Abstract
We developed an X-ray microscope using partially coherent object illumination instead of previously used quasi-incoherent illumination. The design permitted the incorporation of a cryogenic tilt stage, enabling tomography of frozen-hydrated, intact adherent cells. We obtained three-dimensional reconstructions of mouse adenocarcinoma cells at ~36-nm (Rayleigh) and ~70-nm (Fourier ring correlation) resolution, which allowed us to visualize the double nuclear membrane, nuclear pores, nuclear membrane channels, mitochondrial cristae and lysosomal inclusions.
Soft X-ray microscopy is a promising approach to obtain nanoscale three-dimensional (3D) views of intact cells using only the natural contrast afforded by the different absorption of organic matter and water1. This permits entire cells to be examined in their native state without chemical fixation, chemical staining or physical sectioning but only with cryopreservation. Cellular soft X-ray tomography is beginning to be used for quantitative measurements of organelles that are visible by light microscopy2,3, though prior experiments have not yet revealed many of the ultrastructural details that are visible by electron microscopy. Prior analyses have also been limited to cells that could be drawn into a capillary4, making it difficult to examine adherent mammalian cells.
An alternative for visualizing 3D mammalian cell ultrastructure is cryo-electron tomography performed on multiple serial sections, each with a thickness of a micrometer or less, which are then aligned to produce 3D views. This process requires 2–3 weeks per cell5. Much faster nanoscale 3D imaging of intact cells can be performed with fluorescence super-resolution microscopy6. This approach, however, is fundamentally limited to examining the distribution of a few molecular markers per cell and therefore cannot be used to resolve a full spectrum of ultrastructural features. Thus, X-ray microscopy with its potential to reveal 3D ultrastructure in intact cells with a thickness of 5–10 μm could fill the gap in current technologies.
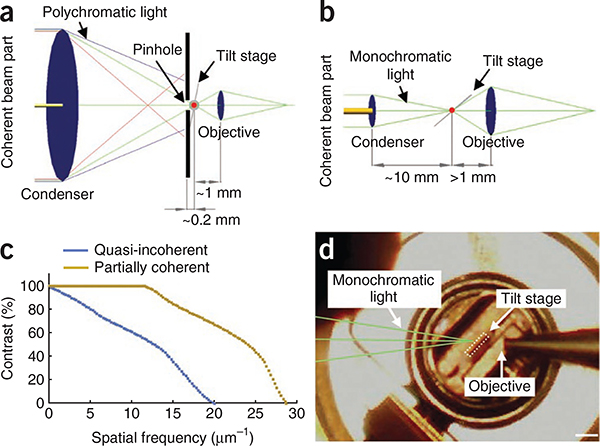
We built a transmission soft X-ray microscope that takes advantage of improved nanofabrication techniques, allowing manufacture of higher-resolution X-ray objectives with smaller outermost zone widths. We combined this higher-resolution objective with partially coherent specimen illumination instead of the lower-resolution objective and quasi-incoherent illumination used in previous designs2–4,7,8 (Fig. 1a,b). Whereas partial coherence lowers the maximal possible resolution compared to incoherence, it provides much larger contrast at medium to higher spatial frequencies9. Thus, combining partial coherence with a better-resolution objective should yield better contrast for fine-scale features. Using Fourier optical theory, we calculated that our microscope design should produce a dramatic increase in contrast for finer-scale structures in the specimen (Fig. 1c).
Figure 1 |.
X-ray microscope designs. (a) Schematic of the design for quasi-incoherent imaging, in which the X-ray source produces a divergent X-ray beam containing small coherently illuminated areas (dim yellow), which are a tiny fraction of the total beam and condenser diameter (~10 mm). The numerical apertures (NA) of the condenser and the objective (both of which are zone plates) were matched (NAcondenser / NAobjective − 1), and a 50-nm zone plate objective is used. (b) Schematic of the design for partially coherent imaging, in which the X-ray source produces a more collimated photon beam containing ~100× more coherent light (bright yellow) with a coherent beam diameter that is a substantial fraction of the condenser diameter (2 mm). The numerical aperture of the objective is more than twice that of the condenser (NAcondenser / NAobjective − 0.43). A 25-nm zone plate objective is used. (c) Contrast transfer functions are plotted for partially coherent and quasi-incoherent microscope designs. The curves were calculated theoretically using the optical parameters of the two designs. (d) Photograph of the tilted flat sample holder (tilt stage) inside the microscope chamber. Scale bar, 1 mm.
An added advantage of our X-ray microscope design is that there is now sufficient space in the object plane to insert a flat sample holder that can be tilted for tomography (Fig. 1b,d). In earlier designs, the sample space had been so small (Fig. 1a) that rotation of the sample could only be achieved with a ~10-μm diameter capillary4,7,8. These cramped conditions arose because a pinhole had to be placed directly in front of the specimen to obtain monochromatic illumination. In our design, we eliminated the need for a pinhole by using a monochromator in front of an elliptically shaped glass capillary condenser to illuminate the specimen with monochromatic light. There is now sufficient space for a flat sample holder to be tilted up to ± 79° (Fig. 1b,d).
We used this partially coherent X-ray microscope to image intact, ~5-μm-thick mouse adenocarcinoma cells10. We grew the cells on cellulose-nitrate–coated gold grids and directly examined them on the X-ray microscope after vitrification by plunge-freezing in liquid ethane. Cells grown on the grids had normal morphology and physiology (Supplementary Figs. 1 and 2).
We acquired X-ray microscope images of these vitrified mammalian cells at tilt angles from −60° to +60° in increments of 1° at a pixel size of either 9.8 nm (25-nm zone plate objective) or 15.6 nm (40-nm zone plate objective). Exposure times for each tilt angle were 2−24 s. The total X-ray exposure (~109 Gy) produced negligible radiation damage, as we detected no difference in image quality between images acquired at the beginning and end of the tilt series (Supplementary Fig. 3). We processed the images using a reciprocal space algorithm11 to generate a 3D tomogram composed of cubic voxels whose side lengths were either 9.8 nm (25-nm zone plate objective) or 15.6 nm (40-nm zone plate objective). Some reconstructions (Supplementary Figs. 4 and 5) were binned by a factor of two for faster processing. We obtained X-ray tomograms of 14 cells, six of which are shown (Fig. 2 and Supplementary Figs. 4 and 5). The principal limitation was poor cryopreservation (Supplementary Fig. 5a).
Figure 2 |.
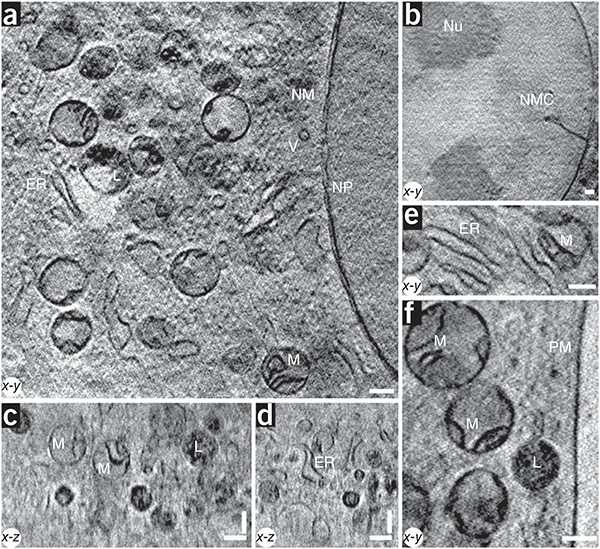
X-ray images of a cell. (a–f) The 3D partially coherent X-ray tomograms of mouse adenocarcinoma cells show many subcellular organelles including mitochondria (M), lyosomes (L), endoplasmic reticulum (ER), vesicles (V), the plasma membrane (PM), the nuclear membrane (NM), nuclear pores (NP), nucleoli (Nu) and nuclear membrane channels (NMC). All images were acquired with a 25-nm zone plate at 510 eV photon energy, except for the image shown in b, which was acquired with a 40-nm zone plate. Pixel sizes and slice thicknesses are 9.8 nm (a,c–f) and 15.6 nm (b). Scale bars, 0.39 μm.
The 3D tomograms of these interphase mouse cells revealed many recognizable subcellular features that had been either undetected or only faintly detected in earlier X-ray tomography studies. We achieved excellent contrast for all membrane-bound structures. We readily identified the plasma membrane, nuclear membrane, nuclear pores, nucleoli, endoplasmic reticulum, vesicles, lysosomes and mitochondria. We could also resolve internal organellar structures, such as mitochondrial cristae, the double nuclear membrane and lysosomal inclusions.
The partially coherent X-ray images exhibited many of the features visible by transmission electron microscopy (TEM) of thin sections of the same cell type (Supplementary Fig. 6). By either modality, mitochondria, lysosomes, endoplasmic reticulum and vesicles predominated in the cytoplasm, whereas the nuclei contained prominent nucleoli but lacked heterochromatin, even at the nuclear periphery. The cells contained very few Golgi structures. The X-ray images frequently revealed nuclear channels that partially traversed the nuclear interior (Figs. 2b and 3a and Supplementary Fig. 4a,b), even though we did not detect comparable channels by TEM (Supplementary Fig. 6a). Similar nuclear channels are found in interphase nuclei of many cell types12, but they are rarely detected by TEM presumably because the channels usually intersect the section plane at glancing angles.
Figure 3 |.
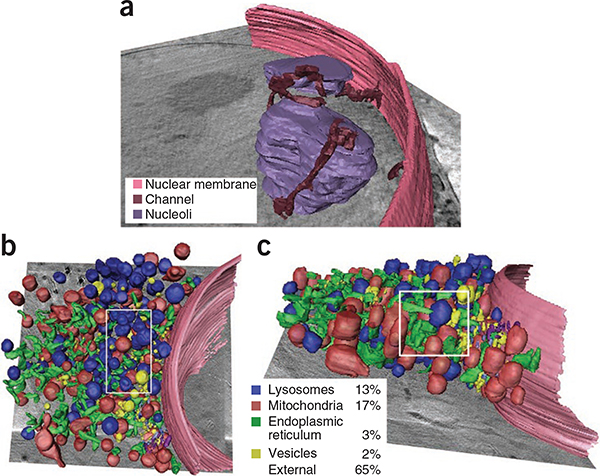
Volumetric rendering of cell cytoplasm. (a) The 3D data corresponding to the image in Figure 2b was segmented to visualize the association of the nuclear membrane channels with the nuclear membrane. (b,c) The 3D data corresponding to the image in Figure 2a were segmented, yielding x-y (b) and x-z (c) views of the cytoplasm. Percentages indicate the volume fraction occupied by different organelles measured in the 3D subvolume delineated by the white rectangles.
We did not detect some structures by X-ray tomography that we detected by TEM, such as ribosomes and the double membrane of the mitochondrial cristae. These probably fall below the current resolution limit (see below). An additional limitation was the restricted tilt angle range (±60°) used in these experiments. This led to poorer resolution in the z dimension, as indicated by a distortion in the 3D shape of some organelles, which appeared more cylindrical in x-z views (Fig. 3b) as well as an inability to obtain face-on views of the nuclear pores (data not shown) or follow the complete circumference of the nuclear membrane (Supplementary Fig. 5b).
The resolution in most previous X-ray tomographic studies of cells is based only on the zone plate objective used1–3, but this may not be a measure of the resolution achieved in the image. The Rayleigh criterion corresponds to the smallest resolvable lateral separation of two high-contrast objects in the image, enabling a useful comparison to resolution reported for super-resolution fluorescence microscopy. Using the 25-nm zone plate objective, we measured a peak-to-peak distance of 36 nm for the nuclear membrane (Supplementary Fig. 7a). This resolution estimate is the first direct demonstration, to our knowledge, that cellular X-ray tomography can substantially surpass conventional light microscope resolution.
Fourier ring correlation (FRC) is a complementary measure of resolution, suitable for comparison to measurements from cryo-electron tomography. In this method, the resolution of each image from the tilt series is estimated by comparing it to a reprojection from a tomogram constructed without that particular image11. For cellular images, the FRC criterion is typically lower than the Rayleigh criterion because it accounts for all structures in the image including lower-contrast structures. At higher tilt angles, the FRC criterion decreases because of increasing thickness of the imaged region and inclusion of structures beyond the tomogram’s reconstruction volume. With a cutoff value commonly used in cryo-electron tomography of 0.5 on the FRC curve, our tomograms had a best resolution of 70 nm (Supplementary Fig. 7b), equivalent to 35 nm in ‘half-pitch resolution’, a measure reported in some X-ray microscopy studies. Together these Rayleigh and FRC measures will enable unbiased comparisons of resolution achieved in future X-ray tomographic images of cells.
The high 3D resolution achieved here requires a synchrotron radiation source, but ‘bench-top’ X-ray microscopes using plasma sources may eventually achieve comparable resolutions13. Other future improvements potentially will include improved X-ray optics that could increase resolution to better than 10 nm. However, better objectives will have smaller depths of focus, necessitating 3D image reconstruction by optical sectioning rather than tomography. Even the present setup with a 25-nm zone plate objective might benefit from alternate image reconstruction procedures. We estimated a depth of focus of 2–3 μm in these 5-μm-thick cells, reflecting an intermediate regime between pure tomography and pure optical sectioning.
A key feature of the X-ray microscope is its ability to generate 3D views of intact cells at nanoscale resolution. The x-z slices through our tomograms reveal cellular organelles in all three dimensions (Fig. 2c,d), thus enabling a comprehensive overview of 3D cellular ultrastructure. To visualize this, we performed a 3D segmentation of all detectable cytoplasmic organelles from one of our 3D views (Fig. 3) and found that organelles accounted for ~35% of the total cytoplasmic volume in that region. This estimated packing density was lower than the maximal value for random close-packing of spheres, which is 63%, but it still represents a crowded environment through which transport processes in the cytoplasm must operate.
The practical workflow we developed for X-ray imaging of adherent cells (Supplementary Protocol) will facilitate broad application of this technology to any specimen that can be grown or placed on a flat sample holder. Future applications include the pairing of partially coherent X-ray microscopy with superresolution fluorescence microscopy6, thereby combining the molecular precision of fluorescence with the ultrastructural landmarks of X-ray tomography.
Cell growth and freezing
The cells were grown on custom-made gold grids coated with a thin layer (0.1 μm) of cellulose nitrate. The grids were placed on the bottom of a Petri dish containing the cells’ usual growth medium. About 1 h before freezing, 2 μl of a solution of 270 nm fiducial markers (silica core with gold shell produced by C. Graf, Free University Berlin) was added to the media in the Petri dish, and the beads were allowed to settle onto the grids.
In preparation for freezing, the grids containing the cells were removed from the Petri dish, briefly rinsed in growth medium and then blotted with Whatman #1 paper to leave a thin layer of fluid (~10–30 μm thick). Immediately after blotting, the grids were rapidly plunge-frozen in liquid ethane at a temperature of around −170 °C. The vitrified cells were either stored in liquid nitrogen or loaded onto the Gatan model 630 holder for examination on the X-ray microscope.
Light microscopy
Cells grown on either cover slips or grids were examined in a Leica DMI6000B microscope in either brightfield or fluorescence mode. Images were acquired with either a 63×, 1.4 numerical apture (NA) objective or a 100×, 1.4 NA objective and recorded on a Leica DFC350FXR2 charge-coupled device (CCD) camera. Before imaging, cells on the grid were overlaid with a coverslip.
Transmission electron microscopy
The cells grown in culture were fixed, processed and embedded by an in situ method. Specifically, cells were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 1 h at room temperature (24 °C), and then overnight at 4 °C. Then the cells were washed three times in buffer and stained for 1 h in 1% osmium tetroxide, followed by a 1-h stain in 0.5% uranyl acetate in 0.1 M acetate buffer. The cells were dehydrated through an ethanol series and finally embedded in epoxy resin. Thin sections (90 nm) were cut and stained with 0.5% uranyl acetate and lead citrate for 2 min. The sections were examined on a Hitachi H7600 electron microscope operating at 80 kV.
X-ray microscopy
Grids with adherent cells were loaded into a Gatan holder under liquid nitrogen and then inserted into the FEI CompuStage in the X-ray microscope. Cells were imaged on the X-ray microscope at −170 °C. The contrast of all images shown in the figures was adjusted by setting the image’s minimum value to zero and its maximum value to 255 (arbitrary units) and then displaying the image with a linear gray scale contrast ramp between these two endpoints using the ImageJ software.
Tomographic reconstruction
Images from the tilt series were processed using the package Bsoft11. An average flat-field image was calculated from ten flat-field images acquired under the same conditions as the tilt series and then divided into each image in the tilt series. To correct for the different exposure times used at higher tilt angles, all images were normalized to an average intensity of zero and an s.d. of one. Camera edge artifacts were eliminated by Gaussian smoothing at the image boundaries. Images from the corrected tilt series were then aligned using the 270-nm beads as fiducial markers. Finally, the tomographic reconstruction was performed using a reciprocal space algorithm14.
Segmentation
The 3D X-ray reconstructions were hand segmented using Amira software (Visage Imaging). The surface rendering software available with this package was used to generate 3D views of the cytoplasm. The same software was used to calculate the volume of every segmented organelle. From these data we calculated average organelle volume as well as the total volume occupied by organelles in a cytoplasmic subvolume.
Calculation of contrast transfer function
For the bending magnet microscope setup (Fig. 1a), the condenser zone plate had 50-nm outermost zone width with zones as large as 121.5 nm and in the center, a beam stop. For the zone plate objective, we assumed 50-nm outermost zone width. In our undulator microscope design (Fig. 1b), the reflective elliptically shaped glass capillary condenser had an equivalent aperture of a zone plate condenser with 58.2-nm outermost and inner zones of 81.7 nm. The X-ray objective had outermost zones of 25 nm corresponding to the experiment described above. Owing to the small NA of the X-ray optics, the contrast transfer functions were calculated numerically with the preceding parameters using a previously developed Fourier optical model15, which assumes a multiple plane wave illumination of the object. Originally, this optical model was based on a simulated object of variable period and thickness and therefore variable contrast at different spatial frequencies. For the calculations reported in this paper we assumed a periodic object with 100% contrast at all spatial frequencies.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Niemann for support and A. Leis for help with learning cryopreservation. This work was funded in part by the Human Frontier Science Program Research grant RGP0053/2005-C, the German Federal Ministry of Education and Research under contract 05KS4BY1/7, the intramural program of the US National Institutes of Health, including the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and the National Cancer Institute, Center for Cancer Research, including Science Applications International Corporations Frederick contract HHSN26120080001E.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Supplementary information is available on the Nature Methods website.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/
References
- 1.McDermott G, Le Gros MA, Knoechel CG, Uchida M & Larabell CA Trends Cell Biol. 19, 587–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larabell CA & Le Gros MA Mol. Biol. Cell 15, 957–962 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchida M et al. Proc. Natl. Acad. Sci. USA 106, 19375–19380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider G et al. Surf. Rev. Lett. 9, 177–183 (2002). [Google Scholar]
- 5.Noske AB, Costin AJ, Morgan GP & Marsh BJ J. Struct. Biol. 161, 298–313 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schermelleh L, Heintzmann R & Leonhardt HJ Cell Biol. 190, 165–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss D et al. Ultramicroscopy 84, 185–197 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Le Gros MA, McDermott G & Larabell CA Curr. Opin. Struct. Biol. 15, 593–600 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Attwood D Soft X-Rays and Extreme Ultraviolet Radiation: Principles and Applications (Cambridge University Press, 1999). [Google Scholar]
- 10.McNally JG, Müller WG, Walker D, Wolford R & Hager GL Science 287, 1262–1265 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Heymann JB, Cardone G, Winkler DC & Steven AC J. Struct. Biol. 161, 232–242 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fricker M, Hollinshead M, White N & Vaux DJ Cell Biol. 136, 531–544 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertilson M, von Hofsten O, Vogt U, Holmberg A & Hertz HM Opt. Express 17, 11057–11065 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Heymann JB & Belnap DM J. Struct. Biol. 157, 3–18 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Schneider G Ultramicroscopy 75, 85–104 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.