Abstract
Mitochondria are vital for cellular energy supply and intracellular signaling after stress. Here, we aimed to investigate how mitochondria respond to acute DNA damage with respect to mitophagy, which is an important mitochondrial quality control process. Our results show that mitophagy increases after DNA damage in primary fibroblasts, murine neurons and Caenorhabditis elegans neurons. Our results indicate that modulation of mitophagy after DNA damage is independent of the type of DNA damage stimuli used and that the protein Spata18 is an important player in this process. Knockdown of Spata18 suppresses mitophagy, disturbs mitochondrial Ca2+ homeostasis, affects ATP production, and attenuates DNA repair. Importantly, mitophagy after DNA damage is a vital cellular response to maintain mitochondrial functions and DNA repair.
INTRODUCTION
In response to DNA damage, an extensive cellular response takes place. This includes efforts to initiate DNA repair, cell cycle arrest or apoptosis to ensure genome stability and integrity (1). Mitochondrial activities and quality are critical to maintain cellular homeostasis because they are the major producers of ATP and other essential factors. Mitochondria respond to physiologic cues by altering mitochondrial content, fusion, fission, and the unfolded protein response (2). These adaptations ensure continuity of the energy supply and promote cellular signaling after stress (2). DNA damage signaling is well-characterized in the nucleus, however, much less is known about how mitochondria respond to stress, even though mitochondria significantly contribute to cell fate decisions.
The importance of mitochondria is underscored by the number and variety of diseases associated with mitochondrial dysfunction. It is important to understand how mitochondria respond to both exogenous and endogenous stressors. During stress events there is crosstalk between the nucleus and mitochondria (3). However, the mechanisms governing this phenomenon remain elusive. Furthermore, it is unclear how mitochondria respond immediately after DNA damage and how these responses affect DNA repair and cell fate.
DNA repair is an energy consuming process. Cells need to maintain proper mitochondrial quality and quantity to ensure sufficient ATP availability (4). It has been reported that mitochondrial mass and mitochondrial DNA, indicators of mitochondrial quantity, are increased after DNA damage (5). Additionally, bioenergetics are enhanced, as indicated by higher oxygen consumption and mitochondrial ATP production after gamma-irradiation (IR) (4). Reactive oxygen species (ROS) are generated as side-products during the oxidative phosphorylation process and their overproduction in turn contributes to mitochondrial stress and damage (6). Thus, proper maintenance of a healthy pool of mitochondrial by quality control mechanisms are hypothesized to be essential, especially after DNA damage.
Mitophagy is an important mitochondrial quality control mechanism because it selectively eliminates unwanted or damaged mitochondria (7). Mitophagy is responsible for basal mitochondrial turnover and elimination of damaged mitochondria under stress (8). Dysfunction of mitophagy is seen in a variety of premature and aging related diseases, including ataxia telangiectasia (9), AD (10), Parkinson's disease (PD) (11) and cancer (12). Cells are continuously exposed to both internal and external DNA damaging sources every day, and these are thought to contribute to aging. Understanding how mitophagy is regulated after DNA damage provides insights into the crosstalk between the nucleus and mitochondria and elucidates aging mechanism by linking genomic instability and mitochondrial dysfunction, which are two major aging hallmarks (13).
Here, we have investigated how mitophagy is regulated in response to acute DNA damage and its association with DNA repair. We monitored mitophagy after various types of damage in different primary fibroblasts, in a mitophagy-reporter mouse model, and in a neuronal mitophagy-reporter in Caenorhabditis elegans after IR. Mitophagy was universally upregulated at later times after DNA damage. Further study indicated that Spata18 was an important protein for mitophagy induction after DNA damage. Spata18, also known as Mieap, is a p53 transcriptional target and was previously reported to contribute to mitochondrial quality control by inducing intramitochondrial lysosome-like organelles after mitochondrial damage (14). Consistently, our study showed that Spata18 KD in primary fibroblast cells significantly suppressed mitophagy induction and was associated with less efficient DNA repair after DNA damage.
MATERIALS AND METHODS
Reagents
Mitophagy Detection Kit (MT02-10; Dojindo Molecular Technologies, INC); Mitomycin (10107409001, sigma), H2O2 (NP0336BOX, Sigma); Rhod2 AM (R1245MP, Invitrogen), ATP measure kit (G7570, Promega); LysoTracker™ Deep Red (L12492, Thermofisher Scientific), MitoSOX™ Red Mitochondrial Superoxide Indicator (M36008, Thermofisher Scientific); MitoTracker™ Green FM (M7514, Thermofisher Scientific); Tetramethylrhodamine, Methyl Ester, Perchlorate (T668, Thermofisher Scientific); PureLink™RNA mini kit (12183025, ThermoFisher); jetPRIME® DNA and siRNA transfection reagent (Polyplus transfection).
Antibodies
Spata18 (HPA036854, Millipore sigma); BNIP3 (#44060S, Cell Signaling); Nix (#12396S, Cell Signaling); PAR (4336-BPC-100, Trevigen); GAPDH (#2118, Cell Signaling); PARP1(#9542S, Cell Signaling); LC3B (NB100-2220, Novus); 53BP1 (NB100-304, Novus), γ-H2AX (05-636, Millipore Sigma); P-ATM (sc-47739, Santa Cruz); P53 (sc-126, Santa Cruz), P21(BD610234, Biosciences); β-actin (sc-1616, Santa Cruz); PINK1 (23274-1-AP, Proteintech); Parkin (14060-1-AP, Proteintech); Rab9a (11420-1-AP, Proteintech); p-ULK1(5869, Cell Signaling); ULK1(6439, Cell Signaling).
Plasmid
ShSpata18 (NO.1-TRCN0000141572, NO.2-TRCN0000141024, Sigma), ShBNIP3 (TRCN0000007833, Sigma), shATM (TRCN0000010299, Sigma); shscramble (#1864, Addgene); psPAX2 (#12260, Addgene); pMD2.G (#12259, Addgene)
Lentivirus
p53 shRNA (h2) Lentiviral Particles (sc-44218-V, Santacruz); Control shRNA Lentiviral Particles-A (sc-108080, Santa Cruz).
Cell culture
Normal human fetal lung fibroblast (IMR-90), abbreviated as HFLF, and normal human fibroblast (GM05757) were from Coriell Institute. Mouse embryonic fibroblast (MEF) were prepared by following standard procedures as described previously (15). Brains were removed from embryonic day 13.5 (E13.5) C57BL/6J mouse embryos and the rest part was digested by trypsin at 37°C for 10 min. Cells were cultured in DMEM (Thermo Fisher Scientific), supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (10 000 U/ml; Thermo Fisher Scientific). Cells were maintained in a 37°C incubator gassed with 5% CO2/95% air. HFLF, normal human fibroblast, and MEF cells utilized in experiments were from passage 16–19, passage 20–23 and passage 2–5, respectively.
Western blot
Cell pellets were lysed in RIPA buffer (Cell Signaling Technology, MA, USA) containing complete mini protease inhibitor (Roche, Basel, Switzerland) and sonicated at power 4 for 10 s on ice by an ultrasonic cell disruptor. Lysates were centrifuged at 14 000 g for 30 min before the supernatants were collected. Protein concentration was measured by BCA assay (ThermoFisher Scientific). Proteins were separated by SDS–PAGE and transferred onto a PVDF membrane. The membrane was incubated with 5% milk (Bio-Rad) in TBST (20 mM Tris–HCl pH7.4, 0.14 mM NaCl, 0.1% Tween® 20) for 1 h at room temperature, followed by overnight incubation with a primary antibody at 4°C. After incubation with the HRP-conjugated secondary antibody for 1 h at room temperature, the signals were detected using a PharosFX plus Molecular Imager (Bio-Rad). Band intensity was determined using FIJI-ImageJ software.
Detection of mitophagy and mitochondria parameters in cells
To detect mitophagy, cells were stained by following the manufacturer's protocol (Dojindo Molecular Technologies, Inc.). Briefly, attached cells were washed with FBS-free medium twice and then incubated in FBS-free medium containing 100 nmol/l Mtphagy dye. After a 30 min incubation at 37°C, in the incubator, the mitophagy dye was removed and cells were washed in FBS-free medium twice before fresh medium was replaced for further treatment. To detect different mitochondrial parameters, trypsinized cells were incubated with different dyes including TMRM (10 nM for 15 min), to detect mitochondrial membrane potential, MitoTracker Green (50 nm for 15 min) for mitochondrial content, and MitoSOX (3 μM for 30 min) for mitochondrial ROS, followed by analysis by flow cytometry (BD FACSCanto™ II). Data was analyzed by Flow Jo V10.
DNA damage exposure
Attached cells were irradiated using gamma irradiation (IR) (either 2 Gy or 6 Gy at a rate of 0.74 Gy/min) or ultraviolet light (UVC 10 J/m2 at rate of 1.13 J/s). To induce oxidative stress damage, cells were cultured with complete medium and 0.25 mM H2O2 for 10 min at 37°C, with 5% CO2. To induce crosslink DNA damage, cells were cultured in complete medium containing 2 μM mitomycin C (MMC) for 48 h. Cells were harvested at the indicated time points after the DNA damage as mentioned in the figures and legends. To inhibit autophagy after 6 Gy IR, irradiated cells were cultured in medium containing DMSO, and 2 nM bafilomycin A1 or 5 mM 3-methyladdenine for 48 h before mitophagy detection.
Mitochondria ATP detection
Cells were plated in 96-well plates 24 h prior to irradiation. ATP levels were measured 1 or 48 h post-irradiation using manufacturer's protocol (Promega) and read on a Synergy II plate reader using the end point protocol settings. The ATP levels were normalized to protein concentrations measured in a parallel plate.
Mitochondria Ca2+ detection
To measure mitochondrial Ca2+, cells were washed with HBSS, followed by incubation with 5 μM dihydro Rhod-2 AM and 0.02% (v/v) Pluronic F-127 for 30 min at 37°C. The fluorescence was observed at ex/em wavelengths of 552/581 nm. Z-stacks were captured with a Zeiss LSM 880 inverted laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) using a 40×/1.3 Plan-NeoFluar oil-immersion objective at 1 μm z-step intervals with lateral pixel dimensions of 0.22 μm. Captured z-series were imported and analyzed using FIJI-ImageJ.
Cell proliferation
Cells were seeded on 96-well plates one day before different treatments. Cells were stained with 5 μM Hoechst 33258 dye for 10 min in the incubator before imaging and counting on the Celigo (Nexcelom Bioscience). Relative cell proliferation was calculated by normalizing cell numbers to that of the cells without any treatment at 24 h.
Ex vivo mitophagy detection using mt-keima mice
The mt-Keima mice were imported from Toren Finkel's lab (16) via Jurrien Dean's lab at National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Mice were maintained at the National Institute on Aging (NIA), Baltimore on a standard NIH diet in a 12 h light/dark cycle. All animal experiments were performed using protocols approved by the appropriate institutional animal care and use committee of the NIA. Both male and female adult mice were included in the experiments. Mice were subjected to whole body IR in a Nordion Gamma cell 40 Exactor Irradiator by placing them in a stereotaxic cube box where they were exposed to IR or not (sham). IR (6 Gy) was delivered at a rate of 0.74 Gy/min. After exposure, mice were returned to home cages and sacrificed by cervical dislocation 48 h after irradiation. To prepare the tissue for confocal imaging, we followed the protocol of Sun et al. (16). Briefly, tissues were immediately dissected out and rinsed with cold PBS. The brain was sliced into 1-mm sections in a pre-chilled Brain Slicer Matrix while the liver and heart were cut into small pieces (around 100 mm3). All tissues were placed onto a 35-mm confocal dishes (VWR) and analyzed immediately under a confocal microscope (Zeiss LSM 880). The principle of mt-Keima for mitophagy detection is based on its pH-dependent fluorescent property that the shorter wavelength (green) excitation predominates at the physiological pH 8.0 while shifting to the longer-wavelength (red) within the acidic lysosome (pH 4.5) after mitophagy. In the present study, emission signals obtained after excitation with the 458-nm laser (green) and 561-nm laser (red) represented signals of mitochondria from a neutral or acidic environment, respectively. Mitophagy was calculated as reported (16) on a pixel-by-pixel basis with pixel-intensity mapping on Zeiss ZEN 3.0 blue edition. Briefly, pixels for both green and red channels in the acquired images were plotted in a scatter diagram on the basis of its fluorescence intensity and the level of mitophagy was calculated by dividing the number of pixels with high red intensity by the total number of mitochondrial pixels. Total number of mitochondrial pixel is defined as the total pixel number after deduction of background pixels.
Mitophagy detection in C. elegans
The neuronal mitophagy reporter C. elegans strain IR1864: N2; Ex001 [punc-119TOMM-20: Rosella; rol-6(su1006) (17) was a gift from the lab of Nektarios Tavernarakis. Synchronized populations of worms were prepared by bleaching (4.45 ml ddH2O, 0.3 ml of standard commercial bleach and 0.25 ml 5 M NaOH) for 5 min with constant shake. Worm pellets were washed with M9 buffer for three times and eggs were hatched in M9 buffer at 15°C before transferring to fresh Escherichia coli (OP50)-seeded NGM agar plate and maintained at 20°C. L4 larvae were either placed in a Nordion Gamma cell 40 Exactor Irradiator and receive 50 Gy irradiation or sham treated. Worms were transferred to a confocal dish and immobilized in 20 mM levamisole solution 24 h post-IR. Images were taken on a confocal microscopy (Zeiss LSM 880). Green Fluorescence emission (490–553 nm) and red fluorescence emission (565–712 nm) images were acquired sequentially upon excitation with 488 nm or 561 nm laser light, respectively. Mitophagy was calculated by determining the ratio of red fluorescent intensity to green fluorescent intensity by using Zeiss ZEN 3.0 blue edition.
Microarray
RNA from mice brain, liver and heart was purified with PureLink™ RNA isolation kit following manufacturer's protocol (Thermo). RNA concentration and quality were measured using the NanoDrop ND-1000 spectrophotometer and a 2100 Bioanalyzer (Agilent Technologies). Microarray analysis was performed by the Gene expression and Genomics core facility (NIA) and analyzed using DIANE 6.0 software. Each group consisted of five mice. Pathways/GO terms were considered significant if they had more than three genes and a t test P value <0.05 with an FDR of not >0.3. The detailed data analysis methods were described previously (18).
Electron microscopy for mitochondrial morphology
Electron microscopy was performed by Electron Microscopy BioServices, MD. Cells were fixed in EM Grade Fixative (2.5% glutaraldehydein PBS) for 10 min, then scraped off and centrifuged at 15 000 RPM for 10 min. Cell pellets in the fixative were refrigerated for another 24 h before they were washed in Millonig's sodium phosphate Buffer and minced into small pieces. Cells were then post-fixed in 1.0% osmium tetroxide, stained with 2.0% aqueous uranyl acetate, dehydrated in a series of graded ethanol and embedded in Spurr's plastic resin. 60 to 80 nm ultra-thin sections were collected and mounted onto 200 mesh copper grids. The grids were then post-stained with Reynold's lead citrate. Sections were examined in a FEI Tecnai Spirit Twin Transmission Electron Microscope, operating at 80 kV. Mitochondria size were analyzed using FIJI-ImageJ software.
Immunofluorescence
Cells were seeded on an eight-well cover slide one day before IR. Cells were fixed in 4% paraformaldehyde for 10 min at RT and then treated with 0.2% Triton X-100 at RT for 5 min. After wash with PBS for two times, cells were incubated in blocking buffer (5% BSA in PBS) for 30 min at RT before incubation with the primary antibody for 53BP1 (Novus, 1:1500 in 5% BSA) overnight at 4°C. Cells were then washed with PBS 3 times, 5 min each time before incubation with the fluorescent probe-conjugated secondary antibody (Thermofisher, 1:1000) for 1 h at RT. Nuclei were counterstained with DAPI. Slides were scanned using Zeiss LSM 880 microscope and analyzed using FIJI-ImageJ software.
Protein knockdown
Knockdown (KD) of P53 in HFLF cells was performed following manufacturer's protocol (Santa Cruz). 5 μg/ml Polybrene were used during infection and cells were selected using complete medium containing 1 μg/ml puromycin 2 days after infection. Cells were used for experiment after at least 2 days’ selection. Knockdown of Spata18 and BNIP3 was performed using lentivirus packaged using the plasmid purchased from Sigma or Addgene. To be specific, lentivirus was packed using transfection reagent (Polyplus transfection) and 10 μg total DNA composed of 4 μg target DNA, 4 ug psPAX2 and 2 ug pMD2.G was used per 100 mm dish in HEK293T cells. Medium was replaced with fresh cell growth medium 4 h after transfection and virus was collected after another 72 h's culture. Viruses were stocked at –80°C after filtration through a 0.45 μm filter. Cell infection was conducted by culturing them with virus-containing medium for 4 h and cells were selected in medium containing 1 μg/ml puromycin 1 day after infection. Cells were used for experiment after at least 2 days’ selection.
Statistical analysis
GraphPad Prism 6.0 was used. Data shown are mean ± SEM. Statistical analysis used in this study was two-tailed unpaired t-test for comparison between two groups, or ANOVA for comparison among multiple groups. Differences were considered statistically significant when P < 0.05.
RESULTS
Mitophagy is induced after DNA damage in primary fibroblasts
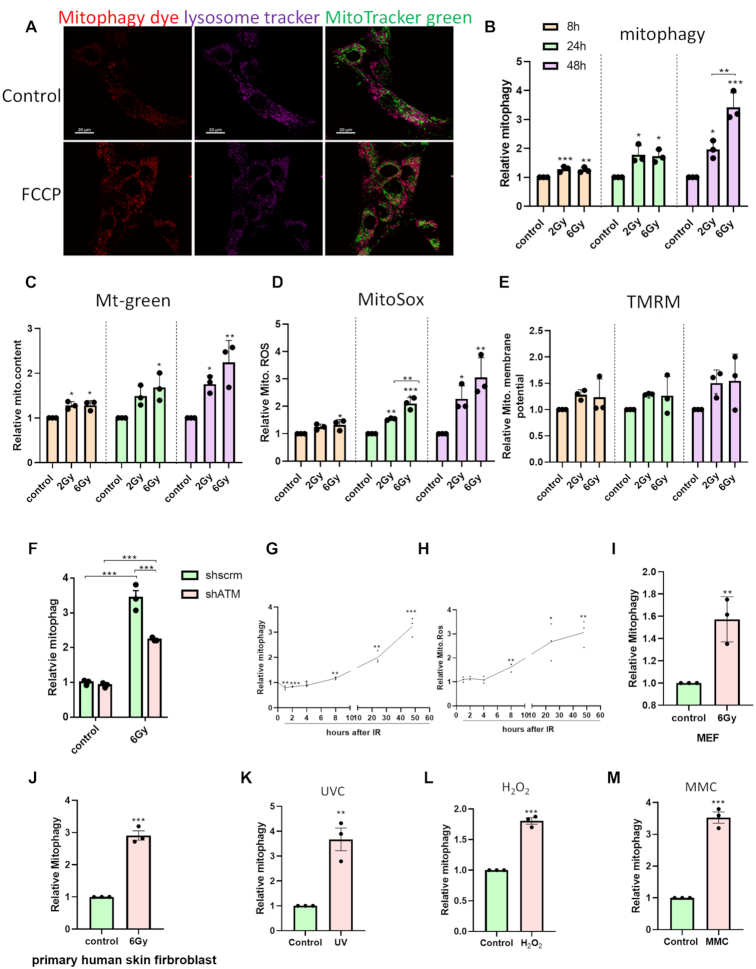
Primary cells were applied to assess mitophagy response after DNA damage. HFLF cells used here were normal primary cells which have been widely used as normal human fibroblast model of DNA damage (19). The cells were exposed to non-lethal doses of IR (2 or 6 Gy), which caused a dose-dependent suppression of cell proliferation (Supplementary Figure S1A), and increased levels of p53 and p21, but there was no cleavage of PARP1 (Supplementary Figure S1B). Dojindo mitophagy dye was employed to detect mitophagy in these cells. To verify its reliability for mitophagy detection, HFLF cells pre-stained with this mitophagy dye were treated with 10 μM Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) for 6 h to induce mitophagy and then co-stained with Lysotracker Deep Red before confocal imaging. As shown in Figure 1A, mitochondria undergoing mitophagy co-localized with the lysosome markers, and, as expected, exposure to FCCP increased mitophagy, indicating reliable detection of mitophagy induction using this method. Mitophagy at the single cell level was then quantified using flow cytometry. Our results show that after irradiation at 2 and 6 Gy, mitophagy increased significantly in comparison to the untreated cells at 8 h. It showed a time-dependent increase between 8 and 48 h (Figure 1B). There was a dose-dependent effect at 48 h. The representative images showing mitophagy induction after IR are shown in Supplementary Figure S1C. Additionally, mitochondrial content (Figure 1C) and ROS production (Figure 1D) both increased in a time-dependent manner while mitochondrial membrane potential (Figure 1E) remained unchanged, indicating a coordinated response of mitochondria after DNA damage.
Figure 1.
DNA damage induces mitophagy in primary fibroblasts. (A) Representative images showing mitophagy staining. HFLF cells were treated with DMSO or 5 μM FCCP for 5h. Mitochondria undergoing mitophagy, lysosomes and normal mitochondria are shown in red, purple, and green, respectively. The scale bars represent 20 μm. (B–E) Quantification of mitophagy by Dojindo mitophagy dye in (B), mitochondrial content by MitoGreen in (C), mitochondrial ROS by MitoSox in (D) and mitochondrial membrane potential by TMRM in (E) via flow cytometer. HFLF cells were irradiated at different doses and mitochondrial parameters were measured at the indicated time points post treatment. Fold change was calculated by normalizing parameters to the controls at the respective time points. (F) Detection of mitophagy in shATM HFLF cells after 6Gy IR. (G,H) Time course of mitophagy in (G) and mitochondria ROS in (H) after 6Gy IR. (I,J) Relative mitophagy level of MEFs in (I) and primary human skin fibroblasts in (J) at 48 h after 6 Gy IR. (K–M) Relative mitophagy of HFLF cells after DNA damage induced by 10 J/m2 UVC in (K), 0.25 mM H2O2 for 10 min in (L) or 2 μM MMC in (M). Mitophagy was detected 48 h post treatment. N = 3, mean ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001.
To check whether induction of mitophagy is dependent on the DNA damage response (DDR), ATM, a central mediator of DDR signaling was depleted in HFLF. The protein level of ATM is shown in Supplementary Figure S1D. The result in Figure 1F showed that mitophagy induction was significantly lower in ATM KD cells than in scrambled cells after IR, but it was not completely abolished. This indicates that induction of mitophagy after DNA damage is at least partially dependent on ATM pathways.
Since DNA repair is initiated immediately after damage, we looked more closely at the time course of mitophagy in HFLF cells starting at 1 h after 6 Gy IR. Interestingly, immediately after IR (1–4 h), mitophagy was slightly lower in treated cells than in control cells (Figure 1G), and then increased over time since 8 h. ROS can be a major contributor to mitochondrial damage so mitochondrial ROS was also quantified as a function of time. Results in Figure 1H showed a consistent trend with the mitophagy time course in Figure 1G that significant increase of ROS was not observed until 8 h post treatment, suggesting an important role of ROS in mitophagy induction after DNA damage.
To investigate whether IR-induced mitophagy showed similar trends in other cells, primary MEFs and primary human skin fibroblasts were irradiated at 6 Gy and analyzed 48 h post-treatment. As shown in Figure 1I and J, Mitophagy increased in a manner similar to the HFLF cells in both MEFs and human skin fibroblasts when compared to the control cells after irradiation, but the extent of change was different, as MEF was less responsive.
To ask whether DNA damage induced by other types of stimuli also impacts mitophagy similarly to IR, HFLF cells were treated with 10 J/m2 UVC, 2 μM MMC and 0.25 mM H2O2. The doses of the different agents were based on previous reports to induce different types of DNA damages (20–22). As shown in Figure 1K–M, these agents all caused significant increases in mitophagy relative to control cells at 48 h. This indicates that the mitophagy increase as a late DNA damage response is not related to a specific source of damage.
Mitophagy is altered in a tissue-specific manner in response to DNA damage induced by IR
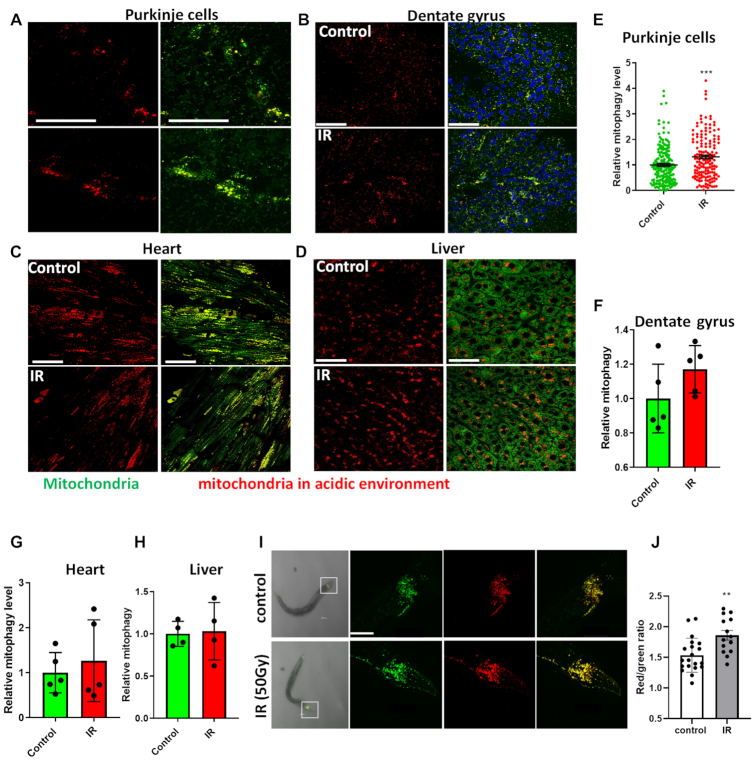
To examine mitophagy induction by IR in mice, the mitophagy reporter strain, mt-Keima mice were used. In this transgenic mouse model, a mitochondrial-targeted (COX VIII) form of the pH-sensitive protein Keima is expressed such that it allows the distinction of mitochondria in an acidic (like lysosome) or normal pH environment (23). Mt-Keima mice were sham-treated or irradiated with 6 Gy and sacrificed 48 h post-treatment. Mitophagy was quantified in various fresh tissues, including the Purkinje cell layer of the cerebellum and the dentate gyrus, which are neuron-rich regions of the brain with high basal mitophagy, as well as in the liver and heart. As shown in Figure 2A–H, among all the tested tissues, a significant increase of mitophagy at 48 h was only observed in the Purkinje cells while cells in the dentate gyrus region showed a slight, but non-significant increase after IR. Mitophagy did not increase significantly in liver or heart cells after irradiation, suggesting a tissue-specific mitophagy response to IR with a preponderance in the central nervous system.
Figure 2.
Ex vivo and in vivo mitophagy after IR damage. Mt-Keima mice were treated with 6 Gy whole body IR and sacrificed 48 h post treatment. After cooling in cold PBS, different fresh tissues were dissected and imaged for mitophagy detection by confocal microscopy. N = 5 for brain and heart tissue, N = 4 for liver tissue per group, with multiple regions imaged per group. (A–D) Representative images showing mitophagy (red) and mitochondria (green) from Purkinje cells in (A), dentate gyrus region in (B), heart in (C) and liver in (D). The scale bars represent 50 μm. (E–H) Quantification of mitophagy level in the Purkinje cells in (E), dentate gyrus region in (F), heart in (G) and liver in (H). (I) Representative images of mitophagy in the head region neurons in mt-Rosella C. elegans. L4 worms were treated with 50 Gy whole body IR and mitophagy was detected 24 h after treatment. The scale bars represent 50 μm. (J) Relative mitophagy level in neurons in the head regions. A total of 14–20 worms from three independent experiments for each group were analyzed. Data: mean ± S.E.M. **P < 0.01, ***P < 0.001.
Gene expression array analysis was performed to get unbiased insights into pathways affected by IR in different tissues and sample quality was confirmed by Principal Component Analysis as shown in Supplementary Figure S2A. Pathways that showed an absolute z-score ≥ 1.5, a P-value ≤ 0.05, and a FDR < 0.3 with a minimum of three changed genes in at least one of the four tissues (as mentioned above) were pursued. Results showed that pathways related to energy and metabolism (Supplementary Figure S2B) and DNA damage responses (Supplementary Figure S2C) were significantly altered in cerebrum, cerebellum, liver, and heart 48 h after irradiation. The expression patterns changed in a similar manner in the cerebellum and the cerebrum, while heart tissue showed substantially less changes and sometimes in the opposite direction in metabolism related pathways. Of note, ATP production and mitochondrial activity-related pathways were up regulated in the cerebrum and cerebellum, suggesting a higher energy demand in neurons after DNA damage and this may explain why neuronal tissues had increased levels of mitophagy.
Similar experiments were carried out using a neural mitophagy reporter strain in N2 C. elegans, mt-Rosella. Mt-rosella is a pH-sensitive fluorescent biosensor containing two tandem fluorescent proteins: a pH-sensitive green fluorescent protein pHluorin and a non-pH sensitive red fluorescent protein DsRed.T3 (24). This has been applied successfully to detect mitophagy in cross species including yeast (25) and worms (26). In the present study, confocal microscopy was conducted on anesthetized live worms 24 h after exposure to 50 Gy IR. Mitophagy was quantified in the neurons of the head ganglia of treated versus untreated worms. As observed in the mouse brains, IR-induced DNA damage promoted an increase in mitophagy in the neurons of the worms at 24 h (Figure 2I and J). Combined, these results suggest that the increased mitophagy following IR-mediated damage is a conserved process across species.
IR-mediated mitophagy is independent of conventional autophagy
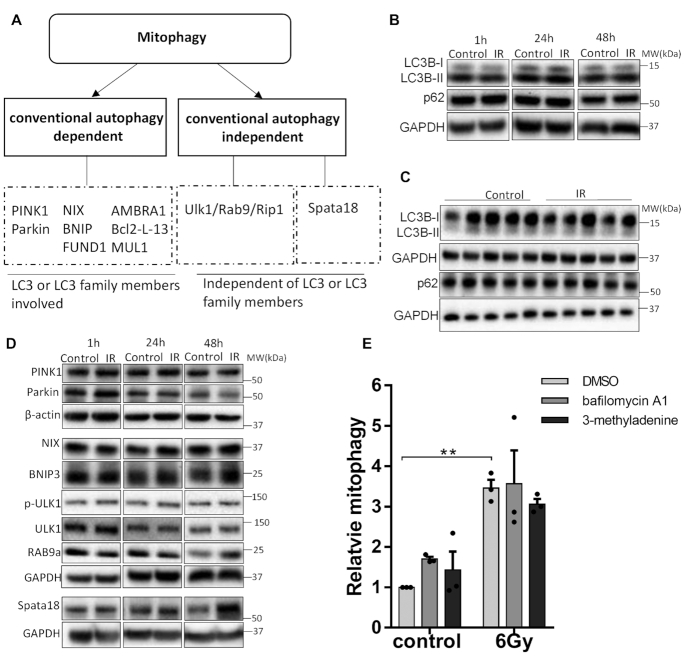
Several pathways have been reported to regulate mitophagy (as shown in Figure 3A) and the most studied is mediated through PINK1/Parkin. In this pathway, PINK1 accumulates on the outer membrane of depolarized mitochondria and recruits Parkin, which subsequently leads to the recruitment of autophagosomes and subsequent engulfment of the mitochondria (27). Mitophagy can also take place in a PINK1/parkin-independent manner by regulation of FUNDC1, NIX/BNIP3, AMBRA1, Bcl2-L-13 or MUL1 (8,28). All the aforementioned proteins mediate mitophagy through pathways involving LC3 family members and are dependent on conventional autophagy (29). There also exists two additional mechanisms whereby mitophagy can take place independently of conventional autophagy machinery. In one scenario, mitophagy is mediated through recruitment of trans-Golgi membrane to damaged mitochondria through Rab9, defined as ‘alternative mitophagy’ (30). In the other, mitochondrial quality control through degradation in an acidic environment is mediated through an accumulation of lysosome-like organelle in mitochondria through Spata18 (14).
Figure 3.
Mitophagy induced by IR is independent of conventional autophagy. (A) Summary of mitophagy pathways. (B, C) Western blot showing the protein level of LC3B and P62 in HFLF cells after 6 Gy IR in (B), and mice cerebellum 48 h after IR in (C). (D) Western blot showing the protein level of PINK1, Parkin, NIX, BNIP3, p-ULK1, ULK1 and RAB9a and Spata18 in HFLF cells after 6 Gy IR. (E) Mitophagy levels of HFLF cells cultured in the presence or DMSO, or 2 nM bafilomycin A1 or 5 mM 3-methyladdenine for 48 h after 6 Gy IR. For western blot data, the result of one out of three independent samples were shown. N = 3, mean ± S.E.M. **P < 0.01.
To dissect the molecular mechanism of mitophagy regulation after DNA damage, the protein levels of LC3 and p62, which are important mediators of conventional autophagy, were measured. Intriguingly, the protein level of LC3II/LC3I and P62 were not significantly changed in both HFLF cells (Figure 3B and Supplementary Figure S3A) and mouse cerebellum after 6 Gy IR (Figure 3C and Supplementary Figure S3B). Consistently, in comparison to the normalized untreated control at different timepoints, there were no significant changes after IR in protein levels of PINK1, Parkin, NIX and BNIP3, which regulate mitophagy through pathways involving LC3, indicating a mechanism independent of conventional autophagy (Figure 3D and Supplementary Figure S3C). The result in Figure 3E also showed that in the presence of two autophagy inhibitors, bafilomycin A1 or 3-methyladdenine, the induction of mitophagy after IR was not significantly affected, further supporting the independence of mitophagy induction on conventional autophagy after IR. We then checked the protein levels of p-ULK1, ULK1, Rab9a and Spata18 at different time points after 6 Gy IR in HFLF cells and found that only the protein level of Spata18 was significantly increased after IR (Figure 3D and Supplementary Figure S3C). These results suggest that IR-mediated mitophagy may involve increased dependence on Spata18 rather than conventional autophagy mediators.
Mitophagy is regulated through the p53-Spata18 axis after DNA damage
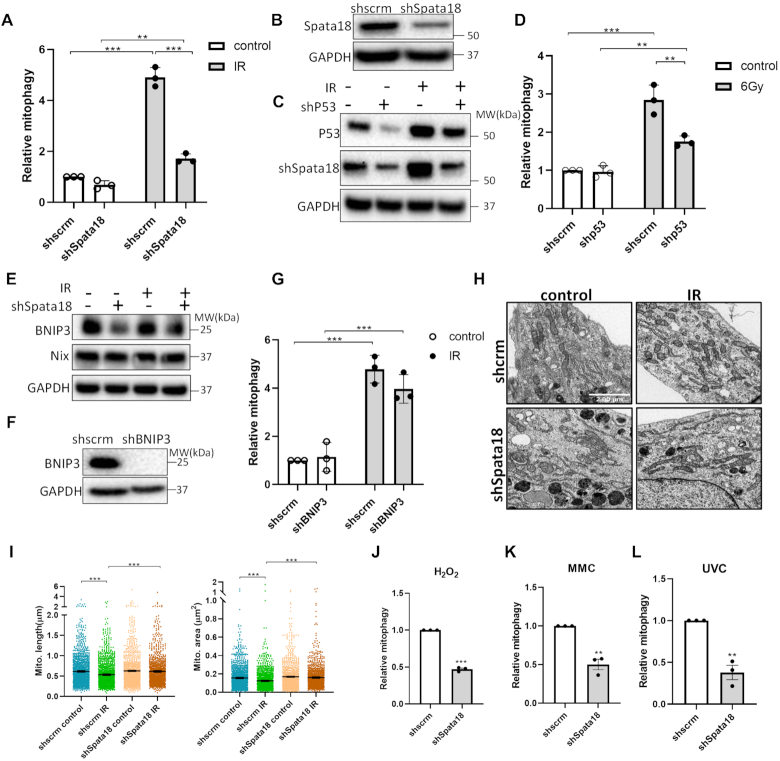
To further investigate whether Spata18 is involved in mitophagy regulation after DNA damage, the mitophagy level of scrambled and shSpata18 HFLF cells were compared 48 h post IR treatment. Relative mitophagy was calculated by normalizing data to that of scrambled cells without IR treatment. As shown in Figure 4A, Spata18 deficiency caused a 3-fold decrease in IR-mediated mitophagy induction in comparison to scramble cells. The knockdown of Spata18 in these cells was confirmed by western blot analysis (Figure 4B). Further, KD of p53, an upstream molecule that mediates the transcription of Spata18, decreased the protein level of Spata18 (Figure 4C and Supplementary Figure S4A) and decreased mitophagy after IR (Figure 4D). Together, these results confirm the important role of Spata18 in mitophagy regulation after IR and suggest a DNA damage–p53–Spata18 pathway.
Figure 4.
Spata18 plays an important role in mitophagy regulation after DNA damage. (A) Mitophagy levels of scrambled and shSpata18 HFLF cells plus or minus IR treatment. (B) Western blot showing the protein level of Spata18. (C) Western blot images showing p53 and Spata18 protein levels in p53 KD and scrambled HFLF cells. (D) Mitophagy level of HFLF cells with p53 KD. (E) Western blot images showing BNIP3 and NIX protein levels in Spata18 KD and scrambled cells. (F) Western blot showing BNIP3 protein levels in shBNIP3 KD cells. (G) Relative mitophagy level of HFLF cells with BNIP3 KD. (H) Representative TEM images showing mitochondria of scrambled and shSpata18 HFLF cells 48 h after 6 Gy IR. The scale bars represent 2 μm. (I) Quantification of mitochondrial size by checking mitochondrial length and area. (J–L) Relative mitophagy level of Spata18 KD HFLF cells after DNA damage induced by 0.25 mM H2O2 in (J), 2 μM MMC in (K) and 10 J/m2 UVC in (L). For (A), (D), (G) and (J–L), mitophagy was detected 48 h after 6 Gy IR. Data was normalized to scrambled cells without radiation. For (C) and (E), extracts were collected 48 h after 6 Gy IR and the result of one out of three independent samples were shown. N = 3, mean ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001.
According to previous research [28], BNIP3 and NIX are critical mediators in Spata18-regulated mitochondrial quality control. We found that BNIP3 was significantly decreased in Spata18 KD cells while NIX was slightly (but not significantly) decreased (Figure 4E and Supplementary Figure S4B). Knockdown of BNIP3 slightly decreased mitophagy after IR (Figure 4F, G). However, its effects were not as significant as that of Spata18 KD cells, suggesting that Spata18 may also mediate induction of mitophagy partially through BNIP3-independent pathways.
Transmission electron microscopy (TEM) analysis of mitochondrial morphology showed that mitochondrial size was significantly decreased in the scrambled cells after radiation, with smaller mitochondrial area and shorter length evident in Figure 4H and I. This is consistent with previous observations that radiation promotes mitochondrial fission (31). When Spata18 was knocked down, mitochondrial size could be not reduced after irradiation. This may explain reduced mitophagy in Spata18-deficient cells, since fission is a prerequisite for mitophagy (32).
Further investigation showed that knockdown of Spata18 also tempered the mitophagy response to other stimuli since mitophagy was attenuated after exposure to H2O2, MMC or UVC in Spata18 KD cells (Figure 4J–L). The role of Spata18 in mitophagy regulation was further confirmed by a second shRNA targeting Spata18 at a different site. The expression of Spata18 in HFLF cells is shown in Supplementary Figure S4C and the new shRNA is marked as shSpata18 (2). Consistent with the results of the first shRNA, knockdown of spata18 by shSpata18 (2) also attenuated mitophagy after DNA damage induced by multiple stimuli (IR, MMC, H2O2 or UVC) when compared with the corresponding controls (Supplementary Figure S4D–G).
Mitophagy suppression by Spata18 KD affects mitochondrial functions and DNA repair
DNA repair is an energy-demanding process and ATP supply determines cell fate by tipping the balance towards survival or apoptosis (33). Mitochondria display increased activity with higher ROS and bioenergetics after DNA damage by IR (4). Modulation of mitophagy to remove damaged mitochondria induced by ROS is hypothesized to be critical in maintaining a healthy mitochondria pool and may also be important for DNA repair.
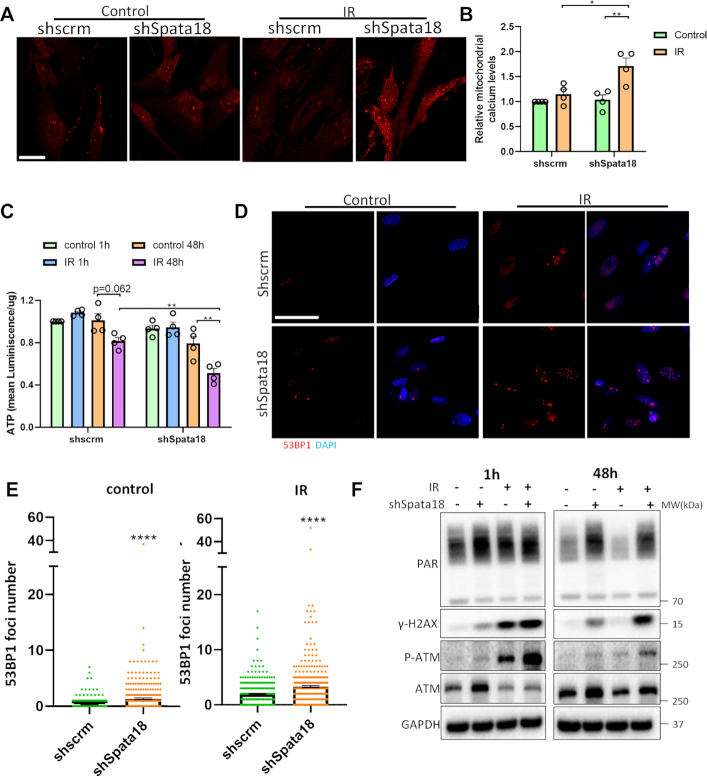
To investigate how mitophagy suppression after Spata18 KD affected mitochondrial health, mitochondrial calcium levels and cellular ATP levels were monitored in the HFLF cells. Results showed that 48 h after IR, mitochondrial calcium remain unchanged in scrambled cells while it increased in shSpata18 KD cells (Figure 5A and B). As Ca2+ overload leads to mitochondrial damage and dysfunction (34), Spata18 KD cells may be more sensitive to IR damage. Consistently, cellular ATP levels in Spata18 KD cells were down regulated by 40% when compared to that of scrambled cells 48 h after IR (Figure 5C).
Figure 5.
Mitophagy suppression due to Spata18 KD affects mitochondrial functions and DNA damage levels. (A) Fluorescent staining indicating mitochondrial calcium level in scrambled and shSpata18 HFLF cells after 6 Gy IR. Live microscopy was performed after staining with Rhod2 dye. Representative images are maximum intensity z-projection of all the slices from z-stacks. Scale bar represents 50 μm. (B) Quantification of maximum fluorescence intensity of z-stacks per slide. (C) Quantification of cellular ATP levels in scrambled and shSpata18 HFLF cells 1 h and 48 h after 6 Gy IR. (D) Representative images showing 53BP1 staining (red) in scrambled and shSpata18 HFLF cells 48 h after radiation. Nucleus was stained with DAPI, blue. A total of 317–437 cells from three independent experiments for each group were analyzed. The scale bars represent 50 μm. (E) Quantification of 53BP1 foci numbers in D. (F) Western blot analyses showing PAR, γ-H2AX, P-ATM, ATM and GAPDH in HFLF extracts collected at different timepoints post radiation at 6 Gy. The result of one out of three independent samples were shown Data: mean ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001.
DNA repair is a critical element of the DNA damage response. To check whether loss of Spata18 impacts DNA damage levels, markers for DNA damage were evaluated. As shown in Figure 5D and E, Spata18 KD cells had significantly higher 53BP1 foci number, a marker for double strand breaks, than scramble cells, indicating higher basal DNA damage. This result was further confirmed by western blot data in Figure 5F and Supplementary Figure S5, which show higher PAR and γ-H2AX levels in Spata18 KD cells without IR treatment. 1 h after IR, no significant change was observed in PAR, however, phospho-ATM (P-ATM) and γ-H2AX were significantly increased in both scramble and Spata18 KD cells, indicating an active DNA damage response. In the irradiated scrambled cells 48 h after IR, γ-H2AX was reduced to a level close to the untreated scrambled cells, indicating resolution of the DNA damage; however, in Spata18 KD cells, γ-H2AX and P-ATM still remained high, indicating less efficient repair of IR DNA damage. The combined results together suggest that Spata18 KD cells have higher basal DNA damage and are less efficient at repairing DNA damage.
DISCUSSION
Cells possess a systematic DNA damage response, which is executed both inside and outside the nucleus. The DNA damage response involves cooperation between the nucleus and mitochondria, and an extensive crosstalk exists between them. As the list of diseases with overlap between deficits in DNA repair and mitochondrial function continues to grow, it underscores the importance of elucidating how these two organelles coordinate their responses to DNA damage and contribute to cell survival (35–37).
In the present study, we found that after non-lethal doses of DNA damage, mitochondria respond in multiple ways. The responses included: an increase in mitochondrial content, ROS, and most intriguingly, changes in mitophagy. Modulation of mitophagy was observed as early as one hour after radiation, when it slightly declined, then gradually increased. It seems that at early timepoints after DNA damage, the cells attempt to preserve as many mitochondria as possible as a means to support energy demand; while at later time points, mitophagy increases and cells clear the superfluous or damaged mitochondria produced during oxidative phosphorylation process. Accumulation of mitochondrial ROS could be a driving force for mitophagy in this process and our preliminary data show that ROS scavenger can decrease mitophagy level (data not shown). Consistently, a similar time course of mitochondrial ROS to that of mitophagy after DNA damage was observed (Figure 1G, H), suggesting an important role of ROS in mitophagy induction after DNA damage. Along with increased mitophagy, new mitochondria were produced through biogenesis to maintain mitochondria homeostasis. This was observed in our results in Figure 1C, and also seen in other reports (38). The increase in mitophagy was a late response to DNA damage and not specific to any certain type of DNA damage since various DNA damaging stimuli increased mitophagy (Figure 1G–M). Importantly, this was a conserved response across species (Figure 2). Further, experiments in mice showed that the mitochondrial response to IR damage varied by tissue type and as such brain tissues (especially Purkinje cells) seemed to be more responsive than non-neuronal tissues, which may be related to their high metabolic activity and demands for energy. This result provides important clues for the explanation of Purkinje cells targeted in Ataxia telangiectasia (A-T), a premature disease induced by the mutation of the ATM gene, an important player in DNA damage response pathways (9).
Mitophagy is a dynamic cellular process and is regulated by different molecular pathways (39). In the case of mitophagy induction after DNA damage, we found that Spata18 had a profound regulatory role. Spata18 is a p53-inducible protein and was reported to induce intramitochondrial lysosome-like organelle after mitochondrial damage, which was independent of conventional autophagy (14). Although higher levels of mitophagy were observed at 24 and 48 h following IR in comparison to the control, we consistently saw no significant changes in LC3BII/LC3BI, p62 or other mitophagy-related proteins, which require the involvement of LC3B (Figure 3). This further supports the idea that mitophagy was induced after DNA damage, which is independent of conventional autophagy. It was reported earlier that Spata18 mediates mitochondrial protein degradation by affecting lysosomal protein translocation from cytoplasm to the mitochondrial matrix through physical interaction with BNIP3 and NIX (40). However, our result showed that when BNIP3 was knocked down, mitophagy was slightly suppressed, but not to the same extent as that produced by the KD of Spata18. This indicates that Spata18 may also mediate induction of mitophagy through pathways that are independent of BNIP3.
To investigate if Spata18 impacts DNA damage and repair, DNA damage was compared between Spata18 KD and scramble cells. DNA damage markers, γ-H2AX, P-ATM and 53BP1 foci, were all increased in cells with Spata18 KD at basal level as well as after IR. It is noteworthy that Spata18 is a cytoplasmic protein which has been found to be recruited to mitochondria after radiation (14). Therefore, we believe it effects nuclear DNA damage and repair indirectly via its role on mitophagy and mitochondrial homeostasis.
The DNA damage exposures applied in the present study will induce nuclear DNA damage and also some mtDNA damage. Though our data (Figure 1F) showed that ATM mediated DDR is important for mitophagy induction, the relative influence of nuclear DNA damage and mtDNA damage on mitophagy induction needs further study.
In conclusion, our results show that mitophagy is modulated after DNA damage. It first slightly decreases and then with time significantly increases. Modulation of mitophagy after DNA damage is consistently observed in primary fibroblasts from different sources and after different DNA damaging stimuli. We also evaluated this process cross species by using mice and C. elegans and showed that mitophagy is altered in a tissue-specific manner ex vivo after IR. Further, we show that the protein Spata18, a p53-inducible protein, is critical in mitophagy regulation after DNA damage. Knockdown of Spata18 did not only impair mitochondrial function but it also altered levels of cellular DNA damage. Our study has provided new insights into the nuclear-mitochondrial signaling axis that responds to acute DNA damage with active mitochondrial quality control.
DATA AVAILABILITY
The GSE number for the microarray data is GSE150273.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Afred May, Tomasz Kulikowicz, Christopher Dunn and Mariana Martinsfor help in the experiments; Elin Lehrmann and Yongqing Zhang for microarray technical support; and Drs Yujun Hou and Louis Christiansen for critical reading of the manuscript.
Contributor Information
Xiuli Dan, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
Mansi Babbar, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
Anthony Moore, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
Noah Wechter, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
Jingyan Tian, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
Joy G Mohanty, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
Deborah L Croteau, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
Vilhelm A Bohr, Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA; Danish Center for Healthy Aging, University of Copenhagen, 2200 Copenhagen, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program, National Institute on Aging. Funding for open access charge: National Institute on Aging, Intramural Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Foster S.S., De S., Johnson L.K., Petrini J.H., Stracker T.H.. Cell cycle- and DNA repair pathway-specific effects of apoptosis on tumor suppression. Proc. Natl Acad. Sci. U.S.A. 2012; 109:9953–9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hill S., Van Remmen H.. Mitochondrial stress signaling in longevity: a new role for mitochondrial function in aging. Redox. Biol. 2014; 2:936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poyton R.O., McEwen J.E.. Crosstalk between nuclear and mitochondrial genomes. Annu. Rev. Biochem. 1996; 65:563–607. [DOI] [PubMed] [Google Scholar]
- 4. Qin L., Fan M., Candas D., Jiang G., Papadopoulos S., Tian L., Woloschak G., Grdina D.J., Li J.J.. CDK1 enhances mitochondrial bioenergetics for Radiation-Induced DNA repair. Cell Rep. 2015; 13:2056–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamamori T., Yasui H., Yamazumi M., Wada Y., Nakamura Y., Nakamura H., Inanami O.. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic. Biol. Med. 2012; 53:260–270. [DOI] [PubMed] [Google Scholar]
- 6. Roberts R.F., Tang M.Y., Fon E.A., Durcan T.M.. Defending the mitochondria: the pathways of mitophagy and mitochondrial-derived vesicles. Int. J. Biochem. Cell Biol. 2016; 79:427–436. [DOI] [PubMed] [Google Scholar]
- 7. Ashrafi G., Schwarz T.L.. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013; 20:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez-Vicente M. Neuronal mitophagy in neurodegenerative diseases. Front. Mol. Neurosci. 2017; 10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang E.F., Kassahun H., Croteau D.L., Scheibye-Knudsen M., Marosi K., Lu H., Shamanna R.A., Kalyanasundaram S., Bollineni R.C., Wilson M.A. et al.. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016; 24:566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang E.F. Mitophagy and NAD+ inhibit Alzheimer disease. Autophagy. 2019; 15:1112–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao F., Yang J., Wang D., Li C., Fu Y., Wang H., He W., Zhang J.. Mitophagy in Parkinson's Disease: Pathogenic and therapeutic implications. Front Neurol. 2017; 8:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang J.Y., Yi H.S., Kim H.W., Shong M.. Dysregulation of mitophagy in carcinogenesis and tumor progression. Biochim. Biophys. Acta Bioenerg. 2017; 1858:633–640. [DOI] [PubMed] [Google Scholar]
- 13. Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G.. The hallmarks of aging. Cell. 2013; 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyamoto Y., Kitamura N., Nakamura Y., Futamura M., Miyamoto T., Yoshida M., Ono M., Ichinose S., Arakawa H.. Possible existence of lysosome-like organella within mitochondria and its role in mitochondrial quality control. PLoS One. 2011; 6:e16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durkin M.E., Qian X., Popescu N.C., Lowy D.R.. Isolation of mouse embryo fibroblasts. Biol. Protoc. 2013; 3:e908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun N., Malide D., Liu J., Rovira I.I., Combs C.A., Finkel T.. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat. Protoc. 2017; 12:1576–1587. [DOI] [PubMed] [Google Scholar]
- 17. Fang E.F., Hou Y., Palikaras K., Adriaanse B.A., Kerr J.S., Yang B., Lautrup S., Hasan-Olive M.M., Caponio D., Dan X. et al.. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat. Neurosci. 2019; 22:401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou Y., Lautrup S., Cordonnier S., Wang Y., Croteau D.L., Zavala E., Zhang Y., Moritoh K., O’Connell J.F., Baptiste B.A. et al.. NAD(+) supplementation normalizes key Alzheimer's features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl Acad. Sci. U.S.A. 2018; 115:E1876–E1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huna A., Salmina K., Jascenko E., Duburs G., Inashkina I., Erenpreisa J.. Self-Renewal signalling in presenescent tetraploid IMR90 cells. J Aging Res. 2011; 2011:103253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gentile M., Latonen L., Laiho M.. Cell cycle arrest and apoptosis provoked by UV radiation-induced DNA damage are transcriptionally highly divergent responses. Nucleic Acids Res. 2003; 31:4779–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee Y.J., Park S.J., Ciccone S.L., Kim C.R., Lee S.H.. An in vivo analysis of MMC-induced DNA damage and its repair. Carcinogenesis. 2006; 27:446–453. [DOI] [PubMed] [Google Scholar]
- 22. Kuehne A., Emmert H., Soehle J., Winnefeld M., Fischer F., Wenck H., Gallinat S., Terstegen L., Lucius R., Hildebrand J. et al.. Acute activation of oxidative pentose phosphate pathway as First-Line response to oxidative stress in human skin cells. Mol. Cell. 2015; 59:359–371. [DOI] [PubMed] [Google Scholar]
- 23. Sun N., Yun J., Liu J., Malide D., Liu C., Rovira I.I., Holmstrom K.M., Fergusson M.M., Yoo Y.H., Combs C.A. et al.. Measuring in vivo mitophagy. Mol. Cell. 2015; 60:685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sargsyan A., Cai J., Fandino L.B., Labasky M.E., Forostyan T., Colosimo L.K., Thompson S.J., Graham T.E.. Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor. Sci. Rep. 2015; 5:12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosado C.J., Mijaljica D., Hatzinisiriou I., Prescott M., Devenish R.J.. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy. 2008; 4:205–213. [DOI] [PubMed] [Google Scholar]
- 26. Fang E.F., Palikaras K., Sun N., Fivenson E.M., Spangler R.D., Kerr J.S., Cordonnier S.A., Hou Y., Dombi E., Kassahun H. et al.. In vitro and in vivo detection of mitophagy in human cells, C. Elegans, and mice. J. Vis. Exp. 2017; doi:10.3791/56301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sekine S., Youle R.J.. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 2018; 16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yun J., Puri R., Yang H., Lizzio M.A., Wu C., Sheng Z.H., Guo M.. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. eLife. 2014; 3:e01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Novak I., Dikic I.. Autophagy receptors in developmental clearance of mitochondria. Autophagy. 2011; 7:301–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saito T., Nah J., Oka S.I., Mukai R., Monden Y., Maejima Y., Ikeda Y., Sciarretta S., Liu T., Li H. et al.. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J. Clin. Invest. 2019; 129:802–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamamori T., Ike S., Bo T., Sasagawa T., Sakai Y., Suzuki M., Yamamoto K., Nagane M., Yasui H., Inanami O.. Inhibition of the mitochondrial fission protein dynamin-related protein 1 (Drp1) impairs mitochondrial fission and mitotic catastrophe after x-irradiation. Mol. Biol. Cell. 2015; 26:4607–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Twig G., Shirihai O.S.. The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox. Signal. 2011; 14:1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kulkarni R., Thomas R.A., Tucker J.D.. Expression of DNA repair and apoptosis genes in mitochondrial mutant and normal cells following exposure to ionizing radiation. Environ. Mol. Mutagen. 2011; 52:229–237. [DOI] [PubMed] [Google Scholar]
- 34. Kaddour-Djebbar I., Choudhary V., Brooks C., Ghazaly T., Lakshmikanthan V., Dong Z., Kumar M.V.. Specific mitochondrial calcium overload induces mitochondrial fission in prostate cancer cells. Int. J. Oncol. 2010; 36:1437–1444. [DOI] [PubMed] [Google Scholar]
- 35. Valentin-Vega Y.A., Maclean K.H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F.C., Cleveland J.L., Green D.R., Kastan M.B.. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012; 119:1490–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang E.F., Scheibye-Knudsen M., Brace L.E., Kassahun H., SenGupta T., Nilsen H., Mitchell J.R., Croteau D.L., Bohr V.A.. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014; 157:882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sugimoto M. A cascade leading to premature aging phenotypes including abnormal tumor profiles in Werner syndrome (review). Int. J. Mol. Med. 2014; 33:247–253. [DOI] [PubMed] [Google Scholar]
- 38. Bartoletti-Stella A., Mariani E., Kurelac I., Maresca A., Caratozzolo M.F., Iommarini L., Carelli V., Eusebi L.H., Guido A., Cenacchi G. et al.. Gamma rays induce a p53-independent mitochondrial biogenesis that is counter-regulated by HIF1alpha. Cell Death Dis. 2013; 4:e663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pickles S., Vigie P., Youle R.J.. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 2018; 28:R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakamura Y., Kitamura N., Shinogi D., Yoshida M., Goda O., Murai R., Kamino H., Arakawa H.. BNIP3 and NIX mediate Mieap-induced accumulation of lysosomal proteins within mitochondria. PLoS One. 2012; 7:e30767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GSE number for the microarray data is GSE150273.