Abstract
Large observational data networks that leverage routine clinical practice data in electronic health records (EHRs) are critical resources for research on coronavirus disease 2019 (COVID-19). Data normalization is a key challenge for the secondary use of EHRs for COVID-19 research across institutions. In this study, we addressed the challenge of automating the normalization of COVID-19 diagnostic tests, which are critical data elements, but for which controlled terminology terms were published after clinical implementation. We developed a simple but effective rule-based tool called COVID-19 TestNorm to automatically normalize local COVID-19 testing names to standard LOINC (Logical Observation Identifiers Names and Codes) codes. COVID-19 TestNorm was developed and evaluated using 568 test names collected from 8 healthcare systems. Our results show that it could achieve an accuracy of 97.4% on an independent test set. COVID-19 TestNorm is available as an open-source package for developers and as an online Web application for end users (https://clamp.uth.edu/covid/loinc.php). We believe that it will be a useful tool to support secondary use of EHRs for research on COVID-19.
Keywords: COVID-19, natural language processing, testing name normalization, LOINC, COVID-19 TestNorm
INTRODUCTION
Coronavirus disease 2019 (COVID-19) was declared a pandemic by the World Health Organization on March 11, 2020; it has become a serious global health crisis since then. As stated by Barton et al.1 in Science “it is more important than ever for scientists around the world to openly share their knowledge, expertise, tools, and technology.” Researchers worldwide have worked diligently to understand the mechanisms of transmission and action for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and to discover effective treatments and interventions. One important data source for COVID-19 research is COVID-19 patients’ clinical data stored in electronic health records (EHRs). Several consortia have been formed to construct large clinical data networks for COVID-19 research, including the National COVID-19 Cohort Collaborative (N3C),2 the international EHR-derived COVID-19 Clinical Course Profiles (4CE),3 and many others.
To efficiently conduct clinical studies across different institutions within a network, one requirement is to normalize clinical data to common data models and standard terminologies. One such example is the Observational Medical Outcomes Partnership common data model maintained by the Observational Health Data Science and Informatics consortium.4 Among different types of clinical data, COVID-19 diagnostic tests are critical for all the following analyses, as they are the primary means to identify the confirmed COVID-19 cases. To address the urgency of the pandemic, individual institutions have created local names and local codes for those new COVID-19 tests in their EHRs. Meanwhile, Logical Observation Identifiers Names and Codes (LOINC), a widely used international standard for lab tests, has responded quickly by developing a new set of standard codes for COVID-19 tests5 to guide standard coding of these tests in clinical settings. Nevertheless, there is a lack of mappings between local COVID-19 testing names and standard LOINC codes, which hampers cross-institutional studies that rely on normalized clinical data at each institution. Existing natural language processing systems such as MetaMap6 or CLAMP7 provide concept mapping functions, but none of them has been updated to accommodate new concepts for COVID-19 tests.
To address this urgent need for reliable mappings, we developed an automated tool—COVID-19 TestNorm—to normalize a local COVID-19 testing name to a standard LOINC code. This tool is available to the community via an open-source package at GitHub and via an online Web application. We believe that COVID-19 TestNorm can be a useful tool for the secondary use of EHRs for research studies on the pandemic.
MATERIALS AND METHODS
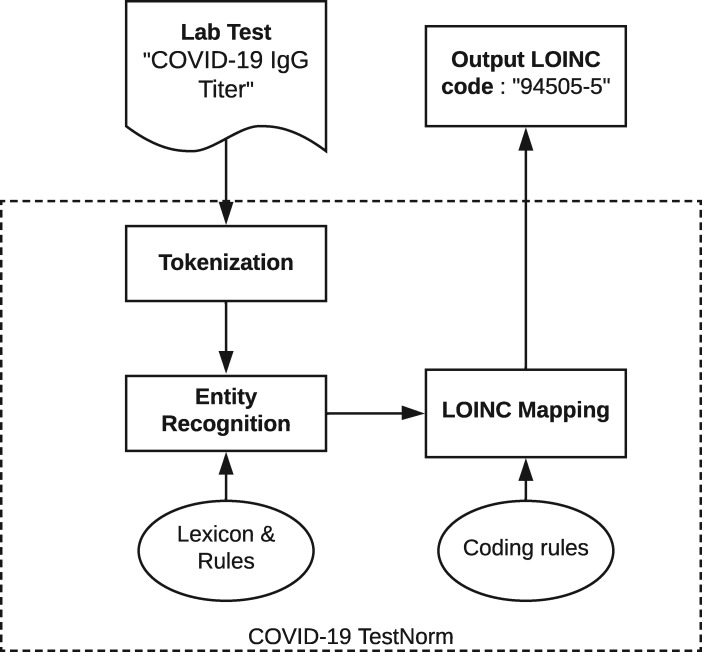
Using COVID-19 testing data collected from 8 healthcare systems, we developed a rule-based system to automatically normalize a local testing name to an LOINC code for COVID-19. Figure 1 shows an overview of the modules of the COVID-19 TestNorm system, mainly including entity recognition and LOINC mapping modules, with inputs from knowledge components such as lexicons and coding rules. The input lab testing names are tokenized first, then specific entities are recognized and appropriate LOINC codes are automatically mapped based on the coding rules.
Figure 1.
An overview of the COVID-19 TestNorm system. COVID-19: coronavirus disease 2019; LOINC: Logical Observation Identifiers Names and Codes.
Dataset
We collected COVID-19 testing data from 8 healthcare systems across the United States, including University of Texas Physicians; Memorial Hermann Health System; University of California, San Diego; Mayo Clinic; University of Florida; University of Minnesota; Columbia University Medical Center; and the national Department of Veterans Affairs (collected from 170 medical centers and 1063 outpatient sites) in April 2020. Data from each institution primarily contained testing names, as well as other fields available in local lab tables, such as specimen information. In total, 568 records were collected from the 8 sources. Although some institutions provided LOINC codes with the names, we manually reviewed all the records and assigned corresponding LOINC codes. Two annotators followed the LOINC COVID-19 coding guideline5 and manually mapped the 568 records to LOINC codes. The Cohen’s kappa agreement8 between the 2 annotators was 99.3%. We then randomly divided the dataset into a development dataset (454 records) and a test dataset (114 records). The COVID-19 TestNorm tool was developed using the development dataset and evaluated on the test dataset.
Entity recognition
LOINC describes each concept using 6 primary axes: Component, System, Method, Time, Property, and Scale,9 some of which were included in our COVID-19 entity categories. Our 5 root categories were Component, System, Method, Quantitative/Qualitative, which defines if a test returns a qualitative or quantitative result, and Institution, which specifies the manufacturer of the testing kit. The LOINC team at Regenstrief has worked with several in vitro diagnostics test kits manufacturers and commercial labs to develop and assign appropriate LOINC codes for their SARS-CoV-2 tests. Some of these mappings are listed on the LOINC website.5
Furthermore, from the manual review of the training set data and coding rules by LOINC,5 we identified that accurate mapping requires more specific values under each root category. For example, for System, which refers to the testing specimen, “Serum or plasma,” “Saliva,” “Nasopharyngeal specimen,” “ANY respiratory specimen,” and “Unspecified specimen” will lead to different LOINC codes, as the corresponding testing methods may vary. In this case, these subcategories of the root category System are essential elements for accurate mapping. This finding also applies to the other root categories. As a result, we divided the 5 root axes into subcategories. Table 1 lists all the detailed entity categories used in our LOINC coding system, as well as corresponding examples. Once entity categories were defined, we further analyzed the development dataset and manually extracted all related terms for each category, which were appended to the lexicon file used for the COVID-19 TestNorm tool. The lexicon file is publicly available together with the COVID-19 TestNorm software package. Potential users can manually revise the lexicon file to further improve COVID-19 TestNorm’s performance on their local data.
Table 1.
Semantic categories used by COVID-19 TestNorm
| LOINC Axes | Fine Entity Types | Example Values |
|---|---|---|
| Component | Covid19 | “COVID-19,” “SARS-COV-2” |
| Covid19_Related | “SARS-related CoV,” “SARS-like CoV” | |
| RNA_Comp | “RNA,” “N gene,” “RdRp gene” | |
| Sequence_Comp | “Whole genome” | |
| Antigen_Comp | “Ag,” “Antigen” | |
| Growth_Comp | “Organism” | |
| Antibody_Comp | “Ab,” “Antibody,” “IgM,” “IgG” | |
| Interpretation_Comp | “Interpretation,” “Recent infection” | |
| System | Blood | “Blood,” “Serum,” “Plasma” |
| Respiratory | “NARES,” “NASAL MUCUS” | |
| NP | “NP,” “Swab,” “NASOPHARYNX” | |
| Saliva | “SALIVA,” “ORAL FLUID” | |
| Other | “UNSPECIFIED,” “UNKNOWN SPECIMEN” | |
| Method | RNA_Method | “Non-probe-based,” “NAA,” “PCR” |
| Sequence_Method | “Sequencing” | |
| Antigen_Method | “Rapid IA,” “Immunoassay,” “IA” | |
| Growth_Method | “Organism specific culture” | |
| Antibody_Method | “Rapid IA,” “Immunoassay,” “IA” | |
| Panel_Method | “Panel,” “Panl” | |
| Quantitative_Qualitative | Quantitative | “Cycle Threshold,” “viral load” |
| Qualitative | “Presence,” “Ord” | |
| Institution | Manufacturer | “Abbott” |
COVID-19: coronavirus disease 2019.
The entity recognition consists of 2 steps: (1) an initial step that combines dictionary lookup and regular expression matching and (2) a disambiguation step that converts the ambiguous tags from the initial step into the final tags according to a set of predefined rules. During the initial step, most information can be captured and tagged to its corresponding category, whereas some ambiguous words need to be further reviewed. For example, the word “IA” can be either mapped to a “method,” which represents the abbreviation of “immunoassay,” or to a “system,” which represents the state “Iowa.” We developed context-based rules to determine the correct semantic categories for those terms.
LOINC mapping
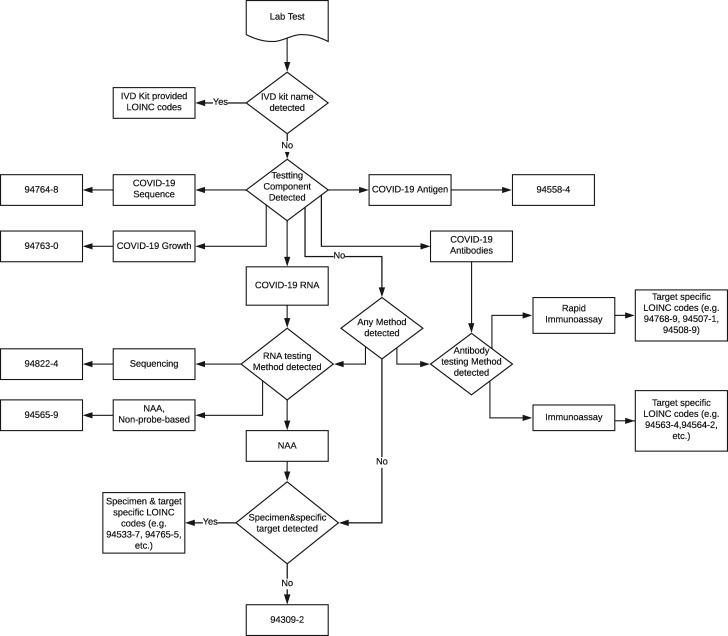
LOINC guidelines for COVID-19 tests (as of May 30, 2020)5 were followed to guide the development of the initial coding rules, which consist of decision-making algorithms based on extracted entities in the previous step. The coding rules were then iteratively updated using the development dataset collected across institutions. Figure 2 shows the overall decision workflow based on the coding rules. It starts with checking of manufacturer information, as specific LOINC codes are assigned to known testing kits by specific manufacturers. If no specific manufacturer information is available, the tool continues the mapping procedure using testing purpose rules. Five testing purpose rules are defined based on the tagged entities for Component, Method and System with the following information: (1) RNA, (2) sequence, (3) antigen, (4) growth, and (5) antibodies. For each testing purpose rule, specific tagged entities for the analyte (Component), specimen (System), Method, or Qualitative/Quantitative are further checked to map to appropriate LOINC codes.
Figure 2.
Coding rules for Logical Observation Identifiers Names and Codes (LOINC) mapping. COVID-19: coronavirus disease 2019; IVD: in vitro diagnostics; NAA: nucleic acid amplification.
Evaluation
We developed the COVID-19 TestNorm tool using the development set (454 records) and evaluated its performance using the independent test set (114 records). We compared the system’s output with the manually annotated gold standard and reported the accuracy of the system (the percentage of correct LOINC codes generated by the system among 114 records).
RESULTS
Table 2 shows the distribution of different COVID-19 tests’ LOINC codes on the full annotated dataset (568 records). LOINC codes 94759-8 (“SARS-CoV-2 (COVID19) RNA [Presence] in Nasopharynx by NAA with probe detection”), 94500-6 (“SARS-CoV-2 (COVID19) RNA [Presence] in Respiratory specimen by NAA with probe detection,” and 94309-2 (“SARS-CoV-2 (COVID19) RNA [Presence] in Unspecified specimen by NAA with probe detection”), were the most frequent codes across institutions, of which 94759-8 is the most frequent one, with over 40% of occurrences in the collected dataset. All 3 codes represent testing for SARS-CoV-2 RNA using nucleic acid (RNA) amplification with a probe-based detection method without specifying the gene or region being tested. The 94500-6 code is used for tests that can be run on a variety of respiratory specimens, 94759-8 is specific for nasopharyngeal specimens, and 94309-2 is for unspecified specimens. Nucleic acid amplification with probe-based detection is the most widely used testing method so far across the 8 sources.
Table 2.
Distribution of mapped LOINC codes
| LOINC Code | Total | Percentage | LOINC Long Common Name |
|---|---|---|---|
| Molecular | |||
| 94759-8 | 240 | 42.25 | SARS-CoV-2 (COVID19) RNA [Presence] in Nasopharynx by NAA with probe detection |
| 94500-6 | 202 | 35.56 | SARS-CoV-2 (COVID19) RNA [Presence] in Respiratory specimen by NAA with probe detection |
| 94309-2 | 75 | 13.20 | SARS-CoV-2 (COVID19) RNA [Presence] in Unspecified specimen by NAA with probe detection |
| 94502-2 | 13 | 2.29 | SARS-related coronavirus RNA [Presence] in Respiratory specimen by NAA with probe detection |
| 94660-8 | 11 | 1.94 | SARS-CoV-2 (COVID19) RNA [Presence] in Serum or Plasma by NAA with probe detection |
| Antibody | |||
| 94563-4 | 10 | 1.76 | SARS-CoV-2 (COVID19) IgG Ab [Presence] in Serum or Plasma by Immunoassay |
| 94564-2 | 4 | 0.70 | SARS-CoV-2 (COVID19) IgM Ab [Presence] in Serum or Plasma by Immunoassay |
| 94762-2 | 2 | 0.35 | SARS-CoV-2 (COVID19) Ab [Presence] in Serum or Plasma by Immunoassay |
| 94504-8 | 2 | 0.35 | SARS-CoV-2 (COVID19) Ab panel - Serum or Plasma by Immunoassay |
| 94505-5 | 2 | 0.35 | SARS-CoV-2 (COVID19) IgG Ab [Units/volume] in Serum or Plasma by Immunoassay |
| 94507-1 | 1 | 0.18 | SARS-CoV-2 (COVID19) IgG Ab [Presence] in Serum, Plasma or Blood by Rapid immunoassay |
| 94508-9 | 1 | 0.18 | SARS-CoV-2 (COVID19) IgM Ab [Presence] in Serum, Plasma or Blood by Rapid immunoassay |
| Other | |||
| 56831-1 | 4 | 0.70 | Problem associated signs and symptoms |
| 90101-7 | 1 | 0.18 | Internal control result |
LOINC: Logical Observation Identifiers Names and Codes.
In addition, we also counted the number of unique COVID-19 testing codes at each participating site. As shown in Figure 3, the number of unique tests at each site varied, with Columbia University Medical Center at the top, probably indicating that many testing methods have been used in this medical center in New York City.
Figure 3.
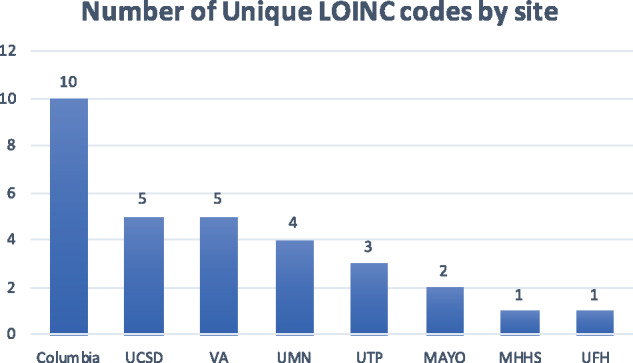
Number of unique Logical Observation Identifiers Names and Codes (LOINC) codes by site. MAYO: Mayo Clinic; MHHS: Memorial Hermann Health System; UCSD: University of California, San Diego; UFH: University of Florida Health; UMN: University of Minnesota; UTP: University of Texas Physicians; VA: Veterans Health Affairs.
The overall accuracy of COVID-19 TestNorm on the development set was 98.9%. When evaluated using the independent test set, the system achieved an accuracy of 97.4%, indicating that the rule-based approach was effective in normalizing COVID-19 testing names to LOINC codes.
The source code of the LOINC TestNorm tool is available at a GitHub repository.10 An online Web application (https://clamp.uth.edu/covid/loinc.php) is also provided so that users can enter local COVID-19 testing names and retrieve mapped LOINC codes automatically.
DISCUSSION
In this study, we collected the lab tests from 8 healthcare systems across the country. We developed a simple but effective normalization system for mapping COVID-19 lab tests to LOINC codes to facilitate rapid research response to the pandemic. The tool is publicly available with source code. For ease of use, we developed a Web application so that end users can easily map their local COVID-19 lab testing names to standardized LOINC codes using the online form, thus improving the efficiency of multicenter data aggregation and global knowledge sharing.
We conducted an error analysis for the mismapped codes. TestNorm achieved 100% accuracy on most of the LOINC codes in the test set, except for codes 94500-6 (2 records) and 56831-1 (1 record). For the 2 errors for 94500-6, one testing name was “UF BKR QUEST OVERALL RESULTS LAB17003” and the other was “CONFIRMATORY TESTING-QUEST.” Both were missed because they do not contain the key entity of COVID-19, which is required by our current coding rules. In the future, we may lift this constraint if we assume that all testing names are about COVID-19. For code 56831-1, the original local testing name “PATIENT SYMPTOM (SARS COV 2)” does not contain any specific testing information, and COVID-19 TestNorm assigned 94309-2 even though the original data came with a specific LOINC code 56831-1, probably owing to additional information available to the local hospital only.
LOINC codes are designed for use in clinical settings, assuming that all information is available. For secondary use scenarios, data submitted by local healthcare facilities do not always contain such detailed information. When the information is incomplete, more general LOINC codes will have to be assigned. For example, when the specimen is unknown, LOINC 94309-2 (“SARS-CoV-2 (COVID19) RNA [Presence] in Unspecified specimen by NAA with probe detection”) will be mapped, which accounts for 13.20% (n = 75 of 568) in our dataset.
One of the limitations of this study is that, even though we collected data from 8 large healthcare systems across the United States, the sample size and data heterogeneity could still be limited. For example, all codes in our dataset are about molecular and antibody tests. With new tests available in the market, the LOINC code sets for COVID-19 are evolving, that is, with weekly updates from Regenstrief, as well as continuous updates from the CDC which maintains a file containing recommended LOINC mappings for test kits currently approved by the Food and Drug Administration (https://www.cdc.gov/csels/dls/sars-cov-2-livd-codes.html). Therefore, it is critical for us to keep updating our tool with new code sets and updated coding rules. When large and diverse samples are accumulated, we will also look into more sophisticated machine learning approaches for this task.
Although we primarily designed COVID-19 TestNorm for secondary use of EHRs for research purposes, the tool could be useful at clinical operational settings or public health agencies as well. Unlike large academic medical centers included in this study, many community hospitals, federally qualified health centers, and nonacademic medical centers and clinics are much less familiar with the difficulties in harmonizing data across multiple systems. Given that the Department of Health and Human Services has just announced more standard reporting for lab testing of COVID-19,11 COVID-19 TestNorm could be a handy tool for improving COVID-19 lab reporting quality for both healthcare providers and public health agencies.
CONCLUSION
Multisite data aggregation and normalization are essential for rapid response to COVID-19 research using clinical data. We developed an automated tool to normalize local COVID-19 testing names to standard LOINC codes. This offers a foundational first step in enabling testing data interoperability for research related to COVID-19.
FUNDING
This project is partially supported by National Center for Advancing Translational Sciences grant nos. UL1TR003167 (UTHealth CTSA, HX, and RM) and 5U01TR002062 (HL, XJ, SP, HX, and KN); National Cancer Institute grant no. U24 CA194215 (HX and HL); National Institute of Health grant nos. R01AG066749 (XJ), R01GM114612 (XJ) and U01TR002062 (XJ); Veterans Affairs Health Services Research RES 13-457 (MM); Cancer Prevention and Research Institute of Texas grant nos. RP170668 (HX), RR180012 (XJ) and RP160015 (XD); the National Science Foundation RAPID grant no. 2027790 (XJ); and Gordon and Betty Moore Foundation grant no. 9639 (LO-M and HX). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Cancer Prevention and Research Institute of Texas, the Department of Veterans Affairs, or the U.S. government.
AUTHOR CONTRIBUTIONS
HX was responsible for the conception and design of the study. HX, XD, and JL designed the natural language processing tool. XD, HX, and JL drafted the manuscript. JL, ES, and HX developed the web API. HX, XD, SA, TC, and JD interpreted the data and results. HX, JB, SLD, EH, HL, KEL, MM, KN, LO-M, SP, RMR, AMS, XJ, and RM contributed to the collection, assembly, and quality control of the data. All authors revised the manuscript critically for important intellectual content and agreed to submit the report for publication.
ACKNOWLEDGMENTS
XD, SLD, MM, KN, RMR, and HX are members of the Observational Health Data Science and Informatics Consortium, Natural Language Processing Working Group.
CONFLICT OF INTEREST STATEMENT
HX and the University of Texas Health Science Center at Houston have research-related financial interests in Melax Technologies, Inc.
REFERENCES
- 1. Barton CM, Alberti M, Ames D, et al. Call for transparency of COVID-19 models. Science 2020; 368 (6490): 482–3. [DOI] [PubMed] [Google Scholar]
- 2. National COVID Cohort Collaborative (N3C). https://covid.cd2h.org/ Accessed June 1, 2020.
- 3. Brat GA, Weber GM, Gehlenborg N, et al. International electronic health record-derived COVID-19 clinical course profiles: the 4CE Consortium. medRxiv: 2020.04.13.20059691; 2020. [DOI] [PMC free article] [PubMed]
- 4. Observational Health Data Sciences and Informatics. https://ohdsi.org/ Accessed June 1, 2020.
- 5.Guidance for mapping to SARS-CoV-2 LOINC terms. https://loinc.org/sars-coronavirus-2/ Accessed June 1, 2020.
- 6. Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc JAMIA 2010; 17 (3): 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soysal E, Wang J, Jiang M, et al. CLAMP—a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc 2018; 25 (3): 331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McHugh ML. Interrater reliability: the kappa statistic. Biochem Med 2012; 22 (3): 276–82. [PMC free article] [PubMed] [Google Scholar]
- 9. Quick Start Guide for Mapping to Laboratory LOINC. https://loinc.org/guides/quick-start/ Accessed June 2, 2020.
- 10. UTHealth-CCB/covid19_testnorm. https://github.com/UTHealth-CCB/covid19_testnorm Accessed June 3, 2020.
- 11. U.S. Department of Health and Human Services. COVID-19 pandemic response, laboratory data reporting: CARES Act section 18115. https://www.hhs.gov/sites/default/files/covid-19-laboratory-data-reporting-guidance.pdf Accessed June 7, 2020.