Supplemental Digital Content is available in the text
Keywords: cetuximab, colorectal cancer, integrin-beta1, signaling pathway
Abstract
Background:
Colorectal cancer is the second commonly seen cancer around the world and accounts for 13% of all human cancers. Among them, 25% of all case were diagnosed with metastasis and 50% occurs metastasis during the development of disease. Cetuximab is a chimeric monoclonal antibody against epidermal growth factor receptor, and is used for treatment of metastatic colorectal cancer alone or combined with chemotherapy or radiation therapy. Integrin-beta 1 (ITGB1), which is also known as CD29, and plays an important role in development of malignant cancers. However, the effect of ITGB1 in promoting the anti-tumor effect of cetuximab is not fully understand.
Methods:
The model of ITGB1 inhibition and overexpression was firstly constructed in LS174T cells, and the viability of cells in each group was detected using CCK-8 assay. The expression of key factors in tumor formation process at transcription level was detected using real-time quantitative polymerase chain reaction method. The expression of key proteins in metastasis process, cell apoptosis and activation of Ras/Raf/MEK signaling pathway was detected using western blotting analysis. And the concentration of key factors of in tumor formation process in cultured medium of LS174T cells were detected using enzyme-linked immunosorbent assay method.
Results:
We found that cetuximab could inhibit the proliferation of LS174T cells, and inhibition of ITGB1 enhanced this effect while overexpression of ITGB1 reduced this effect. We further found that cetuximab could inhibit the expression and secretion of extracellular matrix degradation related molecules in cultured medium and transcription level. Besides, we also found that the expression of key factors in angiogenesis and extracellular matrix degradation related proteins were also reduced after cetuximab treatment. These effects might be mediated by Ras/Raf/MAPK signaling pathway and enhanced after inhibition of ITGB1 expression.
Conclusion:
Inhibition of ITGB1 might be a new therapeutic method in colorectal cancer.
1. Introduction
Colorectal cancer (CRC) is one of the most commonly seen cancers worldwide, which is characterized by high mortality caused by strong metastatic potential. However, the treatment for CRC was still in a slow progression.[1,2] Cetuximab is approved as the first-line treatment for CRC alone or combined with chemotherapy for patients with epidermal growth factor receptor (EGFR) expression or RAS wild-type metastatic colorectal cancer, and presented an effective inhibition effect for CRC.[3] Treatment of cetuximab combined with chemotherapy prolong the survival time for 2.6 months compared with chemotherapy alone.[4] Previous study indicated that integrin-beta 1 (ITGB1) is overexpressed in tumor cells and participate in angiogenesis, tumor progression and metastasis process.[5,6] There is also study indicated that ITGB1 induces the resistance of cancer cells to radiotherapies, chemotherapies and other therapies.[7,8] Based on these findings, researchers has been studied extensively of ITGB1 in terms of biology of tumors, and expression of ITGB1 at protein or RNA level has been indicated to be relater to poor prognosis in lung cancer, breast cancer, and colorectal cancer.[9–11] However, the effect of ITGB1 in promoting the anti-tumor effect of cetuximab was not fully understood. In this study, we firstly established a cell model of ITGB1 inhibition or overexpression in LS174T cells, and found that inhibition of ITGB1 could enhance the inhibition effect of cetuximab on proliferation of cancer cells. We also found that inhibition of ITGB1 also decreased the expression and secretion of extracellular matrix (ECM) degradation and angiogenesis factors, and these effects might be mediated by Ras/Raf/MAPK signaling pathway. We speculated that inhibition of ITGB1 might be a new therapeutic target of CRC.
2. Material and methods
2.1. Reagents
RPMI1640 (12633012) and FBS (10099141) were purchased from Thermo. Cetuximab (A2000) was purchased from Selleck. Human SDF-1α (ab100637), E-Cadherin (ab233611), CXCL9 (ab219047), CXCL10 (ab83700), MMP2 (ab100606) and MMP9 (ab100610) enzyme-linked immunosorbent assay (ELISA) kits were purchase from Abcam. EGFR (ab52894, 175), Ras (ab52939, 21), Raf (ab200653, 68), MEK1 (ab32091, 45), p-MEK1 (ab96379, 45), p-STAT3 (ab 76315, 87), STAT3 (ab 119352, 87), SDF-1 (ab155090, 11), CXCR4 (ab124824, 39), VEGF-α (ab53465, 27), CXCL9 (ab193851), CXCL10 (ab8098), vimentin (ab8978, 57), Fibronectin 1 (ab2413, 285).
2.2. Ethical statement
Our study did not require an ethical board approval because it did not contain human or animal trials.
2.3. Vector construction
Full length of ITGB1 cDNA was obtained using PCR method with following primer: forward: 5’-CCTCTCAGCCTCCAGCGTTG-3’, reverse: 5’-TGCTCTTGCTCACTCACACTCC-3’. Blank Flag-HA-pcDNA3.1 vector and cDNA were firstly digested by BamHI (R3136S, NEB) and KpnI (R3142S, NEB) enzymes. And the ITBG1 overexpression vector was constructed using T4 Ligase (M0202S, NEB). Then, vector was transfected into LS174T cells using Lipofectamine 3000 transfection reagent (L3000015, Invitrogen) for 48 hour. Stable expressed cells were screened by 800 μg/mL G418 for 2 weeks. ITGB1 knock down vector was constructed according to previous study.[12] Briefly, oligos were obtained using T4 PNK (M0201S, NEB) with following primer: forward: 5’-CACCGTGCTGTGTGTTTGCTCAAAC-3’, reverse: 5’-AAACGTTTGAGCAAACACACAGCACGGTGC-3’, and incubated at 37°C for 30 minute followed with incubation at 95°C for 5 minute. Then, lentiCRISPRv2 vector was digested using BsmBI (R0580S, NEB) overnight. Digested vector and oligos were used to construct IGTB1 knockdown vector using Quick Ligase (M2200S, NEB). ITGB1 knockdown vector was firstly transfected into 293T cells (CRL-11268, ATCC) to construct lentivirus vector. Then, LS174T cells were transfected with lentivirus vector and stable expressed cells were screened using 2 ng/mL puromycin.
2.4. Cultured methods and grouping
LS174T cells (CL-188) were purchased from ATCC. Cells were cultured in RPMI (61870044, Thermo) with 10% FBS (10100, Thermo) under 5% CO2 humid atmosphere at 37°C. Cells were divided into 4 groups: normal control group (NC), cetuximab treatment group (CB), cetuximab treatment combined with ITGB1 inhibition group (CI) and cetuximab treatment combined with ITGB1 overexpression group (CO). And in cetuximab treatment groups, cells were firstly incubated with 50 μg/mL cetuximab for 24 hour before performing the following experiments.[13]
2.5. CCK-8 assay
CCK-8 assay was performed according to the protocol of CCK-8 cell proliferation and cytotoxicity assay kit (CA1210, Solarbio). Briefly, cells in each group were seeded into a 96-well plate at a concentration of 1 × 105, and incubated with cetuximab as previous described. After 24 hour incubation, cells were incubated with CCK-8 reagent for 4 hour, then the optical density was measured at 450 nm using Multiskan GO Spectrophotometer (Thermo).
2.6. RNA extraction and reverse transcription
Total RNA extraction was performed according to the protocol of total RNA extraction kit (R1200, Solarbio). Briefly, cells were seeded into a 100 mm plate and cultured until the confluence reached 70% to 80% followed with incubation with cetuximab for 24 hour. Then, cells were lysed with lysis buffer and chloroform for 5 minute at room temperature. After centrifuged at 12000 rpm for 10 minute at 4°C, water phase was removed into an absorption tube. RNA was absorbed onto the absorption tube after the centrifugation and eluted with elution buffer. Concentration of RNA was determined using NanoDrop 3300 (Thermo). cDNA was synthesized using universal HiFiScript gDNA Removal cDNA Synthesis Kit (CW2582, CWBio). Reaction mixture was made up according to the protocol and reaction was performed at 42°C for 15 minute, 85°C for 5 minute. cDNA was stored at –80°C until performing following experiment.
2.7. Real-time quantitative polymerase chain reaction (qPCR)
qPCR was performed according to the protocol of SuperStar Probe One Step RT-qPCR Kit (CW2695, CWBio) with following primers: MMP-2: forward: 5’-CACCTACACCAAGAACTT-3’, reverse: 5’-GGTCCTTGAAGAAGAAGAT-3’; MMP-9: forward: 5’-GCTTAGATCATTCCTCAGT-3’, reverse: 5’-CATTCACGTCGTCCTTAT-3’; caspase-3: forward: 5’-CGAAACTCTTCATCATTCAGGC-3’, reverse: 5’-AGTAAGCATACAGGAAGTCGGC-3’, caspase-9: forward: 5’-GGCTGTCTACGGCACAGATGCA-3’, reverse: 5’-CTGGCTCGGGGTTACTGCCAG-3’; cyclin D1: forward: 5’-GATGCCAACCTCCTCAACGAC-3’, reverse: 5’-CTCCTCGCACTTCTGTTCCTC-3’; CDK4: forward: 5’-GAGGCGACTGGAGGCTTTT-3’, reverse: 5’-GGATGTGGCACAGACGTCC-3’. The reaction mixture was made up as recommended, and reaction was performed with following steps: reverse transcription at 45°C for 20 minute, pre-degeneration at 95°C for 5 minute, repeat these 2 steps for 40 cycles: degeneration at 95°C for 15 second and annealing at 60°C for 45 second. The expression of relative genes was calculated using the 2-ΔΔCt method.[14] Quantification results for each target gene were normalized to GAPDH. Each experiment was repeated for 3 times.
2.8. Western blotting analysis
Cells in each group were treated as previously described and lysed with lysis buffer (CW2333, CWBio) on ice for 30 minute. After centrifugated at 12000 rpm for 10 minute, concentration of proteins was determined using BCA Protein Assay Kit (CW0014, CWBio). 60 μg protein was separated with 10% SDS-PAGE electrophoresis. After electrophoresis, proteins were transferred onto a PVDF membrane using Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Then, membranes were blocked with 5% skim milk at room temperature for 1 hour and incubated with primary antibodies (1:1000) overnight at 4°C. Then, membranes were incubated with secondary antibody (1:5000) for 1 hour at room temperature. Gray value of proteins were detected using iBright CL1500 (Thermo) via chemiluminescent immunoassay. GAPDH was used as an internal control, each experiment was repeated for 3 times independently.
2.9. Enzyme-linked immunosorbent assay
ELISA was performed according to the protocol. Briefly, standard sample and cultured medium sample were firstly added into each well of a 96-well plate followed with incubation overnight at 4°C. After washed with washing buffer, biotinylated CXCL9 detection antibody was added into each well, and after incubated with HRP-streptavidin solution and TMB 1-step substrate reagent, stop solution was added into each well and optical density was measured at 450 nm using Multiskan GO Spectrophotometer (Thermo).
2.10. Statistical analysis
All of data were presented as the mean ± SD. One-way ANOVA was performed to compare the differences between the groups, and followed by Turdey post hoc test. P-value < .05 was set as statistical difference. SPSS software (version 19.0, Inc., Chicago, IL, USA) was used to analyze the data.
3. Results
3.1. Expression of ITGB1 in LS174T cells without cetuximab treatment
As shown in Figure 1A, the expression of ITGB1 in NC, CO and CI groups were 0.69 ± 0.05, 0.93 ± 0.07, and 0.24 ± 0.02. The expression of ITGB1 was significantly increased in CO group and significantly decreased in CI group (P < .05). There results indicating that ITGB1 overexpression and knockdown model were successfully established in LS174T cells.
Figure 1.
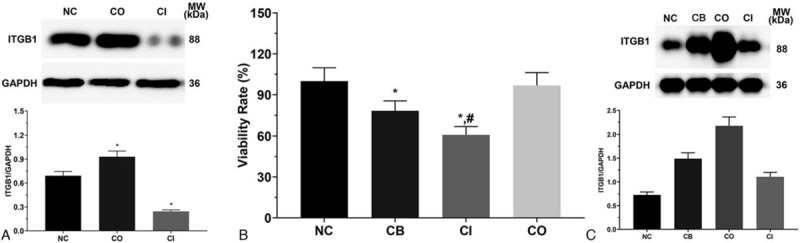
Expression of integrin-beta 1 and cellular proliferation of LS174T cells. (A) Expression of integrin-beta 1 in LS174T cells in each group without cetuximab treatment. (B) Cellular proliferation of LS174T cells in each group under cetuximab treatment. Each experiment was repeated for 3 times independently. Data was presented as mean ± SD. P < .05 was set as a statistic difference. ∗P < .05 compared with normal control group, #P < .05 compared with cetuximab treatment group.
3.2. Effect of cetuximab on proliferation of LS174T cells
As shown in Figure 1B, the proliferation of LS174T cells in CB, CI and CO groups were 78.3 ± 7.2, 60.8 ± 6.1, and 96.8 ± 9.4. The viability rate in CB and CI group was significantly decreased compared with NC group (P < .05), and was significantly decreased in CI group compared with CB group (P < .05).
3.3. Expression of invasion-related gene at transcription level
As shown in Figure 2, the expression of MMP-2 in NC, CB, CI and CO groups were 1.56 ± 0.12, 1.09 ± 0.09, 0.84 ± 0.07 and 1.32 ± 0.10. The expression of MMP-2 was significantly decreased in CB and CI group (P < .05) compared with NC group, and compared with CB group, the expression of MMP-2 was significantly decreased in CI group (P < .05) and significantly increased in CO group (P < .05). The expression of MMP-9 in these groups were 1.35 ± 0.15, 0.91 ± 0.08, 0.62 ± 0.05, and 1.00 ± 0.09. The expression of MMP-9 was significantly decreased in all treatment group (P < .05) compared with NC group, and compared with CB group, the expression of MMP-9 was significantly decreased in CI group (P < .05) compared with CB group. The expression of caspase-3 in these groups were 0.84 ± 0.09, 1.35 ± 0.15, 1.76 ± 0.19, and 1.02 ± 0.11. The expression of caspase-9 in these groups were 0.62 ± 0.04, 0.96 ± 0.09, 1.52 ± 0.13, and 0.71 ± 0.06. Changing in expression of canspase-3 and caspase-9 presented a similar trend, the expression of these 2 molecules was significantly increased in CB and CI group (P < .05) compared with NC group, and compared with CB group, the expression was significantly increased in CI group (P < .05) and significantly decreased in CO group (P < .05). The expression of cyclin D1 in these groups were 1.68 ± 0.18, 1.23 ± 0.11, 0.93 ± 0.07, and 1.26 ± 0.12. The expression of CDK1 in these groups were 1.71 ± 0.15, 1.22 ± 0.13, 0.86 ± 0.09, and 1.30 ± 0.12. Changing in expression of cyclin D1 and CDK1 presented a similar trend, the expression was significantly decreased in all treatment groups compared with NC group (P < .05), and compared with CB group, the expression was significantly decreased in CI group (P < .05) compared with CB group.
Figure 2.
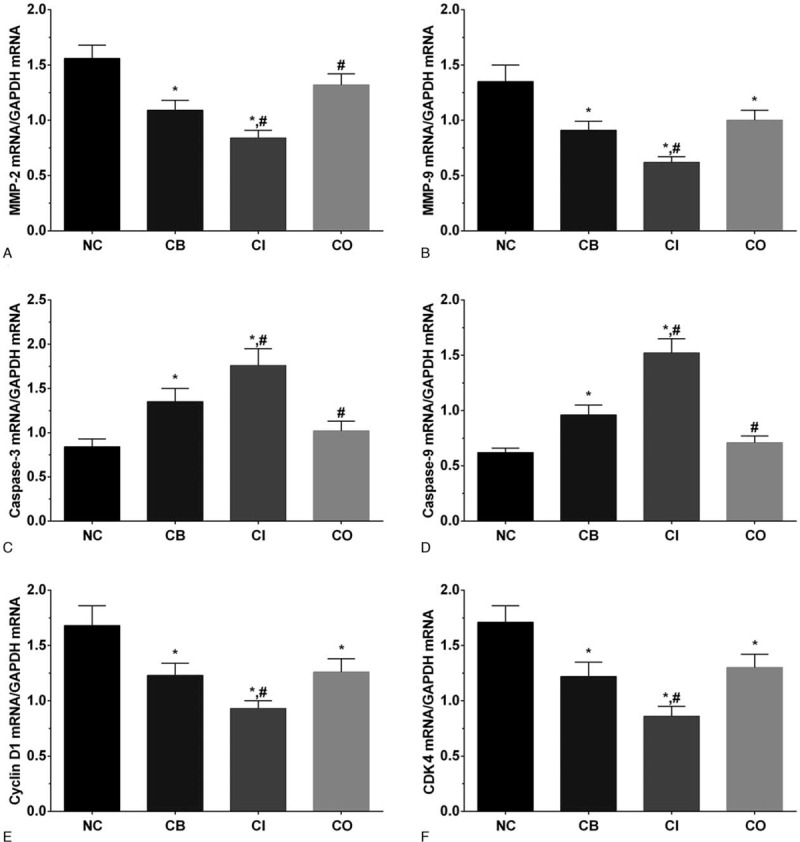
Expression of MMPs, caspases, and cellular cyclin-related genes in LS174T cells of each group. (A) Expression of MMP2 in each group of LS174T cells. (B) Expression of MMP9 in each group of LS174T cells. (C) Expression of caspase-3 in each group of LS174T cells. (D) Expression of caspase-9 in each group of LS174T cells. (E) Expression of cyclin D1 in each group of LS174T cells. (F) Expression of CDK1 in each group of LS174T cells. Each experiment was repeated for 3 times independently. Data was presented as mean ± SD. P < .05 was set as a statistic difference. ∗P < 0.05 compared with normal control group, #P < .05 compared with cetuximab treatment group.
3.4. Concentration of cell invasion related factors in cultured medium
As shown in Figure 3, the concentration of SDF-1α in cultured medium of LS174T cells in NC, CB, CI and CO groups were 1988.2 ± 132.3, 1767.1 ± 126.5, 1502.8 ± 112.4, and 1854.3 ± 131.2 pg/mL. The concentration of SDF-1α was significantly decreased in CI group (P < .05) compared with NC and CB group. The concentration of E-cadherin in these groups was 305.6 ± 27.6, 485.3 ± 31.3, 572.3 ± 35.7, and 421.2 ± 32.4 pg/mL. The concentration of E-cadherin was significantly increased in all treatment groups (P < .05) compared with NC group, and was significantly increased in CI group (P < .05) compared with CB group. The concentration of CXCL9 in these groups was 876.2 ± 68.2, 965.1 ± 83.0, 1201.2 ± 96.3, and 914.7 ± 81.6 pg/mL. The concentration of CXCL10 in these groups was 72.2 ± 6.8, 98.3 ± 8.2, 126.4 ± 11.3, and 86.1 ± 7.5 pg/mL. The expression of CXCL9 and CXCL1 was significantly increased in CI group (P < .05) compared with NC and CB group. The expression of MMP-2 in these groups was 365.2 ± 24.3, 301.4 ± 20.1, 247.3 ± 17.5, and 335.1 ± 22.8. The expression of MMP-2 was significantly decreased in CB group (P < .05) compared with NC group and significantly decreased in CI group (P < .05) compared with NC and CB group. The expression of MMP-9 in these groups was 993.2 ± 83.5, 862.5 ± 73.2, 702.3 ± 56.7, and 920.1 ± 79.4. The expression of MMP-9 was significantly decreased in CI group (P < .05) compared with NC and CB group.
Figure 3.
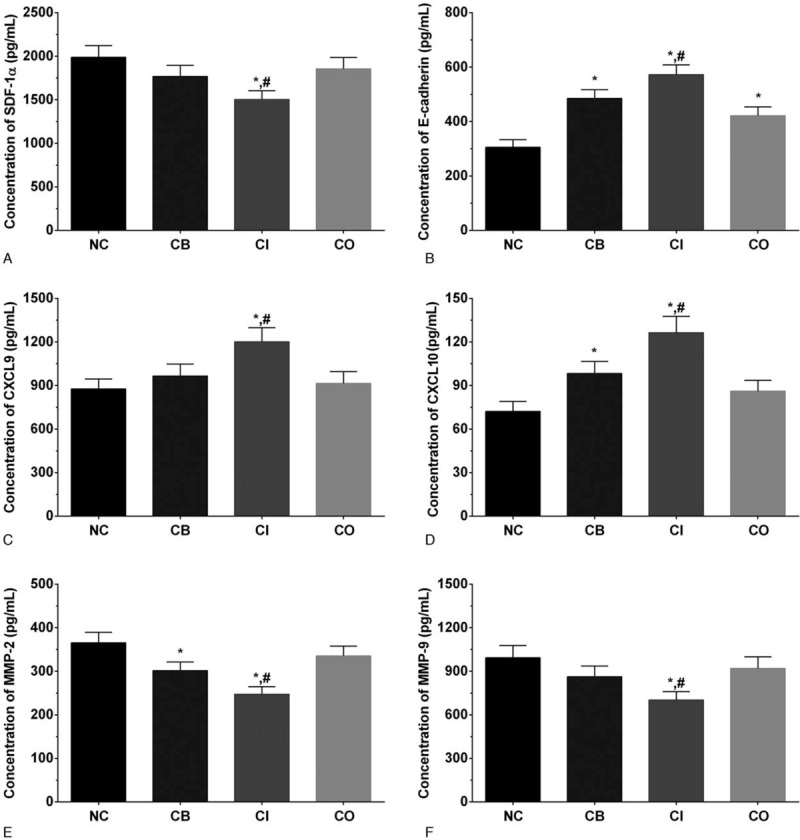
Concentration of invasion related factors in cultured medium. (A) Concentration of SDF-1α in cultured medium. (B) Concentration of E-cadherin in cultured medium. (C) Concentration of CXCL9 in cultured medium. (D) Concentration of CXCL10 in cultured medium. (E) Concentration of MMP-2 in cultured medium. (F) Concentration of MMP-9 in cultured medium. Each experiment was repeated for 3 times independently. Data was presented as mean ± SD. P < .05 was set as a statistic difference. ∗P < .05 compared with normal control group, #P < .05 compared with cetuximab treatment group.
3.5. Expression of metastasis related proteins in each group of LS174T cells
As shown in Figure 4, the expression of SDF-1 in each group of LS174T cells were 1.11 ± 0.09, 1.02 ± 0.08, 0.65 ± 0.07, and 1.51 ± 0.13. The expression of CXCR4 in these group were 0.85 ± 0.07, 0.88 ± 0.07, 0.65 ± 0.05, and 1.50 ± 0.12. The expression of SDF-1 and CXCR4 were significantly decreased in CI group (P < .05) and significantly increased in CO group (P < .05) compared with NC and CB group. The expression of CXCL9 in these group were 0.22 ± 0.02, 0.75 ± 0.06, 1.09 ± 0.09, and 0.70 ± 0.06. The expression of CXCL10 in these group were 0.53 ± 0.04, 0.90 ± 0.08, 1.19 ± 0.10, and 0.83 ± 0.07. The expression of CXCL9/10 was significantly increased in all treatment groups (P < .05) compared with NC group, and was significantly increased in CI group compared with CB group (P < .05). The expression of vimentin in these group were 1.36 ± 0.11, 1.15 ± 0.10, 0.63 ± 0.07, and 1.58 ± 0.13. The expression of fibronectin in these group were 1.34 ± 0.11, 1.15 ± 0.10, 0.60 ± 0.05, and 1.23 ± 0.10. The expression of vimentin and fibronectin was significantly decreased in CI group (P < .05) compared with NC and CB group, and the expression of vimentin was significantly increased in CO group (P < .05) compared with CB group.
Figure 4.
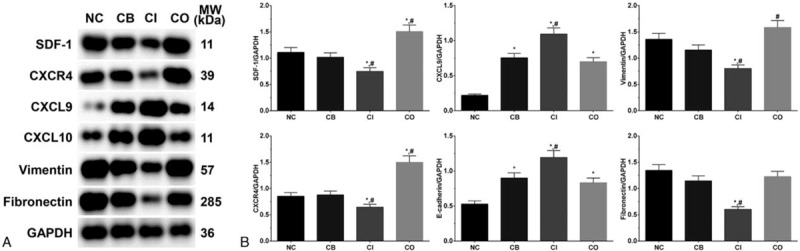
Expression of metastasis related proteins in each group of LS174T cells. (A) Western blotting analysis of each protein. (B) Quantitative analysis of each protein. Each experiment was repeated for 3 times independently. Data was presented as mean ± SD. P < .05 was set as a statistic difference. ∗P < .05 compared with normal control group, #P < .05 compared with cetuximab treatment group.
3.6. Activation of Ras/Raf/MEK signaling pathway in each group of LS174T cells
As shown in Figure 5, the expression of EGFR in each group of LS174T cells were 1.46 ± 0.12, 0.50 ± 0.04, 0.33 ± 0.03, and 1.79 ± 0.15. The expression of EGFR was significantly decreased in CB and CI group (P < .05) and significantly increased in CO group (P < .05) compared with NC group, and was significantly decreased in CI group (P < .05) and significantly increased in CO group (P < .05) compared with CB group. The expression of Ras in these group were 1.67 ± 0.14, 1.36 ± 0.11, 0.80 ± 0.07, and 1.60 ± 0.13. The expression of Ras was significantly decreased in CB and CI group (P < .05) compared with NC group, and was significantly decreased in CI group (P < .05) compared with CB group. The expression of Raf in these group were 1.39 ± 0.12, 1.21 ± 0.10, 0.75 ± 0.06, and 1.89 ± 0.16. The expression of Raf were significantly decreased in CI group (P < .05) and significantly increased in CO group (P < .05) compared with NC and CB group. The ratio of p-MEK1/MEK1 in these group were 2.52 ± 0.21, 2.66 ± 0.22, 2.18 ± 0.18, and 2.97 ± 0.25. The ratio of p-STAT3/STAT3 in these group were 1.15 ± 0.10, 1.00 ± 0.08, 0.68 ± 0.06, and 1.02 ± 0.09. The ratio of p-MEK1/MEK1 and p-STAT3/STAT3 was significantly decreased in CI group (P < .05) compared with NC group, and the ratio of p- STAT3/STAT3 was significantly decreased in CI (P < .05) compared with CB group. The expression of VEGF-α in these group were 1.18 ± 0.13, 1.07 ± 0.12, 0.80 ± 0.11, and 1.59 ± 0.19. The expression of VEGF-α were significantly decreased in CI group (P < .05) and significantly increased in CO group (P < .05) compared with NC and CB group. However, the expression of these key molecules in control, ITGB1 overexpression and ITGB1 inhibition groups was not significantly changed, indicating that changing in ITGB1 expression did not affected the activation of Ras/Raf/MEK signaling pathway (Supplement Figure 1).
Figure 5.
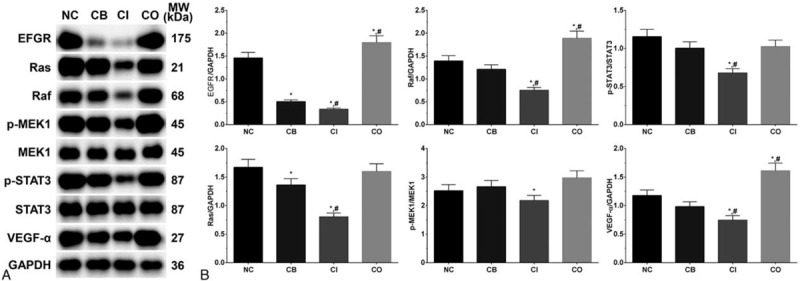
Activation of Ras/Raf/MEK signaling pathway in each group of LS174T cells. (A) Western blotting analysis of each protein. (B) Quantitative analysis of each protein. Each experiment was repeated for 3 times independently. Data was presented as mean ± SD. P < .05 was set as a statistic difference. ∗P < .05 compared with normal control group, #P < .05 compared with cetuximab treatment group.
4. Discussion
Colon cancer is one of the most commonly seen cancers around the world, and almost 1.36 million people were diagnosed with colon cancer each year. Colon cancer is an threaten especially for elder people, median age of diagnosis for colon cancer is 69 years, with 70% of cases were more than 65 years and 40% of cases were over 75 years.[15] Cetuximab is an anti-EGFR agent which has been proved effective for treatment of HNSCC.[16] Cetuximab could bind with the extracellular domain of EGFR as cetuximab is a IgG-subclass monoclonal antibody, inhibiting the activation of the intracellular domain of EGFR and further inhibiting the activation of subsequent signaling transduction pathway.[17] Cetuximab could also stimulate the internalization of EGFR, preventing the interaction with its ligand through removing the receptor from the cell surface.[18] ITGB1 is a useful marker for prognosis of postoperative cancer patients in comparison of tumor size and tumor node metastasis (TNM) staging system.[18] However, the effect of ITGB1 in treatment of CRC combined with cetuximab was not clear.
Highly invasive ability is one of the most important characteristics of cancer, leading to the rapid metastasis and development. MMPs play an important role in degradation of ECM contributes a lot in promoting the invasion of cancer cells. Among the family of MMPs identified, MMP-2 and MMP-9 play a critical role in degradation of type IV collagen and gelation, 2 mainly components of ECM. Increased expression of MMP-2 and MMP-9 was commonly seen in plenty types of tumors, such as breast and prostate tumors,[19] leading to the invasion of cancer cells.[20]
E-cadherin is firstly found as a mediator of cell-cell adhesion in early embryo development.[21] Expression of E-cadherin was lost in Epithelial-Mesenchymal Transition (EMT) process and lead to the increasing in cellular motility of cancer cells.[22] Besides, in EMT process, cancer stem cell features were acquired in cancer cells, resulting in drug resistance and anti-cancer therapies.[23] In presented study, we found that cetuximab inhibition the expression of E-cadherin and MMPs, further inhibit the degradation of ECM and EMT, resulting in inhibition of cellular invasion ability. And inhibition of ITGB1 enhance the anti-tumor effect of cetuximab, decreased the expression of MMPs and E-cadherin. Intrinsic apoptosis is a process characterized by mitochondrial outer membrane permeabilization (MOMP) induced cell death, resulting in activation of caspase-9 and effector caspases. Previous study indicated that caspase-3 cleaved and activated by caspase-9, leading to the activation of apoptosis process.[24] Expression of caspase-9 and caspase-3 were both elevated after cetuximab treatment and even higher after inhibition of ITGB1, indicating that apoptosis process was activation in LS174T cells. Cyclin D1 regulates the G1/G-phase via binding followed with activating of CDK4.[25] Overexpression of cyclin D1 is considered as a collaborative oncogene in plenty types of cancer, and leading to tumorigenesis.[26] Cetuximab inhibits the expression of cyclin D1 and CDK4, and inhibition of ITGB1 would enhance the effect of cetuximab, inhibiting the proliferation of cancer cells. These results revealed that inhibition of ITGB1 could enlarge the effect of cetuximab treatment on reduction of expression of apoptosis and cell cycle related molecules, presented an anti-tumor effect.
Stromal cell-derived factor-1 (SDF-1), also known as CXCL12, is a ligand of CXCR4, and plays an important role in many physiological and pathological processes of cancer.[27] Previous study found that SDF-1 regulates the formation of epithelial-mesenchymal transition (EMT) in multiple types of cancer,[28,29] and also promotes the angiogenesis and metastasis of cancer cells.[30] EGFR is a cell-surface receptor tyrosine kinases and belongs to ERBB family.[31] EGF binding with EGFR triggers the downstream molecules, followed with phosphorylation and activation of downstream effectors including Ras/Raf/MAPK and PI3K/AKT/mTOR signaling pathway.[18] Raf/MAPK signaling pathway is an essential effector pathway. In a animal model, deletion of Ras leading to the inhibition of proliferation and migration, and these effects could be rescued by supplement of Raf, MEK or ERK, but nor PI3K.[32] Expression of Ras could be reduced by deletion of Raf in a mouse model, followed with activation of MAPK signaling pathway.[33] Previous study also found that CXCL12/CXCR4 is critical for cell proliferation via activation of ERK signaling pathway,[34] and there is also study indicated that down-regulation of CXCR4 reduce the proliferation of cancer cells via inhibition of NF-κB signaling pathway with induction of cellular apoptosis.[35] Thus, we speculated that cetuximab targeted on Ras/Raf/MAPK signaling pathway, leading to the reduction in signaling pathway activation. Besides, we found that inhibition of ITGB1 targets on CXCL12/CXCR4, the upstream molecular of Ras/Raf/MAPK signaling pathway, resulting in reduction of cellular proliferation. Besides, there is also study indicated that VEGF expression is also regulated by CXCR4 in cancer cells, promoting the angiogenesis and cancer cell viability.[36] There is also study indicated that VEGF could promote the activation of MAPK signaling pathway.[37] ITGB1 enhance the anti-tumor effect of cetuximab on inhibition of CXCL12/CXCR4 and VEGF further reduce the activation of MAPK signaling pathway, leading to the proliferation of cancer cells.
The expression of CXCL9 and CXCL10 is elevated in CRC model, which in turn led to the recruitment of NK cells to liver, thereby activating immune response, presented an anti-metastatic effect.[38] Previous study also found that knockdown of activator transcription 3 (STAT3) increase the expression of CXCL9 and CXCL10, and improve the outcome of chemotherapy.[39] Another study found that depletion of regulatory T cells (Treg) in CRC mice model leading to the infiltration and proliferation of T cells and up-regulated expression of CXCL9 and CXCL10, improve the effect of immunotherapy.[40] Vimentin is a 57 kDa protein, which is mainly expression in mesenchymal cell types.[41] As a major of intermediate filament protein, vimentin is critical for cellular adhesion, migration and signaling.[42] Vimentin is regarded as a marker of up-regulation of EMT in cancer cells, which promotes the metastasis of cancer cells.[43] Fibronectin is a glycoprotein in ECM of various connective tissues, regulating cellular adhesion, migration and proliferation.[44] Fibronectin not only initiation of assemblies of ECM related protein, but also for the stabilization of these proteins.[45] These proteins functions critical for cellular adhesion and immune response, and we speculated that these effectors might be the downstream molecules of Ras/Raf/MAPK signaling pathway. Inhibition of ITGB1 might enhance the effect of cetuximab on increase the expression of CXCL9/10 and reduce the expression of vimentin and fibronectin, performing the anti-tumor effect.
5. Conclusion
Inhibition of ITGB1 might enhance the anti-tumor effect of cetuximab through inhibition of cellular invasion and metastasis, activation of immune response and inhibition of angiogenesis process. We further noticed that these effects might be mediated by Ras/Raf/MAPK signaling pathway.
Author contributions
Xiaohui Yang performed most of the experiments and wrote the manuscript. Shuai Wang and Weihua Yu collected the samples. Yixiong Zheng performed the statistical analysis. Yulian Wu designed the experiments and revised the manuscript.
Supplementary Material
Footnotes
Abbreviations: CB = cetuximab treatment group, CRC = colorectal cancer, ECM = extracellular matrix, EGFR = epidermal growth factor receptor, ELISA = enzyme-linked immunosorbent assay, ITGB1 = integrin-beta 1, NC = normal control group, qPCR = real-time quantitative polymerase chain reaction.
How to cite this article: Yang X, Wang S, Yu W, Zheng Y, Wu Y. Inhibition of ITGB1 enhance the anti-tumor effect of cetuximab in colorectal cancer cell. Medicine. 2020;99:27(e20944).
These experiments were supported by Basic Public Research Program of Zhejiang Province (No. LGF18H070002) and Zhejiang Education Research Program (No. Y201737989).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
- [1].Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet 2016;7:105–14. [PMC free article] [PubMed] [Google Scholar]
- [2].Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 2018;103:356–87. [DOI] [PubMed] [Google Scholar]
- [3].Jonker DJ, O’Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040–8. [DOI] [PubMed] [Google Scholar]
- [4].Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337–45. [DOI] [PubMed] [Google Scholar]
- [5].Guo G, Gong K, Wohlfeld B, et al. Ligand independent EGFR signaling. Cancer Res 2015;75:3436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hu C, Ni Z, Li BS, et al. hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation. Gut 2017;66:31–42. [DOI] [PubMed] [Google Scholar]
- [7].Fedorenko IV, Abel EV, Koomen JM, et al. Fibronectin induction abrogates the BRAF inhibitor response of BRAF V600E/PTEN-null melanoma cells. Oncogene 2016;35:1225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hirata E, Girotti MR, Viros A, et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin (1/FAK signaling). Cancer Cell 2015;27:574–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zheng W, Jiang C, Li R. Integrin and gene network analysis reveals that ITGA5 and ITGB1 are prognostic in non-small-cell lung cancer. Onco Targets Ther 2016;9:2317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Petricevic B, Vrbanec D, Jakic-Razumovic J, et al. Expression of Toll-like receptor 4 and beta 1 integrin in breast cancer. Med Oncol 2012;29:486–94. [DOI] [PubMed] [Google Scholar]
- [11].Liu QZ, Gao XH, Chang WJ, et al. Expression of ITGB1 predicts prognosis in colorectal cancer: a large prospective study based on tissue microarray. Int J Clin Exp Pathol 2015;8:12802–10. [PMC free article] [PubMed] [Google Scholar]
- [12].Wu TM, Huang JZ, Oung HM, et al. H2O2-based method for rapid detection of transgene-free rice plants from segregating CRISPR/Cas9 genome-edited progenies. Int J Mol Sci 2019;20:3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim S, Kim N, Kang K, et al. Whole transcriptome analysis identifies TNS4 as a key effector of cetuximab and a regulator of the oncogenic activity of KRAS mutant colorectal cancer cell lines. Cells 2019;8:E878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [15].Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med 2008;359:1116–27. [DOI] [PubMed] [Google Scholar]
- [17].Goldstein NI, Prewett M, Zuklys K, et al. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res 1995;1:1311–8. [PubMed] [Google Scholar]
- [18].Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 2008;358:1160–74. [DOI] [PubMed] [Google Scholar]
- [19].Nelson AR, Fingleton B, Rothenberg ML, et al. Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol 2000;18:1135–49. [DOI] [PubMed] [Google Scholar]
- [20].Kleiner DL, Stetler-Stevenson WG. Matrix metalloproteinases and metastasis. Cancer Chemother Pharmacol (Suppl) 1999;43:S42–51. [DOI] [PubMed] [Google Scholar]
- [21].Hyafl F, Babinet C, Jacob F. Cell-cell interactions in early embryogenesis: a molecular approach to the role of calcium. Cell 1981;26:447–54. [DOI] [PubMed] [Google Scholar]
- [22].Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871–90. [DOI] [PubMed] [Google Scholar]
- [23].Barkeer S, Chugh S, Batra SK, et al. Glycosylation of cancer stem cells: function in stemness, tumorigenesis, and metastasis. Neoplasia 2018;20:813–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Slee EA, Harte MT, Kluck RM, et al. Ordering the cytochrome cinitiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol 1999;144:281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fu M, Wang C, Li Z, et al. Minireview: cyclin D1: normal and abnormal functions. Endocrinology 2004;145:5439–47. [DOI] [PubMed] [Google Scholar]
- [26].Casimiro MC, Di Sante G, Crosariol M, et al. Kinase-independent role of cyclin D1 in chromosomal instability and mammary tumorigenesis. Oncotarget 2015;6:8525–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Duan Y, Zhang S, Wang L, et al. Targeted silencing of CXCR4 inhibits epithelial-mesenchymal transition in oral squamous cell carcinoma. Oncol Lett 2016;12:2055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yao CJ, Li PP, Song HS, et al. CXCL12/CXCR4 axis upregulates twist to induce EMT in human glioblastoma. Mol Neurobiol 2016;53:3948–53. [DOI] [PubMed] [Google Scholar]
- [29].Yang P, Wang G, Huo HJ, et al. SDF-1/CXCR4 signaling up-regulates survivin to regulate human sacral chondrosarcoma cell cycle and epithelial-mesenchymal transition via ERK and PI3K/AKT pathway. Med Oncol 2015;32:377. [DOI] [PubMed] [Google Scholar]
- [30].Sun XJ, Charbonneau C, Wei L, et al. CXCR4-targeted therapy inhibits VEGF expression and chondrosarcoma angiogenesis and metastasis. Mol Cancer Ther 2013;12:1163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cataldo VD, Gibbons DL, Perez-Soler R, et al. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med 2011;364:947–55. [DOI] [PubMed] [Google Scholar]
- [32].Yeh P, Chen H, Andrews J, et al. DNA-mutation Inventory to Refine and Enhance Cancer Treatment (DIRECT): a catalog of clinically relevant cancer mutations to enable genome-directed anticancer therapy. Clin Cancer Res 2013;19:1894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Oxnard GR, Miller VA, Robson ME, et al. Screening for germline EGFR T790 M mutations through lung cancer genotyping. J Thorac Oncol 2012;7:1049–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Barbieri F, Bajetto A, Porcile C, et al. CXC receptor and chemokine expression in human meningioma: SDF1/CXCR4 signaling activates ERK1/2 and stimulates meningioma cell proliferation. Ann NY Acad Sci 2006;1090:332–43. [DOI] [PubMed] [Google Scholar]
- [35].Jiang C, Ma S, Hu R, et al. Effect of CXCR4 on apoptosis in osteosarcoma cells via the PI3K/Akt/NF-kappabeta signaling pathway. Cell Physiol Biochem 2018;46:2250–60. [DOI] [PubMed] [Google Scholar]
- [36].Pages G, Pouyssegur J. Transcriptional regulation of the vascular endothelial growth factor gene–a concert of activating factors. Cardiovasc Res 2005;65:564–73. [DOI] [PubMed] [Google Scholar]
- [37].Suarez S, Ballmer-Hofer K. VEGF transiently disrupts gap junctional communication in endothelial cells. J Cell Sci 2001;114:1229–35. [DOI] [PubMed] [Google Scholar]
- [38].Brackett CM, Kojouharov B, Veith J, et al. Toll-like receptor-5 agonist, entolimod, suppresses metastasis and induces immunity by stimulating an NK-dendritic-CD8+ T-cell axis. Proc Natl Acad Sci USA 2016;113:E874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang H, Yamazaki T, Pietrocola F, et al. STAT3 inhibition enhances the therapeutic efficacy of immunogenic chemotherapy by stimulating type 1 interferon production by cancer cells. Cancer Res 2015;75:3812–22. [DOI] [PubMed] [Google Scholar]
- [40].Akeus P, Langenes V, Kristensen J, et al. Treg-cell depletion promotes chemokine production and accumulation of CXCR3(+) conventional T cells in intestinal tumors. Eur J Immunol 2015;45:1654–66. [DOI] [PubMed] [Google Scholar]
- [41].Chung BM, Rotty JD, Coulombe PA. Networking galore: intermediate filaments and cell migration. Curr Opin Cell Biol 2013;25:600–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ivaska J, Pallari HM, Nevo J, et al. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res 2007;313:2050–62. [DOI] [PubMed] [Google Scholar]
- [43].Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 2011;68:3033–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu J, Mosher D. Diagnostic accuracy of glycoproteins in the assessment of liver fibrosis: a comparison between laminin, fibronectin, and hyaluronic acid. Turk J Gastroenterol 2019;30:524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sabatier L, Djokic J, Fagotto-Kaufmann C, et al. Complex contributions of fibronectin to initiation and maturation of microfibrils. Biochem J 2013;456:283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


