Abstract
Individual colorectal adenomas have different propensities to progress to invasive disease. In this study, we explored whether these differences could be explained by gene copy number alterations. We evaluated 18 adenomas of patients without synchronous or subsequent carcinoma (6.5 years follow-up), 23 adenomas of carcinoma patients, and 6 related carcinomas. All samples were measured for their DNA ploidy status. Centromere probes for chromosomes 17 and 18, as well as gene-specific probes for SMAD7, EGFR, NCOA3, TP53, MYC, and RAB20 were assessed by multicolor fluorescence in situ hybridization. An increased genomic instability index of CEP17, SMAD7, and EGFR, as well as TP53 deletions and MYC amplifications defined adenomas of patients with synchronous carcinoma (P<0.05). Diploid NCOA3 signal counts were associated with longer adenoma recurrence-free surveillance (P = 0.042). In addition, NCOA3, MYC, EGFR, and RAB20 amplifications, as well as TP53 deletions correlated with increased DNA stem line values and/or aneuploidy in adenomas (P<0.05). Furthermore, aberrations of NCOA3, MYC, and RAB20 were associated with histopathologically defined high-risk adenomas (P<0.05). RAB20 amplifications were also correlated with high-grade dysplastic adenomas (P = 0.002). We conclude that genomic instability in colorectal adenomas is reflected by EGFR, MYC, NCOA3, and RAB20 amplifications that do correlate with histomorphological features and are indicative for adenoma recurrence and the presence of synchronous carcinomas.
Keywords: amplifications, colorectal adenomas, colorectal carcinogenesis, deletions, genomic instability
Colorectal carcinomas are the third leading cause of death from cancer in industrialized countries.1,2 Most of these tumors are based on genetic alterations of single mucosal cells that develop to invasive carcinomas according to the ‘adenoma–carcinoma sequence’.3 However, not all adenomas necessarily progress to invasive carcinomas, and the risk of malignant transformation depends on, for example, their histopathological subtype.4
Approximately 80% of all colorectal carcinomas show aneuploidy.5 Cytogenetic and molecular cytogenetic analyses have revealed that tumor-specific gains and losses of entire chromosomes or chromosome arms are early genome mutations.6–9 The genomic imbalances show a strikingly conserved recurrent pattern that is characteristic for colorectal carcinomas and is distinct from other solid tumors.6 For a comprehensive overview, please see http://cgap.nci.nih.gov/Chromosomes/Mitelman. Bomme et al10 could show that such specific chromosomal aberrations — especially, a gain of chromosomes 7 and 13 — already exist in adenomas. These aberrations are faithfully conserved in carcinomas and are accompanied by gains of chromosomes and chromosomal arms 6, 8q, and 20, as well as losses of 4q, 8p, 17p, and 18q.6 We could further show that introduction of trisomic chromosomes in an otherwise diploid genome increases the average gene expression level of the resident genes independent of cell type or chromosome.11 This effect was validated in a clinical cohort of colorectal carcinomas in which we could identify a correlation between chromosome amplifications and increased expression levels of the genes resident on the affected chromosomes.6 Therefore, it seems reasonable to assume that genes located on chromosomes that are affected by amplification or deletion do have an important role for colorectal carcinogenesis.
In this study, we report the use of three composite fluorescence in situ hybridization (FISH) panels to identify sequential oncogene amplifications and tumor-suppressor gene deletions during adenoma progression and a correlation of specific copy number changes with general genomic instability. We also queried whether the FISH cocktails would be useful for predicting individual progression risk by analyzing copy number changes in adenoma specimens of 23 patients with synchronous carcinoma and of 18 patients without malignancy. Six related colon carcinomas were analyzed as positive controls.
Materials and methods
Clinical Data
We obtained paraffin-embedded adenoma specimens from 18 patients without synchronous or subsequent carcinoma and from 23 patients with synchronous carcinoma, as well as 6 related carcinoma samples. All patients were diagnosed with at least one colorectal adenoma at the University Clinic Schleswig-Holstein, Campus Lübeck, between 1995 and 2000. The average observation time after polyp extirpation amounted to 6.5 years follow-up period (range 4–9 years; clinical data, Table 1a). All patients could be divided into four groups (Table 1b): patients in group A (n=8) did not present any additional adenoma, whereas patients in group B (n=10) presented at least one additional adenoma (recurrence) within the follow-up period. Group A and B patients did not develop a subsequent carcinoma during follow-up and were summarized as group C (n=18). In contrast, groups D and E included patients also presenting colorectal carcinoma in addition to adenoma. Group D (n=9) patients did not present adenoma recurrence, whereas group E patients (n=14) did. Groups D and E were summarized as group F (n=23).
Table 1a.
Clinical data
| No. | Sex | Age (years) | Group | Adenoma and carcinoma | Adenoma recurrence | Observation time (months) | Grade of dysplasia | Histology | Localization | Size (mm) | Synchronous adenomas |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 57 | A | No | No | 104 | Low grade | Tubular | Transverse | 2 | 2 |
| 2 | M | 70 | A | No | No | 85 | Low grade | Tubular | Left flexur | 2 | No |
| 3 | F | 70 | A | No | No | 71 | Low grade | Tubular | Descendent | 1 | No |
| 4 | M | 69 | A | No | No | 70 | Low grade | Tubulo-villous | Sigmoid | 12 | 4 |
| 5 | F | 82 | A | No | No | 6 | High grade | Tubulo-villous | Sigmoid | 10 | No |
| 6 | F | 78 | A | No | No | 117 | pTis | Tubulo-villous | Rectum | 17 | 3 |
| 7 | M | 79 | A | No | No | 104 | pTis | Tubulo-villous | Cecum | 60 | 3 |
| 8 | F | 67 | A | No | No | 71 | pTis | Tubulo-villous | Sigmoid | 9 | No |
| 9 | F | 65 | B | No | Month 38 | 91 | Low grade | Villous | Rectum | 20 | No |
| 10 | F | 62 | B | No | Month 35 | 92 | Low grade | Tubulo-villous | Sigmoid | 10 | 5 |
| 11 | F | 59 | B | No | Month 39 | 89 | Low grade | Tubulo-villous | Transverse | 7 | 2 |
| 12 | F | 59 | B | No | Month 3 | 73 | Low grade | Tubulo-villous | Sigmoid | 2 | No |
| 13 | F | 64 | B | No | Month 48 | 101 | High grade | Villous | Rectum | 140 | No |
| 14 | F | 76 | B | No | Month 3 | 119 | pTis | Tubulo-villous | Rectum | 20 | No |
| 15 | M | 82 | B | No | Month 18 | 111 | pTis | Tubular | Rectum | 3 | 2 |
| 16 | M | 66 | B | No | Month 14 | 101 | pTis | Tubular | ND | 18 | No |
| 17 | M | 77 | B | No | Months 12 and 21 | 80 | pTis | Tubulo-villous | Rectum | 17 | No |
| 18 | M | 78 | B | No | Month 3 | 68 | pTis | Tubulo-villous | ND | 8 | 3 |
| 19 | F | 85 | D | pT4,N2,MX; G3 | No | 86 | Low grade | Tubulo-villous | ND | 12 | 3 |
| 20 | M | 72 | D | pT4,N1,MX; G2 | No | 86 | Low grade | Tubulo-villous | Rectum | 2 | 2 |
| 21 | F | 61 | D | pT4,N0,MX; G2 | No | 85 | Low grade | Tubulo-villous | ND | 10 | No |
| 22 | M | 65 | D | pT3,N1,MX; G3 | No | 80 | Low grade | Tubular | Right flexur | 6 | No |
| 23 | F | 75 | D | pTis;G3 | No | 96 | Low grade | Tubulo-villous | Sigmoid | 20 | 3 |
| 24 | M | 68 | D | pTis;G3 | No | 3 | Low grade | Tubulo-villous | Transverse | 2 | 2 |
| 25 | F | 75 | D | pTis;G3 | No | 96 | High grade | Tubulo-villous | Right flexur | 3 | 3 |
| 26 | M | 59 | D | pTis; G3 | No | 84 | High grade | Tubulo-villous | Rectum | 18 | 5 |
| 27 | F | 68 | D | pT3,N1,MX; G2 | No | 79 | pTis | Tubulo-villous | ND | 16 | 3 |
| 28 | M | 78 | E | pTis; G3 | Month 36 | 36 | Low grade | Tubular | Sigmoid | 3 | No |
| 29 | M | 56 | E | pT3,N2,MX; G2 | Month 10 | 10 | Low grade | Tubular | Sigmoid | 5 | 2 |
| 30 | M | 78 | E | pT4,N1,MX; G2 | Month 24 | 24 | Low grade | Tubular | ND | 5 | 2 |
| 31 | M | 67 | E | pT2,N1,MX; G2 | Month 44 | 44 | Low grade | Tubular | ND | 4 | No |
| 32 | F | 70 | E | pT3,N1,MX; G2 | Months 11 and 24 | 24 | Low grade | Tubulo-villous | Cecum | 10 | 2 |
| 33 | F | 73 | E | pT1;G2 | Month 10 | 10 | Low grade | Tubulo-villous | Sigmoid | 16 | 2 |
| 34 | F | 77 | E | pT1;G2 | Month 13 | 13 | High grade | Tubulo-villous | Ascendent | 27 | No |
| 35 | F | 44 | E | pTis;G3 | Month 11 | 11 | High grade | Tubulo-villous | Rectum | 30 | 2 |
| 36 | M | 76 | E | pTis;G3 | Month 20 | 20 | High grade | Tubulo-villous | Rectum | 70 | No |
| 37 | M | 76 | E | pTis;G3 | Month 20 | 20 | High grade | Tubulo-villous | Cecum | 30 | 5 |
| 38 | M | 67 | E | pTis;G3 | Month 10 | 10 | High grade | Tubulo-villous | Descendent | 31 | 3 |
| 39 | F | 58 | E | pT1;G2 | Month 24 | 24 | pTis | Tubulo-villous | Rectum | 20 | No |
| 40 | F | 59 | E | pTis,N0,MX;G3 | Months 17 and 18 | 18 | pTis | Tubulo-villous | Sigmoid | 70 | No |
| 41 | M | 75 | E | pT3,N0,MX; G2 | Month 7 | 7 | pTis | Tubulo-villous | Rectum | 28 | No |
Patient groups A, B, C, and D are described in detail in Table 1b.
f, female; m, male; ND, not determined.
Table 1b.
Patient groups
| Group | Definition | Sex |
Age (years) | Adenoma size (mm) | Synchronous adenomas | Recurrence-free timea | |
|---|---|---|---|---|---|---|---|
| Male | Female | ||||||
| A (n = 8) | No carcinoma and no adenoma recurrence | 4 | 4 | 71.5 (57–82) | 14.1 (1–60) | 1.5 (2–4) | 78.5 months (6–117) |
| B (n = 10) | No carcinoma but adenoma recurrence | 4 | 6 | 68.8 (59–82) | 24.5 (2–140) | 1.5 (2–5) | 21.3 months (3–48) |
| C (n = 18) | No carcinoma | 8 | 10 | 70 (57–82) | 18 | 1.5 | 46.7 months (3 –117) |
| D (n = 9) | Synchronous carcinoma but no adenoma recurrence | 4 | 5 | 69.8 (61–85) | 9.9 (2–20) | 2.3 (2–5) | 77.2 months (3–96) |
| E (n = 14) | Synchronous carcinoma and adenoma recurrence | 8 | 6 | 68.1 (44–78) | 24.9 (3–70) | 1.3 (2–5) | 18.4 months (7–44) |
| F (n = 23) | Synchronous carcinoma | 12 | 11 | 68.8 (44–85) | 19 | 1.7 | 41.4 months (3–96) |
Diagnoses of adenoma and/or carcinoma were considered as recurrence.
Histomorphology
Classification of the degree of dysplasia was performed by an experienced pathologist without knowledge of clinical data. The degree of dysplasia was rated as suggested by Riddell et al (none, low grade, and high grade).12
Immunohistochemistry
Immunohistochemistry was performed using the standard peroxidase avidin–biotin–complex technique (Vector, Elite Standard Kit, cat. PK-6100). The sections were deparaffinized, rehydrated, and microwave treated in 0.01 M sodium citrate buffer (pH 6) for 10 min at 500 W. After rinsing in Tris-buffered saline solution (pH 7.6), endogenous peroxidase activity was blocked by slide immersion in 0.5% hydrogen peroxidase for 30 min. Unspecific staining was prevented by application of 1% bovine serum albumin (BSA) for 20 min. For laminin-5 immunohistochemistry, a polyclonal antibody was raised in rabbits against a fusion protein containing the C terminus of the laminin γ2 chain (containing amino-acid residues 1017–1178).13 This antibody was used for overnight incubation at 4°C in 1% BSA 1:200, followed by a biotinylated anti-rabbit IgG (diluted 1:200) for 30 min. Cyclin A analysis was performed using a monoclonal mouse antibody against human cyclin A protein (Novocastra Laboratories, Newcastle Upon Tyne, UK) diluted 1:100 in 1% BSA. This was followed by incubation with the avidin–biotin–peroxidase complex for an additional 30 min. The peroxidase reaction was visualized using DAB (diaminobenzidine tetrahydrochloride, 0.6 mg/ml with 0.03% H2O2 for 6 min). After counterstaining with hematoxylin, slides were dehydrated and mounted with a xylene-soluble mounting medium. As negative controls, primary antibodies were replaced by BSA. Cells were regarded as laminin-5 immunoreactive when a distinct staining of the cytoplasm was visible. Cyclin A immunoreactivity was confined to the cell nuclei. The percentage of stained cells was calculated in each specimen. On the basis of a semiquantitative scoring system, immunoreactivity for both laminin-5 γ2 chain and cyclin A was recorded as follows: category 0 = no specific antibody expression, category 1 = <20%, category 2 = 20–50%, and category 3 = >50% immunoreactivity of all mucosal cells.14 The slides were evaluated independently by two investigators (JKH and GA), and all cases with discrepant scores were reviewed until a conclusive judgment was reached.
Hedley Method and Cytospin Preparation
A modified Hedley method was used for isolating nuclei from formalin-fixed, paraffin-embedded tissues for multicolor FISH analysis on a single-layer preparation.15,16 Two 50-μm-thick sections from each of the formalin-fixed, paraffin-embedded tissues were deparaffinized in xylene, rehydrated in ethanol series and dH2O, and disintegrated in 500 μl of 0.1% protease/1 × phosphate-buffered saline (PBS) (protease: Type XXIV, Bacterial, P8038, Sigma, St Louis, MO, USA; Dulbecco’s 1 × PBS, Life Technologies, Rockville, MD, USA) at 45°C for 45–70 min. The reaction was stopped by adding 500 μl 1 × PBS at room temperature. The samples were filtered through a 30-μm nylon membrane (CN 051, Dako, Glostrup, Denmark), centrifuged, and resuspended in 1 × PBS. Cytospin slides were prepared by the Shanndon Cytospin and fixed in an ethanol series. These single-layer nuclei preparations were then used for ploidy assessment by image cytometry and for multicolor FISH analysis.
Ploidy Assessment
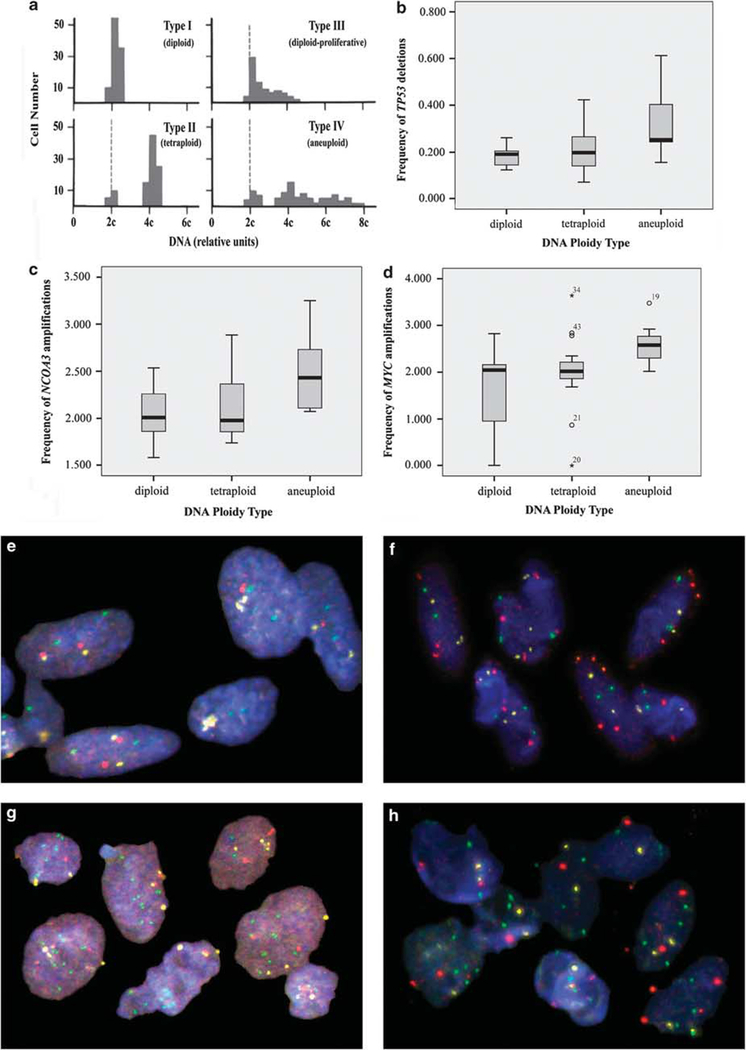
Image cytometry was performed on Feulgen-stained cytospins of paraffin-embedded tissue samples (see the section ‘Hedley Method and Cytospins’). The staining procedure, internal standardization, tumor cell selection, and analysis were based on methods described previously.17 DNA distribution profiles (histograms) were based on 200 interphase nuclei for each specimen. In brief, all DNA values were expressed in relation to the corresponding staining controls, which were given the value 2c, denoting the normal diploid DNA content. DNA profiles were classified according to Auer (Figure 1a).17 Histograms characterized by a single peak in the diploid or near-diploid region (1.5c–2.5c) were classified as type I.
Figure 1.
(a) DNA histograms according to Auer. Type I, II, and III reflect euploid DNA distribution pattern, and type IV characterizes aneuploid cell populations. (b) Frequency of TP53 deletions depending on the ploidy type. (c) Frequency of NCOA3 amplifications depending on the ploidy type. (d) Frequency of MYC amplifications according to ploidy types. Physiological copy numbers in diploid (e) and aberrant copy numbers in aneuploid adenomas (g) of probe panel II: CEP17, yellow/NCOA3, green/TP53, red. Physiological copy numbers in diploid (f) and aberrant copy numbers in aneuploid (h) adenomas of probe panel III: CEP17, yellow/MYC, green/RAB20, red.
The total number of cells with DNA values exceeding the diploid region (>2.5c) was <10%. Type II histograms showed a single peak in the tetraploid region (3.5c–4.5c) or peaks in both the diploid and the tetraploid regions (>90% of the total cell population). The number of cells with DNA values between the diploid and tetraploid regions and those exceeding the tetraploid region (>4.5c) was <10%. Type III histograms represent highly proliferating near-diploid cell populations and are characterized by DNA values ranging between the diploid and the tetraploid regions. Only a few cells (<5%) showed >4.5c. The DNA histograms of types I, II, and III characterize euploid cell populations. Type IV histograms show increased (>5%) and/or distinctly scattered DNA values exceeding the tetraploid region (>4.5c). These histograms reflect aneuploid cell populations. It has to be carefully considered though, that ploidy analysis is a crude measure of nuclear DNA content and that chromosomal instability by DNA cytometry alone can be hampered by balanced translocations or equal amounts of gains and losses, both leading to false-negative results in terms of chromosomal instability assessment. Against this background, genomic instability indices were calculated on signal enumeration of gene-specific probes by dividing the number of different signal patterns by the number of analyzed cells as described below.
Multicolor FISH
Multicolor FISH analysis was performed on each case using three different probe panels: Panel I consisted of one centromere probe specific for chromosome 18 (CEP18, labeled with Spectrum Aqua (SA)), one BAC contig that contains the SMAD7 gene on chromosome 18q12.3 (labeled with Spectrum Green (SG)), and another BAC contig that contains the EGFR gene on chromosome 7p12 (labeled with Spectrum Orange (SO)). In total, 12 562 nuclei were analyzed, 4503 nuclei of 17 adenomas of patients without carcinoma (Ø 265 nuclei per sample) and 6090 nuclei of 23 adenomas of patients with carcinoma (Ø 265 per sample). For 6 adenoma patients, the corresponding colorectal carcinoma was analyzed with a total of 1969 nuclei counts (Ø 328 per sample).
Panel II comprised one probe specific for centromere 17 (CEP17, labeled with SA), one BAC contig that contains the NCOA3 gene on 20q12 (labeled with SG), and another BAC contig that contains the TP53 gene on chromosome 17p13.1 (labeled with SO). In total, 6726 nuclei were analyzed, 2889 nuclei of 17 adenomas of patients without carcinoma (Ø 170 nuclei per sample) and 3139 of 21 adenomas of carcinoma patients (Ø 149 nuclei per sample). For 5 adenoma patients, the corresponding colorectal carcinoma was analyzed with a total of 698 nuclei counts (Ø 140 nuclei per sample).
Panel III summarized one centromeric probe specific for chromosome 17 (CEP17, labeled with SA), one BAC contig that contains the MYC gene on 8q24.12 (labeled with SG), and another BAC contig that contains the RAB20 gene on chromosome 13 (labeled with SO). In total, 10 335 nuclei were analyzed, 4230 nuclei of 17 adenomas of patients without carcinoma (Ø 249 nuclei per sample) and 4032 of 21 adenomas of carcinoma patients (Ø 192 nuclei per sample). For 6 adenoma patients, the corresponding colorectal carcinoma was analyzed with a total of 2073 nuclei counts (Ø 346 nuclei per sample). An overview of the three probe panels including detailed information about the BAC contigs used is shown in Table 2a.
Table 2a.
FISH probe panels
| Probe panel |
Spectrum Aqua |
Spectrum Green (gene probe 1) |
Spectrum Orange (gene probe 2) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Centromere probe | Gene | Location | Entrez Gene ID | BAC clones | Gene | Location | Entrez Gene ID | BAC clones | |
| I | Chromosome 18 (CEP18) | SMAD7 | 18q21.1 | 4092 | 748M14 716L10 15F12 756O18 8L20 |
EGFR | 7p12 | 1956 | 805M4 775L16 |
| II | Chromosome 17 (CEP17) | NCOA3 | 20q12 | 8202 | 456N23 109C3 |
TP53 | 17p13.1 | 7157 | 199F11 186B7 404G1 |
| III | Chromosome 17 (CEP17) | MYC | 8q24.21 | 4609 |
aCTD 2056O22 |
RAB20 | 13q34 | 55647 | 212E4 120J20 |
All BAC clones (Bacterial Artificial Chromosome) were selected through the Human BAC Library of the RPCI11 Rosswell Park Institute and ordered by BACPAC Resources, Oakland, CA, USA.
Ordered from Invitrogen.
The centromeric probes (Vysis/Abbott Laboratories Inc., Downers Grove, IL, USA) were used as internal controls for signal enumeration. The performance of all three probe panels was evaluated on methanol/acetic acid-fixed peripheral lymphocyte cultures derived from karyotypically normal individuals. FISH was performed as follows: In brief, cytospin slides were incubated in a 2 × saline-sodium citrate (SSC) solution at room temperature for 5 min. The slides were digested in 200 μl pepsin (Sigma P6887) in 0.01M HCL at 37°C, washed twice in 1 × PBS, and fixed in an ethanol series. Slides were denatured in 70% deionized formamide/2 × SSC for 4 min at 80°C and dehydrated in an ethanol series at −20°C.
For hybridization, 5 μl of each probe panel was applied to the slides. After overnight hybridization at 37°C, coverslips were removed and slides were washed three times for 5 min in 50% formamide/2 × SSC at 45°C and three times in 0.1 × SSC at 60°C. Followed by a 4 × SSC/0.1% Tween 20 wash (45°C), slides were incubated with 120 μl blocking solution (3%BSA/4 × SSC/0.1% Tween 20) for 30 min at 37°C. Two antibody layers were incubated separately for 45 min at 37°C as follows: the first layer comprised avidin-FITC (1:200) and mouse anti-digoxigenin (Sigma), and the second layer comprised rabbit anti-mouse-TRITC (Sigma). The slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and antifade was added (details of the experimental procedures can be retrieved from http://www.riedlab.nci.nih.gov).
Signal Enumeration
Image analysis was performed using a Leica DMRXA fluorescence microscope (Leica, Wetzlar, Germany) equipped with custom optical filters for DAPI, SA, SG, and SO (Chroma Technologies, Rockingham, VT, USA) and a ×40 Plan Apo (NA 1.25) objective. Images were taken in areas of optimal cell density using the ORCA ER (IEEE1394 I/F) digital camera (Hamamatsu, Bridgewater, NJ, USA). Leica Q-FLUORO was used to acquire multi-focus images for each of the SA, SG, and SO optical filters. A total of 20 images were acquired, and signal enumeration was performed on these digital images for 100–500 nuclei for each probe per case. The counted signals were manually checked and then listed in an Excel-based format for further statistical analysis.
Statistical Analysis
Explorative data analysis was carried out using standard statistical methods such as χ2 test, Wilcoxon rank-sum test, and Spearman’s rank correlation coefficient, depending on the scaling of the 16 observed parameters, including multi-FISH gene-specific targets, ploidy assessment, and clinical data. Survival times of four clinical subgroups were described by Kaplan–Meier methods and compared with log-rank tests. The assumption of normal distribution of genetic instability indices was not violated (Shapiro–Wilks test), such that the increase from diploid over tetraploid to aneuploid cases could be compared by one-sided t-tests. All other statistical tests were two sided. The level of significance was 0.05 and not adjusted for multiple comparisons, thus requiring careful interpretation of analysis data.
Results
Individual colorectal adenomas have different propensities to progress to invasive disease. In this study, we explored whether these differences could be explained by a different mutation spectrum. To study this question and to identify patterns of genome aberrations, we used FISH with probe sets that target commonly aberrant chromosomes, oncogene amplifications, and deletions of tumor-suppressor genes to a series of 18 adenomas of patients without synchronous or subsequent carcinoma, 23 adenomas of carcinoma patients, and 6 matched carcinomas. The probe sets included centromere probes for chromosomes 17 and 18, as well as gene-specific probes for SMAD7 (chromosome band 18q21), EGFR (7p12), NCOA3 (20q12), TP53 (17p13), MYC (8q24), and RAB20 (13q34).
Comparison Between Adenomas with and without Synchronous Carcinoma
No significant differences regarding the two adenoma groups could be observed when assessing individual gene probes for SMAD7 deletions and EGFR amplifications of panel 1 separately. However, an increased genomic instability index (the genomic instability index was measured by dividing the number of different signal patterns by the number of analyzed cells) of CEP18, SMAD7, and EGFR together could be observed in the adenoma samples of patients with carcinoma as compared with adenoma samples of patients without synchronous or subsequent carcinoma (P=0.037; Supplementary Figure 1a). NCOA3 amplifications showed a trend of higher frequency in adenomas of patients with synchronous colorectal cancer (26.9%) as compared with adenomas of patients without malignancy (20.1%, P=0.144). Furthermore, TP53 deletion was more frequently observed in adenoma samples of patients with synchronous carcinoma (P=0.045; Supplementary Figure 1b). TP53 deletions were found in 14.8% in adenomas of patients without and in 23.2% in adenomas of patients with synchronous colorectal cancer. MYC amplifications were found in 21.4% in adenomas of patients without malignancy and in 31.3% in adenomas of patients with synchronous colorectal cancer (P=0.256). No differences could be observed regarding patient groups for RAB20 signal enumeration.
Evaluation of the Prognostic Potential of Gene Probes for Adenoma Recurrence
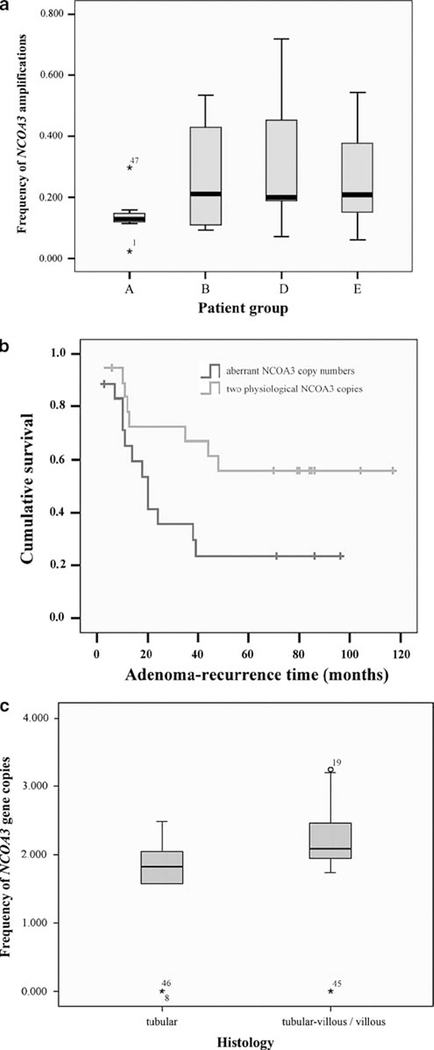
NCOA3 amplifications seemed to indicate patients with adenoma recurrence and/or synchronous carcinoma (P=0.058; Figure 2a). In turn, a diploid signal count for NCOA3 was associated with a longer adenoma recurrence-free observation time (P=0.042; Figure 2b). None of the other gene probes tested showed the prognostic potential for adenoma recurrence.
Figure 2.
(a) Frequency of NCOA3 amplifications according to patient groups (A, patients without adenoma recurrence and without carcinoma; (B) patients with adenoma recurrence but without carcinoma; (D) patients without adenoma recurrence but with carcinoma; (E) patients with adenoma recurrence and with carcinoma). (b) Adenoma recurrence-free survival time depending on NCOA3 copy numbers. (c) Frequency of NCOA3 copy numbers according to the histological subtypes.
Comparison of Adenomas and Synchronous Carcinomas
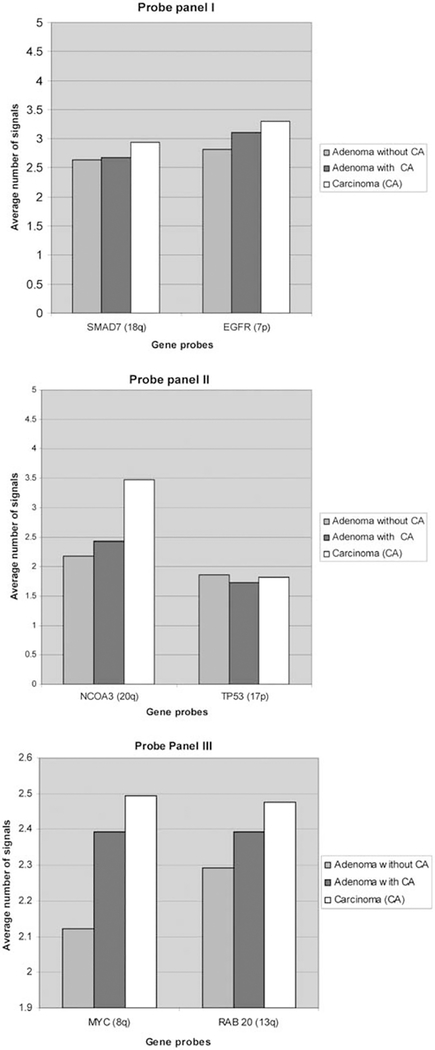
Deletions of SMAD7 did not show any differences between adenoma groups, could be detected in 10.5% of adenomas overall, and were not significantly more frequent in carcinomas (11.3%). In contrast to expected deletions of SMAD7, we observed SMAD7 gains. However, the average signal count of SMAD7 per patient group showed no differences between adenoma groups but was increased in carcinomas (Figure 3, panel I). These data point to an overall low frequency of SMAD7 aberrations during colorectal carcinogenesis and seem to indicate a rather late event during carcinogenesis. EGFR amplifications did not significantly vary between adenoma groups, were present in 36.2% of adenoma cells overall, and were even more frequently observed in carcinomas (44%). The presence of EGFR amplifications in more than one-third of adenomas shows its frequent involvement early during colorectal carcinogenesis. The manifestation of EGFR amplifications during disease progression goes in line with the subsequent increase of average signal counts from adenoma to carcinoma (Figure 3, panel I). NCOA3 amplifications were found in 20.1% in adenomas of patients without malignancy, in 26.9% in adenomas of patients with synchronous colorectal cancer, and in 33.4% of synchronous colorectal carcinomas (P = 0.12/P = 0.278). Although NCOA3 amplifications are already frequently observed in adenomas overall, they seem to be particularly involved in the progression from adenoma to carcinoma. This is in line with the analysis of average NCOA3 signal counts: on average, almost 3.5 NCOA3 gene copies can be detected in carcinomas. This is the highest average copy number alteration in carcinomas of all gene probes analyzed in this study (Figure 3, panel II). TP53 deletions increased from 14.8% in adenomas of patients without malignancy through 23.2% in adenomas of patients with synchronous carcinoma (P = 0.045); however, they were not significantly more frequent in the related carcinomas (20.2%, P = 0.057). TP53 deletions (Figure 3, panel II) seem to drive tumorigenesis mainly from adenoma to carcinoma progression. MYC amplifications were found in 21.4% in adenomas of patients without malignancy vs 31.3% in adenomas of patients with synchronous colorectal cancer (P = 0.256) and in 36.4% in colorectal carcinomas (P = 0.013). Despite their presence in 20% of adenomas, MYC amplifications seem to have a major role in the progression from adenoma to carcinoma. MYC had the second highest number of aberrant copy numbers in carcinomas of all gene probes tested (Figure 3, panel III).
Figure 3.
Average signal counts per gene probe, patient group, and hybridization panel without 2/2/2 signal pattern. For MYC signal counts, one patient with up to 20–30 signals per cell was not included in the visualization to reflect the overall average of MYC signal counts within groups.
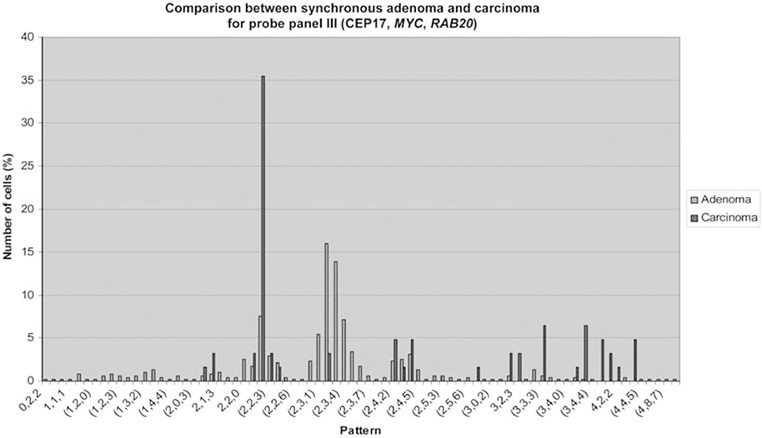
No significant difference could be observed regarding patient groups for RAB20 amplifications, which were found in 34.2% in adenomas overall and in 41.4% in carcinomas. This observation places RAB20 amplifications as a late event during colorectal tumorigenesis. Average signal counts showed a slight increase in RAB20 copy number signals during malignant transformation (Figure 3, panel III). Increasing signal counts for MYC and RAB20 from adenoma to carcinoma within the same patient are exemplarily shown in Figure 4, hereby indicating a certain degree of clonality of aberrant gene signals.
Figure 4.
Example of increasing copy numbers for MYC and RAB20 (panel III) from synchronous adenoma to carcinoma, indicating clonality of cells with aberrant copy number signals.
Correlation of Gene Copy Numbers with DNA Ploidy Status
Overall, EGFR amplifications correlated with SMAD7 deletions (P<0.01) and an increased DNA stem line value (P=0.019). NCOA3 amplifications were more frequently observed in aneuploid adenomas (P=0.04; Figure 1c). This finding is also supported by the fact that increasing NCOA3 gene copy number signals correlated with higher DNA stem line values (P=0.023). An equal copy number count of TP53 as compared with CEP17 reflected genomic stability and was more frequently observed in diploid adenomas (P=0.04; Supplementary Figure 1c). In contrast, deletion of TP53 was more frequently observed in aneuploid adenoma samples overall (P=0.029; Figure 1b). For visualization of probe panel I staining in diploid and aneuploid adenomas, please see Figures 1e and g. MYC amplifications were more frequently observed in adenoma samples with increased DNA stem line values (P<0.01) and in adenoma samples that were assessed to be aneuploid (P=0.029; Figure 1d). In addition, RAB20 amplifications were correlated with increased DNA stem line values (P<0.05). Panel III staining of diploid and aneuploid adenomas is exemplarily shown in Figures 1f and h.
In addition to the correlation of individual gene probes with DNA ploidy status, we also evaluated the genomic instability indices of all three probe panels with respect to ploidy measurements. Genomic instability indices were calculated by dividing the number of observed signal patterns by the number of analyzed cells. This index was calculated for each probe panel separately while excluding the signal pattern 2/2/2. Genomic instability indices for all three probe panels increased from diploid over tetraploid to aneuploid cases (Tables 2b and c). The most prominent increase could be observed comparing the diploid and/or tetraploid cases with the aneuploid cases. The comparison was particularly significant for panel II (CEP17, NCOA3, TP53; diploid vs aneuploid: P=0.0105; tetraploid vs aneuploid: P=0.0145) and panel III (CEP17, MYC, RAB20; diploid vs aneuploid: P=0.017).
Table 2b.
Genomic instability indices and immunohistochemistry
| No. | Sex | Age (years) | Group | Adenoma and carcinoma | Adenoma recurrence | DNA ploidy | Stem line | Genomic instability indexa |
Ln-5 | CyA | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Panel 1 | Panel 2 | Panel 3 | ||||||||||
| 1 | M | 57 | A | No | No | ND | ND | 0.214 | 0.366 | 0.14 | 0 | 1 |
| 2 | M | 70 | A | No | No | ND | ND | 0.232 | 0.23 | 0.247 | 0 | 1 |
| 3 | F | 70 | A | No | No | ND | ND | ND | ND | 0.238 | 0 | 1 |
| 4 | M | 69 | A | No | No | I | 1.83 | 0.158 | 0.195 | 0.277 | 2 | 3 |
| 5 | F | 82 | A | No | No | II | 2.03 | 0.154 | 0.26 | 0.327 | 1 | 2 |
| 6 | F | 78 | A | No | No | I | 2.03 | 0.17 | 0.257 | 0.22 | 1 | 2 |
| 7 | M | 79 | A | No | No | I | 1.91 | 0.226 | 0.229 | 0.25 | 3 | 2 |
| 8 | F | 67 | A | No | No | II | 1.99 | 0.223 | 0.202 | 0.25 | 2 | 1 |
| 9 | F | 65 | B | No | Month 38 | I | 1.86 | 0.165 | 0.292 | 0.192 | 0 | 1 |
| 10 | F | 62 | B | No | Month 35 | II | 1.94 | 0.146 | 0.204 | 0.175 | 1 | 1 |
| 11 | F | 59 | B | No | Month 39 | II | 2.11 | 0.271 | 0.167 | 0.367 | 1 | 1 |
| 12 | F | 59 | B | No | Month 3 | ND | ND | 0.245 | 0.354 | 0.226 | 1 | 2 |
| 13 | F | 64 | B | No | Month 48 | I | 1.96 | 0.291 | 0.247 | 0.231 | 2 | 2 |
| 14 | F | 76 | B | No | Month 3 | III | 3.13 | 0.331 | 0.343 | 0.391 | 2 | 2 |
| 15 | M | 82 | B | No | Month 18 | ND | ND | 0.267 | 0.191 | 0.175 | 1 | 2 |
| 16 | M | 66 | B | No | Month 14 | II | 1.77 | 0.25 | 0.21 | ND | 2 | 2 |
| 17 | M | 77 | B | No | Months 12 and 21 | II | 1.92 | 0.214 | 0.323 | 0.167 | 1 | 2 |
| 18 | M | 78 | B | No | Month 3 | I | 1.86 | 0.284 | 0.274 | 0.322 | 2 | 3 |
| 19 | F | 85 | D | pT4,N2,MX; G3 | No | ND | ND | 0.198 | 0.225 | 0.246 | 1 | 1 |
| 20 | M | 72 | D | pT4,N1,MX; G2 | No | ND | ND | 0.019 | 0.264 | 0.292 | 0 | 1 |
| 21 | F | 61 | D | pT4,N0,MX; G2 | No | I | 1.92 | 0.315 | 0.205 | 0.423 | 1 | 1 |
| 22 | M | 65 | D | pT3,N1,MX; G3 | No | ND | ND | 0.143 | 0.163 | 0.244 | 1 | 1 |
| 23 | F | 75 | D | pTis;G3 | No | III | 3.01 | 0.241 | 0.243 | 0.315 | 1 | 3 |
| 24 | M | 68 | D | pTis;G3 | No | I | 1.89 | 0.216 | 0.296 | 0.276 | 1 | 1 |
| 25 | F | 75 | D | pTis;G3 | No | II | 1.99 | 0.282 | 0.257 | 0.397 | 0 | 2 |
| 26 | M | 59 | D | pTis; G3 | No | III | 3.25 | 0.154 | 0.156 | 0.182 | 1 | 1 |
| 27 | F | 68 | D | pT3,N1,MX; G2 | No | I | 1.83 | 0.168 | 0.178 | ND | 2 | 2 |
| 28 | M | 78 | E | pTis; G3 | Month 36 | ND | ND | ND | ND | 0.208 | 0 | 1 |
| 29 | M | 56 | E | pT3,N2,MX; G2 | Month 10 | I | 1.89 | 0.165 | 0.25 | 0.219 | 0 | 1 |
| 30 | M | 78 | E | pT4,N1,MX; G2 | Month 24 | II | 1.79 | 0.261 | 0.288 | 0.226 | 0 | 1 |
| 31 | M | 67 | E | pT2,N1,MX; G2 | Month 44 | I | 1.82 | 0.14 | 0.116 | 0.188 | 1 | 1 |
| 32 | F | 70 | E | pT3,N1,MX; G2 | Months 11 and 24 | II | 1.85 | 0.268 | 0.216 | 0.311 | 1 | 1 |
| 33 | F | 73 | E | pT1;G2 | Month 10 | I | 2.01 | 0.177 | 0.325 | 0.16 | 2 | 1 |
| 34 | F | 77 | E | pT1;G2 | Month 13 | II | 1.87 | 0.189 | 0.219 | 0.208 | 1 | 2 |
| 35 | F | 44 | E | pTis;G3 | Month 11 | III | 3.47 | 0.333 | 0.344 | 0.292 | 1 | 1 |
| 36 | M | 76 | E | pTis;G3 | Month 20 | I | 1.92 | 0.393 | 0.2 | 0.194 | 1 | 2 |
| 37 | M | 76 | E | pTis;G3 | Month 20 | I | 1.94 | 0.207 | 0.216 | ND | 1 | 1 |
| 38 | M | 67 | E | pTis;G3 | Month 10 | III | 3.42 | 0.267 | 0.322 | 0.387 | 1 | 1 |
| 39 | F | 58 | E | pT1;G2 | Month 24 | III | 3.62 | 0.179 | 0.369 | 0.296 | 2 | 3 |
| 40 | F | 59 | E | pTis,N0,MX;G3 | Months 17 and 18 | ND | ND | 0.6 | ND | 0.75 | 2 | 1 |
| 41 | M | 75 | E | pT3,N0,MX; G2 | Month 7 | III | 3.11 | 0.25 | 0.393 | 0.468 | 1 | 2 |
Cy A, cyclin A; Ln-5, laminin-5 γ2 chain; f, female; m, male; ND, not determined.
The genomic instability index was calculated separately for each probe panel. Here, indices are reported without accounting for the physiologically normal hybridization pattern 2/2/2.
Table 2c.
Correlation between genomic instability indices and DNA ploidy type
| DNA ploidy | Panel I index, average | Panel II index, average | Panel III index, average |
|---|---|---|---|
| Diploid | 0.2196 | 0.2343 | 0.2460 |
| Tetraploid | 0.2258 | 0.2346 | 0.2698 |
| Aneuploid | 0.2507 | 0.3100 | 0.3330 |
For the increase in panel I indices, significance level was not reached.
For panel II indices, differences between diploid and aneuploid (P = 0.0105) and tetraploid and aneuploid (P = 0.0145) were significant.
For panel III indices, the difference between diploid and aneuploid (P = 0.017) was significant, whereas the difference between tetraploid and aneuploid missed significance for a small margin (P = 0.0865).
The correlation of ploidy type and genomic instability indices prompted us to evaluate whether the ploidy status would be dependent on any clinical features. The diameter size of adenomas was on average 29.3 mm of diploid adenomas, 10.5 mm of tetraploid, and 24 mm of aneuploid adenomas. Thus, the size of adenomas did not affect the degree of genomic instability in our data set. When comparing the histological subtype with the averaged instability indices of panels I, II, and III, increased genomic instability could be observed for the tubular/villous and villous subtypes (tubular: x=0.2164; tubular-villous; and villous: x=0.2662). Moreover, a high genomic instability index of probe panel III was significantly associated with the tubular-villous/villous subtype (P=0.015; Supplementary Figure 2a). A similar trend became apparent with high-grade adenomas presenting increased genomic instability.
Correlation of probes across panels
When comparing the signal enumeration of all tested probes across panels, the following correlation pattern became apparent at a significance level of P<0.05.
MYC, NCOA3, and RAB20 amplifications correlated with each other. In addition, NCOA3 amplifications also correlated with those of EGFR. In summary, a close correlation of increasing copy numbers could be identified for those genes that are located on chromosomal regions that are frequently gained in adenomas and colorectal carcinomas. The chromosome arm 18q is frequently deleted in colorectal carcinomas, but seldom in adenomas. We could observe increased gene copy numbers of SMAD7 in larger adenomas (P=0.017) and when NCOA3 amplifications were evident (P<0.01). In contrast, decreased copy numbers of SMAD7 were correlated with increased RAB20 copy numbers (P<0.05) and TP53 deletions (P<0.01). The latter finding was further supported by the fact that signal enumeration for SMAD7 and TP53 showed a close correlation overall (P<0.01).
Correlation of Signal Enumeration and Clinical Data
Size
Larger adenomas were associated with SMAD7 amplifications (P=0.017) and increased DNA stem line values (P=0.044), whereas RAB20 amplifications were more frequently observed in small adenomas (P<0.01).
Histology
The sample cohorts presented in this study comprised 10 tubular, 29 tubular-villous, and 2 villous adenomas. Thus, as villous adenomas were under-represented in our cohorts, villous and tubular-villous adenomas were combined into one group and compared with the tubular subtype. The tubular-villous/villous subtype was associated with NCOA3 amplifications (P=0.024; Figure 2c) and a high genomic instability index of probe panel III (CEP17, MYC, RAB20) (P=0.015; Supplementary Figure 2a).
Grade of dysplasia
Comparing the grade of dysplasia with the histological subtype revealed a strong correlation of high-grade dysplasia with tubular-villous and villous samples (P=0.004; Supplementary Figure 2b): ~61% of tubular-villous and villous adenomas showed high-grade dysplasia or a carcinoma in situ vs 20% of tubular adenomas. Carcinoma in situ was detected in 32.3% of the tubular-villous/villous subgroup as compared with 20% in the tubular entities.
Location
We further assessed whether any dependency of the grade of dysplasia regarding the location of the adenoma could be identified. Therefore, the colorectum was divided into three sections: (1) cecum, ascendent colon, right flexure, and transverse colon; (2) left flexure, descendent colon, sigmoid colon; and (3) rectum. This distinct classification did not show any significant correlation. However, when all colon segments were combined into one group and compared with the rectum, high-grade adenoma and carcinoma in situ were more frequently observed in lesions of the rectum (83.3%; P=0.026; Supplementary Figure 3a). This observation was in line with the finding that adenoma with aneuploid DNA distribution patterns were more frequently detected in the rectum than in the colon (P=0.045; Supplementary Figure 3b).
Proliferation and invasion
Adenoma samples were also assessed immunohistochemically for cyclin A and laminin-5 γ2-chain expression (Table 2b). Increased expression of both markers was observed predominantly in high-grade (P=0.001) and carcinoma in situ adenomas (P=0.002). In addition, low-to-moderate laminin-5 expression (1–50% of cells show immunopositivity) was more commonly detectable in tubulo-villous/villous adenomas (87.1%) as compared with the tubular type (40%) (P=0.01; Supplementary Figure 2c). Strong laminin-5 immunoreactivity (>50% and up to 100% of cells showing expression) was observed only in the tubulo-villous/villous subtype. No difference could be detected between patient groups with and without synchronous colorectal cancer regarding cyclin A and laminin-5 immunoreactivity.
Discussion
Colorectal tumorigenesis is defined by the sequential acquisition of chromosomal aneuploidies. In an attempt to define such patterns and to investigate the dynamics of genome aberrations, we selected FISH probes specific for genes of known and supposedly to be importance in colorectal carcinogenesis (such as EGFR, MYC, TP53, and SMAD7, NCOA3, RAB20), which reside on recurrent aneuploid chromosomes. Copy number alterations of these six genes were then analyzed in interphase nuclei by triple-color FISH using three probe panels, each comprising two of the genes plus one centromere probe for chromosome 17 or 18. These probe cocktails were hybridized to cytospin preparations of adenomas of 41 patients. In 23 cases, synchronous carcinoma was present. A total of 18 patients showed no sign of malignant disease, yet 10 of them presented with recurring adenomas. We also analyzed six matched pairs of adenomas and carcinomas.
Increased size, villous histology, and high-grade dysplasia of the adenoma samples investigated in this study correlated with increased DNA stem line values, an increased genomic instability index of probe panel III, and with amplifications of EGFR, MYC, NCOA3, and RAB20. However, histopathological features did not show any differences or predictive value for patients with increased adenoma recurrence and/or carcinoma risk.
We were then eager to test whether increased proliferation as assessed by cyclin A and/or enhanced malignancy potential by laminin-5 γ2 chain overexpression would indicate individual progression risk. Increased immunoreactivity of cyclin A and laminin-5 was predominantly observed in high-grade dysplastic and carcinoma in situ adenomas. Laminin-5 expression further correlated with villous subtypes. The cyclin A protein is complexed to the kinase subunit Cdk2 during the S phase of the cell cycle.18 It can be used as a proliferation marker for committed cells that will pass through the S and G2 phases.19 Moreover, cyclin A overexpression has been associated with a poor prognosis of colorectal carcinoma patients.20 The laminin-5 isoform has an important role in epithelial cell adhesion to the basement membrane.21 Increased expression has been found not only at the invasion front of colorectal cancers but also in premalignant lesions, such as ulcerative colitis mucosa.14,22 In our data, increased cyclin A and laminin-5 immunoreactivity indicate adenoma cells with increased proliferative and malignancy potential, respectively. However, no differences could be observed for cyclin A or laminin-5 immunoreactivity regarding different patient groups.
We then evaluated the copy numbers of the six genes within three probe panels with respect to the different patient groups. Although individual assessment of SMAD7 and EGFR aberrations did not show any distinction between adenomas with and without synchronous carcinoma, their genomic instability index did. This index was calculated by dividing the number of observed signal patterns (CEP18, SMAD7, EGFR) by the number of analyzed cells. This index was significantly increased in adenoma samples of patients with synchronous colorectal carcinoma. Activation of EGFR, a transmembrane protein with intrinsic tyrosine-kinase activity, leads to proliferation and cell integrity. However, overexpression causes apoptotic resistance and tumor propagation, especially in colorectal carcinogenesis.23,24 Therefore, our finding of EGFR amplifications seems to identify cells that drive tumor formation and propagation. Interestingly, such amplifications were accompanied by deletions of SMAD7.
Adenoma patients with synchronous carcinoma were also indicated by the increased genomic instability index of CEP17, NCOA3, and TP53. Aberrations of NCOA3 and TP53 were also correlated with increased DNA stem line values and overall aneuploid DNA patterns. Deletions of the tumor-suppressor TP53 are known to appear late during colorectal carcinogenesis. They can be detected at early stages in 15% and up to 60% in advanced stage carcinomas.25 A substantial amount of adenoma cells did harbor TP53 deletions in the specimens analyzed in this study, indicating a yet underestimated role of TP53 deletions early during carcinogenesis. NCOA3 has been detected as being amplified in breast cancer (synonym AIB1).26 It has also been described to be amplified in endometrial, stomach, prostate, pancreatic, and hepatocellular carcinomas.27–29 NCOA3 is as transcription coactivator involved in cell proliferation, migration, differentiation, and growth.30 Henke et al31 could show that NCOA3 amplification seems to be an early event during pancreatic cancer development. In colorectal carcinomas, NCOA3 amplifications were detected by different groups in 10–32%.32,33 Interestingly, we could detect NCOA3 amplifications in 20.1% of adenoma cells of patients without malignancy, in 26.9% of adenoma cells of patients with malignancy and in 33.4% of carcinoma cells. NCOA3 amplifications have also been described to correlate with the occurrence of lymph-node and liver metastases.34 Five of our six analyzed carcinoma samples showed lymph-node metastasis. In addition, aberrant NCOA3 copy numbers as compared with CEP17 in the adenoma samples were significantly associated with a shorter time period until adenoma recurrence was diagnosed. Thus, NCOA3 amplifications seem to have a high impact on adenoma development and progression of premalignant stages to malignancy. The importance of NCOA3 amplifications for colorectal carcinogenesis is further corroborated by the fact that NCOA3 amplifications overall correlated with amplifications of EGFR, MYC, and RAB20.
Although no differences between adenomas with and without synchronous carcinoma could be observed for RAB20, MYC amplifications were present more frequently in adenomas of patients with synchronous carcinoma. In addition, MYC amplifications were more frequently observed in aneuploid cells. Amplifications of the oncogene MYC are well known for their impact on colorectal carcinogenesis and are frequently detected in advanced stage carcinomas.35 Our data suggest that MYC amplifications already do seem to be of importance in premalignant lesions and characterize cell clones of increased malignancy potential. RAB proteins belong to the superfamily of RAS-GTPases.36 Their main function is the control of endocytotic vesicle transport. RAB20 is mainly localized at the apical membrane of intestinal cells and is involved in endocytosis and recycling.37,38 RAB20 has not been associated with colorectal carcinogenesis so far. However, we revealed RAB20 amplifications in ~34.2% of adenoma cells, which is comparable with the most frequent gene copy number alteration observed in our data set (gain of EGFR in 36.2% of adenoma cells). The high rate of RAB20 amplifications could cause an increased recycling process for growth factor receptors that are important for proliferation and disintegration during tumor progression.38 This would suggest RAB20 amplification to be an early event during colorectal carcinogenesis.
The results of our probe panel assessments showed that across samples and patient groups, gene-specific copy number alterations could be observed in a substantial number of cells. Interestingly, the most frequently observed alterations were a gain of EGFR (36.2%) and RAB20 (34.2%). Although the frequency of these two aberrations was increased in the carcinoma samples (EGFR, 44%/RAB20, 41.4%), there was no difference between patients with and without malignancy. Therefore, we conclude that amplifications of EGFR and RAB20 could trigger adenoma development. Individual risk assessment for adenoma recurrence and the presence of synchronous carcinoma could be improved through the evaluation of the probe panel I genomic instability index (CEP 18, SMAD7, EGFR) and the frequency of NCOA3 amplifications, MYC amplifications, and TP53 deletions.
In summary, genomic instability in colorectal adenomas is reflected by genomic amplification of the oncogenes EGFR, MYC, NCOA3, and RAB20 that correlate with histomorphological features. These amplifications are indicative for adenoma recurrence and for the presence of synchronous carcinomas. Detection of such amplifications using FISH can contribute to the assessment of individual progression risk.
Supplementary Material
Acknowledgements
We express our gratitude to Timo Gemoll, Buddy Chen and Joseph Cheng for IT support, to Ulla Aspenblad and Inga Maurin for excellent assistance with immunohistochemistry, to Marco Gerling and Gisela Grosser-Pape for excellent assistance with image cytometry, and to Elke Gheribi and Regina Kaatz for clinical sample and data collection. This project was supported by the Intramural Research Program of the NIH, National Cancer Institute and by the Werner und Klara Kreitz-Foundation, Schleswig-Holstein, Germany. CAB received a stipend by the DAAD (Deutscher Akademischer Austauschdienst) and a travel grant by the Boehringer Ingelheim Fond (BIF), Germany. This study was performed in collaboration with the North German Tumorbank Colorectal Cancer (DKH no. 108446) and the Surgical Center for Translational Oncology—Lübeck (SCTO-L).
Footnotes
Disclosure/conflict of interest
The authors declare no conflict of interest.
References
- 1.Weitz J, Koch M, Debus J, et al. Colorectal cancer. Lancet 2005;365:153–165. [DOI] [PubMed] [Google Scholar]
- 2.Malila N, Hakulinen T. Epidemiological trends of colorectal cancer in the Nordic countries. Scand J Surg 2003;92:5–9. [DOI] [PubMed] [Google Scholar]
- 3.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–767. [DOI] [PubMed] [Google Scholar]
- 4.Burgart LJ. Colorectal polyps and other precursor lesions. Need for an expanded view. Gastroenterol Clin North Am 2002;31:959–970. [DOI] [PubMed] [Google Scholar]
- 5.Araujo SE, Bernardo WM, Habr-Gama A, et al. DNA ploidy status and prognosis in colorectal cancer: a meta-analysis of published data. Dis Colon Rectum 2007;50:1800–1810. [DOI] [PubMed] [Google Scholar]
- 6.Habermann JK, Paulsen U, Roblick UJ, et al. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosomes Cancer 2007;46:10–26. [DOI] [PubMed] [Google Scholar]
- 7.Habermann JK, Upender MB, Roblick UJ, et al. Pronounced chromosomal instability and multiple gene amplifications characterize ulcerative colitis-associated colorectal carcinomas. Cancer Genet Cytogenet 2003;147:9–17. [DOI] [PubMed] [Google Scholar]
- 8.Ried T, Knutzen R, Steinbeck R, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer 1996;15:234–245. [DOI] [PubMed] [Google Scholar]
- 9.Geigl JB, Obenauf AC, Schwarzbraun T, et al. Defining ‘chromosomal instability’. Trends Genet 2008;24: 64–69. [DOI] [PubMed] [Google Scholar]
- 10.Bomme L, Bardi G, Pandis N, et al. Clonal karyotypic abnormalities in colorectal adenomas: clues to the early genetic events in the adenoma-carcinoma sequence. Genes Chromosomes Cancer 1994;10:190–196. [DOI] [PubMed] [Google Scholar]
- 11.Upender MB, Habermann JK, McShane LM, et al. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res 2004;64:6941–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riddell RH, Goldman H, Ransohoff DF, et al. Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Hum Pathol 1983;14:931–968. [DOI] [PubMed] [Google Scholar]
- 13.Salo S, Haakana H, Kontusaari S, et al. Laminin-5 promotes adhesion and migration of epithelial cells: identification of a migration-related element in the gamma2 chain gene (LAMC2) with activity in transgenic mice. Matrix Biol 1999;18:197–210. [DOI] [PubMed] [Google Scholar]
- 14.Habermann J, Lenander C, Roblick UJ, et al. Ulcerative colitis and colorectal carcinoma: DNA-profile, laminin-5 gamma2 chain and cyclin A expression as early markers for risk assessment. Scand J Gastroenterol 2001;36:751–758. [DOI] [PubMed] [Google Scholar]
- 15.Castro J, Heiden T, Wang N, et al. Preparation of cell nuclei from fresh tissues for high-quality DNA flow cytometry. Cytometry 1993;14:793–804. [DOI] [PubMed] [Google Scholar]
- 16.Heiden T, Wang N, Tribukait B. An improved Hedley method for preparation of paraffin-embedded tissues for flow cytometric analysis of ploidy and S-phase. Cytometry 1991;12:614–621. [DOI] [PubMed] [Google Scholar]
- 17.Auer GU, Caspersson TO, Wallgren AS. DNA content and survival in mammary carcinoma. Anal Quant Cytol 1980;2:161–165. [PubMed] [Google Scholar]
- 18.Sherr CJ. Mammalian G1 cyclins. Cell 1993;73: 1059–1065. [DOI] [PubMed] [Google Scholar]
- 19.Zindy F, Lamas E, Chenivesse X, et al. Cyclin A is required in S phase in normal epithelial cells. Biochem Biophys Res Commun 1992;182:1144–1154. [DOI] [PubMed] [Google Scholar]
- 20.Handa K, Yamakawa M, Takeda H, et al. Expression of cell cycle markers in colorectal carcinoma: superiority of cyclin A as an indicator of poor prognosis. Int J Cancer 1999;84:225–233. [DOI] [PubMed] [Google Scholar]
- 21.Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin alpha 3 beta 1 in epithelial basement membranes. Cell 1991;65:599–610. [DOI] [PubMed] [Google Scholar]
- 22.Sordat I, Bosman FT, Dorta G, et al. Differential expression of laminin-5 subunits and integrin receptors in human colorectal neoplasia. J Pathol 1998;185:44–52. [DOI] [PubMed] [Google Scholar]
- 23.Roberts RB, Min L, Washington MK, et al. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc Natl Acad Sci USA 2002;99:1521–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grunwald V, Hidalgo M. Development of the epidermal growth factor receptor inhibitor OSI-774. Semin Oncol 2003;30:23–31. [DOI] [PubMed] [Google Scholar]
- 25.Risio M, Casorzo L, Chiecchio L, et al. Deletions of 17p are associated with transition from early to advanced colorectal cancer. Cancer Genet Cytogenet 2003;147: 44–49. [DOI] [PubMed] [Google Scholar]
- 26.Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 1997;277:965–968. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Wu MC, Sham JS, et al. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-through-put tissue microarray. Cancer 2002;95:2346–2352. [DOI] [PubMed] [Google Scholar]
- 28.Zhou G, Hashimoto Y, Kwak I, et al. Role of the steroid receptor coactivator SRC-3 in cell growth. Mol Cell Biol 2003;23:7742–7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghadimi BM, Schrock E, Walker RL, et al. Specific chromosomal aberrations and amplification of the AIB1 nuclear receptor coactivator gene in pancreatic carcinomas. Am J Pathol 1999;154:525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao L, Kuang SQ, Yuan Y, et al. Molecular structure and biological function of the cancer-amplified nuclear receptor coactivator SRC-3/AIB1. J Steroid Biochem Mol Biol 2002;83:3–14. [DOI] [PubMed] [Google Scholar]
- 31.Henke RT, Haddad BR, Kim SE, et al. Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma. Clin Cancer Res 2004;10:6134–6142. [DOI] [PubMed] [Google Scholar]
- 32.Xie D, Sham JS, Zeng WF, et al. Correlation of AIB1 overexpression with advanced clinical stage of human colorectal carcinoma. Hum Pathol 2005;36:777–783. [DOI] [PubMed] [Google Scholar]
- 33.Zeng SD, Xie D, Lin F, et al. [Expression and amplification of steroid receptor coactivator-3 gene in colorectal carcinoma and its clinicopathological significance]. Zhonghua Wei Chang Wai Ke Za Zhi 2005;8:67–70. [PubMed] [Google Scholar]
- 34.Sakakura C, Hagiwara A, Yasuoka R, et al. Amplification and over-expression of the AIB1 nuclear receptor co-activator gene in primary gastric cancers. Int J Cancer 2000;89:217–223. [PubMed] [Google Scholar]
- 35.Platzer P, Upender MB, Wilson K, et al. Silence of chromosomal amplifications in colon cancer. Cancer Res 2002;62:1134–1138. [PubMed] [Google Scholar]
- 36.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev 2001;81:153–208. [DOI] [PubMed] [Google Scholar]
- 37.Lutcke A, Parton RG, Murphy C, et al. Cloning and subcellular localization of novel rab proteins reveals polarized and cell type-specific expression. J Cell Sci 1994;107:3437–3448. [DOI] [PubMed] [Google Scholar]
- 38.Stein MP, Dong J, Wandinger-Ness A. Rab proteins and endocytic trafficking: potential targets for therapeutic intervention. Adv Drug Deliv Rev 2003;55:1421–1437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.