Abstract
Promoter design remains one of the most important considerations in metabolic engineering and synthetic biology applications. Theoretically, there are 450 possible sequences for a 50-nt promoter, of which naturally occurring promoters make up only a small subset. To explore the vast number of potential sequences, we report a novel AI-based framework for de novo promoter design in Escherichia coli. The model, which was guided by sequence features learned from natural promoters, could capture interactions between nucleotides at different positions and design novel synthetic promoters in silico. We combined a deep generative model that guides the search for artificial sequences with a predictive model to preselect the most promising promoters. The AI-designed promoters were optimized based on the promoter activity in E. coli and the predictive model. After two rounds of optimization, up to 70.8% of the AI-designed promoters were experimentally demonstrated to be functional, and few of them shared significant sequence similarity with the E. coli genome. Our work provided an end-to-end approach to the de novo design of novel promoter elements, indicating the potential to apply deep learning methods to de novo genetic element design.
INTRODUCTION
Well-characterized regulatory elements are indispensable for the design of synthetic circuits and metabolic engineering, which offer enormous potential for industrial biotechnology to produce chemical, medical and material products (1,2). Promoters are key elements that regulate gene expression at the level of transcription; hence, the choice of promoter elements is an essential consideration in synthetic biology applications (3). Researchers have proposed several methods to generate novel synthetic promoters (4–6).
Previous studies searching for novel promoters have mainly focused on mutagenesis (4,7,8) or regulatory element combinations (9–11). Methods based on mutagenesis, such as constructing random mutation libraries, were reported to successfully generate novel synthetic promoters (12). For example, Alper et al. used error-prone PCR to mutagenize the bacteriophage PL-λ promoter in Escherichia coli, resulting in a novel library with 22 functional mutants (13). In addition, sequence combination strategies, such as integrating TF binding sites with promoter backgrounds (14,15), combining known short functional components (10,11) and random sequence assembly methods (16–18), have generated some novel promoters for regulating target genes.
Although these experimental approaches are available, these works were constrained by a relatively small library size compared to the large number of all possible sequence combinations. Even for a 50-nt long prokaryotic promoter, the number of all combinations of DNA sequences is 450. The number of possibilities is even much larger for eukaryotic promoters, which have longer promoter lengths and more complex structures. Therefore, it is an interesting question whether one could use computational methods to navigate the vast potential sequence space effectively to find novel promoters.
Recent advances in deep learning methods have provided novel alternative approaches for promoter design. In particular, generative adversarial networks (GANs) (19), which are deep neural network (DNN)-based generative models, offer a promising way to navigate sequence space and thus to generate novel promoters. Based on the minimax adversarial game between two neural networks (the generator and discriminator), GAN can extract essential features from data and automatically generate novel samples. Multiple state-of-the-art image generation methods have been created with GANs (20–22), and GANs have demonstrated the ability to generate novel images with sufficient diversity (23,24). Recently, some variants of GAN have been used to design probes for protein binding microarrays (25), synthetic genes coding for antimicrobial peptides (26), and drug-like molecular structures (27–29).
Here, we proposed a deep learning-based approach for de novo promoter sequence design, and validated the activities of the generated promoters in vivo. A GAN model was trained to extract features from natural promoters, and generated millions of brand new artificial sequences. These AI-generated sequences could mimic key characteristics of natural promoters such as k-mer frequency, –10 and –35 motifs and their spacing constraints. After filtering by a promoter activity predictive model, up to 70.8% of the AI-designed promoters were experimentally demonstrated to be functional, and a number of them showed comparable or even higher activities than most active natural promoters and their strongest mutants. These novel promoters showed low global sequence similarity to E. coli genomic sequences, and noncanonical motifs were found in highly-expressed AI-generated promoters, offering new insight into the design of novel promoters. In conclusion, our method provides a new strategy to effectively design brand new functional promoters.
MATERIALS AND METHODS
Promoter library plasmid construction
The E. coli strain DH5α [F-, ϕ80d, lacZΔ M15, Δ (lacZYA-argF), U169, endA1, recA1, hsdR17(rk-, mk+), sup E44λ, thi-, gyrA96, relA1, phoA] was used as the host organism and cultivated in Luria−Bertani (LB) media supplemented with 100 μg/ml kanamycin at 37°C for promoter activity validation.
Promoter constructs were cloned into the medium-copy-number modified vector pFAB217 with a p15A replicon driving the expression of sfGFP from the reporter gene sfgfp. We also cloned the AI-designed promoters into modified vector pFAB217 using the reporter gene mrfp1. Promoters from two rounds of optimization are displayed in Supplementary Table S1. We used the fixed 5′UTR sequence downstream of promoters, which has been applied in previously works (30,31) and iGEM (international genetically engineered machine competition) parts testing experiments (32). The forward primer designed from the 5′UTR fragments (B0030: GGGCTCTGTAAGATCTATTAAAGAGGAGAAAG) contained an EcoRI site, a BglII site and BamHI site. The putative Shine-Dalgarno sequence in the 5′UTR was placed 7 nt upstream of the original TSS region. Annealing reactions were performed by incubating the complementary oligonucleotides at 95°C for 2 min (2 μl of 100 μM forward and reverse oligonucleotides in sterile water) each cycle for 57 cycles and cooling to 4°C for storage. The annealed oligonucleotides were phosphorylated using T4 polynucleotide kinase (PNK from NEB) with ATP for 1 h. Then, the 5′UTR oligonucleotides were digested by restriction enzymes EcoRI and BamHI and cloned into the EcoRI–BglII sites in modified pFAB217 by T4 DNA ligase. The recombinant plasmids with 5′UTR sequences were verified by sequencing. The modified vector pFAB217 and promoter oligonucleotides were digested with the restriction enzymes EcoRI and BglII, and the designed promoter oligonucleotides flanking the same multiple cloning site were cloned into the new EcoRI-BglII sites. Six positive control promoters, five random baseline promoters (with GC content near 50%) and two blank control plasmids were also tested for promoter activity. All of the reporter plasmids were verified by sequencing.
Assay strains were stored as glycerol stocks (20% glycerol) in sterile centrifuge tubes (1.5 ml). E. coli with target plasmid was picked out using a sterilized metal pinner and grown on plates containing 5 ml of LB medium supplemented with kanamycin. Monoclonal selections were performed overnight (16 hours) in a 96-well U-bottom deep-well plate covered with sterile breathable sealing film (sterile sealing films; Axygen) at 30°C with shaking at 300 rpm on an orbital shaker.
Then the overnight cultures were diluted 1:100 into a final volume of 1.5 ml of fresh medium with the appropriate kanamycin concentration and grown for another 8 h. Then, 200 μl of culture was added to each well of clear bottom black plates, and repeated measurements of the optical density at 600 nm (OD600) and fluorescence (relative fluorescence units [RFU]; excitation at 485 nm and emission at 510 nm) were performed with a microplate reader-incubator-shaker (Thermo). All experiments were repeated at least three times, and the positions of strains in the wells of the microplates were changed during three repeated experiments to avoid any local position effects.
Promoter activity measurement
Five control variants were generated in which the synthetic promoters were replaced by complete random sequences, and the GC content was controlled at near 50%. The resulting expression levels were measured based on the basal expression of the sfgfp gene. This experiment was conducted to test the transcription baseline in our system. Six positive control promoter sequences were also used in the present work, including two different types of wild-type promoters, BBa_J23119 and Ptrc (Trc1), with two of their corresponding mutants (BBa_J23100, BBa_J23102 and Ptrc_m010 (Trc2), Ptrc_m004 (Trc3)), which showed the highest expression in the previously reported mutation library (33). BBa_J23100 and BBa_J23102 were obtained from the BioBrick (34) part in the iGEM Registry of Standard Biological Parts (http://parts.igem.org/Promoters/Catalog/Constitutive). Ptrc (Trc1), Ptrc_m010 (Trc2) and Ptrc_m004 (Trc3) were obtained from previous research (33) and were cultivated in 0.5 ml LB medium with 0.1 mM IPTG. We generated 91 artificial sequences that shared the same –10 and –35 motifs with the wild-type Ptrc promoter, but the other base positions were randomly synthesized with the same possibilities (Supplementary Table S1). Two repeated blank control variants were designed by replacing the synthetic promoter sequences with a 10-nt random sequence (GGGCTCTGTA), which could not provide enough length for RNA polymerase to bind to the upstream sequence of the protein coding region. The promoter strength is calculated as (33):
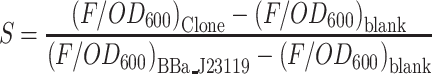 |
Final reported promoter activities were calculated by taking the average of three independent biological experiments and all the artificial sequences with their promoter activity were shown in Supplementary Table S1.
A brief introduction to GAN model
Generative adversarial networks (GANs) (19) have achieved impressive results in the fields like natural image generation (35), image-to-image translation (36), and super resolution image creation (20). GANs contain two ‘adversarial’ networks: the generator 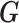 and the discriminator
and the discriminator 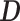 . The generator tries to capture the data distribution and produces artificial samples to fool the discriminator, whereas the discriminator tries to distinguish generated samples from training data. Thus, a minmax game between two networks could be described in the following function:
. The generator tries to capture the data distribution and produces artificial samples to fool the discriminator, whereas the discriminator tries to distinguish generated samples from training data. Thus, a minmax game between two networks could be described in the following function:
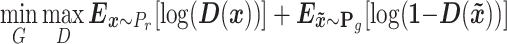 |
where 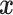 represents training samples from the real distribution, and
represents training samples from the real distribution, and 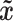 represents the generated samples from the generator model,
represents the generated samples from the generator model,  is implicitly defined by
is implicitly defined by 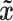
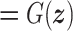 ,
, 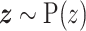 , in which
, in which 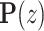 is the latent variable distribution, say a gaussian distribution here. After model training, we could do random sampling from
is the latent variable distribution, say a gaussian distribution here. After model training, we could do random sampling from 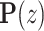 and use the generator network to map them to artificial promoters. We used the WGAN-GP framework which uses Earth Mover's Distance (37) as well as gradient penalty technique (38) to solve the gradient vanishing problem in the original vanilla GAN (39). Besides, the resblock structure (40) is introduced in both the generator and the discriminator model, which helps to solve gradient degradation problem and improves the feature learning ability. The architecture of the WGAN-GP model is shown in Supplementary Figure S1(A).
and use the generator network to map them to artificial promoters. We used the WGAN-GP framework which uses Earth Mover's Distance (37) as well as gradient penalty technique (38) to solve the gradient vanishing problem in the original vanilla GAN (39). Besides, the resblock structure (40) is introduced in both the generator and the discriminator model, which helps to solve gradient degradation problem and improves the feature learning ability. The architecture of the WGAN-GP model is shown in Supplementary Figure S1(A).
In addition, we trained the deep convolutional GAN (DCGAN) model (41) as a control method, which has been widely used for image generation tasks (42). We slightly changed the architecture of the original DCGAN model to adapt to our promoter generation task, which used the 1D convolutional kernel instead of 2D kernel, and decreased the number of convolutional and deconvolutional layers. The architecture of the DCGAN model is shown in Supplementary Figure S2(A).
Model training of GAN models
The training dataset contains a total of 14098 experimentally identified promoters in the E. coli K12 MG1655 genome (43). Most of the promoters recognized in this dataset are 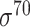 promoters. The promoter sequence is defined as 50 bp upstream of the TSS, which could include key motifs, such as the –10 and –35 regions, but not too long to exclude unnecessary sequences (44).
promoters. The promoter sequence is defined as 50 bp upstream of the TSS, which could include key motifs, such as the –10 and –35 regions, but not too long to exclude unnecessary sequences (44).
We used all the promoter sequences in the dataset described above as the real samples. In the DCGAN model, the input of the generator was the uniform distribution random variable. The batch size was set to 128, the iteration time was set to 100, and we used stochastic gradient descent as the optimization method of our model.
Different from the DCGAN model, in the WGAN-GP model, we sampled from the standard normal distribution as the input random variable of the generator. The batch size was set to 32, and we trained our network for 160 epochs. We found that the best result was approximately 12 epochs, so we selected synthetic promoters from that range of iterations. Note that to obtain the best result from the WGAN-GP model, we trained 5 times for discriminator and one time for generator in each batch training (37). The optimizer used Adam with a learning rate equal to 0.0001, beta1 equal to 0.5 and beta2 equal to 0.9.
The model training of predictive models
For the first round of preselection, we trained a convolutional neural network (CNN) predictive model based on public transcriptome data. The training dataset was from Thomason et al., which contains 14098 promoters with corresponding gene expression levels measured by dRNA-seq (43). The batch size was set to 128, and we used stochastic gradient descent (SGD) as the optimization method. We trained this model with 9000 samples as the training set, 1000 samples as the validation set and others as the testing set, and we achieved a Pearson correlation coefficient (PCC) = 0.25 in the testing set. Notice that we used kernel size equal to 6 to capture the –10 and –35 motifs in the promoter region.
For the second round of preselection, we trained a support vector regression (SVR) model based on the first-round results, which included 83 model-generated promoter sequences and five random sequences. We used the radial basis function (RBF) kernel in the SVR model, i.e.
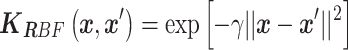 |
Here, the inverse of the standard deviation  was selected by the grid search technique with 5-fold cross validation. To test the performance of the SVR model, we used 66 promoter sequences as the training and validation set and 22 promoter sequences as the testing set. The model achieved a Pearson correlation coefficient of 0.57 in the testing set. We also tested two previous models which predicted the promoter activity by linear SVR model (45) and artificial neural network (ANN) (33), and they achieved Pearson correlation coefficient of 0.514 and 0.480 respectively on our dataset. These results demonstrated the effectiveness of our model.
was selected by the grid search technique with 5-fold cross validation. To test the performance of the SVR model, we used 66 promoter sequences as the training and validation set and 22 promoter sequences as the testing set. The model achieved a Pearson correlation coefficient of 0.57 in the testing set. We also tested two previous models which predicted the promoter activity by linear SVR model (45) and artificial neural network (ANN) (33), and they achieved Pearson correlation coefficient of 0.514 and 0.480 respectively on our dataset. These results demonstrated the effectiveness of our model.
Finding the –10 and –35 motifs by PSSM matrix
We first used the occurrence possibility of each position in the promoter sequences to calculate the PSSM matrix and then calculated the logit function, i.e., 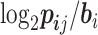 , where
, where 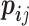 implied the element in the PSSM matrix and
implied the element in the PSSM matrix and 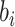 implied the background distribution. Here, we selected the background by calculating the occurrence possibility of T, C, G, A in the whole dataset. The –10 and –35 motif-finding regions were restricted to 1–23 bp and 25–45 bp upstream of TSS, respectively.
implied the background distribution. Here, we selected the background by calculating the occurrence possibility of T, C, G, A in the whole dataset. The –10 and –35 motif-finding regions were restricted to 1–23 bp and 25–45 bp upstream of TSS, respectively.
Novel motifs found by DREME and FIMO
By exploiting our computational and experimental results, we used DREME (46) with e-value threshold 10−50 to find novel motifs from the top 1% highest expression promoters selected by our second-round predictor. Then, the experimental high-expression promoters were scanned by FIMO (47) with P-value threshold 10−3 to search the novel motifs found by DREME. Other parameters for DREME and FIMO were set by default.
BLAST search on experimental functional promoters
BLAST compared 55 AI-designed functional promoters (38 from the first round and 17 from the second round) with E. coli K-12 genome (taxid:83333). Here, we used the default setting in blastn algorithm.
RESULTS
An AI framework for de novo promoter design
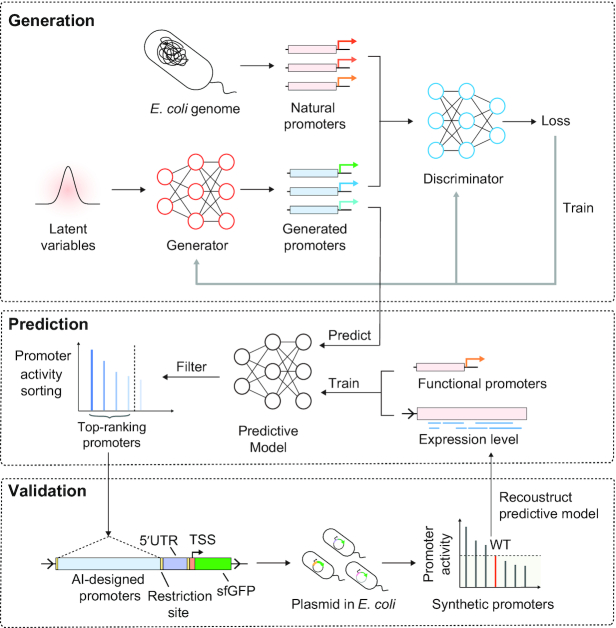
To generate functional synthetic promoters, we introduced an AI-based design workflow (Figure 1), including a GAN network for de novo promoter generation and a predictive model to select promoters with high activity. Then, the generated synthetic promoters were tested by fluorescent protein expression in E. coli.
Figure 1.
The synthetic promoter design approach. In the generation stage, millions of synthetic promoters were generated by a GAN model. By activity prediction, candidate promoters with high potential expression levels were selected and experimentally validated in vivo. The promoter activity of first-round promoters tested in E. coli were used to reconstruct the predictive model for the second-round optimization.
The GAN works as follows: the generator network takes samples from the low-dimensional latent space and maps them to artificial promoters, while the discriminator network evaluates the divergence between the generated promoters and the natural promoters. Based on the minimax game between the discriminator and generator, the GAN model could generate new sequences according to the feature distribution learned from natural promoters. A predictive model trained by gene expression data was introduced to predict sequence activity to preselect the most promising artificial promoters. (Materials and Methods).
AI-designed promoters were cloned into a reporter vector to build a promoter library in E. coli (Supplementary file 1). A fixed 5′UTR region was used to control the influence of interaction effects between core transcriptional elements (30). These promoters were designed to drive the expression of the sfgfp gene (48), and their activities were verified in vivo.
The WGAN-GP model captured essential promoter sequence features
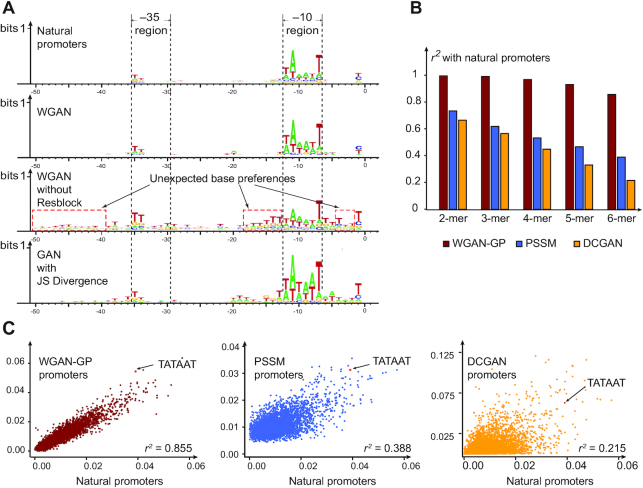
We tried several GAN frameworks to generate promoters and found that the WGAN-GP (38) model (Supplementary Figure S1) could efficiently learn the essential promoter features (Figure 2). Three critical characteristics of the WGAN-GP model (25,38) are important: convolutional layers, resblock structure (40) and the utilization of Earth Mover's Distance (37). The convolutional layers have been reported to extract motif features as well as high-order base pair dependencies (49–51). The resblock network structure was first proposed (40) to handle the gradient degradation problem in DNNs to improve feature learning abilities. We found that the resblock structure helped decrease the biased base occurrence during promoter generation (Figure 2(A)). The Earth Mover's Distance is continuous and provides a usable gradient everywhere (37), which showed more promising results than the Jensen-Shannon (JS divergence) used in the original vanilla GAN model (19) (Figure 2(A)).
Figure 2.
The -10 and -35 motif and k-mer distribution of PSSM, DCGAN and WGAN-GP generated promoters. (A) The sequence logos of natural promoters, artificial sequences generated by WGAN-GP with resblock, without resblock and generated by original vanilla GAN with JS divergence. (B) R-squared value evaluates the correlation of k-mer frequencies (k = 2–6) between natural promoters and artificial sequences designed by WGAN-GP, PSSM and DCGAN. (C) The 6-mer scatter plot of natural promoters and generated promoters. Each point represents a certain 6-mer. The x- and y-axes represent the k-mer frequencies in natural and generated promoters.
To make a comprehensive evaluation of the AI-designed promoters, we analyzed the features of them computationally and experimentally. We firstly analyzed the distribution of the WGAN-GP-generated promoters computationally by the following three aspects: the sequence motif logo, k-mer frequency and the motif spacing constraint. We also tried the deep convolutional GAN (DCGAN)-based network structure (Supplementary Figure S2(A)) as a control method, which was also widely used in image generation tasks (42). The position-specific scoring matrix (PSSM) sampling method was used as another control method; this method independently generated bases according to natural promoter base frequencies.
In terms of the sequence motif logo, the DCGAN model could partly learn sequence motifs in the –10 and –35 regions, but some base preferences appeared in the spacer sequence compared to the base distribution of WGAN-GP promoters (Supplementary Figure S1(B)). We calculated the 2-mer to 6-mer base frequency in promoters generated by the WGAN-GP, PSSM and DCGAN models (Supplementary Figure S3) and found that the occurrence frequency of some common 6-mers, such as TATAAT, increased with the WGAN-GP method, whereas they decreased with the PSSM method (Figure 2(C)), and the correlation of k-mer frequency between natural promoters and DCGAN-generated promoters dropped quickly when the k increased (Figure 2(B)). We also analyzed the top 10 most frequently occurring 6-mers of natural promoters in the WGAN-GP, DCGAN and PSSM promoters (Supplementary Table S2). The WGAN-GP promoters shared five common 6-mers with natural promoters, while DCGAN promoters and PSSM promoters both shared only one 6-mer, indicating that the WGAN-GP model captured important k-mers in natural promoters.
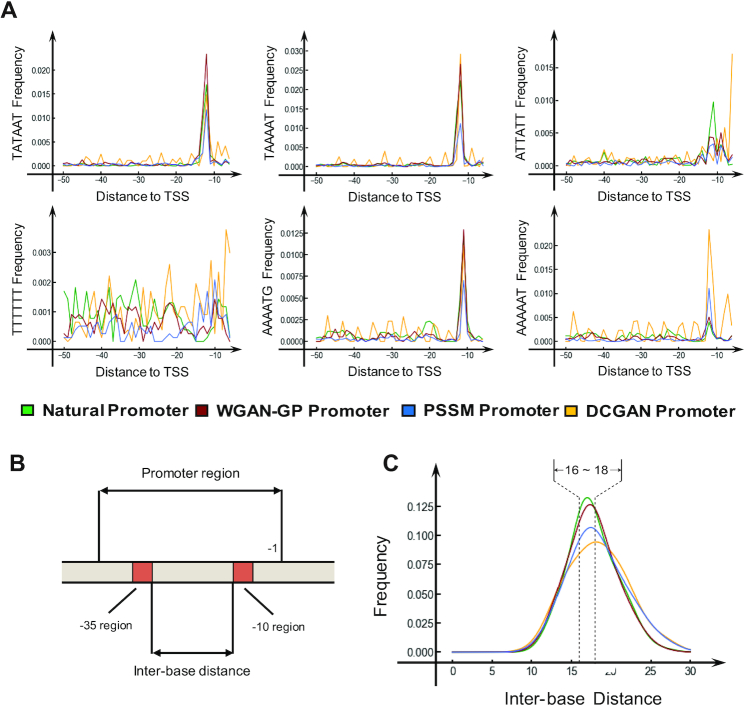
To explore the position distribution of the naturally most frequently occurring 6-mers and the Pribnow box (TATAAT) in these model-generated promoters, their relative distances to the transcriptional start site (TSS) were analyzed. The position distribution of the top 20 most frequently occurring 6-mers in natural promoters is shown in Supplementary Figure S4. As shown in Figure 3(A), the WGAN-GP model outperformed the PSSM method and showed a more similar position distribution with natural promoters, which demonstrated that the WGAN-GP model could learn the k-mer location preference of natural promoters. In addition, the separation between the –10 and –35 regions was also calculated. As a result, the length of the separation between –10 and –35 regions of the WGAN-GP promoters was more centrally distributed in the interval of 16–18 bp compared with those of the DCGAN and PSSM promoters (Figure 3B, C). It was previously reported that a separation of ∼16–18 bp was beneficial for the binding of RNA polymerase (52), thus indicating that the WGAN-GP model-generated promoters offered a better chance for RNA polymerase binding.
Figure 3.
The location of high-frequency k-mers and the separation between the –10 and –35 regions. (A) Pribnow box consensus sequence (TATAAT) and the top five common 6-mers are selected to illustrate the location learning ability of the GAN model. The x-axis represents the location relative to the TSS, and the y-axis represents the frequency of the 6-mer at this location. (B) The definition of inter-base distance between –10 and –35 motifs. (C) The inter-base distance distribution between the –10 and –35 motifs. The natural promoters are marked in green, and the WGAN-GP, PSSM and DCGAN-generated promoters are marked in red, blue, and orange, respectively.
In conclusion, compared with the PSSM and DCGAN models, which cannot learn long-range base pair dependencies, the WGAN-GP model showed the ability to capture higher-order promoter sequence features, including crucial k-mer frequency, k-mer location preference and separation constraint.
The AI-designed promoters showed a high success rate and transcriptional activity
After generating millions of artificial sequences with critical characteristics, a predictive model (Supplementary Figure S5) was trained for selecting the most promising sequences. We then tested 83 WGAN-GP designed sequences with the top predicted activity in E. coli. In addition, six positive controls, including two wild-type promoters (BBa_J23119 and Trc1) and their corresponding strongest mutants (33) (BBa_J23100, BBa_J23102 and Trc2, Trc3) were also tested. Furthermore, five random sequences with controlled GC content were generated as negative control variants (Materials and Methods). The detailed sequences are provided in Supplementary Table S1.
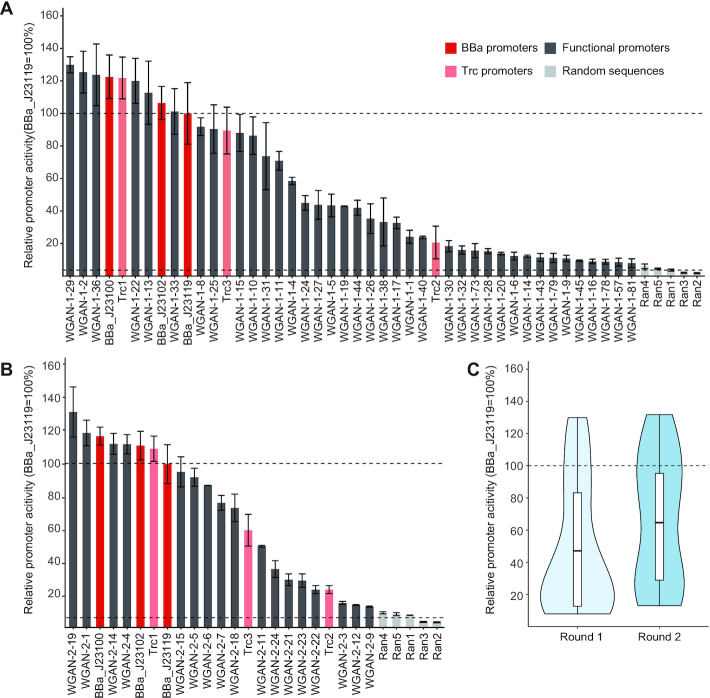
As a result, 45.8% (38 out of 83) of AI-designed promoters showed significantly higher promoter activities than random sequences (t-test with Benjamini Hochberg correction, FDR < 5%). Interestingly, three of the synthetic promoters showed comparable or even higher activities than any of the six positive control promoters (Figure 4 and Supplementary Figure S6). We also tested the promoter activity of 44 WGAN-GP designed promoters using the reporter gene mrfp1 and the results were highly correlated with those measured using sfgfp (Pearson correlation coefficient (PCC) = 0.81, Supplementary Figure S7).
Figure 4.
(A) The promoter activity of 38 first-round AI-designed functional promoters (WGAN-1). BBa_J23119 (dark red) and Trc1 (light red) are wild-type promoters. BBa_J23100, BBa_J23102 and Trc2, Trc3 are their highly-expressed mutants. Ran1–5 represent five random sequences. The error bar represents the standard deviation of three biological replicates. The AI-designed promoters (dark grey) are named by the predicted expression level rank obtained from the CNN model. The dashed lines are the 100% baseline represented by BBa_J23119 (high) and average relative activity of five random sequences (low), respectively. (B) The promoter activity of 24 second-round AI-designed functional promoters (WGAN-2). The second-round AI-designed promoters (dark grey) are named by the predicted expression level rank obtained from the SVR model. (C) The promoter activity distribution of first- and second-round AI-designed functional promoters.
For comparison, we tested the in vivo promoter activity of 20 DCGAN-generated promoters selected by our predictive model (Supplementary Table S1, Supplementary Figure S8). As a result, the successful design rate (20%) was significantly lower than the WGAN-generated promoters (Fisher exact test, P-value < 0.03). We also synthesized 91 artificial sequences containing the same –10 and –35 motifs as the wild-type Trc1 promoter with the other bases randomly synthesized (Supplementary Table S1). Few of these sequences showed promoter activity in E. coli (Supplementary Figure S9), demonstrating that our AI model gained other important features beyond canonical –10 and –35 boxes and generated promoters with high success rates and strong activity.
Iterative optimization significantly improved model performance
The experimental validation of designed promoters demonstrated the feasibility of the AI framework. However, as the genome wide gene expression profile not only depends on promoter activity but also many other regulatory elements, the correlation between the predicted promoter activity and dRNA-seq level in the testing set was moderate (Pearson correlation coefficient (PCC) = 0.25, Supplementary Figure S10). And due to the short half-lives of mRNA in E. coli, it has been shown that the mRNA and protein level could have relatively low correlation (53), the success rate of functional promoters could be limited by the activity prediction model.
To further improve the performance of promoter design strategy, we adopted an iterative strategy by using the first-round promoters to reconstruct the predictive model. In the second round, the reconstructed predictor was trained based on the sfGFP expression level of the first-round WGAN-GP promoters. Considering the relatively small sample size of the first-round tested promoters, we trained a SVR model instead of deep neural networks. This model achieved a much higher PCC of 0.57 (materials and methods, Supplementary Figure S10). Twenty-four promoters were selected in the second round, which were sequences predicted to have the highest expression level by the SVR model. These promoters were cloned into a reporter vector in E. coli to test the promoter activity. As a result, the promoters in the second round showed on average 1.68-fold higher expression activity than those in the first round, and the success rate was significantly improved from 45.8% to 70.8% (Fisher exact test, P-value < 0.05).
AI-model explored new sequence features
To explore the novel features in our AI-designed promoters, we first discovered sequence motifs in the top 1% computationally highly-expressed WGAN-GP generated promoters by DREME (46) (e-value < 10−50). We used FIMO (47) to search these motifs in experimentally validated highly-expressed promoters and found several noncanonical motifs occurring in these promoters (Supplementary Figure S11). As an example, the sequence ‘TACCCT’ from the first noncanonical motif appeared twice in the top 5 most highly-expressed promoters. This sequence differed from the canonical –10 motif (TATAAT) but showed a similar distance (–12 to –7 bp) to TSS, which suggested that AI-based method could capture noncanonical motifs to help design the highly-expressed promoters. Considering these new motif features could be helpful in designing novel synthetic promoters.
To examine how the AI-designed promoters were different from natural promoters, a standard nucleotide BLAST search on experimental functional promoters was conducted against the whole E. coli genome, and no high similarity matches were found. The average e-value obtained for functional AI-designed promoters was 34.98, indicating that the newly designed functional promoters have low similarity with natural E. coli genome (Supplementary Figure S12). The e-value of our functional promoters and random sequences were at the same level and our functional promoters showed lower similarity to the natural genome than the promoters designed by Alper (13) and most constitutive 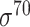 promoters in iGEM BioBrick (34) standard parts (Supplementary Table S3). These results suggested that our framework could design novel synthetic promoters rather than copying the original sequences, indicating that the DNN could effectively explore the sequence space to find novel promoters for E. coli.
promoters in iGEM BioBrick (34) standard parts (Supplementary Table S3). These results suggested that our framework could design novel synthetic promoters rather than copying the original sequences, indicating that the DNN could effectively explore the sequence space to find novel promoters for E. coli.
DISCUSSION
In this study, we conducted de novo promoter design based on the GAN framework and validated synthetic promoter activities in vivo. Our method benefited from recent developments in the deep generative model, which helped us to extract higher-order sequence features and generate millions of novel promoters. In vivo experimental results suggested that 70.8% of the selected novel sequences are functional promoters in E. coli. These generated promoters inherited the key features of E. coli promoters, such as the –10 and –35 regions, and shared similar k-mer frequencies with natural promoters. Meanwhile the promoters showed substantial sequence differences from the genomic sequences, which could help avoid genetic instability due to the lower probability of recombination with the E. coli genome (9).
Naturally occurring promoters have evolved for millions of years but make up only a small subset of the large potential sequence space. While taking advantage of the powerful feature learning ability of deep learning, a great number of synthetic promoters could be automatically designed, which could largely extend functional promoter sequence reservoirs.
Conventional sequence generation methods by which fixing the –10 and –35 regions and randomizing the surrounding nucleotides generated ∼46 functional inducible lac promoters in Lactococcus lactis (54), but this lac promoter selection method could not work in E. coli (55). The mutagenic PCR method used in E. coli by which the Hamming distance to the natural promoter is typically 1–2 base pair (56), and showed that less than 0.1% of colonies could be functional promoters (13). Here, we fixed the –10 and –35 regions of wild-type promoter in E. coli, sampling the functional promoters in the possible combinatorial space of ∼1019 sequences, showing that directly randomizing the surrounding bases of –10 and –35 regions could not find promoters efficiently, indicating that sequence beyond the –10 and –35 regions also contain important information for generating functional promoters.
From the perspective of pattern recognition, transcriptional machinery could be considered as a molecular classifier, which distinguishes real promoter sequences from the other genomic regions to initiate transcription. Thus, synthetic promoters need to have similar properties as natural promoters to recruit the transcriptional machinery. GANs suit this logic well: the discriminator learns to distinguish the real promoters from the artificial ones, mimicking the role of transcriptional machinery. The generator tries to produce artificial sequences that have crucial sequence features similar to those of natural promoters, resembling the mutation process in nature. Unlike the naturally occurring mechanisms, in which promoters are mutated randomly and passively, the AI model could automatically learn to generate optimized sequences, and the model performance could be greatly improved by testing a relatively small number of promoters experimentally, showing its potential ability to reduce the scale of biological screening experiments.
Recently, deep generative models has shown great potential in generating novel images, antimicrobial peptides (26) and small molecular drugs (28), etc. The most powerful aspect of the model is that it could automatically extract crucial features from training samples even without prior knowledge constrains. Thus, we expect that such model frame would also be used to learn different crucial sequence features of other genetic elements. An interesting future attempt could be the generation of synthetic regulatory elements optimized for specific properties. For example, the conditional GAN model (57), which could generate samples with different properties conditioning on additional information, could be implemented to design conditional specific promoters. Combining other recent advances in machine learning like few-shot learning (58), transfer learning (59) and reinforcement learning (60) may help us to learn the model with only a few training samples such as light-response or stress-response promoters.
As our prediction model was trained by a relatively small number of promoters tested experimentally, further improvement could be achieved by high-throughput experiments, such as massively parallel reporter assay, which could test the strength of thousands promoters in a single experiment (61). Combining the high-throughput experiments with AI-based design could help improve the design efficiency of synthetic promoters.
In summary, we proposed an AI-based generative framework for the de novo design of promoter sequences, which showed a high success rate based on experimental validation in vivo. Our work provided new insights into de novo synthetic element design, indicating the potential ability for deep learning approaches to explore the sequence space of synthetic elements.
DATA AVAILABILITY
The computer source code is available from the public GitHub repository (https://github.com/HaochenW/Deep_promoter).
Supplementary Material
ACKNOWLEDGEMENTS
X.W.W., Y.W. and H.C.W. conceived the study. H.C.W. implemented in silico designs. Y.W., W.L. and L.S.L. designed the experiments. Y.W. performed the experiments and analyzed the data. Y.W. and H.C.W. wrote the manuscript. We also thank Huan Fang, Kui Hua and Xianglin Zhang for the positive discussion.
Contributor Information
Ye Wang, Ministry of Education Key Laboratory of Bioinformatics; Center for Synthetic and Systems Biology; Bioinformatics Division, Beijing National Research Center for Information Science and Technology; Department of Automation, Tsinghua University, Beijing 100084, China.
Haochen Wang, Ministry of Education Key Laboratory of Bioinformatics; Center for Synthetic and Systems Biology; Bioinformatics Division, Beijing National Research Center for Information Science and Technology; Department of Automation, Tsinghua University, Beijing 100084, China.
Lei Wei, Ministry of Education Key Laboratory of Bioinformatics; Center for Synthetic and Systems Biology; Bioinformatics Division, Beijing National Research Center for Information Science and Technology; Department of Automation, Tsinghua University, Beijing 100084, China.
Shuailin Li, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Liyang Liu, Ministry of Education Key Laboratory of Bioinformatics; Center for Synthetic and Systems Biology; Bioinformatics Division, Beijing National Research Center for Information Science and Technology; Department of Automation, Tsinghua University, Beijing 100084, China.
Xiaowo Wang, Ministry of Education Key Laboratory of Bioinformatics; Center for Synthetic and Systems Biology; Bioinformatics Division, Beijing National Research Center for Information Science and Technology; Department of Automation, Tsinghua University, Beijing 100084, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [61773230, 61721003]. Funding for open access charge: National Natural Science Foundation of China
Conflict of interest statement. None declared.
REFERENCES
- 1. Lynch S.A., Gill R.T.. Synthetic biology: new strategies for directing design. Metab. Eng. 2012; 14:205–211. [DOI] [PubMed] [Google Scholar]
- 2. Sadeghpour M., Veliz-Cuba A., Orosz G., Josić K., Bennett M.R.. Bistability and oscillations in co-repressive synthetic microbial consortia. Quant. Biol. 2017; 5:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meng H., Ma Y., Mai G., Wang Y., Liu C.. Construction of precise support vector machine based models for predicting promoter strength. Quant. Biol. 2017; 5:90–98. [Google Scholar]
- 4. Guiziou S., Sauveplane V., Chang H.J., Clerte C., Declerck N., Jules M., Bonnet J.. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res. 2016; 44:7495–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Mey M., Maertens J., Lequeux G.J., Soetaert W.K., Vandamme E.J.. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotech. 2007; 7:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilman J., Love J.. Synthetic promoter design for new microbial chassis. Biochem. Soc. Trans. 2016; 44:731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nevoigt E., Kohnke J., Fischer C.R., Alper H., Stahl U., Stephanopoulos G.. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006; 72:5266–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Du J., Yuan Y., Si T., Lian J., Zhao H.. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012; 40:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Portela R.M., Vogl T., Kniely C., Fischer J.E., Oliveira R., Glieder A.. Synthetic core promoters as universal parts for fine-tuning expression in different yeast species. ACS Synth. Biol. 2017; 6:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blazeck J., Liu L., Redden H., Alper H.. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl. Environ. Microbiol. 2011; 77:7905–7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blazeck J., Garg R., Reed B., Alper H.S.. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 2012; 109:2884–2895. [DOI] [PubMed] [Google Scholar]
- 12. Yim S.S., An S.J., Kang M., Lee J., Jeong K.J.. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol. Bioeng. 2013; 110:2959–2969. [DOI] [PubMed] [Google Scholar]
- 13. Alper H., Fischer C., Nevoigt E., Stephanopoulos G.. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:12678–12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vogl T., Ruth C., Pitzer J., Kickenweiz T., Glieder A.. Synthetic core promoters for Pichia pastoris. ACS Synth. Biol. 2014; 3:188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weingarten-Gabbay S., Nir R., Lubliner S., Sharon E., Kalma Y., Weinberger A., Segal E.. Systematic interrogation of human promoters. Genome Res. 2019; 29:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guazzaroni M.-E., Silva-Rocha R.. Expanding the logic of bacterial promoters using engineered overlapping operators for global regulators. ACS Synth. Biol. 2014; 3:666–675. [DOI] [PubMed] [Google Scholar]
- 17. Liu D., Mao Z., Guo J., Wei L., Ma H., Tang Y., Chen T., Wang Z., Zhao X.. Construction, model-based analysis, and characterization of a promoter library for fine-tuned gene expression in Bacillus subtilis. ACS Synth. Biol. 2018; 7:1785–1797. [DOI] [PubMed] [Google Scholar]
- 18. Mohamed H., Chernajovsky Y., Gould D.. Assembly PCR synthesis of optimally designed, compact, multi-responsive promoters suited to gene therapy application. Sci. Rep. 2016; 6:29388–29400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y.. Generative adversarial nets. Adv. Neural Inform. Process. Syst. 2014; 2672–2680. [Google Scholar]
- 20. Ledig C., Theis L., Huszar F., Caballero J., Cunningham A., Acosta A., Aitken A.P., Tejani A., Totz J., Wang Z.. Photo-realistic single image super-resolution using a generative adversarial network. Comput. Vis. Pattern Recognit. 2017; 105–114. [Google Scholar]
- 21. Zhu J.-Y., Park T., Isola P., Efros A.A.. Unpaired image-to-image translation using cycle-consistent adversarial networks. Proc. IEEE Int. Conf. Comput. Vis. 2017; 2223–2232. [Google Scholar]
- 22. Odena A., Olah C., Shlens J.. Conditional image synthesis with auxiliary classifier GANs. Proceedings of the 34th International Conference on Machine Learning. 2017; 70:2642–2651. [Google Scholar]
- 23. Denton E.L., Chintala S., Fergus R.. Deep generative image models using a laplacian pyramid of adversarial networks. Adv. Neural Inform. Process. Syst. 2015; 1486–1494. [Google Scholar]
- 24. Yang J., Kannan A., Batra D., Parikh D.. LR-GAN: layered recursive generative adversarial networks for image generation. Int. Conf. Learn. Represent. 2017; [Google Scholar]
- 25. Killoran N., Lee L.J., Delong A., Duvenaud D., Frey B.J.. Generating and designing DNA with deep generative models. 2017; arXiv doi:17 December 2017, preprint: not peer reviewedhttps://arxiv.org/abs/1712.06148.
- 26. Gupta A., Zou J.. Feedback GAN for DNA optimizes protein functions. Nat. Mach. Intell. 2019; 1:105–111. [Google Scholar]
- 27. Putin E., Asadulaev A., Ivanenkov Y., Aladinskiy V., Sanchez-Lengeling B., Aspuru-Guzik A., Zhavoronkov A.. Reinforced adversarial neural computer for de novo molecular design. J. Chem. Inf. Model. 2018; 58:1194–1204. [DOI] [PubMed] [Google Scholar]
- 28. Kadurin A., Nikolenko S., Khrabrov K., Aliper A., Zhavoronkov A.. druGAN: an advanced generative adversarial autoencoder model for de novo generation of new molecules with desired molecular properties in silico. Mol. Pharm. 2017; 14:3098–3104. [DOI] [PubMed] [Google Scholar]
- 29. De Cao N., Kipf T.. MolGAN: An implicit generative model for small molecular graphs. ICML 2018 workshop on Theoretical Foundations and Applications of Deep Generative Models. 2018; [Google Scholar]
- 30. Davis J.H., Rubin A.J., Sauer R.T.. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011; 39:1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al.. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 32. Kelly J.R., Rubin A.J., Davis J.H., Ajo-Franklin C.M., Cumbers J., Czar M.J., de Mora K., Glieberman A.L., Monie D.D., Endy D.. Measuring the activity of BioBrick promoters using an in vivo reference standard. J Biol Eng. 2009; 3:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meng H., Wang J., Xiong Z., Xu F., Zhao G., Wang Y.. Quantitative design of regulatory elements based on high-precision strength prediction using artificial neural network. PLoS One. 2013; 8:e60288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smolke C.D. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 2009; 27:1099–1102. [DOI] [PubMed] [Google Scholar]
- 35. Zhang H., Goodfellow I., Metaxas D., Odena A.. Self-attention generative adversarial networks. Int. Conf. Mach. Learn. 2019; 7354–7363. [Google Scholar]
- 36. Choi Y., Choi M., Kim M., Ha J.-W., Kim S., Choo J.. StarGAN: unified generative adversarial networks for multi-domain image-to-image translation. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2018; 8789–8797. [Google Scholar]
- 37. Arjovsky M., Chintala S., Bottou L.. Wasserstein generative adversarial networks. Proceedings of the 34th International Conference on Machine Learning. 2017; 70:214–223. [Google Scholar]
- 38. Gulrajani I., Ahmed F., Arjovsky M., Dumoulin V., Courville A.C.. Improved training of wasserstein GANs. Adv. Neural Inform. Process. Syst. 2017; 5767–5777. [Google Scholar]
- 39. Arjovsky M., Bottou L.. Towards principled methods for training generative adversarial networks. Int. Conf. Learn. Represent. 2017; [Google Scholar]
- 40. He K., Zhang X., Ren S., Sun J.. Deep residual learning for image recognition. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2016; 770–778. [Google Scholar]
- 41. Radford A., Metz L., Chintala S.. Unsupervised representation learning with deep convolutional generative adversarial networks. 2015; arXiv doii:07 January 2016, preprint: not peer reviewed,https://arxiv.org/abs/1511.06434.
- 42. Isola P., Zhu J.-Y., Zhou T., Efros A.A.. Image-to-image translation with conditional adversarial networks. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2017; 1125–1134. [Google Scholar]
- 43. Thomason M.K., Bischler T., Eisenbart S.K., Forstner K.U., Zhang A., Herbig A., Nieselt K., Sharma C.M., Storz G.. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2015; 197:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim D., Hong J.S.-J., Qiu Y., Nagarajan H., Seo J.-H., Cho B.-K., Tsai S.-F., Palsson B.Ø. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLos Genet. 2012; 8:e1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiryu H., Oshima T., Asai K.. Extracting relations between promoter sequences and their strengths from microarray data. Bioinformatics. 2005; 21:1062–1068. [DOI] [PubMed] [Google Scholar]
- 46. Bailey T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011; 27:1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S.. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006; 24:79–88. [DOI] [PubMed] [Google Scholar]
- 49. Quang D., Xie X.. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016; 44:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeng H., Edwards M.D., Liu G., Gifford D.K.. Convolutional neural network architectures for predicting DNA–protein binding. Bioinformatics. 2016; 32:i121–i127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kelley D.R., Snoek J., Rinn J.L.. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016; 26:990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harley C.B., Reynolds R.P.. Analysis of E. coli pormoter sequences. Nucleic Acids Res. 1987; 15:2343–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li G.-W., Xie X.S.. Central dogma at the single-molecule level in living cells. Nature. 2011; 475:308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jensen P.R., Hammer K.. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 1998; 64:82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jensen P.R., Hammer K.. Artificial promoters for metabolic optimization. Biotechnol. Bioeng. 2010; 58:191–195. [PubMed] [Google Scholar]
- 56. Pritchard L., Corne D., Kell D., Rowland J., Winson M.. A general model of error-prone PCR. J. Theor. Biol. 2005; 234:497–509. [DOI] [PubMed] [Google Scholar]
- 57. Mirza M., Osindero S.. Conditional generative adversarial nets. 2014; arXiv doi:06 November 2014, preprint: not peer-reviewedhttps://arxiv.org/abs/1411.1784.
- 58. Koch G., Zemel R., Salakhutdinov R.. Siamese neural networks for one-shot image recognition. ICML Deep Learning Workshop. 2015; 2: [Google Scholar]
- 59. Segler M.H., Kogej T., Tyrchan C., Waller M.P.. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Central Sci. 2017; 4:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Popova M., Isayev O., Tropsha A.. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018; 4:eaap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P., Feizi S., Gnirke A., Callan C.G., Kinney J.B. et al.. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012; 30:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The computer source code is available from the public GitHub repository (https://github.com/HaochenW/Deep_promoter).