Abstract
The epidemiology of foot-and-mouth disease (FMD) in Africa is unique in the sense that six of the seven serotypes of FMD viruses (Southern African Territories [SAT] 1, SAT2, SAT3, A, O, and C), with the exception of Asia-1, have occurred in the last decade. Due to underreporting of FMD, the current strains circulating throughout sub-Saharan Africa are in many cases unknown. For SAT1, SAT2, and serotype A viruses, the genetic diversity is reflected in antigenic variation, and indications are that vaccine strains may be needed for each topotype. This has serious implications for control using vaccines and for choice of strains to include in regional antigen banks. The epidemiology is further complicated by the fact that SAT1, SAT2, and SAT3 viruses are maintained and spread by wildlife, persistently infecting African buffalo in particular. Although the precise mechanism of transmission of FMD from buffalo to cattle is not well understood, it is facilitated by direct contact between these two species. Once cattle are infected they may maintain SAT infections without the further involvement of buffalo. No single strategy for control of FMD in Africa is applicable. Decision on the most effective regional control strategy should focus on an ecosystem approach, identification of primary endemic areas, animal husbandry practices, climate, and animal movement. Within each ecosystem, human behavior could be integrated in disease control planning. Different regions in sub-Saharan Africa are at different developmental stages and are thus facing unique challenges and priorities in terms of veterinary disease control. Many science-based options targeting improved vaccinology, diagnostics, and other control measures have been described. This review therefore aims to emphasize, on one hand, the progress that has been achieved in the development of new technologies, including research towards improved tailored vaccines, appropriate vaccine strain selection, vaccine potency, and diagnostics, and how it relates to the conditions in Africa. On the other hand, we focus on the unique epidemiological, ecological, livestock farming and marketing, socioeconomic, and governance issues that constrain effective FMD control. Any such new technologies should have the availability of safe livestock products for trade as the ultimate goal.
Keywords: vaccine, foot-and-mouth disease virus, vaccine matching, new-generation vaccine, diagnostic tests
Introduction
Foot-and-mouth disease (FMD), of which FMD virus (FMDV) is the causative agent, is a contagious viral disease which affects cloven hoofed animals such as cattle, pigs, sheep, goats, and other artiodactyl species.1 FMD ranks as one of the most economically important infectious diseases of animals according to the World Organisation for Animal Health (OIE). The occurrence of the disease not only affects international trade in livestock and animal products but also results in damaging consequences for the livelihoods of local farmers due to impacts upon productivity, food security, and losses of income. The disease is widely distributed in the developing world, in particular Africa, Asia, and South America. In these regions, livestock farming forms the backbone of rural economies that supports approximately 70% of the world’s poor. FMD outbreaks particularly affect vulnerable individuals such as women and children since approximately 75% of livestock in Africa are raised under the communal smallholder, communal-grazing, or pastoral systems that sustain livelihoods of these groups.2–4 The lack of veterinary infrastructure, human resources, movement controls, and appropriate vaccines render many developing countries particularly exposed to the spread of FMD.5–7
The epidemiology of FMD in Africa is influenced by two different patterns (ie, a cycle involving wildlife), in particular the African buffalo (Syncerus caffer), and an independent cycle maintained within domestic animals.8–12 Another unique feature of FMD epidemiology in Africa is the presence of the three Southern African Territories (SAT) serotypes (ie, SAT1, SAT2, and SAT3), which are maintained within the African buffalo populations.12–15 The presence of large numbers of African buffalo provides a potential source of sporadic infection to domestic livestock and other wildlife species.16–18 Although the precise mechanism of transmission of FMD from buffalo to cattle is not well understood, it is facilitated by direct contact between these two species. Once cattle are infected they may maintain SAT infections without the further involvement of buffalo.12,19–21 Sub-Saharan Africa is endowed with an abundance of wildlife, which has been preserved within national parks and game reserves.22 In communities neighboring these parks, the livestock/wildlife interface presents unique challenges to livestock disease control.12,23,24 In addition, the creation and expansion of transfrontier conservation areas in southern and eastern Africa presents a particular challenge to the management of FMD.
In response to this unique epidemiological situation, certain southern (eg, Botswana, Namibia, and South Africa) and North African (such as Egypt) countries20 have invested in regular livestock vaccination programs to manage the disease and facilitate access to international and regional trade markets in livestock and livestock products. However, in southern Africa, the incidence of the disease has increased appreciably over the last decade, and since the eradication of the disease in Africa is unlikely in the near future, more flexible ways of managing FMD are required to obviate clashes between conservation-based and livestock-based initiatives aimed at rural development. Despite systematic use of vaccination, numerous outbreaks of FMD have been recorded, and there is evidence of sustained virus circulation in vaccinated cattle populations in southern and eastern Africa since 2000,25,26 as well as in North Africa where exotic incursions of FMD (serotypes A, SAT2, and O) have caused widespread outbreaks.20,27
Effective control and prevention of FMD relies largely on the implementation of strategies such as physical separation of wildlife and livestock, repeated vaccination of cattle herds exposed to wildlife, control of animal movements, and careful assessment of the risk of FMDV introduction into disease-free areas.12,23,25 The current inactivated vaccines have proven effective in reducing clinical disease in FMD-endemic areas and have been critical to the success of FMD control programs in South America and Europe.28 In Africa, the diversity of circulating field strains of FMDV makes the selection of sufficiently cross-protective FMD vaccines a challenge. Therefore, local and regional programs of surveillance to monitor FMDVs circulating in wildlife and livestock populations are a crucial component of vaccine control, to provide vaccine matching data and access to appropriate viral strains that can be used in the development of new vaccines. There is need also for risk-based surveillance to be able to determine primary endemic areas and factors that influence disease dissemination, to assist the design of targeted, area-wide, or ecosystem-based disease control strategies, as African regions embark on the Food and Agriculture Organization of the United Nations (FAO)-OIE Progressive Control Pathway (PCP) for the Control of FMD.29 The success of any FMD control campaign ultimately depends on the abundant supply of vaccine of the appropriate strain composition and proven potency, adequate vaccine coverage, rapid vaccine development, overall planning and management by a well-resourced veterinary service, and the involvement and cooperation of the livestock farmer.30
In summary, animal diseases, in particular transboundary animal diseases such as FMD, severely constrain the development of competitive livestock enterprises in developing countries.31,32 The aim of this review is to identify the current limitations that are experienced in the control of FMD in endemic settings in Africa caused by gaps in knowledge of epidemiology, vaccinology, and diagnostics. We also emphasize the progress that has been achieved in the development of new technologies, including research towards improved tailored vaccines, vaccine matching, and diagnostics, and how this relates to the conditions in Africa. Furthermore, we advocate applied research into vaccination and disease control strategies to enable fit-for-purpose approaches to FMD control in Africa.
Epidemiological patterns in Africa
The current global burden of FMDV infection is maintained within three continental reservoirs in Asia, Africa, and South America, which can be further subdivided into seven major virus pools of infection.29,33 Each of these contains at least three serotypes of virus, and because virus circulation is mainly within these regional reservoirs, strains have evolved which are specific to the region and which often (in the case of type A and SAT viruses) require tailored diagnostics and vaccines for control.34
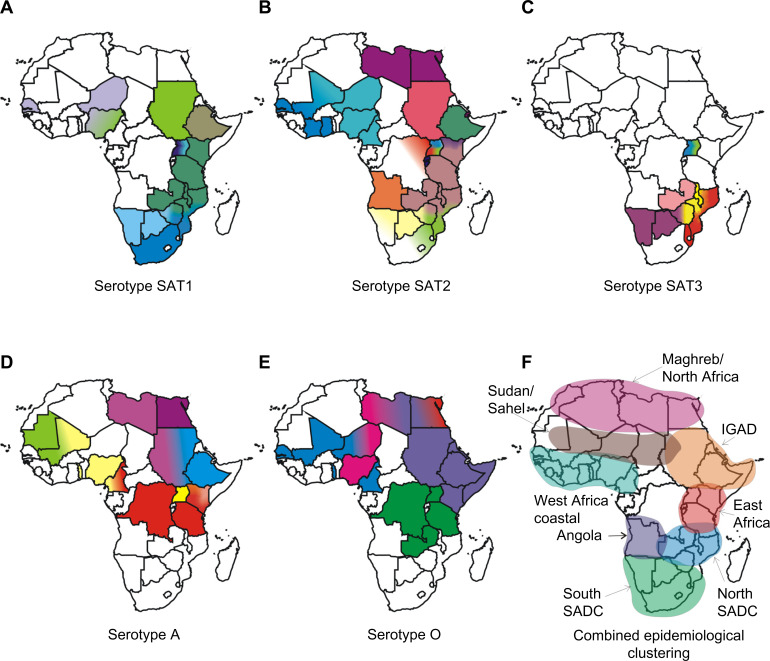
In Africa, the FMDV serotypes are not uniformly distributed, and each serotype results in different epidemiological patterns. The cumulative incidence of FMDV serotypes show that six of the seven serotypes of FMD (O, A, C, SAT1, SAT2, and SAT3) have occurred in Africa.35,36 The distribution of five serotypes and the different topotypes are shown in Figure 1A–E. Based on the genetic characterization of the virus and antigenic relationship of FMDV in Africa, the virus distribution has been divided into three virus pools: namely, pool 4 covering East and North Africa, with predominance of serotypes A, O, SAT1, and SAT2; pool 5 restricted to West and northern Africa, with serotypes O, A, SAT1, and SAT2; and pool 6 restricted mainly to South Africa, with SAT1, SAT2, and SAT3 serotypes. Recent studies in East and southern Africa have revealed genetic differences between viruses isolated at different times and places.37–39 Periodically, there have been incursions of types SAT1 and SAT2 from Africa into the Middle East, probably as a result of animal movement.35,36 The most recent reports include the spread of viruses of the SAT2 serotype to Yemen in 1990, to Kuwait and Saudi Arabia in 2000, and to the Palestinian Autonomous Territories and Bahrain in 2012.40 Overall, the geographic and genetic clustering of FMDVs suggest ecological adaptation and/or separation, but in many endemically affected areas, the temporal and spatial dynamics of infection still need to be much more accurately determined by analysis of host animal distributions and contact opportunities, serosurveys to estimate weight of infection, and use of the latest available techniques in genetic tracing of FMDV incursions into disease-free regions.41 Generally, many of these factors are driven by climatic factors and socioeconomic changes centered on human behavior. Also, findings regarding the epidemiology of FMD involving wildlife within a particular ecosystem of Africa may not be applicable to other ecosystems because of ecological, host, and viral variability differences.42 Understanding of how these risk factors are clustered and associated in space and time may assist in effective disease control planning.43,44
Figure 1.
Maps of Africa showing the serotype and topotype distribution.
Notes: The topotypes are color coded. The epidemiological clustering is indicated. The epidemiological clusters shown in the maps (A–F) do not necessarily indicate political borders of the countries.
Abbreviations: IGAD, Intergovernmental Authority on Development; SADC, Southern African Development Community; SAT, Southern African Territories.
To understand the complexity of FMD epidemiology in Africa and to assist decision making and improve the continental control of FMD, it is important to further divide the virus pools into epidemiological clusters.29 Rweyemamu et al29 proposed eight epidemiological clusters for Africa (Figure 1F) based on the distribution of serotypes and topotypes in different regions in Africa (Figure 1A–E), animal movement patterns, impact of wildlife, and farming systems. Here, we revisited the epidemiological clusters in light of the latest information regarding outbreaks in the region. The epidemiological clusters for Africa have the following characteristics.
Indian Ocean Island Countries (Madagascar, Mauritius, and Seychelles) are free of FMD, with a recognized status of FMD freedom without vaccination.
The South Southern African Development Community (SADC) cluster includes Swaziland, Lesotho, South Africa, Botswana, and Namibia, the southern and western part of Zimbabwe, and the southern part of Mozambique. The commercial livestock sectors of South SADC countries, with the exception of Zimbabwe and Mozambique, are free from FMD and meet the conditions of the OIE for zonal or country freedom from FMD without vaccination. Over the last 5 years, the region has suffered from an increasing number of outbreaks in cattle, most of which has been caused by SAT2 viruses. Cross-border epidemiological events have occurred on a number of occasions in South SADC, and in some cases, outbreaks were caused by viruses from different topotypes (Figure 1). The epidemiology of FMD in this region is characterized by virus circulation between the wildlife host, the African buffalo, and domestic animals, as well as spread among domestic animals, without the involvement of wildlife.18 In some of these countries, there are segregated wildlife areas that harbor African buffalo known to be infected, asymptomatically, with FMDV serotypes SAT1, SAT2, and SAT3. These wildlife parks are segregated from livestock through a system of game-proof fencing and vigorous surveillance. In these countries, game ranching or other wildlife conservation activities with FMD-infected African buffalo are not allowed within FMD-free zones. However, for this epidemiological cluster, the primary source of FMD seems to be the risk posed by the wild buffalo herds,45,46 as evidenced by many outbreaks in or near transfrontier conservation areas (TFCAs), such as the Kavango-Zambezi TFCA.47
The North SADC cluster comprises the northern part of Zimbabwe, Zambia, northern Mozambique, Malawi, and southern Tanzania. The North SADC cluster countries have to deal with at least four serotypes of the virus (A, O, SAT1, and SAT2), and maybe even five (SAT3), each with multiple subtypes in the region (Figure 1). This may require the incorporation of more than one strain of a given serotype into a single vaccine to allow effective control in this region. Viral diversity and thus antigenic diversity is a complicating factor in effective vaccination against FMD in this cluster. Cross-border spread of the disease is common, and SAT1 and/or SAT2 outbreaks in Mozambique, Malawi, and Zambia between 2002 and 2013 were either because of outbreaks spreading from neighboring countries or to internal buffalo–cattle contact. Northern Malawi and Northern Zambia are under constant threat of FMD spread from southern Tanzania.29,38,39,48
The Angola cluster may also include the western Democratic Republic of Congo (DRC). Very little is known about the true incidence of FMD within this cluster, and no official information is available on the isolation of FMDV from Angola since 1974. However, an FMD outbreak has been recorded in Angola in 2009, although no virus could be isolated. The southern part of Angola forms part of the Kavango-Zambezi TFCA, and it may be appropriate to include it within the South SADC cluster.
The East African Community cluster is comprised of Tanzania, Uganda, Kenya, Rwanda, and Burundi, plus the eastern part of the DRC (Figure 1). In addition to large livestock populations, this cluster has the highest concentration of wildlife in the world. The transmission and maintenance of FMD in this region is complex, as farming practices, trade, and wildlife contribute to the maintenance and spread of the virus. Farming is dominated by agro-pastoral and pastoral communities and is characterized by communal grazing and migrations. Eastern DRC is heavily dependent on trade in livestock from Uganda, Tanzania, Rwanda, and Burundi. The cluster probably contains several FMD primary endemic foci, and cross-border epidemiological events suggest that animal movement plays an important role in virus dissemination.39 At least four serotypes (A, O, SAT1, and SAT2) are endemic in this cluster,37–39,49–51 with serological evidence for a fifth serotype (C)52 (Figure 1). A sixth serotype (SAT3) was isolated in wildlife (African buffalo) in Uganda in 1970,10 although it has never been isolated from livestock in this cluster. SAT3 was also reported in Uganda in 199753 and in the DRC in 2005, but was not genotyped.54 Isolates of serotypes A, O, SAT1, and SAT2 from Tanzania and Kenya (2004–2009) were genetically related.38,39 Similarly, viruses from Uganda and Kenya (1998–2008) were related.51 FMDV isolates belonging to serotypes SAT1 (topotype IV) and SAT2 (topotype X) have been isolated from African buffalo.15 As discussed above, there are also wide genetic and antigenic variations in the virus strains in this epidemiological cluster. The role of the African buffalo in the maintenance and transmission of FMD serotypes (eg, A and O)55 that occur in this cluster has not been systematically studied.
The Intergovernmental Authority on Development (IGAD) cluster comprises Sudan, South Sudan, Eritrea, Ethiopia, Djibouti, Somalia, Northern Kenya, and Northern Uganda (Figure 1). Similar to the East African Community cluster, this cluster probably harbors major FMD primary endemic foci. Ethiopia and Sudan have the highest cattle populations in Africa.29 Historically, isolates of serotypes A, O, SAT1, and SAT2 from Sudan and Ethiopia were genetically related to isolates from Uganda, Kenya, and Tanzania, most likely as a result of cross-border movement, a situation that has not changed.
The Soudan/Sahel cluster comprises Western Sudan, Niger, Chad, Burkina Faso, Mali, Northern Nigeria, Senegal, and Mauritania. The farming system in this ecosystem is predominantly pastoral, characterized by long-distance movement of livestock due to either transhumance or trade. This cluster probably also contains important FMD primary endemic areas, and at least four serotypes (A, O, SAT1, and SAT2) of the virus have been found. Furthermore, it may be an important disease-corridor cluster, linking the IGAD cluster with West Africa and probably West Africa with North Africa. The 1999 FMD strain in Algeria was found to be related to the West African type O topotype.29 Similarly, isolates of serotype O from Niger (2007) and Nigeria (2007 and 2009) were genetically related to viruses found in Eritrea (2004 and 2011), Ethiopia (2005, 2006, 2008, and 2010–2012), and Sudan (2005, 2008–2011).56 Viruses belonging to serotype A were isolated from cattle samples from Togo (2005), Nigeria (2009) in West Africa, and Cameroon (2005) in Central Africa, which had close genetic relationships with viruses from Eritrea (1998) and Sudan (2006 and 2011) in East Africa56,57 (Figure 1).
Although the epidemiology of FMD in the coastal belt countries of West and Central Africa has not been deeply studied, it seems that this cluster probably gets infected from the Soudan/Sahel cluster. It could therefore be described as secondary endemic.
North Africa/Maghreb cluster countries Morocco, Algeria, and Tunisia have not reported FMD since 1999, most likely because of routine preventive vaccination and other measures. Libya and Egypt have sporadic FMD, and take routine preventive vaccination. Libya reported a SAT2 outbreak in 2003 (topotype VII) (Figure 1), probably as a result of live animal introductions from neighboring countries to the south, breaching the Sahara barrier. The virus was genetically related to outbreaks in cattle in Saudi Arabia in 2000 and Eritrea in 1998.35 In 2012, Libya experienced another SAT2 outbreak (topotype VII), this time genetically related to isolates from Sudan (2007) and Nigeria (2008). Egypt also reported a SAT2 outbreak in 2012, the first occurrence of this serotype since 1950, and at least three sub-lineages (one Libyan and two Egyptian)20 were identified. Egypt also reported African type A viruses in 2006, 2007, 2009, and 2012, as well as ME-SA (Middle East–South Asia) types O and A. Yemen reported EA (East Africa)-3 type O in 2007 and 2009. Thus, North African countries will remain at risk from the south and east, but across the majority of their territories, and at-risk populations should effectively maintain FMD freedom.
Antigenic diversity of FMDV: implication for the selection of vaccine strains
The selection of FMD vaccine candidates is complicated by the wide spectrum of genetic and antigenic variability of the FMDV and the continuous emergence of new mutants from populations that escape the host immune response.58–61 Most of the impacts of this variation derive from changes within the three major surface-exposed capsid proteins of the virus (ie, VP1, VP2, and VP3). At least 30%–50% of the residues that constitute the capsid proteins are exposed on the virus surface, many of which encompass neutralizing epitopes.62–67 It has been shown that the majority of FMDV-neutralizing antibodies are directed against conformational epitopes located on the β-barrel connecting loops projecting from the surface of the virus, especially the highly mobile βG–βH loop in VP1.62,68–70 It is important to recognize that the SAT1 and SAT2 viruses display greater antigenic variation compared with the Euro-Asian serotypes (O, A, C, and Asia-1).61,66,71 The variation is not random, but tends to be concentrated at the surface-exposed β-barrel connecting loops. Therefore, knowledge of the amino acid residues that comprise the antigenic determinants of FMDV, and those that function as protective epitopes in particular, will greatly improve our understanding of virus neutralization in vivo.72,73
Several studies have been carried out to delineate the neutralizing antigenic sites of representative viruses from serotypes A,74–76 O,68,77 C,78 and Asia-1.79,80 In these studies, monoclonal antibodies (MAbs) have been pivotal in identifying critical amino acid residues of the different neutralizing antigenic sites. In addition, mapping the topography of the mutations on the X-ray crystallographic structure of FMDV62,63,67 and predicting B-cell epitopes using computational algorithms81 have resolved several other antigenic sites on the capsid of FMDV. Although the overall role and importance of these antigenic sites in induction of protective immunity and cross-reactivity in target species is still poorly understood, it appears that animals vaccinated with FMDV do not elicit a predominant antibody response against a single antigenic site, but rather utilize a broad repertoire of epitopes on the viral capsid.82–84
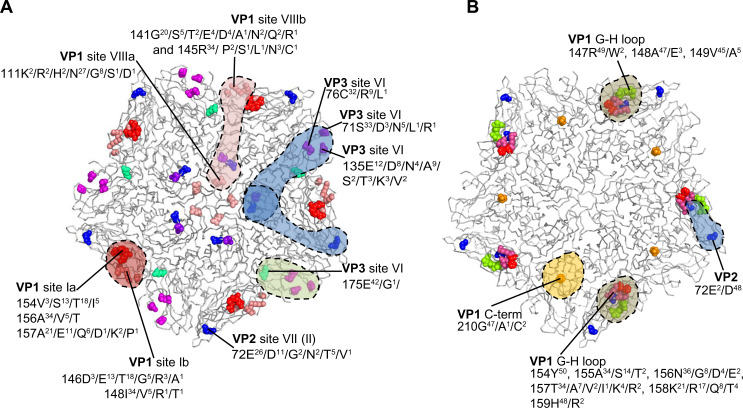
In a study to investigate the antigenic structure of FMDV serotypes SAT1 and SAT2, unique critical MAb contact residues were found on VP3 and VP1 (site VI), VP2 and VP1 (site VII), or VP1 residue 111 (site VIII) of a SAT1 virus.85 Similarly, for a SAT2 virus, two epitopes have been mapped, one encompassing the βG–βH loop of VP1 and the other involving residue 210 in the C-terminus of VP1.85–87 These antigenic sites are summarized in Figure 2. The residue positions where substitutions occurred in MAb-resistant mutants are variable, with high entropy or uncertainty in SAT1 and SAT2 capsid protein alignments (Figure 2). For both serotypes, the variable regions were clustered around the fivefold and threefold axes of symmetry of the virion. A single amino acid change can lead to the abrogation of antibody-binding, which emphasizes the structural flexibility of the surface-exposed loops.
Figure 2.
The antigenic structure of foot-and-mouth disease SAT1 (A) and SAT2 (B) viruses are depicted.
Notes: The amino acid substitutions observed in SAT1 and SAT2 monoclonal antibody-resistant (MAR) mutants are shown as spheres on the grey ribbon backbone of the pentamer (five copies of VP1, VP2, and VP3) unit, and the potential antibody footprints are stipulated. The variation in amino acid residues at each antigenic residue position, in a complete capsid protein alignment of 43 SAT1 and 50 SAT2 viruses available on the genetic sequence database (GenBank) are indicated. The viruses in the alignments include isolates from South Africa, Zimbabwe, Mozambique, Namibia, Botswana, Zambia, Angola, Malawi, Tanzania, Kenya, Uganda, Rwanda, Zaire, Nigeria, Senegal, Ghana, Eritrea, Saudi Arabia, and Sudan. Adapted from Grazioli S, Moretti M, Barbieri I, Crosatti M, Brocchi E. Use of monoclonal antibodies to identify and map new antigenic determinants involved in neutralization of FMD viruses type SAT 1 and SAT 2 In: European Commission for the Control of Foot-and-Mouth Disease: International control of Foot-and-Mouth disease: Tools, Trends and perspectives. Paphos, Cyprus: Food and Agriculture Organization; 2006:287–297.85
Abbreviation: SAT, Southern African Territories.
Vaccine matching: the selection of vaccine strains for specific geographic, endemic regions
The two important determinants that will affect the efficacy of a vaccine and determine whether it will protect or not are 1) the ability of the vaccine strain to elicit antibodies that will cross-react and protect against the field or outbreak virus in question (defined as the vaccine or antigenic match), and 2) the potency of the vaccine to elicit a strong and long-lasting immune response. The quality and quantity of the antigen in the vaccine as well as the formulation of the vaccines and inclusion of immune-stimulating adjuvants are all factors that will influence and contribute to the overall potency of the vaccine.88 In addition to vaccine efficacy, the number of animals vaccinated in the target population during a vaccination campaign will determine effective protection at herd level and should be taken into consideration. Basic capability to undertake vaccine matching tests on a routine basis in diagnostic laboratories in African countries is severely limited, and therefore current advice regarding the selection of the best vaccine to be used in these settings is normally provided by regional (ARC-Onderstepoort Veterinary Institute, South Africa; Botswana Veterinary Institute, Botswana) and international reference centers.
The OIE/FAO FMD Reference Laboratory Network reports over the last five years have revealed a gap in the vaccine strains available to match against circulating SAT1 and especially SAT2 viruses. The urgent requirement for the development of new SAT vaccine strains with good immunogenicity for use in Africa was also highlighted at the recent Global FMD Research Alliance congress (Arusha, Tanzania in October 2013). For the African continent (FMD endemic pools 4, 5, and 6), at least five vaccine strains are available for SAT1, and seven vaccine strains are described for SAT2 viruses. However, not all these vaccine strains are of recent derivation or are currently used in production, and it is therefore imperative that outbreak samples are properly matched to the vaccine strains that are available for use in control programs.
The direct assessment of the degree of protection induced by a vaccine against the field virus is performed using animal challenge studies and is the most accurate way to determine whether a vaccine will cross-protect against a particular field virus.82,89,90 However, routine heterologous challenge studies are rarely performed, as this is time consuming, requires access to expensive bio-containment facilities, and raises serious animal welfare issues when large numbers of animals need to be vaccinated and challenged. If small groups of animals are used, these challenge studies can suffer from low precision.91
In South America, a variation of this direct method has been developed, based on the probability that cattle will be protected against a challenge of 10,000 infective doses of test virus following a single or boosted vaccination.92 In order for the expected percentage of protection method to be effective, the vaccine needs to be extensively tested in many (hundreds) cattle by previous challenge tests with homologous virus.92 Although this method has been widely used in South America,90,93 the availability of good datasets and sera and the need for diverse vaccine strains are limiting factors for use of this approach in the African context.
Therefore, in vitro alternatives to measure cross-reaction between sera elicited by a vaccine and a particular field/outbreak isolate is recommended, supporting the need to accurately predict vaccine matching without the involvement of animals.94 Traditionally, antigenic characterization of a field or outbreak virus is performed indirectly using the in vitro virus neutralization test (VNT), which measures the ability of sera from vaccinated animals to cross-react with the field virus.61,82,89,90 The neutralization titers are used to calculate r1-values to determine antigenic relationships.95 However, interpretation of the test is plagued by limitations, including the uncertainty as to how well the in vitro matching data actually correlates to in vivo cross-protection, the impact of vaccine potency on protection, and the availability of reference reagents.88 Furthermore, the use of r1-values to estimate cross-protection relies on having sufficient repeated measures to overcome the inherent variability of the neutralization titers.96 In a recent study with SAT1 viruses, we found a number of factors to impair reproducibility in one-way relationships, such as the operator, batch variability of reagents, day-to-day variation in the cells, and variation in individual cattle sera. It is also known that measuring the titer ratio to a known control is not sufficient to eliminate the inter-experiment variability, highlighting the necessity for time-consuming duplicate tests to be undertaken on separate and independent occasions to compensate for day-to-day variations.96 A novel way to quantify and visualize antigenic relationships is antigenic cartography.97,98 However, the combination of genetic sequencing and antigenic profiling of the outbreak virus are still useful methods to identify newly emerging or re-emerging virus strains and whether available vaccine strains are likely to provide protection against the outbreak virus or not.
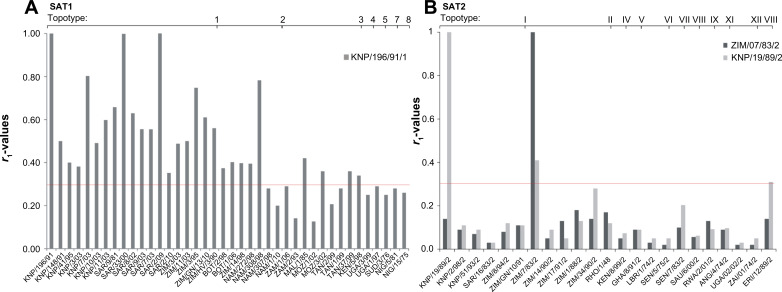
Using this in vitro approach in vaccine matching, we have shown that the SAT1 vaccine strains SAR/9/81 and KNP/196/91 are antigenically relevant for South Africa, Zimbabwe, Mozambique, Botswana, Namibia, Zambia, and Tanzania.61 The r1-values determined in assays with antisera from cattle that had been vaccinated with SAT1/KNP/196/91, and a panel of SAT1 viruses, was greater than 0.3 for 75% (n=30) of viruses (Figure 3A). This result suggests that efficient cross-protection may be induced in animals following vaccination with these strains. Similarly, a vaccine based on NIG/5/81 could most likely protect against many strains circulating in West Africa (pool 5 viruses), while a vaccine based on BOT/1/06 virus alone or in combination with another strain from East Africa will effectively cross-react to control most pool 4 viruses.99 Thus, a high potency or bivalent SAT1 vaccine could most likely be effective across topotypes. However, there is a dearth of knowledge about the antibody response of animals vaccinated with a multivalent vaccine containing a combination of intra-serotype strains to cross-react against field viruses. Furthermore, it has been shown for serotype A viruses that a high potency vaccine provides protection against heterologous challenge, despite low r1-values.100 In the past, the combination of SAT1/SAR/9/81 and SAT1/KNP/196/91 in a tetravalent vaccine, containing also a SAT2 and a SAT3 strain, were able to protect against SAT1 outbreaks in southern Africa.101 The antigenic relationship of SAT2 viruses against two SAT2 vaccine strains showed poor cross-reactivity, indicative of low level of cross-neutralization in vivo (Figure 3B). Notably, a single type O vaccine strain was efficacious in controlling the circulation of different topotypes in Kenya, even though temporal fluctuation in the prevalence of a particular type was observed.50
Figure 3.
Chart to indicate the r1-values of various SAT1 and SAT2 foot-and-mouth disease virus isolates from different topotypes compared with antiserum prepared from the vaccine strains (A) SAT1/KNP/196/91 (topotype 1) and (B) SAT2/KNP19/89/2 (topotype I) and SAT2/ZIM7/83 (topotype II).
Notes: The antigenic relationships of the field isolates were determined using cross-neutralization assays on IB-RS-2 cells, carried out as described by the OIE.207 The r1-values were calculated as: r = serum titer against heterologous virus/serum titer against homologous virus, and interpreted as proposed by Samuel et al.104 Viruses from the Kruger National Park (South Africa) (KNP), South Africa (SAR), Ghonerazhou National Park (Zimbabwe) (GN), Zimbabwe (ZIM or RHO), Botswana (BOT), Namibia (NAM), Mozambique (MOZ), Angola (ANG), Zambia (ZAM), Malawi (MAL), Tanzania (TAN), Kenya (KEN), Uganda (UGA), Rwanda (RWA), Sudan (SUD), Zaire (ZAI) Ghana (GHA), Nigeria (NIG), Senegal (SEN), Eritrea (ERI), Saudi Arabia (SAU) were included in the study.
Abbreviations: OIE, World Organisation for Animal Health; SAT, Southern African Territories.
Alternatively, serological cross-reactivity can be estimated using a liquid-phase blocking enzyme-linked immunosorbent assay (ELISA) procedure for vaccine matching reviewed by Paton et al88 and Kitching et al.102 In common with the VNT method, this approach is based on the reactivity of bovine serum raised against the available vaccine strains to the field virus and the homologous vaccine strain.103 The r1-value is calculated as the heterologous titer divided by the homologous titer and interpreted as proposed by Samuel et al.104 The r1-value calculated by the two assays differ, because the VNT approach is based on the ability of the antisera to neutralize the virus, whereas the liquid-phase blocking ELISA method measures binding of the antisera to the virus or viral components. Therefore, there is still a need for a faster and more reliable method that is as effective as the VNT at evaluating vaccine matching.
Since the antigenic variability, reflected in the VNT titers, is a measure of whether the sites with which the immune system reacts to neutralize the virus remain sufficiently similar between a homologous and heterologous virus to be recognized,105 we have described a new approach, using linear mixed-effect models, to estimate antigenic matching.66 In principle, the amino acid variation and in vitro cross-protection titers from VNTs were combined with crystallographic structural data to generate antigenic matching information indirectly.66 Examining SAT1 and SAT2 viruses, we identified a correlation between genetic distance and antigenic relatedness (r1-values), but more significantly, we could identify areas on the surface of the capsid where mutations were strong predictors of antigenic distance. These were consistent within serotypes, and were found to match some of the independently identified antigenic sites in other serotypes. Applying the linear mixed-effect model to SAT1 viruses, two surface-exposed regions of the capsid (ie, VP1 βG–βH (132–174) and VP3 βH–βI (191–202) loops were identified as better predictors of cross-reactivity between the field virus and vaccine strain66 than serologically calculated r1-values. Both regions were identified by studying MAb-resistant mutant viruses.85 For SAT2 viruses, amino acid residues in the VP1 C-terminus (200–224), the VP2 βB–βC loop (70–82), and residue 178 in the VP1 βH–βI loop were found to be good predictors of cross-reactivity.66 However, the aim in endemic settings is to identify potentially new vaccine viruses when existing vaccine strains are found to not cross-react to the outbreak virus sufficiently. The linear mixed-effect approach was found useful to assess the likely cross-reaction of new vaccine strains against a group of field viruses.66 Taken together, this approach will assist in the future in the selection of potential new vaccine strains for sub-Saharan Africa.
Simple antibody recognition measures do not always correctly predict the ability of a vaccine to protect against an outbreak virus.106 The antibody isotype, the avidity of the antibody to the virus in question, and the type of immune response elicited are also important factors to consider.107–109 In a recent study comparing the accuracy of traditional and novel serological assays to predict cross-protection, it was found that the use of VNT titers and r1-values are inaccurate indicators of protection.110 However, when the VNT titers were combined with the IgG1 titer, a more accurate estimate of FMD vaccine protection against the heterologous virus for serotype A was achieved. To date, the correlation of in-parallel serological data, like VNT and IgG1, IgG1/IgG2, or antibody avidity in cross-protection in the case of SAT viruses is unknown.
Control of FMD by vaccination
The existing vaccines against FMD consist of complete, chemically inactivated virions combined with an adjuvant.7 The adjuvant used in the vaccine formulation has undeniably a huge effect on the efficacy and potency of the vaccine and has been reviewed elsewhere.7,111 Despite successful application in the developed world, the effective administration and optimal induction of protective immunity are hampered by several factors in developing countries. In addition to the vaccine-matching constraints that have been discussed in the previous section, some viruses are very difficult to adapt to cell culture, slowing the introduction of new vaccine strains, reducing vaccine yield, and potentiating through prolonged passage the selection of undesirable antigenic changes.112,113 Vaccination does not induce sterile immunity, and animals may still be able to infect non-vaccinated animals and may also become persistently infected.7,114–117 The presence of contaminating non-structural proteins in some vaccine formulations makes it problematic to distinguish between vaccinated and convalescent animals, impacting on the ability to export from FMD-controlled regions. In addition, the hot climate in many African regions calls for vaccines with improved stability and which are less reliant on a cold-chain. During production, the manufacturer also has to compensate for this instability by increasing the quantity of antigen per vaccine dose, which is expensive and reduces vaccine yield. Based on the findings of Doel and Bacarini,118 it is believed that unstable vaccines are less immunogenic due to degradation before and after inoculation. Therefore, FMD vaccines require frequent booster vaccinations in order to be effective. Lastly, the current vaccines are relatively expensive, especially for the small and subsistence farmer.
Vaccines used in the control of FMD in endemic regions are mostly used for mass prophylactic application. Such vaccines are multivalent to provide protection against multiple serotypes, and should have a potency of at least 3 PD50 per dose.119 Generally, prophylactic vaccines incorporate 146S particles combined with saponin-alhydrogel or oil-adjuvant.119 Oil-adjuvanted vaccines have been used successfully in FMD-eradication campaigns in South America.5,120,121 A study evaluating different adjuvants for SAT vaccines has shown that a double water-in-oil-in-water adjuvant, ISA206, elicited protective antibody responses against SAT2 serotype in cattle.122 Inactivated vaccines induce short-lived immunity, and it is recommended that naïve animals receive two initial vaccinations (a primary and secondary dose) 3–4 weeks apart, followed by re-vaccination every 4–6 months101,122 to prevent spread of disease within populations. However, in the African environment, this may differ for different manufacturers, depending on the potency of the vaccine, and some manufacturers recommend five vaccinations per annum. There is a definite need to assess whether different adjuvants may enhance the duration of immunity against SAT antigens. For these reasons vaccination campaigns should be performed regularly, based on 1) the epidemiological circumstances and risk of disease spread, 2) the value and life expectancy of species, and 3) the economic status of the country. The interval between vaccinations is critical to prevent a “window of susceptibility” and where the continuous or sporadic presence of virus in carrier animals is present.
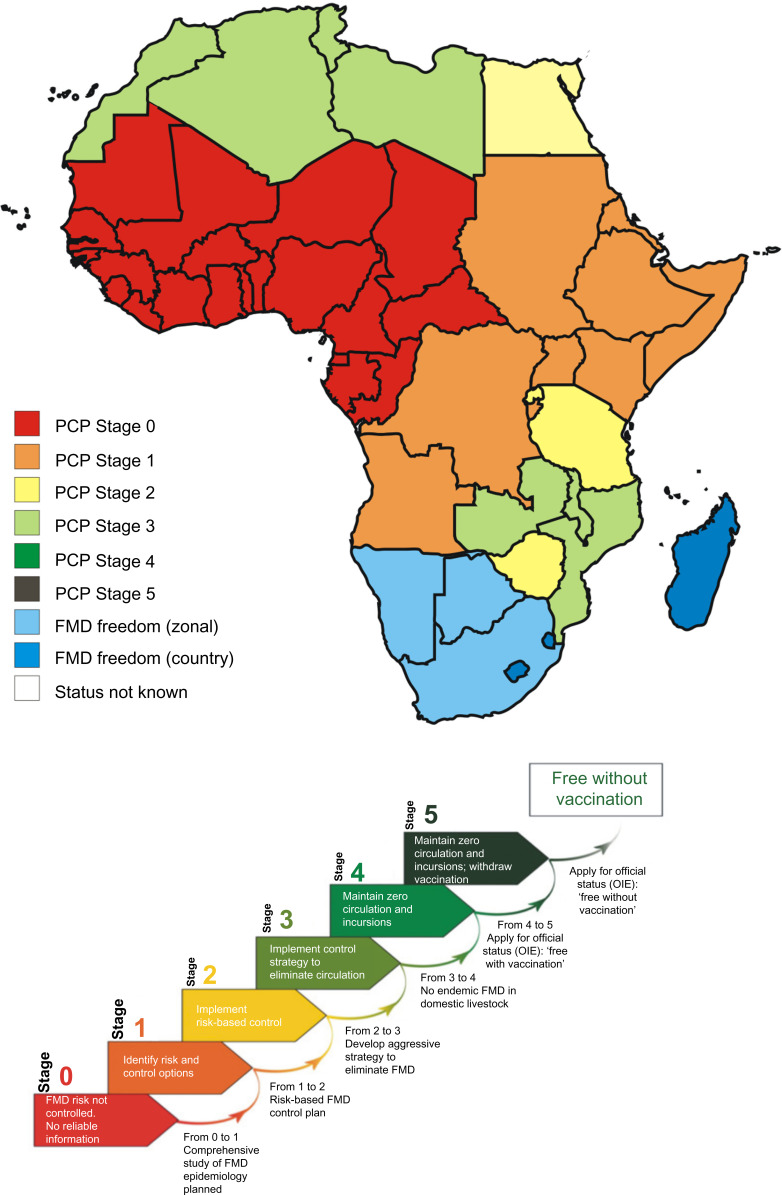
The PCP is the strategy proposed by OIE and FAO to control and ultimately eradicate FMD from endemic countries. Different regions in sub-Saharan Africa are at different developmental stages of control and are thus facing unique challenges and priorities in terms of FMD control (Figure 4). In many African endemic countries, there are various knowledge gaps, such as disease occurrence and mechanisms of virus maintenance and transmission, and therefore no routine vaccination campaigns are implemented (PCP Stage 1 countries; Figure 4). In other African endemic countries, even where surveillance is conducted to provide knowledge about high-risk populations, often implementation of effective, scheduled vaccination campaigns still does not take place (PCP Stage 2 countries; Figure 4). There are various reasons why governments do not subsidize FMD vaccines, leading to individuals needing to carry the cost and implement their own vaccine schedules. Additionally, individuals would need to source vaccines without knowledge of the current circulating strains in their region, leading to a poor vaccine match. This often leads to no or ineffective control in endemic African regions. The development of new vaccines against FMD in endemic countries in Africa should therefore take into account the ecosystem-based synchronization as FMD control strategies employed in these regions.30
Figure 4.
Map indicating the different PCP stages of countries in southern and eastern Africa.
Notes: The results are based on self-assessment and aspirations from the member states, with the goal to make progress on the PCP for FMD.208–210 The FMD PCP consists of six stages ranging from zero (0), when there is continuous FMD virus circulation with no reporting or control actions, to five (5), where a country is ready to be officially recognized by the OIE as free without vaccination. Currently, the OIE recognizes only three categories for countries with regard to FMD: 1) countries not free from FMD (PCP stages 0–3), 2) FMD-free countries or zones practicing vaccination (PCP stage 4), and 3) FMD-free countries or zones where vaccination is not practiced (PCP stage 5). This figure was adapted with permission from the Food and Agriculture Organization of the United Nations. © FAO 2011. Report of the 39th Session of EuFMD, Rome 2011. 1–31. http://www.fao.org/ag/againfo/commissions/eufmd/commissions/eufmd-home/reports/general-sessions/2011-39th-session/en/. Accessed July 25, 2014.211
Abbreviations: FMD, foot-and-mouth disease; OIE, World Organisation for Animal Health; PCP, Progressive Control Pathway.
Design of improved inactivated vaccines
Some of the abovementioned limitations of current vaccines are being addressed by the development of reverse genetic approaches. Additionally, new alternative vaccines that do not require infectious virus as well as efforts to understand the role of innate immunity and cytokines to induce protection and boost the immune response offer tremendous potential for the control of FMD in endemic regions.
Genome-length viral RNA (vRNA) derived from complementary DNA (cDNA) clones of FMDV is infectious when transfected into suitable mammalian cells.123–126 These cDNA clones are readily amenable to genetic engineering to introduce changes to the virus genome that allows for the replacement of the external capsid-coding region or structural, surface-exposed antigenic loops with the corresponding regions of an emerging virus. The outcome of such a chimeric virus is the transfer of the spectrum of neutralizing epitopes from the etiological agent to the recombinant virus123–127 and the ability to antigenically simulate the outbreak virus and induce protective immunity in host animals.123,128,129 Furthermore, it has been shown that inter-serotype chimeric vaccines with the capsid proteins of SAT1 within a SAT2 background128 and O serotype within an A serotype background123,130 confer protective immunity.
However, capsid swapping may transfer other undesirable traits such as capsid instability and poor cell culture adaptation, which are limitations that can be overcome by site-directed mutagenesis of the amino acid(s) associated with improved performance as vaccine candidates.129
The development of new vaccine FMDV strains relies strongly on virus growth and high antigen yields of the new strain in the production cell line.7,131,132 We have demonstrated that SAT-type viruses, previously impossible to adapt to cell culture, can be structurally modified by introducing an adaptation phenotype which is able to interact with sulfated glycosaminoglycans, enabling improved vaccine production.129 Several glycosaminoglycan-binding sites have been identified for the various serotypes that can be used for this purpose.112,129,133–138 Furthermore, alternative cell entry pathways exist and can be applied in future to improve cell adaptation.134,135,139 Continued reverse genetic approaches to rapidly adapt African FMDV to BHK-21 cell culture will greatly enhance the ability to produce region-specific vaccines tailored to current circulating strains.
The stability of vaccines is of crucial importance in Africa, where the logistical process to get the vaccine from the manufacturer to the animal may take months and in many remote regions is in the absence of a cold-chain. Vaccines with improved stability and less reliant on a cold-chain are needed and could improve the longevity of immune responses elicited in animals.118 FMD is known to be unstable, especially for O and SAT2 serotypes,118 in mildly acidic pH conditions or at elevated temperatures, leading to dissociation of the capsid (146S particle) and loss of immunogenicity. The residues at the capsid inter-pentamer interfaces, and their interactions, are important for the infectivity and stability of the virion,140,141 and mutations adjacent to these interfaces have an effect on the conformational stability of FMDV.142–145 However, experimental studies on the relative importance of residues and molecular interactions in viral capsid assembly, disassembly, and/or stability are still very limited. Recent research has compared more thermostable serotype A viruses with unstable O and SAT2 viruses, together with crystallography structures, sequence data, and in silico calculations of stability, to predict residue substitutions that could increase stability at inter-pentameric interfaces. With reverse genetic approaches, stabilizing mutations have been introduced into infectious copy clones. SAT2 and O viruses with improved stability have been developed, and their antigens are currently being tested in animal models for potential use as vaccines (unpublished observation).
Alternative vaccine strategies: subunit and live viral-vector vaccines
Due to the limitations of inactivated vaccines, alternative strategies for vaccine development have focused initially on the use of VP1-proteins and peptides either isolated from FMDV or produced by recombinant DNA;146,147 VP1-derived peptides148 or chemically synthesized VP1 peptides;149–152 vectors expressing VP1 fusion proteins;153–155 inoculation with DNA expressing VP1 epitopes alone156 or with DNA encoding IL-2;157 and transgenic plants or recombinant tobacco mosaic virus expressing VP1.158,159 However, they rarely achieved protection against virus challenge in livestock,151,160,161 or as a result of a limited subset of epitopes, selected for antigenic variants that escaped from protection.162 The reduced level of protection may be due to a lack of T-cell epitopes.163
Subviral particles or virus-like particles which are assembled as empty particles lack RNA (noninfectious), but are immunogenic, as their antigenic surface is indistinguishable from the complete virus.164 Previous attempts to express the P1, leader, and 3C regions in recombinant baculovirus systems has yielded limited amounts of empty capsid due to toxicity in insect cells165 or in Escherichia coli resulted in inefficient capsid assembly.164 Major advances have been made, firstly in developing methods to efficiently express empty capsids in vaccinia virus or baculovirus expression systems by lowering viral protease activity, which is toxic to cells, and secondly by enhancing capsid stability.166 Cattle vaccinated with such an A-serotype construct have shown sustained VNT titers and protection from challenge 34 weeks post-vaccination.166 This approach has several advantages over conventional vaccines, by reducing production costs, eliminating risk of infectivity, and enhancing stability.166 However, producing vaccines using insect cells requires highly skilled staff, and scaling up of the manufacturing process is difficult and expensive to optimize, which poses disadvantages for the African market. More studies are needed to assess whether this strategy would be effective in endemic regions of Africa for SAT serotypes and when applied to the field and to find out whether the production process would be economically viable.
Alternative strategies which incorporate sections of the FMDV genome inserted in a live viral vector have been investigated with bovine rhinotracheitis virus,154 and the G–H loop region has been incorporated in a chimeric poliovirus;155 capsid and/or non-structural proteins in vaccinia virus,167,168 fowlpox virus,169 or pseudorabies virus170,171 provides partial protection in pigs and guinea pigs. A chimeric bamboo mosaic virus containing 1D epitopes induces humoral and cell-mediated responses and protection in pigs.172 The experimental vaccines so developed have several disadvantages, such as requirement for multiple doses, low-level antigen expression, and uncertain safety. Additionally, repeated vaccination with a viral-vector vaccine, as is necessary in endemic regions, leads to immunity against the vector and reduced protection. The most successful advances have arisen from incorporating FMDV genes into replication-defective human adenovirus (Ad) vectors.173,174 The resulting vaccine has several advantages over conventional vaccines: 1) it can induce protection within 7 days and within 1 day if combined with interferon-α expression, particularly useful for emergency use; 2) it is fully compatible with test systems that allow to differentiate infected from vaccine animals; 3) incomplete inactivation problems are not a concern; and 4) it has excellent immunogenicity, attributed to its natural adjuvant properties, high transduction efficiency of targeted cells, and ability to induce antiviral cytotoxic T-cells.174 However, whether animals vaccinated with Ad-FMD vaccines would be protected from developing a carrier state is unclear, as is the significance of pre-existing immunity in the field. Finally, it is of major importance to determine the protective immunity and breadth of antigenic coverage against evolving variants in the field.
To be optimal for use in Africa, new vaccines should be easily delivered to the animal, proven to be more effective and induce longer lasting immunity than conventional inactivated vaccines, safe in production, easily made, and inexpensive. This is especially important for African producers that need to design vaccines tailored to African continental control, focusing on circulating serotypes and subtypes in endemic regions where eradication is difficult to attain due to the presence of maintenance hosts and continuous wildlife–livestock interfaces.
Current and novel diagnostic tests for field application in endemic regions
The accurate diagnosis of FMDV infection is of utmost importance for the control and eradication of the disease in endemic regions. The initial diagnosis of FMD is normally based on clinical signs, but this can easily be confused with other vesicular diseases.175 Hence, it is vital that the recognition of signs of the disease by the farmer is promptly conveyed to the relevant veterinary authorities to verify clinical symptoms, and suspect samples should then be sent to the reference laboratory for confirmation. Rapid and precise data generated by laboratories provides vital support to FMD control and vaccination programs. However, in many African countries, samples received by the laboratory can be of poor quality due to an ineffective cold-chain and long transport periods. These factors make laboratory diagnosis challenging, and it is evident that sub-Saharan Africa requires diagnostic tools that are fit for purpose in these settings to allow for rapid diagnosis and the appropriate measures taken for control.
Existing diagnostic techniques for the detection of FMD are mainly based on the following principles:
The identification of the infectious agent by virus isolation involving propagation on susceptible cell cultures176
The detection of viral antigen by ELISA systems using FMDV-specific antibody or capturing reagents177–179
Molecular detection of viral nucleic acid by reverse-transcription polymerase chain reaction (RT-PCR) and the genetic analysis of the nucleotide sequence, mostly of the VP1-coding region180
Detection of FMDV-specific antibody in animals previously exposed to the virus. The VNT is usually used as a confirmatory test for sera found positive by ELISA.88
These techniques are primarily suited for well-equipped laboratories which are usually either national or regional reference laboratories.30 The virus cell culture system, for example, requires careful handling of specimens to prevent environmental and cross-contamination, trained personnel, and a BSL3 (biosecurity level 3) laboratory. The success of virus isolation is dependent on the sample quality and requires special transport conditions from the sampling point to the laboratory.176 Both the solid-phase competition ELISA and the liquid-phase blocking ELISA for serological detection of FMDV-specific antibodies against structural proteins are relatively simple procedures and easily implementable in diagnostic laboratories in endemic regions.181–183 The VNT on the other hand, requires technical skill to be performed accurately and is dependent on cell culture facilities,184,185 which may not be conducive for laboratories in endemic regions.
Molecular techniques, like RT-PCR and real-time RT-PCR have the advantage that a wide range of samples (eg, esophageal/pharyngeal scrapings, tissue, and serum) can be tested rapidly186,187 and are now widely considered to be front-line diagnostic tests for the detection of FMD.188 Furthermore, RT-PCR is the first step to determine the nucleotide sequences and contributes to molecular epidemiology studies and provides a tool to support regional and country-wide FMD control programs in an outbreak situation. To various degrees, these technologies are being implemented and applied in many diagnostic laboratories across the African continent, although these approaches are sometimes still too expensive to be implemented as routine across the whole of the African environment.
One of the limitations in the early diagnosis of FMD in an endemic situation is that outbreaks often occur in remote areas where the nearest veterinarian and national laboratory can be many kilometers away.30 This can delay the laboratory results for a month or longer. Lateral flow devices (LFDs) are immuno-chromatographic tests that allow the diagnosis of FMDV at the site of a suspected outbreak. Different LFDs have been developed and evaluated and are either serotype specific189 or can detect all seven FMDV serotypes.190,191 The LFD can utilize vesicular fluid or vesicular epithelial suspensions but not nasal swabs or sera.189,191 The method makes use of capture and detection MAbs or specific polyclonal antisera on a strip test, and studies thus far have shown the test to be as sensitive and specific as the ELISA; however, the sensitivity of the strip test may differ for the various FMDV strains.192 The use of LFDs will allow for immediate determination of an FMDV-positive or -negative result and thus will facilitate decisions to be undertaken immediately for FMDV control in case of a positive result. Furthermore, only those samples that test positive on the field devices can be sent to the laboratory for further confirmatory tests, subtyping, and genetic characterization.
Portable real-time PCR platforms offer many advantages in endemic point-of-disease situations.193–195 These platforms, which can be utilized by non-specialists, are designed to perform all the steps of an RT-PCR test (eg, capable of nucleic acid extraction and performing RT-PCR). The Enigma FL field laboratory platform (Enigma Diagnostics, Wiltshire, UK) is one such system capable of nucleic acid extraction, PCR thermocycling, and analysis of data without the requirement for user intervention and has been tested for FMD diagnosis.193,196 The question remains whether field-based assays can ensure the detection of new viruses as they continue to evolve in sub-Saharan Africa.
An alternative molecular detection technique is loop-mediated isothermal amplification (LAMP) assay, which has been widely used for the detection of RNA and DNA viruses that infect livestock.197,198 This approach is an autocycling, strand-displacement DNA-synthesis method performed by the large fragment of Bst DNA polymerase at a single temperature. To facilitate the LAMP assay in the field and because the RT-LAMP products are generated in abundance, the results can be visualized by the naked eye, either in the form of visual turbidity or visual fluorescence. The diagnostic performance of real-time RT-PCR and RT-LAMP are governed by the nucleotide variability of the genome signatures between the different serotypes and genetic lineages of the virus. Since the primer recognition sites used for RT-LAMP have a larger footprint than RT-PCR assays, designing assays that can accommodate the large range of sequence variability of FMDV can be challenging, particularly for SAT strains. There have been many studies that have shown the high specificity and high amplification efficiency of RT-LAMP.198–200 Furthermore, Yamazaki et al201 developed the first application of a multiplex RT-LAMP approach to accommodate the high sequence variability encountered in RNA virus genomes and found the analytical sensitivity to be comparable to the singleplex RT-LAMP assays.198 The combination of a simple method to prepare template RNA and RT-LAMP can prove useful for sub-Saharan Africa in the field or laboratory where expensive equipment may not be readily available.
Additional novel diagnostic assays such as biosensors,202 microarrays,203 gold nanoparticle improved immuno-PCR,204 and nucleic acid sequence-based amplification205,206 have been shown to enable rapid and reliable diagnosis, surveillance screening, and strain typing for FMDV. Although these assays have promising capabilities for sub-Saharan Africa and can improve many of the current problems faced, there are also many limiting factors that prevent the routine use of certain assays. For example, many of the novel assays are still to be optimized for the FMDV SAT serotypes where a high degree of sequence variability exists, the costs involved per test will determine how widely these assays will be used, the field-based novel assays will require training of personnel, and some laboratory-based tests require specialized equipment which is not readily available and personnel capable of correctly interpreting and analyzing the datasets produced.
Thus, open communication between national and international reference labs becomes important as a support system to endemic regions. Also, the possibility of diagnostic banks where diagnostic kits become readily available in outbreak situations can alleviate many of the problems faced by endemic regions.
Conclusion
Most countries in Africa are ill equipped to control transboundary animal diseases such as FMD because of the lack of infrastructure and financial resources, ineffective animal health authorities, civil unrest, and even military conflict. Furthermore, most governments ascribe low priority to the control of animal diseases in the face of many other pressing problems like human health and education. Even in countries that controlled FMD successfully in the past, like South Africa, Namibia, Botswana, and Zimbabwe, an increase in the amount of FMD outbreaks has been observed over the last two decades. However, for other countries, this may reflect the situation since the 1960s. In many others, the current situations regarding their FMD status is unknown, and the majority of outbreaks remain unrecorded. There are several reasons for this: 1) countries may not be involved in intercontinental trade in animals and animal products and therefore have little incentive to report FMD outbreaks; 2) in many regions where pastoral systems predominate, surveillance systems are inadequate or nonexistent; 3) transporting suitable material from the field to a suitably equipped laboratory to confirm and type the occurrence of FMDV is logistically complicated and expensive; and 4) very few laboratories in Africa have the means to diagnose FMD adequately.
Nevertheless, there have been successes in parts of Africa – for example, Botswana, Namibia, and South Africa have FMD-free zones certified by the OIE and protected by a vaccination zone surrounding high-risk areas. Furthermore, Morocco, Algeria, and Tunisia have OIE-endorsed control plans. These successes indicate that FMD control is possible in Africa and can be extended.
More effective control of FMD in Africa can be achieved if primary endemic areas and factors that influence disease dissemination are known to assist in the design of appropriate control strategies. This will assist the design of targeted, area-wide or ecosystem-based disease control strategies, which may include more effective movement control, improve decisions on appropriate vaccine strains, and improved vaccines and point-of-disease diagnostic assays. Various new and exciting technologies to improve vaccinology, vaccine matching, and diagnostic tests have seen the light. However, for these technologies to make an impact in the control of FMD in Africa, they should be tailored to the unique environment and needs of Africa.
Many countries are embarking on the stepwise PCP approach to improve their FMD control capacity in a sustainable manner. Different regions in sub-Saharan Africa are at different developmental stages of control and thus face unique challenges and priorities in terms of FMD control. Progressive control of FMD may be achieved if founded on sound epidemiological understanding of the disease that is ecosystem specific.
Acknowledgments
The authors are grateful to Dr L Rotherham for logistical input in the manuscript and Dr Keith Sumption from the EuFMD for information on countries, PCP stages. The work reported here was partly funded by the Wellcome Trust Grant WT087546MA to Southern African Centre for Infectious Disease Surveillance.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Grubman MJ, Baxt B. Foot-and-mouth disease. Clin Microbiol Rev. 2004;17(2):465–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scoones I, Bishi A, Mapitse N, et al. Foot-and-mouth disease and market access: challenges for the beef industry in southern Africa. Pastoralism. 2010;1(2):135–164. [Google Scholar]
- 3.Miguel E, Grosbois V, Caron A, et al. Wildlife-livestock contacts: frequency of interactions contact and foot-and-mouth disease dynamic in cattle populations at the periphery of Transfrontier Conservation Areas in Southern Africa. Ecosphere. 2013;4(4):art51. [Google Scholar]
- 4.Ferguson KJ, Cleaveland S, Haydon DT, et al. Evaluating the potential for the environmentally sustainable control of foot and mouth disease in sub-Saharan Africa. Ecohealth. 2013;10(3):314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutmoller P, Barteling SS, Olascoaga RC, Sumption KJ. Control and eradication of foot-and-mouth disease. Virus Res. 2003;91(1):101–144. [DOI] [PubMed] [Google Scholar]
- 6.Perry BD, Rich KM. Poverty impacts of foot-and-mouth disease and the poverty reduction implications of its control. Vet Rec. 2007;160(7):238–241. [DOI] [PubMed] [Google Scholar]
- 7.Doel TR. FMD vaccines. Virus Res. 2003;91(1):81–99. [DOI] [PubMed] [Google Scholar]
- 8.Condy JB, Hedger RS, Hamblin C, Barnett IT. The duration of the foot-and-mouth disease virus carrier state in African buffalo i) in the individual animal and ii) in a free-living herd. Comp Immunol Microbiol Infect Dis. 1985;8(3–4):259–265. [DOI] [PubMed] [Google Scholar]
- 9.Hedger RS, Condy JB, Golding SM. Infection of some species of African wild life with foot-and-mouth disease virus. J Comp Pathol. 1972;82(4):455–461. [DOI] [PubMed] [Google Scholar]
- 10.Hedger RS. Foot-and-mouth disease and the African buffalo (Syncerus caffer). J Comp Pathol. 1972;82(1):19–28. [DOI] [PubMed] [Google Scholar]
- 11.Bengis RG, Kock RA, Fischer J. Infectious animal diseases: the wildlife/livestock interface. Rev Sci Tech. 2002;21(1):53–65. [DOI] [PubMed] [Google Scholar]
- 12.Thomson GR, Vosloo W, Bastos AD. Foot and mouth disease in wildlife. Virus Res. 2003;91(1):145–161. [DOI] [PubMed] [Google Scholar]
- 13.Vosloo W, Bastos AD, Kirkbride E, et al. Persistent infection of African buffalo (Syncerus caffer) with SAT-type foot-and-mouth disease viruses: rate of fixation of mutations, antigenic change and interspecies transmission. J Gen Virol. 1996;77:1457–1467. [DOI] [PubMed] [Google Scholar]
- 14.Vosloo W, Bastos AD, Sangare O, Hargreaves SK, Thomson GR. Review of the status and control of foot-and-mouth disease in sub-Saharan Africa. Rev Sci Tech. 2002;21(3):437–449. [DOI] [PubMed] [Google Scholar]
- 15.Ayebazibwe C, Mwiine FN, Tjornehoj K, et al. The role of African buffalos (Syncerus caffer) in the maintenance of foot-and-mouth disease in Uganda. BMC Vet Res. 2010;6:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawe PS, Flanagan FO, Madekurozwa RL, et al. Natural transmission of foot-and-mouth disease virus from African buffalo (Syncerus caffer) to cattle in a wildlife area of Zimbabwe. Vet Rec. 1994;134(10):230–232. [DOI] [PubMed] [Google Scholar]
- 17.Bastos AD, Boshoff CI, Keet DF, Bengis RG, Thomson GR. Natural transmission of foot-and-mouth disease virus between African buffalo (Syncerus caffer) and impala (Aepyceros melampus) in the Kruger National Park, South Africa. Epidemiol Infect. 2000;124(3):591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vosloo W, Thomson GR. Natural habitats in which foot-and-mouth disease viruses are maintained In: Domingo E, Sobrino F, editors. Natural Habitats in Which Foot-and-Mouth Disease Viruses are Maintained. Norfolk, United Kingdom: Horizon Bioscience; 2004: 384–410. [Google Scholar]
- 19.Thomson GR. Foot-and-mouth disease In: Coetzer JAW, Thomson GR, Tustin RC, editors. Foot-and-Mouth Disease. Cape Town: Oxford University Press; 1994: 825–852. [Google Scholar]
- 20.Ahmed HA, Salem SA, Habashi AR, et al. Emergence of foot-and-mouth disease virus SAT 2 in Egypt during 2012. Transbound Emerg Dis. 2012;59(6):476–481. [DOI] [PubMed] [Google Scholar]
- 21.Thomson GR, Bengis RG, Brown CC. Picornavirus infections In: Williams ES, Barker IK, editors. Infectious Diseases of Wild Mammals. Iowa: Iowa State University Press; 2001: 119–130. [Google Scholar]
- 22.Chardonnet P, des Clers B, Fischer J, et al. The value of wildlife. Rev Sci Tech. 2002;21(1):15–51. [DOI] [PubMed] [Google Scholar]
- 23.Brückner GK, Vosloo W, Du Plessis BJ, et al. Foot-and-mouth disease: the experience of South Africa. Rev Sci Tech. 2002;21(3):751–764. [DOI] [PubMed] [Google Scholar]
- 24.de Garine-Wichatitsky M, Miguel E, Mukamuri B, et al. Coexisting with wildlife in transfrontier conservation areas in Zimbabwe: cattle owners’ awareness of disease risks and perceptions of the role played by wildlife. Comp Immunol Microbiol Infect Dis. 2013;36(3):321–332. [DOI] [PubMed] [Google Scholar]
- 25.Jori F, Vosloo W, Du Plessis B, et al. A qualitative risk assessment of factors contributing to foot and mouth disease outbreaks in cattle along the western boundary of the Kruger National Park. Rev Sci Tech. 2009;28(3):917–931. [DOI] [PubMed] [Google Scholar]
- 26.WAHID Interface [database on the Internet]. 2013. Available from: http://www.oie.int/wahis_2/public/wahid.php/Wahidhome/Home. Accessed June 14, 2014.
- 27.Knowles NJ, Wadsworth J, Reid SM, et al. Foot-and-mouth disease virus serotype A in Egypt. Emerg Infect Dis. 2007;13(10):1593–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown F. The history of research in foot-and-mouth disease. Virus Res. 2003;91(1):3–7. [DOI] [PubMed] [Google Scholar]
- 29.Rweyemamu M, Roeder P, Mackay D, et al. Epidemiological patterns of foot-and-mouth disease worldwide. Transbound Emerg Dis. 2008;55(1):57–72. [DOI] [PubMed] [Google Scholar]
- 30.Rweyemamu MM, Garland AJM. The design of vaccines and diagnostics for use in endemic FMD settings In: Global Endemic FMD roadmap workshop; Agra: 2006. [Google Scholar]
- 31.Perry B, Sones K. Science for development. Poverty reduction through animal health. Science. 2007;315(5810):333–334. [DOI] [PubMed] [Google Scholar]
- 32.Knight-Jones TJ, Rushton, J. The economic impacts of foot and mouth disease – what are they, how big are they and where do they occur? Prev Vet Med. 2013;112(3–4):161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tully DC, Fares MA. The tale of a modern animal plague: tracing the evolutionary history and determining the time-scale for foot and mouth disease virus. Virology. 2008;382(2):250–256. [DOI] [PubMed] [Google Scholar]
- 34.Paton DJ, Sumption KJ, Charleston B. Options for control of foot-and-mouth disease: knowledge, capability and policy. Philos Trans R Soc Lond B Biol Sci. 2009;364(1530):2657–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valarcher J-F, Knowles N, Fernandez R, et al. Global FMD situation 2003–2004 In: Report of the Session of the Research Group of the Standing Technical Committee of the European Commission for the Control of Foot-and-Mouth. Chania, Greece; 2004: 137–148. [Google Scholar]
- 36.Donaldson A. The global status of foot-and-mouth disease and its relevance to control and eradication efforts in South-East Asia Report of 33rd session of the European Commission for the Control of Foot-and-Mouth Disease (EUFMD Commission); April 7–9, 1999; Rome. [Google Scholar]
- 37.Sangula AK, Belsham GJ, Muwanika VB, et al. Co-circulation of two extremely divergent serotype SAT 2 lineages in Kenya highlights challenges to foot-and-mouth disease control. Arch Virol. 2010;155(10):1625–1630. [DOI] [PubMed] [Google Scholar]
- 38.Kasanga CJ, Sallu R, Kivaria F, et al. Foot-and-mouth disease virus serotypes detected in Tanzania from 2003 to 2010: conjectured status and future prospects. Onderstepoort J Vet Res. 2012;79(2):462. [DOI] [PubMed] [Google Scholar]
- 39.Kasanga CJ, Wadsworth J, Mpelumbe-Ngeleja CA, et al. Molecular characterization of foot-and-mouth disease viruses collected in Tanzania between 1967 and 2009. Transbound Emerg Dis. Epub January 25, 2014. [DOI] [PubMed]
- 40.Hall MD, Knowles NJ, Wadsworth J, Rambaut A, Woolhouse ME. Reconstructing geographical movements and host species transitions of foot-and-mouth disease virus serotype SAT 2. MBio. 2013;4(5): e00591–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cottam EM, Thebaud G, Wadsworth J, et al. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proc Biol Sci. 2008;275(1637):887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vosloo W, Thompson PN, Botha B, Bengis RG, Thomson GR. Longitudinal study to investigate the role of impala (Aepyceros melampus) in foot-and-mouth disease maintenance in the Kruger National Park, South Africa. Transbound Emerg Dis. 2009;56(1–2): 18–30. [DOI] [PubMed] [Google Scholar]
- 43.Molla B, Ayelet G, Asfaw Y, et al. Epidemiological study on foot-and-mouth disease in cattle: seroprevalence and risk factor assessment in South Omo zone, south-western Ethiopia. Transbound Emerg Dis. 2010;57(5):340–347. [DOI] [PubMed] [Google Scholar]
- 44.Megersa B, Beyene B, Abunna F, et al. Risk factors for foot and mouth disease seroprevalence in indigenous cattle in Southern Ethiopia: the effect of production system. Trop Anim Health Prod. 2009;41(6): 891–898. [DOI] [PubMed] [Google Scholar]
- 45.Sutmoller P, Thomson GR, Hargreaves SK, Foggin CM, Anderson EC. The foot-and-mouth disease risk posed by African buffalo within wildlife conservancies to the cattle industry of Zimbabwe. Prev Vet Med. 2000;44(1–2):43–60. [DOI] [PubMed] [Google Scholar]
- 46.Hargreaves SK, Foggin CM, Anderson EC, et al. An investigation into the source and spread of foot and mouth disease virus from a wildlife conservancy in Zimbabwe. Rev Sci Tech. 2004;23(3):783–790. [DOI] [PubMed] [Google Scholar]
- 47.AHEAD. Special FMD bulletin: FMD occurrence in southern Africa shows a worrying upward trend with unusual patterns of transboundary spread. 2011. Available from: http://www.afrivip.org/education/livestock/disease-bulletins/fmd-bulletin/2014/materials. Accessed July 24, 2014.
- 48.Sinkala Y, Pfeiffer D, Kasanga C, et al. Foot-and-mouth disease control in Zambia: a review of the current situation. Onderstepoort J Vet Res. 2012;79(2):109. [Google Scholar]
- 49.Sangula AK, Belsham GJ, Muwanika VB, et al. Evolutionary analysis of foot-and-mouth disease virus serotype SAT 1 isolates from east Africa suggests two independent introductions from southern Africa. BMC Evol Biol. 2010;10:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balinda SN, Sangula AK, Heller R, et al. Diversity and transboundary mobility of serotype O foot-and-mouth disease virus in East Africa: implications for vaccination policies. Infect Genet Evol. 2010;10(7): 1058–1065. [DOI] [PubMed] [Google Scholar]
- 51.Balinda SN, Belsham GJ, Masembe C, et al. Molecular characterization of SAT 2 foot-and-mouth disease virus from post-outbreak slaughtered animals: implications for disease control in Uganda. Epidemiol Infect. 2010;138(8):1204–1210. [DOI] [PubMed] [Google Scholar]
- 52.Sangula AK, Siegismund HR, Belsham GJ, et al. Low diversity of foot-and-mouth disease serotype C virus in Kenya: evidence for probable vaccine strain re-introductions in the field. Epidemiol Infect. 2011;139(2):189–196. [DOI] [PubMed] [Google Scholar]
- 53.Bastos AD, Anderson EC, Bengis RG, et al. Molecular epidemiology of SAT3-type foot-and-mouth disease. Virus Genes. 2003;27(3): 283–290. [DOI] [PubMed] [Google Scholar]
- 54.Sumption K, Rweyemamu M, Wint W. Incidence and distribution of foot-and-mouth disease in Asia, Africa and South America; combining expert opinion, official disease information and livestock populations to assist risk assessment. Transbound Emerg Dis. 2008;55(1):5–13. [DOI] [PubMed] [Google Scholar]
- 55.Ayebazibwe C, Mwiine FN, Balinda SN, et al. Antibodies against foot-and-mouth disease (FMD) virus in African buffalos (Syncerus caffer) in selected National Parks in Uganda (2001–2003). Transbound Emerg Dis. 2010;57(4):286–292. [DOI] [PubMed] [Google Scholar]
- 56.wrlfmd.org [homepage on the Internet]. Ref Lab Reports; 2005–2011. Available from: http://www.wrlfmd.org/ref_labs/fmd_ref_lab_reports.htm Accessed June 15, 2014.
- 57.Bronsvoort BM, Radford AD, Tanya VN, et al. Molecular epidemiology of foot-and-mouth disease viruses in the Adamawa province of Cameroon. J Clin Microbiol. 2004;42(5):2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haydon DT, Samuel AR, Knowles NJ. The generation and persistence of genetic variation in foot-and-mouth disease virus. Prev Vet Med. 2001;51(1–2):111–124. [DOI] [PubMed] [Google Scholar]
- 59.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. [DOI] [PubMed] [Google Scholar]
- 60.Holland J, Spindler K, Horodyski F, et al. Rapid evolution of RNA genomes. Science. 1982;215(4540):1577–1585. [DOI] [PubMed] [Google Scholar]
- 61.Maree FF, Blignaut B, Esterhuysen JJ, et al. Predicting antigenic sites on the foot-and-mouth disease virus capsid of the South African Territories types using virus neutralization data. J Gen Virol. 2011;92(Pt 10):2297–2309. [DOI] [PubMed] [Google Scholar]
- 62.Acharya R, Fry E, Stuart D, et al. The three-dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature. 1989;337(6209):709–716. [DOI] [PubMed] [Google Scholar]
- 63.Logan D, Abu-Ghazaleh R, Blakemore W, et al. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature. 1993;362(6420):566–568. [DOI] [PubMed] [Google Scholar]
- 64.Mateu MG, Camarero JA, Giralt E, Andreu D, Domingo E. Direct evaluation of the immunodominance of a major antigenic site of foot-and-mouth disease virus in a natural host. Virology. 1995;206(1): 298–306. [DOI] [PubMed] [Google Scholar]
- 65.Usherwood EJ, Nash AA. Lymphocyte recognition of picornaviruses. J Gen Virol. 1995;76:499–508. [DOI] [PubMed] [Google Scholar]
- 66.Reeve R, Blignaut B, Esterhuysen JJ, et al. Sequence-based prediction for vaccine strain selection and identification of antigenic variability in foot-and-mouth disease virus. PLoS Comput Biol. 2010;6(12): e1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lea S, Abu-Ghazaleh R, Blakemore W, et al. Structural comparison of two strains of foot-and-mouth disease virus subtype O1 and a laboratory antigenic variant, G67. Structure. 1995;3(6):571–580. [DOI] [PubMed] [Google Scholar]
- 68.Kitson JD, McCahon D, Belsham GJ. Sequence analysis of monoclonal antibody resistant mutants of type O foot-and-mouth disease virus: evidence for the involvement of the three surface exposed capsid proteins in four antigenic sites. Virology. 1990;179(1):26–34. [DOI] [PubMed] [Google Scholar]
- 69.Thomas AA, Woortmeijer RJ, Barteling SJ, Meloen RH. Evidence for more than one important, neutralizing site on foot-and-mouth disease virus. Brief report. Arch Virol. 1988;99(3–4):237–242. [DOI] [PubMed] [Google Scholar]
- 70.Xie QC, McCahon D, Crowther JR, Belsham GJ, McCullough KC. Neutralization of foot-and-mouth disease virus can be mediated through any of at least three separate antigenic sites. J Gen Virol. 1987;68: 1637–1647. [DOI] [PubMed] [Google Scholar]
- 71.Esterhuysen JJ, Thomson GR, Ashford WA, et al. The suitability of a rolled BHK-21 monolayer system for the production of vaccines against the SAT types of foot-and-mouth disease virus. I. Adaptation of virus isolates to the system, immunogen yields achieved and assessment of subtype cross reactivity. Onderstepoort J Vet Res. 1988;55(2): 77–84. [PubMed] [Google Scholar]
- 72.Dunn CS, Samuel AR, Pullen LA, Anderson J. The biological relevance of virus neutralisation sites for virulence and vaccine protection in the guinea pig model of foot-and-mouth disease. Virology. 1998;247(1):51–61. [DOI] [PubMed] [Google Scholar]
- 73.Juleff N, Windsor M, Lefevre EA, et al. Foot-and-mouth disease virus can induce a specific and rapid CD4+ T-cell-independent neutralizing and isotype class-switched antibody response in naïve cattle. J Virol. 2009;83(8):3626–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baxt B, Vakharia V, Moore DM, Franke AJ, Morgan DO. Analysis of neutralizing antigenic sites on the surface of type A12 foot-and-mouth disease virus. J Virol. 1989;63(5):2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolwell C, Clarke BE, Parry NR, et al. Epitope mapping of foot-and-mouth disease virus with neutralizing monoclonal antibodies. J Gen Virol. 1989;70:59–68. [DOI] [PubMed] [Google Scholar]
- 76.Thomas AA, Woortmeijer RJ, Puijk W, Barteling SJ. Antigenic sites on foot-and-mouth disease virus type A10. J Virol. 1988;62(8): 2782–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crowther JR, Farias S, Carpenter WC, Samuel AR. Identification of a fifth neutralizable site on type O foot-and-mouth disease virus following characterization of single and quintuple monoclonal antibody escape mutants. J Gen Virol. 1993;74:1547–1553. [DOI] [PubMed] [Google Scholar]
- 78.Mateu MG, Martinez MA, Capucci L, et al. A single amino acid substitution affects multiple overlapping epitopes in the major antigenic site of foot-and-mouth disease virus of serotype C. J Gen Virol. 1990;71: 629–637. [DOI] [PubMed] [Google Scholar]
- 79.Sanyal A, Venkataramanan R, Pattnaik B. Antigenic features of foot-and-mouth disease virus serotype Asia1 as revealed by monoclonal antibodies and neutralization-escape mutants. Virus Res. 1997;50(2): 107–117. [DOI] [PubMed] [Google Scholar]
- 80.Grazioli S, Fallacara F, Brocchi E. Mapping of antigenic sites of foot-and-mouth disease virus serotype Asia 1 and relationships with sites described in other serotypes. J Gen Virol. 2013;94:559–569. [DOI] [PubMed] [Google Scholar]
- 81.Borley DW, Mahapatra M, Paton DJ, et al. Evaluation and use of in-silico structure-based epitope prediction with foot-and-mouth disease virus. PLoS One. 2013;8(5):e61122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aggarwal N, Barnett PV. Antigenic sites of foot-and-mouth disease virus (FMDV): an analysis of the specificities of anti-FMDV antibodies after vaccination of naturally susceptible host species. J Gen Virol. 2002;83:775–782. [DOI] [PubMed] [Google Scholar]
- 83.Fowler VL, Knowles NJ, Paton DJ, Barnett PV. Marker vaccine potential of a foot-and-mouth disease virus with a partial VP1 G-H loop deletion. Vaccine. 2010;28(19):3428–3434. [DOI] [PubMed] [Google Scholar]
- 84.Mahapatra M, Hamblin P, Paton DJ. Foot-and-mouth disease virus epitope dominance in the antibody response of vaccinated animals. J Gen Virol. 2012;93:488–493. [DOI] [PubMed] [Google Scholar]
- 85.Grazioli S, Moretti M, Barbieri I, Crosatti M, Brocchi E. Use of monoclonal antibodies to identify and map new antigenic determinants involved in neutralization of FMD viruses type SAT 1 and SAT 2 In: European Commission for the Control of Foot-and-Mouth Disease: International control of Foot-and-Mouth disease: Tools, Trends and perspectives. Paphos, Cyprus: Food and Agriculture Organization; 2006: 287–297.
- 86.Crowther JR, Rowe CA, Butcher R. Characterization of monoclonal antibodies against a type SAT 2 foot-and-mouth disease virus. Epidemiol Infect. 1993;111(2):391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Opperman PA, Maree FF, Van Wyngaardt W, Vosloo W, Theron J. Mapping of antigenic determinants on a SAT2 foot-and-mouth disease virus using chicken single-chain antibody fragments. Virus Res. 2012;167(2):370–379. [DOI] [PubMed] [Google Scholar]
- 88.Paton DJ, Valarcher JF, Bergmann I, et al. Selection of foot-and-mouth disease vaccine strains-a review. Rev Sci Tech. 2005;24(3):981–993. [PubMed] [Google Scholar]
- 89.Cox SJ, Voyce C, Parida S, et al. Protection against direct-contact challenge following emergency FMD vaccination of cattle and the effect on virus excretion from the oropharynx. Vaccine. 2005;23(9): 1106–1113. [DOI] [PubMed] [Google Scholar]
- 90.Mattion N, Konig G, Seki C, et al. Reintroduction of foot-and-mouth disease in Argentina: characterisation of the isolates and development of tools for the control and eradication of the disease. Vaccine. 2004;22(31–32):4149–4162. [DOI] [PubMed] [Google Scholar]
- 91.Goris N, Merkelbach-Peters P, Diev VI, et al. European Pharmacopoeia foot-and-mouth disease vaccine potency testing in cattle: between test variability and its consequences. Vaccine. 2007;25(17): 3373–3379. [DOI] [PubMed] [Google Scholar]
- 92.Alonso Fernandez A, Casas Olascoaga R, Astudillo VM, Söndahl MS, Gomes I, Vianna Filho YL. Updating of foot-and-mouth disease virus strains of epidemiological importance in South America. Bol Centr Panam Fiebre Aftosa. 1987;53:11–18. [Google Scholar]
- 93.Mattion N, Goris N, Willems T, et al. Some guidelines for determining foot-and-mouth disease vaccine strain matching by serology. Vaccine. 2009;27(5):741–747. [DOI] [PubMed] [Google Scholar]
- 94.oie.int [homepage on the Internet]. Manual of diagnostic tests and vaccines for terrestrial animals. Paris: Office International des Epizooties; 2012. Available from: http://www.oie.int/manual-of-diagnostic-tests-and-vaccines-for-terrestrial-animals. Accessed June 6, 2014. [Google Scholar]
- 95.Rweyemamu MM, Booth JC, Head M, Pay TW. Microneutralization tests for serological typing and subtyping of foot-and-mouth disease virus strains. J Hyg (Lond). 1978;81(1):107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rweyemamu MM. Antigenic variation in foot-and-mouth disease: studies based on the virus neutralization reaction. J Biol Stand. 1984;12(3):323–337. [DOI] [PubMed] [Google Scholar]
- 97.Ludi A, Rodriguez L. Novel approaches to foot-and-mouth disease vaccine development. Dev Biol (Basel). 2013;135:107–116. [DOI] [PubMed] [Google Scholar]
- 98.Ludi AB, Horton DL, Li Y, et al. Antigenic variation of foot-and-mouth disease virus serotype A. J Gen Virol. 2014;95:384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maree F, Chitray M, Blignaut B, et al. Report to FAO: improved control of FMD in SADC. (MTF/INT/003/EEC). 2010. Available from: http://www.fao.org/ag/againfo/commissions/docs/research_group/erice/APPENDIX_17.pdf. Accessed July 24, 2014.
- 100.Brehm KE, Kumar N, Thulke HH, Haas B. High potency vaccines induce protection against heterologous challenge with foot-and-mouth disease virus. Vaccine. 2008;26(13):1681–1687. [DOI] [PubMed] [Google Scholar]
- 101.Hunter P. Vaccination as a means of control of foot-and-mouth disease in sub-Saharan Africa. Vaccine. 1998;16(2–3):261–264. [DOI] [PubMed] [Google Scholar]
- 102.Kitching RP, Rendle R, Ferris NP. Rapid correlation between field isolates and vaccine strains of foot-and-mouth disease virus. Vaccine. 1988;6(5):403–408. [DOI] [PubMed] [Google Scholar]
- 103.Ferris NP, Donaldson AI. The World Reference Laboratory for Foot and Mouth Disease: a review of thirty-three years of activity (1958–1991). Rev Sci Tech. 1992;11(3):657–684. [DOI] [PubMed] [Google Scholar]
- 104.Samuel AR, Ouldridge EJ, Arrowsmith AE, Kitching RP, Knowles NJ. Antigenic analysis of serotype O foot-and-mouth disease virus isolates from the Middle East, 1981 to 1988. Vaccine. 1990;8(4):390–396. [DOI] [PubMed] [Google Scholar]
- 105.Golding SM, Hedger RS, Talbot P. Radial immuno-diffusion and serum-neutralisation techniques for the assay of antibodies to swine vesicular disease. Res Vet Sci. 1976;20(2):142–147. [PubMed] [Google Scholar]
- 106.McCullough KC, De Simone F, Brocchi E, et al. Protective immune response against foot-and-mouth disease. J Virol. 1992;66(4): 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mulcahy G, Reid E, Dimarchi RD, Gale C, Doel TR. Maturation of functional antibody affinity in animals immunised with synthetic foot-and-mouth disease virus. Res Vet Sci. 1992;52(2):133–140. [DOI] [PubMed] [Google Scholar]
- 108.Capozzo AV, Wilda M, Bucafusco D, et al. Vesicular stomatitis virus glycoprotein G carrying a tandem dimer of foot-and-mouth disease virus antigenic site A can be used as DNA and peptide vaccine for cattle. Antiviral Res. 2011;92(2):219–227. [DOI] [PubMed] [Google Scholar]
- 109.Lavoria MA, Di-Giacomo S, Bucafusco D, et al. Avidity and subtyping of specific antibodies applied to the indirect assessment of heterologous protection against foot-and-mouth disease virus in cattle. Vaccine. 2012;30(48):6845–6850. [DOI] [PubMed] [Google Scholar]
- 110.Brito BP, Perez AM, Capozzo AV. Accuracy of traditional and novel serology tests for predicting cross-protection in foot-and-mouth disease vaccinated cattle. Vaccine. 2014;32(4):433–436. [DOI] [PubMed] [Google Scholar]
- 111.Kitching RP. Vaccination of calves against FMD in the presence of maternally derived antibody In: European Commission for the Control of Foot-and-Mouth Disease; Israel;1997:191–195. [Google Scholar]
- 112.Sa-Carvalho D, Rieder E, Baxt B, et al. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J Virol. 1997;71(7):5115–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao Q, Pacheco JM, Mason PW. Evaluation of genetically engineered derivatives of a Chinese strain of foot-and-mouth disease virus reveals a novel cell-binding site which functions in cell culture and in animals. J Virol. 2003;77(5):3269–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Salt JS. The carrier state in foot and mouth disease – an immunological review. Br Vet J. 1993;149(3):207–223. [DOI] [PubMed] [Google Scholar]
- 115.Sutmoller P, Gaggero A. Foot-and mouth diseases carriers. Vet Rec. 1965;77(33):968–969. [DOI] [PubMed] [Google Scholar]
- 116.Sutmoller P, Cottral GE, McVicar JW. A review of the carrier state in foot-and-mouth disease. Proc Annu Meet U S Anim Health Assoc. 1967;71:386–395. [PubMed] [Google Scholar]
- 117.Mackay DK, Forsyth MA, Davies PR, Salt JS. Antibody to the nonstructural proteins of foot-and-mouth disease virus in vaccinated animals exposed to infection. Vet Q. 1998;20 Suppl 2:S9–S11. [DOI] [PubMed] [Google Scholar]
- 118.Doel TR, Baccarini PJ. Thermal stability of foot-and-mouth disease virus. Arch Virol. 1981;70(1):21–32. [DOI] [PubMed] [Google Scholar]
- 119.Rweyemamu M, Roeder P, MacKay D, et al. Planning for the progressive control of foot-and-mouth disease worldwide. Transbound Emerg Dis. 2008;55(1):73–87. [DOI] [PubMed] [Google Scholar]
- 120.Bahneman HG, Mesquita JA. Oil-adjuvanted vaccine against foot and mouth disease. Bol Centro Panam Aftosa. 1987;53:25. [Google Scholar]
- 121.Dora JFP, Coelho Nunes JC, et al. Epidemic of foot-and-mouth disease in Bage, RS, Brazil, 1980. Evaluation of two systems of vaccination. Bol Centr Panam Fiebre Aftosa. 1984;49–50:11–17. [Google Scholar]
- 122.Cloete M, Dungu B, Van Staden LI, Ismail-Cassim N, Vosloo W. Evaluation of different adjuvants for foot-and-mouth disease vaccine containing all the SAT serotypes. Onderstepoort J Vet Res. 2008;75(1):17–31. [DOI] [PubMed] [Google Scholar]
- 123.Rieder E, Baxt B, Lubroth J, Mason PW. Vaccines prepared from chimeras of foot-and-mouth disease virus (FMDV) induce neutralizing antibodies and protective immunity to multiple serotypes of FMDV. J Virol. 1994;68(11):7092–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zibert A, Maass G, Strebel K, Falk MM, Beck E. Infectious foot-and-mouth disease virus derived from a cloned full-length cDNA. J Virol. 1990;64(6):2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Almeida MR, Rieder E, Chinsangaram J, et al. Construction and evaluation of an attenuated vaccine for foot-and-mouth disease: difficulty adapting the leader proteinase-deleted strategy to the serotype O1 virus. Virus Res. 1998;55(1):49–60. [DOI] [PubMed] [Google Scholar]
- 126.Van Rensburg HG, Henry TM, Mason PW. Studies of genetically defined chimeras of a European type A virus and a South African Territories type 2 virus reveal growth determinants for foot-and-mouth disease virus. J Gen Virol. 2004;85:61–68. [DOI] [PubMed] [Google Scholar]
- 127.Van Rensburg HG, Mason PW. Construction and evaluation of a recombinant foot-and-mouth disease virus: implications for inactivated vaccine production. Ann N Y Acad Sci. 2002;969:83–87. [DOI] [PubMed] [Google Scholar]
- 128.Blignaut B, Visser N, Theron J, Rieder E, Maree FF. Custom-engineered chimeric foot-and-mouth disease vaccine elicits protective immune responses in pigs. J Gen Virol. 2011;92:849–859. [DOI] [PubMed] [Google Scholar]
- 129.Maree FF, Blignaut B, de Beer TA, Visser N, Rieder EA. Mapping of amino acid residues responsible for adhesion of cell culture-adapted foot-and-mouth disease SAT type viruses. Virus Res. 2010;153(1):82–91. [DOI] [PubMed] [Google Scholar]
- 130.Fowler VL, Paton DJ, Rieder E, Barnett PV. Chimeric foot-and-mouth disease viruses: evaluation of their efficacy as potential marker vaccines in cattle. Vaccine. 2008;26(16):1982–1989. [DOI] [PubMed] [Google Scholar]
- 131.Preston KJ, Owens H, Mowat GN. Sources of variations encountered during the selection and production of three strains of FMD virus for the development of vaccine for use in Nigeria. J Biol Stand. 1982;10(1):35–45. [DOI] [PubMed] [Google Scholar]
- 132.Pay TWF, Rweyemamu MM, O’Reilly KJ. Experiences with Type SAT 2 foot-and-mouth disease vaccines in Southern Africa. In: XVth Conference of the Office International Des Epizzoties Permanent Commision on foot-and-mouth disease. 1978: 1–25.
- 133.Rieder E, Baxt B, Mason PW. Animal-derived antigenic variants of foot-and-mouth disease virus type A12 have low affinity for cells in culture. J Virol. 1994;68(8):5296–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Baranowski E, Ruiz-Jarabo CM, Sevilla N, et al. Cell recognition by foot-and-mouth disease virus that lacks the RGD integrin-binding motif: flexibility in aphthovirus receptor usage. J Virol. 2000;74(4):1641–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baranowski E, Sevilla N, Verdaguer N, et al. Multiple virulence determinants of foot-and-mouth disease virus in cell culture. J Virol. 1998;72(8):6362–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fry EE, Lea SM, Jackson T, et al. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 1999;18(3):543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fry EE, Newman JW, Curry S, et al. Structure of foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation. J Gen Virol. 2005;86:1909–1920. [DOI] [PubMed] [Google Scholar]
- 138.Jackson T, Ellard FM, Ghazaleh RA, et al. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70(8):5282–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Berryman S, Clark S, Kakker NK, et al. Positively charged residues at the five-fold symmetry axis of cell culture-adapted foot-and-mouth disease virus permit novel receptor interactions. J Virol. 2013;87(15): 8735–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mateo R, Diaz A, Baranowski E, Mateu MG. Complete alanine scanning of intersubunit interfaces in a foot-and-mouth disease virus capsid reveals critical contributions of many side chains to particle stability and viral function. J Biol Chem. 2003;278(42): 41019–41027. [DOI] [PubMed] [Google Scholar]
- 141.Mateo R, Luna E, Rincon V, Mateu MG. Engineering viable foot-and-mouth disease viruses with increased thermostability as a step in the development of improved vaccines. J Virol. 2008;82(24): 12232–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Filman DJ, Syed R, Chow M, et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8(5):1567–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Airaksinen A, Roivainen M, Hovi T. Coxsackievirus A9 VP1 mutants with enhanced or hindered A particle formation and decreased infectivity. J Virol. 2001;75(2):952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mateo R, Luna E, Mateu MG. Thermostable variants are not generally represented in foot-and-mouth disease virus quasispecies. J Gen Virol. 2007;88(Pt 3):859–864. [DOI] [PubMed] [Google Scholar]
- 145.Twomey T, Newman J, Burrage T, et al. Structure and immunogenicity of experimental foot-and-mouth disease and poliomyelitis vaccines. Vaccine. 1995;13(16):1603–1610. [DOI] [PubMed] [Google Scholar]
- 146.Bachrach HL, Moore DM, McKercher PD, Polatnick J. Immune and antibody responses to an isolated capsid protein of foot-and-mouth disease virus. J Immunol. 1975;115(6):1636–1641. [PubMed] [Google Scholar]
- 147.Kleid DG, Yansura D, Small B, et al. Cloned viral protein vaccine for foot-and-mouth disease: responses in cattle and swine. Science. 1981;214(4525):1125–1129. [DOI] [PubMed] [Google Scholar]
- 148.Strohmaier K, Franze R, Adam KH. Location and characterization of the antigenic portion of the FMDV immunizing protein. J Gen Virol. 1982;59:295–306. [DOI] [PubMed] [Google Scholar]
- 149.Bittle JL, Houghten RA, Alexander H, et al. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature. 1982;298(5869):30–33. [DOI] [PubMed] [Google Scholar]
- 150.Francis MJ, Hastings GZ, Brown F, et al. Immunological evaluation of the multiple antigen peptide (MAP) system using the major immunogenic site of foot-and-mouth disease virus. Immunology. 1991;73(3): 249–254. [PMC free article] [PubMed] [Google Scholar]
- 151.DiMarchi R, Brooke G, Gale C, et al. Protection of cattle against foot-and-mouth disease by a synthetic peptide. Science. 1986; 232(4750): 639–641. [DOI] [PubMed] [Google Scholar]
- 152.Nargi F, Kramer E, Mezencio J, et al. Protection of swine from foot-and-mouth disease with one dose of an all-D retro peptide. Vaccine. 1999;17(22):2888–2893. [DOI] [PubMed] [Google Scholar]
- 153.Clarke BE, Brown AL, Currey KM, et al. Potential secondary and tertiary structure in the genomic RNA of foot-and-mouth disease virus. Nucleic Acids Res. 1987;15(17):7067–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kit M, Kit S, Little SP, Di Marchi RD, Gale C. Bovine herpesvirus-1 (infectious bovine rhinotracheitis virus)-based viral vector which expresses foot-and-mouth disease epitopes. Vaccine. 1991;9(8): 564–572. [DOI] [PubMed] [Google Scholar]
- 155.Kitson JD, Burke KL, Pullen LA, Belsham GJ, Almond JW. Chimeric polioviruses that include sequences derived from two independent antigenic sites of foot-and-mouth disease virus (FMDV) induce neutralizing antibodies against FMDV in guinea pigs. J Virol. 1991;65(6):3068–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wong HT, Cheng SC, Chan EW, et al. Plasmids encoding foot-and-mouth disease virus VP1 epitopes elicited immune responses in mice and swine and protected swine against viral infection. Virology. 2000;278(1):27–35. [DOI] [PubMed] [Google Scholar]
- 157.Wong HT, Cheng SC, Sin FW, et al. A DNA vaccine against foot-and-mouth disease elicits an immune response in swine which is enhanced by co-administration with interleukin-2. Vaccine. 2002;20(21–22):2641–2647. [DOI] [PubMed] [Google Scholar]
- 158.Wigdorovitz A, Perez Filgueira DM, Robertson N, et al. Protection of mice against challenge with foot and mouth disease virus (FMDV) by immunization with foliar extracts from plants infected with recombinant tobacco mosaic virus expressing the FMDV structural protein VP1. Virology. 1999;264(1):85–91. [DOI] [PubMed] [Google Scholar]
- 159.Wigdorovitz A, Carrillo C, Dus Santos MJ, et al. Induction of a protective antibody response to foot and mouth disease virus in mice following oral or parenteral immunization with alfalfa transgenic plants expressing the viral structural protein VP1. Virology. 1999;255(2):347–353. [DOI] [PubMed] [Google Scholar]
- 160.Mulcahy G, Gale C, Robertson P, et al. Isotype responses of infected, virus-vaccinated and peptide-vaccinated cattle to foot-and-mouth disease virus. Vaccine. 1990;8(3):249–256. [DOI] [PubMed] [Google Scholar]
- 161.Mulcahy G, Pullen LA, Gale C, DiMarchi RD, Doel TR. Mouse protection test as a predictor of the protective capacity of synthetic foot-and-mouth disease vaccines. Vaccine. 1991;9(1):19–24. [DOI] [PubMed] [Google Scholar]
- 162.Taboga O, Tami C, Carrillo E, et al. A large-scale evaluation of peptide vaccines against foot-and-mouth disease: lack of solid protection in cattle and isolation of escape mutants. J Virol. 1997;71(4): 2606–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rodriguez A, Saiz JC, Novella IS, Andreu D, Sobrino F. Antigenic specificity of porcine T cell response against foot-and-mouth disease virus structural proteins: identification of T helper epitopes in VP1. Virology. 1994;205(1):24–33. [DOI] [PubMed] [Google Scholar]
- 164.Grubman MJ, Lewis SA, Morgan DO. Protection of swine against foot-and-mouth disease with viral capsid proteins expressed in heterologous systems. Vaccine. 1993;11(8):825–829. [DOI] [PubMed] [Google Scholar]
- 165.Rooisen J, Belsham GJ, Ryan MD, King AMQ, Vlak JM. Synthesis of foot-and-mouth disease virus capsid proteins in insect cells using baculovirus expression vectors. J Gen Virol. 1990;71:1703–1711. [DOI] [PubMed] [Google Scholar]
- 166.Porta C, Kotecha A, Burman A, et al. Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen. PLoS Pathog. 2013;9(3):e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sanz-Parra A, Sobrino F, Ley V. Infection with foot-and-mouth disease virus results in a rapid reduction of MHC class I surface expression. J Gen Virol. 1998;79:433–436. [DOI] [PubMed] [Google Scholar]
- 168.Berinstein A, Roivainen M, Hovi T, Mason PW, Baxt B. Antibodies to the vitronectin receptor (integrin αVβ3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J Virol. 1995;69(4): 2664–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zheng M, Jin N, Zhang H, et al. Construction and immunogenicity of a recombinant fowlpox virus containing the capsid and 3C protease coding regions of foot-and-mouth disease virus. J Virol Methods. 2006;136(1–2):230–237. [DOI] [PubMed] [Google Scholar]
- 170.Li X, Liu R, Tang H, et al. Induction of protective immunity in swine by immunization with live attenuated recombinant pseudorabies virus expressing the capsid precursor encoding regions of foot-and-mouth disease virus. Vaccine. 2008;26(22):2714–2722. [DOI] [PubMed] [Google Scholar]
- 171.Yao Q, Qian P, Cao Y, et al. Synergistic inhibition of pseudorabies virus replication by porcine alpha/beta interferon and gamma interferon in vitro. Eur Cytokine Netw. 2007;18(2):71–77. [DOI] [PubMed] [Google Scholar]
- 172.Yang CD, Liao JT, Lai CY, et al. Induction of protective immunity in swine by recombinant bamboo mosaic virus expressing foot-and-mouth disease virus epitopes. BMC Biotechnol. 2007;7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rodriguez LL, Gay CG. Development of vaccines toward the global control and eradication of foot-and-mouth disease. Expert Rev Vaccines. 2011;10(3):377–387. [DOI] [PubMed] [Google Scholar]
- 174.Grubman MJ, Moraes MP, Schutta C, et al. Adenovirus serotype 5-vectored foot-and-mouth disease subunit vaccines: the first decade. Future Virol. 2010;5(1):51–64. [Google Scholar]
- 175.Remond M, Kaiser C, Lebreton F. Diagnosis and screening of foot-and-mouth disease. Comp Immunol Microbiol Infect Dis. 2002;25(5–6):309–320. [DOI] [PubMed] [Google Scholar]
- 176.Jamal SM, Belsham GJ. Foot-and-mouth disease: past, present and future. Vet Res. 2013;44:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Abu Elzein EM, Crowther JR. Enzyme-labelled immunosorbent assay techniques in foot-and-mouth disease virus research. J Hyg (Lond). 1978;80(3):391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ferris NP, Dawson M. Routine application of enzyme-linked immunosorbent assay in comparison with complement fixation for the diagnosis of foot-and-mouth and swine vesicular diseases. Vet Microbiol. 1988;16(3):201–209. [DOI] [PubMed] [Google Scholar]
- 179.Ferris NP, Abrescia NG, Stuart DI, et al. Utility of recombinant integrin αvβ6 as a capture reagent in immunoassays for the diagnosis of foot-and-mouth disease. J Virol Methods. 2005;127(1):69–79. [DOI] [PubMed] [Google Scholar]
- 180.Di Nardo A, Knowles NJ, Paton DJ. Combining livestock trade patterns with phylogenetics to help understand the spread of foot and mouth disease in sub-Saharan Africa, the Middle East and Southeast Asia. Rev Sci Tech. 2011;30(1):63–85. [DOI] [PubMed] [Google Scholar]
- 181.Paiba GA, Anderson J, Paton DJ, et al. Validation of a foot-and-mouth disease antibody screening solid-phase competition ELISA (SPCE). J Virol Methods. 2004;115(2):145–158. [DOI] [PubMed] [Google Scholar]
- 182.King DP, Madi M, Mioulet V, et al. New technologies to diagnose and monitor infectious diseases of livestock: challenges for sub-Saharan Africa. Onderstepoort J Vet Res. 2012;79(2):456. [DOI] [PubMed] [Google Scholar]
- 183.Mackay DK, Bulut AN, Rendle T, Davidson F, Ferris NP. A solid-phase competition ELISA for measuring antibody to foot-and-mouth disease virus. J Virol Methods. 2001;97(1–2):33–48. [DOI] [PubMed] [Google Scholar]
- 184.Mackay DKJ, Rendle T, Kitching RP. FAO collaborate study on FMD antibody detection In: European Commision for the Control of Foot-and-Mouth Disease. Aldershot, UK: 1998: 148–157. [Google Scholar]
- 185.Donaldson AI, Kitching RP, Barnett PV. Foot and mouth disease, Part 2, Section 2.1., OIE List A diseases, Chapter 2.1.1. In: OIE Standards Commission, editors Manual of Standards for Diagnostic Tests and Vaccines. 4th ed. Paris;2000:77–92. [Google Scholar]
- 186.Amaral-Doel CM, Owen NE, Ferris NP, Kitching RP, Doel TR. Detection of foot-and-mouth disease viral sequences in clinical specimens and ethyleneimine-inactivated preparations by the polymerase chain reaction. Vaccine. 1993;11(4):415–421. [DOI] [PubMed] [Google Scholar]
- 187.Marquardt O, Straub OC, Ahl R, Haas B. Detection of foot-and-mouth disease virus in nasal swabs of asymptomatic cattle by RT-PCR within 24 hours. J Virol Methods. 1995;53(2–3):255–261. [DOI] [PubMed] [Google Scholar]
- 188.Reid SM, Ebert K, Bachanek-Bankowska K, et al. Performance of real-time reverse transcription polymerase chain reaction for the detection of foot-and-mouth disease virus during field outbreaks in the United Kingdom in 2007. J Vet Diagn Invest. 2009;21(3): 321–330. [DOI] [PubMed] [Google Scholar]
- 189.Yang M, Goolia M, Xu W, Bittner H, Clavijo A. Development of a quick and simple detection methodology for foot-and-mouth disease virus serotypes O, A and Asia 1 using a generic RapidAssay Device. Virol J. 2013;10:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reid SM, Ferris NP, Bruning A, et al. Development of a rapid chromatographic strip test for the pen-side detection of foot-and-mouth disease virus antigen. J Virol Methods. 2001;96(2):189–202. [DOI] [PubMed] [Google Scholar]
- 191.Ferris NP, Nordengrahn A, Hutchings GH, et al. Development and laboratory validation of a lateral flow device for the detection of foot-and-mouth disease virus in clinical samples. J Virol Methods. 2009;155(1):10–17. [DOI] [PubMed] [Google Scholar]
- 192.Oem JK, Ferris NP, Lee KN, et al. Simple and rapid lateral-flow assay for the detection of foot-and-mouth disease virus. Clin Vaccine Immunol. 2009;16(11):1660–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.King DP, Dukes JP, Reid SM, et al. Prospects for rapid diagnosis of foot-and-mouth disease in the field using reverse transcriptase-PCR. Vet Rec. 2008;162(10):315–316. [DOI] [PubMed] [Google Scholar]
- 194.Shirley MW, Charleston B, King DP. New opportunities to control livestock diseases in the post-genomics era. J Agric Sci. 2010;149: 115–121. [Google Scholar]
- 195.Sammin D, Ryan E, Ferris NP, et al. Options for decentralized testing of suspected secondary outbreaks of foot-and-mouth disease. Transbound Emerg Dis. 2010;57(4):237–243. [DOI] [PubMed] [Google Scholar]
- 196.Madi M, Hamilton A, Squirrell D, et al. Rapid detection of foot-and-mouth disease virus using a field-portable nucleic acid extraction and real-time PCR amplification platform. Vet J. 2012;193(1):67–72. [DOI] [PubMed] [Google Scholar]
- 197.James HE, Ebert K, McGonigle R, et al. Detection of African swine fever virus by loop-mediated isothermal amplification. J Virol Methods. 2010;164(1–2):68–74. [DOI] [PubMed] [Google Scholar]
- 198.Dukes JP, King DP, Alexandersen S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch Virol. 2006;151(6):1093–1106. [DOI] [PubMed] [Google Scholar]
- 199.Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin Chem. 2001;47(9):1742–1743. [PubMed] [Google Scholar]
- 200.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–229. [DOI] [PubMed] [Google Scholar]
- 201.Yamazaki W, Mioulet V, Murray L, et al. Development and evaluation of multiplex RT-LAMP assays for rapid and sensitive detection of foot-and-mouth disease virus. J Virol Methods. 2013;192(1–2): 18–24. [DOI] [PubMed] [Google Scholar]
- 202.Sanchez-Aparicio MT, Rosas MF, Ferraz RM, et al. Discriminating foot-and-mouth disease virus-infected and vaccinated animals by use of beta-galactosidase allosteric biosensors. Clin Vaccine Immunol. 2009;16(8):1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Baxi MK, Baxi S, Clavijo A, Burton KM, Deregt D. Microarray-based detection and typing of foot-and-mouth disease virus. Vet J. 2006;172(3):473–481. [DOI] [PubMed] [Google Scholar]
- 204.Ding YZ, Liu YS, Zhou JH, et al. A highly sensitive detection for foot-and-mouth disease virus by gold nanopariticle improved immuno-PCR. Virol J. 2011;8:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Collins RA, Ko LS, Fung KY, et al. A method to detect major serotypes of foot-and-mouth disease virus. Biochem Biophys Res Commun. 2002;297(2):267–274. [DOI] [PubMed] [Google Scholar]
- 206.Lau LT, Reid SM, King DP, et al. Detection of foot-and-mouth disease virus by nucleic acid sequence-based amplification (NASBA). Vet Microbiol. 2008;126(1–3):101–110. [DOI] [PubMed] [Google Scholar]
- 207.World Organisation for Animal Health (OIE). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris: Office International des Epizooties; 2009. [Google Scholar]
- 208.Mtei B, Mapitse N, Knopf L, Mokopasetso M, Bastiaensen P. Progressing towards Foot and Mouth Disease (FMD) Control and OIE recognized status of SADC Member States. Botswana: Regional Animal Health Centre for Southern Africa; 2011. Available from: http://www.oie.int/doc/ged/D10576.PDF. Accessed July 24, 2014. [Google Scholar]
- 209.Development of a Long Term Roadmap for the Progressive Control of FMD in Eastern Africa. Nairobi: Report of a Workshop held in Nairobi, Kenya March 5–6 2012. Convened by FAO, OIE, and AU-IBAR as a joint meeting and workshop; 2012 Available from: http://www.fao.org/fileadmin/user_upload/eufmd/docs/PCP_nairobi/Report.pdf. Accessed July 24, 2014. [Google Scholar]
- 210.Workshop on the development of a long term action plan (roadmap) for improved survelliance and control of foot-and-mouth disease in Africa. Nairobi: 26–30th January 2009. Available from: http://www.fao.org/ag/againfo/commissions/docs/Regional_FMD_meetings/Report_workshopNairobi1.pdf. Accessed July 24, 2014. [Google Scholar]
- 211.Report of the 39th Session of EuFMD, Rome 2011. Rome: Food and Agricultural Organization of the United Nations; 2011. Available from: http://www.fao.org/ag/againfo/commissions/eufmd/commissions/eufmd-home/reports/general-sessions/2011–39th-session/en/. Accessed July 25, 2014. [Google Scholar]