Abstract
Despite their denaturing properties, detergents are used to purify and study membrane proteins. Herein, we demonstrated a polymer-based detergent-free extraction of the membrane protein cytochrome-b5 along with E. coli lipids. Nuclear magnetic resonance experiments revealed the suitability of using nanodiscs for high-resolution studies and revealed the type of native lipids associated with the protein.
Graphical Abstract
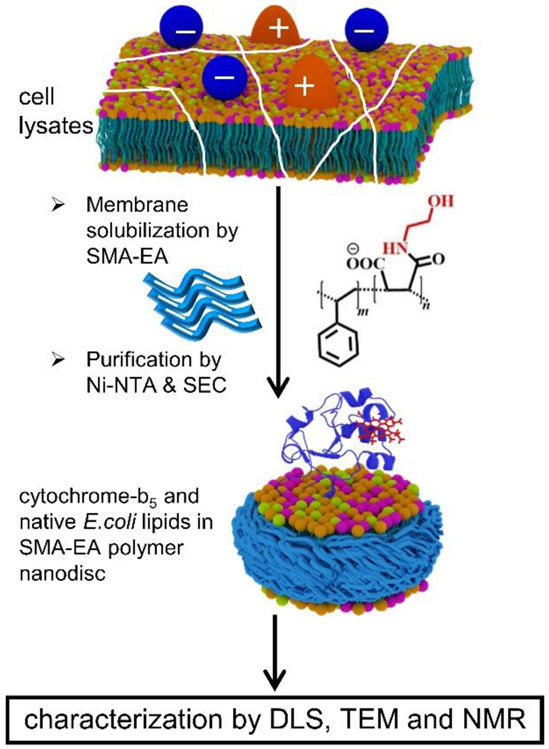
Despite the need for high-resolution structures of membrane proteins to better understand their numerous functions, only <3% of the structures reported in the Protein Data Bank are of membrane proteins. Although a combination of a variety of biophysical techniques is used to overcome the challenges to investigate high-resolution structures and dynamics of membrane proteins,1, 2 the key issue is related to the preparation of samples exhibiting stable and natively-folded structures that retain the function of the membrane proteins.3, 4 Although detergents, and micelles as membrane mimetics, are continued to be used in the structural studies, mainly the X-ray crystallography studies, it is known that detergents destabilize protein structures and disrupt their functions.5 Bicelles, although considered to be better membrane mimetic systems, have also been shown to distort protein structures and functions due to the diffusion of the detergent molecules from the bicelle rim to the planar lipid bilayer region.6 These difficulties have been well demonstrated for the cytochrome proteins containing large soluble domains.6 To overcome these difficulties, recent reported works have focused on the feasibility of directly extracting membrane proteins from cell lysates by using synthetic polymers7-10 without the use of any detergents and reconstituting the proteins in polymer-based nanodiscs.11-13 In this study, we demonstrated the feasibility of a direct extraction of a ~16-kDa cytochrome-b5 from E. coli using a synthetic styrene-co-maleic acid-ethanolamine (SMA-EA) polymer and reconstituting the cytochrome-b5 protein in polymer nanodiscs along with native lipids from E. coli (Fig. 1).
Figure 1.
A schematic representation of the direct extraction of a 16-kDa rabbit cytochrome-b5 in native E. coli lipids using a negatively charged SMA-EA polymer. + and − indicate the net charge of a membrane protein. SEC: size-exclusion chromatography.
Recent studies have reported low-molecular-weight styrene-maleic acid (SMA)14 polymer derivatives such as anionic SMA-EA15 and cationic SMA-QA16 that have been successfully used in the formation of nanodiscs with synthetic lipids and the ability to vary the size of the lipid nanodisc by simply changing the lipid-to-polymer ratio. The large nanodiscs (>20 nm, called macro-nanodiscs) have been shown to align in the presence of an external magnetic field, and therefore can be used in solid-state nuclear magnetic resonance (NMR)-based structural studies under static conditions.15-17 In the current study, we showed that these polymer variants can be used for extracting cytochrome-b5 directly from E. coli without the use of any detergent and also showed the feasibility of carrying out structural studies of this protein using well-established solution NMR and other biophysical techniques. Since the anionic transmembrane protein cytochrome-b5 contains a large soluble domain with a large number of negatively charged residues, we used this protein to further demonstrate the need to avoid any non-specific interaction between the membrane protein to be extracted and the polymer used in order to successfully stabilize the native structure, folding, and function of a membrane protein in nanodiscs. The feasibility of identifying and quantifying the lipids, directly extracted along with cytochrome-b5 from E. coli, by performing simple 1D 31P NMR experiments was also demonstrated.
Cytochrome-b5 is a single transmembrane protein containing heme as a prosthetic group.18 This protein functions by donating the second electron to the membrane-bound cytochrome-P450 enzyme, which requires two electrons to perform the metabolism of drugs and xenobiotics. Cytochrome-b5 also delivers electrons to various other proteins that regulate fatty acid metabolism.19, 20 The 16-kDa cytochrome-b5 consists of an N-terminal heme-binding domain (residues 1-89), a 15-residue flexible linker region (residues 90-104), and a hydrophobic C-terminal domain (residues 105-134).18
Cytochrome-b5 is a negatively charged protein with a net charge of −8.8 at pH 7.4;21 and, therefore, a negatively charged synthetic SMA-EA polymer (Figs. 1 and S1) was used for the direct extraction to avoid non-specific interactions between the protein and polymer. The 7.4 pH was maintained throughout the purification protocol. Cytochrome-b5 was expressed in E. coli C41 cells as reported previously22 and briefly explained in Supporting Information. The membranes containing cytochrome-b5 were collected by subjecting cell lysate to centrifugation, and were resuspended in potassium phosphate buffer (pH 7.4); and the concentration of SMA-EA polymer was adjusted to have a 1:1 membrane-to-polymer ratio (w/w). The cytochrome-b5 in SMA-EA nanodiscs containing native E. coli lipids was purified using Ni-NTA affinity chromatography and then size-exclusion chromatography (SEC).
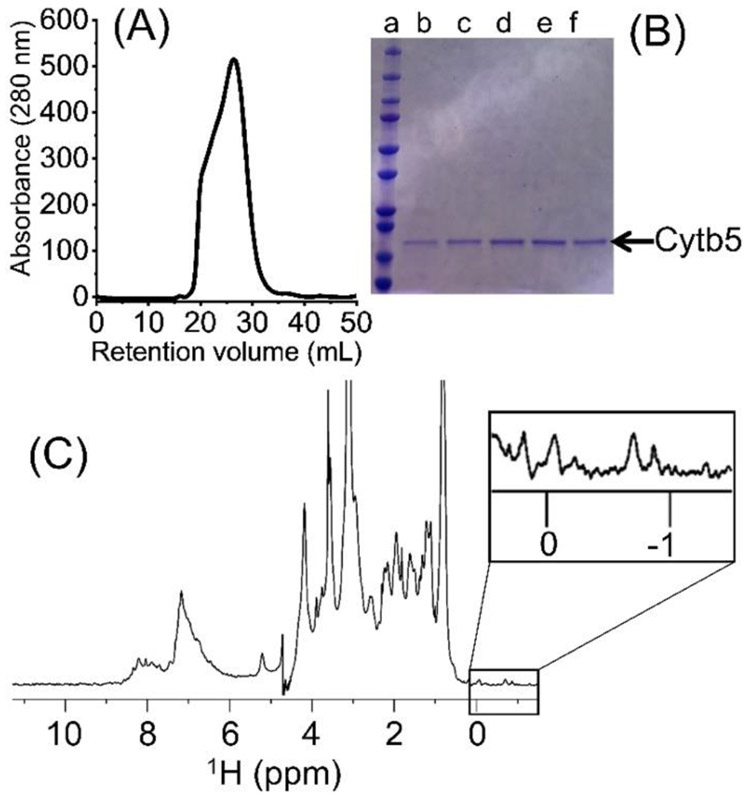
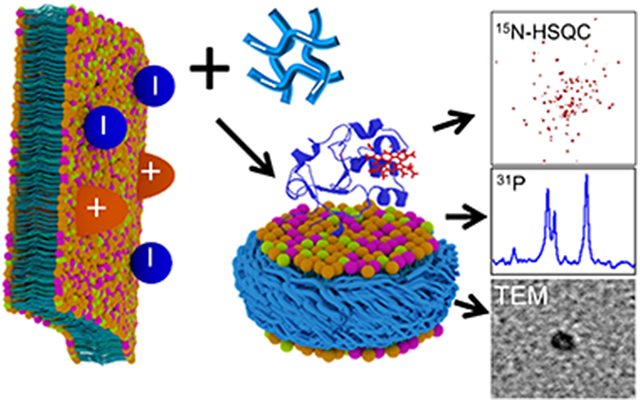
The directly extracted cytochrome-b5 in SMA-EA polymer nanodiscs containing native E. coli lipids was experimentally examined using SEC, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), dynamic light scattering (DLS), transmission electron microscopy (TEM) and NMR spectroscopy as shown in Figures 2, 3 and 4. The SEC elution volume for cytochrome-b5 was between 21 and 26 mL, much lower than the expected elution volume for cytochrome-b5 alone (MW ~16 kDa), indicating the presence of a complex larger than cytochrome-b5 alone (Fig. 2A). The protein showed a single band (MW ~16 kDa) on the SDS-PAGE gel, indicating the high purity of the detergent-free extracted cytochrome-b5 (Fig. 2B). The final protein yields were about 2.5 to 3.0 mg per L culture. UV-visible absorption spectra were recorded in the absence and presence of sodium dithionite. Shifts from 409 nm (oxidized form) to 424, 526 and 556 nm were observed upon the addition of sodium dithionite (Fig. S2). These characteristics are typical of the oxidized state of cytochrome-b5.23 A 1H NMR spectrum showed peaks corresponding to lipids (0.84 and 3.14 ppm), SMA-EA polymer (6.5 to 7.4 ppm from the styrene group) and protein (amide-proton resonances in the 7-9 ppm region), which further confirmed the presence of cytochrome-b5 in the SMA-EA native E. coli lipid nanodiscs (Fig. 2C). The nanodiscs were further characterized using DLS, which showed a hydrodynamic radius of ~5 nm (Fig. 3A), and by acquiring TEM images of them (Fig. 3B), which revealed the presence of nanodiscs having dimensions of ~10 nm.
Figure 2.
(A) Size-exclusion chromatogram (SEC) of the extracted cytochrome-b5. (B) SDS-PAGE analysis of the directly extracted cytochrome-b5. Lane a includes the protein markers and lanes b-f the SEC protein fractions. (C) 1H NMR spectrum of the directly extracted cytochrome-b5. Methyl resonances in the upfield region (boxed) indicating the folded conformation of the protein. The protein sample was prepared in 20 mM potassium phosphate buffer pH 7.4, 50 mM NaCl. The NMR spectrum data were collected at 25 °C using a Bruker 500 MHz NMR spectrometer equipped with a TXI probe.
Figure 3.
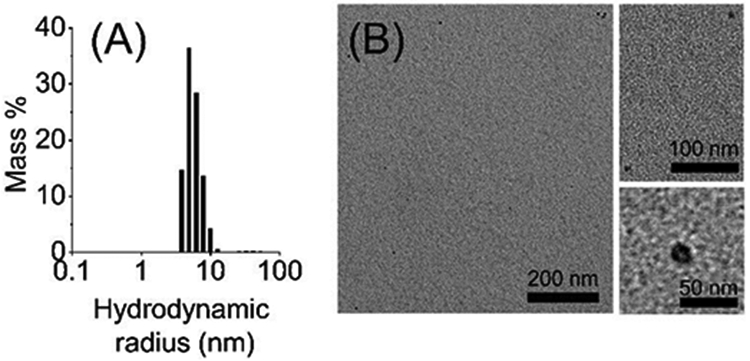
(A) Dynamic light scattering profile of the nanodiscs showing the presence of a large complex with a hydrodynamic radius of ~5 nm. (B) Transmission electron microscopy images showing SMA-EA-based nanodiscs containing directly extracted cytochrome-b5 and E. coli lipids; enlarged images are shown on the right side of the panel for clarity.
Figure 4.
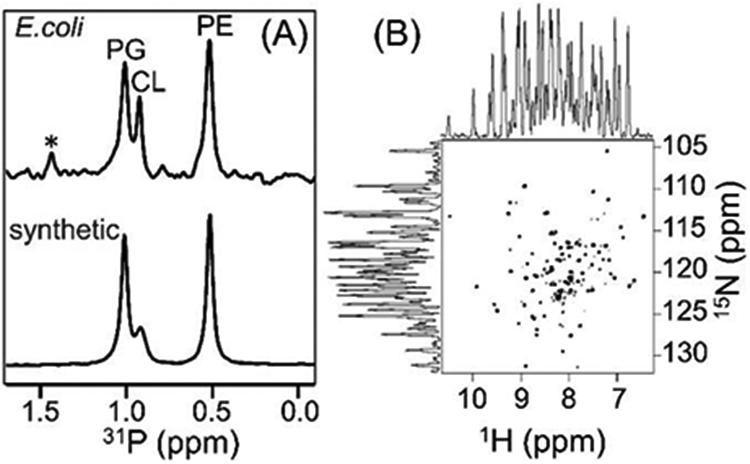
(A) 31P NMR spectra of native E. coli lipids present in the polymer nanodiscs and of the artificially reconstituted synthetic lipids (a reference sample) acquired under by decoupling protons. The protein sample was prepared in 20 mM HEPES buffer pH 7.4 containing 50 mM NaCl and 100 mM sodium cholate. The spectra were recorded using a 500 MHz Bruker NMR spectrometer equipped with a room-temperature 1H/31P/2H BB reverse probe. The synthetic lipids (PE: phosphatidylethanolamine, CL: cardiolipin, and PG: phosphatidylglycerol) were prepared in a PE:CL:PG molar ratio of 7:1.2:4.3. * indicate the uncharacterized E. coli lipids. (B) 2D 1H/15N TROSY-HSQC NMR spectrum of 65 μM 15N-labelled cytochrome-b5 in native nanodiscs. The 1H and 15N line widths were measured to be 27-35 Hz and ~18 Hz, respectively. The protein sample was prepared in 20 mM potassium phosphate buffer pH 7.4, 50 mM NaCl. The 2D NMR spectrum was recorded at 25 °C using a Bruker Avance III 800-MHz spectrometer equipped with a cryogenically cooled triple-resonance probe.
Native E. coli lipids present in the nanodiscs were analyzed using 31P NMR. The 31P NMR spectrum exhibited a very low signal-to-noise ratio with broad spectral lines (with a line width of ~250 Hz) for the extracted native lipids in the SMA-EA nanodiscs. The observed line broadening can be attributed to the unaveraged 1H-1H dipolar couplings, due to the slow tumbling of the ~10-nm-sized nanodiscs, contributing to 31P spin-spin relaxation (Fig. S3). In addition, the applied weak radio-frequency irradiation for proton decoupling during 31P signal acquisition may not have been sufficient to completely suppress 1H-31P dipolar couplings. In order to resolve the overlapped NMR peaks and hence identify the lipids directly extracted by the polymer from E. coli, the nanodiscs were dissolved by adding the non-denaturing detergent sodium cholate at a concentration of 100 mM, and the resultant solution was used to record the 31P NMR spectrum. A reference 31P NMR spectrum was also recorded from commercially available synthetic lipids in the same buffer conditions (Fig. S3). Three well-resolved resonances were observed in the 31P NMR spectrum for the extracted native lipids, as shown in Figure 4A. Based on the spectrum of the reference sample (Fig. 4A), the major peaks observed in the 31P NMR spectrum for the extracted lipids were assigned to phosphatidylethanolamine (PE), phosphatidylglycerol (PG) and cardiolipin (CL). The unassigned low-intensity peak (denoted with an asterisk) appearing in the downfield region of the spectrum was likely due to uncharacterized lipids of the E. coli membrane. The observation of all types of E. coli native lipids in the polymer nanodiscs indicated that cytochrome-b5 does not have a preference to interact with any E. coli lipids. This observation was found to be in agreement with our recent study that utilized the lipid exchange process in peptide-based nanodiscs to report a non-specific preference of cytochrome-b5 for lipids.24
The polymer nanodiscs consisting of the directly extracted lipids and cytochrome-b5 are small (~10 nm); therefore, the isotropic nature of the nanodiscs should enable the application of solution NMR experiments to study the structure and dynamics of cytochrome-b5. To demonstrate this feasibility, uniformly 15N-labelled cytochrome-b5 was expressed in E. coli and directly extracted in native E.coli lipids using the polymer as described above, and the resultant nanodiscs were characterized using the above-mentioned biophysical and NMR experiments. A 2D 1H/15N TROSY-HSQC NMR spectrum of cytochrome-b5 reconstituted in native E. coli lipids was then acquired, and is shown in Fig. 4B. This spectrum exhibited well-dispersed resonances both in the 1H and 15N dimensions, indicating the correctly folded conformation of the protein, and in good agreement with previous studies.18 The spectrum also showed narrow spectral lines, which were attributed to the highly dynamic nature of the large soluble domain of cytochrome-b5, as reported in the previous studies. On the other hand, broad spectral lines invisible in this 2D 1H/15N TROSY-HSQC solution NMR spectrum have been observed in an 19F NMR spectrum25 and also according to solid-state NMR experiments,26 and were attributed to the residues in the transmembrane domain undergoing a slow motion, on the milliseconds or slower time scale. The narrow spectral lines observed for the soluble domain residues in the 2D 1H/15N TROSY-HSQC NMR spectrum confirmed the native-like mobility of residues in the soluble domain of cytochrome-b5 reconstituted in the nanodiscs and also proved the absence of any interaction between the polymer belt and the residues in the reconstituted protein. Taken together, these results suggested that SMA-EA polymer-lipid nanodiscs with cytochrome-b5 were intact under the purification conditions employed in this study.
A direct extraction of cytochrome-b5 was also attempted using a positively charged SMA-QA polymer (Fig. S1) in order to determine the effect of both polymer and protein charges on the solubilization and purification of membrane proteins. The E. coli membranes were solubilized using the SMA-QA polymer under conditions otherwise identical to those described above for the negatively charged SMA-EA polymer. The SMA-QA polymer-protein fraction from the Ni-NTA chromatography column showed, on an SDS-PAGE gel, a protein band with an intensity much lower than that obtained for the SMA-EA protein-fraction (Fig. S4). Unlike the case for SMA-EA, when the membranes were solubilized with SMA-QA, most of the cytochrome-b5 was in the insoluble portion of the membranes and buffer flow-through, indicating the role played by the interaction of the positively charged SMA-QA polymer with the negatively charged cytochrome-b5. Therefore, the charge of the polymer must be the same as that of the membrane protein under investigation to avoid any non-specific interaction between the polymer and the membrane protein.27, 28
Despite the recent advances in the direct extraction of membrane proteins using synthetic polymers, the results reported in this study demonstrated the need to consider a few limitations for achieving a successful membrane protein extraction and reconstitution. As demonstrated for cytochrome-b5, the charge of the polymer used needs to be carefully chosen for a successful reconstitution/extraction of a membrane protein from the cell membrane — in agreement with a recent study on the effect of polymer charge on the stability and functional folding of the reconstituted protein.27, 28 As demonstrated in the current study, to avoid any charge-charge interaction between the polymer and cytochrome-b5, we chose the negatively charged SMA-EA polymer to purify the negatively charged cytochrome-b5. Note that due to the presence of carboxylic groups in the SMA-EA polymer, the non-specific binding of the polymer to Ni-NTA beads required extensive washing steps, which reduced the final yield of the cytochrome-b5 (Fig. S5). Therefore, the use of other purification tags29 in place of the 6His-tag would be helpful to overcome such non-specific interactions.
In conclusion, the membrane-anchored heme-containing cytochrome-b5 was successfully extracted from E. coli and reconstituted in native E. coli lipids by using a negatively charged synthetic SMA-EA polymer. The type of native E. coli lipids present in the nanodiscs was identified using 31P NMR. These results demonstrated that 31P NMR can be used to identify the type of native lipids, and their ratio, associated with the directly extracted membrane protein. The information obtained on the lipids from 31P NMR experiments can also be useful to optimize the conditions/procedure used to successfully reconstitute the membrane proteins extracted using detergents when following the well-established protocols. Our results revealed that the directly extracted 15N-labelled cytochrome-b5 was of high purity as indicated by the 2D 1H/15N TROSY-HSQC NMR spectrum, which also demonstrated the correctly folded state of the protein and the feasibility to carry out structural and dynamical measurements using the well-established multidimensional solution NMR techniques. Although both SMA-QA and SMA-EA polymers were observed to behave similarly in solubilizing membranes, the polymer charge was found to have a significant effect on the protein purification. The attempts to purify the cytochrome-b5 using the positively charged polymer SMA-QA failed due to the charge-charge interactions between the protein and the polymer. Therefore, our results suggested that the polymer charge should be seriously considered based on the net charge of the desired protein that is to be purified at a given pH. We expect the electrostatic interactions between the polymer and protein to be highly significant for most membrane proteins containing a large soluble domain, such as single-pass and double-pass transmembrane proteins.
Supplementary Material
Acknowledgements
This study was supported by NIH (GM084018 to A.R.).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Hagn F, Nasr ML and Wagner G, Nat Protoc, 2018, 13, 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang B and Tamm LK, Nat. Struct. Mol. Biol, 2016, 23, 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang S, Munro RA, Shi L, Kawamura I, Okitsu T, Wada A, Kim S-Y, Jung K-H, Brown LS and Ladizhansky V, Nature Methods, 2013, 10, 1007–1012. [DOI] [PubMed] [Google Scholar]
- 4.Denisov IG and Sligar SG, Nat. Struct. Mol. Biol, 2016, 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, Huang R, Ackermann R, Im S-C, Waskell L, Schwendeman A and Ramamoorthy A, 2016, 55, 4497–4499. [DOI] [PubMed] [Google Scholar]
- 6.Dürr UHN, Soong R and Ramamoorthy A, Prog. Nucl. Magn. Reson. Spectrosc, 2013, 69, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall SCL, Tognoloni C, Charlton J, Bragginton EC, Rothnie AJ, Sridhar P, Wheatley M, Knowles TJ, Arnold T, Edler KJ and Dafforn TR, Nanoscale, 2018, 10, 10609–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T and Overduin M, J Am Chem Soc, 2009, 131, 7484–7485. [DOI] [PubMed] [Google Scholar]
- 9.Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi J.-i. and Ramamoorthy A, J. Am. Chem. Soc, 2017, 139, 18657–18663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison KA, Akram A, Mathews A, Khan ZA, Patel JH, Zhou C, Hardy DJ, Moore-Kelly C, Patel R, Odiba V, Knowles TJ, Javed M.-u.-H., Chmel NP, Dafforn TR and Rothnie AJ, Biochem. J, 2016, 473, 4349–4360. [DOI] [PubMed] [Google Scholar]
- 11.Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EAW, Dafforn TR, Baldus M and Killian JA, Proc Natl Acad Sci, 2014, 111, 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP and Dafforn TR, Nat Protoc, 2016, 11, 1149–1162. [DOI] [PubMed] [Google Scholar]
- 13.Barniol-Xicota M and Verhelst SHL, J. Am. Chem. Soc, 2018, 140, 14557–14561. [DOI] [PubMed] [Google Scholar]
- 14.Bada Juarez JF, Harper AJ, Judge PJ, Tonge SR and Watts A, Chem Phys Lipids, 2019, 221, 167–175. [DOI] [PubMed] [Google Scholar]
- 15.Ravula T, Ramadugu SK, Di Mauro G and Ramamoorthy A, Angew Chem Int Ed Engl, 2017, 56, 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 16.Ravula T, Hardin NZ, Ramadugu SK, Cox SJ and Ramamoorthy A, Angew Chem Int Ed Engl, 2018, 57, 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 17.Radoicic J, Park SH and Opella SJ, Biophys J, 2018, 115, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahuja S, Jahr N, Im SC, Vivekanandan S, Popovych N, Le Clair SV, Huang R, Soong R, Xu J, Yamamoto K, Nanga RP, Bridges A, Waskell L and Ramamoorthy A, J Biol Chem, 2013, 288, 22080–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Napier JA, Michaelson LV and Sayanova O, Prostaglandins, Leukotrienes and Essential Fatty Acids, 2003, 68, 135–143. [DOI] [PubMed] [Google Scholar]
- 20.Guillou H, D'Andrea S, Rioux V, Barnouin R, Dalaine S, Pedrono F, Jan S and Legrand P, 2004, 45, 32–40. [DOI] [PubMed] [Google Scholar]
- 21.Kozlowski LP, Nucleic Acids Res, 2017, 45, D1112–D1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulrooney SB and Waskell L, Prot. Expr Purif, 2000, 19, 173–178. [DOI] [PubMed] [Google Scholar]
- 23.Guzov VM, Houston HL, Murataliev MB, Walker FA and Feyereisen R, 1996, 271, 26637–26645. [DOI] [PubMed] [Google Scholar]
- 24.Ravula T, Ishikuro D, Kodera N, Ando T, Anantharamaiah GM and Ramamoorthy A, Chem. Mater, 2018, 30, 3204–3207. [Google Scholar]
- 25.Bai J, Wang J, Ravula T, Im S-C, Anantharamaiah GM, Waskell L and Ramamoorthy A, Biochimica et Biophysica Acta (BBA) - Biomembranes, 2020, 1862, 183194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandey MK, Vivekanandan S, Yamamoto K, Im S, Waskell L and Ramamoorthy A, J. Magn. Reson, 2014, 242, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopf AH, Dorr JM, Koorengevel MC, Antoniciello F, Jahn H and Killian JA, Biochim Biophys Acta Biomembr, 2020, 1862, 183125. [DOI] [PubMed] [Google Scholar]
- 28.Ravula T, Hardin NZ, Bai J, Im SC, Waskell L and Ramamoorthy A, Chem Commun (Camb), 2018, 54, 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimple ME, Brill AL and Pasker RL, Curr Protoc Protein Sci, 2013, 73, 9 9 1–9 9 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.