Abstract
Functional crosstalk between histone modifications and chromatin remodeling has emerged as a key regulatory mode of transcriptional control during cell fate decisions, but the underlying mechanisms are not fully understood. Here we discover an HRP2–DPF3a–BAF epigenetic pathway that coordinates methylated histone H3 lysine 36 (H3K36me) and ATP-dependent chromatin remodeling to regulate chromatin dynamics and gene transcription during myogenic differentiation. Using siRNA screening targeting epigenetic modifiers, we identify hepatoma-derived growth factor-related protein 2 (HRP2) as a key regulator of myogenesis. Knockout of HRP2 in mice leads to impaired muscle regeneration. Mechanistically, through its HIV integrase binding domain (IBD), HRP2 associates with the BRG1/BRM-associated factor (BAF) chromatin remodeling complex by interacting directly with the BAF45c (DPF3a) subunit. Through its Pro-Trp-Trp-Pro (PWWP) domain, HRP2 preferentially binds to H3K36me2. Consistent with the biochemical studies, ChIP-seq analyses show that HRP2 colocalizes with DPF3a across the genome and that the recruitment of HRP2/DPF3a to chromatin is dependent on H3K36me2. Integrative transcriptomic and cistromic analyses, coupled with ATAC-seq, reveal that HRP2 and DPF3a activate myogenic genes by increasing chromatin accessibility through recruitment of BRG1, the ATPase subunit of the BAF complex. Taken together, these results illuminate a key role for the HRP2-DPF3a-BAF complex in the epigenetic coordination of gene transcription during myogenic differentiation.
INTRODUCTION
Higher organisms comprise a myriad of phenotypically and functionally distinctive cell types that contain almost the same genomic DNA. Epigenetic mechanisms, such as histone modifications and ATP-dependent chromatin remodeling, utilize identical genomic information to establish unique chromatin structures and gene expression patterns that cells acquire in fate specification during development or in response to environmental perturbations (1,2). Crosstalk between histone modifications and chromatin remodeling that precisely controls chromatin dynamics during transcription has emerged as a key regulatory mode in cell fate decisions (3,4), and its misregulation is implicated in neurodegenerative diseases, diabetes and many cancers (5–7). However, the underlying mechanisms governing the coordination of these two aspects of epigenetic regulation are not fully understood.
Histone modifications alter the direct contacts between histones and DNA and thus generate docking sites for effector proteins, or readers (8). Binding of readers to histones recruits components of the transcriptional machinery and chromatin remodeling complexes to chromatin, regulates gene expression and determines the functional outcome of the corresponding histone modifications (9,10). The Pro-Trp-Trp-Pro (PWWP) domain belongs to the Tudor domain ‘Royal family’ and functions as a histone methylation reader (11), and is noteworthy for its affinity for methylated histone 3 lysine 36 (H3K36me) (12). PWWP proteins are involved in numerous nuclear processes such as gene transcription (13–16), DNA methylation (17–19), mRNA splicing (20), chromatin remodeling (21) and DNA damage repair (22–24). Hepatoma-derived growth factor-related protein 2 (HRP2) belongs to the HRP family and is a PWWP domain-containing histone reader protein (12,25). The function of HRP2 has previously been reported to be linked to chromatin dynamics. HRP2 facilitates HIV-1 integration by interacting with and tethering the HIV integrase to active transcription sites (26,27). Through recruitment of the homologous recombination repair machinery, HRP2 has also been shown to promote DNA repair (23).
Myogenesis, the formation of muscular tissue, occurs during embryonic development, and is recapitulated in adult skeletal muscle in response to injury and disease. During myogenesis, activated muscle progenitors withdraw from the cell cycle, express lineage-specific genes and terminally differentiate to multinucleated myotubes (28–30). Epigenetic regulation such as histone modification and chromatin remodeling ensures the correct integration of developmental signals at gene regulatory regions and creates a permissive chromatin environment for RNA Pol II binding and transcription in myogenesis (31–34). These features make myogenesis a classical model for studying the role of epigenetic regulation in cell fate decisions.
In this study, we find that HRP2 is indispensable for myogenic differentiation and that its deficiency impairs post-injury muscle regeneration in mice. HRP2 preferentially binds to H3K36me2 and recruits the BRG1/BRM-associated factor (BAF) chromatin remodeling complex, by interacting directly with its DPF3a subunit, to myogenic gene promoters. Thus, the HRP2-DPF3a-BAF complex coordinates histone modification and chromatin remodeling and activates muscle-specific gene expression during myogenic differentiation.
MATERIALS AND METHODS
Cell culture
C2C12 cells were maintained in growth medium (DMEM) with 20% fetal bovine serum (FBS). After reaching to 90% confluence, growth medium were replaced with differentiation medium containing 2% horse serum to induce differentiation. HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). All cell lines were maintained at 37°C incubator containing 5% CO2.
Generation of C2C12-E3MCK-luc M21 reporter cell line
To generate the C2C12-E3MCK-luc M21 reporter cell line, MCK promoter and enhancer were inserted into pGL4.16 [luc2CP/Hygro] vector and transfected into C2C12 cells. Stable integrated clone that could monitor myogenesis though luciferase reporter activity was picked up after hygromycin selection, referred to as C2C12-E3MCK-luc M21.
siRNA screening
Epigenetics siRNA library targeting 220 epigenetic modifiers (four siRNAs targeting the same gene) was purchased from Dharmacon. C2C12-E3MCK-luc M21 was seeded in a 24-well plate overnight and then transfected with siRNAs using Lipofectamine RNAiMAX (Invitrogen) for 48 h before differentiation. The 2nd transfection was performed after 24 h of differentiation in differentiation medium. The luciferase activity was measured after 96 h of differentiation. The luciferase raw data was calculated and ranked by relative log2 fold change compared to control group. Heatmaps were generated in R using the pheatmap package.
siRNA knockdown assay
All siRNAs were transfected twice using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's recommendations. The final concentration of the siRNA is 20 nM. C2C12 myoblast cells were seeded on day 1. Twenty-four hours later, the first transfection was performed (day 2) when the cell confluence reached to 40%. Forty-eight hours later, the myoblast cells were induced to differentiation with 2% horse serum (day 4). The second transfection was performed in the differentiated myotube cells 24 h after the induction of differentiation (day 5). The siRNA sequences used were as follows: siDPF3a-#1 (5′-CACGUACCACGGAGGACUU-3′), siDPF3a-#2 (5′-CUGACUCUGGUCAUUGUUC-3′), siDPF3a-#3 (5′-GUCAUUGUUCUAGUUCUGA-3′), siHRP2-#1 (5′-GGUACAAGGCCAACAAGGA-3′), siHRP2-#2 (5′-GUGAAGAAGCGUAGCCGAA-3′), siBRG1-#1 (5′-CUGAUUCCUCCGCAACCAA-3′), siBRG1-#2 (5′-CUGAUGAUGAGACCGUCAA-3′), siMYOD-#1 (5′-GGUCUCAGGUGUAACAGGU-3′), siMYOD-#2 (5′-CGAACCAGCGGCUACCCAA-3′) siMYOD-#3 (5′-GCUCUGAUGGCAUGAUGGA-3′). siNSD2-#1 (5′-GAUGUAUGUGGCAAACCUU-3′). and siNSD2-#2 (5′-GAAUUGCUUGGGUCCCAGU-3′).
Immunoblotting
Tissue samples and cells were lysed with RIPA buffer supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche). After adding SDS loading buffer, samples were boiled for 10 min. Protein samples were resolved by SDS-PAGE gels and then transferred to nitrocellulose membranes. The membranes were blocked and incubated with primary antibodies. Antibodies used were TY1 (Sigma, SAB4800032), HRP2 (Proteintech, 15134-1-AP), MHC (Developmental Studies Hybridoma Bank, MF20), Myog (Santa Cruz, SC-12732), GAPDH (Millipore, MAB347), FLAG-HRP (Sigma, F7425), DPF3a (Custom made), P-DPF3a (Custom made), Pan-DPF3a (Custom made), GST (Santa Cruz, sc-459) and MYOD (Santa Cruz, SC-304).
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 10 min and permeabilized with 1% Triton X-100 for 5 min at room temperature. Then cells were blocked in 5% BSA for 1 h at room temperature and incubated with primary antibody overnight at 4°C. Cells were washed with PBS, stained with goat anti-mouse Alexa Fluor 488 secondary antibody (Thermo Fisher Scientific) for 1 h at at room temperature and counter-stained with DAPI to detect nuclei. Images were captured using a fluorescence microscope (DMi8, LEICA). MHC (Developmental Studies Hybridoma Bank, MF20) antibody was used.
Mass spectrometry
To identification of HRP2-interacting proteins, HEK293T cells were transfected with plasmid expressing FLAG-tagged HRP2. Cells were resuspended in NP-40 lysis buffer (150mM sodium chloride, 1% NP-40, 50 mM Tris PH 8.0) with protease inhibitor cocktail (Roche) and centrifuged at 12 000 g for 20 min at 4°C. The supernatants were incubated with anti-FLAG M2 affinity gel at 4°C overnight. After washing four times with NP-40 lysis buffer, FLAG protein complex was eluted with FLAG peptide (Sigma). The elutes were resolved on NuPAGE 4–12% Bis–Tris gel (Invitrogen) and stained with a silver staining kit (Pierce). The protein bands were cut out and analysed by liquid chromatography–tandem mass spectrometry (LC–MS/MS).
Co-immunoprecipitation (Co-IP)
Cells were resuspended in ice-cold NP-40 lysis buffer (150mM sodium chloride, 1% NP-40, 50 mM Tris PH 8.0) with protease inhibitor cocktail (Roche) and centrifuged at 12 000 g for 20 min at 4°C. For FLAG-tagged proteins, the supernatants were incubated with anti-FLAG M2 affinity gel at 4°C overnight. For other immunoprecipitations, the supernatants were incubated with the indicated antibodies overnight at 4°C. Dynabeads Protein G were added for 4 h at 4°C. After washing four times with NP-40 lysis buffer, SDS loading buffer was added and boiled for 10 min. The supernatants were subjected to immunoblotting. Antibodies used were TY1 (Sigma, SAB4800032), FLAG-HRP (Sigma, F7425), BRG1 (Proteintech, 21634-1-AP), BAF155 (Proteintech, 17722-1-AP), BAF60A (Proteintech, 10998-2-AP), DPF3 (GeneTex, GTX122249), Myc (Cell Signaling Technology, 2278) and DPF3a (Custom made).
Protein purification and pulldown
S-tag-DPF3a, GST-HRP2 and truncated GST-HRP2 fusion proteins were expressed and purified from Escherichia coli BL21 (DE3). After induction with IPTG overnight at 16°C, cells were resuspended in GST lysis buffer (100 mM NaCl, 50 mM Tris–HCl pH 8.0) supplemented with 1% Triton X-100, 1 mM dithiothreitol (DTT), protease inhibitor cocktail (Roche) and 0.2 mM PMSF, sonicated and centrifuged. The supernatants were mixed with S-protein agarose or glutathione agarose. The beads were washed with lysis buffer. S-tag-DPF3a were eluted with elution buffer. For pulldown assay, GST-HRP2 and truncated GST-HRP2 fusion proteins immobilized on glutathione agarose were incubated with HEK293T cell lysis transfected with myc-tagged DPF3a. GST-HRP2 immobilized on glutathione agarose were incubated with S-tag-DPF3a. After washing four times, the beads were boiled for 10 min. The supernatants were analyzed by Coomassie blue staining and immunoblotting. Antibodies used were Myc (Cell Signaling Technology, 2278) and S-protein HRP Conjugate (Millipore, 69047).
CRISPR–Cas9 mediated gene knockdown of DPF3a
LentiCas9-Blast (#52962) construct was purchased from Addgene (Watertown, MA, USA). To generate stable cells expressing cas9, HEK293T cells were co-transfected with lentiCas9-Blast and the packaging plasmids (PMD2G and PSPAX2). After transfection for 6 h, the medium containing the transfection reagent was removed and replaced with fresh complete DMEM. The lentiviral particles were harvested from HEK293T cells after another 48 h. Cells were then infected with appropriate amounts of lentiviral particles for 12 h, and then virus-containing medium was removed and replaced with fresh medium for 48 h. Stably transduced cells were selected by blasticidin. DPF3a-KD cell line was generated by cloning gRNA targeting exon of DPF3a into the gRNA cloning vector lentiGuide-Puro (Addgene, #52963). sgRNAs were designed using sgRNA Designer (Broad Institute). Stable cells expressing cas9 was infected and selected with puromycin. The expression of DPF3a were detected via immunoblotting. The sgRNA sequences used were as follows: sgRNA 1F (5′-GGAACCGAATAAGTCCTCCG-3′), sgRNA 1R (5′-CGGAGGACTTATTCGGTTCC-3′), sgRNA 2F (5′-GGCCGCAGCACGTACCACGG-3′), sgRNA 2R (5′-CCGTGGTACGTGCTGCGGCC-3′), sgRNA 3F (5′-TGACGTGTCACTTTCTGACG-3′) and sgRNA 3R (5′-CGTCAGAAAGTGACACGTCA-3′).
Rescue assay
C2C12 stable line expressing siRNA-resistant FLAG-tagged WT and mutated HRP2 (R527A/R528A) was seeded in a six-well plate overnight and then transfected with siRNAs against Hrp2 or control using Lipofectamine RNAiMAX (Invitrogen) for 36 h before differentiation. The second transfection was performed after 24 h of differentiation. The expression of myogenic differentiation markers were detected by immunoblotting after 96 h of differentiation.
Protein–protein ChIP assays
Cells were transfected with FLAG-tagged HRP2 or mutated HRP2 (W21A). Then cells were treated with 1% formaldehyde for 15 min. The reaction was quenched with 125 mM glycine. Then cells were washed twice with PBS, resuspended in 400 μl ChIP lysis buffer (50 mM Tris–HCl pH 7.5, 140 mM NaCl, 1% Triton X-100, 1 mM EDTA and 0.1% sodium deoxycholate) supplemented with protease inhibitor cocktail (Roche) and sonicated to DNA fragment sizes of 500 bp to 2 kb. Samples were centrifuged at 20 000 g at 4°C for 20 min. The supernatants were incubated with anti-FLAG M2 affinity gel at 4°C overnight. After washing four times with ChIP washing buffer, SDS loading buffer was added and boiled for at least 30 min to reverse the crosslinks. The supernatant were subjected to immunoblotting. Antibodies used were FLAG-HRP (Sigma, F7425), Histone H3 (Abcam, ab1791), H3K36me2 (Abcam, ab9049), H3K36me3 (Abcam, ab9050), H3K4me3 (Millipore, 05-745), H3K9me3 (Millipore, 07-442) and H3K27me3 (Abcam, ab6002).
Native ChIP assays
293T cells were transfected with FLAG-tagged HRP2 or mutated HRP2 (W21A). 24 hours later, nuclei were prepared and resuspended in 10 mM Tris–HCl pH 7.5, containing 10 mM NaCl, 3 mM MgCl2, 3 mM CaCl2, 0.1 mM PMSF and chromatin was released by digestion with micrococcal nuclease (Sigma) at 37°C for 10 min. Digestion was optimized to produce primarily mono-nucleosomes. Digestion was stopped by addition of 20 mM EDTA. Nuclear debris was removed by centrifugation and the soluble chromatin was incubated with anti-FLAG M2 affinity gel at 4°C overnight. After washing four times, SDS loading buffer was added and boiled for 10 min. The supernatant were subjected to immunoblotting. Antibodies used were FLAG-HRP (Sigma, F7425), Histone H3 (Abcam, ab1791), H3K36me2 (Abcam, ab9049), H3K36me3 (Abcam, ab9050), H3K4me3 (Millipore, 05-745), H3K9me3 (Millipore, 07-442) and H3K27me3 (Abcam, ab6002).
RNA extraction and qRT-PCR
Total RNAs were extracted from TA tissues or cells using TRIzol (Invitrogen). 2 μg of total RNA was reverse-transcribed into cDNA by the cDNA Synthesis Kit (Roche). Primers used for amplification were listed in Supplementary Table S1.
Peptide pulldown assays
Biotinylated peptides were purchased from ChinaPeptides. Briefly, 1 μg, 5 μg or 10 μg of peptides were incubated with GST-HRP2 (IBD) fusion proteins overnight at 4°C. Streptavidin agarose beads (Millipore) were added for 6 h at 4°C. After washing four times with GST lysis buffer, SDS loading buffer was added and boiled for 10 min. The supernatant were subjected to immunoblotting. The sequences of biotinylated peptides used were as follows: Biotin-IBM (Biotin-LFGSTSESDTSTFHGFDEDD) and Biotin-p-IBM [Biotin-LFGSTSESDT(p-S)TFHGFDEDD].
RNA-seq
Total RNAs from cells were isolated using TRIzol (Invitrogen) following the procedures described previously (35). Libraries were constructed and sequenced on the BGISEQ-500 platform (BGI Group, Shenzhen, P.R. China).
RNA-seq data analysis
The raw sequencing reads were checked using FastQC (version 0.11.8). Cutadapt (v2.0) was used for adapter trimming and low-quality filtering to get the clean data. The genome sequence and gene annotation of Mus musculus (mm10, GRCm38) from GENCODE (Release M17) were used. The gene expression was quantified by Salmon (version 0.8.0). Based on the quantification, differential expressed genes (DEGs) between sample groups were generated by DESeq2 (R version 3.3.2). DEGs were filtered using the cutoff with fold change ≥1.5 and adjust P-value ≤0.05.
ChIP-seq and ChIP-qPCR
ChIP experiments were performed following the procedures described previously (36). For TY1, Hrp2 and Dpf3a ChIP, cells were double crosslinked by incubation with DMA (Sangon Biotech, Shanghai, China) for 1 h followed by treatment with 1% formaldehyde for 10 min at room temperature. For H3K36me2 and Brg1 ChIP, cells were crosslinked with 1% formaldehyde. The crosslink was quenched by adding 125 mM glycine at RT for 5 min. Cells were washed three times with PBS and lysed using ChIP lysis buffer supplemented with protease inhibitor cocktail (Roche). The chromatin were then sonicated to DNA fragment sizes of 300–500 bp with Bioruptor Sonicator. Immunoprecipitation was performed with TY1 (Sigma, SAB4800032), Hrp2 (Proteintech, 15134-1-AP), Dpf3a (Custom made), H3K36me2 (Abcam, ab9049) and Brg1 (Santa Cruz, sc-17796 X). Input was used as control for ChIP-seq. After elution and reversal cross-linking, samples were treated with RNase A at 37°C for 30 min. For ChIP-seq, libraries were constructed and sequenced on Illumina HiSeq X ten. For ChIP-qPCR, 1 μl purified DNA was used per qPCR reaction. Primers for ChIP-qPCR were listed in Supplementary Table S2.
ChIP-seq data analysis
FastQC (version 0.11.8) and cutadapt (v2.0) were applied to raw sequencing reads from ChIP and input libraries for quality control and pre-processing to get the clean reads, respectively. The reads were mapped to M. musculus genome (mm10) by Bowtie2 (version 2.2.6). The duplicated reads were removed using SAMtools (version 1.9). Then the peak finding algorithm from MACS2 (version 2.1.1) was used to detect regions with significant enrichment of ChIP signals. The ENCODE Processing Pipeline for transcription factor and histone modification ChIP-seq was refered to set the parameters of reads mapping and peak calling. To associate the peaks with nearby genes and genomic regions, annotatePeaks.pl from HOMER (v4.10) was used. The deepTools suite (version 3.2.0) was used to produce the binding profiles and heatmaps.
ATAC-seq
100 000 cells were lysed in lysis buffer (10 mM Tirs–HCl, PH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% (v/v) IGPAL CA-630) for 10 min on ice before centrifuged at 500g for 5 min. The nuclei were added with 50 μl transposition reaction buffer (5 μl TruePrep Tagment Enzyme, 10 μl TruePrep Tagment Buffer L and 35 μl ddH2O from Vazyme TD501-01) and followed with incubation at 37°C for 30 min. After tagmentation, VAHTS DNA Clean Beads were used to stop the reaction and DNA was purified for final library construction (TruePrepTM DNA Library Prep Kit V2 for Illumina) before paired-end high-throughput sequencing using HiSeq XTen.
ATAC-seq data analysis
Similar to the ChIP-seq data analysis, clean reads from ATAC-seq were processed according to ENCODE Processing Pipeline for ATAC-seq Data. Generally, ATAC-seq reads were mapped to M. musculus genome (mm10) by Bowtie2 (version 2.2.6), The deepTools suite (version 3.2.0) was used to produce the binding profiles and heatmaps.
Generation of Hrp2 knockout mice
Hrp2 knockout mice were generated by Shanghai Model Organisms Center, Inc. through CRISPR/Cas9-mediated non-homologous end joining (NHEJ).
CTX injury and histological analysis
Nine-week-old male WT and Hrp2-KO mice were anaesthetized and injected intramuscularly into tibialis anterior (TA) muscles using 50 μl of 20 μM CTX in saline to induce muscle regeneration. Mice were sacrificed at 3, 5, 7 and 14 days post-injection by cervical dislocation. TA muscles were harvested, immediately frozen in liquid nitrogen and stored at −80°C. TA muscles were embedded in 100% optimal cutting temperature (O.C.T.). 8-μm sections from TA muscle were stained with hematoxylin and eosin. The CSA of myofibers (n > 1000) in TA muscles and number of nuclei within regenerating fibers (n > 300) were determined using the Image J program from four animals (for CTX day 7). The distributions of fiber sizes were analysed (n > 1000) from four animals (for CTX day 7).
Study approval
Mice were maintained under a 12 h light/12 h dark cycle and fed with fresh water and rodent diet. Animal studies were approved by the Ethical Committee of Tianjin Medical University (permit number: SYXK 2009-0001).
Statistical analysis
Significance was calculated using the two-tailed unpaired t-test to compare two groups of independent samples using GraphPad Prism Software. P < 0.05 was considered statistically significant. Results were presented as means ± SD for at least three independent experiments.
RESULTS
siRNA screening identifies HRP2 as a key epigenetic regulator of myogenesis
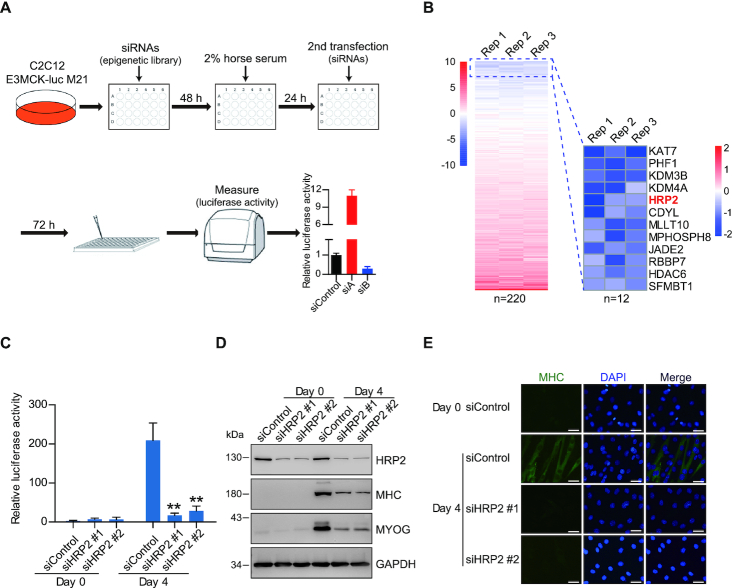
To systematically identify epigenetic factors that regulate myogenesis, we first developed a high-throughput screening assay in the C2C12 myoblast cell line (Figure 1A). We generated an engineered C2C12 cell line carrying a muscle creatine kinase (MCK) enhancer-driven luciferase reporter gene (37), hereafter referred to as C2C12-E3MCK-luc M21 (Supplementary Figure S1A). MCK expression is induced upon C2C12 differentiation (Supplementary Figures S1B and D); therefore, myogenesis can be monitored by the expression of the MCK-luc reporter gene. To validate this system, we first tested the role of MYOD, a key transcription factor for myogenesis (38,39). As expected, MYOD knockdown resulted in decreased expression of myogenic markers, such as myogenin and MHC (Supplementary Figures S1C and E). Notably, MYOD knockdown also caused a reduction in reporter activity (Supplementary Figure S1C), supporting the effectiveness of this reporter system for evaluating myogenesis. Next, we conducted siRNA screening in the engineered C2C12-E3MCK-luc M21 cells using a customized siRNA library that targets 220 epigenetic modifiers, including histone modification writers, erasers and readers. Consistent with previous reports (40), we found that depletion of the Kdm4a gene strongly reduced the MCK-luc reporter activity, thereby further validating our screening approach (Figure 1B).
Figure 1.
Identification of HRP2 as a key epigenetic regulator in myogenesis. (A) Schematic of screening procedure using an siRNA library targeting 220 epigenetic regulators in C2C12-E3MCK-luc M21 cells. (B) Heatmap showing changes in luciferase activity of C2C12-E3MCK-luc M21 myotubes transfected with an epigenetic siRNA library (left). Heatmap of the 12 epigenetic candidates yielding lowest luciferase activity in C2C12-E3MCK-luc M21 myotubes upon their knockdown (right). This experiment was performed in 3 replicates. (C) Luciferase activity quantification of C2C12-E3MCK-luc M21 myoblasts (Day 0) and myotubes (Day 4) transfected with scramble (siControl) and two different Hrp2 siRNAs. Data represent as means ± SD of three replicates. ** P < 0.01. (D) Western blotting analysis of MYOG, MHC and GAPDH in C2C12 myoblasts and myotubes transfected with scramble and two different HRP2 siRNAs. (E) Immunofluorescence analysis of MHC and DAPI in C2C12 myoblasts and myotubes transfected with scramble and two different HRP2 siRNAs. Scale bars, 25 μm. DAPI, 4′,6-diamidino-2-phenylindole.
HRP2, an H3K36me reader protein, was among the top hits in the screen. Several H3K36 methyltransferases, including SET domain containing 2 (Setd2) (41), Setd3 (42), nuclear receptor binding SET domain protein 2 (NSD2) (43) and absent, small or homeotic 1-like (Ash1L) (44), have previously been shown as essential regulators of myogenesis, although how the alteration of H3K36me impairs muscle differentiation remains unclear. Thus, we chose HRP2 for further investigation. Using another two siRNAs, we found that knockdown of HRP2 greatly reduced the activity of MCK-luc reporter and muscle differentiation marker genes (Figure 1C–E), confirming an essential role for HRP2 in myogenesis.
HRP2 interacts with the BAF chromatin remodeling complex
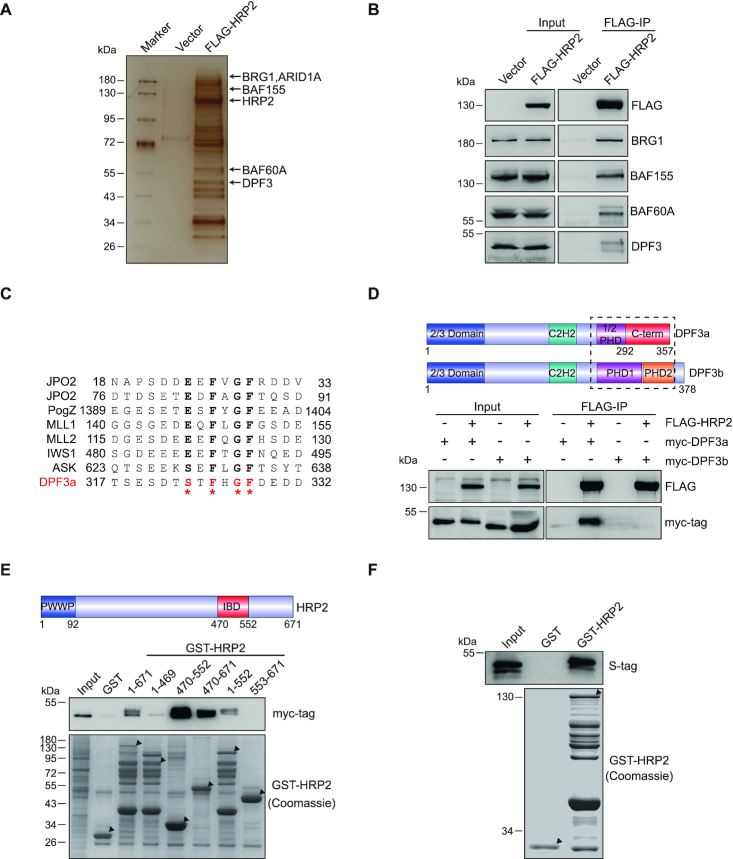
HRP2 does not possess any enzymatic domain; therefore, it may exert its function by interacting with other transcriptional machineries. To test this hypothesis, we carried out a proteomics analysis to identify HRP2 binding proteins. We performed HRP2 immunoprecipitation (IP) using FLAG antibody with extracts from FLAG-HRP2-expressing cells. Mass spectrometry analysis identified a set of BAF chromatin remodeling complex subunits as enriched proteins in HRP2-IP (Figure 2A), including BRG1, BAF155, BAF60A, ARID1A and DPF3 (45,46). The associations between HRP2 and BAF subunits were further confirmed by FLAG-HRP2 IP followed by immunoblotting analysis using antibodies recognizing individual subunits (Figure 2B). Together, these results demonstrate that HRP2 associates with the BAF complex.
Figure 2.
HRP2 directly interacts with DPF3a. (A) 293T cells were transfected with FLAG-HRP2 and whole cell lysates were subjected to co-IP. Isolated HRP2 binding proteins were visualized by silver staining. (B) Co-IP analysis of the interactions between FLAG-HRP2 and BAF complex subunits in 293T cells. (C) Alignment of DPF3a with other IBD binding protein sequences shows that DPF3a possesses a typical IBD binding motif (IBM). (D) Schematic representation of the DPF3 isoforms. DPF3a contains a half PHD finger and a specific C-terminus, whereas DPF3b contains a double PHD finger. Blue: 2/3 domain; teal: C2H2-Krüppel-like zinc finger; purple: first plant homeodomain; orange: second plant homeodomain; red: DPF3a-specific C-terminus (upper). Co-IP analysis of the interaction between FLAG-HRP2 and myc-DPF3a or myc-DPF3b in 293T cells (lower). (E) HRP2 IBD domain is essential for DPF3a binding. Schematic representation of HRP2. Amino acid positions are depicted (upper). GST pull-down assays examining the interactions between GST-HRP2 (WT and its truncated mutants as indicated) and myc-DPF3a shows that the IBD domain of HRP2 is indispensable for DPF3a binding (lower). (F) S-tag-tagged DPF3a was purified in bacteria and subjected to GST pull-down assay together with GST-HRP2. The result shows that HRP2 interacts with DPF3a directly in vitro.
HRP2 associates with the BAF complex through direct interaction with DPF3a
To gain more molecular insights into the association between HRP2 and the BAF complex, we next sought to identify the specific BAF complex subunit(s) that mediate the interaction. HRP2 protein contains two known functional domains: a PWWP domain and an HIV integrase binding domain (IBD). The IBD domain is mainly responsible for the interactions of HRP2 with its binding partners, including interacts-with-Spt6 (IWS1), JPO2 (also known as CDCA7L, RAM2 and R1), CDC7-activator of S-phase kinase complex (ASK) and pogo transposable element with zinc finger domain (PogZ) (47). Alignment of the amino acid sequences of these known HRP2-interacting proteins revealed a consensus motif (S/EXFXGF) termed the IBD binding motif (IBM) (47,48). Interestingly, we found that DPF3a, but not other BAF subunits, possesses an IBM in its C-terminus region (Figure 2C), suggesting that DPF3a is the subunit which mediates the association between HRP2 and the BAF complex.
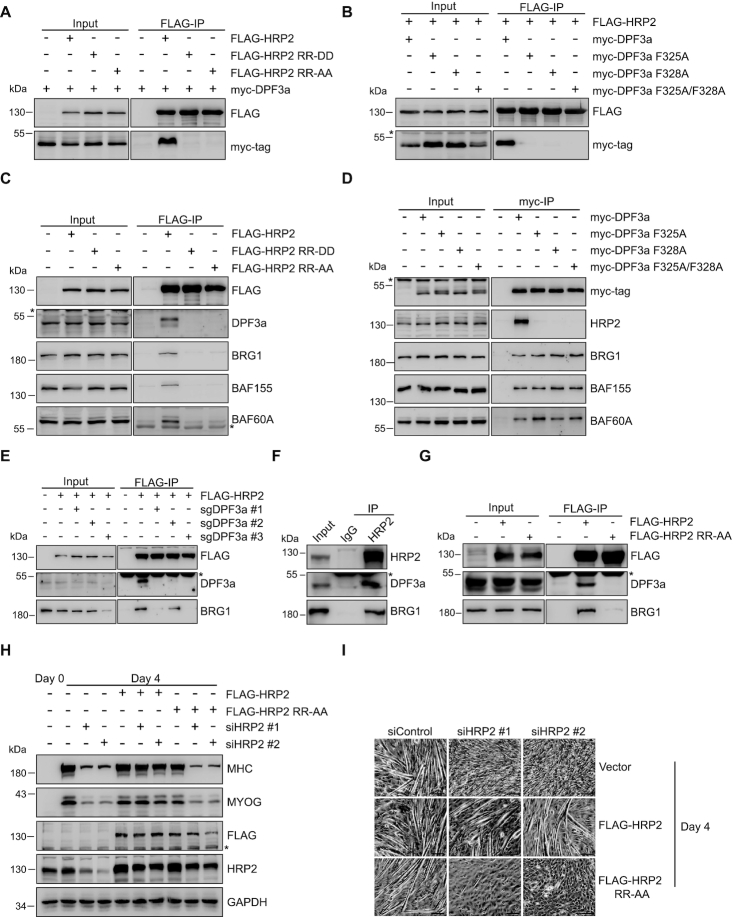
DPF3, also called BAF45C, consists of two distinct isoforms (49). DPF3b contains tandem plant homeodomain (PHD) fingers that can bind to acetylated and methylated histone lysine residues (50,51). In contrast, DPF3a possesses a half PHD finger and a distinct C-terminus of unknown function. As shown in Figure 2D, HRP2 interacted with isoform DPF3a, but not DPF3b. To examine whether HRP2 directly binds to DPF3a, we performed a GST pull-down assay with purified full-length and truncated HRP2. As shown in Figure 2E and F, HRP2 directly interacts with DPF3a and deletion of the IBD abolished this interaction. We further identified the critical residues that mediate this interaction. As illustrated in Figure 3A and B, mutations in either the IBD (R527D/R528D or R527A/R528A) of HRP2 or the IBM (F325A or F328A or F325A/F328A) of DPF3a effectively abolished the interaction between these two proteins. Collectively, these results demonstrate that HRP2 directly interacts with DPF3a through the IBD-IBM binding interface.
Figure 3.
HRP2 associates with the BAF complex by interacting with DPF3a. (A) Co-IP analysis of the interactions between FLAG-HRP2 (WT or mutants) and myc-DPF3a in 293T cells. (B) Co-IP analysis of the interactions between FLAG-HRP2 and myc-DPF3a (WT or mutants) in 293T cells. (C) Co-IP analysis of the interactions between FLAG-HRP2 (WT or mutants) and BAF complex subunits in 293T cells. ‘*’ indicates nonspecific bands. (D) Co-IP analysis of the interactions between myc-DPF3a (WT or mutants) and BAF complex subunits in 293T cells. (E) 293T cells were transfected with three sgRNAs targeting DPF3a, and stable cells were obtained by puromycin selection. Co-IP assay examining the interactions of FLAG-HRP2 and BRG1. ‘*’ indicates nonspecific bands. (F) Co-IP analysis of the interactions between HRP2 and BAF complex subunits in C2C12 myotubes. ‘*’ indicates nonspecific bands. (G) Co-IP analysis of the interactions between FLAG-HRP2 (WT or mutants) and BAF complex subunits in C2C12 myotubes. ‘*’ indicates nonspecific bands. (H) C2C12 cells were transfected with empty vector, FLAG-HRP2 and FLAG-HRP2 mutant, and stable cells were obtained by puromycin selection. These stable cells were transfected with scramble (siControl) and two different HRP2 siRNAs as indicated and then subjected to differentiation. Western blot analysis of MHC, MYOG, FLAG, HRP2 and GAPDH in myoblasts and myotubes shows that overexpression of WT, but not the mutant, HRP2 rescued the myogenic defects of HRP2-depleted cells. (I) C2C12 cells were transfected with empty vector, FLAG-HRP2 and FLAG-HRP2 mutant, and stable cells were obtained by puromycin selection. These stable cells were transfected with scramble (siControl) and two different HRP2 siRNAs as indicated and then subjected to differentiation. Phase contrast microscopy images show that WT, but not the mutant, HRP2 rescued the myogenic defects of HRP2-depleted cells. Scale bars, 100 μm.
To investigate whether DPF3a could bridge HRP2 to the BAF complex, we performed a co-IP assay with wild type (WT) and IBD-mutant HRP2. As shown in Figure 3C, WT HRP2 co-immunoprecipitated with DPF3a and BAF core components, whereas the IBD mutants that did not interact with DPF3a failed to immunoprecipitate with other BAF subunits. These results suggest that the HRP2-BAF interaction depends on DPF3a. In contrast, DPF3a IBM mutants, which lost the interaction with HRP2, retained the ability to interact with other BAF components (Figure 3D), suggesting that HRP2 is not required for the association between DPF3a and other BAF complex subunits. To confirm this, DPF3a was depleted by CRISPR-mediated gene editing. As shown in Figure 3E, in the absence of DPF3a, HRP2 failed to interact with the core BAF component BRG1. To investigate the association of HRP2 with the BAF complex in muscle cells, we performed immunoprecipitation analysis using anti-HRP2 antibody to enrich endogenous HRP2. As shown in Figure 3F, endogenous HRP2 effectively co-immunoprecipitates with DPF3a and BRG1 in differentiated C2C12 cells. Furthermore, the R527A/R528A mutant of HRP2 failed to interact with the BAF complex, in contrast to WT HRP2 (Figure 3G). These data indicate that HRP2 interacts with the BAF complex in muscle cells. Altogether, these results demonstrate that DPF3a works as a hub linking HRP2 and the BAF complex.
To explore the functional importance of the interaction between HRP2 and DPF3a in myogenesis, we transfected siRNA-resistant WT and mutated HRP2 (R527A/R528A) expression constructs into endogenous HRP2-depleted myotube cells. As shown in Figure 3H and I, overexpression of WT, but not the mutant, HRP2 rescued the myogenic defects of HRP2-depleted cells. These data indicate that the regulation of myogenesis by HRP2 depends on its association with DPF3a.
DPF3a is up-regulated during myogenesis and is essential for muscle differentiation
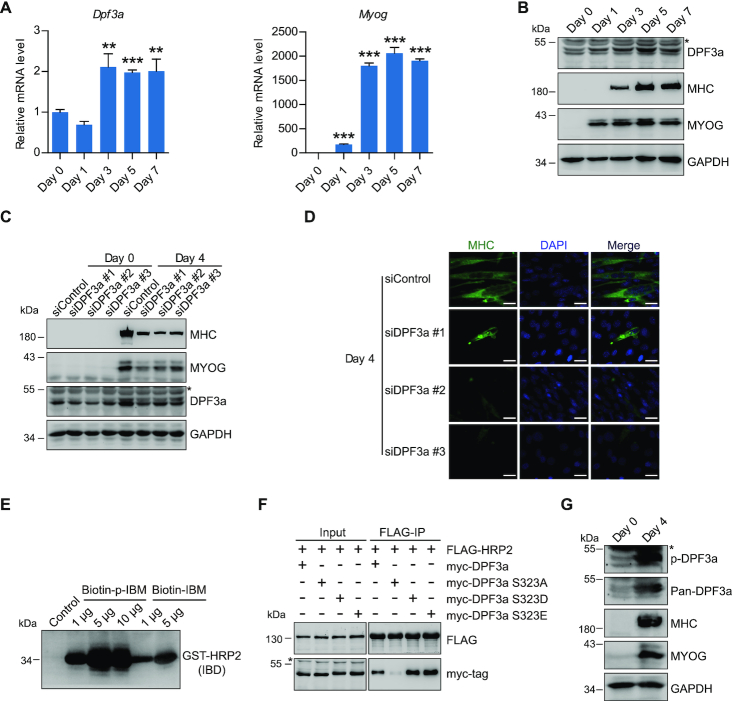
Previous studies have shown that morpholino-mediated knockdown of dpf3 in zebrafish led to severely reduced cardiac contractility, incomplete cardiac looping and defective organization of cardiac and skeletal muscle fibers (50). Consistent with the predicted role of DPF3a in myogenesis, we detected an increase of DPF3a mRNA during myogenic differentiation (Figure 4A). To confirm this increase of DPF3a expression, we generated an antibody specifically recognizing DPF3a (Supplementary Figure S2A). As shown in Figure 4B, the abundance of DPF3a protein also increased upon myoblast differentiation. Moreover, depletion of DPF3a dramatically diminished myotube formation following differentiation (Figure 4C and D). These results, together with previous reports, support a key role for DPF3a in myogenesis.
Figure 4.
DPF3a is critical for C2C12 differentiation, and elevation of DPF3a expression and phosphorylation in myotubes enhances its binding to HRP2. (A) RT–qPCR analysis for Dpf3a (left) and Myog (right) transcripts of myoblasts and myotubes at various differentiation stages as indicated. Data represent as means ± SD of three replicates. ** P < 0.01 and *** P < 0.001. (B) Western blotting analysis of DPF3a, MYOG, MHC and GAPDH in myoblasts and myotubes at various differentiation stages as indicated. (C) Western blotting analysis of MYOG, MHC, DPF3a and GAPDH in myoblasts and myotubes transfected with scramble and three DPF3a-specific siRNAs as indicated. (D) Immunofluorescence analysis of MHC and DAPI in myotubes transfected with scramble and three different DPF3a siRNAs. Scale bars, 25 μm. DAPI, 4′,6-diamidino-2-phenylindole. (E) Biotin-labeled DPF3a or p-DPF3a peptide was incubated with GST-IBD. Peptide pull-down assay shows that phosphorylation of DPF3a enhanced its affinity for HRP2. (F) Co-IP assay examining the interaction of FLAG-HRP2 and indicated myc-DPF3a (WT or mutants). (G) Western blotting analysis of DPF3a, p-DPF3a, MHC, MYOG and GAPDH in myoblasts and myotubes showing that phosphorylation of DPF3a increased upon cell differentiation.
Phosphorylation of DPF3a enhances its interaction with HRP2
Unlike other IBM-containing proteins, DPF3a and ASK possess a serine (S) instead of a glutamic acid (E) as the first amino acid of the canonical IBM (Figure 2C). Glutamic acid is a negatively charged amino acid that can mimic serine phosphorylation. Since phosphorylation is important for protein interaction, we speculated that phosphorylation of DPF3a at serine 323 (S323) might affect its interaction with HRP2. Cross-species amino acid sequence alignment of the DPF3a C-terminus shows that DPF3a S323 is evolutionarily conserved (Supplementary Figure S2B). Pulldown assays showed that S323-phosphorylated peptide exhibited higher affinity for recombinant GST-IBD compared to unphosphorylated peptide (Figure 4E and Supplementary Figure S2C). As shown in Figure 4F, the amounts of HRP2 coprecipitated with phospho-mimetic DPF3a mutants (S323D or S323E) markedly increased, while the S323A phospho-dead DPF3a mutant (S323A) failed to bind to HRP2. To further analyze DPF3a phosphorylation in vivo, we generated a phospho-specific DPF3a antibody (anti-p-DPF3a) against the epitope containing the phospho-S323 residue. As shown in Supplementary Figure S2D, anti-p-DPF3a antibody recognized the phosphorylated peptide with markedly higher selectivity over the unphosphorylated peptide. The p-DPF3a antibody failed to recognize the phospho-dead S323A mutant, further validating the specificity of this antibody (Supplementary Figure S2E). Importantly, both total and phosphorylated DPF3a abundance increased upon myoblast differentiation (Figure 4G). Collectively, these results demonstrate that phosphorylation of DPF3a enhances the association between HRP2 and DPF3a.
Characterization of HRP2 genome-wide distribution during myogenesis
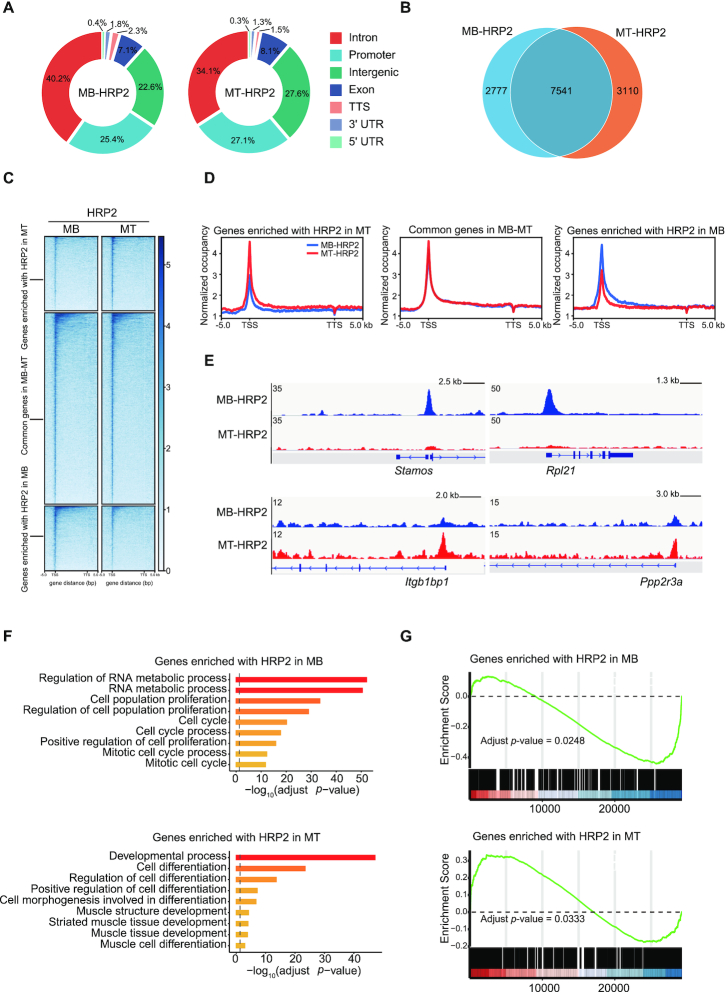
Methylation of H3K36 has long been implicated in transcriptional regulation (52,53). Since HRP2 is an H3K36me reader protein and is required for myogenesis, we sought to explore the regulatory mechanisms of HRP2 in gene expression during myogenic differentiation. To do this, we first mapped the genome-wide distribution of HRP2 by chromatin immunoprecipitation sequencing (ChIP-seq) in undifferentiated C2C12 myoblasts (MB) and differentiated myotubes (MT). As shown in Figure 5A, 25% of total HRP2 peaks in MB and 27% in MT were detected within gene promoter regions, indicating an enrichment of HRP2 at promoter regions in both MB and MT. To validate the genome-wide distribution of HRP2 in muscle cells, we generated cells stably expressing an HRP2 construct with a TY1 tag at its N-terminus. Western blotting analysis showed that the expression of exogenous TY1-HRP2 was comparable to that of endogenous HRP2 (Supplementary Figure S3A). We then performed ChIP-seq using TY1 antibody in myoblasts and myotubes, and bioinformatics analysis revealed a high correlation between HRP2 and TY1-HRP2 ChIP-seq datasets (Supplementary Figure S3B). Based on the ratio of HRP2 enrichment on these genes in MB versus MT, we categorized HRP2 target genes into three subgroups: HRP2-enriched genes in MB, common genes in MB and MT, and HRP2-enriched genes in MT (Figure 5B–E). Gene ontology analysis was performed with HRP2-enriched genes in MB and in MT, and revealed that these two categories of genes were enriched for cell proliferation and muscle cell differentiation, respectively (Figure 5F). We performed RNA-seq analysis and identified genes that were differentially expressed during myogenic differentiation. Gene set enrichment analysis (GSEA) showed that HRP2-enriched genes in MB and HRP2-enriched genes in MT were enriched for down-regulated genes and up-regulated genes upon differentiation, respectively (Figure 5G). Taken together, these findings demonstrate a redistribution of HRP2 on the genome during myogenesis.
Figure 5.
Characterization of HRP2 distribution in myoblasts and myotubes. (A) Genome-wide distribution of HRP2 in C2C12 myoblasts and myotubes. (B) HRP2 binding genes were categorized into three subgroups: HRP2-enriched genes in MB, common genes in MB and MT, and HRP2-enriched genes in MT. Venn diagram showing the number of genes in these three groups. (C) Heatmaps showing the distribution of HRP2 binding in the three groups of genes indicated in (B) for the region −5 kb from the TSS to +5 kb from the TTS. Genes on the heatmaps are ordered according to decreasing total level of HRP2. (D) Average genome-wide HRP2 occupancies are shown for the three groups of genes indicated in (B). Sequences 5 kb upstream of the TSS and 5 kb downstream of the TTS are also included. (E) Representative ChIP-seq tracks of HRP2-enriched genes in MB and MT. (F) Gene ontology enrichment analysis of two groups of genes: HRP2-enriched genes in MB and HRP2-enriched genes in MT, as indicated in (B). (G) GSEA analysis combining the gene sets indicated in (F) with RNA sequencing profiles upon myoblast differentiation. HRP2-enriched genes in MB were enriched for down-regulated genes upon myoblast differentiation (upper). HRP2-enriched genes in MT were enriched for up-regulated genes upon myoblast differentiation (lower).
HRP2 genome recruitment depends on the H3K36me2 mark
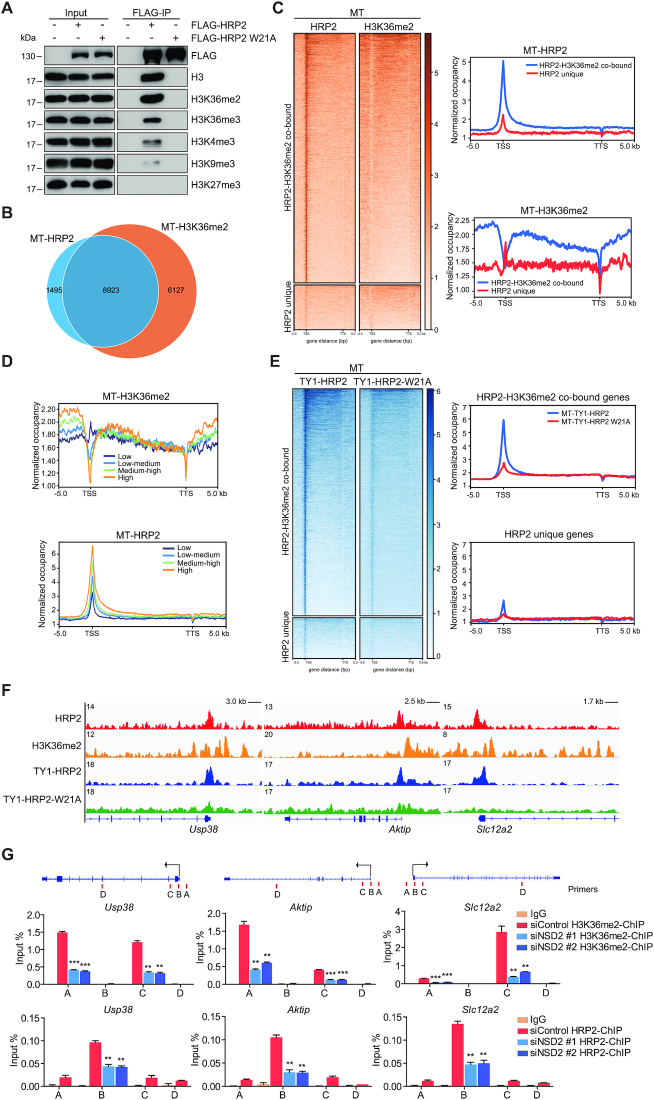
HRP2 has been reported to interact with various methylated histone peptides in vitro (12), and we therefore sought to examine its binding preference in vivo. A protein-protein ChIP assay showed that WT HRP2 strongly associated with chromatin containing H3K36me2, while its binding to H3K36me3 and other histone methylation marks was much weaker. Mutation of a conserved aromatic residue (W21A) in the hydrophobic cavity of HRP2 PWWP (11,12) abolished these bindings (Supplementary Figure S3D). Similar results were obtained from native ChIP assay with mono-nucleosomes (Figure 6A and Supplementary Figure S3C). These data indicate that HRP2 preferentially binds to chromatin regions marked by H3K36me2. To further investigate whether genome recruitment of HRP2 depends on the H3K36me2 mark, we performed H3K36me2 ChIP-seq in MT and observed that almost 80% of HRP2 occupied genes that overlapped with H3K36me2-enriched genes (Figure 6B). Bioinformatics analysis showed that the normalized tag density of HRP2 ChIP-seq was higher in HRP2/H3K36me2-co-bound genes than in HRP2-unique genes (Figure 6C). Furthermore, we categorized HRP2/H3K36me2-co-bound genes into four groups according to H3K36me2 normalized tag density around TSS. As shown in Figure 6D, genes with higher H3K36me2 enrichment also exhibited higher enrichment of HRP2 at the TSS (Figure 6D), indicating that HRP2 genome occupancy correlates with the H3K36me2 mark. Notably, we observed a shift ∼1.5 kb between the maxima of HRP2 and H3K36me2 signals (Supplementary Figure S3E). To test if the recruitment of HRP2 to chromatin depends on H3K36me2, we performed ChIP-sequencing with TY1 antibody in C2C12 cells stably expressing TY1-HRP2 and the TY1-HRP2 (W21A) mutation. Metagene analysis and representative gene tracks showed that the TSS enrichment of the W21A mutant decreased substantially relative to that of WT HRP2 (Figure 6E and F), which indicates that the loss of H3K36me2 binding abolishes the recruitment of HRP2 to target gene TSS. Zhuang et al. reported that the H3K36me2 methyltransferase NSD2 is important for muscle development in mice (43). ChIP-qPCR analysis of several HRP2-H3K36me2-co-bound genes revealed that depletion of NSD2 specifically decreased H3K36me2 signals around TSS, together with a marked reduction of HRP2 binding at the TSS (Figure 6G and Supplementary Figure S3F). Taken together, these findings demonstrate that HRP2 genome recruitment depends on the H3K36me2 histone mark.
Figure 6.
HRP2 genome recruitment depends on the H3K36me2 mark. (A) 293T cells expressing FLAG-tagged HRP2 (WT or W21A mutant) were subjected to native ChIP assay. Nucleosomes pulled down with anti-FLAG antibody were analyzed by western blotting using antibodies against various histone marks. (B) Venn diagram showing the number of overlaps between genes bound by HRP2 and H3K36me2 in C2C12 myotubes. (C) Heatmaps showing the distribution of HRP2 and H3K36me2 on HRP2-H3K36me2-co-bound genes and HRP2-unique genes for the region −5 kb from the TSS to +5 kb from the TTS. Genes on the heatmaps are ordered according to decreasing total level of HRP2. (left). Average genome-wide H3K36me2 and HRP2 occupancies are shown for HRP2-H3K36me2-co-bound genes and HRP2-unique genes. Sequences 5 kb upstream and 5 kb downstream of the TTS are also included (right). (D) HRP2-H3K36me2-co-bound genes are categorized into four groups according to H3K36me2-normalized tag density around TSS. Average genome-wide H3K36me2 and HRP2 occupancies are shown for these four gene sets. Sequences 5 kb upstream of the TSS and 5 kb downstream of the TTS are also included. (E) Heatmaps showing the distribution of TY1-HRP2 and TY1-HRP2-W21A binding on HRP2-H3K36me2-co-bound genes and HRP2-unique genes for the region -5 kb from the TSS to +5 kb from the TTS. Genes on the heatmaps are ordered according to decreasing total level of TY1-HRP2 (left). Average genome-wide TY1-HRP2 and TY1-HRP2-W21A occupancies are shown for HRP2-H3K36me2-co-bound genes and HRP2-unique genes. Sequences 5 kb upstream of the TSS and 5 kb downstream of the TTS are also included (right). (F) Gene tracks showing representative ChIP-Seq profiles for the indicated proteins and histone marks at the Usp38, Aktip and Slc12a2 gene loci in myotubes. (G) ChIP-qPCR of H3K36me2 and HRP2 at Usp38, Aktip and Slc12a2 gene loci in MT C2C12 cells transfected with siRNA as indicated. PCR primers were designed according to ChIP-seq peaks of corresponding proteins on these gene loci. Schematic representation of PCR primer design is provided. Enrichment values (Y axis) are expressed as fold change normalized to input chromatin. Data are represented as means ± SD of three replicates. ** P < 0.01 and *** P < 0.001.
HRP2 colocalizes with DPF3a at gene promoters.
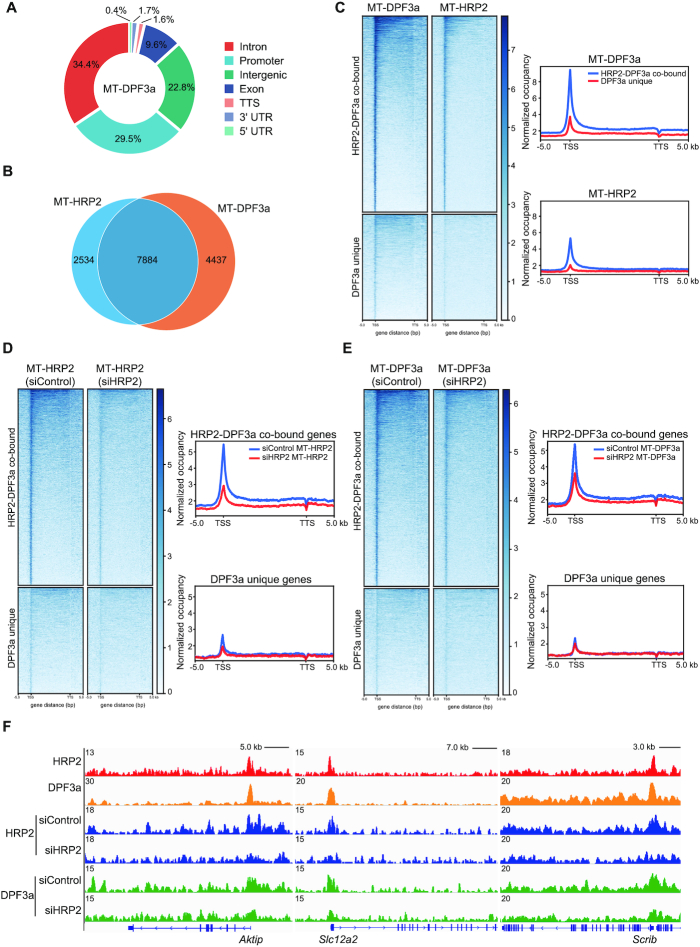
Since HRP2 directly interacts with DPF3a, we next tested whether HRP2 acted in concert with DPF3a to regulate gene expression. As shown in Figure 4B, the expression of DPF3a was low in MB but markedly increased in MT. Thus, we performed ChIP-seq with a focus on examining the genome-wide site occupancy by DPF3a in MT. As shown in Figure 7A, nearly 30% of total DPF3a peaks in MT occurred within gene promoters. Importantly, >60% of DPF3a-bound genes overlapped with HRP2-occupied genes (Figure 7B). Furthermore, the distribution of DPF3a-bound peaks resembled the genomic distribution of HRP2 (Supplementary Figure S4D). Metagene analysis revealed that higher DPF3a occupancy occurred in HRP2-DPF3a-co-bound genes than in DPF3a-unique genes (Figure 7C), indicating that DPF3a genome recruitment correlates with that of HRP2. Further bioinformatics analysis showed that HRP2/DPF3a-co-bound genes exhibit longer gene length, higher expression level and enrichment for muscle-related genes (Supplementary Figure S4A–C), which is consistent with the roles of HRP2 and DPF3a in muscle cell differentiation.
Figure 7.
Genome-wide colocalization of DPF3a and HRP2 in myotubes. (A) Genome-wide distribution of DPF3a binding in C2C12 myotubes. (B) Venn diagram showing the number of overlaps between genes bound by HRP2 and DPF3a in C2C12 myotubes. (C) Heatmaps showing the distribution of DPF3a and HRP2 on HRP2-DPF3a-co-bound genes and DPF3a-unique genes for the region -5 kb from the TSS to +5 kb from the TTS. Genes on the heatmaps are ordered according to decreasing total level of DPF3a (left). Average genome-wide DPF3a and HRP2 occupancies are shown for HRP2-DPF3a-co-bound genes and DPF3a-unique genes. Sequences 5 kb upstream of the TSS and 5 kb downstream of the TTS are also included (right). (D) Heatmaps showing the distribution of HRP2 on HRP2-DPF3a-co-bound genes and DPF3a-unique genes in WT and HRP2 knockdown cells for the region from -5 kb to +5 kb from the TTS. Genes on the heatmaps are ordered according to decreasing total level of HRP2 (left). Average genome-wide HRP2 occupancies are shown for HRP2-DPF3a-co-bound genes and DPF3a-unique genes in WT and HRP2 knockdown cells. Sequences 5 kb upstream of the TSS and 5 kb downstream of the TTS are also included (right). (E) Heatmaps showing the distribution of DPF3a on HRP2-DPF3a-co-bound genes and DPF3a-unique genes in WT and HRP2 knockdown cells for the region −5 kb from the TSS to +5 kb from the TTS. Genes on the heatmaps are ordered according to decreasing total level of DPF3a (left). Average genome-wide DPF3a occupancies are shown for HRP2-DPF3a-co-bound genes and DPF3a-unique genes in WT and HRP2 knockdown cells. Sequences 5 kb upstream of the TSS and 5 kb downstream of the TTS are also included (right). (F) Gene tracks showing representative ChIP-Seq profiles for the indicated proteins in various cells as indicated at the Aktip, Slc12a2 and Scrib gene loci.
To further investigate whether DPF3a binding to chromatin depends on HRP2, we performed ChIP-seq in control and HRP2-depleted C2C12 cells. Metagene analysis and representative gene tracks showed that the loss of HRP2 effectively reduced the promoter binding of HRP2 on HRP2/DPF3a-co-bound genes, together with a marked reduction of DPF3a enrichment around the TSS (Figure 7D–F). Consistent with this, ChIP-qPCR on several HRP2-DPF3a-co-bound genes revealed that depletion of HRP2 binding concurrently reduced the DPF3a occupancy around the TSS (Supplementary Figure S4E). Collectively, these findings demonstrate that HRP2 is required for the recruitment of DPF3a to target gene TSS.
Identification of common target genes of HRP2 and DPF3a during myogenic differentiation
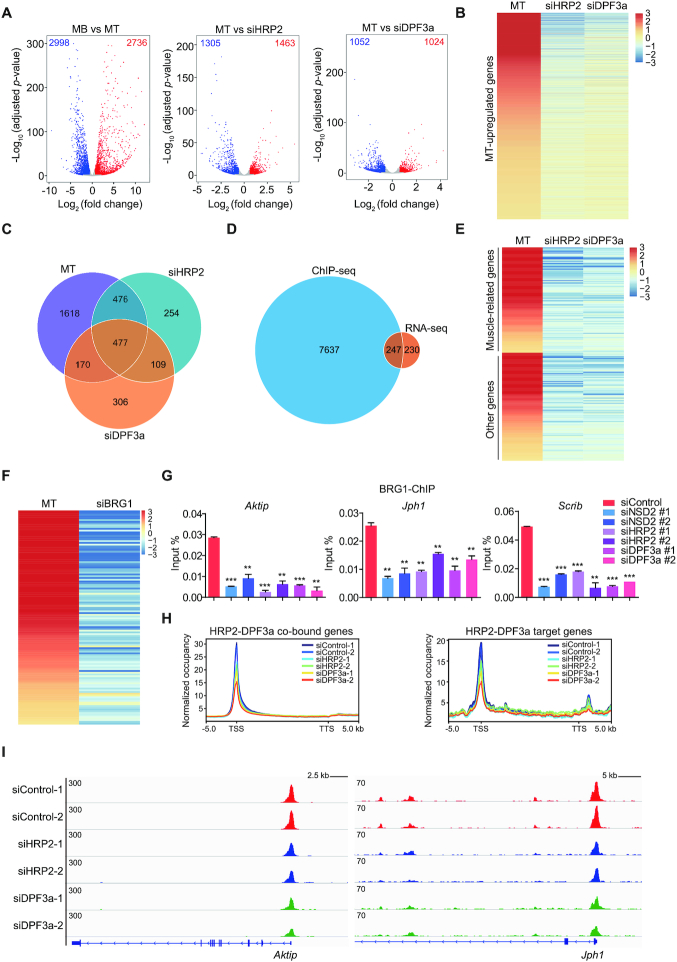
Since HRP2 colocalizes with DPF3a, and both are required for myogenesis, we proceeded to identify their co-regulated genes that are responsible for myogenic differentiation. First, we performed RNA-seq analyses in MB and MT with depletion of HRP2 or DPF3a. We observed 2736 genes up-regulated upon MB differentiation, while 1305 genes and 1052 genes were down-regulated by HRP2 and DPF3a knockdown, respectively (Figure 8A and B). Combining these three RNA-seq datasets, we identified 477 genes regulated by HRP2 and DPF3a (Figure 8C). Furthermore, among these 477 genes, more than half (247/477) overlapped with HRP2/DPF3A-co-bound genes (Figure 8D). Importantly, ∼50% of these 247 genes (122) belonged to categories of myogenesis (Figure 8E). These data suggest that HRP2 and DPF3a modulate muscle differentiation by regulating myogenic gene expression.
Figure 8.
HRP2 and DPF3a regulate chromatin accessibility of myogenic gene loci through recruitment of the BAF complex. (A) Volcano plots of differentially expressed genes in three groups: MB versus MT (left), siControl vs. siHRP2 (middle) in myotubes and siControl versus siDPF3a in myotubes (right). (B) Representative heatmaps of genes up-regulated in myotubes (compared to myoblasts) and down-regulated upon DPF3a or HRP2 knockdown in myotubes. (C) Venn diagram showing the number of overlaps between the three genes sets indicated in (A). (D) Venn diagram showing the number of overlaps between HRP2-DPF3a-co-bound genes and common genes of three RNA-seq datasets indicated in (A). (E) Representative heatmap of 247 overlapped genes indicated in (D). Of these, 122 can be sub-categorized into muscle-related genes. (F) Representative heatmaps of 122 muscle-related genes down-regulated upon BRG1 knockdown in myotubes. (G) ChIP-qPCR of BRG1 at Aktip, Jph1 and Scrib gene loci in MT C2C12 cells transfected with siRNA as indicated. PCR primers were designed at ‘B’ region as indicated in Figure 6G. Enrichment values (Y axis) are expressed as fold change normalized to input chromatin. Data are represented as means ± SD of three replicates. ** P < 0.01 and *** P < 0.001. (H) Average genome-wide ATAC signals are shown for HRP2-DPF3a-co-bound genes and 247 overlapped genes in myotubes, with transfection as indicated. Sequences 5 kb upstream of the TSS and 5 kb downstream of the TTS are also included. (I) Gene tracks showing representative ATAC-seq profiles in cells, with transfection as indicated, at the Aktip and Jph1 gene loci.
HRP2 and DPF3a recruit BRG1 and increase chromatin accessibility at myogenic genes
Since HRP2 associates with the BAF complex through direct interaction with DPF3a, we next tested whether BRG1, the enzymatic subunit of BAF, also regulates HRP2-DPF3a target genes. As shown in Figure 8F, the expression of the 122 myogenic genes commonly regulated by HRP2-DPF3a also markedly decreased upon BRG1 knockdown. Consistent with this observation, BRG1 knockdown abolished myotube formation following differentiation (Supplementary Figure S5A). Furthermore, as analyzed by ChIP-qPCR, the binding of BRG1 to myogenic genes was decreased upon HRP2, DPF3a and NSD2 knockdown (Figure 8G and Supplementary Figure S5B). To examine chromatin accessibility, ATAC-sequencing (ATAC-seq) was performed in MT with HRP2 and DPF3a knockdown. Metagene analysis and representative gene tracks showed that chromatin accessibility decreased upon HRP2 and DPF3a knockdown at HRP2-DPF3a-co-bound genes as well as at the 247 co-regulated muscle genes (Figure 8H, I and Supplementary Figure S5C). Taken together, these data indicate that HRP2 and DPF3a regulate myogenic genes by recruiting the BAF complex and remodeling the corresponding chromatin loci.
HRP2 is essential for muscle regeneration in mice
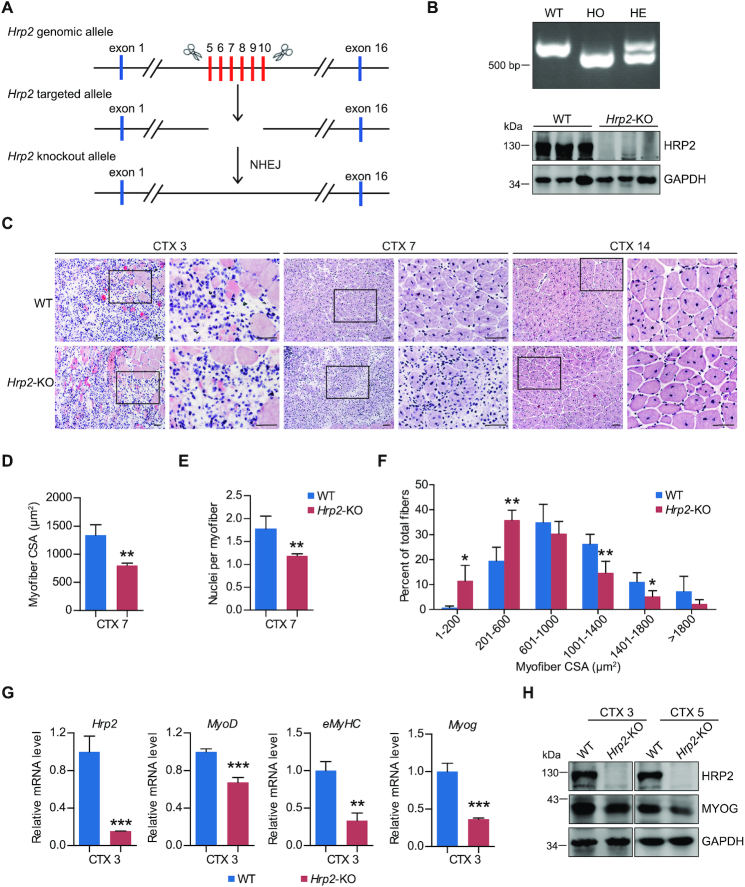
To further investigate the functions of HRP2 in vivo, we generated Hrp2 knockout (KO) mice (Figure 9A and B). Consistent with a previous report (54), no apparent developmental defects were observed in Hrp2 KO mice. To examine the roles of HRP2 in muscle regeneration, the tibialis anterior (TA) muscles from both WT and Hrp2 KO were degenerated by cardiotoxin (CTX) treatment. As shown in Supplementary Figure S6A, on Day 5 after CTX injury both WT and Hrp2 KO muscles contained fibers with centralized nuclei, indicating the occurrence of muscle regeneration at the point of maximal injury. A compact assembly of centrally nucleated regenerating fibers was observed at the point of maximal injury in the WT muscles on Day 7 after injury; however, the regenerating fibers in the Hrp2 KO muscles at Day 7 appeared more sparsely organized, and quantification of regenerating fibers revealed that muscle regeneration is more severely impaired in Hrp2 KO muscles than in WT muscles (Figure 9C–F). Furthermore, we observed that the expression of HRP2, MYOD, MYOG and eMyHC, and of six representative HRP2 and DPF3a common target genes (Supplementary Figure S6B), was substantially decreased in Hrp2 KO mice, indicating delayed muscle regeneration (Figure 9G and H). Taken together, these results revealed that Hrp2 depletion in mice led to impaired muscle regeneration, further emphasizing the importance of HRP2 in myogenesis.
Figure 9.
Muscle regeneration is impaired in Hrp2 KO mice treated with CTX. (A) Schematic outlining the generation of Hrp2 knockout mice using the CRISPR/Cas9 system. The targeting sites of Hrp2 are shown. (B) Hrp2−/− KO male mice were established by breeding Hrp2+/− males and females. The targeted fragment of Hrp2 was amplified by PCR using genomic DNA templates (upper). HRP2 abundance in WT and Hrp2 knockout mice was examined by western blot (lower). HO: homozygous, HE: heterozygous. (C) Representative H&E-stained cross-sections of TA muscles from male WT and Hrp2−/− mice. Sections were obtained from CTX-injured muscles at Days 3, 7 and 14 post-treatment. Boxed areas are enlarged in the panels to their right. Scale bar, 50 μm. (D) Mean myofiber cross-sectional area (CSA) of regenerating muscles in male WT and Hrp2−/− mice (number of fibers counted > 1000) at Day 7 post-CTX-induced injury (n = 4 per group). (E) Numbers of nuclei per myofiber in TA muscle from WT or Hrp2−/− mice at Day 7 post-CTX-induced injury (n = 4 per group). (F) Myofiber CSA distribution in WT and Hrp2−/− mice 7 days after CTX injection. (G) mRNA levels of embryonic MyHC, Myog, MyoD and Hrp2 were analyzed in TA muscle of WT or Hrp2−/− mice at Day 3 post-CTX-induced injury (n = 3 per group). (H) Western blot analysis of MYOG and GAPDH protein levels in TA muscle of WT or Hrp2−/− mice at Days 3 and 5 post-CTX-induced injury. Data are represented as means ± SD of 4 (D–F) or 3 (G) replicates. ** P < 0.01 and *** P < 0.001.
DISCUSSION
Chromatin dynamics plays a crucial role in the regulation of gene transcription by providing the transcriptional machinery with dynamic access to genomic DNA. Histone post-translational modifications and ATP-dependent chromatin remodeling represent two primary mechanisms that typically work in concert to control chromatin dynamics and function. Targeting a chromatin remodeling complex to a particular genomic region through site-specific histone modifications represents a key regulatory mode for this functional crosstalk (3,4). Several components of the BAF chromatin remodeling complex possess unique motifs that mediate their binding to various histone marks. For instance, the bromodomain of BRG1/BRM recognizes histone lysine acetylation, thus helping the recruitment and retention of the BAF complex onto chromatin templates (55–57). The DPF3b isoform of BAF45C contains two PHD fingers that are responsible for recruiting the BAF complex to H3K14ac-marked nucleosomes (50,51). It has recently been shown that DPF3b also interacts with monomethylated histone H3K4 and thereby enhances BAF complex binding to enhancers (58). However, experimental evidence of the interplay between the BAF complex and other histone modifications is lacking. Both isoforms of DPF3 can associate with the BAF complex. Unlike DPF3b, the DPF3a isoform contains a single truncated PHD finger, which does not bind to histone, raising the question of whether and how DPF3a recruits the BAF complex to specific gene loci. In this study, we find that the PWWP domain of HRP2 preferentially binds to H3K36me2. HRP2 directly interacts with DPF3a and recruits the BAF complex to genomic regions depending on the binding of HRP2 to H3K36me2. These findings therefore indicate that HRP2-DPF3a acts as a ‘bridging’ adaptor to connect H3K36me2 and the BAF complex, and establish H3K36me2 as a histone mark that coordinates with remodeling enzymes to regulate chromatin dynamics and gene expression.
Methylation of H3K36 plays important roles at multiple stages of transcriptional regulation. Its ultimate effect on transcription depends on the degree of methylation, the location of this specific modification and the binding of reader proteins (52,53). ChIP-seq analyses reveal that H3K36me3 often appears within the gene bodies of actively transcribed genes, while H3K36me2 displays a marked enrichment at promoter or intergenic regions (13,14,18,59,60). The gene body distribution of H3K36me3 is widely considered a hallmark of gene activation; however, the functions and mechanisms of H3K36me2 in transcription are not well defined. Zhu et al. showed that LEDGF, a PWWP domain-containing protein, recognizes Ash1L-catalyzed H3K36me2 and recruits mixed-lineage leukemia (MLL) protein to activate leukemia target genes (13). Our biochemical analyses show that HRP2 preferentially binds to H3K36me2 mark. Further ChIP analyses indicate that (i) HRP2 genome occupancy correlates with the H3K36me2 mark; (ii) W21A mutation of HRP2, which abolishes HRP2 binding to H3K36me2, reduces the recruitment of HRP2 to target genes and (iii) depletion of NSD2 leads to decreased H3K36me2 signals and a marked reduction of HRP2 binding to chromatin. Collectively, these findings demonstrate that HRP2 genome recruitment depends on the H3K36me2 histone mark. Consistent with our work, LeRoy et al. recently reported that HRP2 binds to H3K36me2/3 and facilitates RNA Pol II transcription elongation by relieving the nucleosome-induced barrier (14). Together, these findings illustrate an epigenetic mechanism by which H3K36me2 contributes to transcriptional activation.
Importantly, we observed that the maxima of H3K36me2 and HRP2 binding signals do not coincide perfectly. A multi-step mechanism may be responsible for this location shift. The HRP2/DPF3a/BAF complex is initially recruited to target genes by direct binding of HRP2 to the H3K36me2 mark upstream/downstream of the TSS region. The BAF complex interacts with other proteins, such as MYOD (61,62), which are located at the TSS and may facilitate the relocation and stabilization of the BAF complex at the TSS. HRP2 is then dragged along with the BAF complex to the TSS and thus dissociates from the H3K36me2 mark. Future investigations are needed to prove the above multi-step mechanism (or some other mechanisms) underlying this location shift.
A prominent function of epigenetic regulation is to control cell fate and differentiation. Several lines of evidence indicate that H3K36 methylation is required for myogenic differentiation. Yi et al. reported that Setd2 is essential for the proliferation and differentiation of myoblasts (41). It has been shown that NSD2 is important for muscle development in mice (43). Likewise, Ash1L has been shown to activate myogenic genes and promote myoblast fusion (44). We find that the chromatin binding of HRP2 undergoes a switch from cell proliferation to cell differentiation genes during myogenic differentiation. HRP2 colocalizes and cooperates with DPF3a to regulate the myogenic gene expression program and MB differentiation. Loss of function studies in mice underscore a key role for HRP2 in injury-induced muscle regeneration, further supporting its function in muscle cell fate specification. The PWWP domain of HRP2 displays low-affinity binding to methylated histone peptides in vitro (12,23). In contrast, cistromic analysis in our study and others reveals that HRP2 binds to H3K36me2 at specific genomic regions (14). The underlying mechanism for precise and selective binding of HRP2 to distinct target genes in MB and MT is currently unknown. One possibility is that HRP2 acts in concert with associated proteins to enhance the binding affinity and specificity of HRP2 for chromatin. During myogenic differentiation, both the abundance and phosphorylation level of DPF3a increase, which leads to a marked increase in the association between HRP2 and DPF3a. The enhanced interaction between these two proteins further promotes recruitment of the BAF complex. Several components of the BAF complex contain histone modification reader domains. Thus, combinations of multiple effectors may work synergistically to increase the binding affinity and specificity of the HRP2–DPF3a–BAF complex assemblage at target loci. Through its IBD domain, HRP2 interacts with various IBM-containing proteins. Therefore, HRP2 may associate with other factors to drive cell proliferation gene expression in MB. The switch of binding partners during myogenesis represents a plausible explanation for the observed specific HRP2 distribution and function.
Protein phosphorylation functions as a key regulatory mode for signal transduction and cellular responses to external cues (63). In response to hypertrophic stimuli, DPF3a is phosphorylated by casein kinase 2 at S348, which is critical for the activation of cardiac fetal genes through the recruitment of BRG1 (64). Here we show that DPF3a is also phosphorylated at S323 upon myogenic differentiation, which enhances its association with HRP2 and facilitates recruitment of the BAF complex to target genes. Thus, DPF3a phosphorylation at different sites may generate sensors that mediate cellular responses to various environmental changes, which eventually leads to the modulation of chromatin dynamics and the specification of cell fate. The kinase responsible for S323 phosphorylation of DPF3a is currently unknown, however, and further investigation is therefore needed to identify this kinase and define its role in epigenetic regulation and muscle cell fate specification.
DATA AVAILABILITY
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq, ChIP-seq and ATAC-seq data can be found at the Gene Expression Omnibus database under accession numbers GSE141407.
Supplementary Material
Contributor Information
Xu Zhu, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Bingxue Lan, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Xianfu Yi, School of Biomedical Engineering and Technology, Tianjin Medical University, Tianjin 300070, China.
Chaoran He, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Lin Dang, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Xingquan Zhou, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Yumei Lu, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Yongzhan Sun, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Zhiheng Liu, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Xue Bai, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Kai Zhang, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Bing Li, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Mulin Jun Li, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Department of Pharmacology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Yupeng Chen, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Lirong Zhang, 2011 Collaborative Innovation Center of Tianjin for Medical Epigenetics, Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31571336 to L.Z., 31601111 to X.Z., 31871327 to M.J.L, 81770658 and 31571337 to Y.C.]; National Key Research and Development Program of China [2017YFA0504102 to Y.C.]; Natural Science Foundation of Tianjin [19JCJQJC63800 to Y.C., 19JCQNJC09000 to X.Y., 18JCZDJC34700 to M.J.L.]; Excellent Talent Project of Tianjin Medical University (to M.J.L. and Y.C.). Funding for open access charge: National Key Research and Development Program of China [2017YFA0504102 to Y.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jenuwein T., Allis C.D.. Translating the histone code. Science. 2001; 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 2. Chen T., Dent S.Y.. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014; 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petty E., Pillus L.. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013; 29:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swygert S.G., Peterson C.L.. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim. Biophys. Acta. 2014; 1839:728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dawson M.A., Kouzarides T.. Cancer epigenetics: from mechanism to therapy. Cell. 2012; 150:12–27. [DOI] [PubMed] [Google Scholar]
- 6. Reddy M.A., Zhang E., Natarajan R.. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2015; 58:443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lardenoije R., Iatrou A., Kenis G., Kompotis K., Steinbusch H.W., Mastroeni D., Coleman P., Lemere C.A., Hof P.R., van den Hove D.L. et al.. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 2015; 131:21–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venkatesh S., Workman J.L.. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015; 16:178–189. [DOI] [PubMed] [Google Scholar]
- 9. Yun M., Wu J., Workman J.L., Li B.. Readers of histone modifications. Cell Res. 2011; 21:564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Musselman C., Lalonde M., Côté J., Kutateladze T.. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012; 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qin S., Min J.. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 2014; 39:536–547. [DOI] [PubMed] [Google Scholar]
- 12. Wu H., Zeng H., Lam R., Tempel W., Amaya M.F., Xu C., Dombrovski L., Qiu W., Wang Y., Min J.. Structural and histone binding ability characterizations of human PWWP domains. PLoS One. 2011; 6:e18919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu L., Li Q., Wong S.H., Huang M., Klein B.J., Shen J., Ikenouye L., Onishi M., Schneidawind D., Buechele C. et al.. ASH1L links Histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 2016; 6:770–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. LeRoy G., Oksuz O., Descostes N., Aoi Y., Ganai R.A., Kara H.O., Yu J.R., Lee C.H., Stafford J., Shilatifard A. et al.. LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription in differentiated cells. Sci. Adv. 2019; 5:eaay3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pradeepa M.M., Grimes G.R., Taylor G.C., Sutherland H.G., Bickmore W.A.. Psip1/Ledgf p75 restrains Hox gene expression by recruiting both trithorax and polycomb group proteins. Nucleic Acids Res. 2014; 42:9021–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J. et al.. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014; 508:263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schubeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 18. Weinberg D.N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K.N., Horth C., McGuire J.T., Xu X., Nikbakht H. et al.. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019; 573:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heyn P., Logan C.V., Fluteau A., Challis R.C., Auchynnikava T., Martin C.A., Marsh J.A., Taglini F., Kilanowski F., Parry D.A. et al.. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet. 2019; 51:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo R., Zheng L., Park J.W., Lv R., Chen H., Jiao F., Xu W., Mu S., Wen H., Qiu J. et al.. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell. 2014; 56:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smolle M., Venkatesh S., Gogol M.M., Li H., Zhang Y., Florens L., Washburn M.P., Workman J.L.. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012; 19:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li F., Mao G., Tong D., Huang J., Gu L., Yang W., Li G.M.. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013; 153:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baude A., Aaes T.L., Zhai B., Al-Nakouzi N., Oo H.Z., Daugaard M., Rohde M., Jaattela M.. Hepatoma-derived growth factor-related protein 2 promotes DNA repair by homologous recombination. Nucleic Acids Res. 2016; 44:2214–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daugaard M., Baude A., Fugger K., Povlsen L.K., Beck H., Sorensen C.S., Petersen N.H., Sorensen P.H., Lukas C., Bartek J. et al.. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012; 19:803–810. [DOI] [PubMed] [Google Scholar]
- 25. Izumoto Y., Kuroda T., Harada H., Kishimoto T., Nakamura H.. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 1997; 238:26–32. [DOI] [PubMed] [Google Scholar]
- 26. Wang H., Jurado K.A., Wu X., Shun M.C., Li X., Ferris A.L., Smith S.J., Patel P.A., Fuchs J.R., Cherepanov P. et al.. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012; 40:11518–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schrijvers R., De Rijck J., Demeulemeester J., Adachi N., Vets S., Ronen K., Christ F., Bushman F.D., Debyser Z., Gijsbers R.. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012; 8:e1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chal J., Pourquie O.. Making muscle: skeletal myogenesis in vivo and in vitro. Development. 2017; 144:2104–2122. [DOI] [PubMed] [Google Scholar]
- 29. Bentzinger C.F., Wang Y.X., Rudnicki M.A.. Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012; 4:a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wagers A.J., Conboy I.M.. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell. 2005; 122:659–667. [DOI] [PubMed] [Google Scholar]
- 31. Saccone V., Puri P.L.. Epigenetic regulation of skeletal myogenesis. Organogenesis. 2010; 6:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toto P.C., Puri P.L., Albini S.. SWI/SNF-directed stem cell lineage specification: dynamic composition regulates specific stages of skeletal myogenesis. Cell. Mol. Life Sci. 2016; 73:3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson D.C.L., Dilworth F.J.. Epigenetic regulation of adult myogenesis. Curr. Top. Dev. Biol. 2018; 126:235–284. [DOI] [PubMed] [Google Scholar]
- 34. Perdiguero E., Sousa-Victor P., Ballestar E., Munoz-Canoves P.. Epigenetic regulation of myogenesis. Epigenetics. 2009; 4:541–550. [DOI] [PubMed] [Google Scholar]
- 35. Cao X., Dang L., Zheng X., Lu Y., Lu Y., Ji R., Zhang T., Ruan X., Zhi J., Hou X. et al.. Targeting super-enhancer-driven oncogenic transcription by CDK7 inhibition in anaplastic thyroid carcinoma. Thyroid. 2019; 29:809–823. [DOI] [PubMed] [Google Scholar]
- 36. Sun Y., Liu Z., Cao X., Lu Y., Mi Z., He C., Liu J., Zheng Z., Li M.J., Li T. et al.. Activation of P-TEFb by cAMP-PKA signaling in autosomal dominant polycystic kidney disease. Sci. Adv. 2019; 5:eaaw3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ozturk-Kaloglu D., Hercher D., Heher P., Posa-Markaryan K., Sperger S., Zimmermann A., Wolbank S., Redl H., Hacobian A.. A noninvasive in vitro monitoring system reporting skeletal muscle differentiation. Tissue Eng. Part C Methods. 2017; 23:1–11. [DOI] [PubMed] [Google Scholar]
- 38. Berkes C.A., Tapscott S.J.. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005; 16:585–595. [DOI] [PubMed] [Google Scholar]
- 39. Sartorelli V., Puri P.L.. Shaping gene expression by landscaping chromatin architecture: lessons from a master. Mol. Cell. 2018; 71:375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verrier L., Escaffit F., Chailleux C., Trouche D., Vandromme M.. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 2011; 7:e1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yi X., Tao Y., Lin X., Dai Y., Yang T., Yue X., Jiang X., Li X., Jiang D.S., Andrade K.C. et al.. Histone methyltransferase Setd2 is critical for the proliferation and differentiation of myoblasts. Biochim. Biophys. Acta Mol. Cell Res. 2017; 1864:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eom G.H., Kim K.B., Kim J.H., Kim J.Y., Kim J.R., Kee H.J., Kim D.W., Choe N., Park H.J., Son H.J. et al.. Histone methyltransferase SETD3 regulates muscle differentiation. J. Biol. Chem. 2011; 286:34733–34742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhuang L., Jang Y., Park Y.K., Lee J.E., Jain S., Froimchuk E., Broun A., Liu C., Gavrilova O., Ge K.. Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat. Commun. 2018; 9:1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castiglioni I., Caccia R., Garcia-Manteiga J.M., Ferri G., Caretti G., Molineris I., Nishioka K., Gabellini D.. The Trithorax protein Ash1L promotes myoblast fusion by activating Cdon expression. Nat. Commun. 2018; 9:5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alfert A., Moreno N., Kerl K.. The BAF complex in development and disease. Epigenet. Chromatin. 2019; 12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hargreaves D.C., Crabtree G.R.. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011; 21:396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tesina P., Cermakova K., Horejsi M., Prochazkova K., Fabry M., Sharma S., Christ F., Demeulemeester J., Debyser Z., Rijck J. et al.. Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif. Nat. Commun. 2015; 6:7968. [DOI] [PubMed] [Google Scholar]
- 48. Blokken J., De Rijck J., Christ F., Debyser Z.. Protein-protein and protein-chromatin interactions of LEDGF/p75 as novel drug targets. Drug Discov. Today Technol. 2017; 24:25–31. [DOI] [PubMed] [Google Scholar]
- 49. Ninkina N.N., Mertsalov I.B., Kulikova D.A., Alimova-Kost M.V., Simonova O.B., Korochkin L.I., Kiselev S.L., Buchman V.L.. Cerd4, third member of the d4 gene family: expression and organization of genomic locus. Mamm. Genome. 2001; 12:862–866. [DOI] [PubMed] [Google Scholar]
- 50. Lange M., Kaynak B., Forster U.B., Tonjes M., Fischer J.J., Grimm C., Schlesinger J., Just S., Dunkel I., Krueger T. et al.. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008; 22:2370–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zeng L., Zhang Q., Li S., Plotnikov A.N., Walsh M.J., Zhou M.M.. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010; 466:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li J., Ahn J.H., Wang G.G.. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell. Mol. Life Sci. 2019; 76:2899–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wagner E.J., Carpenter P.B.. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012; 13:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang H., Shun M.C., Dickson A.K., Engelman A.N.. Embryonic lethality due to arrested cardiac development in Psip1/Hdgfrp2 Double-Deficient mice. PLoS One. 2015; 10:e0137797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen W., Xu C., Huang W., Zhang J., Carlson J.E., Tu X., Wu J., Shi Y.. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007; 46:2100–2110. [DOI] [PubMed] [Google Scholar]
- 56. Hassan A.H., Prochasson P., Neely K.E., Galasinski S.C., Chandy M., Carrozza M.J., Workman J.L.. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002; 111:369–379. [DOI] [PubMed] [Google Scholar]
- 57. Agalioti T., Chen G., Thanos D.. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002; 111:381–392. [DOI] [PubMed] [Google Scholar]
- 58. Local A., Huang H., Albuquerque C.P., Singh N., Lee A.Y., Wang W., Wang C., Hsia J.E., Shiau A.K., Ge K. et al.. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 2018; 50:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al.. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kuo A.J., Cheung P., Chen K., Zee B.M., Kioi M., Lauring J., Xi Y., Park B.H., Shi X., Garcia B.A. et al.. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell. 2011; 44:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de la Serna I.L., Ohkawa Y., Berkes C.A., Bergstrom D.A., Dacwag C.S., Tapscott S.J., Imbalzano A.N.. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 2005; 25:3997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Forcales S.V., Albini S., Giordani L., Malecova B., Cignolo L., Chernov A., Coutinho P., Saccone V., Consalvi S., Williams R. et al.. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 2012; 31:301–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pawson T., Scott J.D.. Protein phosphorylation in signaling–50 years and counting. Trends Biochem. Sci. 2005; 30:286–290. [DOI] [PubMed] [Google Scholar]
- 64. Cui H., Schlesinger J., Schoenhals S., Tonjes M., Dunkel I., Meierhofer D., Cano E., Schulz K., Berger M.F., Haack T. et al.. Phosphorylation of the chromatin remodeling factor DPF3a induces cardiac hypertrophy through releasing HEY repressors from DNA. Nucleic Acids Res. 2016; 44:2538–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq, ChIP-seq and ATAC-seq data can be found at the Gene Expression Omnibus database under accession numbers GSE141407.