Hedgehog signaling involves the dynamic movement of receptors and effectors in and out of cilia. Desai et al. found that the dynamics of Smo are regulated by ubiquitination, which modulates its interaction with the intraflagellar transport system to control ciliary levels of this receptor.
Abstract
In the absence of Hedgehog ligand, patched-1 (Ptch1) localizes to cilia and prevents ciliary accumulation and activation of smoothened (Smo). Upon ligand binding, Ptch1 is removed from cilia, and Smo is derepressed and accumulates in cilia where it activates signaling. The mechanisms regulating these dynamic movements are not well understood, but defects in intraflagellar transport components, including Ift27 and the BBSome, cause Smo to accumulate in cilia without pathway activation. We find that in the absence of ligand-induced pathway activation, Smo is ubiquitinated and removed from cilia, and this process is dependent on Ift27 and BBSome components. Activation of Hedgehog signaling decreases Smo ubiquitination and ciliary removal, resulting in its accumulation. Blocking ubiquitination of Smo by an E1 ligase inhibitor or by mutating two lysine residues in intracellular loop three causes Smo to aberrantly accumulate in cilia without pathway activation. These data provide a mechanism to control Smo’s ciliary level during Hedgehog signaling by regulating the ubiquitination state of the receptor.
Introduction
Primary cilia play critical roles in development by monitoring the extracellular environment and transmitting this information to the cell body and nucleus, allowing the cell to coordinate its physiology with surrounding cells. In mammals and other vertebrates, ciliary dysfunction leads to a variety of structural birth defects in the brain, heart, lungs, kidneys, and other organs, along with craniofacial and other skeletal abnormalities. While numerous receptors are localized in the cilia, Hedgehog signaling is the best studied ciliary pathway. This pathway plays fundamental roles during development, and many of the developmental defects caused by ciliary dysfunction can be attributed to abnormal Hedgehog signaling. All the key components of the Hedgehog pathway are enriched in the cilium (Corbit et al., 2005; Haycraft et al., 2005; Ocbina and Anderson, 2008; Rohatgi et al., 2007), and their localization changes dynamically in response to the activity of the pathway. In the off state, patched-1 (Ptch1), the Hedgehog ligand receptor, accumulates in cilia and prevents ciliary accumulation and activation of smoothened (Smo). Upon binding of the ligand, Ptch1 is removed from the cilium, and Smo is derepressed and now accumulates in the cilium. Smo subsequently activates downstream signaling, which results in the accumulation of the Gli transcription factors at the ciliary tip before their modification and translocation to the nucleus, where they modulate expression of target genes.
The mechanisms underlying the trafficking of Hedgehog components to and within the cilium are not well understood. Part of the movement is facilitated by intraflagellar transport (IFT; Eguether et al., 2018; Eguether et al., 2014; Keady et al., 2012), and perturbing IFT disrupts Hedgehog signaling (Huangfu et al., 2003; Liem et al., 2012). IFT, which is critical for the assembly and maintenance of cilia, involves motor-driven transport of large complexes called IFT particles along the cilia. These particles are composed of at least 30 proteins organized in IFT-A, IFT-B, and BBSome subcomplexes. The IFT particles serve as motor adaptors connecting various proteins that need to be moved into or out of cilia to kinesin and dynein motors. We previously showed that Ift25 and Ift27, which are subunits of IFT-B, are not required for ciliary assembly. Instead, these two IFTs work with the adaptor protein Lztfl1 and the BBSome to regulate Hedgehog signaling by facilitating the removal of Smo from cilia at the basal state and Ptch1 after pathway activation (Eguether et al., 2018; Eguether et al., 2014; Keady et al., 2012).
In this work, we explore the mechanism underlying the dynamics of Smo localization to cilia and find that the removal of Smo from cilia is regulated by ubiquitination. Ubiquitination involves the covalent attachment of the 76–amino acid peptide ubiquitin (Ub) to cellular proteins. Ub is usually added to lysine residues, and it can be further ubiquitinated on one of its seven lysine residues to produce polyubiquitinated proteins. The type of Ub modification on a protein determines its cellular fate. For instance, K48 polyubiquitination drives degradation by the proteasome, K11 polyubiquitination drives degradation during specific points in the cell cycle, and K63 polyubiquitination often regulates complex formation and signaling (Swatek and Komander, 2016). The addition of Ub results from a cascade of activity whereby an E1 enzyme activates Ub and passes it to an E2 enzyme, which then either passes the Ub to an E3 enzyme for attachment to the protein of interest or works with an E3 to modify that protein. The process is reversible by the action of deubiquitinating enzymes (Swatek and Komander, 2016). Prior work identified components of the Ub system in cilia (Huang et al., 2009; Pazour et al., 2005; Raman et al., 2015), where it has been implicated in disassembly (Huang et al., 2009; Wang et al., 2019). The Ub system is also known to regulate a number of steps in Hedgehog signaling (Hsia et al., 2015). In this work, we found that ubiquitination of Smo facilitates its interaction with the IFT system for efficient ciliary removal/export when the Hedgehog pathway is suppressed.
Results
Ubiquitinated Smo is retained in Ift27−/−, Lztfl1−/−, and Bbs2−/− cilia but not in wild-type cilia
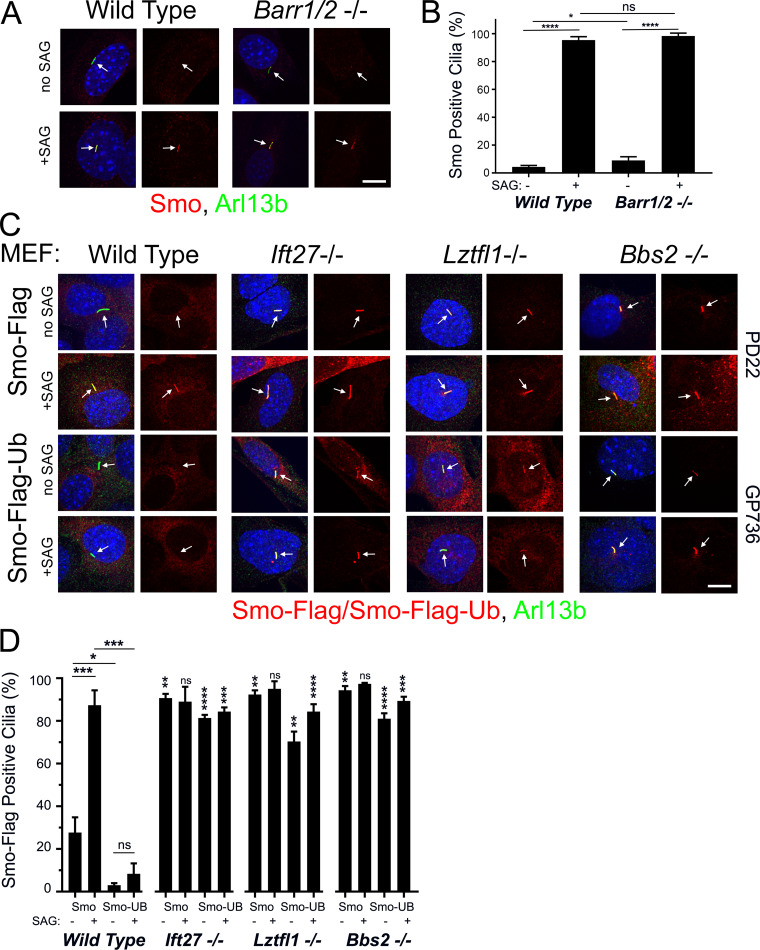
Our previous work indicated that Ift25 and Ift27 are required for the regulated removal of Smo from cilia (Eguether et al., 2014; Keady et al., 2012). This suggests that Hedgehog signaling regulates the interaction between Smo and the IFT particle, but the underlying mechanism is unknown. Structurally, Smo is a member of the G protein–coupled receptor (GPCR) seven transmembrane protein family. Many GPCRs cycle between the cell surface and the endomembrane system depending upon ligand binding. The dynamics are often driven by β-arrestin binding to activated receptor, causing the receptor to associate with clathrin for internalization (Tian et al., 2014). Alternatively, the receptor may be ubiquitinated and internalized via the endosomal sorting complexes required for transport (ESCRT) machinery (Skieterska et al., 2017). While a published report indicates that Smo delivery to cilia required β-arrestin (Kovacs et al., 2008), we were not able to observe any Smo trafficking defects in mouse embryonic fibroblasts (MEFs) lacking β-arrestin 1 and 2 (Barr1/2; Fig. 1, A and B).
Figure 1.
Smo-Ub is removed from wild-type MEF cilia but not from Ift27, Lztfl1, or Bbs2 mutant cilia. (A) Wild-type and Barr1/Barr2 double mutant MEFs were serum starved and stimulated with SAG for 24 h before being fixed and stained for endogenous Smo (red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Left image of each pair is a three-color composite; right image shows only the red Smo channel. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Scale bar is 10 μm and applies to all images in the panel. (B) Presence of endogenous Smo in cilia was quantitated from the cells described in A. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; *, P < 0.05; ns, not significant by two-way ANOVA. Error bars indicate SD. (C) Wild-type, Ift27, Lztfl1, and Bbs2 mutant MEFs were transfected with PD22 (Smo-Flag) or GP736 (Smo-Flag-Ub) and selected with Bsd. Confluent cells were serum starved and stimulated with SAG for 24 h before being fixed and stained for Smo or Smo-Ub (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Left image of each pair is a three-color composite; right image shows only the red Flag channel. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Scale bar is 10 μm and applies to all images in the panel. (D) Presence of ciliary Smo or Smo-Ub was quantitated from the cells described in C. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. For the mutants, significance is shown with respect to same condition in wild-type by three-way ANOVA. Error bars indicate SD.
To explore the role of Ub in regulating the ciliary localization of Smo, we fused a single Ub open reading frame to the intracellular C-terminal end of Smo. This approach has been used to study the role of Ub in trafficking of GPCRs (Shih et al., 2000; Terrell et al., 1998). Cells expressing Smo-Flag from a cytomegalovirus (CMV) promoter show ∼25% Flag-positive cilia without pathway activation, and the numbers are elevated to ∼90% positive cilia upon pathway activation by Smoothened Agonist (SAG). The addition of Ub to the C-terminus greatly reduces the amount of ciliary Smo under basal conditions and notably also blocks the accumulation usually associated with pathway activation by addition of SAG (Fig. 1, C and D) or Sonic Hedgehog (SHH)-conditioned medium (Fig. S1, A and B). As we and others have previously observed for endogenous Smo (Eguether et al., 2014; Seo et al., 2011), Smo-Flag is retained in Ift27−/−, Lztfl1−/−, and Bbs2−/− MEF cilia at both the basal state and after SAG induction. In contrast to the depletion of Smo-Flag-Ub from cilia that we saw in control cells, Smo-Flag-Ub is highly enriched in Ift27−/−, Lztfl1−/−, and Bbs2−/−cilia (Fig. 1, C and D; and Fig. S2). Importantly, the expression of Smo-Flag-Ub does not interfere with the trafficking of endogenous Smo (Fig. S1 C). These data suggest that the IFT/Bardet-Biedl syndrome (BBS) system targets ubiquitinated Smo for ciliary removal.
Figure S1.
Related to Fig. 1: Endogenous Smo is trafficked normally in Smo-Flag-Ub–expressing cells. (A) Wild-type MEFs were transfected with PD22 (Smo-Flag) or GP736 (Smo-Flag-Ub) and selected with Bsd. Confluent cells were serum starved and stimulated with SHH-conditioned medium for 24 h before being fixed and stained for Smo or Smo-Ub (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Left image of each pair is a three-color composite; right image shows only the red Flag channel. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Scale bar is 10 μm and applies to all images in the panel. (B) Presence of Smo or Smo-Ub in cilia was quantitated from the cells described in A. N is 3 replicates. ****, P < 0.0001; ns, not significant by two-way ANOVA. Error bars indicate SD. (C) Wild-type MEFs were transfected with GP736 (Smo-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 24 h and then stimulated with SAG for 24 h before being fixed and stained for endogenous Smo (red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Left image of each pair is a three-color composite; right image shows only the red Smo channel. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Scale bar is 10 μm and applies to all images in the panel. 3.67% ± 3.2% of cells were Smo positive without SAG stimulation, and 92.7% ± 3.2% of cells were Smo positive after SAG stimulation (P < 0.0001; Student’s t test).
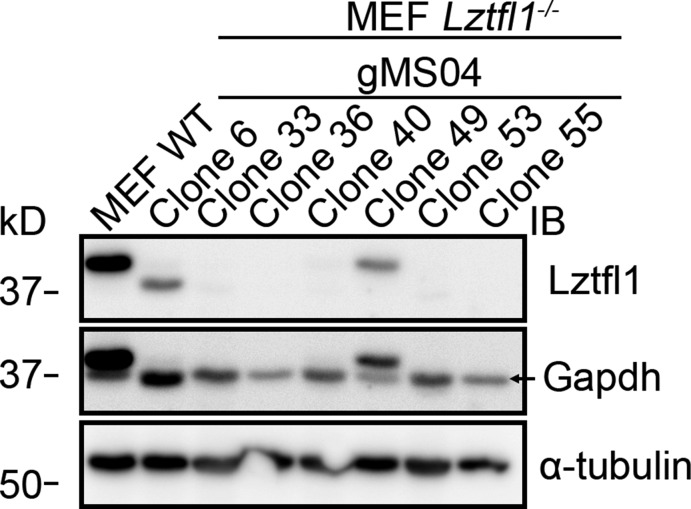
Figure S2.
Related to Fig. 1: Genome-edited cells. Western blot to document loss of Lztfl1 in edited MEFs. The Lztfl1 blot was reprobed with GAPDH and α-tubulin antibodies to ensure relatively even loading. Note that the Lztfl1 bands in wild-type, clone 6, and clone 49 are visible in the GAPDH blot. Clone 36 was used in Fig. 1.
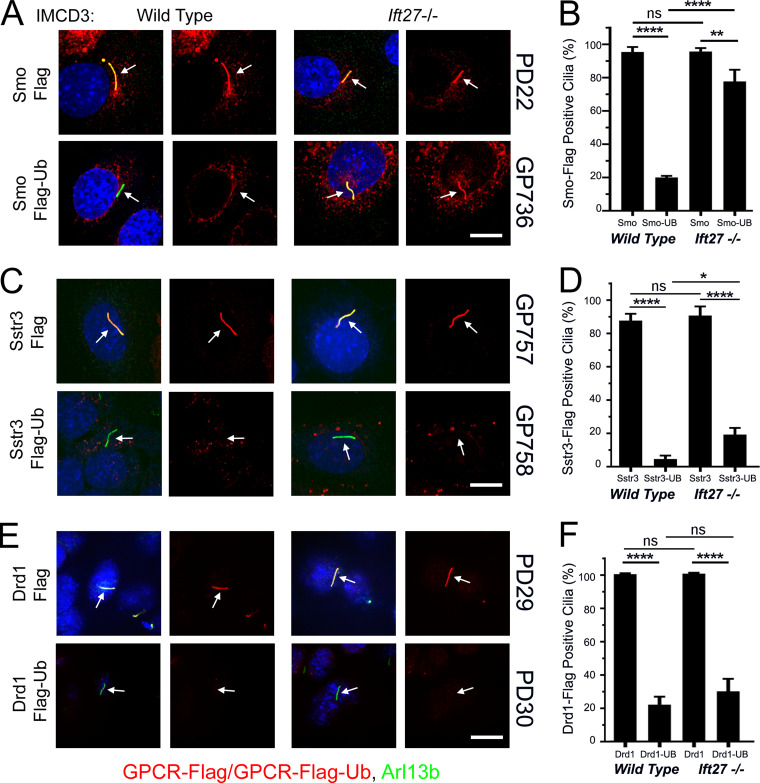
To confirm our finding that Smo-Ub was retained in Ift27−/− mutant cilia but not in control cilia, we examined IMCD3 control and IMCD3Ift27−/− cells. In these kidney epithelial cells, Smo-Flag is retained in wild-type cilia without pathway activation (Fig. 2, A and B). Confirming the MEF studies, Smo-Flag-Ub does not accumulate in control cilia but does accumulate in IMCD3Ift27−/− cilia (Fig. 2, A and B).
Figure 2.
GPCR trafficking in IMCD3 cells reveals diversity in ciliary retrieval mechanisms. (A) Wild-type and Ift27 mutant IMCD3 cells were transfected with PD22 (Smo-Flag) or GP736 (Smo-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 48 h before being fixed and stained for Smo or Smo-Ub (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. Scale bar is 10 μm and applies to all images in the panel. (B) Presence of ciliary Smo or Smo-Ub was quantitated from cells in A. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; **, P < 0.01; ns, not significant by two-way ANOVA. Error bars indicate SD. (C) Wild-type and Ift27 mutant IMCD3 cells were transfected with GP757 (Sstr3-Flag) or GP758 (Sstr3-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 48 h before being fixed and stained for Sstr3 or Sstr3-Ub (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. Scale bar is 10 μm and applies to all images in the panel. (D) Presence of ciliary Sstr3 or Sstr3-Ub was quantitated from cells in C. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; *, P < 0.05; ns, not significant by two-way ANOVA. Error bars indicate SD. (E) Wild-type and Ift27 mutant IMCD3 cells were transfected with PD29 (Drd1-Flag) or PD30 (Drd1-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 48 h before being fixed and stained for Drd1 or Drd1-Ub (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. Scale bar is 10 μm and applies to all images in the panel. (F) Presence of ciliary Drd1 or Drd1-Ub was quantitated from cells in C. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ns, not significant by two-way ANOVA. Error bars indicate SD.
To determine how other GPCRs behave, we tagged the somatostatin receptor Sstr3 with Flag and Ub. Sstr3-Flag is highly enriched in both control and IMCD3Ift27−/− cilia, while Sstr3-Flag-Ub is not highly enriched in cilia of either cell line (Fig. 2, C and D). Similar results were obtained for the Ddr1 dopamine receptor (Fig. 2, E and F), suggesting that an Ift27-independent mechanism exists for the removal of some ubiquitinated cargos from cilia.
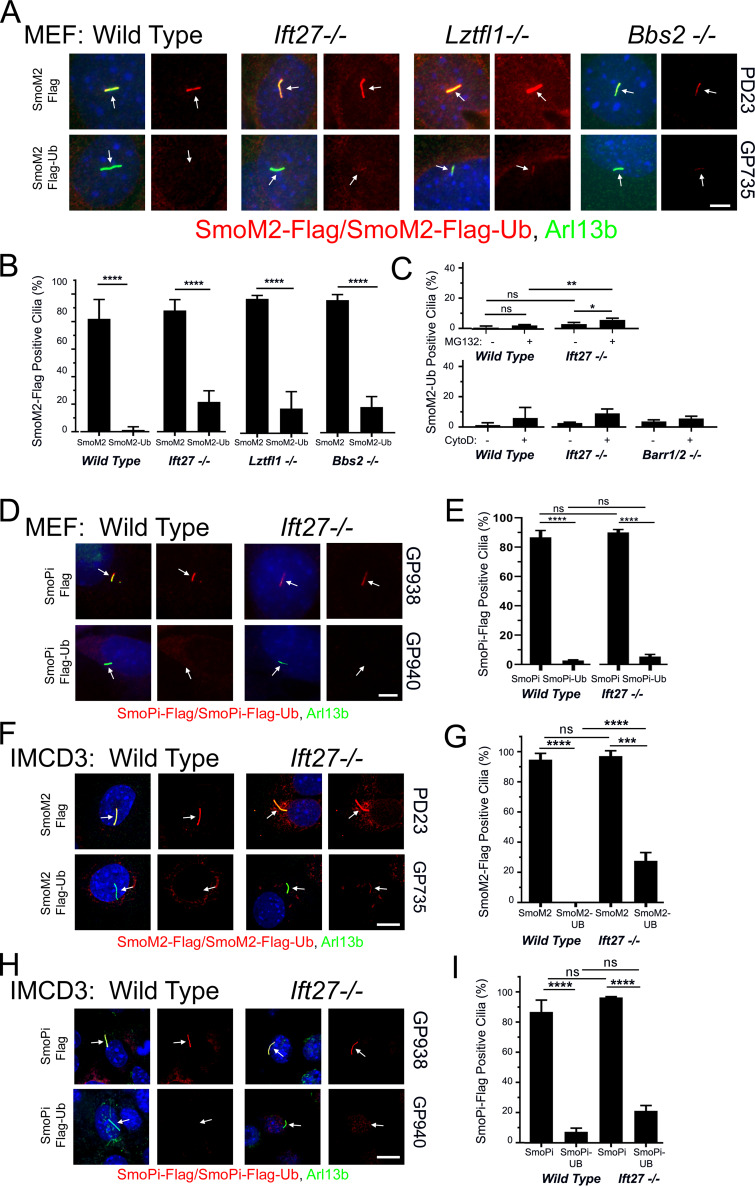
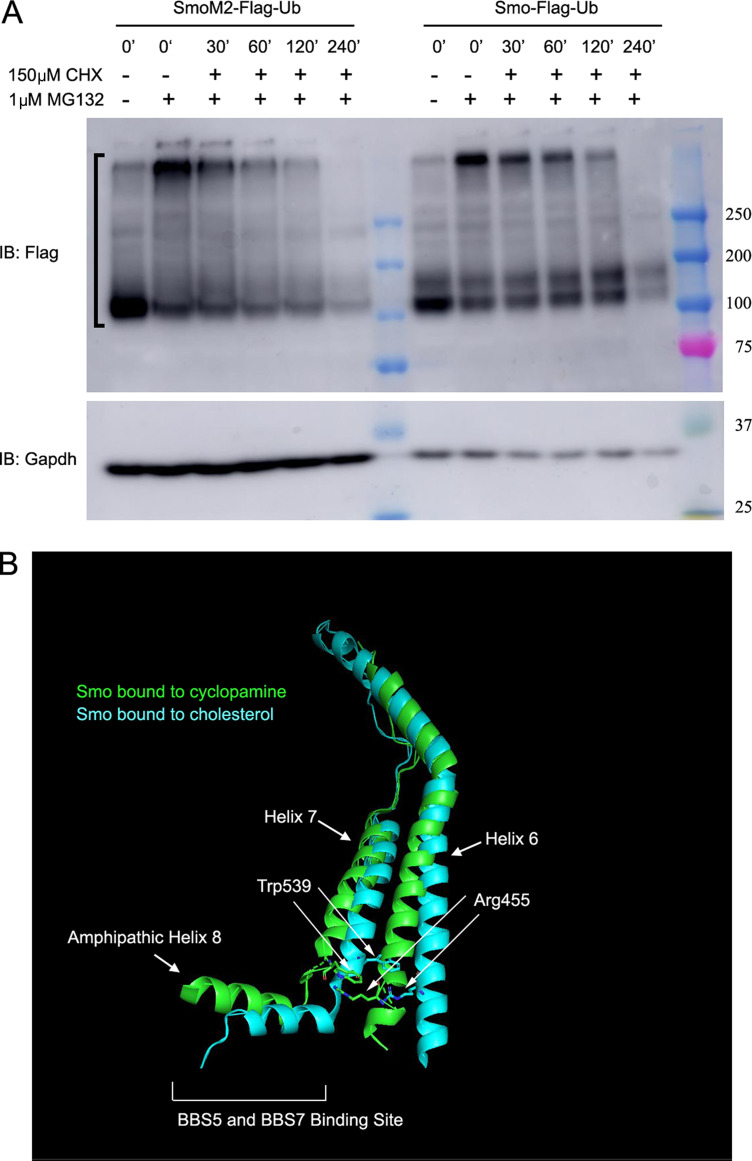
SmoM2 is an oncogenic form of Smo that contains a Trp to Leu mutation (MmSmoW539L) at the cytoplasmic face of the last transmembrane domain (Xie et al., 1998). This mutation constitutively activates Hedgehog signaling, and SmoM2 accumulates in cilia regardless of whether Hedgehog ligand is present or not. Tagging SmoM2 with Ub removed it from wild-type cilia as expected, but unexpectedly SmoM2-Ub was observed to be barely above background in Ift27−/−, Lztfl1−/−, and Bbs2−/− MEF cilia (Fig. 3, A and B). Similar results were obtained in Ift27−/− IMCD3 cells (Fig. 3, F and G). One possibility for the failure of SmoM2-Ub to accumulate in Ift27−/−, Lztfl1−/−, and Bbs2−/− mutant cells is that perhaps SmoM2-Ub is highly unstable and degraded before it is able to traffic to cilia. To compare the relative stability of Smo-Ub and SmoM2-Ub, we treated cells expressing these proteins with MG132 for 4 h to build up levels of Smo-Ub or SmoM2-Ub, then added cycloheximide to prevent new synthesis, and removed the MG132 to allow proteasomal degradation to proceed. MG132 treatment caused significant buildup of Smo-Ub and SmoM2-Ub in cells, which decayed with similar rates, suggesting that it is not the stability of SmoM2-Ub that prevents its accumulation in Ift27−/− cilia (Fig. S3 A). The large buildup of SmoM2-Ub caused by MG132 treatment was not sufficient to cause SmoM2-Ub to accumulate in Ift27−/− cilia (Fig. 3 C). Recently, ectocytosis was proposed as an alternative pathway for clearing some GPCRs from cilia (Nager et al., 2017). This pathway is activated in Bbs mutants, suggesting that perhaps ectocytosis can remove SmoM2-Ub from cilia on Ift27−/−, Lztfl1−/−, and Bbs2−/− mutant cells. However, ectocytosis does not appear to be the explanation, as SmoM2-Ub did not accumulate when ectocytosis was blocked with cytochalasin D (Fig. 3 C). β-arrestin–driven retrieval is another possible mechanism for removal of SmoM2-Ub, but this does not appear to be the mechanism as loss of Barr1/2 did not cause accumulation of SmoM2-Ub in cilia (Fig. 3 C).
Figure 3.
SmoM2 is different from Smo. (A) Wild-type, Ift27, Lztfl1, and Bbs2 mutant MEFs were transfected with PD23 (SmoM2-Flag) or GP735 (SmoM2-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 48 h before being fixed and stained for SmoM2 or SmoM2-Ub (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Left image of each pair is a three-color composite; right image shows only the red Flag channel. Scale bar is 5 μm and applies to all images in the panel. (B) Presence of SmoM2 or SmoM2-Ub in cilia was quantitated from the cells described in A. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ns, not significant. Mutants were not significantly different from the same condition in wild-type by two-way ANOVA. Error bars indicate SD. (C) Wild-type, Ift27, and Barr1/2 mutant SmoM2-Ub cells treated as in B except that 1 µM MG132 for 4 h or 500 nM cytochalasin D (CytoD) was added for 24 h before fixation. **, P < 0.01; *, P < 0.05; ns, not significant. No differences in CytoD treatment were significant by two-way ANOVA. Error bars indicate SD. (D) Wild-type and Ift27 mutant MEFs were transfected with GP938 (SmoPi-Flag) or GP940 (SmoPi-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 48 h before being fixed and stained for SmoPi or SmoPi-Ub (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Left image of each pair is a three-color composite; right image shows only the red Flag channel. Scale bar is 5 μm and applies to all images in the panel. (E) Presence of SmoPi or SmoPi-Ub in cilia was quantitated from the cells described in D. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ns, not significant by two-way ANOVA. Error bars indicate SD. (F) Wild-type and Ift27 mutant IMCD3 cells were transfected with PD23 (SmoM2-Flag) or GP735 (SmoM2-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 48 h before being fixed and stained for Flag (red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. Scale bar is 10 μm and applies to all images in the panel. (G) Presence of ciliary SmoM2 or SmoM2-Ub was quantitated from cells described in F. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ***, P < 0.001; ns, not significant. Error bars indicate SD. (H) Wild-type and Ift27 mutant IMCD3 cells were transfected with GP938 (SmoPi-Flag) or GP940 (SmoPi-Flag-Ub) and selected with Bsd. Confluent cells were serum starved for 48 h before being fixed and stained for Flag (red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. Scale bar is 10 μm and applies to all images in the panel. (I) Presence of ciliary SmoPi or SmoPi-Ub was quantitated from IMCD3 cells described in H. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ns, not significant by two-way ANOVA. Error bars indicate SD.
Figure S3.
Related to Fig. 3: Structure of Smo. (A) MEF cells expressing either SmoM2-Flag-Ub or Smo-Flag-Ub were treated with (+) or without (−) 1 μM MG132 for 4 h. After 4 h, 0-min time points were collected, the MG132 was washed out, and 150 μM cycloheximide was added to the remaining cells. Samples were then collected at 30, 60, 120, and 240 min after the removal of MG132 and the addition of cycloheximide. The half-life of Smo-Flag-Ub and SmoM2-Ub was 110 min and 181 min, respectively (N is 2 replicates). Bracket marks the region of the gel that was used to calculate Smo decay. IB, immunoblot. (B) Ribbon diagram of Smo showing the location of SmoM2 (W539) in transmembrane 7 and SmoPi (R455) residues in transmembrane 6 (redrawn by Pymol [PyMOL.org] from data in Huang et al., (2018). Blue green is cholesterol-bound human Smo (5L7I; Byrne et al., 2016), and green is cyclopamine-bound Xenopus Smo (6D32; Huang et al., 2018). The two states predict the transitions from inactive (blue green) to active (green) when the Pi bond is broken. The breaking of the Pi bond is accompanied by shifts in the position of transmembrane helixes 6 and 7 and amphipathic helix 8, which is the proposed binding site of BBS5 and BBS7 (Seo et al., 2011).
The observation that SmoM2 is retained in cilia in spite of the repressive activity of Ptch1 suggests that SmoM2 is in a conformation that is not a substrate for ubiquitination and removal by Ift27 and the BBSome. Huang et al. (2018) recently proposed a model for how the SmoM2 mutation shifts the structure of Smo toward the activated state. Their model proposes that in the inactive state, Trp539 at the end of helix 7 forms a Pi bond with Arg455 in helix 6. The SmoM2 mutation, which converts Trp539 to a Leu, would break this bond, shifting the positions of transmembrane helixes 6 and 7 and the amphipathic helix that follows helix 7 (Fig. S3 B; Huang et al., 2018). SmoPi-Flag (Arg455Ala) mimicked SmoM2-Flag in that the receptor accumulated in cilia without the need for pathway activation, and SmoPi-Flag-Ub was removed from cilia independent of Ift27 (Fig. 3, D and E). Similar results were obtained in Ift27−/− IMCD3 cells (Fig. 3, H and I). This finding suggests that Smo’s conformation influences the mechanism of removal from cilia, with inactive Smo using an Ift27-dependent mechanism and activated SmoM2 and SmoPi using a retrieval pathway similar to the one used by Sstr3 and other GPCRs that are constitutively localized in cilia.
Blocking ubiquitination causes Smo to accumulate in cilia
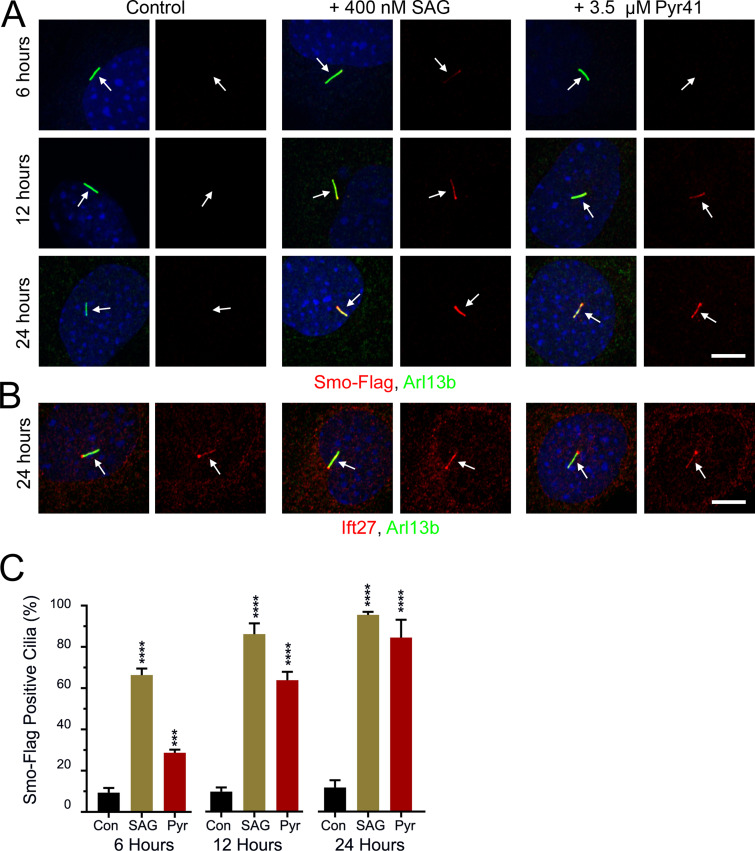
The ligation of Ub to a substrate requires a cascade starting with an E1 Ub-activating enzyme, followed by an E2 Ub-conjugating enzyme, and lastly an E3 Ub ligase. Pyr41 inhibits E1 enzymes (Yang et al., 2007), and we reasoned that if ubiquitination of Smo was required for its removal from cilia, Pyr41 should cause Smo to accumulate in cilia. While blocking an E1 activating enzyme is expected to affect many processes, we found that cells treated with 3.5 µM Pyr41 appeared healthy, remained ciliated, and accumulated Smo in cilia. The Smo accumulation in cilia was detectable within 6 h of treatment and at 24 h was similar to the accumulation driven by SAG treatment (Fig. 4, A and C). Pyr41 did not appear to affect general IFT, as Ift27 staining was similar in Pyr41-treated cells and controls (Fig. 4 B).
Figure 4.
E1 inhibition elevates ciliary Smo levels. (A) Wild-type MEFs were transfected with Smo-Flag (PD22), and a single-cell clone (11479.6T PD22 Clone3) was selected that showed low ciliary Smo-Flag at the basal level and high ciliary Smo-Flag after SAG stimulation. These cells were serum starved and not treated (Con), treated with 400 nM SAG (SAG), or treated with 3.5 µM Pyr41 (Pyr). Coverslips were removed at the indicated times, fixed, and stained for Smo (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Scale bar is 10 μm and applies to all images. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. (B) Cells described in A were fixed at 24 h and stained for Ift27 (red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Scale bar is 10 μm and applies to all images. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Ift27 channel. (C) Presence of ciliary Smo-Flag was quantitated from cells in A. N is 3 replicates with at least 100 cells counted per condition. ***, P < 0.001; ****, P < 0.0001 compared with control at each time point by one-way ANOVA. Error bars indicate SD.
Lysines in Smo’s third intracellular loop are required for regulated ciliary localization
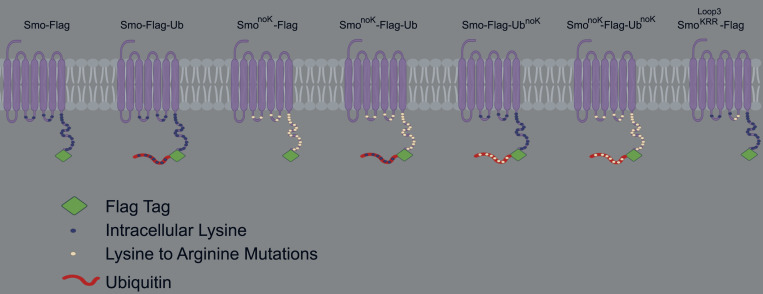
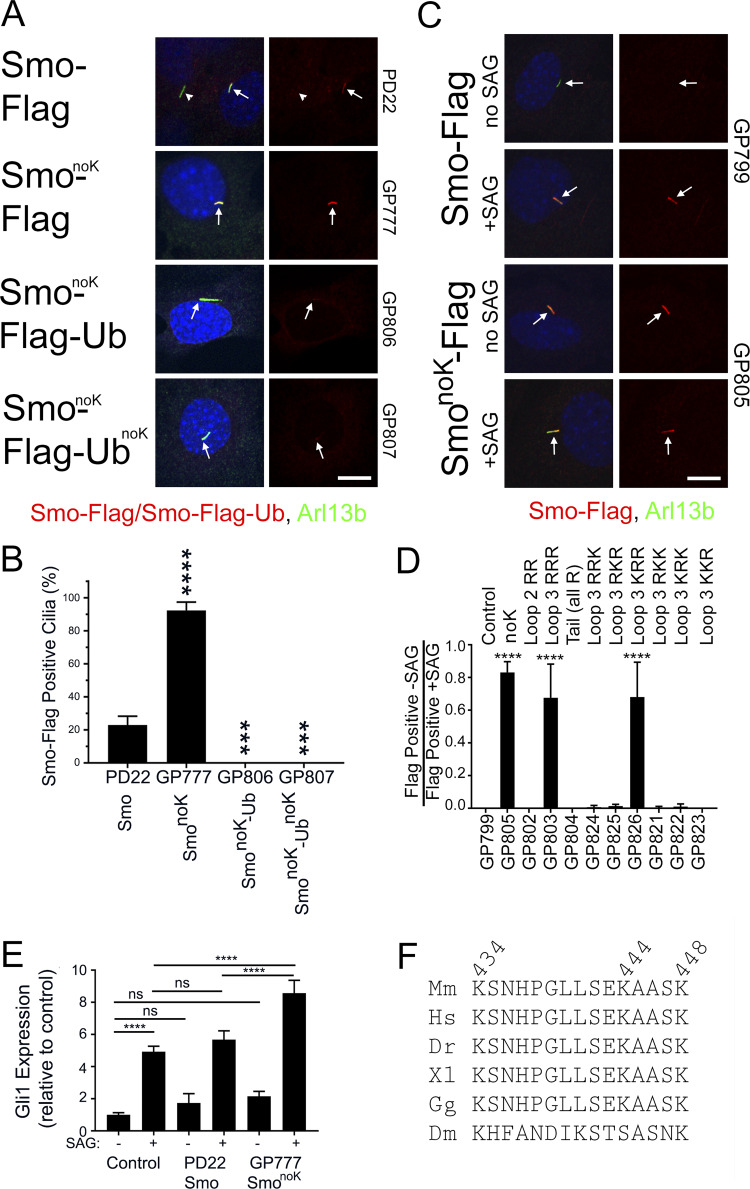
If blocking ubiquitination with Pyr41 is sufficient to cause Smo to accumulate in cilia, then mutating the site of ubiquitination should also cause Smo to accumulate. It is expected that a cytoplasmic lysine residue would be ubiquitinated, and mouse Smo has two lysines in cytoplasmic loop two, three in cytoplasmic loop three, and 16 in the C-terminal tail (Fig. S4). Ubiquitination predictors gave inconsistent results without overlap of predicted sites of ubiquitination, so SmonoK was generated where all 21 cytoplasmic lysines were mutated to arginine (GP777). SmonoK accumulated to high levels in cilia without pathway activation, supporting the hypothesis that Ub is needed for ciliary removal (Fig. 5, A and B). To ensure that SmonoK is still capable of being removed, we attached Ub and found that SmonoK-Ub was efficiently removed from the cilia. UbnoK (all seven lysines of Ub are mutated to arginine) that is not a substrate for polyubiquitin was also effective in removing SmonoK, indicating that a single Ub is sufficient to remove Smo from the cilium (Fig. 5, A and B).
Figure S4.
Related to Fig. 5: Diagram of Smo constructs. Experiments used mouse Smo tagged with a Flag epitope (green diamond) on the C-terminal end. Cytoplasmic lysines are marked with dark blue circles, which were changed to white circles when the lysine was mutated to arginine. In some constructs, Ub (red ribbon) was fused to the C-terminal end of Smo. Ub contains seven lysines marked with dark blue circles, which were changed to white circles when the lysines were mutated to arginine.
Figure 5.
Smo loop three lysines are required for regulated ciliary localization. (A) Wild-type MEFs transfected with Smo-Flag (PD22), SmonoK-Flag (GP777), SmonoK-Flag-Ub (GP806), or SmonoK–Flag-UbnoK (GP807) were serum starved, fixed, and stained for Smo (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Scale bar is 10 μm and applies to all images. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. (B) Presence of ciliary Smo-Flag or Smo-Flag-Ub was quantitated from cells in A. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ***, P < 0.001 compared with control by one-way ANOVA. Error bars indicate SD. (C) Wild-type MEFs transfected with GgCryD1-Bsd-IRES-Smo-Flag (GP799), GgCryD1-Bsd-IRES-SmonoK-Flag (GP805) were serum starved, treated ±SAG, fixed, and stained for Smo (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Scale bar is 10 μm and applies to all images. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. (D) Ciliary Smo was quantitated from the cells described in C as well as a series of mutants designed to identify the lysines important for regulated localization of Smo to cilia. Results are reported as a ratio of the percentage of cells with Smo-positive cilia without SAG divided by the percentage of cells with Smo-positive cilia after treatment with SAG. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001 compared with control (GP799) by one-way ANOVA. Error bars indicate SD. (E) Wild-type MEFs untransfected (control) or transfected with Smo-Flag (PD22) or SmonoK-Flag (GP777) were serum starved and treated ±SAG for 24 h. RNA was collected, reverse transcribed, and quantitated using real-time PCR. N is 3 replicates. Data are reported with respect to wild-type unstimulated (−SAG). ****, P < 0.0001; ns, not significant by two-way ANOVA. Error bars indicate SD. (F) Sequence from Smo cytoplasmic loop three showing complete conservation in vertebrate models. Drosophila melanogaster (Dm) retained three lysines but was otherwise divergent. Dr, Danio rerio; Hs, Homo sapiens; Gg, Gallus gallus; Mm, Mus musculus; Xl, Xenopus laevis.
To understand the importance of Smo ubiquitination to signaling, we measured activation of the pathway (based on level of Gli1 expression) in response to SAG in untransfected cells compared with ones transfected with Smo-Flag or SmonoK-Flag. Smo-Flag cells were similar to control cells, but SmonoK-Flag cells had a higher level of pathway activation, suggesting that ubiquitination of Smo dampens the pathway (Fig. 5 E).
The observation that SmonoK accumulates in cilia suggested that we could identify the specific lysine residue(s) that is important for Smo removal by adding lysines back to SmonoK. This experiment is complicated by the fact that 20%–25% of cilia were positive when wild-type Smo was transfected into cells. We reasoned that this was due to the high level of expression of Smo driven by the CMV promoter saturating the ciliary removal system. To circumvent this problem, we created a construct with low Smo expression by replacing the CMV promoter with a weak GgCryD1 promoter, placing the Smo open reading frame downstream of an internal ribosome entry site (IRES) and adding an out-of-frame methionine upstream of the Smo start codon (GP799). Cells expressing this construct did not show ciliary Smo until the pathway was activated (Fig. 5, C and D). However, drug selection did not work well for this construct, so the percentage of transfection was variable. To circumvent the problem of variable transfection, we ratioed the percentage of Smo-Flag positive without SAG treatment to the percentage positive after SAG treatment (Fig. 5 D). Confirming the results with the CMV promoter (Fig. 5, A and B), SmonoK expressed from the GgCryD1 promoter (GP805) accumulated in cilia without pathway activation (Fig. 5, C and D). Further analysis showed that the lysines in the C-terminal tail and loop two are not needed for removal of Smo-Flag from cilia, but at least one of the three lysines in loop three (GP803) is critical. Further mutation of loop three indicated that when both Lys444 and Lys448 are mutated to arginine (GP826), Smo accumulates in cilia without pathway activation. Mutation of either of these lysines on its own was not enough to retain Smo (Fig. 5 D), suggesting that either residue can serve the regulatory role. This region of loop three is highly conserved in vertebrates, with no substitutions in any of the main model organisms. In Drosophila, the region is not conserved except that it contains three lysines (Fig. 5 F).
Smo ubiquitination is reduced by pathway activation
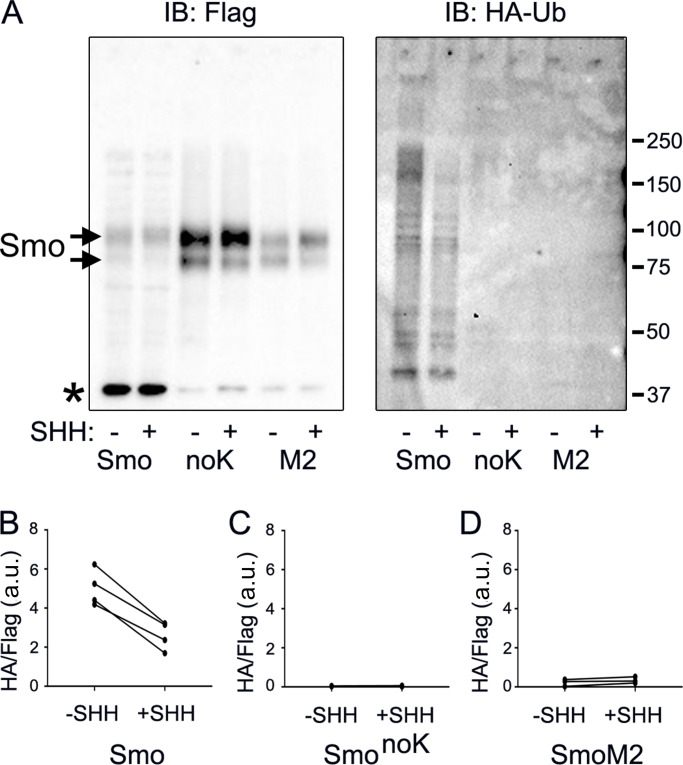
To explore the ubiquitination state of Smo during pathway activation, stable cell lines expressing Smo-Flag, SmonoK-Flag, or SmoM2-Flag and HA-Ub (Kamitani et al., 1997) from different promoters were grown to confluence, serum starved for 24 h, and then treated with or without Hedgehog-conditioned medium for an additional 24 h. After 20 h of SHH treatment, 1 µM MG132 was added for 4 h to block proteasomal degradation, and then the cells were harvested for immunoprecipitation. Smo, SmonoK, and SmoM2 were precipitated with Flag antibody beads. Probing the immunoprecipitates for Flag showed that approximately equal amounts of Smo were precipitated under both conditions (Fig. 6 A). Probing for HA showed a smear up the gel as expected for ubiquitinated proteins and showed that significantly more HA-Ub precipitated with Smo from control cells compared with cells treated with SHH-conditioned medium (Fig. 6, A and B). No HA-Ub was detected after immunoprecipitation of SmonoK, indicating that the HA signal was due to ligation of Ub onto Smo (Fig. 6, A and C). Interestingly, we did not detect SmoM2 ubiquitination at either basal or induced state (Fig. 6, A and D). This suggests that the ubiquitination of SmoM2 is blocked by the conformation change that accompanies its activation.
Figure 6.
Ubiquitination of Smo is reduced by pathway activation. (A) Wild-type MEFs transfected with Smo-Flag (PD22), SmonoK-Flag (GP777), and SmoM2-Flag (PD23) along with HA-Ub (PD14) were grown to confluence, serum starved 24 h, treated with or without SHH–conditioned medium for 24 h, treated with 1 μM MG132 for the last 4 h, and immunoprecipitated with anti-Flag beads. Precipitates were probed for Smo-Flag and HA-Ub. * marks an unrelated endogenous protein detected by the Flag antibody. Smo bands are marked with arrows, with the upper band being a glycosylated form. Both panels were run on the same gel, but different exposures were used for Flag and HA. IB, immunoblot. (B) Quantification of decreased Ub incorporation in Smo-Flag after treatment with Hedgehog-conditioned medium. N is 4 replicates. P = 0.003 with paired t test. (C) Quantification of Ub incorporation into SmonoK after treatment with Hedgehog-conditioned medium. N is 4 replicates. Differences are not significant. (D) Quantification of Ub incorporation into SmoM2 after treatment with Hedgehog-conditioned medium. N is 3 replicates. Differences are not significant.
Discussion
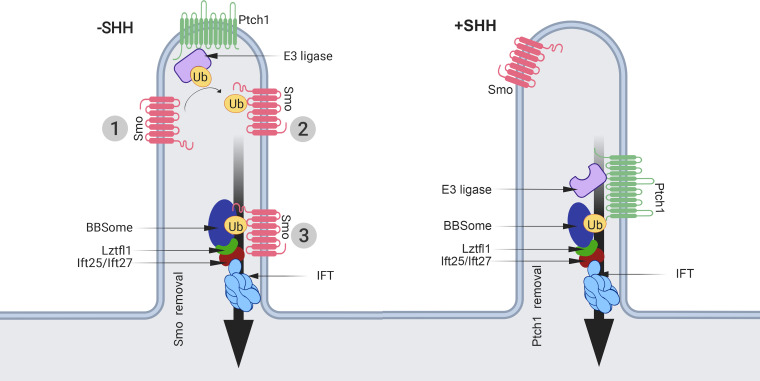
During vertebrate Hedgehog signaling, receptors and other components of the pathway are moved into and out of cilia. While these movements are critical to the proper regulation of the pathway, little is known about the molecular mechanisms driving them. In this work, we show that the addition of Ub to the C-terminal end of Smo prevents its accumulation in wild-type cilia in response to pathway activation. However, Smo-Ub still accumulates in cilia on cells lacking Ift27, Lztfl1, or Bbs2, suggesting that Ub couples Smo to the IFT system for removal from cilia. In support of this observation, blocking ubiquitination with an E1 inhibitor or by removing two critical lysines from Smo caused Smo to inappropriately accumulate in cilia in the off state. We further show that activation of Hedgehog signaling reduces the amount of Ub on Smo. Taken together, these data suggest a model where an E3 ligase ubiquitinates Smo in the off state, which promotes its interaction with the IFT/BBSome for removal from cilia. Upon pathway activation, we expect this E3 ligase will be inactivated, and there may also be concomitant activation of a Smo deubiquitinase, thus allowing Smo to remain in cilia (Fig. 7).
Figure 7.
Model for the mechanism regulating ciliary levels of Smo. Our data and data from the literature support a model where the ubiquitination state of Smo regulates its ciliary localization. In the simplest form of the model, Smo that enters cilia at the basal state (1) is ubiquitinated by unknown E3 ligase (2) on lysine residues in intracellular loop three, making it cargo for removal from the cilium by the IFT system (3). Activation of the pathway by SHH binding to Ptch1 would suppress the activity of the E3 ligase or, if the E3 is ciliary localized, remove it from the cilium. This would allow Smo that enters the cilium to remain, become activated, and trigger downstream signaling events. A simple mechanism for regulating the ciliary localization of an E3 ligase could involve binding the ligase to Ptch1, which is known to bind E3 ligases. By attaching the ligase to Ptch1, the ligase would be ciliary localized at the basal state and be removed upon pathway activation. While we have no data on the role of Ub in regulating Ptch1, a similar mechanism whereby Ptch1 is ubiquitinated by pathway activation could serve to keep ciliary Ptch1 levels low in the activated state.
The removal of ubiquitinated GPCRs from cilia appears to be a general phenomenon as the addition of Ub to Sstr3 prevented it from accumulating in cilia. However, in contrast to Smo-Ub, the removal of Sstr3-Ub was not dependent on Ift27. Unexpectedly, oncogenic SmoM2 behaved like Sstr3 rather than nononcogenic Smo, as SmoM2-Ub did not accumulate in cilia lacking Ift27. This suggests that an alternative mechanism exists to remove certain GPCRs from cilia. The removal was not blocked by loss of β-arrestin or by inhibiting ectocytosis, making it likely to be an IFT-dependent process. If it is IFT dependent, then subunits of either IFT-A or IFT-B (besides Ift25 and Ift27) must be able to bind ubiquitinated GPCRs to remove them from cilia. Smo and SmoM2 differ by a single amino acid, Trp539Leu, located at the end of transmembrane 7. Trp539 is thought to interact with Arg455 to form a Pi lock that keeps Smo in the inactive configuration. Activation of Smo or mutating of either Trp539 or Arg455 is thought to open the Pi lock, shifting transmembrane helixes 6 and 7 and causing amphipathic helix 8 in the C-terminal cytoplasmic tail to shift and partially unwind (Huang et al., 2018). Our finding that SmoPi-Ub is removed from Ift27 mutant cilia such as SmoM2-Ub suggests that activation of Smo changes its conformation such that ubiquitinated forms no longer depend on the BBSome for removal. The detailed mechanism of how Smo interacts with the BBSome remains to be determined, but a short region just downstream of the SmoM2 mutation is reported to bind to BBS5 and BBS7 (Seo et al., 2011). This region contains the amphipathic helix that is shifted and partially unwound by the activation of Smo (Huang et al., 2018). It is possible that the SmoM2 and SmoPi mutations change the structure of this helix and the tail so it can be bound by other components of the IFT system for removal from cilia.
The role of Ub in ciliary biology and Hedgehog signaling has not been extensively studied, although several studies indicate that Ub is critical to both cilia and Hedgehog signaling. In Chlamydomonas reinhardtii, Ub and isoforms of E1, E2, and E3 ligases are present in the cilium (Huang et al., 2009; Long et al., 2016; Pazour et al., 2005). Ciliary proteins become ubiquitinated while cilia are disassembling, and ubiquitinated cargos accumulate in cilia when IFT is disrupted (Huang et al., 2009). In mammalian cells, valosin-containing protein (VCP), which uses ATP to segregate Ub from binding partners, is complexed with IFT-B though Ubxn10. Both VCP and Ubxn10 are required for ciliogenesis (Raman et al., 2015). In Trypanosoma brucei, Ub copurifies with the BBSome, and BBSome mutations affect cellular localization of ubiquitinated receptors (Langousis et al., 2016). In Caenorhabditis elegans, attachment of Ub to the ciliary membrane protein Pkd2 reduced its level in cilia, and similar to our findings, the BBSome appeared to be required (Hu et al., 2007; Xu et al., 2015).
Ub has been implicated in a number of steps of the Hedgehog pathway, ranging from the production of Hedgehog ligand to regulating the proteolytic conversion of Gli3 full length to Gli3 repressor (Hsia et al., 2015). Less is known about the role of Ub in regulating the dynamics of Ptch1 and Smo, although previous studies support a function for Ub in this process. In Drosophila, Smo is ubiquitinated in the off state, and this drives sequestration in cytoplasmic vesicles (Ma et al., 2016; Xia et al., 2012; Zhou et al., 2018). The E3 ligase modifying Drosophila Smo is reported to be Herc4 (Jiang et al., 2019; Sun et al., 2019), and other studies suggest that the Usp8 and Uchl5 deubiquitinases regulate cellular localization of Smo. Herc4 was identified as a genetic modifier of Hedgehog signaling, and its loss stabilized Smo protein (Jiang et al., 2019; Sun et al., 2019). Usp8 was identified in an RNAi screen as a Smo deubiquitinase, and its loss increased Smo ubiquitination (Xia et al., 2012). Uchl5 stabilizes Drosophila Smo and promotes its localization on the cell surface. The human homologue UCH37 was reported to promote Smo localization to cilia (Zhou et al., 2018). However, we used CRISPR to knock out Herc4 along with its close paralog Herc3, Usp8, and Uchl5/UCH37 and did not observe any effects on Smo localization, suggesting that they are not the critical enzymes regulating Smo removal from mammalian cilia.
Ptch1 is also likely to be regulated by ubiquitination, as the E3 ligases Smurf1 and Smurf2 ubiquitinate Ptch1 and promote its degradation (Yue et al., 2014). While this work focused on the degradation of Ptch1 by ubiquitination, it showed that knockdown of Smurf1 and Smurf2 increased the intensity of Ptch1-GFP in mammalian cilia (Yue et al., 2014). Another relevant study found two PY motifs, which are binding sites for HECT family ubiquitinases, in Ptch1 and showed that mutating these binding sites caused Ptch1 to remain in cilia after pathway activation (Kim et al., 2015). Proteomic analysis indicates that numerous E3 ligases bind Ptch1 (Yamaki et al., 2016).
Two recent large-scale CRISPR screens of genes involved in Hedgehog signaling identified a number of Ub-related enzymes regulating Hedgehog signaling (Breslow et al., 2018; Pusapati et al., 2018). Genes identified included E1, E2, and E3 ligases along with deubiquitinases and adaptor proteins. Pusapati et al. (2018) identified three genes, Mgrn1, Megf8, and Atthog, that all increased ciliary levels of Smo at the basal state. Of these, the loss of Megf8 and Atthog drove ciliary Smo to high levels that appear similar to what we observed with the loss of Ift27, whereas Mgrn1 loss was more variable, with some cilia showing strong enrichment and others similar to controls. Currently, it is not known if Megf8 and Atthog are involved in ubiquitination, whereas Mgrn1 encodes an E3 ligase. This makes Mgrn1 a strong candidate to be the enzyme that regulates the ciliary localization of Smo, but the variable amount of Smo accumulation in cilia suggests that there are likely other enzymes involved.
In summary, our work uncovers a mechanism for regulating the dynamic distribution of Smo during Hedgehog signaling (Fig. 7). In this model, Hedgehog signaling regulates the activity or subcellular distribution of E3 ligases (and perhaps deubiquitinases) that modify Smo. At the basal state, the E3 ligase is active against Smo, causing Smo to become ubiquitinated. The simplest model would place the E3 ligase in the cilium, but it is possible that Smo is ubiquitinated outside the cilium before it enters. Ciliary localized ubiquitinated Smo would be a substrate for removal from cilia by the IFT/BBSome. It is likely that the BBSome binds both Ub and Smo, as a Smo binding site was identified in BBS7 (Seo et al., 2011). Upon activation of Hedgehog signaling, the E3 ligase is inhibited and/or removed from the cilium, or alternatively, the activation of Smo may change its confirmation such that it is no longer a substrate for ubiquitination. Thus, Smo that enters the cilium will not be ubiquitinated and will not interact with the IFT/BBS complex; instead, it will remain in the cilium. The simplest mechanism for regulating the ciliary localization of an E3 ligase is to bind it to Ptch1, which is known to be removed from cilia upon pathway activation. As discussed previously, Ptch1 contains two PY motifs and is known to bind HECT family E3 ligases, making this a plausible mechanism. In addition to regulating Smo distribution, Ub could more generally regulate the ciliary levels of other receptors, the most likely being Ptch1. Perhaps the binding of ligand to Ptch1 activates an E3 ligase that promotes Ptch1 removal from cilia.
Materials and methods
Plasmids
Plasmids were assembled by Gibson assembly (NEB) into the pHAGE lentiviral backbone (Wilson et al., 2008). All inserts are derived from mouse unless otherwise stated. Mutations were generated by PCR amplification with mutated primers and the products Gibson assembled. For mutation of the 16 lysines in the C-terminal tail of Smo, the tail sequence was chemically synthesized (gBlock; IDT) and PCR amplified for Gibson assembly. All inserts were fully sequenced and match the National Center for Biotechnology Information reference sequence or expected mutant forms. Plasmids are described in Table 1 and Fig. S4. SnapGene files will be provided upon request.
Table 1. Plasmids.
| Name | Description | Promoter | Insert | Tags | Selection |
|---|---|---|---|---|---|
| MS03 | SHH Expression Plasmid | CMV | HsSHH | None | Bsd |
| GP734 | Flag-Ub | CMV | Ub | Flag, Ub | Bsd |
| PD19 | Flag-UbnoK (7 Lys mutated to Arg) | CMV | UbnoK | Flag, UbnoK | Bsd |
| PD14 | HA-Ub | CMV | Ub | HA, Ub | Puro |
| PD13 | HA-UbnoK (7 Lys mutated to Arg) | CMV | UbnoK | HA, UbnoK | Puro |
| PD22 | Smo-Flag | CMV | Smo | Flag | Bsd |
| GP736 | Smo-Flag-Ub | CMV | Smo | Flag, Ub | Bsd |
| GP777 | SmonoK-Flag | CMV | SmonoK | Flag | Bsd |
| GP806 | SmonoK-Flag-Ub | CMV | SmonoK | Flag, Ub | Bsd |
| GP807 | SmonoK-Flag-UbnoK | CMV | SmonoK | Flag, UbnoK | Bsd |
| PD23 | SmoM2-Flag | CMV | SmoW539L | Flag | Bsd |
| GP735 | SmoM2-Flag-Ub | CMV | SmoW539L | Flag, Ub | Bsd |
| GP938 | SmoPi-Flag | CMV | SmoR455A | Flag | Bsd |
| GP940 | SmoPi-Flag-Ub | CMV | SmoR455A | Flag, Ub | Bsd |
| GP757 | Sstr3-Flag | CMV | Sstr3 | Flag | Bsd |
| GP758 | Sstr3-Flag-Ub | CMV | Sstr3 | Flag, Ub | Bsd |
| PD29 | Drd1-Flag | CMV | Drd1 | Flag | Bsd |
| PD30 | Drd1-Flag-Ub | CMV | Drd1 | Flag, Ub | Bsd |
| GP799 | Smo-Flag | GgCryD1 | Smo | Flag | Bsd |
| GP805 | Like GP799 but with the 21 cytoplasmic lysines of Smo mutated to Arg | ||||
| GP802 | Like GP799 but with the two lysines in loop two of Smo mutated to Arg | ||||
| GP803 | Like GP799 but with the three lysines (KKK to RRR) in loop three of Smo mutated to Arg | ||||
| GP804 | Like GP799 but with the 16 lysines in C-terminal tail of Smo mutated to Arg | ||||
| GP824 | Like GP799 but with two lysines (KKK to RRK) in loop three of Smo mutated to Arg | ||||
| GP825 | Like GP799 but with two lysines (KKK to RKR) in loop three of Smo mutated to Arg | ||||
| GP826 | Like GP799 but with two lysines (KKK to KRR) in loop three of Smo mutated to Arg | ||||
| GP821 | Like GP799 but with one lysine (KKK to RKK) in loop three of Smo mutated to Arg | ||||
| GP822 | Like GP799 but with one lysine (KKK to KRK) in loop three of Smo mutated to Arg | ||||
| GP823 | Like GP799 but with one lysine (KKK to KKR) in loop three of Smo mutated to Arg | ||||
Cell culture
Wild-type, Ift27−/−, and Bbs2−/− MEFs were derived from E14 embryos and immortalized with SV40 Large T antigen. Barr1−/−/Barr2−/− double knockout MEFs were obtained from R. Lefkowitz (Duke University, Durham, NC) and immortalized with SV40 Large T antigen. Lztfl1−/− cells (Fig. S2) were obtained by genome editing of immortalized wild-type MEFs using guide (gMS04: GCTCGATCAAGAAAACCAAC) cloned into pLentiCRISPRv2 (Addgene plasmid #52961; Sanjana et al., 2014). All MEFs were cultured in 95% DMEM (4.5 g/L glucose), 5% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin (all from Gibco-Invitrogen).
IMCD3 cells (Rauchman et al., 1993) were cultured in 47.5% DMEM (4.5 g/liter glucose), 47.5% F12, 5% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin (all from Gibco-Invitrogen). IMCD3Ift27−/− cells were obtained from Max Nachury (University of California, San Francisco, San Francisco, CA).
For SAG experiments, MEFs were plated at near-confluent densities and serum starved (same culture medium as described above but with 0.25% fetal bovine serum) for 48 h before treatment to allow ciliation. SAG (Calbiochem) was used at 400 nM.
Sonic Hedgehog (SHH)-conditioned medium was generated from HEK cells transfected with a human (Hs) SHH expression construct (MS03). Cells stably expressing MS03 were grown to confluency in 90% DMEM (4.5 g/liter glucose), 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin; medium was then replaced with low serum medium (0.25% fetal bovine serum) and grown for 48 h. Medium was collected, and the filter was sterilized and titered for ability to relocate Smo to cilia. Dilutions similar in effect to 400 nM SAG were used for experiments.
Lentivirus production
Lentiviral packaged pHAGE-derived plasmids (Wilson et al., 2008) were used for transfection. These vectors were packaged by a third-generation system comprising four distinct packaging vectors (Tat, Rev, Gag/Pol, and VSV-g) using HEK 293T cells as the host. DNA (Backbone: 5 µg; Tat: 0.5 µg; Rev: 0.5 µg; Gag/Pol: 0.5 µg; VSV-g: 1 µg) was delivered to the HEK cells using calcium phosphate precipitates. After 48 h, supernatant was harvested, filtered through a 0.45-μm filter, and added to subconfluent cells. After 24 h, cells were selected with puromycin (Puro; 1 µg/ml) or blasticidin (Bsd; 60 µg/ml for CMV promoter or 20 µg/ml for GgCryD1 promoter).
Immunofluorescence
Cells were fixed with 2% paraformaldehyde for 15 min, permeabilized with 0.1% Triton-X-100 for 2 min, and stained as described (Follit et al., 2006). In some cases, fixed cells were treated with 0.05% SDS for 5 min before prehybridization to retrieve antigens. The primary antibodies are described in Table 2.
Table 2. Antibodies.
| Primary antibody | Clone or antibody name | Concentration | Supplier |
|---|---|---|---|
| Flag | F1804 | 1:1,000 | Sigma |
| Arl13b | N295B/66 | 1:1,000 | Davis/National Institutes of Health NeuroMab |
| Arl13b | 17711-1-AP | 1:1,000 | Proteintech |
| Smo | E5 | 1:100 | Santa Cruz |
| IFT27 | 719 | 1:250 | Keady et al., 2012 |
| IFT88 | 448 | 1:250 | Pazour et al., 2002 |
| Bbs5 | B-11 | 1:500 | Santa Cruz |
| Bbs2 | A-12 | 1:100 | Santa Cruz |
| Bbs2 | 11188-2-AP | 1:500 | Proteintech |
| Lztfl1 | 17073-1-AP | 1:500 | Proteintech |
| α-Tubulin | B-5-1-2 | 1:5,000 | Sigma |
| GAPDH | 14C10 3683S | 1:5,000 | Cell Signaling |
Confocal images were acquired with an inverted microscope (TE-2000E2; Nikon) equipped with a Solamere Technology–modified spinning-disk confocal scan head (CSU10; Yokogawa). Three-image Z stacks were acquired at 0.2-μm intervals and converted to single planes by maximum projection with MetaMorph software (MDS Analytical Technologies). Widefield images were captured using an Orca ER or a FLIR camera on a Zeiss Axiovert 200M microscope equipped with a 100× Zeiss objective. Contrast adjustment and cropping were done in ImageJ or Photoshop.
Protein and mRNA analysis
For Western blots, MEFs were lysed directly into denaturing gel loading buffer (Tris-HCl 125 mM, pH 6.8, glycerol 20% vol/vol, SDS 4% vol/vol, β-mercaptoethanol 10% vol/vol, and bromophenol blue). Western blots were developed by chemiluminescence (Super Signal West Dura; Pierce Thermo) and imaged using an Amersham Imager 600 or Bio-Rad ChemiDoc XRS+.
For immunoprecipitations, cells were serum starved for 48 h, and proteins were extracted with lysis buffer (Cell Lytic Solution; C2978; Sigma) with 0.1% CHAPSO, 0.1% NP-40, and protease inhibitor (Complete EDTA-Free; Roche). Insoluble components were removed by centrifugation at 20,000 g. Flag beads (Anti Flag M2 Affinity Gel; A2250; Sigma) were added to the cell extract and the mixture was incubated for 2 h at 4°C. After centrifugation, beads were washed 3X with 0.1% Tween20/TBS and 3X with TBS before elution with 3XFLAG peptide (F4799; Sigma). Gel loading buffer was added to the eluates for SDS-PAGE electrophoresis and Western blotting analysis.
Isolation of mRNA and quantitative mRNA analysis were performed as previously described (Jonassen et al., 2008) using the primers described in Table 3.
Table 3. Quantitative RT-PCR primers.
| Primer | Genbank accession no. | Sequence | Tm | Length (bp) |
|---|---|---|---|---|
| MmGAPDHExon3for | NM_008084 | 5′-GCAATGCATCCTGCACCACCA-3′ | 61.1 | |
| MmGAPDHExon4rev | 5′-TTCCAGAGGGGCCATCCACA-3′ | 61.1 | 138 | |
| MmGli1Exon4for | NM_010296 | 5′-CCAGGGTTATGGAGCAGCCAGA-3′ | 61.2 | |
| MmGli1Exon5rev | 5′-CTGGCATCAGAAAGGGGCGAGA-3′ | 61.5 | 135 |
Statistics
Data groups were analyzed as described in the figure legends using GraphPad Prism. Differences between groups were considered statistically significant if P < 0.05. Statistical significance is denoted with asterisks (*, P = 0.01–0.05; **, P = 0.01–0.001; ***, P < 0.001 to 0.0001; ****, P < 0.0001). Error bars are all SD.
Online supplemental material
Fig. S1 shows that Smo-Flag and Smo-Flag-Ub cells respond to SHH and SAG similarly and that endogenous Smo is trafficked normally in Smo-Flag-Ub–expressing cells. Fig. S2 documents the generation of Lztfl1 genome edited cells. Fig. S3 shows degradation of SmoM2-Flag-Ub compared with that of Smo-Flag-Ub and ribbon diagrams of the Smo structure. Fig. S4 contains diagrams of the various Smo, Ub, and no lysine versions used throughout the paper.
Supplementary Material
Acknowledgments
We thank Drs. K. Bellve, L. Lifshitz, and K. Fogarty (University of Massachusetts Medical School Biomedical Imaging Group) for assistance with microscopy; Dr. R. Wang (University of Massachusetts Medical School, Worcester, MA) for assistance with statistical analysis; and Dr. W. Tsang (Institut de Recherches Cliniques de Montréal, Quebec, Canada) for sharing HA-Ub and HA-UbnoK constructs. We thank Drs. J. Chen (Stony Brook University Medicine, Stony Brook, NY), Thibaut Eguether (Faculte de Medecine Pierre et Marie Curie, Paris, France), William Monis (Sanofi, Paris, France), and Abigail Smith (University of Massachusetts Medical School, Worcester, MA) for critical reading of this manuscript and for valuable input during its production.
This work was supported by the National Institutes of Health (GM060992 and DK103632 to G.J. Pazour).
The authors declare no competing financial interests.
Author contributions: Conceptualization, P.B. Desai and G.J. Pazour; Formal Analysis, P.B. Desai, M.W. Stuck, B. Lv, and G.J. Pazour; Funding Acquisition, G.J. Pazour; Investigation, P.B. Desai, M.W. Stuck, B. Lv, and G.J. Pazour; Methodology, P.B. Desai, M.W. Stuck, B. Lv, and G.J. Pazour; Resources, P.B. Desai, M.W. Stuck, B. Lv, and G.J. Pazour; Supervision, G.J. Pazour; Validation, P.B. Desai, M.W. Stuck, B. Lv, and G.J. Pazour; Visualization, P.B. Desai and G.J. Pazour; Writing/Original Draft Preparation, P.B. Desai and G.J. Pazour; and Writing/Review and Editing, P.B. Desai, M.W. Stuck, B. Lv, and G.J. Pazour.
References
- Breslow D.K., Hoogendoorn S., Kopp A.R., Morgens D.W., Vu B.K., Kennedy M.C., Han K., Li A., Hess G.T., Bassik M.C., et al. . 2018. A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat. Genet. 50:460–471. 10.1038/s41588-018-0054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne E.F.X., Sircar R., Miller P.S., Hedger G., Luchetti G., Nachtergaele S., Tully M.D., Mydock-McGrane L., Covey D.F., Rambo R.P., et al. . 2016. Structural basis of Smoothened regulation by its extracellular domains. Nature. 535:517–522. 10.1038/nature18934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., and Reiter J.F.. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Eguether T., San Agustin J.T., Keady B.T., Jonassen J.A., Liang Y., Francis R., Tobita K., Johnson C.A., Abdelhamed Z.A., Lo C.W., et al. . 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell. 31:279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T., Cordelieres F.P., and Pazour G.J.. 2018. Intraflagellar transport is deeply integrated in hedgehog signaling. Mol. Biol. Cell. 29:1178–1189. 10.1091/mbc.E17-10-0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., and Pazour G.J.. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792. 10.1091/mbc.e06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C.J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E.J., and Yoder B.K.. 2005. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1 e53 10.1371/journal.pgen.0010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia E.Y., Gui Y., and Zheng X.. 2015. Regulation of Hedgehog signaling by ubiquitination. Front. Biol. (Beijing). 10:203–220. 10.1007/s11515-015-1343-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wittekind S.G., and Barr M.M.. 2007. STAM and Hrs down-regulate ciliary TRP receptors. Mol. Biol. Cell. 18:3277–3289. 10.1091/mbc.e07-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Diener D.R., and Rosenbaum J.L.. 2009. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 186:601–613. 10.1083/jcb.200903066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Zheng S., Wierbowski B.M., Kim Y., Nedelcu D., Aravena L., Liu J., Kruse A.C., and Salic A.. 2018. Structural Basis of Smoothened Activation in Hedgehog Signaling. Cell. 174:312–324.e16. 10.1016/j.cell.2018.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., and Anderson K.V.. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 426:83–87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Jiang W., Yao X., Shan Z., Li W., Gao Y., and Zhang Q.. 2019. E3 ligase Herc4 regulates Hedgehog signalling through promoting Smoothened degradation. J. Mol. Cell Biol. 11:791–803. 10.1093/jmcb/mjz024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen J.A., San Agustin J., Follit J.A., and Pazour G.J.. 2008. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J. Cell Biol. 183:377–384. 10.1083/jcb.200808137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T., Nguyen H.P., and Yeh E.T.. 1997. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J. Biol. Chem. 272:14001–14004. 10.1074/jbc.272.22.14001 [DOI] [PubMed] [Google Scholar]
- Keady B.T., Samtani R., Tobita K., Tsuchya M., San Agustin J.T., Follit J.A., Jonassen J.A., Subramanian R., Lo C.W., and Pazour G.J.. 2012. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell. 22:940–951. 10.1016/j.devcel.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Hsia E.Y.C., Brigui A., Plessis A., Beachy P.A., and Zheng X.. 2015. The role of ciliary trafficking in Hedgehog receptor signaling. Sci. Signal. 8:ra55 10.1126/scisignal.aaa5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs J.J., Whalen E.J., Liu R., Xiao K., Kim J., Chen M., Wang J., Chen W., and Lefkowitz R.J.. 2008. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 320:1777–1781. 10.1126/science.1157983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langousis G., Shimogawa M.M., Saada E.A., Vashisht A.A., Spreafico R., Nager A.R., Barshop W.D., Nachury M.V., Wohlschlegel J.A., and Hill K.L.. 2016. Loss of the BBSome perturbs endocytic trafficking and disrupts virulence of Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 113:632–637. 10.1073/pnas.1518079113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem K.F. Jr., Ashe A., He M., Satir P., Moran J., Beier D., Wicking C., and Anderson K.V.. 2012. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J. Cell Biol. 197:789–800. 10.1083/jcb.201110049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Zhang F., Xu N., Liu G., Diener D.R., Rosenbaum J.L., and Huang K.. 2016. Comparative Analysis of Ciliary Membranes and Ectosomes. Curr. Biol. 26:3327–3335. 10.1016/j.cub.2016.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Li S., Han Y., Li S., Yue T., Wang B., and Jiang J.. 2016. Regulation of Smoothened Trafficking and Hedgehog Signaling by the SUMO Pathway. Dev. Cell. 39:438–451. 10.1016/j.devcel.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nager A.R., Goldstein J.S., Herranz-Perez V., Portran D., Ye F., Garcia-Verdugo J.M., and Nachury M.V.. 2017. An Actin Network Dispatches Ciliary GPCRs into Extracellular Vesicles to Modulate Signaling. Cell. 168:252–263.e14. 10.1016/j.cell.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina P.J., and Anderson K.V.. 2008. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev. Dyn. 237:2030–2038. 10.1002/dvdy.21551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Baker S.A., Deane J.A., Cole D.G., Dickert B.L., Rosenbaum J.L., Witman G.B., and Besharse J.C.. 2002. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157:103–113. 10.1083/jcb.200107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., and Witman G.B.. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–114. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusapati G.V., Kong J.H., Patel B.B., Krishnan A., Sagner A., Kinnebrew M., Briscoe J., Aravind L., and Rohatgi R.. 2018. CRISPR Screens Uncover Genes that Regulate Target Cell Sensitivity to the Morphogen Sonic Hedgehog. Dev. Cell. 44:113–129.e8. 10.1016/j.devcel.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M., Sergeev M., Garnaas M., Lydeard J.R., Huttlin E.L., Goessling W., Shah J.V., and Harper J.W.. 2015. Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat. Cell Biol. 17:1356–1369. 10.1038/ncb3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauchman M.I., Nigam S.K., Delpire E., and Gullans S.R.. 1993. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. Am. J. Physiol. 265:F416–F424. 10.1152/ajprenal.1993.265.3.F416 [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., and Scott M.P.. 2007. Patched1 regulates hedgehog signaling at the primary cilium. Science. 317:372–376. 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Zhang Q., Bugge K., Breslow D.K., Searby C.C., Nachury M.V., and Sheffield V.C.. 2011. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 7 e1002358 10.1371/journal.pgen.1002358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S.C., Sloper-Mould K.E., and Hicke L.. 2000. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 19:187–198. 10.1093/emboj/19.2.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skieterska K., Rondou P., and Van Craenenbroeck K.. 2017. Regulation of G Protein-Coupled Receptors by Ubiquitination. Int. J. Mol. Sci. 18:923 10.3390/ijms18050923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Sun B., Cui M., and Zhou Z.. 2019. HERC4 exerts an anti-tumor role through destabilizing the oncoprotein Smo. Biochem. Biophys. Res. Commun. 513:1013–1018. 10.1016/j.bbrc.2019.04.113 [DOI] [PubMed] [Google Scholar]
- Swatek K.N., and Komander D.. 2016. Ubiquitin modifications. Cell Res. 26:399–422. 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell J., Shih S., Dunn R., and Hicke L.. 1998. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell. 1:193–202. 10.1016/S1097-2765(00)80020-9 [DOI] [PubMed] [Google Scholar]
- Tian X., Kang D.S., and Benovic J.L.. 2014. β-arrestins and G protein-coupled receptor trafficking. Handb. Exp. Pharmacol. 219:173–186. 10.1007/978-3-642-41199-1_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Peng Z., Long H., Deng X., and Huang K.. 2019. Polyubiquitylation of α-tubulin at K304 is required for flagellar disassembly in Chlamydomonas. J. Cell Sci. 132 jcs229047 10.1242/jcs.229047 [DOI] [PubMed] [Google Scholar]
- Wilson A.A., Kwok L.W., Hovav A.H., Ohle S.J., Little F.F., Fine A., and Kotton D.N.. 2008. Sustained expression of alpha1-antitrypsin after transplantation of manipulated hematopoietic stem cells. Am. J. Respir. Cell Mol. Biol. 39:133–141. 10.1165/rcmb.2007-0133OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Jia H., Fan J., Liu Y., and Jia J.. 2012. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol. 10 e1001238 10.1371/journal.pbio.1001238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Murone M., Luoh S.M., Ryan A., Gu Q., Zhang C., Bonifas J.M., Lam C.W., Hynes M., Goddard A., et al. . 1998. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 391:90–92. 10.1038/34201 [DOI] [PubMed] [Google Scholar]
- Xu Q., Zhang Y., Wei Q., Huang Y., Li Y., Ling K., and Hu J.. 2015. BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci. Rep. 5:11855 10.1038/srep11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki Y., Kagawa H., Hatta T., Natsume T., and Kawahara H.. 2016. The C-terminal cytoplasmic tail of hedgehog receptor Patched1 is a platform for E3 ubiquitin ligase complexes. Mol. Cell. Biochem. 414:1–12. 10.1007/s11010-015-2643-4 [DOI] [PubMed] [Google Scholar]
- Yang Y., Kitagaki J., Dai R.M., Tsai Y.C., Lorick K.L., Ludwig R.L., Pierre S.A., Jensen J.P., Davydov I.V., Oberoi P., et al. . 2007. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 67:9472–9481. 10.1158/0008-5472.CAN-07-0568 [DOI] [PubMed] [Google Scholar]
- Yue S., Tang L.Y., Tang Y., Tang Y., Shen Q.H., Ding J., Chen Y., Zhang Z., Yu T.T., Zhang Y.E., et al. . 2014. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. eLife. 3 e02555 10.7554/eLife.02555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Yao X., Pang S., Chen P., Jiang W., Shan Z., and Zhang Q.. 2018. The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating Smoothened. J. Mol. Cell Biol. 10:243–257. 10.1093/jmcb/mjx036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.