Abstract
The nucleolus is a membrane-less nuclear structure that disassembles when cells undergo mitosis. During mitosis, nucleolar factors are thus released from the nucleolus and dynamically change their subcellular localization; however, their functions remain largely uncharacterised. Here, we found that a nucleolar factor called nucleolar protein 11 (NOL11) forms a protein complex with two tryptophan-aspartic acid (WD) repeat proteins named WD-repeat protein 43 (WDR43) and Cirhin in mitotic cells. This complex, referred to here as the NWC (NOL11-WDR43-Cirhin) complex, exists in nucleoli during interphase and translocates to the periphery of mitotic chromosomes, i.e., perichromosomal regions. During mitotic progression, both the congression of chromosomes to the metaphase plate and sister chromatid cohesion are impaired in the absence of the NWC complex, as it is required for the centromeric enrichment of Aurora B and the associating phosphorylation of histone H3 at threonine 3. These results reveal the characteristics of a novel protein complex consisting of nucleolar proteins, which is required for regulating kinetochores and centromeres to ensure faithful chromosome segregation.
INTRODUCTION
The nucleolus is a large membrane-less subnuclear compartment assembled around tandemly repeated clusters of ribosomal RNA (rRNA) genes (rDNA) in interphase nucleus (1,2). The nucleolus primarily serves as a site of ribosome biogenesis, i.e., rRNA transcription, pre-rRNA processing, and ribosomal subunit assembly, but it also seems to be involved in other cellular functions, such as mitotic progression, senescence, and stress responses (1). Consistent with these diversified nucleolar functions, a bioinformatic-based classification indicates that many of the ∼4500 nucleolar proteins potentially participate in processes other than ribosome biogenesis (3,4).
The nucleolus exhibits phase-separated, liquid droplet-like properties (5–7), and its structure changes in response to various external stimuli, such as deprivation of growth factors, perturbation of rRNA transcription, inhibition of casein kinase II and cyclin-dependent kinase, and exposure to factors that result in cell injury (4,8–10), in a manner that depends on the RNA levels in the nucleolus. In response to changes in the nucleolar structure, proteins diffuse out from the nucleolus (11) and execute several cellular functions, such as stress responses (1,8,9).
The nucleolar structure is also altered during the cell cycle, most prominently during mitosis in higher eukaryotes (1,12–14). The nucleolus disassembles at the onset of mitosis, when rRNA transcription is shut down, and it reassembles as the cell exits mitosis and rRNA transcription resumes. As the nucleolus disassembles, a portion of the rRNA transcription machinery, such as the upstream binding factor (UBF), remains associated with the rDNA repeats (1), but a number of nucleolar factors translocate from the nucleolus to the cytoplasm. Among these factors, Ki67, nucleophosmin/B23, fibrillarin, nucleolin, and pre-rRNA are known to bind to the surface area of condensed mitotic chromosomes, the layer called the perichromosomal region (PR) (1,13–15). The PR surrounds mitotic chromosomes throughout, except at centromeric regions, where centromeric proteins (CENPs) are enriched (1,15,16). Recent studies have revealed that Ki67 is involved in the individualisation and integrity of mitotic chromosomes (17,18). Interestingly, Ki67 seems to recruit several nucleolar proteins to PRs, but the relevance of their translocation to PRs is not well understood (19–21).
It was previously reported that a nucleolar factor called nucleolar protein 11 (NOL11), which is essential for optimal pre-rRNA transcription and processing (22), is required for nucleolar integrity during interphase, which relates to the timely activation of cyclin-dependent kinase 1 (Cdk1) and mitotic entry (23). There seemed more to NOL11 than controlling mitotic entry, as NOL11 depletion affected mitotic transitions reflected by an obvious increase of the mitotic index (23). In the present study, we found that NOL11 forms a protein complex with two characteristic tryptophan-aspartic acid (WD) repeat proteins named WD-repeat protein 43 (WDR43) and Cirhin. WDR43 and Cirhin are reportedly nucleolar components of the ribosomal t-UTP/UTPA subcomplex of the small-subunit (SSU) processome (22,24) and are involved in the normal development of vertebrates (25–30). WDR43 binds to several gene promoters to facilitate transcriptional elongation (31).This complex, referred to here as the NWC (NOL11–WDR43–Cirhin) complex, translocates from interphase nucleoli to the PRs of mitotic chromosomes and promotes Aurora B enrichment at centromeres.
MATERIALS AND METHODS
Drug treatments
Mitotic cells were collected by adding 75 to 330 nM nocodazole followed by a mitotic shake-off. S phase cells were collected by adding 2 mM thymidine for 24 h. Cells were synchronised in metaphase by adding 10 μM MG132 for 2 h. To perform monastrol release assay, cells were treated with 100 μM monastrol (Sigma) for 4 h and then released into fresh medium containing 10 μM MG132.
Plasmids
cDNAs encoding full-length NOL11 and Cirhin were amplified by polymerase chain reaction (PCR) and subcloned into the pcDNA3 plasmid (Invitrogen) containing sequences coding for FLAG/HA.
FLAG/HA-NOL11 or FLAG/HA-Cirhin stably expressing HeLa cells
HeLa cells were transfected with PvuI-digested pcDNA3-FLAG/HA-NOL11 or pcDNA3-FLAG/HA-Cirhin plasmid. One day after transfection, cells were treated with 1000 μg/ml geneticin (G418) and cultured until G418-resistant colonies were apparent. Individual colonies were maintained in growth medium containing 1000 μg/ml G418. Cells were screened by immunoblotting to obtain the cell lines where FLAG/HA-NOL11 or Cirhin was highly expressed.
Immunofluorescence
Cells were grown on poly-L-lysine-coated coverslip. For immunostaining using anti-α-tubulin antibodies, cells were rinsed with pre-warmed phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, and 8.1 mM Na2HPO4), then pre-fixed in pre-warmed 4% paraformaldehyde (PFA) in PBS for 5 s, and permeabilised in 1% Triton X-100 in PBS for 5 min and then fixed with 4% PFA for 15 min. For immunofluorescence using the other antibodies, cells were fixed with 4% PFA in PBS for 15 min and permeabilised with 0.1% Triton X-100 in PBS for 3 min. For pre-extraction immunostaining, coverslips were incubated with 0.1% (v/v) Triton X-100 in PBS for 2 min before fixation.
Fixed cells were blocked with 3% (w/v) bovine serum albumin in PBS for 1 h at room temperature. Then, cells were incubated with the indicated primary antibodies at room temperature for 1 h or 4°C overnight in humidity box, stained with Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 1 h, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min, and mounted with Vectashield (Vector Laboratories). Images were captured on a BioRevo microscope (BZ-9000, Keyence) equipped with a 40× objective lens (Plan Apochromat, NA 0.95, Nikon) or 100× oil objective lens (Plan Apochromat VC, NA 1.4) or confocal microscope (LSM 700, Carl Zeiss) equipped with a 100× oil objective lens (Plan Apochromat, NA 1.4, Carl Zeiss) and a CCD camera (Axio Cam MRm, Carl Zeiss). Observed images were processed and analysed ImageJ software.
Small interfering RNA (siRNA) transfection
All siRNAs were purchased from Invitrogen. siRNAstreansfection was performed using Lipofectamine RNAiMAX according to the manufacturer's protocol. The sequences of siRNA duplexes were as follows: NOL11-#1, 5′-CCAAACGCAUGUGCUUUCUUACAGU-3′; NOL11-#2, 5′-GUCUACUUCUGGAUGGGAAUU-3′; WDR43-#1, 5′-GAGCAGUACAUGACCGGUUACUUAA-3′; WDR43-#2, 5′-GGAAGACCUCCAGACGAAUAGCUUU-3′; Cirhin-#1, 5′-UGUGAUAGGAGUAUCCCUUUGGAGC-3′; Cirhin-#2, 5′-AUGAUUGUGGACAGGCAGUAUAUGG-3′; and TIF-IA, 5′-CGACACCGUGGUUUCUCAUGCCAAU-3′; MAD2L1, 5′-AGAAUUGGUAAUAAACUGUGGUCCC-3′.
Stealth™ RNAi Luciferase Reporter Control was used as a negative control throughout this paper.
Chromosome spreads
For Giemsa staining, mitotic cells were collected by shake-off and swelled in a hypotonic solution of 40% PBS/MilliQ for 5 min at room temperature. The swollen cells were fixed by dropwise addition of fixing solution (methanol/acetic acid, 3:1), dropped onto glass slides, and stained with Giemsa staining solution.
For immunostaining, chromosome spreads was prepared as described previously (32) with modification. Mitotic cells were incubated in 40% PBS/MilliQ for 5 min 30 s on ice or in hypotonic buffer (75 mM KCl/0.8% sodium citrate/H2O, 1:1:1) containing 100 nM okadaic acid (Wako) at room temperature for 15 min. Then, swollen cells were cytospun onto ethanol-prewashed glass slides at 113 × g for 5 min (Cytospin 4), fixed with PBS containing 2% PFA 15 min, and permeabilised with PBS containing 0.1% Triton X-100 for 3 min. The subsequent procedure was conducted as described in immunofluorescence.
Immunoprecipitation
For immunoprecipitation of the endogenous proteins, HeLa cells were lysed in CSK buffer [10 mM PIPES (pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 1 mM EGTA] containing 1 mM DTT, 250 μM phenylmethylsulfonyl fluoride (PMSF), complete protease inhibitor cocktail, 0.3% Triton X-100 and 1 mM ATP at 4°C for 30 min. The cleared lysate was incubated for 1 h with 0.4 μg antibodies, added with 10 μl protein G Sepharose (GE Healthcare), and rotated for 1 h. After washing four times with the same buffer, immunoprecipitants were examined by immunoblotting.
For sequential immunoprecipitation, mitotic HeLa cells or those stably expressing FLAG/HA-NOL11 were collected and lysed in TNE buffer [20 mM Tris–HCl (pH 7.5), 150 mM NaCl, and 2 mM EDTA] containing 1% NP-40 and complete protease inhibitor cocktail at 4°C for 30 min. The cleared lysate was incubated for 4 h with anti-FLAG M2-agarose beads. After washing three times with TNE buffer containing 1% NP-40, the bound proteins were eluted with 0.5 mg/ml FLAG peptide in TNE buffer containing 1% NP-40 at 4°C (first elute). The eluates were incubated with 0.4 μg anti-Cirhin antibodies for 1 h, added with 10 μl protein G Sepharose (GE Healthcare), and incubated on a rotating wheel for 1 h. The beads were washed four times with the same buffer, immunoprecipitants were examined by immunoblotting (second IP).
Statistical analysis
The quantified data are shown as the mean ± standard error of the mean (SEM). Statistical significances were tested by one-tailed unpaired Welch's t-test. Significant differences are shown as follows: *P < 0.05, **P < 0.01 and ***P < 0.001.
RESULTS
NOL11 forms a protein complex with WDR43 and Cirhin and associates with mitotic chromosomes
To elucidate the prospective mitotic function(s) of NOL11, the subcellular localisation of NOL11 during mitosis was first examined. Immunofluorescence staining showed that NOL11 localised in the nucleolus during interphase, subsequently dispersed into the cytoplasm, and concentrated around the chromosomes in mitosis (Figure 1A; Supplementary Figure S1). When the cytoplasmic NOL11 signals were extracted by detergent treatment, the chromosome periphery signals persisted (Figure 1B). Immunofluorescence microscopy of chromosome spreads revealed that NOL11 localized at Ki67-positive chromosome periphery (PR) but did not co-localise with ACA, marking centromeres, or condensin subunit SMC2, marking the chromosome axis (Figure 1C). A fractionation experiment conducted on mitotic cell extracts also showed that NOL11 was present in the chromosome-enriched fraction, whereas TIF-IA, another nucleolar protein and rRNA transcription factor, was barely detected (Figure 1D). These results indicated that NOL11 localized at the PR, which consists of subsets of nucleolar proteins (1,3,15,20,33).
Figute 1.
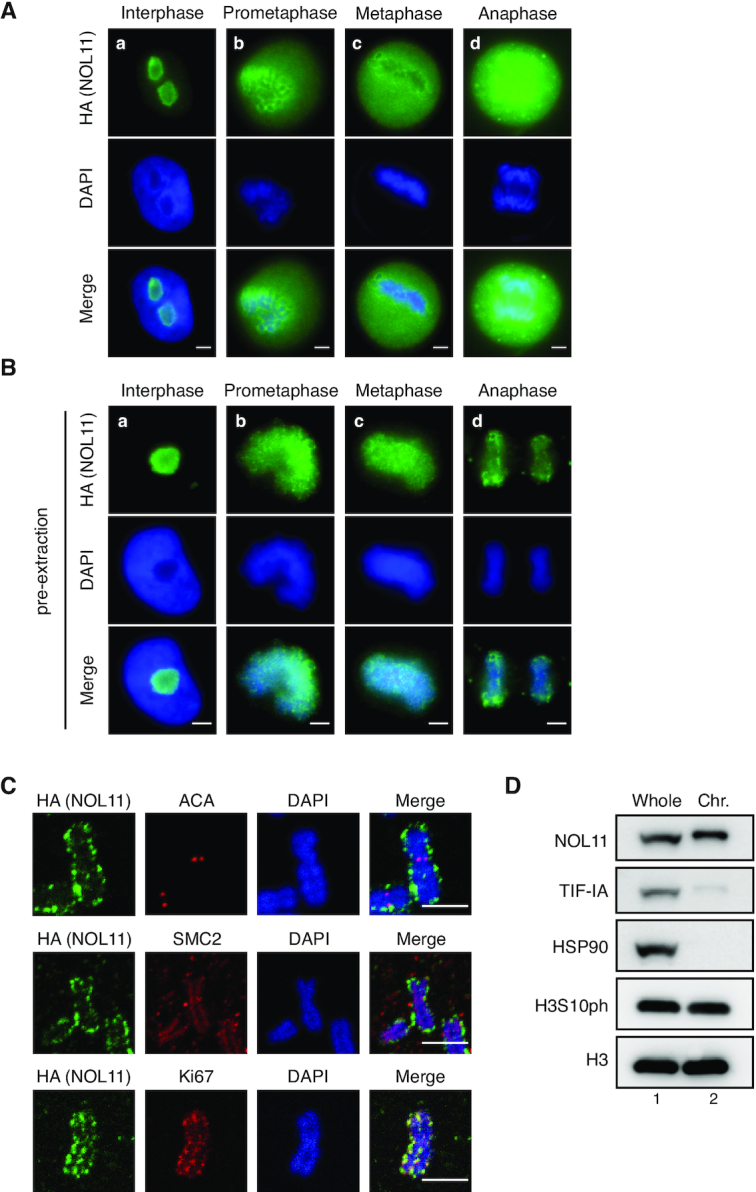
NOL11 localises at the chromosome periphery. (A–C) Localisation of NOL11 at the PR. HeLa cells that stably expressed FLAG- and HA-tagged NOL11 (FLAG/HA-NOL11 HeLa cells) were fixed with 4% PFA after pre-extraction (B) or without pre-extraction (A) and stained with anti-HA antibodies (NOL11; green) and DAPI (blue). Representative cells in interphase (a), prometaphase (b), metaphase (c), and anaphase (d) are shown. Scale bar, 5 μm. (C) Chromosome spreads prepared from FLAG/HA-NOL11 HeLa cells were co-stained with anti-HA antibodies (NOL11; green) and ACA (red), anti-SMC2 antibodies (red), or anti-Ki67 antibodies (red) and then DAPI (blue). Scale bar, 2 μm. (D) Localisation of NOL11 in the chromosomal fraction of mitotic cells. HeLa cells were synchronised in mitosis and fractionated. Whole-cell extracts (Whole) and chromosomal fractions (Chr.) were immunoblotted with the indicated antibodies. HSP90 is a marker for cytosolic proteins and H3 is a marker for chromosomal proteins. Note that mobility of NOL11 in the chromosomal fraction appears slower than that in the Whole-cell extracts.
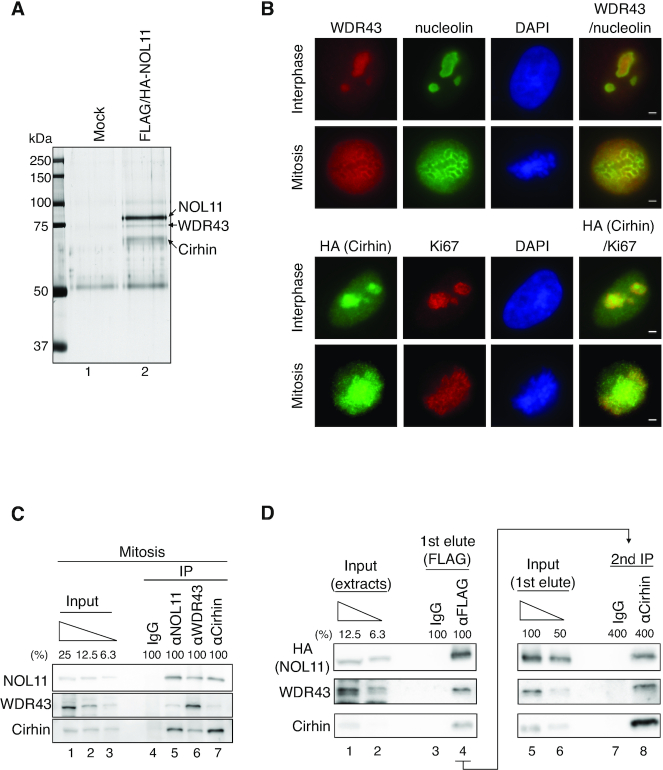
As no functional motifs in NOL11 could be found, one possibility was that NOL11 is involved in the mitotic control as a component of a protein complex. To examine this, the tandem affinity purification approach was chosen to identify NOL11-interacting proteins from mitotic HeLa cells that stably express FLAG- and HA-tagged NOL11 (FLAG/HA-NOL11). We examined several discernible species in a sample from FLAG/HA-NOL11-expressing cell extracts by mass spectrometry analysis and only WDR43 and Cirhin were identified as candidates for NOL11-interacting proteins (Figure 2A). Immunofluorescence staining revealed that WDR43 and Cirhin localized in the nucleolus during interphase and associated with the PR during mitosis in a manner similar to NOL11 (Figure 2B). Importantly, an interaction between the endogenous proteins in mitotic cells was found by co-immunoprecipitation (co-IP) experiments (Figure 2C).
Figure 2.
NOL11, WDR43, and Cirhin comprise the NWC complex. (A) Identification of NOL11-interacting proteins. Extracts prepared from nocodazole-treated HeLa cells (Control) or those stably expressing FLAG/HA-NOL11 were incubated with anti-FLAG antibody-conjugated agarose beads, and bound proteins were eluted using the FLAG peptide. The eluates were subsequently immunoprecipitated with anti-HA antibody-conjugated protein G Sepharose beads. Bound proteins were resolved through SDS-PAGE, visualised by silver staining, and analysed via MS. (B) Localization of WDR43 and Cirhin. Interphase (top row) or mitotic (second row) HeLa cells were co-stained with anti-WDR43 antibodies (red) and anti-Nucleolin antibodies (green), followed by DAPI staining (blue). Alternatively, interphase (third row) or mitotic (bottom row) HeLa cells stably expressing FLAG/HA-Cirhin were co-stained with anti-HA antibodies (green) and anti-Ki67 antibodies (red), followed by DAPI staining (blue). Scale bar, 2 μm. (C) Endogenous interactions among NOL11, WDR43, and Cirhin. Endogenous NOL11, WDR43, or Cirhin was immunoprecipitated from mitotic HeLa cell extracts and then immunoblotted with the indicated antibodies. (D) Complex formation by NOL11, WDR43 and Cirhin. Mitotic extracts prepared from HeLa cells stably expressing FLAG/HA-NOL11 were mixed with normal mouse IgG or anti-FLAG antibody-conjugated agarose beads, and bound proteins were eluted using the FLAG peptides (1st elute). Then, the eluates from anti-FLAG antibody-conjugated agarose beads were immunoprecipitated using anti-Cirhin antibodies (second IP). Proteins were analysed using the indicated antibodies.
Next, we conducted a sequential immunoprecipitation to confirm that NOL11, WDR43 and Cirhin were present in the same protein complex (Figure 2D). Extracts prepared from mitotic HeLa cells stably expressing FLAG/HA-NOL11 were mixed with control or FLAG agarose beads followed by FLAG peptide elution (Figure 2D, left). WDR43 and Cirhin in the fraction were found to be eluted by FLAG peptides (Figure 2D, lane 4). Then, the eluates that contained proteins bound to FLAG/HA-NOL11 were immunoprecipitated with anti-Cirhin antibodies (Figure 2D, right). Immunoblot analysis showed the presence of WDR43 in the final immunoprecipitants (Figure 2D, lane 8). These observations indicate that NOL11 forms a protein complex with WDR43 and Cirhin in mitotic cells, and we hereafter called this complex the NWC complex. We also performed Co-IP experiments using interphase-synchronised extracts, and found that NOL11 interacted with WDR43 and Cirhin even in interphase cells (Supplementary Figure S2). These results implied that the NWC complex is assembled throughout the cell cycle
It should be noted that NOL11, WDR43 and Cirhin are reportedly the components of the t-UTP/UTPA complex with three other proteins (hUTP10, hUTP15 and hUTP17) in human cells (24), raising the possibility that the NWC complex is included in the t-UTP/UTPA complex. To test this, we immunoprecipitated the NWC complex using antibodies for each subunit of this complex from either mitotic or interphase HeLa cell extracts; we then analyzed the immunoprecipitants by immunoblotting for other known components of the t-UTP/UTPA complex, hUTP10 and hUTP15. In all cases, the amounts of hUTP10 and 15 were beyond the detection limits, when NOL11, WDR43 and Cirhin were efficiently immunoprecipitated, suggesting that the majority of the NWC complexes do not comprise the t-UTP/UTPA complex (Supplementary Figure S3).
All components of the NWC complex are required for chromosomal association of this complex
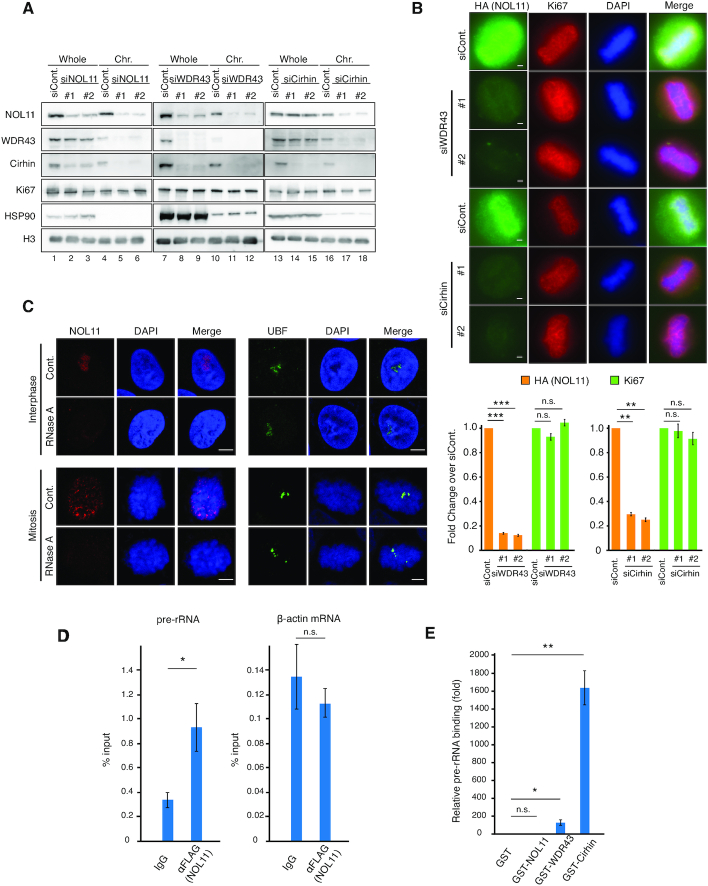
To address how protein stability and chromosomal association may depend on each component, the fractionation experiment was conducted after siRNA-mediated knockdown experiments for NOL11, WDR43, or Cirhin (Figure 3A). The total amount of Cirhin was decreased when WDR43 or NOL11 was depleted (Figure 3A, lanes 2, 3, 8 and 9), and the amount of NOL11 was reduced by depleting WDR43 but not Cirhin (Figure 3A, lanes 8, 9, 14 and 15), but the amount of WDR43 was unaffected by depleting NOL11 or Cirhin (Figure 3A, lanes 2, 3, 14 and 15). It was obvious that these effects were mediated at the protein level, as quantitative RT-PCR showed that depletion of any of the component did not grossly affect the mRNA levels (Supplementary Figure S4). These data suggest that the protein stability of the NWC components is interdependent, implicating that NOL11, WDR43 and Cirhin form a functional complex in cells.
Figure 3.
All components of the NWC complex are required for chromosomal association of this complex. (A) Effect of NOL11, WDR43, or Cirhin knockdown on the chromosomal association of the NWC complex. HeLa cells transfected with luciferase-specific siRNA (siCont.), NOL11-specific siRNA (siNOL11), WDR43-specific siRNA (siWDR43), or Cirhin-specific siRNA (siCirhin) were synchronised in mitosis and fractionated. Whole-cell extracts and chromosomal fractions were immunoblotted with the indicated antibodies. (B) Dissociation of NOL11 from the PR through knockdown of WDR43 or Cirhin. FLAG/HA-NOL11 HeLa cells transfected with siCont., siWDR43, or siCirhin were fixed with 4% PFA after pre-extraction and co-stained with anti-HA antibodies (NOL11; green) or anti-Ki67 antibodies (red) and then DAPI (blue). Representative cells in metaphase are shown. Scale bar, 2 μm. For each condition, the relative intensity of NOL11 and Ki67 signals at the PR was measured and shown in the graph. Values are mean ± SEM (n = 3), **P < 0.01, and ***P < 0.001, n.s. not statistically different (one-tailed Welch's t-test). Chromosomes from >25 mitotic cells were measured. (C) Translocation of the NOL11 from the nucleolus or mitotic chromosomes in the presence of RNase A. HeLa cells in interphase (top and second rows) and mitosis (third and bottom rows) were permeabilised and incubated without (Cont.) or with RNase A. Samples were subsequently fixed and stained with anti-NOL11 antibodies (red), anti-UBF antibodies (green), and DAPI (blue). Scale bar, 5 μm. (D) Association between pre-RNA and the NWC complex. The NWC complex was immunoprecipitated from mitotic HeLa cells that stably expressed FLAG/HA-NOL11 using anti-FLAG antibodies, and associated RNA was analysed using quantitative RT-PCR using specific primer sets for pre-rRNA (5′ETS regions) or β-actin mRNA. Values are mean ± SEM (n = 3). *P < 0.05, n.s. not statistically different (one-tailed Welch's t-test). (E) RNA-binding activity of Cirhin. RNA purified from HeLa cells was incubated with glutathione Sepharose-immobilised GST (lane 1) or GST fusion proteins of each component of the NWC complex (lanes 2–4). After extensive washing, bound RNA was analysed using quantitative RT-PCR using specific primer sets for pre-rRNA (5′ETS regions). Results were normalized to those for GST. Values are mean ± SEM (n = 3). *P < 0.05, **P < 0.01, n.s. not statistically different (one-tailed Welch's t-test).
The fractionation assay notably indicated that all components of the NWC complex are required for chromosomal association of the complex (Figure 3A, lanes 4–6, 10–12, and 16–18). In the chromosome-enriched fraction, the residual amounts of NOL11 and WDR43 were reduced in the absence of Cirhin (Figure 3A, lanes 17 and 18), and WDR43 was undetectable in the absence of NOL11 (Figure 3A, lanes 11 and 12). In contrast, the chromosomal association of Ki67, a major PR protein, was little affected in the absence of any of the NWC complex (Figure 3A). Consistent with these results, immunofluorescence staining indicated that NOL11 was displaced from the PR, but Ki67 remained in the PR of mitotic chromosomes when cells were depleted of WDR43 or Cirhin (Figure 3B).
Recent studies have indicated that the newly synthesised pre-rRNA associates with the PR and acts as a binding scaffold for several pre-rRNA processing proteins (20,33). Consistent with this notion, treatment with RNase A reduced the NOL11 signals from the PR (Figure 3C), unlike UBF that directly binds to the rDNA promoter region (Figure 3C). Of note, RNA immunoprecipitation assays showed that the NWC complex could interact with pre-rRNA but not with β-actin mRNA in mitotic cells (Figure 3D). Thus, pre-rRNA may act as a binding scaffold for the NWC complex in mitotic chromosomes. Consistent with this notion, NOL11 signals in the PR were reduced by the treatment of low-dose actinomycin D, an inhibitor of RNA polymerase I, but hardly affected by the treatment of amanitin, an inhibitor of RNA polymerase II (Supplementary Figure S5). In contrast, Ki67 signals in the PR were not affected by these inhibitors (Supplementary Figure S5) as reported previously (20). To determine which components possess RNA-binding activity, GST fusion proteins of each component of the NWC complex were purified and tested for their ability to bind to pre-rRNA. Although NOL11 did not exhibit RNA-binding activity (Figure 3E), Cirhin showed significant RNA-binding activity (Figure 3E). WDR43 showed much lower RNA-binding activity than Cirhin (Figure 3E). Thus, it is possible that the NWC complex binds to the mitotic chromosome periphery by virtue of RNA-binding ability of Cirhin.
The NWC complex is involved in mitotic function to regulate microtubule-kinetochore (MT-KT) attachments
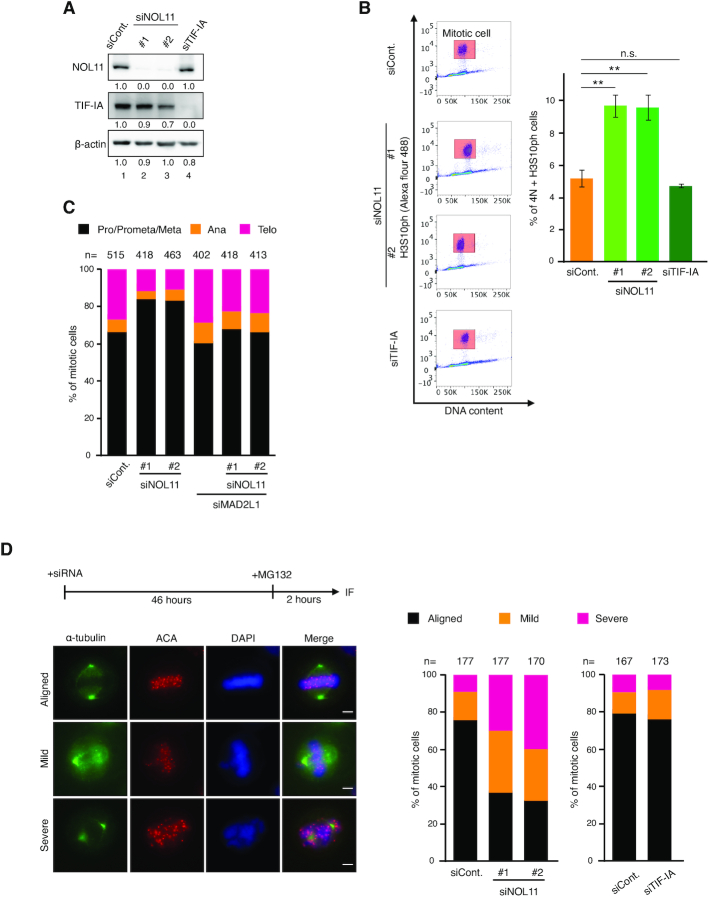
To examine the mitotic functions of the NWC complex, we conducted RNA interference-mediated knockdown experiments in HeLa cells using two different siRNAs (Figure 4A). Fluorescence-activated cell sorting (FACS) analysis showed that the mitotic index was increased by more than twofold in NOL11-depleted cells compared to that observed in control cells (Figure 4B). In contrast, TIF-IA depletion, which also caused nucleolar disruption and delayed mitotic entry similarly to NOL11 depletion (23), had a limited effect on the mitotic index (Figure 4B). Mitotic cells depleted of NOL11 appeared to spend a longer time progressing from prophase to metaphase as indicated by their increased population (Figure 4C). Co-depletion of MAD2L1, an essential spindle checkpoint protein (34), was found to largely restore the mitotic delay caused by NOL11 depletion, indicating that the spindle-assembly checkpoint remained active for a longer time in the absence of NOL11 (Figure 4C). The depletion of WDR43 and Cirhin also showed similar results (Supplementary Figure S6A).
Figure 4.
NOL11 depletion delays progression through mitosis and caused chromosome misalignment. (A) Efficiency of depletion. HeLa cells were transfected with siCont., two distinct siNOL11, or TIF-IA-specific siRNA (siTIF-IA). Forty-eight hours after transfection, whole-cell extracts were immunoblotted using the indicated antibodies. β-actin was used as a loading control. Numbers below the panel of immunoblots indicated the relative intensity of signals. (B) Mitotic arrest in NOL11-depleted cells. The mitotic index was determined by dual-colour FACS analysis using propidium iodide and anti-H3S10ph antibody 48 h after siRNA transfection. Mitotic cells were gated for H3S10ph-positive cells with ≃4N DNA content (4th column, red rectangle). Values are mean ± SEM (n = 3). **P < 0.01, n.s. not statistically different (one-tailed Welch's t-test). (C) Mitotic arrest by NOL11 depletion and rescue by MAD2L1 depletion. HeLa cells transfected with combination of siCont., siNOL11 (#1 and #2), or MAD2L1-specific siRNA (siMAD2L1) for 48 h were analysed. Mitotic cells were classified as prophase to metaphase (Pro/Prmeta/Meta), anaphase (Ana), and telophase (Telo). The distribution of mitotic cells in each phase is shown. (D) Defective chromosome alignment in NOL11-depleted cells. After treatment with 10 μM MG132 for 2 h, cells transfected with siCont., two distinct siNOL11, or siTIF-IA were stained with anti-α-tubulin antibodies (green), ACA (red), and DAPI (blue). Scale bar, 5 μm. The percentage of mitotic cells with different degrees of chromosome alignment is presented (right). The classification of chromosome alignment was as follows: full alignment (Aligned), one to five misaligned chromosomes (Mild), and more than six misaligned chromosomes (Severe).
Upon close inspection of those mitotic cells in the absence of NOL11, or of WDR43 and Cirhin, it was found that chromosome alignment on the metaphase plate was severely compromised (Figure 4D; Supplementary Figure S6B), which would explain why these cells sustained the checkpoint and caused mitotic delay (Figure 4C; Supplementary Figure S6A). In contrast, the depletion of TIF-IA did not show a gross effect on chromosome alignment (Figure 4D).
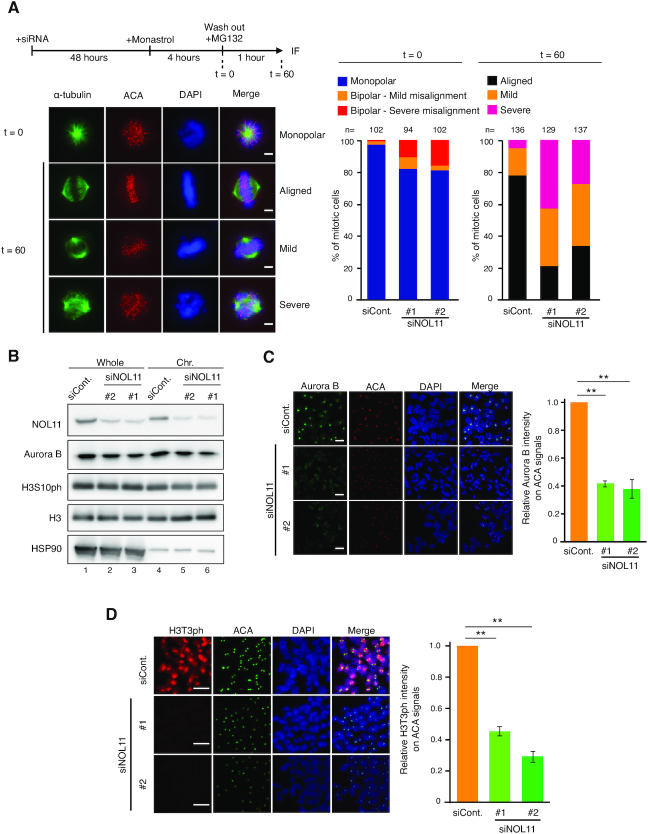
As an efficient chromosome alignment depends on the regulated interaction between MT-KT and the establishment of bipolar orientation on the spindle (35–37), the following question was whether the NWC complex might be involved in these processes. To address this, the monastrol release assay was used, in which the ability to regulate MT-KT attachments can be quantitatively measured by the correction rate from monopolar to bipolar spindle configuration. Treatment with monastrol, an Eg5 inhibitor, accumulates cells with monopolar spindles, causing the formation of syntelically attached sister kinetochore pairs (38). The removal of monastrol quickly converts monopolar to bipolar spindles, replacing syntelic attachments with amphitelic attachments by releasing syntelic MT-KT attachments and correcting the orientation. Whereas chromosome alignment was established shortly after the removal of monastrol in control experiments, it was severely perturbed in cells depleted of NOL11, WDR43 or Cirhin (Figure 5A; Supplementary Figure S7), indicating that the regulated interaction of MT-KT attachments became defective by depleting the NWC complex.
Figure 5.
NOL11 depletion causes enrichment failure of Aurora B at centromeres. (A) Defect of chromosome alignment after release from monastrol arrest in NOL11-depleted cells. HeLa cells were cultured for 48 h after transfection with siCont. or siNOL11, treated with 100 μM monastrol for 4 h, and subsequently transferred in medium containing 10 μM MG132. Immunofluorescence and classification of chromosome alignment were performed as shown in Figure 4D. Representative images of cells 0 or 60 min after the removal of monastrol are shown. Scale bar, 5 μm. (B) Aurora B and H3S10ph levels on mitotic chromosomes are not detectably affected by depleting NOL11. HeLa cells transfected with siCont. or siNOL11 were synchronised in mitosis and fractionated. Whole-cell extracts and chromosomal fractions were immunoblotted with the indicated antibodies. (C) Reduced localisation of Aurora B at centromeres in NOL11-depleted cells. Mitotic chromosome spreads were prepared 48 h after siRNA transfection and stained with anti-Aurora B antibodies (green), ACA (red), and DAPI (blue). Scale bar, 2 μm. For each condition, the relative intensity of Aurora B signals at centromeres was measured (right). Values are mean ± SEM (n = 3). Chromosomes from more than 15 mitotic cells were measured. **P < 0.01 (one-tailed Welch's t-test). (D) Reduced localisation of H3T3ph at centromeres in NOL11-depleted cells. Mitotic chromosome spreads were prepared 48 h after siRNA transfection and stained with anti-H3T3ph antibodies (red), ACA (green) and DAPI (blue). Scale bar, 2 μm. For each condition, the relative intensity of H3T3ph signals at centromeres was measured (right). Values are mean ± SEM (n = 3). Chromosomes from more than 15 mitotic cells were measured. **P < 0.01 (one-tailed Welch's t-test).
Consistent with this idea, NOL11 depletion severely compromised the kinetochore association of a plus end-directed microtubule motor CENP-E (Supplementary Figure S8), which plays an essential role in congressing chromosomes to the equatorial plane (39,40). In contrast, NOL11 depletion did not largely affect the level of Hec1 (Supplementary Figure S8), the kinetochore protein that directly mediates microtubule interaction (41,42). These results suggest that the NWC complex is involved in the regulation of MT-KT attachment, rather than having a role in kinetochore assembly.
The NWC complex is required to enrich the histone mark that recruits Aurora B at centromeres
The phenotypes induced by the depletion of the NWC subunits bore some resemblance to those expected upon the inhibition of Aurora B. This mitotic kinase forms the chromosomal passenger complex along with inner centromere protein (INCENP), survivin, and borealin/dasra and is involved in multiple mitotic events, including the regulation of bipolar MT-KT attachments and maintenance of centromeric cohesion (37,43–46). To accomplish these functions, the centromeric enrichment of the activity of Aurora B is essential. Specifically, the kinetochore localisation of CENP-E is known to depend on the activity of Aurora B (39). Thus we examined the effects of NOL11 depletion on the localisation of Aurora B.
Immunoblotting experiments showed that NOL11 depletion had little effect on the total amount of Aurora B, as well as on the chromosome-bound Aurora B (Figure 5B). In addition, NOL11 depletion did not detectably affect the levels of H3S10ph on mitotic chromosomes, implying that the overall Aurora B activity remained unchanged (Figure 5B). However, in immunofluorescence staining, NOL11 depletion noticeably caused a reduction of Aurora B-specific signal intensities at centromeres (Figure 5C). Similar effects on Aurora B were also found after the depletion of WDR43 or Cirhin (Supplementary Figure S9A and B). Thus, the loss of centromeric enrichment of Aurora B seems to account for the mitotic defects associated with the depletion of the NWC complex. Supporting this idea, we found that treatment of an Aurora B inhibitor ZM447439 caused a similar degree of chromosome misalignment between control and the NOL11-depleted cells (Supplementary Figure S10).
To address the mechanism by which the NWC complex concentrates Aurora B to centromeres, we first asked if NOL11 may directly regulate Aurora B localisation. In immunoprecipitation experiments, we found that NOL11 did not co-immunoprecipitate with Aurora B (Supplementary Figure S11A and B), discounting such possibility. As H3T3ph is known to be the histone mark to recruit Aurora B at centromeres by serving as the direct binding sites for survivin (47–49), we addressed whether NOL11 depletion influenced this modification. Immunofluorescence staining revealed that NOL11 depletion markedly reduced H3T3ph-specific signal intensities at centromeres (Figure 5D), the total levels of H3T3ph were not detectably affected in immunoblotting analysis (Supplementary Figure S11C). These seemingly inconsistent results (Figure 5D; Supplementary Figure S11C) allowed us to speculate that the NWC complex is required for the centromeric concentration of H3T3ph to recruit Aurora B at centromeres. NOL11 did not co-immunoprecipitate with Haspin (Supplementary Figure S11B), the kinase that phosphorylates H3T3 (50–52), indicating that the function of the NWC complex is involved in specifying the H3T3ph at centromeres by means other than interacting with Haspin.
The NWC complex is involved in maintaining sister chromatid cohesion
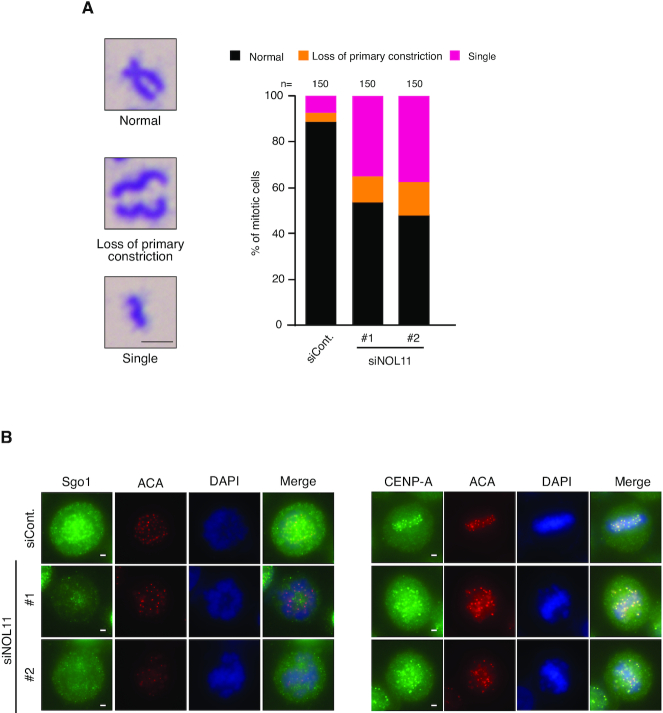
Finally, we addressed whether NWC depletion influenced sister chromatid cohesion, which also depends on centromeric Aurora B activity. In Giemsa staining of spread chromosomes, we found that NOL11, WDR43, or Cirhin depletion led to defects in mitotic chromosome cohesion, including loss of primary constrictions and separated single chromatids (Figure 6A; Supplementary Figure S12), suggesting that centromere organization for the maintenance of sister chromatid cohesion requires the NWC complex in mitosis. Of note, NOL11 depletion caused the delocalisation of Sgo1 from centromeres (Figure 6B), which should also contribute to these defects.
Figure 6.
NOL11 depletion disrupts sister chromatid cohesion. (A) Defect in sister chromatid cohesion by NOL11 depletion. Mitotic chromosome spreads were prepared from HeLa cells transfected with siCont. or siNOL11 and stained with Giemsa stain. (Left) Typical examples of normal (top), loss of primary constriction (middle), or single chromosomes (bottom), respectively. Scale bar, 2 μm. (Right) Percentages of normal and aberrant chromosomes. (B) Delocalisation of Sgo1 from centromeres. HeLa cells transfected with siCont. or siNOL11 were fixed with 4% PFA after pre-extraction and co-stained with anti-Sgo1 (left, green) and anti-CENP-A (right, green) and ACA antibodies (red) and then DAPI (blue). Representative cells are shown. Scale bar, 2 μm.
We furthermore examined the effect of NOL11 depletion on the localisation of Ki67 and a condensin subunit SMC2 (Supplementary Figure S13), major PR and axial proteins, respectively, both of which are required for the integrity of chromosomal arms. These proteins show little if any changes in their localisation on chromosomes, supporting the conclusion that the maintenance of centromeric function is the one of the prime function of the NWC complex in mitosis (Figure 6; Supplementary Figure S13).
DISCUSSION
Identification of the NWC complex that comprises nucleolar proteins during mitosis
In this study, we found that NOL11 formed a protein complex with WDR43 and Cirhin in HeLa cells (Figure 2; Supplementary Figure S2). This complex, the NWC complex, is associated with the PR of mitotic chromosomes (Figures 1 and 2B; Supplementary Figure S1) and required for centromere functions (Figures 4–6; Supplementary Figures S6, S7 and S12). Importantly, these three nucleolar proteins comprise a functional complex, as all subunits were indispensable for its chromosomal localisation and mitotic function (Figure 3A and B). It is likely that the NWC complex associates with the PR depending on the direct interaction of Cirhin with pre-rRNA (Figure 3C–E; Supplementary Figure S5). The finding that Cirhin has an ability to bind to RNAs purified from mitotic cells seems reasonable, as Cirhin depletion caused the dissociation of the other components from mitotic chromosomes without affecting their protein stability (Figure 3A and B). This mode of interaction is consistent with the report that newly synthesised pre-rRNA on the PR serves as a binding scaffold (33) and also with our previous observation that the pre-rRNA links nucleolar proteins at the outer layer of PRs (20).
We also found that NOL11 associated with WDR43 and Cirhin in interphase cells (Supplementary Figure S2), which is consistent with reports describing that these proteins are components of the t-UTP/UTPA complex of the SSU processome that is required for both optional pre-rRNA transcription and processing (22,24). The t-UTP/UTPA complex contains three other subunits (hUTP10, hUTP15, and hUTP17) in addition to the NWC complex in human cells. However, our results indicate that the majority of the NWC complexes do not comprise the t-UTP/UTPA complex (Supplementary Figure S3). Notably, in a recent report, it was described that WDR43 binds to several gene promoters through promoter-associated noncoding/nascent RNAs and is implicated in the regulation of pluripotency. In this mode of action, WDR43 appears also to be dissociated from the t-UTP/UTPA complex. Thus, it is possible that NOL11, WDR43 and Cirhin form several different complexes, with different protein compositions.
The NWC complex is involved in both mitotic entry and mitotic progression
We previously found that NOL11 depletion caused precocious nucleolar disruption during interphase, which in turn suppressed the activation of Cdk1 and delayed mitotic entry (23). It has been known that treating cells with low-dose Cdk1 inhibitor RO-3306 causes several mitotic chromosomal defects and polyploidy (53,54). Therefore, it was formally possible that an insufficient activity of Cdk1 underlies the mitotic phenotypes of NWC depletion, including metaphase arrest, chromosome misalignment, concentration defect of Aurora B at centromeres, defect in sister chromatid cohesion, and delocalisation of several centromere/kinetochore proteins (Figure 4C and D, 5C, and 6; Supplementary Figures S6, S7, S8 and S12). However, it is unlikely that this is the case for NOL11 depletion, as the level of the Cdk1 activity eventually reached the level attained in unperturbed mitosis, indicated by Cdk1-dependent MPM2 phosphoepitope (23). Depletion of TIF-IA, another protein involved in rRNA transcription at nucleoli, also caused nucleolar disruption in interphase nuclei and delayed Cdk1 activation (23), supporting the idea that delayed mitotic entry is a common consequence after nucleolar disruption. However, as TIF-IA depletion caused neither mitotic arrest (Figure 4B) nor mitotic chromosome misalignment (Figure 4D), having a role in mitotic chromosome organisation may be a unique feature of the NWC complex.
The NWC complex facilitates the enrichment of Aurora B at centromeres
Our results showed that the depletion of the NWC complex impaired the enrichment of Aurora B at centromeres (Figure 5C; Supplementary Figure S9), and a reduction of the centromeric histone mark H3T3ph provides binding sites for the Aurora B complex (Figure 5D) (47–49). We found that the mitotic delay caused by NWC complex depletion was associated with defects in chromosome alignment, mitotic spindle assembly, and chromosome biorientation (Figures 4D and 5A; Supplementary Figures S6B and S7).
These mitotic phenotypes can be stemmed from a defective regulation of microtubule attachments at kinetochore, in which Aurora B plays a direct role (37,44,45,55). In line with this idea, NOL11 depletion severely impaired the kinetochore association of CENP-E, a plus end-directed microtubule motor required for microtubule attachment (39,40). Moreover, the chromosome spread analysis indicated that sister chromatid cohesion is perturbed in cells depleted of each NWC complex (Figure 6A; Supplementary Figure S12), which might be a consequence of the delocalisation of Sgo1 from centromeres (Figure 6B). As cohesin-mediated sister chromatid cohesion at centromeres serves as a mechanical foundation to establish stable KT-MT attachments and to promote chromosome biorientation, loss of sister chromatid cohesion seems to provide an alternative explanation of the mitotic phenotypes.
An obvious following question is how the NWC complex facilitates the centromeric concentration of Aurora B through H3T3ph. One possibility is that the NWC complex directly recruits Aurora B or Haspin, a mitotic kinase that phosphorylates the H3T3 site of H3 (50–52), to the centromeres of mitotic chromosomes. However, the findings of this study discounted this possibility (Supplementary Figure S11B).
Notably, the finding that NOL11 depletion did not affect the total levels of H3T3ph (Supplementary Figure S11C) allows us to raise an alternative possibility that the NWC complex is required to prevent the histone mark, H3T3ph, from spreading throughout chromosome arms. Once it is confined to centromeres, Aurora B will be accumulated through a positive feedback circuit formed between Aurora B and Haspin (52).
The centromeric Aurora B is known to protect cohesin, along with Haspin and Sgo1 (32,49,55); conversely, cohesin indirectly recruits the Aurora B complex to centromeres (55), suggesting the interdependency between the centromeric association of Aurora B and the establishment of sister chromatid cohesion. Thus, a second positive feedback regulation between Aurora B and cohesin may also contribute in building the centromere architecture.
Mechanistically how the NWC complex control the histone mark awaits future investigation, but one appealing hypothesis can be predicted based on the amino acid sequence of the Cirhin subunit of the NWC complex. Cirhin contains the conserved RVXF motif at its N-terminal end (amino acids 8–11), a consensus that indicates a direct binding to PP1. This RVXF motif is thought to be present in the targeting subunits of PP1 (56–58), which determines the intracellular localisation and substrate specificities of PP1. In fact, we found that the NWC complex interacted with PP1 in mitotic cell extracts (Supplementary Figure S14). One of the subclass of PP1, PP1γ, has been implicated in the dephosphorylation of H3T3ph on chromosome arms (58–60), limiting H3T3ph at centromeres (61). Thus, the balanced activities of Haspin and PP1γ underlie the centromeric confinement of H3T3ph and Aurora B (61), which may require the tethering of PP1γ to the chromosome periphery by the NWC complex along the arms of mitotic chromosomes.
CONCLUSIONS
This work identified the NWC complex that participates in regulation of MT-KT attachment and centromere assembly in human cells. The complex associates with the nucleolus during interphase and translocates to the periphery of mitotic chromosomes at the onset of mitosis when the nucleolus disassembles. There appear to exist several sets of proteins that dynamically move between nucleoli and PRs in a cell cycle-dependent manner. Whereas many of the nucleolar-perichromosome shuttling proteins seem to be involved in a timely transition into mitosis from interphase, the NWC complex is specifically involved in mitotic chromosome function through facilitating the enrichment of Aurora B at centromeres, presumably by generating different biochemical environments for chromosome arms and centromeres.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Sae Higashiura, Masanori Maki, Reina Yagishita, and Makiko Yanagida for technical assistance and Enago (www.enago.jp) for the English language review.
Authors contributions: A.F., Y.H., K.K. and K.K. designed the experiment. A.F., Y.H., K.K., Y.K., H.S. and K.K. conducted the experiments. A.F., Y.H., K.K., T.H., T.D., A.F. and K.K. conducted the data interpretation. A.F., Y.H., K.K., T.H. and K.K. wrote the manuscript.
Notes
Present address: Akiko Fujimura, Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
Present address: Yuki Hayashi, Cell Biology and Biophysics Unit, European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
Contributor Information
Akiko Fujimura, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Yuki Hayashi, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan; Life Science Center for Survival Dynamics, Tsukuba Advanced Research Alliance, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Kazashi Kato, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Yuichiro Kogure, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Mutsuro Kameyama, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Haruka Shimamoto, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Hiroaki Daitoku, Life Science Center for Survival Dynamics, Tsukuba Advanced Research Alliance, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Akiyoshi Fukamizu, Life Science Center for Survival Dynamics, Tsukuba Advanced Research Alliance, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
Toru Hirota, Cancer Institute of the Japanese Foundation for Cancer Research, Division of Experimental Pathology, 3-8-1 Ariake, Koto-ku, Tokyo 135-8550, Japan.
Keiji Kimura, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan; Life Science Center for Survival Dynamics, Tsukuba Advanced Research Alliance, University of Tsukuba, 1-1-1 Tenno-dai, Tsukuba Science City, Ibaraki 305-8577, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research, KAKENHI [25290064, 19H03257 to K.K., 15H02365, 15H05977 to T.H., 15J00599 to Y.H.] from the Japan Society for the Promotion of Sciences; The Novartis Foundation (Japan) for the Promotion of Science (to K.K.); Takeda Science Foundation (to K.K.). Funding for open access charge: Japan Society for the Promotion of Science [19H03257].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I.. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007; 8:574–585. [DOI] [PubMed] [Google Scholar]
- 2. Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D.. Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 2008; 129:13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmad Y., Boisvert F.M., Gregor P., Cobley A., Lamond A.I.. NOPdb: Nucleolar Proteome Database–2008 update. Nucleic Acids Res. 2009; 37:D181–D184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grummt I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma. 2013; 122:487–497. [DOI] [PubMed] [Google Scholar]
- 5. Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P.. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5237–E5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyman A.A., Simons K.. Cell biology. Beyond oil and water–phase transitions in cells. Science. 2012; 337:1047–1049. [DOI] [PubMed] [Google Scholar]
- 7. Zhu L., Brangwynne C.P.. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 2015; 34:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I.. The nucleolus under stress. Mol. Cell. 2010; 40:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. James A., Wang Y., Raje H., Rosby R., DiMario P.. Nucleolar stress with and without p53. Nucleus. 2014; 5:402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Sluis M., McStay B.. Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 2017; 46:81–86. [DOI] [PubMed] [Google Scholar]
- 11. Nunez Villacis L., Wong M.S., Ferguson L.L., Hein N., George A.J., Hannan K.M.. New roles for the nucleolus in health and disease. BioEssays. 2018; 40:e1700233. [DOI] [PubMed] [Google Scholar]
- 12. Dimario P.J. Cell and molecular biology of nucleolar assembly and disassembly. Int. Rev. Cytol. 2004; 239:99–178. [DOI] [PubMed] [Google Scholar]
- 13. Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011; 2:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zharskaia O.O., Zatsepina O.V.. [Dynamics and mechanisms of the nucleolus reorganization during mitosis]. Tsitologiia. 2007; 49:355–369. [PubMed] [Google Scholar]
- 15. Booth D.G., Earnshaw W.C.. Ki-67 and the chromosome periphery compartment in mitosis. Trends Cell Biol. 2017; 27:906–916. [DOI] [PubMed] [Google Scholar]
- 16. Gautier T., Robert-Nicoud M., Guilly M.N., Hernandez-Verdun D.. Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J. Cell Sci. 1992; 102:729–737. [DOI] [PubMed] [Google Scholar]
- 17. Cuylen S., Blaukopf C., Politi A.Z., Muller-Reichert T., Neumann B., Poser I., Ellenberg J., Hyman A.A., Gerlich D.W.. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016; 535:308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takagi M., Ono T., Natsume T., Sakamoto C., Nakao M., Saitoh N., Kanemaki M.T., Hirano T., Imamoto N.. Ki-67 and condensins support the integrity of mitotic chromosomes through distinct mechanisms. J. Cell Sci. 2018; 131:jcs212092. [DOI] [PubMed] [Google Scholar]
- 19. Booth D.G., Takagi M., Sanchez-Pulido L., Petfalski E., Vargiu G., Samejima K., Imamoto N., Ponting C.P., Tollervey D., Earnshaw W.C. et al.. Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. eLife. 2014; 3:e01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayashi Y., Kato K., Kimura K.. The hierarchical structure of the perichromosomal layer comprises Ki67, ribosomal RNAs, and nucleolar proteins. Biochem. Biophys. Res. Commun. 2017; 493:1043–1049. [DOI] [PubMed] [Google Scholar]
- 21. Sobecki M., Mrouj K., Camasses A., Parisis N., Nicolas E., Lleres D., Gerbe F., Prieto S., Krasinska L., David A. et al.. The cell proliferation antigen Ki-67 organises heterochromatin. eLife. 2016; 5:e13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freed E.F., Prieto J.L., McCann K.L., McStay B., Baserga S.J.. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet. 2012; 8:e1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayashi Y., Fujimura A., Kato K., Udagawa R., Hirota T., Kimura K.. Nucleolar integrity during interphase supports faithful Cdk1 activation and mitotic entry. Sci. Adv. 2018; 4:eaap7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prieto J.L., McStay B.. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007; 21:2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sondalle S.B., Baserga S.J., Yelick P.C.. The contributions of the ribosome biogenesis protein Utp5/WDR43 to craniofacial development. J. Dent. Res. 2016; 95:1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilkins B.J., Lorent K., Matthews R.P., Pack M.. p53-mediated biliary defects caused by knockdown of cirh1a, the zebrafish homolog of the gene responsible for North American Indian Childhood Cirrhosis. PLoS One. 2013; 8:e77670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao C., Andreeva V., Gibert Y., LaBonty M., Lattanzi V., Prabhudesai S., Zhou Y., Zon L., McCann K.L., Baserga S. et al.. Tissue specific roles for the ribosome biogenesis factor Wdr43 in zebrafish development. PLoS Genet. 2014; 10:e1004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chagnon P., Michaud J., Mitchell G., Mercier J., Marion J.F., Drouin E., Rasquin-Weber A., Hudson T.J., Richter A.. A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am. J. Hum. Genet. 2002; 71:1443–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griffin J.N., Sondalle S.B., Del Viso F., Baserga S.J., Khokha M.K.. The ribosome biogenesis factor Nol11 is required for optimal rDNA transcription and craniofacial development in Xenopus. PLoS Genet. 2015; 11:e1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sondalle S.B., Baserga S.J.. Human diseases of the SSU processome. Biochimi. Biophys. Acta. 2014; 1842:758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bi X., Xu Y., Li T., Li X., Li W., Shao W., Wang K., Zhan G., Wu Z., Liu W. et al.. RNA targets ribogenesis factor WDR43 to chromatin for transcription and pluripotency control. Mol. Cell. 2019; 75:102–116. [DOI] [PubMed] [Google Scholar]
- 32. Dai J., Sullivan B.A., Higgins J.M.. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell. 2006; 11:741–750. [DOI] [PubMed] [Google Scholar]
- 33. Sirri V., Jourdan N., Hernandez-Verdun D., Roussel P.. Sharing of mitotic pre-ribosomal particles between daughter cells. J. Cell Sci. 2016; 129:1592–1604. [DOI] [PubMed] [Google Scholar]
- 34. Musacchio A., Salmon E.D.. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007; 8:379–393. [DOI] [PubMed] [Google Scholar]
- 35. Cheeseman I.M., Desai A.. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008; 9:33–46. [DOI] [PubMed] [Google Scholar]
- 36. Tanaka T.U., Desai A.. Kinetochore-microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 2008; 20:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carmena M., Wheelock M., Funabiki H., Earnshaw W.C.. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012; 13:789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kapoor T.M., Mayer T.U., Coughlin M.L., Mitchison T.J.. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 2000; 150:975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S.. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003; 161:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A.. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006; 311:388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hara M., Fukagawa T.. Kinetochore assembly and disassembly during mitotic entry and exit. Curr. Opin. Cell Biol. 2018; 52:73–81. [DOI] [PubMed] [Google Scholar]
- 42. Joglekar A.P., Bloom K.S., Salmon E.D.. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr. Opin. Cell Biol. 2010; 22:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cimini D. Detection and correction of merotelic kinetochore orientation by Aurora B and its partners. Cell Cycle. 2007; 6:1558–1564. [DOI] [PubMed] [Google Scholar]
- 44. Hengeveld R.C.C., Vromans M.J.M., Vleugel M., Hadders M.A., Lens S.M.A.. Inner centromere localization of the CPC maintains centromere cohesion and allows mitotic checkpoint silencing. Nat. Commun. 2017; 8:15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lampson M.A., Cheeseman I.M.. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011; 21:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruchaud S., Carmena M., Earnshaw W.C.. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007; 8:798–812. [DOI] [PubMed] [Google Scholar]
- 47. Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., Funabiki H.. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010; 330:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., Higgins J.M.. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010; 330:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamagishi Y., Honda T., Tanno Y., Watanabe Y.. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010; 330:239–243. [DOI] [PubMed] [Google Scholar]
- 50. Dai J., Sultan S., Taylor S.S., Higgins J.M.. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005; 19:472–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Markaki Y., Christogianni A., Politou A.S., Georgatos S.D.. Phosphorylation of histone H3 at Thr3 is part of a combinatorial pattern that marks and configures mitotic chromatin. J. Cell Sci. 2009; 122:2809–2819. [DOI] [PubMed] [Google Scholar]
- 52. Wang F., Ulyanova N.P., van der Waal M.S., Patnaik D., Lens S.M., Higgins J.M.. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 2011; 21:1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McCloy R.A., Rogers S., Caldon C.E., Lorca T., Castro A., Burgess A.. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014; 13:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Voets E., Marsman J., Demmers J., Beijersbergen R., Wolthuis R.. The lethal response to Cdk1 inhibition depends on sister chromatid alignment errors generated by KIF4 and isoform 1 of PRC1. Sci. Rep. 2015; 5:14798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Trivedi P., Stukenberg P.T.. A Centromere-Signaling network underlies the coordination among mitotic events. Trends Biochem. Sci. 2016; 41:160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cohen P.T. Protein phosphatase 1–targeted in many directions. J. Cell Sci. 2002; 115:241–256. [DOI] [PubMed] [Google Scholar]
- 57. Egloff M.P., Johnson D.F., Moorhead G., Cohen P.T., Cohen P., Barford D.. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997; 16:1876–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trinkle-Mulcahy L., Andersen J., Lam Y.W., Moorhead G., Mann M., Lamond A.I.. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol. 2006; 172:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bollen M., Gerlich D.W., Lesage B.. Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 2009; 19:531–541. [DOI] [PubMed] [Google Scholar]
- 60. Wurzenberger C., Gerlich D.W.. Phosphatases: providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 2011; 12:469–482. [DOI] [PubMed] [Google Scholar]
- 61. Qian J., Lesage B., Beullens M., Van Eynde A., Bollen M.. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr. Biol. 2011; 21:766–773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.