Abstract
False-negative severe acute respiratory syndrome coronavirus 2 test results can negatively impact the clinical and public health response to coronavirus disease 2019 (COVID-19). We used droplet digital polymerase chain reaction (ddPCR) to demonstrate that human DNA levels, a stable molecular marker of sampling quality, were significantly lower in samples from 40 confirmed or suspected COVID-19 cases that yielded negative diagnostic test results (ie, suspected false-negative test results) compared with a representative pool of 87 specimens submitted for COVID-19 testing. Our results support suboptimal biological sampling as a contributor to false-negative COVID-19 test results and underscore the importance of proper training and technique in the collection of nasopharyngeal specimens.
Keywords: COVID-19, ddPCR, false negative, nasopharyngeal swab, sample quality
Improper nasopharyngeal swab collection could compromise COVID-19 diagnosis. In support of this, suspected false-negative specimens from confirmed or suspected COVID-19 cases contained less human DNA (a molecular marker of sampling quality) compared with a representative specimen pool submitted for testing.
Accurate coronavirus disease 2019 (COVID-19) diagnosis is critical to a successful clinical and public health response. Current COVID-19 tests detect 1 or more targets in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ribonucleic acid (RNA) genome, usually by real-time reverse-transcriptase polymerase chain reaction (RT-PCR), and nasopharyngeal swabs have been the preferred sample for testing to date [1]. Although PCR-based tests are highly sensitive, false-negative COVID-19 test results do occur [2, 3], although reported rates vary. A recent large retrospective study estimated the clinical sensitivity of SARS-CoV-2 molecular assays to be between 58% and 96% [4], whereas another reported a 67% SARS-CoV-2 RNA detectability rate in respiratory samples taken within 7 days of hospitalization for COVID-19 [5]. Various factors other than molecular technology contribute to test sensitivity, including the timing of sample collection with respect to infection stage [6, 7] as well as specimen storage and transport [2]. Improper specimen collection could also contribute to false-negative COVID-19 test results. Although nasopharyngeal swabs are routinely ordered for respiratory viruses, the collection of a high-quality specimen requires training and expertise because it involves insertion of the swab to the posterior nasopharynx, a depth of approximately 7 cm, followed by rotation and withdrawal of the swab [8]. To investigate suboptimal sample collection as a possible cause of false-negative test results, we quantified human deoxyribonucleic acid (DNA) levels recovered on nasopharyngeal swabs submitted to a single laboratory for COVID-19 testing, hypothesizing that human DNA could serve as a stable molecular marker of specimen collection quality.
METHODS
The St. Paul’s Hospital Virology laboratory is 1 of 5 provincially designated SARS-CoV-2 diagnostic laboratories in British Columbia, Canada. COVID-19 testing on nasopharyngeal swabs (Copan UTM collection kit or BD universal viral transport system) was performed by total nucleic acid extraction from 500 μL medium on the Roche MagNA Pure 96 followed by real-time RT-PCR using the Roche LightMix 2019 novel coronavirus (2019-nCoV) real-time RT-PCR assay, which uses E-Sarbeco primers/probes [9], or using the Roche cobas SARS-CoV-2 test.
Between March and May 2020, we identified 40 suspected false-negative nasopharyngeal swab test results from presumed or confirmed COVID-19 cases for which >1 mL medium remained for retesting. These included 23 negative samples from individuals who recorded a positive test within ±12 days of the negative test (in which the median time elapsed between negative and positive tests was 4 days, with an interquartile range of 1–6 days) and 17 samples from individuals who tested negative but for whom there was high clinical suspicion of infection by the treating physician with no alternate diagnosis established. A convenience sample of 87 consecutively submitted nasopharyngeal swabs served as a comparison dataset. Remnant specimens were stored at −20°C until retesting. To standardize nucleic acid extraction across all specimens and to maximize viral RNA recovery, 1 mL medium was extracted on the BioMérieux EasyMag and eluted in 35 μL buffer. SARS-CoV-2 detection in suspect false-negative samples was reattempted using a nested RT-PCR and sequencing protocol targeting conserved regions in open reading frame (ORF)-1a and Spike [10], in which the lower limit of detection of this assay was estimated in-house using serial dilutions of synthetic SARS-CoV-2 RNA standards (Exact Diagnostics).
Human DNA levels were quantified using droplet digital PCR (ddPCR), a technique in which each sample is fractionated into 20 000 nL-sized water-in-oil droplets before PCR amplification with sequence-specific primers and fluorescent probes, and in which Poisson statistics are used to calculate input template concentrations at reaction endpoint. Nucleic acid extracts were combined with primer/probe sets targeting 2 regions in the human RPP30 gene ~8 kb apart, ddPCR Supermix for Probes (no dUTPs) (BioRad), XhoI restriction enzyme (New England Biolabs), and nuclease free water. Primers and probes are as follows: RPP30 Forward-GATTTGGACCTGCGAGCG, RPP30 Probe- VIC-CTGACCTGA-ZEN-AGGCTCT- 3IABkFQ, RPP30 Reverse Primer- GCGGCTGTCTCCACAAGT; RPP30 Shear Forward Primer- CCAATTTGCTGCTCCTTGGG, RPP30 Shear Probe- FAM- AAGGAGCAA-ZEN-GGTTCTATTGTAG- 3IABkFQ, RPP30 Shear Reverse Primer- CATGCAAAGGAGGAAGCCG (Integrated DNA Technologies; ZEN = internal ZEN quencher; 3IABkFQ = 3’ Iowa Black Fluorescent Quencher). Droplets were generated using an Automated Droplet Generator (BioRad) and cycled at 95°C for 10 minutes; 40 cycles of (94°C for 30 seconds, 53°C for 1 minute) and 98°C for 10 minutes and analyzed on a QX200 Droplet Reader (BioRad) using QuantaSoft software (version 1.7.4; BioRad). Measured copies of RPP30/reaction are averaged across primer/probe sets, divided by 2 (as each human cell carries 2 RPP30 copies), and normalized to input volume to determine cells/μL extract.
A convenience panel of 91 remnant nucleic acid extracts performed on the Roche MagNA Pure 96 as part of COVID-19 testing was also assessed for human RNAseP RNA levels using the US Centers for Disease Control and Prevention (US-CDC) protocol on a Roche Lightcycler 480 [11] and human RPP30 DNA levels as described above.This study was approved by the Providence Health Care/University of British Columbia and Simon Fraser University Research Ethics Boards.
RESULTS
We began by validating a SARS-CoV-2-nested RT-PCR/sequencing protocol targeting ORF1a and Spike [10] on a blinded test panel of 24 SARS-CoV-2-negative and -positive samples extracted on the EasyMag, and confirming 100% concordance with results obtained using the Roche LightMix 2019-nCoV real-time RT-PCR assay (in which positives in the latter assay yielded Ct values of 21.5 to 35.4; data not shown). Using synthetic SARS-CoV-2 RNA standards, we further estimated the in-house lower limit of detection (LLOD) of the nested RT-PCR assay to be ~1.5 copies/reaction (data not shown), which is lower than the reported LLOD of ~10 copies/reaction from the Roche LightMix 2019-nCoV real-time RT-PCR kit package insert. Reasoning that the nested RT-PCR assay might therefore offer increased SARS-CoV-2 detection sensitivity compared with the original real-time RT-PCR assay, we retested the 40 suspected false-negative specimens by nested RT-PCR. However, all suspect false-negative samples again tested negative. This indicated that the original negative results were not likely attributable to suboptimal real-time RT-PCR assay performance, but rather suggested that SARS-CoV-2 RNA was exceedingly low or absent in these samples.
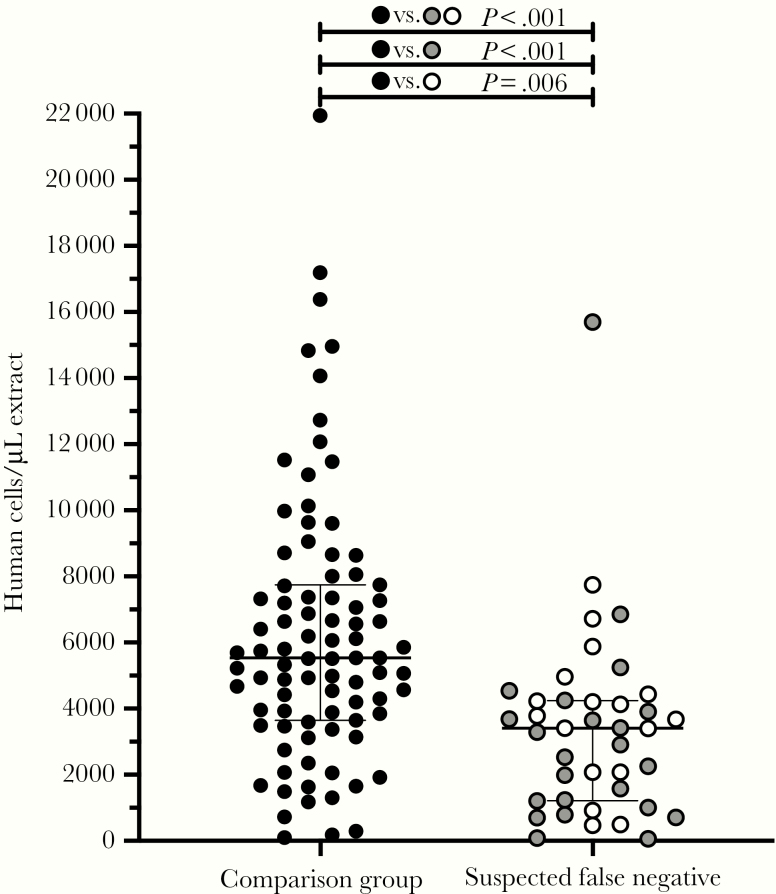
We then investigated whether human DNA recovered on the nasopharyngeal swab could serve as a molecular marker of specimen collection quality, reasoning that DNA (by virtue of its stability) would be well preserved in remnant clinical specimens. We used a sensitive, multiplexed ddPCR protocol for absolute human RPP30 gene copy number quantification [12]. Overall, we observed significantly lower human DNA levels in the suspected false-negative nasopharyngeal swab samples compared with a panel of consecutive samples submitted for testing during the same period, although overlap between groups was still substantial (P < .001) (Figure 1). Specifically, suspected false-negative specimens harbored a median 3409 (interquartile range [IQR], 1213–4242) human cells/μL extract, whereas samples in the comparison group harbored a median 5539 (IQR, 3649–7744) human cells/μL. Further stratification of the false-negative samples by type (ie, negative tests from individuals with a confirmed positive test within ±12 days, and negative tests from individuals with high clinical suspicion of infection) revealed that both subgroups harbored significantly less human DNA compared with the control group (P < .001 and P = .006, respectively). This supports suboptimal biological sampling as a contributing cause of false-negative COVID-19 test results.
Figure 1.
Suspected false-negative coronavirus disease 2019 test samples contained significantly lower human deoxyribonucleic acid (DNA) levels compared with a representative pool of specimens submitted for testing. Human DNA levels (RPP30 gene target) were measured using droplet digital polymerase chain reaction (ddPCR) in nasopharyngeal extracts as a molecular marker of biological sampling quality. “Suspect false-negatives” included 23 negative samples from individuals who recorded a positive test within ±12 days of the negative test (gray) and 17 samples from individuals with high clinical suspicion of being infected but never molecularly confirmed (white). The comparison dataset was a consecutive set of 87 samples submitted for testing in April 2020 to the same laboratory (black). P values report the significance level between the comparison dataset and the suspect false-negative group as a whole (black), between the comparison dataset and the negative samples from individuals who reported a positive test within ±12 days (gray) and between the comparison dataset and the negative samples from individuals with high clinical suspicion (white).
We chose human DNA as a molecular marker of sampling quality because of its stability. However, the US-CDC 2019-nCoV real-time RT-PCR diagnostic panel includes a human RNAseP RNA-specific primer/probe set, in part to assess sample quality [11]. To investigate the relationship between human cells as measured by RPP30 gene copy number using ddPCR, and RNAseP RNA Ct values measured by real time RT-PCR, we retrospectively quantified both molecules in a convenience panel of 91 remnant nasopharyngeal swab nucleic acid extracts generated on the Roche MagNA Pure 96. The measurements correlated strongly: RNAseP RNA Ct values ranged from 19.65 to 27.77, whereas human cells/μL extract measured by ddPCR ranged from 82 to 32 498 (median, 1454; IQR, 667–4100) human cells/μL extract, yielding a Spearman’s ρ = −0.97 (P < .0001).
DISCUSSION
Our results underscore the importance of proper training and technique in the collection of high-quality nasopharyngeal specimens. They also highlight the potential utility of including a molecular marker of sampling quality in SARS-CoV-2 diagnostic assays that could serve as an endogenous control. Although the major commercial assays (eg, Roche cobas SARS-CoV-2; https://www.fda.gov/media/136049) include an internal RNA control for nucleic acid extraction and RT-PCR amplification, these do not provide a measure of biological sampling quality. Although the US-CDC 2019-nCoV real-time RT-PCR diagnostic panel does feature a human RNAseP RNA-specific primer/probe set, in part to assess sample quality [11], our findings suggest that the interpretation criteria for this control may be too liberal. Specifically, the US-CDC’s instructions for use, issued on March 15, 2020, state that failure to detect RNAseP within 40 PCR cycles can indicate insufficient biological material in the sample or other assay problems. In our retrospective test panel of 91 remnant nasopharyngeal nucleic acid extracts, however, the 90th percentile Ct value for RNAseP was 25.9 (range 19.65 to 27.77; see Results). The observation that even the lowest decile of samples in terms of RNAseP RNA levels (possibly representing those for which sampling was the least robust) still amplified well before Ct <40 suggests that this threshold may be insufficient to identify suboptimally collected samples.
Some limitations of our study merit mention. Our use of a convenience sample of 87 consecutively submitted nasopharyngeal swabs may not represent an ideal control group, because there is no guarantee that these samples were collected using appropriate or consistent technique. Indeed, the wide range of human DNA levels observed in this group, and the substantial overlap with the suspected false-negative group, corroborate this notion. However, this limitation should only serve to reduce our study’s statistical power. Moreover, approximately 40% of our suspected false-negative tests derived from patients with high clinical suspicion of SARS-CoV-2 infection but whose diagnosis was never confirmed. Because diagnoses may not have been made in a consistent manner across treating physicians, these samples may be less likely to represent false-negative results. However, our observation that human DNA levels in both subcategories of the false-negative group were significantly lower than in the comparison group suggests that this limitation may be minimal. It is also important to note that our study was not designed to identify a threshold of human DNA (or RNA) that could define a properly collected SARS-CoV-2 nasopharyngeal swab. Future studies attempting to do so would need to consider that recovery efficiency (and thus total yield) of different types of nucleic acid may differ by extraction platform (eg, human DNA levels recovered in the present study differed between BioMérieux and MagNA Pure platforms; see Results), and possibly by swab type. It is also important to note that, although human DNA levels can serve as a surrogate marker of the amount of biological material collected, sampling the correct anatomical location is also critical, particularly for nasopharyngeal swabs.
CONCLUSIONS
Our observations strongly support suboptimal biological sampling, but not PCR sensitivity for SARS-CoV-2 RNA detection, as a contributing cause of false-negative COVID-19 test results.
Notes
Acknowledgments. We thank Dr. Christopher Sherlock for helpful discussions. We also thank the laboratory teams at the St. Paul’s Hospital Virology Laboratory and the BC Centre for Excellence in HIV/AIDS for technical assistance.
Financial support. This work was funded by a Genome BC COVID-19 Rapid Response grant (number COV-115) awarded to C. F. L. and Z. L. B. as co-principal investigators. N. N. K. holds a Vanier Canada Graduate Scholarship from the Canadian Institutes for Health Research. Z. L. B. holds a Scholar Award from the Michael Smith Foundation for Health Research.
Potential conflicts of interest. N. N. K. reports grants from Vanier Canada Graduate Scholarships, Canadian Institutes for Health Research, during the conduct of the study. G. R. reports grants from Genome BC, during the conduct of the study. C. J. B. reports grants from Genome BC, during the conduct of the study and personal fees from Gilead Science, Canada, outside the submitted work. R. B. J. reports personal fees from Abbvie Inc., outside the submitted work. J. S. G. M. reports grants from Genome BC, during the conduct of the study, as well as grants from the Public Health Agency of Canada, the BC Ministry of Health, the US National Institutes of Health, Gilead Sciences, Merck, and ViiV Healthcare, all paid to his institution outside the submitted work. V. L. reports grants from Genome BC during the conduct of the study. M. G. R. reports grants from Genome BC during the conduct of the study. A. S. reports grants from Genome BC during the conduct of this study. N. M. reports grants from Genome BC during the conduct of the study. C. F. L. reports grants from Genome BC during the conduct of the study. Z. L. B. reports grants from Genome BC and the Michael Smith Foundation for Health Research during the conduct of the study. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. US Centers for Disease Control and Prevention. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19). 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed 23 March 2020. [Google Scholar]
- 2. Prinzi A. False Negatives and Reinfections: the Challenges of SARS-CoV-2 RT-PCR Testing Available at: https://asm.org/Articles/2020/April/False-Negatives-and-Reinfections-the-Challenges-of. Accessed 3 May 2020.
- 3. Woloshin S, Patel N, Kesselheim AS. False negative tests for SARS-CoV-2 infection - challenges and implications. N Engl J Med 2020. doi: 10.1056/NEJMp2015897 [DOI] [PubMed] [Google Scholar]
- 4. Green DA, Zucker J, Westblade LF, et al. Clinical performance of SARS-CoV-2 molecular testing. J Clin Microbiol 2020. doi: 10.1128/JCM.00995-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 7. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med 2020. doi: 10.7326/M20-1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marty FM, Chen K, Verrill KA. How to obtain a nasopharyngeal swab specimen. N Engl J Med 2020; 382(22):e76. [DOI] [PubMed] [Google Scholar]
- 9. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nao N, Shirato K, Katano H, Matsuy S, Takeda M.. Detection of second case of 2019-nCoV infection in Japan, 2020. Available at: https://www.niid.go.jp/niid/en/2019-ncov-e/9334-ncov-vir3-2.html. Accessed 23 March 2020. [Google Scholar]
- 11. US Centers for Disease Control and Prevention. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel Instructions for use. 2020. Available at: https://www.fda.gov/media/134922/download. Accessed 23 March 2020. [Google Scholar]
- 12. Bruner KM, Wang Z, Simonetti FR, et al. A quantitative approach for measuring the reservoir of latent human immunodeficiency virus-1 proviruses. Nature 2019; 566:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]