Abstract
Bacteria often carry “extra DNA” in the form of plasmids in addition to their chromosome. Many plasmids have a copy number greater than one such that the genes encoded on these plasmids are present in multiple copies per cell. This has evolutionary consequences by increasing the mutational target size, by prompting the (transitory) co-occurrence of mutant and wild-type alleles within the same cell, and by allowing for gene dosage effects. We develop and analyze a mathematical model for bacterial adaptation to harsh environmental change if adaptation is driven by beneficial alleles on multicopy plasmids. Successful adaptation depends on the availability of advantageous alleles and on their establishment probability. The establishment process involves the segregation of mutant and wild-type plasmids to the two daughter cells, allowing for the emergence of mutant homozygous cells over the course of several generations. To model this process, we use the theory of multitype branching processes, where a type is defined by the genetic composition of the cell. Both factors—the availability of advantageous alleles and their establishment probability—depend on the plasmid copy number, and they often do so antagonistically. We find that in the interplay of various effects, a lower or higher copy number may maximize the probability of evolutionary rescue. The decisive factor is the dominance relationship between mutant and wild-type plasmids and potential gene dosage effects. Results from a simple model of antibiotic degradation indicate that the optimal plasmid copy number may depend on the specific environment encountered by the population.
Keywords: rapid adaptation, antibiotic resistance, bacterial evolution, plasmid copy number, dominance
PLASMIDS are extrachromosomal DNA elements that can be transmitted vertically or be transferred horizontally between cells and are commonly found in bacteria. Plasmids usually do not carry essential genes and are sometimes depicted as parasites that infect and exploit the bacterial cell, imposing a cost on its host. However, genes on plasmids can code for traits, such as resistance to antibiotics or the ability to metabolize rarely encountered carbon sources, that are beneficial in specific environments (Eberhard, 1989, 1990; MacLean and San Millan 2015).
While some (usually large) plasmids exist only in a single copy within the bacterial cell, many plasmids are present in several copies per cell, some even in several hundreds (Friehs 2004). The importance of plasmid copy number for the evolutionary dynamics of genes on plasmids, and the consequences for bacterial adaptation have recently started to gain increased attention (San Millan et al. 2016; Rodriguez-Beltran et al. 2018; Ilhan et al. 2019).
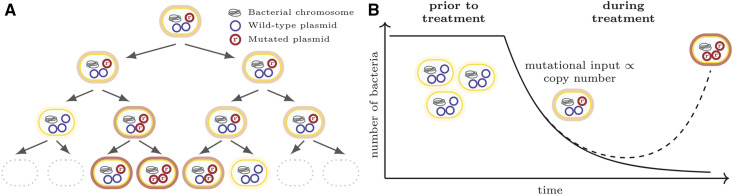
There are several important differences between carrying a gene on a multicopy plasmid and carrying it on a single-copy plasmid or a haploid chromosome. First, the mutational target size is multiplied by the copy number, making the appearance of mutations more likely (San Millan et al. 2016). Second, novel alleles appearing through mutation or novel genes acquired through transformation are initially present only on one (or few) of the plasmid copies. At cell division, as sketched in Figure 1A, the segregation of plasmids to the daughter cells generates cells with a plasmid composition different from that of the mother cell, and, thereby, cells with a higher fraction of mutant plasmids may arise (San Millan et al. 2016; Halleran et al. 2019). Yet, through this segregation process, alleles on plasmids are subject to an extra layer of drift, termed “segregational drift” by Ilhan et al. (2019). This affects, in particular, the establishment probability of novel alleles on plasmids. On the other hand, the coexistence of mutant and wild-type plasmids within the same cell may allow the cell to escape from fitness trade-offs if these different plasmid variants confer different benefits (Rodriguez-Beltran et al. 2018). Last, gene dosage effects can lead to an amplification of a gene’s effect, such as increased levels of resistance if carried on a multicopy plasmid, i.e., cells with a larger number of plasmids carrying this gene display higher fitness (Martinez and Baquero 2000; San Millan et al. 2016; Santos-Lopez et al. 2017).
Figure 1.
Bacterial adaptation on multicopy plasmids. (A) Random segregation of plasmids at cell division and establishment of the resistance mutation [figure adapted from San Millan et al. (2016)]. (B) Evolutionary rescue through de novo mutations on a multicopy plasmid.
Each of these effects has been demonstrated in evolution experiments. San Millan et al. (2016) exposed Escherichia coli populations that carried a blaTEM-1 β-lactamase gene either on the chromosome or on a multicopy plasmid to increasing levels of ceftazidime. Different alleles of the blaTEM-1 β-lactamase gene confer resistance to different antibiotics, and the original allele inserted by San Millan et al. (2016) conferred resistance to ampicillin but not to ceftazidime . Mutations can, however, generate an allele that provides ceftazidime resistance (see also Blazquez et al. 1995). San Millan et al. (2016) found that high levels of resistance were more likely to evolve when the gene was present on a multicopy plasmid due to (1) the increased mutational input, (2) the following increase of the fraction of mutant plasmids per cell through segregation, and (3) gene dosage effects. Using the same experimental system, Rodriguez-Beltran et al. (2018) showed that the existence of heterozygous cells, carrying wild-type and mutant plasmids, allowed populations to escape from fitness trade-offs, since these cells were resistant to both antibiotics. In a different setup, in the study by Santos-Lopez et al. (2017), the coexistence of a mutated plasmid and a wild-type plasmid allowed for a higher total plasmid copy number and resistance to higher antibiotic concentrations than found for either plasmid type on its own, combining again two benefits of multicopy plasmids (the possibility of heterozygosity and gene dosage effects). Ilhan et al. (2019) finally studied the rate of evolution on two multicopy plasmids, one with a low and the other one with a high copy number. The accumulation of new mutations was lower than expected by the mutational target size, reflecting the decreased establishment probability of mutations on multicopy plasmids (at least in the absence of dosage effects).
Theoretical studies that quantify and disentangle the described effects are still rare. Some experiments are complemented by models and computer simulations (Rodriguez-Beltran et al. 2018; Ilhan et al. 2019). In addition, Halleran et al. (2019) show that random segregation of plasmid variants into the daughter cells speeds up adaptation compared to an equal partition that leads to exact copies of the mother cell. Beyond these studies, which focus specifically on plasmids, models exist for chromosomal adaptation in polyploid prokaryotes, which has similarities to adaptation on multicopy plasmids (Markov and Kaznacheev 2016; Sun et al. 2018).
In this article, we develop a mathematical framework to study bacterial adaptation driven by novel alleles on nontransmissible multicopy plasmids. To be specific, we will often refer to the evolution of antibiotic resistance throughout the manuscript. Yet, the results apply equally to other beneficial alleles. In our model, wild-type and mutant plasmid copies are distributed randomly to daughter cells at cell division, but each daughter cell receives the same number of plasmid copies. This resembles early models used to investigate plasmid incompatibility and segregation rates of incompatible plasmids (Ishii et al. 1978; Novick and Hoppensteadt 1978; Cullum and Broda 1979). Using the mathematical theory of multitype branching processes, we determine the establishment probability of a novel allele that initially arises on a single plasmid copy within a single cell. Using these results, we calculate the probability of evolutionary rescue, i.e., the probability that the bacterial population escapes extinction following environmental change through adaptive evolution, if the novel allele appears through mutation in an existing plasmid-carried gene. This probability depends on the mutational input and on the establishment probability of mutations. The plasmid copy number often has an antagonistic effects on these two factors, making it nonobvious whether a higher or lower copy number leads to a higher probability of rescue. We especially explore how the relationship between the plasmid composition within a cell and bacterial fitness influences the results. As an example, we apply the framework to a simple model for antibiotic resistance through enzymatic antibiotic degradation, in which we derive this relationship mechanistically. We finally extend the modeling framework to account for adaptation from the standing genetic variation.
The Model
We consider a bacterial population of initially cells. Each bacterial cell contains exactly n copies of a nontransmissible plasmid. We distinguish two variants of this plasmid: the wild-type plasmid and the mutant plasmid. Initially, all cells are homozygous for the wild-type plasmid and sensitive to an antibiotic in their environment. The population size therefore declines due to the drug pressure, and the population will go extinct unless resistance evolves (see Figure 1B). A resistance allele can appear through a single mutation of a gene on the multicopy plasmid (mutant plasmid). As explained above, this is possible if, for example, the wild-type plasmid carries a resistance gene that confers resistance to some antibiotic but not to the one currently present (see San Millan et al. 2016). The level of resistance of a cell depends on its plasmid composition. Further below, we extend our model to include standing genetic variation.
We model the population dynamics by a birth–death process with birth (cell division) and death rates and for a cell with n plasmids and i mutated plasmids. We denote the Malthusian fitness of a bacterial cell by For cells that are homozygous for the wild-type plasmid (i = 0), and, hence, sensitive to the antibiotic, it holds that With an increasing number of mutant plasmids in a cell, the birth rate increases and/or the death rate decreases We normally assume that at least the cell type with only mutated plasmids (i = n) is resistant, so that Otherwise the bacterial population could not escape ultimate extinction. The shape of and as a function of i reflects the dominance relationship between the mutant and the wild-type allele. Gene dosage effects can lead to a higher fitness of mutant homozygotes for bacteria with a higher plasmid copy number (i.e., or ). On the other hand, plasmids can impose a cost on the cell, and it is plausible to assume that this cost increases with the copy number. We implement plasmid costs as an increase in the death rate that depends linearly on n, e.g., However, for the most part, we ignore plasmid costs (c = 0) in order to disentangle the cost-independent effects of n on rescue. While the analytical results are derived for arbitrary and we choose (or = 1 + cn) and for all numerical examples presented in the figures, such that only the net growth rate depends on the cell type. We set a hard carrying capacity of cells. When the population size exceeds a random cell is removed.
We assume that plasmid replication and cell division are perfectly synchronized. Right before cell division, each plasmid is replicated exactly once. At plasmid replication, mutation from the wild-type to the mutant plasmid occurs with probability u per plasmid. We neglect back mutations. Then the cell divides, and each daughter cell receives exactly n plasmids. The distribution of wild-type and mutant plasmids to the daughter cells, however, is random. The probability that a cell that contains i mutant plasmids before and 2i + x mutant plasmids after plasmid replication (x denotes here the number of newly mutated wild-type plasmids) divides into daughter cells with k and mutant plasmids, respectively, is given by
| (1) |
For example, disregarding mutation (x = 0), a cell with two plasmids (n = 2), one of which is mutated (i = 1), divides into two heterozygous daughter cells with probability and in a homozygous wild-type and a homozygous mutant cell with probability .
The chosen model for plasmid replication and segregation is mathematically convenient and captures the essence of the process by introducing stochasticity in the number of mutant plasmids inherited by each daughter cell. However, it contains several simplifying assumptions. For example, plasmids are normally replicated after cell division. Thereby, some plasmids may be copied several times, and others not at all (Nordström 2006). An equal distribution of the plasmid copies contained in the mother cell to the daughter cells is an idealization, reflecting a perfect partitioning system. How segregation occurs varies across plasmids, and, moreover, differs between low-copy and high-copy plasmids (Million-Weaver and Camps 2014). Given the variation in segregation mechanisms and differences between plasmids with low and high copy numbers, no model fits them all, and we here choose one extreme. To test the robustness of our results, we set up and analyze two alternative models for plasmid replication and segregation in the supplemental information, section S1, in which we relax some of the assumptions. In these two alternative models, plasmid copies are replicated after cell division. In alternative model (1), we keep the restriction that plasmid copies are distributed to daughter cells in (nearly) equal numbers. If the plasmid copy is even, n/2 plasmid copies are segregated to both daughter cells, and, otherwise, daughter cells receive and copies, respectively. In the alternative model (2), we relax the assumption of an equal distribution of plasmid copies. Each daughter cell receives at least one plasmid copy to avoid plasmid loss, but otherwise plasmid copies are distributed randomly to the two daughter cells. In both models, we consider plasmid replication to happen iteratively after cell division, i.e., a plasmid copy is selected from the pool to replicate and put back to the pool before the next plasmid replicates until the carrying capacity of n plasmids is reached [cf. the models on plasmid incompatibility by Ishii et al. (1978), Novick and Hoppensteadt (1978), and Cullum and Broda (1979) that also consider either replication of each copy as in our main model or iterative replication as in our alternative models].
Analysis
Stochastic computer simulations
We perform stochastic computer simulations that exactly implement the model. The code is written in the C++ programming language, following the Gillespie algorithm (Gillespie 1976) and making use of the GNU Scientific library (Galassi et al. 2009). We track the bacterial population until it has either gone extinct or until the mutant homozygote has reached a critical number Nc above which the probability of extinction of the homozygote population is <1%. We determine this threshold mathematically through where is the probability that a single mutant homozygous individual establishes long-term lineage of offspring. We provide an expression for in Equation (A.6).
Mathematical approach
Whether the bacterial population successfully adapts or goes extinct depends on two factors: the mutational supply and the establishment probability of the resistance mutation once it has appeared. New resistance mutations appear approximately at rate during population decline, where is the number of wild-type homozygous cells at time t. (The approximation assumes that within any one cell, at most one plasmid copy acquires a mutation during replication.) The mutational supply hence linearly increases with the plasmid copy number n. Since the initial population size is large, we describe the dynamics of wild-type homozygous cells deterministically (cf. for example Orr and Unckless 2008; Alexander and Bonhoeffer 2012; Martin et al. 2013; Tazzyman and Bonhoeffer 2014; Uecker et al. 2014; Uecker and Hermisson 2016; Uecker 2017). Hence:
| (2) |
Yet, for evolutionary rescue to occur, it is not sufficient that a resistance mutation appears before the wild-type population goes extinct. It also needs to escape stochastic loss. It is hence not sufficient to compute the rate of appearance of resistance mutations. Instead, we need to compute the rate of appearance of successful resistance mutations. Any resistance mutation first appears in a single plasmid copy within a single cell, and we denote by its establishment probability. Then, the rate of appearance of successful resistance mutations is given by To determine we use the mathematical theory of multitype branching processes, where a type is determined by the plasmid composition of the cell. Branching process theory is a classic approach to approximate establishment probabilities of beneficial alleles (Fisher 1922; Haldane 1927; for a review see Patwa and Wahl 2008). The underlying idea is that cells carrying the beneficial allele spread independently from each other as long as they are rare and the fate of the beneficial allele uncertain. Details of how to apply the approach to the current problem are given in Appendix A.
With this, the probability of evolutionary rescue by de novo mutations can be approximated by
| (3) |
(see Orr and Unckless 2008; Alexander and Bonhoeffer 2012; Martin et al. 2013; Tazzyman and Bonhoeffer 2014; Uecker et al. 2014; Uecker and Hermisson 2016; Uecker 2017; Anciaux et al. 2018). The exponential function is the zeroth term of a Poisson distribution and gives the probability that no successful resistance mutation appears in time, i.e., that the population goes extinct. From Equation (3), we see that
in the absence of plasmid costs ( and independent of n).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental Material (File S1) and the simulation code and the scripts used for the numerical analysis (File S2) are available at figshare: https://doi.org/10.25386/genetics.12357731
Results
Analytical solutions for one and two plasmid copies per cell
In a first step, we consider plasmids with copy numbers n = 1 and n = 2. Note that, within our approach, rescue on a single-copy plasmid is identical to rescue on a haploid chromosome (assuming equal mutation probabilities), which means that any comparison of n > 1 to n = 1 is simultaneously a comparison of adaptation on a multicopy plasmid to adaptation on a haploid chromosome. Detailed derivations of all results can be found in SI section S2.
For single-copy plasmids, establishment of the resistance mutation is determined simply by the dynamics of cell division and death, while for two-copy plasmids, segregation plays a role since heterozygous cells can either have two heterozygous or two homozygous daughter cells. The potential production of a daughter cell that is homozygous for the maladaptive wild-type allele a priori opposes establishment of the resistance mutation on a two-copy plasmid. On the other hand, gene dosage effects may outweigh this disadvantage. Moreover, the mutational input is twice as high on a two-copy plasmid (assuming that the plasmid copy number does not affect the fitness of wild-type homozygotes).
For n = 1 and n = 2, simple analytical solutions for exist [from Equation (A.2)]:
| (4) |
| (5) |
This allows us to analyze in detail how these effects play out. Under which conditions is the establishment probability of a beneficial allele higher on a two-copy than on a single-copy plasmid? And when is evolutionary rescue more likely?
We distinguish two cases: (1) There are no gene dosage effects, and the level of resistance depends on the relative number of mutated plasmids in the cell, hence and , i.e., the fitness of mutant homozygous cells is independent of the total plasmid copy number n. (2) There are gene dosage effects, and the level of resistance depends on the absolute number of mutated plasmids in the cell, hence and In both cases, we assume that there is no cost associated with a higher plasmid copy number (i.e., and
It is intuitive and can also be derived from a comparison of the formulas for and that in scenario (1), the establishment probability is always higher if the cell carries one plasmid than if it carries two, i.e., irrespective of the birth and death rates of all cell types. However, with two plasmid copies, the mutational input is higher, which may outweigh the lower establishment probability. Inserting Equations (4) and (5) into Equation (3) and comparing the rescue probabilities shows that
| (6) |
where λhet and μhet are the birth and death rates of the heterozygous two-copy cell and The fitness of heterozygous cells hence needs to be sufficiently high, otherwise the higher mutational input for n = 2 is insufficient to outweigh the lower establishment probability. In particular, evolutionary rescue is less likely on a two-copy plasmid than on a single-copy plasmid if heterozygous two-copy cells have a negative Malthusian fitness λhet - μhet. In the special case for all n and i and λhom = 1+s and , condition (Equation 6) simplifies to
| (7) |
The Malthusian fitness of a heterozygous cell hence needs to be at least one-third that of a homozygous mutant cell (if s is small).
The variable α should not be confounded with the dominance coefficient of the mutation, since s is the Malthusian fitness of the mutant homozygous cell and not the effect size of the mutation. The dominance coefficient is given by Analogous to Equation (7), we can derive a threshold for the dominance coefficient above which rescue is more likely on a two-copy than on a single-copy plasmid:
| (8) |
The threshold depends both on λ0 and on the beneficial effect sE of the mutation, and is a decreasing function in both. The higher λ0 and the more beneficial the mutation, the lower is the required dominance coefficient.
In scenario (2) with gene dosage effects, unlike in scenario (1), the establishment probability can be higher with two plasmid copies than with one:
| (9) |
For the probability of evolutionary rescue, we find that two plasmid copies are always advantageous, i.e.,
| (10) |
irrespective of the birth and death rates of all types. Remember though that we assumed that the second plasmid copy did not impose any additional cost. An additional cost, if strong enough, can make the second copy disadvantageous despite gene dosage effects.
Arbitrary copy numbers
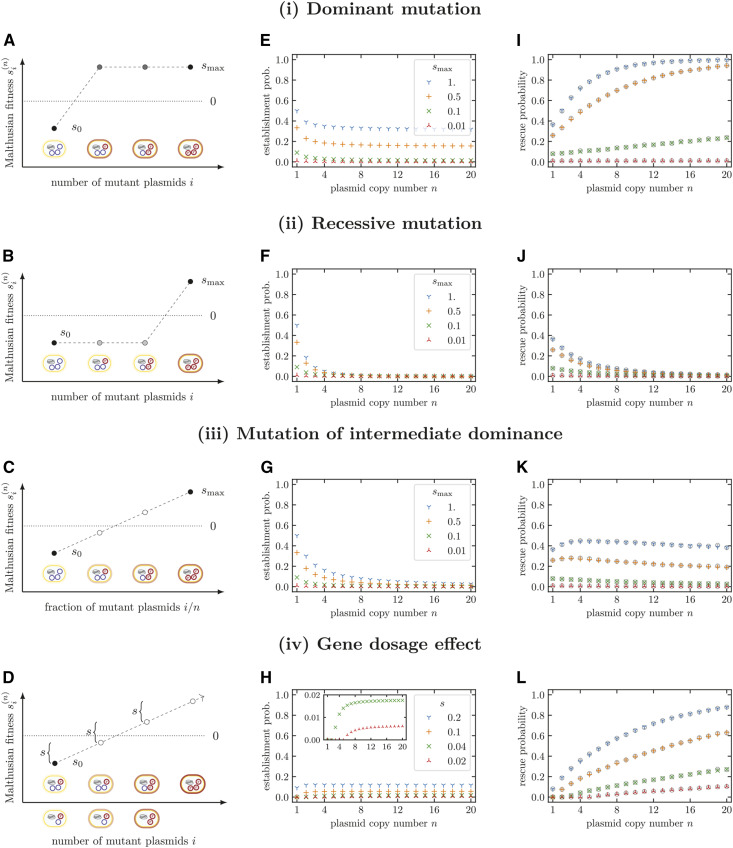
In the previous section, we saw that—as is also intuitively expected—the fitness of heterozygous cells and potential gene dosage effects determine whether a copy number of one or two is advantageous. Here, we proceed to investigate for arbitrary copy numbers how the relationship between the plasmid composition of a cell, i.e., the number of mutant plasmids i and wild-type plasmids (n – i), and the cell’s fitness influences the form of In the absence of gene dosage effects (i.e., if the fitness of mutant homozygotes is independent of n), we refer to this relationship as the dominance function of a mutant allele. In order to determine the establishment probabilities we solve Equation (A.5) numerically [and then use Equation (A.1)]. Throughout this section, we set for all n and i, and only the birth rate depends on n and i. The Malthusian fitness of wild-type cells is set to independent of n. We consider examples of three different types of plasmid mutations with different shapes of dominance functions: (i) dominant mutations (Figure 2A), (ii) recessive mutations (Figure 2B), (iii) mutations of intermediate dominance (Figure 2C), and (iv) mutations with a gene dosage effect (Figure 2D). For (i)–(iii), is independent of n, while it increases with n in case (iv).
Figure 2.
The influence of the dominance function of a plasmid mutation and of gene dosage effects on the establishment of resistance-conferring plasmid variants and the probability of evolutionary rescue. We consider three generic types of dominance functions for plasmid mutations (i–iii) and gene dosage effects (iv). (A–D) Illustration of the relation between cell types and their Malthusian fitness. (E–H) Establishment probabilities of a plasmid mutation arising on a single plasmid copy. (I–L) Probability of evolutionary rescue from de novo mutations. Without gene dosage effects, the establishment probability decreases with the plasmid copy number. Yet, the probability of evolutionary rescue may still increase with n if the decline in the establishment probability is compensated by the increased mutational supply. Results for the establishment probabilities were obtained numerically by using Equation (A.5) with Equation (A.1). These were inserted into Equation (3) to obtain the probability of evolutionary rescue. Parameters: s0 = −0.1, Open circles show averages over 104 stochastic simulations. Error bars indicate twofold SE.
For n = 1, the fate of the mutant plasmid is determined entirely by the birth and death dynamics of the mutant cell. In contrast, for higher copy plasmid numbers, segregation of mutant plasmids during cell division of heterozygous cells leads in general to one daughter cell with a lower number of mutant plasmids, stretching the establishment process of the mutant plasmid over a larger number of cell generations. If the fitness of mutant homozygotes is independent of n [cases (i)–(iii)], establishment of the mutant plasmid therefore becomes less likely with increasing copy number n irrespective of the concrete dominance function (Figure 2, E–G). How strongly the establishment probability drops with n, however, depends on the dominance function, which we illustrate by a set of numerical examples. While strictly speaking, we only describe results for the concrete parameter sets and do not give a general proof, nothing about the chosen examples is special, and they are most likely representative. In Supplemental Material, Figure S3.1, we show results for several other parameter sets, observing the same behaviors.
For dominant mutations [case (i)], cells only need to carry one mutant plasmid to be resistant. Formally, dominant mutations are defined by for i > 0. Then, decreases only moderately with n and eventually converges to a constant (Figure 2E). We find that the increase in the mutational supply with higher copy numbers exceeds the decrease in the establishment probability, such that the probability of evolutionary rescue increases with the plasmid copy number (Figure 2I).
For recessive plasmid mutations [case (ii)], only cells where all plasmid copies are mutated are resistant. Cells with a heterozygous plasmid composition are, just as wild-type cells, fully susceptible ( for i < n). Establishment probabilities for this plasmid type fall sharply with increasing plasmid copy numbers to very low values (Figure 2F). Since heterozygous cells are susceptible to the antibiotic and are less likely to divide than to die, it is unlikely that a resistant homozygous mutant cell establishes if many cell divisions are required to generate it. Unlike for dominant mutations, rescue probabilities decrease with plasmid copy number (Figure 2J). The drop in the establishment probability is so dramatic that it cannot be compensated for by the higher supply of new mutations for higher plasmid copy numbers.
For intermediate dominance [case (iii)], we assume a Malthusian fitness that increases linearly with the relative number of mutant plasmids, i.e., The behavior of lies between the two extremes of dominant and recessive mutations (Figure 2G). If the fitness of homozygous mutant cells smax is large, we observe a slight maximum in the rescue probability as a function of the plasmid copy number (Figure 2K). This is the result of the antagonistic effects of the plasmid copy number on the establishment probability (decrease with n) and on the mutational input (increase with n). This maximum is shifted to higher plasmid copy numbers with increasing smax (and with increasing s0, see Figure S3.1). If the fitness benefit of the mutation is small, Prescue decreases monotonically with n, which is consistent with the comparison of the rescue probabilities for n = 1 and n = 2 in the previous section.
Finally, we discuss the dependence of establishment and rescue probabilities on the plasmid copy number for mutations with a gene dosage effect [case (iv)]. In this case, fitness of a bacterium depends on the absolute number of mutated plasmids within a cell, and cells with different plasmid copy numbers need the same absolute number of mutated plasmid copies to become resistant. Here, we implement a linear increase of the fitness with n, i.e., In that case, we find that the establishment probability increases with the plasmid copy number since the negative effect of segregational drift on establishment of the mutant plasmid is outweighed by the higher fitness of mutant homozygotes (Figure 2H). In the limit of high copy numbers, the establishment probability reaches a constant value. If the increase in fitness per beneficial plasmid s is small and the plasmid copy number low, mutant homozygotes do not have a positive Malthusian fitness, and the establishment probability of the mutant plasmid is hence zero (see inset in Figure 2H). Obviously, since both the mutational input and increase with n under the implemented gene dosage effects, rescue is more likely for high than for low copy number plasmids (Figure 2L).
In the Supplementary information, section S1, we show the rescue probabilities under the two alternative models of plasmid replication and segregation, and compare the results with those presented here. General trends are robust. Yet, especially for recessive mutations, differences appear. The decline of Prescue with n is much less strong under the alternative schemes, and low but intermediate copy numbers may even be best (though only by a very slight margin) for the bacterial population.
Example: the influence of antibiotic concentration
So far, we have fixed the relationship between the plasmid composition of a cell and its fitness. Yet, this relationship derives from the specific resistance mechanism, and likely also from the environmental conditions. Here, we use a simple model to demonstrate how the above formalism can be applied to concrete situations of resistance evolution. Using a basic mechanistic model for cell resistance, we establish how the dominance function depends on the external antibiotic concentration to which the bacterial population is exposed. We then determine the concentration-dependent probability of evolutionary rescue for a range of plasmid copy numbers.
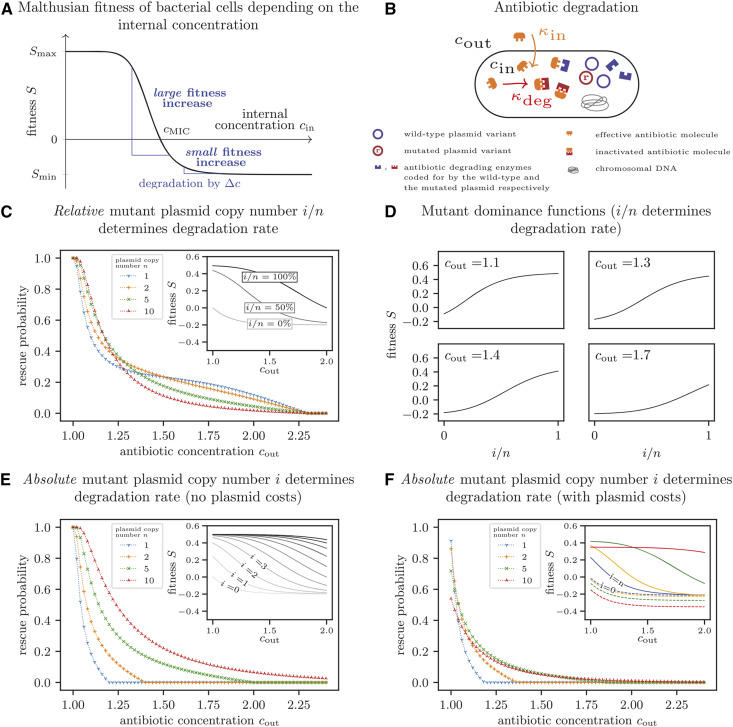
We assume that the fitness of a bacterial cell depends on the antibiotic concentration, cin, within the cell, and take the corresponding dose-response curve, S(cin), as given [Figure 3A and Equation (B.1)]. For the wild type, the internal concentration cin equals the antibiotic concentration in the environment cout. A decrease in the internal concentration occurs through enzymes that degrade the antibiotic within the cell [Equation (B.3)]. The corresponding gene is located on the plasmid and carries a mutation in the mutant plasmid. The mutation does not impose a fitness cost. While both plasmid variants, wild-type and mutant, code for the enzyme, only the mutated version can degrade the specific antibiotic (Figure 3B). The rate of antibiotic degradation increases with the number of mutated plasmids within the bacterial cell. Consequently, the internal concentration decreases with i, and bacterial fitness increases, as determined by the shape of the dose response curve (Figure 3A). Details on the model can be found in Appendix B.
Figure 3.
Dependence of the optimal plasmid copy number on the antibiotic concentration. (A) Malthusian fitness S of a bacterial cell as a function of the antibiotic concentration within the cell. (B) Illustration of a simple model to determine the antibiotic concentration within the cell. Antibiotic molecules diffuse into the cell, where they become inactivated due to reactions with the mutant version of a plasmid-encoded enzyme. We assume that inflow of antibiotic molecules kin and degradation kdeg are in equilibrium. (C) Probability of evolutionary rescue if the relative number of mutant plasmids within the cell determines the degradation rate. The enzyme production increases linearly with the relative number of mutated plasmid copies, and the cell fitness is thus determined by the fraction i/n of mutated plasmids within the cell (inset). (D) Mutant dominance functions for various antibiotic concentrations if the relative number of mutant plasmids i/n within a cell determines the degradation rate. With increasing concentration, the shape changes from concave to convex. (E) Probability of evolutionary rescue if the absolute number of mutant plasmids within the cell determines the degradation rate. The enzyme production increases linearly with the absolute number of mutated plasmid copies, corresponding to a gene dosage effect (see inset for the concentration-dependent fitness of each cell type). Rescue probabilities increase with higher plasmid copy numbers. (F) Probability of evolutionary rescue if the absolute number of mutant plasmids within the cell determines the degradation rate and plasmids impose a copy-number-dependent cost. Unlike in (E), plasmid costs increase bacterial death rates by c = 0.015 per plasmid copy. For very low antibiotic concentrations, the probability is lowest for high copy numbers due to the high associated costs. For high concentrations, the disadvantage of a higher burden is outweighed by the advantage of more efficient antibiotic degradation, and the rescue probability increases with the copy number. In the inset of (F), dashed and solid lines show the fitness of wild-type and mutant homozygous cells, respectively. Results for the rescue probabilities were obtained as in Figure 2. The fitnesses are calculated from Equation (B.1) and Equation (B.7) with either Equation (B.4) (for C and D) or Equation (B.5) (for E and F). Parameters:
We distinguish two limiting cases. In the first case, we assume that the resources or molecules (e.g., ribosomes) needed for the production of the degrading enzyme are limited, and a single plasmid copy is enough to obtain the maximum possible amount of enzyme. The limiting factor that determines the rate of mutant enzyme production is not the number of mutated plasmid copies but the available resources for which wild-type and mutant plasmids are competing. In that case, the rate of mutated enzyme production, and, thus, the antibiotic degradation rate depend on the relative number of mutated plasmids i/n per cell (see inset in Figure 3C for the fitness of different cell types as a function of cout). Concretely, we assume that the enzyme production increases linearly with i/n [Equation (B.4)]. We find that the optimal plasmid copy number n changes with the antibiotic concentration (Figure 3C). The reason is that the dominance function changes along the antibiotic gradient (Figure 3D). For low concentrations, a small change in the internal antibiotic concentration (i/n small) leads to a large fitness increase (Figure 3A) such that the mutation has a concave dominance function (cout = 1.1 in Figure 3D). This resembles the dominant mutation from the previous section. For these concentrations, a high plasmid copy number maximizes the rescue probability. For high concentrations, in contrast, a high fraction of mutant plasmids is required to achieve an appreciable increase in fitness (Figure 3A). Then, the dominance function is convex, resembling a recessive mutation (cout = 1.6 in Figure 3D). In this regime, a low copy number leads to the highest probability of evolutionary rescue. For intermediate concentrations, the dominance function has an inflection point and an intermediate copy number is most advantageous for the bacterial population (cout = 1.3 and cout = 1.4 in Figure 3D).
In the second case that we consider, the enzyme production and the degradation rate depend on the absolute number of mutated plasmids i, i.e., it is not the resources needed for enzyme production that are the limiting factor but the number of mutant plasmids. This reflects a gene dosage effect. We assume here that the enzyme production increases linearly with i [Equation (B.5)]. The Malthusian fitness of cell types with mutated plasmid copies as a function of the external antibiotic concentration is shown in the inset of Figure 3E, assuming, as in the previous section, that plasmids do not impose a fitness cost. At all concentrations, rescue is more likely for higher n (Figure 3E).
This simple picture changes if plasmids have a high fitness cost, which increases the death rate by c per copy number in our model. In that case, the fitness of wild-type cells decreases with the plasmid copy number, and, even for mutant homozygotes, a higher copy number does not always imply a higher fitness (see the inset of Figure 3F, where dashed lines indicate the wild type and solid lines the mutant homozygote). In contrast to the results without plasmid costs (Figure 3E) where the highest copy number always led to the highest chance of evolutionary rescue, the optimal copy number now depends on the antibiotic concentration. For low concentrations, the high burden imposed by a high copy number leads to a low probability of rescue. However, for higher concentrations, where efficient antibiotic degradation is important, rescue is more likely with higher plasmid copy numbers even though carrying plasmids is costly.
The mutant frequency in the standing genetic variation
So far, we have focused on evolutionary rescue from de novo mutations that arise during the decline of the bacterial population after the change in the environment, i.e., in the presence of antibiotics. Yet, mutated plasmids may be present in the population prior to environmental change. Here, we extend our model to account for rescue from the standing genetic variation. We assume that mutant plasmids that confer resistance in the antibiotic environment are disadvantageous in the absence of antibiotics and segregate in mutation-selection balance in the population. We denote by −σ the selection coefficient for mutant homozygous cells for mutations, where the relative proportion of mutated plasmids determines the cell fitness, and by −σGDE the deleterious effect per plasmid copy if there is a gene dosage effect. To determine the frequency of mutant plasmids in the absence of antibiotics, we adapt a multitype Moran model that describes the dynamics of the various cell types in a bacterial population of constant size where Ni denotes the number of cells with i mutated plasmid copies. From the Moran model, we derive a set of ordinary differential equations, from which we numerically obtain the numbers of mutant homozygous and heterozygous cells in deterministic mutation-selection equilibrium. From those, we estimate the contribution of the standing genetic variation to the probability of evolutionary rescue. Since the population faces ultimate extinction if all resistance mutations become extinct, the rescue probability can be approximated by
| (11) |
where Qi denotes the probability that a cell of type i does not leave a long-term lineage of descendants. A detailed description of the model is given in Appendix C. The deterministic approach assumes an infinite population size and neglects stochastic fluctuations in Ni. We therefore additionally perform simulations of the stochastic model to account for finite populations.
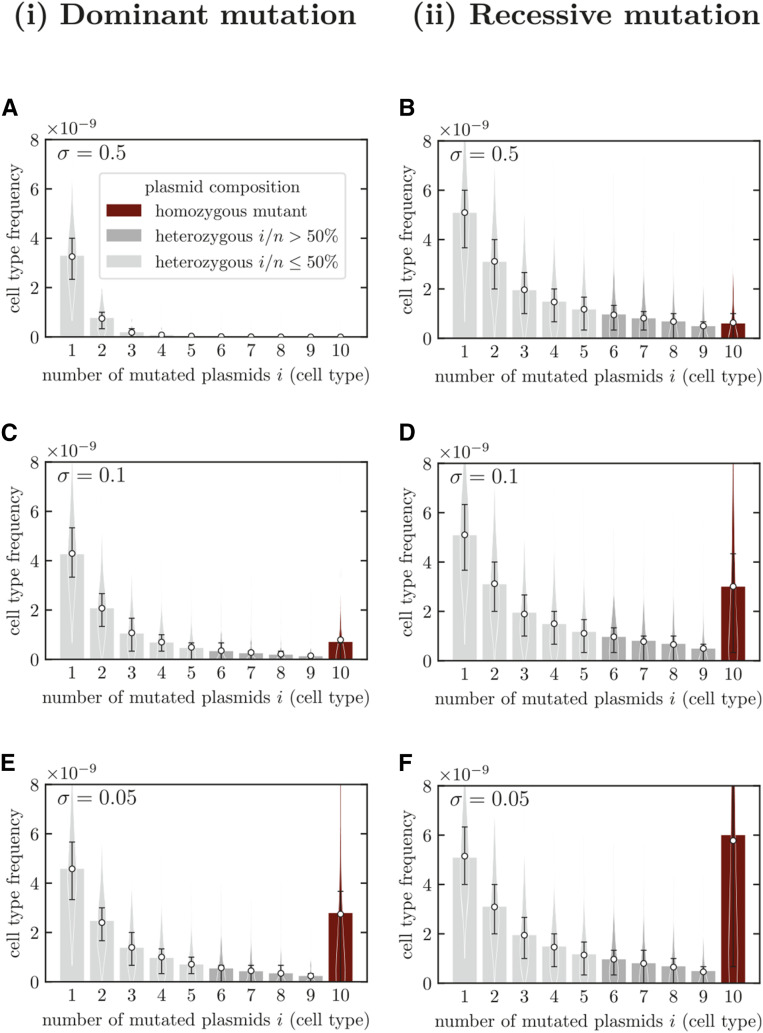
Before considering rescue probabilities, we briefly discuss the distribution of the frequencies of the various cell types in the standing genetic variation, comparing dominant and recessive mutations. Figure 4 shows, in an exemplary manner for a bacterial population with n = 10 plasmid copies per cell, the expected frequencies for different strengths of selection against the mutant plasmid. As expected, the frequency of cells containing mutated plasmids is higher for a recessive than for a dominant mutation, since in that case the mutation is exposed to selection only if it occurs in a mutant homozygous cell (cf. also Figure 5, A and B). Since cells with a heterozygous plasmid composition are not subject to selection for recessive mutations, their frequencies do not visibly change with the strength of selection. The frequency of homozygous mutant cells, however, increases strongly with decreasing purifying selection, and mutant homozygotes become the most frequent mutant class for weak selection. Unlike heterozygous cell types, mutant homozygotes have only daughter cells of their own type (up to mutation), allowing them to rise to higher frequencies than heterozygous cells if selection is weak. For weak selection against the mutant plasmid, the difference between recessive and dominant mutations shows mainly in classes with high numbers i of mutant plasmids and most prominently in the number of mutant homozygotes. Generally, the variance in the cell frequencies is high (see violin plots in Figure 4 and Figure S4.2). We will discuss consequences of this high variance below.
Figure 4.
Distribution of mutant cell type frequencies in the standing genetic variation for dominant and recessive mutations. Mutant plasmids are deleterious prior to antibiotic treatment with a selection coefficient of σ (see Appendix C). Bar plots show frequencies obtained from the equilibrium of the deterministic model, i.e., by numerically integrating Equations (C.5), (C.6) and (C.7). Violin plots show frequencies obtained by 103 stochastic simulations. Open markers and error bars show the mean and the interquartile range respectively. Parameters: n = 10, Figure S4.2 shows the same plot as in F with the full extent of the interquartile range.
Figure 5.
Dependence of mutant frequencies on the dominance function and the influence of standing genetic variation on the probability of evolutionary rescue. (A–D) Deterministic frequencies of mutant cells for a bacterial population in the standing genetic variation (SGV) for a range of plasmid copy numbers. Mutant plasmids are deleterious with a selection coefficient of σ = 0.1 (dominant, recessive, intermediate). For the gene dosage effect (iv), each mutant plasmid decreases the selective fitness of its host cell by σGDE = 0.01. (E–H) Probabilities of evolutionary rescue from SGV for those bacterial populations in an antibiotic environment. Closed markers show the approximation using deterministic frequencies for the cell type frequencies in the SGV. Open markers show averages over 103 stochastic simulations. Error bars indicate twofold SE. (I–L) Total probabilities of evolutionary rescue, including rescue from SGV and rescue from de novo mutations. For comparison, small markers indicate rescue probabilities from de novo mutations only. After the switch to the antibiotic environment, wild-type cells have a birth rate The fitness of resistant bacteria is increased to for (i–iii) and by s per mutated plasmid for (iv) (analogous to Figure 2). Frequencies (A–D) are obtained from the equilibrium of the deterministic model, i.e., by numerically integrating Equations (C.5), (C.6) and (C.7). Rescue probabilities from SGV (E–H) are obtained from Equation (11) and Equation (A.5). Total rescue probabilities including de novo mutations (E–H) are obtained from Equation (12), which is based on the deterministic frequencies for rescue from SGV. Other parameters:
As for rescue from de novo mutations, we are interested in how the probability of rescue from the standing genetic variation depends on the plasmid copy number n. For this, we assume that the dominance function remains the same upon the environmental change, e.g., mutations that are dominant prior to the change are also dominant afterward. Likewise, gene dosage effects persist, such that a mutant homozygous cell is particularly fit in the new environment but particularly unfit in the old environment.
Figure 5 shows the expected cell type frequencies, the probability of rescue from the standing genetic variation, and the total rescue probability as a function of the plasmid copy number. Both for dominant and for recessive mutations, as well as for mutations of intermediate dominance, the expected overall frequency of mutant cells increases with the plasmid copy number n due to the increased mutational supply (Figure 5, A–C). The effect is particularly strong for recessive mutations, where the deleterious mutation is masked in heterozygous cells. For recessive mutations, we moreover observe that the frequency of mutant homozygotes Nn is (nearly) independent of the plasmid copy number (Figure 5B). Generally for high n, irrespective of the dominance function, most cells harbor only few (<50%) mutated plasmids. Especially for recessive mutations, or those with intermediate dominance, these cells are not very likely to leave a long-term lineage of descendants after the environmental change. Therefore, the probability of evolutionary rescue increases only slightly (if at all) with n (Figure 5, E–G). If there is a gene dosage effect, i.e., if selection against the mutation increases with the absolute number of mutant plasmids, there is a minimum in the frequency of mutant cells for intermediate plasmid copy numbers n, stemming from the antagonistic effects of the plasmid copy number on the mutational supply and on the strength of selection against the mutant plasmid (Figure 5D). Yet, since the establishment probability strongly increases with n, rescue from the standing genetic variation still increases with plasmid copy number for the chosen parameter set (Figure 5H). Further analysis shows that, if the mutation is highly beneficial after the environmental change, a slight minimum in as a function of n may exist (e.g., for λ0 = 0.95, σGDE = 0.1, s = 2, N = 3 × 108, u = 3 × 10−10).
Unless the selective advantage of the mutant plasmid is very weak after the environmental change, the deterministic approximation for Ni leads to an overestimation of the rescue probabilities from the standing genetic variation (see Figure 5E for and Figure 5H for ). The error gets larger for smaller population sizes (Figure S4.1). The reason for the deviation between the analytical theory and the stochastic simulations is the large variance in Ni. Since is strongly concave in Ni for strong selection, the negative and positive effects of fluctuations in Ni below or above the average do not cancel. Due to these stochastic effects, in Figure 5F (recessive mutation), the probability of evolutionary rescue decreases for high copy numbers in contrast to the deterministic approximation, which predicts a monotonic increase. It seems plausible that, for high n, stochastic fluctuations in the numbers of cells with high i are stronger, causing this decrease. To probe this intuition, in Appendix S4, we investigate this behavior in some more detail for recessive mutations and a lethal wild type in the new environment. In that case, rescue relies entirely on mutant homozygotes in the standing genetic variation. Prescue then decreases with n if stochasticity in Nn is taken into account but remains constant under the deterministic approximation of Nn (Figure S4.3). Nevertheless, in most cases, the trend of is correctly estimated based on the deterministic approximation of the cell type frequencies in the standing genetic variation.
Finally, we combined rescue from the standing genetic variation and from de novo mutations to determine the total probability of evolutionary rescue (Figure 5, I–L):
| (12) |
For mutations where the relative number of mutant plasmids determines the cell fitness [cases (i)–(iii)], the effect of the dominance function on total rescue probabilities is much weaker than its effect on rescue by de novo mutations alone. In all three cases, rather similar total rescue probabilities are observed.
Discussion
Adaptation depends fundamentally on two factors, the presence of adaptive alleles and their probability to escape stochastic loss while rare. For plasmid-carried genes, the plasmid copy number influences both of these factors. On one hand, the mutational target size increases with the number of plasmid copies per cell. On the other hand, unlike for alleles on haploid genomes, it takes several cell divisions until a cell with a large fraction of mutant plasmids is generated, negatively affecting the mutant plasmid’s chances to establish. We here developed a framework to model bacterial adaptation driven by novel alleles on multicopy plasmids. We determined the probability of evolutionary rescue, taking into account new mutations and mutations from the standing genetic variation.
The influence of the dominance function
It is no surprise that the dominance of the mutant allele is key for whether a low or a high copy number maximizes the probability of evolutionary rescue. We studied examples of dominant and recessive mutations and mutations of intermediate dominance and mutations in genes with a gene dosage effect, always assuming that bacterial fitness is increasing (or at least nondecreasing) with the number of mutated plasmid copies. Only with a gene dosage effect do both the mutational input and the establishment probability increase with plasmid copy number, while in all other cases, the effects are antagonistic. That is, unless there are gene dosage effects, the plasmid copy number augments the mutational input but decreases the establishment probability. We study the outcome of this interplay of effects by means of numerical examples, which—while being no universal proof—indicate trends that likely hold for the entire biologically relevant range of parameters. We observe that, for dominant mutations, the benefits of a high per-cell mutation rate outweigh the negative effects on the establishment probability such that the rescue probability increases with plasmid copy number. This holds true both for de novo rescue and for rescue from the standing genetic variation. For recessive mutations, there is a phenotypic delay of several generations until the allele can be picked up by selection. The establishment probability therefore drops much more strongly with plasmid copy number than for dominant mutations. In that case, the probability of de novo rescue decreases with n. On the other hand, in an environment where the allele is deleterious, its effect is masked in all heterozygous cells, leading to a high number of mutant cells in the standing genetic variation. In a deterministic analysis of the standing genetic variation, this leads to an increase in the probability of rescue from the standing genetic variation with n, even for recessive mutations (for consequences of stochasticity see below). In sum, for recessive mutations, the total probability of rescue remains constant as n increases.
These results are in line with the findings of Sun et al. (2018) for a model of bacterial adaptation on the chromosome. While bacteria are usually treated as haploid, making dominance inconsequential, Sun et al. (2018) take into account that bacterial chromosomes can be polyploid during growth, such that the dominance of mutations strongly affects the adaptive process. The segregation mechanism is different from the one in the present model, since the chromosome copies are not distributed randomly to the daughter cells at cell division. Rather, all chromosomes carrying the mutation go into the same daughter cell such that homozygosity is reached in the smallest possible number of steps. That both models make consistent predictions shows that the results are (at least to some extent) robust to the segregation mechanism (but see also the discussion further below). Indeed, we study two alternative models for plasmid replication and segregation, and these mostly confirm the conclusions except for a very slight maximum in the rescue probability for low copy numbers if the mutation is recessive. In these alternative models, we assume that plasmid copies are chosen at random for replication rather than each copy being replicated exactly once (see Model). Therefore, homozygous mutant cells can arise more quickly than in our original model. The negative effect of a high copy number on establishment of the mutant plasmid is therefore less strong, leading—in the interplay with the copy-number-dependent mutational input—to the observed slight maximum in Prescue for recessive mutations.
Based on a deterministic analysis of the cell numbers in the standing genetic variation, both the present study and that of Sun et al. (2018) find that the mean number of mutant homozygous cells is independent of the plasmid copy number for recessive mutations. Stochastic simulations confirm that this holds for their mean number. Yet, the variance is large and increases with n. Stochasticity in genotype frequencies prior to environmental change affects adaptation from the standing genetic variation, since there is, on the one hand, a chance that only a few (or even no) beneficial mutants are present, and, on the other, the possibility that there are more than needed for successful adaptation [see, for example, also Figure 2 in Hermisson and Pennings (2005) and Figure 3A in Hermisson and Pennings (2017)]. Therefore, the probability of rescue from the standing genetic variation starts to decrease with large copy numbers—an effect that was not observed previously.
In most of our examples, we assume that the dominance function remains the same across environments. But, for diploid organisms, for which dominance has been studied more intensely, it has been found that the dominance coefficient can be different in different environments (Gerstein et al. 2014). Indeed, within a simple model of antibiotic degradation, we find that the dominance function is concentration—and hence environment—dependent.
Empirical dominance relationships
In this latter model, we derive the dominance relationship between mutated and wild-type plasmids mechanistically. In all other parts of the manuscript, however, we treat the dominance of mutations as a parameter. This raises the question about the nature of naturally occurring resistance determinants, and we here discuss a few examples that represent (partially) dominant or (partially) recessive mutations or resistance genes with gene dosage effects. Macrolide antibiotics such as erythromycin bind to the 23S rRNA, ultimately interfering with protein synthesis (Vester and Douthwaite 2001). Mutations in rrn genes, altering the ribosomal RNA, can confer resistance but these mutations have low dominance. Therefore, a sufficiently high fraction of gene copies need to be mutated to yield a resistant phenotype (Sigmund and Morgan 1982; Mark et al. 1983; Weisblum 1995; Sander et al. 1997; Vester and Douthwaite 2001). Consequently, bacterial species that carry only one or two copies of the respective gene on the chromosome such as Helicobacter pylori or Mycobacterium smegmatis can acquire resistance through mutations in rrn genes while this is not possible in E. coli that carries seven copies of the rrn operon (Sander et al. 1997; Vester and Douthwaite 2001). Yet, Sigmund and Morgan (1982) isolated mutations conferring erythromycin resistance in rrn genes on a multicopy plasmid and demonstrated that E. coli carrying the mutant plasmid were resistant. Interestingly, Sigmund and Morgan (1982) discuss the possibility of resistance evolution via this pathway (i.e., de novo mutation of a rrn gene on a multicopy plasmid, followed by segregation to reach a high fraction of mutated copies in the cell). In the examples in our article, we do not consider genes that are present both in the chromosome and on the plasmid, e.g., essential genes such as the rrn genes that are additionally carried on a plasmid. This would lead to a more complex function for the type-dependent bacterial fitness that would depend on the number of chromosomal gene copies. While we did not explicitly study such a case, it is straightforward to implement. For dominant mutations, mutations in gene copies on the chromosome or on the plasmid could lead to resistance, while recessive mutations would have no chance to establish. For mutations of intermediate dominance, the maximum possible fitness that can be achieved through mutations on plasmids would increase with the plasmid copy number (even without gene dosage effects). Therefore, the optimal plasmid copy number would probably be higher than in the absence of gene copies on the chromosome. While mutations in rrn genes are partially recessive, mutations in loci that affect methylation of rRNA have been shown to be dominant (Heiser and Davies 1972; Weisblum 1995; Vester and Douthwaite 2001). As another example, let us consider resistance to trimethoprim—an antibiotic that prevents DNA synthesis by binding to an enzyme (dihydrofolate reductase) in the DNA synthesis pathway. Resistance to trimethoprim is conferred by a plasmid carrying a mutated drf gene that codes for a mutated enzyme to which the antibiotic cannot bind and therefore provides a bypass (Hall and Partridge 2004; van Hoek et al. 2011). This indicates at least a certain degree of dominance. Last, as already pointed out, some resistance genes display a gene dosage effect. This is especially true for genes coding for antibiotic-degrading enzymes such as β-lactamases (Martinez and Baquero 2000; San Millan et al. 2016). Notably, negative gene dosage effects also exist: it has been found that E. coli displays lower levels of resistance to tetracycline if the resistance-conferring transposable element Tn10 is carried on a multicopy plasmid than if it is present in a single copy on the chromosome (Moyed et al. 1983). Note also that, in all the examples discussed here (except maybe for the last one), new resistance could occur through mutation in existing genes that can be plasmid-encoded and are hence relevant to the present study.
Plasmid replication and segregation
We developed a mathematical framework to describe the dynamics of multicopy plasmids that allows for a clear and easy separation of the mutational input and the establishment probability of mutant alleles. We apply this model across the entire range of copy numbers. Obviously, this neglects large parts of the complexity of plasmid dynamics. Here, we briefly discuss some of the assumptions.
In the model presented in the main text, we assume that plasmid replication is initiated immediately before the host cell divides. In reality, plasmids replicate throughout the cell cycle (Bogan et al. 2001; Nordström 2006). Fully accounting for this would require use of a full nested model that describes both the within-cell dynamics of plasmids and the birth–death dynamics of bacterial cells. This, in turn, would require knowledge of the period during the cell cycle during which the drug is effective, the number of plasmids that are present during that period, and the level of resistance they confer. Such a nested model would be appropriate to describe a specific system in detail but is less suitable to serve as a general model.
The second, more crucial assumption of our model of plasmid replication is that each plasmid copy reproduces exactly once. While (ideally) every plasmid replicates once per cell cycle on average, individual plasmid copies are picked for replication at random from the pool of all existing copies (Nordström 2006). This implies especially that a cell with more than one mutated plasmid copy can appear from one cell generation to the next. We therefore also tested alternative models where plasmid replication is carried out iteratively (see Supplemental information section S1). In these alternative models, we moreover inverted the order of plasmid replication and segregation. As discussed above, a comparison with the main text model showed that the trends in the probability of evolutionary rescue are mostly similar, independent of the assumed scheme of plasmid replication.
The other set of assumptions concerns the segregation of the plasmid copies to the daughter cells. For the main text model, we assume that each daughter cell receives the exact same number of plasmid copies. Most low-copy-number plasmids possess active partitioning systems that force plasmid copies within a bacterial cell to opposite directions, leading to two (or few) clusters and at least partially balancing plasmid numbers in both daughter cells (Million-Weaver and Camps 2014). High-copy-number plasmids do not possess any known active partitioning systems, although their existence has been discussed (Million-Weaver and Camps 2014; Wang 2017). Indeed, active partitioning systems are, in that case, less relevant, since, with increasing copy number, it becomes less and less likely that the plasmid becomes lost, even for a random distribution of plasmid copies to the daughter cells. Early models for the segregation of high-copy-number plasmids assume that the plasmids diffuse freely within the cell cytoplasm and are distributed randomly in space (Summers 1991). However, it was later found that high-copy-number plasmids form clusters within the cell (e.g., Pogliano et al. 2001). To explain the random distribution of high-copy-number plasmids to daughter cells despite clusters, it has been hypothesized that for replication, single plasmids become detached from the clusters and are transported to the mid-cell, where they are replicated; afterwards, the two sibling plasmids diffuse in space and attach to existing clusters or become the founders of new ones (Nordström and Gerdes 2003). Another (compatible) explanation is the observed existence of single plasmid copies in addition to clusters (Wang 2017). Assuming a random distribution of high-copy-number plasmids to daughter cells, the relative deviation from an equal share decreases with copy number. Overall, our assumption of an equal split of all plasmids seems to be a reasonable approximation for a baseline model. For one of the alternative models (no. 2) in the supplementary information, we have loosened the restriction of equally distributed plasmids.
The most critical assumption in all our model variants is the random distribution of wild-type and mutant plasmids at cell division. For high-copy-number plasmids, this assumption is generally considered as true (San Millan et al. 2016; Ilhan et al. 2019). Yet, for low-copy-number plasmids with active partitioning systems, it may be less well justified. The common view is that plasmids are recruited for replication to the center of the cell; afterwards, in the presence of active partitioning systems, the two replicates are pushed (or pulled) toward the two cell poles, thus separating them such that, at cell division, they will end up one in each of the daughter cells (Nordström and Gerdes 2003). Hence, in this case, our assumption might distort the chance of resistance evolution. If mutations are recessive, for example, a random distribution allows for an efficient accumulation of mutated plasmids, which is needed to generate phenotypically resistant cells. With active partitioning of individual plasmid clones, homozygous mutant cells (resistant) are generated at a much lower rate or—in the absence of errors—in our main text model with replication of each plasmid copy not at all, leading to a much lower chance of resistance evolution. The opposite effect happens for dominant mutations, leading to a higher chance of resistance evolution due to the active partitioning of mutated plasmids to both daughter cells (cf. also Markov and Kaznacheev 2016). While such a deterministic partitioning of replicate plasmids is often proposed, there is also some evidence that partitioning could be more random. First, different plasmid types with the same partitioning system are not stably inherited together. To explain this, it has been suggested that there is a time delay between plasmid replication and partitioning, during which heterologous plasmid pairs (i.e., pairs that consist of one copy of each type) may be formed in the cell center; these two plasmids (rather than replicate plasmid) are then partitioned into the two daughter cells (Nordström 2006). For our model, heterozygous plasmid pairs (consisting of a wild-type and a mutant plasmid) would correspond to the heterologous plasmid pairs of two different plasmid types. Second, while in some cases, immobilization of plasmid clusters has been observed (Derman et al. 2008), others describe a much more chaotic behavior, in which plasmids (or plasmid clusters) diffuse in the cell and are, over the course of the cell cycle, repeatedly repelled by filaments that dynamically form and dissolve (Campbell and Mullins 2007; Sengupta et al. 2010). This would presumably lead to a mixup of plasmid replicates within the cell. Interestingly, based on this observation, Anand and Khan (2010) explicitly point to the “possibility of random assortment of daughter plasmids (sisters and nonsisters) during cell division”. In that case, our assumption of random segregation of wild-type and mutant plasmids would be reasonable.
As a last remark, we would like to stress that a copy number of one (or two, three…) is usually meant to correspond to one plasmid copy per genome equivalent, and, often, more than one copy is present in the cell even for single-copy plasmids. We ignore this in our model in the same way as most models for adaptation on the bacterial chromosome assume that bacteria are strictly haploid.
Further limitations and extensions
As a caveat, we would like to stress that, for the most part, we ignored plasmid costs in this manuscript. The aim was to disentangle the immediate consequences of plasmid copy number on dynamics without conflating factors. The burden imposed by plasmids on their bacterial host is expected to increase with plasmid copy number, and, at some point, outweighs the benefits. Hence, for dominant mutations, where we found rescue probabilities to increase with plasmid copy number, we can conclude that, in the presence of plasmid costs, an intermediate number of plasmids is most beneficial. The same holds true for adaptation in genes with a gene dosage effect, since the positive effect of a larger number of plasmids in reality saturates (unlike in our model, which approximates the fitness benefits of additional plasmid copies by a linear function). However, it is important to note that plasmid costs are often alleviated by compensatory mutations such that they can be very low, undetectable, or even absent (although it is hard to imagine that they could be zero with an ever increasing copy number); see e.g., San Millan et al. (2014), Harrison et al. (2015), Loftie-Eaton et al. (2017).
In our model of evolutionary rescue, we considered only two environments and an abrupt switch between them. Yet, in the context of resistance evolution, it would be highly relevant to include the pharmacokinetics of the drug and to study effects of treatment factors, such as the frequency of drug administration, on resistance evolution. While the general approach is suitable to deal with more complex scenarios, the mathematical analysis is restricted to constant environments. Considering time-varying drug pressures would require to derive time-dependent establishment probabilities based on time-inhomogeneous branching processes.
Importantly, we assumed that the plasmids are nontransmissible. For transmissible plasmids, horizontal gene transfer could potentially have a non-negligible effect on the establishment of mutated plasmid copies. Conjugative plasmids usually have rather low copy numbers (<10) since the genes required for conjugation are costly (Norman et al. 2009). Transmissible plasmids with higher copy numbers could still cotransfer with other plasmids or phages (Eberhard 1990; Barry et al. 2019). Horizontal transfer is relevant mainly if the population consists of plasmid-carrying and plasmid-free cells since conjugative transfer of plasmids into cells that already contain plasmids of the same incompatibility group (i.e., especially copies of its own type) is unstable or is even prohibited through surface exclusion proteins (Eberhard 1990).
Last, throughout the article, we assumed that novel beneficial alleles arise by mutation. If, instead, genes are picked up by transformation and integrated into one of the plasmid copies, the rate of acquisition of beneficial traits is independent of the copy number. Hence, it is only the establishment probability that determines which copy number maximizes the probability of rescue. From our results for the establishment probability, it is straightforward to see that, in that case, rescue becomes less likely with increasing copy numbers unless there are gene dosage effects. This result is supported also by simulations performed by Ilhan et al. (2019) comparing the fixation dynamics of beneficial alleles on multicopy plasmids. In their study, the proportions of populations where the beneficial plasmid allele gets lost by random fluctuations is higher for 100 plasmids per cell compared to 10 plasmids per cell given the total number of mutated plasmids is the same at the beginning (compare the results for p10 × 105, copy number 10; population of 105 cells—with those for p100 × 105, copy number 100; population of 105 cells—in Figure 3 in Ilhan et al. (2019)).
Conclusion
Multicopy plasmids are pervasive in bacteria, influencing their ecology and evolution. To capture the population genetics of bacteria, theory therefore needs to account for the contribution of plasmids that are present in the bacterial cell in several copies. Such theory needs to describe the segregation process and take the—possibly environment-dependent—dominance of plasmid-carried alleles into consideration. Here, we developed and analyzed a model for evolutionary rescue through mutations on multicopy plasmids. Our analysis provides insight into the factors governing the adaptive process and how they balance for various dominance functions and gene dosage effects. We find that the probability of evolutionary rescue may increase or decrease with plasmid copy number, depending on the nature of the mutation and the environment. Overall, the results demonstrate the relevance of the plasmid copy number for the dynamics of plasmid-carried alleles and for plasmid-mediated bacterial adaptation.
Acknowledgments
The authors thank Sebastian Bonhoeffer, Tal Dagan, and Anne Kupczok for helpful discussions, Alvaro San Millan for providing valuable insights into the biology of multicopy plasmids, Javier Lopez-Garrido for pointing out the existence of negative gene dosage effects, and two anonymous referees for many helpful comments. M.S. is a member of the International Max Planck Research School for Evolutionary Biology and gratefully acknowledges the benefits provided by the program.
Appendix A: Calculating the establishment probability from branching process theory
To determine the establishment probability of the resistance mutation, we use the mathematical theory of multitype branching processes. A “type” is determined by the plasmid composition of the cell. With a plasmid copy number of n, there are hence n + 1 types, since cells can contain 0, 1, …, n mutated plasmids. In the following, we speak of cells of type i if a cell contains i mutant plasmids. At cell division, a cell produces cells of types that may differ from its own. Only homozygous cells exclusively produce cells of their own types (up to mutations). As a consequence, unless the branching process goes extinct, a permanent lineage of mutant homozygotes will establish. We denote by the probability that a cell lineage that is founded by a single cell of type i goes extinct. Since the alternative to extinction is establishment of the resistance mutation, it holds
| (A.1) |
While our model is formulated in continuous time, we consider a discrete-time branching process. This is possible since within our framework, with time-independent birth and death rates, it does not matter for establishment or loss of the allele when cells divide but only if they divide. It is therefore sufficient to focus on whether the next event in the life of a cell is cell division or cell death. The respective probabilities of cell division and cell death for a cell of type i are given by and Given the cell divides, it produces cells of type k and with probability as defined in Equation (1). Transitions between types in the branching process therefore occur with probabilities Since the mutation probability u is small, we can neglect new mutations during the establishment process. We therefore set x = 0 in the branching process approximation.
Following these considerations, it is intuitive to derive a set of (coupled) equations for the probabilities The lineage founded by a single cell of type i goes extinct either if the cell dies or if the lineages founded by its two daughter cells both go extinct. Thus:
| (A.2) |
Note that since we ignore new mutations during the establishment process, we have
In the following, we provide a brief formal summary of how to determine the vector This follows standard theory on multitype branching processes, and we refer to textbooks for details (Mode 1971; Jagers 1975; Sewastjanow 1975; Haccou et al. 2005).
We denote the probability generating function of the multitype branching process by
where the vector components are given by
| (A.3) |
The vector is a dummy variable without biological meaning. The probability generating function can be used in two different ways to determine In our model analysis below, we will make use of both methods.
First, the vector Q(n) of extinction probabilities is a fixpoint of the probability generating function (e.g. Sewastjanow 1975), i.e.
| (A.4) |
This exactly corresponds to the set of equations (A.2). We use the fixpoint equation to obtain the extinction probabilities for n = 1 and n = 2.
Second, it holds that [Equation (7.4) in Mode (1971)]
| (A.5) |
We apply this to numerically obtain the extinction probabilities for n > 2.
From Equation (A.2), one immediately obtains for the establishment probability of a process that is initiated by a single mutant homozygote (i.e. a cell of type i = n):
| (A.6) |
Appendix B: Model linking the dominance function to the antibiotic concentration
In this section, we provide a detailed presentation of the model that connects the fitness of a specific cell type to the antibiotic concentration in the environment and that underlies Figure 3 in the main text.
For the bacterial response to a given antibiotic concentration within the cell, , we use a pharmacodynamic function (cf. Regoes et al. 2004). We assume that the net growth of a bacterial population in dependence of the internal antibiotic concentration is given by
| (B.1) |
The four parameters denote maximal and minimal net growth (Smax and Smin), the antibiotic concentration level cMIC where the net growth is zero and the Hill coefficient κ, which determines the slope of the curve. As we will see below, the concentration within a cell depends on its type, i.e., . We set the death rate for all n and i and the birth rate as
The rate of antibiotic inflow kin into the cell is proportional to the difference in the concentrations outside and inside the cell cout and cin, i.e.,
| (B.2) |
where γ denotes the diffusion coefficient.
Within the cell, the antibiotic can be degraded by enzymes that are encoded by the mutant plasmid. The degradation rate kdeg depends on the antibiotic concentration in the cell and the amount of degrading enzymes that can break antibiotic molecules and that depends on n and i,
| (B.3) |
In the main text, we consider two cases. In one case, the amount of available enzymes is proportional to the relative number of mutant plasmids,
| (B.4) |
In the other case, it is proportional to the absolute number of mutant plasmids,
| (B.5) |
We assume that inflow of antibiotic molecules into the cell and degradation of antibiotics are in equilibrium:
| (B.6) |
From this, we obtain the concentration within cells that carry i mutant and n − i wild-type plasmids, given an external concentration cout:
| (B.7) |
Substituting equation (B.7) in (B.1), we obtain the net growth as a function of the plasmid composition.
Appendix C: Modeling the cell type frequencies in the standing genetic variation
To obtain the cell type frequencies in the standing genetic variation, we set up a multitype Moran model that describes a bacterial population of constant size prior to antibiotic treatment. When a cell divides, the daughter cells replace the parental cell as well as another randomly chosen cell from the population. Wild-type cells replicate at rate . Since resistance mutations may cause a cost in the antibiotic-free environment, we assume that replication of cells carrying mutated plasmids occurs at a lower rate. We set for i > 0:
| (C.1) |
Mutation, plasmid replication, and plasmid segregation happen in the same way as described in the main text for the phase after the environmental change.
For the stochastic simulations, the model is implemented in a straightforward way. The initial population consists of wild-type cells. Transitions between states may happen when a cell of type i divides, another cell of type k is replaced and two daughter cells enter the population. We denote the types of the first daughter cell by j1 and of the second daughter cell by j2 [note that the cells are ordered now, unlike in Equation (1)]. The reaction can then be expressed by
| (C.2) |
where is the vector of the numbers of all cell types and denote unit vectors.
The rate of this reaction is given by
| (C.3) |
where denotes the probability that the first daughter cell of a type i cell is of type j1 and the second one of type j2:
| (C.4) |
We simulate this process, using the Gillespie algorithm (Gillespie 1976), which we implemented in the Python programming language. At simulation time tsim = 1000, the switch in the environment occurs and we continue the simulations according to the model dynamics in the stressful environment described in the main text to determine the probability of evolutionary rescue.In order to obtain the frequencies of the various cell types in the standing genetic variation, we record those at time tsim = 1000. Increasing the simulation time to tsim = 2000 did not change the results, showing that the simulation time tsim = 1000 chosen is large enough.
We next derive a set of ordinary differential equations that describes the dynamics of the system deterministically. As in the approximation for the establishment process, we consider only new mutations at replication of wild-type cells (i = 0). For other cells, the dynamics of mutant plasmids are governed by segregation, and new mutations can be neglected. Furthermore, within one cell division it is highly unlikely that more than one mutation occurs. Consequently, only the dynamics of cells with i = 0 or i = 1 mutated plasmids are affected by mutations. We approximate the probability that no mutation occurs by 1 − un, and the probability that one mutation occurs by un. From now on, we use the short-hand notation as given in Equation (1) (here, daughter cells are not ordered). Table C.1 summarizes all events that involve a cell of type i. Note that ; the probability of new mutations is accounted for by extra factors un and .
Table C.1. Reactions in the Moran model that involve cells of type i.
| Parental cell-type k | Daughter cell-types {j, j′} | Cell type of replaced cell l | Change in Ni | Rate of reaction |
|---|---|---|---|---|
| excluding k = 0, Ni = N0 or Ni = N1 | ||||
| k = i | l = i | 0 | ||
| k = i | l ≠ i | |||
| k = i | , | l = i | ||
| k = i | , | l ≠ i | ||
| k ≠ i | l = i | 0 | ||
| k ≠ i | l ≠ i | |||
| k ≠ i | , | l = i | ||
| k ≠ i | , | l ≠ i | 0 | |
| , | ||||
| k = 0 | l = 0 | 0 | ||
| k = 0 | l ≠ 0 | |||
| k = 0 | l = 0 | |||
| k = 0 | l ≠ 0 | 0 | ||
| , | ||||
| k = 0 | l = 1 | |||
| k = 0 | 0 | |||
| k = 0 | l = 1 | 0 | ||
| k = 0 | l ≠ 1 | |||
Note that it is not possible that a cell of type i has only one daughter cell of type i and one of another type with the exception of wild-type cells (i = 0) that may have daughter cells of type 0 and of type 1. Similarly, if the parent is of type k ≠ i, it is not possible that both daughter cells are of type i since there are 2k mutant plasmids before cell division.
Putting everything together, we obtain for the dynamics of cells of type i with :
| (C.5) |
For cells of type , we obtain:
| (C.6) |
where we used that . And for cells of type i = 1:
| (C.7) |
Approximating and , we obtain for the relative frequencies , in the standing genetic variation:
| (C.8) |
where δ denotes the Kronecker delta.
We numerically integrate the nonlinear system given by Equation (C.8) starting with x0 = 1 and xi = 0 for i > 0, using the solve_ivp method from the SciPy package in Python. The equilibrated frequencies ( for all i) are used to estimate the average number of cells of type i for a given population size
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12357731.
Communicating editor: J. Masel
Literature Cited
- Alexander H. K., and Bonhoeffer S., 2012. Pre-existence and emergence of drug resistance in a generalized model of intra-host viral dynamics. Epidemics 4: 187–202. 10.1016/j.epidem.2012.10.001 [DOI] [PubMed] [Google Scholar]
- Anand S. P., and Khan S. A, 2010. Plasmid segregation: Birds of a feather try not to flock together. J. Bacteriol. 192: 1171–1174. 10.1128/JB.01551-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anciaux Y., Chevin L.-M., Ronce O., and Martin G., 2018. Evolutionary rescue over a fitness landscape. Genetics 209: 265–279. 10.1534/genetics.118.300908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry K. E., Wailan A. M., Sheppard A. E., Crook D., Vegesan K. et al. , 2019. Don’t overlook the little guy: an evaluation of the frequency of small plasmids co-conjugating with larger carbapenemase gene containing plasmids. Plasmid 103: 1–8. 10.1016/j.plasmid.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Blazquez J., Morosini M.-I., Negri M.-C., Gonzalez-Leiza M., and Baquero F., 1995. Single amino-acid replacements at positions altered in naturally occurring extended-spectrum TEM β-lactamases. Antimicrob. Agents Chemother. 39: 145–149. 10.1128/AAC.39.1.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan J. A., Grimwade J. E., Thornton M., Zhou P., Denning G. D., and Helmstetter C. E., 2001. P1 and NR1 plasmid replication during the cell cycle of Escherichia coli. Plasmid 45: 200–208. 10.1006/plas.2000.1512 [DOI] [PubMed] [Google Scholar]
- Campbell C. S., and Mullins R. D., 2007. In vitro visualization of type II plasmid segregation: bacterial actin filaments pushing plasmids. J. Cell Biol. 179: 1059–1066. 10.1083/jcb.200708206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullum J., and Broda P., 1979. Rate of segregation due to plasmid incompatibility. Genet. Res. 33: 61–79. 10.1017/S0016672300018176 [DOI] [Google Scholar]
- Derman A. I., Lim-Fong G., and Pogliano J., 2008. Intracellular mobility of plasmid DNA is limited by the ParA family of partitioning systems. Mol. Microbiol. 67: 935–946. 10.1111/j.1365-2958.2007.06066.x [DOI] [PubMed] [Google Scholar]
- Eberhard W. G., 1990. Evolution in bacterial plasmids and levels of selection. Q. Rev. Biol. 65: 3–22. 10.1086/416582 [DOI] [PubMed] [Google Scholar]
- Eberhard W. G., 1989. Why do bacterial plasmids carry some genes and not others? Plasmid 21: 167–174. 10.1016/0147-619X(89)90040-1 [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1922. On the dominance ratio. Proc. R. Soc. Edinb. 42: 321–341. 10.1017/S0370164600023993 [DOI] [Google Scholar]
- Friehs K., 2004. Plasmid copy number and plasmid stability, pp. 47–82 in New Trends and Developments in Biochemical Engineering, edited by Scheper T. Springer, Berlin, Heidelberg: 10.1007/b12440 [DOI] [PubMed] [Google Scholar]
- Galassi M., Davies J., Theiler J., Gough B., Jungman G. et al. , 2009. GNU Scientific Library Reference Manual, Ed. 3rd Network Theory Ltd., Bristol, UK. [Google Scholar]
- Gerstein A. C., Kumzin A. and Otto S. P., 2014. Loss of heterozygosity facilitates passage through Haldanes sieve for Saccharomyces cerevisiae undergoing adaptation. Nat. Commun. 5: 3819 10.1038/ncomms4819 [DOI] [PubMed] [Google Scholar]
- Gillespie D. T., 1976. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. of Computational Physics, 22: 403–434. [Google Scholar]
- Haccou P., Jagers P., and Vatutin V. A., 2005. Branching Processes: Variation, Growth, and Extinction of Populations. Cambridge University Press, Cambridge, England: 10.1017/CBO9780511629136 [DOI] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. v. selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. 10.1017/S0305004100015644 [DOI] [Google Scholar]
- Hall R., and Partridge S., 2004. Evolution of multiple antibiotic resistance by acquisition of genes, pp. 37–51 in Antibiotic Development and Resistance, edited by Hughes D., and Andersson D. I.. Taylor and Francis, New York. [Google Scholar]
- Halleran A. D., Flores-Bautista E., and Murray R. M., 2019. Quantitative characterization of random partitioning in the evolution of plasmid-encoded traits. bioRxiv (Preprint posted March 31, 2019). 10.1101/594879 [DOI] [Google Scholar]
- Harrison E., Guymer D., Spiers A. J., Paterson S., and Brockhurst M. A., 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25: 2034–2039. 10.1016/j.cub.2015.06.024 [DOI] [PubMed] [Google Scholar]
- Heiser T. L., and Davies J. E., 1972. Mechanism of Kasugamycin resistance in Escherichia coli. Nat. New Biol. 235: 6–9. 10.1038/newbio235006a0 [DOI] [PubMed] [Google Scholar]
- Hermisson J., and Pennings P. S., 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. 10.1534/genetics.104.036947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J., and Pennings P. S., 2017. Soft sweeps and beyond: understanding the patterns and probabilities of selection footprints under rapid adaptation. Methods Ecol. Evol. 8: 700–716. 10.1111/2041-210X.12808 [DOI] [Google Scholar]
- Ilhan J., Kupczok A., Woehle C., Wein T., Nils F Hülter P. R. et al. , 2019. Segregational drift and the interplay between plasmid copy number and evolvability. Mol. Biol. Evol. 36: 472–486. 10.1093/molbev/msy225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Hashimoto-Gotoh T., and Matsubara K., 1978. Random replication and random assortment model for plasmid incompatibility in bacteria. Plasmid 1: 435–445. 10.1016/0147-619x(78)90002-1 [DOI] [PubMed] [Google Scholar]
- Jagers P., 1975. Branching Processes With Biological Applications, John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- Loftie-Eaton W., Bashford K., Quinn H., Dong K., Millstein J. et al. , 2017. Compensatory mutations improve general permissiveness to antibiotic resistance plasmid. Nat. Ecol. Evol. 1: 1354–1363. 10.1038/s41559-017-0243-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean R. C., and San Millan A., 2015. Microbial Evolution: towards resolving the plasmid paradox. Curr. Biol. 25: R764–R767. 10.1016/j.cub.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Mark L. G., Sigmund C. D., and Morgan E. A., 1983. Spectinomycin resistance due to a mutation in an rrna operon of Escherichia coli. J. Bacteriol. 155: 989–994. 10.1128/JB.155.3.989-994.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov A. V., and Kaznacheev I. S., 2016. Evolutionary consequences of polyploidy in prokaryotes and the origin of mitosis and meiosis. Biol. Direct 11: 28 10.1186/s13062-016-0131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Aguilée R., Ramsayer J., Kaltz O., and Ronce O., 2013. The probability of evolutionary rescue: towards a quantitative comparison between theory and evolution experiments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120088 10.1098/rstb.2012.0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L., and Baquero F., 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44: 1771–1777. 10.1128/AAC.44.7.1771-1777.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million-Weaver S., and Camps M., 2014. Mechanisms of plasmid segregation: have multicopy plasmids been overlooked? Plasmid 75: 27–36. 10.1016/j.plasmid.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mode C. J., 1971. Multitype Branching Processes: Theory and Applications, American Elsevier Publishing Company, New York. [Google Scholar]
- Moyed H. S., Nguyen T. T., and Bertrand K. P., 1983. Multicopy Tn10 tet plasmids confer sensitivity to induction of tet gene expression. J. Bacteriol. 155: 549–556. 10.1128/JB.155.2.549-556.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., 2006. Plasmid R1–replication and its control. Plasmid 55: 1–26. 10.1016/j.plasmid.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Nordström K., and Gerdes K., 2003. Clustering versus random segregation of plasmids lacking a partitioning function: a plasmid paradox? Plasmid 50: 95–101. 10.1016/S0147-619X(03)00056-8 [DOI] [PubMed] [Google Scholar]
- Norman A., Hansen L. H., and Sørensen S. J., 2009. Conjugative plasmids: vessels of the communal gene pool. Proc. Biol. Sci. 364: 2275–2289. 10.1098/rstb.2009.0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., and Hoppensteadt F. C., 1978. On plasmid incompatibility. Plasmid 1: 421–434. 10.1016/0147-619X(78)90001-X [DOI] [PubMed] [Google Scholar]
- Orr H. A., and Unckless R. L., 2008. Population extinction and the genetics of adaptation. Am. Nat. 172: 160–169. 10.1086/589460 [DOI] [PubMed] [Google Scholar]
- Patwa Z., and Wahl L. M., 2008. The fixation probability of beneficial mutations. J. R. Soc. Interface 5: 1279–1289. 10.1098/rsif.2008.0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J., Ho T. Q., Zhong Z., and Helinski D. R., 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98: 4486–4491. 10.1073/pnas.081075798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Beltran J., Hernandez-Beltran J. C. R., DelaFuente J., Escudero J. A., Fuentes-Hernandez A. et al. , 2018. Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat. Ecol. Evol. 2: 873–881. 10.1038/s41559-018-0529-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes R. R., Wiuff C., Zappala R. M., Garner K. N., Baquero F., and Levin B. R., 2004. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob. Agents and Chemother. 48: 3670–3676. 10.1128/AAC.48.10.3670-3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander P., Prammananan T., Meier A., Frischkorn K., and Böttger E. C., 1997. The role of ribosomal RNAs in macrolide resistance. Mol. Microbiol. 26: 469–480. 10.1046/j.1365-2958.1997.5811946.x [DOI] [PubMed] [Google Scholar]
- San Millan A., Peña-Miller R., Toll-Riera M., Halber Z. V., McLean A. R. et al. , 2014. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5: 5208 10.1038/ncomms6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan A., Escudero J. A., Gifford D. R., Mazel D., and MacLean R. C., 2016. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat. Ecol. Evol. 7: 10 10.1038/s41559-016-0010 [DOI] [PubMed] [Google Scholar]
- Santos-Lopez A., Bernabe-Belas C., Ares-Arroyo M., Ortega-Huedo R., Hoefer A. et al. , 2017. A naturally occurring single nucleotide polymorphism in a multicopy plasmid produces a reversible increase in antibiotic resistance. Antimicrob. Agents Chemother. 61: e01735–e16. 10.1128/AAC.01735-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M., Nielsen H. J., Youngren B., and Austin S., 2010. P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation. J. Bacteriol. 192: 1175–1183. 10.1128/JB.01245-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewastjanow B. A., 1975. Verzweigungsprozesse, Oldenbourg, Munich; 10.1002/mana.19750672208 [DOI] [Google Scholar]
- Sigmund C. D., and Morgan E. A., 1982. Erythromycin resistance due to a mutation in a ribosomal RNA operon of Escherichia coli. Proc. Natl. Acad. Sci. USA 79: 5602–5606. 10.1073/pnas.79.18.5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. K., 1991. . The kinetics of plasmid loss. Trends Biotechnol. 9: 273–278. 10.1016/0167-7799(91)90089-Z [DOI] [PubMed] [Google Scholar]
- Sun L., Alexander K. H., Bogos B., Kiviet D. J., Ackermann M., and Bonhoeffer S., 2018. Effective polyploidy causes phenotypic delay and influences bacterial evolvability. PLoS Biol. 16: e2004644 10.1371/journal.pbio.2004644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazzyman S. J., and Bonhoeffer S., 2014. Plasmids and evolutionary rescue by drug resistance. Evolution 68: 2066–2078. 10.1111/evo.12423 [DOI] [PubMed] [Google Scholar]
- Uecker H., 2017. Evolutionary rescue in randomly mating, selfing, and clonal populations. Evolution 71: 845–858. 10.1111/evo.13191 [DOI] [PubMed] [Google Scholar]
- Uecker H., and Hermisson J., 2016. The role of recombination in evolutionary rescue. Genetics 202: 721–732. 10.1534/genetics.115.180299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uecker H., Otto S. P., and Hermisson J., 2014. Evolutionary rescue in structured populations. Am. Nat. 183: E17–E35. 10.1086/673914 [DOI] [PubMed] [Google Scholar]
- van Hoek A. H. A. M., Mevious D., Guerra B., Mullany P., Roberts A. P. et al. , 2011. Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2: 1–27. 10.3389/fmicb.2011.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B., and Douthwaite S., 2001. Mocrolide resistance conferred by base substitutions in 23s rRNA. Antimicrob. Agents Chemother. 45: 1–12. 10.1128/AAC.45.1.1-12.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., 2017. Spatial distribution of high copy number plasmids in bacteria. Plasmid 91: 2–8. 10.1016/j.plasmid.2017.02.005 [DOI] [PubMed] [Google Scholar]
- Weisblum B., 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39: 577–585. 10.1128/AAC.39.3.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental Material (File S1) and the simulation code and the scripts used for the numerical analysis (File S2) are available at figshare: https://doi.org/10.25386/genetics.12357731