SUMMARY
Fast purinergic signaling is mediated by ATP and ATP-gated ionotropic P2X receptors (P2XRs), and it is implicated in pain-related behaviors. The properties exhibited by P2XRs vary between those expressed in heterologous cells and in vivo. Several modulators of ligand-gated ion channels have recently been identified, suggesting that there are P2XR functional modulators in vivo. Here, we establish a genome-wide open reading frame (ORF) collection and perform functional screening to identify modulators of P2XR activity. We identify TMEM163, which specifically modulates the channel properties and pharmacology of P2XRs. We also find that TMEM163 is required for full function of the neuronal P2XR and a pain-related ATP-evoked behavior. These results establish TMEM163 as a critical modulator of P2XRs in vivo and a potential target for the discovery of drugs for treating pain.
Graphical Abstract
In Brief
Fast purinergic signaling is mediated by ATP-gated P2X receptors. Salm et al. perform functional screening with a genome-wide ORF collection and identify TMEM163, which modulates the channel properties and pharmacology of P2XRs and ATP-evoked behavior. They propose TMEM163 as a critical modulator of fast purinergic signaling.
INTRODUCTION
ATP and purinergic P2X receptors (P2XRs) mediate pain (Basbaum et al., 2009; Bleehen and Keele, 1977; Burnstock, 1977, 2008; Chizh and Illes, 2001; North and Jarvis, 2013). Thus, P2XRs are considered as a target for the discovery of drugs for treating pain. However, it is difficult to design drugs that function across P2XR subtypes because there are seven P2XR subtypes with properties that overlap and properties that are distinct (Khakh and North, 2012; Li et al., 2013). For example, the half-maximal effective concentrations (EC50) of ATP-evoked currents are varied from 0.1 μM to 4 mM among the P2XR subtypes (Coddou et al., 2011). In addition, the kinetics of P2XR subtypes differ: there are both fast- and slow-desensitizing P2XR subtypes (Khakh and North, 2012; Li et al., 2013). Native P2XRs showed diverse responses that are dependent on cell types (Chizh and Illes, 2001; North, 2002). Due to the diversity in the properties of P2XR subtypes and the expression of multiple subtypes in a single cell, it is difficult to conclude whether P2XR subtypes by themselves can explain the properties of native P2XRs. The pharmacology of native and recombinant P2XRs shows variation. For example, ivermectin was initially shown to activate invertebrate glutamate-gated chloride channels (Cully et al., 1994) and later found to potentiate ATP-evoked activity of heterologously expressed P2X4R but not heterologously expressed P2X2R and neuronal P2XRs from embryonic hippocampus and the brainstem trigeminal mesencephalic nucleus (MNB) (Cully et al., 1994; Khakh et al., 1999; Priel and Silberberg, 2004; Silberberg et al., 2007). These differences could be due to a difference in P2XR subtypes expressed in vivo and in heterologous cells. Alternatively, it is possible functional modulators are missing in heterologous cell systems.
Many ion channels require modulatory molecules or auxiliary subunits specific to each particular channel type for their biogenesis and appropriate activity in vivo. For example, the ionotropic AMPA-type glutamate receptor forms a stable complex with TARPs and CNIHs (Kato et al., 2010; Schwenk et al., 2009; Tomita et al., 2003; Vandenberghe et al., 2005) and requires PORCN for its maturation (Erlenhardt et al., 2016). Likewise, the ionotropic nicotinic-type acetylcholine receptor requires NACHO (encoded by the TMEM35 gene) for its surface expression (Gu et al., 2016). Finally, the ionotropic GABA receptor forms a stable complex with GARLH4 (encoded by the LHFPL4 gene) and neuroligin (Yamasaki et al., 2017) and requires clptm1 for its maturation or surface expression (Ge et al., 2018). Furthermore, some of these molecules modulate receptor pharmacology in vivo and are potential targets for drug discovery. Indeed, a specific modulator of the AMPA receptor, its TARPγ-8 auxiliary subunit, has been used for drug discovery. With the incorporation of TARPγ-8 into a compound screening system, compounds suitable for treating epilepsy were identified (Kato et al., 2016; Maher et al., 2017; Maher et al., 2016). These examples also suggest that P2XRs may have as-yet unidentified specific functional modulators.
Here, we utilized a genome-wide open reading frame (ORF) collection to identify TMEM163 as a specific modulator of P2XRs. TMEM163 co-expression enhances the ATP-evoked current in heterologous cells expressing exogenous P2X3R and P2X4R. Further, TMEM163 modulates the channel properties of P2X3R and P2X4R, which alter their ATP potency and pharmacological efficacy. Finally, TMEM163 is required for ATP-evoked currents in the cerebellar granule cells and dorsal root ganglion (DRG) neurons including those that convey pain-related information from peripheral tissue. Taken together, our results establish a critical system of regulation of P2XRs and ATP-evoked behavior.
RESULTS
Functional ORF-Based Screening Identifies TMEM163 as a P2X Receptor Modulator
The correct functioning of ion channels is facilitated by functional modulators and these can be any type of protein, including membrane, cytosolic, and secreted ones. To identify functional modulators of P2XRs, we performed an unbiased, high-throughput screening using a genome-wide human ORF collection, established here, and an automated analysis (Figures 1A and 1B). We have established a mammalian genome-wide ORF collection containing 17,284 non-redundant ORFs including 817 OriGene transmembrane ORFs, 13,193 CCSB-Broad Human ORFs, and our 3,274 ORFs cloned in this study. This collection was constructed with either a C-terminal V5 tag or a native stop codon in various mammalian expression vectors with a CMV promoter. The HUGO database defines 19,224 distinct protein-coding genes as of March 2019 (https://www.genenames.org/), and our ORFs correspond to 90% of these human protein-coding genes.
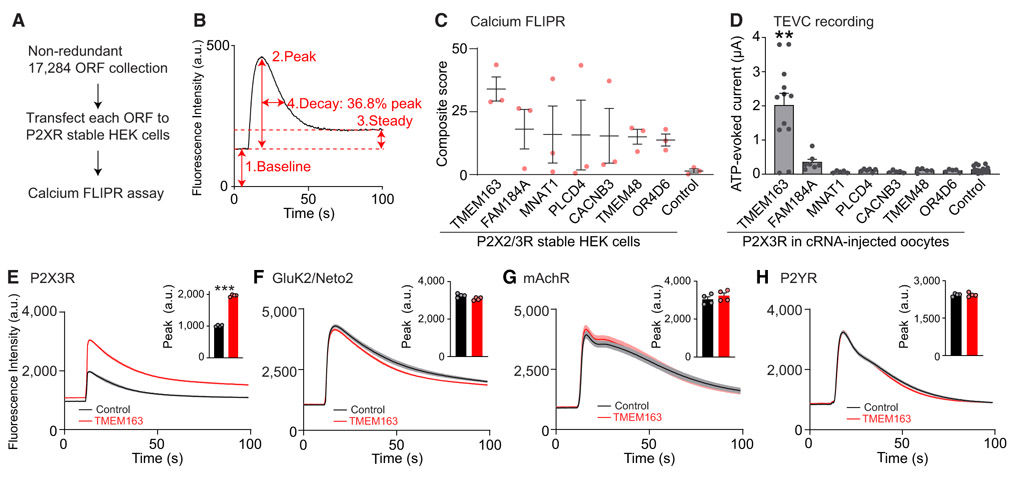
Figure 1. Genome-wide ORF-Based FLIPR Screening Identifies a P2X3R Modulator.
(A) Scheme of genome-wide ORF-based FLIPR screening.
(B) Analysis of each FLIPR response. Individual calcium FLIPR traces were analyzed using four different factors shown on the example trace: (1) baseline, (2) peak, (3) steady state, and (4) decay, as the time to reach 36.8% of the peak from the peak time, as well as the ratios of peak and baseline (5) and steady state and baseline (6) as baseline-normalized values. The effect of these factors were then multiplied together, creating a composite score for hit ORF prioritization.
(C) The top seven P2XR-specific ORFs and RFP (control) are listed based on composite score (n = 3).
(D) ATP (300 nM)-evoked currents were measured with two-electrode voltage clamp (TEVC) recording (Vh = −30 mV) from oocytes co-injected with 100 pg of P2X3R cRNA and 2 ng of cRNA for each ORF or Neto2 as a control (n = 5–20).
(E–H) Modulation of agonist-evoked calcium FLIPR responses by TMEM163 co-expression. HEK cells or HEK cells stably expressing P2X3R or GluK2 and Neto2 were transiently transfected with TMEM163 or pcDNA3 (control). Agonist-induced calcium FLIPR responses were measured from transfected cells pre-incubated with 0.3 U/mL apyrase for 1 h. Summary bar graphs of peak amplitude are shown in insets (n = 4). TMEM163 enhanced peak and steady-state responses of the P2X3R-stable HEK cells in response to α,β-MeATP (55.5 μM, E). TMEM163 did not affect the calcium FLIPR responses of GluK2/Neto2-stable HEK cells in response to 3.33 μM kainate (F), of HEK cells to 1 mM acetylcholine probing endogenous muscarinic acetylcholine receptor (mAchR) (G), and of HEK cells to 100 μM ATP probing endogenous P2Y receptors (P2YRs) (H).
Data are mean ± SEM. Kruskal-Wallis with Dunn’s post-test (D), two-way ANOVA followed by Bonferroni-Dunn’s post-test for (E)–(H); **p < 0.01; ***p < 0.001.
We next established HEK cell lines stably expressing both P2X3R and P2X2R as representative P2XRs (presumably a mixture of P2X2R and P2X3R heteromers, P2X2R and P2X3R homomers) with α,β-MeATP EC50 of 656.5 ± 88.7 nM (Figure S1A). Here, we used α,β-MeATP instead of ATP to distinguish the activity from endogenous P2Y receptors in HEK cells. Then, we individually transfected each of the ORFs into stable P2XR cells plated in individual wells on 384-well plates, with a transfection efficiency around 80%–90% as estimated by RFP (red fluorescent protein) (Figure S1B). Two days after transfection, we measured 1 μM α,β-MeATP-evoked P2XR activity with the FLIPR calcium 4 dye using the 384-well FLIPR Tetra system (Figure 1A). In this study, we used an EC70 dose (α,β-MeATP, 1 μM) for the screening, which provides a reasonable separation of the signal and background (Z′ = 0.61 for the agonist-evoked responses and SNR [the ratio of signal mean to STD of the background] = 39.5) (Zhang et al., 1999). The FLIPR results were automatically analyzed using a custom analytical program that examined six factors (baseline, peak amplitude, steady-state amplitude, decay, ratio of peak and baseline, and ratio of steady-state and baseline) (Figure 1B), and ORFs with any factor greater than 3 times the SD above the mean of the plate were selected for validation. Then, we validated the top 309 ORFs using a similar assay with three replicates on P2XRs and ranked all ORFs by a composite score or the multiplication of all six factors. By comparing the positive ORFs across other channels, we established a list of 60 ORFs as specific modulators of α,β-MeATP-evoked calcium responses in the P2XR-stably expressing HEK cells (top seven clones in Figure 1C).
Changes in calcium responses could be due to the modulation of P2XR or other molecules. To identify ORFs that alter P2XR ion channel activity, we tested the top seven ORFs by measuring ATP-evoked currents from Xenopus laevis oocytes injected with cRNAs of P2X3R and each ORF with a fast perfusion system and two-electrode voltage clamp (TEVC) recording. Here, we expressed only a single P2X3R subtype, instead of the two P2XR subtypes (P2X2R and P2X3R), to be used for screening because the expression of both P2X2R and P2X3R might complicate data interpretations due to heterogeneity in receptor compositions. As a result, we identified TMEM163 as a modulator of P2X3R activity (Figure 1D).
We further validated the specificity of TMEM163 on P2X3Rs via the calcium FLIPR assay in transfected HEK cells. We transiently transfected TMEM163 into P2X3R-stably expressing HEK cells and pre-treated them with an ATP hydrolyzer, apyrase, for 1 h to reduce extracellular ATP, which desensitizes P2XRs before stimulation. The results showed that TMEM163 specifically enhanced agonist-evoked calcium FLIPR response from cells stably expressing P2X3R (Figure 1E), but not the kainate-type glutamate receptor (GluK2 and Neto2) (Figure 1F). Furthermore, TMEM163 expression did not alter the GPCR-dependent calcium signaling tested by the endogenous muscarinic acetylcholine receptor (mAchR) and endogenous ATP-dependent P2Y receptor signaling (Figure 1G and H), establishing TMEM163 as a modulator of P2X3Rs.
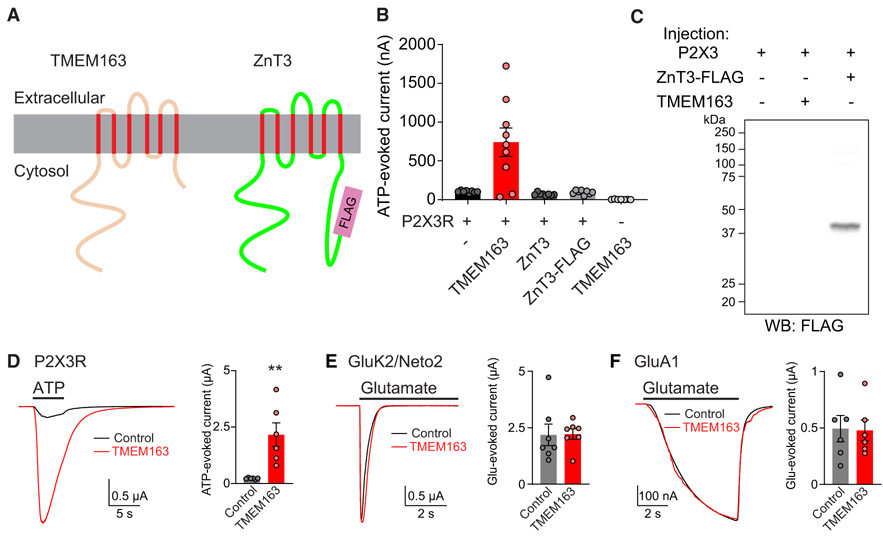
TMEM163 is a 289-amino-acid protein with six transmembrane domains in humans and shares structural homology with zinc transporters, such as ZnT3 (Figure 2A). TMEM163 was previously identified as a component of synaptic vesicles and named SV31 (Burré et al., 2007). It was also revealed to be a zinc transporter (Barth et al., 2011; Cuajungco et al., 2014; Waberer et al., 2017). We thus examined whether zinc transporter activity plays a role in modulating P2XR activity by examining ZnT3. We injected P2X3R cRNA with cRNA of either ZnT3, ZnT3 with C-terminal FLAG tag, or TMEM163 into oocytes and compared the 300 nM ATP-evoked currents (Figure 2B). Although TMEM163 co-injection robustly enhanced ATP-evoked currents, there was no difference in ATP-evoked currents upon the co-injection of ZnT3 or ZnT3-FLAG (Figure 2B). One possible explanation for this is that ZnT3 or ZnT3-FLAG was not expressed in cRNA-injected oocytes, but we confirmed expression of ZnT3-FLAG with the anti-FLAG antibody in lysate from oocytes (Figure 2C). Notably, TMEM163 alone did not show any ATP-dependent activity in cRNA-injected oocytes (Figure 2B). Moreover, in experiments performed in parallel under similar conditions in which TMEM163 enhanced P2X3R activity (Figure 2D), TMEM163 co-expression did not alter glutamate-evoked currents from oocytes injected with kainate receptor components, GluK2 and Neto2, or with AMPA receptor, GluA1 (Figures 2E and 2F). Meanwhile, at both −30 mV and −70 mV holding potentials, TMEM163 co-expression significantly enhanced 300 nM ATP-evoked currents from oocytes expressing P2X3R (Figure S2). These results suggest that TMEM163 specifically enhances P2X3R activity in heterologous systems.
Figure 2. TMEM163 Specifically Enhances P2XR Activity.
(A) Domain architecture of TMEM163 with its structural homolog, ZnT3, with FLAG epitope tag at the C terminus of ZnT3 (ZnT3-FLAG).
(B) 300 nM ATP-evoked currents were measured from oocytes injected with combinations of cRNAs as indicated (P2X3R 25 pg; all others 2 ng) using TEVC recording (Vh = −30 mV, n = 7–9).
(C) Expression of ZnT3-FLAG was confirmed at the expected molecular weight by western blotting (WB) of cRNA-injected oocyte lysate with anti-FLAG antibody.
(D–F) Representative traces and quantification of agonist-evoked currents in cRNA-injected oocytes. cRNAs of 100 pg of P2X3R, 500 pg of GluK2 and 2 ng of Neto2, 1 ng of GluA1, 2 ng of TMEM163, and 2 ng of control (Neto2 for P2XR and GluA1 and ZnT3 for GluK2) were injected individually. TMEM163 co-expression enhanced 300 nM ATP-evoked currents of P2X3R (D), but not 500 μM glutamate-evoked currents of the kainate receptor, GluK2/Neto2 (E), or 10 μM glutamate with 50 μM cyclothiazide-evoked currents of the AMPA receptor, GluA1 (F) (n = 6–7).
Data are mean ± SEM. Mann-Whitney U-test (D). **p < 0.01.
Neuronal P2X Receptor Function Requires TMEM163
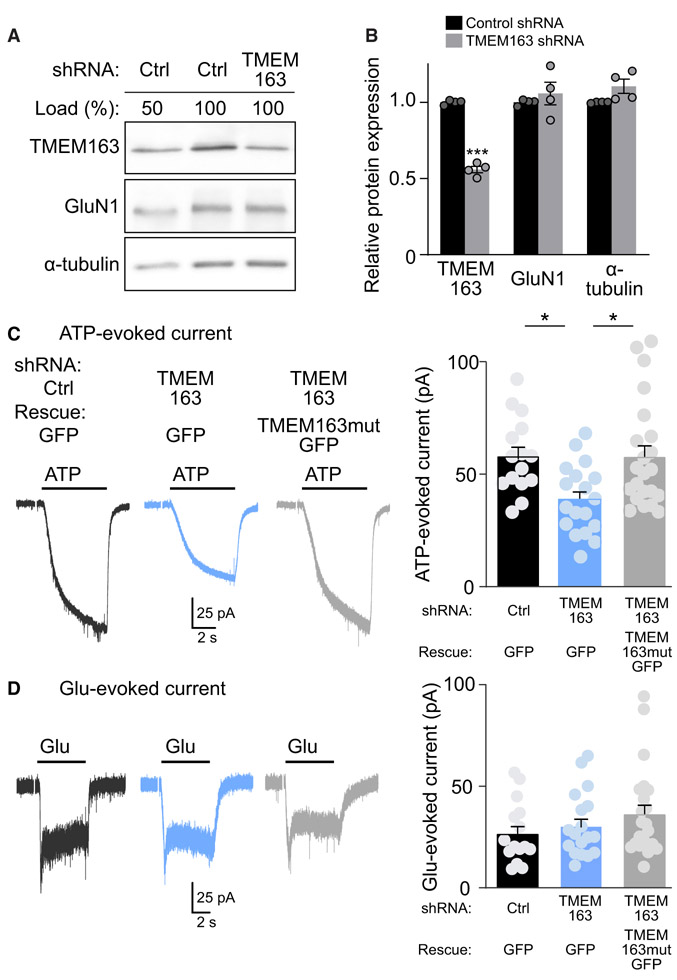
We next examined the roles of TMEM163 in neurons. In situ hybridization showed the expression of TMEM163 in mouse cerebellar granule cells (http://mouse.brain-map.org/) (Figure S3A). Thus, we prepared a lentivirus carrying TMEM163 short hairpin RNA (shRNA) and infected mouse primary cerebellar granule neurons with it. We detected TMEM163 protein with an anti-TMEM163 antibody generated for this study, which recognized a single band consistent with the expected molecular weight of 31 kDa in mouse cerebellum (Figure S3B). We confirmed a reduction of the TMEM163 level by 44.1% ± 2.2% by TMEM163 shRNA compared to the level with control shRNA without altering other unrelated proteins, GluN1 and α-tubulin (Figures 3A and 3B).
Figure 3. TMEM163 Potentiates P2XR Function in Cerebellar Granule Cells.
Primary cerebellar granule neurons were treated with lentivirus carrying TMEM163 shRNA or a random sequence in parental pFUGW (Ctrl) as well as C-terminally GFP-tagged TMEM163 mutant resistant to the shRNA (TMEM163mutGFP).
(A) Total protein amounts of TMEM163 and other proteins were measured in primary cerebellar granule neurons treated with shRNA lentivirus. For comparison, we loaded 50% of the control (Ctrl) lysate.
(B) Quantitation of total protein amounts. TMEM163 shRNA significantly reduced TMEM163 protein compared with control shRNA. Total protein amounts of GluN1 and α-tubulin were unaltered (n = 4).
(C and D) Agonist-evoked whole-cell currents were measured under whole-cell configuration (Vh = −70 mV) with the cell capacitance between 4.0–4.3 pF. Representative traces and quantitation of peak amplitudes of 100 μM ATP-evoked currents (C) or 100 μM Glu-evoked currents (D) upon 6-s application on neurons with a GFP signal from GFP or TMEM163mutGFP (n = 15–22). Artifacts of a 150 ms/2 mV pulse for cell capacitance measurement were omitted from traces shown.
Data are mean ± SEM. Student’s t test (B) and one-way ANOVA, followed by Bonferroni’s post-test (C); *p < 0.05; ***p < 0.001.
We examined whether TMEM163 plays a role in modulating ATP-evoked P2XR currents. Under the whole-cell configuration, we measured ATP-evoked P2XR currents from neurons expressing GFP, representing lentivirus infection. Under these conditions, we confirmed that a P2XR inhibitor, TNP-ATP (10 μM), blocked all ATP-evoked currents in cerebellar granule cells (23.55 ± 6.96 pA from untreated and 0.53 ± 0.25 pA from TNP-ATP-treated cells, n = 3). We identified an ~33% reduction in ATP-evoked currents in neurons treated with TMEM163 shRNA compared with currents observed in those treated with control shRNA (Figure 3C). Moreover, this reduction was restored by co-expression of C-terminal GFP-tagged TMEM163 mutant resistant to TMEM163 shRNA (Figure 3C). Glutamate-evoked currents in neurons treated with various lentiviruses were unaltered (Figure 3D). The similar extents of shRNA-mediated reduction in TMEM163 protein expression and ATP-evoked P2XR activity suggest that TMEM163 is involved in neuronal P2XR activity.
TMEM163 Modulates Channel Properties of P2XRs
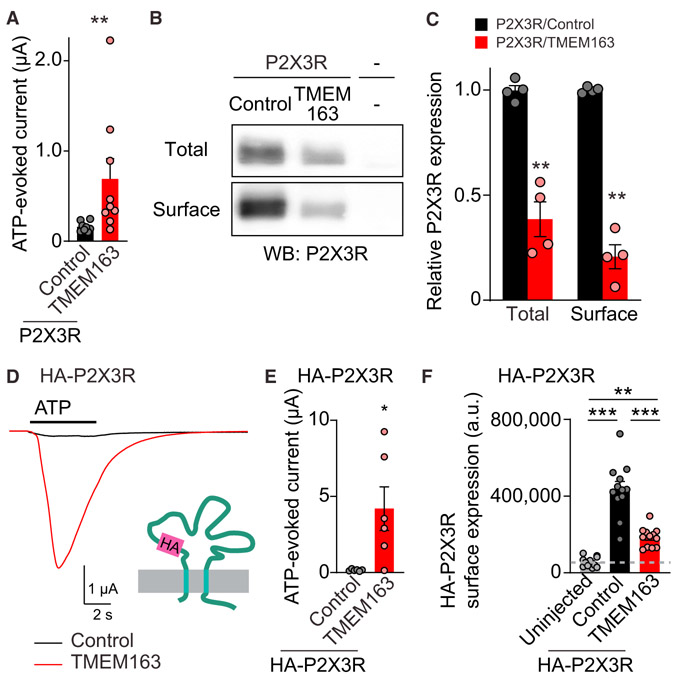
TMEM163 increases ATP-evoked P2X3R activity, which could be due to an increase in the number of P2X3Rs at the cell surface or an increase in each channel’s activity. We examined these possibilities in cRNA-injected oocytes by measuring ATP-evoked currents and protein surface expression in parallel. We expressed P2X3R with either TMEM163 or control protein (Neto2) and confirmed that TMEM163 increased ATP-evoked currents (Figure 4A). We also biotinylated proteins at the surface of oocytes with cell-impermeable sulfo-NHS-SS-biotin and prepared membrane lysate from the biotinylated oocytes. The surface and total proteins were isolated from membrane lysate with neutravidin and anti-P2X3R antibody, respectively (Figure 4B). The results revealed that the co-expression of TMEM163 significantly reduced both total and surface protein amounts of P2X3R to similar extents (Figure 4C), suggesting that TMEM163 affects P2X3R expression. We also confirmed that TMEM48, a negative ORF in Figure 1D, did not affect the surface expression of HA-P2X3Rs and P2X3R (Figures S4A and S4B). Despite the reduction in the surface expression of P2X3R by TMEM163 co-expression (Figures 4B and 4C), TMEM163 co-expression increased ATP-evoked P2X3R current (Figure 4A). These results indicate that TMEM163 enhances the activity of each P2X3R channel.
Figure 4. TMEM163 Enhances ATP-Evoked P2XR Channel Activity.
(A–C) Surface expression of P2X3R was biochemically measured in oocytes injected with cRNAs of 100 pg of P2X3R and 2 ng of TMEM163 or Neto2 (control). (A) TEVC recording (Vh = −30 mV, n = 8–9) showed an increase in 300 nM ATP-evoked currents by TMEM163 co-expression. (B) Total and surface expression of P2X3R in the oocytes. Proteins at the oocyte surface were biotinylated. The “Surface” and “Total” proteins were isolated with neutravidin and anti P2X3R antibody, respectively. (C) Quantification of P2X3R expression. P2X3R was detected in oocytes only injected with P2X3R cRNA, but not in “uninjected” oocytes, and co-injection of TMEM163 cRNA reduced both total and surface expression of P2X3Rs (n = 4).
(D–F) Agonist-evoked currents and chemiluminescently detected surface expression of extracellularly HA-tagged P2X3R (HA-P2X3R) were measured in cRNA-injected oocytes (cRNA amount injected: 1 ng of HA-P2X3; 2 ng of TMEM163 or Neto2 indicated as a control). (D and E) HA epitope was inserted into the extracellular domain of P2X3R (HA-P2X3R). TMEM163 robustly enhanced ATP (1 μM)-evoked currents of HA-P2X3R. Representative traces (D) and summary bar graph (E) are shown (n = 6). (F) Surface expression of HA-P2X3R in oocytes was measured chemiluminescently. HA proteins at the oocyte surface were detected with anti-HA antibody under non-permeabilized conditions as chemiluminescence signal (n = 10). The signal from uninjected oocytes indicates background in this assay.
Data are mean ± SEM. Mann-Whitney U-test (A and E), unpaired t test (C), one-way ANOVA with Bonferroni’s post-test (F); *p < 0.05; **p < 0.01; ***p < 0.001.
We further confirmed this by applying an alternative method of chemiluminescent detection of cell surface proteins using extracellularly HA-tagged P2X3R as previously reported (Zhang et al., 2009). We inserted an HA epitope in the extracellular loop of P2X3R between asparagine and arginine at positions 72 and 73, respectively (HA-P2X3R) (Figure 4D) and confirmed TMEM163 increased ATP-evoked currents of HA-P2X3R (Figures 4D and 4E). We next labeled HA-P2X3R at the oocyte surface with an anti-HA antibody without cell permeabilization and detected surface expression of HA-P2X3R as a luminescent signal from the combination of an HRP-conjugated secondary antibody and substrate (Zerangue et al., 1999). HA-P2X3Rs were detected at the surface in oocytes with and without TMEM163 co-expression compared to uninjected oocytes (Figure 4F). Surface expression of HA-P2X3R was reduced by TMEM163 co-expression (Figure 4F), which is consistent with the results from biotinylated proteins with untagged receptors (Figures 4A-4C). These results confirm that TMEM163 can increase channel activity of P2X3Rs and reduce surface expression of P2X3Rs in heterologous cells.
We showed that TMEM163 modulates P2X3R with two opposing effects, reducing P2X3R surface expression and increasing P2X3R channel activity. Toward testing this modulation on other P2XRs, we cloned all of the other human P2XR subtypes (P2XR1, 2, 4, 5, 6, and 7) and measured ATP-evoked currents from oocytes injected with each P2XR (Figures 6A and S4C). As shown previously (Lê et al., 1997; Soto et al., 1996), oocytes expressing human P2XR1, 2, 4, or 7, but not human P2X5R and P2X6R, showed ATP-evoked currents (Figures 6A and S4C). Then, we examined the effects of TMEM163 on P2XR1, 2, 4, and 7. We found that TMEM163 enhanced and reduced ATP-evoked currents from oocytes expressing P2X4R and P2X1R or P2X7R, respectively, but not P2X2R (Figures 6A and S4C). Because TMEM163 modulates P2X3R both positively and negatively by distinct mechanisms, we postulated that the balance of positive and negative modulation is different among P2XR subtypes. Thus, we generated HA-tagged P2X7R and examined the effects of TMEM163 on surface expression of HA-P2X7R in cRNA-injected oocytes. As a result, we found that TMEM163 reduced surface expression of HA-P2X7R as well as ATP-evoked current (Figures S4D and S4E). Again, TMEM48 did not show any effects (Figure S4E). These results indicate that TMEM163 modulates surface expression and properties of P2XRs in a subtype specific manner.
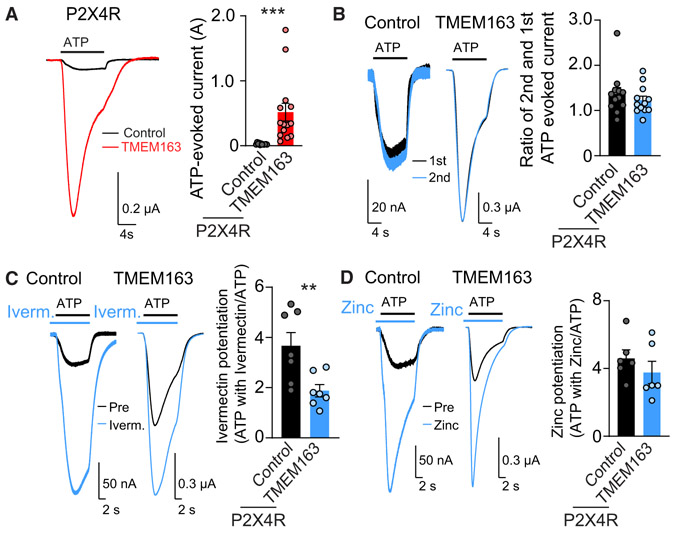
Figure 6. TMEM163 Modulates P2XR Pharmacology.
ATP-evoked responses were measured using TEVC recording (Vh = −30 mV) with various P2XR pharmacological reagents from oocytes co-injected with cRNAs of 25 pg of P2X4R and 2 ng of TMEM163 or Neto2 as a control.
(A) TMEM163 enhanced the ATP-evoked currents of P2X4Rs. ATP (1.3 μM)-evoked responses were measured. Representative traces and quantification of the peak amplitudes (n = 12–14).
(B) ATP (1.3 μM ATP)-evoked responses were measured at a 2-min interval. Representative traces and the ratio of peak amplitudes of the second to first ATP-evoked currents (n = 7). P2X4Rs both with and without TMEM163 recovered fully within 2 min after stimulation.
(C and D) TMEM163 co-expression reduced ivermectin-mediated, but not zinc-mediated, potentiation of ATP-evoked P2X4R currents. ATP (1.3 μM)-evoked currents were measured, then 10 μM ivermectin (C) or 10 μM ZnCl2 (D) was applied for 10 s before stimulation with ATP (1.3 μM) along with each drug as shown by bars above traces. Representative traces and the ratios of ATP-evoked currents with and without ivermectin (C, n = 7) or ZnCl2 (D, n = 6).
Data are mean ± SEM. Mann-Whitney U-test (A, C); **p < 0.01; ***p < 0.001.
We next examined whether TMEM163 modulates the potency of the endogenous agonist ATP by comparing the EC50 dose of ATP on P2X3R with and without TMEM163. We first examined this using the calcium FLIPR assay in P2X3R-stably expressing HEK cells with transient TMEM163 expression. Co-expression of TMEM163 shifted the dose-response curve of P2X3R-stably expressing HEK cells to the left (Figure 5A). However, the calcium responses are from both P2X3R- and endogenous P2 receptors including both P2X and P2Y receptors. Indeed, HEK cells (Mock) responded to ATP at concentrations above 1 μM, and TMEM163 alone did not alter this response (Figure S5A).
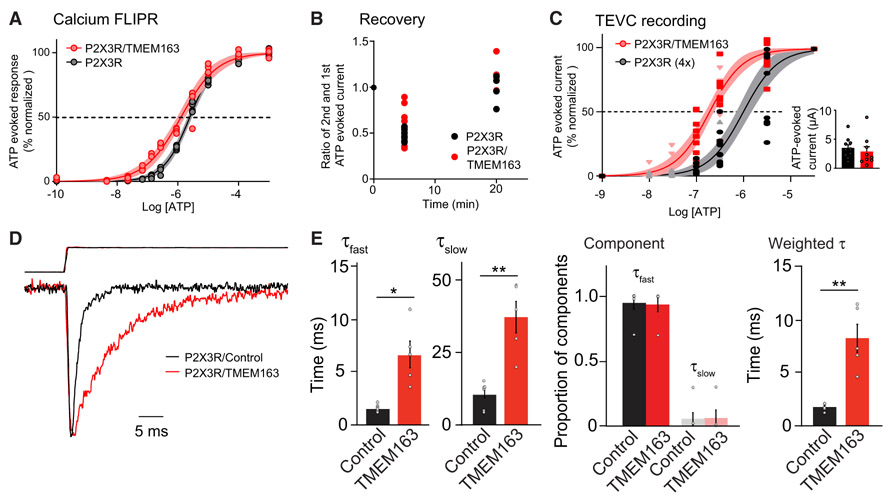
Figure 5. TMEM163 Shifts EC50 of ATP for P2XRs and Slows Decay Kinetics.
(A) Calcium FLIPR response at various concentrations of ATP was measured in HEK cells stably expressing P2X3R transfected transiently with TMEM163 or pcDNA3. Apyrase was pre-incubated for 1 h before stimulation to reduce extracellular ATP. P2X3R-stably expressing HEK cells responded to ATP, and co-expression of TMEM163 shifted the ATP EC50 (EC50: 1.27 ± 0.11 μM for P2X3R with TMEM163 and 2.50 ± 0.09 μM for P2X3R, p = 0.0013; Hill coefficient: 0.73 ± 0.04 for P2X3R with TMEM163 and 0.99 ± 0.03 for P2X3R p = 0.0023, n = 4). Data are mean ± SEM. Student’s t test.
(B and C) ATP-evoked currents from cRNA-injected oocytes were measured by TEVC recording (Vh = −30 mV). (B) TMEM163 did not affect recovery of ATP-evoked response of P2X3Rs at various intervals. ATP (10 μM)-evoked currents were measured at 5- or 20-min intervals, and the ratios of first and second ATP-evoked currents were calculated (n = 8). ATP-evoked responses were fully recovered with a 20-min interval. (C) TMEM163 shifted the ATP EC50 of P2X3R lower. To evaluate dose-response curves similarly, we adjusted the peak amplitudes of ATP-evoked currents of two conditions by injecting different amounts of P2X3R cRNAs: 25 pg of P2X3R with 2 ng of TMEM163 or 100 pg of P2X3R (4×) (small inset). We then measured ATP-evoked responses of various concentrations with a 20-min interval between stimulations for full recovery (n = 9). The estimated EC50 values were 0.26 ± 0.03 μM for P2X3R with TMEM163 and 1.30 ± 0.23 μM for P2X3R (4×). Hill coefficient: 0.98 ± 0.15 for P2X3R with TMEM163 and 1.04 ± 0.14 for P2X3R.
(D and E) TMEM163 slows the decay kinetics of P2X3R. We transiently transfected TMEM163-IRES2-EGFP (TMEM163) or IRES2-EGFP (control) in P2X3R-stably expressing HEK cells and applied 100 μM ATP to outside-out patch membranes of transfected cells using a piezoelectric device for ultrafast agonist application. (D) Superimposed traces of 100 μM ATP-evoked, peak-normalized responses (bottom) and open tip response (top) from outside-out parch membranes expressing P2X3R with either control (black, n = 6) or TMEM163 (red, n = 5). (E) The decays of the currents were fitted with bi-exponential curves, and fast and slow decay time constants (τfast and τslow) estimated (see STAR Methods for details). TMEM163 slows both fast and slow decay components without changing a proportion of each component (n = 6 for control, n = 5 for TMEM163). As a result, TMEM163 expression significantly slowed the weighted decay time constant (τ).
Data are mean ± SEM. Mann-Whitney U-test (E); *p < 0.05; **p < 0.01; ***p < 0.001.
To test the modulation of ionotropic P2XR channel properties by TMEM163 directly, we measured ATP-evoked currents from P2X3R cRNA-injected oocytes using TEVC recording. Some P2XR subtypes are known to desensitize rapidly, and its recovery takes a significant amount of time (Cook and McCleskey, 1997; Garcia-Guzman et al., 1997). We first examined the potential effects of TMEM163 on the recovery kinetics of P2X3R in cRNA-injected oocytes. We compared the peak amplitude of 10 μM ATP-evoked currents at a 5-or 20-min interval. We found no difference in amplitude recovery with or without TMEM163 co-expression (Figure 5B). We observed ~50% of the peak amplitude of the first ATP application (48.7% ± 2.3% for P2X3R alone and 56.3% ± 6.1% for P2X3R with TMEM163) at 5 min after stimulation, suggesting that some of the P2X3R was still desensitized. On the other hand, at 20 min after stimulation, similar responses to initial ATP application were observed, suggesting that the receptors had fully recovered from desensitization and that percent of desensitized P2XR was consistent among experiments.
We then determined the ATP dose–response curves of P2X3R alone and with TMEM163. When we injected the same amount (25 pg) of P2X3R, ATP-evoked currents at saturating ATP concentration (33 μM) from oocytes co-injected with TMEM163 were substantially larger than those from oocytes expressing P2X3R alone (Figure S5B). Because the different maximum amplitude of ATP-evoked responses may provide amplitude-dependent effects independent from TMEM163 co-expression, we adjusted the maximal ATP-evoked currents from each condition to similar levels by altering amounts of cRNA injected (100 pg P2X3R alone and 25 pg P2X3R and 2 ng TMEM163) (Figures 5C, inset, and S5B). We then measured ATP-evoked currents at various ATP concentrations with a 20-min interval to avoid effects of desensitization (Figure 5C). EC50 values estimated by fitting dose responses with a sigmoid curve showed that TMEM163 co-expression shifted the EC50 of P2X3R from 1.3 ± 0.23 μM to 261 ± 27.2 nM with a Hill equation value (1.04 ± 0.14 for P2X3R and 0.98 ± 0.15 for P2X3R with TMEM163). Thus, we concluded that TMEM163 modulates the channel properties of P2X3R.
P2X3R opens and desensitizes on the order of milliseconds, which is faster than the temporal resolution of our TEVC recording of cRNA-injected oocytes or the whole-cell recording of neurons that detected the enhancement of ATP-evoked P2X3R currents by TMEM163 co-expression. To examine TMEM163 modulation of P2X3R at a higher temporal resolution, we used outside-out patch membranes with ultra-fast solution exchange provided by a piezo electric device (Yan et al., 2013). We transiently transfected and expressed TMEM163-IRES2-EGFP or IRES2-EGFP (control) into P2X3R-stable HEK cells. We confirmed the interaction of TMEM163 with P2X3R in transfected HEK cells using co-immunoprecipitation (Figure S5C). We next prepared outside-out membranes from GFP-positive cells and applied 100 μM ATP with the piezo. The solution exchange rate was estimated by measuring the open tip potential at the end of each recording by zapping membranes. Then, experiments with an open tip potential rise time within 0.4 ms were analyzed. We found the mean rise time of open tip potential was 0.24 ± 0.005 ms for P2X3R and 0.25 ± 0.009 ms for P2X3R with TMEM163, and the mean rise time of ATP-evoked currents was 0.43 ± 0.042 ms for P2X3R and 0.50 ± 0.061 ms for P2X3R with TMEM163. Because the rise time of the open tip (solution exchange rate) is significantly faster than the rise time of ATP-evoked currents (p = 0.005 for P2X3R and p = 0.014 for TMEM163/P2X3R), we concluded that we had sufficient resolution to evaluate channel kinetics. Despite no significant change in rise time of ATP-evoked p2X3R currents by TMEM163 co-expression, we observed that TMEM163 co-expression robustly slowed decay kinetics of ATP-evoked P2X3R currents (Figure 5D). The decay kinetics were estimated by fitting the decay of each current with bi-exponential curves. TMEM163 co-expression slowed both fast and slow components (τfast and τslow) without changing the proportion of each component (Figure 5E). As a result, TMEM163 co-expression slowed the weighted τ of P2X3R by a fourth. These results showed that TMEM163 modulates the decay kinetics of P2X3Rs, suggesting that TMEM163 increases the open probability of P2X3R at a single channel level.
TMEM163 Modulates Channel Pharmacology of P2XRs
A discrepancy was identified between the pharmacology of native and recombinant P2XRs. As ivermectin potentiates P2X4R expressed in heterologous cells better than in neuronal P2XRs (Khakh et al., 1999; Priel and Silberberg, 2004; Silberberg et al., 2007), we decided to examine P2X4Rs. Because a molecule that modulates channel properties often modulates channel pharmacology, we postulated that TMEM163 may modulate the pharmacology of P2X4Rs. We first examined modulation of P2X4R by TMEM163 and found that TMEM163 co-expression significantly enhanced ATP-evoked currents from P2X4R cRNA-injected oocytes (Figure 6A). We next examined ivermectin potentiation of P2X4R with and without TMEM163 by pre-incubating each oocyte with 10 μM ivermectin for 10 s, followed by an application of ATP (1.3 μM) and ivermectin (Figure 6C). We observed that ivermectin potentiated ATP-evoked currents of P2X4R alone 3.7 ± 0.5 times, whereas it potentiated those of P2X4R with TMEM163 1.9 ± 0.2 times (Figure 6C). TMEM163 enhanced and ivermectin potentiated ATP-evoked currents of P2X4R. However, with ivermectin, TMEM163 did not enhance ATP-evoked currents of P2X4R as much as P2X4R alone, suggesting that TMEM163 enhancement and ivermectin potentiation share a similar mechanism.
Ivermectin potentiates P2XR activity through the transmembrane domains of P2X4R (Jelínkova et al., 2008; Silberberg et al., 2007). On the other hand, zinc potentiates P2X4R activity through its interaction with the extracellular domain of P2X4R (Kasuya et al., 2016). Because of the distinct action sites of zinc and ivermectin, we examined zinc potentiation of ATP-evoked currents of P2X4R and P2X4R co-expressed with TMEM163. But we did not identify any significant differences in zinc potentiation of peak amplitudes (Figure 6D). These results suggest that TMEM163 modulates the pharmacology of P2X4R by modifying P2XR transmembrane domains but not the extracellular domain.
TMEM163 Modulates ATP-Evoked Behavior and P2XR Activity In Vivo
ATP induces pain in humans (Bleehen and Keele, 1977; Coutts et al., 1981; Hamilton et al., 2000), and P2X3R knockout (KO) mice were reported to exhibit a reduction in ATP-evoked behaviors (Cockayne et al., 2000; Souslova et al., 2000). Therefore, we next tested the role of TMEM163 in regulating ATP-evoked behaviors. To this end, we generated TMEM163 KO mice through the CRISPR/Cas9 system. We designed a guide1 RNA to disrupt exon 2 of TMEM163 encoding the 1st transmembrane domain (Figure S6A) and established three independent mouse lines (1, 2, and 5). Deletion of either 70 or 200 bp, including exon 2, was confirmed by Sanger sequencing of genomic PCR products from lines 1 or 2 and 5, respectively (Figure S6B). We backcrossed all three TMEM163 KO mouse lines to C57/B6J at least three times independently and maintained them separately. All three lines of TMEM163 KO mice were viable and fertile. Loss of TMEM163 proteins was confirmed by western blotting of DRG lysate from TMEM163 KO mice with the anti-TMEM163 antibody (Figure S6C). The total protein amount of P2X3R was unaltered as determined with anti-P2X3R antibody (Figure S6C). This anti-P2X3R antibody recognized a band at the expected molecular weight only in DRG among the various nervous tissues tested (Figure S6D). Specific detection of a 60 kDa band in DRG is consistent with strong expression of P2X3R mRNA in DRG (Chen et al., 1995; Lewis et al., 1995).
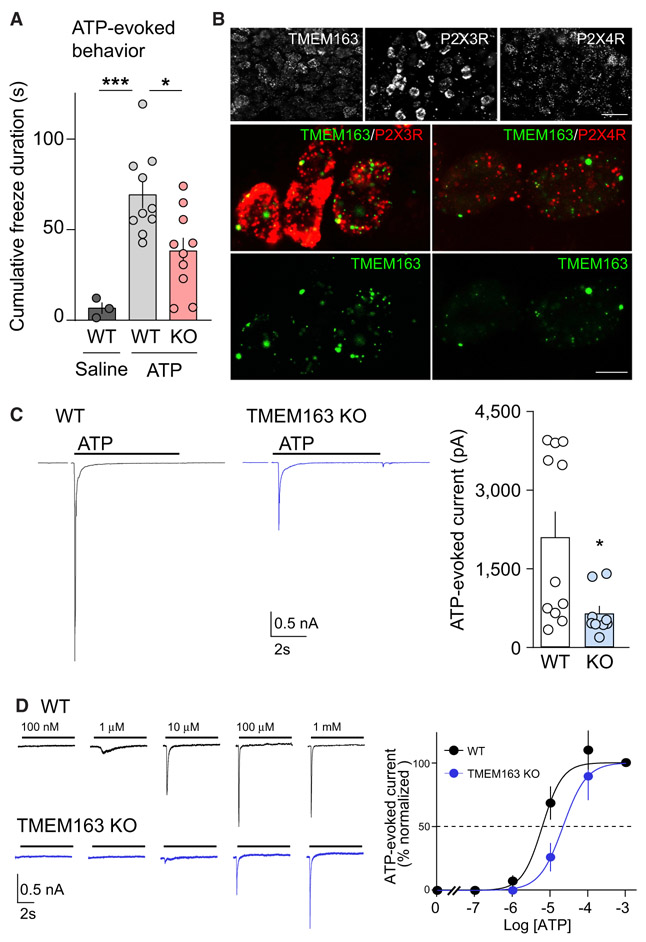
We examined ATP-evoked behavioral responses in a genotype-blinded manner. Mice were habituated to experimental conditions for 5 days before examining behaviors induced by an injection of ATP into the hind paw. Upon ATP injection into the hind paw of TMEM163 KO and wild-type (WT) littermate mice, we observed changes in a pain-related measure of paw lifting and mouse freezing behavior (Dogishi et al., 2015; Cockayne et al., 2000; Souslova et al., 2000).
Although P2X3R KO mice showed no ATP-evoked currents in DRG neurons and 50% reduction in ATP-induced paw lifting (Cockayne et al., 2000), ATP injection-induced paw lifting was unaltered between WT and TMEM163 KO mice (Figure S6E), suggesting residual P2XR activity in DRG neurons in TMEM163 KO mice. The time of occurrence and duration of each freezing behavior was recorded for at least 4 min after injection for each mouse (Figure S6F). The mean cumulative duration of freezing behaviors after ATP injection in WT mice was significantly greater than that after injection of the saline vehicle injection 74 ± 7.1 s versus 6.7 ± 3.2 s, respectively. In comparison with WT mice, TMEM163 KO mice exhibited a significant reduction in the mean cumulative duration of freezing upon ATP injection (Figures 7A and S6F). These results show that TMEM163 KO affects one, but not all, pain behaviors (i.e., due to residual current). The reduction in the freezing duration of the TMEM163 KO mice suggests that TMEM163 modulates this ATP-evoked behavior.
Figure 7. TMEM163 Is a Modulator of ATP-Evoked Behavior and P2XR Activity In Vivo.
(A) ATP-injected responses were evaluated for at least 4 min after injection of 3 μmol ATP into mouse hind paw. The mean cumulative durations of freezing behavior of wild-type (WT) and knockout (KO) mice were each significantly greater in response to ATP than to saline. On the other hand, the TMEM163 KO mice froze significantly less than WT mice in response to ATP, indicating that TMEM163 modulates this pain-related behavior (n = 10 for ATP-injected mice and 3 for saline-injected mice).
(B) The mRNA expression of TMEM163 and P2XRs in mouse dorsal root ganglion (DRG) detected by fluorescence in situ hybridization (FISH). Low-magnification images (top) showed a heterogeneous expression of TMEM163, P2X3R, and P2X4R across DRG neurons. Middle: double-labeling FISH for TMEM163 (green) and P2X3R or P2X4R (red) showed TMEM163 mRNA was expressed in both P2X3R- and P2X4R-positive cells.
Scale bars, 50 μm (top panels); 10 μm (middle and bottom panels).
(C and D) ATP-evoked whole-cell responses were measured in primary DRG neurons from adult WT and TMEM163 KO mice under whole-cell configuration (Vh = −70 mV) with an inter-stimulus interval longer than 3 min for full recovery. (C) Representative traces and quantitation of peak amplitudes of 100 μM ATP-evoked currents (n = 11). Artifacts from 2 mV step for 150 ms for access resistance measurement were omitted from traces shown. (D) Representative traces and dose-response curves of peak amplitudes normalized at 1 mM (n = 5 each). The estimated EC50 values were 5.92 ± 1.17 μM for WT and 31.0 ± 15.1 μM for TMEM163 KO, and these EC50 values were significantly different (p = 0.006). The Hill co-efficient values were unaltered (1.52 ± 0.15 for WT and 1.93 ± 0.37 for TMEM163 KO).
Data are mean ± SEM; one-way ANOVA with Bonferroni’s post-test (A), unpaired t test (C and D); *p < 0.05, ***p < 0.001.
ATP-evoked signaling in the hind paw is mediated by DRG neurons. Thus, we postulated that the TMEM163 KO mice may exhibit a reduction in P2XR activity in DRG neurons. We examined whether TMEM163 and P2XRs are expressed in the same DRG neurons by using double fluorescence in situ hybridization (FISH) with probe sequences available from Allen Brain Institute. We found that almost all DRG neurons expressed TMEM163 and P2X4R and that DRG neurons: small- and medium-sized diameters expressed P2X3R strongly (Figure 7B). High-magnification images showed the co-expression of TMEM163 with P2X3R or P2X4R (Figure 7B).
If TMEM163 modulates P2XR activity in vivo, the FISH results presented in Figure 7B imply that ATP-evoked currents will vary in DRG neurons because of varied expression levels of P2XR subtypes. However, DRG neurons from TMEM163 KO mice might be expected to show a reduction in the overall averaged ATP-evoked activity because of the corresponding expression of TMEM163 in DRG neurons. To directly test this, we prepared primary DRG neurons from TMEM163 KO mice and WT littermates. ATP-evoked currents under whole-cell configuration (Vh = −70 mV) were recorded from DRG neurons with diameters around 30 μm to ensure consistencies of voltage-clamp. In DRG neurons from TMEM163 KO mice, we found a reduction in peak amplitude of ATP-evoked currents (Figure 7C). Because TMEM163 co-expression shifted the ATP dose-response of P2X3R to the left in cRNA-injected oocytes (Figure 5C), we examined whether the loss of TMEM163 shifts the ATP dose-response of P2XRs in DRG neurons. We measured ATP-evoked whole-cell currents (Vh = −70 mV) of P2XRs in DRG neurons from WT and TMEM163 KO mice at various ATP concentrations. The ATP dose-response curves were estimated as the relative ratio of peak amplitudes measured at each concentration to the peak amplitude at saturated ATP concentration (1 mM). We found that the ATP dose-response curve was shifted to the right in TMEM163 KO DRG neurons (Figure 7D). The estimated EC50 values were 5.92 ± 1.17 μM for WT and 31.0 ± 15.1 μM for TMEM163 KO and were significantly different (p = 0.006). These results indicate that TMEM163 regulates neuronal P2XR properties, consistent with the findings from recombinant systems co-expressed with TMEM163.
DISCUSSION
This study identifies TMEM163 as a modulator of P2XR channel properties in vivo and of ATP-evoked behavior. We performed a functional screen for P2XR modulators with a genome-wide ORF collection before measuring the effect of potential modulators on P2XR channel activity. Our results show that TMEM163 plays essential roles in regulating channel activity and pharmacology of P2X3R and P2X4R, as well as in mouse behavior evoked by ATP, an agonist implicated in pain in humans.
P2XRs Require TMEM163 for Their Function In Vivo
Our results show that TMEM163 is required in mice for a full manifestation of the fundamental channel properties of P2XRs and an ATP-evoked pain-related behavior. Because P2XRs have seven subtypes with distinct properties (Khakh and North, 2012; Li et al., 2013), it is difficult to draw conclusions as to whether differences in properties of native and recombinant P2XRs is due to differences in P2XR subtypes expressed or the lack of TMEM163 expression in heterologous cells. We showed that TMEM163 modulates the EC50 of ATP-evoked P2XR currents in DRG neurons. In the future, it should be investigated whether TMEM163 modulates other properties of P2XRs in vivo.
TMEM163 has been identified as a component of the synaptic vesicle, called SV31, and is known to interact with TRPML1 (Burré et al., 2007; Cuajungco et al., 2014). Based on the homology of TMEM163 with zinc transporters, TMEM163 was tested for zinc activity and shown to regulate zinc in heterologous systems. However, this has yet to be shown in vivo (Waberer et al., 2017). Because P2XR activity can be modulated by zinc, TMEM163 may modulate P2XR activity through zinc regulation. However, we think that this is unlikely because of two lines of evidence: (1) TMEM163 modulates P2XRs in heterologous cells, whereas structurally homologous zinc transporter (ZnT3) does not, and (2) if TMEM163 acts on P2XR activity through its potential ability to regulate zinc, additional zinc should not show comparable modulation of P2XRs co-expressed with TMEM163. However, we detected similar levels of zinc-dependent potentiation of ATP-evoked P2X4R currents on P2X4R alone and P2X4R with TMEM163 (Figure 6), suggesting that zinc and TMEM163 act on P2X4Rs independently.
TMEM163 Modulates Channel Properties of P2XRs
TMEM163 slows the decay kinetics of P2X3Rs as evaluated on a millisecond timescale. P2XR channel activity is determined by the open probability and conductance of single channels. Our discovery of the slower decay kinetics induced by TMEM163 indicates that TMEM163 increases the open probability of P2X3R. A similar phenomenon was previously observed in other neuro-transmitter receptors, including AMPA- and kainate-type glutamate receptors. Their auxiliary subunits, TARPs and Neto1 and Neto2, slow the decay kinetics of glutamate-evoked currents of AMPA and kainate receptors, respectively (Tomita et al., 2005; Zhang et al., 2009). In this case, both TARP and Neto2 increase both the open probability and single channel conductance. At this stage, it remains unclear whether TMEM163 changes the single channel conductance of P2X3R. Further analysis of single channel behavior of P2XRs with and without TMEM163 will reveal biophysical mechanism of TMEM163 modulation of P2XR.
TMEM163 modulates the potency with which ATP affects P2XRs. Agonist potency is determined by ligand-binding and pore-gating, suggesting that TMEM163 alters structures surrounding the ligand-binding domain or gating machinery. We found that TMEM163 reduced ivermectin potentiation of P2X4Rs, whereas TMEM163 and zinc potentiated P2X4Rs independently. Interestingly, ivermectin and zinc act on distinct sites on P2XRs: the transmembrane and extracellular domains, respectively. Because ivermectin potentiation was occluded by TMEM163 modulation of P2X4R activity, ivermectin and TMEM163 may affect P2XR activity through a similar mechanism. Ivermectin was shown to increase the open probability of P2X4R (Priel and Silberberg, 2004). Because our data suggest that TMEM163 increases open probability of P2X3R, TMEM163 may increase the open probability of P2X4R as well. These findings support the idea that TMEM163 modulates the transmembrane domains of P2XRs similar to ivermectin and alters their gating machinery.
In addition, P2XRs as well as TRPV1 are modulated through interactions of their cytoplasmic domains with phosphoinositides (Fujiwara and Kubo, 2006; Prescott and Julius, 2003; Zhao et al., 2007). A phosphoinositide binding protein, Pirt, can also modulate the activity of P2X3R and TRPV1 as well as TRPM8 (Gao et al., 2015; Kim et al., 2008; Tang et al., 2013). Because TMEM163 is similar to zinc transporters, it is unlikely to regulate the cellular levels of phosphoinositide levels. The molecular structures of several P2XR subtypes in different states have been reported (Hattori and Gouaux, 2012; Kasuya et al., 2017; Kawate et al., 2009; Mansoor et al., 2016). A comparison of the structures of P2XR alone and P2XR co-expressed with TMEM163 should reveal its mechanism of action at an atomic resolution.
Genome-wide ORF Collection
Although we have been designing and expanding our ORF collection since 2009 from OriGene’s 4,000 transmembrane ORF collection to a genome-wide ORF collection supported by NIH, a private sector group has performed a similar screening and successfully identified NACHO (encoded by the TMEM35 gene) and several other molecules as modulators of ionotropic nicotinic acetylcholine receptors (Gu et al., 2016, 2019; Rex et al., 2017). The successful identification of specific modulators for acetylcholine and P2XRs strongly suggests the utility of this ORF-based functional screening for identifying functional modulators.
There are several factors critical to the success of the screening, including the size and quality of genome-wide ORF collections in expression vectors. In principle, it is best to test all ORFs present in vivo and confirm their expression in heterologous cells. At the time of writing, HUGO listed 19,224 protein-coding genes (https://www.genenames.org/). In this study, using a combination of three ORF collection resources (Figure 1A), we could assemble 17,284 non-redundant ORF-encoding genes, which cover 90% of human protein-coding genes. However, this collection does not include splicing isoforms or proteins with RNA-edited forms. Thus, the collection of all potential ORFs should be increased. Genome-wide ORF collections have been generated by both academia and industry, for example, by the Broad Institute and OriGene (Yang et al., 2011). However, there is a need for standardization, because no consistent approach has been taken among organizations. It would be beneficial if a society were to organize an initiative to establish one ORF collection that covers a minimum set of human non-redundant ORF-encoding genes in expression vectors and another ORF collection that covers all ORF variants, including splicing isoforms with verified protein expression. Ideally, the NIH or a non-profit organization could support this activity further to establish such ORF collections for the application of ORF-based screening in the near future, as extensive efforts have already been made for KO-based screening.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Susumu Tomita (Susumu.Tomita@yale.edu).
Materials Availability
There are restrictions to the availability of the ORF collection used in this study due to the MTA with the commercial sources. All other reagents are available upon reasonable requests.
Data and Code Availability
The published article includes all dataset and code generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal handling was in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Yale University. Animal care and housing were provided by the Yale Animal Resource Center (YARC), in compliance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 1996). Animals were maintained in a 12 hr light/dark cycle with ad libitum access to food and water. Wild-type (C57BL/6J, Stock# 000664) were obtained from the Jackson Laboratory. Female Xenopus laevis frogs were obtained from Nasco and used for oocyte preparation. Both sexes of mice were used for this study (P7 for primary cerebellar granule neurons, around P30 for primary DRG neurons, and 2-3 months for behavior and biochemistry).
Generation of TMEM163 knockout mice
TMEM163 knockout mice were generated using the CRISPR/Cas approach at the Yale Genome Editing Center (https://medicine.yale.edu/compmed/ags/). Four sgRNAs were designed for the 5′ and 3′ ends of the exon 2 of TMEM163 based on a CRISPR Design Tool (http://zlab.bio/guide-design-resources). Combinations of Two sgRNAs together with Cas9 were injected into C57BL/6J zygotes. Chimeric mice were identified by genomic PCR and backcrossed to C57BL/6J (Stock# 000664) at least three generations. Each mouse lines were maintained separately. TMEM163 homozygous knockout mice were obtained by mating heterozygous mice.
Target sequences were as follows.
Guide #3, GGGCTTCATGAGGTTTCAGCCGG.
Guide #12, GTTTACTAGAAAGCAGCACCCGG.
Guide #9, ACTGAAGGCGGCCACTGCCAGGG.
Guide #11, CAGGGCCAGGGTGACAATAATGG.
Cells
HEK293 (ATCC, CRL-1573) and HEK cells expressing exogenous genes stably were cultured in advanced DMEM supplemented with 4% FBS, GlutaMAX, and penicillin/streptomycin. All cells were kept in a 37°C, 5% CO2 incubator. Stable HEK cell lines were created by transfection and subsequent selection for 2 weeks with appropriate antibiotics, G418 and Hygromycin B. Single colonies were isolated and tested for receptor activity using the calcium FLIPR. Lines with non-saturated activity were selected for further analysis.
Antibodies
The following antibodies were used at the indicated dilutions: rabbit polyclonal antibodies to TMEM163 (1:1000-3000 for WB), P2X3 (Alomone; 1:1000-4000 for WB): mouse monoclonal antibodies to alpha tubulin (Sigma; 1:100,000 for WB), NR1/GluN1 (BD Biosciences; 1:1000 for WB), FLAG (Sigma; 1:1000 for WB): rat monoclonal antibodies to HA (Roche; 1:400 for surface expression). Polyclonal antisera to TMEM163 proteins were raised by injecting rabbits with a GST-TMEM163 fusion protein encoding the first 88 amino acids of TMEM163. Antisera were affinity purified on Affi-gel columns (Bio-Rad) containing the His-tagged TMEM163 fusion proteins.
METHOD DETAILS
Construction of Genome-wide non-redundant ORF collection
Three ORF libraries (ORFeome Collaboration Complete Human Collection, hORFeome v8.11 OC Supplemental Library, CCSB-Broad Human Lentiviral Expression Library) were obtained from ThermoFisher Scientific. The CCSB-Broad Lentiviral Expression Library contains 15,743 ORFs in pLX304, and the other two ORF libraries contain total 23,644 ORFs in pENTR221/223. Sequences of the ORF libraries were obtained from the NCBI database, and one ORF of each unique gene was selected for constructing the non-redundant ORF collection: 13,193 ORFs in pLX304 and 3,647 ORFs in pENTR221/223. Among them, 3,274 ORFs in pENTR were cloned successfully into pcDNA3.1 with a c-terminal V5 tag (pcDNA3.1-cV5) using Gateway cloning. In addition, 817 transmembrane ORFs from the 4,000 OriGene transmembrane collection were added. As a result, we constructed a Genome-wide non-redundant ORF collection with 17,284 ORFs, of which most contain a c-terminal V5 tag in pLX304 or in pcDNA3.1-cV5.
Transfection
Transient transfection of each ORF into cells was prepared using Fugene 6. Briefly, a mixture of Fugene 6 and a plasmid was incubated in a 384-well plate, followed by plating cells at 4-6x104 cells per well. Cells were used for experiments two days after transfection.
Calcium FLIPR Assay
We followed the manufacturer’s protocol. Briefly, cells were washed with 1x HBSS with 5 mM CaCl2, 0.49 mM MgCl2, 0.41 mM MgSO2, 20 mM HEPES pH7.4 once and incubated with the FLIPR Calcium 4 reagent (Molecular Devices) with 1.25 mM probenecid for one hour. The plates were loaded into the FLIPR Tetra system (Molecular Devices) and fluorescence signal was measured (Total 110 s recording with an agonist application at 20 s). For ATP dose response experiments, MgATP was used.
Xenopus laevis oocyte preparation
Oocytes were harvested from Xenopus laevis frogs as done previously (Tomita et al., 2004). Briefly, oocytes were extracted surgically from anesthetized frogs, treated with collagenase, and selected manually for cRNA injection. The cRNAs were transcribed in vitro using the T7 mMessage mMachine kit.
Two-electrode voltage clamp (TEVC) recording
Two-electrode voltage clamp (TEVC) recording was performed at room temperature in a recording solution (90 mM NaCl, 1 mM KCl, 2 mM CaCl2, and 10 mM HEPES pH7.4). The membrane potential was held at −30 mV (P2XR and GluK2/Neto2) or −70 mV (GluA1) using GeneClamp 500B (Axon Instruments). For P2X3R and GluK2/Neto2 recordings, agonist was applied using an air pressure perfusion system (AutoMate Scientific). Data were acquired and analyzed by LabChart (ADInstruments). MgATP was used for all ATP additions.
Protein biotinylation at the oocyte surface
Xenopus laevis oocytes were incubated in 1.5 mg/mL NHS SS Biotin in ND96 for 30 minutes on ice. Oocyte membranes were then solubilized with 0.5% SDS and 1% Triton X-100 in homogenization buffer (50 mM Tris-Cl pH7.6, 2 mM EDTA, 2 mM EGTA 100 mM NaCl). Biotinylated proteins were isolated with Neutravidin plus beads (Pierce).
Oocyte chemiluminescent surface expression
Measurement of protein surface expression in oocytes was performed as previously described (Tomita et al., 2004). Oocytes expressing extracellularly HA-tagged P2X3R were blocked in 1% BSA ND96, followed by labeling with anti-HA rat antibody and HRP conjugated anti-rat antibody under a non-permeabilized condition at 4°C. HRP activity was measured with SuperSignal ELISA Femto Maximum Sensitivity Substrate using a 96-well plate reader (Glomax Multi+ Detection System, Promega).
Primary cell culture preparation
Primary cerebellar granule cell cultures were prepared as described previously (Yan et al., 2013). Briefly, cerebella were dissected from P7 mice, digested with trypsin, and plated 1.5x106 cells in 10% FBS BME media supplemented with B27 and glutamine on a PDL-coated 24-well. Cells were maintained at 37°C and 5% CO2 and used at DIV11-14. Lentivirus was added at DIV1. At DIV3, the media was changed to MEM supplemented with glutamine, cytosine arabinoside, and insulin-transferrin-sodium selenite.
Primary dorsal root ganglion (DRG) neurons were prepared as described previously with minor alterations (Matos-Cruz et al., 2017). Briefly, DRGs were harvested from vertebrae in the thoracic and lumbar regions in HBSS with 20 mM HEPES. DRGs were digested in 1 mg/mL collagenase P for 45 minutes and 0.25% trypsin EDTA for 15 minutes. Cells were plated in F12 culture media supplemented with 50 mg/mL NGF onto PDL and Matrigel-coated coverslips in a 24-well plate.
Preparation of lentivirus carrying shRNAs
TMEM163 shRNA (CCCAGAACTACAGGAAGAA) was inserted into FUGW-H1 vector (a gift from Dr. Sally Temple through Addgene, #25870) (Fasano et al., 2007). The “TMEM163 mutant” resistant to the shRNA carries six mutations (GCACAAAATTATAGAAAA), which did not alter the amino acids encoded. The TMEM163 mutant was fused with EGFP at its C terminus and replaced EGFP in FUGW-H1.
Lentivirus was prepared as described previously (Zhang et al., 2009). Briefly, three plasmids of pDelta8.9, pVSVG (Stewart et al., 2003) using and pFUGW-H1 encoding TMEM163 shRNAs were co-transfected into 293FT cells using the calcium phosphate method. 36-40 hr later supernatant was collected, filtered, and concentrated by ultracentrifugation (120,000 g for 1.5 hours at 4°C). The viral pellet was resuspended in PBS and stored at −80°C. 293FT cells were maintained in D-MEM supplemented with 10% FBS, 0.1 mM MEM non-essential amino acids, 2 mM glutamine, 1mM sodium pyruvate, 1% penicillin/streptomycin, and 500 μg/mL geneticin.
Preparation of mouse homogenate
Tissues were homogenized in 20 mM Tris/1 mM EDTA pH8.0 with the HALT protease inhibitor. SDS was added to final 0.5% and samples were incubated for 5 minutes on ice. After sonication, samples were spun at 14,000 g for 10 minutes, and supernatant was collected. Protein concentrations were measured using a BCA protein assay (Pierce).
Mouse behavioral analysis
Mice (2-3-month-old) were habituated to handling and to the test apparatus daily for 5 days before administering the first injection on day 6. Mice were placed into clear plastic containers (9 × 9 × 13 cm) with four angled mirrors surrounding them in a soundproof room with stable temperature. Behavior was recorded from above with a camcorder (Sony Model DCR-DVD 300) with the mirrors providing side views of the mice (Shimada and LaMotte, 2008). 30 μL of ATP (total 3 μmols in saline) or saline alone was injected into the mouse hindpaw, and behavior was recorded for four minutes after the injection (Cockayne et al., 2005). The time of occurrence and duration of each “freezing” behavior (Dogishi et al., 2015), defined as a cessation of movement for 1 s or longer, was scored manually from video recordings. The cumulative duration of freezing behaviors for each mouse was found to be the most reliable indicator of behavioral responses to the injection. These behaviors were not triggered by external events such as sounds within or outside the testing room as supported by our analyses of the video recordings. Experimenters were blinded to each genotype. Mice were randomized during treatment. For analyses of knockout mice, littermates of the same sex were randomly assigned to experimental group, and no mice were excluded from the analysis.
Whole-cell recording
Cerebellar granule cell recording. Whole-cell recordings were performed between DIV10–15 with the external solution containing (in mM): 140 NaCl, 2.5 KCl, 2.5 CaCl2, 2.7 MgCl2, 1.3 MgSO4, 10 D-glucose, 0.001 TTX, and 10 HEPES (301 mOsm, pH7.4 adjusted with NaOH). Patch pipette was filled with the internal solutions containing (in mM): 125 CsMeSO4, 15 CsCl, 5 TEA-Cl, 0.4 EGTA, 20 HEPES, 2.5 Mg-ATP, 0.25 Na-GTP, 1 QX-314, (280 mOsm, pH7.2 adjusted with CsOH). The MgATP solution was made in Ca2+ and Mg2+ free extracellular solution (301 mOsm, adjusted with sucrose). We confirmed elimination of P2XR-mediated current by co-application of MgATP and 10 μM TNP-ATP (Tocris), a high affinity P2XR antagonist. To confirm access resistance of each recording, 2mV step for 150 ms was given before agonist applications.
Recordings from dissociated dorsal root ganglion neuron in acute culture were performed on DIV 2 and DIV 3. Neurons with diameters around 30 μm were recorded in the external solution containing (in mM): 140 NaCl, 4.7 KCl, 2.5 CaCl2, 1.3 MgCl2, 5.6 D-glucose, 0.001 TTX and 10 HEPES pH7.4. For agonist stimulation, MgATP was prepared in the external solution. The patch pipette was filled with internal solution containing (in mM): 125 CsMeSO4, 20 TEA-Cl, 5 EGTA, 5 HEPES, 2.5 Mg-ATP, 0.2 Na-GTP (280 mOsm, pH7.2 adjusted with CsOH).
All experiments were performed at room temperature, and cells were voltage clamped under −70 mV (Multiclamp 700B amplifier, Axon Instruments). A 3–6 MΩ patch pipette was adopted accordingly. Under the whole-cell configuration, series resistance was continuously monitored to keep under 40 MΩ. Solution exchange was done by a Valvelink 8.2 perfusion system (AutoMate Scientific Inc.) and the 360 μm glass pipette was placed 40 μm above cell body. Signals were sampled at 50 kHz, and low-pass filtered at 2 kHz. Offline analysis was performed using pCLAMP10 (Molecular Devices).
Fluorescent in situ hybridization (FISH)
Generation of riboprobes and hybridization procedures were performed as previously described (Yamasaki et al., 2010). In brief, riboprobes were designed according to the Allen Brain Atlas. Mouse cDNA fragments of TMEM163 (1085–1896 bp; GenBank accession number, NM_028135.2), P2X3R (1002–1953 bp, NM_145526.1), and P2X4R (921–1828 bp, NM_001310720.1), were subcloned into the pCR4-TOPO plasmid vector (Invitrogen), and digoxigenin (DIG)- or fluorescein-labeled cRNA probes were transcribed in vitro using DIG or fluorescein RNA Labeling Mix (Roche Diagnostics). For tissue preparation to perform double-labeling FISH, mice were killed by cervical dislocation and lumbar DRGs were dissected, rapidly frozen with powdered dry ice, and embedded in OCT compound. Cryosections (20 μm in thickness) were collected on Superfrost Excell slides (Fisher Scientific) and allowed to dry for 10 min at room temperature. After fixation with 4% paraformaldehyde and acetylation, sections were hybridized with a mixture of DIG- or fluorescein-labeled cRNA probes. After a stringent post-hybridization wash, DIG and fluorescein were detected using a two-step method as follows: the first detection was performed using a peroxidase-conjugated anti-fluorescein antibody (Roche Diagnostics) and FITC-TSA plus amplification kit (PerkinElmer). After inactivation of residual peroxidase activity by dipping sections in 1% H2O2 for 30 min, the second detection was performed by peroxidase-conjugated anti-DIG antibody (Roche Diagnostics), and Cy3-TSA plus amplification kit (PerkinElmer). Images of FISH were captured using a confocal laserscanning microscope LSM800 (Carl Zeiss).
Outside-out patch combined with a piezo electrical device
The P2X3 stable cell line were transiently transfected with pIRES2-EGFP (Clontech) or TMEM163/pIRES2-EGFP using Fugene 6 (Promega). Recordings from outside-out patch membranes were performed 1 day after transfection at room temperature. The external solution contained (in mM): 140 NaCl, 4.7 KCl, 10 D-glucose and 10 HEPES (305 mOsm, pH 7.3 adjusted with NaOH). The Patch pipette was filled with an internal solution containing (in mM): 130 Cs-Gluconate, 10 TEA-Cl, 2 MgCl2, 10 EGTA and 10 HEPES (280 mOsm, pH 7.2 adjusted with CsOH). The external solution and 100 uM MgATP in the external solution were constantly running through each barrel of the two-barrel theta glass capillaries (Warner Instruments, TG200-4). The tip diameter of the theta glass pipette was ~100 um pulled from a flaming/brown micropipette puller (Sutter Instrument, P-1000) directly. The position of the theta glass tube was controlled by a piezo driven electrical device (Siskiyou) to apply ATP to outside-out patch membranes on the patch pipette. At the end of measurements of ATP-evoked responses of each membrane, the membrane on the patch pipette was zapped and the open tip potential was measured with diluted extracellular solution to monitor the solution exchange without moving the patch pipette and the theta tube. The open tip potentials with a 10%–90% rise time of less than 400 μs were used for analysis.
Co-immunoprecipitation
HEK cells were transfected with P2X3R and TMEM163 or GFP in mammalian expression vectors. Two days post-transfection transfected cells were suspended in lysis buffer (25 mM Tris-Cl, pH 8.0, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 1% CHAPS) and centrifuged. The supernatants were incubated with 1 μg of the anti TMEM163 antibody and 10 μl of protein-A Sepharose beads (GE Health). The beads were then washed five times with lysis buffer and the proteins retained on the beads were analyzed by western blotting.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical details of experiments can be found in the figure legends or Method Details section. Statistical analyses were performed using GraphPad Prism 8.0 software (GraphPad Software). All data are given as mean ± s.e.m., and 1% outliers were excluded for whole-cell recordings. Data were analyzed for normality using Shapiro-Wilkes test. When normality was confirmed, statistical analysis was performed using One-way ANOVA followed by Bonferroni’s post-test, Paired t test, or Unpaired t test based on the number of conditions. When normality was not confirmed, we used Kruskal-Wallis test followed by Dunn’s post-test or Mann-Whitney U-test based on the number of conditions. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOCURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Tubulin | Sigma | Cat#: T9026; RRID AB_477593 |
| Mouse monoclonal anti-FLAG | Sigma | Cat#: F1804; RRID: AB_262044 |
| Rat monoclonal anti-HA | Roche | Cat#: 11867423001; RRID:AB_390918 |
| Rabbit polyclonal anti-P2X3 | Alomone | Cat#: APR-016; RRID:AB_2313760 |
| Mouse monoclonal anti-NR1 | BD Biosciences | Cat#: 556308; RRID: AB_396353 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Halt protease inhibitor cocktail | ThermoFisher Scientific | Cat#: 78446 |
| GlutaMax | GIBCO | 35050–061 |
| L-Glutamine | GIBCO | 25030–081 |
| Penicillin Streptomycin | GIBCO | 15140–122 |
| poly-D-lysine | Sigma | Cat#: P6407 |
| Protein A Sepharose CL4B | GE Healthcare | Cat#: 17-0780-01 |
| Sulfo-NHS-SS-Biotin | Thermo Scientific | Cat#: 21331 |
| NeutraAvidin Plus Ultralink Resin | Thermo Scientific | Cat#: 53151 |
| FuGENE® 6 Transfection Reagent | Promega | Cat#: E2691 |
| Trypsin-EDTA (0.05%), phenol red | GIBCO | Cat#: 25300054 |
| Advanced DMEM | GIBCO | Cat#: 12491023 |
| Adenosine 5′-triphosphate magnesium salt | Sigma | Cat#: A9187 |
| TNP-ATP triethylammonium salt | Tocris | Cat#: 2464 |
| α, β-Methyleneadenosine 5′-triphosphate lithium salt | Sigma | Cat#: M6517 |
| Ham’s F-12 Nutrient Mix | GIBCO | Cat#: 11765054 |
| NGF 2.5S Native Mouse Protein | GIBCO | Cat#: 13257019 |
| Collagenase P | Roche | Cat#: 11213857001 |
| Fetal Bovine Serum, qualified, USDA-approved regions | GIBCO | Cat#: 10437028 |
| Trypsin-EDTA (0.25%), phenol red | GIBCO | Cat#: 25200056 |
| L-Glutamic acid monosodium salt hydrate | Sigma | Cat#: G5889 |
| (−)-(α)-Kainic Acid (hydrate) | Cayman Chemical Company | Cat#: 78050 |
| Acetylcholine chloride | Sigma | Cat#: A5625 |
| Cyclothiazide | Sigma | Cat#: C9847 |
| HBSS, no calcium, no magnesium, no phenol red | GIBCO | Cat#: 14175-095 |
| MEM Non-Essential Amino Acids Solution (100X) | GIBCO | Cat#: 11140050 |
| Gentamycin | GIBCO | Cat#: 11811-031 |
| Sodium Pyruvate | Sigma | Cat#: P5280-25G |
| Hygromycin B (50 mg/mL) | GIBCO | Cat#: 10687010 |
| Gateway LR Clonase II Enzyme mix | Invitrogen | Cat#: 11791-020 |
| Minimum Essential Medium | GIBCO | Cat#: 11090-081 |
| Collagenase from Clostridium histolyticum | Sigma | Cat#: C9891-1G |
| Trypsin from bovine pancreas | Sigma | Cat#: T1426-1G |
| Deoxyribonuclease I from bovine pancreas | Sigma | Cat#: D4527 |
| Cytosine β-D-arabinofuranoside hydrochloride | Sigma | Cat#: C6645-25MG |
| Insulin-transferrin-sodium selenite media supplement | Sigma | Cat#: I1884-1VL |
| 2-Mercaptoethanol | Sigma | Cat#: M3148-25ML |
| Dithiothreitol | American Bioanalytical | Cat#: AB00490-00010 |
| Basal Eagle’s medium | GIBCO | Cat#: 21010-046 |
| Geneticin | GIBCO | Cat#: 15710-064 |
| Critical Commercial Assays | ||
| mMESSAGE mMACHINE T7 Transcription Kit | Ambion | Cat#: AM1344 |
| Pierce BCA Protein Assay Kit | Pierce | Cat#: 23225 |
| 5Prime Perfectprep® Plasmid 96 VAC Base Kit | 5Prime | Cat#: 2300320 |
| SuperSignal ELISA Femto Maximum Sensitivity Substrate | Thermo Scientific | Cat#: 37074 |
| Experimental Models: Cell Lines | ||
| HEK293 | ATCC | Cat#: CRL-1573 |
| 293FT | Life Technologies | Cat#: R70007 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Wild-type (C57BL/6J) | The Jackson Laboratory | Stock#: 000664 |
| Recombinant DNA | ||
| Plasmid: pGEM-HE | Liman et al., 1992 | N/A |
| Plasmid: pcDNA3.1(+) | Invitrogen | V790-20 |
| Plasmid: FUGW-H1 | Addgene | #25870 |
| Plasmid: pVSVg | Stewart et al., 2003 | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| LabChart | ADInstruments | https://www.adinstruments.com/products/labchart |
Highlights.
Genome-wide ORF-based functional screening identifies a P2X receptor modulator
TMEM163 modulates P2X receptor channel properties and pharmacology
Neuronal P2X receptor function requires TMEM163
TMEM163 regulates ATP-evoked behavior
ACKNOWLEDGMENTS
The authors thank Drs. Elena Gracheva, Marina Picciotto, Michael Crair, Slav Bagriantsev, Steven Shimada, Lydia Hoffstaetter, Erika Hoyos-Ramirez, the late Dr. Jim Howe, and members of the Tomita lab for insightful comments and technical advice. We thank the Yale Genome Editing Core for helping us to generate TMEM163 KO mice and Addgene for the plasmids listed in the STAR Methods. This work was supported by NIH/NINDS (RC1 NS068966), NIH/NIMH (U01 MH104984), Yale University (to S.T.), the Gruber Foundation, the Kavli Foundation, the NIH Cellular and Molecular Biology Training Grant (T32-GM007223), the NIH Neurobiology of Cortical Systems Training Grant (T32-GNS007224) (to E.J.S.), the Grant-in-Aid for Scientific Research on Innovative Areas (17H06313), and the Fund for the Promotion of Joint International Research (17KK0160) from MEXT, Japan (to M.Y. and T.M.).
Footnotes
DECLARATION OF INTERESTS
The research being reported in this publication was supported by materials purchased from OriGene Technologies. The terms of this purchase have been reviewed and approved by Yale University in accordance with its policy on objectivity in research.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107704.
REFERENCES
- Barth J, Zimmermann H, and Volknandt W (2011). SV31 is a Zn2+-binding synaptic vesicle protein. J. Neurochem 118, 558–570. [DOI] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G, and Julius D (2009). Cellular and molecular mechanisms of pain. Cell 139, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleehen T, and Keele CA (1977). Observations on the algogenic actions of adenosine compounds on the human blister base preparation. Pain 3, 367–377. [DOI] [PubMed] [Google Scholar]
- Burnstock G (1977). The purinergic nerve hypothesis. Ciba Found. Symp (48), 295–314. [DOI] [PubMed] [Google Scholar]
- Burnstock G (2008). Purinergic signalling and disorders of the central nervous system. Nat. Rev. Drug Discov 7, 575–590. [DOI] [PubMed] [Google Scholar]
- Burré J, Zimmermann H, and Volknandt W (2007). Identification and characterization of SV31, a novel synaptic vesicle membrane protein and potential transporter. J. Neurochem 103, 276–287. [DOI] [PubMed] [Google Scholar]
- Chen CC, Akopian AN, Sivilotti L, Colquhoun D, Burnstock G, and Wood JN (1995). A P2X purinoceptor expressed by a subset of sensory neurons. Nature 377, 428–431. [DOI] [PubMed] [Google Scholar]
- Chizh BA, and Illes P (2001). P2X receptors and nociception. Pharmacol. Rev 53, 553–568. [PubMed] [Google Scholar]
- Cockayne DA, Hamilton SG, Zhu QM, Dunn PM, Zhong Y, Novakovic S, Malmberg AB, Cain G, Berson A, Kassotakis L, et al. (2000). Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X3-deficient mice. Nature 407, 1011–1015. [DOI] [PubMed] [Google Scholar]
- Cockayne DA, Dunn PM, Zhong Y, Rong W, Hamilton SG, Knight GE, Ruan HZ, Ma B, Yip P, Nunn P, et al. (2005). P2X2 knockout mice and P2X2/P2X3 double knockout mice reveal a role for the P2X2 receptor subunit in mediating multiple sensory effects of ATP. J. Physiol 567, 621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coddou C, Yan Z, Obsil T, Huidobro-Toro JP, and Stojilkovic SS (2011). Activation and Regulation of Purinergic P2X Receptor Channel. Pharmacol. Rev 63, 641–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SP, and McCleskey EW (1997). Desensitization, recovery and Ca(2+)-dependent modulation of ATP-gated P2X receptors in nociceptors. Neuropharmacology 36, 1303–1308. [DOI] [PubMed] [Google Scholar]
- Coutts AA, Jorizzo JL, Eady RA, Greaves MW, and Burnstock G (1981). Adenosine triphosphate-evoked vascular changes in human skin: mechanism of action. Eur. J. Pharmacol 76, 391–401. [DOI] [PubMed] [Google Scholar]
- Cuajungco MP, Basilio LC, Silva J, Hart T, Tringali J, Chen CC, Biel M, and Grimm C (2014). Cellular zinc levels are modulated by TRPML1-TMEM163 interaction. Traffic 15, 1247–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully DF, Vassilatis DK, Liu KK, Paress PS, Van der Ploeg LH, Schaeffer JM, and Arena JP (1994). Cloning of an avermectin-sensitive glutamate-gated chloride channel from Caenorhabditis elegans. Nature 371, 707–711. [DOI] [PubMed] [Google Scholar]
- Dogishi K, Kodera M, Oyama S, Shirakawa H, Nakagawa T, and Kaneko S (2015). Long-lasting pain-related behaviors in mouse chronic cystitis model induced by a single intravesical injection of hydrogen peroxide. J. Pharmacol. Sci 129, 244–246. [DOI] [PubMed] [Google Scholar]
- Erlenhardt N, Yu H, Abiraman K, Yamasaki T, Wadiche JI, Tomita S, and Bredt DS (2016). Porcupine Controls Hippocampal AMPAR Levels, Composition, and Synaptic Transmission. Cell Rep. 14, 782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, and Temple S (2007). shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 1, 87–99. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, and Kubo Y (2006). Regulation of the desensitization and ion selectivity of ATP-gated P2X2 channels by phosphoinositides. J. Physiol 576, 135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XF, Feng JF, Wang W, Xiang ZH, Liu XJ, Zhu C, Tang ZX, Dong XZ, and He C (2015). Pirt reduces bladder overactivity by inhibiting purinergic receptor P2X3. Nat. Commun 6, 7650. [DOI] [PubMed] [Google Scholar]
- Garcia-Guzman M, Stühmer W, and Soto F (1997). Molecular characterization and pharmacological properties of the human P2X3 purinoceptor. Brain Res. Mol. Brain Res 47, 59–66. [DOI] [PubMed] [Google Scholar]
- Ge Y, Kang Y, Cassidy RM, Moon K, Lewis R, Wong ROL, Foster LJ, and Craig AM (2018). Clptm1 Limits Forward Trafficking of GABAA Receptors to Scale Inhibitory Synaptic Strength. Neuron 97, 596–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Matta JA, Lord B, Harrington AW, Sutton SW, Davini WB, and Bredt DS (2016). Brain α7 Nicotinic Acetylcholine Receptor Assembly Requires NACHO. Neuron 89, 948–955. [DOI] [PubMed] [Google Scholar]
- Gu S, Matta JA, Davini WB, Dawe GB, Lord B, and Bredt DS (2019). alpha6-Containing Nicotinic Acetylcholine Receptor Reconstitution Involves Mechanistically Distinct Accessory Components. Cell Rep. 26, 866–874. [DOI] [PubMed] [Google Scholar]
- Hamilton SG, Warburton J, Bhattacharjee A, Ward J, and McMahon SB (2000). ATP in human skin elicits a dose-related pain response which is potentiated under conditions of hyperalgesia. Brain 123, 1238–1246. [DOI] [PubMed] [Google Scholar]
- Hattori M, and Gouaux E (2012). Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature 485, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelínkova I, Vávra V, Jindrichova M, Obsil T, Zemkova HW, Zemkova H, and Stojilkovic SS (2008). Identification of P2X(4) receptor transmembrane residues contributing to channel gating and interaction with ivermectin. Pflugers Arch. 456, 939–950. [DOI] [PubMed] [Google Scholar]
- Kasuya G, Fujiwara Y, Takemoto M, Dohmae N, Nakada-Nakura Y, Ishitani R, Hattori M, and Nureki O (2016). Structural Insights into Divalent Cation Modulations of ATP-Gated P2X Receptor Channels. Cell Rep. 14, 932–944. [DOI] [PubMed] [Google Scholar]
- Kasuya G, Yamaura T, Ma XB, Nakamura R, Takemoto M, Nagumo H, Tanaka E, Dohmae N, Nakane T, Yu Y, et al. (2017). Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel. Nat. Commun 8, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, and Bredt DS (2010). Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68, 1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Burris KD, Gardinier KM, Gernert DL, Porter WJ, Reel J, Ding C, Tu Y, Schober DA, Lee MR, et al. (2016). Forebrain-selective AMPA-receptor antagonism guided by TARP γ-8 as an antiepileptic mechanism. Nat. Med 22, 1496–1501. [DOI] [PubMed] [Google Scholar]
- Kawate T, Michel JC, Birdsong WT, and Gouaux E (2009). Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature 460, 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, and North RA (2012). Neuromodulation by extracellular ATP and P2X receptors in the CNS. Neuron 76, 51–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, Proctor WR, Dunwiddie TV, Labarca C, and Lester HA (1999). Allosteric control of gating and kinetics at P2X(4) receptor channels. J. Neurosci 19, 7289–7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AY, Tang Z, Liu Q, Patel KN, Maag D, Geng Y, and Dong X (2008). Pirt, a phosphoinositide-binding protein, functions as a regulatory subunit of TRPV1. Cell 133, 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê KT, Paquet M, Nouel D, Babinski K, and Séguéla P (1997). Primary structure and expression of a naturally truncated human P2X ATP receptor subunit from brain and immune system. FEBS Lett. 418, 195–199. [DOI] [PubMed] [Google Scholar]
- Lewis C, Neidhart S, Holy C, North RA, Buell G, and Surprenant A (1995). Co-expression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature 377, 432–435. [DOI] [PubMed] [Google Scholar]
- Li M, Silberberg SD, and Swartz KJ (2013). Subtype-specific control of P2X receptor channel signaling by ATP and Mg2+. Proc. Natl. Acad. Sci. USA 110, E3455–E3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher MP, Wu N, Ravula S, Ameriks MK, Savall BM, Liu C, Lord B, Wyatt RM, Matta JA, Dugovic C, et al. (2016). Discovery and Characterization of AMPA Receptor Modulators Selective for TARP-γ8. J. Pharmacol. Exp. Ther 357, 394–414. [DOI] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, and Hess P (1992). Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9, 861–871. [DOI] [PubMed] [Google Scholar]
- Maher MP, Matta JA, Gu S, Seierstad M, and Bredt DS (2017). Getting a Handle on Neuropharmacology by Targeting Receptor-Associated Proteins. Neuron 96, 989–1001. [DOI] [PubMed] [Google Scholar]
- Mansoor SE, Lü W, Oosterheert W, Shekhar M, Tajkhorshid E, and Gouaux E (2016). X-ray structures define human P2X(3) receptor gating cycle and antagonist action. Nature 538, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos-Cruz V, Schneider ER, Mastrotto M, Merriman DK, Bagriantsev SN, and Gracheva EO (2017). Molecular Prerequisites for Diminished Cold Sensitivity in Ground Squirrels and Hamsters. Cell Rep. 21, 3329–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (1996). Guide for the Care and Use of Laboratory Animals (Washington, DC: The National Academies Press; ). [Google Scholar]
- North RA (2002). Molecular physiology of P2X receptors. Physiol. Rev 82, 1013–1067. [DOI] [PubMed] [Google Scholar]
- North RA, and Jarvis MF (2013). P2X receptors as drug targets. Mol. Pharmacol 83, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott ED, and Julius D (2003). A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science 300, 1284–1288. [DOI] [PubMed] [Google Scholar]
- Priel A, and Silberberg SD (2004). Mechanism of ivermectin facilitation of human P2X4 receptor channels. J. Gen. Physiol 123, 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex EB, Shukla N, Gu S, Bredt D, and DiSepio D (2017). A Genome-Wide Arrayed cDNA Screen to Identify Functional Modulators of alpha7 Nicotinic Acetylcholine Receptors. SLAS Discov. 22, 155–165. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, and Klöcker N (2009). Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319. [DOI] [PubMed] [Google Scholar]
- Shimada SG, and LaMotte RH (2008). Behavioral differentiation between itch and pain in mouse. Pain 139, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg SD, Li M, and Swartz KJ (2007). Ivermectin Interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron 54, 263–274. [DOI] [PubMed] [Google Scholar]
- Soto F, Garcia-Guzman M, Karschin C, and Stühmer W (1996). Cloning and tissue distribution of a novel P2X receptor from rat brain. Biochem. Biophys. Res. Commun 223, 456–460. [DOI] [PubMed] [Google Scholar]
- Souslova V, Cesare P, Ding Y, Akopian AN, Stanfa L, Suzuki R, Carpenter K, Dickenson A, Boyce S, Hill R, et al. (2000). Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X3 receptors. Nature 407, 1015–1017. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Kim A, Masuch T, Park K, Weng H, Wetzel C, and Dong X (2013). Pirt functions as an endogenous regulator of TRPM8. Nat. Commun 4, 2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, and Bredt DS (2003). Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J. Cell Biol 161, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, and Bredt DS (2005). Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature 435, 1052–1058. [DOI] [PubMed] [Google Scholar]
- Tomita S, Fukata M, Nicoll RA, and Bredt DS (2004). Dynamic Interaction of Stargazin-Like TARPs With Cycling AMPA Receptors at Synapse. Science 303, 1508–1511. [DOI] [PubMed] [Google Scholar]
- Vandenberghe W, Nicoll RA, and Bredt DS (2005). Stargazin is an AMPA receptor auxiliary subunit. Proc. Natl. Acad. Sci. USA 102, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waberer L, Henrich E, Peetz O, Morgner N, Dötsch V, Bernhard F, and Volknandt W (2017). The synaptic vesicle protein SV31 assembles into a dimer and transports Zn2. J. Neurochem 140, 280–293. [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Matsui M, and Watanabe M (2010). Preferential localization of muscarinic M1 receptor on dendritic shaft and spine of cortical pyramidal cells and its anatomical evidence for volume transmission. J. Neurosci 30, 4408–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Hoyos-Ramirez E, Martenson JS, Morimoto-Tomita M, and Tomita S (2017). GARLH family proteins stabilize GABAA receptors at synapses. Neuron 93, 1138–1152.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Yamasaki M, Straub C, Watanabe M, and Tomita S (2013). Homeostatic control of synaptic transmission by distinct glutamate receptors. Neuron 78, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. (2011). A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, and Jan LY (1999). A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22, 537–548. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, and Oldenburg KR (1999). A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang W, St-Gelais F, Grabner CP, Trinidad JC, Sumioka A, Morimoto-Tomita M, Kim KS, Straub C, Burlingame AL, Howe JR, and Tomita S (2009). A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron 61, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Logothetis DE, and Séguéla P (2007). Regulation of ATP-gated P2X receptors by phosphoinositides. Pflugers Arch. 455, 181–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all dataset and code generated or analyzed during this study.