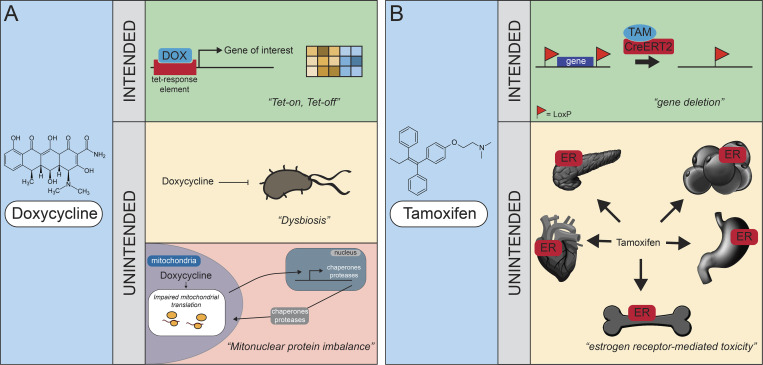
This Reproducibility Viewpoint discusses confounding factors of Tet-On/Tet-Off and Cre/loxP systems, including doxycycline-induced microbiome alterations, mitochondrial dysfunction, and tamoxifen-induced toxicity.
Abstract
Spatiotemporally regulated targeted gene manipulation is a common way to study the effect of gene variants on phenotypic traits, but the Cre/loxP and Tet-On/Tet-Off systems can affect whole-organism physiology and function due to off-target effects. We highlight some of these adverse effects, including whole-body endocrinology and disturbances in the gut microbiome and in mitochondrial and metabolic function.
Gain- and loss-of-function strategies to study gene function
Linking the effect of gene variants with phenotypes can be accomplished in various ways. Natural population variation can be assessed in humans using genome-wide association studies or in animal reference populations (Williams and Auwerx, 2015). The development of genetically modified animals has allowed researchers to elucidate the function of proteins of interest in the context of an intact organism. Overexpressing a gene of interest or introducing deletions or mutations with homologous recombination strategies (e.g., cyclization recombination [Cre]/locus of crossing over of bacteriophage P1 [loxP]–mediated excision) or CRISPR/Cas9 editing has led to valuable insights into the function of many genes. Unfortunately, the deletion of a gene that is required during embryogenesis can result in embryonic or perinatal lethality, making it impossible to study the effects of the gene ablation at later developmental ages. Genetic redundancy (Nowak et al., 1997), compensatory mechanisms, and/or toxic effects of byproducts can also make interpretation of data obtained from whole-body genetically modified animals difficult.
The use of inducible expression systems overcomes some of the limitations associated with transgenic and targeted mutagenesis studies. Here, we mainly focus on developments in mammalian inducible systems as applied in the mouse. Several strategies have been developed to achieve inducible spatiotemporal control of gene expression in mice. Temporal control often requires responsiveness to an exogenously added inducer, critically allowing one to understand the consequences of genetic deletion of key proteins in animals for which whole-organism and/or developmental knockout is lethal. Two prototypical examples of such temporal control are the use of chimeric Cre fusion recombinase proteins that can be activated/inhibited by small-molecule ligands (Utomo et al., 1999) and the Tet-On/Tet-Off system, which relies on the use of tetracycline or its derivatives such as doxycycline (Gossen and Bujard, 1992).
These spatiotemporal models are currently widely used and are more precise than the development of germline whole-body knockout/knock-in models that can suffer from premature death and often poorly understood comorbidities. This has opened new opportunities to understand the role of a gene of interest in a tissue of interest and has allowed for better delineation of the cause of (dys)function of the gene and compensatory mechanisms by altered signaling pathways. Obviously, these inducible systems have proven to be important in the understanding of gene function as well as organ and whole-body function.
Although these models are now standard methods in the toolbox of molecular biologists, they come with limitations. For instance, unexpected transient expression of Cre recombinase in the germline or during early development can occur. Appropriate genotyping and careful monitoring can deal with these confounding factors. However, it has become apparent that these ligand-based models for spatial and temporal control of gene expression are prone to secondary effects caused by the inducing agent, introducing inherent bias in the experimental setup. Here, we discuss these different inducible systems, the confounding factors of the inducing agents, and possible alternative approaches to circumvent part of these confounding factors.
Temporal regulation of gene expression using the Tet-On/Tet-Off system
The Tet-On/Tet-Off system has been widely adopted for temporal regulation of gene expression and uses tetracycline antibiotics to up- or down-regulate gene expression (Fig. 1 A; Gossen and Bujard, 1992). A tetracycline resistance gene is constitutively repressed by tetracycline repressor, a protein that binds specifically to tetracycline operator sequences within the promoter, silencing gene transcription. This system relies on a promoter that is responsive to tetracycline-type antibiotics and induces gain- or loss-of-function (Gossen and Bujard, 1992). The system can be used in two ways: (1) in Tet-On mode, whereby tetracycline binds the tetracycline operator transactivator and activates expression of the gene of interest, and (2) in Tet-Off mode, whereby tetracycline binding in fact represses target gene expression and tetracycline removal reactivates the expression (Gossen and Bujard, 1992). The Tet-On/Tet-Off system therefore allows for exquisite flexibility to study gene function. The most widely used effector for tetracycline-controlled transactivation is doxycycline, the exemplar used here, but other tetracyclines, such as 9-tert-butyl-doxycycline (Zhu et al., 2007), can exert similar effects. However, while toxicological analyses revealed a detrimental effect of doxycycline on cell viability even at a low dose (Ermak et al., 2003), only few researchers consider the potential detrimental effects of the use of tetracyclines in mammalian systems. Doxycycline is part of the widely used tetracycline antibiotic class that blocks protein synthesis in bacteria and thereby limits bacterial growth (Boynton et al., 2017). Importantly, doxycycline acutely accumulates in all major tissues to much higher levels than observed in blood serum (Blanchard et al., 1975).
Figure 1.
Intended and unintended effects of spatiotemporal gene expression regulation. (A) The antibiotic doxycycline (DOX) causes disturbances in microbiota and mitochondrial dysfunction. Doxycycline is used to alter the tetracycline (Tet) response element and thereby activates (Tet-On) or deactivates (Tet-Off) expression of the gene of interest. Side effects of this antibiotic-induced temporal gene alteration include those affecting microbiota and those impairing mitochondrial function. In Tet-On/Tet-Off experiments, these adverse effects could cause a second hit that blurs the interpretation of the function of the gene of choice. (B) Tamoxifen (TAM) induces unintended estrogen receptor–mediated toxicity. Various studies have indicated that unwarranted side effects via estrogen receptor–mediated toxicity affect whole-body physiology and blur the interpretation of the function of the gene of choice. Some examples, such as the effects of tamoxifen on pancreatic, cardiac, bone, gastrointestinal, and fat function, are discussed in the text. ER, estrogen receptor.
Off-target effects in Tet-On/Tet-Off models
The first thing to consider as a side effect of doxycycline antibiotics is the disturbance in the composition and function of the skin, pulmonary, mouth, vaginal, and gut microbiota (Fig. 1 A). For instance, the gut microbiota play an important role in many physiological processes such as energy balance and metabolism. As expected, doxycycline significantly alters the gut microbiome after even 7 d of exposure (Boynton et al., 2017), impacting whole-body metabolic flexibility (Smith et al., 2018). If proper controls are not included in the study design, antibiotic-mediated alterations in the microbiome and subsequent whole-body metabolism, cognition, and immune function should be considered as confounding factors in animal experiments with the Tet-On/Tet-Off system.
Another effect of the use of doxycycline comes from a completely different angle. Mitochondria are derived from the endosymbiosis of an α-proteobacterium with a host amitochondriate cell, the evolutionary predecessor of the modern eukaryotic cell (Lang et al., 1999). They still retain their own bacterial-like, circular DNA (mitochondrial DNA), encoding the mitochondrial 16S and 12S ribosomal RNA, 22 tRNAs, and 13 core subunits of the oxidative phosphorylation (OXPHOS) system. Because the remaining OXPHOS subunits are encoded by nuclear DNA, the stoichiometric balance between both genomes is tightly regulated (Houtkooper et al., 2013). Because of the evolutionary similarities between the bacterial and mitochondrial protein synthesis machinery, it is not surprising that doxycycline can also inhibit mitochondrial translation (Moullan et al., 2015; Fig. 1 A). In fact, that classes of antibiotics aimed at inhibiting bacterial protein translation also inhibit mitochondrial function was observed in the 1960s (Clark-Walker and Linnane, 1966), and this has been of groundbreaking importance for our current understanding of the evolutionary origins of mitochondria.
Doxycycline inhibition of mitochondrial protein synthesis results in reduced expression of subunits of the respiratory complexes encoded by the mitochondrial DNA (such as MTCO1 in complex IV), while nuclear-encoded proteins (such as subunit SDHA in complex II) are not directly affected (Moullan et al., 2015). This mitonuclear protein imbalance results in a lower OXPHOS capacity and renders mitochondria more fragmented. On a whole-body level, doxycycline increases motility in worms and flies and can even extend the life span in worms (Houtkooper et al., 2013; Moullan et al., 2015). At the same time, however, developmental delay and physiological impairment reflected by reduced body size and fertility have been observed in worms and flies (Moullan et al., 2015). Even plants (Arabidopsis thaliana) treated with doxycycline show mitochondrial dysfunction and growth retardation (Moullan et al., 2015). These findings clearly demonstrate that doxycycline can cause widespread effects on mitochondrial metabolism and whole-body homeostasis through a mechanism that is conserved between the plant and animal kingdoms (Fig. 1 A).
Considering the role of mitochondria for optimal cell and organ physiology, it is no surprise that mitochondrial dysfunction has been implicated in virtually all multifactorial age-related diseases, such as type 2 diabetes mellitus, heart failure, neurodegeneration, and cancer (Smith et al., 2018). Therefore, it is likely that doxycycline-induced mitochondrial dysfunction can impact the cellular and whole-body effects of the primary gene manipulation. This two-hit model could seriously interfere with the interpretation of Tet-On/Tet-Off studies. Mitochondrial alterations following doxycycline treatment hence have the possibility to compromise whole-cell and organ function, particularly in energy-demanding tissue such as the brain and cardiac muscle.
In conclusion, treatment with doxycycline (or with other tetracyclines) by itself has the potential to severely affect the microbiome, as well as causing whole-body mitochondrial dysfunction. These add-on effects can cause additional cellular, organ, and whole-body adjustments that are not primarily attributed to the manipulation of the gene of choice.
Cre/loxP models
The Cre/loxP system offers another way to regulate gene expression in a spatial manner (Feil et al., 2009). The method requires an essential exon of the gene of interest to be flanked—or floxed—with two 34-bp DNA recognition sites named loxP sites (by homologous recombination in embryonic stem cells). Both loxP sites are in the same orientation, and the site-specific Cre recombination removes the DNA between these two sites, leaving a single loxP site behind (Fig. 1 B). Crossing floxed mice with mice harboring a Cre recombinase gene driven by tissue-specific promoters leads to a selective excision of the gene of interest in the tissue of interest. As a further development of this technology, inducible Cre recombinases have been designed that are inactive in the basal state but can be activated by addition of their cognate ligands, such as the synthetic estrogen receptor antagonist tamoxifen and its active metabolite 4-hydroxytamoxifen for CreERT2, where noninduced background recombination is minimal; other Cre systems have been developed using different ligand-receptor combinations (Feil et al., 2009; Fig. 1 B).
Off-target effects in inducible Cre/loxP models
It has become evident, however, that the addition of hormones in itself can have widespread detrimental effects on cellular and organismal physiology. Here, we mainly focus on the ligand tamoxifen, as this system is most widely used (Fig. 1 B). For instance, substantial gastric toxicity was observed after administration of a single dose of tamoxifen to multiple mouse strains in both sexes, with apoptosis of gastric parietal cells and metaplasia of zymogenic chief cells as early as 3 d after administration (Huh et al., 2012). This can have consequences for studying gastric and intestinal physiology.
With the growing emphasis on sex as a biological variable, the use of tamoxifen-based systems to understand sex-based differences is clearly problematic. Estrogens are active in numerous tissues, and as such, tamoxifen can have other unexpected side effects (Ye et al., 2015). Tamoxifen remained detectable in adipose tissue after cessation of treatment, suggesting that tamoxifen remains bioavailable for weeks after administration (Ye et al., 2015). The systemic and washout effects of tamoxifen on fat and glucose homeostasis blur the effects of the primary gene knockout on various organs (Al Batran et al., 2018). The reproductive system and bone homeostasis are also affected by tamoxifen treatment.
Together, these examples suggest that even a single dose of the recombination inducers can have off-target effects that negatively affect systemic metabolism and endocrinology. Consequently, the combination of the altered expression of a gene of interest together with the systemic off-target effects of tamoxifen can aggravate the combined overall phenotype of the animal and makes the physiological interpretation from these models sometimes difficult. This double-hit model of tamoxifen-induced altered physiology is hence a major drawback of the Cre/loxP strategy to control gene expression and can only be partially circumvented by the incorporation of proper control groups.
Conclusions and perspectives
The development of inducible gene expression models, such as the Cre/loxP and Tet-On/Tet-Off systems, has allowed for a more detailed functional study of gene function without the adverse effects associated with germline gain- or loss-of-function mutants. Here, we warn of potential confounding factors of both tamoxifen and doxycycline treatment in these inducible mouse models. We propose a “two-hit model” whereby the resultant effects of the combined altered gene expression and of the inducible compounds (tamoxifen or doxycycline) can result in additive/synergistic/antagonistic phenotypes that can confound data interpretation. Obviously, incorporating control groups with a similar dose and exposure time of tamoxifen or doxycycline should be used. However, this does not completely remove the risks of hitting two targets, complicating inferences about the function of the gene of interest.
There is an opportunity to further improve the Cre/loxP model to avoid some of these limitations. Lower concentrations and shorter tamoxifen treatment, for instance, would be beneficial. Clearly, the efficiency of tamoxifen-inducible Cre recombinase is sensitive to the dose and route of administration of tamoxifen due to differences in the genetic background of the animals, and as such, careful monitoring of recombination efficiency and optimal (i.e., minimal) dose is warranted (Gridley and Groves, 2014). Alternative tissue-specific delivery methods such as filled nanoparticles with tissue-specific receptors could provide a new approach. Other inducers for the Cre system, such as RU-486 or ecdysone, might provide an alternative strategy to alter spatiotemporal gene expression, but can have similar liabilities (Alexander et al., 2007). Another alternative is the use of raloxifene, a weaker estrogen analogue that typically does not produce as robust a recombination as tamoxifen and does not induce tamoxifen-induced organ dysfunction (Huh et al., 2012; Koitabashi et al., 2009). However, it remains to be studied if these estrogen analogues allow for a clearer interpretation of the temporally altered expression of the gene of interest.
Similarly, the development of more sensitive models of doxycycline-induced (in)activation of gene expression is needed to reduce doxycycline concentration and exposure time. The recently developed, highly sensitive, doxycycline-inducible adeno-associated virus requires less doxycycline, reducing side effects but also allowing gene control in tissues where lower doxycycline concentrations are achieved (e.g., in the brain due to the blood–brain barrier; Das et al., 2016).
Other models of spatiotemporal gene (in)activation are currently under development. Optogenetics has the potential to circumvent some of the above-mentioned shortcomings of the inducers used to activate or silence gene expression in mice. Optogenetics makes use of genetically encoded light-sensitive proteins to control the behavior of living cells and organisms. Some LightOn systems are based on a genetically encoded light sensor that uses flavin adenine dinucleotide as a photon acceptor (Wang et al., 2012). Various technical challenges remain with the use of this technique. One of these is the use of light with excitation wavelengths from 450 to 600 nm, which significantly damage mitochondrial DNA and induce a loss of mitochondrial respiratory activity in cell culture (Godley et al., 2005). Also, further research is necessary to establish whether optical absorption by specific mitochondrial proteins (such as cytochromes) interferes with other cellular processes.
Given the risks associated with using inducible systems, we stress that tamoxifen- and doxycycline-treated control experiments are critically important and should be rigorously used to ensure proper interpretation of the data without overestimating the (patho)physiological effect of the gene expression of interest. This not only applies for scientists investigating (mitochondrial) metabolism specifically, but given the central role of mitochondria in cellular physiology, it is also apparent in other fields, including cardiac physiology, endocrinology, immunology, and cancer.
Acknowledgments
R.H. Houtkooper is financially supported by a VIDI grant from ZonMw (no. 91715305). J. Auwerx is supported by the Ecole Polytechnique Federale de Lausanne.
The authors declare no competing financial interests.
References
- Al Batran R., Gopal K., Martin M.D., Ho K.L., Almutairi M., Aburasayn H., Eaton F., Campbell J.E., and Ussher J.R.. 2018. Skeletal muscle-specific Cre recombinase expression, controlled by the human α-skeletal actin promoter, improves glucose tolerance in mice fed a high-fat diet. Diabetologia. 61:1849–1855. 10.1007/s00125-018-4643-x [DOI] [PubMed] [Google Scholar]
- Alexander H.K., Booy E.P., Xiao W., Ezzati P., Baust H., and Los M.. 2007. Selected technologies to control genes and their products for experimental and clinical purposes. Arch. Immunol. Ther. Exp. (Warsz.). 55:139–149. 10.1007/s00005-007-0025-7 [DOI] [PubMed] [Google Scholar]
- Blanchard P., Rudhardt M., and Fabre J.. 1975. Behaviour of doxycycline in the tissues. Chemotherapy. 21(Suppl 1):8–18. 10.1159/000221886 [DOI] [PubMed] [Google Scholar]
- Boynton F.D.D., Ericsson A.C., Uchihashi M., Dunbar M.L., and Wilkinson J.E.. 2017. Doxycycline induces dysbiosis in female C57BL/6NCrl mice. BMC Res. Notes. 10:644 10.1186/s13104-017-2960-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G.D., and Linnane A.W.. 1966. In vivo differentiation of yeast cytoplasmic and mitochondrial protein synthesis with antibiotics. Biochem. Biophys. Res. Commun. 25:8–13. 10.1016/0006-291X(66)90631-0 [DOI] [PubMed] [Google Scholar]
- Das A.T., Tenenbaum L., and Berkhout B.. 2016. Tet-On Systems For Doxycycline-inducible Gene Expression. Curr. Gene Ther. 16:156–167. 10.2174/1566523216666160524144041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermak G., Cancasci V.J., and Davies K.J.. 2003. Cytotoxic effect of doxycycline and its implications for tet-on gene expression systems. Anal. Biochem. 318:152–154. 10.1016/S0003-2697(03)00166-0 [DOI] [PubMed] [Google Scholar]
- Feil S., Valtcheva N., and Feil R.. 2009. Inducible Cre mice. Methods Mol. Biol. 530:343–363. 10.1007/978-1-59745-471-1_18 [DOI] [PubMed] [Google Scholar]
- Godley B.F., Shamsi F.A., Liang F.Q., Jarrett S.G., Davies S., and Boulton M.. 2005. Blue light induces mitochondrial DNA damage and free radical production in epithelial cells. J. Biol. Chem. 280:21061–21066. 10.1074/jbc.M502194200 [DOI] [PubMed] [Google Scholar]
- Gossen M., and Bujard H.. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 89:5547–5551. 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridley T., and Groves A.K.. 2014. Overview of genetic tools and techniques to study Notch signaling in mice. Methods Mol. Biol. 1187:47–61. 10.1007/978-1-4939-1139-4_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., and Auwerx J.. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497:451–457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.J., Khurana S.S., Geahlen J.H., Kohli K., Waller R.A., and Mills J.C.. 2012. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology. 142:21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koitabashi N., Bedja D., Zaiman A.L., Pinto Y.M., Zhang M., Gabrielson K.L., Takimoto E., and Kass D.A.. 2009. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ. Res. 105:12–15. 10.1161/CIRCRESAHA.109.198416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B.F., Gray M.W., and Burger G.. 1999. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33:351–397. 10.1146/annurev.genet.33.1.351 [DOI] [PubMed] [Google Scholar]
- Moullan N., Mouchiroud L., Wang X., Ryu D., Williams E.G., Mottis A., Jovaisaite V., Frochaux M.V., Quiros P.M., Deplancke B., et al. 2015. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell Rep. 10:1681–1691. 10.1016/j.celrep.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A., Boerlijst M.C., Cooke J., and Smith J.M.. 1997. Evolution of genetic redundancy. Nature. 388:167–171. 10.1038/40618 [DOI] [PubMed] [Google Scholar]
- Smith R.L., Soeters M.R., Wüst R.C.I., and Houtkooper R.H.. 2018. Metabolic Flexibility as an Adaptation to Energy Resources and Requirements in Health and Disease. Endocr. Rev. 39:489–517. 10.1210/er.2017-00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utomo A.R., Nikitin A.Y., and Lee W.H.. 1999. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat. Biotechnol. 17:1091–1096. 10.1038/15073 [DOI] [PubMed] [Google Scholar]
- Wang X., Chen X., and Yang Y.. 2012. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods. 9:266–269. 10.1038/nmeth.1892 [DOI] [PubMed] [Google Scholar]
- Williams E.G., and Auwerx J.. 2015. The Convergence of Systems and Reductionist Approaches in Complex Trait Analysis. Cell. 162:23–32. 10.1016/j.cell.2015.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Wang Q.A., Tao C., Vishvanath L., Shao M., McDonald J.G., Gupta R.K., and Scherer P.E.. 2015. Impact of tamoxifen on adipocyte lineage tracing: Inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Mol. Metab. 4:771–778. 10.1016/j.molmet.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Aller M.I., Baron U., Cambridge S., Bausen M., Herb J., Sawinski J., Cetin A., Osten P., Nelson M.L., et al. 2007. Silencing and un-silencing of tetracycline-controlled genes in neurons. PLoS One. 2 e533 10.1371/journal.pone.0000533 [DOI] [PMC free article] [PubMed] [Google Scholar]