Abstract
Streptomyces phage ϕC31 integrase (Int)—a large serine site-specific recombinase—is autonomous for phage integration (attP x attB recombination) but is dependent on the phage coded gp3, a recombination directionality factor (RDF), for prophage excision (attL x attR recombination). A previously described activating mutation, E449K, induces Int to perform attL x attR recombination in the absence of gp3, albeit with lower efficiency. E449K has no adverse effect on the competence of Int for attP x attB recombination. Int(E449K) resembles Int in gp3 mediated stimulation of attL x attR recombination and inhibition of attP x attB recombination. Using single-molecule analyses, we examined the mechanism by which E449K activates Int for gp3-independent attL x attR recombination. The contribution of E449K is both thermodynamic and kinetic. First, the mutation modulates the relative abundance of Int bound attL-attR site complexes, favoring pre-synaptic (PS) complexes over non-productively bound complexes. Roughly half of the synaptic complexes formed from Int(E449K) pre-synaptic complexes are recombination competent. By contrast, Int yields only inactive synapses. Second, E449K accelerates the dissociation of non-productively bound complexes and inactive synaptic complexes formed by Int. The extra opportunities afforded to Int(E499K) in reattempting synapse formation enhances the probability of success at fruitful synapsis.
INTRODUCTION
Serine recombinases (SRs) of the small and large types are both characterized by similar small amino-terminal catalytic domains, but differ considerably in their carboxyl-terminal domains (CTDs) (1,2). The well characterized resolvases and invertases of the small SR family contain a typical helix-turn-helix motif in the DNA binding carboxyl-terminus. In the large SR family—which includes phage-coded integrases—the CTD is much larger (300–500 amino acids), and likely harbor regulatory peptide motifs involved in the control of recombination in vivo. Aside from their utility in unveiling mechanisms and regulation of recombination, serine integrases have been developed as tools for genome engineering (3,4), for creating binary genetic switches in synthetic biology (5), for building biological computers that count and record input stimuli (6–8) and for assembling DNA fragments in vitro to reconstitute gene clusters encoding metabolic pathways (9,10).
In contrast to tyrosine family phage integrases, their serine family counterparts mediate integration without accessory proteins by exchanging short (<50 bp long) phage and bacterial attachment sites (attP and attB, respectively) (1,11–12) (Figure 1A). The reaction follows the classical serine recombination mechanism: (i) concerted double strand breaks within synapsed att sites, each bound by an Int dimer, (ii) concomitant covalent linkage of the catalytic serine residues to the 5′-phosphate ends of the broken DNA, (iii) 180° relative rotation of the cleaved DNA–protein complex and (iv) rejoining of the broken strands by the chemical reversal of cleavage (2). Unlike integration, excision of the prophage by recombination between flanking attL and attR sites requires, in addition to the integrase, a phage coded recombination directionality factor (RDF) (13–16) (Figure 1B). Stoichiometric binding of RDF to the integrase is thought to trigger the lysogeny-to-lysis switch by promoting assembly of the excision synapse (attL-attR), and also by inhibiting formation of the integration synapse (attP-attB) (14–15,17).
Figure 1.
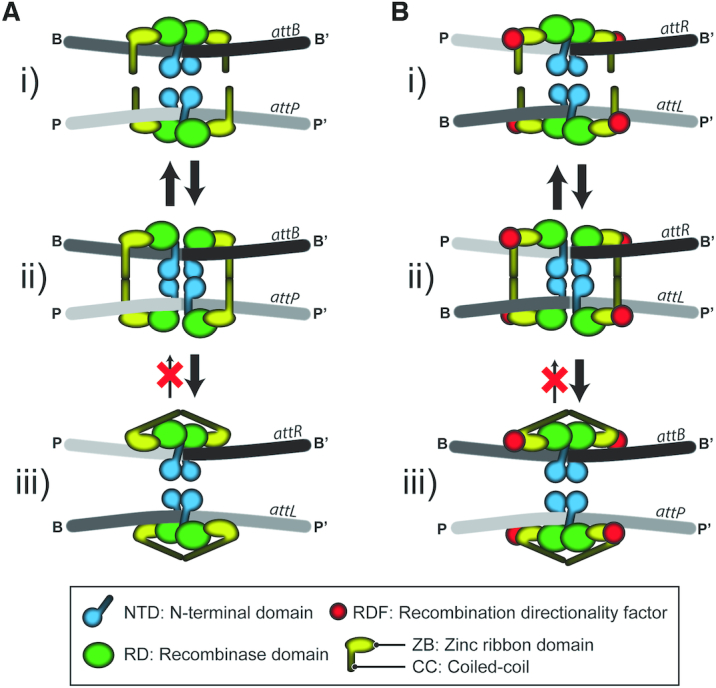
Phage integration (attP x attB → attL + attR) and excision (attL x attR → attP + attB) mediated by serine integrases; control of recombination directionality by RDF. The integration and excision reactions are schematically diagrammed in panels (A) and (B), respectively. The pre-chemical steps of the reaction involve binding of Int dimers to att sites (I) followed by synapsis of the bound sites (II). In the functional synapse, the sites are paired in a complementary fashion, that is, a P-type half-site is paired with a B-type half-site between the synaptic partners. The ensuing chemical/conformational steps include DNA cleavage, 180° rotation of the cleaved complex and relegation of the broken strands (III). The reaction is irreversible because of the auto-inhibited state of the post-recombination Int-att site complex. In the schematic representation of Int, the amino-terminal domain (NTD) is distinguished from the CTD comprised of a tripartite motif, namely, a recombinase domain (RD), a zinc ribbon domain (ZD) and a coiled coil (CC) region. RDF regulates the directionality of recombination by modulating the CC trajectories. The presence of RDF promotes the auto-inhibited state of Int bound attP-attB, while the absence of RDF has a similar effect on Int bound attL-attR. This figure follows the model for the recombination synapse proposed from the structure of the L1 integrase CTD bound to the A118 attP half-site (23).
A subset of the phage/prophage coded Integrase-RDF pairs identified so far has provided clues to the possible mechanisms for controlling recombination directionality (14–15,17–21). Particularly useful insights have come from the crystal structure of a complex between the CTD of the L1 (Listeria innocua) prophage integrase and a half-attP target site of the 98% identical Listeria phage A118 integrase (22,23). The structure shows extensive interactions between attP and two DNA binding subdomains within CTD—a recombinase domain (RD) and connected to it an unusual zinc ribbon domain (ZD). A coiled coil (CC) motif nestled between helices within ZD projects away from the main body of the protein–DNA assembly. A structural model built from the derived interactions of the CC motifs at juxtaposed attP and attB half-sites suggests that attL and attR bound ZDs form an auto-inhibited complex in which the CC interactions occur within integrase dimers bound to each att site (intra-dimer) (Figure 1A, iii; B, i). RDF is thought to reconfigure these interactions into the inter-dimer format, between an integrase dimer bound to attL and one bound to attR (Figure 1B, ii). This model is consistent with DNA binding and recombination results obtained using att site and integrase mutants (23–26). More recent data suggest that the RDF (gp44) of phage A118 binds to the CC motif to foster the Int tetramer conformation required for productive attL-attR synapsis (17).
While phage integrases have strong sequence homology, their RDFs are highly variable (8,21). The RDFs are generally mono-specific in integrase interaction, raising questions about a common mechanism for activation of attL-attR recombination among the different integrase-RDF systems. A recent study identified key amino acids in the integrase of the Streptomyces phage ϕC31 (hereafter referred to as Int) and in its RDF (the phage coded gp3) required for their interaction (27). Mutations that disrupt the interaction result in both defective excision and integration. The findings are consistent with gp3 binding to a hinge region at the base of the CC within Int, and hinge flexibility being important for integration as well as excision. Mutational analyses of the A118 Int-gp44 system suggest the involvement of two separate contacts between the integrase and its RDF in determining recombination directionality (17). Both interactions are required for activating attL x attR recombination; however, the one mediated by the extreme amino-terminal region of gp44 is dispensable in suppressing attP × attB recombination.
Self-activated variants of Int have been isolated that perform attL × attR recombination without gp3, but at relatively low efficiency (26). Fully competent in attP × attB recombination, they are capable of attL × attL or attR × attR recombination when the central dinucleotide exchanged during the reaction is symmetrized (5′TA3′) in artificial substrates or kept asymmetric (5′AA3′) as in the native substrates. Symmetrizing relieves the homology/base pairing constraint on recombination. In theory, either ‘normal’ or ‘contrary’ site alignment within the recombination synapse could result in product formation. In the normal alignment, referred to as complementary pairing (26), the synaptic interaction is between an Int subunit bound to a P-type (P or P’) half-site and one bound to a B-type half-site (B or B’), P-B, P’-B, P-B’ or P’-B’ (Figure 1). In the contrary (non-complementary) synapsis, the synaptic interaction is P-to-P or B-to-B type. Combined results from pairwise combinations of attLAA, attRAA, attLTA and attRTA sites demonstrate that recombination by one of the self-activated variants, Int(E449K), occurs from both normal and contrary site alignments, with a strong bias for the former. Consistent with a central role for the CC motif in determining the functional status of the synapse (22–23,28), the strongest among the activating mutations map to the same helical phase of the CC motif (26). The E449K mutation chosen for the present analysis is located at the start of the CC region.
Our previous analyses of wild-type Int using single-molecule tethered particle motion (TPM) revealed differences in Brownian motion (BM) amplitudes among recombination competent or incompetent attP-attB and attL-attR synaptic structures (29). In broad terms, the functional synapse formed by head-to-tail att sites has a slightly higher BM amplitude than the non-functional synapse, which would be consistent with the active ‘parallel’ and inactive ‘anti-parallel’ arrangement of the paired sites (Figure 1A and B; Supplementary Figure S1). Furthermore, the attP-attB synapse formed in the presence of Int plus gp3 and the attL-attR synapse formed by Int alone (both synapses are non-functional in recombination) fit a bi-modal Gaussian pattern of BM amplitudes. The similar difference in the mean BM amplitudes between the Gaussian pairs for the attP-attB and attL-attR synapses suggest that, depending on the att site partners, the presence or absence of gp3 induces analogous inactive synaptic conformations.
The present side-by-side analysis of Int and Int(E449K) by single-molecule TPM reveals a bipartite thermodynamic and kinetic role for E449K in the activation of attL × attR recombination. In its thermodynamic role, E449K modulates the landscape of Int-bound att site complexes and the synaptic conformations assembled from them. The fraction of bound complexes (BC) that proceed to synapsis is higher for Int(E449K) compared to Int. While a subset of the synaptic structures formed by Int(E449K) complete recombination, none do so in the case of Int. In its kinetic role, E449K promotes the rapid dissociation of non-productively bound Int-att site complexes and of abortive wayward synaptic complexes. The opportunity for reattempts at productive attL-attR synapsis, with the likelihood of success, is thereby enhanced. This dual role of E449K partially recapitulates that of the native RDF, gp3. The thermodynamic effect of gp3 far exceeds that of E449K, while the two are nearly indistinguishable in their kinetic effect.
MATERIALS AND METHODS
Proteins
The ϕC31 integrase (Int), its variant Int(E499K) and gp3 proteins were expressed in Escherichia coli, and purified according to published procedures (15,30). The Int and Int(E499K) stocks were stored in aliquots at −70°C in 20 mM Tris–HCl (pH 7.5), 2 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 0.3 M NaCl and 10% glycerol. Aliquots of gp3 were stored at −70°C in10 mM sodium phosphate (pH 7.4), 2.7 mM KCl, 300 mM NaCl and 5% glycerol. For standard biochemical assays, the proteins were diluted in the reaction buffer (10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 100 mM NaCl, 5 mM DTT, 5 mM spermidine, 4.5% glycerol and 0.5 mg/ml bovine serum albumin (BSA)) on ice just prior to use. The reaction buffer in TPM and FCS (fluorescence correlation spectroscopy) assays was 10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 4.5% glycerol, 5 mM DTT and 1 mg/ml BSA (29).
DNA substrates
The 1303 att sites containing linear DNA substrates utilized for the TPM assays have been described previously (29). The spacing between the cross-over points within the two att sites was 752 bp for head-to-tail and head-to-head att sites.
The plasmid substrates used in ensemble assays contained the attP-attB or attL-attR pair of sites cloned into the pUC19 vector. First, an ∼600 bp DNA fragment was amplified from a plasmid template using two synthetic DNA primers that included the desired att site sequence. The primers were designed to generate an EcoRI site and a BamHI site close to the termini of the polymerase chain reaction (PCR) product. The amplified DNA was digested with EcoRI plus BamHI, and was inserted between the EcoRI and BamHI sites in pUC19. The ∼500 bp DNA sequence sandwiched between attL and attR was derived from the TRP1 gene of Saccharomyces cerevisiae.
The 849 bp linear DNA substrate for the FCS experiments was prepared by PCR amplification of the relevant region of a plasmid containing attP and attB in head-to-tail orientation. One of the primers used in the PCR reaction was labeled with the fluorophore TMR (trimethylrhodamine). The 91 bp DNA fragment containing a single attL site, mimicking the linear excision product (of the same size and sequence), was also obtained by PCR amplification from a template plasmid using two primers, one of which was tagged with TMR at the 5′ end.
DNA substrates employed in the various assays, circular plasmids isolated from bacterial cultures and the linear DNA molecules prepared by PCR amplification of appropriate templates, were purified by phenol-chloroform extraction and ethanol precipitation.
Single-molecule TPM analysis
The TPM assays were carried out essentially as previously described for wild-type ϕC31 integrase (29) with a few modifications. The preparation of reaction chambers and tethering surfaces to prevent non-specific interactions now employed a PEG treatment protocol (31,32) instead of the earlier procedure of washing with buffer containing casein or BSA (33). The ratio of maleimide-PEG5000 to biotin-PEG5000 (Laysan Bio, Inc.) in the surface preparation buffer was 181:1. The mean diameter (240 nm) of the polystyrene beads (Bangs Laboratories, Inc.) used in the present study was slightly larger than that (200 nm) of beads previously used. The acquisition of single molecule BM amplitude time traces, estimation of dwell times from these traces and fitting the dwell time plots to a single exponential algorithm were performed as described in published studies (29,34–35). The data set for each assay (Figures 3–6) was derived from molecules observed in five to six separate TPM experiments and cumulatively depicted in Supplementary Figures S2–5.
Figure 3.
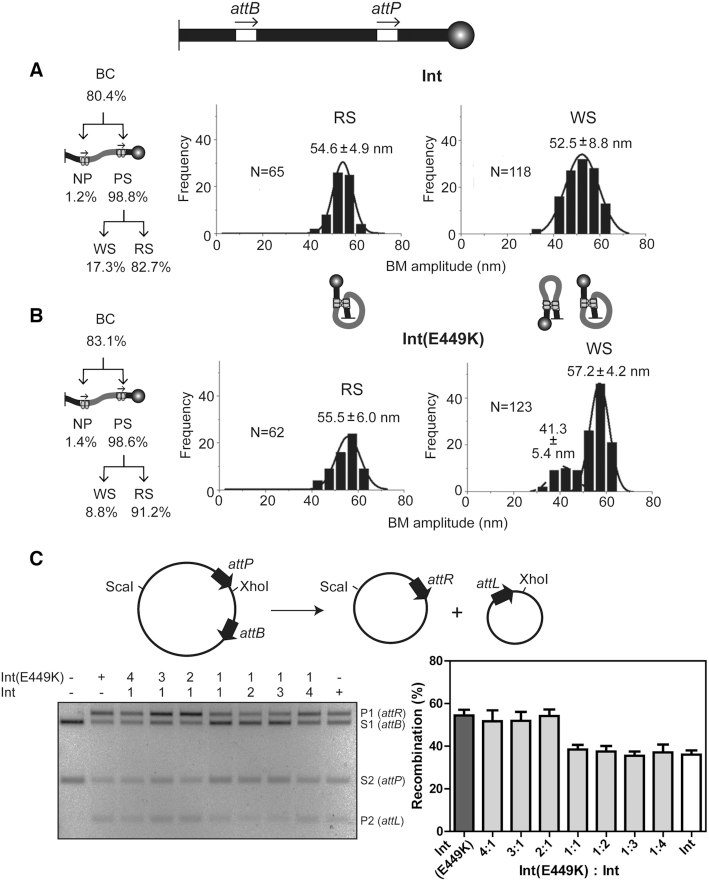
Int and Int(E449K) mediated attP × attB recombination. (A and B) The DNA substrate containing attB and attP sites in head-to-tail orientation is schematically drawn at the top with the tethering surface and the polystyrene bead indicated as in Figure 2. The partitioning of the total BC into individual complexes is indicated at the left. The plots of the BM amplitude distributions of the functional (RS) and non-functional (WS) synaptic complexes are based on the data from Supplementary Figure S2A. N refers to the number of these two synaptic states (or the number of transition events generating them) captured in single molecule time traces. For example, the trace III shown in Figure 2 would have contributed N = 2 toward WS events. N is larger for the WS plot as one molecule can potentially form an abortive synapse in a recurrent fashion. An RS synapse by definition is a unique event for a molecule, resulting in completion of recombination. In this figure (and in Figures 4–6), the schematics of synapsed molecules are meant to indicate that the arrangement of the att sites in the WS complexes may be heterogeneous, parallel (lower BM amplitude species) or anti-parallel (higher amplitude species). In the RS complex, the arrangement is parallel. (C) The circular substrate used in the ensemble assays and the products of recombination are shown schematically at the top. Reactions contained Int or Int(E449K), or the indicated molar ratios of the two. Recombination was assayed by ScaI plus XhoI digestion of DNA, and agarose gel electrophoresis. The bands resulting from the substrate are labeled S1 (attB), S2 (attP); those resulting from the products are labeled P1 (attR), P2 (attL). The mean recombination efficiencies (±SD) from three separate assays are plotted.
Figure 6.
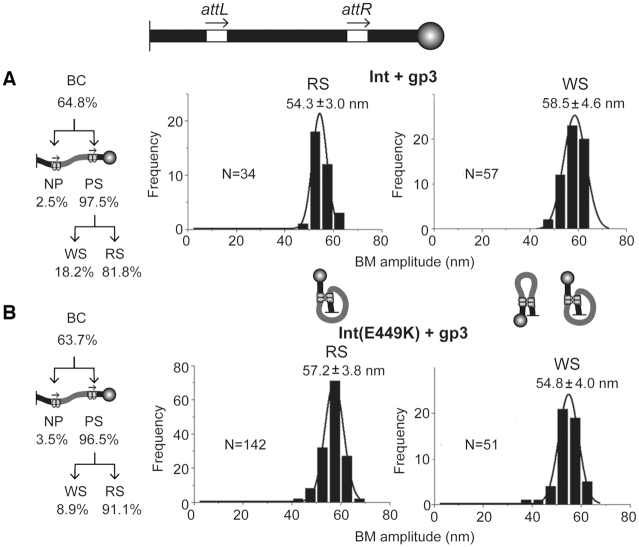
Activation of Int and stimulation of Int(E449K) in attL x attR recombination by gp3. (A and B) The only difference between the analysis shown here and that presented in Figure 5 is that reactions were initiated by adding a mixture of Int or Int(E449K) and gp3 (Supplementary Figure S5).
FCS analysis of attP X attB recombination by Int
FCS was carried out using a confocal system assembled using a Nikon Ti Eclipse microscope. The 561 nm laser was directed via a dichroic mirror (Semrock, Di01-R405/488/561/635) and focused with a Nikon Apochromat 100 × NA 1.4 oil immersion objective to excite reaction samples containing the TMR-labeled DNA substrate. Fluorescence emission was collected using a Semrock NF03-405/488/561/635E-25 notch filter and recorded by a set of avalanche photodiodes (Picoquant MPD-5C5T). FCS measurements were performed on 100 μl samples placed in a glass chamber. A 60 s fluorescence intensity fluctuation was acquired at 1 min intervals over a 60 min time course. In parallel assays, the fluorescence emission data was collected after stopping the reactions with sodium dodecyl sulphate (SDS) (0.05%) addition at various time points. FCS curves were built in Symphotime (Picquant), and were fitted to a two-component 3D free diffusion model:
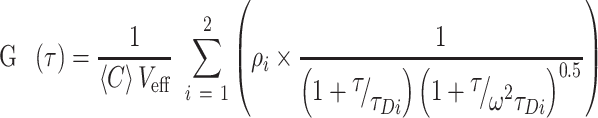 |
V eff is the excitation volume, C is the average concentration of molecules in the observation volume, τ is the correlation time and τD is the diffusion time, which represents the average lateral transition time of the particle through the observation volume with an axial (z0) to lateral (r0) dimension ratio w = (z0/r0). The ‘structural parameter’ w and Veff were determined by calibrating the experimental system using a standard dye, R6G (D = 414 μm2/s) (36). All calculations, including the evaluation of the autocorrelation curves, were performed using symphotime (Picoquant).
Quantitation of recombination by standard biochemical assays
The ensemble recombination reactions were carried out under buffer conditions similar to those described previously (26). The plasmid DNA was present at 4.62 nM together with Int, Int(E449K) or mixtures of the two and gp3 at 330 nM (estimated as protein monomers). After incubation at 30°C for 2 h, reactions were stopped by transfer to 80°C for 10 min. Samples were digested with ScaI and XhoI, and were fractionated by electrophoresis in 1% agarose gels to separate the substrate and recombinant product bands. Ethidium bromide stained DNA bands were quantitated using Quantity One software (Bio-Rad). The recombination efficiency was estimated as the sum of the intensities of the product bands divided by the sum of the intensities of the product plus unreacted substrate bands.
RESULTS
The rationale for the TPM analysis of recombination between two att sites by Int
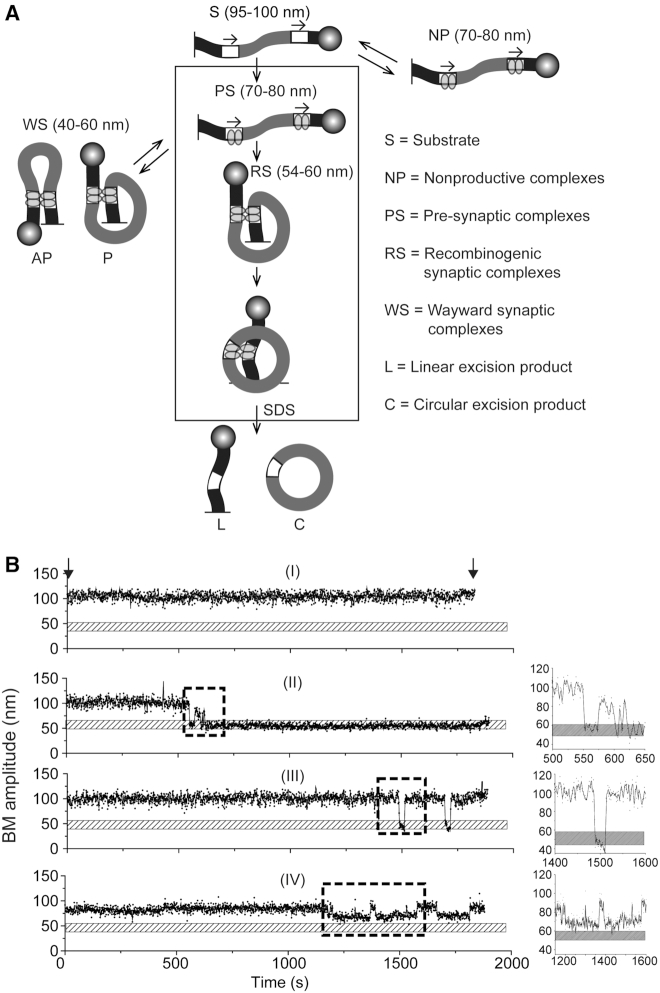
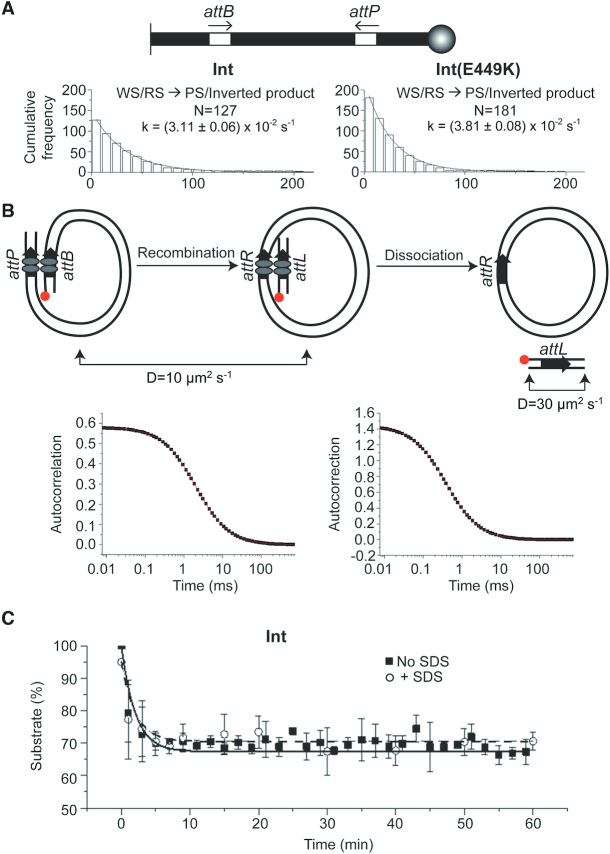
In TPM analysis, the individual steps of recombination from free substrate → protein bound intermediate complexes → product(s) is reported by finite differences in the BM amplitudes of a polystyrene bead attached to one end of the linear substrate DNA molecule whose other end is tethered to a glass slide (29,34,37–38). An unbound substrate molecule has a ‘high’ BM amplitude, one occupied by Int dimers at the att sites present within it has an ‘intermediate’ BM amplitude, and one in which the bound att sites have synapsed by looping of the intervening DNA has a ‘low’ BM amplitude (Figure 2 and Supplementary Figure S1). In the case of att sites oriented in a head-to-tail fashion, the linear product of the deletion reaction will retain the low BM amplitude of the synapse. Successful recombination can be distinguished from an unsuccessful synapse by treatment with SDS, which dissociates the bound Int. Abortively synapsed molecules are restored to the high BM amplitude of the substrate. For att sites in the head-to-head orientation, SDS addition cannot distinguish between recombination or the lack of it within a synapse, as the substrate and the inversion product have the same length. However, it is possible to kinetically identify recombination, provided the dissociation of the abortive synapse and that of the product synapse differ sufficiently in their rate constants (35).
Figure 2.
TPM analysis of Int-mediated recombination. (A) The schematic diagram represents the recombination reaction attP × attB → attL + attR for sites in head-to-tail orientation. The tethered and bead-attached ends of the substrate DNA molecule (S) are indicated by the short vertical line at the left and the sphere at the right, respectively. The Int dimers bound to att sites are shown as twin ovals. The vertical series of arrows denote the conversion of S via the PS complex and the recombinogenic synaptic complex (RS) to the products L (linear; tethered) and C (circular; free). Futile binding events (NP; non-productive complex) and futile synapsis (WS; wayward synaptic complex) are placed outside the box enclosing the functional reaction path. NP and WS may join the vertical sequence of events following their dissociation. The approximate BM amplitudes characteristic of the individual DNA species are indicated in parentheses (see also Supplementary Figure S1). The RS complex contains the att sites in their functional parallel (P) geometry. The WS complexes may contain the sites in parallel or anti-parallel (AP) geometry (or conformations that approximate these geometries), thus broadening their BM amplitude distribution. The sum of NP + PS (WS + RS) gives the total bound complexes, indicated by BC in Figures 3–6 and Supplementary Figure S2B. (B) Typical single molecule BM amplitude time traces indicating unbound DNA substrate, Int binding to DNA and transitions of the bound DNA–protein complexes are shown. The stippled horizontal bar depicts the BM amplitude of synapsed molecules. The left and right short vertical arrows indicate the addition of Int (0 min) and SDS (30 min), respectively. (i) A molecule that failed to bind during the observation period (S); (ii) a molecule that formed a transient abortive synapse (WS), dissociated (PS), synapsed productively (RS) and completed recombination; (iii) a molecule that made two synaptic attempts (both futile; WS) and returned each time to the substrate state (S) by protein dissociation; (iv) A molecule that oscillated between the unbound (S) and non-productively bound (NP) states without succeeding to form synapsis (WS or RS). The BM amplitude changes that report specific transition events are enclosed in the dashed boxes, and are accentuated in the panels at the right with the expanded Y-axis. The number of molecules observed by time trace records in individual sets of experiments and their BM amplitude patterns are shown in Supplementary Figures S2–5. The synaptic complexes identified from these time traces, and classified into the RS and WS groups, were the source of the data for Figures 3–6.
The recombination pathway characterized previously for Int, and diagrammed schematically in Figure 2 for a pair of head-to-tail att sites, provides a frame of reference for the present analysis. The reaction proceeds through the following progressive conversion steps: S (substrate) → PS (pre-synaptic complexes) → RS (recombinogenic synaptic complexes) → L + C (linear plus circular deletion products). The ‘off-pathway’ complexes NP (non-productively bound complexes; formed from S) and WS (wayward synaptic complexes; formed from PS) may gain entry to the reaction path after dissociation to S and PS, respectively. In order to register a consistent initial decrease in BM amplitude from that of S, both the att sites in the substrate DNA have to be bound by Int dimers (NP or PS in Figure 2) Int occupancy of only one of the two att sites is not reliably reported by BM amplitude, and is a limitation of the TPM analysis. The situation is different for tyrosine recombinases (YRs) such as Cre and Flp whose occupancy of a single target site bends the DNA sufficiently to cause a drop in BM amplitude (34,35).
The experimental results described below (Figures 3–9) were obtained using 1303 bp DNA molecules containing a pair of att sites with a spacing of 752 bp between their cross-over points (29) (Supplementary Figure S1). In interpreting the data presented, it is helpful to keep in mind the following BM amplitude values for the relevant DNA species derived from published work (29) and the present study: S (unbound), 95–100 nm; NP and PS (Int bound but unsynapsed), 70–80 nm; RS and WS (synapsed, productively or abortively), 40–60 nm; L (tethered linear product from deletion), 40–45 nm (Supplementary Figure S1). The rather broad BM amplitude range of the synapsed molecules would be consistent with the presence of heterogeneous conformations within the synaptic structures. In a subset of the cases, these BM amplitude distributions for WS could be split into two overlapping Gaussians. Small variations of BM amplitude in the bound DNA were noticed depending on whether a reaction contained Int alone or Int plus gp3. However, such variations do not preclude the identification of the individual complexes of interest.
Figure 9.
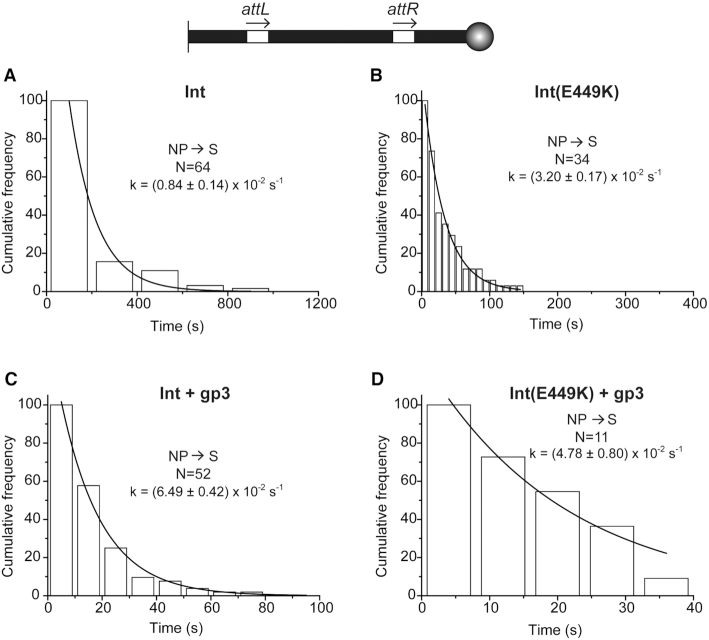
Dissociation kinetics of NP assembled in the attL-attR (head-to-tail) DNA substrate. (A–D). The dwell times of the NP complexes before dissociating to S were determined from single molecule time traces, and were plotted as in Figure 8 with the ordinate at time zero normalized as ‘100’. A single exponential algorithm was employed to derive k-NPd. N = the total number of NP complexes observed.
TPM analysis of Int (29), unlike prior biochemical studies (15,39), is able to identify synaptic structures between attL and attR formed by Int and between attP and attB formed by Int plus gp3. Presumably, these futile synaptic structures are unable to survive the conditions of gel electrophoresis used to probe them. The TPM terminology of WS encompasses all of the synaptic structures that fail to produce recombinants in attP × attB and attL × attR reactions with Int alone or with Int plus gp3. Our analysis cannot directly address whether a subset of the WS complexes are chemically competent to execute strand cleavage even though they cannot complete recombination. In our observations, SDS addition to TPM reactions almost never separates the bead from the tethered DNA segment whereas cutting the DNA with a restriction enzyme readily untethers the bead (29). Thus, strand cleavage is either absent within the WS complexes, or is readily reversible and therefore highly transient.
Int(E449K)-mediated recombination between attP and attB sites
Previous ensemble biochemical assays showed that Int(E449K) is capable of carrying out attP × attB recombination efficiently, in addition to its ability to recombine attL × attR weakly (26). In the present study, we wished to examine at the single molecule level what effects the E449K mutation has on the individual steps of the reaction. The general strategy, as in our previous TPM investigations of YRs and SRs (29,34–35), was to observe the BM amplitude responses of individual DNA molecules to Int/Int(E449K) (either alone or together with gp3) over a 30 min interval. At 30 min, the change in the BM amplitude distribution resulting from SDS addition was recorded (Supplementary Figures S2–5). Furthermore, the BM amplitude transitions of BC and the time dependence of such events were derived from the 0–30 min single molecule traces.
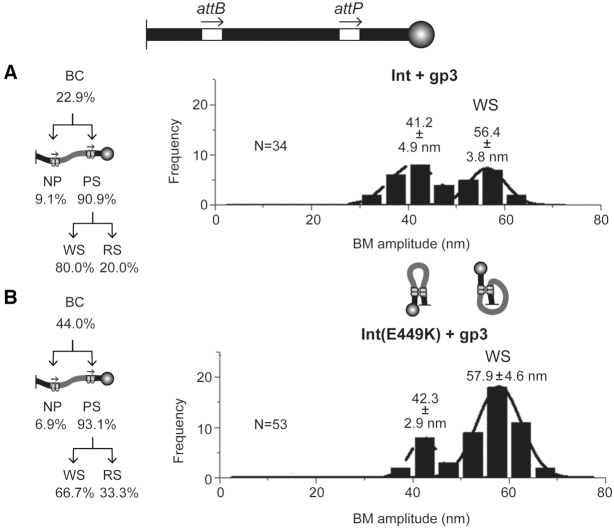
In the head-to-tail orientation of attP-attB, Int and Int(E449K) were nearly identical in the formation of BC and their partitioning into NP and PS (Figure 3A and B; Supplementary Figure S2A). Nearly all binding events were subsequently fruitful in forming synapsis (∼99% PS), with few non-productively bound molecules (≤1.4% NP). The PS → RS conversion was slightly higher for Int(E449K), with a corresponding lower PS → WS conversion, than Int (Figure 3A, B). Int and Int(E449K) converted ∼66% and ∼75% of the substrate into recombinant products, respectively. When normalized to BC, recombination by Int was ∼82% and that by Int(E449K) was ∼90%. In ensemble assays containing a mixture of Int and Int(E449K), the activities of the hetero-tetramers assembled on attP-attB were consistent with the activities of the homo-tetramers of the individual proteins (Figure 3C). Thus, the E449K substitution per se is innocuous in the attP X attB reaction in the context of an [Int-Int(E449K)]-hetero-tetramer.
The mean BM amplitudes of the RS complexes for Int (54.6 ± 4.9 nm) and Int (E449K) (55.0 ± 6.0 nm) (Figure 3A, B) are consistent with a reactive synapse containing the attP-attB sites in parallel geometry. The broader distribution of the WS complexes formed by Int contained a subpopulation shifted toward lower BM amplitudes suggestive of sites paired in an anti-parallel fashion (Figure 3A and B). The histograms for the Int(E449K) WS complexes showed a bi-modal pattern with mean BM amplitudes at 41.3 ± 5.4 nm and 57.2 ± 4.2 nm (Figure 3B). The inactive synaptic structures formed by Int and Int(E449K) appear to span a range of conformations—from ‘parallel-like’ to ‘anti-parallel-like’.
The closely matching behavior between Int and Int(E449K) in head-to-tail attP × attB recombination is seen for head-to-head attP × attB recombination as well (Supplementary Figure S2B). However, in this case, it is not possible to separate the PS complexes into WS and RS complexes. Dissociation of the synapse without recombination (WS) or after DNA inversion (RS)—prior to or following SDS treatment—cannot not be distinguished because the resultant BM amplitude is the same for both events. The two types of dissociation events are also not distinguishable kinetically because of their similar rate constants (29) (see also results shown in Figure 7A).
Figure 7.
Recombination kinetics analyzed by TPM and FCS. (A) The substrate for the TPM analysis contained attP-attB in head-to-head orientation. The histogram plots depict the dwell times of BC in the low BM amplitude synapsed state (WS/RS; as recorded by the single molecule time traces). Their dissociation kinetics followed a single exponential model. N = the set of all synapsed molecules. (B) In the substrate containing head-to-tail attP-attB sites used for the FCS assay, the position of the TMR-fluorophore is indicated. The recombination reaction is schematically diagrammed to show the size difference between the fluorophore containing DNA species before (849 bp) and after (91 bp) the reaction. To determine the diffusion coefficient of the short linear product, a PCR amplified DNA of identical length with the fluorophore at the same location was used. The dissociation of the product synapse (with or without SDS addition) was assayed by the generation of the fast diffusing short fluorophore containing DNA. For a fast dissociating product synapse, the SDS effect will be minimal or absent. (C) The progress of recombination with and without SDS challenge is plotted as the decrease in the slow diffusing substrate with time.
Effect of gp3 on attP × attB recombination by Int and Int(E449K)
Consistent with its physiological role of promoting ϕC31 excision from the prophage state, while preventing reintegration of the excised phage, gp3 strongly inhibits attP × attB recombination by Int in vitro as well (15). Int associates with gp3 in solution and in the att-bound state in apparently stoichiometric fashion (15,40). A chimeric protein in which gp3 is fused to the carboxyl-terminus of Int mimics the effects of gp3 included in Int recombination reactions (41). Int(E449K) is also subject to inhibition by gp3 in the attP × attB reaction (15), suggesting that the substitution does not alleviate the gp3 induced block in assembling the attP-attB recombination synapse. We have now utilized TPM to parse out potential differences between Int and Int(E449K) in their responses to gp3, even though the overall recombination output in ensemble assays is affected similarly for the wild-type and mutant proteins.
The presence of gp3 in addition to Int or Int(E449K) in the attP-attB head-to-tail reaction caused a marked reduction in the formation of BC (the sum of NP and PS (WS plus RS)) by both proteins over the 30 min observation period (Figure 4 and Supplementary Figure S3). This effect was more marked for Int (from ∼80% to ∼23%) than for Int(E449K) (from ∼83% to ∼44%) (Figures 3A-B and 4A-B). In sharp contrast to reactions lacking gp3, the vast majority of the PS complexes formed by Int or Int(E449K) in the presence of gp3 produced the WS-type synaptic complexes. However, Int(E449K) was more efficient than Int in forming the smaller fraction of RS complexes (∼33% versus 20%).
Figure 4.
Inhibition of attP × attB recombination by gp3: differential effects on Int and Int(E449K). (A B). The data shown here correspond to TPM assays in which Int or Int(E449K) plus gp3 were added to the attP-attB (head-to-tail) substrate (see Supplementary Figure S3). The relative abundance of the individual complexes and the BM amplitude distributions of the WS complexes are arranged as in Figure 3.
The attP–attB WS complexes formed in the presence of gp3 could be split into two Gaussian BM amplitude distributions (Figure 4A and B). For Int, the lower (41.2 ± 4.9 nm) and higher (56.4 ± 3.8 nm) amplitude distributions were nearly equal (with a slight bias in favor of the lower amplitude distribution). For Int(E449K), the higher amplitude distribution (57.9 ± 4.6 nm) was dominant over the lower one (42.3 ± 2.9 nm). These results are consistent with a higher abundance of ‘anti-parallel-like’ attP-attB synapses induced by Int plus gp3 compared to Int alone (Figures 3A and 4A). Such a marked difference due to gp3 was not noticed for Int(E449K) (Figures 3B and 4B). Thus, the attP-attB sites appear to preferentially assume a ‘parallel-like’ geometry with Int(E449K) even in the presence of gp3, although the synaptic structures containing them are incompetent in recombination.
The TPM results bring to light differences between Int and Int(E449K), not revealed by bulk assays, in how gp3 modulates their association with attP-attB as well as the synaptic interactions that follow binding. In the presence of gp3, the efficiency of total BC formation by Int(E449K) is nearly twice that of Int. The fraction of bound substrate in the PS form is almost equal for the two proteins. The partitioning of the synaptic complexes strongly favors the inactive WS over the active RS for Int and Int(E449K). However, the yield of RS is ∼1.7-fold higher for Int(E449K). The combined effects produce a large overall inhibition of recombination, the residual activity for Int(E449K) plus gp3 being 3- to 4-fold more than that for Int plus gp3. When normalized to the fraction of Int or Int(E449K) bound attP-attB sites, the recombination yields in the presence of gp3 are ∼18% for Int and ∼31% for Int(E449K).
Current TPM results are consistent with our prior observations (29) that the inclusion of gp3 does not significantly alter the shift in BM amplitude caused by the binding of Int or Int(E449K) to an att site. This earlier work also found that the addition of gp3 in the absence of Int does not change the mean BM amplitude of DNA molecules containing head-to-tail attP-attB or attL-attR sites, indicating a lack of direct interaction between gp3 and an att site. The additional mass from gp3 present at an Int-bound att site may not produce a BM amplitude change sufficient to be reliably detected in the TPM assay.
Int and Int(E449K) activities on attL-attR sites oriented head-to-tail
The absolute dependence of Int on gp3 for ϕC31 excision in vivo is recapitulated by attL × attR reactions in vitro (15). By contrast, Int(E449K) alone shows detectable recombination activity on attL-attR in standard biochemical assays (26). This low basal activity is further stimulated in the presence of gp3 (15). In attempts to potentially delineate the step(s) influenced by E449K and gp3 in the attL-attR reaction path, we followed the TPM behaviors of the head-to-tail attL-attR containing DNA substrate when bound by Int or Int(E449K), with and without added gp3.
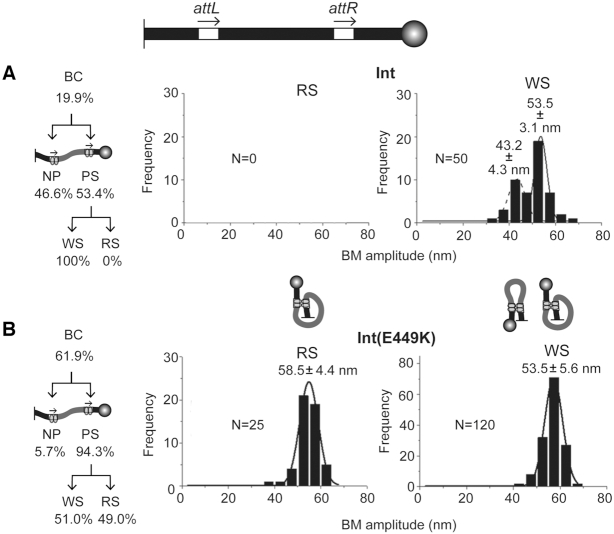
The cumulative formation of attL-attR BC in the absence of gp3, indicated by the lowering of substrate BM amplitude, was considerably higher for Int(E449K) (∼62%) compared to Int ∼20%) (Figure 5A and B; Supplementary Figure S4). For Int, the bound molecules were almost equally distributed into NP (46.6%) and PS (53.4%) complexes, and the latter gave rise exclusively to WS complexes yielding no recombination (Figure 5A). For Int(E449K), nearly all the bound att sites (∼94%) proceeded to synapsis (PS → WS + RS) (Figure 5B). The WS and RS complexes were roughly equimolar, giving a net recombination efficiency of ∼29% with respect to the input substrate DNA and ∼46% with respect to the attL-attR bound molecules (Figure 5B).
Figure 5.
Int and Int(E449K) activities on head-to-tail oriented attL-attR sites. The TPM results from reactions of the head-to-tail attL-attR substrate with Int or Int(E449K) were used to obtain the relative amounts of BC formed and the BM amplitude distributions of the WS and RS complexes (see Supplementary Figure S4).
The BM amplitude peak for the Int(E449K) RS complexes was 58.5 ± 4.4 nm, and was slightly downshifted for the WS complexes to 53.5 ± 5.6 nm (Figure 5B). The WS complexes formed by Int on attL-attR conformed to a bimodal distribution with mean BM amplitudes of 53.5 ± 3.1 and 43.2 ± 4.3 nm (Figure 5A). This bimodality is reminiscent of the WS complexes formed by Int plus gp3 and Int(E449K) plus gp3 on head-to-tail attP-attB sites (Figure 4A and B). The qualitative similarities of these WS complexes suggest that attP-attB synapse formed by Int or Int(E449K) in the presence of gp3 and attL-attR synapse generated by Int alone are populated by analogous inactive conformations that include both parallel-like and anti-parallel-like att site arrangements. The contrasting uni-modal distribution of the attL-attR WS complexes of the higher BM amplitude assembled by Int(E449K) is consistent with the ability of the self-activating mutation to promote principally parallel-like att site orientation even in those synaptic structures that do not execute recombination.
Similar recombination efficiencies of Int and Int(E449K) on head-to-tail attL-attR in the presence of gp3
Addition of gp3 to in vitro reactions restores attL × attR recombination activity to Int, and strongly stimulates the basal activity of Int(E449K) in this reaction (15). Results of gel mobility shift assays suggest that gp3 promotes the formation of the excision synaptic complex—and suppresses the formation of the integration synaptic complex—by both Int and Int(E449K). By the structural model (22–23,28), the role of gp3 would be to reconfigure the trajectories of the CC regions of Int differently depending on the att sites to which it is bound. The E449K mutation is only partially effective in establishing the functional CC trajectory on attL-attR, and requires gp3 assistance to elicit the full activity of Int(E449K) in attL × attR recombination. To test how gp3 modulates the activities of Int and Int(E449K) in finer detail, we carried out TPM assays analogous to the attL × attR reactions depicted in Figure 5, but in the presence of gp3.
In contrast to the reactions lacking gp3, those containing gp3 showed very similar behaviors of Int and Int(E449K) (Figure 6A and B; Supplementary Figure S5). The yield of BC was nearly the same for the two proteins (64–65%). They were predominantly PS, with very little non-productive binding (NP). In the case of Int, these complexes populated ∼82% active synaptic structures (RS) and ∼18% inactive ones (WS). For Int(E449K), the corresponding values were ∼91% RS complexes and ∼9% WS complexes. The conversion of BC into recombinant products was high for both proteins—∼80% for Int and ∼88% for Int(E449K). The recombination yields based on the input substrate were ∼52% for Int and ∼56% for Int(E449K).
The RS complexes formed by Int and Int(E449K) fell into single Gaussian distributions with mean BM amplitudes of 54.3 ± 3.0 and 57.2 ± 3.8 nm, respectively (Figure 6A and B). The distributions of the WS complexes were also uni-modal with peaks at 58.5 ± 4.6 nm for Int and 54.8 ± 4.0 nm for Int(E449K) (Figure 6A and B). The slight displacement in the location of the BM amplitude peaks of synaptic complexes formed by the two proteins was not anticipated, given their similar recombination efficiencies on attL-attR in the presence of gp3. The differences may indicate that E449K modulates the interactions of Int with att site DNA and/or gp3 to cause small perturbations in the conformations of synaptic structures without affecting their functional status.
The uni-modal distribution of the WS complexes is strikingly different from the bi-modality of these complexes formed by Int in the absence of gp3 (Figure 5A). The disappearance of the Gaussian centered at ∼43 nm, with retention of the one centered at 53–58 nm, suggests that the overall conformations of the synaptic structures induced in the presence of gp3 are not grossly different between their active (RS) and inactive (WS) forms (parallel-like).
Kinetics of recombination by Int: product dissociation is not rate limiting
The dissociation of the synapsed recombination target sites has two components to it—dissociation of the substrate synapse without recombination (in WS complexes) and of the product synapse following recombination (in RS complexes). The two cannot be distinguished for sites in head-to-head orientation, as the BM amplitude change is the same in both cases. If the kinetics of the two events are significantly different, it is possible to separate the two rate constants by fitting the dwell times of the WS complexes to a two-exponential algorithm (34). Applying this rationale to YRs, our TPM studies revealed a slower rate constant for product dissociation relative to WS dissociation (34,35). In a different approach, FCS analysis of Cre-mediated recombination between head-to-tail loxP sites revealed two kinetically distinct modes of product formation, depending on whether reactions were SDS-quenched or not. From the slower rate in the absence of SDS, the dissociation of the product synapse could be assigned as the rate limiting step (42).
In principle, the rate of WS dissociation (k-WSd)—and also NP dissociation, k-NPd—relative to the rate of recombination (krec = k-RSd) can have a significant impact on the overall reaction output. If the NP and WS complexes dissociate slowly compared to conversion of the RS complex into product(s), they would behave as dead-end complexes. If their dissociation rates are faster than—or comparable to—the rate of recombination, they will contribute to recombination by starting over from the PS or the S state. As the first step in probing the potential kinetic effects of E449K, we utilized TPM-dwell time and FCS analyses to characterize key kinetic features of Int-mediated recombination.
The dwell times of WS/RS complexes resulting from the association of Int with head-to-head attP-attB sites best fit a single exponential model with a rate constant for dissociation kd = (3.11 ± 0.06) × 10−2 s−1 for Int and (3.81 ± 0.08) × 10−2 s−1 for Int(E449K) (Figure 7A). One interpretation of this observation is that the rate constants of WS and RS dissociation are nearly the same (krec (k-RSd) = k-WSd), and cannot be separated by their BM amplitude dwell times. In order to verify this possibility, we employed FCS to monitor product release during head-to-tail attP X attB recombination by Int. The rationale of the assay is based on the slow and fast diffusion rates of the substrate and the short linear product resulting from it, respectively (Figure 7B). The placement of the fluorophore in the substrate is such that it will be carried by the linear product following recombination. The diffusion coefficient of this short DNA fragment will be determined by whether it remains associated with the larger circular product or dissociates from it.
The reaction utilized a TMR (tetramethylrhodamine)-labeled 849 bp long linear DNA containing head-to-tail attP-attB sites placed near its ends. Recombination would generate a 91 bp linear DNA carrying the fluorophore and a 758 bp non-fluorescent circular DNA. The diffusion coefficient of the substrate was estimated as 10 μm2/s−1. The corresponding value for a TMR-labeled 91 bp DNA (identical in sequence to the recombination product) was 30 μm2/s−1 (Figure 7B). The dissociation of the RS complex (post-recombination) was monitored over a 60 min time course after addition of Int. The data were fit to a two-component 3D diffusion model, and plotted as the fraction of unreacted substrate with progression of time. In parallel, aliquots of the reaction were quenched with SDS to follow total product formation with time (associated with and dissociated from the synapse combined). The native reactions and the SDS-quenched reactions displayed single exponential decay curves with nearly identical rate constants of (1.90 ± 0.5) × 10−2 s−1 and (1.80 ± 0.3) × 10−2 s−1, respectively (Figure 7C). This finding is in good agreement with the value of kd for synapsed molecules (WS/RS) = (1.50 ± 0.1) × 10−2 s−1 derived by earlier TPM analysis of head-to-head attP-attB (29) and (3.11 ± 0.06 × 10−2 s−1) obtained in this study (Figure 7A).
The FCS results, in conjunction with the single exponential decay of synapsed head-to-head attP-attB sites (Figure 7A) (29), establish that dissociation of the product synapse is not rate liming during Int mediated recombination. If the release of the fluorophore tagged 91 bp recombinant molecule were slow, SDS addition would have made dissociation instantaneous. In this case, the FCS data should have been distinct between a reaction treated with SDS and an untreated one (which is contrary to experimental findings; Figure 7C). Rapid product release is consistent with the nearly identical kinetics for the dissociation of WS and for recombination within RS.
The E449K substitution mimics gp3 in promoting faster dissociation of non-productively bound Int from attL-attR and of abortive attL-attR synapses
Prior studies have designated functional synapsis as the central regulatory step in determining the directionality of recombination by Int or Int plus gp3 (39,43). Results from the TPM and FCS assays (Figure 7A and B) now suggest that the dissociation of WS complexes will be kinetically relevant to recombination, provided a subset of the reassembled synapses are of the recombination competent RS type (WS → PS → synapsis [RS/WS])). Similarly, dissociation of the NP complexes could also influence synapse formation: NP → S → PS → synapsis [RS/WS]. In the case of attL-attR sites in the presence of Int alone, synapse assembly following NP and WS dissociation can only produce WS complexes. In the case of Int(E449K) (or Int plus gp3), the outcome will be WS as well as RS complexes. To test whether E449K modulates the dissociation kinetics of WS and/or NP, we determined the rate constants k-WSd and k-NPd for head-to-tail attL-attR treated with Int or Int(E449K) in the presence and absence gp3.
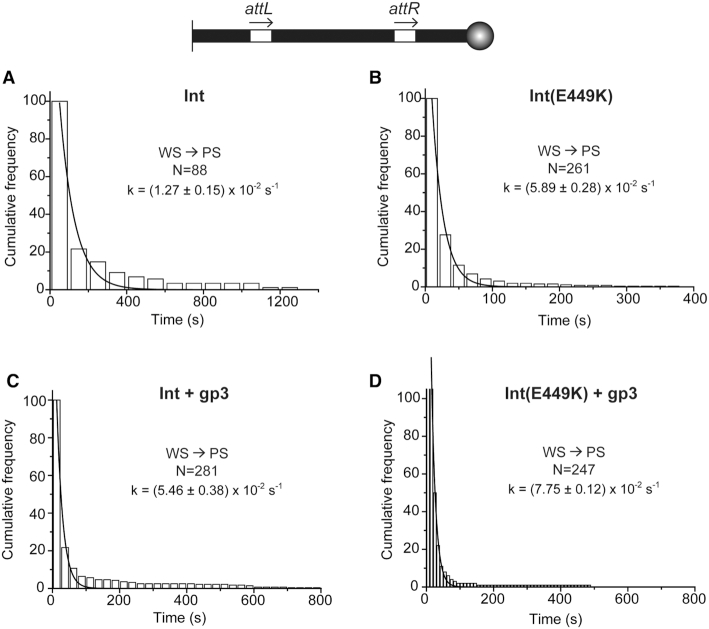
The dwell time histograms for the WS synaptic state of the attL-attR (head-to-tail) substrate were fit to a single exponential algorithm to obtain the rate constants for WS dissociation (Figure 8). The estimated k-WSd for Int, (1.27 ± 0.15) × 10−2 s−1, (Figure 8A) was 3- to 4-fold lower than that for Int(E449K), k-WSd = (5.89 ± 0.28) × 10−2 s−1 (Figure 8B). Inclusion of gp3 in the reaction allowed Int to nearly catch up with Int(E449K) in k-WSd, (5.46 ± 0.38) × 10−2 s−1 (Figure 8C). The k-WSd for Int(E449K) was also increased—but only slightly compared to Int—by gp3, k-WSd = (7.75 ± 0.12) × 10−2 s−1 (Figure 8D).
Figure 8.
Dissociation kinetics of abortive synaptic complexes assembled in the attL-attR (head-to-tail) DNA substrate. (A–D). The dwell times of the WS complexes before their dissociation to PS complexes were collected from single molecule time traces. For easy comparison between individual panels, the ordinate at time zero was normalized as ‘100’ in all plots. The rate constants for dissociation k-WSd was obtained by fitting the histogram plots to a single exponential algorithm. N = the total number of WS complexes observed.
A similar kinetic analysis of the NP complexes (Figure 9) gave a ∼4-fold lower k-NPd for Int, (0.84 ± 0.14) × 10−2 s−1 (Figure 9A) than for Int(E449K), k-NPd = (3.20 ± 0.17) × 10−2 s−1 (Figure 9B). This dissociation—like WS dissociation—was also accelerated by gp3, the effect being markedly stronger for Int, k-NPd = (6.49 ± 0.42) × 10−2 s−1 (Figure 9C) than for Int(E449K), k-NPd = (4.78 ± 0.80) × 10−2 s−1 (Figure 9D).
Thus, the E449K substitution and the addition of gp3 to the reaction have nearly the same effect in accelerating the dissociation of a non-functional attL-attR synapse or a non-productively occupied attL-attR, and affording it the chance to reattempt synapsis in two steps (WS → PS → synapsis) or three (NP → S → PS → synapsis), respectively. However, the lower yields of RS complexes by Int(E449K) compared to Int plus gp3 or Int (E449K) plus gp3 (Figures 5 and 6) suggest that E449K is less effective than gp3 in how successful such attempts are going to be in producing RS complexes rather than forming WS complexes.
DISCUSSION
Single molecule analysis of site-specific DNA recombination and DNA transposition have raised the mechanistic analysis of these reactions to a level of resolution that is not attainable by standard ensemble biochemical studies (37–38,42,44–48). The analytical tools employed include magnetic tweezers, TPM, TFM (tethered fluorophore motion), TFM-FRET (fluorescence resonance energy transfer) and PIFE (protein induced fluorescence enhancement). TPM, because of its simplicity of implementation and sensitivity to small changes in DNA length, has been particularly useful in characterizing the pre-chemical and chemical steps performed by YRs λ Int, Flp, Cre and XerC/D (34–35,38,45). More recently we applied TPM to the stepwise analysis of the large SR ϕC31 integrase (Int) (29). The present TPM analysis of Int(E449K) was designed to understand how this Int variant is able to overcome, to a certain degree, the gp3 requirement for attL × attR recombination. The TPM data report qualitative and quantitative differences in the conformations of the pre-chemical BC formed by Int and Int (E449K). Furthermore, non-productively bound attL-attR and abortive synaptic structures formed by Int(E449K) have faster dissociation kinetics, thereby increasing the number of chances they get to reassemble synapses, a subset of which would be functional in recombination.
Our findings can be accommodated by the structural model (22–23,28) suggesting a role for RDF in inducing an inter-dimer open conformation of the CC motifs when Int is bound to attL-attR. They are also consistent with the proposal, based on attP binding affinity differences between Listeria integrase and its isolated CTD as well as the crystal structure of CTD-attP half-site, that the opening of the integrase dimer for attP binding requires free energy expenditure (49). It is conceivable that E449K in ϕC31 integrase lowers the energy cost for the conformational transition during attL/attR binding by the Int dimer. This substitution may concurrently promote, albeit weakly, the CC trajectory that confers recombination competence on the attL-attR synapse.
Conformational differences in synaptic complexes organized by Int and Int(E449K) on attL-attR sites
Int(E449K) differs from Int in the relative partitioning of attL-attR bound DNA molecules into NP and PS complexes, and in the conversion of PS complexes into WS and RS complexes (Figure 5). The fraction of PS relative to NP is much higher for Int(E449K). Int, unlike Int(E449K), fails to generate RS complexes from PS. Int(E449K), by contrast, converts PS complexes into RS and WS complexes in roughly equal amounts. The WS complexes generated by the two proteins differ in the synaptic conformations they harbor, as reported by their BM amplitudes. The lower BM amplitude distribution observed for Int (Figure 5A), suggestive of incorrectly oriented sites within the synapse (presumably in an anti-parallel-like geometry), is almost absent in the case of Int(E449K) (Figure 5B). The higher BM amplitude distribution of WS complexes seen for both proteins likely contains synaptic conformations (presumably parallel-like) that are more akin to the functional RS complexes (Figure 5).
The presence of gp3 in the attL × attR reaction almost completely eliminates the differences between Int and Int(E449K) in the total amounts and relative proportions of the NP, PS, WS and RS complexes produced by them (Figure 6A and B). Furthermore, the WS complexes from the gp3 added reactions fit a single Gaussian distribution of the higher mean BM amplitude for both Int and Int(E449K) (Figure 6A and B). The disappearance of the lower BM amplitude distribution seen for Int in the absence of gp3 suggests that RDF promotes proper orientation of att sites (parallel-like) even when the synapse that harbors these sites is non-functional.
Functional and non-functional attP-attB complexes formed by Int and Int(E449K)
In the absence of gp3, Int and Int(E449K) are similar in the amount, as well as the relative abundance, of NP, PS, WS and RS complexes that they yield in the attP × attB reaction (Figure 3A and B). The broad distribution of the WS complexes formed by Int is suggestive of both correctly and incorrectly oriented sites (parallel-like and anti-parallel-like) in the inactive synaptic structures (Figure 3A). Such a mixture appears to be the case with Int(E449K) as well, although here a shallow lower BM amplitude peak (anti-parallel-like) can be distinguished from the more prominent higher BM amplitude distribution (parallel-like) (Figure 3B). The inclusion of gp3 demarcates these WS conformations into two distinct distributions, the ratio of the higher amplitude population (parallel-like) to the lower amplitude one (anti-parallel-like) being higher for Int (E449K) (Figure 4A and B). The overall reduction in the bound attP-attB complexes (as indicated by the fraction of substrate molecules shifted to lower BM amplitudes) due to gp3 is greater for Int compared to Int(E449K) (Figures 3A-B and 4A-B). Furthermore, the decrease in the formation of RS (in favor of WS) complexes from PS complexes in the presence of gp3 is slightly more for Int than Int(E449K) (Figures 3A-B and 4A-B).
Overall, the quantitative differences between Int and Int(E449K) in the parallel-like and anti-parallel-like synaptic conformations organized among the recombination-incompetent WS complexes may be due to E449K-mediated modulations in Int-att site binding interactions as well as in interactions between Int and gp3 in solution and/or on the att site DNA. The preference for the parallel-like conformation in WS complexes formed by Int(E449K) would be consistent with its partially activated state for attL-attR recombination, which would occur from the parallel synaptic conformation assembled in RS complexes.
Activation of gp3-independent attL x attR recombination by E449K: thermodynamic contribution
The activation of attL × attR recombination by Int(E449K), as interpreted according to the currently accepted structural models (22–23,28), results from the ability of this mutation to promote inter-dimer CC trajectories across attL-attR sites occupied by Int (Figure 10). The extent of self-activation by E449K is modest, and normal levels of attL × attR recombination requires roughly the same stoichiometry of gp3 for Int and Int(E449K). A reasonable explanation for these observations is that E449K populates a small fraction of the activated Int dimer in solution or upon its occupancy of an attL or attR site. This self-activated (or activation-competent) form is in equilibrium with an excess of the inactive dimer. Only when both att site bound dimers are in the activated state, do they become capable of forming a functional synapse (RS complexes). In the presence of gp3, the equilibrium is shifted strongly toward the active (or activation-competent) dimer to give efficient attL × attR recombination. The distinction between E449K and gp3 in the conversion of pre-activated Int into its activated form during attL-attR reaction is schematically illustrated in Supplementary Figure S6.
Figure 10.
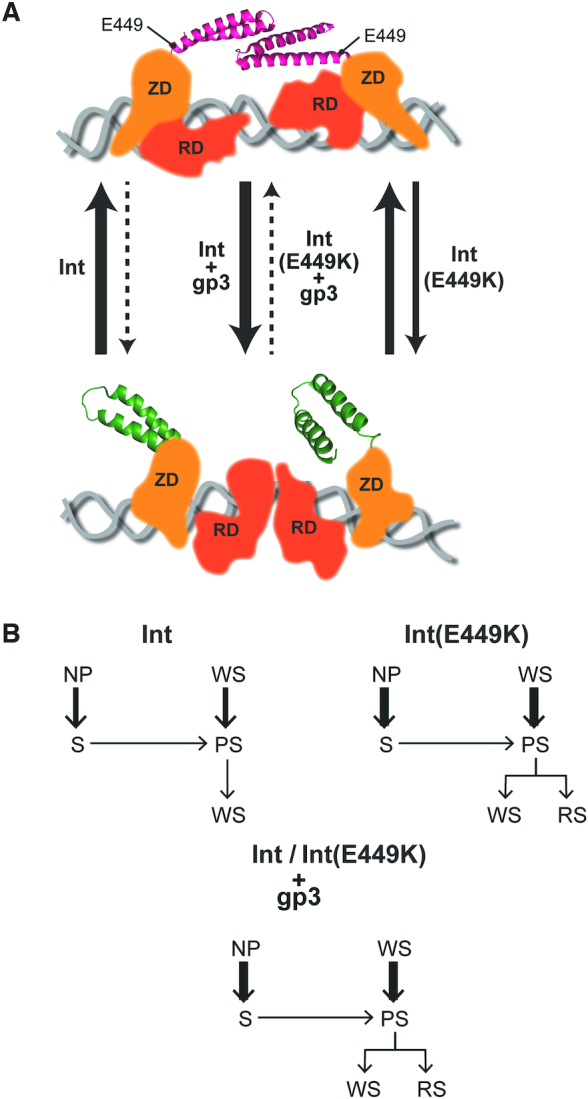
Comparison of E449K with gp3 in the activation of Int for attL × attR recombination. (A) The closed arrangement of the CC motifs (shown in red) within the Int dimer bound to attL (or attR), which proscribes functional synapsis, is schematically diagrammed at the top. RDF (gp3) shifts the equilibrium toward the open conformation of the coiled coils (shown in green), fostering inter-dimer interactions and productive synapsis (bottom schematic). E449K mimics gp3 in its thermodynamic effect; however, the magnitude of the effect is considerably lower for E449K. The representations follow the proposed structural model for the L1 integrase (23,28). The CC trajectories in the bottom and top panels correspond to the CC1 and CC2 designations, respectively, in the structure. The relative thickness of the arrows qualitatively indicates the direction of the equilibrium between synapsis-competent and synapsis-incompetent conformations of Int and Int(E449K) with and without gp3. As the ϕC31 Int structure has not been solved, the location of E449 shown here at the base of CC is only an approximation. RD = recombinase domain; ZD = zinc finger domain. (B) The kinetic re-routing of the WS and NP complexes toward synaptic complexes is schematically diagrammed. The thickness of the arrows representing k-WSd and k-NPd qualitatively conveys the speed of WS and NP dissociation, respectively, for Int and Int(E449K) with and without gp3.
Interestingly, the amounts of BC formed on attP-attB in the presence of gp3 and on attL-attR in its absence are higher for Int(E449K) than Int (Figures 4 and 5). By contrast, both Int and Int(E449K) give nearly the same amounts of BC with attP-attB when gp3 is absent and with attL-attR when gp3 is present (Figures 3 and 6). The observed differences in binding affinities, depending on the individual att site pairs and the presence or absence of gp3, may be relevant to the activation of Int by E449K for gp3-independent attL × attR recombination. The low basal level of constitutive self-activation in Int(E449K) may account for its lower inhibition, compared to Int, by gp3 in attP × attB recombination (Figure 4). Under the gp3-inhibited condition, recombination is completed by roughly one third of the synapses formed by Int(E449K); the success rate is only one fifth for Int.
The kinetic component of attL × attR recombination by Int(E449K)
The combination of TPM and FCS analyses suggests that the kinetics of recombination (RS → product) and of the dissociation of non-functional synaptic complexes (WS → PS) are quite similar for Int. A dissociated WS ( = PS) can enter the reaction path by re-forming the synapse within the time scale of the TPM assays. A dissociated NP ( = S) can do the same by first forming PS. While the re-formed attL-attR synapses are all inactive (WS) in the case of Int, ∼50% of such synapses are active (RS) in the case of Int(E449K). The rate constants for WS and NP dissociation for head-to-tail attL-attR are ∼4-fold higher for Int(E449K) than Int in the absence of gp3 (Figures 8A-B and 9A-B). While WS or NP dissociation for Int(E449K) is more or less unaffected (or is only modestly increased) by gp3, gp3 hastens WS and NP dissociations for Int to rates comparable to those of Int(E449K) (Figures 8C-D and 9C-D). Thus, there is a significant kinetic component to the activation mechanism by E449K. Furthermore, the kinetic effects of E449K and gp3 on attL × attR recombination via NP or WS dissociation are nearly the same.
The rate constants for WS dissociation (WS → PS) and for synapse formation (PS → WS or PS → RS) are of the same order (10−2 s−1) (29). Thus, the advantage of faster WS dissociation will not be rate limited by the speed with which the dissociated molecules reform synapses (of the WS and RS types for Int(E449K) and only of the WS type for Int when gp3 is absent). Given the association rate constant of ∼106 M−1 s−1 for S → PS (29), provided the Int concentration is not insufficient, the dissociated NP molecules will not be rate limited in PS formation. The 30 min time traces of individual molecules reveal multiple dissociation events for NP and PS (Supplementary Figure S7). A subset of these molecules succeeds in RS assembly and recombination, provided the reaction conditions do not proscribe this outcome (time trace I in Supplementary Figure S7).
A bipartite role for E449K in the activation of gp3-independent attL × attR recombination
In summary, E449K plays a bipartite, thermodynamic and kinetic, role in activating Int to perform attL × attR recombination in the absence of gp3, albeit with modest efficiency. The kinetic effect of E449K in disassembling and potentially redirecting futile associations of Int with attL-attR toward the productive pathway is comparable to that of gp3. However, the thermodynamic contribution of E449K toward populating the recombination competent state among the distinct Int BC is much stronger with gp3. These relative contributions are schematically summarized in Figure 10. Mutations other than E449K have been identified that also license attL × attR recombination by Int without gp3, and these map to the same face of the CC region (39). We are now testing whether combinations of such mutations are synergistic in the activation of attL × attR recombination. Strongly self-activated variants of Int would be ideal candidates for TPM analyses in verifying the mechanisms proposed here for the action of E449K.
From a broader perspective, the E449K and gp3 mediated stimulation of recombination by Int has parallels to the modulation in the activities of regulatory or enzymatic proteins effected by simple amino acid modifications and by non-covalent or covalent interactions of effector proteins, respectively. The underlying principle is the utilization of alternative mechanisms to manipulate the free energies of the intermediates and transition state along a reaction path.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Professor Margaret M Smith for critical reading of the manuscript.
Contributor Information
Hsiu-Fang Fan, Institute of Medical Science and Technology, National Sun Yat-sen University, Sizihwan, Kaohsiung 804, Taiwan; Department of Chemistry, National Sun Yat-sen University, Sizihwan, Kaohsiung 804, Taiwan; Aerosol Science Research Center, National Sun Yat-sen University, Sizihwan, Kaohsiung 804, Taiwan.
Bo-Yu Su, Department of Life Sciences and Institute of Genome Sciences, National Yang-Ming University, Taipei 112, Taiwan.
Chien-Hui Ma, Department of Molecular Biosciences, UT Austin, Austin, TX 78712, USA.
Paul A Rowley, Department of Biological Sciences, University of Idaho, Moscow, ID 83844, USA.
Makkuni Jayaram, Department of Molecular Biosciences, UT Austin, Austin, TX 78712, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [MOST 108-2113-M-010-002 to H.F.F.]; Welch Foundation [F-1274 to M.J.]; National Science Foundation [MCB 1049925, 1949821 to M.J.]; National Institutes of Health [P20 GM104420 to Center for Modeling Complex Interactions, University of Idaho (P.A.R.)]. Funding for open access charge: Welch Foundation [F-1274 to M.J.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Smith M.C.M. Phage-encoded serine integrases and other large serine recombinases. Microbiol. Spectr. 2015; 3:doi:10.1128/microbiolspec.MDNA3-0059-2014. [DOI] [PubMed] [Google Scholar]
- 2. Stark W.M. The serine recombinases. Microbiol Spectr. 2014; 2:doi:10.1128/microbiolspec.MDNA3-0046-2014. [DOI] [PubMed] [Google Scholar]
- 3. Chavez C.L., Calos M.P.. Therapeutic applications of the φC31 integrase system. Curr. Gene Ther. 2011; 11:375–381. [DOI] [PubMed] [Google Scholar]
- 4. Groth A.C., Calos M.P.. Phage integrases: biology and applications. J. Mol. Biol. 2004; 335:667–678. [DOI] [PubMed] [Google Scholar]
- 5. Bonnet J., Subsoontorn P., Endy D.. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl Acad. Sci. U.S.A. 2012; 109:8884–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonnet J., Yin P., Ortiz M.E., Subsoontorn P., Endy D.. Amplifying genetic logic gates. Science. 2013; 340:599–603. [DOI] [PubMed] [Google Scholar]
- 7. Siuti P., Yazbek J., Lu T.K.. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 2013; 31:448–452. [DOI] [PubMed] [Google Scholar]
- 8. Yang L., Nielsen A.A., Fernandez-Rodriguez J., McClune C.J., Laub M.T., Lu T.K., Voigt C.A.. Permanent genetic memory with >1-byte capacity. Nat. Methods. 2014; 11:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Colloms S.D., Merrick C.A., Olorunniji F.J., Stark W.M., Smith M.C., Osbourn A., Keasling J.D., Rosser S.J.. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 2014; 42:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L., Zhao G., Ding X.. Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination. Sci. Rep. 2011; 1:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghosh P., Kim A.I., Hatfull G.F.. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol. Cell. 2003; 12:1101–1111. [DOI] [PubMed] [Google Scholar]
- 12. Smith M.C., Thorpe H.M.. Diversity in the serine recombinases. Mol. Microbiol. 2002; 44:299–307. [DOI] [PubMed] [Google Scholar]
- 13. Bibb L.A., Hancox M.I., Hatfull G.F.. Integration and excision by the large serine recombinase φRv1 integrase. Mol. Microbiol. 2005; 55:1896–1910. [DOI] [PubMed] [Google Scholar]
- 14. Ghosh P., Wasil L.R., Hatfull G.F.. Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol. 2006; 4:e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khaleel T., Younger E., McEwan A.R., Varghese A.S., Smith M.C.. A phage protein that binds φC31 integrase to switch its directionality. Mol. Microbiol. 2011; 80:1450–1463. [DOI] [PubMed] [Google Scholar]
- 16. Zhang L., Zhu B., Dai R., Zhao G., Ding X.. Control of directionality in Streptomyces phage φBT1 integrase-mediated site-specific recombination. PLoS One. 2013; 8:e80434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mandali S., Gupta K., Dawson A.R., Van Duyne G.D., Johnson R.C.. Control of recombination directionality by the Listeria phage A118 protein Gp44 and the coiled-coil motif of Its serine integrase. J. Bacteriol. 2017; 199:e00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abe K., Takamatsu T., Sato T.. Mechanism of bacterial gene rearrangement: SprA-catalyzed precise DNA recombination and its directionality control by SprB ensure the gene rearrangement and stable expression of spsM during sporulation in Bacillus subtilis. Nucleic Acids Res. 2017; 45:6669–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bibb L.A., Hatfull G.F.. Integration and excision of the Mycobacterium tuberculosis prophage-like element, φRv1. Mol. Microbiol. 2002; 45:1515–1526. [DOI] [PubMed] [Google Scholar]
- 20. Breuner A., Brondsted L., Hammer K.. Novel organization of genes involved in prophage excision identified in the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 1999; 181:7291–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fogg P.C.M., Haley J.A., Stark W.M., Smith M.C.M.. Genome integration and excision by a new Streptomyces bacteriophage, φJoe. Appl. Environ. Microbiol. 2017; 83:e02767-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta K., Sharp R., Yuan J.B., Li H., Van Duyne G.D.. Coiled-coil interactions mediate serine integrase directionality. Nucleic Acids Res. 2017; 45:7339–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rutherford K., Yuan P., Perry K., Sharp R., Van Duyne G.D.. Attachment site recognition and regulation of directionality by the serine integrases. Nucleic Acids Res. 2013; 41:8341–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta M., Till R., Smith M.C.. Sequences in attB that affect the ability of φC31 integrase to synapse and to activate DNA cleavage. Nucleic Acids Res. 2007; 35:3407–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu S., Ma J., Wang W., Zhang M., Xin Q., Peng S., Li R., Zhu H.. Mutational analysis of highly conserved residues in the phage φC31 integrase reveals key amino acids necessary for the DNA recombination. PLoS One. 2010; 5:e8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rowley P.A., Smith M.C., Younger E., Smith M.C.. A motif in the C-terminal domain of φC31 integrase controls the directionality of recombination. Nucleic Acids Res. 2008; 36:3879–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fogg P.C.M., Younger E., Fernando B.D., Khaleel T., Stark W.M., Smith M.C.M.. Recombination directionality factor gp3 binds φC31 integrase via the zinc domain, potentially affecting the trajectory of the coiled-coil motif. Nucleic Acids Res. 2018; 46:1308–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rutherford K., Van Duyne G.D.. The ins and outs of serine integrase site-specific recombination. Curr. Opin. Struct. Biol. 2014; 24:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan H.F., Hsieh T.S., Ma C.H., Jayaram M.. Single-molecule analysis of φC31 integrase-mediated site-specific recombination by tethered particle motion. Nucleic Acids Res. 2016; 44:10804–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thorpe H.M., Wilson S.E., Smith M.C.. Control of directionality in the site-specific recombination system of the Streptomyces phage φC31. Mol. Microbiol. 2000; 38:232–241. [DOI] [PubMed] [Google Scholar]
- 31. Chandradoss S.D., Haagsma A.C., Lee Y.K., Hwang J.H., Nam J.M., Joo C.. Surface passivation for single-molecule protein studies. J. Vis. Exp. 2014; 86:e50549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsu K.W., Chow S.Y., Su B.Y., Lu Y.H., Chen C.J., Chen W.L., Cheng M.Y., Fan H.F.. The synergy between RSC, Nap1 and adjacent nucleosome in nucleosome remodeling. Biochim. Biophys. Acta Gene Regul. Mech. 2019; 1862:129–140. [DOI] [PubMed] [Google Scholar]
- 33. Fan H.F., Li H.W.. Studying RecBCD helicase translocation along Chi-DNA using tethered particle motion with a stretching force. Biophys. J. 2009; 96:1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fan H.F. Real-time single-molecule tethered particle motion experiments reveal the kinetics and mechanisms of Cre-mediated site-specific recombination. Nucleic Acids Res. 2012; 40:6208–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan H.F., Ma C.H., Jayaram M.. Real-time single-molecule tethered particle motion analysis reveals mechanistic similarities and contrasts of Flp site-specific recombinase with Cre and lambda Int. Nucleic Acids Res. 2013; 41:7031–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Culbertson C.T., Jacobson S.C., Michael Ramsey J.. Diffusion coefficient measurements in microfluidic devices. Talanta. 2002; 56:365–373. [DOI] [PubMed] [Google Scholar]
- 37. Fan H.F., Ma C.H., Jayaram M.. Single-molecule tethered particle motion: stepwise analyses of site-specific DNA recombination. Micromachines (Basel). 2018; 9:E216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mumm J.P., Landy A., Gelles J.. Viewing single lambda site-specific recombination events from start to finish. EMBO J. 2006; 25:4586–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rowley P.A., Smith M.C.. Role of the N-terminal domain of φC31 integrase in attB-attP synapsis. J. Bacteriol. 2008; 190:6918–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pokhilko A., Zhao J., Ebenhoh O., Smith M.C., Stark W.M., Colloms S.D.. The mechanism of φC31 integrase directionality: experimental analysis and computational modelling. Nucleic Acids Res. 2016; 44:7360–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Olorunniji F.J., McPherson A.L., Rosser S.J., Smith M.C.M., Colloms S.D., Stark W.M.. Control of serine integrase recombination directionality by fusion with the directionality factor. Nucleic Acids Res. 2017; 45:8635–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pinkney J.N., Zawadzki P., Mazuryk J., Arciszewska L.K., Sherratt D.J., Kapanidis A.N.. Capturing reaction paths and intermediates in Cre-loxP recombination using single-molecule fluorescence. Proc. Natl Acad. Sci. U.S.A. 2012; 109:20871–20876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McEwan A.R., Rowley P.A., Smith M.C.. DNA binding and synapsis by the large C-terminal domain of φC31 integrase. Nucleic Acids Res. 2009; 37:4764–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bai H., Sun M., Ghosh P., Hatfull G.F., Grindley N.D., Marko J.F.. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proc. Natl Acad. Sci. U.S.A. 2011; 108:7419–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Diagne C.T., Salhi M., Crozat E., Salome L., Cornet F., Rousseau P., Tardin C.. TPM analyses reveal that FtsK contributes both to the assembly and the activation of the XerCD-dif recombination synapse. Nucleic Acids Res. 2014; 42:1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fournes F., Crozat E., Pages C., Tardin C., Salome L., Cornet F., Rousseau P.. FtsK translocation permits discrimination between an endogenous and an imported Xer/dif recombination complex. Proc. Natl Acad. Sci. U.S.A. 2016; 113:7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. May P.F., Zawadzki P., Sherratt D.J., Kapanidis A.N., Arciszewska L.K.. Assembly, translocation, and activation of XerCD-dif recombination by FtsK translocase analyzed in real-time by FRET and two-color tethered fluorophore motion. Proc. Natl Acad. Sci. U.S.A. 2015; 112:E5133–E5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pouget N., Turlan C., Destainville N., Salome L., Chandler M.. IS911 transpososome assembly as analysed by tethered particle motion. Nucleic Acids Res. 2006; 34:4313–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H., Sharp R., Rutherford K., Gupta K., Van Duyne G.D.. Serine Integrase attP Binding and Specificity. J. Mol. Biol. 2018; 430:4401–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.