Here, Serrano-Saiz et al. describe the cis-regulatory logic of how neurotransmitter identity is imposed onto individual, distinct neuron types...
Keywords: C. elegans, cis-regulatory control, neurotransmitter, transcription factors
Abstract
We explore here the cis-regulatory logic that dictates gene expression in specific cell types in the nervous system. We focus on a set of eight genes involved in the synthesis, transport, and breakdown of three neurotransmitter systems: acetylcholine (unc-17/VAChT, cha-1/ChAT, cho-1/ChT, and ace-2/AChE), glutamate (eat-4/VGluT), and γ-aminobutyric acid (unc-25/GAD, unc-46/LAMP, and unc-47/VGAT). These genes are specifically expressed in defined subsets of cells in the nervous system. Through transgenic reporter gene assays, we find that the cellular specificity of expression of all of these genes is controlled in a modular manner through distinct cis-regulatory elements, corroborating the previously inferred piecemeal nature of specification of neurotransmitter identity. This modularity provides the mechanistic basis for the phenomenon of “phenotypic convergence,” in which distinct regulatory pathways can generate similar phenotypic outcomes (i.e., the acquisition of a specific neurotransmitter identity) in different neuron classes. We also identify cases of enhancer pleiotropy, in which the same cis-regulatory element is utilized to control gene expression in distinct neuron types. We engineered a cis-regulatory allele of the vesicular acetylcholine transporter, unc-17/VAChT, to assess the functional contribution of a “shadowed” enhancer. We observed a selective loss of unc-17/VAChT expression in one cholinergic pharyngeal pacemaker motor neuron class and a behavioral phenotype that matches microsurgical removal of this neuron. Our analysis illustrates the value of understanding cis-regulatory information to manipulate gene expression and control animal behavior.
ONLY a small fraction of an animal’s genome is transcribed into messenger, regulatory, or structural RNAs. Much of the remainder of the genome carries cis-regulatory information in the form of binding sites for trans-acting regulatory factors that dictates patterns of gene expression (Davidson 2006). Genomic binding profiles have been determined for many transcriptional regulators, but it is far from clear how reliably such binding profiles predict the presence of functionally relevant cis-regulatory elements (Ecker et al. 2012; Doolittle 2013; Eddy 2013; Brown and Celniker 2015). De novo prediction of functional cis-regulatory elements by bioinformatic analysis has also proven to be challenging, at least in part due to the fact that binding sites of trans-acting regulatory factors are often small and degenerate. An alternative approach to define and decode cis-regulatory control elements employs transgenic reporter gene analysis, which can reveal the functionality of cis-regulatory elements based on their ability to direct reporter expression in specific cell types (Yáñez-Cuna et al. 2013).
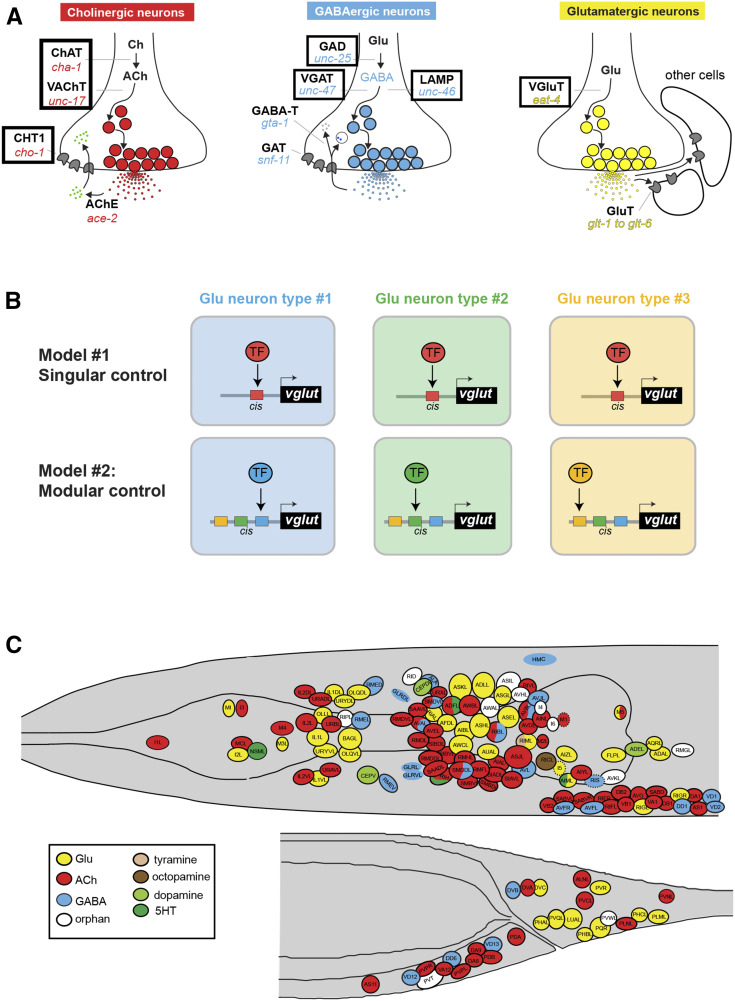
The identification of cis-regulatory elements by reporter gene analysis has the potential to reveal insights into the regulatory logic of cell type-specific gene expression. Genes are often expressed in multiple distinct cell types. This could indicate that distinct cell types may utilize the same transcription factor that operates through a singular cis-regulatory element to turn on expression of a given target gene in distinct cell types. Alternatively, a gene could be expressed in different cell types by harboring multiple cis-regulatory elements that are responsive to distinct, cell type-specific transcription factors. This problem is particularly evident in the nervous system, in which distinct neuron types choose to express usually one of a discrete number of different neurotransmitter systems, thereby acquiring a specific neurotransmitter identity. Neurotransmitter identity is determined by specific enzymes and transporters that synthesize and synaptically package a given neurotransmitter (“neurotransmitter pathway genes”) (Figure 1A). The neurotransmitter identity of a specific neuron can be considered a phenotypic trait. This trait (e.g., the usage of glutamate as neurotransmitter) is shared by many different neuron types. Hence, the question arises as to whether such a shared trait is (1) induced by the same transcriptional regulatory mechanism that operates through the same cis-regulatory elements in all neuron types that share this neurotransmitter phenotype (“singular control”), or (2) is independently controlled by a different transcription factor or transcription factor combinations in distinct neuron types through distinct cis-regulatory elements (“modular control”) (Figure 1B). The analysis of cis-regulatory control regions of neurotransmitter pathway genes has the potential to distinguish between these two possibilities. If the phenotypic trait of a specific neurotransmitter identity is regulated separately in distinct neuron types, cis-regulatory information should be organized in a modular manner. That is, discrete cis-regulatory elements will be responsible for driving expression of the same neurotransmitter pathway gene in different neuron types.
Figure 1.
Introduction to the problem of neurotransmitter identity specification. (A) Neurotransmitter pathway genes. In this paper, fluorescence reporters were generated for cis-regulatory elements contained in the regulatory regions of the genes highlighted in boxes. (B) Conceptual models for how the shared phenotypic trait of neurotransmitter identity could be regulated in different neuron types. Previous studies as well as this paper corroborate the modular control model. However, as we will show here in this paper, there are cases where the same transcription factor (TF) is used in multiple different neuron types (seemingly in support of the singular model), but this is usually because a given TF usually operates in distinct combinations in different cell types, as discussed in the text. For simplicity, these two models just focus on the question of whether a specific neurotransmitter identity is controlled by single TFs. (C) Neurotransmitter atlas of the C. elegans adult hermaphrodite head and tail. Adapted from Gendrel et al. (2016).; ChAT, choline acetyltransferase; VAChT, vesicular acetylcholine (ACh) transporter; CHT1: choline reuptake transporter, AChE: acetylcholinesterase, GAD: glutamic acid decarboxylase; VGAT, vesicular GABA transporter; GABA-T, GABA transaminase GAT, GABA transporter; LAMP, lysosomal associated membrane protein; GluT, Glutamate transporter; 5HT, 5-hydroxytryptamine; ACh, acetylcholine; Ch, choline; GABA, γ-aminobutyric acid; Glu, glutamate; TF, transcription factor.
There is presently ample evidence to support such modular cis-regulatory organization, particularly in the nematode Caenorhabditis elegans (Eastman et al. 1999; Wenick and Hobert 2004; Flames and Hobert 2009; Kratsios et al. 2011; Serrano-Saiz et al. 2013; Lloret-Fernandez et al. 2018). We further extend this concept here via analysis of the cis-regulatory control regions of several neurotransmitter pathway genes. However, we also uncover additional complexities. For a few neuron classes we have not been able to define independent cis-regulatory elements, indicating that the same transcription factor may act in different cell types, but is assisted by different coregulators in distinct cell types. We discuss our findings in the context of phenotypic convergence, a concept aiming to explain how a phenotypic trait shared by distinct neuron types (neurotransmitter identity) can be specified by distinct regulatory means (Konstantinides et al. 2018; Kratsios and Hobert 2018). We also report on evidence for the existence of enhancer pleiotropy, in which similar cis-regulatory elements can drive expression in distinct neuron classes.
Besides providing mechanistic insights into gene regulation, substantial methodological benefits can be gained from the dissection of cis-regulatory control regions. Cis-regulatory elements that drive expression in a small subset of cell types provide “genetic access” to these cell types. That is, these elements can be used as drivers to express genetic tools that allow the visualization or isolation of these cells or, in the context of the nervous system, the optogenetic activation or inhibition of these cells.
Materials and Methods
Transgenes
A list of all transgenic strains used in this study is shown in Supplemental Material, Table S1. Strains were generated by injecting the transgene mix as simple or complex arrays. Briefly, complex arrays contained the fosmid (for otIs388, otIs518, otIs354, otEx6066, and otEx4431) linearized by restriction enzyme digestion or obtained as a PCR product (for unc-17, cho-1, ace-2, unc-25, unc-46, and unc-47 reporters; Table S2). The reporters were mixed with bacterial genomic DNA from Escherichia coli OP50 (previously sonicated and purified with a Gentra Puregene kit; QIAGEN, Valencia, CA) and a co-injection marker plasmid, also linearized by restriction enzymes. For simple arrays, plasmids were mixed directly with the co-injection marker plasmid. For each construct, two to three transgenic lines and at least 10 worms per line were analyzed.
DNA constructs
A list of all reporter constructs and sequences can be found in Table S2. Reporters were generated by different methods: (1) cloning into pDP95.75, pOH569, or pPD95.67 using standard molecular biology cloning techniques; (2) by PCR fusion, as described in Hobert (2002); or (3) fosmid recombineering, as described in Tursun et al. (2009).
Clustered regularly interspaced short palindromic repeats/Cas9 genome engineering
The unc-17 regulatory allele ot1025 was generated using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome engineering, as described previously (Dokshin et al. 2018). The E3 motif was replaced with a PmeI restriction site with the following oligo repair template: tttttctgtctcaaaacggagctcgagcccatgacgtgtGTTTAAACtccgcccaattccttca (capital letters indicate the introduced PmeI site replacing the E3 motif). The engineering was done in unc-17(ot907[unc-17::mKate2::3xFLAG]IV) animals (Pereira et al. 2019) (strain OH15568), generating strain OH16363 unc-17(ot907ot1025).
Neuron identification
Neuronal identification was performed by colocalization of the reporter constructs with landmark strains. Specifically, individual reporter transgene-expressing rfp or mChopti strains were crossed with a green transgenic landmark strain, otIs354 [cho-1fosmid::sl2::yfp::h2b] (Pereira et al. 2015) or otIs388 [eat-4fosmid::sl2::yfp::h2b] (Serrano-Saiz et al. 2013), and gfp or yfp transgene-expressing lines were crossed with either otIs544 [cho-1fosmid::sl2::mChopti::h2b], otIs518 [eat-4fosmid::sl2::mChopti::h2b], or otIs564 [unc-47fosmid::sl2::h2b::mChopti (Gendrel et al. 2016).
Worms were anesthetized using 100 mM of sodium azide (NaN3) and mounted on 5% agarose on glass slides. All images were acquired using a Zeiss ([Carl Zeiss], Thornwood, NY) confocal microscope (LSM880). Representative images are shown following orthogonal projection of 2–10 z-stacks. Images shown in Figure 8 were taken using an automated fluorescence microscope (AXIO Imager Z1 Stand; Zeiss). Acquisition of several z-stack images was performed with the Micro-Manager software. Representative images are shown using the maximum intensity projection type. Image reconstruction was performed using ImageJ software (Schneider et al. 2012).
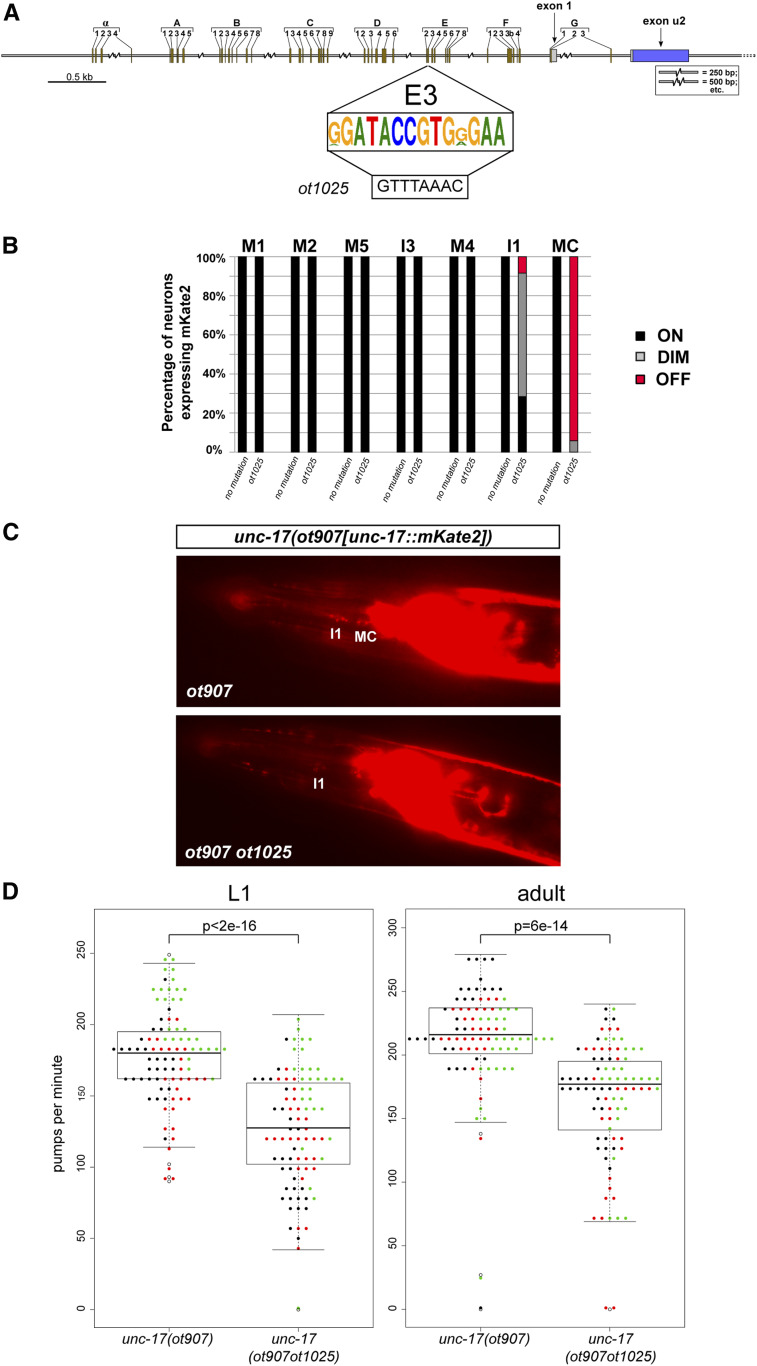
Figure 8.
Cis-regulatory alleles of unc-17, generated by clustered regularly interspaced short palindromic repeats/Cas9 genome engineering. (A) Box E3 is the consensus sequence of multiple Caenorhabditis species, as shown in Figure 3. The E3 box was replaced with a PmeI site (ot1025 allele) in the background of the ot907 allele, which designates the mKate2 tag on the unc-17 locus. (B) Scoring of fluorescent reporter expression of unc-17(ot907) and unc-17(ot907ot1025) animals in cholinergic pharyngeal neurons. The E3 box deletion affects unc-17 expression in the bilaterally symmetric MC neuron class and, mildly, the I1 neuron class. The percentages of neurons that show the indicated phenotypes are indicated in the bar chart. Representative images are shown in panel C Animals were scored at the L4 or young adult stage, n > 50. (D) Pumping rates in worms of different genotypes were compared at the L1 and adult stages. Three different cohorts of worms were tested on different days (each dot color represents a worm from a different assay day). The ANOVA test was used to make a model to fit a linear mixed-effects model to the obtained data. Then, pairwise comparisons of genotypes were done with Tukey’s correction. Statistical significance indicated for L1 comparison P < 2e–16, and for adult comparison P = 6e–14.
Scoring of pharyngeal pumping
Pumping assays were performed at room temperature. Household bleach-mediated egg preparations were used to synchronize C. elegans, and animals were then transferred from an uncrowded nonstarved plate to a fresh NGM plate with a uniform thin layer of OP50. After 5 min recovery, they were observed under a Nikon (Garden City, NY) Eclipse E400 upright microscope equipped with DIC optics using a 50× objective lens for scoring L1-stage animals and a 20× objective for adult animals. The number of pumps in a 20-sec period was measured by focusing on a single worm and then this number was multiplied by three to obtain pumps per minute (ppm). For each stage, ppm was counted for 30 worms.
Phylogenetic analysis
Genomic sequences spanning the cho-1 and unc-17 loci of C. elegans and 12 closely related Caenorhabditis species (C. brenneri, C. briggsae, C. doughertyi, C. inopinata, C. latens, C. nigoni, C. remanei, C. sinica, C. tribulationis, C. tropicalis, C. wallacei, and C. zanzibari) were identified by Basic Local Alignment Search Tool search, and then downloaded from WormBase.org or Caenorhabditis.org. Sequences were aligned using MAFFT or ClustalW and, if necessary, adjusted by inspection; sequence logos were generated by LogOddsLogo (https://www.ncbi.nlm.nih.gov/CBBresearch/Yu/logoddslogo/).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12354695.
Results
In previous work, we used fosmid (large genomic clone)-based reporters and/or CRISPR/Cas9-engineered reporter alleles to comprehensively assign neurotransmitter identities throughout the entire C. elegans nervous system (Serrano-Saiz et al. 2013; Pereira et al. 2015; Gendrel et al. 2016) (Figure 1C). Cholinergic neurons are marked by two fosmid-based reporters: (1) the unc-17 locus, which encodes the C. elegans ortholog of the vesicular acetylcholine (ACh) transporter (VAChT) (Alfonso et al. 1993) and reports on transcription of both unc-17/VAChT and cha-1/ChAT, which shares its first noncoding exon with unc-17 (Alfonso et al. 1994); and (2) cho-1, the choline reuptake transporter (Okuda et al. 2000; Mullen et al. 2007), which marks most, but not quite all, cholinergic neurons (Pereira et al. 2015). The expression pattern of the fosmid-based unc-17 reporter is mimicked by a CRISPR/Cas9-engineered reporter insertion into the unc-17 locus (Pereira et al. 2015). A fosmid-based reporter for ace-2/AChE, one of several C. elegans acetylcholinesterase-encoding genes, is expressed in a subset of cholinergic neurons (Pereira et al. 2015). All glutamatergic neurons are marked by a fosmid-based reporter of the vesicular transporter for glutamate, eat-4/VGLUT (Lee et al. 1999; Serrano-Saiz et al. 2013). Lastly, γ-aminobutyric acid (GABA)-ergic neurons are marked by CRISPR/Cas9-engineered reporter alleles of the unc-25 locus, which encode glutamic acid decarboxylase (GAD; unc-25) (Jin et al. 1999; Gendrel et al. 2016), and by fosmid-based reporters for the unc-47 and unc-46 loci (Gendrel et al. 2016), which encode the vesicular GABA transporter unc-47/VGAT (McIntire et al. 1997) and an associated cofactor (Schuske et al. 2007), respectively. We collectively refer to this cohort of genes as neurotransmitter pathway genes (Figure 1A).
Armed with the knowledge of the precise expression pattern of those fosmid-based and/or CRISPR/Cas9-engineered reporters, we generated a series of reporter constructs that contained distinct DNA fragments of the respective loci. We generated transgenic C. elegans animals carrying these reporters and analyzed their expression patterns in the nervous system with single-cell resolution. This analysis was motivated by a specific hypothesis. Based on previous studies of cis-regulatory control regions of neurotransmitter pathway genes, as well as extensive analysis of genetic mechanisms that control neurotransmitter specification, a theme of modular control of neurotransmitter identity has been emerging (Eastman et al. 1999; Wenick and Hobert 2004; Kratsios et al. 2011; Serrano-Saiz et al. 2013; Pereira et al. 2015; Gendrel et al. 2016; Lloret-Fernandez et al. 2018). To test whether the theme of modularity broadly applies to multiple neurotransmitter pathway genes, we carried out an extensive dissection of their cis-regulatory control regions.
Cholinergic pathway genes: regulation of the cholinergic locus (unc-17/VAChT and cha-1/ChAT)
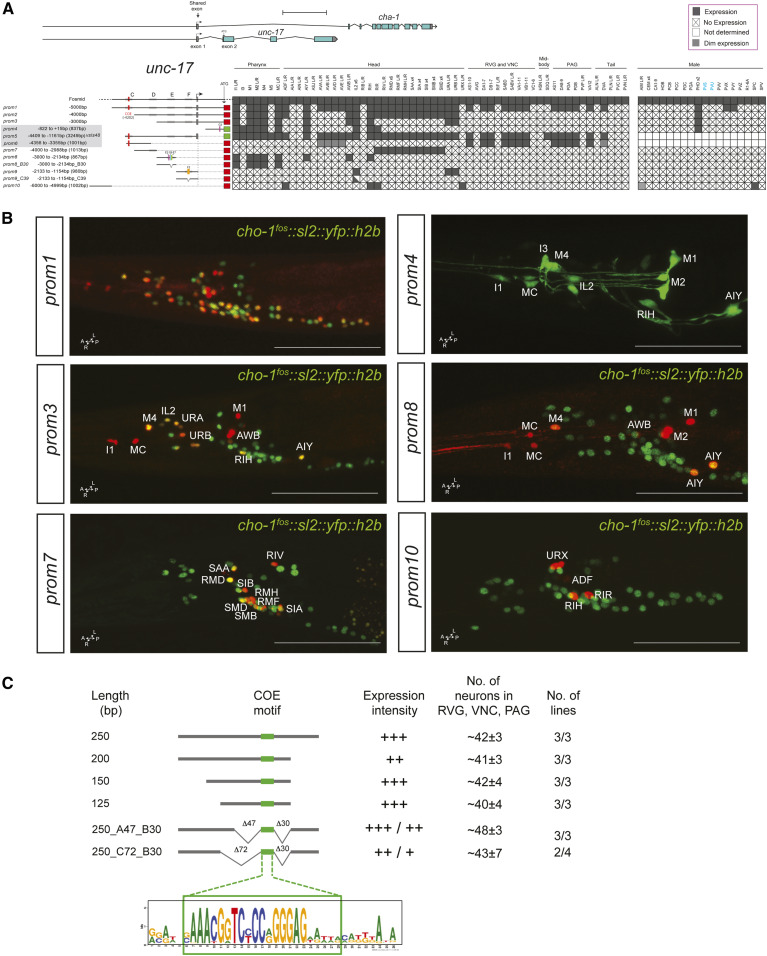
In animal species ranging from worms to vertebrates, the enzymes that synthesize (choline acetyltransferase; cha-1) and package ACh into synaptic vesicles (VAChT; unc-17) are organized into an operon-like conserved structure, termed the cholinergic locus (Alfonso et al. 1994; Eiden 1998). 5 kb upstream of the first protein-coding exon of unc-17 drive reporter gene expression in the majority, but clearly not all, cholinergic neurons (prom1 in Figure 2A,B). Within the male-specific nervous system, only a very small fraction of neurons express this reporter. A 1-kb fragment preceding this 5-kb fragment captures a number of additional cholinergic neurons, in both the sex-shared as well as male-specific nervous system (unc-17prom10, Figure 2A,B).
Figure 2.
Analysis of cis-regulatory elements of the unc-17/cha-1 locus. (A) Overview of the unc-17/cha-1 locus, reporter constructs, and summary of sites of expression. The constructs are referred to the unc-17/cha-1 locus to scale. Expression of the fosmid reporter is from Pereira et al. (2015). unc-17prom4, prom5 worms were shown in previous papers (Wenick and Hobert 2004; Kratsios et al. 2011) but expressing cells were not identified. Within the matrix, the gray color indicates expression, “X” indicates no expression, and white color indicates “not determined.” Scale bar = 2 kb. (B) Representative images of several constructs in the adult head (lateral views). In the images, the unc-17 reporters are shown in red and the cho-1 fosmid (otIs354) is also shown in green as a landmark. Cells in the unc-17prom1 worm are not labeled because there is complete colocalization with the cho-1 fosmid. Usually two or three lines were scored for each reporter and > 10 animals/line. Bar, 50 μm. (C) Analysis of an unc-3 responsive element from the unc-17 locus required for expression in the RVG, VNC, and PAG. This small 250-bp element is also expressed in a few unidentified head neurons, but we only consider here the expression in the RVG, VNC, and PAG, which has previously been shown to require the indicated COE motif and unc-3 (Kratsios et al. 2011). No., number; PAG, preanal ganglion; RVG, retrovesicular ganglion; VNC, ventral nerve cord.
Reporter expression is eliminated in cholinergic motor neurons of the ventral nerve cord (VNC), the flanking retrovesicular ganglion (RVG), and the preanal ganglion (PAG), as well as the command interneurons by removal of distal sequences within the 5-kb fragment (compare unc-17prom1 vs. unc-17prom2). These distal sequences contain a binding site for the UNC-3 transcription factor (“COE motif”), the terminal selector of ventral cord motor neuron and command interneuron identity (Kratsios et al. 2011; Pereira et al. 2015). Conversely, a construct containing only distal sequences and the COE motif (unc-17prom6) is sufficient to drive reporter gene expression in cholinergic motor neurons in the VNC, RVG, and PAG (Kratsios et al. 2011). Further shortening of the 5 kb fragment (unc-17prom3) leads to consecutive loss of expression in more neuron classes (Figure 2A).
Reporter animals carrying 3249 bp of cis-regulatory information upstream of the noncoding first exon shared by unc-17 and cha-1 (unc-17prom5) yielded similar but not identical expression as the 5-kb construct that contained the first intron of the locus (unc-17prom1). We broke this 3249-bp upstream region into several nonoverlapping pieces. One small piece, which contains the COE motif, is sufficient to drive expression in VNC motor neurons (unc-17prom6), as previously described (Kratsios et al. 2011). Three additional, nonoverlapping ∼1-kb fragments (unc-17prom7 to prom9) show distinct patterns of expression in largely (but not completely) nonoverlapping neuron classes. One fragment (unc-17prom7) drives expression exclusively in head motor neuron classes (RMD, SMD, SMB, SIA, SIB, RMF, and RMH) as well as two ring interneuron classes (RIR and RIV). Another, distinct ∼900-bp element captures expression in pharyngeal inter- and motor neuron classes (unc-17prom8). Notably, very similar expression is observed from a different regulatory element in the first intron of the unc-17 locus (unc-17prom4). The ∼900-bp upstream pharyngeal element is also expressed in a few extrapharyngeal neuron classes (AIY, AWB, and RIH). Lastly, the third ∼1-kb fragment is exclusively expressed in three neuron classes of the anterior ganglion (IL2, URA, and URB) (unc-17prom9).
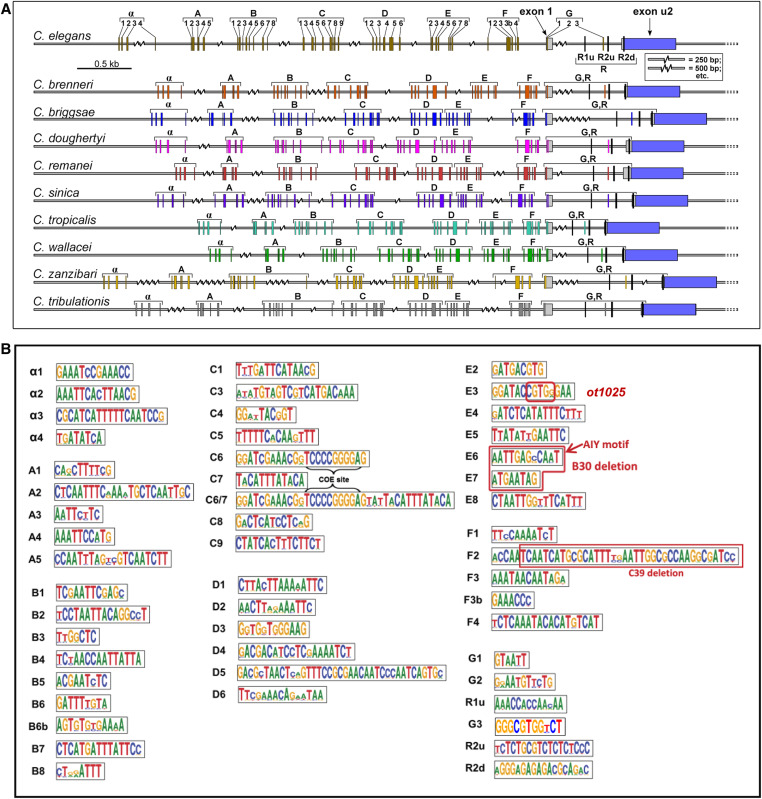
We sought to increase the granularity of our cis-regulatory analysis of the unc-17/cha-1 cholinergic locus by phylogenetic footprinting. Examining 10 different nematode species, we identified a host of small conserved elements (Figure 3A). We examined the functional relevance of two small clusters of these elements in the context of two separate ∼1-kb fragments (Figure 2, A and B). One is the ∼900-bp element (unc-17prom8) that drives expression in a few pharyngeal and extrapharyngeal neuron classes (AIY, AWB, and RIH). This element contains a small motif (“E6”) that we previously identified to be a binding site for the terminal selector-type TTX-3/CEH-10 homeodomain complex (“AIY motif”). We found this motif to be required for unc-17 expression and hence the acquisition of cholinergic identity of the AIY interneuron (Wenick and Hobert 2004). An adjacent element (“E7”) matches the consensus binding site for the UNC-86 POU homeodomain transcription factor and we previously found UNC-86 to be required for cholinergic identity acquisition of the RIH neuron (Pereira et al. 2015). We find that combined deletion of E6 and E7 results in the expected loss of expression of this element, both in AIY and RIH (unc-17prom8_B30; Figure 2A). Expression in AWB, which expresses neither ttx-3, ceh-10, nor unc-86, is also lost, suggesting that a related homeodomain transcription factor that recognizes the E6 and/or E7 motifs may control AWB cholinergic identity.
Figure 3.
Identification of conserved motifs upstream of the cholinergic locus (unc-17/cha-1) of 10 nematode species. (A) Locations of 51 conserved motifs in unc-17 promoters derived from sequence alignments, as described in the Materials and Methods. The R1 and R2 motifs have been described previously, and regulate the alternate splicing of unc-17 and cha-1 transcripts (Mathews et al. 2015). (B) Sequence logos of the conserved promoter motifs. The small element designated E6 = AIY motif (TTX-3/CEH-10-binding site) has been functionally validated in C. elegans to be required to drive unc-17 expression in AIY by Wenick and Hobert (2004). The element designated E7 contains the consensus binding site for the UNC-86 transcription factor. The combined deletion of E6 and E7 (designated unc-17prom8_B30 in Figure 2) results in the loss of expression of the reporter in AIY and RIH. The red box in E3 indicates the nucleotides that were replaced in ot1025 (Figure 8). The red box in F2 indicates the deleted nucleotides in unc-17prom9_C39 (Figure 2).
The ∼1-kb element, that is exclusively expressed in the IL2, URA, and URB neuron classes (unc-17prom9), contains a stretch of conserved sequence motifs (termed F2 in Figure 3B) including another predicted binding site for the UNC-86 POU homeodomain transcription factor, previously shown to be required for cholinergic identity acquisition of the IL2, URA, and URB neuron classes (Zhang et al. 2014). Deletion of the F2 motif affects expression in all these neuron classes (unc-17prom9_C39; Figure 2A). In summary, this cis-regulatory analysis indicates that: (1) the terminal selector UNC-86 exerts its effect on cholinergic identity specification in distinct neuron classes, likely via direct binding and activation of the unc-17/cha-1 cholinergic locus, and (2) that UNC-86 operates through distinct regulatory elements in different cholinergic neuron cell types. These points are further supported by a distal cis-regulatory element that is expressed in four different neuron classes: ADF, RIH, RIR, and URX (unc-17prom10; Figure 2, A and B). Three of these neuron classes express UNC-86 and at least two of them (RIH and URX) have been shown to require UNC-86 for proper unc-17 expression (Pereira et al. 2015).
Lastly, we sought to define the minimal cis-regulatory element upstream of unc-17 that is sufficient to drive unc-3-dependent expression in cholinergic motor neuron classes in the RVG, VNC, and PAG. We focused on unc-17prom6 (1001 bp) (Figure 2A), which contains the UNC-3-binding site (COE motif) previously shown to be required for reporter gene expression in unc-3-dependent neuron classes in the RVG, VNC, and PAG (Kratsios et al. 2011). Shorter reporter versions of 250, 200, 150, and 125 bp that contain the COE motif are sufficient to drive expression in motor neuron classes of the RVG, VNC, and PAG (Figure 2C). Through the introduction of a number of deletions surrounding the COE motif, we found no evidence for the existence of additional required cis-regulatory elements (Figure 2C). While this may indicate that UNC-3 is the sole factor required for expression of unc-17 in motor neuron classes of the RVG, VNC, and PAG, we note that there are highly conserved sequences directly adjacent to the COE motif. Whether these sequences are covered by the UNC-3 protein (which contacts DNA beyond the COE motif (Treiber et al. 2010) or may represent a binding site for a cofactor is presently not clear.
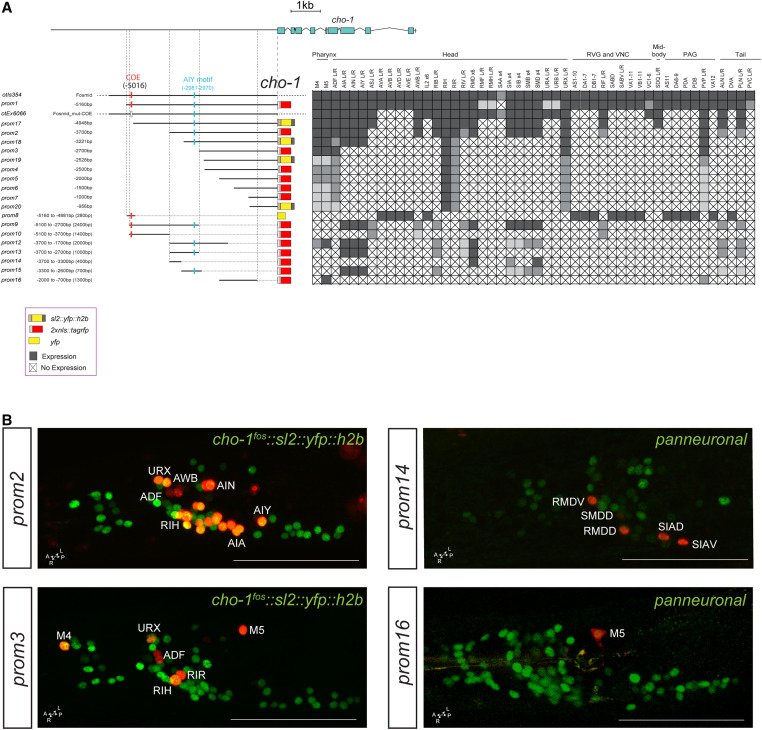
Cholinergic pathway genes: regulation of the cho-1 locus
Dissection of the cho-1/ChT locus reveals a similar modular organization of cis-regulatory elements. A ∼5-kb fragment upstream of the cho-1 locus recapitulates almost the entire expression pattern of the fosmid-based reporter (cho-1prom1; Figure 4A). Successive deletions from the 5′ end result in sequentially increasing losses of expression in multiple cholinergic neuron types. The 2.7-kb fragment (cho-1prom3) directly upstream of the start site of the gene only contains regulatory information for six classes of head and tail neurons, including the only two pharyngeal cholinergic neuron classes that express cho-1. Expression in the pharyngeal neurons can be separated from the remaining few head and tail neurons through a deletion from the 3′ end of this construct (compare cho-1prom3 with cho-1prom11, prom12, and prom16). As is the case for the unc-17 locus, there appear to be two distinct regions of the cho-1 locus that can drive expression in pharyngeal neurons (Figure 4). Another striking similarity to unc-17 regulation is that we isolated an element from both cho-1 and unc-17 that is active in the same set of extrapharyngeal neuron classes: ADF, RIH, RIR, and URX. As mentioned above, three of these neuron classes express UNC-86 and two have been shown to require UNC-86 for both unc-17 and cho-1 expression (Pereira et al. 2015).
Figure 4.
Analysis of cis-regulatory elements of the cho-1 locus. (A) Overview of the cho-1 locus. Schematic of the cho-1 reporters and their expression in hermaphrodite. The constructs are referred to the cho-1 locus to scale. otIs354 (fosmid reporter) and otEx6066 reporters were shown in previous papers (Pereira et al. 2015; Stefanakis et al. 2015), but expressing cells were not identified. Within the matrix, gray color indicates expression, “X” indicates no expression, and white indicates not determined. (B) Representative images of several constructs in the adult head (lateral views). In the images the cho-1 reporters are shown in red and the cho-1 fosmid (otIs354) is also shown in green as a landmark. Usually two or three lines were scored for each reporter and > 10 animals/line. Bar, 50 μm. PAG, preanal ganglion; RVG, retrovesicular ganglion; VNC, ventral nerve cord; yfp, yellow fluorescent protein.
Successive deletions of constructs from the 3′ end also separate expression into distinct neuron classes (Figure 4A). Most notable is the previously reported 280-bp distal element that contains the unc-3-binding COE motif (cho-1prom8). The expression of this element precisely matches all neurons throughout the C. elegans nervous system that require UNC-3 for their cholinergic identity specification. Removal of 60 bp from the 5′ end of this element, leaving the COE motif intact, eliminates expression in the UNC-3(+) neurons (compare prom8 and prom9), suggesting that these 60 bp contain a binding site for another factor that may collaborate with UNC-3. Such a factor is expected to exist because UNC-3 alone is not sufficient to induce cholinergic fate [e.g., UNC-3 is expressed in the noncholinergic ASI neurons (Prasad et al. 1998)].
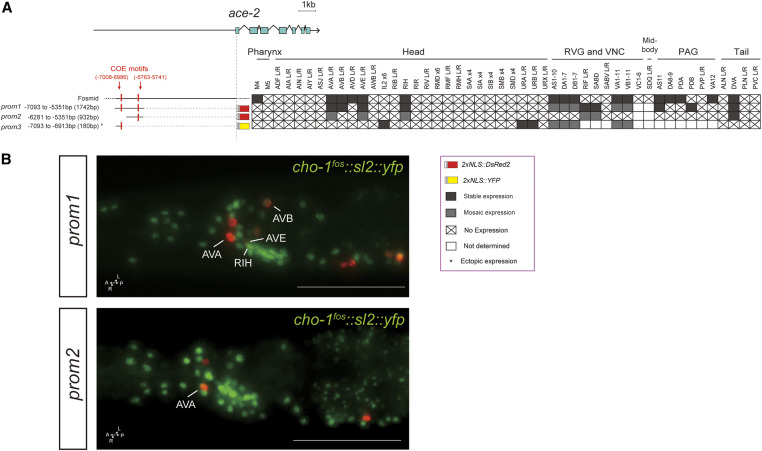
Cholinergic pathway genes: regulation of ace-2.
The ace-2 gene is an ortholog of human acetylcholinesterase (ACHE), an enzyme that catalyzes the breakdown of ACh (Culotti et al. 1981). Using a fosmid-based reporter, we previously determined that ace-2 is expressed in some, but not all, cholinergic neuron classes (Pereira et al. 2015) (Figure 5). A distal element of 1.8 kb (prom1) contains two COE motifs and is sufficient for expression in cholinergic VNC motor neurons, as previously described (Kratsios et al. 2011). We find this element to also be expressed in command interneuron classes (AVA, AVB, and AVE), as well as the RIH and DVA neuron classes, almost completely recapitulating the ace-2 fosmid expression pattern; Figure 5). Supporting the notion of modularity, a shorter 1-kb version of this 1.8-kb element (prom2) drives reporter expression only in AVA, AVB, and DVA, and a limited number of VNC motor neuron classes (Figure 5). A small 180-bp element (prom3) at the distal end of the 1.8-kb fragment drives expression in VNC motor neuron classes, as well as a dozen head (IL2D/V, URA, and URB) and tail (PLM and PHC) neuron classes that normally do not express ace-2 (based on a fosmid-based reporter). This is one of the rare cases where an isolated cis-regulatory element showed derepression in ectopic neuron types.
Figure 5.
Analysis of cis-regulatory elements of the ace-2 locus. (A) Overview of the ace-2 locus. Schematic of ace-2 reporters and their expression in hermaphrodites. The constructs are referred to the ace-2 locus to scale. (B) In the images two ace-2 reporters are shown in red and the cho-1 fosmid (otIs354) is also shown in green as a landmark. Expression of the ace-2 fosmid was characterized in Kratsios et al. (2011) and Pereira et al. (2015). Constructs prom1 and prom2 were described in Kratsios et al. (2011), but cell identification in the head and tail neurons was not performed. Within the matrix, gray color indicates expression, “X” indicates no expression, and white indicates not determined. Some representative images of two reporter constructs are shown (also containing a cho-1 fosmid landmark reporter). Usually two or three lines were scored for each reporter and > 10 animals/line. Bar, 50 μm. PAG, preanal ganglion; RVG, retrovesicular ganglion; VNC, ventral nerve cord.
Glutamatergic identity: regulation of eat-4/VGluT expression
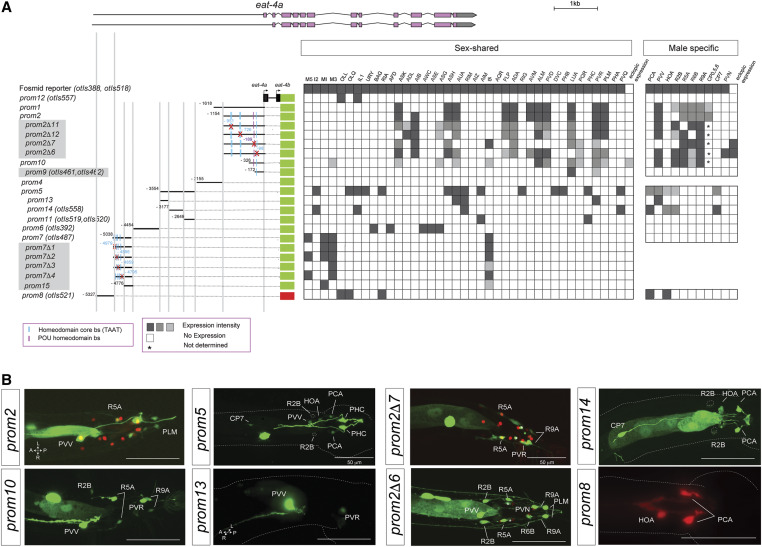
In previous work, we had already mapped cis-regulatory elements that drive expression of eat-4 in distinct eat-4(+) neuron classes in the hermaphrodite nervous system (Serrano-Saiz et al. 2013). While we had examined eat-4 expression in the male nervous system as well (Serrano-Saiz et al. 2017b), we had not examined cis-regulatory elements that drive expression in the male nervous system. We now find that such elements are located in distinct regions of the intergenic sequence upstream of the eat-4 locus (Figure 6A). One element, the 377-bp eat-4prom13 element, drives reporter gene expression exclusively in the male-specific PVV neuron (Figure 6B). This element will be a useful driver to visualize and manipulate PVV neuron anatomy and function. Some of the elements of the eat-4 locus appear to act redundantly, i.e., separate regions drive expression in the same male-specific neuron classes. For example, two nonoverlapping elements are expressed in PVV (eat-4prom1 and eat-4prom13) (Figure 6).
Figure 6.
Analysis of cis-regulatory elements of the eat-4 locus. (A) Overview of the eat-4 locus. Schematic of eat-4 gfp reporter constructs and their expression in hermaphrodite and male nervous systems. The constructs are referred to the eat-4 locus to scale. Gray boxed reporters have not been published before, while the information for the other constructs had been partially shown in Serrano-Saiz et al. (2013), but only in the context of the hermaphrodite and not in the male nervous system (Table S1). The red cross indicates the position of the deletion in each construct. Within the matrix, the gray intensity of each box indicates relative GFP intensity; white boxes indicate no expression. (*) CP1-6 expression was not determined. (B) Representative images of several constructs in the adult male tail. In the images for eat-4prom2 and eat-4prom2Δ7, the eat-4 fosmid (otIs518) is also shown in red as a landmark. Usually two or three lines were scored for each reporter and > 10 animals/line. Bar, 50 μm.
We also undertook a more fine-grained analysis of the previously identified regulatory elements that drive expression in multiple glutamatergic neuron classes with the goal of identifying putative transcription factor-binding sites. Given the preponderance of homeobox genes in controlling neurotransmitter identity (Jin et al. 1994; Serrano-Saiz et al. 2013; Zhang et al. 2014; Pereira et al. 2015; Gendrel et al. 2016), we mutated a number of predicted homeodomain-binding sites (TAAT) as well as a predicted and highly conserved POU homeodomain site in various cis-regulatory elements of the eat-4 locus. We found that several of these deletions lead to losses of expression of the respective eat-4 reporter in very specific neuron classes. For example, deletion of distinct, predicted homeodomain-binding sites eliminates expression in individual neuron classes of the pharynx within the eat-4prom7 element (Figure 6A). Similarly, deletion of distinct homeodomain-binding sites eliminates expression in distinct ray sensory neurons and deletion of a conserved POU homeodomain-binding site specifically eliminates expression in the male-specific PVV neuron (eat-4prom2Δ7; Figure 6). Taken together, this cis-regulatory analysis confirms and extends our previous conclusion that the eat-4 locus, and hence glutamatergic neurotransmitter identity, is controlled in a modular manner through distinct cis-regulatory elements that likely respond to neuron type-specific homeodomain transcription factors (Serrano-Saiz et al. 2013).
GABAergic pathway
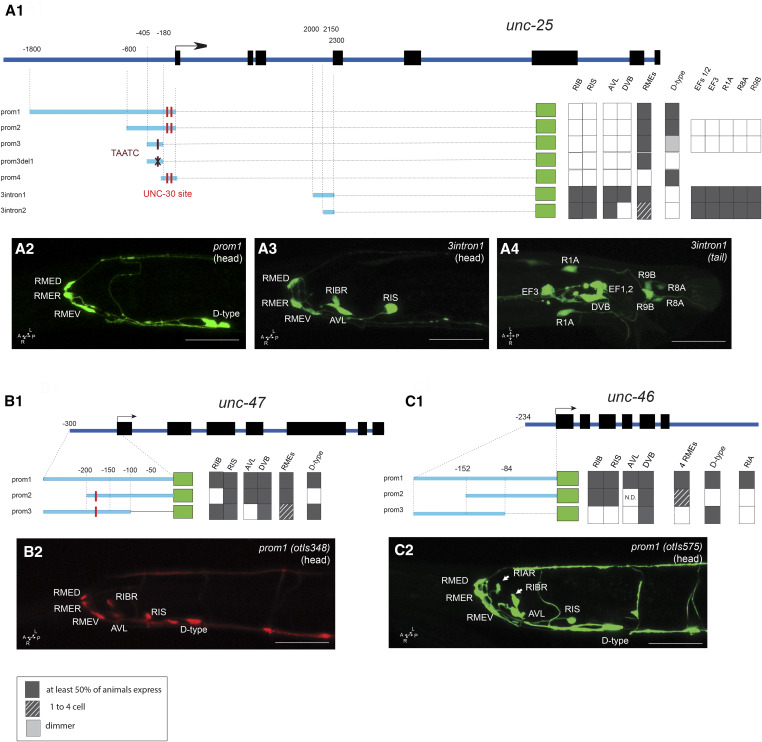
The cis-regulatory region of the unc-25/GAD locus that directs expression in all GABAergic neuron classes, except the RIB neuron class whose GABAergic identity was discovered recently (Gendrel et al. 2016), had been previously localized to < 1 kb upstream of the first exon of unc-25 (Eastman et al. 1999; Jin et al. 1999). We sought to locate the cis-regulatory elements for the previously unrecognized RIB neurons, as well as for expression in the male-specific nervous system (Figure 7). We found that 600-bp upstream of the unc-25 locus drove reporter gene expression exclusively in the RME motor neurons and the D-type motor neurons (unc-25prom2; Figure 7), consistent with this element containing a previously characterized binding site for the unc-30 transcription factor, a terminal selector that controls unc-25 expression (Eastman et al. 1999). However, this element did not show expression in other GABAergic neuron classes in the sex-shared or sex-specific nervous system. Rather, we found those elements to reside within an intron of the unc-25 locus. Specifically, we found that 300 bp (3intron1) revealed expression in RIS, AVL, and DVB, as well as the newly identified GABAergic RIB neuron and the GABAergic neuron classes of the male-specific nervous system (Figure 7A, 3 and 4). Similar to the upstream 600-bp element (prom3), the intronic 300-bp element also drives expression in the RME neurons. Further deletion of this 300-bp element (3intron2) eliminated reporter gene expression in the DVB motor neuron, but retained expression in the RME neuron class (Figure 7A1).
Figure 7.
Analysis of cis-regulatory elements of GABA pathway genes. Analysis of the unc-25/GAD (A1), unc-47/VGAT (B1), and unc-46/LAMPG (C1) loci. (A1, B1, and C1) Schematic of the unc-47, unc-25, and unc-46 GABA pathway genes. The constructs are referred to their respective loci to scale. Dark gray boxes indicate that in at least two lines, > 50% of the worms show gfp expression in the cell. Dashed lined color indicates that, in at least two lines, > 50% of the worms had gfp expression in at least one RME (if there are no dashed lines, gfp expression is always present in the four RMEs). The black cross indicates the position of the deletion in unc-25prom3del1 (TAATC). The red line indicates the position of the UNC-30-binding site sequence (TAATCC). Note that there is no TAATCC sequence in the unc-46 promoter. (A2, A3, A4, B2, and C2) Representative confocal images of adult animals for unc-25prom1 head (A2), unc-25_3intron1 [head (A3) and tail (A4)], unc-47prom1 head (B2), and unc-46prom1 head (C2). Usually two or three lines were scored for each reporter and > 10 animals/line. Bar, 50 μm. GABA, γ-aminobutyric acid.
A previous analysis of the unc-47/VGAT locus revealed that cis-regulatory elements required for expression in GABAergic neurons also reside in a few hundred base pairs upstream of the start of the gene (Eastman et al. 1999). We corroborated these findings with a smaller 300-bp element (prom1) that also reveals expression in the RIB neurons, which were not identified in the original unc-47 studies as GABAergic (McIntire et al. 1993; Eastman et al. 1999) (Figure 7B). Further dissection of these 300 bp separated expression in D-type motor neurons from the remainder of the GABAergic neuron classes (Figure 7B1).
The cis-regulatory regions for unc-46 expression in GABA neuron classes were previously found to reside somewhere upstream of the unc-46 start site (Schuske et al. 2007). We narrowed down this region to a 246-bp element (unc-46prom1), which recapitulates the expression in the entire GABAergic nervous system, including the RIB interneurons (Figure 7C), as well as in the RIA non-GABAergic neuron. As with unc-47, two overlapping subfragments again separate expression in D-type motor neurons from the remainder of the GABAergic neuron classes.
In conclusion, in all GABAergic pathway genes, the unc-30-dependent regulatory element that drives expression in VNC GABAergic (D-type) motor neurons can generally be separated easily from elements driving expression in other GABAergic neuron classes, but expression in other neurons is harder to separate from one another. This is not surprising because the RME, AVL, RIS, and DVB neuron classes appear to share several common trans-acting factors for their identity specification, namely the orphan nuclear hormone receptor nhr-67 and the LIM homeobox gene lim-6 (Hobert et al. 1999; Sarin et al. 2009; Gendrel et al. 2016).
Assessment of potentially redundant activity of distinct cis-regulatory modules in the native genome context
In most neurotransmitter pathway genes, we noted some incidences of distinct, nonoverlapping cis-regulatory regions driving expression in the same neuron type. For example, in the unc-17 locus, two distinct cis-regulatory elements (unc-17prom4 and unc-17prom8 in Figure 2) produce the exact same expression in five pharyngeal neuron classes (I1, M1, M2, M4, and MC). At first sight, a sufficiency of two distinct elements to direct expression in these neurons would argue for redundancy, i.e., one would predict that mutations in either element in the context of the genomic locus should not result in expression defects because the other element should be able to compensate for the mutated one. To test whether such redundancy does indeed exist, we used CRISPR/Cas9 genome engineering to mutate a highly conserved motif within the unc-17prom8 element (Figure 2A and Figure 8A). This motif, the E3 box, is completely conserved in 10 nematode species (Figure 3A). The E3 box mutation (Figure 8A) was engineered in a strain in which the endogenous unc-17 locus had been tagged with mKate2 [the ot907 allele (Pereira et al. 2019)]. We found that this mutation unc-17(ot907ot1025) resulted in an almost fully penetrant loss of unc-17/VAChT expression in the MC neuron class and a much milder effect on expression in I1 (Figure 8, B and C). This observation argues that, at least for expression in the MC neuron, the existence of the regulatory elements present in the unc-17prom4 fragment cannot compensate for disruption of the unc-17prom8 element.
The unc-17/VAChT and cha-1/ChaT genes share a common first exon, and are thought to be jointly regulated (Alfonso et al. 1994) (Figure 2A). Therefore, unc-17(ot907ot1025) animals are predicted to be unable to synthesize ACh (due to loss of the CHA-1 protein) and unable to package ACh into synaptic vesicles (due to loss of the UNC-17 protein) specifically in the MC neurons. The two MC neurons are the key pacemaker neurons of the pharynx (Avery and Horvitz 1989; Raizen et al. 1995; Trojanowski et al. 2014). Hence, we tested whether animals carrying the regulatory allele unc-17(ot907ot1025) display pharyngeal pumping defects akin to those observed upon laser ablation or genetic silencing of the MC neurons. We indeed find that unc-17(ot907ot1025) animals, which appear otherwise behaviorally indistinguishable from wild-type animals, indeed pump less (Figure 8D). Reductions in pumping are also observed in animals treated with nicotinic ACh receptor (nAChR) antagonist or in animals that lack the AChR subunit EAT-2 that is expressed in MC target tissue (Raizen et al. 1995; McKay et al. 2004). Our finding of neuron type-specific removal of the unc-17/VAChT therefore provides further evidence for a cholinergic signal from MC neurons to target tissues being essential for pharyngeal pumping behavior.
Discussion
Past work has defined the modular nature of neurotransmitter identity specification in C. elegans from two different perspectives. First, distinct terminal selector-type transcription factors were found to be required to specify the same neurotransmitter identity in different neuron types [reviewed in Hobert (2016)]. In several cases, it was explicitly shown that this specificity is achieved through the binding of the respective transcription factor to defined cis-regulatory elements within neurotransmitter pathway genes (Eastman et al. 1999; Wenick and Hobert 2004; Kratsios et al. 2011). Second, reporter constructs that capture distinct regions of the loci that encode for neurotransmitter pathway genes were shown to be selectively expressed in distinct subsets of ACh, glutamate, GABA, or monoaminergic neurons, respectively (Eastman et al. 1999; Wenick and Hobert 2004; Flames and Hobert 2009; Kratsios et al. 2011; Serrano-Saiz et al. 2013; Lloret-Fernandez et al. 2018). We have expanded here the analysis of cis-regulatory control regions of neurotransmitter pathway genes, thereby corroborating the largely modular control of neurotransmitter identity (Figure 1B). However, not every neuron type is controlled by an independent, dedicated module, since the same cis-regulatory element can be employed by the same transcription factor in different cell types, likely by acting in the context of a distinct combinatorial code of transcription factors. Hence, cell type-specific gene expression profiles are controlled by combinations of different types of regulatory constellations, as illustrated in Figure 9. We discuss these points in more detail below.
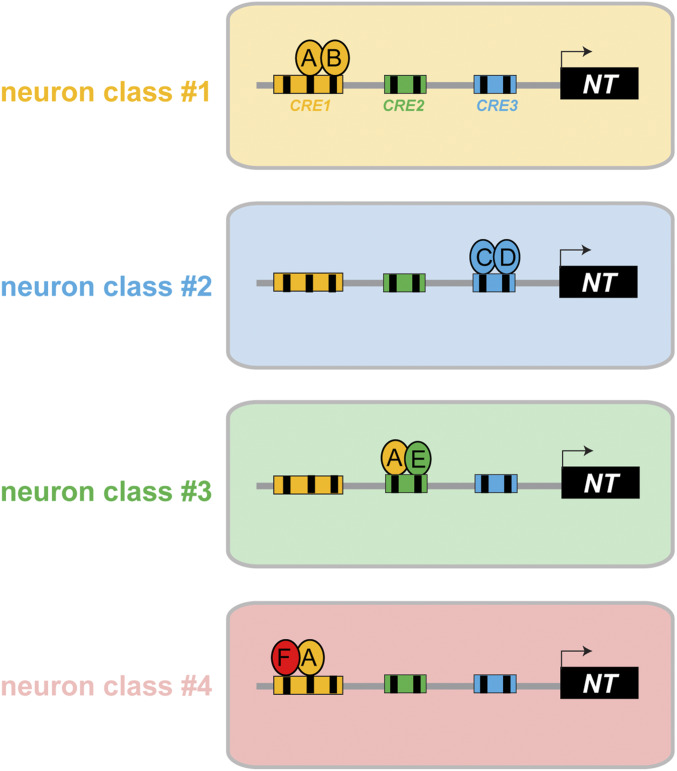
Figure 9.
Illustrations of the organization of cis-regulatory elements for neuron type-specific gene expression. A neurotransmitter pathway gene (NT) is controlled by different sets of transcription factors (A–F) in four different neuron classes. Reporter gene analysis (“promoter bashing”) has revealed distinct cis-regulatory elements in the NT locus that drive expression in four different neuron classes. The indicated transcription factors act through specific binding sites (black boxes) within a given cis-regulatory element. The following scenarios are illustrated: comparing neuron class 1 and class 2 illustrates the simplest scenario of modularity where different combinations of neuron type-specific transcription factors (A + B or C + D) control NT expression in different neuron classes. Comparing neuron class 1 and 3 illustrates that the same transcription factor A can be used in different neuron types, but it operates through distinct, separable cis-regulatory elements that each contain a cognate binding site for this transcription factor. One example for this are the two distinct elements from the unc-17/VAChT locus that drive expression in distinct sets of UNC-86-dependent neurons (RIH vs. IL2/URA/URB), with each of these elements containing validated UNC-86-binding sites (see Results). Comparing neuron class 1 and 4 illustrates a cis-regulatory element that drives expression in multiple distinct neuron classes, and for both neuron classes the same transcription factor-binding site and the same cognate transcription factor are required (e.g., unc-17 element driving UNC-86-dependent expression in IL2, URA, and URB). In such a case, UNC-86 may interact with different transcription factors in neuron 1 vs. neuron 4, but it will not be possible to generate reporters that separate expression in neuron 1 and neuron 4. Hence, the modularity of cis-regulatory control is not manifested by distinct separable elements, but by distinct transcription factor-binding sites that are used in a modular and combinatorial manner.
This and past analysis clearly shows that distinct, modular regulatory inputs control the expression of neurotransmitter pathway systems in distinct neuron types (Eastman et al. 1999; Jin et al. 1999; Wenick and Hobert 2004; Flames and Hobert 2009; Kratsios et al. 2011; Serrano-Saiz et al. 2013; Stefanakis et al. 2015; Lloret-Fernandez et al. 2018) (illustrated in Figure 9). Apart from using transgenic reporter gene assays, we provide here a striking example of this modularity by engineering a cis-regulatory allele of the unc-17/VAChT locus in which we mutated a small, phylogenetically conserved motif. We find that this manipulation not only leads to a selective loss of unc-17/VAChT expression in a single neuron class, but also causes the phenotype that one would expect from dysfunctionality of this neuron.
Nevertheless, past work, as well as the work presented here, also demonstrates that regulatory elements that drive expression in distinct neuron types can be difficult to disentangle, i.e., even small cis-regulatory elements still drive expression in multiple neuron classes. Does this observation contrast with the modular control model and support the singular control model shown in Figure 1B? Theoretically, a more finely grained mutational analysis may still separate distinct elements for distinct cell types. However, there are already clear cases where we know that the same transcription factor acts in distinct cells to specify, for example, their cholinergic identity, seemingly in support of the singular model. However, in many if not all cases, this singular transcription factor may act in combination with different sets of cell type-specific partner proteins (illustrated in Figure 9, compare neurons 1 and 4). For example, the COE-type transcription factor unc-3 controls unc-17/VAChT and cho-1/ChT expression in VNC motor neurons and their presynaptic command interneurons, which are all cholinergic and specified by unc-3 (Kratsios et al. 2011; Pereira et al. 2015). Mutation of the unc-3-binding site leads to loss of expression in all these neuron classes. Similarly, a regulatory element from the unc-17/VAChT locus shows expression in the IL2, URA, and URB neuron classes, all of which require unc-86 for their proper neurotransmitter identity acquisition (Zhang et al. 2014). Likewise, a regulatory element from the eat-4 locus is expressed in the AIM and PHC neuron classes, both of which require unc-86 for their glutamatergic identity specification. Cis-regulatory elements that control expression of GABA pathway genes in the RIS and AVL neuron classes (both controlled by the nhr-67 and lim-6 transcription factors) can also not be easily separated, while cis-regulatory elements for GABAergic VNC motor neurons (which are dependent on the unc-30 transcription factor) can be separated from RIS and AVL. These examples demonstrate that the modularity of cis-regulatory control does not need to manifest itself by the existence of distinct, completely separable cis-regulatory elements that drive expression in distinct neuron types. Rather, the same cis-regulatory element and its cognate transcription factor may be deployed in different cell types because transcription factors operate in distinct, cell type-specific combinations.
Enhancers that are employed to drive expression in different cell types have been termed “pleiotropic enhancers” (Sabarís et al. 2019). Such pleiotropy has been mostly inferred from genome-wide binding profiles of histone modification marks that indicate (but do not prove) enhancer activity at a relatively coarse level. Specifically, marks of enhancer activity can be found in the same location in distinct tissue types (McKay and Lieb 2013). The underlying mechanistic basis for enhancer pleiotropy is often not clear. The cis-regulatory analysis conducted in this and previous papers (Kratsios et al. 2011; Serrano-Saiz et al. 2013, 2017a) not only supports the concept of enhancer pleiotropy in an orthogonal and more granular manner, but also strongly hints at its mechanistic basis. Different neuron types utilize the same cis-regulatory element because this regulatory element contains binding sites for the same trans-acting factor (Figure 9; compare neurons 1 and 4).
While the unc-3 case provides the clearest evidence for the same cis-regulatory motif to be employed in different neuron types, our analysis also reveals that a transcription factor required for acquisition of the same neurotransmitter identity in distinct neuron types does not necessarily always act through the same cis-regulatory element. For example, UNC-86 operates in more than a dozen different neuron types to specify neurotransmitter identity (Sze et al. 2002; Serrano-Saiz et al. 2013; Zhang et al. 2014; Pereira et al. 2015). It appears that UNC-86 functions through separable cis-regulatory elements in the unc-17 locus to enable cholinergic gene expression in different cell types. These distinctive types of engagement of UNC-86 with its target promoters are a likely reflection of distinct molecular partnerships that UNC-86 may engage in (Figure 9, compare neurons 1 and 3).
In several neurotransmitter pathway genes, we noted the existence of separable elements that drive expression in the same neuron types. For example, RME motor neuron expression is controlled by separable cis-regulatory elements in the unc-25/GAD locus. This is akin to the “shadow enhancers” that have been observed to drive the expression of developmental control genes in Drosophila (Hong et al. 2008; Lagha et al. 2012). One immediate question that arises from such a constellation is whether this is an indication of true enhancer redundancies. Complete redundancy would predict that deletion of one of these elements would result in no expression defect because of the existence of the other element. Alternatively, the sufficiency of isolated elements to drive expression could be a result of the multicopy nature of concatamerized C. elegans transgenes (Mello et al. 1991). In the natural genomic context, several separable cis-regulatory elements may need to cooperate to turn on expression in a specific cell type. We tested this notion here explicitly through mutating a small conserved motif that is located in one of two separate cis-regulatory elements of the unc-17 locus, which each drive expression in the same set of pharyngeal neurons. We find completely penetrant defects in unc-17 expression in a single neuron class, thereby indicating that, in their endogenous context, a cis-regulatory element may be required even if another element produces, in the context of a multicopy array, the same pattern of expression. This finding is not without precedent. We had previously defined two separate cis-regulatory elements in the cog-1 homeobox gene locus that are alone sufficient to drive expression in the ASE neuron (O’Meara et al. 2009). Nevertheless, we isolated a cis-regulatory allele of cog-1 from a mutant screen in which only one of those two elements is mutated. These examples illustrate an important limitation for the interpretation of reporter gene analysis: the expression of isolated elements may demonstrate that an element is sufficient to produce an expression pattern, but they do not prove that such an element is required in its normal genomic context.
Pioneering work by Eric Davidson and colleagues in the sea urchin embryo revealed that “nuts and bolts” differentiation genes, i.e., genes that do not encode regulatory factors, but rather “function” proteins that endow a cell with specific anatomical or functional properties, are largely controlled by positive regulatory inputs, i.e., by transcriptional activators (Davidson 2006). Our cis-regulatory analysis of neurotransmitter pathway genes provides further support for this notion. If the expression patterns of neurotransmitter pathway genes were frequently shaped by repressive mechanisms, our successive deletion of regulatory elements should have revealed ectopic expression of reporter genes. While we observed such derepression in some rare cases, repression does not appear to be a major mechanism in shaping the expression of neurotransmitter pathway genes. The cis-regulatory analysis of other neuronal differentiation genes, undertaken by our laboratory over the years (Wenick and Hobert 2004; Etchberger et al. 2007, 2009; Kratsios et al. 2011; Doitsidou et al. 2013; Zhang et al. 2014; Serrano-Saiz et al. 2018), also provides only limited evidence in favor of repression being an important mechanism to shape neuron type-specific gene expression profiles. This is not to say that transcriptional repression plays no role in shaping the terminal differentiation program. Several studies have revealed that transcriptional repression is used in a number of distinct cellular contexts (sensory neurons and motor neurons) to establish subtle, but important molecular differences between members of the same neuron class [e.g., differences between left vs. right ASE neurons (Etchberger et al. 2009) or differences between different VNC motor neuron classes (Kerk et al. 2017)].
Lastly, the dissection of cis-regulatory elements of genes that define specific phenotypic traits of distinct neuron types (i.e., neurotransmitter identities) highlights the underlying mechanistic basis of phenotypic convergence. In evolutionary theory, phenotypic convergence refers to independent evolution of similar traits in different organisms (Rosenblum et al. 2014). This terminology has recently been applied to the acquisition of the phenotypic trait of neurotransmitter identity (Konstantinides et al. 2018). In both C. elegans and Drosophila, distinct transcription factors control the same phenotypic trait in distinct neuron types (Serrano-Saiz et al. 2013; Konstantinides et al. 2018; Kratsios and Hobert 2018). Cis-regulatory analyses like the ones described here demonstrate that it is the cis-regulatory control regions of neurotransmitter pathway genes that integrate distinct regulatory inputs to turn on a common neurotransmitter trait in distinct neuron types.
Acknowledgments
We thank Chi Chen for expert assistance in generating transgenic strains, Kelsey Roberts for generating constructs, Surojit Sural for advice on pumping assays, and Steven Cook for help with statistics. This work was supported by the Howard Hughes Medical Institute. E.S.-S. has been supported by the Ramon y Cajal program (RYC-2016-20537), and M.G. was supported by the European Molecular Biology Organization and Human Frontier Science Program postdoctoral fellowships.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12354695.
Communicating editor: V. Reinke
Literature Cited
- Alfonso A., Grundahl K., Duerr J. S., Han H. P., and Rand J. B., 1993. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261: 617–619. 10.1126/science.8342028 [DOI] [PubMed] [Google Scholar]
- Alfonso A., Grundahl K., McManus J. R., Asbury J. M., and Rand J. B., 1994. Alternative splicing leads to two cholinergic proteins in Caenorhabditis elegans. J. Mol. Biol. 241: 627–630. 10.1006/jmbi.1994.1538 [DOI] [PubMed] [Google Scholar]
- Avery L., and Horvitz H. R., 1989. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron 3: 473–485. 10.1016/0896-6273(89)90206-7 [DOI] [PubMed] [Google Scholar]
- Brown J. B., and Celniker S. E., 2015. Lessons from modENCODE. Annu. Rev. Genomics Hum. Genet. 16: 31–53. 10.1146/annurev-genom-090413-025448 [DOI] [PubMed] [Google Scholar]
- Culotti J. G., Von Ehrenstein G., Culotti M. R., and Russell R. L., 1981. A second class of acetylcholinesterase-deficient mutants of the nematode Caenorhabditis elegans. Genetics 97: 281–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., 2006. The Regulatory Genome. Academic Press, San Diego. [Google Scholar]
- Doitsidou M., Flames N., Topalidou I., Abe N., Felton T. et al. , 2013. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes Dev. 27: 1391–1405. 10.1101/gad.217224.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin G. A., Ghanta K. S., Piscopo K. M., and Mello C. C., 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210: 781–787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F., 2013. Is junk DNA bunk? A critique of ENCODE. Proc. Natl. Acad. Sci. USA 110: 5294–5300. 10.1073/pnas.1221376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman C., Horvitz H. R., and Jin Y., 1999. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 19: 6225–6234. 10.1523/JNEUROSCI.19-15-06225.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker J. R., Bickmore W. A., Barroso I., Pritchard J. K., Gilad Y. et al. , 2012. Genomics: ENCODE explained. Nature 489: 52–55. 10.1038/489052a [DOI] [PubMed] [Google Scholar]
- Eddy S. R., 2013. The ENCODE project: missteps overshadowing a success. Curr. Biol. 23: R259–R261. 10.1016/j.cub.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Eiden L. E., 1998. The cholinergic gene locus. J. Neurochem. 70: 2227–2240. 10.1046/j.1471-4159.1998.70062227.x [DOI] [PubMed] [Google Scholar]
- Etchberger J. F., Lorch A., Sleumer M. C., Zapf R., Jones S. J. et al. , 2007. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21: 1653–1674. 10.1101/gad.1560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger J. F., Flowers E. B., Poole R. J., Bashllari E., and Hobert O., 2009. Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development 136: 147–160. 10.1242/dev.030064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N., and Hobert O., 2009. Gene regulatory logic of dopamine neuron differentiation. Nature 458: 885–889. 10.1038/nature07929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M., Atlas E. G., and Hobert O., 2016. A cellular and regulatory map of the GABAergic nervous system of C. elegans. Elife 5: e17686 10.7554/eLife.17686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. 10.2144/02324bm01 [DOI] [PubMed] [Google Scholar]
- Hobert O., 2016. A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscip. Rev. Dev. Biol. 5: 474–498. 10.1002/wdev.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., Tessmar K., and Ruvkun G., 1999. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126: 1547–1562. [DOI] [PubMed] [Google Scholar]
- Hong J. W., Hendrix D. A., and Levine M. S., 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Hoskins R., and Horvitz H. R., 1994. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372: 780–783. 10.1038/372780a0 [DOI] [PubMed] [Google Scholar]
- Jin Y., Jorgensen E., Hartwieg E., and Horvitz H. R., 1999. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J. Neurosci. 19: 539–548. 10.1523/JNEUROSCI.19-02-00539.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk S. Y., Kratsios P., Hart M., Mourao R., and Hobert O., 2017. Diversification of C. elegans motor neuron identity via selective effector gene repression. Neuron 93: 80–98. 10.1016/j.neuron.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N., Kapuralin K., Fadil C., Barboza L., Satija R. et al. , 2018. Phenotypic convergence: distinct transcription factors regulate common terminal features. Cell 174: 622–635.e13. 10.1016/j.cell.2018.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P., and Hobert O., 2018. Nervous system development: flies and worms converging on neuron identity control. Curr. Biol. 28: R1154–R1157. 10.1016/j.cub.2018.08.032 [DOI] [PubMed] [Google Scholar]
- Kratsios P., Stolfi A., Levine M., and Hobert O., 2011. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15: 205–214. 10.1038/nn.2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M., Bothma J. P., and Levine M., 2012. Mechanisms of transcriptional precision in animal development. Trends Genet. 28: 409–416. 10.1016/j.tig.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y., Sawin E. R., Chalfie M., Horvitz H. R., and Avery L., 1999. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in Caenorhabditis elegans. J. Neurosci. 19: 159–167. 10.1523/JNEUROSCI.19-01-00159.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret-Fernandez C., Maicas M., Mora-Martinez C., Artacho A., Jimeno-Martin A. et al. , 2018. A transcription factor collective defines the HSN serotonergic neuron regulatory landscape. Elife 7: e32785 10.7554/eLife.32785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews E. A., Mullen G. P., Manjarrez J. R., and Rand J. B., 2015. Unusual regulation of splicing of the cholinergic locus in Caenorhabditis elegans. Genetics 199: 729–737. 10.1534/genetics.114.173765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire S. L., Jorgensen E., and Horvitz H. R., 1993. Genes required for GABA function in Caenorhabditis elegans. Nature 364: 334–337. 10.1038/364334a0 [DOI] [PubMed] [Google Scholar]
- McIntire S. L., Reimer R. J., Schuske K., Edwards R. H., and Jorgensen E. M., 1997. Identification and characterization of the vesicular GABA transporter. Nature 389: 870–876. 10.1038/39908 [DOI] [PubMed] [Google Scholar]
- McKay D. J., and Lieb J. D., 2013. A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27: 306–318. 10.1016/j.devcel.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J. P., Raizen D. M., Gottschalk A., Schafer W. R., and Avery L., 2004. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics 166: 161–169. 10.1534/genetics.166.1.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., and Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen G. P., Mathews E. A., Vu M. H., Hunter J. W., Frisby D. L. et al. , 2007. Choline transport and de novo choline synthesis support acetylcholine biosynthesis in Caenorhabditis elegans cholinergic neurons. Genetics 177: 195–204. 10.1534/genetics.107.074120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T., Haga T., Kanai Y., Endou H., Ishihara T. et al. , 2000. Identification and characterization of the high-affinity choline transporter. Nat. Neurosci. 3: 120–125. 10.1038/72059 [DOI] [PubMed] [Google Scholar]
- O’Meara M. M., Bigelow H., Flibotte S., Etchberger J. F., Moerman D. G. et al. , 2009. Cis-regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog-1 affect neuronal. Dev. Genet. 181: 1679–1686. 10.1534/genetics.108.097832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Kratsios P., Serrano-Saiz E., Sheftel H., Mayo A. E. et al. , 2015. A cellular and regulatory map of the cholinergic nervous system of C. elegans. Elife 4: e12432 10.7554/eLife.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Aeschimann F., Wang C., Lawson H., Serrano-Saiz E. et al. , 2019. Timing mechanism of sexually dimorphic nervous system differentiation. Elife 8: e42078 10.7554/eLife.42078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B. C., Ye B., Zackhary R., Schrader K., Seydoux G. et al. , 1998. unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 125: 1561–1568. [DOI] [PubMed] [Google Scholar]
- Raizen D. M., Lee R. Y., and Avery L., 1995. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141: 1365–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum E. B., Parent C. E., and Brandt E. E., 2014. The molecular basis of phenotypic convergence. Annu. Rev. Ecol. Evol. Syst. 45: 203–226. 10.1146/annurev-ecolsys-120213-091851 [DOI] [Google Scholar]
- Sabarís G., Laiker I., Preger-Ben Noon E., and Frankel N., 2019. Actors with multiple roles: pleiotropic enhancers and the paradigm of enhancer modularity. Trends Genet. 35: 423–433. 10.1016/j.tig.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Sarin S., Antonio C., Tursun B., and Hobert O., 2009. The C. elegans Tailless/TLX transcription factor nhr-67 controls neuronal identity and left/right asymmetric fate diversification. Development 136: 2933–2944. 10.1242/dev.040204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., and Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuske K., Palfreyman M. T., Watanabe S., and Jorgensen E. M., 2007. UNC-46 is required for trafficking of the vesicular GABA transporter. Nat. Neurosci. 10: 846–853. 10.1038/nn1920 [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E., Poole R. J., Felton T., Zhang F., de la Cruz E. D. et al. , 2013. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155: 659–673. 10.1016/j.cell.2013.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Oren-Suissa M., Bayer E. A., and Hobert O., 2017a Sexually dimorphic differentiation of a C. elegans hub neuron is cell autonomously controlled by a conserved transcription factor. Curr. Biol. 27: 199–209. 10.1016/j.cub.2016.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Pereira L., Gendrel M., Aghayeva U., Battacharya A. et al. , 2017b A neurotransmitter atlas of the Caenorhabditis elegans male nervous system reveals sexually dimorphic neurotransmitter usage. Genetics 206: 1251–1269. 10.1534/genetics.117.202127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Leyva-Díaz E., De La Cruz E., and Hobert O., 2018. BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr. Biol. 28: 2813–2823.e2. 10.1016/j.cub.2018.06.045 [DOI] [PubMed] [Google Scholar]
- Stefanakis N., Carrera I., and Hobert O., 2015. Regulatory logic of pan-neuronal gene expression in C. elegans. Neuron 87: 733–750. 10.1016/j.neuron.2015.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Zhang S., Li J., and Ruvkun G., 2002. The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development 129: 3901–3911. [DOI] [PubMed] [Google Scholar]
- Treiber T., Mandel E. M., Pott S., Gyory I., Firner S. et al. , 2010. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity 32: 714–725. 10.1016/j.immuni.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Trojanowski N. F., Padovan-Merhar O., Raizen D. M., and Fang-Yen C., 2014. Neural and genetic degeneracy underlies Caenorhabditis elegans feeding behavior. J. Neurophysiol. 112: 951–961. 10.1152/jn.00150.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I., and Hobert O., 2009. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One 4: e4625 10.1371/journal.pone.0004625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenick A. S., and Hobert O., 2004. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell 6: 757–770. 10.1016/j.devcel.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Yáñez-Cuna J. O., Kvon E. Z., and Stark A., 2013. Deciphering the transcriptional cis-regulatory code. Trends Genet. 29: 11–22. 10.1016/j.tig.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Zhang F., Bhattacharya A., Nelson J. C., Abe N., Gordon P. et al. , 2014. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development 141: 422–435. 10.1242/dev.099721 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12354695.