Abstract
Heat-shock protein (Hsp) 90 assists in the folding of diverse sets of client proteins including kinases and growth hormone receptors. Hsp70 plays a major role in many Hsp90 functions by interacting and modulating conformation of its substrates before being transferred to Hsp90s for final maturation. Each eukaryote contains multiple members of the Hsp70 family. However, the role of different Hsp70 isoforms in Hsp90 chaperoning actions remains unknown. Using v-Src as an Hsp90 substrate, we examined the role of each of the four yeast cytosolic Ssa Hsp70s in regulating Hsp90 functions. We show that the strain expressing stress-inducible Ssa3 or Ssa4, and the not constitutively expressed Ssa1 or Ssa2, as the sole Ssa Hsp70 isoform reduces v-Src-mediated growth defects. The study shows that although different Hsp70 isoforms interact similarly with Hsp90s, v-Src maturation is less efficient in strains expressing Ssa4 as the sole Hsp70. We further show that the functional distinction between Ssa2 and Ssa4 is regulated by its C-terminal domain. Further studies reveal that Ydj1, which is known to assist substrate transfer to Hsp70s, interacts relatively weakly with Ssa4 compared with Ssa2, which could be the basis for poor maturation of the Hsp90 client in cells expressing stress-inducible Ssa4 as the sole Ssa Hsp70. The study thus reveals a novel role of Ydj1 in determining the functional distinction among Hsp70 isoforms with respect to the Hsp90 chaperoning action.
Keywords: Hsp70, Hsp40, Hsp90, v-Src
HEAT-SHOCK protein (Hsp) 90 is a highly conserved chaperone across all eukaryotes (Chen et al. 2006). Vertebrates and lower eukaryotes contain multiple highly homologous cytosolic Hsp90 isoforms with only partial functional redundancy (Voss et al. 2000; Li et al. 2012). Hsp90 is involved in the folding of various key cellular proteins and is thus essential for cellular survival in eukaryotes. Its client proteins include transcription factors, kinases, telomerase, and many viral proteins (Rajapandi et al. 2000; Citri et al. 2006; Kim et al. 2008; Srisutthisamphan et al. 2018). Several Hsp90 clients also include those involved in carcinogenesis such as p53, and thus the chaperone has been extensively studied for its role in cancer biology (Boysen et al. 2019; Dahiya et al. 2019). Although Hsp90 influences the maturation of large numbers of cellular proteins, the requirements vary with the substrates. For some substrates, such as steroid hormone receptors, it is essential for both maturation and maintenance, and for others such as kinases, the chaperone is required only during the synthesis and folding into the native state (Picard et al. 1990; Xu et al. 1999). The diversity of Hsp90 functions is believed to be due to its interaction with various cochaperones. In spite of extensive research, no sequence or structural motif conserved across different client proteins has been identified, and thus how Hsp90 binds and assists in the folding of diverse sets of substrates continues to be under intense investigation (Taipale et al. 2012).
Hsp90 is a homodimeric protein and each protomer consists of three domains, namely, the N-terminal domain that binds to ATP, the client-binding middle domain that also interacts with other cochaperones, and the MEEVD motif, which contains C-terminal domains required for dimerization and interaction with tetratricopeptide repeat (TPR) domains containing cochaperones (Minami et al. 1994; Prodromou et al. 1999; Li et al. 2012). The client proteins are known to interact with both the middle and the N-terminal domains of Hsp90 (Sato et al. 2000; Karagöz et al. 2014). ATP binding to the N-terminal domain leads to conformational changes in Hsp90 (Graf et al. 2009); through an intermediary stage, the chaperone forms a closed state in which the N-terminal domain is dimerized. In this structurally compact state, the ATP is hydrolyzed, which leads to dissociation of the N-terminal domains and the release of ADP followed by a transition of Hsp90 back to the original open conformation. The Hsp90 reaction cycles are regulated by dynamic associations with various cochaperones, which are broadly divided into TPR and non-TPR-containing proteins such as Sti1, Cpr7, and Aha1 (Chang et al. 1997; Mayr et al. 2000; Panaretou et al. 2002).
Hsp70 is one of the other major cellular chaperones that plays a central role in maturation of Hsp90 client proteins such as transcription factors and protein kinases (Kirschke et al. 2014; Roy et al. 2015). Hsp70 has its own chaperoning activity and its role in the Hsp90 reaction cycle is for the early folding of Hsp90 client proteins. Any defect in the Hsp70-Hsp90 folding cycle results in ubiquitination and degradation of Hsp90 client proteins (Leu et al. 2011; Rodina et al. 2013; Roy et al. 2015). Hsp70 interacts with Hsp90 through an adaptor molecule (e.g., Sti1 and Hop in yeast and mammals, respectively) that acts as a bridge between the two chaperones. Sti1 bridges the two chaperones via its helical TPR domains that bind to the C-terminal EEVD motif present in Hsp70 and Hsp90 (Schmid et al. 2012; Röhl et al. 2015). In addition to being bridged by the adaptor molecules, the two proteins also exhibit direct interactions (Kravats et al. 2018). The substrate first interacts with Hsp70 and the partially folded substrate is then transferred to Hsp90 for further maturation. The Hsp90 cycle progresses by its interaction with other cochaperones such as peptidyl prolyl cis/trans isomerases (Duina et al. 1996; Warth et al. 1997) and Sba1 (e.g., homologous to mammalian p23) (Sullivan et al. 2002; McLaughlin et al. 2006), followed by dissociation of Hsp90 from the Hsp70-Hsp90 complex. The exact role of Hsp70 in regulating Hsp90 functions is not clearly understood. It is believed that the role of Hsp70 in the Hsp90 chaperone machinery is to stabilize protein substrates in a configuration that can be recognized and bound by Hsp90 (Karagöz et al. 2014).
Eukaryotes contain multiple, highly homologous members of cytosolic Hsp70, e.g., the genetically tractable Saccharomyces cerevisiae harbors four cytosolic SSA (Stress-Seventy subfamily A) Hsp70 isoforms (Ssa1–4) (Werner-Washburne et al. 1987). Ssa1 and Ssa2 are constitutively expressed, whereas Ssa3 and Ssa4 are expressed only under stress conditions such as high temperature and oxidative stress (Werner-Washburne et al. 1989). Previous studies have shown that although they are highly homologous, the Ssa isoforms possess both redundant as well as distinct functions (Lotz et al. 2019). It has been shown that cells expressing Ssa2 but not Ssa1 stably propagate one of the yeast prions (URE3) (Tibor Roberts et al. 2004). It has been similarly shown that the presence of Ssa3 under heat-shock conditions is required to suppress α-synuclein-mediated toxicity (Flower et al. 2005; Gupta et al. 2018). Although various studies have examined the roles of different Hsp70 isoforms in substrate folding, not much is known about their significance for Hsp90 functions. It is also not clear whether different members of the Hsp70 family function similarly or distinctly in the Hsp90 chaperoning pathway. As Hsp70 is required for the Hsp90 chaperoning function, the differences in the actions of these different Hsp70 isoforms could affect the fates of Hsp90 client proteins.
In the present study, using v-Src as a model Hsp90 client protein, we have investigated the roles of different Ssa Hsp70s in the Hsp90 chaperoning pathway. Our results show that although Ssa1 and Ssa2 stabilize v-Src, its maturation is inhibited in strains expressing Ssa3 or Ssa4 as the sole source of Ssa Hsp70. We show that different Ssa Hsp70 isoforms interact similarly with Hsp90 and that their distinct role in the Hsp90 pathway is due to their different affinities with Hsp40 Ydj1. The present study thus reveals that Ydj1 is required not only to stimulate the activities of Hsp90s, but also in defining their functional specificity in Hsp90 chaperoning activity.
Materials and Methods
Strains and plasmids
The strains and plasmids used in the study are described in Table 1 and Table 2, respectively.
Table 1. List of strains used in the present study.
| Strain | Genotype | Reference |
|---|---|---|
| SY187 | MATa, kar 1-1, PDAL5::ADE2, his3Δ202, leu2Δ1, trp1Δ63, ura3-52 | Sharma and Masison (2011) |
| SY135 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS315PSSA2-SSA1 | Sharma and Masison (2008) |
| SY136 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS315PSSA2-SSA2 | Sharma and Masison (2008) |
| SY143 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS315PSSA2-SSA3 | Sharma and Masison (2008) |
| SY211 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS315PSSA2-SSA4 | Sharma and Masison (2008) |
| A42 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS315PSSA2-SSA42 | This study |
| A24 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS315PSSA2-SSA24 | This study |
| Asc200 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS416PGPD-His6SSA2 | Gupta et al. (2018) |
| Asc400 | MATa, PDAL5::ADE2, ssa1::Kan, ssa2::HIS3, ssa3::TRP1, ssa4:: ura3-2f /pRS416PGPD-His6SSA4 | Gupta et al. (2018) |
Table 2. List of plasmids used in the present study.
| Plasmid | Marker | Reference |
|---|---|---|
| pRS316PGAL1-FLAG-vSrc | URA3 | This study |
| pRS316PGAL1-GFP | URA3 | This study |
| PRE-lacZ | URA3 | Morano and Thiele (1999) |
| pRS315PSSA2-SSA1 | LEU2 | Sharma and Masison (2008) |
| pRS315PSSA2-SSA2 | LEU2 | Sharma and Masison (2008) |
| pRS315PSSA2-SSA3 | LEU2 | Sharma and Masison (2008) |
| pRS315PSSA2-SSA4 | LEU2 | Sharma and Masison (2008) |
| pRS315PSSA2-SSA42 | LEU2 | This study |
| pRS315PSSA2-SSA24 | LEU2 | This study |
| pRS416PGPD-His6SSA2 | URA3 | Gupta et al. (2018) |
| pRS416PGPD-His6SSA4 | URA3 | Gupta et al. (2018) |
| pPROEXHTV-YDJ1 | Ampicillin | Sharma and Masison (2011) |
| pET29bHTV-HSP82 | Kanamycin | This study |
| pET29bHTV-STI1 | Kanamycin | This study |
Plasmid pRS316PGAL1-GFP is a URA3-based single-copy vector with GFP under a GAL1 promoter. For protein purification, HSP82 and STI1 were subcloned into the pET29bHTV plasmid using PCR-based amplification of the respective genes, and further digestion using BamH1 and Xho1 to generate pET29bHTV-HSP82 and pET29bHTV-STI. The plasmid encodes (in the 5′ to 3′ direction) a Hexa-His-tag, a TEV protease recognition site, and HSP82 or STI1.
Media and growth conditions
Media composition is as described before (Kumar et al. 2014). Synthetic-defined (SD) media is composed of yeast nitrogen base (233520; BD) with ammonium sulfate (0.67%) and 2% dextrose (50-99-7; Fisher Scientific). SGal media is similar to SD media, except it is supplemented with 2% raffinose (R0250; Sigma [Sigma Chemical], St. Louis, MO) and 2% galactose (G0625; Sigma) instead of dextrose. YPAD media is composed of 1% yeast extract (212750; BD), 2% peptone (244620; BD), and 2% dextrose supplemented with 0.005% adenine (A9126; Sigma). Cells were grown at 30° unless specifically mentioned. Amino acids were supplemented as required.
Immunoblot analysis
Cells grown in liquid media were harvested by centrifugation, and lysed using glass beads. The lysate was further fractionated into supernatant and pellet. The proteins were separated by SDS-PAGE, transferred onto PVDF membranes, and probed using the desired antibodies. The primary antibodies used in the study were as follows: anti-FLAG (F3165; Sigma), anti-phosphotyrosine (05-321; Millipore, Bedford, MA), Anti-GFP (MA5-15256; Thermo Fisher Scientific), anti-Sse1 (a kind gift from J. Brodsky), anti-Ydj1 (SAB5200007; Sigma), anti-Hsc82 (ab30920; Abcam), anti-Hsp70 (ADI-SPA-822-F; Enzo Lifesciences), and anti-Pgk1 (catalog number 459250; Invitrogen, Carlsbad, CA). Primary antibodies were used at dilution of 1:5000 and incubated with an immunoblot for 1 hr at 25°. Lysate for A4 was loaded at three times the amount of A2 lysate to correct for the lower v-Src level in A4 strain, and also for the lower anti-Hsp70 antibody affinity for Ssa4 than Ssa2. Densitometric analysis of immunoblots was performed using UN-SCAN-IT software.
Chase experiments
Yeast strains were grown in selective liquid SD media at 30° until OD600nm reached 0.8–1. Cells were collected using centrifugation and washed three times with sterile water. The v-Src expression was induced by diluting cells into selective liquid SGal media to 0.5 OD600nm. Cells were further grown for 12 hr at 30°. Galactose-mediated protein expression was terminated by shifting cells to growth media containing dextrose instead of galactose. To monitor v-Src degradation, the culture aliquots were collected at the mentioned time points. Cells were harvested and lysed using mechanical disruption. The cellular lysates were probed on an immunoblot using anti-FLAG antibody (F3165; Sigma).
Protein purification
Ydj1 was purified as described earlier (Sharma and Masison 2011).
For Hsp82 and Sti1 purification, plasmids pET29bHTV-HSP82 and pET29bHTV-STI1 were transformed in Escherichia coli Rosetta DE3 (Invitrogen) strains. Cultures were grown in LB broth until OD600nm reached 0.7 and protein expression was induced with 0.3 mM IPTG at 37° for 3 hr. Cells were lysed, and His6-tagged Hsp82 was purified from cellular lysates through cobalt-based Talon metal affinity resin. To remove the His6-tag, purified His6-Hsp82 was incubated with His6-TEV (Tobacco etch virus). The cleaved His6-tag, along with the TEV protease recognition site and His6-TEV protease, was further removed using a metal affinity column.
Sti1 was purified using a process similar to that described above for Hsp82.
Ssa2 and Ssa4 were purified using a process similar to that described for Ssa2 (Gupta et al. 2018). Briefly, strains harboring plasmid pRS416PGPD-His6SSA2 or pRS416PGPD-His6SSA4 as a sole source of Ssa Hsp70 were grown in liquid YPAD media for 24 hr at 30°. Cells were harvested and resuspended in 20 mM HEPES, 150 mM NaCl, 20 mM KCl, and 20 mM MgCl2 pH 7.4 buffer (buffer A) containing protease inhibitor cocktail (Pierce Chemical, Rockford, IL). Cell lysis was carried out using glass beads followed by sonication. Protein purification was carried out using cobalt-based metal affinity resin. The N-terminal His6 tag was removed using TEV protease as described above for Hsp82. The His6 tag cleaved purified protein was further incubated with ATP-agarose resin for 4 hr and eluted with buffer A containing 7 mM ATP and 1 mM DTT (Gupta et al. 2018). Protein purity was confirmed on 10% SDS-PAGE.
The hybrid proteins Ssa24 and Ssa42 were purified using a process similar to that described above for Ssa2.
Immunoprecipitation and pull down
For immunoprecipitation studies, cells were resuspended in 20 mM Tris pH 7.5 buffer containing 150 mM NaCl, 0.5 mM EDTA, and 1 mM PMSF, and lysed by mechanical disruption using glass beads. The lysate was incubated overnight with anti-FLAG antibody-attached resin (A220; Sigma). The unbound fraction in the supernatant was removed by centrifugation at 4000 × g for 1 min. The beads were washed with buffer containing 150 mM NaCl, 0.1% Triton X-100, and 20 mM Tris pH 7.5, and immunoprecipitated proteins were further analyzed on immunoblots using various antibodies.
The pull-down studies were performed as described earlier (Kumar et al. 2015). Briefly, purified His6-Ydj1 or His6-Hsp82 was bound to cobalt-based affinity resin, followed by incubation with yeast lysate. The resin was further washed to remove unbound proteins. The bound fraction was eluted using 20 mM EDTA and probed with the desired antibodies.
Quantitative real-time PCR
The cells were harvested and total RNA was purified using a HiPurA Yeast RNA Purification Kit (MB611 from HiMedia) following the manufacturer’s protocol. About 100 ng of isolated RNA was used to prepare complementary DNA (cDNA) using a cDNA synthesis kit (Verso from AB1453B; Thermo Fisher Scientific). Then, 50 ng of cDNA was used as template for quantitative real-time PCR (qRT-PCR) using a DyNAmoColorFlash SYBR green PCR kit (FNZ416L; Thermo Fisher Scientific) on the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA).
Luciferase refolding assay
The luciferase refolding assay was carried following a procedure similar to that described previously (Kravats et al. 2018), with a few modifications. Briefly firefly luciferase (80 nM) from Promega (Madison, WI) was denatured in the presence of 1 mM ATP at 45° for 10 min. The denatured luciferase (40 nM) was refolded in the presence of 0.3 μM Ydj1 with or without 0.5 μM Hsp70 (Ssa2 or Ssa4). The reaction was incubated at 25° and refolding was initiated by the addition of 1 mM ATP. Refolding was measured as an increase in luminescence with time. To examine the effect of Hsp90 on luciferase refolding, denatured luciferase (40 nM) was incubated in the presence of 0.3 μM Ydj1, 0.5 μM Hsp70 (Ssa2 or Ssa4), 2.4 μM Sti1, and 0.9 μM Hsp82, and the refolding was monitored as stated above.
Ste11 kinase assay
Yeast cells were transformed with plasmid pheromone response element (PRE)-lacZ with the lacZ gene driven by PREs (Morano and Thiele 1999). The cells were grown until OD600nm reached 1 and further treated with α-factor (5 μM) for 6 hr. The β-galactosidase activity was measured as described before (Morano and Thiele 1999). Briefly, OD600nm 1 cells were permeabilized using the freeze–thaw method. The cells were incubated with 200 μl of ONPG (o-nitrophenyl-β-D-galactopyranoside) (4 mg/ml) for 15 min followed by the addition of 1 M Na2CO3. The cells were separated using centrifugation, and the supernatant was used for measuring absorbance at 420 nm.
Biolayer interferometry
Biolayer interferometry studies were performed using an Octet K2 instrument (ForteBio) to monitor the interaction between Ydj1 with different Hsp70 isoforms at 30°. Ydj1 was immobilized on amine-reactive second-generation (AR2G) biosensors, activated using a 1:1 ratio of 0.1 M N-hydroxysuccinimide and 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide, to a response signal of 1 nm. Postimmobilization, the biosensor was blocked with 1 M ethanolamine. The reference biosensor was activated similarly except that assay buffer (25 mM HEPES pH 7.4, 150 mM NaCl, 20 mM MgCl2, and 20 mM KCl) lacking Ydj1 was used for immobilization on AR2G biosensors. The immobilized Ydj1 and reference biosensors were dipped into assay buffer containing increasing concentrations (0.25–1 μM) of Ssa Hsp70 isoforms in a 96-well plate, and binding was monitored as an increase in binding response for 200 sec. Dissociation was monitored in buffer alone for 200 sec. The nonspecific signal obtained from reference biosensor was subtracted from the corresponding response signal of Ydj1 and Hsp70 interaction. The obtained binding sensograms were analyzed using software “Data Analysis 9.0” available from ForteBio.
Significance test
Three biological replicates were performed for each experiment. To compare significance among the groups, the Student’s t-test was used. P-values are shown as follows: *P < 0.05, **P < 0.01, and ***P < 0.001, with P < 0.05 considered statistically significant.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12136941.
Results
Hsp90 chaperoning action varies with its partner Ssa Hsp70 isoforms
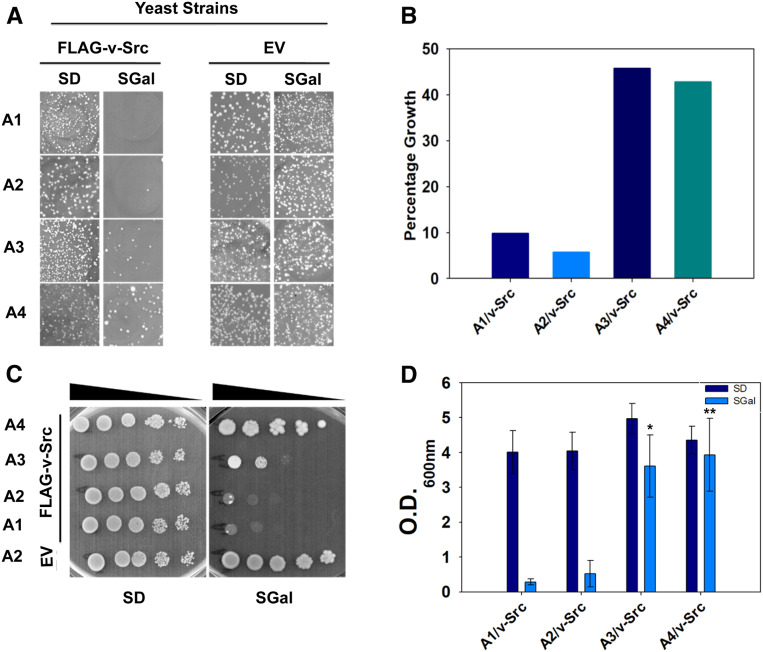
The oncogene v-Src of the Rous sarcoma virus is one of the most well-studied Hsp90 clients in the yeast S. cerevisiae. Although S. cerevisiae does not encode for v-Src kinase, its heterologous expression and subsequent maturation leads to tyrosine phosphorylation of many cellular proteins (Brugge et al. 1987). The uncontrolled phosphorylation activity induces cellular growth arrest in the yeast. The poor growth of the yeast cells upon v-Src overexpression is thus indicative of native folding and maturation of the kinase to its active form. We first confirmed v-Src-mediated cellular growth arrest in a wild-type strain encoding all four cytosolic Ssa Hsp70 isoforms (Supplemental Material, Figure S1). To examine the role of different Ssa Hsp70 isoforms in Hsp90 chaperoning functions, we overexpressed v-Src from a galactose-inducible promoter in yeast strains that express only one of the desired isoforms in the absence of all four chromosomally encoded Hsp70s. To achieve similar expression levels of the expressed Ssa Hsp70s, all isoforms are expressed under the same native constitutive Ssa2 promoter. Figure 1 shows the growth phenotypes of cells harboring either empty plasmid or plasmid-encoding v-Src under a galactose-inducible promoter. In the absence of v-Src expression, all strains expressing individual Ssa Hsp70 isoforms grew similarly (Figure 1, A and C). The v-Src overexpression in strains expressing Ssa1 (A1) or Ssa2 (A2) as the sole source of Hsp70 led to growth defects, as is evident from their poor growth onto SGal solid media. Interestingly, cells overexpressing v-Src with Ssa3 (A3) or Ssa4 (A4) as the sole Ssa Hsp70 grew better than A1 or A2. The v-Src-mediated growth defects were further examined using spot dilution assays onto solid growth media (Figure 1C). As observed for the primary transformants, the A1 and A2 strains showed higher v-Src-mediated toxicity than strains expressing stress-inducible Ssa Hsp70s. Similar results were obtained when A1–A4 strains expressing v-Src were grown at 30° for 72 hr in liquid SD media or with galactose (SGal) as the carbon source (Figure 1D). As the A1–A4 strains are isogenic except for the presence of different Ssa Hsp70 isoforms, the data suggest that stress-inducible Ssa Hsp70s function differently than the constitutive isoforms in promoting the maturation of Hsp90 substrate v-Src.
Figure 1.
Strains expressing Ssa3 or Ssa4 showed reduction in v-Src toxicity. (A) S. cerevisiae strains A1–A4 were transformed with pRS316 (EV) or pRS316PGAL1-FLAG-v-Src (FLAG-v-Src). Growth of transformants is shown on SD and SGal solid media after 3 and 5 days, respectively, at 30°. (B) Graph represents percentage numbers of colonies grown on SGal growth media. Equal numbers of cells were plated onto SD and SGal media, and percentage growth on SGal was calculated with respect to SD media. (C) Cells grown in selective liquid SD media were washed and serially diluted onto SD and SGal media. Growth is shown after 4 days of incubation at 30°. (D) First, 5–6 transformants were pooled and grown in selective liquid SD media. Cells were reinoculated at OD600nm 0.02 into SD or SGal media. Shown is the OD600nm of each cell culture after 72 hr of incubation at 30°. Error bar represents SD from three different biological replicates. P-values were calculated using the Student’s t-test and A2 as a control. EV, empty vector; SGal, SD media supplemented with 2% raffinose, 2% galactose and 2% dextrose respectively.
To examine whether the distinct effect of stress-inducible Hsp70s is specific to v-Src or is more general for other Hsp90 clients, we further explored the maturation of an another Hsp90 client, Ste11, in the A2 and A4 strains. The Ste11 kinase is required for the activation of Ste12, which regulates PREs. Ste11 maturation can thus be widely monitored by the activity of β-galactosidase expressed under the control of a PRE. The plasmid encoding PRE-lacZ was transformed into the A2 or A4 strains. The transformants were grown on liquid SD media until OD600nm reached 1 and were treated with α-factor for the induction of Ste11 expression. Next, β-galactosidase activity was monitored as described in the Materials and Methods section. Compared with A2, the β-galactosidase activity was found to be lower in the A4 cells (Figure S2). These results suggest that, similar to v-Src, Ste11 maturation is also reduced in A4 cells.
The reduction in v-Src toxicity is due to faster degradation of v-Src
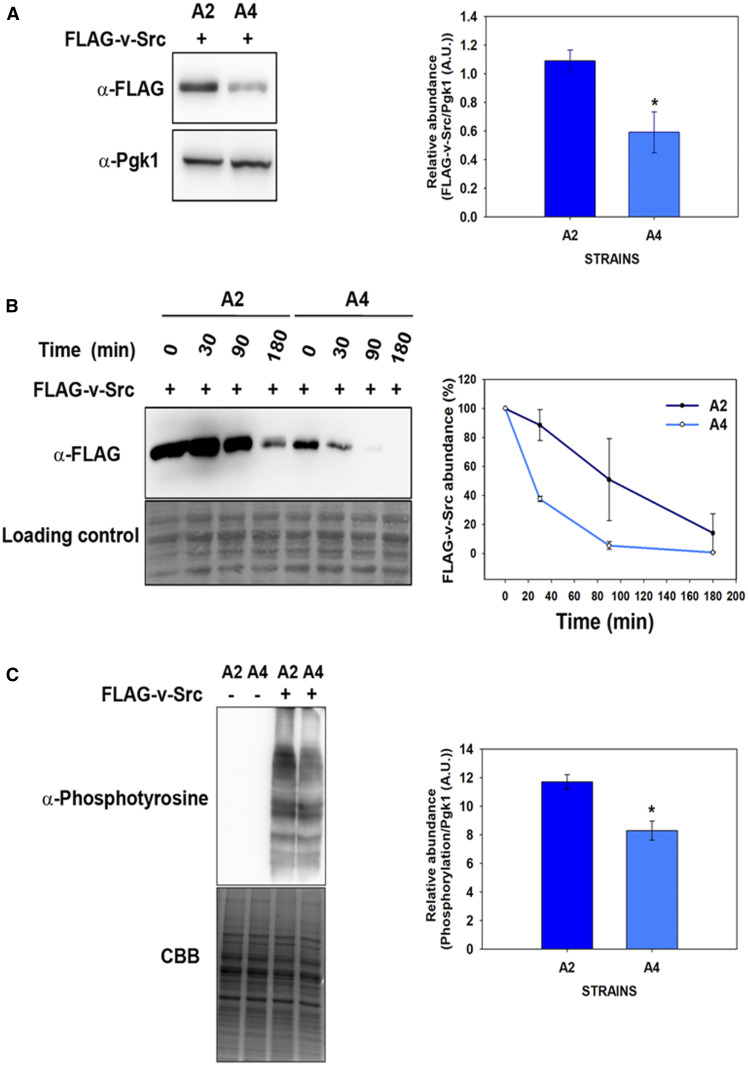
The above results show that strains with stress-inducible Ssa3 or Ssa4 as the sole Hsp70 reduce v-Src-mediated toxicity. Furthermore, a comparison of the growth phenotypes of A3 vs. A4 cells shows that v-Src overexpression is less detrimental in cells expressing Ssa4 than in those expressing Ssa3. To further explore the mechanism of reduced v-Src toxicity, we used A2 and A4 as representative members of constitutive and stress-inducible Hsp70 chaperones. v-Src kinase requires Hsp90 chaperone machinery to fold it to the native state, and if folding fails the kinase is targeted for degradation (An et al. 2000; Kundrat and Regan 2010). Thus, we examined v-Src abundances in the A2 and A4 strains to explore whether v-Src toxicity in the A4 strain is reduced due to a defect in its maturation. The cells expressing FLAG-tagged v-Src from a galactose-inducible promoter were grown in liquid growth media containing galactose for 12 hr and the cellular lysate was then fractionated onto 12% SDS-PAGE before probing with the anti-FLAG antibody. As shown in Figure 2A, the v-Src expression was ∼2–2.5-fold lower in A4 than in the A2 strain.
Figure 2 .
v-Src activity and its degradation in the A4 and A2 strains. (A) v-Src expression was induced for 12 hr, and its abundance was monitored in cellular lysate with the anti-FLAG antibody. (B) v-Src expression was induced for 12 hr in SGal media. Cells were then shifted to noninducible SD liquid media and v-Src abundance was chased at the indicated time intervals with anti-FLAG antibody. (C) Equal amounts of cellular lysate from the A2 or A4 strains expressing v-Src were probed with anti-phosphotyrosine antibody. Panel toward right depicts quantification of respective immunoblots. Error bars represents SD from three different biological replicates. P-values were calculated using the Student’s t-test and A2 as a control. A.U., arbitrary units; SGal, SD media supplemented with 2% raffinose, 2% galactose and 2% dextrose respectively.
To examine whether the relatively reduced abundance of v-Src in the A4 strain is due to an effect of the galactose-inducible promoter, GFP was used as a reporter gene to monitor the strength of the GAL1 promoter in the A1–A4 strains. The gene encoding GFP was subcloned under the GAL1 promoter, and its expression was monitored using immunoblot analysis with anti-GFP antibodies and with fluorescence microscopy. Both the immunoblot analysis with the anti-GFP antibody (Figure S3A) and fluorescence microscopy (Figure S3B) showed that the GFP levels were similar in the A1–A4 strains, thus suggesting that the galactose promoter’s strength remains independent of variations in the Hsp70 isoforms.
We further examined the degradation rates of v-Src in cells expressing Ssa2 or Ssa4 as the sole Ssa Hsp70 source. The cells were grown under inducible conditions for 12 hr and were then shifted to repressible media to suppress v-Src expression. The abundances of preformed v-Src were then monitored at different time intervals (Figure 2B). As was observed, although even after 30 min of chase, there was no significant change in the v-Src levels in the A2 cells and >60% of v-Src was found to be degraded in the A4 cells. After ∼90 min of growth in repressible media, most of the preformed v-Src was degraded in the A4 cells whereas >50% of the v-Src was still present in the A2 cells. The enhanced degradation in the A4 strain was specific to v-Src as both GFP and other cellular proteins showed similar abundances in the A2 and A4 strains (Figure S4). Collectively, the above data suggest that v-Src degradation rates are higher in the A4 strain than in the A2 strain.
The constitutively active mature v-Src randomly phosphorylates most of the tyrosine-containing proteins of the yeast proteome (Brugge et al. 1987). To examine the tyrosine kinase activity of v-Src, cells harboring a v-Src expression plasmid were grown in inducible growth media. The cells were then lysed and the lysate was immunoblotted with an anti-phosphotyrosine antibody. As expected, most of cellular proteins were detected with the anti-phosphotyrosine antibody (Figure 2C,) but the phosphorylation levels varied between the A2 and A4 strains. Overall, the band intensity, which is a reflection of v-Src kinase activity, was found to be higher in the lysate obtained from the A2 cells than that obtained from the A4 cells, thus suggesting relatively higher v-Src maturation levels in cells expressing Ssa2 than those expressing Ssa4 as the sole Ssa Hsp70 source. Thus, the v-Src kinase activities in the A2 and A4 cells paralleled the growth defects observed in these strains. Overall, these results show that v-Src maturation is significantly reduced in strains expressing Ssa4 Hsp70 as the partner Hsp90 protein; these results indicate that different Hsp70 isoforms function differently in the Hsp90 chaperoning pathway.
The A2 and A4 strains expressing v-Src show similar abundances of other major chaperones
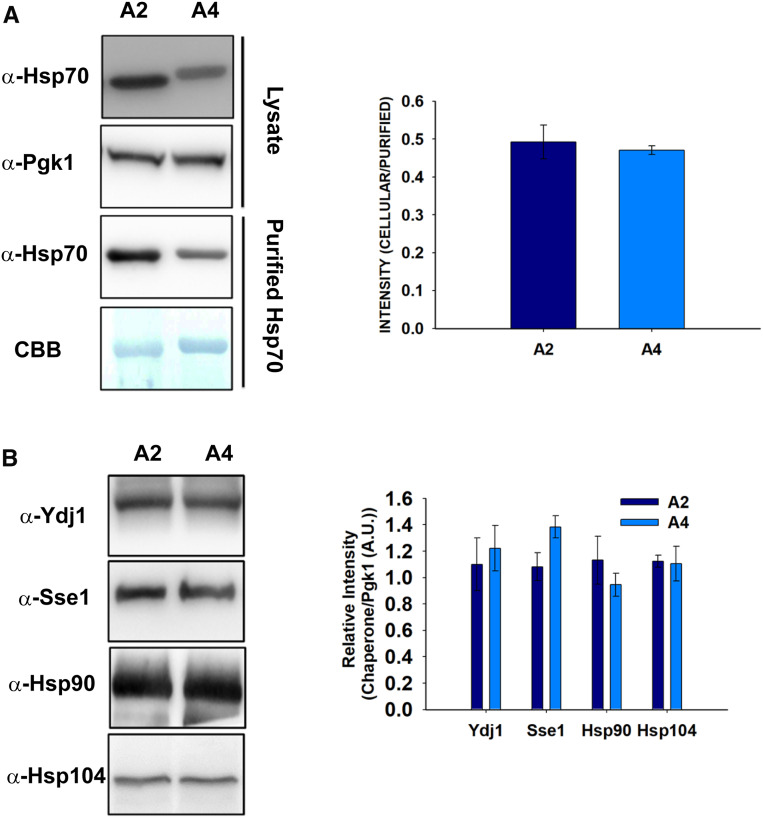
To explore whether the observed differences in v-Src maturation are related to altered abundances of the major heat-shock proteins involved in client protein maturation, we examined the expression levels of Hsp70, Ydj1, Sse1, Hsp104, and Hsp90 in both the A2 and A4 strains. Both Ydj1 and Sse1 are known to affect v-Src maturation, suggesting that both cochaperones play important roles in Hsp90-dependent functions (Dey et al. 1996; Goeckeler et al. 2002). Similarly, Hsp104 is known to interact with Hsp90 cochaperones as well as with Hsp70 (Abbas-Terki et al. 2001; Reidy and Masison 2010).
We first examined the Ssa2 and Ssa4 levels in the A2 and A4 strains, respectively. Since the Hsp70 antibody recognizes Ssa2 with three times higher affinity than Ssa4 (Gupta et al. 2018), the cellular abundances of the Ssa Hsp70 isoforms were measured by normalizing the amount of Hsp70 detected in the whole-cell lysate with respect to that observed using in vitro purified Hsp70s (Figure 3A). In agreement with our previous study, similar Ssa2 and Ssa4 levels were observed from the A2 and A4 strains, respectively, suggesting that the variations in kinase maturation are not due to varying Hsp70s levels.
Figure 3.
A2 and A4 strains show similar abundance of major chaperones. Yeast lysate was prepared from the indicated strains overexpressing v-Src for 12 hr. (A) Equal amounts of the cellular lysate (upper panel) or purified His6-tagged Ssa Hsp70 (Lower panel) were loaded onto 10% SDS-PAGE gels and probed with anti-Hsp70 antibody. CBB was used to stain purified Hsp70s as loading control. (B) Equal amounts of cellular lysates from indicated strains were probed with antibodies against Ydj1, Sse1, Hsp90, or Hsp104. Right panel shows quantification of respective immunoblots. Error bars represent SD from three different biological replicates. A.U., arbitrary units; CBB, Coomassie Brilliant Blue
To monitor Ydj1 abundance, the cellular lysates from the A2 and A4 cells were normalized for the total protein amount, and were further probed on an immunoblot with antibodies against Ydj1 (Figure 3B). As seen in Figure 3B, Ydj1 was found to be similar in both the A2 and A4 strains. As v-Src is an Hsp90 substrate, we further examined Hsp90 abundances similar to those described above for Ydj1. We first examined the specificity of the anti-Hsp90 antibody for either of the two Hsp90 isoforms, namely, Hsc82 and Hsp82. As shown in Figure S5, the antibody detects both of the Hsp90 isoforms. The immunoblot results with the anti-Hsp90 antibodies showed that Hsp90 is expressed similarly in both the A2 and A4 strains (Figure 3B). Similarly, no significant differences were observed for Hsp104 and Sse1 in the A4 strain vs. the A2 strain. These results generally suggest that decreased v-Src toxicity in the A4 strain is not related to altered levels of the major heat-shock proteins.
v-Src interaction with Ssa4 is lower than with Ssa2
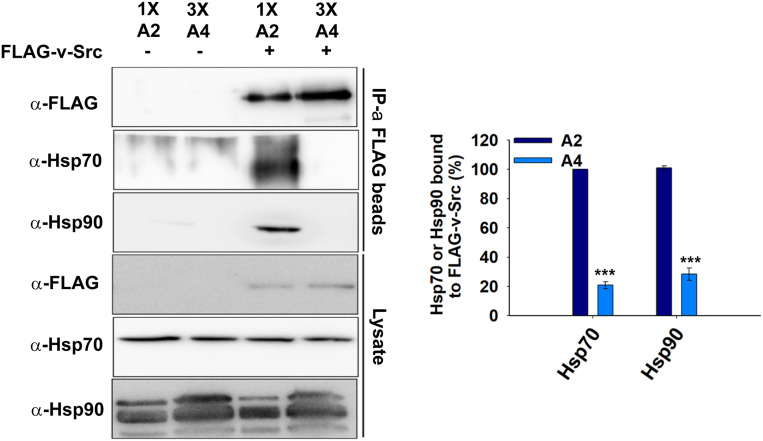
The reduced tyrosine phosphorylation activity and associated toxicity of v-Src in the A4 strain indicates a defect in its maturation to a native folded state. As v-Src is an Hsp90 substrate and this interaction facilitates v-Src maturation, we conducted immunoprecipitation assays to examine the v-Src interactions with Hsp90 in both the A2 and A4 strains. Cells expressing FLAG-v-Src were grown for 12 hr and the cellular lysates were incubated with immobilized beads of anti-FLAG antibodies. As the v-Src steady-state level is lower in A4 cells, for immunoprecipitation studies greater quantities of cellular lysates from A4 cells (3x) were incubated with beads coated with anti-FLAG antibodies to capture equal amounts of v-Src from the A2 and A4 strains. The similar levels of immobilized v-Src were confirmed on an immunoblot with an anti-FLAG antibody (Figure 4, upper panel). The co-immunoprecipitated proteins were further probed with the anti-Hsp90 antibody. As shown in Figure 4, although similar levels of v-Src were detected, more Hsp90 was obtained from A2 than from the A4 strain, thus suggesting that less of the v-Src interacts with Hsp90 in the A4 strain.
Figure 4.
The v-Src interaction with Hsp70 and Hsp90 using immunoprecipitation studies. The indicated strains were grown in selective SGal media for v-Src expression. The cells were lysed, and lysates were incubated with anti-FLAG antibody immobilized beads. The immunoprecipitated proteins were probed with indicated antibodies. Right panel shows quantification of respective immunoblots. Error bar represents SD from three different biological replicates. P-values were calculated using the Student’s t-test and A2 as a control. IP, immunoprecipitated; SGal, SD media supplemented with 2% raffinose, 2% galactose and 2% dextrose respectively.
It is well known that many Hsp90 substrates first interact with Hsp70 before being transferred to Hsp90 (Arlander et al. 2006; Cintron and Toft 2006). To explore whether reduced levels of the v-Src-Hsp90 complex in the A4 strain were due to altered upstream interactions of v-Src with Hsp70s, we examined its interactions with Ssa2 and Ssa4. Similar to that described above for Hsp90, the co-immunoprecipitated proteins with FLAG-v-Src were further probed with anti-Hsp70 antibodies. As seen for Hsp90, although both Ssa2 and Ssa4 were detected at similar levels in the cellular lysates, less Ssa4 was found in the co-immunoprecipitated protein sample (Figure 4). These results suggest that although Ssa2 or Ssa4 are highly homologous, their binding properties with v-Src differ significantly in vivo.
Both the Ssa2 and Ssa4 isoforms interact similarly with Hsp90
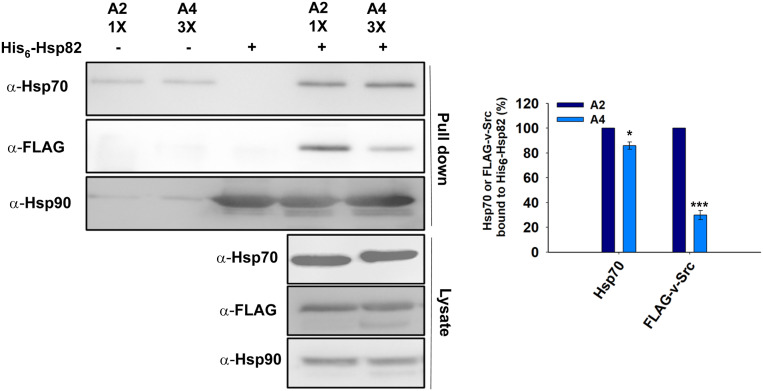
We further examined the ability of Ssa2 and Ssa4 to interact with Hsp90. His6-Hsp82 was used as the immobilized bait for the Hsp70s present in the cellular lysates from the A2 and A4 strains. Equal amounts of purified hexa-His-tagged Hsp82 (His6-Hsp82) were bound over a cobalt-based metal affinity resin. The cellular lysates obtained from the A2 and A4 cells expressing FLAG-v-Src were passed through His6-Hsp82-bound beads. The beads were subsequently washed and the bound proteins were eluted using 20 mM EDTA. The eluted proteins were then immunoblotted with anti-Hsp70, anti-Hsp90, and anti-FLAG tag antibodies. The eluted fractions from the A2 and A4 cellular lysates, when probed with the anti-Hsp70 antibody, showed similar levels of Hsp70s, thus suggesting that the two Hsp70 isoforms bind with similar affinities to Hsp90 (Figure 5). Furthermore, lower amounts of v-Src were found in the eluted fraction from A4 than in the eluted fraction with A2 cells; this result is in agreement with the above data and shows that v-Src binds with lower affinity to Hsp90 in A4 (Figure 5). Overall, the above data show that both A2 and A4 bind with similar affinity to Hsp90 and A4 cells, but that less of the v-Src interacts with Hsp90, which could be due its relatively poor interaction with Ssa4 compared with that with Ssa2.
Figure 5.
The Hsp82 interaction is similar with Ssa2 and Ssa4 isoforms of Hsp70. The purified His6-Hsp82 was adsorbed onto cobalt metal affinity resin and further incubated with yeast lysate from A2 or A4 strains expressing FLAG-v-Src. The bound fractions were probed for Ssa Hsp70s, v-Src, or Hsp82 with anti-Hsp70 antibody, anti-FLAG antibody, or anti-Hsp90 antibody, respectively. Panel toward right depicts quantification of respective western blots. Error bar represents SD from three different biological replicates. P-values were calculated using the Student’s t-test and A2 as a control.
Ydj1 interacts poorly with Ssa4 compared with Ssa2
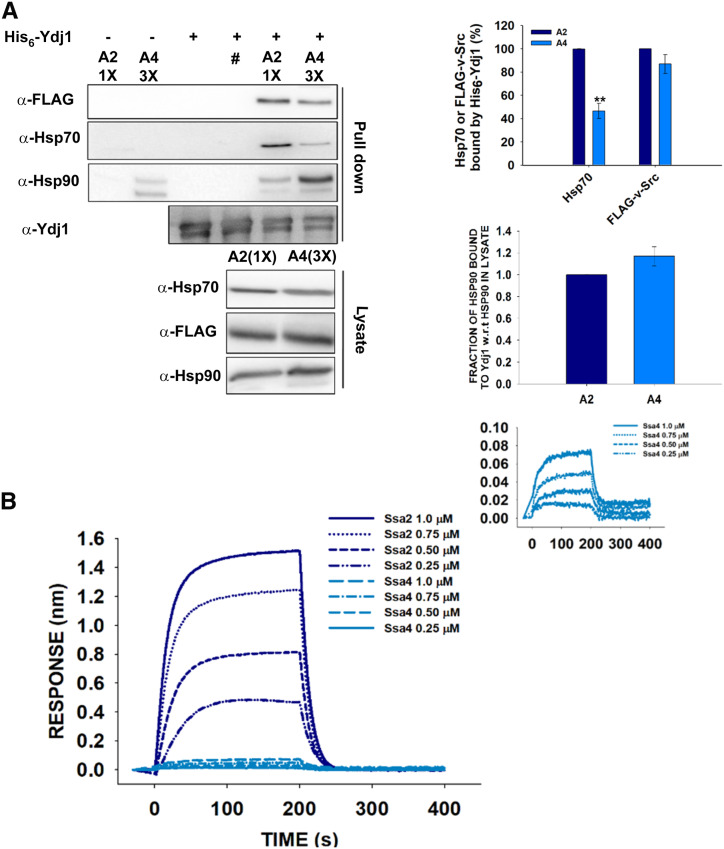
As Ydj1 is known to play an important role in substrate transfer to Hsp70s (Dey et al. 1996), we next explored the ability of v-Src to interact with Ydj1 in A2 and A4 cells expressing FLAG-tagged v-Src. The interactions were examined using a pull-down assay with Ydj1 as the bait protein and the bound fractions were probed with anti-FLAG antibodies. As shown in Figure 6A, roughly similar amounts of v-Src were detected in the A2 and A4 strains, suggesting that Ydj1 binds with similar affinity to v-Src from either the A2 or A4 strain.
Figure 6.
The Ydj1 interaction is stronger with Ssa2 than Ssa4. (A) The His6-Ydj1 immobilized beads were incubated with yeast lysate from A2 (1X) or A4 (3X) cells expressing FLAG-tagged v-Src. The bound proteins were probed with anti-FLAG, anti-Hsp70, anti-Hsp90, or anti-Ydj1 antibody. Relatively higher amount of Hsp70 from the A2 strain was detected indicating a stronger interaction of Ydj1 with Ssa2 than Ssa4. Panel toward right depicts quantification of respective western blot. #A mutant of Ydj1 [His6-Ydj1(H34Q)] was used as negative control. (B) The biolayer interferometry sensogram showing interaction of Ydj1 with increasing concentrations of Hsp70 isoforms (Ssa2 or Ssa4) as analytes. A much stronger interaction of Ydj1 with Ssa2 than Ssa4 was observed at similar concentrations of the two Hsp70 isoforms. Inset shows the zoomed-in view of Ydj1 interaction with Ssa4. The labels are represented as the same order of magnitude of the curve. Error bars represent SD from three different biological replicates. P-values were calculated using the Student’s t-test and A2 as a control. w.r.t is defined as with respect to.
We next determined the interactions between Ydj1 and Ssa2 or Ssa4 using methods similar to those mentioned above for its interaction with v-Src. The eluted fractions from Ydj1-bound beads were probed with anti-Hsp70 or anti-Hsp90 antibodies. Figure 6A shows that compared with Ssa2, significantly lower amounts of Ssa4 were detected in the eluted fraction, thus suggesting that Ydj1 binds with relatively weaker affinity to Ssa4 than to Ssa2. Interestingly, Hsp90 from the A2 and A4 cells was found to interact similarly with Ydj1.
To further examine the Ydj1 interactions with the Ssa Hsp70 isoforms, we conducted biolayer interferometry studies with purified chaperones as described in the Materials and Methods section. Biolayer interferometry is an optical label-free method that is extensively used to monitor biomolecular interactions in real time. The Ydj1-loaded biosensor tips were immersed in solutions containing ATP (5 mM) and with increasing concentrations of one of the Hsp70 isoforms (Ssa2 or Ssa4). Figure 6B shows the sensograms for the binding of Ydj1 with the Ssa Hsp70 isoforms. As seen, incubation of the Ydj1-coated biosensor tips with solutions containing Ssa2 or Ssa4 led to increases in the biolayer interferometry signals. The binding response increased with increasing concentrations of the Ssa Hsp70 isoforms. Furthermore, at similar concentrations, the binding response was much stronger for Ssa2 than for Ssa4, suggests that the Ydj1 binding affinity is higher for Ssa2 than for Ssa4, which is in agreement with the above pull-down assays that showed relatively stronger binding of Ydj1 with Ssa2.
Ydj1-assisted substrate refolding is more efficient with Ssa2 than with Ssa4
As Ydj1 assists Hsp70 in stimulating ATPase activity as well as in substrate transfer, any variation in its interaction with Ssa2 vs. Ssa4 might affect the downstream maturation of Hsp70 or Hsp90 substrates. Therefore, we further explored the Ydj1-assisted refolding of thermally denatured luciferase, which is a well-known Hsp70 substrate. Luciferase unfolds upon incubation at higher temperatures and its refolding back to a native state requires the presence of Hsp70. This refolding is further enhanced when Hsp90 is co-incubated with Hsp70s in a refolding buffer.
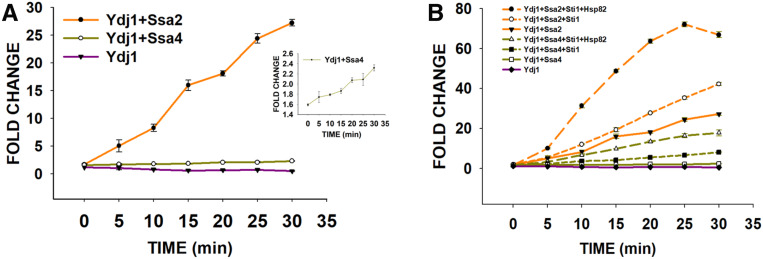
Luciferase was denatured by incubation at 45° for 10 min. Refolding was initiated by incubating denatured luciferase with Ydj1 in both the presence and absence of Ssa2 or Ssa4 at 25° for different time intervals. As seen in Figure 7A, the presence of Ydj1 alone in the refolding buffer was not able to refold luciferase. As expected, the fraction of refolded luciferase, as measured by the increase in luminescence, increased when Ssa2 or Ssa4 was added into the refolding buffer containing Ydj1. The refolding level increased with increasing incubation time and saturated at ∼30 min. Although luciferase refolded in the presence of either Ssa2 or Ssa4, the fraction of refolding was found to be significantly greater for Ssa2 than for Ssa4 (Figure 7A). After 30 min of incubation with Ssa2:Ydj1, the luciferase refolding level was ∼25-fold, compared with only 2.5-fold with Ssa4:Ydj1. Overall, the data suggest that the Ssa2:Ydj1 complex is more efficient than Ssa4:Ydj1 for the refolding of denatured luciferase. These results are in agreement with the above pull-down assay, which showed weaker Ydj1 interaction with Ssa4 than with Ssa2.
Figure 7.
Ssa2 has higher luciferase refolding activity compared to Ssa4. (A) Luciferase was denatured at 45°, and refolded in the presence of Ssa2 or Ssa4 and Ydj1. As shown, fraction of luciferase that refolded was higher in the presence of A2 than A4. (B) The denatured luciferase (40 nM) was incubated in the presence of 0.3 μM Ydj1, 0.5 μM Hsp70 (Ssa2 or Ssa4), 2.4 μM Sti1, and 0.9 μM Hsp82, and the refolding was monitored by measuring the increase in luminescence. The luciferase refolding curves for Ydj1:Ssa2, Ydj1:Ssa4, and Ydj1 alone in (B) are adapted from (A) for comparison. The labels are represented as the same order of magnitude of curve. Error bars represent SD from three different biological replicates.
We next examined the effect of Hsp90 on the refolding of denatured luciferase. The bridge protein Sti1 was added to the refolding reaction containing either Ssa2 or Ssa4 isoforms and Ydj1, and the refolding was monitored in both the presence and absence of an Hsp82 isoform of Hsp90. As expected, the addition of Hsp90 further enhanced luciferase refolding and relatively more so in reactions containing Ssa2 rather than Ssa4 (Figure 7B). After 30 min of incubation, the luciferase refolding was ∼66-fold in the reaction containing Ssa2:Ydj1:Sti1:Hsp82, compared with a level of only 16-fold with Ssa4:Ydj1:Sti1:Hsp82. Overall, these studies suggest that Ydj1:Ssa2:Hsp90 has higher refolding activity than Ydj1:Ssa4:Hsp90.
The above results suggest that Ydj1 association with Ssa Hsp70 is crucial for substrate refolding. To confirm whether Ydj1 is also crucial for the maturation of v-Src in vivo, we examined wild-type and ydj1Δ strains for v-Src-mediated growth defects. The cells were transformed with the plasmid-encoding FLAG-v-Src under a galactose-inducible promoter and the transformants were further monitored for growth onto media containing dextrose or galactose as the carbon source. As shown in Figure S6, in the presence of v-Src, cells lacking Ydj1 grew significantly more than wild-type cells, thus suggesting the critical role of Ydj1 in v-Src maturation.
The functional distinction between Ssa2 and Ssa4 is governed by their C-terminal domains
Ydj1 interacts with Hsp70 at its ATPase, as well as the C-terminal, domain (Gong et al. 2018). Although Ydj1 interaction with the ATPase domain is crucial for stimulating Hsp70 ATPase activity, its coordination with the C-terminal domain facilitates substrate transfer (Demand et al. 1998).
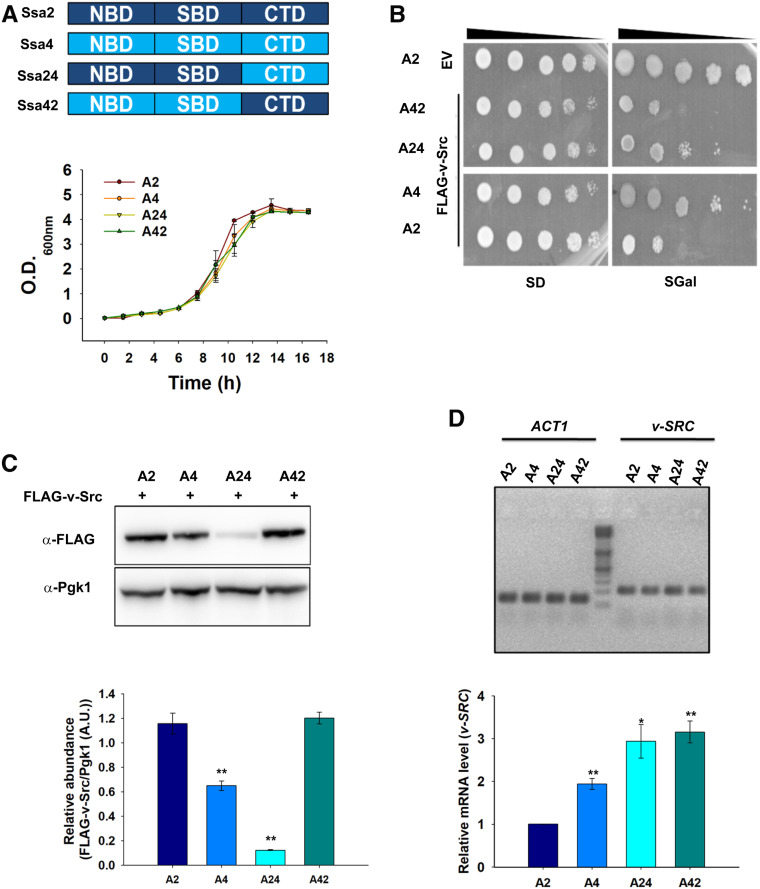
To examine the role of nucleotide-binding domain (NBD) and C-terminal domain in determining the functional distinction of the Ssa Hsp70 isoforms, we swapped these domains and constructed two hybrid proteins based upon Ssa2 and Ssa4 as the parent proteins (Figure 8A). The hybrid Ssa24 encodes the NBD and SBD (substrate binding domain) of Ssa2, and the C-terminal domain of Ssa4. Similarly, Ssa42 encodes the NBD and SBD of Ssa4, and the C-terminal domain of Ssa2. Strains expressing Ssa24 or Ssa42 (e.g., A24 or A42, respectively) as the sole Ssa Hsp70 source were constructed and examined for v-Src toxicity. In the absence of v-Src, the strains expressing the hybrid proteins showed similar growth on liquid YPAD as well as on solid SD media, thus suggesting that designed hybrid proteins are well folded and functionally active (Figure 8, A and B). Furthermore, the hybrid proteins were expressed at similar levels as those of wild-type Ssa2 or Ssa4 (Figure S7). As seen before, the toxicity is reduced in the Ssa4 strain compared with Ssa2. For strains expressing hybrid proteins, we found that the v-Src overexpression was less toxic in strains expressing Ssa24 than in those expressing Ssa42 (Figure 8B); this result suggests that the C-terminal domain governs the functional distinction between Ssa2 and Ssa4 for the activities required for maturation of the Hsp90 client protein v-Src. The reduced toxicity of the Ssa24 strain is not due to altered expression of other chaperones such as Ydj1 and Hsp90, and suggests a direct role of the hybrid chaperone in the maturation of v-Src kinase (Figure S7). v-Src maturation was further confirmed by measuring its steady-state level in the Ssa24- and Ssa42-expressing strains. As shown in Figure 8C, the v-Src levels were found to be much lower in Ssa24 than in Ssa42; this result suggests its higher degradation in Ssa24. To further explore whether lower levels of v-Src in A4 and A24 are related to lower amounts of v-Src transcription, we conducted qRT-PCR with primers specific for v-Src. As shown in Figure 8D, no significant differences in v-Src messenger RNA levels were observed in A4 and A24 compared with A2 and A42, respectively.
Figure 8.
C-terminal domain of Hsp70 governs its specificity for v-Src maturation. (A) Upper panel shows schematics of designed hybrid Hsp70 proteins. Amino acid sequences at hybrid junction in Ssa24 and Ssa42 are 537–538 of Ssa2 and 538–539 of Ssa4, respectively. Lower panel shows growth curve of indicated strains in liquid YPAD media. (B) Shown is the growth of indicated strains on solid SD or SGal media after 5 days of incubation at 30°. (C) Immunoblot showing steady-state expression of FLAG-tagged v-Src or Pgk1 (as control) in indicated strains. (D) The relative abundance of mRNA encoding v-Src in indicated strains as measured using qRT-PCR. The qRT-PCR was carried out using primers specific for v-SRC or ACT1 (as control). Lower panels (C and D) show relative quantification. Error bar represents SD from three different biological replicates. P-values were calculated using the Student’s t-test and A2 as a control. A.U. arbitrary units; CTD, C-terminal domain; EV, empty vector; mRNA, messenger RNA; NBD, nucleotide-binding domain; SBD, substrate binding domain; qRT-PCR, quantitative real-time PCR; SGal, SD media supplemented with 2% raffinose, 2% galactose and 2% dextrose respectively. YPAD, 1% yeast extract, 2% peptone, and 2% dextrose supplemented with 0.005% adenine.
We further measured the Ydj1 interactions with hybrid Ssa24 and Ssa42 using a pull-down assay with His6-tagged Ydj1 as the bait protein. His6-Ydj1 was bound over cobalt metal affinity beads, and the cellular lysates from the A24 and A42 cells were passed through the Ydj1-bound beads (Figure S8). Similar to the results shown above, Ydj1 binds with relatively higher affinity to Ssa2 than to Ssa4. For the hybrids, the Ydj1 interaction with Ssa24 is stronger than with Ssa42, thus suggesting that the NBD plays a dominant role in regulating Ydj1 interactions with Ssa Hsp70; this is expected based on previous findings (Jiang et al. 2007).
Overall, these results point toward a role of the C-terminal domain of the Hsp70 isoforms in determining the functional distinction between Ssa2 and Ssa4 in the Hsp90 pathway.
Discussion
Hsp70 and Hsp90, along with their cochaperones, form two major classes of cellular chaperone machinery. Hsp70-mediated substrate refolding is independent of Hsp90 actions; however, Hsp90 requires Hsp70 for the maturation of many of its client proteins. Therefore, insight into how Hsp70 coordinates with Hsp90 is critical to further enhance our understanding of Hsp90 function. As eukaryotes carry multiple cytosolic members of the Hsp70 family, the role of each of these highly homologous members in Hsp90 activity is not clear. Although the members of the Hsp70 or Hsp90 families are very homologous within each family, they function distinctly in many cellular processes. The current study thus dissects the role of each of the Hsp70 isoforms in the Hsp90 pathway and shows that different isoforms behave differently with Hsp90s, and that the distinction is primarily governed by the C-terminal domain of Hsp70s.
The results showing maturation of the Hsp90 clients v-Src and Ste11 in strains with different Ssa Hsp70 isoforms reveal clear distinctions between the roles of the constitutive and stress-inducible Hsp70s in the Hsp90 chaperoning pathway. The growth of A3 and A4 cells, even with v-Src overexpression, could either be due to a cytoprotective action of the stress-inducible Ssa Hsp70 isoforms or lack of the function required for folding of the Hsp90 substrate to its active conformation that is critical for uncontrolled protein tyrosine phosphorylation. Several lines of evidence suggest that a lack of v-Src folding protects cells from growth arrest. First, the level of protein phosphorylation is significantly lower in the A4 than in the A2 strain, and suggests reduced accumulation of active v-Src kinase. Second, the higher degradation rate of v-Src in A4 than in A2 cells is also an indication of its reduced maturation. Third, the wild-type strain expressing all four Ssa Hsp70 isoforms when grown under heat-stress conditions is unable to reduce v-Src toxicity, and suggests a recessive role for Ssa3 or Ssa4 with regard to v-Src-associated toxicity (Figure S9). These results thus suggest that constitutively expressed Ssa Hsp70s cooperate better than their stress-inducible members with Hsp90s for folding of their client proteins. It is likely that the evolutionary pressure on Hsp70s to coordinate with Hsp90s might have been more selective toward the constitutively present Ssa Hsp70s rather than for those isoforms that are expressed only under stress. Thus, these results suggest that in addition to their redundant roles, the constitutive and stress-inducible Hsp70 members also evolved with more specialized distinct functions, such as their role in the Hsp90 pathway. This agrees with our previous results showing that A3 cells more effectively reduce α-synuclein toxicity than A2 cells; this cytoprotective effect is primarily mediated through autophagy (Gupta et al. 2018).
Hsp90 coordinates with Hsp70 for many of its cellular functions. Interaction studies using immunoprecipitation of FLAG-v-Src have revealed that the client interacts poorly with Hsp90s in A4 compared with the A2 strain. Since the same Hsp90 isoforms are present in the A2 and A4 strains, it is intriguing to note that the v-Src interaction with Hsp90 is strain-dependent. Both the A2 and A4 strains are isogenic except for the presence of Ssa2 and Ssa4, respectively, and thus the distinct v-Src-Hsp90 interactions must be regulated by the Hsp70 isoforms or their interacting cochaperones. The Hsp70 interactions with Hsp90 are known to be mediated through the nucleotide-binding domain as well as the EEVD motif present at the C-termini of these proteins (Kravats et al. 2017). Our results from the pull-down assay show that both Ssa2 and Ssa4 interact similarly with Hsp82, and thus the relatively weaker interaction of v-Src with Hsp82 in the A4 strain might be due to a cellular process upstream of the Hsp70-Hsp90 collaboration.
Next, we find that the v-Src interaction with Ssa4 is relatively weaker than the interaction with Ssa2, which could be the basis for the relatively poor interaction observed between v-Src and Hsp90 in the A4 strain. Similar to other known substrates, v-Src could interact with Hsp70s either directly or through the Hsp70 cochaperone Ydj1 (Dey et al. 1996). The pull-down study using His6-tagged Ydj1 shows that the cochaperone interaction with v-Src is similar in both the A2 and A4 strains. Furthermore, the pull-down study revealed that the Ydj1 interaction with Ssa2 is stronger than with Ssa4. The weaker interaction of Ydj1 with Ssa4 was further confirmed using purified chaperones in vitro. The relatively weaker interaction of Ydj1 with Ssa4 is in agreement with the lower luciferase refolding activity seen in the reactions containing Ydj1 and Ssa4 instead of Ssa2. Ssa2 was able to promote luciferase refolding at levels 25–30 times greater than for Ssa4. Similarly, higher activity was observed with Ssa2 than with Ssa4 when luciferase refolding assays were carried out in the presence of Hsp82, with either of the Hsp70 isoforms and Ydj1. This study generally reveals a novel distinction in the interactions of Ydj1 with Ssa2 vs. the interactions with Ssa4. As the Ssa4 interaction with Ydj1 is relatively weaker, it is possible that this leads to poor substrate transfer from Ydj1 to Ssa4, resulting in lower abundance of the v-Src-Ssa4 complex, as observed in the immunoprecipitated complex against v-Src.
Hsp40 proteins are known to interact at the NBD (Jiang et al. 2007) or the C-terminal domain of Hsp70 (Demand et al. 1998; Gong et al. 2018), and influence Hsp70 function (Sluder et al. 2018). The C-terminal domains of Ssa2 and Ssa4 are more divergent (i.e., a sequence identity of 42%) than their NBDs (i.e., a sequence identity of 87%). The interaction of Ydj1 with the NBD stimulates the ATPase activity of Hsp70 (Laufen et al. 1999; Jiang et al. 2007), whereas those at the C-terminal domain are known to facilitate substrate transfer (Freeman et al. 1995; Demand et al. 1998). The pull-down assay shows that the Ydj1 interaction with Ssa24 is stronger than it is with Ssa42 and suggests that the Ydj1-Ssa interaction is primarily mediated by the N-terminal domain. Our study used the hybrid Ssa chaperones Ssa24 and Ssa42, showing that cells expressing Ssa24 reduce v-Src toxicity better than those expressing Ssa42, and reveals that the C-terminal domain mediates the functional distinction of Ssa proteins in the Hsp90 chaperoning pathway. It is possible that, although Ydj1 interacts strongly with Ssa24 through its N-terminal domain, its interaction with the less-conserved C-terminal domain of Ssa4 remains compromised and results in poor v-Src transfer to the hybrid Ssa Hsp70, and leads to relatively lower maturation of the kinase. This is in agreement with the lower abundance of the v-Src-Ssa4 complex in the A4 strain (Figure 4), although Ydj1 interacts similarly with v-Src in both the A2 and A4 strains. Our results suggest that although the N-terminal domain of Hsp70 governs its interaction with Ydj1, the C-terminal domain determines the functional specificity between Ssa2 and Ssa4 with respect to v-Src toxicity.
The pull-down experiments using Ydj1 further reveal its interaction with Hsp90. The Ydj1 interaction with Hsp90 could either be direct or indirect through Hsp70. Since Ydj1 shows similar interactions with Hsp90 in spite of the different affinities toward the Ssa Hsp70 isoforms, the interaction of Ydj1 with Hsp90 (as seen in the pull-down assay) is more likely to be a direct interaction. Furthermore, as Ydj1 interacts similarly with Hsp90 in the A2 and A4 strains, the distinct v-Src activity and related toxicity might not be due to any activity associated with the formation of the Ydj1-Hsp90 complex in these strains.
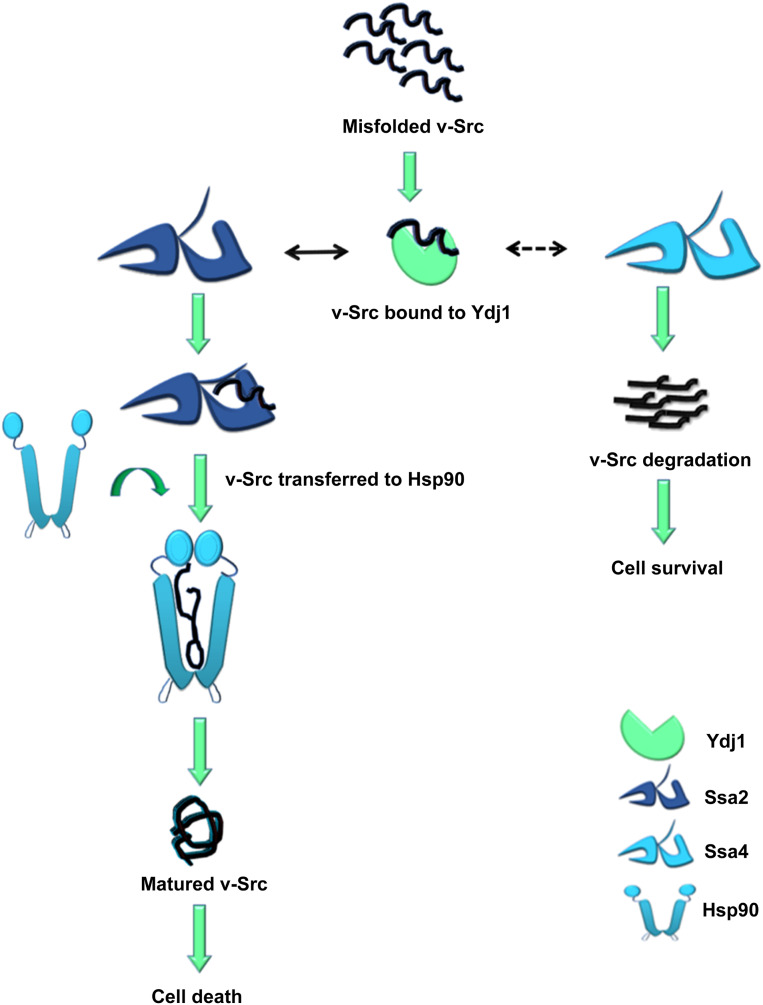
In summary, our data suggest that v-Src interacts similarly with Ydj1 in the A2 and A4 strains, but that the client transfer from Ydj1 to Hsp70 is less efficient for Ssa4 than for Ssa2. The lack of v-Src-Ssa4 interaction leads to substrate inactivation and degradation. In contrast, v-Src is transferred efficiently to Ssa2. Hsp90 then interacts with v-Src and promotes its maturation to an active kinase (Figure 9).
Figure 9.
Model of how Ydj1 regulates activity of Ssa2 and Ssa4. Misfolded v-Src interacts with Ydj1. Ydj1 further recruits v-Src to Hsp70. Solid black arrow represents transfer of v-Src to Ssa2 while dashed arrow represents the fact that Ydj1 is not able to transfer v-Src to Ssa4. Lack of Ydj1-Ssa4 interaction results in degradation of v-Src and cell survival in Ssa4 background. Hsp90 interacts with Ssa2, thus v-Src is transferred to Hsp90 and gets matured. v-Src maturation through Hsp90 results in cell death in Ssa2 background.
Hsp70s are involved in a variety of cellular functions. Although different Hsp70 isoforms perform many redundant functions, functional distinctions among these are also known. What governs the functional specificity among highly homologous Hsp70s is not clear and the role of the cochaperones is generally believed to the underlying basis of specificity. The current study shows that different Ssa Hsp70 isoforms function distinctly in the Hsp90 chaperoning functions. Interestingly, the functional distinctions of the Ssa Hsp70 isoforms lie upstream of their interactions with the cochaperone Ydj1. Thus, the results provided here show that Hsp40s are not only required to activate the Hsp70 reaction cycle, but also influence its function and thus provide functional diversity within members of the Hsp70 family.
Acknowledgments
We thank Daniel Masison for critical reading of the manuscript, Kevin Morano for providing the PRE-lacZ plasmid as a kind gift, and Jeffrey Brodsky for providing the anti-Sse1 antibody as a kind gift. The work was supported by the Council of Scientific and Industrial Research, India and a Rapid Grant for Young Investigators, Department of Biotechnology, India grant (BT/PR6463/GBD/27/420/2012). The authors declare that they have no conflicts of interest.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12136941.
Communicating editor: O. Cohen-Fix
Literature Cited
- Abbas-Terki T., Donzé O., Briand P. A., and Picard D., 2001. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 21: 7569–7575. 10.1128/MCB.21.22.7569-7575.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W. G., Schulte T. W., and Neckers L. M., 2000. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 11: 355–360. [PubMed] [Google Scholar]
- Arlander S. J. H., Felts S. J., Wagner J. M., Stensgard B., Toft D. O. et al. , 2006. Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J. Biol. Chem. 281: 2989–2998. 10.1074/jbc.M508687200 [DOI] [PubMed] [Google Scholar]
- Boysen M., Kityk R., and Mayer M. P., 2019. Hsp70- and hsp90-mediated regulation of the conformation of p53 DNA binding domain and p53 cancer variants. Mol. Cell 74: 831–843.e4. 10.1016/j.molcel.2019.03.032 [DOI] [PubMed] [Google Scholar]
- Brugge J. S., Jarosik G., Andersen J., Queral-Lustig A., Fedor-Chaiken M. et al. , 1987. Expression of Rous sarcoma virus transforming protein pp60v-src in Saccharomyces cerevisiae cells. Mol. Cell. Biol. 7: 2180–2187. 10.1128/MCB.7.6.2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. C., Nathan D. F., and Lindquist S., 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17: 318–325. 10.1128/MCB.17.1.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Zhong D., and Monteiro A., 2006. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics 7: 156 10.1186/1471-2164-7-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cintron N. S., and Toft D., 2006. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J. Biol. Chem. 281: 26235–26244. 10.1074/jbc.M605417200 [DOI] [PubMed] [Google Scholar]
- Citri A., Harari D., Shohat G., Ramakrishnan P., Gan J. et al. , 2006. Hsp90 recognizes a common surface on client kinases. J. Biol. Chem. 281: 14361–14369. 10.1074/jbc.M512613200 [DOI] [PubMed] [Google Scholar]
- Dahiya V., Agam G., Lawatscheck J., Rutz D. A., Lamb D. C. et al. , 2019. Coordinated conformational processing of the tumor suppressor protein p53 by the Hsp70 and Hsp90 chaperone machineries. Mol. Cell 74: 816–830.e7. 10.1016/j.molcel.2019.03.026 [DOI] [PubMed] [Google Scholar]
- Demand J., Lüders J., and Höhfeld J., 1998. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol. 18: 2023–2028. 10.1128/MCB.18.4.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B., Caplan A. J., and Boschelli F., 1996. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol. Biol. Cell 7: 91–100. 10.1091/mbc.7.1.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A. A., Chang H.-C. J., Marsh J. A., Lindquist S., and Gaber R. F., 1996. A cyclophilin function in hsp90-dependent signal transduction. Science 274: 1713–1715. 10.1126/science.274.5293.1713 [DOI] [PubMed] [Google Scholar]
- Flower T. R., Chesnokova L. S., Froelich C. A., Dixon C., and Witt S. N., 2005. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson’s disease. J. Mol. Biol. 351: 1081–1100. 10.1016/j.jmb.2005.06.060 [DOI] [PubMed] [Google Scholar]
- Freeman B. C., Myers M. P., Schumacher R., and Morimoto R. I., 1995. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14: 2281–2292. 10.1002/j.1460-2075.1995.tb07222.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler J. L., Stephens A., Lee P., Caplan A. J., and Brodsky J. L., 2002. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1–151 thermosensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell 13: 2760–2770. 10.1091/mbc.02-04-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Hu W., Xu L., Wu H., Wu S. et al. , 2018. The C-terminal GGAP motif of Hsp70 mediates substrate recognition and stress response in yeast. J. Biol. Chem. 293: 17663–17675. 10.1074/jbc.RA118.002691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf C., Stankiewicz M., Kramer G., and Mayer M. P., 2009. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 28: 602–613. 10.1038/emboj.2008.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Puri A., Singh P., Sonam S., Pandey R. et al. , 2018. The yeast stress inducible Ssa Hsp70 reduces α-synuclein toxicity by promoting its degradation through autophagy. PLoS Genet. 14: e1007751 10.1371/journal.pgen.1007751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Maes E. G., Taylor A. B., Wang L., Hinck A. P. et al. , 2007. Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28: 422–433. 10.1016/j.molcel.2007.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagöz G. E., Duarte A. M. S., Akoury E., Ippel H., Biernat J. et al. , 2014. Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell 156: 963–974. 10.1016/j.cell.2014.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. H., Kim R., Chen W., Hu S., Shin K.-H. et al. , 2008. Association of hsp90 to the hTERT promoter is necessary for hTERT expression in human oral cancer cells. Carcinogenesis 29: 2425–2431. 10.1093/carcin/bgn225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke E., Goswami D., Southworth D., Griffin P. R., and Agard D. A., 2014. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157: 1685–1697. 10.1016/j.cell.2014.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravats A. N., Doyle S. M., Hoskins J. R., Genest O., Doody E. et al. , 2017. Interaction of E. coli Hsp90 with DnaK involves the DnaJ binding region of DnaK. J. Mol. Biol. 429: 858–872. 10.1016/j.jmb.2016.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravats A. N., Hoskins J. R., Reidy M., Johnson J. L., Doyle S. M. et al. , 2018. Functional and physical interaction between yeast Hsp90 and Hsp70. Proc. Natl. Acad. Sci. USA 115: E2210–E2219. 10.1073/pnas.1719969115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Gaur D., Masison D. C., and Sharma D., 2014. The BAG homology domain of Snl1 cures yeast prion [URE3] through regulation of Hsp70 chaperones. G3 (Bethesda) 4: 461–470. 10.1534/g3.113.009993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Gaur D., Gupta A., Puri A., and Sharma D., 2015. Hsp90-associated immunophilin homolog Cpr7 is required for the mitotic stability of [URE3] prion in Saccharomyces cerevisiae. PLoS Genet. 11: e1005567 10.1371/journal.pgen.1005567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrat L., and Regan L., 2010. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry 49: 7428–7438. 10.1021/bi100386w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A. et al. , 1999. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. USA 96: 5452–5457. 10.1073/pnas.96.10.5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J. I. J., Pimkina J., Pandey P., Murphy M. E., and George D. L., 2011. HSP70 inhibition by the small-molecule 2-phenylethyne‐sulfonamide impairs protein clearance pathways in tumor cells. Molecular cancer research. MCR 9: 936–947. 10.1158/1541-7786.MCR-11-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Soroka J., and Buchner J., 2012. The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta 1823: 624–635. [DOI] [PubMed] [Google Scholar]
- Lotz S. K., Knighton L. E., Nitika G. W. Jones, and Truman A. W., 2019. Not quite the SSAme: unique roles for the yeast cytosolic Hsp70s. Curr. Genet. 65: 1127–1134. 10.1007/s00294-019-00978-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Richter K., Lilie H., and Buchner J., 2000. Cpr6 and Cpr7, two closely related hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 275: 34140–34146. 10.1074/jbc.M005251200 [DOI] [PubMed] [Google Scholar]
- McLaughlin S. H., Sobott F., Yao Z., Zhang W., Nielsen P. R. et al. , 2006. The Co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 356: 746–758. 10.1016/j.jmb.2005.11.085 [DOI] [PubMed] [Google Scholar]
- Minami Y., Kimura Y., Kawasaki H., Suzuki K., and Yahara I., 1994. The carboxy-terminal region of mammalian HSP90 is required for its dimerization and function in vivo. Mol. Cell. Biol. 14: 1459–1464. 10.1128/MCB.14.2.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano K. A., and Thiele D. J., 1999. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J. 18: 5953–5962. 10.1093/emboj/18.21.5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J. K. et al. , 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone Aha1. Mol. Cell 10: 1307–1318. 10.1016/S1097-2765(02)00785-2 [DOI] [PubMed] [Google Scholar]
- Picard D., Khursheed B., Garabedian M. J., Fortin M. G., Lindquist S. et al. , 1990. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 348: 166–168. 10.1038/348166a0 [DOI] [PubMed] [Google Scholar]
- Prodromou C., Siligardi G., O’Brien R., Woolfson D. N., Regan L. et al. , 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18: 754–762. 10.1093/emboj/18.3.754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapandi T., Greene L. E., and Eisenberg E., 2000. The molecular chaperones Hsp90 and Hsc70 are both necessary and sufficient to activate hormone binding by glucocorticoid receptor. J. Biol. Chem. 275: 22597–22604. 10.1074/jbc.M002035200 [DOI] [PubMed] [Google Scholar]
- Reidy M., and Masison D. C., 2010. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell. Biol. 30: 3542–3552. 10.1128/MCB.01292-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodina A., Patel P. D., Kang Y., Patel Y., Baaklini I. et al. , 2013. Identification of an allosteric pocket on human hsp70 reveals a mode of inhibition of this therapeutically important protein. Chem. Biol. 20: 1469–1480. 10.1016/j.chembiol.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl A., Wengler D., Madl T., Lagleder S., Tippel F. et al. , 2015. Hsp90 regulates the dynamics of its cochaperone Sti1 and the transfer of Hsp70 between modules. Nat. Commun. 6: 6655 10.1038/ncomms7655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J., Mitra S., Sengupta K., and Mandal A. K., 2015. Hsp70 clears misfolded kinases that partitioned into distinct quality-control compartments. Mol. Biol. Cell 26: 1583–1600. 10.1091/mbc.E14-08-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Fujita N., and Tsuruo T., 2000. Modulation of Akt kinase activity by binding to Hsp90. Proc. Natl. Acad. Sci. USA 97: 10832–10837. 10.1073/pnas.170276797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A. B., Lagleder S., Gräwert M. A., Röhl A., Hagn F. et al. , 2012. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31: 1506–1517. 10.1038/emboj.2011.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., and Masison D. C., 2008. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics 179: 1301–1311. 10.1534/genetics.108.089458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., and Masison D. C., 2011. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc. Natl. Acad. Sci. USA 108: 13665–13670. 10.1073/pnas.1107421108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder I. T., Nitika L. E. Knighton, and Truman A. W., 2018. The Hsp70 co-chaperone Ydj1/HDJ2 regulates ribonucleotide reductase activity. PLoS Genet. 14: e1007462 10.1371/journal.pgen.1007462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisutthisamphan K., Jirakanwisal K., Ramphan S., Tongluan N., Kuadkitkan A. et al. , 2018. Hsp90 interacts with multiple dengue virus 2 proteins. Sci. Rep. 8: 4308 10.1038/s41598-018-22639-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W. P., Owen B. A. L., and Toft D. O., 2002. The Influence of ATP and p23 on the Conformation of hsp90. J. Biol. Chem. 277: 45942–45948. 10.1074/jbc.M207754200 [DOI] [PubMed] [Google Scholar]
- Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K. D. et al. , 2012. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150: 987–1001. 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor Roberts B., Moriyama H., and Wickner R. B., 2004. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast 21: 107–117. 10.1002/yea.1062 [DOI] [PubMed] [Google Scholar]
- Voss A. K., Thomas T., and Gruss P., 2000. Mice lacking HSP90beta fail to develop a placental labyrinth. Development 127: 1. [DOI] [PubMed] [Google Scholar]
- Warth R., Briand P. A., and Picard D., 1997. Functional analysis of the yeast 40 kDa cyclophilin Cyp40 and its role for viability and steroid receptor regulation. Biol. Chem. 378: 381–391. 10.1515/bchm.1997.378.5.381 [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M., Stone D. E., and Craig E. A., 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 2568–2577. 10.1128/MCB.7.7.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M., Becker J., Kosic-Smithers J., and Craig E. A., 1989. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol. 171: 2680–2688. 10.1128/JB.171.5.2680-2688.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Singer M. A., and Lindquist S., 1999. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc. Natl. Acad. Sci. USA 96: 109–114. 10.1073/pnas.96.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12136941.