-splicing (Blumenthal 2012) (Figure 4). Trans-splicing precisely joins exons from two discontinuous primary transcripts (Figure 4A), whereas cis-splicing precisely joins two exons from the same primary transcript (Figure 4B) (discussed in detail below).
Figure 4.
Schematic representations of trans-splicing and cis-splicing in C. elegans. (A) Spliced leader trans-splicing. A 2,2,7-trimethylguanosine (TMG)-capped 22-nt spliced leader (SL) sequence derived from an SL snRNA in SL snRNP replaces a 7-methylguanosine (m7G)-capped outron in a pre-mRNA. A y-shaped outron is excised. (B) Cis-splicing. A lariat-shaped intron is excised and the upstream and downstream exons are ligated. The 5′-splice site (5′SS), 3′-splice site (3′SS) and branch point (BP) are recognized by U1 snRNP (U1), U2 auxiliary factor (U2AF) and U2 snRNP (U2), respectively. Boxes represent exons and solid lines indicate introns and outrons. Dashed lines connect exons that are ligated in the splicing reactions. Cap structures and branch points are indicated with green and black circles, respectively. Almost invariable nucleotide sequences of the splice sites are indicated.
Operons and trans-splicing
mRNAs of >84% of C. elegans protein-coding genes begin with a spliced leader (SL), one of two common extragenically derived 22 nt sequences (SL1 or SL2) (Allen et al. 2011; Tourasse et al. 2017). The SL is donated by a ∼100 nt RNA, SL1 or SL2 RNA, in a process termed spliced leader trans-splicing (Figure 4A). The SL RNA forms a small nuclear ribonucleoprotein particle (snRNP), which is structurally and functionally similar to the U snRNAs (U1, U2, U4, U5, and U6) that play key roles in cis-splicing or intron removal (Van Doren and Hirsh 1988). The trans-splicing event is very closely related to cis-splicing: the 5′ splice site (5′ SS) is on the SL RNA and the 3′ splice site (3′ SS) is the trans-splice site or the site of SL addition on the pre-mRNA (Figure 4A). Unlike the U snRNAs, the SL RNA is consumed in every trans-splicing event (Van Doren and Hirsh 1988). The C. elegans genome contains 110–150 tandem repeats of SL1 RNA gene loci (Krause and Hirsh 1987; Yoshimura et al. 2019) and 18 SL2 RNA genes, including a variety of variant SL2 RNAs at dispersed loci (Evans et al. 1997). As the SL RNAs have a 2,2,7-trimethylguanosine (TMG) cap, mRNAs processed by trans-splicing also have a TMG cap rather than the usual 7-methylguanosine (m7G) cap (Figure 4) (Van Doren and Hirsh 1988). The TMG cap is considered to stimulate translation of the trans-spliced mRNAs (Maroney et al. 1995; Lall et al. 2004; Wallace et al. 2010). Detailed mechanisms of trans-splicing in C. elegans are documented in a previous article in WormBook (Blumenthal 2012).
More than half of pre-mRNAs are subject to SL1 trans-splicing, which trims off the 5′ ends of pre-mRNAs and replaces them with the SL1 sequence. The region between the transcription start site (TSS) or the 5′ cap and the trans-splice site is called the outron. As trans-splicing is very efficient, pre-mRNAs with outron sequences are hardly detected, which makes it difficult to determine the TSSs for trans-spliced genes in C. elegans. RNA-seq analysis of captured 5′ ends of nuclear RNAs revealed a collection of TSSs for 7351trans-spliced genes, often with multiple TSS clusters per gene (Kruesi et al. 2013; Saito et al. 2013). Lengths of the outrons range from <10 to >3000 nt, with a median of 369 nt (Saito et al. 2013).
The other splice leader, SL2, is trans-spliced to mRNAs derived from downstream genes in operons (Spieth et al. 1993; Blumenthal et al. 2002; Allen et al. 2011). In C. elegans, there are 1255 verified operons containing 3193 genes, representing ∼15% of all protein-coding genes (Allen et al. 2011; Blumenthal 2012). Half of the operons contain only two genes, whereas the other half contain three to eight genes (Allen et al. 2011). The distance between the genes in an operon is typically ∼100 bp, but can be up to >2 kb-pairs (kb) (Morton and Blumenthal 2011a). First genes in the operons are either trans-spliced to SL1 or not trans-spliced. Downstream genes are trans-spliced predominantly to SL2, but some of them are also trans-spliced to SL1 at the same trans-splice site as SL2, and the ratio of SL2 is negatively correlated with the intergenic distance (Allen et al. 2011; Tourasse et al. 2017). Some downstream genes in operons are predominantly trans-spliced to SL1 due to transcription from intergenic promoters, and such operons are termed “hybrid” operons (Huang et al. 2007; Allen et al. 2011). Further variations in the structure of gene clusters and detailed lists of their examples are summarized in a previous article in WormBook (Blumenthal et al. 2015). Information about the positions of the trans-splice sites, as well as the ratio of SL1/SL2 trans-splicing for each gene, derived from a compendium of 1682 publicly available C. elegans RNA-seq data sets are now available (Tourasse et al. 2017). Operons appear to be highly stable in the genus Caenorhabditis; 96% of C. elegans operons are conserved in Caenorhabditis briggsae (Stein et al. 2003). Features of trans-splicing and operons in nematodes and other organisms are summarized in a previous review article (Lasda and Blumenthal 2011).
cis-splicing
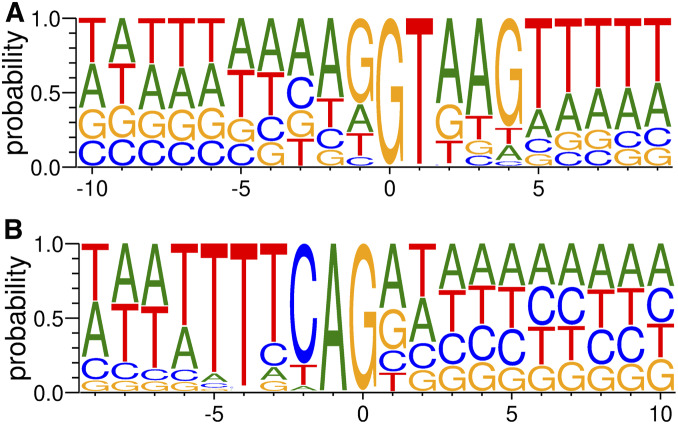
Similar to other multicellular organisms, C. elegans has an intron-rich genome, and intron excision from pre-mRNAs by the spliceosome—a process referred to as cis-splicing—is a fundamental step of gene expression (Figure 4B) (reviewed in a previous WormBook article (Zahler 2012)). A notable peculiarity of C. elegans introns is that many of them are relatively short compared to other metazoan introns; around half of C. elegans introns are <65 nt long, with 47 nt being the most commonly observed intron length (Lander et al. 2001; Spieth et al. 2014). Similar to other eukaryotes, cis-splicing in C. elegans involves base-pairing between the U1 snRNA and the 5′ splice donor site (Figure 4B) (Thomas et al. 1990; Zahler et al. 2004) with a consensus sequence of AG/GUAAGUU (where / indicates the intron/exon boundary) (Figure 5A). It is important to note that in C. elegans (as in humans) a small portion of introns (<1%) begins with GC instead of GU (Farrer et al. 2002) (Burset et al. 2001). In addition, at least one intron begins with GA (H.K., unpublished data). The consensus sequence of the C. elegans 3′ splice acceptor site—UUUUCAG/R (Figure 5B)—is recognized by U2 auxiliary factor (U2AF) (Figure 4B) (Zorio and Blumenthal 1999a; Hollins et al. 2005). U2AF is a heterodimer composed of U2AF65 and U2AF35 (UAF-1 and UAF-2, respectively, in C. elegans) (Zorio et al. 1997; Zorio and Blumenthal 1999b). In most metazoans, U2AF65 recognizes the polypyrimidine tract separated from the 3′SS (Sickmier et al. 2006; Mackereth et al. 2011). In contrast, C. elegans lacks the polypyrimidine tract (Blumenthal and Steward 1997; Schwartz et al. 2008) and UAF-1 and UAF-2 recognize the U stretch and the AG dinucleotide, respectively, in the conserved octamer sequence of the 3′SS (Zorio and Blumenthal 1999a; Hollins et al. 2005). It is assumed that lariat formation occurs during splicing in C. elegans like in other eukaryotes, but few studies experimentally addressed the branchpoints (Zahler 2012; Ragle et al. 2015) due to the very short and AU-rich nature of the introns. Unlike in budding yeast, branchpoint consensus is not found in sequence analysis of C. elegans introns, even though branchpoint binding protein ortholog SFA-1 is essential for embryonic development (Mazroui et al. 1999).
Figure 5.
Sequence motifs of the 5′ (A) and 3′ (B) splice sites. Probability of the nucleotides at each position is displayed by using Weblogo version 3.7.1 (Crooks et al. 2004). The sequences of the introns are derived from 114,417 (5′) and 114,006 (3′) unique splice sites in 20-nt or longer introns annotated in WormBase (WS254). Position 0 indicates the beginning (A) and the end (B) of the introns.
A compendium of the publicly available C. elegans RNA-seq data sets from 96 individual studies, including >6.6 billion exon-exon junction reads, identified as many as 667,779 junctions in the genome (Tourasse et al. 2017). However, 78.8% of these splice junctions were detected only rarely (<100 reads over the datasets). In contrast, 97.6% of the reads came from 63,156 robustly detected junctions (>10,000 reads). Rare splice junctions (including those predicted in gene models in WormBase) may be derived from alternative splicing (discussed below), while some of these transcripts may represent sequencing errors or biological noise. In support of the latter idea, the total number of detected junctions per gene increases with the gene expression level (top 5% genes having on average ∼70 junctions) and “rare” junctions are less evolutionarily conserved than more frequently used ones (Tourasse et al. 2017).
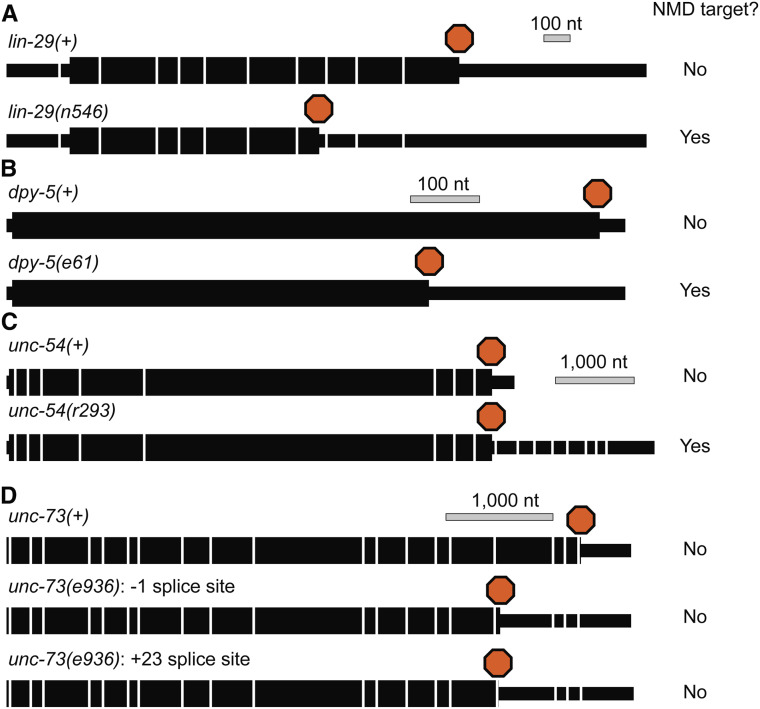
It is generally assumed that homologs of human and yeast splicing machinery components also function in splicing in C. elegans (Table 2). As cis-splicing is a critical step of pre-mRNA processing, crucial components of the splicing machinery are essential for embryonic development (Hebeisen et al. 2008). Forward and reverse genetic screens revealed that loss of function of many splicing machinery proteins leads to the Masculinization of Germline (Mog) phenotype (Puoti and Kimble 1999, 2000; Kasturi et al. 2010; Zanetti et al. 2011), germline overproliferation (Kerins et al. 2010), or distal tip cell migration phenotypes (Doherty et al. 2014). Recent extensive forward and reverse genetic studies of cryptic splicing in the unc-73(e936) allele that harbor a 5′ SS mutation demonstrated functional roles for a conserved 27 kDa component of U4/U6-U5 tri-snRNP-specific proteins (SNRP-27) and a conserved U5 snRNP protein PRP-8 in maintaining the position of the 5′ SS defined by U1 snRNA during rearrangements of the spliceosome (Zahler et al. 2018; Mayerle et al. 2019).
Table 2 . C. elegans orthologs of human spliceosome component proteins.
| C. elegans | Human Ortholog | Domains | Description |
|---|---|---|---|
| snr-2 | SNRPB/Sm B/B’ | LSm | Sm protein (Common in U1, U2, U4 and U5 snRNPs) |
| snr-3 | SNRPDl/Sm D1 | LSm | Sm protein (Common in U1, U2, U4 and U5 snRNPs) |
| snr-4 | SNRPD2/Sm D2 | LSm | Sm protein (Common in U1, U2, U4 and U5 snRNPs) |
| snr-1 | SNRPD3/Sm D3 | LSm | Sm protein (Common in U1, U2, U4 and U5 snRNPs) |
| snr-6 | SNRPE/Sm E | LSm | Sm protein (Common in U1, U2, U4 and U5 snRNPs) |
| snr-5 | SNRPF/Sm F | LSm | Sm protein (Common in U1, U2, U4 and U5 snRNPs) |
| snr-7 | SNRPG/Sm G | LSm | Sm protein (Common in U1, U2, U4 and U5 snRNPs) |
| gut-2 | LSM2/Lsm2 | LSm | LSm protein (U6 snRNP) |
| lsm-3 | LSM3/Lsm3 | LSm | LSm protein (U6 snRNP) |
| lsm-4 | LSM4/Lsm4 | LSm | LSm protein (U6 snRNP) |
| lsm-5 | LSM5/Lsm5 | LSm | LSm protein (U6 snRNP) |
| lsm-6 | LSM6/Lsm6 | LSm | LSm protein (U6 snRNP) |
| lsm-7 | LSM7/Lsm7 | LSm | LSm protein (U6 snRNP) |
| lsm-8 | LSM8/Lsm8 | LSm | LSm protein (U6 snRNP) |
| rnp-7 | SNRNP70/U1-70K | U1 snRNP70, RRM | U1 snRNP |
| rnp-2 | SNRPA/U1A | RRM | U1 snRNP |
| snrp-3 | SNRPC/U1C | U1 Zn finger | U1 snRNP |
| prp-40 | PRPF40A/FBP11 | WW, FF | U1 snRNP |
| rbm-25 | RBM25/S164 | RRM, PWI | U1 snRNP |
| ddx-17 | DDX5/p68 | DEAD-box helicase | U1 snRNP |
| tcer-1, tcer-2 | TCERG1/CA150 | WW, FF | U1 snRNP |
| mog-2 | SNRPA1/U2A’ | Leucine-rich repeat | U2 snRNP |
| rnp-3 | SNRPB2/U2B” | RRM | U2 snRNP |
| prp-21 | SF3A1/SF3a120 | SWAP, PRP21-like, Ubiquitin | U2 snRNP |
| repo-1 | SF3A2/SF3a66 | C2H2 Zn finger | U2 snRNP |
| prp-9 | SF3A3/SF3a60 | SF3a60 binding | U2 snRNP |
| sftb-1 | SF3B1/SF3b155 | Sf3b1 | U2 snRNP |
| sftb-2 | SF3B2/SF3b150 | Proline-rich | U2 snRNP |
| teg-4 | SF3B3/SF3b130 | MMS1, CPSF A subunit region | U2 snRNP |
| sap-49 | SF3B4/SF3b49 | RRM | U2 snRNP |
| moa-2 | SF3B5/SF3b10 | SF3b10 | U2 snRNP |
| sftb-6 | SF3B6/SF3b14a | RRM | U2 snRNP |
| phf-5 | PHF5A/SF3b14b | PHD-finger | U2 snRNP |
| snu-13 | SNU13/hSnu13 | L7Ae | U4/U6 snRNP |
| prp-31 | PRPF31/hPrp31 | snoRNA binding, Prp31 | U4/U6 snRNP |
| prp-3 | PRPF3/hPrp3 | PWI, PRP3 | U4/U6 snRNP |
| prp-4 | PRPF4/hPrp4 | PRP4-like, WD40 repeat | U4/U6 snRNP |
| cyn-11 | PPIH/hCypH | Peptidyl-prolyl cis-trans isomerase | U4/U6 snRNP |
| prp-8 | PRPF8/hPrp8 | PROCN, RRM, U5-snRNA binding, U6-snRNA interacting, PRP8 domain IV, MPN, PROCT | U5 snRNP |
| snrp-200 | SNRNP200/hBrr2 | DEAD/DEAH box helicase | U5 snRNP |
| eftu-2 | EFTUD2/hSnu114 | GTP-binding, Elongation factor Tu, Elongation factor G | U5 snRNP |
| snrp-40.1, snrp-40.2 | SNRNP40/U5-40K | WD40 repeat | U5 snRNP |
| prp-6 | PRPF6/hPrp6 | PRP1, TPR | U5 snRNP |
| teg-1 | CD2BP2/hLin1 | GYF | U5 snRNP |
| dib-1 | TXNL4A/hDib1 | DIM1 | U5 snRNP |
| ddx-23 | DDX23/hPrp28 | DEAD-box helicase | U5 snRNP |
| snrp-27 | SNRNP27/U4/U6.U5-27K | U4/U6.U5 tri-snRNP | |
| usp-39 | USP39/hSad1 | Ubiquitin-hydrolases Zn finger, Ubiquitin carboxyl-terminal hydrolase | U4/U6.U5 tri-snRNP |
| sart-1 | SART1/hSnu66 | SART-1 | U4/U6.U5 tri-snRNP |
| rbm-42 | RBM42 | RRM | U4/U6.U5 tri-snRNP |
| uaf-1 | U2AF2/U2AF65 | RRM | U2 related |
| uaf-2 | U2AF1/U2AF35 | CCCH Zn finger, RRM | U2 related |
| rnp-6 | PUF60 | RRM | U2 related |
| smr-1 | SMNDC1/SPF30 | SMN | U2 related |
| dnj-30 | DNAJC8/SPF31 | DnaJ | U2 related |
| rbm-17 | RBM17/SPF45 | G-patch, RRM | U2 related |
| tag-65 | CHERP | SWAP, CTD-binding, G-patch | U2 related |
| sap-140 | U2SURP/SR140 | RRM, SWAP, cwf21 | U2 related |
| ddx-15 | DHX15/PRP43 | DEAH-box helicase | U2 related |
| ddx-46 | DDX46 | DEAD-box helicase | U2 related |
| hel-1 | DDX39B/UAP56 | DExD/H-box helicase | Transcription and export (TREX) complex |
| aly-1, aly-2, aly-3 | ALYREF/Aly/REF | RRM, Fop | Transcription and export (TREX) complex |
| thoc-1 | THOC1 | Thoc1, Death domain | Transcription and export (TREX) complex |
| thoc-2 | THOC2 | Thoc2 | Transcription and export (TREX) complex |
| thoc-3 | THOC3 | WD40 repeat, WD40- like beta propeller repeat | Transcription and export (TREX) complex |
| luc-7L | LUC7L | LUC7 | A complex protein |
| prp-39 | PRPF39 | PRP39 | A complex protein |
| tiar-1, tiar-2, tiar-3 | TIA1 | RRM, Q-rich | A complex protein |
| bub-3 | BUB3 | WD40 repeat | A complex protein |
| - | TRIR/MGC2803 | TRIR | A complex protein |
| - | SUGP1/SF4 | SWAP, G-patch | A complex protein |
| ccar-1 | CCAR1/FLJ10839 | S1-like RNA binding, DBC1, SAP | A complex protein |
| cdk-11.1, cdk-11.2 | CDK11A/CDC2L2 | Protein kinase | A complex protein |
| cus-2 | HTATSF1/Tat SF1 | RRM | A complex protein |
| fust-1 | FUS/TLS | RRM, RanBP Zn finger | A complex protein |
| rbm-5 | RBM5 | RRM, RanBP Zn finger, G-patch | A complex protein |
| rbm-5 | RBM10 | RRM, RanBP Zn finger, G-patch | A complex protein |
| sfa-1 | SF1 | KH, CCHC Zn finger | A complex protein |
| bud-13 | BUD13/MGC13125 | Bud13 | RES complex |
| pmlr-1 | SNIP1 | FHA | RES complex |
| rbmx-2 | RBMX2/CGI-79 | RRM | RES complex |
| snu-23 | ZMAT2/hSnu23 | dsRNA-binding Zn finger | B complex protein |
| prp-38 | PRPF38A/hPrp38 | PRP38 | B complex protein |
| mfap-1 | MFAP1 | MFAP1 | B complex protein |
| ubl-5 | UBL5 | Ubiquitin | B complex protein |
| smu-2 | IK/RED | RED-like | B complex protein |
| smu-1 | SMU1 | WD40 repeat | B complex protein |
| multiple genes | HSPB1/HSP27 | Hsp20 | B complex protein |
| dxbp-1 | KIN/HsKin17 | Kin17 curved DNA-binding | B complex protein |
| mtr-4 | MTREX/Skiv2L2 | DEAD/DEAH box helicase | B complex protein |
| prpf-4 | PRPF4B/hPrp4 kinase | Protein kinase | B complex protein |
| pqbp-1.1, pqbp-1.2 | PQBP1/NPW38 | WW | B complex protein |
| wbp-11 | WBP11/NPW38BP | WBP11 | B complex protein |
| wbp-4 | WBP4/FBP21 | U1 Zn finger, WW | B complex protein |
| mog-4 | DHX16/hPrp2 | DEAD/DEAH box helicase | Bact complex protein |
| let-858 | CWC22/KIAA1604 | MIF4G, MA3 | Bact complex protein |
| rnf-113 | RNF113A | CCCH Zn finger, RING finger | Bact complex protein |
| cyn-16 | CWC27/NY-CO-10 | Peptidyl-prolyl cis-trans isomerase | Bact complex protein |
| cyn-4 | PPIL2 | Rtf2 RING finger, Peptidyl-prolyl cis-trans isomerase | Bact complex protein |
| ccdc-12 | CCDC12 | cwf18 | Bact complex protein |
| cyn-10 | PPIL3/PPIL3b | Peptidyl-prolyl cis-trans isomerase | Bact complex protein |
| gkow-1 | GPKOW/hSPP2 | G-patch, KOW | Bact complex protein |
| prp-19 | PRPF19/Prp19 | U-box, PRP19-like, WD40 repeat | Prp19 complex (nineteen complex, NTC) |
| cdc-5L | CDC5L/CDC5 | Myb-like DNA-binding, Cdc5p/Cef1 | Prp19 complex (nineteen complex, NTC) |
| bcas-2 | BCAS2/SPF27 | BCAS2 | Prp19 complex (nineteen complex, NTC) |
| syf-1 | XAB2/hSyf1 | TPR | Prp19 complex (nineteen complex, NTC) |
| syf-2 | SYF2/GCIP p29 | SYF2 | Prp19 complex (nineteen complex, NTC) |
| syf-3 | CRNKL1/hSyf3 | HAT repeat | Prp19 complex (nineteen complex, NTC) |
| isy-1 | ISY1/hIsy1 | Isy1-like | Prp19 complex (nineteen complex, NTC) |
| ctnb-1 | CTNNBL1/catenin beta like 1 | Catenin-beta-like | Prp19 complex (nineteen complex, NTC) |
| hsp-1, hsp-70, F44E5.4, F44E5.5, F11F1.1 | HSPA8/Hsp73 | Hsp70 | Prp19 complex (nineteen complex, NTC) |
| rbm-22 | RBM22 | RRM | NTC-related (NTR) complex |
| skp-1 | SNW1/SKIP | SKIP/SNW | NTC-related (NTR) complex |
| bud-31 | BUD31/G10 | G10 | NTC-related (NTR) complex |
| cyn-12 | PPIL1 | Peptidyl-prolyl cis-trans isomerase | NTC-related (NTR) complex |
| cwc-15 | CWC15/AD-002 | Cwf15/Cwc15 | NTC-related (NTR) complex |
| plrg-1 | PLRG1/PRL1 | WD40 repeat | NTC-related (NTR) complex |
| emb-4 | AQR/Aquarius | AAA | NTC-related (NTR) complex |
| sel-13 | ZNF830/CCDC16 | C2H2 Zn finger | Intron-binding complex (IBC) |
| cyn-13 | PPIE/CypE | RRM, Peptidyl-prolyl cis-trans isomerase | Intron-binding complex (IBC) |
| cwf-19L2 | CWF19L2 | CwfJ | Intron-binding complex (IBC) |
| F33D11.10, Y65B4A.6 | EIF4A3/eIF4A3 | DEAD/DEAH box helicase | Exon junction complex (EJC) |
| mag-1 | MAGOH | Mago nashi | Exon junction complex (EJC) |
| rnp-4 | RBM8A/Y14 | RRM | Exon junction complex (EJC) |
| casc-3 | CASC3/MLN51 | Btz | Exon junction complex (EJC) |
| mog-1 | DHX38/hPrp16 | DEAH-box helicase | Step 1 factor |
| yju-2 | YJU2/CCDC94 | YJU2 | Step 1 factor |
| mog-3 | CWC25/CCDC49 | CIR, CWC25 | Step 1 factor |
| sacy-1 | DDX41/Abstrakt | DEAD-box helicase | C complex protein |
| cacn-1 | CACTIN | Cactin | C complex protein |
| ddx-35 | DHX35/DDX35 | DEAH-box helicase | C complex protein |
| gpch-1 | GPATCH1/Q9BRR8 | G-patch | C complex protein |
| cyn-15 | PPWD1 | WD40 repeat, Peptidyl-prolyl cis-trans isomerase | C complex protein |
| Y66D12A.8 | CXorf56 | UPF0428 | C complex protein |
| T23G11.4 | C9orf78 | HCA59 | C complex protein |
| cyn-8 | PPIG | Peptidyl-prolyl cis-trans isomerase | C complex protein |
| C08H9.16 | FRA10AC1 | Fra10Ac1 | C complex protein |
| frg-1 | FRG1 | FRG1-like | C complex protein |
| wdr-83 | WDR83/MORG1 | WD40 repeat | C complex protein |
| ess-2 | ESS2/DGCR14 | Es2 | C complex protein |
| R05G6.4 | NOSIP | NOSIP | C complex protein |
| sde-2 | SDE2/C1orf55 | Sde2 | C complex protein |
| C47E8.4 | FAM50A, FAM50B | XAP5 | C complex protein |
| K01G5.8 | FAM32A | FA32A | C complex protein |
| rsr-2 | SRRM2/SRm300 | cwf21, SRRM | C complex protein |
| prp-17 | CDC40/hPrp17 | WD40 repeat | Step 2 factor |
| prp-18 | PRPF18/hPrp18 | PRP18 | Step 2 factor |
| mog-5 | DHX8/hPrp22 | DEAH-box helicase | Step 2 factor |
| sluh-7 | SLU7 | Pre-mRNA splicing Prp18-interacting factor | Step 2 factor |
| F37A4.2 | PRKRIP1 | PRKRIP1 | Step 2 factor |
This table describes C. elegans orthologs or closest homologs of human spliceosomal protein components described in Wahl and Luhrmann (2015) with some updates.
Mutations in splicing machinery genes can cause human diseases with specific symptoms such as autosomal dominant retinitis pigmentosa and spinal muscular atrophy (Daguenet et al. 2015; Carey and Wickramasinghe 2018), and C. elegans has been utilized as a model organism to study pathogenesis and potential therapeutics for such diseases (Briese et al. 2009; Sleigh et al. 2011; Gao et al. 2014, 2019; Rubio-Peña et al. 2015; Wheway et al. 2015; Dimitriadi et al. 2016). RNA-seq analyses have identified many genes affected in such conditions, though how reduction of splicing machinery function leads to such specific phenotypes in C. elegans and other organisms is unknown.
There is an increasing appreciation for roles for pre-mRNA splicing homeostasis in aging. Expression of fluorescent ret-1 splicing reporter minigenes (Kuroyanagi et al. 2013b) is deregulated with age, and dietary restriction (which extends lifespan) prolongs proper splicing regulation (Heintz et al. 2017). In addition, sfa-1, which encodes the C. elegans ortholog of branchpoint binding protein [BBP, also known as splicing factor 1 (SF1)], is required for maintenance of youthful splicing of the ret-1 reporters as well as lifespan extension by dietary restriction (Heintz et al. 2017). RNA-seq analyses of endogenous mRNAs confirmed age-induced splicing deregulation, which is suppressed by dietary restriction in an sfa-1-dependent manner (Heintz et al. 2017). Although SFA-1 is essential for embryonic development, and considered to be involved in branchpoint recognition in C. elegans cis-splicing (Mazroui et al. 1999), it is unclear whether SFA-1 is globally required for intron excision because consensus sequences of the yeast/vertebrate branchpoint have not been found in C. elegans introns (Blumenthal and Steward 1997; Schwartz et al. 2008). Indeed, RNA-seq analyses revealed that genes whose splicing is affected by sfa-1 knockdown upon dietary restriction are functionally enriched for metabolic processes, including lipid catabolism and carbohydrate transport (Heintz et al. 2017). Although the link between the longevity signals upon dietary restriction and SFA-1 function is still unclear, this study provided functional relevance of splicing homeostasis and healthy aging.
Alternative splicing
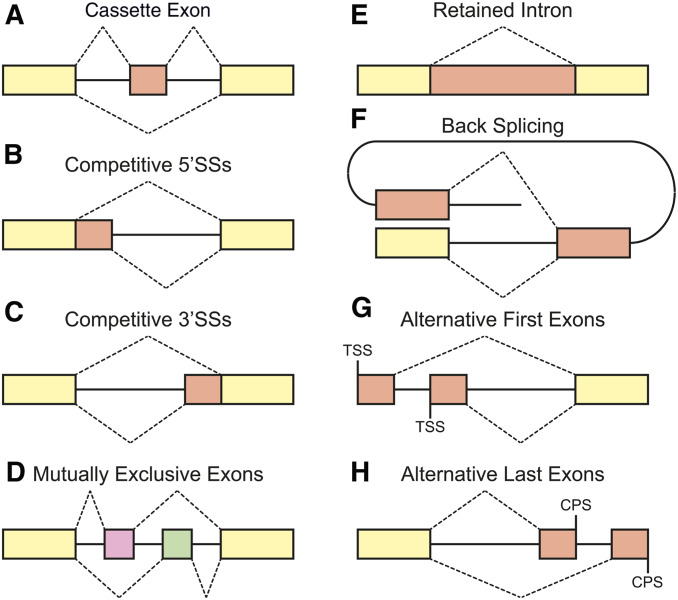
Alternative pre-mRNA splicing is a process by which the exons of primary transcripts are joined in different combinations of splice sites to produce structurally and functionally distinct mRNA and protein variants and enables organisms to generate vast protein diversity from a limited number of genes (Matlin et al. 2005; Nilsen and Graveley 2010). In humans, ∼95% of multi-exon genes undergo alternative splicing, most of which are regulated in a tissue- or cell-type-specific manner (Pan et al. 2008; Wang et al. 2008). Elementary alternative splicing events can be classified into several types (Figure 6). A cassette exon is a discrete exon and can be either included in, or excluded from, an mRNA (Figure 6A); this is the most widely appreciated type of alternative splicing in humans as well as in C. elegans (Wang et al. 2008; Ramani et al. 2011). One splice site may be selected from two or more competitive 5′ (Figure 6B) or 3′ (Figure 6C) SSs in an exon. For mutually exclusive exons, only one out of two or more discrete exons is selected at a time in a mutually exclusive manner (Figure 6D). An intron can be either excised from, or included in, an mRNA (Figure 6E). Back splicing generates a circular RNA by ligating a 5′ SS of an exon to a 3′ SS of the same or an upstream exon (Figure 6F). Alternative first exons (Figure 6G) are regulated by transcription start site selection, and alternative last exons (Figure 6H) are coupled with alternative polyadenylation (discussed below). Multiple elementary alternative splicing events within a gene, and even within an exon, can combinatorially expand the number of isoforms potentially produced by a single gene.
Figure 6.
Schematic representations of elementary alternative splicing events. (A) Cassette exon. (B and C) Competitive 5′- (B) and 3′- (C) splice sites. (D) Mutually exclusive exons. (E) Retained intron. (F) Back splicing. (G) Alternative first exons. The first exon of an mRNA is selected by alternative promoters. TSS, transcription start site. (H) Alternative last exons. The last exon of an mRNA is selected in conjunction with alternative polyadenylation sites. CPS, cleavage and polyadenylation site.
Alternative splicing occurs in ∼23.5–35% of C. elegans genes, depending on the read cutoffs used for detection (Ramani et al. 2011; Tourasse et al. 2017). To visualize the relative expression levels of possible splice variants, Tourasse et al. (2017) diagrammed the abundance of supporting reads for all splicing junctions for each of the 20,335 protein-coding genes in C. elegans (Tourasse et al. 2017). For instance, usage of each of four mutually exclusive exons in the mrp-1 gene (Yabe et al. 2005) is supported by at least 7% of junction reads (Tourasse et al. 2017). In contrast, only one isoform is considered to be expressed for the ant-1.1 gene, one of the most highly expressed genes in C. elegans, even though >50 isoforms were predicted in WormBase (WS251) and 170 other junctions were actually detected (Tourasse et al. 2017). RNA-seq analysis of C. briggsae suggested limited conservation of the alternative splicing events between C. elegans and C. briggsae (Uyar et al. 2012), although this result could also be explained readily by differences in the read depth and annotation quality between C. elegans and C. briggsae.
Early findings about splicing regulatory factors in C. elegans came from genetic suppressor screenings that unexpectedly identified mutations in U1 snRNA genes (Zahler et al. 2004) and RBP genes (Lundquist et al. 1996; Spike et al. 2001, 2002; Spartz et al. 2004; Dassah et al. 2009) as allele-specific suppressors. Our current understanding of factors and elements that influence specific alternative splicing events in C. elegans came from a number of technical advantages, such as the fact that trans-acting factors and cis-acting elements can be identified by genetic analysis with fluorescent reporter animals (Kuroyanagi et al. 2007, 2013a; Ohno et al. 2008), and the evolutionary conservation of trans-acting factors and their binding sequences (Kabat et al. 2006; Ray et al. 2013; Soufari and Mackereth 2017). The nonessential nature of many splicing regulators allows for analysis of partially spliced pre-mRNAs and insight into the specific order of intron excision (Ohno et al. 2008, 2012; Kuroyanagi et al. 2013a). RNA-seq analysis of splicing regulator mutants, as well as crosslinking and immunoprecipitation coupled with deep sequencing (CLIP-seq) analysis of the regulators, allow a global search for alternative splicing events and target genes in C. elegans (Kuroyanagi et al. 2013b; Norris et al. 2014; Ragle et al. 2015; Chen et al. 2016). With these advantages, it is now clear that alternative splicing events are coordinately regulated by multiple splicing regulators (Barberan-Soler et al. 2011; Amrane et al. 2014; Kuwasako et al. 2014; Norris et al. 2014; Tomioka et al. 2016; Tan and Fraser 2017).
Regulation of alternative splicing events and/or isoform-specific functions in C. elegans was discussed extensively in previous review articles (Zahler 2012; Gracida et al. 2016; Wani and Kuroyanagi 2017). Some highlights of those reviews include that regulatory mechanisms for tissue-specific mutually exclusive exons vary from gene to gene (Kuroyanagi et al. 2006, 2007, 2013a, 2014; Ohno et al. 2008), a fraction of the alternative splicing events are coupled with nonsense-mediated mRNA decay (NMD) (see below for details) (Barberan-Soler et al. 2011; Takei et al. 2016; Muir et al. 2018), a weak GC splice site can be used in alternative splicing regulation (Farrer et al. 2002; Ohno et al. 2008), and germlines tend to utilize proximal 3′ splice sites that are not accompanied by pyrimidine stretches (Ragle et al. 2015). Here, we summarize alternative splicing regulation of genes that are not discussed in previous review articles.
Splice site usage can be affected by core components of the splicing machinery, and such events help illuminate mechanisms of splice site selection fidelity. tos-1 (target of splicing-1) was originally identified as a gene whose pre-mRNA splicing pattern was altered in a temperature-sensitive mutant of uaf-1 encoding U2AF large subunit U2AF65; UAF-1 and SFA-1 facilitate recognition of weak 3′ SSs of intron 1 and intron 2 of the tos-1 gene (Ma et al. 2011). A B-complex-specific protein homolog MFAP-1 (Table 2) also affects splicing of tos-1 intron 2 and exon 3 (Ma et al. 2012). Despite detailed analysis of its alternative splicing, the function of the tos-1 gene is still unknown. Aging-induced changes in alternative splicing of the tos-1 gene were used as a readout of SFA-1-mediated pre-mRNA splicing homeostasis (Heintz et al. 2017). Because overexpression of SFA-1 extends lifespan (Heintz et al. 2017), identification of critical target(s) for SFA-1 in alternative or constitutive splicing is of particular interest. Aging-induced splicing changes were confirmed for ret-1 exon 5, lipl-7 intron 4, slo-2 cassette exon, and lea-1 exon 10 (Heintz et al. 2017), yet their relevance to longevity is yet to be elucidated.
Neuron-type-specific alternative splicing can be achieved by specific combinations of splicing regulatory proteins that are regulated by transcription factors specifying the fate of the neurons. sad-1 (synapses of amphids defective-1) encodes a conserved neuronal protein serine/threonine kinase that regulates axonal identity and synapse formation (Crump et al. 2001; Hung et al. 2007). Inclusion of sad-1 exon 15, which carries a termination codon, leads to truncation of the SAD-1 C-terminus that includes the PDZ domain binding sequence (Hung et al. 2007). Only full-length SAD-1 protein, SAD-1(L), can physically interact with an F-actin binding scaffolding protein Neurabin (NAB-1) to control neuronal polarity, and with a pseudokinase STRD-1 to mediate synaptogenesis (Hung et al. 2007; Kim et al. 2010a,b). A dichromatic fluorescent reporter revealed that sad-1 exon 15 is regulated in a neuron-type specific manner; motor neurons in the ventral nerve cord express both isoforms, a touch-sensing neuron ALM expresses only the inclusion isoform, and its sister neuron BDU expresses only the skipped isoform (Thompson et al. 2019). The neuron-type-specific alternative splicing of sad-1 exon 15 is mediated by differential expression of multiple RBPs that are regulated by multiple neuron-type-specific transcription factors; co-expression of transcription factors UNC-86, MEC-3, and ALR-1 specifies the identity of ALM and controls expression of MEC-8 and a Muscleblind homolog MBL-1, leading to complete inclusion of exon 15 (Thompson et al. 2019). In excitatory motor neurons, MBL-1, and not MEC-8, is expressed, and exon 15 of sad-1 is partially included, whereas in inhibitory motor neurons, another RBP, a Musashi homolog, MSI-1, is responsible for the partial inclusion of sad-1 exon 15 (Thompson et al. 2019).
Although alternative splicing can, in theory, generate a large number of isoforms, depending on the locus, only a handful of isoforms may be observed. lev-11 (levamisole resistant-11) is the sole gene encoding tropomyosin, an evolutionarily conserved actin-binding protein that influences actomyosin contractility and actin filament dynamics. It has two tissue-specific promoters, and two, three, two, and three mutually exclusive splicing events for exons 4, 5, 7, and 9, respectively (Kagawa et al. 1995; Anyanful et al. 2001). With this complex gene structure, the lev-11 locus can potentially produce up to 72 distinct mRNAs/proteins and WormBase (WS271) predicts >20 distinct gene models. Extensive analyses of RT-PCR products, however, detected only six isoforms (Watabe et al. 2018). Dichromatic and trichromatic fluorescent splicing reporters revealed tissue-specific expression patterns; body wall muscles in the main body express lev-11a and lev-11d; pharynx and an excretory cell express lev-11e; the intestine and neurons express lev-11c; head muscles express lev-11o; and an unidentified tissue expresses lev-11t (Barnes et al. 2018; Watabe et al. 2018). We expect that future work using long-read RNA sequencing will clarify the observable populations in genes such as lev-11 where only a subset of possible mRNA isoforms is expressed.
CircRNAs have recently been appreciated as common products of many eukaryotic protein-coding genes (Wilusz 2018). They are produced by a kind of alternative splicing termed back splicing (Figure 6F), which is facilitated by short intronic repeat sequences flanking an upstream 3′ SS and a downstream 5′ SS (Liang and Wilusz 2014). CircRNAs are highly stable and accumulate in cells (Zhang et al. 2016) because they are resistant to exonucleases. Certain vertebrate circRNAs have multiple binding sites for specific miRNAs, and, thus, negatively regulate miRNA function by serving as a sponge (Hansen et al. 2013; Memczak et al. 2013). In C. elegans, at least 1166 circRNAs from 797 genes have been identified, and some of them accumulate with age (Memczak et al. 2013; Cortés-López et al. 2018), yet their functions remain to be elucidated.
Some alternative splicing events are conserved between C. elegans and mammals. PTB-1 is the sole C. elegans homolog of mammalian hnRNP proteins PTBP1 (also known as PTB) and PTBP2 (also known as neural PTB, nPTB). PTBP1 negatively auto-regulates its own expression by repressing splicing of the 34-nt exon 11 to elicit NMD (Wollerton et al. 2004), and negatively cross-regulates splicing of its paralogue PTBP2 by repressing a paralogous 34-nt exon (Boutz et al. 2007; Spellman et al. 2007). In C. elegans, PTB-1 represses inclusion of its own 34 nt orthologous exon to elicit NMD (Tomioka et al. 2016), demonstrating conservation of the gene structure and autoregulation of the PTB family. While it is known that PTB family proteins have four RNA-recognition motif (RRM) domains and binds to UC-rich sequences (Oberstrass et al. 2005), cis-elements for the ptb-1 autoregulation remain to be identified.
These examples illustrate that alternative splicing in C. elegans is regulated in a cell-type-specific manner by multiple RBPs. The RBPs responsible may vary from gene to gene in the same cell type, as well as from cell type to cell type for the same gene. In order to elucidate the entire network of RBPs that regulates all the alternative pre-mRNA processing events in all the cell types in this organism, high-throughput analysis of cell-type-specific transcriptomes, as well as systematic characterization of conditional RBP mutant strains, will be needed. High-throughput genome-editing (Norris et al. 2017), deep single-cell RNA sequencing (Cao et al. 2017), isolation of intact RNAs from single tissues (Kunitomo et al. 2005; Spencer et al. 2011; Kaletsky et al. 2016), and long-read RNA sequencing will contribute to this ultimate goal.
Processing and 3′ End Formation of mRNAs
Eventually, an elongating RNA polymerase II (Pol II) transcription complex reaches the end of the gene and transcription termination must occur. The 3′ end of eukaryotic mRNAs are not formed simply by transcription termination, but rather by RNA processing. These RNA processing events include cleavage of the nascent transcript and poly(A) tail formation, both of which typically precede transcription termination. The poly(A) tails of eukaryotic mRNAs are important elements for nuclear export, translation, and stability (Mangus et al. 2003; Goldstrohm and Wickens 2008; Roy and Jacobson 2013). As with splicing, 3′ end formation and polyadenylation can be regulated to generate mRNAs with different 3′ UTRs, which can impact post-transcriptional gene regulation. While our understanding of the molecular mechanisms underlying 3′ end formation and polyadenylation are more completely worked out in mammalian and yeast systems, genetic screens, reporter analysis, and transcriptomics in C. elegans have revealed important insights into these regulatory steps, and will be the focus of this section.
mRNA 3′ end formation and polyadenylation machinery and genetics
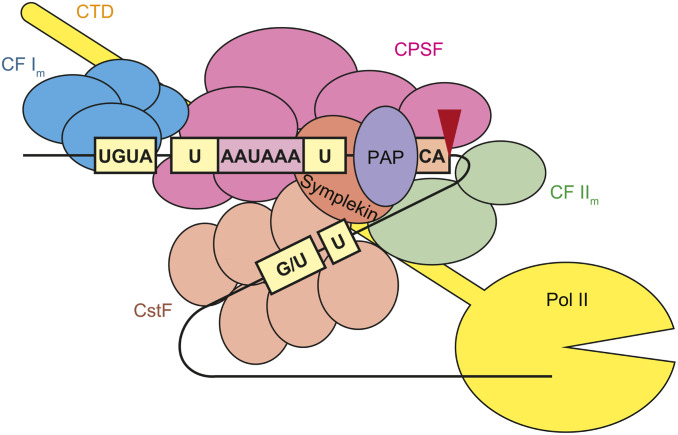
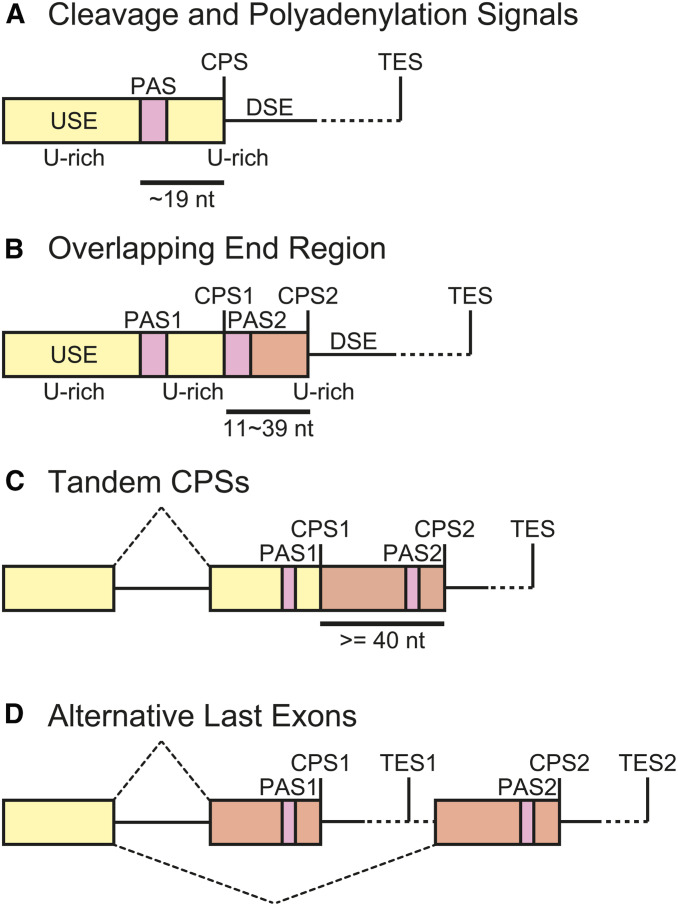
An evolutionarily conserved protein complex forms the poly(A) tail through endonucleolytic cleavage and template-independent polyadenylation of nascent pre-mRNAs (Table 3) (Chan et al. 2011; Shi and Manley 2015). Although there is considerable deviation in individual genes, the current consensus sequences for cleavage and polyadenylation in mammals consist of an upstream sequence conforming to the consensus UGUA, a U-rich upstream element (USE), AAUAAA or similar sequences 15–30 nt upstream of the cleavage and polyadenylation site (CPS), a CA dinucleotide immediately 5′ to the CPS, and the U/GU-rich downstream element (DSE) (Figure 7) (Proudfoot 2011; Gruber et al. 2014; Shi and Manley 2015). Cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF) synergistically bind to the USE-AAUAAA hexamer and the DSE, respectively (MacDonald et al. 1994; Schönemann et al. 2014; Casañal et al. 2017; Clerici et al. 2018). Cleavage factor I (CF Im) binds to the UGUA motif (Brown and Gilmartin 2003). These factors directly bind to the pre-mRNA to form a core complex, which, in turn, recruits other factors, including cleavage factor II (CFIIm), a scaffolding protein symplekin, and the poly(A) polymerase (PAP) to assemble the active 3′ end processing complex (Chan et al. 2011; Shi and Manley 2015). The C-terminal domain (CTD) of the largest subunit of Pol II, which comprises tandem repeats of YSPTSPS heptads, facilitates cotranscriptional assembly of these factors (Hirose and Manley 2000; Proudfoot et al. 2002; Bentley 2005). Recently, a minimal machinery for cleavage and polyadenylation in yeast was reconstituted in vitro, and the cryo-EM structure of the complex was solved (Hill et al. 2019).
Table 3. Brief summary of factors considered to act in cleavage and polyadenylation in C. elegans.
| C. elegans | Human Ortholog | Domains | Description |
|---|---|---|---|
| Cleavage and polyadenylation specificity factor (CPSF) | |||
| cpsf-1 | CPSF160 | MMS1_N, CPSF_A | Cleavage and polyadenylation specificity factor 160 kDa subunit |
| cpsf-2 | CPSF100 | Lactamase_B, Beta-Casp, RMMBL, CPSF100_C | Cleavage and polyadenylation specificity factor 100 kDa subunit, homologous to CPSF73 |
| cpsf-3 | CPSF73 | Lactamase_B, Beta-Casp, RMMBL, CPSF73-100_C | Cleavage and polyadenylation specificity factor 73 kDa subunit, considered to be an endonuclease that cleaves pre-mRNAs at CPSs |
| cpsf-4 | CPSF30 | zf-CCCH, zf-CCHC | Cleavage and polyadenylation specificity factor 30 kDa subunit, ZF2 and ZF3 directly recognize AAUAAA |
| fipp-1 | Fip1 | Fip1 | Pre-mRNA 3′-end-processing factor FIP1, binds to U-rich RNA |
| pfs-2 | WDR33 | six WD40 domains | Directly recognizes AAUAAA |
| symk-1 | Symplekin | DUF3453, Symplekin C | Scaffold protein that functions as a component of a multimolecular complex involved in histone mRNA 3′-end processing. Is involved in pre-mRNA polyadenylation. |
| Cleavage stimulatory factor (CstF) | |||
| cpf-1 | CstF-50 | CSTF1 dimer, five WD40 domains | Cleavage stimulation factor 50 kDa subunit |
| cpf-2 | CstF-64 | RRM, CSTF2 hinge, CSTF C | Cleavage stimulation factor 64 kDa subunit, recognizes DSE |
| suf-1 | CstF-77 | Suf | Cleavage stimulation factor 77 kDa subunit, stimulates CstF-64 |
| Cleavage factor Im (CF Im) | |||
| cfim-1 | CFIm 25 | NUDIX | Cleavage factor Im complex 25 kDa subunit |
| cfim-2 | CFIm 68, CFIm59 | RRM | Cleavage factor Im complex 68 kDa subunit, Cleavage factor Im complex 59 kDa subunit |
| Cleavage factor IIm (CF IIm) | |||
| pcf-11 | PCF11 | CTD bind | Enhances transcription termination and 3′ end processing, genome-wide in human cells |
| clpf-1 | CLP1 | CLP1 N, CLP1 P, Clp1 | Polyribonucleotide 5′-hydroxyl-kinase |
| Poly(A) polymerase (PAP) | |||
| pap-1 | PAP-alpha, beta, gamma | PAP central, NTP transf 2, PAP RNA-bind | Poly(A) Polymerase |
| pap-2 | PAP-alpha, beta, gamma | PAP central, NTP transf 2, PAP RNA-bind | Poly(A) Polymerase |
| pap-3 | PAP-alpha, beta, gamma | PAP central, NTP transf 2, PAP RNA-bind | Poly(A) Polymerase |
This table describes C. elegans orthologs of human proteins involved in 3′ end processing.
Figure 7.
Schematic representations of core sequence elements and factors involved in cleavage and polyadenylation of mRNAs in mammals. Red arrowhead indicates CPS. CA, CA dinucleotide immediately 5′ to the CPS; CTD, C-terminal domain of Pol II; CF Im, mammalian cleavage factor I; CF IIm, mammalian cleavage factor II; CPSF, cleavage and polyadenylation specificity factor; CstF, cleavage stimulation factor; PAP, poly(A) polymerase; U, U-rich upstream element (USE); UGUA, upstream elements with UGUA consensus.
Factors functionally involved in cleavage and/or polyadenylation in C. elegans were first identified in an RNAi screen for suppressors of a Synthetic Multi-Vulva (SynMuv) phenotype of the lin-15AB (n765) mutant (Cui et al. 2008). The Lin phenotype of the n765 allele is caused by premature transcription termination in a transposon integrated into the lin-15AB operon. Suppressors that restored expression of lin-15A (the downstream gene of the operon) were recovered and proposed to function by compromising transcription termination within the inserted transposon (Cui et al. 2008). This screen identified functional homologs of much of the eukaryotic cleavage and polyadenylation machinery, including CPSF subunit genes: cpsf-1, cpsf-2, and cpsf-4; CstF subunit genes: cpf-1 and cpf-2; a symplekin gene symk-1; and a poly(A) polymerase gene pap-1 (Table 3) (Cui et al. 2008). This screen also identified another group of genes, cids-1, cids-2, and nrd-1, encoding proteins with a Pol II CTD interacting domain (CID), although their function in 3′ end formation remains to be elucidated. A dpy-13 transcriptional readthrough reporter that carries a GFP-fused tubulin cDNA downstream from the CPS was also used to identify cleavage and polyadenylation factors (Miki et al. 2017). Efficient repression of the dpy-13 reporter readthrough requires the CPSF subunit genes cpsf-1, cpsf-2, cpsf-3, cpsf-4, fipp-1, and pfs-2, CstF subunit genes cpf-2 and suf-1, a CFIIm subunit gene pcf-11, and symk-1, likely due to the functions of these proteins in 3′ end cleavage (Miki et al. 2017). Chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) analysis revealed peaks of CstF subunit proteins CPF-1 and CPF-2 almost overlapping with that of Pol II with Ser2-phosphorylated CTD at 0.5–0.6 kb downstream from the CPSs (Garrido-Lecca et al. 2016). These results indicated that functions of the factors involved in the 3′ end processing of pre-mRNAs and termination of Pol II transcription are conserved, and that these processes are also coupled in C. elegans.
Termination of RNA polymerase II transcription
In eukaryotes, termination of Pol II transcription is not necessary for mRNA 3′ end formation. Rather, efficient transcription termination requires 3′ end processing of pre-mRNAs because these processes are functionally coupled. Here, we summarize the current models of this mechanism and the fate of pre-mRNAs downstream from the CPS. Two, nonmutually exclusive models for transcription termination have been proposed in mammals and yeast. In the “torpedo” model, pre-mRNA is first cleaved at the CPS (Kim et al. 2004b). For a subset of genes, the pre-mRNA is cleaved within the cotranscriptional cleavage (CoTC) site located downstream from the CPS (Teixeira et al. 2004; Nojima et al. 2013). Then, a processive 5′-to-3′ exoribonuclease XRN2 (Rat1p in budding yeast) degrades the nascent RNA from a new 5′ end and catches up with and disassembles the transcription elongation complex (Kim et al. 2004b; Teixeira et al. 2004; West et al. 2004; Nojima et al. 2013). In the alternative “conformational change” model, the transcription elongation complex is disassembled by factors associated with, and/or dissociated from, the Pol II CTD in a polyadenylation signal (PAS)-dependent, yet cleavage-independent, manner (Kim et al. 2004a; Zhang et al. 2005, 2015). As phosphorylation status of the YSPTSPS heptad repeats in the Pol II CTD is regulated dynamically during transcription cycles, CTD phosphatases are implicated in transcription termination (see below).
Experiments in C. elegans support both models of transcription termination. ChIP-seq analysis, revealed that Pol II with Ser2-phosphorylated CTD peaked ∼0.5 kb downstream from the CPS (Garrido-Lecca et al. 2016), suggesting that Pol II pauses in this region to facilitate 3′ end formation and transcription termination. Consistent with the “torpedo” model, temperature-sensitive alleles of xrn-2 (an ortholog of human XRN2) were recovered in a forward genetic screen for mutants that allow transcriptional readthrough of the dpy-13 reporter (described above) (Miki et al. 2017). However, additional experiments revealed two types of genes: those that depend on xrn-2 for transcription termination and those that do not (Miki et al. 2017). When xrn-2 was inhibited, pre-mRNAs of some genes, including dpy-13, were cleaved at the CPSs, yet Pol II did not terminate transcription, and some downstream genes in the same direction were processed into mature mRNAs (Miki et al. 2017). In the same experiments, other genes exhibited no transcription termination defect. ChIP-seq analysis of XRN-2::GFP suggested that, in both types of genes, XRN-2 is recruited to promoter regions, travels along the gene bodies, and pauses just downstream from the CPSs together with Pol II (Miki et al. 2017). Fragment swapping of readthrough reporter minigenes suggested that it is the promoter regions that determine whether or not the transcription termination depends on XRN-2 (Miki et al. 2017), although detailed mechanisms are yet to be determined. Interestingly, transcription termination of transcripts from XRN-2-independent genes requires a CPS downstream element of unknown composition (Miki et al. 2017).
A unique property of pre-mRNA processing of operon genes is that 3′ end formation of the upstream genes is coupled with trans-splicing of the downstream genes and not with transcription termination (Blumenthal 2012; Blumenthal et al. 2015). Briefly, cleavage and polyadenylation of the upstream mRNA occurs by a conventional mechanism (Garrido-Lecca et al. 2016), and the resulting 5′-phosphate end of the nascent RNA is subjected to XRN-2-mediated degradation (Miki et al. 2016). The downstream pre-mRNA is, however, protected from XRN-2 by a U-rich (Ur) element proposed to recruit SL2 snRNP for trans-splicing (Graber et al. 2007; Lasda et al. 2010). Recently, it has been shown that expression of xrn-2, a downstream gene of the rpl-43∼xrn-2 operon, is negatively auto-regulated by XRN-2 activity in vivo (Miki et al. 2016); XRN-2 may compete with transcribing Pol II and/or the trans-splicing machinery to terminate transcription by the “torpedo” mechanism without affecting expression of the upstream rpl-43 gene (Miki et al. 2016). XRN-2 activity also affects expression of downstream genes in other operons, including clpf-1, by the same mechanism (Miki et al. 2016).
Alternative polyadenylation
Selection of an appropriate CPS can regulate gene expression. A diversity of techniques have been used to annotate C. elegans CPSs genome-wide, which has led to a more complete picture of cleavage and polyadenylation than would be possible from single reporter analyses (Mangone et al. 2010; Jan et al. 2011). At least one representative CPS has been identified in 83% of RefSeq mRNAs so far, and most CPSs in individual mRNAs are clustered within 4 nt of the representative CPS (Mangone et al. 2010; Jan et al. 2011). Searches for the most likely PAS within 50 nt upstream of each of the CPSs identified the canonical PAS motif AAUAAA in 39% of the CPSs and many PAS variants that differ by 1–2 nt in 48% of the CPSs (Mangone et al. 2010). The positions of both the canonical PAS motif and the PAS variants peaked 19 nt upstream of the CPS and were embedded within a U-rich region that extends ∼20 nt beyond the CPS (Mangone et al. 2010; Jan et al. 2011) (Figure 8A). Surprisingly, ∼13% of the CPSs lacked a detectable PAS motif (Mangone et al. 2010); such “no PAS” CPSs are within a U-rich region (Jan et al. 2011). The canonical PAS motifs were much more frequently found in nontrans-spliced (43% of 5131 CPSs) than in trans-spliced transcripts (30% of 14,873 CPSs), and more frequently in SL1-spliced nonoperon genes (32% of 10,879 CPSs) than in SL1-spliced operon genes (22% of 1409 sites), implying some correlation between 5′- and 3′-end processing.
Figure 8.
Schematic representations of cis-elements for 3′ end processing of mRNAs in C. elegans. (A) Typical cleavage and polyadenylation signals. AAUAAA or related sequences embedded in a U-rich region functions as a polyadenylation signal (PAS). CPS, cleavage and polyadenylation site; DSE, downstream element; TES, transcription end site; USE, upstream element. (B) Overlapping end region (ORE). Two closely located (11–39 nt apart) CPSs have their own PAS and share a U-rich region as either DSE or USE. (C) Tandem cleavage and polyadenylation sites (CPSs). Two or more PASs and CPSs are located in the same last exon but are ≥ 40 nt apart. (D) Alternative last exons (ALEs). Choice of the CPS is coupled with choice of the last exon of an mRNA. Boxes represent exons and solid lines indicate introns or 3′ flanking regions.
About half of genes have more than one CPS (Mangone et al. 2010; Jan et al. 2011). Two or more closely spaced (usually 12–22 nt) CPSs may form an overlapping end region (OER) (Figure 8B), in which each of the CPSs contains a PAS motif; such PAS motifs are separated by a U-rich region that can serve as either an upstream or a downstream element depending on the CPS used (Jan et al. 2011). In total, 17,596 CPSs from 7116 OERs represent the largest currently known class of alternative mRNA isoforms in C. elegans (Jan et al. 2011). The end regions of 2448 genes even overlap with those of convergent genes, contributing to genome compaction without significantly impacting regulatory autonomy (Jan et al. 2011).
Multiple CPSs in a single gene may be separated by 40 nt or more; 11,285 such upstream CPSs were identified in 31% of the Entrez genes with sequencing information (Jan et al. 2011) (Figure 8C). Among them, 8148 CPSs (72%) are within the same last exon as the 3′-most CPSs to form tandem CPSs (Jan et al. 2011). As miRNAs recognize complementary elements in 3′ UTRs of their target mRNAs (Zisoulis et al. 2010), alternative choice of tandem CPSs may affect regulation by miRNAs (Jan et al. 2011; Blazie et al. 2017). The 3′-most or distal CPSs prefer a common PAS, whereas the proximal CPSs more often show no PAS (Mangone et al. 2010; Jan et al. 2011), consistent with the fact that the cleavage and polyadenylation machinery sometimes bypasses the proximal CPSs. Some of the upstream “no PAS” CPSs may be used upon physical constraint, such as queuing of Pol II (Mangone et al. 2010). In the rest of the multi-CPS genes, choice of the CPS is coupled with choice of alternative last exons (ALEs), often with distinct termination codons, thereby affecting the function of the protein products (Figure 8D). So far, 1398 ALEs have been identified across 1277 Entrez genes (Jan et al. 2011). Recent polyA-tagging and sequencing (PAT-Seq) analysis of mRNAs from eight somatic tissues revealed widespread tissue-specific alternative polyadenylation (Blazie et al. 2015, 2017). Regulation mechanisms for the alternative polyadenylation events coupled with ALEs are reported for unc-60, unc-64, and cha-1/unc-17; tissue-specific RBPs and/or RNA secondary structure have been shown to play switch-like roles (Ohno et al. 2012; Kuroyanagi 2013; Mathews et al. 2015; Chen et al. 2016).
Regulators of alternative polyadenylation have been identified in a suppressor screen for synaptogenesis and axon development phenotypes of a sydn-1 null mutant (Van Epps et al. 2010; Chen et al. 2015). The neuronal phenotypes of the sydn-1 mutant were suppressed by loss-/reduction-of-function or knockdown of pfs-2, cpf-1, cpf-2, cpsf-2, pap-2 (T15H9.6), cpsf-4, zfp-3 (encoding a zinc finger protein that can interact with CPF-2), ssup-72 (encoding an ortholog of human and yeast CTD phosphatase SSU72), symk-1, cids-1, and cids-2 (Van Epps et al. 2010; Chen et al. 2015), indicating that these genes are specifically required for the sydn-1 neuronal phenotypes. Pol II occupancy analysis by ChIP-seq and reporter analysis revealed that inhibition of SSUP-72 activity by a nuclear protein, SYDN-1, at a strong internal PAS region of the endogenous unc-44 locus is required for producing a neuron-specific isoform of unc-44 (unc-44f) that utilizes the 3′-most PAS (Chen et al. 2015). Paradoxically, SYDN-1 inhibition of SSUP-72 promotes neuron-specific expression of an isoform of the dlk-1 gene (dlk-1S) that utilizes a weak intronic PAS (Chen et al. 2015), indicating that the function of SSUP-72 is context-dependent. A dlk-1 null mutation suppressed the axonal phenotype of the sydn-1 null mutant (Chen et al. 2015), suggesting that this sydn-1 phenotype is due to dysregulation of dlk-1 alternative polyadenylation. This study thus revealed mechanisms for neuron-specific alternative polyadenylation as well as the functions of neuron-specific protein isoforms.
Does length matter? Insights into poly(A) tails from genome-wide studies
Since poly(A) tails of eukaryotic mRNAs are important for nuclear export, translation, and stability, it has long been believed that longer poly(A) tails contribute more to mRNA stabilization and efficient translation, and that is true at pregastrulation stages of frog, zebrafish, and fruit fly embryos (Subtelny et al. 2014; Eichhorn et al. 2016; Lim et al. 2016). However, recent poly(A) tail profiling with TAIL-seq or PAL-seq protocols in eukaryotes, including C. elegans, reveal that short poly(A) tails are a feature of abundant and well-translated mRNAs (Chang et al. 2014; Subtelny et al. 2014; Park et al. 2016; Lima et al. 2017). For example, at the L4 stage, 90% of mRNA molecules had tail lengths between 26 and 132 nt, with the most abundant species of poly(A) tails being 33–34 nt (Lima et al. 2017); 33–34 nt is the size of one PABP footprint (Lima et al. 2017). In addition, a phasing pattern was observed, with peaks at the poly(A) tail lengths consistent with serial PABP binding (Lima et al. 2017), suggesting that unprotected 3′ adenosines are trimmed. When looking at median overall poly(A) lengths of 13,601 individual protein-coding genes, the most frequent median length was 82 nt, with 90% of mRNAs having median tails ranging from 53 to 115 nt (Lima et al. 2017). Gene ontology (GO) analysis revealed that short-tailed transcripts were highly enriched for genes involved in translation, nucleosome components, and cuticular collagens, whereas long-tailed transcripts were enriched for genes with regulatory functions, such as transcription factors, signal transduction proteins, mediators of neuronal activity, and hormone receptors (Lima et al. 2017).
Processing of histone mRNAs
In metazoans, mRNAs of canonical, replication-dependent histone genes (H2a, H2b, H3, and H4) are not polyadenylated, but end in a 26-nt conserved structure with a stem-loop (Marzluff et al. 2008). During formation of the 3′ end, the stem-loop is recognized by stem-loop binding protein (SLBP), and the pre-mRNAs are cleaved by the endonuclease subunit CPSF73 in CPSF with the help of the U7 snRNP, which binds to the histone downstream element (HDE) located downstream from the cleavage site (Strub and Birnstiel 1986; Mowry and Steitz 1987; Wang et al. 1996; Dominski et al. 2005; Kolev and Steitz 2005).
C. elegans has 64 histone genes that all harbor conserved 3′ end sequences with a stem-loop structure and a highly conserved AATCC element immediately followed by at least one canonical PAS element (Pettitt et al. 2002; Keall et al. 2007). The majority of C. elegans histone mRNAs end 3–6 nt downstream from the hairpin structure and lack a poly(A) tail, as revealed by poly(A) selection, RNase protection assays, and sequencing analysis (Keall et al. 2007). The sole SLBP homolog in C. elegans, CDL-1, specifically binds to the conserved 16-nt hairpin sequence (Michel et al. 2000; Kodama et al. 2002). While knockdown of cdl-1 does not significantly affect histone mRNA levels, it does severely deplete histone protein levels (Pettitt et al. 2002; Keall et al. 2007), consistent with the role of CDL-1 in post-transcriptional control of histone gene expression. Surprisingly, the C. elegans genome lacks a U7 snRNA gene (Davila Lopez and Samuelsson 2008). Instead, CSR-1-bound endogenous small interfering RNAs (endo-siRNAs) produced by the RNA-dependent RNA polymerase (RdRP) EGO-1 and the dicer-related helicase DRH-3 are proposed to be involved in the processing of histone mRNAs (Avgousti et al. 2012).
Even though the majority of C. elegans histone mRNAs lack poly(A) tails (Keall et al. 2007), poly(A) profiling analyses revealed polyadenylated transcripts for 57 of the 64 histone genes (Mangone et al. 2010; Jan et al. 2011). These observations lend support to models in which the canonical PAS elements ensure transcription termination, i.e., C. elegans histone mRNAs are initially 3′-end-processed via cleavage and polyadenylation at the conserved PAS sites, followed by further processing to remove sequences downstream of the stem-loop (Mangone et al. 2010). Recently, it has been shown that total and polyadenylated mRNAs are upregulated for most of the replication-dependent and -independent histone genes in an smn-1 null mutant (Gao et al. 2019). C. elegans smn-1 is the sole ortholog of mammalian SMN proteins, which facilitate assembly of snRNPs, implying that some snRNPs may be involved in histone mRNA processing.
mRNA Quality Control: Counteracting Errors and Regulating Gene Expression
There is ample room for errors during mRNA processing and maturation. For example, the frequency of transcriptional errors in C. elegans has been estimated at ∼4 × 10−6 (Gout et al. 2013). While this error frequency may seem low, the number and size of mRNA molecules produced in each cell guarantees some number of errors. For example, a cell with 10,000 mRNAs of an average length of 1500 nt and a transcriptional error frequency of 4 × 10−6 would be expected to harbor ∼60 mRNAs with errors from transcription alone. The spliceosome is also not perfect: a meta-analysis of over 1000 C. elegans RNA-seq samples identified a persistent, low level of novel splice junctions (deemed “biological noise”), especially in highly transcribed mRNAs, where sensitivity to detect low-abundance isoforms would be expected to be greatest (Tourasse et al. 2017). As with errors that arise during DNA replication (i.e., mutations), errors during mRNA production may be benign, or they may alter the encoded information, leading to gene products deleterious to the organism.
To ensure the fidelity of gene expression, and to mitigate the deleterious consequences of mistakes in mRNA production, C. elegans has quality control mechanisms that act on the intermediates and products of gene expression. The intermediates (mRNAs) and products (proteins) of gene expression are tested for certain properties, and a molecule that fails a test is often destroyed. In this manner, cells constantly scrutinize gene expression, prune errors, and effectively heighten the fidelity of gene expression. Many of these quality control pathways are conserved throughout eukaryotes. Here, we will focus on quality control pathways relating to protein-coding genes, though we note there is also quality control of ncRNAs, including ribosomal RNAs, tRNAs, and telomerase RNA (for reviews, see de la Cruz et al. 2015; Hopper and Huang 2015; Zinder and Lima 2017).
There are several distinct quality control pathways that collectively attenuate the levels and effects of a wide variety of errors. For example, mRNAs that are inefficiently spliced are often retained in the nucleus, effectively preventing their translation (e.g., Shiimori et al. 2013). Misfolded proteins are recognized by chaperones, and either refolded or degraded (reviewed in Voisine et al. 2010). Double-stranded RNAs are produced by some viruses, selfish genetic elements, and repetitive sequences and are edited or silenced, altering the encoded protein or preventing protein production altogether (Fire et al. 1998; Morse et al. 2002). Many quality control processes act cell-autonomously, though some can communicate across tissues (Prahlad et al. 2008; Sun et al. 2011). While these pathways collectively capture the products of a great diversity of errors, they are not perfect and they do have blind spots: some proteins escape or overwhelm quality control, misfold, and accumulate, leading to cellular and organismal dysfunction (Satyal et al. 2000; Parker et al. 2001).
It should be noted that, while many quality control processes have defined roles in mitigating the production and consequences of errors in gene expression, they serve other conserved functions as well, including regulation of endogenous gene expression. For example, NMD (discussed in detail in the next section) affects mRNA expression for a large fraction of the genome, including many apparently “normal” (i.e., “error-free”) transcripts (Morrison et al. 1997; Barberan-Soler et al. 2009; Muir et al. 2018). In fungi, mammals, and plants, some quality control components and pathways function as antiviral factors, to help cells distinguish between “self” and “nonself” and fight invasions (Toh et al. 1978; Balistreri et al. 2014; Garcia et al. 2014). This remains an area of active research, and C. elegans will continue to contribute to the appreciation of the biological functions that quality control pathways provide.
One important class of quality control pathways are those acting at the level of translation. Collectively, these pathways are known as translation surveillance and involve communication between a ribosome and mRNA decay machinery. A schematic of the translational surveillance pathways we will overview in this chapter are depicted in Figure 9, and the names, domains, and functions of specific factors are tabulated (Table 4). These pathways prevent protein production from mRNAs with an early stop codon (NMD), mRNAs that lack a stop codon (Nonstop Decay), or mRNAs with a block to translation elongation (No-Go Decay). A theme of all three pathways is that failure of a ribosome to transit an mRNA in a timely or “normal” manner is coupled to mRNA decay. This has led to the view of translational surveillance pathways as enforcing a “translate or perish” rule on mRNAs (Brogna et al. 2016).
Figure 9.
Translational surveillance pathways under consideration. (A) Translation of a normal mRNA. Ribosomes (gray) load near the trimethylguanosine cap (m2,2,7G) and locate a start codon (green stoplight). Ribosomes elongate until they terminate at a stop codon (red octagon). Upon termination, protein is released, and ribosomes are recycled for further rounds of translation. (B) Nonsense-mediated mRNA decay (NMD) arises from translation termination at a premature stop codon. The mRNA is destabilized through the action of at least seven SMG proteins. (C) Nonstop Decay arises from translation to the 3′ end of an mRNA, which can arise from mRNA cleavage (left) or polyadenylation upstream of a stop codon (right). The mRNA and nascent protein are degraded, and the ribosome is rescued. (D) No-Go Decay arises when a ribosome stalls as a result of a roadblock during elongation (yellow triangle). Such roadblocks include RNA hairpins, rare codons, and polybasic (Arg or Lys) amino acid runs. As with Nonstop Decay, the mRNA and nascent protein are degraded, and the ribosome is rescued.
Table 4. Brief summary of factors known to act in C. elegans translational surveillance.
| C. elegans | Human Ortholog | Domains | Description |
|---|---|---|---|
| Nonsense-mediated decay | |||
| smg-1 | SMG1 | PI3Kinase | SMG-1 encodes a huge protein with a PI3K domain required for NMD, presumably via phosphorylation of SMG-2 |
| smg-2 | UPF1 | RNA helicase, AAA | Binds mRNAs and is dynamically phosphory-lated/dephosphorylated, with the phosphorylated form exhibiting a preference for NMD targets. Binding observed throughout 3′UTRs. |
| smg-3 | UPF2 | MIF4G, Upf2 | SMG-2/3/4 interact and are thought to constitute a “core” NMD complex conserved in most eukaryotes. |
| smg-4 | UPF3A and UPF3B | Upf3 | SMG-2/3/4 interact and are thought to constitute a “core” NMD complex conserved in most eukaryotes. |
| smg-5 | SMG5 | PIN (catalytically inactive) | With SMG-7, required for efficient dephosphorylation of SMG-2 |
| smg-6 | SMG6 | EST1, PIN | SMG-6 contains a PIN endoribonuclease domain required for NMD |
| smg-7 | SMG7 | EST1 | With SMG-5, required for efficient dephosphorylation of SMG-2 |
| smgl-1 | NBAS | WD40 | Identified alongside smgl-2; loss-of-function via RNAi stabilizes some NMD targets. Complete loss-of-function thought to be lethal. |
| smgl-2 | DHX34 | DExH-box helicase | Identified alongside smgl-1; loss-of-function via RNAi stabilizes some NMD targets. Complete loss-of-function thought to be lethal. |
| Nonstop/no-go decay | |||
| skih-2 | SKIV2L | DEAD-box helicase, rRNA processing Arch | Catalytic subunit of the SKI RNA helicase and is required for 3′>5′ decay of Nonstop mRNAs |
| ttc-37 | TTC37 | Tetratricopeptide repeat | Scaffolding subunit of the SKI RNA helicase and is required for 3′>5′ decay of Nonstop mRNAs |
| pelo-1 | PELO | eRF1 domains 1, 2, and 3 | Ribosome rescue factor required for release of ribosomes from Nonstop mRNAs; homologous to eRF1 |
| nonu-1 | N4BP2 | P-loop Kinase, Cue, Smr | Putative endoribonuclease required for repression of Nonstop and No-Go mRNAs |
This table describes factors currently known to act in translational surveillance in C. elegans. This list is not exhaustive, esp. for Nonstop/No-Go Decay where genetic screens for factors are far from saturated and several more factors are known in systems other than C. elegans. There is an extensive literature for many of these individual factors, and readers are referred to references and reviews for further reading (see main text).
Nonsense-mediated mRNA decay discovery and genetics
NMD in C. elegans was discovered as an allele-specific, gene-nonspecific informational suppression phenomenon (Hodgkin et al. 1989). This origin is reflected in the naming of proteins that carry out NMD: Suppressor with Morphogenetic effect on Genitalia (smg, pronounced “smug”) are genes that, when inactivated, suppress certain alleles in unrelated genes (dpy-5, lin-29, unc-54, tra-1, tra-2) and also exhibit abnormal genitalia (a protruding vulva in the hermaphrodite and a swollen bursa in males). At least some smg-suppressible alleles are mutations of the 3′ UTR downstream of an otherwise normal ORF (Hodgkin et al. 1989; Pulak and Anderson 1993). At the time of its discovery, this distinguished smg from other informational suppression mechanisms, all of which directly altered the nature of information transfer in coding regions (e.g., nonsense suppressor tRNAs that recode some stop codons to an amino acid). Instead, it was suggested that SMGs act in mRNA metabolism.
This turned out to be the case, with an interesting twist: the SMG machinery acts to destabilize certain mRNAs in a translation-dependent manner (Losson and Lacroute 1979; Daar and Maquat 1988; Peltz et al. 1993; Pulak and Anderson 1993). smg-suppressible alleles produce mRNAs that encode functional proteins, but the mRNA is destabilized by the SMG machinery. In such cases, the mutant phenotype arises from decreased levels of functional protein rather than an mRNA that encodes a nonfunctional protein. Loss of a SMG protein allows for derepression of the mRNA (i.e., normal mRNA levels), and sufficient protein production for suppression of the mutant phenotype. A distinguishing feature of many smg-suppressible alleles is the presence of a premature stop codon, which can arise from mutation, frameshifts, and/or an abnormally long 3′ UTR, to name but a few (Pulak and Anderson 1993; Longman et al. 2007). In such cases, the C-terminally truncated protein is apparently still functional. This is where the name NMD comes from (which we will hereafter use interchangeably with SMG): NMD is an mRNA decay process acting on certain nonsense (early stop codon) alleles. In at least one case, phenotypic suppression by smg may yield full-length protein via spontaneous readthrough of an early stop codon (tra-3 (Hodgkin et al. 1989)); loss of NMD leads to an increase in mRNA levels and readthrough of stop codons in S. cerevisiae (Wang et al. 2001; Keeling et al. 2004). We note that while many early stop codon mutations will trigger degradation by NMD, not all such early stop codons will be phenotypically smg-suppressible; phenotypic smg suppression requires the truncated polypeptide be functional [for several examples of this, see unc-54 (Pulak and Anderson 1993)].
A surprising feature of C. elegans is that its NMD system is nonessential, in contrast to other metazoans where loss of NMD is lethal (Medghalchi et al. 2001; Metzstein and Krasnow 2006; Wittkopp et al. 2009). This fact allows C. elegans’ smg suppressors to be easily isolated via genetic screens or as spontaneous suppressors, both of which contributed to the isolation of smg genes in C. elegans (Hodgkin et al. 1989; Cali et al. 1999). It is not uncommon to unintentionally obtain spontaneous smg suppressors when growing large populations of mutant animals, where a mutation can suppress the mutant phenotype and improve growth and/or health of the animal. It is an outstanding question why NMD is essential in most animals but not in C. elegans; recent work suggests many metazoans (but not C. elegans) contain an apoptosis-promoting factor (GAD45Beta) that NMD must constitutively repress for viability (Nelson et al. 2016). Whatever the cause, the nonessential nature of C. elegans’ NMD has had many positive outcomes, including:
(1) The ability to study and characterize functional effectors of NMD to learn how the NMD pathway works (see next section).
(2) A view of mRNA metabolism in the absence of NMD, useful for analyzing more directly the products of transcription and splicing divorced from cellular efforts to erase errors in these processes (Morrison et al. 1997; Mitrovich and Anderson 2000; Barberan-Soler et al. 2009; Tourasse et al. 2017; Muir et al. 2018).
(3) Reverse genetic tools to dissect gene function and expression, by combining smg mutants with construction of smg-suppressible alleles of a gene-of-interest (Wilkinson et al. 1994; Getz et al. 1997; Maher et al. 2013).
Many of the above studies have served as a genesis for models and studies in other organisms, and also benefited from discoveries in other species—a trend that we expect will continue into the future.
Mechanism of NMD
Soon after its discovery, it was appreciated that NMD is a widely conserved process throughout eukaryotes, including humans (Chang and Kan 1979; Losson and Lacroute 1979; Maquat et al. 1981; Hodgkin et al. 1989). The observation that a large fraction (∼11–20%) of human inherited genetic diseases results from mutations that create an early stop codon pointed to NMD as an important player in a diverse swath of human health and disease (Mort et al. 2008). This realization stimulated much interest in understanding how NMD (Figure 9B) identifies target mRNAs and destroys them. However, there still is no consensus on many questions one might consider fundamental to an understanding of NMD. Below we describe observations and models in C. elegans and other systems, highlighting areas for future work.
One question central to NMD is how SMG proteins identify their targets. Translation is required for NMD, and all known NMD targets contain an early stop codon. These two observations spatiotemporally align translation termination and NMD and have led to the idea that some stop codons are deemed “premature” and are targeted by NMD. Consistent with the idea that there are qualitative or quantitative differences in recognition of premature and normal stop codons, ribosomal toeprints in S. cerevisiae and mammalian extracts are slightly longer on at least some NMD targets (Amrani et al. 2004; Peixeiro et al. 2012), and ribosomes in C. elegans protect different footprint sizes on NMD targets compared to non-NMD targets (Arribere and Fire 2018). While the physical basis for such effects remains unclear, these studies provide evidence that differences between premature and normal stop codons indeed exist at the level of translation.
One approach to address how premature stop codons are defined has been to analyze NMD targets and mutate features of interest. These studies point to the 3′ UTR as being critical for NMD target definition, and we provide some exemplary alleles that do or do not elicit NMD in Figure 10.
Figure 10.
Schematic of mRNAs that do, or do not, trigger NMD. For each gene, the wild-type allele is diagrammed above a mutant allele. In the diagrams, exons are indicated by boxes, coding regions by thicker boxes, stop codons by red octagons and exon-exon junctions by white gaps. The scale bar shows 100 or 1000 nt for each mRNA. The far-right column indicates whether the allele is an NMD target or not. Examples were chosen to illustrate different classes of NMD targets and are not exhaustive (see text for further examples). Annotations were taken from Ensembl, with 3′ UTR annonations from (Jan et al. 2011). (A) An example of a premature stop codon upstream of exon-exon junctions. lin-29(n546) is a smg-suppressible allele of lin-29 (Hodgkin et al. 1989) that encodes an Arg > Stop mutation in the lin-29 ORF (Rougvie and Ambros 1995). (B) An example of an NMD target from a mono-exonic gene. dpy-5(e61) is a Gly > Stop mutation (Thacker et al. 2006) that confers a smg-suppressible Dpy phenotype (Hodgkin et al. 1989). (C) An example of an allele that converts a normal stop codon to a premature stop codon. unc-54(r293) is a 256 bp deletion spanning the unc-54 3′ UTR and poly(A) site that results in a fusion of the unc-54 transcript with the downstream aex-5 transcript. The unc-54(r293) allele is smg-suppressible (Hodgkin et al. 1989). The exact positions of the exon-exon junctions in the unc-54(r293) transcript are not known; the diagram represents a “best guess” based on the work of Pulak and Anderson (1993), Loepold and Ahmed (2014) and the aex-5 transcript. (D) An example of an allele that creates premature stop codons but does not elicit NMD. unc-73(e936) is a splice site mutation that leads to usage of two cryptic splice sites (−1 and +23 nt relative to the normal splice site) generating out-of-frame premature stop codons. Neither transcript is an NMD target, though the reasons for this are not known (Roller et al. 2000).
One model for premature stop codon recognition is that NMD detects stop codons with downstream splicing events. In this model, translation termination upstream of an exon junction complex [deposited ∼20–24 nt upstream of an exon junction (Le Hir et al. 2000)] gives rise to NMD. While this model has proven useful for understanding NMD targeting of some human mRNAs (Cheng et al. 1990; Zhang et al. 1998a,b), it cannot explain NMD in S. cerevisiae, which is thought to lack an exon junction complex. Furthermore, it cannot explain all NMD targeting in C. elegans, as NMD in C. elegans has been shown to act on mono-exonic transcripts [where splicing is not thought to happen, e.g., dpy-5 (Figure 10B)], and splicing downstream of a stop codon is neither necessary (Longman et al. 2007) nor sufficient (Figure 10D) (Roller et al. 2000) for NMD.
A second model from NMD substrate recognition is that premature stop codons are distinguished from normal stop codons by virtue of the length of the downstream 3′ UTR. It is unclear what factors might sense 3′ UTR length. Two overlapping possibilities are: (1) one or more SMG proteins detect 3′ UTR length, and (2) 3′ UTR length is sensed by interactions between a terminating ribosome and the PABP (Behm-Ansmant et al. 2007; Ivanov et al. 2008; Silva et al. 2008). However, an argument against a role for the PABP is that NMD can occur even on transcripts that lack a poly(A) tail (Meaux et al. 2008). Furthermore, in humans, there is a relatively poor correlation between 3′ UTR length and susceptibility to NMD genome-wide, with many long 3′ UTRs apparently resistant to NMD (Hurt et al. 2013; Toma et al. 2015). Interestingly, SMG-2 binds throughout the 3′ UTR of NMD targets in mammals, though binding also occurs throughout the 3′ UTRs of normal mRNAs, suggesting that the mere presence of SMG-2 is insufficient to discriminate NMD targets from nontargets (Hogg and Goff 2010; Hurt et al. 2013; Zund and Muhlemann 2013; Kurosaki et al. 2014).
Cellular identification of NMD targets requires the action of each of at least seven different SMG proteins, SMG-1 through SMG-7 (Hodgkin et al. 1989; Cali et al. 1999). SMG-1 through SMG-7 have functional homologs throughout metazoans, and at least SMG-2, SMG-3, and SMG-4 are conserved to S. cerevisiae (Upf1, Upf2, Upf3, respectively). Loss of any one of the seven smg genes yields nearly identical phenotypes with respect to NMD, consistent with the idea that action of all seven SMG proteins is required for the NMD pathway (Pulak and Anderson 1993). Curiously, complete knockout of smg-6 appears to be lethal, though the reasons for this are currently unknown (Cali and Anderson 1998). One possibility is that SMG-6 has essential functions outside of NMD; at least some SMG proteins have additional functions outside the NMD pathway [e.g., SMG-2/Upf1 in Staufen-mediated decay (reviewed in Kim and Maquat 2019)]. Additionally, there may be other genes with a smg phenotype (Longman et al. 2007, 2013; Yamashita et al. 2009; Hug and Caceres 2014; Melero et al. 2016), but the role of at least some of these factors in NMD remains controversial (Rosains and Mango 2012).
The SMG proteins include several conserved domains that are known to act in RNA metabolism and in protein–protein interactions, suggesting SMGs act directly on RNA and interact with one another. Targeted studies of particular SMG proteins have provided glimpses of the NMD pathway in C. elegans. SMG-1 has a PIK3 kinase domain that is required for NMD, possibly via phosphorylation of SMG-2 (Page et al. 1999; Grimson et al. 2004). SMG-5/7 interact with SMG-2 and are required for its efficient dephosphorylation (Anders et al. 2003). Phospho-SMG-2 is bound preferentially to NMD targets (Johns et al. 2007; Muir et al. 2018), though it is unclear how this binding relates to other steps in the pathway (coming before/after NMD target identification) (Grimson et al. 2004). In humans, the phosphorylation state of SMG-2 changes during NMD and affects interactions with other SMG proteins and translational components, and it has been proposed that phospho-SMG-2 may serve as a binding site for mRNA decay effectors (Ohnishi et al. 2003; Kashima et al. 2006; Okada-Katsuhata et al. 2012). For more information on SMG proteins, their domains, and their functions, see Karousis et al. (2016).
Eventually, the SMG proteins bring about the demise of an NMD target mRNA by stimulating RNA decay. There are several possible mRNA decay pathways, with many of them reported as active on NMD targets in diverse eukaryotes including 3′>5′ exonucleolytic decay, decapping and 5′>3′ exonucleolytic decay, and endonucleolytic cleavage (e.g., Muhlrad and Parker 1994; Mitchell and Tollervey 2003; Gatfield and Izaurralde 2004). One technical hurdle to the study of NMD target decay mechanisms is the essential nature of much of the decay machinery: the exonucleolytic machinery implicated in NMD target removal is essential for mRNA decay generally, and inactivating mutations are largely lethal across metazoans, confounding experimental interpretation.
There is at least one RNA decay factor that is clearly genetically required for NMD target degradation: SMG-6. Fly and mammalian SMG-6 proteins contain a PIN endonuclease domain with catalytic activity required for NMD, and it has been suggested that SMG-6 directly cleaves NMD targets in those systems (Gatfield and Izaurralde 2004; Glavan et al. 2006; Huntzinger et al. 2008; Eberle et al. 2009). The SMG-6 protein of C. elegans also contains a PIN domain, and catalytic residues of C. elegans SMG-6 are also required for NMD (J.A., unpublished data). Cleavage activity of SMG-6 is thus a central conserved feature of metazoan NMD, and it is possible this is an initiating event in degradation of NMD targets. We note that this is in contrast to S. cerevisiae, where the dominant model has long been that NMD triggers decapping followed by 5′>3′ degradation of mRNA targets (Muhlrad and Parker 1994). However, other work identified a homolog of SMG-6 in S. cerevisiae (Nmd4) that acts during NMD of at least some transcripts (He and Jacobson 1995; Dehecq et al. 2018). After cleavage, NMD target mRNA fragments are further digested by exonucleases. As part of this in C. elegans and flies, the RNA helicase SKI and 3′>5′exosome are thought to degrade the mRNA 3′>5′, and, with the help of the ribosome rescue factor PELO-1, dissociate any upstream straggling ribosomes (see section of endogenous roles for Nonstop/No-Go; Hashimoto et al. 2017; Arribere and Fire 2018).
Endogenous roles for NMD
Soon after its discovery, the idea emerged that NMD is a quality control pathway to prevent the accumulation of deleterious, and potentially dominantly acting, truncated proteins. Consistent with this idea, NMD changes the mode of inheritance of some premature stop codon-containing alleles from dominant to recessive. For example, in an NMD-deficient background (smg-(−)), certain nonsense mutations in unc-54 are dominant over wild-type unc-54, whereas, in an NMD-competent background (smg-(+)), the same unc-54 mutations are recessive (Cali and Anderson 1998). The effect depends on the position of the premature stop codon in relation to the domains of the UNC-54 protein, as would be expected if the mechanism depended on the nature of the encoded protein. An explanation for these observations is that alleles encoding truncated proteins can be dominant negatives (antimorphs) and are normally kept in check by NMD-dependent destabilization of their mRNA. This model has proven useful to understand the mode of inheritance of certain disease-causing premature stop codon-containing alleles in humans (Miller and Pearce 2014).
A major source of NMD targets in wild-type organisms is pseudogenes. Pseudogenes are generally thought to be nonfunctional and imperfect copies of normal genes, and accumulate inactivating mutations and rearrangements through genetic drift. Many such mutations in pseudogenes shift the reading frame or introduce stop codons, making the pseudogene an NMD target. There are ∼1600 annotated pseudogenes in C. elegans (Yates et al. 2016), with at least 131 pseudogene mRNAs destabilized in a smg-dependent manner (Mitrovich and Anderson 2005; Muir et al. 2018). Thus, a major function of NMD is to buffer cells from pseudogene-encoded products that might otherwise interfere with the functions of normal genes.
In addition to truncating mutations and pseudogenes, NMD affects expression of mRNAs from ∼20% of genes in C. elegans (Ramani et al. 2009). The fraction of genes with mRNAs affected by NMD is similar in other eukaryotes (He et al. 2003; Mendell et al. 2004). A major challenge in analyzing gene expression differences in NMD-deficient strains is identifying what mRNA changes are the direct result of NMD vs. secondary effects. A better understanding of the NMD pathway will facilitate the identification of direct NMD targets from indirect effects, with recent work leveraging an evolving understanding of SMG-2 binding and RNA decay processes toward this end (Arribere and Fire 2018; Muir et al. 2018).
The consensus from both genome-wide and individual gene studies is that NMD targets include many endogenous mRNAs that have an upstream ORF (uORF), a long 3′ UTR, one or more 3′ UTR-contained introns, or a splicing event that introduces a premature stop codon (Ramani et al. 2009). In such cases, NMD can serve a regulatory role. For example, mRNA expressed from gna-2 contains two uORFs that elicit NMD (Lee and Schedl 2004). gna-2 mRNA is also a target of the RNA binding protein GLD-1, which can act as a translational repressor. Upon GLD-1 binding, translation of the gna-2 mRNA is blocked, NMD is prevented, and the mRNA is thus stabilized. In this example, NMD enables GLD-1-dependent mRNA stabilization of gna-2 mRNA.
An important class of endogenous NMD targets are mRNAs with premature stop codons introduced by splicing. Such transcripts can arise from errors in splicing, and the ability to knockout the NMD pathway has led to a more complete picture of these events (Barberan-Soler et al. 2009; Tourasse et al. 2017). Spliced transcripts with premature stop codons can also be programmed, intentional, alternative splicing events. RBPs such as some ribosomal proteins and mRNA splicing factors can regulate the splicing of a premature stop-codon-containing exon in their own transcripts as a means to autoregulate functional protein levels (Morrison et al. 1997; Mitrovich and Anderson 2000; Takei et al. 2016). The model of alternative splicing coupled to NMD has proven mutually beneficial to understand the regulation, substrates, and functions of both splicing and NMD in C. elegans and throughout metazoans (Lareau et al. 2007; Barberan-Soler et al. 2009; Hansen et al. 2009; Tourasse et al. 2017).
Organisms have co-opted NMD to regulate several endogenous mRNAs, involving NMD in many biological functions. While some examples are understood in molecular detail (e.g., Lee and Schedl 2004), there are several known functions of NMD with an incompletely understood molecular basis. For example, smg mutants exhibit a protruding vulva (Pvl) phenotype, often associated with cellular proliferation defects, though the basis for this is unknown (Hodgkin et al. 1989). Work has shown that NMD is influenced by, or required for, normal aging and insulin signaling (Son et al. 2017). At least some SMG proteins also act in pathways other than NMD [SMG-2/UPF1 in staufen-mediated mRNA decay (Kim and Maquat 2019)]. We expect that the experimental tractability of C. elegans in general, and its NMD system in particular, coupled with its powerful research tools and a rich literature, will continue to enable insight on the intersection of NMD and organism function and physiology.
No-Go/Nonstop mRNA decay
While NMD is the most well-studied translational surveillance pathway, it is not the only one, and here we will discuss two additional pathways: Nonstop and No-Go Decay (Figure 9, C and D). Because of the close relationship between No-Go and Nonstop Decay, we will discuss them together. To date, C. elegans has sat on the sidelines while much of the discovery and characterization of No-Go and Nonstop Decay pathways has taken place in yeast and mammalian systems. We will describe the current models of No-Go and Nonstop Decay developed from these systems, and mention what is known about C. elegans in particular at the end. There has been a flood of molecular detail of No-Go/Nonstop discovered even since relatively recent reviews (Joazeiro 2017).
Nonstop Decay refers to the mRNA and protein loss that results from ribosomal stalling at the 3′ end of an mRNA (Frischmeyer et al. 2002). No-Go Decay refers to the mRNA and protein loss that results from ribosomal stalling during translation elongation, e.g., from a stretch of rare codons, polybasic amino acids, or structured RNAs (Doma and Parker 2006). No-Go and Nonstop Decay overlap substantially. For example, translation of the poly(A) tail during Nonstop Decay gives rise to polylysine, which can stall ribosomes and elicit No-Go Decay (Ito-Harashima et al. 2007). Both No-Go and Nonstop Decay elicit mRNA degradation as well as ubiquitination and degradation of the nascent protein chain (Dimitrova et al. 2009; Bengtson and Joazeiro 2010). A commonality between No-Go and Nonstop Decay is a failure of the ribosome to leave the mRNA in a timely fashion. In such cases, the ribosome does not fall off but remains bound at the stall site, even if the stall is simply a free 3′ end. The stalled ribosome::mRNA complex marks the mRNA, and recruits cellular machinery to the site to degrade the mRNA and nascent peptide.
How does a cell distinguish a terminally stalled ribosome from a ribosome that has transiently paused? One idea that has recently emerged from S. cerevisiae is that it is not a stalled ribosome that is recognized but rather a pile-up of ribosomes on an mRNA (Simms et al. 2017). As translation can occur on polyribosomes (multiple ribosomes bound on a single mRNA), if one ribosome stalls, the ribosomes that lie upstream will collide with it, forming a ribosomal traffic jam. Recent work supports the idea that the interface between collided ribosomes may recruit cellular decay machinery, with Cryo-EM structures of collided diribosomes highlighting a conserved interface between the small ribosomal subunits (Juszkiewicz and Hegde 2018; Ikeuchi et al. 2019).
After recognition of ribosomal stalling, three things take place (the temporal relationship of the first two processes is still unclear):
(1) The mRNA is degraded. Both No-Go and Nonstop are thought to involve endonucleolytic cleavage in the vicinity of stalled ribosomes, as well as 3′>5′ exonucleolytic decay facilitated by the SKI RNA helicase and the 3′>5′ exosome (van Hoof et al. 2002; Doma and Parker 2006; Tsuboi et al. 2012; D’Orazio et al. 2019).
(2) The ribosome is rescued. A specialized ribosome rescue factor (Dom34/Pelota with Hbs1) recognizes the stalled ribosome and recruits ABCE-1 for ribosomal splitting (Shoemaker et al. 2010; Pisareva et al. 2011; Young et al. 2015).
(3) The nascent peptide chain is degraded in a process deemed “Ribosome Quality Control.” After ribosomal subunit splitting, the peptidyl-tRNA remains bound to the 60S. Factors elongate the nascent chain with alanine and threonine, ubiquitinate the nascent peptide, and eventually degrade the protein (Bengtson and Joazeiro 2010; Shao and Hegde 2014; Shen et al. 2015; Kostova et al. 2017).
An initial study in C. elegans demonstrated that Nonstop Decay exists in this organism, though the machinery that executes repression was not able to be identified (Parvaz and Anderson 2007). At least one reason for this is that, in C. elegans, translation of many 3′ UTRs is sufficient to repress protein expression through a poorly understood mechanism independent of No-Go and Nonstop Decay (Arribere et al. 2016). This added layer of repression may have caused early screens for Nonstop Decay factors to fail (Parvaz and Anderson 2007). An appreciation of the repressive effect of translation of the 3′ UTR allowed subsequent work to remove the effect, leading to identification of functional homologs of the SKI RNA helicase (skih-2 and ttc-37) as well as the pelota ribosome rescue factor (pelo-1) (Arribere and Fire 2018). Knockout of skih-2 stabilizes Nonstop mRNA reporter levels, and knockout of pelo-1 leads to an accumulation of stalled ribosomes on the same Nonstop mRNA reporter. It thus appears that C. elegans contains a Nonstop mRNA Decay pathway mechanistically similar to that in yeast and mammals. Recent work also suggests C. elegans has a No-Go mRNA Decay pathway, and has identified a putative endoribonuclease (nonu-1) acting in both Nonstop and No-Go mRNA Decay (Glover et al. 2020), similar to work in S. cerevisiae (D’Orazio et al. 2019). There have been no reports to date on the fate of nascent peptides during No-Go/Nonstop Decay in C. elegans, though C. elegans contains homologs of factors known to act in these pathways in yeast and mammals.
Endogenous roles for No-Go/Nonstop Decay
As with NMD, cells have co-opted No-Go/Nonstop Decay to repress and regulate endogenous mRNAs. Because at least some mutants of Nonstop Decay factors are viable in C. elegans, translation can be examined in their absence and used to define endogenous Nonstop Decay targets (as is done with smg mutants to define targets of that pathway). While NMD targets can be examined by looking for changes in RNA-seq between smg and wild-type strains, with No-Go/Nonstop Decay targets, a modified ribosome footprint profiling technique is used to capture the ribosomes that stall in the absence of skih-2 and pelo-1 (Ingolia et al. 2009; Guydosh and Green 2014; Arribere and Fire 2018). This approach has revealed that endogenous Nonstop Decay targets include mRNAs that are endonucleolytically cleaved, for example the mRNA xbp-1/hac1 in C. elegans/S. cerevisiae that regulates the unfolded protein response (Guydosh and Green 2014; Arribere and Fire 2018). mRNAs undergoing RNAi would also be expected to experience mRNA cleavage within their ORF, and recent work has shown these mRNAs are also targeted by Nonstop Decay in C. elegans (Pule et al. 2019), consistent with earlier findings in flies and plants (Orban and Izaurralde 2005; Hashimoto et al. 2017; Szádeczky-Kardoss et al. 2018). These studies illustrate that one function of Nonstop Decay is to efficiently dismantle ribosomes on an mRNA undergoing cotranslational degradation.
One function of the Nonstop Decay pathway in C. elegans and Drosophila melanogaster is to clear the degradation products of NMD (Hashimoto et al. 2017; Arribere and Fire 2018). In animals deficient for Nonstop Decay factors (lacking the RNA helicase skih-2 and the ribosome rescue factor pelo-1), ribosomes accumulate at premature stop codons. The link between NMD and Nonstop Decay occurs after a committed step of mRNA decay in NMD: mutations in Nonstop factors do not appreciably derepress steady state levels of NMD targets, and smg mutations are epistatic to ribosomal stalling at premature stop codons (Arribere and Fire 2018). This work suggests that NMD funnels into Nonstop Decay, an axis that may inform an understanding of the NMD pathway as well as functions of Nonstop Decay. The relationship between NMD and Nonstop Decay is conserved between C. elegans and D. melanogaster (Hashimoto et al. 2017).
The picture attained so far shows important roles for Nonstop Decay in C. elegans. However, the study of endogenous functions and targets for No-Go/Nonstop Decay has lagged behind similar studies of NMD targets. There are a number of technical issues that have stymied the study of endogenous No-Go/Nonstop functions, including:
Nonstop Decay targets have been systematically ignored by annotation software. Virtually all gene annotation programs require protein coding genes to have start and stop codons. Nonstop mRNA targets would violate this rule as they lack a stop codon. Historically, techniques to annotate polyadenylation sites suffered from high false positive rates inside coding regions [due to internal priming artifacts (Nam et al. 2002)], leading to skepticism about the existence of bona fide premature polyadenylation. However, other work has shown that Nonstop Decay targets can arise from premature polyadenylation in a coding region, generating stop-codon-less transcripts (Sparks and Dieckmann 1998; Ozsolak et al. 2010). The absence of a stop codon would cause Nonstop Decay targets to be either discarded or annotated as nonprotein-coding. An appreciation of this effect has led to manual annotation of Nonstop Decay mRNA targets in humans, but such efforts have not yet occurred in C. elegans.
There is still a relatively poor understanding of which endogenous mRNAs elicit No-Go Decay. This is in part because most No-Go Decay reporters include translational stalls, which are hyperbolic examples of what is observed in the genome. For example, No-Go Decay reporters in yeast and mammals often include 12 consecutive rare codons encoding basic amino acids, though such runs are rare in normal proteins. As No-Go Decay is not yet well studied in C. elegans, it remains unclear what structures or sequences stall ribosomes in a reporter context, let alone endogenous mRNAs. There are a few endogenous genes known to trigger No-Go Decay in S. cerevisiae (e.g., Brandman et al. 2012), and we expect that as an understanding of No-Go Decay evolves so too will the understanding of how No-Go acts on endogenous mRNAs.
Currently the experimental identification of endogenous No-Go/Nonstop Decay targets is more difficult than the identification of NMD targets. For NMD, a smg mutation confers a large-fold increase in the levels of NMD targets, as much as 10- to 20-fold, which can be detected readily by any number of techniques (RT-qPCR, northern, RNA-seq). For No-Go/Nonstop Decay, there are multiple redundant mechanisms acting to repress mRNA levels, and loss of any one mechanism has little, if any, detectable effect on total mRNA levels for its targets. For this reason, No-Go/Nonstop Decay targets are defined by monitoring the ribosomes that stall on them. As the understanding of No-Go/Nonstop Decay improves, it may become easier to experimentally identify targets.
Once these issues are resolved, we expect there will be greater insight into the role of No-Go/Nonstop Decay in normal animal physiology and function.
Concluding Remarks and Perspective
In the above sections, we have briefly outlined what is known about mRNA editing, splicing, and quality control in C. elegans. Historically, the rich literature and physiology of C. elegans, its vibrant research community, and the genetic tractability of C. elegans has proven to be powerful assets toward attaining insight into mRNA processing and regulation in vivo. We expect this trend to continue. We now have a greater appreciation for the complexity of mRNA processing and quality control, and expect that this view will facilitate deeper insight into the individual pathways of mRNA metabolism, as well as their combined effects on all mRNAs in the cell. Moving forward, the development and application of novel enrichment and high throughput techniques will be an exciting area with new discoveries. The genetic tractability, well-annotated cell types, and rich physiology of C. elegans will provide an important setting in which to apply such techniques, and to rigorously explore the role of mRNA metabolism in the normal health and life of an organism.
Acknowledgments
Work in the laboratory of J.A.A. is supported by the National Institutes of Health (NIH) (R01GM131012), the Searle Scholars Foundation, and startup funds from the University of California, Santa Cruz. Work in the laboratory of H.K. is supported by KAKENHI from Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) or the Japan Society for the Promotion of Science (JSPS) [grant numbers JP20112004, JP25118506, JP26291003, JP15H01350, JP15H01467, JP15KK0252, JP17H03633, JP17H05596, JP20H03181, JP221S0002 and JP16H06279 (PAGS)], Precursory Research for Embryonic Science and Technology (PRESTO) from Japan Science and Technology Agency (JST) and Nanken-Kyoten from Tokyo Medical and Dental University (TMDU). Work in the laboratory of H.A.H. is supported by the American Cancer Society (RSG-15-051 RMC), the National Science Foundation (Award Number 1917050) and the NIH (R01GM130759).
Footnotes
Communicating editor: J. Kim
Literature Cited
- Allen M. A., Hillier L. W., Waterston R. H., and Blumenthal T., 2011. A global analysis of C. elegans trans-splicing. Genome Res. 21: 255–264. 10.1101/gr.113811.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrane S., Rebora K., Zniber I., Dupuy D., and Mackereth C. D., 2014. Backbone-independent nucleic acid binding by splicing factor SUP-12 reveals key aspects of molecular recognition. Nat. Commun. 5: 4595 10.1038/ncomms5595 [DOI] [PubMed] [Google Scholar]
- Amrani N., Ganesan R., Kervestin S., Mangus D. A., Ghosh S. et al. , 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]
- Anders K. R., Grimson A., and Anderson P., 2003. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 22: 641–650. 10.1093/emboj/cdg056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyanful A., Sakube Y., Takuwa K., and Kagawa H., 2001. The third and fourth tropomyosin isoforms of Caenorhabditis elegans are expressed in the pharynx and intestines and are essential for development and morphology. J. Mol. Biol. 313: 525–537. 10.1006/jmbi.2001.5052 [DOI] [PubMed] [Google Scholar]
- Arribere J. A., and Fire A. Z., 2018. Nonsense mRNA suppression via nonstop decay. eLife 7: pii: e33292. 10.7554/eLife.33292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Cenik E. S., Jain N., Hess G. T., Lee C. H. et al. , 2016. Translation readthrough mitigation. Nature 534: 719–723. 10.1038/nature18308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgousti D. C., Palani S., Sherman Y., and Grishok A., 2012. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 31: 3821–3832. 10.1038/emboj.2012.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R., Yan C., Wan R., Lei J. and Shi Y., 2017. Structure of the post-catalytic spliceosome from Saccharomyces cerevisiae. Cell 171: 1589–1598.e8. 10.1016/j.cell.2017.10.038 [DOI] [PubMed] [Google Scholar]
- Bai R., Wan R., Yan C., Lei J., and Shi Y., 2018. Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. Science 360: 1423–1429. 10.1126/science.aau0325 [DOI] [PubMed] [Google Scholar]
- Balistreri G., Horvath P., Schweingruber C., Zund D., McInerney G. et al. , 2014. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe 16: 403–411. 10.1016/j.chom.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Barberan-Soler S., Lambert N. J., and Zahler A. M., 2009. Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans. RNA 15: 1652–1660. 10.1261/rna.1711109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S., Medina P., Estella J., Williams J., and Zahler A. M., 2011. Co-regulation of alternative splicing by diverse splicing factors in Caenorhabditis elegans. Nucleic Acids Res. 39: 666–674. 10.1093/nar/gkq767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D. E., Watabe E., Ono K., Kwak E., Kuroyanagi H. et al. , 2018. Tropomyosin isoforms differentially affect muscle contractility in the head and body regions of the nematode Caenorhabditis elegans. Mol. Biol. Cell 29: 1075–1088. 10.1091/mbc.E17-03-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B., Hundley H., Li J. B., Peng Z., Pickrell J. et al. , 2012. The difficult calls in RNA editing. Interviewed by H Craig Mak. Nat. Biotechnol. 30: 1207–1209. 10.1038/nbt.2452 [DOI] [PubMed] [Google Scholar]
- Bass B. L., 2006. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb. Symp. Quant. Biol. 71: 285–292. 10.1101/sqb.2006.71.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L., and Weintraub H., 1987. A developmentally regulated activity that unwinds RNA duplexes. Cell 48: 607–613. 10.1016/0092-8674(87)90239-X [DOI] [PubMed] [Google Scholar]
- Bass B. L., and Weintraub H., 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55: 1089–1098. 10.1016/0092-8674(88)90253-X [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Gatfield D., Rehwinkel J., Hilgers V., and Izaurralde E., 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 26: 1591–1601. 10.1038/sj.emboj.7601588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson M. H., and Joazeiro C. A., 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467: 470–473. 10.1038/nature09371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17: 251–256. 10.1016/j.ceb.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Bertram K., Agafonov D. E., Dybkov O., Haselbach D., Leelaram M. N. et al. , 2017a Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170: 701–713.e11. 10.1016/j.cell.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Bertram K., Agafonov D. E., Liu W. T., Dybkov O., Will C. L. et al. , 2017b Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. 10.1038/nature21079 [DOI] [PubMed] [Google Scholar]
- Billi A. C., Fischer S. E., and Kim J. K., 2014. Endogenous RNAi pathways in C. elegans (May 7, 2014), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.170.1, http://www.wormbook.org. 10.1895/wormbook.1.170.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie S. M., Babb C., Wilky H., Rawls A., Park J. G. et al. , 2015. Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol. 13: 4 10.1186/s12915-015-0116-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie S. M., Geissel H. C., Wilky H., Joshi R., Newbern J. et al. , 2017. Alternative polyadenylation directs tissue-specific miRNA targeting in Caenorhabditis elegans somatic tissues. Genetics 206: 757–774. 10.1534/genetics.116.196774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., 2012. Trans-splicing and operons in C. elegans (November 20, 2012), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.5.2, http://www.wormbook.org. [Google Scholar]
- Blumenthal T., and Steward K., 1997. RNA processing and gene structure in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., and Priess J. R.. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Blumenthal T., Evans D., Link C. D., Guffanti A., Lawson D. et al. , 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854. 10.1038/nature00831 [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Davis P., and Garrido-Lecca A., 2015. Operon and non-operon gene clusters in the C. elegans genome (April 28, 2015), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.175.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack K. E., Hobartner C., and Bohnsack M. T., 2019. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel) 10: pii: E102. 10.3390/genes10020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz P. L., Stoilov P., Li Q., Lin C. H., Chawla G. et al. , 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21: 1636–1652. 10.1101/gad.1558107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C. et al. , 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151: 1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese M., Esmaeili B., Fraboulet S., Burt E. C., Christodoulou S. et al. , 2009. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet. 18: 97–104. 10.1093/hmg/ddn320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S., McLeod T., and Petric M., 2016. The meaning of NMD: translate or perish. Trends Genet. 32: 395–407. 10.1016/j.tig.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Brown K. M., and Gilmartin G. M., 2003. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell 12: 1467–1476. 10.1016/S1097-2765(03)00453-2 [DOI] [PubMed] [Google Scholar]
- Burset M., Seledtsov I. A., and Solovyev V. V., 2001. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 29: 255–259. 10.1093/nar/29.1.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali B. M., and Anderson P., 1998. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet. 260: 176–184. 10.1007/s004380050883 [DOI] [PubMed] [Google Scholar]
- Cali B. M., Kuchma S. L., Latham J., and Anderson P., 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell Z. T., Bhimsaria D., Valley C. T., Rodriguez-Martinez J. A., Menichelli E. et al. , 2012. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 1: 570–581. 10.1016/j.celrep.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Packer J. S., Ramani V., Cusanovich D. A., Huynh C. et al. , 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357: 661–667. 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey K. T., and Wickramasinghe V. O., 2018. Regulatory potential of the RNA processing machinery: implications for human disease. Trends Genet. 34: 279–290. 10.1016/j.tig.2017.12.012 [DOI] [PubMed] [Google Scholar]
- Casañal A., Kumar A., Hill C. H., Easter A. D., Emsley P. et al. , 2017. Architecture of eukaryotic mRNA 3′-end processing machinery. Science 358: 1056–1059. 10.1126/science.aao6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella G., Tsitsipatis D., Abdelmohsen K., and Gorospe M., 2019. mRNA methylation in cell senescence. Wiley Interdiscip. Rev. RNA 10: e1547 10.1002/wrna.1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S., Choi E. A., and Shi Y., 2011. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip. Rev. RNA 2: 321–335. 10.1002/wrna.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Lim J., Ha M., and Kim V. N., 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53: 1044–1052. 10.1016/j.molcel.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Chang J. C., and Kan Y. W., 1979. beta 0 thalassemia, a nonsense mutation in man. Proc. Natl. Acad. Sci. USA 76: 2886–2889. 10.1073/pnas.76.6.2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Fogel-Petrovic M., and Maquat L. E., 1990. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol. 10: 5215–5225. 10.1128/MCB.10.10.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu Z., Zhou B., Wei C., Zhou Y. et al. , 2016. CELF RNA binding proteins promote axon regeneration in C. elegans and mammals through alternative splicing of Syntaxins. eLife 5: pii: e16072. 10.7554/eLife.16072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhou Y., Qi Y. B., Khivansara V., Li H. et al. , 2015. Context-dependent modulation of Pol II CTD phosphatase SSUP-72 regulates alternative polyadenylation in neuronal development. Genes Dev. 29: 2377–2390. 10.1101/gad.266650.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C., Ronai D., Than M. T., Walker C. J., Sewell A. K. et al. , 2016. Nucleotide levels regulate germline proliferation through modulating GLP-1/Notch signaling in C. elegans. Genes Dev. 30: 307–320. 10.1101/gad.275107.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Faini M., Muckenfuss L. M., Aebersold R., and Jinek M., 2018. Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nat. Struct. Mol. Biol. 25: 135–138 (erratum: Nat. Struct. Mol. Biol. 25: 355). 10.1038/s41594-017-0020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés-López M., Gruner M. R., Cooper D. A., Gruner H. N., Voda A. I. et al. , 2018. Global accumulation of circRNAs during aging in Caenorhabditis elegans. BMC Genomics 19: 8 10.1186/s12864-017-4386-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covelo-Molares H., Bartosovic M., and Vanacova S., 2018. RNA methylation in nuclear pre-mRNA processing. Wiley Interdiscip. Rev. RNA 9: e1489 10.1002/wrna.1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E., 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. G., Zhen M., Jin Y., and Bargmann C. I., 2001. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron 29: 115–129. 10.1016/S0896-6273(01)00184-2 [DOI] [PubMed] [Google Scholar]
- Cui M., Allen M. A., Larsen A., Macmorris M., Han M. et al. , 2008. Genes involved in pre-mRNA 3′-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc. Natl. Acad. Sci. USA 105: 16665–16670. 10.1073/pnas.0807104105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar I. O., and Maquat L. E., 1988. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol. Cell. Biol. 8: 802–813. 10.1128/MCB.8.2.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daguenet E., Dujardin G., and Valcarcel J., 2015. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 16: 1640–1655. 10.15252/embr.201541116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassah M., Patzek S., Hunt V. M., Medina P. E., and Zahler A. M., 2009. A genetic screen for suppressors of a mutated 5′ splice site identifies factors associated with later steps of spliceosome assembly. Genetics 182: 725–734. 10.1534/genetics.109.103473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila Lopez M., and Samuelsson T., 2008. Early evolution of histone mRNA 3′ end processing. RNA 14: 1–10. 10.1261/rna.782308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffit S. N., Yee B. A., Manning A. C., Rajendren S., Vadlamani P. et al. , 2017. The C. elegans neural editome reveals an ADAR target mRNA required for proper chemotaxis. eLife 6: pii: e28625. 10.7554/eLife.28625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehecq M., Decourty L., Namane A., Proux C., Kanaan J. et al. , 2018. Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes. EMBO J. 37: pii: e99278. 10.15252/embj.201899278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Karbstein K., and Woolford J. L. Jr, 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 84: 93–129. 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezi V., Ivanov C., Haussmann I. U., and Soller M., 2016. Nucleotide modifications in messenger RNA and their role in development and disease. Biochem. Soc. Trans. 44: 1385–1393 [corrigenda: Biochem. Soc. Trans. 47: 957 (2019)]. 10.1042/BST20160110 [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., and Goldstein B., 2016. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202: 885–901. 10.1534/genetics.115.182162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadi M., Derdowski A., Kalloo G., Maginnis M. S., O’Hern P. et al. , 2016. Decreased function of survival motor neuron protein impairs endocytic pathways. Proc. Natl. Acad. Sci. USA 113: E4377–E4386. 10.1073/pnas.1600015113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova L. N., Kuroha K., Tatematsu T., and Inada T., 2009. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284: 10343–10352. 10.1074/jbc.M808840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty M. F., Adelmant G., Cecchetelli A. D., Marto J. A., and Cram E. J., 2014. Proteomic analysis reveals CACN-1 is a component of the spliceosome in Caenorhabditis elegans. G3 (Bethesda) 4: 1555–1564. 10.1534/g3.114.012013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K., and Parker R., 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564. 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang X. C., and Marzluff W. F., 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123: 37–48. 10.1016/j.cell.2005.08.002 [DOI] [PubMed] [Google Scholar]
- D’Orazio K. N., Wu C. C., Sinha N., Loll-Krippleber R., Brown G. W. et al. , 2019. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. eLife 8: pii: e49117.. 10.7554/eLife.49117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle A. B., Lykke-Andersen S., Muhlemann O., and Jensen T. H., 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 16: 49–55. 10.1038/nsmb.1530 [DOI] [PubMed] [Google Scholar]
- Eichhorn S. W., Subtelny A. O., Kronja I., Kwasnieski J. C., Orr-Weaver T. L. et al. , 2016. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 5: pii: e16955. 10.7554/eLife.16955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E., and Levanon E. Y., 2018. A-to-I RNA editing - immune protector and transcriptome diversifier. Nat. Rev. Genet. 19: 473–490. 10.1038/s41576-018-0006-1 [DOI] [PubMed] [Google Scholar]
- Evans D., Zorio D., MacMorris M., Winter C. E., Lea K. et al. , 1997. Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc. Natl. Acad. Sci. USA 94: 9751–9756. 10.1073/pnas.94.18.9751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer T., Roller A. B., Kent W. J., and Zahler A. M., 2002. Analysis of the role of Caenorhabditis elegans GC-AG introns in regulated splicing. Nucleic Acids Res. 30: 3360–3367. 10.1093/nar/gkf465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica S. M., Oubridge C., Galej W. P., Wilkinson M. E., Bai X. C. et al. , 2017. Structure of a spliceosome remodelled for exon ligation. Nature 542: 377–380. 10.1038/nature21078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica S. M., Oubridge C., Wilkinson M. E., Newman A. J., and Nagai K., 2019. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science 363: 710–714. 10.1126/science.aaw5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E. et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Fischer S. E., 2010. Small RNA-mediated gene silencing pathways in C. elegans. Int. J. Biochem. Cell Biol. 42: 1306–1315. 10.1016/j.biocel.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Frischmeyer P. A., van Hoof A., O’Donnell K., Guerrerio A. L., Parker R. et al. , 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261. 10.1126/science.1067338 [DOI] [PubMed] [Google Scholar]
- Frye M., Harada B. T., Behm M., and He C., 2018. RNA modifications modulate gene expression during development. Science 361: 1346–1349. 10.1126/science.aau1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej W. P., Wilkinson M. E., Fica S. M., Oubridge C., Newman A. J. et al. , 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. S., Ben-Asher N., Manning A. C., Deffit S. N., Washburn M. C. et al. , 2019. Disruption in A-to-I editing levels affects C. elegans development more than a complete lack of editing. Cell Rep. 27: 1244–1253.e4. 10.1016/j.celrep.2019.03.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Teng Y., Luo J., Huang L., Li M. et al. , 2014. The survival motor neuron gene smn-1 interacts with the U2AF large subunit gene uaf-1 to regulate Caenorhabditis elegans lifespan and motor functions. RNA Biol. 11: 1148–1160. 10.4161/rna.36100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Xu J., Chen H., Xue D., Pan W. et al. , 2019. Defective expression of mitochondrial, vacuolar H+-ATPase and histone genes in a C. elegans model of SMA. Front. Genet. 10: 410 10.3389/fgene.2019.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Garcia S., and Voinnet O., 2014. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16: 391–402. 10.1016/j.chom.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Lecca A., Saldi T., and Blumenthal T., 2016. Localization of RNAPII and 3′ end formation factor CstF subunits on C. elegans genes and operons. Transcription 7: 96–110. 10.1080/21541264.2016.1168509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D., and Izaurralde E., 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578. 10.1038/nature02559 [DOI] [PubMed] [Google Scholar]
- Getz S., Xu S., and Fire A., 1997. Termperature sensitive smg mutations as a tool to engineer conditional expression….a progress report. Worm Breed. Gaz. 14: 26. [Google Scholar]
- Glavan F., Behm-Ansmant I., Izaurralde E., and Conti E., 2006. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 25: 5117–5125. 10.1038/sj.emboj.7601377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover M. L., Burroughs A. M., Monem P. C.,Egelhofer T. A., Pule M. N. et al. , 2020. NONU-1 encodes a conserved endonuclease required for mRNA translation surveillance. Cell Rep. 30: 4321–4331.e4. 10.1016/j.celrep.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., Agranat-Tamir L., Light D., Ben-Naim Zgayer O., Fishman A. et al. , 2017. A-to-I RNA editing promotes developmental stage-specific gene and lncRNA expression. Genome Res. 27: 462–470. 10.1101/gr.211169.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm A. C., and Wickens M., 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9: 337–344. 10.1038/nrm2370 [DOI] [PubMed] [Google Scholar]
- Gott J. M., and Emeson R. B., 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34: 499–531. 10.1146/annurev.genet.34.1.499 [DOI] [PubMed] [Google Scholar]
- Gout J. F., Thomas W. K., Smith Z., Okamoto K., and Lynch M., 2013. Large-scale detection of in vivo transcription errors. Proc. Natl. Acad. Sci. USA 110: 18584–18589. 10.1073/pnas.1309843110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber J. H., Salisbury J., Hutchins L. N., and Blumenthal T., 2007. C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA 13: 1409–1426. 10.1261/rna.596707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracida X., Norris A. D., and Calarco J. A., 2016. Regulation of tissue-specific alternative splicing: C. elegans as a model system. Adv. Exp. Med. Biol. 907: 229–261. 10.1007/978-3-319-29073-7_10 [DOI] [PubMed] [Google Scholar]
- Grimson A., O’Connor S., Newman C. L., and Anderson P., 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol. Cell. Biol. 24: 7483–7490. 10.1128/MCB.24.17.7483-7490.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R., Martin G., Keller W., and Zavolan M., 2014. Means to an end: mechanisms of alternative polyadenylation of messenger RNA precursors. Wiley Interdiscip. Rev. RNA 5: 183–196. 10.1002/wrna.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh N. R., and Green R., 2014. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156: 950–962. 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. D., Lareau L. F., Blanchette M., Green R. E., Meng Q. et al. , 2009. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 5: e1000525 10.1371/journal.pgen.1000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B. et al. , 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Haselbach D., Komarov I., Agafonov D. E., Hartmuth K., Graf B. et al. , 2018. Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172: 454–464.e11. 10.1016/j.cell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Takahashi M., Sakota E., and Nakamura Y., 2017. Nonstop-mRNA decay machinery is involved in the clearance of mRNA 5′-fragments produced by RNAi and NMD in Drosophila melanogaster cells. Biochem. Biophys. Res. Commun. 484: 1–7. 10.1016/j.bbrc.2017.01.092 [DOI] [PubMed] [Google Scholar]
- Hebeisen M., Drysdale J., and Roy R., 2008. Suppressors of the cdc-25.1(gf)-associated intestinal hyperplasia reveal important maternal roles for prp-8 and a subset of splicing factors in C. elegans. RNA 14: 2618–2633. 10.1261/rna.1168408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz C., Doktor T. K., Lanjuin A., Escoubas C., Zhang Y. et al. , 2017. Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature 541: 102–106 (erratum: Nature 547: 476). 10.1038/nature20789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., and Jacobson A., 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9: 437–454. 10.1101/gad.9.4.437 [DOI] [PubMed] [Google Scholar]
- He F., Li X., Spatrick P., Casillo R., Dong S. et al. , 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452. 10.1016/S1097-2765(03)00446-5 [DOI] [PubMed] [Google Scholar]
- Helm M., and Motorin Y., 2017. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18: 275–291. 10.1038/nrg.2016.169 [DOI] [PubMed] [Google Scholar]
- Hill C. H., Boreikaite V., Kumar A., Casanal A., Kubik P. et al. , 2019. Activation of the endonuclease that defines mRNA 3′ ends requires incorporation into an 8-subunit core cleavage and polyadenylation factor complex. Mol. Cell 73: 1217–1231.e11. 10.1016/j.molcel.2018.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., and Manley J. L., 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., and Anderson P., 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg J. R., and Goff S. P., 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389. 10.1016/j.cell.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollins C., Zorio D. A., MacMorris M., and Blumenthal T., 2005. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA 11: 248–253. 10.1261/rna.7221605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., and Huang H. Y., 2015. Quality Control Pathways for Nucleus-Encoded Eukaryotic tRNA Biosynthesis and Subcellular Trafficking. Mol. Cell. Biol. 35: 2052–2058. 10.1128/MCB.00131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough R. F., Lingam A. T., and Bass B. L., 1999. Caenorhabditis elegans mRNAs that encode a protein similar to ADARs derive from an operon containing six genes. Nucleic Acids Res. 27: 3424–3432. 10.1093/nar/27.17.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Pleasance E. D., Maydan J. S., Hunt-Newbury R., O’Neil N. J. et al. , 2007. Identification and analysis of internal promoters in Caenorhabditis elegans operons. Genome Res. 17: 1478–1485. 10.1101/gr.6824707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S. M., van Delft P., Mendil L., Bachman M., Smollett K. et al. , 2015. Formation and abundance of 5-hydroxymethylcytosine in RNA. ChemBioChem 16: 752–755. 10.1002/cbic.201500013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N., and Caceres J. F., 2014. The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep. 8: 1845–1856. 10.1016/j.celrep.2014.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley H. A., and Bass B. L., 2010. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 35: 377–383. 10.1016/j.tibs.2010.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley H. A., Krauchuk A. A., and Bass B. L., 2008. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA 14: 2050–2060. 10.1261/rna.1165008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W., Hwang C., Po M. D., and Zhen M., 2007. Neuronal polarity is regulated by a direct interaction between a scaffolding protein, Neurabin, and a presynaptic SAD-1 kinase in Caenorhabditis elegans. Development 134: 237–249. 10.1242/dev.02725 [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Kashima I., Fauser M., Sauliere J., and Izaurralde E., 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617. 10.1261/rna.1386208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt J. A., Robertson A. D., and Burge C. B., 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 23: 1636–1650. 10.1101/gr.157354.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K., Tesina P., Matsuo Y., Sugiyama T., Cheng J. et al. , 2019. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38: pii: e100276. 10.15252/embj.2018100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T., Ghaemmaghami S., Newman J. R., and Weissman J. S., 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O., and Maquat L. E., 2007. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21: 1833–1856. 10.1101/gad.1566807 [DOI] [PubMed] [Google Scholar]
- Ito-Harashima S., Kuroha K., Tatematsu T., and Inada T., 2007. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21: 519–524. 10.1101/gad.1490207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A., Memczak S., Wyler E., Torti F., Porath H. T. et al. , 2015. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10: 170–177. 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Ivanov P. V., Gehring N. H., Kunz J. B., Hentze M. W., and Kulozik A. E., 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27: 736–747. 10.1038/emboj.2008.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G., and Bartel D. P., 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. 10.1038/nature09616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C. A. P., 2017. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol. 33: 343–368. 10.1146/annurev-cellbio-111315-125249 [DOI] [PubMed] [Google Scholar]
- Johns L., Grimson A., Kuchma S. L., Newman C. L., and Anderson P., 2007. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol. Cell. Biol. 27: 5630–5638. 10.1128/MCB.00410-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S., and Hegde R. S., 2018. Quality control of orphaned proteins. Mol. Cell 71: 443–457. 10.1016/j.molcel.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat J. L., Barberan-Soler S., McKenna P., Clawson H., Farrer T. et al. , 2006. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput. Biol. 2: e86 10.1371/journal.pcbi.0020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H., Sugimoto K., Matsumoto H., Inoue T., Imadzu H. et al. , 1995. Genome structure, mapping and expression of the tropomyosin gene tmy-1 of Caenorhabditis elegans. J. Mol. Biol. 251: 603–613. 10.1006/jmbi.1995.0459 [DOI] [PubMed] [Google Scholar]
- Kaletsky R., Lakhina V., Arey R., Williams A., Landis J. et al. , 2016. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 529: 92–96. 10.1038/nature16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R., Yao V., Williams A., Runnels A. M., Tadych A. et al. , 2018. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14: e1007559 10.1371/journal.pgen.1007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karousis E. D., Nasif S., and Muhlemann O., 2016. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA 7: 661–682. 10.1002/wrna.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R. et al. , 2006. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20: 355–367. 10.1101/gad.1389006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi P., Zanetti S., Passannante M., Saudan Z., Muller F. et al. , 2010. The C. elegans sex determination protein MOG-3 functions in meiosis and binds to the CSL co-repressor CIR-1. Dev. Biol. 344: 593–602. 10.1016/j.ydbio.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Keall R., Whitelaw S., Pettitt J., and Muller B., 2007. Histone gene expression and histone mRNA 3′ end structure in Caenorhabditis elegans. BMC Mol. Biol. 8: 51 10.1186/1471-2199-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L., Khan A., Vukic D., and O’Connell M., 2017. ADAR RNA editing below the backbone. RNA 23: 1317–1328. 10.1261/rna.060921.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K. M., Lanier J., Du M., Salas-Marco J., Gao L. et al. , 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703. 10.1261/rna.5147804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerins J. A., Hanazawa M., Dorsett M., and Schedl T., 2010. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation vs. meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev. Dyn. 239: 1555–1572. 10.1002/dvdy.22274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Ahn S. H., Krogan N. J., Greenblatt J. F., and Buratowski S., 2004a Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23: 354–364. 10.1038/sj.emboj.7600053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Hung W., Narbonne P., Roy R., and Zhen M., 2010a C. elegans STRADalpha and SAD cooperatively regulate neuronal polarity and synaptic organization. Development 137: 93–102. 10.1242/dev.041459 [DOI] [PubMed] [Google Scholar]
- Kim J. S., Hung W., and Zhen M., 2010b The long and the short of SAD-1 kinase. Commun. Integr. Biol. 3: 251–255. 10.4161/cib.3.3.11455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N. J., Vasiljeva L., Rando O. J., Nedea E. et al. , 2004b The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [corrigenda: Nature 433: 661 (2005)]. 10.1038/nature03041 [DOI] [PubMed] [Google Scholar]
- Kim U., Wang Y., Sanford T., Zeng Y., and Nishikura K., 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. USA 91: 11457–11461. 10.1073/pnas.91.24.11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., and Maquat L. E., 2019. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25: 407–422. 10.1261/rna.070136.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S. W., and Bass B. L., 2002. The role of RNA editing by ADARs in RNAi. Mol. Cell 10: 809–817. 10.1016/S1097-2765(02)00649-4 [DOI] [PubMed] [Google Scholar]
- Kodama Y., Rothman J. H., Sugimoto A., and Yamamoto M., 2002. The stem-loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development 129: 187–196. [DOI] [PubMed] [Google Scholar]
- Kolev N. G., and Steitz J. A., 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19: 2583–2592. 10.1101/gad.1371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N., and Arakawa K., 2019. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 61: 316–326. 10.1111/dgd.12608 [DOI] [PubMed] [Google Scholar]
- Kostova K. K., Hickey K. L., Osuna B. A., Hussmann J. A., Frost A. et al. , 2017. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357: 414–417. 10.1126/science.aam7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., and Hirsh D., 1987. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell 49: 753–761. 10.1016/0092-8674(87)90613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi W. S., Core L. J., Waters C. T., Lis J. T., and Meyer B. J., 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2: e00808 10.7554/eLife.00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H., Uesugi H., Kohara Y., and Iino Y., 2005. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 6: R17 10.1186/gb-2005-6-2-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Li W., Hoque M., Popp M. W., Ermolenko D. N. et al. , 2014. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 28: 1900–1916. 10.1101/gad.245506.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., 2013. Switch-like regulation of tissue-specific alternative pre-mRNA processing patterns revealed by customized fluorescence reporters. Worm 2: e23834 10.4161/worm.23834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., Kobayashi T., Mitani S., and Hagiwara M., 2006. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat. Methods 3: 909–915. 10.1038/nmeth944 [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H., Ohno G., Mitani S., and Hagiwara M., 2007. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol. Cell. Biol. 27: 8612–8621. 10.1128/MCB.01508-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., Ohno G., Sakane H., Maruoka H., and Hagiwara M., 2010. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat. Protoc. 5: 1495–1517. 10.1038/nprot.2010.107 [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H., Watanabe Y., and Hagiwara M., 2013a CELF family RNA-binding protein UNC-75 regulates two sets of mutually exclusive exons of the unc-32 gene in neuron-specific manners in Caenorhabditis elegans. PLoS Genet. 9: e1003337 10.1371/journal.pgen.1003337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., Watanabe Y., Suzuki Y., and Hagiwara M., 2013b Position-dependent and neuron-specific splicing regulation by the CELF family RNA-binding protein UNC-75 in Caenorhabditis elegans. Nucleic Acids Res. 41: 4015–4025. 10.1093/nar/gkt097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., Takei S., and Suzuki Y., 2014. Comprehensive analysis of mutually exclusive alternative splicing in C. elegans. Worm 3: e28459 10.4161/worm.28459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwasako K., Takahashi M., Unzai S., Tsuda K., Yoshikawa S. et al. , 2014. RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing. Nat. Struct. Mol. Biol. 21: 778–786. 10.1038/nsmb.2870 [DOI] [PubMed] [Google Scholar]
- Lall S., Friedman C. C., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E. et al. , 2004. Contribution of trans-splicing, 5′ -leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J. Biol. Chem. 279: 45573–45585. 10.1074/jbc.M407475200 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C. et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lareau L. F., Brooks A. N., Soergel D. A., Meng Q., and Brenner S. E., 2007. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 623: 190–211. http://compbio.berkeley.edu/people/brenner/pubs/lareau-2007-eurekah-nmd.pdf [DOI] [PubMed] [Google Scholar]
- Lasda E. L., and Blumenthal T., 2011. Trans-splicing. Wiley Interdiscip. Rev. RNA 2: 417–434. 10.1002/wrna.71 [DOI] [PubMed] [Google Scholar]
- Lasda E. L., Allen M. A., and Blumenthal T., 2010. Polycistronic pre-mRNA processing in vitro: snRNP and pre-mRNA role reversal in trans-splicing. Genes Dev. 24: 1645–1658. 10.1101/gad.1940010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., and Schedl T., 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18: 1047–1059. 10.1101/gad.1188404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde E., Maquat L. E., and Moore M. J., 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860–6869. 10.1093/emboj/19.24.6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., and Wilusz J. E., 2014. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28: 2233–2247. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Lee M., Son A., Chang H., and Kim V. N., 2016. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 30: 1671–1682. 10.1101/gad.284802.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima S. A., Chipman L. B., Nicholson A. L., Chen Y. H., Yee B. A. et al. , 2017. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 24: 1057–1063. 10.1038/nsmb.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Li X., Zhang L., Jiang J., Hill R. C. et al. , 2017. Structure of the yeast spliceosomal postcatalytic P complex. Science 358: 1278–1283. 10.1126/science.aar3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loepold L., and Ahmed S., 2014. Surveillance and Targeting of Aberrant Transcripts in Caenorhabditis elegans. UNC, Chapel Hill, NY. [Google Scholar]
- Longman D., Plasterk R. H., Johnstone I. L., and Caceres J. F., 2007. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 21: 1075–1085. 10.1101/gad.417707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Hug N., Keith M., Anastasaki C., Patton E. E. et al. , 2013. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res. 41: 8319–8331. 10.1093/nar/gkt585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., and Lacroute F., 1979. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. USA 76: 5134–5137. 10.1073/pnas.76.10.5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist E. A., Herman R. K., Rogalski T. M., Mullen G. P., Moerman D. G. et al. , 1996. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122: 1601–1610. [DOI] [PubMed] [Google Scholar]
- Ma L., Tan Z., Teng Y., Hoersch S., and Horvitz H. R., 2011. In vivo effects on intron retention and exon skipping by the U2AF large subunit and SF1/BBP in the nematode Caenorhabditis elegans. RNA 17: 2201–2211. 10.1261/rna.027458.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Gao X., Luo J., Huang L., Teng Y. et al. , 2012. The Caenorhabditis elegans gene mfap-1 encodes a nuclear protein that affects alternative splicing. PLoS Genet. 8: e1002827 10.1371/journal.pgen.1002827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C. C., Wilusz J., and Shenk T., 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14: 6647–6654. 10.1128/MCB.14.10.6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth C. D., Madl T., Bonnal S., Simon B., Zanier K. et al. , 2011. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475: 408–411. 10.1038/nature10171 [DOI] [PubMed] [Google Scholar]
- Maher K. N., Catanese M., and Chase D. L., 2013. Large-scale gene knockdown in C. elegans using dsRNA feeding libraries to generate robust loss-of-function phenotypes. J. Vis. Exp. 79: e50693 10.3791/50693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T. et al. , 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. 10.1126/science.1191244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D. A., Evans M. C., and Jacobson A., 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Kinniburgh A. J., Rachmilewitz E. A., and Ross J., 1981. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell 27: 543–553. 10.1016/0092-8674(81)90396-2 [DOI] [PubMed] [Google Scholar]
- Maroney P. A., Denker J. A., Darzynkiewicz E., Laneve R., and Nilsen T. W., 1995. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA 1: 714–723. [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Wagner E. J., and Duronio R. J., 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9: 843–854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews E. A., Mullen G. P., Manjarrez J. R., and Rand J. B., 2015. Unusual regulation of splicing of the cholinergic locus in Caenorhabditis elegans. Genetics 199: 729–737. 10.1534/genetics.114.173765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A. J., Clark F., and Smith C. W., 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6: 386–398. 10.1038/nrm1645 [DOI] [PubMed] [Google Scholar]
- Mayerle M., Yitiz S., Soulette C., Rogel L. E., Ramirez A. et al. , 2019. Prp8 impacts cryptic but not alternative splicing frequency. Proc. Natl. Acad. Sci. USA 116: 2193–2199. 10.1073/pnas.1819020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Puoti A., and Kramer A., 1999. Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA 5: 1615–1631. 10.1017/S1355838299991872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux S., van Hoof A., and Baker K. E., 2008. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol. Cell 29: 134–140. 10.1016/j.molcel.2007.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi S. M., Frischmeyer P. A., Mendell J. T., Kelly A. G., Lawler A. M. et al. , 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10: 99–105. 10.1093/hmg/10.2.99 [DOI] [PubMed] [Google Scholar]
- Melero R., Hug N., Lopez-Perrote A., Yamashita A., Caceres J. F. et al. , 2016. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat. Commun. 7: 10585 10.1038/ncomms10585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J. et al. , 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Mendell J. T., Sharifi N. A., Meyers J. L., Martinez-Murillo F., and Dietz H. C., 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36: 1073–1078 (erratum: Nat. Genet. 36: 1238). 10.1038/ng1429 [DOI] [PubMed] [Google Scholar]
- Metzstein M. M., and Krasnow M. A., 2006. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2: e180 10.1371/journal.pgen.0020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Schumperli D., and Muller B., 2000. Specificities of Caenorhabditis elegans and human hairpin binding proteins for the first nucleotide in the histone mRNA hairpin loop. RNA 6: 1539–1550. 10.1017/S135583820000056X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T. S., Carl S. H., Stadler M. B., and Grosshans H., 2016. XRN2 autoregulation and control of polycistronic gene expresssion in Caenorhabditis elegans. PLoS Genet. 12: e1006313 10.1371/journal.pgen.1006313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T. S., Carl S. H., and Grosshans H., 2017. Two distinct transcription termination modes dictated by promoters. Genes Dev. 31: 1870–1879. 10.1101/gad.301093.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. N., and Pearce D. A., 2014. Nonsense-mediated decay in genetic disease: friend or foe? Mutat. Res. Rev. Mutat. Res. 762: 52–64. 10.1016/j.mrrev.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., and Tollervey D., 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11: 1405–1413. 10.1016/S1097-2765(03)00190-4 [DOI] [PubMed] [Google Scholar]
- Mitrovich Q. M., and Anderson P., 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 14: 2173–2184. 10.1101/gad.819900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovich Q. M., and Anderson P., 2005. mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 15: 963–967. 10.1016/j.cub.2005.04.055 [DOI] [PubMed] [Google Scholar]
- Morrison M., Harris K. S., and Roth M. B., 1997. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 94: 9782–9785. 10.1073/pnas.94.18.9782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D. P., Aruscavage P. J., and Bass B. L., 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. USA 99: 7906–7911. 10.1073/pnas.112704299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M., Ivanov D., Cooper D. N., and Chuzhanova N. A., 2008. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 29: 1037–1047. 10.1002/humu.20763 [DOI] [PubMed] [Google Scholar]
- Morton J. J., and Blumenthal T., 2011a Identification of transcription start sites of trans-spliced genes: uncovering unusual operon arrangements. RNA 17: 327–337. 10.1261/rna.2447111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J. J., and Blumenthal T., 2011b RNA processing in C. elegans. Methods Cell Biol. 106: 187–217. 10.1016/B978-0-12-544172-8.00007-4 [DOI] [PubMed] [Google Scholar]
- Mowry K. L., and Steitz J. A., 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science 238: 1682–1687. 10.1126/science.2825355 [DOI] [PubMed] [Google Scholar]
- Muhlrad D., and Parker R., 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581. 10.1038/370578a0 [DOI] [PubMed] [Google Scholar]
- Muir V. S., Gasch A. P., and Anderson P., 2018. The Substrates of Nonsense-Mediated mRNA Decay in Caenorhabditis elegans. G3 (Bethesda) 8: 195–205. 10.1534/g3.117.300254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam D. K., Lee S., Zhou G., Cao X., Wang C. et al. , 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl. Acad. Sci. USA 99: 6152–6156. 10.1073/pnas.092140899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., and Frokjaer-Jensen C., 2019. The Caenorhabditis elegans transgenic toolbox. Genetics 212: 959–990. 10.1534/genetics.119.301506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. O., Moore K. A., Chapin A., Hollien J., and Metzstein M. M., 2016. Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. eLife 5: pii: e12876. 10.7554/eLife.12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W., and Graveley B. R., 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., 2016. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17: 83–96. 10.1038/nrm.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T., Dienstbier M., Murphy S., Proudfoot N. J., and Dye M. J., 2013. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Rep. 3: 1080–1092. 10.1016/j.celrep.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Gao S., Norris M. L., Ray D., Ramani A. K. et al. , 2014. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell 54: 946–959. 10.1016/j.molcel.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Gracida X., and Calarco J. A., 2017. CRISPR-mediated genetic interaction profiling identifies RNA binding proteins controlling metazoan fitness. eLife 6: pii: e28129. 10.7554/eLife.28129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass F. C., Auweter S. D., Erat M., Hargous Y., Henning A. et al. , 2005. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309: 2054–2057. 10.1126/science.1114066 [DOI] [PubMed] [Google Scholar]
- Ohnishi T., Yamashita A., Kashima I., Schell T., Anders K. R. et al. , 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12: 1187–1200. 10.1016/S1097-2765(03)00443-X [DOI] [PubMed] [Google Scholar]
- Ohno G., Hagiwara M., and Kuroyanagi H., 2008. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev. 22: 360–374. 10.1101/gad.1620608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno G., Ono K., Togo M., Watanabe Y., Ono S. et al. , 2012. Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans. PLoS Genet. 8: e1002991 10.1371/journal.pgen.1002991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H., Fujiwara M., Ohshima Y., and Ishihara T., 2008. ADBP-1 regulates an ADAR RNA-editing enzyme to antagonize RNA-interference-mediated gene silencing in Caenorhabditis elegans. Genetics 180: 785–796. 10.1534/genetics.108.093310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada-Katsuhata Y., Yamashita A., Kutsuzawa K., Izumi N., Hirahara F. et al. , 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 40: 1251–1266. 10.1093/nar/gkr791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban T. I., and Izaurralde E., 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11: 459–469. 10.1261/rna.7231505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F., Kapranov P., Foissac S., Kim S. W., Fishilevich E. et al. , 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029. 10.1016/j.cell.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. F., Carr B., Anders K. R., Grimson A., and Anderson P., 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19: 5943–5951. 10.1128/MCB.19.9.5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L. J., Frey B. J., and Blencowe B. J., 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415 [corrigenda: Nat. Genet. 41: 762 (2009)]. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Park J. E., Yi H., Kim Y., Chang H., and Kim V. N., 2016. Regulation of Poly(A) Tail and Translation during the Somatic Cell Cycle. Mol. Cell 62: 462–471. 10.1016/j.molcel.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Parker J. A., Connolly J. B., Wellington C., Hayden M., Dausset J. et al. , 2001. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc. Natl. Acad. Sci. USA 98: 13318–13323. 10.1073/pnas.231476398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvaz J. B., and Anderson P., 2007. Analysis of Nonstop mRNA Decay in Caenorhabditis elegans. University of Wisconsin, Madison, WI. [Google Scholar]
- Pasquinelli A. E., 2018. A rADAR defense against RNAi. Genes Dev. 32: 199–201. 10.1101/gad.313049.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer E., Rechavi G., and Dominissini D., 2017. Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr. Opin. Chem. Biol. 41: 93–98. 10.1016/j.cbpa.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Peixeiro I., Inacio A., Barbosa C., Silva A. L., Liebhaber S. A. et al. , 2012. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 40: 1160–1173. 10.1093/nar/gkr820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Brown A. H., and Jacobson A., 1993. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 7: 1737–1754. 10.1101/gad.7.9.1737 [DOI] [PubMed] [Google Scholar]
- Pettitt J., Crombie C., Schumperli D., and Muller B., 2002. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone gene expression and is essential for embryonic and postembryonic cell division. J. Cell Sci. 115: 857–866. [DOI] [PubMed] [Google Scholar]
- Pisareva V. P., Skabkin M. A., Hellen C. U., Pestova T. V., and Pisarev A. V., 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30: 1804–1817. 10.1038/emboj.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C., Lin P. C., and Nagai K., 2017. Structure of a pre-catalytic spliceosome. Nature 546: 617–621. 10.1038/nature22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T., and Morimoto R. I., 2008. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320: 811–814. 10.1126/science.1156093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., 2011. Ending the message: poly(A) signals then and now. Genes Dev. 25: 1770–1782. 10.1101/gad.17268411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Furger A., and Dye M. J., 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. 10.1016/S0092-8674(02)00617-7 [DOI] [PubMed] [Google Scholar]
- Pulak R., and Anderson P., 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7: 1885–1897. 10.1101/gad.7.10.1885 [DOI] [PubMed] [Google Scholar]
- Pule M. N., Glover M. L., Fire A. Z., and Arribere J. A., 2019. Ribosome clearance during RNA interference. RNA 25: 963–974. 10.1261/rna.070813.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A., and Kimble J., 1999. The Caenorhabditis elegans sex determination gene mog-1 encodes a member of the DEAH-Box protein family. Mol. Cell. Biol. 19: 2189–2197. 10.1128/MCB.19.3.2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A., and Kimble J., 2000. The hermaphrodite sperm/oocyte switch requires the Caenorhabditis elegans homologs of PRP2 and PRP22. Proc. Natl. Acad. Sci. USA 97: 3276–3281. 10.1073/pnas.97.7.3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragle J. M., Katzman S., Akers T. F., Barberan-Soler S., and Zahler A. M., 2015. Coordinated tissue-specific regulation of adjacent alternative 3′ splice sites in C. elegans. Genome Res. 25: 982–994. 10.1101/gr.186783.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendren S., Manning A. C., Al-Awadi H., Yamada K., Takagi Y. et al. , 2018. A protein-protein interaction underlies the molecular basis for substrate recognition by an adenosine-to-inosine RNA-editing enzyme. Nucleic Acids Res. 46: 9647–9659. 10.1093/nar/gky800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A. K., Nelson A. C., Kapranov P., Bell I., Gingeras T. R. et al. , 2009. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 10: R101 10.1186/gb-2009-10-9-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A. K., Calarco J. A., Pan Q., Mavandadi S., Wang Y. et al. , 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 21: 342–348. 10.1101/gr.114645.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R., Fabrizio P., Dybkov O., Hartmuth K., Pena V. et al. , 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. 10.1126/science.aag1906 [DOI] [PubMed] [Google Scholar]
- Ray D., Kazan H., Cook K. B., Weirauch M. T., Najafabadi H. S. et al. , 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati M. R., and Melton D. A., 1987. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell 48: 599–605. 10.1016/0092-8674(87)90238-8 [DOI] [PubMed] [Google Scholar]
- Reich D. P., and Bass B. L., 2019. Mapping the dsRNA world. Cold Spring Harb. Perspect. Biol. 11: pii: a035352. 10.1101/cshperspect.a035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D. P., Tyc K. M., and Bass B. L., 2018. C. elegans ADARs antagonize silencing of cellular dsRNAs by the antiviral RNAi pathway. Genes Dev. 32: 271–282. 10.1101/gad.310672.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller A. B., Hoffman D. C., and Zahler A. M., 2000. The allele-specific suppressor sup-39 alters use of cryptic splice sites in Caenorhabditis elegans. Genetics 154: 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosains J., and Mango S. E., 2012. Genetic characterization of smg-8 mutants reveals no role in C. elegans nonsense mediated decay. PLoS One 7: e49490 10.1371/journal.pone.0049490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal J. J., 2015. The emerging role of RNA editing in plasticity. J. Exp. Biol. 218: 1812–1821. 10.1242/jeb.119065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., and Ambros V., 1995. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development 121: 2491–2500. [DOI] [PubMed] [Google Scholar]
- Roundtree I. A., Evans M. E., Pan T., and He C., 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B., and Jacobson A., 2013. The intimate relationships of mRNA decay and translation. Trends Genet. 29: 691–699. 10.1016/j.tig.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. J., Stuart J. M., Lund J., and Kim S. K., 2002. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418: 975–979 [corrigenda: Nature 450: 128 (2007)]. 10.1038/nature01012 [DOI] [PubMed] [Google Scholar]
- Rubio-Peña K., Fontrodona L., Aristizabal-Corrales D., Torres S., Cornes E. et al. , 2015. Modeling of autosomal-dominant retinitis pigmentosa in Caenorhabditis elegans uncovers a nexus between global impaired functioning of certain splicing factors and cell type-specific apoptosis. RNA 21: 2119–2131. 10.1261/rna.053397.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. L., Hashimoto S., Gu S. G., Morton J. J., Stadler M. et al. , 2013. The transcription start site landscape of C. elegans. Genome Res. 23: 1348–1361. 10.1101/gr.151571.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T. K., Ash P. E., Wilson G., Gonzales P., Garrido-Lecca A. et al. , 2014. TDP-1, the Caenorhabditis elegans ortholog of TDP-43, limits the accumulation of double-stranded RNA. EMBO J. 33: 2947–2966. 10.15252/embj.201488740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter J. D., Bennett R. P., and Smith H. C., 2016. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 41: 578–594. 10.1016/j.tibs.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C. E., 2019. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses. J. Biol. Chem. 294: 1710–1720. 10.1074/jbc.TM118.004166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin L. P., and Leidel S. A., 2014. Modify or die?–RNA modification defects in metazoans. RNA Biol. 11: 1555–1567. 10.4161/15476286.2014.992279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal S. H., Schmidt E., Kitagawa K., Sondheimer N., Lindquist S. et al. , 2000. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97: 5750–5755. 10.1073/pnas.100107297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva Y. A., Rieder L. E., and Reenan R. A., 2012. The ADAR protein family. Genome Biol. 13: 252 10.1186/gb-2012-13-12-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönemann L., Kuhn U., Martin G., Schafer P., Gruber A. R. et al. , 2014. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 28: 2381–2393. 10.1101/gad.250985.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. H., Silva J., Burstein D., Pupko T., Eyras E. et al. , 2008. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 18: 88–103. 10.1101/gr.6818908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P., Montano M., Puca A., Solovieff N., Kojima T. et al. , 2009. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS One 4: e8210 10.1371/journal.pone.0008210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., and Hegde R. S., 2014. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell 55: 880–890. 10.1016/j.molcel.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P. S., Park J., Qin Y., Li X., Parsawar K. et al. , 2015. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347: 75–78. 10.1126/science.1259724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wei J., and He C., 2019. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 74: 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., 2017. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18: 655–670. 10.1038/nrm.2017.86 [DOI] [PubMed] [Google Scholar]
- Shi Y., and Manley J. L., 2015. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 29: 889–897. 10.1101/gad.261974.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiimori M., Inoue K., and Sakamoto H., 2013. A specific set of exon junction complex subunits is required for the nuclear retention of unspliced RNAs in Caenorhabditis elegans. Mol. Cell. Biol. 33: 444–456. 10.1128/MCB.01298-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C. J., Eyler D. E., and Green R., 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330: 369–372. 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier E. A., Frato K. E., Shen H., Paranawithana S. R., Green M. R. et al. , 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 23: 49–59. 10.1016/j.molcel.2006.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. L., Ribeiro P., Inacio A., Liebhaber S. A., and Romao L., 2008. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576. 10.1261/rna.815108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms C. L., Yan L. L., and Zaher H. S., 2017. Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68: 361–373.e5. 10.1016/j.molcel.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh J. N., Buckingham S. D., Esmaeili B., Viswanathan M., Cuppen E. et al. , 2011. A novel Caenorhabditis elegans allele, smn-1(cb131), mimicking a mild form of spinal muscular atrophy, provides a convenient drug screening platform highlighting new and pre-approved compounds. Hum. Mol. Genet. 20: 245–260. 10.1093/hmg/ddq459 [DOI] [PubMed] [Google Scholar]
- Son H. G., Seo M., Ham S., Hwang W., Lee D. et al. , 2017. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nat. Commun. 8: 14749 10.1038/ncomms14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufari H., and Mackereth C. D., 2017. Conserved binding of GCAC motifs by MEC-8, couch potato, and the RBPMS protein family. RNA 23: 308–316. 10.1261/rna.059733.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks K. A., and Dieckmann C. L., 1998. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 26: 4676–4687. 10.1093/nar/26.20.4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A. K., Herman R. K., and Shaw J. E., 2004. SMU-2 and SMU-1, Caenorhabditis elegans homologs of mammalian spliceosome-associated proteins RED and fSAP57, work together to affect splice site choice. Mol. Cell. Biol. 24: 6811–6823. 10.1128/MCB.24.15.6811-6823.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R., Llorian M., and Smith C. W., 2007. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell 27: 420–434. 10.1016/j.molcel.2007.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L. et al. , 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21: 325–341. 10.1101/gr.114595.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., McWhirter R., Miller T., Strasbourger P., Thompson O. et al. , 2014. Isolation of specific neurons from C. elegans larvae for gene expression profiling. PLoS One 9: e112102 10.1371/journal.pone.0112102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J., Brooke G., Kuersten S., Lea K., and Blumenthal T., 1993. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell 73: 521–532. 10.1016/0092-8674(93)90139-H [DOI] [PubMed] [Google Scholar]
- Spieth J., Lawson D., Davis P., Williams G., and Howe K., 2014. Overview of gene structure in C. elegans (October 29, 2014), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.65.2, http://www.wormbook.org. 10.1895/wormbook.1.65.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Shaw J. E., and Herman R. K., 2001. Analysis of smu-1, a gene that regulates the alternative splicing of unc-52 pre-mRNA in Caenorhabditis elegans. Mol. Cell. Biol. 21: 4985–4995. 10.1128/MCB.21.15.4985-4995.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Davies A. G., Shaw J. E., and Herman R. K., 2002. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development 129: 4999–5008. [DOI] [PubMed] [Google Scholar]
- Stein L. D., Bao Z., Blasiar D., Blumenthal T., Brent M. R. et al. , 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: E45 10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., and Birnstiel M. L., 1986. Genetic complementation in the Xenopus oocyte: co-expression of sea urchin histone and U7 RNAs restores 3′ processing of H3 pre-mRNA in the oocyte. EMBO J. 5: 1675–1682. 10.1002/j.1460-2075.1986.tb04411.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny A. O., Eichhorn S. W., Chen G. R., Sive H., and Bartel D. P., 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Singh V., Kajino-Sakamoto R., and Aballay A., 2011. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332: 729–732. 10.1126/science.1203411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szádeczky-Kardoss I., Csorba T., Auber A., Schamberger A., Nyiko T. et al. , 2018. The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res. 46: 4632–4648. 10.1093/nar/gky279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei S., Togo-Ohno M., Suzuki Y. and Kuroyanagi H., 2016. Evolutionarily conserved autoregulation of alternative pre-mRNA splicing by ribosomal protein L10a. Nucleic Acids Res. 44: 5585–5598. 10.1093/nar/gkw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. H., and Fraser A. G., 2017. The combinatorial control of alternative splicing in C. elegans. PLoS Genet. 13: e1007033 10.1371/journal.pgen.1007033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq A., and Jantsch M. F., 2012. Transcript diversification in the nervous system: a to I RNA editing in CNS function and disease development. Front. Neurosci. 6: 99 10.3389/fnins.2012.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A., Tahiri-Alaoui A., West S., Thomas B., Ramadass A. et al. , 2004. Autocatalytic RNA cleavage in the human beta-globin pre-mRNA promotes transcription termination. Nature 432: 526–530. 10.1038/nature03032 [DOI] [PubMed] [Google Scholar]
- Thacker C., Sheps J. A., and Rose A. M., 2006. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell. Mol. Life Sci. 63: 1193–1204. 10.1007/s00018-006-6012-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J., Lea K., Zucker-Aprison E., and Blumenthal T., 1990. The spliceosomal snRNAs of Caenorhabditis elegans. Nucleic Acids Res. 18: 2633–2642. 10.1093/nar/18.9.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M., Bixby R., Dalton R., Vandenburg A., Calarco J. A. et al. , 2019. Splicing in a single neuron is coordinately controlled by RNA binding proteins and transcription factors. eLife 8: pii: e46726. 10.7554/eLife.46726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh E. A., Guerry P., and Wickner R. B., 1978. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 136: 1002–1007. 10.1128/JB.136.3.1002-1007.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma K. G., Rebbapragada I., Durand S., and Lykke-Andersen J., 2015. Identification of elements in human long 3′ UTRs that inhibit nonsense-mediated decay. RNA 21: 887–897. 10.1261/rna.048637.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M., Naito Y., Kuroyanagi H., and Iino Y., 2016. Splicing factors control C. elegans behavioural learning in a single neuron by producing DAF-2c receptor. Nat. Commun. 7: 11645 10.1038/ncomms11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin L. A., and Bass B. L., 2003. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science 302: 1725 10.1126/science.1091340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin L. A., Saccomanno L., Morse D. P., Brodigan T., Krause M. et al. , 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 21: 6025–6035. 10.1093/emboj/cdf607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse N. J., Millet J. R. M., and Dupuy D., 2017. Quantitative RNA-seq meta-analysis of alternative exon usage in C. elegans. Genome Res. 27: 2120–2128. 10.1101/gr.224626.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E. et al. , 2012. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46: 518–529. 10.1016/j.molcel.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Uyar B., Chu J. S., Vergara I. A., Chua S. Y., Jones M. R. et al. , 2012. RNA-seq analysis of the C. briggsae transcriptome. Genome Res. 22: 1567–1580. 10.1101/gr.134601.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Delft P., Akay A., Huber S. M., Bueschl C., Rudolph K. L. M. et al. , 2017. The Profile and Dynamics of RNA Modifications in Animals. ChemBioChem 18: 979–984. 10.1002/cbic.201700093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren K., and Hirsh D., 1988. Trans-spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature 335: 556–559. 10.1038/335556a0 [DOI] [PubMed] [Google Scholar]
- Van Epps H., Dai Y., Qi Y., Goncharov A., and Jin Y., 2010. Nuclear pre-mRNA 3′-end processing regulates synapse and axon development in C. elegans. Development 137: 2237–2250. 10.1242/dev.049692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P. A., Dietz H. C., and Parker R., 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264. 10.1126/science.1067272 [DOI] [PubMed] [Google Scholar]
- Vesely C., Tauber S., Sedlazeck F. J., von Haeseler A., and Jantsch M. F., 2012. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 22: 1468–1476. 10.1101/gr.133025.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisine C., Pedersen J. S., and Morimoto R. I., 2010. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol. Dis. 40: 12–20. 10.1016/j.nbd.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M. C., and Luhrmann R., 2015. SnapShot: spliceosome dynamics I. Cell 161: 1474-e1 10.1016/j.cell.2015.05.050 [DOI] [PubMed] [Google Scholar]
- Wallace A., Filbin M. E., Veo B., McFarland C., Stepinski J. et al. , 2010. The nematode eukaryotic translation initiation factor 4E/G complex works with a trans-spliced leader stem-loop to enable efficient translation of trimethylguanosine-capped RNAs. Mol. Cell. Biol. 30: 1958–1970. 10.1128/MCB.01437-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R., Yan C., Bai R., Huang G., and Shi Y., 2016. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science 353: 895–904. 10.1126/science.aag2235 [DOI] [PubMed] [Google Scholar]
- Wan R., Yan C., Bai R., Lei J. and Shi Y., 2017. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell 171: 120–132.e12. 10.1016/j.cell.2017.08.029 [DOI] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L. et al. , 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kimble J., and Wickens M., 2004. Tissue-specific modification of gld-2 mRNA in C. elegans: likely C-to-U editing. RNA 10: 1444–1448. 10.1261/rna.7570804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Czaplinski K., Rao Y., and Peltz S. W., 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20: 880–890. 10.1093/emboj/20.4.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. F., Whitfield M. L., Ingledue T. C. 3rd, Dominski Z., and Marzluff W. F., 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10: 3028–3040. 10.1101/gad.10.23.3028 [DOI] [PubMed] [Google Scholar]
- Wani S., and Kuroyanagi H., 2017. An emerging model organism Caenorhabditis elegans for alternative pre-mRNA processing in vivo. Wiley Interdiscip. Rev. RNA 8:pmid:28703462. 10.1002/wrna.1428 [DOI] [PubMed] [Google Scholar]
- Warf M. B., Shepherd B. A., Johnson W. E., and Bass B. L., 2012. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 22: 1488–1498. 10.1101/gr.134841.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. C., and Hundley H. A., 2016. Trans and cis factors affecting A-to-I RNA editing efficiency of a noncoding editing target in C. elegans. RNA 22: 722–728. 10.1261/rna.055079.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. C., Kakaradov B., Sundararaman B., Wheeler E., Hoon S. et al. , 2014. The dsRBP and inactive editor ADR-1 utilizes dsRNA binding to regulate A-to-I RNA editing across the C. elegans transcriptome. Cell Rep. 6: 599–607. 10.1016/j.celrep.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe E., Ono S., and Kuroyanagi H., 2018. Alternative splicing of the Caenorhabditis elegans lev-11 tropomyosin gene is regulated in a tissue-specific manner. Cytoskeleton (Hoboken) 75: 427–436. 10.1002/cm.21489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Gromak N., and Proudfoot N. J., 2004. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525. 10.1038/nature03035 [DOI] [PubMed] [Google Scholar]
- Wheway G., Schmidts M., Mans D. A., Szymanska K., Nguyen T. T. et al. , 2015. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat. Cell Biol. 17: 1074–1087. 10.1038/ncb3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple J. M., Youssef O. A., Aruscavage P. J., Nix D. A., Hong C. et al. , 2015. Genome-wide profiling of the C. elegans dsRNAome. RNA 21: 786–800. 10.1261/rna.048801.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson H. A., Fitzgerald K., and Greenwald I., 1994. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79: 1187–1198. 10.1016/0092-8674(94)90010-8 [DOI] [PubMed] [Google Scholar]
- Wilkinson M. E., Fica S. M., Galej W. P., Norman C. M., Newman A. J. et al. , 2017. Postcatalytic spliceosome structure reveals mechanism of 3′-splice site selection. Science 358: 1283–1288. 10.1126/science.aar3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. E., 2018. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA 9: e1478 10.1002/wrna.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp N., Huntzinger E., Weiler C., Sauliere J., Schmidt S. et al. , 2009. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 29: 3517–3528. 10.1128/MCB.00177-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M. C., Gooding C., Wagner E. J., Garcia-Blanco M. A., and Smith C. W., 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13: 91–100. 10.1016/S1097-2765(03)00502-1 [DOI] [PubMed] [Google Scholar]
- Wu D., Lamm A. T., and Fire A. Z., 2011. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat. Struct. Mol. Biol. 18: 1094–1101. 10.1038/nsmb.2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T., Suzuki N., Furukawa T., Ishihara T., and Katsura I., 2005. Multidrug resistance-associated protein MRP-1 regulates dauer diapause by its export activity in Caenorhabditis elegans. Development 132: 3197–3207. 10.1242/dev.01909 [DOI] [PubMed] [Google Scholar]
- Yamashita A., Izumi N., Kashima I., Ohnishi T., Saari B. et al. , 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 23: 1091–1105. 10.1101/gad.1767209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Wan R., Bai R., Huang G., and Shi Y., 2016. Structure of a yeast activated spliceosome at 3.5 A resolution. Science 353: 904–911. 10.1126/science.aag0291 [DOI] [PubMed] [Google Scholar]
- Yan C., Wan R., Bai R., Huang G., and Shi Y., 2017. Structure of a yeast step II catalytically activated spliceosome. Science 355: 149–155. 10.1126/science.aak9979 [DOI] [PubMed] [Google Scholar]
- Yates A., Akanni W., Amode M. R., Barrell D., Billis K. et al. , 2016. Ensembl 2016. Nucleic Acids Res. 44: D710–D716. 10.1093/nar/gkv1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura J., Ichikawa K., Shoura M. J., Artiles K. L., Gabdank I. et al. , 2019. Recompleting the Caenorhabditis elegans genome. Genome Res. 29: 1009–1022. 10.1101/gr.244830.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. J., Guydosh N. R., Zhang F., Hinnebusch A. G., and Green R., 2015. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell 162: 872–884. 10.1016/j.cell.2015.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman E. M., and Claycomb J. M., 2014. From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front. Genet. 5: 416 10.3389/fgene.2014.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., 2012. Pre-mRNA splicing and its regulation in Caenorhabditis elegans (March 21, 2012), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.31.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., Rogel L. E., Glover M. L., Yitiz S., Ragle J. M. et al. , 2018. SNRP-27, the C. elegans homolog of the tri-snRNP 27K protein, has a role in 5′ splice site positioning in the spliceosome. RNA 24: 1314–1325. 10.1261/rna.066878.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., Tuttle J. D., and Chisholm A. D., 2004. Genetic suppression of intronic +1G mutations by compensatory U1 snRNA changes in Caenorhabditis elegans. Genetics 167: 1689–1696. 10.1534/genetics.104.028746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti S., Meola M., Bochud A., and Puoti A., 2011. Role of the C. elegans U2 snRNP protein MOG-2 in sex determination, meiosis, and splice site selection. Dev. Biol. 354: 232–241. 10.1016/j.ydbio.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Zhan X., Yan C., Zhang X., Lei J., and Shi Y., 2018a Structure of a human catalytic step I spliceosome. Science 359: 537–545. 10.1126/science.aar6401 [DOI] [PubMed] [Google Scholar]
- Zhan X., Yan C., Zhang X., Lei J., and Shi Y., 2018b Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res. 28: 1129–1140. 10.1038/s41422-018-0094-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rigo F., and Martinson H. G., 2015. Poly(A) signal-dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the Poly(A) Site. Mol. Cell 59: 437–448. 10.1016/j.molcel.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun X., Qian Y., LaDuca J. P., and Maquat L. E., 1998a At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 18: 5272–5283. 10.1128/MCB.18.9.5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun X., Qian Y., and Maquat L. E., 1998b Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4: 801–815. 10.1017/S1355838298971849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yan C., Hang J., Finci L. I., Lei J. et al. , 2017. An atomic structure of the human spliceosome. Cell 169: 918–929.e14. 10.1016/j.cell.2017.04.033 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yan C., Zhan X., Li L., Lei J. et al. , 2018. Structure of the human activated spliceosome in three conformational states. Cell Res. 28: 307–322. 10.1038/cr.2018.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhan X., Yan C., Zhang W., Liu D. et al. , 2019. Structures of the human spliceosomes before and after release of the ligated exon. Cell Res. 29: 274–285. 10.1038/s41422-019-0143-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xue W., Li X., Zhang J., Chen S. et al. , 2016. The biogenesis of nascent circular RNAs. Cell Rep. 15: 611–624. 10.1016/j.celrep.2016.03.058 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fu J., and Gilmour D. S., 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 19: 1572–1580. 10.1101/gad.1296305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. Q., Zhang P., Gao H., He X., Dou Y. et al. , 2015. Profiling the RNA editomes of wild-type C. elegans and ADAR mutants. Genome Res. 25: 66–75. 10.1101/gr.176107.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder J. C., and Lima C. D., 2017. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31: 88–100. 10.1101/gad.294769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis D. G., Lovci M. T., Wilbert M. L., Hutt K. R., Liang T. Y. et al. , 2010. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 17: 173–179. 10.1038/nsmb.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D. A., and Blumenthal T., 1999a Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. 10.1038/45597 [DOI] [PubMed] [Google Scholar]
- Zorio D. A., and Blumenthal T., 1999b U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5: 487–494. 10.1017/S1355838299982225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D. A., Cheng N. N., Blumenthal T., and Spieth J., 1994. Operons as a common form of chromosomal organization in C. elegans. Nature 372: 270–272. 10.1038/372270a0 [DOI] [PubMed] [Google Scholar]
- Zorio D. A., Lea K., and Blumenthal T., 1997. Cloning of Caenorhabditis U2AF65: an alternatively spliced RNA containing a novel exon. Mol. Cell. Biol. 17: 946–953. 10.1128/MCB.17.2.946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zund D., and Muhlemann O., 2013. Recent transcriptome-wide mapping of UPF1 binding sites reveals evidence for its recruitment to mRNA before translation. Translation (Austin) 1: e26977 10.4161/trla.26977 [DOI] [PMC free article] [PubMed] [Google Scholar]