Abstract
Background
The coronavirus disease 2019 (COVID-19) pandemic has put tremendous pressure on the healthcare system worldwide. Diagnostic testing remained one of the limiting factors for early identification and isolation of infected patients. This study aimed to evaluate posterior oropharyngeal saliva (POPS) for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection among patients with confirmed or suspected COVID-19.
Methods
The laboratory information system was searched retrospectively for all respiratory specimens and POPS requested for SARS-CoV-2 RNA detection between 1 February 2020 and 15 April 2020. The agreement and diagnostic performance of POPS against NPsp were evaluated.
Results
A total of 13772 specimens were identified during the study period, including 2130 POPS and 8438 nasopharyngeal specimens (NPsp). Two hundred and twenty-nine same-day POPS-NPsp paired were identified with POPS and NPsp positivity of 61.5% (95% confidence interval [CI] 55.1–67.6%) and 53.3% (95% CI 46.8–59.6%). The overall, negative and positive percent agreement were 76.0% (95% CI 70.2–80.9%), 65.4% (95% CI 55.5–74.2%), 85.2% (95% CI 77.4–90.8%). Better positive percent agreement was observed in POPS-NPsp obtained within 7 days (96.6%, 95% CI 87.3–99.4%) compared with after 7 days of symptom onset (75.0%, 95% CI 61.4–85.2%). Among the 104 positive pairs, the mean difference in Cp value was 0.26 (range: 12.63 to −14.74), with an overall higher Cp value in NPsp (Pearson coefficient 0.579). No significant temporal variation was noted between the 2 specimen types.
Conclusions
POPS is an acceptable alternative specimen to nasopharyngeal specimen for the detection of SARS-CoV-2.
Keywords: COVID-19, severe acute respiratory syndrome coronavirus 2, saliva, mass screening, pandemic
INTRODUCTION
Nasopharyngeal specimens (NPsp), for example, nasopharyngeal swabs (NPS) and aspirates (NPA) are the recommended specimen types for the investigation of viral respiratory infections [1]. Currently, the World Health Organization and Centers for Disease Control and Prevention recommended an upper respiratory specimen (NPA or NPS with throat swab) and/or a lower respiratory specimen for the diagnosis of coronavirus disease 2019 (COVID-19) [2, 3]. However, the collection of NPsp is relatively invasive with significant patient discomfort and is contraindicated in patients with recent nasal trauma or surgery, or severe thrombocytopenia [4]. In addition, obtaining NPsp required trained healthcare workers (HCW) and has infection control implications mandating use of personal protective equipment (PPE), including N95 respirator or equivalent, gloves, faceshield/eye protection, and gown [3, 4]. The procedure should also be performed in a well-ventilated room, or a negative pressure airborne infection isolation room (AIIR) due to the potential generation of aerosols. Hence, these requirements might become limiting factors in providing adequate capacity for diagnosing infections during the COVID-19 pandemic. Moreover, there is a surge in global demand for flocked swabs and viral transport medium (VTM) used for collecting NPsp, resulting in substantial pressure and uncertainty over the reliable supply of these essential material [5, 6].
Saliva has previously been shown to be a useful specimen type for other respiratory virus detections, such as influenza and human metapneumovirus, with comparable sensitivity [7] and agreement [8–11] with NPsp. Preliminary studies showed persistent detectable severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in saliva from patients with confirmed COVID-19 [12, 13]. Viral load was highest in posterior oropharyngeal saliva (POPS) during the first week of symptom onset, with median viral load of first available saliva specimen being around 106 copies/mL [14]. On 13 April 2020, the Food and Drug Administration granted accelerated emergency use authorization for the use of saliva, in addition to other respiratory specimen types, in a SARS-CoV-2 assay to facilitate mass screening, based on an evaluation of 60 specimens obtained with the Spectrum Solutions LLC SDNA-1000 Saliva Collection Device [15]. Thus far, studies on saliva as diagnostic specimens for SARS-CoV-2 were small and varied in saliva specimen collection methods (Supplementary Table 1) [16–20]. Hence, to understand the performance of saliva specimens in a field situation, we performed a retrospective comparison of saliva against NPsp in a larger population of patients with suspected or confirmed COVID-19.
MATERIAL AND METHODS
Setting
Because of the announcement of clustered pneumonia of unknown origin by the Wuhan Municipal Health commission [21], the Government of the Hong Kong Special Administrative Region (HKSAR) and Hospital Authority (HA), a governing body that manage public hospitals in Hong Kong, have worked together to strengthen SARS-CoV-2 surveillance in Hong Kong. The surveillance strategies implemented during the study period are summarized in Table 1 [22–25]. To maximize SARS-CoV-2 testing capacity in Hong Kong, the workload from the various surveillance programs were shared between the Public Health Laboratory Service Branch and across the 7 HA hospital clusters. Queen Elizabeth Hospital is a major acute hospital in the Kowloon Central Cluster, and SARS-CoV-2 real-time reverse transcription polymerase chain reaction (RT-PCR) testing was made available since 1 February 2020. Testing is provided for diagnosis of suspected cases and serial monitoring of viral load in confirmed patients. According to local clinical practice guideline at the time of the study, all confirmed cases must be hospitalized and could only be released from isolation when 2 clinical specimens were tested negative for SARS-CoV-2, taken at least 24 hours apart [26], and the consensus was all previous positive sites needed to meet the criteria. Hence, repeated specimens of various types were taken from these patients during the study period to assess if they could be discharged from AIIR.
Table 1.
Laboratory Surveillance and Monitoring Programs in Hong Kong and in the Kowloon Central Cluster Hospitals
| Surveillance/Monitoring Program | Specimen Type(s) | Target Population | Launch Date | |
|---|---|---|---|---|
| Hong Kong wide surveillance | Tier 1 (inpatient) | Upper ± lower respiratory specimens | Symptomatic patients with epidemiological risk factors. | 31 December 2019 |
| Tier 2 (inpatient) | Upper ± lower respiratory specimens | Enhanced laboratory surveillance for patients with pneumonia with (i) unknown cause, or (ii) requiring intensive care support; or (iii) occurring in cluster, or (iv) in healthcare workers. | 13 January 2020 | |
| Tier 3 (inpatient) | Upper ± lower respiratory specimens | Enhanced laboratory surveillance in all patients admitted to hospital with pneumonia. | 31 January 2020 | |
| Tier 4 (outpatient) | Posterior oropharyngeal saliva | AED or general outpatient clinic attendees with fever or acute onset mild respiratory symptoms. | 19 February 2020 | |
| Tier 5 (outpatient) | Posterior oropharyngeal saliva | Private outpatient clinic attendees with fever or acute onset mild respiratory symptoms. | 6 March 2020 | |
| Tier 6 (outpatient) | Posterior oropharyngeal saliva | Asymptomatic returned traveler | 25 March 2020 | |
| Temporary testing centers for symptomatic returned travelers (outpatient) | NPS/T | Symptomatic returned travelers arriving Hong Kong International Airport. | 20 March 2020 | |
| Triage and test (T&T, outpatient) | NPS/T | Symptomatic patients with epidemiological factors attending AED. | 28 March 2020 | |
| Cluster-based surveillance in KCC hospitals | Patient undergoing nonemergency AGP (in and outpatient) |
Posterior oropharyngeal saliva | Asymptomatic patients undergoing nonemergency AGP. | 10 February 2020 |
| Staff with travel history (outpatient) |
Posterior oropharyngeal saliva | Asymptomatic hospital staff with travel history | 16 March 2020 | |
| Confirmed patient monitoring | Confirmed COVID-19 patients (inpatient) |
Respiratory specimens and/or posterior oropharyngeal saliva | Monitoring of patients with confirmed COVID-19 infection in hospital AIIR until discharge criteriaa is achieved. | 13 February 2020 |
Abbreviations: AED, accident and emergency department; AGP, aerosol generating procedure; AIIR, airborne infection isolation room; COVID-19, coronavirus disease 2019; KCC, Kowloon Central Cluster; NPS/T, pooled nasopharyngeal swab and throat swab.
aThe local clinical practice guideline at the time of the study advised that all confirmed cases must be hospitalized and could only be released from isolation when two clinical specimens were tested negative for SARS-CoV-2, taken at least 24 hours apart, and the consensus was all previous positive sites needed to meet the criteria.
Specimen Collection
For the collection of POPS, standard instructions prepared by HA were given to patients (https://www.ha.org.hk/haho/ho/cc/Information_sheet_en_txt.pdf), an online video was also available (https://www.youtube.com/watch?v = rZ3oNsJGBxo). In brief, patients were asked to clear saliva from back of the throat into a sterile container as soon as possible after waking up, before any eating, drinking, or teeth brushing. For NPsp, patients were instructed to blow their nose to clear the nostrils, and both NPS and NPA were collected by a HCW wearing full PPE. NPS was collected by insertion of a flock swab into the nostril parallel to the palate with a rotatory motion to a depth equal to the distance from the nostril to the tragus [4]. The flock swab was left in the position for a few seconds before removal with a rotatory motion. The swab was then placed in 1 mL of VTM for transportation. NPS was collected using a catheter connected one end to a mucus trap and the other end to a vacuum source, which is then inserted into the nasopharynx similar to NPS to the nasopharynx for aspiration of nasopharyngeal secretion into the mucus trap. One mL of VTM was added to the secretion before transportation. If POPS and NPSp were taken at the same time, POPS was always obtained before NPsp. Lower respiratory samples were always preserved in 1 mL of VTM. Other conventional respiratory specimens such as sputum, tracheal aspirate, and broncheoalveolar lavage were collected following usual practice.
Real-time Reverse Transcription Polymerase Chain Reaction Assay for SARS-CoV-2
Upper respiratory tract specimens (eg, NPA or NPS with or without pooled throat swabs), lower respiratory tract specimens (eg, sputum, tracheal aspirate or bronchoalveolar lavage), and POPS were acceptable specimen types for the detection of SARS-CoV-2 RNA in our laboratory. All respiratory specimens, and POPS from inpatients, were received in 1 mL of VTM. POPS collected from outpatients were sent to our laboratory as neat, and 1 mL of VTM was added by the laboratory staff if the specimen was too viscous for accurate pipetting. All specimens were processed within 24 hours of collection. Total nucleic acid extraction was performed using MagNA Pure LC 2.0 (Roche, Switzerland) or MagNA Pure 96 (Roche, Switzerland). Real-time RT-PCR was performed using LightMix® Modular SARS and Wuhan CoV E-gene kit with EAV RNA extract control (TIB-MOLBIOL, Berlin, Germany) on a Cobas z480 real-time PCR analyzer (Roche Diagnostics, Mannheim, Germany) according to manufacturer’s instructions. Briefly, each 20 μL reaction mixture contained 4.0 μL of Roche Master, 0.1 μL of RT enzyme, 0.5 μL of reagent mix, 5.4 μL of water and 10 μL of extracted RNA. RT-PCR was performed under the following conditions: RT step at 55°C for 5 minutes and 95°C for 5 minutes, then 45 thermal cycling at 95°C for 3 seconds, 60°C for 12 seconds, and 72°C for 3 seconds, followed by cooling at 40°C for 30 seconds. SARS-CoV-2 RNA was considered present if the Cp value of the reaction was ≤40 and a sigmoidal amplification curve was observed.
Data Collection and Analysis
Test data for SARS-CoV-2 RNA detection in all respiratory and POPS specimens between 1 February 2020 and 15 April 2020 were retrieved from the laboratory information system retrospectively. Demographic and clinical information of corresponding patients were retrieved from the Clinical Management System. Ethical approval was obtained from the Research Ethics Committee of the Kowloon Central/Kowloon East Cluster, HA.
POPS and NPsp from the same patient on the same day (POPS-NPsp) were paired for further analysis. Only 1 POPS and 1 NPsp were included per patient per day. Analyses of diagnostic performance, agreement, and temporal variation of result discordance were performed.
Statistical Analysis
Sample size for the study was calculated to detect the difference between a true kappa of 0.5 and a kappa of 0.7 under the null hypothesis, at α = 0.05 and power = 0.80, assuming positivity rates for the 2 sample types are 0.5 and 0.6. In the absence of a “gold standard” specimen type, test performance of SARS-CoV-2 RNA detection in paired POPS and NPsp was assessed by percent agreement. The correlation of Cp values between POPS and NPsp was assessed using the Pearson correlation coefficient. Student t-test and Mann-Whitney U tests were performed for comparison where appropriate. A P value of < .05 was considered statistically significant. All statistical analysis was performed using R version 3.6.3.
RESULTS
Patient Characteristics
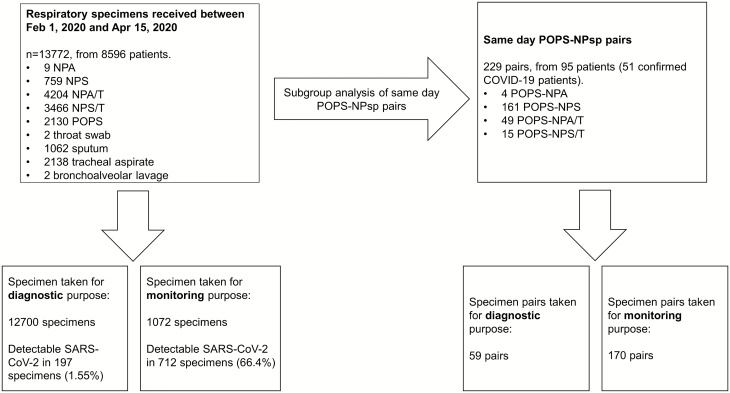
A total of 13772 POPS and respiratory specimens from 8596 patients were received for SARS-CoV-2 RNA detection between 1 February 2020 and 15 April 2020, of which 12700 were performed for diagnostic purpose, with an overall positive rate of 1.55%. In particular, the positive rate of specimens sent from “Tier 1” patients and testing centers for symptomatic returned travelers were 8.34% and 6.18%, respectively. The number of specimens per patient ranged from 1 to 41, with a median of 1. In additional to monitoring in COVID-19 patients, more than 1 specimen was sent from some of the patients due to the paucity of data on the optimal specimen type and concerns with inter-specimen variation. The calculated sample size required for the study was 109 pairs, and we identified 229 same-day POPS-NPsp from 95 patients, with 51 confirmed COVID-19 patients, 7 of whom (13.7%) had asymptomatic infection (Figure 1). The mean and median age of the 95 patients with POPS-NPsp were 39 and 36 years (range 4–92 years), and 57 were male. Fourteen patients had more than one NPsp taken on the same day, and the NPsp with the longer time interval from POPS were excluded from pairing. The mean and median time difference between the POPS-NPsp were 140 and 42 minutes (range, 0–1257 minutes). For POPS, only 128/229 (55.9%) were collected between 0400 and 1200 despite instruction of early morning sample taking, 75/128 showed detectable SARS-CoV-2 RNA.
Figure 1.
Breakdown of respiratory specimens received during the study period. Between 1 February 2020 and 15 April 2020, a total of 13772 specimens for SARS-CoV-2 testing were received from inpatients in the Kowloon Central Cluster, as well as ambulatory patients and asymptomatic patients from different surveillance programs and clinical settings. Subgroup head-to-head comparison of posterior oropharyngeal saliva with nasopharyngeal specimens were performed on the 229 posterior oropharyngeal saliva when concurrent nasopharyngeal specimen taken on the same day from the same patient. Abbreviations: COVID-19, coronavirus disease 2019; NPA, nasopharyngeal aspirates; NPA/T: nasopharyngeal aspirate pooled with throat swab; NPS, nasopharyngeal swabs; NPS/T: nasopharyngeal swab pooled with throat swab; POPS, posterior oropharyngeal saliva; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Comparison of SARS-CoV-2 RNA Detection Between POPS-NPsp
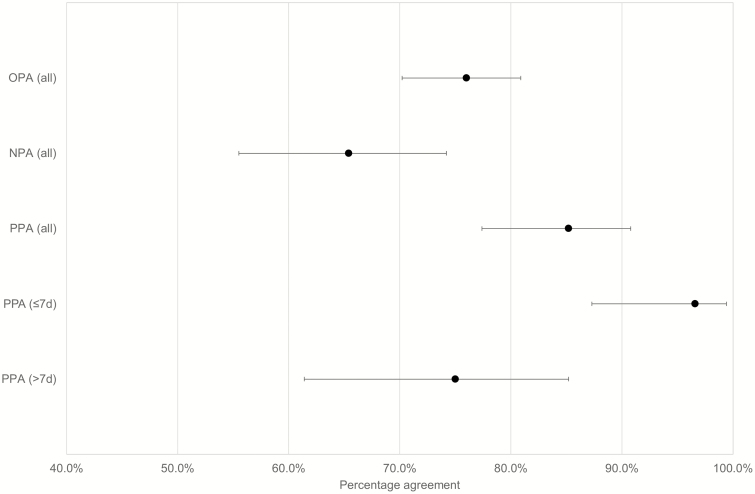
Of the 229 POPS-NPsp identified, the majority (70.3%) of the NPsp were NPS (Figure 1). The results of the POP-NPsp are shown in Table 2. In this subgroup, the positivity of POPS and NPsp was 61.6% (95% confidence interval [CI] 55.1–67.6%) and 53.3% (95% CI 46.8–59.6%), respectively. The positive percent agreement (PPA) and negative percent agreement (NPA) were 85.2% (95% CI 77.4–90.8%) and 65.4% (95% CI 55.5–74.2%) respectively (Figure 2). Overall percent agreement (OPA) was 76.0% (95% CI 70.2–80.9%), and Cohen’s kappa was 0.512 (95% CI .401–.623), suggesting moderate agreement.
Table 2.
Results From Posterior Oropharyngeal Saliva and Nasopharyngeal Specimens in Same-day Matched Pairs (n = 229)
| SARS-CoV-2 RNA | Posterior oropharyngeal Saliva | ||
|---|---|---|---|
| Detected | Not Detected | ||
| Nasopharyngeal specimens | Detected | 104 | 18 |
| Not detected | 37 | 70 | |
Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 2.
Percent agreement of the 229 posterior oropharyngeal saliva-nasopharyngeal pair. Overall negative and positive percent agreement of the 229 pairs and positive percent agreement of the symptomatic patients according to symptom onset with the corresponding 95% confidence interval. Abbreviations: NPA, negative percent agreement; OPA, overall percent agreement; PPA, positive percent agreement.
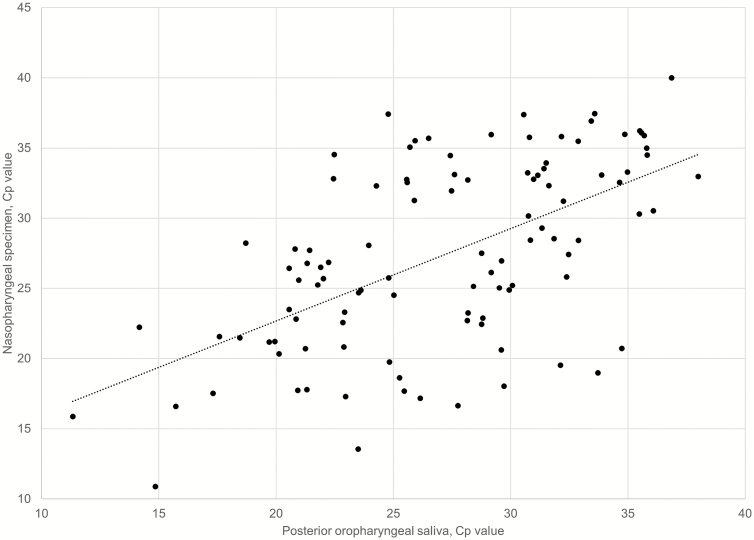
The median and mean of the difference in Cp value among the 104 positive pairs (from 43 patients) were 0.71 and 0.26 (range, 12.63 to −14.74), where a positive difference represents higher Cp, that is, a lower viral load, in the NPsp. Moderate correlation was noted between the Cp values of the 2 specimen types (Pearson correlation coefficient: 0.579) (Figure 3). There was no correlation between the difference in Cp value and the difference in collection time among the POPS-NPsp (Supplementary Figure 1) or between the collection time and result concordance/discordance with the paired NPsp (P = .889).
Figure 3.
Comparison of Cp values of same-day posterior oropharyngeal saliva-nasopharyngeal specimen pairs. Cp values from 104 posterior oropharyngeal saliva and nasopharyngeal specimen pairs collected from 43 patients with detectable SARS-CoV-2 were included for comparison (Pearson correlation coefficient: 0.579). Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
POPS-NPsp Among Symptomatic Patients With Confirmed COVID-19
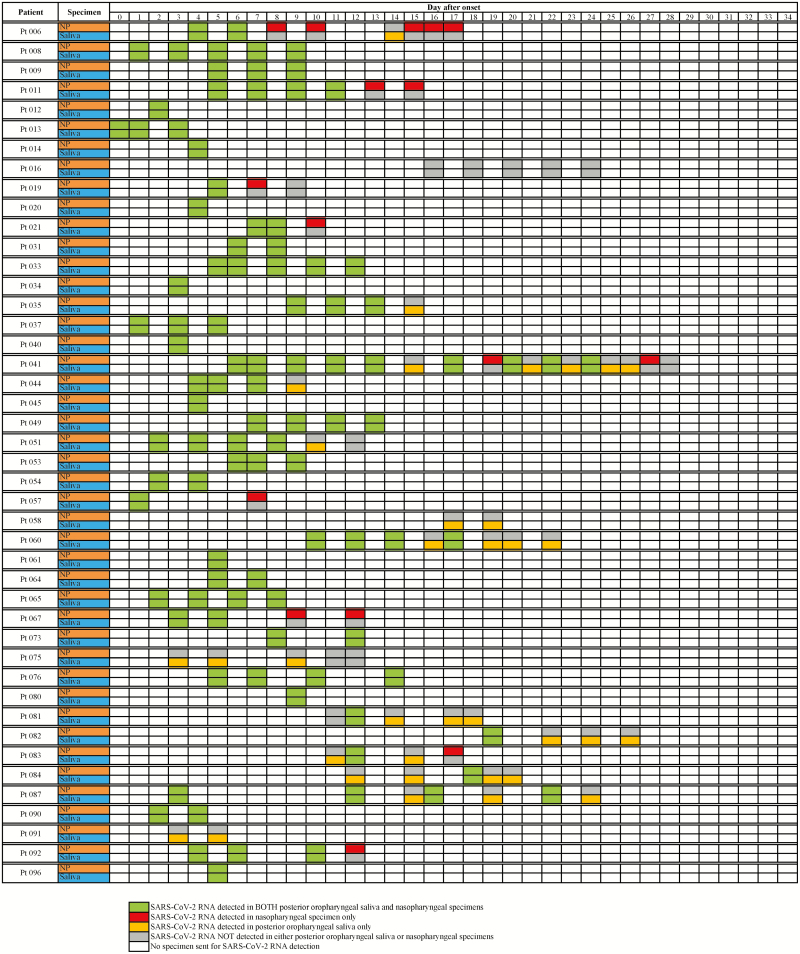
Of the 229 POPS-NPsp, 161 were collected from 44 symptomatic COVID-19 patients. Sixty-three pairs (from 34 patients) were obtained within 1 week ( ≤7 days), and 98 pairs (from 29 patients) were obtained after 1 week from symptom onset. The PPA of POPS-NPsp collected from COVID-19 patients within 7 days of symptom onset was high at 96.6% (95% CI 87.3–99.4%), as compared with the PPA and NPA of 75.0% (95% CI 61.4–85.2%) and 26.2% (95% CI 14.4–42.3%) in POPS-NPsp collected more than 7 days from symptom onset (Figure 2). Of the discordant POPS-NPsp, SARS-CoV-2 RNA was more frequently detected in POPS compared with NPsp. Four of the 6 discordant results within the first week of symptoms onset week were POPS+/NPsp− with a relatively low Cp value of 22.63–28.45 in POPS, compared with the 2 POPS−/NPsp + with a higher Cp values of 34.32 and 37.11 (Supplementary Table 2). A longitudinal analysis of these symptomatic patients showed that intermittent positive results among POPS and NPsp are often seen during the later course of illness (Figure 4). Notably, POPS was the predominant specimen type with detectable SARS-CoV-2 RNA in patient 58, 75, 81, 82, 84, and 87, all of whom were symptomatic and young with age ranged from 17 to 36 years (mean 22 years), suffering from mild respiratory symptoms and no documented hypogeusia or dry mouth.
Figure 4.
Longitudinal SARS-CoV-2 RNA test results from the 44 patients with confirmed COVID-19. Forty-four symptomatic COVID-19 patients were included with SARS-CoV-2 RNA results from posterior oropharyngeal saliva and nasopharyngeal specimens during their stay in the airborne infection isolation facilities in the hospitals of the Kowloon Central Cluster, with the day of symptom onset being day 0 on the chart. Only days where both posterior oropharyngeal saliva and nasopharyngeal specimen types were obtained were included on the diagram. Asymptomatic COVID-19 patients with paired POPS-NPsp specimens were not included in this figure. Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
POPS-NPsp Among Pediatric Patients
Only 21 POPS-NPsp from 7 pediatric patients were identified (age range: 4–18 years) (Supplementary Table 3). Six of the 7 patients have confirmed COVID-19, 4 of whom were asymptomatic. Eight of the 13 concordant POPS-NPsp showed detectable SARS-CoV-2 RNA in both specimen types, with a mean and median Cp value difference of −1.09 and −1.32 (range 2.93 to −5.18), where a negative difference represents higher Cp, that is, lower viral load, in POPS. Nonetheless, 5 of the 8 discordant pairs were POPS+/NPsp−.
DISCUSSION
Hong Kong was the first region in the world to adopt POPS for mass screening in the COVID-19 pandemic. “Deep throat saliva” was the official term used by the government of HKSAR and HA in the promotion materials, but we have used POPS here as per previous publications that better describe the anatomical origin (posterior oropharynx) of the specimen [14]. The first POPS-based enhanced laboratory SARS-CoV-2 surveillance “Tier 4” program was launched on 19 February 2020 [23]. Up to 19 April 2020, over 35000 “Tier 4” POPS were screened Hong Kong wide, facilitated identification of >100 COVID-19 patients, including the uncovering of at least 2 local clusters [27, 28].
We performed a head-to-head comparison of POPS-NPsp, many of which were collected from COVID-19 patients during their course of illness. POPS showed a fair OPA with NPsp, and more discordance pairs were POPS+/NPsp−. The PPA was particularly good between specimens collected within 7 days of symptom onset. Our finding might be explained by several reasons. First, POPS specimens might contain both bronchopulmonary secretions and nasopharyngeal secretions [14], that is, a mixed upper and lower respiratory specimen, hence increasing the detection probability of SARS-CoV-2 RNA which affects both upper and lower respiratory tracts. Compared with saliva straight from salivary glands, posterior oropharyngeal saliva appeared to have higher sensitivity [12–14]. Second, POPS collected in this study were not routinely mixed with VTM, unlike NPsp that are transported in VTM, which minimized any potential dilution effect. Third, epithelial cells lining salivary gland ducts were previously shown to be an early target for SARS-CoV in a rhesus macaque model [29], and a more recent study demonstrated high expression of ACE2 receptors, a receptor used by SARS-CoV-2 and SARS-CoV, on the mucosa of oral cavity [30]; thus, POPS might theoretically contain more SARS-CoV-2 virions during early illness. As expected, the percent agreement of POPS-NPsp is higher during the first 7 days of illness [12, 14, 31, 32] which adds to the support that POPS is suitable for the diagnosis of patients presenting during early infection. Moreover, no difference in categorical agreement was noted between POPS collected during early morning compared with other times of the day, implying that “early morning” collection of POPS may not be mandatory. However, it must be noted that all patients included had refrained from eating, drinking and teeth brushing for at least 2 hours before obtaining POPS regardless of actual collection time. Finally, POPS may also be an acceptable alternative specimen type for SARS-CoV-2 RNA detection in children as fair categorical concordance among POPS-NPsp were also seen in this group. However, more data are required to confirm our finding due to the small number of pediatric patients in our cohort.
There are several limitations in our study. First, specimen subtypes among NPsp were rather heterogeneous, and it could not be excluded that 1 or more NPsp subtype would be superior to POPS for SARS-CoV-2 RNA detection. However, this candidly reflects the logistic difficulties in standardizing the specimen collection protocol among inpatients, ambulatory patients, and asymptomatic patients from different surveillance programs and settings, particularly during the present pandemic. Second, not all POPS were mixed with VMT, which could affect the overall sensitivity of POPS due to dilution effect. However, this should only lead to a bias against POPS and would not change the overall conclusion in favor of POPS specimens in the present study. Finally, due to the retrospective nature of the study, there is considerable heterogeneity in sampling of the patient population, as both suspected and confirmed cases were included and tested at different frequencies. This is further complicated by the absence of a diagnostic gold standard, which precluded the calculation of diagnostic performance like test sensitivity and specificity. Hence, our results may not apply in other settings with a different mix of patients or disease prevalence.
Our result suggested that POPS is a satisfactory alternative specimen type for SARS-CoV-2 RNA detection, with acceptable agreement of test results when compared with NPsp. In addition to the ease of collection and lesser patient discomfort, the cost savings for POPS collection is also considerable compared with conventional NPSp. Our calculation based on the latest costs of equipment required was USD$8.24 per 100 POPS specimen collection, compared with USD$104.87 and USD$165.38 per 100 NPS and NPA specimen collection, respectively. In light of the ongoing COVID-19 pandemic, mass screening program may be undertaken by more territories to facilitate control of ongoing transmission. POPS would be a desirable specimen type in such settings, where patients can collect the specimen by themselves at home, without assistance from HCW or requirements for PPE, at a lower cost compared with the conventional NPsp.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the Molecular Laboratory of Department of Pathology, Queen Elizabeth Hospital, for their tireless support in performing molecular testing for SARS-CoV-2 RNA for patients with suspected or confirmed COVID-19.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Charlton CL, Babady E, Ginocchio CC, et al. Practical guidance for clinical microbiology laboratories: viruses causing acute respiratory tract infections. Clin Microbiol Rev 2019; 32 (1) e00042–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. World Health Organization.2020. Available at: https://apps.who.int/iris/handle/10665/331329. Accessed 6 May 2020.
- 3. Centers for Disease Control and Prevention. Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (covid-19), 14 April 2020 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed 27 April 2020.
- 4. Marty FM, Chen K, Verrill KA. How to obtain a nasopharyngeal swab specimen. N Engl J Med 2020. April 17. doi: 10.1056/NEJMvcm2010260. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 5. Mufson S, Timberg C, Tiku N. When these Boston doctors ran out of virus-testing swabs, they mobilized an army of 3-D printers. The Washington Post.2020. Available at: https://www.washingtonpost.com/climate-environment/2020/04/22/nasal-swabs-shortage-coronavirus/. Accessed 27 April 2020.
- 6. Webber L, Jewett C. Testing swabs run in short supply as makers try to speed up production. National Public Radio2020. Available at: https://www.npr.org/sections/health-shots/2020/03/18/817801222/testing-swabs-run-in-short-supply-as-makers-try-to-speed-up-production. Accessed 27 April 2020.
- 7. Kim YG, Yun SG, Kim MY, et al. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J Clin Microbiol 2017; 55:226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. To KK, Lu L, Yip CC, et al. Additional molecular testing of saliva specimens improves the detection of respiratory viruses. Emerg Microbes Infect 2017; 6:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robinson JL, Lee BE, Kothapalli S, Craig WR, Fox JD. Use of throat swab or saliva specimens for detection of respiratory viruses in children. Clin Infect Dis 2008; 46:e61–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. To KKW, Yip CCY, Lai CYW, et al. Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect 2019; 25:372–8. [DOI] [PubMed] [Google Scholar]
- 11. Yoon J, Yun SG, Nam J, Choi SH, Lim CS. The use of saliva specimens for detection of influenza A and B viruses by rapid influenza diagnostic tests. J Virol Methods 2017; 243:15–9. [DOI] [PubMed] [Google Scholar]
- 12. To KK, Tsang OT, Chik-Yan Yip C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 2020; pii: ciaa149. doi: 10.1093/cid/ciaa149. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen L, Zhao J, Peng J, et al. Detection of 2019-nCoV in saliva and characterization of oral symptoms in COVID-19 patients. Lancet Infect Dis 2020. [Preprint]. 19 March 2020. Accessed 11 May 2020. Available at SSRN: https://ssrn.com/abstract=3556665 or doi: 10.2139/ssrn.3556665. [DOI] [Google Scholar]
- 14. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. U.S. Food and Drug Administration. Accelerated emergency use authorization (EUA) summary: SARS-CoV-2 assay, 2020 April 13.2020. Available at: https://www.fda.gov/media/136875/download. Accessed 27 April 2020.
- 16. Zheng S, Yu F, Fan J, et al. Saliva as a diagnostic specimen for SARS-CoV-2 by a PCR-based assay: a diagnostic validity study. Lancet Infect Dis 2020. [Preprint]. 28 February 2020. Available at SSRN: https://ssrn.com/abstract=3543605 or doi: 10.2139/ssrn.3543605. [DOI] [Google Scholar]
- 17. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect 2020; pii: S0163–4453(20)30213–9. doi: 10.1016/j.jinf.2020.04.005. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kojima N, Turner F, Slepnev V, et al. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for COVID-19 detection. MedRxIV [Preprint] 2020. doi: 10.1101/2020.04.11.20062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol 2020; pii: JCM.00776–20. doi: 10.1128/JCM.00776-20. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs.]MedRxIV [Preprint] 2020. doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
- 21. Wuhan municipal health commission. Report of clustering pneumonia of unknown etiology in Wuhan City. Wuhan, China: Wuhan Municipal Health Commission; 2019http://wjw.whuhan.gov.cn/front/web/showDetail/2019123108989. 31 Dec 2019 Accessed 28 February 2020. (In Chinese). [Google Scholar]
- 22. news.gov.hk. Virus test centres in operation. The Government of the Hong Kong Special Administrative Region.2020. Available at: https://www.news.gov.hk/eng/2020/03/20200320/20200320_165632_459.html Accessed 27 April 2020.
- 23. Hospital Authority. HA enhanced laboratory surveillance programme extends to outpatients with mild symptoms.2020. Available at: https://www.info.gov.hk/gia/general/202002/18/P2020021800437.htm. Accessed 27 April 2020.
- 24. Centre of Health Protection. Enhanced laboratory surveillance with testing for COVID-19 at clinics of private medical practitioners. Department of Health, The Government of the Hong Kong Special Administrative Region 6 March 2020. Available at: https://www.chp.gov.hk/files/pdf/letters_to_doctors_20200306.pdf Accessed 27 April 2020.
- 25. Department of Health. CHP to further extend enhanced laboratory surveillance programme. the government of the Hong Kong special administrative region.2020. March 24. Available at: https://www.info.gov.hk/gia/general/202003/24/P2020032400828.htm Accessed 27 April 2020.
- 26.HA Central Committee on Infectious Diseases and Emergency Response (CCIDER). Interim Recommendation on Clinical Management of Adult Cases with Coronavirus Disease 2019 (COVID-19). Hospital Authority. 13 February 2020. Accessed 28 April 2020.
- 27. Centre of Health Protection. CHP investigates four additional cases of COVID-19. Department of Health, The Government of the Hong Kong Special Administrative Region 4 March2020. Available at: https://www.info.gov.hk/gia/general/202003/04/P2020030400780.htm Accessed 27 April 2020.
- 28. Centre of Health Protection. CHP investigates five additional cases of COVID-19. Department of Health, The Government of the Hong Kong Special Administrative Region. 10 March 2020. Available at: https://www.info.gov.hk/gia/general/202003/10/P2020031000782.htm. Accessed 27 April 2020.
- 29. Liu L, Wei Q, Alvarez X, et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol 2011; 85:4025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci 2020; 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020. Apr 1. doi: 10.1038/s41586-020-2196-x. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 32. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020. Mar 11. doi: 10.1001/jama.2020.3786. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.