The ubiquitin proteasome system plays an essential role in the regulation of flowering time, a precisely timed developmental transition necessary for plant fitness. However, the full impact...
Keywords: protein degradation, flowering time, dominant-negative strategies, plant biology
Abstract
Targeted degradation of proteins is mediated by E3 ubiquitin ligases and is important for the execution of many biological processes. Redundancy has prevented the genetic characterization of many E3 ubiquitin ligases in plants. Here, we performed a reverse genetic screen in Arabidopsis using a library of dominant-negative U-box-type E3 ubiquitin ligases to identify their roles in flowering time and reproductive development. We identified five U-box decoy transgenic populations that have defects in flowering time or the floral development program. We used additional genetic and biochemical studies to validate PLANT U-BOX 14 (PUB14), MOS4-ASSOCIATED COMPLEX 3A (MAC3A), and MAC3B as bona fide regulators of flowering time. This work demonstrates the widespread importance of E3 ubiquitin ligases in floral reproductive development. Furthermore, it reinforces the necessity of dominant-negative strategies for uncovering previously unidentified regulators of developmental transitions in an organism with widespread genetic redundancy, and provides a basis on which to model other similar studies.
FLOWERING is the first committed step in the plant reproductive process, leading to the production of reproductive organs and eventually offspring. Plants use highly complex gene networks that integrate a wide array of internal and external signals to regulate flowering. In the model plant Arabidopsis thaliana, six pathways have been identified to control flowering time. Four of these six pathways regulate the production of the florigen, the protein FLOWERING LOCUS T (FT), while the remaining two bypass FT to promote flowering in a more direct manner (Srikanth and Schmid 2011). In addition, abiotic and biotic stress can modulate flowering time by altering the function of one or multiple flowering pathways (Park et al. 2016; Takeno 2016).
In Arabidopsis, the transition to flowering is an irreversible decision that the plant makes in response to external and internal signals. In order for the response to occur at the appropriate time, plants need to promote the activity of floral activators and repress the activity of floral repressors. One way that plants accomplish this is by leveraging the ubiquitin proteasome system to accurately degrade floral regulator proteins (Nelson et al. 2000; McGinnis et al. 2003; Imaizumi et al. 2005; Sawa et al. 2007; Hu et al. 2014). E3 ubiquitin ligases provide substrate specificity for the ubiquitin proteasome system and mediate the ubiquitylation of target proteins (Vierstra 2009). E3 ubiquitin ligases play central roles in the regulation of the photoperiodic, vernalization, and gibberellin (GA) flowering-time pathways (Nelson et al. 2000; McGinnis et al. 2003; Imaizumi et al. 2005; Sawa et al. 2007; Jang et al. 2008; Lazaro et al. 2012; Hu et al. 2014), demonstrating their important functions in this critical developmental decision.
Despite these well-characterized roles, it is likely that E3 ubiquitin ligases that regulate flowering have not been identified due to the numerous genome duplications that have resulted in widespread gene redundancy (Risseeuw et al. 2003; Yee and Goring 2009; Navarro-Quezada et al. 2013). To overcome these issues, we previously created and validated a library of transgenic plants expressing E3 ubiquitin ligase decoys (Feke et al. 2019). E3 ubiquitin ligase decoys are forms of E3 ubiquitin ligases that lack the protein domain that coordinates substrate ubiquitylation but retain other protein domains. Thus, E3 ubiquitin ligase decoys can still interact with substrate proteins but can no longer promote ubiquitylation and proteasomal degradation (Lee and Feke et al. 2018). In the case of the U-box family of E3 ubiquitin ligases, the U-box domain coordinates substrate ubiquitylation (Azevedo et al. 2001; Finn et al. 2016). A U-box decoy protein lacks the U-box domain but retains the N- and/or C-terminal protein regions that typically contain protein–protein interaction domains (Feke et al. 2019).
Here, we employ the decoy library to identify U-box-type E3 ubiquitin ligases that control flowering time and reproductive development. We focus on four metrics of reproductive development: the number of rosette leaves, the age of the plant in days when 1-cm bolting occurs, the first occurrence of anthesis, and the rate of stem elongation. Using these metrics, we uncover six U-box proteins that regulate 1-cm bolting, six U-box proteins that regulate rosette leaf number, four U-box proteins that control stem elongation, and one U-box protein that controls anthesis.
We perform focused genetic studies on three U-box genes: PLANT U-BOX 14 (PUB14), MAC3A, and MAC3B. We confirm their roles in flowering-time regulation by observing delayed flowering phenotypes in three transfer DNA (T-DNA) insertion mutants, pub14-1 (SALK_118095C), mac3a, and mac3b mutants (Monaghan et al. 2009). We also perform immunoprecipitation mass spectrometry (MS) with the PUB14 decoy, similar to what we did previously for MAC3B (Feke et al. 2019), and find a list of proteins involved in the regulation of flowering time. These findings demonstrate the widespread importance of E3 ubiquitin ligases in the regulation of reproductive development and illustrate the strength of the decoy technique to quickly identify novel E3 ubiquitin ligases in diverse biological processes.
Materials and Methods
Phenotypic screening
The construction of the decoy library, the MAC3B-OX, and the MAC3B-WD was described previously (Feke et al. 2019). Control pCCA1∷Luciferase and decoy seeds were surface sterilized in 70% ethanol and 0.01% Triton X-100 for 20 min prior to being sown on 1/2 MS plates [2.15 g/liter Murashige and Skoog medium, pH 5.7 (catalog number MSP01; Cassion Laboratories) and 0.8% bacteriological agar (catalog number AB01185; AmericanBio)] with or without appropriate antibiotics [15 μg/ml ammonium glufosinate (catalog number 77182-82-2; Santa Cruz Biotechnology) for vectors pB7-HFN and pB7-HFC, or 50 μg/ml kanamycin sulfate (AmericanBio) for pK7-HFN]. Seeds were stratified for 2 days at 4°, transferred to 12 hr light/12 hr dark conditions for 7 days, and then to constant light conditions for 7 days to enable screening for circadian clock studies shown in Feke et al. (2019). Seedlings were then transferred to soil (Fafard II) and grown at 22° in inductive 16 hr light/8 hr dark conditions with a light fluence rate of 135 μmol m−2 s−1. Plants were monitored daily for flowering status, with the dates upon which each individual reached 1-cm inflorescence height, 10-cm inflorescence height, and the first occurrence of anthesis recorded. Additionally, leaf number at 1-cm inflorescence height was recorded.
Homozygous pub14-1, mac3a, mac3b, and mac3a/mac3b mutant seeds were surface sterilized and sown on 1/2 MS plates without antibiotics as described above. Seeds were stratified for 3 days at 4°, then transferred to 12 hr light/12 hr dark conditions for 2 weeks prior to transfer to soil and growth under inductive conditions as described above. Plants were monitored daily for flowering status as described above.
Data normalization and statistical analysis
As the age at anthesis depends on the initiation of flowering, we used anthesis delay as a measurement of anthesis. Anthesis delay was calculated by taking the age at anthesis and subtracting the age at 1-cm inflorescence height. Similarly, the age at 10-cm inflorescence height depends on the initiation of flowering. Thus, we calculated the stem elongation period by subtracting the age at 1-cm inflorescence height from the age at 10-cm inflorescence height. These modified metrics were used for all analyses.
To allow for comparison across independent experiments, data were normalized to the individual wild-type control performed concurrently. The average value of the wild-type control plants was calculated for every experiment, then this average was subtracted from the value of each individual T1 insertion or control wild-type plant done concurrently. These normalized values were used for statistical analyses and are presented in Supplemental Material, Table S1.
Welch’s t-test was used to compare each normalized T1 insertion plant population or subpopulation to the population of all normalized control plants. To decrease the number of false positives caused by multiple testing, we utilized a Bonferroni-corrected α as the P-value threshold. The α applied differed between experiments, and is noted throughout.
Measurement of gene expression in U-box mutants
Homozygous mac3a/mac3b mutant plants in the Col-0 background were generated previously (Monaghan et al. 2009). Col-0, pub14-1, mac3a, mac3b, and mac3a/mac3b seeds were stratified on 1/2 MS plates at 4° for 2 days prior to growth in 16 hr light/8 hr dark conditions at a fluence rate of 130 μmol m−2 s−1 at 22°. Ten-day-old seedlings were collected in triplicate every 4 hr for 1 day starting at zeitgeber time 0 (ZT0) and snap-frozen using liquid nitrogen, then ground using the Mixer Mill MM400 system (Retsch). Total RNA was extracted from ground seedlings using an RNeasy Plant Mini Kit and treated with RNase-Free DNase (catalog numbers 74904 and 79254, respectively; QIAGEN, Valencia, CA) following the manufacturer’s protocols. Complementary DNA (cDNA) was prepared from 1 μg total RNA using iScript Reverse Transcription Supermix (catalog number 1708841; Bio-Rad, Hercules, CA), then diluted 10-fold and used directly as the template for quantitative real-time RT-PCR (qRT-PCR). The qRT-PCR was performed using 3.5 μl of diluted cDNA and 5.5 μM primers listed in Table 1 (Wu et al. 2008; Lee and Thomashow 2012) using iTaq Universal SYBR Green Supermix (catalog number 1725121; Bio-Rad) with the CFX 384 Touch Real-Time PCR Detection System (Bio-Rad). The qRT-PCR began with a denaturation step of 95° for 3 min, followed by 45 cycles of denaturation at 95° for 15 sec, and primer annealing at 53° for 15 sec. Relative expression was determined by the comparative CT method using IPP2 (AT3G02780) as an internal control. The relative expression levels represent the mean values of 2-ΔΔCT from three biological replicates, where ΔCT = CT of FT – CT of IPP2 and the reference is Col-0 replicate number 1. When measuring FT expression, the time point of peak expression (ZT16) was used as the reference point.
Table 1. Primers used in this study.
| Name | Sequence | Reference |
|---|---|---|
| qPCR IPP2 F | ATTTGCCCATCGTCCTCTGT | Lee and Thomashow (2012) |
| qPCR IPP2 R | GAGAAAGCACGAAAATTCGGTAA | Lee and Thomashow (2012) |
| qPCR PUB14-1 F | ATTGTTGTTCCCACGAGGAG | This manuscript |
| qPCR PUB14-1 R | TCGAAGAAAGGGCTGAGAAG | This manuscript |
| qPCR PUB14-2 F | CGGTTAATGGAGGAAGCAAG | This manuscript |
| qPCR PUB14-2 R | CCACTGTCATGTCACGGAAC | This manuscript |
| qPCR PUB14-3 F | CGCAAAATCAAGGGAGCTGTAG | This manuscript |
| qPCR PUB14-3 R | AGTACCGTTGGCCAATTTCTCT | This manuscript |
| qPCR PUB14-4 F | CATGGAAGCTAGAGAGAACGCT | This manuscript |
| qPCR PUB14-4 R | CCCTTGATTTGTTCCCCTGGTA | This manuscript |
| qPCR FT F | ATCTCCATTGGTTGGTGACTGATA | Wu et al. (2008) |
| qPCR FT R | GCCAAAGGTTGTTCCAGTTGTAG | Wu et al. (2008) |
F, forward; R, reverse.
Immunoprecipitation and MS of PUB14 decoy plants
Individual T1 pB7-HFN-PUB14 transgenic plants in a Col-0 background and control Col-0 and pB7-HFC-GFP were grown as described for phenotype analysis. Seven-day-old seedlings were transferred to soil and grown under 16 hr light/8 hr dark at 22° for 2–3 weeks. Prior to harvest, plants were entrained to 12 hr light/12 hr dark at 22° for 1 week. Approximately 40 mature leaves from each background were collected and snap-frozen in liquid nitrogen, such that each sample was a mixture of leaves from multiple individuals to reduce the effects of expression-level fluctuations. Tissue samples were ground in liquid nitrogen using the Mixer Mill MM400 system (Retsch). Immunoprecipitation was performed as described previously (Lu et al. 2010; Huang et al. 2016a,b). Briefly, protein from 2 ml tissue powder was extracted in SII buffer (100 mM sodium phosphate pH 8.0, 150 mM NaCl, 5 mM EDTA, and 0.1% Triton X-100) with cOmplete EDTA-free Protease Inhibitor Cocktail (catalog number 11873580001; Roche), 1 mM PMSF, and a PhosSTOP tablet (catalog number 04906845001; Roche) by sonification. Anti-FLAG antibodies were cross-linked to Dynabeads M-270 Epoxy (catalog number 14311D; Thermo Fisher Scientific) for immunoprecipitation. Immunoprecipitation was performed by incubation of protein extracts with beads for 1 hr at 4° on a rocker. Beads were washed with SII buffer three times, then twice in F2H buffer (100 mM sodium phosphate pH 8.0, 150 mM NaCl, and 0.1% Triton X-100). Beads were eluted twice at 4° and twice at 30° in F2H buffer with 100 μg/mL FLAG peptide, incubated with TALON magnetic beads (catalog number 35636; Clontech) for 20 min at 4°, and then washed twice in F2H buffer and three times in 25 mM ammonium bicarbonate. Samples were subjected to trypsin digestion (0.5 µg, catalog number V5113; Promega, Madison, WI) at 37° overnight, then vacuum dried using a SpeedVac before being dissolved in 5% formic acid/0.1% trifluoroacetic acid (TFA). Protein concentration was determined by nanodrop measurement (A260/A280) (Thermo Scientific Nanodrop 2000 UV-Vis Spectrophotometer). An aliquot of each sample was further diluted with 0.1% TFA to 0.1 µg/µl and 0.5 µg was injected for liquid chromatography (LC)-MS/MS analysis at the Keck MS and Proteomics Resource Laboratory at Yale University.
LC-MS/MS analysis was performed on a Thermo Scientific Orbitrap Elite mass spectrometer equipped with a Waters nanoACQUITY ultra performance LC (UPLC) system utilizing a binary solvent system (buffer A: 0.1% formic acid and buffer B: 0.1% formic acid in acetonitrile). Trapping was performed at 5 µl/min 97% Buffer A for 3 min using a Waters Symmetry C18 180 µm × 20 mm trap column. Peptides were separated using an ACQUITY UPLC PST (BEH) C18 nanoACQUITY column 1.7 µm, 75 µm × 250 mm (37°), and eluted at 300 nl/min with the following gradient: 3% buffer B at initial conditions, 5% buffer B at 3 min, 35% buffer B at 140 min, 50% buffer B at 155 min, 85% B at 160–165 min, and then returned to initial conditions at 166 min. MS was acquired in the Orbitrap in profile mode over the 300–1700 m/z range using one microscan, 30,000 resolution, automatic gain control (AGC) target of 1E6, and a full maximum ion time of 50 ms. Up to 15 MS/MS were collected per MS scan using collision-induced dissociation on species with an intensity threshold of 5000 and charge states of two and above. Data-dependent MS/MS were acquired in centroid mode in the ion trap using one microscan, AGC target of 2E4, full max ion time of 100 ms, 2.0 m/z isolation window, and normalized collision energy of 35. Dynamic exclusion was enabled with a repeat count of 1, repeat duration of 30 sec, exclusion list size of 500, and exclusion duration of 60 sec.
The MS/MS spectra were searched by the Keck MS and Proteomics Resource Laboratory at Yale University using MASCOT (Perkins et al. 1999). Data were searched against the SwissProt_2015_11.fasta A. thaliana database with oxidation set as a variable modification. The peptide mass tolerance was set to 10 ppm, the fragment mass tolerance to 0.5 Da, and the maximum number of allowable missed cleavages was set to 2.
Data availability
The authors affirm that all other data necessary for confirming the conclusions of the article are present within the article, figures, and tables or as supplemental materials uploaded to figshare. The seeds and plasmids are available upon request and will be available from the Arabidopsis Biological Resource Center. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12245753
Results
The role of U-box decoys in flowering time
Protein degradation through the ubiquitin proteasome system plays an essential role in flowering-time pathways (Imaizumi et al. 2003, 2005; McGinnis et al. 2003; Park et al. 2007; Jang et al. 2008; Sun 2011; Lazaro et al. 2012; Hu et al. 2014). However, the extent to which the ubiquitin proteasome system regulates flowering is not fully known. To identify E3 ubiquitin ligases that regulate flowering time, we screened the U-box decoy library. This is a subset of the larger decoy library described in our previous manuscript (Feke et al. 2019).
Parental control and T1 transgenic seedlings expressing the decoys were transferred to soil and grown under long-day (16 hr light and 8 hr dark) conditions. By analyzing a population of T1 transgenics, we avoid the problems that may arise from following a single insertion that may not be representative of the entire population.
To monitor the initiation of flowering, we measure the number of leaves at 1-cm bolting. This is a common flowering-time measurement that informs on the developmental stage of the plant at the vegetative- to reproductive-phase transition. We also measure the age of the plant in days when bolting occurs, as indicated by a 1-cm long inflorescence. This allows us to determine how much time the plant spends in the vegetative stage.
In addition to floral initiation, we also measure two metrics of reproductive development. We measure the first occurrence of anthesis, or the opening of the floral bud. We also measure stem elongation by recording the age of the plant in days when the inflorescence is 10-cm long. As anthesis and stem elongation are dependent on the initiation of flowering, we calculated the delay, or number of days after 1-cm bolting at which anthesis occurs or the stem reaches 10 cm in length, and used these value for our analyses. By measuring all four metrics, we are able to categorize any candidate floral regulator by which aspects of floral development are impacted.
Flowering time and reproductive development can differ between experiments due to uncharacterized variations in growth conditions. To compare across the entire decoy library, we calculated the flowering time difference for each individual decoy transgenic. This value was calculated by determining the average rosette leaf number for the control population in each experiment, then subtracting this value from the rosette leaf number for each individual decoy transgenic (Figure 1). We generated the 1-cm bolting time difference (Figure 2), anthesis delay difference (Figure S1), and stem elongation period difference (Figure S2) in the same manner. To see the variation within experiments, the individual control plants were normalized against the other control plants in the same experiment, as described for the decoy plants above (Figure S3). We perform Welch’s t-test with a Bonferroni-corrected α of 1.25 × 10−3 on these difference values. In this way, we were able to confidently assess whether a decoy population was different from the control in any of our metrics.
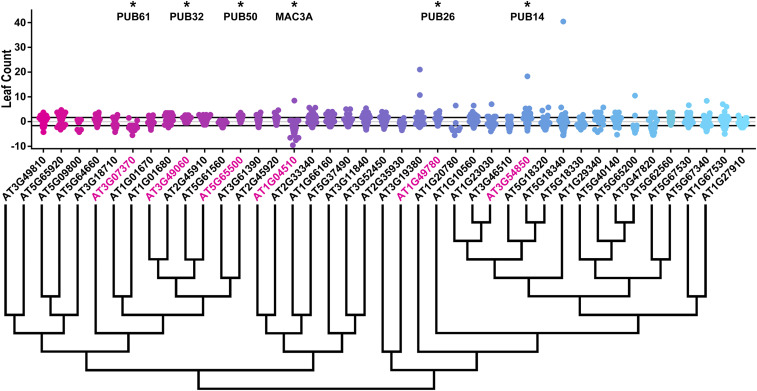
Figure 1.
Leaf-count distributions of U-box decoy plants. Values presented are the differences between the rosette leaf count of the individual decoy plant and the average rosette leaf count of the parental control in the accompanying experiment. The gray line is at the average control value and the black lines are ± SD of the control plants. Genes are ordered by closest protein homology using Phylogeny.Fr, (Dereeper et al. 2008), and a tree showing that homology is displayed beneath the graph. * and pink gene names indicate that the entire population differs from wild-type with a Bonferroni-corrected P < 1.25 × 10−3.
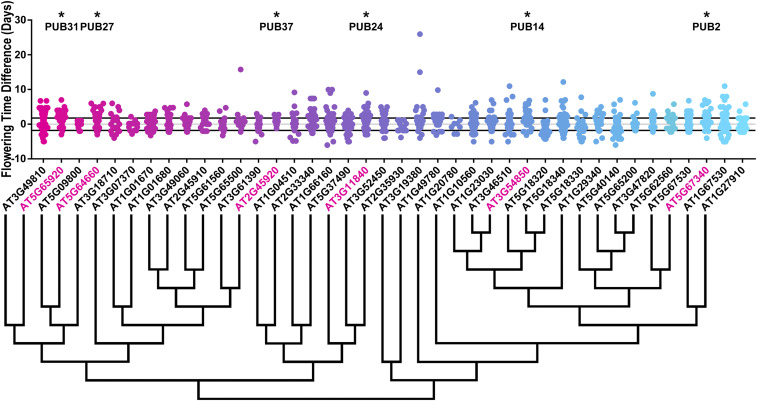
Figure 2.
The 1-cm bolting age distributions of U-box decoy plants. Values presented are the differences between the age at 1-cm inflorescence of the individual decoy plant and the average age at 1-cm inflorescence of the parental control in the accompanying experiment. The gray line is at the average control value and the black lines are ± SD of the control plants. Genes are ordered by closest protein homology using Phylogeny.Fr, (Dereeper et al. 2008), and a tree showing that homology is displayed beneath the graph. * and pink gene names indicate that the entire population differs from wild-type with a Bonferroni-corrected P < 1.25 × 10−3.
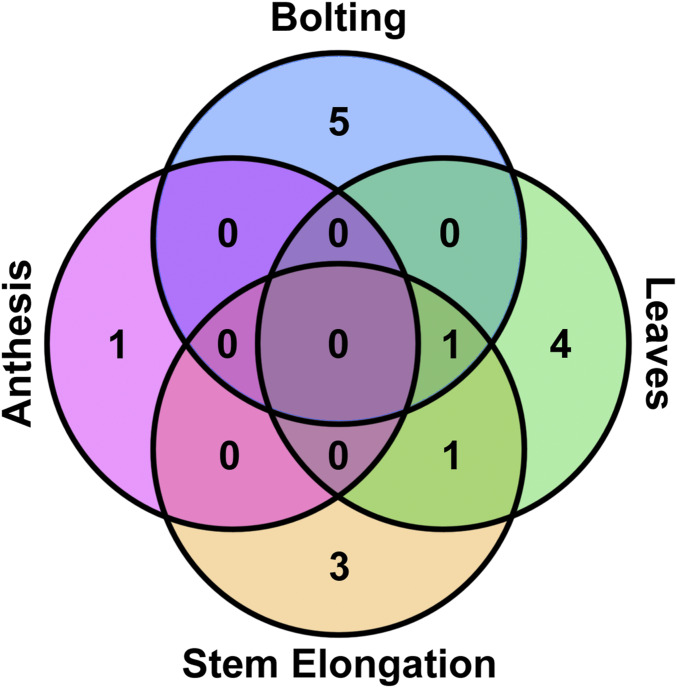
Of the 40 decoy populations assayed, six populations demonstrated a statistically altered age at 1-cm bolting, six had altered rosette leaf number, one had altered anthesis, and four had altered stem elongation time. Most effects on flowering time were minor, which we define as having less than two leaves different from wild-type at 1-cm bolting or < 2 days different from wild-type in the age-based metrics. Correspondingly, we define “major” as more than two leaves or > 2 days different from wild-type, respectively. To identify candidates for detailed genetic follow-up studies, we focused on decoy populations that had any effect on multiple flowering criteria or had a major effect on one criterion.
There were three decoy populations that had a major effect on one flowering criterion. Expressing the PUB31 decoy had a large effect on 1-cm bolting, delaying it by an average of 2.4 days. Expressing the MAC3A and PUB61 decoys caused altered rosette leaf number with 2.9 and 2.2 fewer leaves, respectively. The magnitudes of the phenotypes observed in these populations make them high-priority candidate flowering-time regulators. MAC3A had the greatest magnitude change in rosette leaf number, was previously noted to have a flowering-time defect (Monaghan et al. 2009), and was part of our focused circadian clock genetic studies previously (Feke et al. 2019), making it a major candidate for focused genetic studies with regard to its role in flowering time.
To identify additional major candidate flowering-time regulators, we determined which decoy populations caused defects in multiple flowering-time parameters (Figure 3). Expressing the PUB26 decoy shortened the stem elongation period by 0.75 days and resulted in 1.4 more leaves at flowering time. PUB14 also affected multiple flowering parameters. It flowered with more leaves (1.3 leaves), delayed 1-cm bolting (1.8 days), and also shortened the stem elongation period (0.56 days shorter), with all three parameters reaching statistical significance. Many classic flowering-time regulators affect both rosette leaf number and days to 1-cm bolting (Page et al. 1999; Nelson et al. 2000; Wang et al. 2011), making PUB14 a strong candidate for follow-up studies.
Figure 3.
Overlap between candidate flowering-time regulators for each metric. The statistically significant regulators from Figure 1, Figure 2, and Figures S1 and S2 were categorized based on which metrics were affected.
PUB14 regulates flowering time
PUB14 was the only candidate flowering-time regulator identified in our screen that impacted both rosette leaf number and 1-cm bolting age. To understand the function and regulation of PUB14, we mined publicly available expression data and the literature. PUB14 has the U-box domain centrally located and possesses five ARMADILLO repeats. It has no known genetic function, although it was used as a “prototypical” PUB gene in a structural study on U-box function (Andersen et al. 2004). PUB14 is closely related to PUB13 (E-value of 0), which has been implicated in the control of flowering time, immunity, cell death, and hormone responses (Li et al. 2012a,b; Kong et al. 2015; Zhou et al. 2015, 2018; Liao et al. 2017). Mutants of PUB13 have accelerated flowering time in long-day conditions (Li et al. 2012a,b; Zhou et al. 2015), in contrast to the delayed flowering we observed with the PUB14 decoys. Although these data suggest that PUB14 and PUB13 affect flowering time differently, the identification of a close homolog of a characterized flowering-time regulator provides strength to the hypothesis that PUB14 could also regulate flowering time. Genes that regulate flowering time are often regulated by the circadian clock or diel light cycles, and their expression may differ between inductive (long-day) and noninductive (short-day) conditions. Thus, we attempted to determine whether PUB14 is regulated by the circadian clock or various daily light cycles. To assay for rhythmicity, we queried publicly available microarray data and determine the correlation value, a measure of the similarity between the expression data and the hypothesized cycling pattern (Mockler et al. 2007). If this correlation value is greater than the standard correlation cutoff of 0.8 then it is considered rhythmic. While PUB14 expression does not cycle under circadian, long-day (12 hr light/12 hr dark), or floral inductive long-day (16 hr light/8 hr dark) conditions, it does cycle under noninductive short-day (8 hr light/16 hr dark) conditions, peaking in the evening (19 hr after dawn) (Mockler et al. 2007). Furthermore, many flowering-time genes are regulated by stress, temperature, or hormones. For this reason, we mined expression data using the eFP browser for treatments that effect PUB14 expression (Winter et al. 2007). While PUB14 expression is unaffected by most treatments, it is upregulated when leaves are exposed to Pseudomonas syringae (Winter et al. 2007).
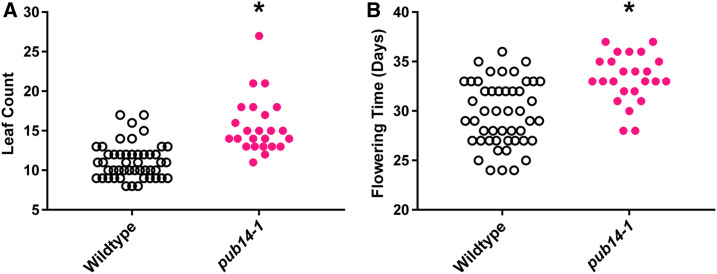
PUB14 is closely related to a gene that regulates flowering time and expressing the decoy causes delayed flowering. We isolated an Arabidopsis mutant with a SALK T-DNA insertion located in the 5′ UTR of PUB14, which we named pub14-1. While a 5′ UTR insertion may have many different effects, we find that expression of the N-terminal portion of the PUB14 gene is increased in the pub14-1 mutant background (Figure S4). We analyzed flowering time in the pub14-1 mutant and compared it to the wild type. We observed that 1-cm bolting was delayed by 3.6 days in the pub14-1 mutant and that it flowered with 4.5 more leaves on average (P < 0.0125; Figure 4), similar to the PUB14 decoy population. Interestingly, we did not recapitulate the stem elongation defect observed in the PUB14 decoy population, but did observe that anthesis was advanced by 1.6 days relative to wild-type (P < 0.0125; Figure S5). The similarity in the phenotypes of the PUB14 decoy population and the pub14-1 mutant suggests that the PUB14 is a bona fide regulator of flowering time, although additional experiments with a true knockout of PUB14 would be beneficial for confirming its role in positive or negative regulation of flowering time.
Figure 4.
Flowering-time analyses of pub14-1 mutants. (A) Rosette leaf number. (B) Age at 1-cm bolting. * represents a significant difference from wild-type with a Bonferroni-corrected P < 0.0125.
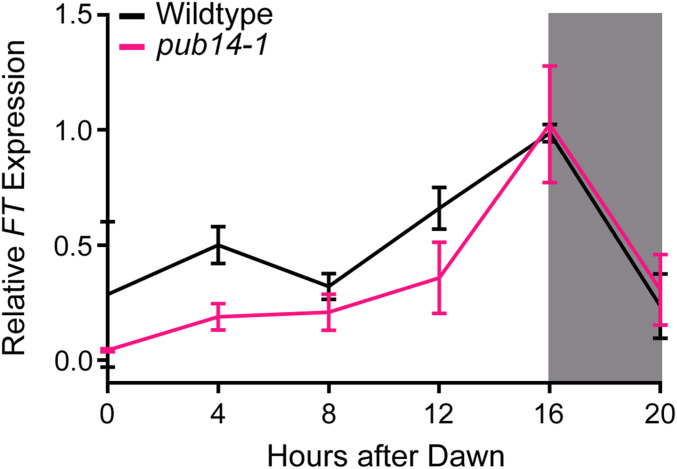
Reduction in FT expression levels is a hallmark of many late-flowering mutants, although some mutants delay flowering independently of FT (Han et al. 2008; Leijten et al. 2018). To determine whether the pub14-1 mutant delays flowering in an FT-dependent manner, we measured FT expression in wild-type and pub14-1 seedlings grown under long-day conditions (Figure 5). In the wild-type plants, we observed patterns of FT expression corresponding to those observed previously (Suárez-López et al. 2001; Wu et al. 2008; Song et al. 2012). However, in the pub14-1 mutant seedlings we observed a reduction of FT expression from ZT0 to ZT12. These results suggest that PUB14 functions upstream of FT in flowering time regulation. The morning peak in FT expression is more prominent in Arabidopsis grown outdoors than under standard laboratory conditions (Song et al. 2018). The drivers of this early day expression peak are the naturally occurring reduced red to far red ratio and temperature cycles (Song et al. 2018). This peak is decreased in the pub14-1 mutant, making it possible that PUB14 plays a role in the control of FT expression by controlling the stability of negative regulators involved in the far red and temperature input pathways that drive the morning peak of FT expression.
Figure 5.
qRT-PCR of FT expression in pub14-1 mutants. FT expression was measured using qRT-PCR in wild-type or homozygous pub14-1 mutants grown under long-day (16 hr light/8 hr dark) conditions. Quantifications are the average of three biological replicates with error bars showing SD. qRT-PCR, quantitative real-time PCR.
PUB14 interacts with flowering-time regulators
Our data suggest that PUB14 is a bona fide regulator of flowering time. However, it is unclear in which flowering time pathways PUB14 functions. To better understand the biochemical function of PUB14, we performed immunoprecipitation followed by MS on tissue expressing the 3XFLAG-6XHIS-tagged PUB14 decoy and searched for flowering-time regulators (Table S2). We included tissue from plants expressing 3XFLAG-6XHIS-tagged GFP as a control for proteins that bind to the tag and wild-type parental plants as a control for proteins that bind to the beads. We identified four proteins with known functions in flowering-time regulation as potential interactors of PUB14 (Table 2). We identified peptides corresponding to SPLAYED, a SWI/SNF ATPase that represses flowering time under short-day conditions, possibly by modulating activity of the floral activator LEAFY (Wagner and Meyerowitz 2002). We also identified peptides corresponding to SNW/SKI-Interacting Protein (SKIP), a component of the spliceosomal-activating complex that represses flowering time by activating FLC (Cao et al. 2015; Cui et al. 2017). Similarly, we identify a potential interaction with ACTIN-RELATED PROTEIN 6, a repressor of flowering that is required for FLC expression (Choi et al. 2005; Deal et al. 2005; Martin-Trillo et al. 2006). Finally, we identify TOPLESS, a protein that acts as a weak repressor of flowering time, potentially by forming a complex with CONSTANS (CO) to repress FT activation (Causier et al. 2012; Graeff et al. 2016). While additional work is required to verify these interactions and test whether they are ubiquitylation targets of PUB14, the identification of flowering-time repressors as putative interacting partners of PUB14 may explain the late-flowering phenotype we observe in the pub14-1 mutant and PUB14 decoy-expressing plants.
Table 2. Selected immunoprecipitation-MS results from the PUB14 decoy.
| Locus | Protein name | Total spectral counts | |
|---|---|---|---|
| PUB14 decoy | Combined controls | ||
| AT3G54850 | PUB14 | 754 | 9 |
| AT1G60780 | SYD | 38 | 0 |
| AT3G14750 | SKIP | 19 | 0 |
| AT1G01090 | ARP6 | 18 | 0 |
| AT2G36170 | TPL | 61 | 17 |
| AT4G37920 | PUB10 | 16 | 0 |
| AT4G13430 | PUB12 | 2 | 0 |
PUB14 decoy peptide hits are from one immunoprecipitation-mass spectrometry experiment using the PUB14 decoy as the bait. Combined control peptide hits are summed from the independent control experiments of wild-type Col-0 and 35S::His-FLAG-GFP-expressing plants. PUB, PLANT U-BOX; SYD, SPLAYED; SKIP, SNW/SKI-Interacting Protein; ARP6, ACTIN-RELATED PROTEIN 6; TLP, TOPLESS.
We have previously observed that E3 ligases interact with close homologs (Lee and Feke et al. 2018; Feke et al. 2019). Thus, we searched our immunoprecipitation-MS data for other U-box genes that interact with PUB14. We did not identify peptides corresponding to PUB13, the closest homolog of PUB14; however, we did identify peptides corresponding to two other close homologs of PUB14, PUB12 (E-value 3 × 10−163) and PUB10 (E-value 8 × 10−142). Interestingly, we did not identify peptides corresponding to the other members of this small subfamily, PUB15 (E–value 6 × 10−134) and PUB11 (E–value 4 × 10−142). While the importance of these interactions has not been verified, these data suggest that interaction between homologs is a common feature of E3 ligase complexes, and that PUB10 and PUB12 may also be involved in flowering-time regulation.
MAC3A and MAC3B regulate flowering time in a partially redundant manner
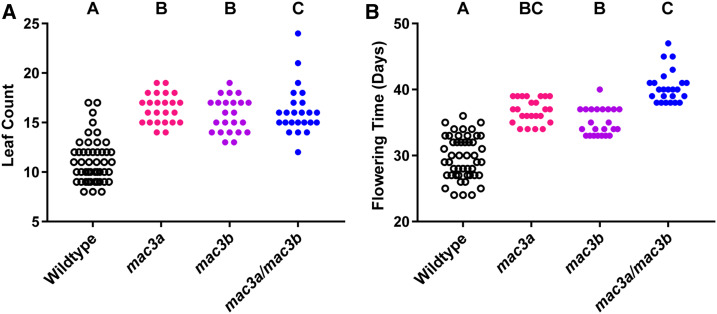
Expression of the MAC3A decoy leads to the greatest magnitude change that we observed in our screen (2.9 more leaves than wild-type, Figure 1). MAC3A and MAC3B can act as fully or partially redundant regulators of processes controlled by the plant spliceosomal activating complex (Monaghan et al. 2009; Jia et al. 2017; Li et al. 2018; Feke et al. 2019). It was previously noted that MAC3A and MAC3B could regulate flowering time (Monaghan et al. 2009). We have established genetic tools to further investigate the genetic interaction of MAC3A and MAC3B in flowering time. We grew the single and double mutants in an inductive photoperiod, and measured our four flowering time parameters (Figure 6 and Figure S6). We observed a statistically significant difference in rosette leaf number between all three mutant backgrounds and the wild-type, but observed no difference between the mutant backgrounds (5.3, 4.0, and 5.1 more leaves than wild-type in the mac3a, mac3b, and mac3a/mac3b mutants, respectively; Figure 6A). Genetically this indicates that these two genes are in series or function together for this aspect of flowering-time control. For the number of days to 1-cm bolting (flowering time; Figure 6B), we observe a statistical difference from wild-type in all three backgrounds, with the double mutant being the most delayed (10.7 days), the mac3b mutant being the least delayed (5.4 days), and the mac3a mutant having an intermediate delay in flowering (7.1 days). The increase in severity of the double mutant indicates that MAC3A and MAC3B can act redundantly for 1-cm bolting. The anthesis delay is shorter in the single mutants than in the wild type (1.5 and 1.3 days for the mac3a and mac3b mutants, respectively; Figure S6A), but is indistinguishable from wild-type in the mac3a/mac3b double mutant. The stem elongation time is also shorter in the mac3a single mutant when compared to the wild type (0.8 days; Figure S6B), but longer in the mac3a/mac3b double mutant (0.8 days). We observe no statistical difference in stem elongation between the mac3b single mutant and the wild type. What is clear from these data is that MAC3A and MAC3B are necessary for the plant to properly time developmental transitions. This experiment also confirms what has previously been seen, i.e., that MAC3A and MAC3B can act partially redundantly and possibly together to control important biological processes.
Figure 6.
Flowering-time analyses of mac3a, mac3b, and mac3a/mac3b mutants. (A) Rosette leaf number. (B) Age at 1-cm bolting. Letters represent statistical groups as defined by a Kruskal–Wallis test with post hoc Dunn’s multiple comparisons test, with statistical difference defined as P < 0.05.
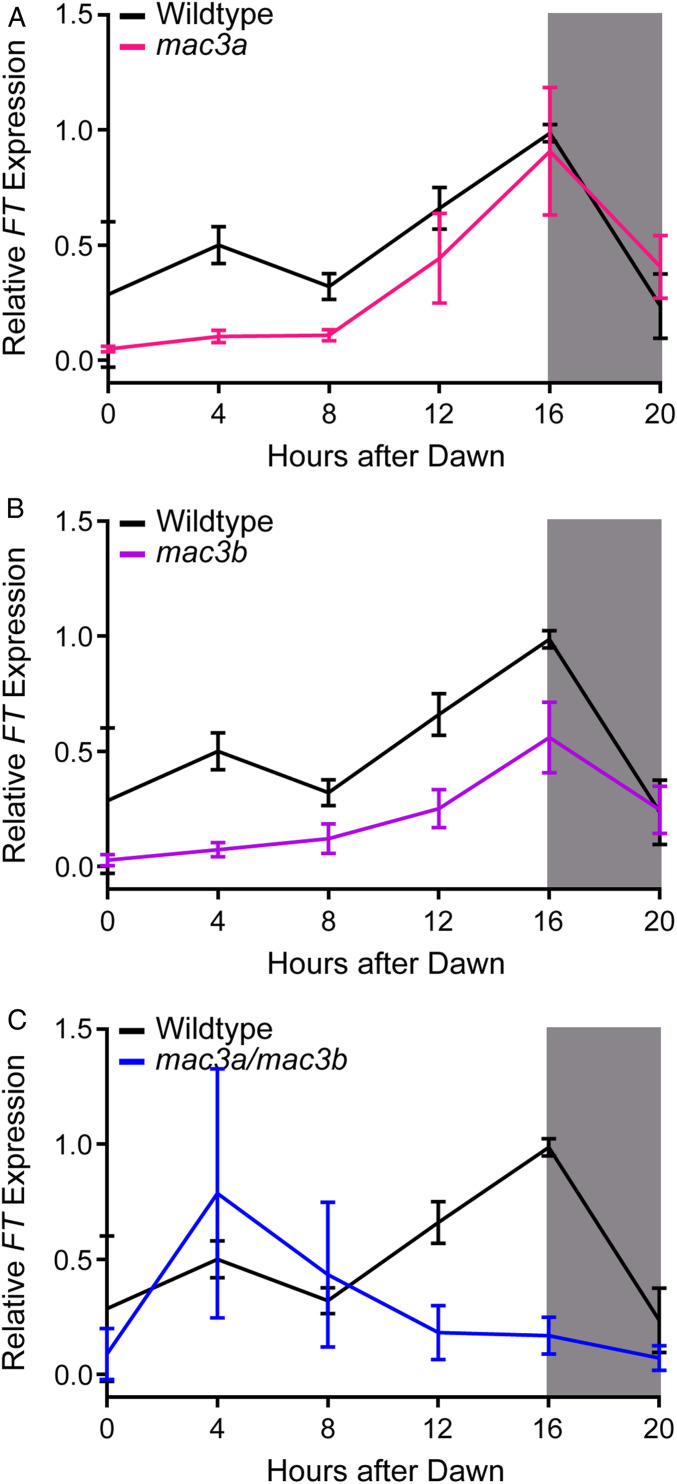
To determine whether the flowering time delays we observe in the mac3a, mac3b, and mac3a/mac3b mutants are due to an FT-dependent or -independent process, we measured FT expression in these plants using qRT-PCR (Figure 7). We observed a decrease in FT expression in all three mutant backgrounds. The decrease in FT expression was maximal from ZT0 to ZT8 in the single mac3a and mac3b mutants, although mac3b also had decreased FT expression at the ZT16 peak. In contrast, the maximal decrease in FT levels in the mac3a/mac3b double mutant occurred at ZT16. The mac3a/mac3b double mutant showed an even greater decrease in FT expression at the peak, which may explain the greater delay in flowering time in this background. These results strongly indicate that MAC3A and MAC3B are functioning upstream of FT to control flowering. In particular, the striking impact on the evening peak of FT expression suggests that the photoperiodic regulation of FT may be impacted, as the ability of this pathway to regulate this peak of FT expression is well understood, and the mac3a/mac3b mutant has a known circadian clock defect (Shim et al. 2017; Feke et al. 2019).
Figure 7.
qRT-PCR of FT expression in mac3A, mac3B, and mac3A/mac3B mutants. FT expression was measured using qRT-PCR in wild-type or homozygous (A) mac3a, (B) mac3b, and (C) mac3a/mac3b mutants grown under long-day (16 hr light/8 hr dark) conditions. Quantifications are the average of three biological replicates with error bars showing SD. qRT-PCR, quantitative real-time PCR.
Ubiquitylation dependency of MAC3B on flowering-time control
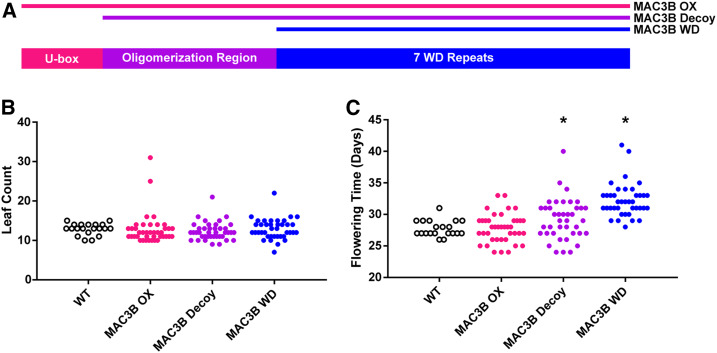
MAC3A and MAC3B are N-terminal U-box proteins that utilize seven WD40 repeats for protein–protein interaction, and are capable of oligomerizing through a canonical coiled-coil domain (Ohi et al. 2005; Grote et al. 2010). We have previously shown that MAC3A and MAC3B can form a heterodimer complex in plants in the absence of the U-box domain (Feke et al. 2019). To test the role of MAC3A/MAC3B dimerization in flowering-time regulation, we created a MAC3B decoy construct that consists of only the annotated WD40 repeats and lacks the coiled coil domain (Feke et al. 2019). We were unable to generate similar constructs for MAC3A due to unknown technical constraints, as described previously (Feke et al. 2019). We conducted the flowering-time assays with plants expressing the MAC3B WD construct and included MAC3B decoy plants as control. The transgenic plants expressing the MAC3B WD delayed 1-cm bolting by 4.5 days (P = 3.8 × 10−12) compared to the decoy-expressing plants, which delayed flowering by 1.45 days (P = 0.016) (Figure 8). Interestingly, this was not the case in our other flowering-time metrics, as rosette leaf number, anthesis delay, and stem elongation period were identical to the wild type in the MAC3B WD population (Figure 8 and Figure S7). The MAC3B decoy had similar trends in anthesis delay and elongation-period defects that we observed in our initial screen, delaying flowering by 0.95 (P = 3 × 10−3) and 0.88 days (P = 0.039), respectively, but did not have any effects on rosette leaf number. Taken together, this suggests that the anthesis delay and stem elongation defects that we observed in the MAC3B decoy are dependent on its ability to form protein dimers, while the delayed 1-cm bolting is a dominant-negative effect.
Figure 8.
Flowering-time analyses of MAC3B overexpression constructs. Flowering time was measured in T1 MAC3B full-length (OX), MAC3B decoy, and MAC3B WD insertion plants. (A) Schematic presenting which domains are present in each construct. (B) Age at 1-cm inflorescence. (C) Leaf number at 1-cm inflorescence. * represents a significant difference from WT with a Bonferroni-corrected P < 0.017. WT, wild-type.
We have previously reported that precise regulation of MAC3B expression is essential for maintaining periodicity in the circadian clock (Feke et al. 2019). However, it is unclear if flowering time is also sensitive to MAC3B expression levels. Thus, we overexpressed the full-length MAC3B and assayed its effects on flowering time. Interestingly, we did not observe any alteration in flowering time in the full-length MAC3B overexpression plants (Figure 8 and Figure S7). This suggests that the role of MAC3B in the regulation of flowering time relies on its ability to ubiquitylate substrates and not on precise regulation of MAC3B expression levels.
Discussion
We have previously demonstrated the utility of the decoy technique to overcome redundancy and identify E3 ligases that regulate the circadian clock (Lee et al. 2018; Feke et al. 2019). Here, we demonstrate that the decoy technique is capable of identifying E3 ligases involved in developmental processes, specifically flowering-time regulation. We were able to identify 5 major candidates and 10 minor candidates for flowering-time regulators, and performed follow-up experiments to validate 2 of the major candidates. While one of these candidates, MAC3A, and its homolog, MAC3B, have been suggested to control flowering time previously (Monaghan et al. 2009), we directly demonstrate flowering defects in single and double mutants, and reveal the complicated partial redundancy between the two genes. This study, and our previous study on the role of MAC3A and MAC3B in clock function, will likely help to clarify the genetic roles of MAC3A and MAC3B in the myriad biological processes that they control (Monaghan et al. 2009; Jia et al. 2017; Li et al. 2018; Feke et al. 2019). Furthermore, a second candidate revealed by our screen, PUB14, had not been identified for its role in any biological process previously, but has sequence similarity to the characterized flowering-time regulator PUB13 (Li et al. 2012a,b; Zhou et al. 2015).
PUB14 is a novel flowering-time regulator
PUB14 was the only candidate flowering-time regulator found in our screen that affected both rosette leaf number and 1-cm bolting age, two hallmarks of flowering time (Koornneef et al. 1991). Although the biochemical structure of the PUB14 protein has been studied (Andersen et al. 2004), to our knowledge no phenotypes have previously been associated with mutations in this gene. Here, we validate PUB14 as a regulator of flowering time. Both PUB14 decoys and the pub14-1 T-DNA insertion mutant delayed 1-cm bolting and increased rosette leaf number, Furthermore, FT expression was reduced in the pub14-1 mutant. As we found that the pub14-1 mutant had increased expression of a portion of PUB14, the similarity in phenotype between the PUB14 decoy and the pub14-1 mutant suggests that the decoy could be acting in a dominant-positive manner in this case, rather than dominant negative. We have previously observed dominant-positive effects with the other decoys in relation to the regulation of the circadian clock and flowering time (Lee and Feke et al. 2018; Feke et al. 2019).
The closest homolog of PUB14 is PUB13, which has previously been implicated in stress responses and the control of flowering time (Li et al. 2012a,b; Zhou et al. 2015). Mutants of PUB13 have accelerated flowering time under long-day growth conditions, suggesting that PUB13 acts as a repressor of photoperiodic flowering (Li et al. 2012a,b; Zhou et al. 2015). pub13 mutants also have elevated levels of the defense hormone salicylic acid (SA), suggesting that PUB13 is a negative regulator of immunity (Li et al. 2012a). SA activates flowering (Martínez et al. 2004; Li et al. 2012a), indicating that PUB13 acts as a repressor of flowering time and immunity through negatively regulating SA levels. Correspondingly, the advanced flowering time in the pub13 mutant is dependent on SA (Li et al. 2012a; Zhou et al. 2015). We were unable to recapitulate the pub13 mutant flowering phenotype with the PUB13 decoy, although we did see a trend toward advanced flowering that did not reach our statistical cutoff (Figure 2). However, the high protein sequence similarities in the substrate recognition domains of PUB14 and PUB13 (83% similar), and the similarities of their functions, suggest that these two homologous genes may share the same targets. In addition to its potential role in stress-regulated flowering, we also identified a suite of potential flowering-time regulators that interact with PUB14 in our immunoprecipitation-MS experiments. Direct interaction studies such as yeast two-hybrid or co-immunoprecipitation are required to verify that these putative substrates interact with PUB14. Further genetic and molecular studies can determine whether they are targets or regulatory partners of PUB14 (Lee et al. 2018, 2019).
MAC3A regulates flowering time
MAC3A [also known as PLANT U-BOX 59 (PUB59) or PRE-mRNA PROCESSING FACTOR 19 A (PRP19A)] and its close homolog MAC3B (PUB60/PRP19B) are the core components of a large, multifunctional protein complex known as the NineTeen complex (NTC) (Monaghan et al. 2009). In plants, the NTC, and MAC3A and MAC3B in particular, has been implicated in splicing, (microRNA) miRNA biogenesis, immunity, and the circadian clock (Monaghan et al. 2009; Jia et al. 2017; Li et al. 2018; Feke et al. 2019). For this reason, we do not believe that MAC3A and MAC3B would be interacting with and ubiquitylating proteins that regulate flowering, but would rather alter processes through splicing or other NTC processes. Interestingly, another component of the NTC, SKIP, has been implicated in flowering-time regulation previously (Cao et al. 2015; Cui et al. 2017). Here, we identified MAC3A as the U-box decoy with the greatest magnitude effect on flowering time, and validated that MAC3A and MAC3B are bona fide regulators of flowering time. We do observe different phenotypes between the MAC3A decoy, MAC3B decoy, and mac3a/mac3b mutants, which suggests a complex relationship with flowering time. However, it is clear that both genes are essential for proper flowering-time control.
The precise methods through which MAC3A and MAC3B alter flowering time are not yet understood and are likely multifactorial. MAC3A and MAC3B are involved in the regulation of splicing, miRNA biogenesis, immunity, and the circadian clock (Monaghan et al. 2009; Jia et al. 2017; Li et al. 2018; Feke et al. 2019). Interestingly, all of these factors are involved in the regulation of flowering time (Yanovsky and Kay 2002; Imaizumi et al. 2003; Chen 2004; Wu and Poethig 2006; Yamaguchi et al. 2009; Wu et al. 2009; Yant et al. 2010; Lyons et al. 2015; Cui et al. 2017; Gil et al. 2017). Alterations in the circadian clock lead to defects in photoperiodic flowering time, similar to what we observe in the mac3a/mac3b double mutant (Nakamichi et al. 2007). Likewise, increased resistance to pathogens, like what is observed in the mac3a/mac3b double mutant, is positively correlated with a delay in flowering time (Korves and Bergelson 2003; Monaghan et al. 2009; Lyons et al. 2015). miRNAs play an essential role in the regulation of flowering time through the aging pathway, with miRNAs having both activating and repressive activity within this pathway (Chen 2004; Wu and Poethig 2006; Yamaguchi et al. 2009; Wu et al. 2009; Yant et al. 2010). However, interpretation of the relationship between MAC3A and MAC3B and this pathway is complicated by the fact that both the repressive and activating miRNAs are likely affected by these genes (Jia et al. 2017; Li et al. 2018). Finally, splicing also plays a role in the regulation of flowering, as both the photoperiodic floral activator CO and the ambient temperature floral repressor FLM are alternatively spliced (Lee et al. 2013; Posé et al. 2013; Gil et al. 2017). In addition, our results suggest that the ubiquitylation activity of MAC3B is essential for its ability to regulate flowering time. In our truncation studies, we observed an anticorrelation between the presence of the U-box domain and the proper regulation of flowering time, with no effect on flowering time observed in plants overexpressing full-length MAC3B and the largest impact on flowering time in plants expressing the putative substrate interaction domain alone. Future investigation into the relationships between the diverse functions of MAC3A and MAC3B and flowering time will likely prove fruitful.
Additional flowering time candidates connect stress to flowering time
Stress is a well-known regulator of flowering time in Arabidopsis (Takeno 2016). Correspondingly, all of our remaining high-priority candidate floral regulators have established roles in stress responses. PUB26 is a negative regulator of immunity and pub26 mutants exhibit elevated levels of immunity (Wang et al. 2018). As resistance to pathogens and delayed flowering are positively correlated (Lyons et al. 2015), we would expect that flowering time would be delayed in these mutants, in concordance with our observations of flowering time in the PUB26 decoy population. Like biotic stresses, abiotic stresses such as salt stress can delay flowering time (Kim et al. 2007). In accordance with this, we observe delayed flowering with the PUB31 decoy, which leads to mild sensitivity to salt stress when mutated (Zhang et al. 2017). In contrast, the correlation between the known stress phenotypes of PUB61, also known as CARBOXYL TERMINUS OF HSC70-INTERACTING PROTEIN (CHIP), is less easily interpretable. We observe early flowering with the CHIP decoy. CHIP was previously identified to alter sensitivity to heat, cold, salt, and abscisic acid, but the system is complicated because mutants and overexpression lines are both sensitive to these stresses (Luo et al. 2006; Zhou et al. 2014; Wei et al. 2015). In this case, use of the decoy may help to untangle the complex relationships between CHIP, stress, and flowering time.
Conclusions
A multitude of factors, ranging from light conditions and temperature to the effects of stress, contribute to the regulation of flowering time. We only selected one condition, the floral-inductive long-day condition, to perform our screen, and due to the labor intensiveness of this screen, we chose to only investigate the U-box library. Despite using these limited conditions, we were able to identify five novel regulators of flowering time and validated two by mutant analysis. This demonstrates the likely magnitude of undiscovered flowering-time regulators within the E3 ligases as a whole, and demonstrates the necessity of targeted, dominant-negative screens to characterize members of these complex gene classes. Our experimental procedures and results provide a model for future studies of the roles of E3 ligases in flowering time and other developmental processes, and solidify the usefulness of the decoy technique as a screening platform for identifying plant E3 ligase functions.
Acknowledgments
We thank Christopher Adamchek, Cathy Chamberlin, Suyuna Eng Ren, Brandon Williams, Milan Sandhu, Annie Jin, and Skylar McDermott for their technical support; Adam Saffer for his critical reading of the manuscript; Chris Bolick, Eileen Williams, and the staff at Marsh Botanical Gardens for their support in maintaining plant growth spaces; Sandra Pariseau and Denise George for their administrative support; the Keck Proteomics Facility at Yale for their assistance with proteomics; and Xin Li for providing the mac3a and mac3a/mac3b double mutants. This work was supported by funding from the National Science Foundation (EAGER #1548538) and the National Institutes of Health (R35 GM-128670) to J.M.G.; by a Forest B.H. and Elizabeth D.W. Brown Fund Fellowship to W.L.; and by funding from the National Institutes of Health (T32 GM-007499), the Gruber Foundation, and the National Science Foundation (GRFP DGE-1122492) to A.M.F. The authors declare no competing interests.
Author contributions: A.M.F. and J.M.G. designed the experiments. A.M.F. performed the experiments and experimental analyses. A.M.F., J.H., and W.L. were involved in the generation of the U-box decoy library. A.M.F. and J.M.G. wrote the manuscript.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12245753.
Communicating editor: T. Juenger
Literature Cited
- Andersen P., Kragelund B. B., Olsen A. N., Larsen F. H., Chua N.-H. et al. , 2004. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279: 40053–40061. 10.1074/jbc.M405057200 [DOI] [PubMed] [Google Scholar]
- Azevedo C., Santos-Rosa M. J., and Shirasu K., 2001. The U-box protein family in plants. Trends Plant Sci. 6: 354–358. 10.1016/S1360-1385(01)01960-4 [DOI] [PubMed] [Google Scholar]
- Cao Y., Wen L., Wang Z., and Ma L., 2015. SKIP interacts with the Paf1 complex to regulate flowering via the activation of FLC transcription in Arabidopsis. Mol. Plant 8: 1816–1819. 10.1016/j.molp.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., and Davies B., 2012. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. 10.1104/pp.111.186999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. 10.1126/science.1088060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kim S., Kim S. Y., Kim M., Hyun Y. et al. , 2005. SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17: 2647–2660. 10.1105/tpc.105.035485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Tong A., Huo Y., Yan Z., Yang W. et al. , 2017. SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 15: 80 [corrigenda: BMC Biol. 17: 25 (2019)]. 10.1186/s12915-017-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R. B., Kandasamy M. K., McKinney E. C., and Meagher R. B., 2005. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646. 10.1105/tpc.105.035196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S. et al. , 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feke A., Liu W., Hong J., Li M. W., Lee C. M. et al. , 2019. Decoys provide a scalable platform for the identification of plant E3 ubiquitin ligases that regulate circadian function. Elife 8: e44558 10.7554/eLife.44558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J. et al. , 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44: D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil K.-E., Park M.-J., Lee H.-J., Park Y.-J., Han S.-H. et al. , 2017. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 89: 128–140. 10.1111/tpj.13351 [DOI] [PubMed] [Google Scholar]
- Graeff M., Straub D., Eguen T., Dolde U., Rodrigues V. et al. , 2016. Microprotein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet. 12: e1005959 10.1371/journal.pgen.1005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M., Wolf E., Will C. L., Lemm I., Agafonov D. E. et al. , 2010. Molecular architecture of the human Prp19/CDC5L complex. Mol. Cell. Biol. 30: 2105–2119. 10.1128/MCB.01505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., García-Ponce B., Fonseca-Salazar G., Alvarez-Buylla E. R., and Yu H., 2008. AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J. 55: 253–265. 10.1111/j.1365-313X.2008.03499.x [DOI] [PubMed] [Google Scholar]
- Huang H., Alvarez S., Bindbeutel R., Shen Z., Naldrett M. J. et al. , 2016a Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15: 201–217. 10.1074/mcp.M115.054064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Alvarez S., and Nusinow D. A., 2016b Data on the identification of protein interactors with the Evening Complex and PCH1 in Arabidopsis using tandem affinity purification and mass spectrometry (TAP-MS). Data Brief 8: 56–60. 10.1016/j.dib.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Kong X., Wang C., Ma L., Zhao J. et al. , 2014. Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell Online 26: 4763–4781. 10.1105/tpc.114.132738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Tran H. G., Swartz T. E., Briggs W. R., and Kay S. A., 2003. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306. 10.1038/nature02090 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T. F., Harmon F. G., Ho L. A., and Kay S. A., 2005. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. 10.1126/science.1110586 [DOI] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K. C. S., Wenkel S., Soppe W. et al. , 2008. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. 10.1038/emboj.2008.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T., Zhang B., You C., Zhang Y., Zeng L. et al. , 2017. The Arabidopsis MOS4-associated complex promotes MicroRNA biogenesis and precursor messenger RNA splicing. Plant Cell 29: 2626–2643. 10.1105/tpc.17.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-G., Kim S.-Y., and Park C.-M., 2007. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226: 647–654. 10.1007/s00425-007-0513-3 [DOI] [PubMed] [Google Scholar]
- Kong L., Cheng J., Zhu Y., Ding Y., Meng J. et al. , 2015. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6: 8630 10.1038/ncomms9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., and van der Veen J. H., 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. MGG 229: 57–66. 10.1007/BF00264213 [DOI] [PubMed] [Google Scholar]
- Korves T. M., and Bergelson J., 2003. A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 133: 339–347. 10.1104/pp.103.027094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Piñeiro M., and Jarillo J. A., 2012. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999. 10.1105/tpc.110.081885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-M., and Thomashow M. F., 2012. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 15054–15059. 10.1073/pnas.1211295109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-M., Feke A., Li M.-W., Adamchek C., Webb K. et al. , 2018. Decoys untangle complicated redundancy and reveal targets of circadian clock F-box proteins. Plant Physiol. 177: 1170–1186. 10.1104/pp.18.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Li M. W., Feke A., Liu W., Saffer A. M. et al. , 2019. GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat. Commun. 10: 3750 10.1038/s41467-019-11769-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Ryu H.-S., Chung K. S., Posé D., Kim S., et al. , 2013. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632. 10.1126/SCIENCE.1241097 [DOI] [PubMed] [Google Scholar]
- Leijten W., Koes R., Roobeek I., and Frugis G., 2018. Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae crop species. Plants (Basel) 7: 111 10.3390/plants7040111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu K., Zhou B., Li M., Zhang S. et al. , 2018. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell 30: 481–494. 10.1105/tpc.17.00953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ahn I.-P., Ning Y., Park C.-H., Zeng L. et al. , 2012a The U-box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159: 239–250. 10.1104/pp.111.192617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Dai L., and Wang G.-L., 2012b PUB13, a U-box/ARM E3 ligase, regulates plant defense, cell death, and flowering time. Plant Signal. Behav. 7: 898–900. 10.4161/psb.20703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D., Cao Y., Sun X., Espinoza C., Nguyen C. T. et al. , 2017. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 214: 1646–1656. 10.1111/nph.14472 [DOI] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K. et al. , 2010. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61: 259–270. 10.1111/j.1365-313X.2009.04048.x [DOI] [PubMed] [Google Scholar]
- Luo J., Shen G., Yan J., He C., and Zhang H., 2006. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 46: 649–657. 10.1111/j.1365-313X.2006.02730.x [DOI] [PubMed] [Google Scholar]
- Lyons R., Rusu A., Stiller J., Powell J., Manners J. M. et al. , 2015. Investigating the association between flowering time and defense in the Arabidopsis thaliana-Fusarium oxysporum interaction. PLoS One 10: e0127699 10.1371/journal.pone.0127699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C., Pons E., Prats G., and León J., 2004. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 37: 209–217. 10.1046/j.1365-313X.2003.01954.x [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M., Lázaro A., Poethig R. S., Gómez-Mena C., Piñeiro M. A. et al. , 2006. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133: 1241–1252. 10.1242/dev.02301 [DOI] [PubMed] [Google Scholar]
- McGinnis K. M., Thomas S. G., Soule J. D., Strader L. C., Zale J. M. et al. , 2003. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130. 10.1105/tpc.010827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T. C., Michael T. P., Priest H. D., Shen R., Sullivan C. M. et al. , 2007. The diurnal project: diurnal and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72: 353–363. 10.1101/sqb.2007.72.006 [DOI] [PubMed] [Google Scholar]
- Monaghan J., Xu F., Gao M., Zhao Q., Palma K. et al. , 2009. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5: e1000526 10.1371/journal.ppat.1000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Niinuma K., Ito S., Yamashino T. et al. , 2007. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 48: 822–832. 10.1093/pcp/pcm056 [DOI] [PubMed] [Google Scholar]
- Navarro-Quezada A., Schumann N., and Quint M., 2013. Plant F-box protein evolution is determined by lineage-specific timing of major gene family expansion waves. PLoS One 8: e68672 10.1371/journal.pone.0068672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. C., Lasswell J., Rogg L. E., Cohen M. A., and Bartel B., 2000. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340. 10.1016/S0092-8674(00)80842-9 [DOI] [PubMed] [Google Scholar]
- Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Ren L., Hirsch J. P. et al. , 2005. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell. Biol. 25: 451–460. 10.1128/MCB.25.1.451-460.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page T., Macknight R., Yang C. H., and Dean C., 1999. Genetic interactions of the Arabidopsis flowering time gene FCA, with genes regulating floral initiation. Plant J. 17: 231–239. 10.1046/j.1365-313X.1999.00364.x [DOI] [PubMed] [Google Scholar]
- Park B. S., Sang W. G., Yeu S. Y., Do Choi Y., Paek N.-C. et al. , 2007. Post-translational regulation of FLC is mediated by an E ubiquitin ligase activity of SINAT5 in Arabidopsis. Plant Sci. 173: 269–275. 10.1016/j.plantsci.2007.06.001 [DOI] [Google Scholar]
- Park H. J., Kim W.-Y., Pardo J. M., and Yun D.-J., 2016. Molecular interactions between flowering time and abiotic stress pathways. Int. Rev. Cell Mol. Biol. 327: 371–412. 10.1016/bs.ircmb.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S., 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567. [DOI] [PubMed] [Google Scholar]
- Posé D., Verhage L., Ott F., Yant L., Mathieu J. et al. , 2013. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417. 10.1038/nature12633 [DOI] [PubMed] [Google Scholar]
- Risseeuw E. P., Daskalchuk T. E., Banks T. W., Liu E., Cotelesage J. et al. , 2003. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34: 753–767. 10.1046/j.1365-313X.2003.01768.x [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D. A., Kay S. A., and Imaizumi T., 2007. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265. 10.1126/science.1146994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. S., Kubota A., and Imaizumi T., 2017. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173: 5–15. 10.1104/pp.16.01327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Smith R. W., To B. J., Millar A. J., and Imaizumi T., 2012. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. 10.1126/science.1219644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Kubota A., Kwon M. S., Covington M. F., Lee N. et al. , 2018. Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat. Plants 4: 824–835. 10.1038/s41477-018-0253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., and Schmid M., 2011. Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F. et al. , 2001. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. 10.1038/35074138 [DOI] [PubMed] [Google Scholar]
- Sun T.-P., 2011. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. 10.1016/j.cub.2011.02.036 [DOI] [PubMed] [Google Scholar]
- Takeno K., 2016. Stress-induced flowering: the third category of flowering response. J. Exp. Bot. 67: 4925–4934. 10.1093/jxb/erw272 [DOI] [PubMed] [Google Scholar]
- Vierstra R. D., 2009. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. 10.1038/nrm2688 [DOI] [PubMed] [Google Scholar]
- Wagner D., and Meyerowitz E. M., 2002. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12: 85–94. 10.1016/S0960-9822(01)00651-0 [DOI] [PubMed] [Google Scholar]
- Wang J., Grubb L. E., Wang J., Liang X., Li L. et al. , 2018. A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69: 493–504.e6. 10.1016/j.molcel.2017.12.026 [DOI] [PubMed] [Google Scholar]
- Wang W., Yang D., and Feldmann K., 2011. EFO1 and EFO2, encoding putative WD-domain proteins, have overlapping and distinct roles in the regulation of vegetative development and flowering of Arabidopsis. J. Exp. Bot. 62: 1077–1088. 10.1093/jxb/erq336 [DOI] [PubMed] [Google Scholar]
- Wei J., Qiu X., Chen L., Hu W., Hu R. et al. , 2015. The E3 ligase AtCHIP positively regulates Clp proteolytic subunit homeostasis. J. Exp. Bot. 66: 5809–5820. 10.1093/jxb/erv286 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V. et al. , 2007. An “electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., and Poethig R. S., 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. 10.1242/dev.02521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J.-W., Weigel D. et al. , 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-F., Wang Y., and Wu S.-H., 2008. Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 148: 948–959. 10.1104/pp.108.124917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.-F., Yang L., Wu G., Poethig R. S. et al. , 2009. The MicroRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278. 10.1016/j.devcel.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M. J., and Kay S. A., 2002. Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312. 10.1038/nature00996 [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T. T., Ott F., Lanz C. et al. , 2010. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. 10.1105/tpc.110.075606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D., and Goring D. R., 2009. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60: 1109–1121. 10.1093/jxb/ern369 [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhao J., Li L., Gao Y., Zhao L. et al. , 2017. The Arabidopsis U-box E3 ubiquitin ligase PUB30 negatively regulates salt tolerance by facilitating BRI1 kinase inhibitor 1 (BKI1) degradation. Plant Cell Environ. 40: 2831–2843. 10.1111/pce.13064 [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhang Y., Qi J., Chi Y., Fan B. et al. , 2014. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10: e1004116 10.1371/journal.pgen.1004116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lu D., Xu G., Finlayson S. A., He P. et al. , 2015. The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J. Exp. Bot. 66: 3353–3366. 10.1093/jxb/erv148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu D., Wang P., Ma X., Lin W. et al. , 2018. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 115: E1906–E1915. 10.1073/pnas.1712251115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all other data necessary for confirming the conclusions of the article are present within the article, figures, and tables or as supplemental materials uploaded to figshare. The seeds and plasmids are available upon request and will be available from the Arabidopsis Biological Resource Center. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12245753