Abstract
The universal L-shaped tertiary structure of tRNAs is maintained with the help of nucleotide modifications within the D- and T-loops, and these modifications are most extensive within hyperthermophilic species. The obligate-commensal Nanoarchaeum equitans and its phylogenetically-distinct host Ignicoccus hospitalis grow physically coupled under identical hyperthermic conditions. We report here two fundamentally different routes by which these archaea modify the key conserved nucleotide U54 within their tRNA T-loops. In N. equitans, this nucleotide is methylated by the S-adenosylmethionine-dependent enzyme NEQ053 to form m5U54, and a recombinant version of this enzyme maintains specificity for U54 in Escherichia coli. In N. equitans, m5U54 is subsequently thiolated to form m5s2U54. In contrast, I. hospitalis isomerizes U54 to pseudouridine prior to methylating its N1-position and thiolating the O4-position of the nucleobase to form the previously uncharacterized nucleotide m1s4Ψ. The methyl and thiol groups in m1s4Ψ and m5s2U are presented within the T-loop in a spatially identical manner that stabilizes the 3′-endo-anti conformation of nucleotide-54, facilitating stacking onto adjacent nucleotides and reverse-Hoogsteen pairing with nucleotide m1A58. Thus, two distinct structurally-equivalent solutions have evolved independently and convergently to maintain the tertiary fold of tRNAs under extreme hyperthermic conditions.
INTRODUCTION
Stable tRNA and rRNA molecules function effectively to translate the genetic code into proteins only after a variety of posttranscriptional modifications have been added to their structures (1–5). Some RNA modifications are specific to certain groups of organisms, whereas others are highly conserved throughout the three domains of life. One such conserved modification is the 5-methyluridine at position 54 (m5U54) in the T-loop of tRNAs. This nucleotide is universally conserved and is methylated in most members of the Bacteria and Eukarya (2,3) with only a few exceptions that include some members of the Mycoplasmatales (6–8). In archaeal tRNAs, m5U54 has only been observed in the Thermococcales group (9), where it is typically modified further by thiolation to form m5s2U (10,11), as is also seen in some thermophilic bacteria (12–15). Uridine-54 is one of several nucleotides that are modified to facilitate the folding and stabilization of the core L-shaped conformation of tRNA (16,17). The density of such modifications is generally highest in thermophilic organisms that must maintain the structural integrity of their tRNAs under demanding environmental conditions (16,18,19).
In most organisms, the m5U54 modification is catalyzed by an enzyme belonging to COG2265 (20), a family of pyrimidine methyltransferases that use S-adenosyl-L-methionine (AdoMet) as the methyl group donor. These m5U54 methyltransferases are present in all three domains of life (21), and are exemplified by the bacterial homolog TrmA of Escherichia coli (22–24), the eukaryotic enzyme Trm2p seen in Saccharomyces cerevisiae (25), and the euryarchaeota homolog PAB0719 (TrmU54) from Pyrococcus abyssi (9,26). The importance of U54 methylation is underlined by the existence of an alternative and phylogenetically-unrelated mechanism directed by the COG1206 flavoprotein TrmFO (27–30), which catalyzes m5U54 formation using N5,N10-methylenetetrahydrofolate as the methyl group donor (31).
In hyperthermophilic organisms where m5U54 is modified further to form m5s2U, the bulky sulphur atom constrains the ribose pucker in the 3′-endo configuration to avoid a steric clash with the 2′-hydroxyl (32) favouring the anti-orientation of the U54 base. This in turn promotes U54 stacking onto adjacent nucleotides and its reverse-Hoogsteen pairing with A58 within the T-loop (33), the net effect of which is to increase the thermostability of the tRNA (14,32,34,35). However, nucleotide m5s2U is absent in the tRNAs of hyperthermophilic archaea of the Crenarchaeota phylum (10), and these organisms modified uridine-54 via an alternative route to form m1Ψ (36). Nucleotide m1Ψ54 engages in similar interactions within the T-loop, including a reverse-Hoogsteen pairing with A58 (37). The extent to which the different U54 modifications in Crenarchaeota and the Thermococcales branch of the Euryarchaeota are physiologically equivalent has not been previously known, and this question is addressed in the present study.
The hyperthermophilic archaea Ignicoccus hospitalis and Nanoarchaeum equitans grow physically attached as a commensal pair under identical environmental conditions (38). I. hospitalis is a member of the Crenarchaeota, whereas N. equitans is closer to the Euryarchaeota (39,40) and has more recently been placed in the Nanoarchaeota branch of the DPANN superphylum (41). I. hospitalis has a genome encoding almost 1500 open reading frames (ORFs) (42) and grows perfectly well as a pure culture, whereas N. equitans has a considerably smaller genome of 552 ORFs (43), and proliferates only when physically attached to its I. hospitalis host (38,39). The genomes of these two archaea encode different complements of RNA modification enzymes. Here we use liquid chromatography with mass spectrometry to map the modifications at and around nucleotide U54 to determine how the two archaea stabilize their tRNA tertiary structures. We show that despite the coupled growth of N. equitans and I. hospitalis under identical hyperthermic conditions, the two archaea modify their tRNAs in distinctly different ways. While N. equitans modifies U54 to form m5s2U, I. hospitalis synthesizes the previously uncharacterized nucleotide m1s4Ψ54, which stabilizes the tRNA T-loop in a structurally equivalent manner.
MATERIALS and METHODS
In silico screening of the I. hospitalis and N. equitans genomes
Previously characterized m5U RNA methyltransferases were used as BLAST queries (44) against open reading frames (ORFs) in I. hospitalis and N. equitans. These included the AdoMet-dependent RlmD (UniProt accession number P55135) and TrmA (P23003) methyltransferases from E. coli K12, the P. abyssi methyltransferases PAB0719 (Q9UZR7) and PAB0760 (Q9UZK1), and the folate-dependent enzymes TrmFO (P39815) from Bacillus subtilis, and RlmFO (Q2SS13) from Mycoplasma capricolum (45).
The I. hospitalis and N. equitans genomes were also screened for homologs of TtuA and TtuB that direct thiolation of m5U54 (46), and the Pus10/TrmY combination that produces m1Ψ54 (47). BLAST queries included the P. abyssi and Pyrococcus furiosus homologs PAB1092 and PF0273 (TtuA), PF1758 (TtuB), PAB2391 and PF1139 (Pus10) and PAB1866 (TrmY).
Growth of I. hospitalis/N. equitans
I. hospitalis was grown on its own in pure culture and in co-culture with N. equitans as previously described (39). Cells were harvested at late log phase by centrifugation, and N. equitans cells were enriched from the co-cultures by differential centrifugation (39). Cells were run twice through a French press, followed by addition of 4 ml of TRIzol® (Life Technologies). After 5 min at 20°C, 0.8 ml chloroform was added, and the samples were centrifuged at 12,000 g for 5 min. The aqueous phase was extracted with phenol/chloroform, before collecting the nucleic acids by ethanol precipitation.
Analyses of tRNAs nucleosides
The total RNA mixtures from I. hospitalis (pure culture) or N. equitans (enriched from co-cultures) were extracted with Nucleobond® RNA/DNA 400 kits (Macherey-Nagel) to isolate the tRNA fractions. The supernatant fractions containing tRNAs and other soluble RNAs were passed through Microcon YM-100 columns (Millipore) to reduce the amount of impurities. The supernatant samples were digested to completion to form nucleosides (48) and analyzed using an Agilent Technologies 1100 HPLC with a Hypercarb column (150 mm × 0.3 mm) containing 5 μm PGC pore size of 250 Å (Thermo Scientific) and run at 0.5 μl/min. Nucleosides were separated using gradients of 0–90% acetonitrile in 0.1% formic acid and were detected by absorbance at 260 nm.
Nucleoside structures were subsequently determined by liquid chromatography (LC) linked with electrospray ionization (ESI) quadrupole time of flight mass spectrometry (qToF-MS) using Agilent 6530B equipment. Total tRNA nucleosides, prepared as above, were lyophilized, resuspended in 15 μl 0.1% formic acid and centrifuged at 16,000 g for 5 min. The supernatant, in 4 μl portions, was injected into an Agilent Technologies 1290 Infinity HPLC system equipped with an Agilent Zorbax Eclipse Plus C18 column (2.1 × 150 mm, 1.8 μm) and a 50 mm guard-column at 40°C. A chromatographic gradient was formed from 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) by increasing the proportion of solvent B from 3 to 95% over 10 min at a flow rate of 400 μl/min. Elutants were analyzed by qToF-MS scanning three time per second from m/z 50 to m/z 1050 at a gas temperature of 325°C with drying gas at 8 l/min, nebulizer at 35 psig, sheath gas temp 350°C, sheath gas flow at 11 min/l, VCap at 3500 V, fragmentor at 125 V and skimmer at 65 V. Spectra were calibrated from the signals of purine and Hexakis (1H,1H,3H-tetrafluoropropoxy) phosphazine delivered through a second needle in the ion source by an isocratic pump (flow rate 20 μl/min). Nucleosides were compared to chemically synthesized standards (Um, m5U, s2U, s4U, m5s2U, Ψ, s2Ψ, m1Ψ, m3Ψ and m1m3Ψ). Tandem MS fragmentation (MS/MS) was carried out in positive and negative ion modes at collision energies of 35 V and 15V, respectively. In order to promote fragmentation of s2U and s4U for comparison to compound #8, the fragmentor voltage was increased to 190V before selecting the protonated nucleobases for MS/MS (pseudo-MS3). Ion chromatograms were extracted using 5 ppm mass tolerance, and spectra were analyzed using Masshunter Qualitative Analysis B.08.00 (Agilent). Oligonucleotide masses and isotope distribution were calculated using the software at https://www.envipat.eawag.ch/.
In the experiments where recombinant NEQ053 was expressed in E. coli, tRNA nucleoside masses and fragmentation patterns were determined after LC separation by a different analytical set-up using ion-trap ESI-MS, as previously reported (48).
Expression and purification of recombinant NEQ053
The NEQ053 open reading frame was amplified by PCR using N. equitans chromosomal DNA as template. This region of the N. equitans chromosome encodes an ATG sequence 27 codons upstream and in-frame with a second ATG and, by alignment with pyrococcal and other homologs, we judged the second ATG to be the true start of the NEQ053 ORF. Our PCR product, extending from this second ATG, was inserted into the E. coli expression vector pQE-80L (QIAGEN) to encode a protein with an N-terminal tag of six histidine residues. Purification of the NEQ053 enzyme to near homogeneity was achieved by chromatography on Ni-NTA resin (QIAGEN) and a HiTrap™ Heparin column (GE Healthcare) (9). Enzyme fractions were pooled and run over a PD10 column (GE Healthcare) equilibrated with 25 mM Tris-HCl pH 7.5, 300 mM NaCl, 10% glycerol and 2 mM DTT. The NEQ053 enzyme was stored in aliquots at –80°C. The methyltransferase function of the recombinant NEQ053 was also tested in vivo in E. coli after expressing from a plasmid (49) in an rlmC, rlmD, trmA null-strain lacking all endogenous m5U RNA modifications (50).
In vitro transcription and methylation of N. equitans tRNAThr
A double-stranded DNA template for N. equitans tRNAThr (anticodon GGU) was produced by PCR using the Vent DNA polymerase (New England Biolabs) and two oligodeoxynucleotides with overlapping ends where one of these primers contained the T7 promoter sequence (51). The tRNAThr transcript was purified on a denaturing gel (52), and 500 nM was incubated with 20 nM of recombinant NEQ053 methyltransferase for 15 min at 80°C in 25 μl methylation buffer consisting of 25 mM Tris–Cl, pH 7.5, 50 mM ammonium acetate, 2 mM dithiotreitol, 10 mM MgCl2, 0.1 mg/ml RNase-free bovine serum albumin (Roche Applied Science) and 80 μM AdoMet (Sigma). Reactions were stopped by extracting with phenol/chloroform and the tRNA was recovered by ethanol precipitation.
Analysis of rRNA and tRNA sequences by MALDI-ToF
In vitro methylation by NEQ053 in the N. equitans tRNAThr was analyzed directly by MALDI-ToF after digestion with RNase A or T1. MALDI analyses of the much larger rRNAs required prior isolation of fragments of approximately 50 nucleotides using DNA oligonucleotides complementary to the I. hospitalis and N. equitans 23S rRNA sequences 720–770, 1890–1945 and 1910–1960. DNA oligos at 1 nmol were mixed in 62.5 mM HEPES, pH 7.0 and 125 mM KCl with 300 pmol of total RNA from pure cultures of I. hospitalis or N. equitans enriched from co-culture and were heated at 90°C for 5 min followed by slow cooling to 37°C. The same rRNA regions were isolated from the E. coli rlmC, rlmD, trmA null-strain recombinants (50,53). The oligonucleotide-rRNA hybrids were digested with RNase A and mung bean nuclease, and protected rRNA fragments of approximately the same length as the oligonucleotides were isolated on gels (54,55). RNase cleavage at any mismatched sites in the rRNA-oligonucleotide hybrids effectively separated any I. hospitalis rRNAs impurities in the N. equitans samples (49). Sites of modification in the rRNA fragments and in tRNA transcripts were identified by digesting with RNase A or T1 and analyzing the resultant oligonucleotides using MALDI-TOF mass spectrometry (UltrafleXtreme, Bruker Daltonics) collecting data in positive ion mode (54,55).
Primer extension
Primer extension was used to investigate sites of 2′-O-methylation in N. equitans 23S rRNA that had been indicated by MALDI-MS. 3 pmol Cy5-labelled DNA primers of 18–20 nucleotides were hybridized to complementary sequences within N. equitans 23S rRNA (4 pmol) by incubating at 80°C for 2 min in 4.5 μl of 56 mM HEPES pH 7.0 and 112 mM KCl followed by slow cooling to 42°C. Extension and sequencing reactions were performed with 1.5 U AMV reverse transcriptase (Thermo Scientific) per reaction (56) and analyzed on 13% polyacrylamide-urea gels. Bands were visualized on a Typhoon FLA 9500 from GE Healthcare.
RESULTS
Screening for candidate m5U methyltransferases
In silico analyses of the genome sequences showed that NEQ053 from N. equitans was the only ORF to score an E-value that is compatible with an RNA m5U methyltransferase. In comparison with methyltransferase homologs for which the RNA targets have already been empirically characterized, the best score for NEQ053 was against P. abyssi PAB0719 (TrmU54) with an E-value of 10−101, while the paralogous enzyme PAB0760, which is specific for U747 in 23S rRNA, gave an E-value of 10−80. However, a different pattern was seen when compared with bacterial enzymes, where E. coli RlmD specific for U1939 in 23S rRNA was more similar to NEQ053 (E-value 10−35) than its U54-specific counterpart TrmA (E-value 10−15). No ORF with significant similarity to the folate-dependent m5U methyltransferases RrmFO or TrmFO was detected. No gene encoding a homolog of any previously characterized RNA m5U methyltransferase was found in the I. hospitalis genome.
Verification of tRNA m5U by HPLC
The bioinformatics findings were tested by assaying for m5U in the archaeal tRNAs. Nucleosides hydrolysates were generated from bulk tRNAs isolated from pure cultures of I. hospitalis and from co-cultures after enrichment of N. equitans, and were analyzed by HPLC (Figure 1). Consistent with the absence of an RNA m5U methyltransferase, no m5U was present in nucleosides isolated from pure cultures of I. hospitalis cells. In the N. equitans tRNA nucleosides, m5U was present (Figure 1C) but constituted only about 3% of the total amount of uridine. Based on their average uridine content, this indicates that approximately one quarter of N. equitans tRNAs contained this single modification.
Figure 1.
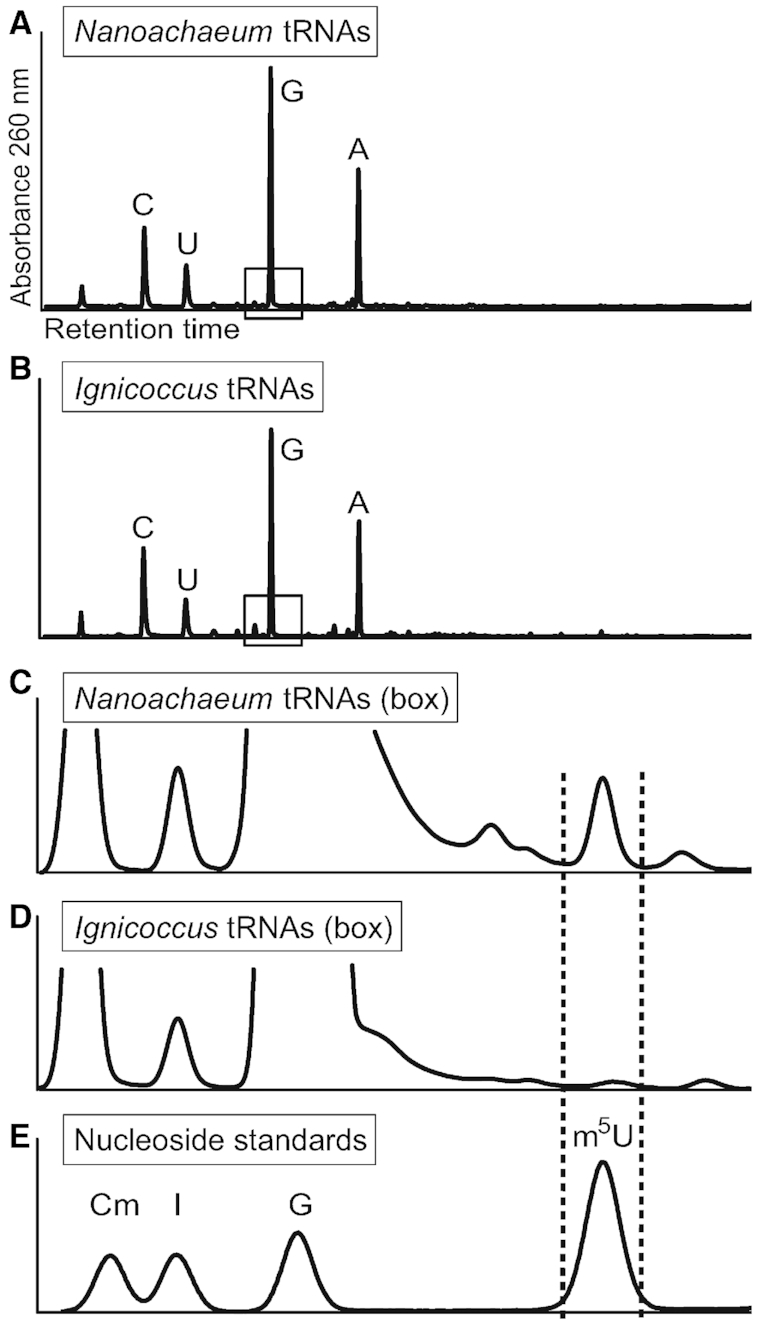
HPLC profiles of tRNA nucleosides from Nanoarchaeum and Ignicoccus cells. (A) Nucleosides from the enriched Nanoarchaeum culture, and (B) from Ignicoccus cultured alone. (C) Enlargement of the boxed region in panel A containing m5U, and (D) the same region from panel B showing the absence of m5U. (E) Nucleoside standardization mixture, indicating the fraction corresponding to the retention time for m5U.
NEQ053 is responsible for tRNA m5U54 modification
An in vitro system with a recombinant version of NEQ053 and an unmodified transcript of N. equitans tRNAThr was used to establish whether this enzyme was responsible for adding m5U and to identify the location of the modification in the tRNA structure. This tRNA substrate generates RNase digestion products that are easily distinguishable by MALDI-MS. After incubation with NEQ053 and digestion with RNase A, a spectral shift occurred in the unique fragment GGGUp containing nucleotide U54 due to a mass increase of 14.0 Da (Figure 2). This mass change is consistent with substitution of a hydrogen atom with a methyl group. The same increase in the overlapping RNase T1 fragment UUCGp established the exact location of the methylation at nucleotide U54 and confirmed that modification here was stoichiometric.
Figure 2.
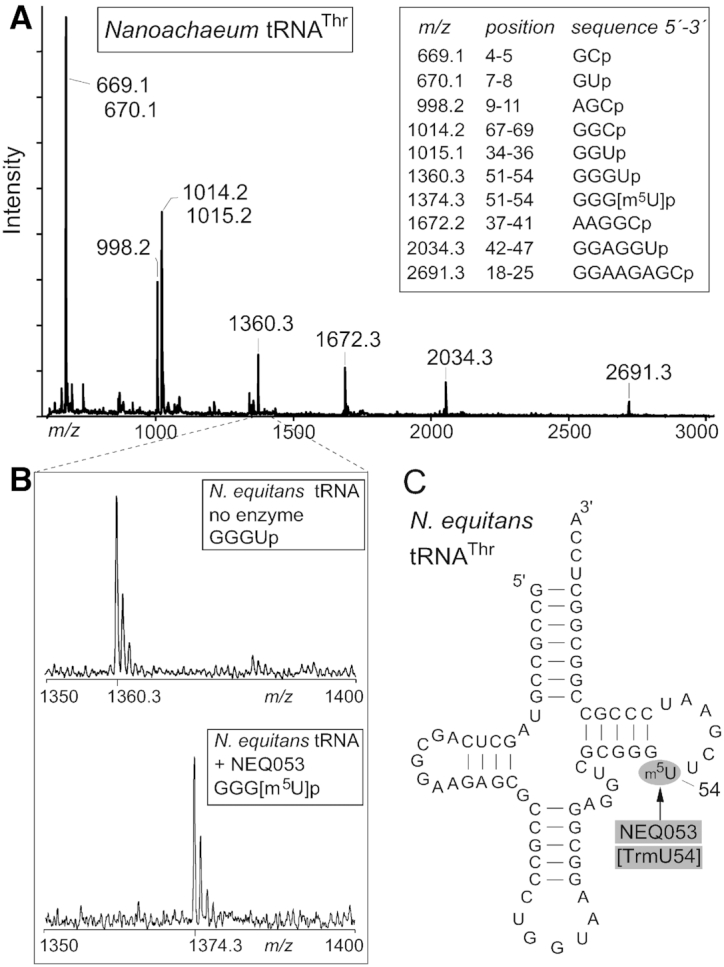
MALDI-MS spectra showing methylation in vitro by NEQ053 at U54 in N. equitans tRNAThr. (A) Spectrum of oligonucleotides formed by RNase A digestion of unmodified tRNAThr. The measured mass/charge (m/z) of each fragment is shown above the peak with the theoretical m/z values (in box). (B) Enlargement of the spectral region containing the GGGUp fragment from nucleotides 51–54 with an m/z of 1360.3 when derived from the unmodified tRNAThr, and with m/z of 1374.3 after incubation of the tRNA in vitro with NEQ053 prior to digestion. (C) Schematic of the N. equitans tRNAThr secondary structure. The target site of NEQ053 indicates that the enzyme could be renamed as TrmU54.
Expression of NEQ053 in E. coli and screening for additional m5U targets
An E. coli strain in which rlmC, rlmD and trmA had been inactivated, and thus lacked m5U modifications in its rRNAs and tRNAs, was used as a host for expressing the recombinant NEQ053. HPLC analysis of bulk tRNAs showed that NEQ053 added m5U to the tRNAs within the E. coli null-strain (Supplementary Figure S1). Generation of a nucleoside with a mass corresponding to methylated uridine was confirmed by ESI-MS (Supplementary Figure S1), and its collision-induced fragmentation generated a pattern of masses that unambiguously matched an m5U standard (Supplementary Figure S1C), confirming the site of methyl group addition on the C5-atom of the uracil base.
Isolation of the nucleotide regions around E. coli 23S rRNA U747 and U1939 and their analysis by MALDI-MS showed that there was no rescue of modification at these other nucleotide targets by NEQ053 (not shown). Similar analyses also ruled out the presence of m5U modification at the equivalent locations in N. equitans 23S rRNA (Supplementary Figure S2). In the course of these studies, numerous sites of 2′-O-methylation within the N. equitans 23S rRNA structures around nucleotide 1939 were revealed by MS and verified by primer extension (Supplementary Figure S2).
In silico screening for supplementary enzymes modifying U54
The relatively low amount of m5U in tRNAs extracted from N. equitans cells (Figure 1) was clearly inconsistent with the activity of this enzyme when tested in vitro (Figure 2) and in vivo in the heterologous E. coli host (Supplementary Figure S1). We reasoned that the low yield of m5U in N. equitans cells could indicate that it represented a precursor that would subsequently be converted into a hypermodified form. In silico screening of the archaeal genomes for ORFs encoding additional uridine modification enzymes revealed homologs of TtuA and TtuB that thiolate the 2-position of m5U in tRNAs. The N. equitans ORF NEQ283 showed high similarity to PF0273, a TtuA homolog from P. furiosus (E-value = 10−53), while NEQ523 displayed close resemblance to the TtuB homolog PF1758 (E-value = 10−73).
A database search for enzymes that might modify U54 in I. hospitalis tRNAs revealed Igni_0342, a homolog of the pyrococcal enzyme Pus10 that isomerizes this nucleotide to Ψ (E-values of 10−77 against PAB2391 and PF1139). In a subsequent modification step seen in many archaeal species, the methyltransferase TrmY converts the pseudouridine to m1Ψ. A likely candidate for catalyzing this reaction was the I. hospitalis protein Igni_0291, which shows an E-value of 10−27 against the P. abyssi TrmY homolog PAB1866. No homologs of Pus10 or TrmY were found in N. equitans.
Database searches also showed that the I. hospitalis genome encodes orthologs of the two putative U54 thiolation enzymes, TtuA and TtuB, where Igni_0707 resembles NEQ283 (E-value = 10−31) and Igni_0506 is similar to NEQ523 (E-value = 10−42). In the absence of the modification pathway through m5U, this raised the possibility that these I. hospitalis enzymes might thiolate m1Ψ54 forming a previously unseen hypermodified version of this nucleotide.
Uridine hypermodification in the archaeal tRNAs
The putative functions of the enzymes predicted by the bioinformatics searches described above were tested by fractionating nucleosides from the N. equitans and I. hospitalis tRNAs by liquid chromatography and defining their masses using ESI-MS. Nucleoside structures were determined from their collision-induced fragmentation and comparison of the fragment patterns with those of known uridine derivatives.
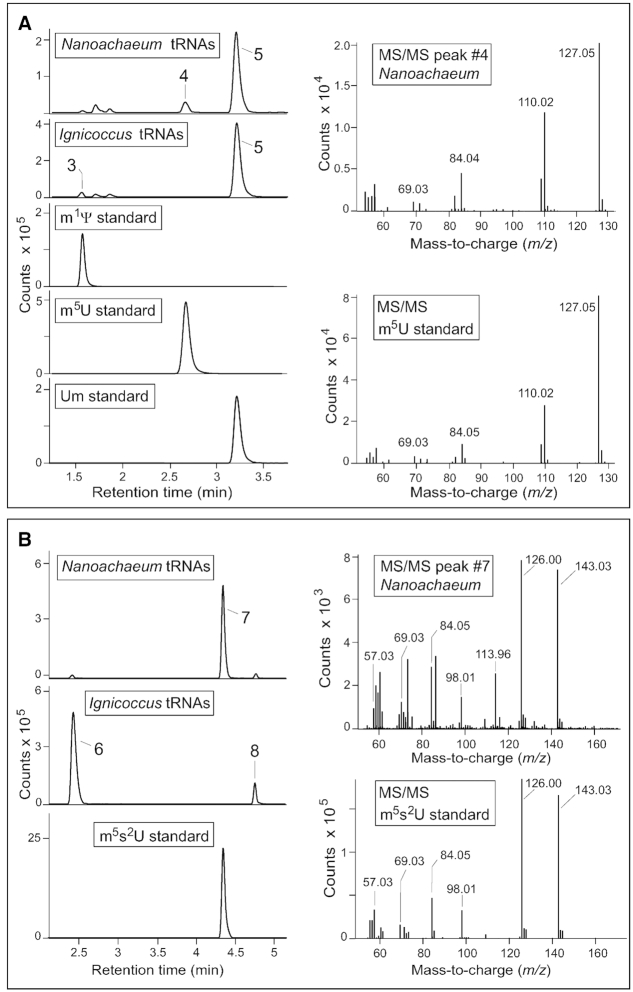
Eleven distinct derivatives of uridine were isolated from the two sets of archaeal tRNAs (Table 1). With a couple of notable exceptions, these matched the retention times, masses and fragmentation patterns of known standards, thus making identification straightforward and unequivocal. Unmodified uridine was the predominant component and eluted immediately after pseudouridine on the LC; both nucleotides (compounds #1 and #2, Table 1) have the same monoisotopic mass of 244.07 Da (m/z 245.08 in positive ion mode; monoisotopic masses are used throughout). Three nucleosides of mass 258.08 Da (m/z 259.09 in positive ion mode) were identified as uridines with a single methyl group. The first of these to elute, m1Ψ (compound #3), was in small amounts and specific to I. hospitalis; the next, m5U (compound #4), was also in small amounts and specific to N. equitans; while the third nucleoside with this mass, Um (compound #5), gave a prominent peak for both organisms (Figure 3A and Supplementary Figure S3).
Table 1.
Uridine nucleoside derivatives identified in the tRNAs of N. equitans and I. hospitalis
| Compound notation and retention time (RT in min) | Archaeon | m/z (negative/positive ion modes) | Molecular composition | Nucleoside | Nucleoside standards for comparison | Notes | |
|---|---|---|---|---|---|---|---|
| #1 | 1.2 | I. hospitalis & N. equitans | 243.062/245.077 | C9H12N2O6 | Ψ | Ψ | a |
| #2 | 1.5 | I. hospitalis & N. equitans | 243.062/245.077 | C9H12N2O6 | U | U | a |
| #3 | 1.5 | I. hospitalis | 257.078/259.093 | C10H14N2O6 | m1Ψ | m1Ψ | a,b |
| #4 | 2.6 | N. equitans | 257.078/259.093 | C10H14N2O6 | m5U | m5U | a,b |
| #5 | 3.2 | I. hospitalis & N. equitans | 257.078/259.093 | C10H14N2O6 | Um | Um | a |
| #6 | 2.5 | I. hospitalis | 273.055/275.070 | C10H14N2O5S | m1s4Ψ | Ψ, m1Ψ, s2Ψ | c |
| #7 | 4.4 | N. equitans | 273.055/275.070 | C10H14N2O5S | m5s2U | m5s2U | a |
| #8 | 4.8 | I. hospitalis | 273.055/275.070 | C10H14N2O5S | s2Um | m5s2U, s2U, s4U, Um | c |
| #9 | 1.6 | N. equitans | 259.039/261.054 | C9H12N2O5S | s4Ψ | Ψ, s2Ψ | b,c |
| #10 | 3.0 | I. hospitalis & N. equitans | 259.039/261.054 | C9H12N2O5S | s2U | s2U | a |
| #11 | 3.7 | I. hospitalis & N. equitans | 259.039/261.054 | C9H12N2O5S | s4U | s4U | a |
| #12 | 3.0 | I. hospitalis | 271.094/273.108 | C11H16N2O6 | m1Ψm | m1Ψ, m3Ψ, m1U | c |
aStructures confirmed by mass determination, chromatographic retention time (RT) and tandem MS (MS/MS) fragmentation analyses of compound and comparison with identical chemically synthesized standard.
bPresent in small amounts compared to the other modified nucleosides.
cNo identical synthetic standard available. Structures deduced from mass determination, isotope distribution, tandem MS analyses and comparison with related (nonidentical) standards to identify and exclude known nucleoside fragments.
Figure 3.
LC/ESI-MS characterization of uridine nucleosides from N. equitans and I. hospitalis tRNAs. (A) Analyses of uridines modified with one methyl group and registered at m/z 259.09 in positive ion mode. On left, retention times during liquid chromatography compared to known methyluridine standards. On right, fragmentation of N. equitans compound #4 confirming its structure from comparison with the m5U standard. The identities of compound #3 (m1Ψ) and compound #5 (Um) were verified by the same methods (Supplementary Figure S3). Minor amounts of non-nucleoside compounds can be seen migrating at retention times close to that of m1Ψ. (B) Uridines modified with a methyl plus a thiol group registered at m/z 275.07 in positive ion mode. Compound #7 is specific to N. equitans and matches the LC profile of the m5s2U standard as well as its MS/MS fragmentation pattern (right). No standards with the same LC migration were available for compounds #6 and #8, and these were identified respectively as m1s4Ψ and s2Um by comparing their fragmentation patterns to a series of nucleosides containing these individual modifications (Figures 4 and 5). Small quantities of these compounds were observed in some preparations of the enriched N. equitans cells (as seen here) and are contaminants resulting from incomplete removal of all the I. hospitalis cells.
Three nucleosides were observed at m/z 275.07, which fits the mass of a methylated uridine with a thiol group (Figure 3B). One of these (compound #7, Table 1) was specific to N. equitans and was unambiguously identified as m5s2U by fragmentation and comparison to a chemically synthesized standard (Figure 3B).
The other two nucleosides (#6 and #8) originated in I. hospitalis and have the same mass as m5s2U (within the accuracy of the instrumentation used here), but with LC retention times that indicate distinctly different structures. Our spectrometric set-up has a mass accuracy better than 5 ppm and this, together with analysis of isotope patterns, is a strong predictor of atomic composition. The five top candidates that match the observed mass/isotope pattern of compound #6 are listed in Supplementary Table S1. From these, the atomic composition C10N2O5SH14 is the best candidate, and fits with the structure of a methylated and thiolated uridine/pseudouridine. The second-best candidate (C11N6OSH10) is very close in score and also in its predicted isotope distribution (Supplementary Figure S4), however, this composition has only a single oxygen making it incompatible with a nucleoside. Apart from m5s2U, no standards with the composition C10N2O5SH14 are available for direct experimental comparison, and therefore the structure of compound #6 was deduced by piecing together distinguishing fragments (and ruling out others) from the set of uridine derivatives in Table 1.
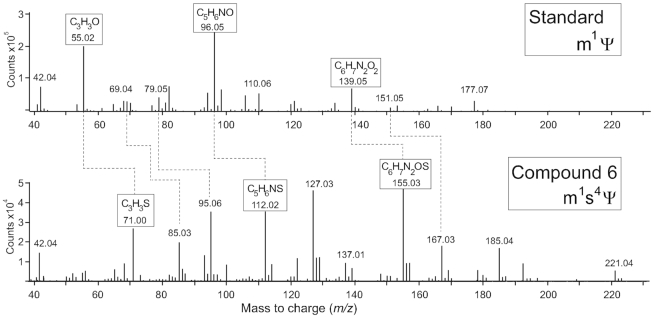
First, it was established that compound #6 is a derivative of pseudouridine. The main positive-ion fragmentation pathway for all nucleosides, except pseudouridine and its derivates, is loss of the sugar moiety. Compound #6 has a stable glycosidic bond (Figure 4) and produces no fragments corresponding to loss of ribose- or methylated ribose-derivatives (theoretically, at m/z 129.01 and 143.03, respectively). Further, there is significant overlap with the m1Ψ fragmentation pattern, indicating that compound #6 is methylated on the N1-position. In addition, some of these fragments show distinctive mass shifts corresponding to the substitution of an oxygen atom with sulphur (Figure 4). Finally, the position of this thiol group at the 4-carbonyl rather than the 2-carbonyl position was deduced from similarities and differences to aspects of s2Ψ and m1Ψ negative ion-mode fragmentation (Figure 5). These fragmentation data, together with the atomic composition derived from the accurate mass and the isotope distribution (Table S1 and Supplementary Figure S4), indicate that compound #6 is m1s4Ψ.
Figure 4.
Tandem MS in positive ion mode of compound #6 comparing with the fragmentation pattern of m1Ψ. Key fragments are indicated in boxes, and are seen to increase in mass by 16 Da showing O- to S- substitution. The fragmentation pattern is consistent with compound #6 being m1s4Ψ. The positions of the methyl group at N1 and the sulphur on C4 of the pseudouridine were conclusively verified by fragmentation in the negative ion mode and comparison with additional standards (Figure 5).
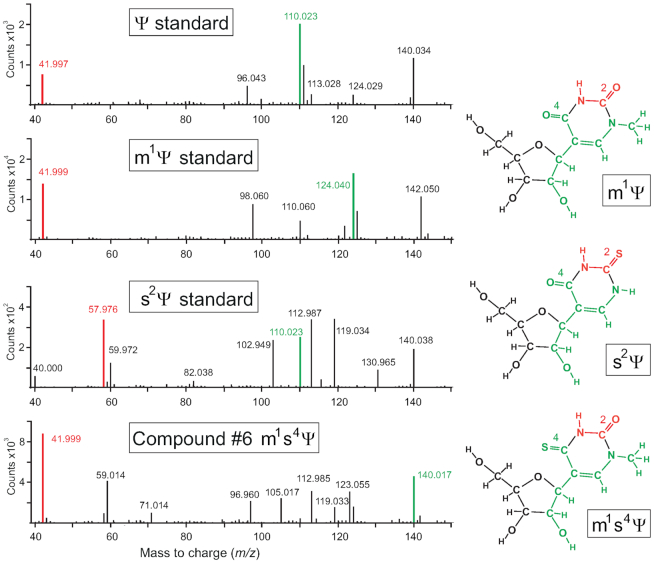
Figure 5.
Tandem MS spectrum of nucleoside compound #6 in negative ion mode, comparing the measured m/z values of its fragmentation pattern to those of the Ψ, m1Ψ and s2Ψ standards. The fragment containing the uracil C2 and N3 atoms (red) has a theoretical m/z of 41.999 in negative ion mode when containing a C2-carbonyl group (the values measured here were m/z 41.997 for Ψ, and m/z 41.999 for m1Ψ and compound #6). This fragment shifts to m/z 57.976 when C2 is thiolated (as for s2Ψ, and not for compound #6). The fragment (in green) spanning from the C4-carbonyl and the N1 atom through the glycosidic bond to the 2′-hydroxyl is seen at m/z 110.023 when unmodified (e.g. for Ψ and s2Ψ); this fragment increases to m/z 124.040 when there is an N1-methyl group (as in m1Ψ); and with a further increase to m/z 140.017 when the C4-position is thiolated (as in compound #6). With the instrumentation used here, the m/z measurement of 140.017 is of sufficient accuracy to distinguish the composition of this fragment as C6H7NOS, as opposed to the m/z 140.034 and 142.050 fragments, which fit the compositions C6H7NO3 and C6H9NO3, respectively. The data are fully consistent with compound #6 being m1s4Ψ.
The minor product compound #8 was identified through the same mass accuracy/isotope considerations used for compound #6, and additionally by comparison of its fragmentation pattern against s2U, s4U and Um standards (Supplementary Figure S5). The characteristic neutral loss of 146.06 Da, corresponding to a methylated ribose-derivative, and a base-fragmentation pattern identical to that of s2U, allow us to conclude that compound #8 is s2Um.
Three uridines with a single thiol and no methyl group (compounds #9, #10 and #11) were identified. Compounds #10 and #11 are present in both archaea and were identified as s2U and s4U, respectively, in a straightforward comparison with known standards (Supplementary Figure S5). Compound #9 was present only in N. equitans, and in low quantities. Here, no standard with the same mass and retention time was available. Fragment data indicate that the structure is most likely to be s4Ψ (Supplementary Figure S6), but with a lower degree of certainty than for the other nucleosides in Table 1.
Compound #12 was found only in I. hospitalis tRNAs (Table 1). The mass of this compound and the resilience of its glycosidic bond under fragmentation showed it to be dimethyl pseudouridine. Comparison with the m1m3Ψ standard ruled out this structure, while similarity to m1Ψ, with a 14 Da increase in the masses of fragments that include the 2′-ribose position, indicate that compound #12 is m1Ψm.
DISCUSSION
N. equitans grows physically attached to the surface of I. hospitalis, and the two archaea thus experience identical environmental conditions where temperatures can exceed 95°C. These organisms are, however, phylogenetically distinct with I. hospitalis belonging to the Crenarchaeota while N. equitans is closer to the Euryarchaeota (40) within the Nanoarchaeota phylum of the DPANN superphylum (41). Both organisms possess highly truncated genomes and while I. hospitalis is self-sufficient, N. equitans lacks many metabolic enzymes and compensates by scavenging small essential compounds from I. hospitalis over their fused cell membranes (38,39). It seems unlikely, however, that larger molecules such as RNA modification enzymes are transported between the two organisms (49), and raises the question of how their tRNA molecules are stabilized to enable them to survive and thrive together under the same extreme conditions.
The N. equitans enzyme NEQ053 was a clear candidate for an m5U methyltransferase with an E-value of 10−101 against PAB0719 that has previously been shown to add the m5U54 modification in P. abyssi tRNAs (9). However, in addition to tRNA-specific methyltransferases, the COG2265 group also contains the closely related enzymes RlmC and RlmD, which respectively add m5U modifications at U747 and U1939 in bacterial 23S rRNA (53,57). When compared with the bacterial enzymes, the NEQ053 sequence shows greater similarity to RlmD than its U54-specific counterpart TrmA, and is furthermore highly similar to the P. abyssi paralog PAB0760 that is responsible for m5U747 modification in 23S rRNA (51). Such ambiguity in bioinformatics-based prediction of pyrimidine C5-methylation targets has already been noted in previous studies, where empirical testing was required to identify the nucleotide target (51,58) and in one case revealed dual RNA modification sites (50). We therefore performed a set of experiments to find the main methylation target of NEQ053 and to see whether any additional sites were modified.
The location of the m5U at position 54 within the tRNA was established in vitro using a recombinant version of the NEQ053 enzyme with a transcript of the N. equitans tRNAThr as its substrate (Figure 2). Under the in vitro conditions used, methylation by the recombinant enzyme was stoichiometric (Figure 2C). Expressing the enzyme in an E. coli strain where rlmC, rlmD and trmA had been inactivated (50), and thus lacked m5U modifications in its rRNAs and tRNAs, led to m5U modification of the tRNAs (Supplementary Figure S1). Notably, this modification was introduced into E. coli tRNAs in vivo at 37 °C, despite this growth temperature being considerably below that at which NEQ053 operates within its natural host. This is reminiscent of a 16S rRNA-specific recombinant I. hospitalis methyltransferase that was previously shown to function in E. coli cells (49).
Analysis of the nucleotide regions around 23S rRNA U747 and U1939 by MALDI-MS (53) in the E.coli null strain showed that there was no rescue of these modifications by the recombinant NEQ053. Analysis of the equivalent positions within N. equitans 23S rRNA similarly showed that there was no m5U modification (Supplementary Figure S2). Taken together with the tRNA modification data, this suggests that the activity of the recombinant NEQ053 in the heterologous host reflects how the enzyme functions in N. equitans.
While ruling out m5U modifications in the N. equitans 23S rRNA, the MS analyses in combination with primer extension unearthed numerous sites of 2′-O-methylation within the structures around nucleotide 1939 (Supplementary Figure S2). Extensive 2′-O-methylation and a paucity of nucleobase modifications has previously been noted in the 3′-minor domain of N. equitans 16S rRNA and could represent a means of facilitating ribosomal subunit maturation and function (49). In the present case, this would suggest that the role of m5U1939 in 23S rRNA might also be substituted by other modifications at neighbouring nucleotides.
All these findings are consistent with the exclusive function of NEQ053 being to add the m5U54 modification in N. equitans tRNAs, and this enzyme can thus be renamed as TrmU54 with the same classification as the PAB0719 enzyme (9,26). Assuming the presence of one m5U54 per fully matured tRNA, as seen in most organisms (2,3,59,60), the proportion of m5U in N. equitans tRNAs would be around 12% of the total uridine content (Figure 2C) rather than the 3% observed here. This small proportion of N. equitans tRNAs containing m5U54 was subsequently shown to represent a transient stage of their maturation process, prior to hypermodification of this nucleotide position (Figure 6A). In silico screening revealed homologs of TtuA and TtuB that thiolate the 2-position of m5U in the tRNAs of some thermophilic bacteria (46), where TtuA acts as a 2-thio synthetase and TtuB as a sulphur carrier/donor (61,62). The N. equitans ORF NEQ283 showed high similarity to PF0273 (TtuA homolog) from P. furiosus, while NEQ523 displayed even closer resemblance to the TtuB homolog PF1758. Genes encoding homologous enzymes are also apparent in the genomes of other Nanoarchaeota. LC/ESI-MS analyses of the N. equitans tRNAs unequivocally showed the presence of m5s2U (Figure 4). Nucleotide m5s2U at tRNA position 54 has been noted in other hyperthermophiles, where the relative proportions of m5U that is thiolated to m5s2U varies with environmental conditions and in particular the growth temperature (10,11,13,15). This modified nucleotide is absent in the I. hospitalis tRNAs (Table 1).
Figure 6.
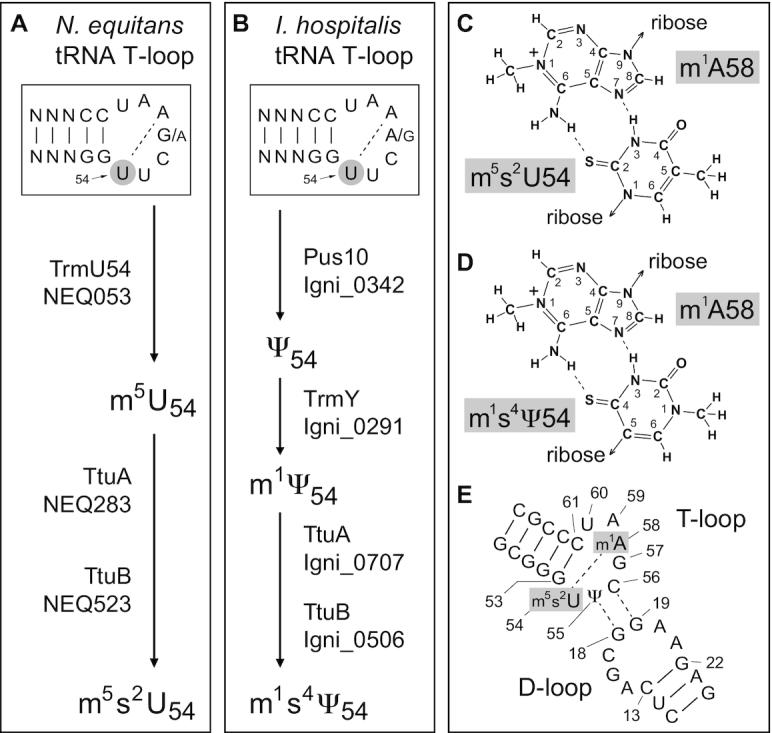
Modification pathways of nucleotide U54 within the tRNAs of (A) N. equitans and (B) I. hospitalis. Modified U54 makes a reverse Hoogsteen interaction with nucleotide 58 in the T-loops of both archaea where (C) the m5s2U54 to m1A58 interaction in N. equitans is functionally equivalent to (D) the m1s4Ψ54 to m1A58 pair found in I. hospitalis. (E) Putative interaction of the archaeal tRNA D- and T-loops based on the yeast tRNAPhe structure (33). The N. equitans and I. hospitalis genomes respectively encode the m1A methyltransferase TrmI homologs Igni_1131 and NEQ337. The presence of m1A was confirmed by LC (retention time of 3.6 min) and by ESI-MS/MS (not shown) in the tRNAs of both archaea; m1I (LC retention time of 3.6 min) was also found in both species, and presumably is formed at position 57 via an m1A intermediate in tRNAs that have an adenosine at this position (67,68). MS fragmentation confirmed the structures of Ψ (Figure 5), m5s2U54 (Figure 3) and m1s4Ψ54 (Figures 4 and 5).
A search for alternative U54 modification pathways in I. hospitalis tRNAs revealed Igni_0342, which is a homolog of the P. abyssi enzyme Pus10 that isomerizes both this nucleotide and its neighbour U55 to Ψ (47). In a subsequent modification step seen in many archaeal species, Ψ54 is converted to m1Ψ by the TrmY methyltransferase (47). Bioinformatics analysis indicated that the methylation reaction is most likely carried out by Igni_0291, which resembles PAB1866, the P. abyssi homolog of TrmY. Not surprisingly, no homologs of Pus10 or TrmY are present in N. equitans. Intriguingly, however, the I. hospitalis genome encodes orthologs of the two putative U54 thiolation enzymes, where Igni_0707 resembles NEQ283, and Igni_0506 is similar to NEQ523, raising the possibility that I. hospitalis might thiolate m1Ψ54. LC fractionation and ESI-MS analyses confirmed that this is indeed the case, and I. hospitalis tRNAs contain the previously uncharacterized nucleoside m1s4Ψ (Figures 4 and 5). The enzymatic steps involved in m1s4Ψ synthesis are summarized in Figure 6B, and homologs of these enzymes are evident in other Crenarchaeota including the close relative, Ignicoccus islandicus.
Modification of U54 helps maintain the highly conserved L-shaped tertiary structure of tRNAs, facilitating interaction with elongation factors and ribosomal binding sites during the course of mRNA translation (63,64). Here, the reverse Hoogsteen interaction between nucleotides 54 and 58 plays a key role in maintaining the internal structure of the T-loop and its contacts with the D-loop (33). Nucleotide 54, hypermodified as m5s2U in N. equitans and m1s4Ψ in I. hospitalis would be conformed in an essentially identical manner with the 2-thiol group in m5s2U (Figure 6C) and 4-thio group of m1s4Ψ (Figure 6D) in the same relative position holding the ribose of nucleotide-54 in a C3′-endo-pucker and the glycosidic bond in the anti-configuration to avoid steric clash with the 2′-hydroxyl. This, in conjunction with the 5-methyl of m5s2U and the equivalently-positioned 1-methyl of m1s4Ψ, would strengthen nucleotide-54 stacking between nucleotides 53 and 55.
Uridine 55 is isomerized to pseudouridine in most tRNAs, and Ψ is present in both N. equitans and I. hospitalis tRNAs (Table 1). The stacked arrangement optimally positions both m5s2U54 and m1s4Ψ54 to make the reverse Hoogsteen pairing with A58, which is itself stacked between G57 and C61. Extrapolating from the yeast tRNAPhe crystal structure, this arrangement facilitates the interaction of Ψ55 and C56 with the D-loop nucleotides 19 and 18 (Figure 6E), respectively, and enables nucleotides 59 and 60 to coordinate a magnesium ion with the D-loop (33). The identities of yeast tRNAPhe nucleotides 59 and 60 are different than in the archaeal tRNAs (Figure 2) where the D-loops lack dihydrouridine (65), and while metal ion coordination between these archaeal positions remains structurally feasible, it remains to be demonstrated. The T-loop structure is further strengthened by m1A58 modification in species of Archaea, Bacteria and Eukarya (66). Nucleoside m1A was identified in both the I. hospitalis and N. equitans tRNAs (Figure 6E legend).
Further screening of the archaeal tRNAs revealed a total of eleven modified forms of uridine (Table 1). Both archaea contained unmodified uridine, Ψ, Um, s2U and s4U in their tRNAs, while nucleotides m5U, m5s2U and a thiolated pseudouridine (compound #9) are specific to N. equitans, and m1Ψ, m1s4Ψ, m1Ψm and s2Um are specific to I. hospitalis. In a study of tRNA modifications in fourteen diverse archaeal species, s2Um was confined to the hyperthermophilic Crenarchaeota branch of the Archaea and absent from both the Euryarchaeota and Bacteria (10). Our findings are consistent with this previous study, and we note that although N. equitans lacks s2Um in its tRNAs, it does produce both Um and s2U.
In conclusion, we demonstrate here that while N. equitans and I. hospitalis grow physically attached under identical hyperthermic conditions, their modifications at U54 in their tRNAs are determined by their phylogenetic origins. These two organisms thus possess structural solutions that have evolved independently and convergently to ensure tRNA function within this extreme environment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Basma El Yacoubi and Valérie de Crécy-Lagard, University of Florida in Gainsville for advice on cloning of NEQ053, Kenneth Holst Seistrup for help with bioinformatics analyses. The Villum Center for Bioanalytical Sciences is thanked for providing access to mass spectrometric equipment.
Contributor Information
Simon Rose, Department of Biochemistry & Molecular Biology, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Sylvie Auxilien, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
Jesper F Havelund, Department of Biochemistry & Molecular Biology, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Finn Kirpekar, Department of Biochemistry & Molecular Biology, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Harald Huber, Lehrstuhl für Mikrobiologie und Archaeenzentrum, Universität Regensburg, Universitätsstraße 31, D-93053 Regensburg, Germany.
Henri Grosjean, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
Stephen Douthwaite, Department of Biochemistry & Molecular Biology, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Research Agency [FNU rammebevilling 10-084554 to S.D.]; Deutsche Forschungsgemeinschaft [Förderkennzeichen HU701/2 to H.H.]. Funding for open access charge: FNU-rammebevilling [10-084554 to S.D.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ofengand J., Del Campo M.. Böck A., Curtis R., Kaper J.B., Neidhardt T., Nyström T., Squires C.. Modified nucleotides of E. coli ribosomal RNA. Escherichia coli and Salmonella. 2005; Washington, DC: ASM Press. [Google Scholar]
- 2. Björk G.R., Hagervall T.G.. Böck A., Curtis R., Kaper J.B., Neidhardt T., Nyström T., Squires C.. Escherichia coli and Salmonella. 2005; Washington DC: ASM Press. [Google Scholar]
- 3. Grosjean H. Grosjean H. Nucleic Acids are not boring long polymers of only four types of nucleotides. 2009; Austin, Texas: 1–18.DNA and RNA modification enzymes: Structure, mechanism, function and evolutionLandes Biosciences. [Google Scholar]
- 4. Agris P.F., Narendran A., Sarachan K., Vare V.Y.P., Eruysal E.. The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity. Enzymes. 2017; 41:1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A. et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hsuchen C.C., Dubin D.T.. Methylation patterns of mycoplasma transfer and ribosomal ribonucleic acid. J. Bacteriol. 1980; 144:991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson L., Hayashi H., Söll D.. Isolation and properties of a transfer ribonucleic acid deficient in ribothymidine. Biochemistry. 1970; 9:2823–2831. [DOI] [PubMed] [Google Scholar]
- 8. Grosjean H., Breton M., Sirand-Pugnet P., Tardy F., Thiaucourt F., Citti C., Barre A., Yoshizawa S., Fourmy D., de Crecy-Lagard V. et al.. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet. 2014; 10:e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Urbonavičius J., Auxilien S., Walbott H., Trachana K., Golinelli-Pimpaneau B., Brochier-Armanet C., Grosjean H.. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol. Microbiol. 2008; 67:323–335. [DOI] [PubMed] [Google Scholar]
- 10. Edmonds C.G., Crain P.F., Gupta R., Hashizume T., Hocart C.H., Kowalak J.A., Pomerantz S.C., Stetter K.O., McCloskey J.A.. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol. 1991; 173:3138–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kowalak J.A., Dalluge J.J., McCloskey J.A., Stetter K.O.. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994; 33:7869–7876. [DOI] [PubMed] [Google Scholar]
- 12. Watanabe K., Oshima T., Saneyoshi M., Nishimura S.. Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GTΨC in tRNA of an extreme thermophile. FEBS Lett. 1974; 43:59–63. [DOI] [PubMed] [Google Scholar]
- 13. Watanabe K., Oshima T., Hansske F., Ohta T.. Separation and comparison of 2-thioribothymidine-containing transfer ribonucleic acid and the ribothymidine-containing counterpart from cells of Thermus thermophilus HB 8. Biochemistry. 1983; 22:98–102. [DOI] [PubMed] [Google Scholar]
- 14. Shigi N., Suzuki T., Tamakoshi M., Oshima T., Watanabe K.. Conserved bases in the TΨC loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J. Biol. Chem. 2002; 277:39128–39135. [DOI] [PubMed] [Google Scholar]
- 15. Shigi N., Suzuki T., Terada T., Shirouzu M., Yokoyama S., Watanabe K.. Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 2006; 281:2104–2113. [DOI] [PubMed] [Google Scholar]
- 16. Grosjean H., Oshima T.. Gerday C., Glansdorff N.. How Nucleic Acids cope with high temperature. Physiology and Biochemistry of Extremophiles. 2007; ASM Press; 39–56. [Google Scholar]
- 17. Lorenz C., Lunse C.E., Morl M.. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Phillips G., de Crecy-Lagard V.. Biosynthesis and function of tRNA modifications in Archaea. Curr. Opin. Microbiol. 2011; 14:335–341. [DOI] [PubMed] [Google Scholar]
- 19. Hori H., Kawamura T., Awai T., Ochi A., Yamagami R., Tomikawa C., Hirata A.. Transfer RNA modification enzymes from thermophiles and their modified nucleosides in tRNA. Microorganisms. 2018; 6:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N. et al.. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003; 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anantharaman V., Koonin E.V., Aravind L.. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002; 30:1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ny T., Lindstrom H.R., Hagervall T.G., Björk G.R.. Purification of transfer RNA m5U54-methyltransferase from Escherichia coli. Association with RNA. Eur. J. Biochem. 1988; 177:467–475. [DOI] [PubMed] [Google Scholar]
- 23. Gu X., Ivanetich K.M., Santi D.V.. Recognition of the T-arm of tRNA by tRNA m5U54 -methyltransferase is not sequence specific. Biochemistry. 1996; 35:11652–11659. [DOI] [PubMed] [Google Scholar]
- 24. Alian A., Lee T.T., Griner S.L., Stroud R.M., Finer-Moore J.. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc. Natl Acad. Sci. U.S.A. 2008; 105:6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nordlund M.E., Johansson J.O., von Pawel-Rammingen U., Byström A.S.. Identification of the TRM2 gene encoding the tRNA (m5U54) methyltransferase of Saccharomyces cerevisiae. RNA. 2000; 6:844–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walbott H., Leulliot N., Grosjean H., Golinelli-Pimpaneau B.. The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)-methyltransferase provides insights into its tRNA specificity. Nucleic. Acids. Res. 2008; 36:4929–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delk A.S., Rabinowitz J.C.. Biosynthesis of ribosylthymine in the transfer RNA of Streptococcus faecalis: a folate-dependent methylation not involving S-adenosylmethionine. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Urbonavičius J., Brochier-Armanet C., Skouloubris S., Myllykallio H., Grosjean H.. In vitro detection of the enzyme activity of folate-dependent tRNA(U54, C5)-methyltransferase. Methods Enzymol. 2007; 425:103–119. [DOI] [PubMed] [Google Scholar]
- 29. Urbonavičius J., Skouloubris S., Myllykallio H., Grosjean H.. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria-evolutionary implications. Nuclec. Acids Res. 2005; 33:3955–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamagami R., Yamashita K., Nishimasu H., Tomikawa C., Ochi A., Iwashita C., Hirata A., Ishitani R., Nureki O., Hori H.. The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO). J. Biol. Chem. 2012; 287:42480–42494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamdane D., Argentini M., Cornu D., Golinelli-Pimpaneau B., Fontecave M.. FAD/folate-dependent tRNA methyltransferase: flavin as a new methyl-transfer agent. J. Am. Chem. Soc. 2012; 134:19739–19745. [DOI] [PubMed] [Google Scholar]
- 32. Watanabe K., Yokoyama S., Hansske F., Kasai H., Miyazawa T.. CD and NMR studies on the conformational thermostability of 2-thioribothymidine found in the TΨC loop of thermophile tRNA. Biochem. Biophys. Res. Commun. 1979; 91:671–677. [DOI] [PubMed] [Google Scholar]
- 33. Shi H., Moore P.B.. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000; 6:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horie N., Hara-Yokoyama M., Yokoyama S., Watanabe K., Kuchino Y., Nishimura S., Miyazawa T.. Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry. 1985; 24:5711–5715. [DOI] [PubMed] [Google Scholar]
- 35. Davanloo P., Sprinzl M., Watanabe K., Albani M., Kersten H.. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 1979; 6:1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pang H., Ihara M., Kuchino Y., Nishimura S., Gupta R., Woese C.R., McCloskey J.A.. Structure of a modified nucleoside in archaebacterial tRNA which replaces ribosylthymine. 1-Methylpseudouridine. J. Biol. Chem. 1982; 257:3589–3592. [PubMed] [Google Scholar]
- 37. Romby P., Carbon P., Westhof E., Ehresmann C., Ebel J.P., Ehresmann B., Giege R.. Importance of conserved residues for the conformation of the T-loop in tRNAs. J. Biomol. Struct. Dyn. 1987; 5:669–687. [DOI] [PubMed] [Google Scholar]
- 38. Jahn U., Gallenberger M., Paper W., Junglas B., Eisenreich W., Stetter K.O., Rachel R., Huber H.. Nanoarchaeum equitans and Ignicoccus hospitalis: new insights into a unique, intimate association of two archaea. J. Bacteriol. 2008; 190:1743–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huber H., Hohn M.J., Rachel R., Fuchs T., Wimmer V.C., Stetter K.O.. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002; 417:63–67. [DOI] [PubMed] [Google Scholar]
- 40. Forterre P., Gribaldo S., Brochier-Armanet C.. Happy together: genomic insights into the unique Nanoarchaeum/Ignicoccus association. J. Biol. 2009; 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.F., Darling A., Malfatti S., Swan B.K., Gies E.A. et al.. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013; 499:431–437. [DOI] [PubMed] [Google Scholar]
- 42. Podar M., Anderson I., Makarova K.S., Elkins J.G., Ivanova N., Wall M.A., Lykidis A., Mavromatis K., Sun H., Hudson M.E. et al.. A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 2008; 9:R158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waters E., Hohn M.J., Ahel I., Graham D.E., Adams M.D., Barnstead M., Beeson K.Y., Bibbs L., Bolanos R., Keller M. et al.. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:12984–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 45. Lartigue C., Lebaudy A., Blanchard A., Yacoubi B.E., Rose S., Grosjean H., Douthwaite S.. The flavoprotein Mcap0476 (RlmFO) catalyzes m5U1939 modification in Mycoplasma capricolum 23S rRNA. Nucleic Acids Res. 2014; 42:8073–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shigi N., Sakaguchi Y., Suzuki T., Watanabe K.. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 2006; 281:14296–14306. [DOI] [PubMed] [Google Scholar]
- 47. Chatterjee K., Blaby I.K., Thiaville P.C., Majumder M., Grosjean H., Yuan Y.A., Gupta R., de Crecy-Lagard V.. The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA. 2012; 18:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giessing A.M., Jensen S.S., Rasmussen A., Hansen L.H., Gondela A., Long K., Vester B., Kirpekar F.. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009; 15:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seistrup K.H., Rose S., Birkedal U., Nielsen H., Huber H., Douthwaite S.. Bypassing rRNA methylation by RsmA/Dim1 during ribosome maturation in the hyperthermophilic archaeon Nanoarchaeum equitans. Nucleic Acids Res. 2017; 45:2007–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Desmolaize B., Fabret C., Bregeon D., Rose S., Grosjean H., Douthwaite S.. A single methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939 modifications in Bacillus subtilis 23S rRNA. Nucleic Acids Res. 2011; 39:9368–9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Auxilien S., Rasmussen A., Rose S., Brochier-Armanet C., Husson C., Fourmy D., Grosjean H., Douthwaite S.. Specificity shifts in the rRNA and tRNA nucleotide targets of archaeal and bacterial m5U methyltransferases. RNA. 2011; 17:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Grosjean H., Droogmans L., Roovers M., Keith G.. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabelled tRNA substrates. Methods Enzymol. 2007; 425:57–101. [DOI] [PubMed] [Google Scholar]
- 53. Madsen C.T., Mengel-Jorgensen J., Kirpekar F., Douthwaite S.. Identifying the methyltransferases for m5U747 and m5U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003; 31:4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Andersen T.E., Porse B.T., Kirpekar F.. A novel partial modification at 2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004; 10:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Douthwaite S., Kirpekar F.. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007; 425:3–20. [DOI] [PubMed] [Google Scholar]
- 56. Stern S., Moazed D., Noller H.F.. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988; 164:481–489. [DOI] [PubMed] [Google Scholar]
- 57. Agarwalla S., Kealey J.T., Santi D.V., Stroud R.M.. Characterization of the 23S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 2002; 277:8835–8840. [DOI] [PubMed] [Google Scholar]
- 58. Purta E., O’Connor M., Bujnicki J.M., Douthwaite S.. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 2008; 383:641–651. [DOI] [PubMed] [Google Scholar]
- 59. Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J.. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic. Acids. Res. 2009; 37:D159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barraud P., Tisné C.. To be or not to be modified: Miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB Life. 2019; 71:1126–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Arragain S., Bimai O., Legrand P., Caillat S., Ravanat J.L., Touati N., Binet L., Atta M., Fontecave M., Golinelli-Pimpaneau B.. Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:7355–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen M., Asai S.I., Narai S., Nambu S., Omura N., Sakaguchi Y., Suzuki T., Ikeda-Saito M., Watanabe K., Yao M. et al.. Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:4954–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 64. Zhang J., Ferre-D’Amare A.R.. The tRNA Elbow in Structure, Recognition and Evolution. Life (Basel). 2016; 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Noon K.R., Guymon R., Crain P.F., McCloskey J.A., Thomm M., Lim J., Cavicchioli R.. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23 degrees C) and Stetteria hydrogenophila (Topt 95 degrees C). J. Bacteriol. 2003; 185:5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guelorget A., Roovers M., Guerineau V., Barbey C., Li X., Golinelli-Pimpaneau B.. Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase. Nucleic Acids Res. 2010; 38:6206–6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Grosjean H., Constantinesco F., Foiret D., Benachenhou N.. A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res. 1995; 23:4312–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roovers M., Wouters J., Bujnicki J.M., Tricot C., Stalon V., Grosjean H., Droogmans L.. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004; 32:465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.