Abstract
Background
Remdesivir is a prodrug of the nucleoside analogue GS-441524 and is under evaluation for treatment of SARS-CoV-2-infected patients.
Objectives
To evaluate the pharmacokinetics of remdesivir and GS-441524 in plasma, bronchoalveolar aspirate (BAS) and CSF in two critically ill COVID-19 patients.
Methods
Remdesivir was administered at 200 mg loading dose on the first day followed by 12 days of 100 mg in two critically ill patients. Blood samples were collected immediately after (C0) and at 1 (C1) and 24 h (C24) after intravenous administration on day 3 until day 9. BAS samples were collected on Days 4, 7 and 9 from both patients while one CSF on Day 7 was obtained in one patient. Remdesivir and GS-441524 concentrations were measured in these samples using a validated UHPLC-MS/MS method.
Results
We observed higher concentrations of remdesivir at C0 (6- to 7-fold higher than EC50 from in vitro studies) and a notable decay at C1. GS-441524 plasma concentrations reached a peak at C1 and persisted until the next administration. Higher concentrations of GS-441524 were observed in the patient with mild renal dysfunction. Mean BAS/plasma concentration ratios of GS-441524 were 2.3% and 6.4% in Patient 1 and Patient 2, respectively. The CSF concentration found in Patient 2 was 25.7% with respect to plasma. GS-441524 levels in lung and CNS suggest compartmental differences in drug exposure.
Conclusions
We report the first pharmacokinetic evaluation of remdesivir and GS-441524 in recovered COVID-19 patients. Further study of the pharmacokinetic profile of remdesivir, GS-441524 and the intracellular triphosphate form are required.
Introduction
On 24 March 2020, the WHO confirmed 375 498 cases of infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 causes coronavirus disease 2019 (COVID-19), which has a clinical picture ranging from mild to severe, including death.1 There is no specific antiviral treatment recommended for COVID-19. Patients with COVID-19 should receive supportive care to help relieve symptoms.2 Therefore, testing the efficacy of existing drugs used for other viruses against SARS-CoV-2 is a fundamental goal in order to develop a specific therapy rapidly. Small molecules, such as nucleoside analogues or protease inhibitors, are candidate drugs as treatments for COVID-19.3 Among these nucleoside analogues, remdesivir (GS-5734), a diastereomer monophosphophoramidate prodrug of the adenine nucleoside analogue GS-441524, is under evaluation. GS-441524 undergoes metabolic conversion in cells and tissues into the pharmacologically active triphosphate form (GS-443902) that inhibits viral RNA polymerases, but does not affect host RNA or DNA polymerases.4 Remdesivir showed a broad-spectrum antiviral activity with potent in vitro efficacy against multiple genetically unrelated RNA viruses similar to SARS-CoV-2, such as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV). According to several in vitro studies, remdesivir appears to be the most promising candidate for COVID-19 treatment.5–8 In Vero cells infected with SARS-CoV-2, remdesivir (molecular weight, 602.6 g/mol) showed an EC50 of 460 ng/mL.5 However, for GS-441524 (molecular weight, 291.3 g/mol) EC50 data from cells infected with SARS-CoV-2 have not been reported. In humans, remdesivir showed moderate protein binding (80%–90%) and GS-441524 exhibited very low protein binding in plasma (<20%).9 Preliminary data from human healthy donors confirm that remdesivir is extensively metabolized by cytochrome P450 enzyme (CYP) (CYP2C8, CYP2D6 and CYP3A4). Specific metabolism data on GS-441524 have not been reported. The half-lives are about 0.89 and 25 h for remdesivir and GS-441524, respectively. Remdesivir is mainly excreted in urine (about 74%). The predominant species detected in urine were GS-441524, followed by remdesivir and other metabolites.9
In clinical practice, a SARS-CoV-2-infected patient recovered after receiving intravenous remdesivir for compassionate use in the USA.10 Currently, 10 clinical trials (1 suspended, 1 terminated, 8 recruiting) have been initiated to evaluate intravenous remdesivir in adult patients with mild, moderate and severe COVID-19 (ClinicalTrials.gov). To date, there are no pharmacokinetic (PK) data for SARS-CoV-2-infected patients. The only published PK data were obtained from observations of Ebola treatment in monkeys.8 The aim of our study was to investigate remdesivir and GS-441524 plasma concentrations in two critically ill COVID-19 patients treated with intravenous remdesivir. Moreover, compartmental concentrations were evaluated, in bronchoalveolar aspirate (BAS) in both patients and in a CSF sample from one of them.
Patients and methods
Ethics
The study was conducted in accordance with the Declaration of Helsinki as well as with national and institutional standards. Informed consent was obtained from family members. All data were collected anonymously.
Patients and treatment
The two patients were a female (Patient 1, PT1) and a male (Patient 2, PT2) 66 and 67 years old, respectively. Both were residents of the city of Wuhan, China, who had travelled to Italy.
In January, they developed fever with respiratory symptoms. Subsequently they were admitted to the National Institute of Infectious Diseases ‘Lazzaro Spallanzani’, in Rome, Italy. Nasopharyngeal and oropharyngeal swabs from both patients were positive for SARS-CoV-2 by RT–PCR assay. Both patients developed progressive respiratory failure and clinical evidence of severe adult respiratory distress syndrome and required mechanical ventilation support in the ICU.11
As it was recommended for compassionate use, and in the absence of any clinical trial available at that time, we started remdesivir treatment on request from the responsible physicians owing to clinical worsening. Gilead Science Inc. provided remdesivir and instructions for preparation and administration for intravenous administration. The remdesivir dosing regimen was similar to that reported in the Ebola clinical trial and therefore was used for 13 days (PALM Consortium Study).12 The manufacturer-recommended adult dosage and duration of remdesivir were 200 mg loading dose on the first day followed by 12 days of 100 mg once-daily maintenance doses to be administered intravenously over 1 h. Both patients concluded 13 days of remdesivir treatment, despite transient renal impairment in PT1. Efficacy and safety evaluations were performed during the study and included virology and clinical laboratory tests (haematology, serum chemistry), vital sign monitoring, and physical examinations were evaluated daily. Monitoring was performed from baseline until the end of treatment.
Bioanalytical method and PK assessments of remdesivir and GS-441524
Blood samples were collected immediately after (C0) and at 1 (C1) and 24 h (C24) after remdesivir intravenous administration on Day 3 until Day 9 for the determination of plasma concentrations of remdesivir and GS-441524. For compartmental distribution, BAS samples were collected on Days 4, 7 and 9 from both patients and a CSF sample was obtained on Day 7 from PT2. Immediately after collection, the samples were kept at 4°C and subsequently centrifuged and stored at −20°C until analysis. Concentrations of remdesivir and the parent nucleoside analogue GS-441524 were measured in the different matrices using a validated UHPLC-MS/MS method.13 Briefly, 50 μL of the sample was mixed with 600 μL of acetonitrile:methanol (50:50, v:v) containing internal standard. Three hundred microlitres of supernatant was diluted with 600 μL of HPLC-MS grade water and 10 μL was injected into the column (Waters Acquity UPLC HSS T3, 1.8 μm, 2.1 × 50 mm). Using this method remdesivir and GS-441524 were measured simultaneously with a limit of quantification (LOQ) of 5.86 ng/mL for remdesivir and 1.96 ng/mL for GS-441524. In order to decrease the plasma esterase activity all steps were performed on ice.
Statistical analysis
Normally distributed continuous data were expressed as mean (SD). The AUC from the time of finish of infusion to the next infusion (AUC0–24) was calculated by Phenix 8.1 with non-compartmental analysis (linear log trapezoidal rule). Pharmacokinetic data were compared using the Mann–Whitney U-test. A P value of <0.05 was considered statistically significant. Statistical analysis was performed using the GraphPad Prism package, version 8.4.0 (GraphPad Software, San Diego, CA, USA).
Results
Patients
The characteristics of the two COVID-19 patients before, during and after treatment are shown in Table S1 (available as Supplementary data at JAC Online). Before starting remdesivir treatment in PT1 a transient renal impairment was observed (creatinine 203.3 μmol/L). Due to the critical condition of PT1 and the absence of effective alternative treatment, remdesivir was administered at the same dose. Moreover, during and after treatment renal function improved in PT1 (creatinine during treatment, 188.2 μmol/L; creatinine after treatment, 123.7 μmol/L). Nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 RT-PCR detection were negative for both patients 1 week after completing remdesivir treatment.
Pharmacokinetics of remdesivir
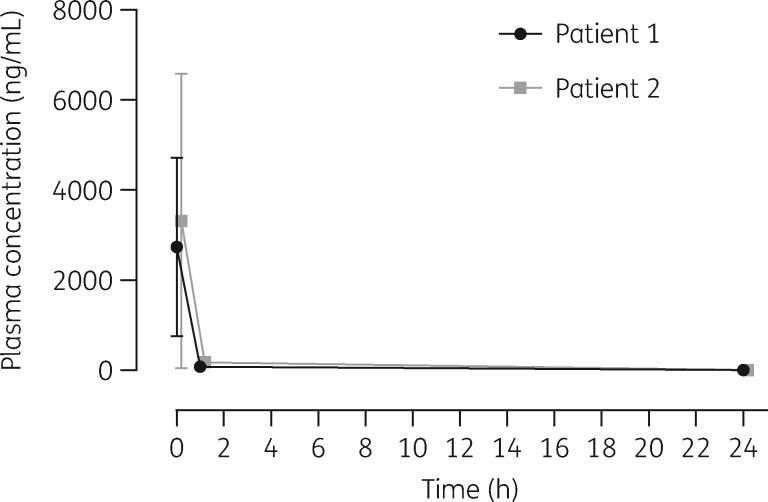
The mean plasma concentration–time profiles of remdesivir after intravenous administration of remdesivir in PT1 and in PT2 are presented in Figure 1. Immediately after remdesivir infusion, peak serum concentrations were observed in both patients. Median (SD) C0 values were 2737 (1985) and 3317 (3280) ng/mL in PT1 and in PT2, respectively; rapid plasma concentration decay was observed 1 h after infusion, to 80.7 (10.7) ng/mL and 171 (75.2) ng/mL, respectively. At 24 h after intravenous administration, the concentrations of remdesivir were below the LOQ in both patients. AUC24 in PT1 and PT2 was 2915 and 4042 ng·h/mL, respectively.
Figure 1.
Pharmacokinetics of remdesivir following intravenous administration of multiple doses of remdesivir in critically ill patients (Patient 1 with renal impairment and Patient 2 without renal impairment; mean ± SD measured 3–9 days after remdesivir initiation).
No significant differences in the mean concentrations were observed at any timepoint between the two patients (P > 0.05). Remdesivir exposure did not appear to be meaningfully affected by renal impairment. Concentrations of remdesivir in all BAS samples and in the CSF were under the LOQ.
Pharmacokinetics of GS-441524
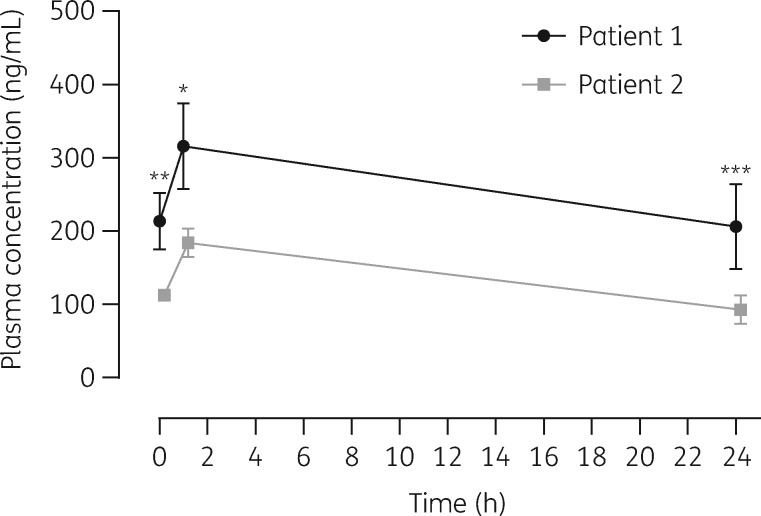
The mean plasma concentration–time profiles of GS-441524 after intravenous administration of remdesivir in PT1 and in PT2 are presented in Figure 2. In PT1, mean (SD) GS-441524 plasma concentrations immediately after and 1 and 24 h after remdesivir infusion were 214 (38.4), 316 (58.5) and 206 (58.1) ng/mL, respectively. In PT2 the GS-441524 plasma concentrations were 113 (6.84), 184 (19.7) and 92.6 (19.6) ng/mL at C0, C1 and C24, respectively. Data demonstrated that 1 h after remdesivir infusion a GS-441524 peak serum concentration was present in both patients. As shown in Figure 2, significant differences in the mean concentrations were observed between the two patients at all timepoints (C0, P = 0.0079; C1, P = 0.0286; C24, P ≤ 0.0001). AUC0–24 was 6131 ng·h/mL in PT1 versus 3117 ng·h/mL in PT2. GS-441524 AUC0–24 was increased 1.96-fold, C0 1.89-fold, C1 1.71-fold and C24 2.20-fold, comparing PT1 with PT2. Therefore, higher concentrations were found in the patient with renal dysfunction.
Figure 2.
Pharmacokinetics of nucleoside analogue GS-441524 following intravenous administration of multiple doses of remdesivir in two critically ill patients (mean ± SD measured 3–9 days after remdesivir initiation). Statistical comparison of GS-441524 plasma concentrations at pharmacokinetic timepoints in Patient 1 (with renal impairment) versus Patient 2 (without renal impairment): *P < 0.05; **P < 0.01; ***P < 0.001 (Mann–Whitney test; P < 0.05 was considered significant).
In BAS, concentrations of GS-441524 on day 4 were 8.6 versus 9.2 ng/mL, those on day 7 were 2.7 versus 3.3 ng/mL and those on day 9 were 3.0 versus 6.1 ng/mL, and the mean BAS/plasma ratio was 2.3% versus 6.4% in PT1 and PT2, respectively. GS-441524 CSF concentration in PT2 on day 7 was 22.1 ng/mL, representing a CSF/plasma ratio of 25.7%.
Discussion
Remdesivir is a nucleotide analogue RNA polymerase inhibitor that showed decreased viral load and improved pulmonary function in an animal model of SARS-CoV and MERS-CoV infection.6,7 An in vitro study showed that remdesivir inhibited SARS-CoV-2 virus in a human cell line.5
These data, together with the addition of human safety profile data for intravenous remdesivir therapy obtained from a randomized trial in the context of Ebola virus disease,12 are the basis for the use of intravenous remdesivir as a treatment for COVID-19 patients.14
We report, for the first time, remdesivir PK data in COVID-19 patients. Data were obtained from two critically ill Chinese patients with severe adult respiratory distress syndrome.11 After intravenous administration, in both patients remdesivir showed a peak at the end of infusion and a half-life of 1 h, while GS-441524 reached a peak 1 h after infusion and then remained detectable until the next remdesivir administration. Similar data have been shown in rhesus monkeys after daily intravenous administration of remdesivir, which was rapidly converted into the nucleoside analogue GS-441524 and appeared to reach sustained intracellular concentrations of the active triphosphate form above its EC90 for SARS-CoV.5,8,14
Furthermore, we found higher GS-441524 plasma concentrations in the patient with renal impairment, indicating that renal excretion was a major route of elimination. There are no specific studies conducted with remdesivir in patients with renal and/or hepatic impairment; during the PALM and MEURI protocol no renal or hepatic abnormalities were attributed to remdesivir.11 Given the benefit–risk calculation in patients with severe SARS-CoV infection and renal impairment, the use of remdesivir should be evaluated. Nevertheless, placebo-controlled clinical trials are needed to determine the balance between safety and efficacy in these patients.15
Remdesivir was undetectable in the compartments evaluated, whereas the GS-441524 metabolite was detected in BAS, but at a lower concentration compared with CSF. Compartment differences in drug exposure could be due to different blood supply or protein binding properties (remdesivir has moderate protein binding while GS-441524 exhibits very low protein binding),9 which are the major factors influencing drug diffusion in cells and tissue. In addition, active efflux by different specific protein transporters in lung and in CNS, such as P-glycoprotein (P-gp), could potentially be involved.9,16,17 Remdesivir is a substrate for P-gp, while no data are yet available on GS-441524, requiring further studies.
Another hypothesis could be the different drug uptake and metabolism in lung cells, where a high number of virus-infected cells are present.18 Evaluation of the triphosphate form in lung cells is necessary to confirm this hypothesis.
These preliminary observations increase knowledge about the PK and use of remdesivir for treatment of COVID-19 patients. However, further studies, with higher numbers of patients, and clinical trials, are needed confirm these preliminary results.
Supplementary Material
Acknowledgements
Members of COVID 19 INMI Study Group
Maria Alessandra Abbonizio, Chiara Agrati, Fabrizio Albarello, Gioia Amadei, Alessandra Amendola, Andrea Antinori, Mario Antonini, Raffaella Barbaro, Barbara Bartolini, Martina Benigni, Nazario Bevilacqua, Licia Bordi, Veronica Bordoni, Marta Branca, Paolo Campioni, Maria Rosaria Capobianchi, Cinzia Caporale, Ilaria Caravella, Fabrizio Carletti, Rita Casetti, Concetta Castilletti, Roberta Chiappini, Carmine Ciaralli, Eleonora Cimini, Francesca Colavita, Angela Corpolongo, Massimo Cristofaro, Salvatore Curiale, Alessandra D’Abramo, Cristina Dantimi, Alessia De Angelis, Giada De Angelis, Rachele Di Lorenzo, Federica Di Stefano, Gianpiero D’Offizi, Federica Ferraro, Lorena Fiorentini, Andrea Frustaci, Paola Gallì, Gabriele Garotto, Maria Letizia Giancola, Filippo Giansante, Emanuela Giombini, Germana Grassi, Maria Cristina Greci, Giuseppe Ippolito, Eleonora Lalle, Simone Lanini, Daniele Lapa, Luciana Lepore, Andrea Lucia, Franco Lufrani, Manuela Macchione, Alessandra Marani, Luisa Marchioni, Andrea Mariano, Maria Cristina Marini, Micaela Maritti, Giulia Matusali, Silvia Meschi, Francesco Messina, Chiara Montaldo, Silvia Murachelli, Emanuele Nicastri, Roberto Noto, Claudia Palazzolo, Emanuele Pallini, Fabrizio Palmieri, Virgilio Passeri, Federico Pelliccioni, Antonella Petrecchia, Ada Petrone, Nicola Petrosillo, Elisa Pianura, Maria Pisciotta, Silvia Pittalis, Costanza Proietti, Vincenzo Puro, Gabriele Rinonapoli, Martina Rueca, Alessandra Sacchi, Francesco Sanasi, Carmen Santagata, Silvana Scarcia, Vincenzo Schininà, Paola Scognamiglio, Laura Scorzolini, Giulia Stazi, Francesco Vaia, Francesco Vairo, Maria Beatrice Valli.
Funding
This study was supported by funds from the Italian Ministry of Health (Ricerca Corrente, Linea 1).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online.
References
- 1. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 2. https://www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html.
- 3. Li G, De Clercq E.. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov 2020; 19: 149–50. [DOI] [PubMed] [Google Scholar]
- 4. Siegel D, Hui HC, Doerrfler E. et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J Med Chem 2017; 60: 1648–61. [DOI] [PubMed] [Google Scholar]
- 5. Wang M, Cao R, Zhang L. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020; 30: 269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sheahan TP, Sims AC, Leist SR. et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun 2020; 11: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheahan TP, Sims AC, Graham RL. et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017; 9: eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Warren TK, Jordan R, Lo MK. et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016; 531: 381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. https://www.ema.europa.eu/en/documents/other/summary-compassionate-use-remdesivir-gilead_en.pdf.
- 10. Holshue ML, DeBolt C, Lindquist S. et al. First case of 2019 novel coronavirus in the United States. N Engl J Med 2020; 382: 929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albarello F, Pianura E, Di Stefano F. et al. 2019-novel coronavirus severe adult respiratory distress syndrome in two cases in Italy: an uncommon radiological presentation. Int J Infect Dis 2020; 93: 192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mulangu S, Dodd LE, Davey RT Jr. et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 2019; 381: 2293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avataneo V, de Nicolò A, Cusato J, et al. Development and validation of a UHPLC-MS/MS method for quantification of the prodrug remdesivir and its metabolite GS-441524: a tool for clinical pharmacokinetics of SARS-CoV-2/COVID-19 and Ebola virus disease. J Antimicrob Chemother 2020; 10.1093/jac/dkaa152. [DOI] [PMC free article] [PubMed]
- 14. Ko WC, Rolain JM, Lee NY. et al. Arguments in favor of remdesivir for treating SARS-CoV-2 infections. Int J Antimicrob Agents 2020; 5: 105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grein J, Ohmagari N, Shin D. et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med 2020; 10: doi:10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rizk ML, Zou L, Savic RM. et al. Importance of drug pharmacokinetics at the site of action. Clin Transl Sci 2017; 10: 133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borst P, Schinkel AH.. P-glycoprotein ABCB1: a major player in drug handling by mammals. J Clin Invest 2013; 123: 4131–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taneva E, Crooker K, Park SH. et al. Differential mechanisms of tenofovir and tenofovir disoproxil fumarate cellular transport and implications for topical preexposure prophylaxis. Antimicrob Agents Chemother 2015; 60: 1667–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.