Abstract
Vibrio fischeri is a non-pathogenic organism related to pathogenic Vibrio species that can be readily grown and stored with common laboratory equipment. In this article, protocols for routine growth, storage, and phenotypic assessment of V. fischeri as well as recipes for useful media are included. Specifically, this article describes procedures and considerations for growth of this microbe in complex and minimal media. It also describes assays for biofilm formation, motility, and bioluminescence, three commonly assessed phenotypes of Vibrio fischeri.
Keywords: Biofilm, Vibrio, Vibrio fischeri, Symbiosis, Bacterial Cultivation, Motility, Luminescence
INTRODUCTION
Vibrio fischeri (also classified as Aliivibrio fischeri) is a Gram-negative, rod-shaped, bioluminescent marine bacterium (Urbanczyk, Ast, Higgins, Carson, & Dunlap, 2007). The bacterial genome consists of two chromosomes with low G-C content and often includes one large and/or one or more small plasmids (Boettcher & Ruby, 1994; Ruby et al., 2005). V. fischeri was originally studied for its bioluminescent properties and now is studied most intensively with respect to its symbiosis with the Hawaiian bobtail squid, Euprymna scolopes. V. fischeri is also capable of colonizing other squid species as well as fish species such as Monocentris japonica (Nishiguchi, 2001; Ruby & Nealson, 1976). In the squid symbioses, the bacteria produce bioluminescence, which protects their host from predation, while the host provides nutrients and a protected niche for the bacteria (Graf & Ruby, 1998; Jones & Nishiguchi, 2004).
Much work has been done to identify key factors that promote the interactions between V. fischeri and E. scolopes, from the perspectives of both the bacteria and their host. The squid hatch in an aposymbiotic state – i.e., lacking their symbiont - and acquire V. fischeri from the surrounding seawater (Nyholm, Stabb, Ruby, & McFall-Ngai, 2000; Stabb & Visick, 2013). Bacteria-containing seawater is flushed through the squid’s mantle cavity and across the surface of the symbiotic light organ, where ciliary movements direct bacteria toward sheltered zones (McFall-Ngai & Ruby, 1991; Nawroth et al., 2017). There, V. fischeri cells form a bacterial aggregate or biofilm on the organ surface from which they subsequently disperse (Nyholm et al., 2000). Using motility and chemotaxis, the bacteria enter into pores and migrate through non-permissive spaces to reach deep crypt spaces where they multiply and produce light (Aschtgen et al., 2019; Graf, Dunlap, & Ruby, 1994). Once the symbiosis is established, the bacteria are maintained through a daily cyclic expulsion of ~95% of the bacteria from the light organ into the surrounding seawater and a regrowth of the remaining ~5% population (Lee & Ruby, 1994). In these different stages, the nutritional availability and therefore growth of V. fischeri is thought to shift (Wier et al., 2010). It is hypothesized that, during the day, the V. fischeri symbiont utilizes glycerol-3-phosphate from host-derived lipids, while at night and prior to expulsion, host-derived chitin is the primary nutrient.
V. fischeri is a non-pathogenic, biosafety level 1 (BSL-1) organism that is related to pathogenic Vibrio species like Vibrio cholerae. This low safety level and a continually expanding genetic toolbox make V. fischeri an excellent bacterium to utilize at any level of research. V. fischeri serves as an important model organism to study a variety of processes including the following: requirements for symbiotic colonization, evolutionary adaptations between bacterium and host, quorum sensing, bioluminescence, host-associated biofilm formation, and motility.
STRATEGIC PLANNING
Strain Considerations
V. fischeri strains have been isolated from a variety of environmental locations. Historically, strain ES114 (K. J. Boettcher & Ruby, 1990), which was isolated from the E. scolopes light organ, has been used as an experimental wild type strain for symbiosis experiments. Thus, most of the laboratory techniques for growth and genetic and phenotypic characterizations were developed and/or optimized for this strain.
More recently, a number of additional symbiotic and environmental strains have been isolated and are beginning to be characterized (e.g., MB13B2 and FQ-A001) (Bongrand & Ruby, 2019; Koehler et al., 2018; Sun et al., 2016). These strains exhibit substantial genotypic and phenotypic differences from ES114. For example, MB13B2 (Bongrand et al., 2016) and FQ-A001 (Bultman, Cecere, Miyashiro, Septer, & Mandel, 2019) contain additional sequences (250 kb and 54 kb, respectively) not present in ES114. In experiments designed to test superior symbiotic competence, MB13B2 or FQ-A001 outcompeted ES114, resulting in majority or exclusive colonization of the squid. As a result, they have been termed “dominant” strains. However, other recent isolates termed “sharing” will co-exist with ES114 within a light organ (Bongrand et al., 2016), a result that indicates that ES114 remains intact as a wild-type strain and hasn’t necessarily become “lab-adapted”.
Different isolates also vary in several other phenotypes. For example, dominant strain MB13B2 is vastly different relative to ES114 in its ability to form biofilms in symbiosis and in laboratory culture (Koehler et al., 2018). While ES114 can successfully form small aggregates on the squid light organ, it forms biofilms poorly in laboratory culture. Due to an incomplete understanding of signals present in the squid that promote these phenotypes, genetic manipulations are generally required to induce biofilm formation by ES114. In contrast, MB13B2 readily produces robust biofilm pellicles in static liquid cultures in the test tube and both produces larger cellular aggregates on the light organ and migrates to the crypts earlier (Bongrand & Ruby, 2019; Koehler et al., 2018). Dominant strain FQ-A001 displays reduced migration in motility assays and increased bioluminescence relative to ES114 (Sun et al., 2016). This strain is also capable of attacking and killing ES114 via a type VI secretory apparatus that ES114 lacks (Speare et al., 2018).
Given the significant genotypic and phenotypic variation of these dominant and sharing strains, comparative analyses have tremendous power to address questions of the evolutionary advantages and trade-offs between bacteria with different colonization strategies. The specific study goal(s) will necessarily dictate the choice of strain(s) and the corresponding expectations for the outcome. One additional consideration for the choice of strain lies in the ease with which a given strain can be genetically manipulated. Our experience to date with MB13B2 suggests that this organism may prove challenging to manipulate. Thus, certain questions may require the genetic manipulability of a strain like ES114 to facilitate dissection of complex phenotypes.
Since much of the research investigating V. fischeri growth and behavior has been based on ES114, the experimental procedures described herein were developed/optimized using this isolate. However, it is likely that some V. fischeri isolates will exhibit different behaviors under the growth conditions described here, and thus these protocols may need alterations or may be wholly ineffective. For these strains, these protocols should serve as a first step but may require significant alterations to achieve the best results.
Growth Conditions
As V. fischeri is a bacterium found in the ocean, its optimum growth in the laboratory reflects those conditions. Optimal growth for V. fischeri occurs at 24–28°C in a high (~20 g/L) salt environment. High temperatures (above 34°C for some strains) prove lethal for the bacterium (Cohen, Mashanova, Rosen, & Soto, 2019). Similarly, V. fischeri will readily lyse in fresh water. Traditionally, growth media for this organism have included glycerol, and many researchers continue to include this carbon source, even in complex media that does not require its addition for V. fischeri growth. However, V. fischeri readily metabolizes glycerol, resulting in the acidification of the growth medium that causes bacterial death. While buffering the medium can reduce this effect, medium acidification is a prime reason why overnight cultures of V. fischeri fail to re-grow, or re-grow poorly, following subculturing. Furthermore, recovery of a V. fischeri culture after overnight growth is reduced proportional to the duration of stationary phase. A shorter overnight incubation time (e.g., ~14 h) is optimal for the recovery of bacteria from frozen stocks and for use in experimental cultures.
Genetic manipulation of V. fischeri typically depends on antibiotics to select for strains with insertions or deletions of genetic material and to obtain and maintain plasmids of interest. Table 1 lists the antibiotic concentrations necessary to achieve selection for the common laboratory strain ES114. While this strain grows optimally on a high salt medium such as LBS, which contains 20 g/L of sodium chloride, certain antibiotics do not work well under these high salt conditions. Therefore, for some antibiotics, LB medium, made with 10 g/L of sodium chloride, is used instead of LBS; the growth of ES114 is not substantially impaired under these lower salt conditions. Alternatively, higher concentrations of antibiotics are required for selection in media like SWT or TMM compared to LBS. Other V. fischeri strains may exhibit altered antibiotic resistance profiles and should be individually assessed to determine the optimal concentrations of antibiotics.
Table 1.
Antibiotic Concentrations and Medium for Selection of V. fischeri
| Antibiotic1,2 | Solvent | Stock Concentration (mg/mL) | Working Concentration (μg/mL) | Medium3 |
|---|---|---|---|---|
| Chloramphenicol | 100% EtOH | 1 | 1 | LBS |
| 5 | TMM or high copy # plasmid in LBS | |||
| Erythromycin | 100% EtOH | 2.5 | 2.5 | LBS |
| Gentamicin | ddH2O | 10 | 10 | LB |
| Kanamycin | ddH2O | 20 | 100 | LBS or TMM |
| Spectinomycin | ddH2O | 40 | 200 | LB |
| Tetracycline | 100% EtOH | 5 | 2.5 | LBS |
| 30 | SWT or TMM | |||
| Trimethoprim | 90% EtOH | 2 | 10 | LBS |
V. fischeri is highly resistant to ampicillin.
Zeocin has been used at a working concentration of 10 μg/mL in LB, but further optimization is necessary to enhance selectivity (Visick, Hodge-Hanson, Tischler, Bennett, & Mastrodomenico, 2018).
Optimal growth medium for different antibiotics was determined empirically.
BASIC PROTOCOL 1: GROWTH OF V. fischeri FROM FROZEN STOCKS
V. fischeri can be recovered from a frozen stock in a glycerol solution (See Basic protocol 3) at −80°C and remains viable on petri plates for about one week. The bacteria can be streaked onto LBS plates, and single colonies can then be used to inoculate liquid cultures. See Figure 1 for an example of a streak plate and expected colony appearance.
Figure 1. Wild-type V. fischeri strain ES114 streak plate.
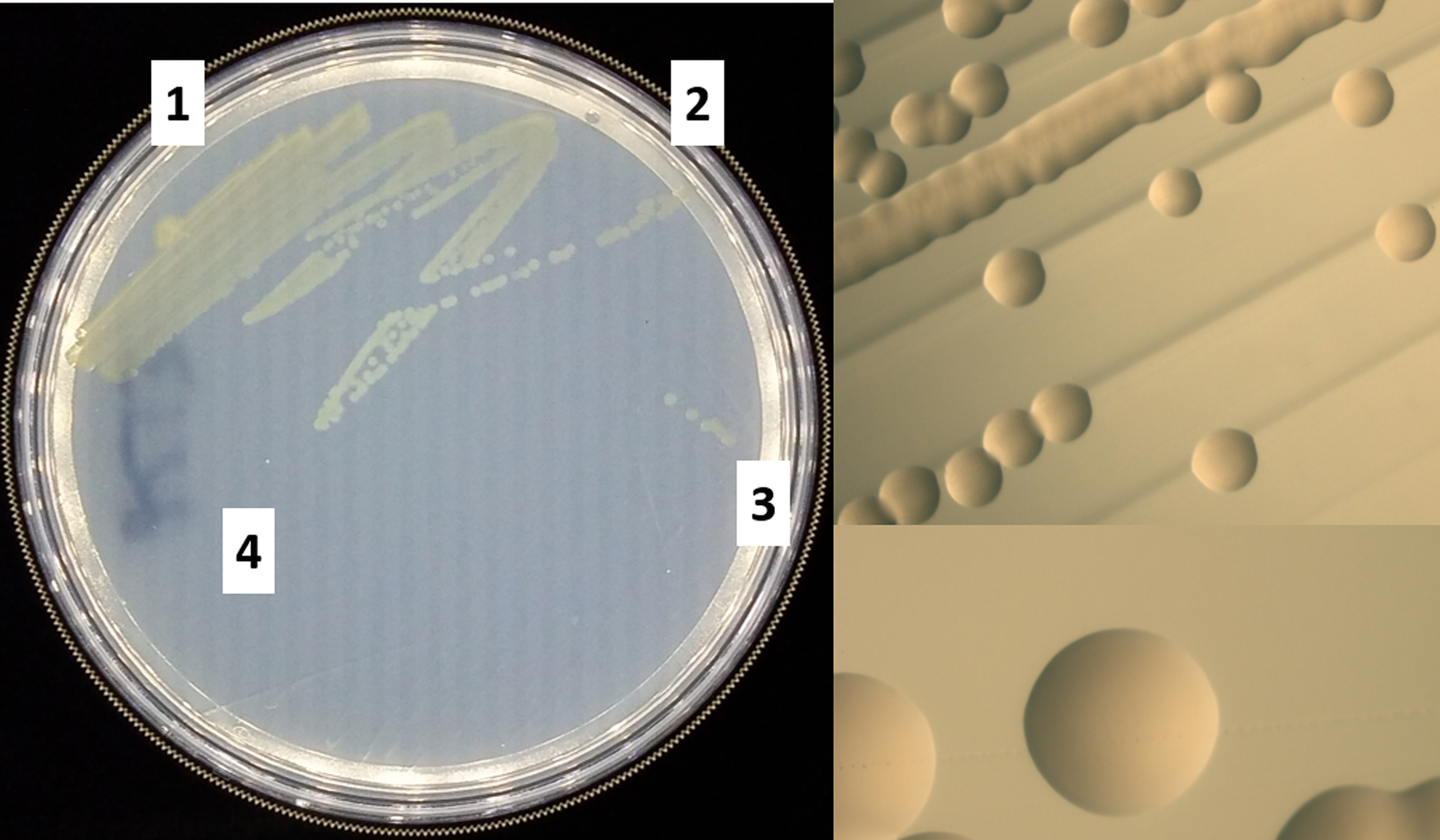
Left, the cells were streaked from a frozen culture with four independent streaks and incubated at 28°C for 20 hours. Top right, magnified image of single colonies from a streak. Bottom right, further magnified image of a single V. fischeri colony.
Materials
Vibrio fischeri frozen glycerol stock in cryovial
LBS plate (see recipe)
Sterile applicator sticks
28°C incubator
Use a sterile applicator stick to scrape a small amount of V. fischeri cells from the frozen stock in the cryovial. Transfer the bacteria and make a heavy streak on a small area of the surface of an LBS plate.
With a second sterile applicator stick, make a second streak by gently passing it through the heavy streak once. Repeat this twice more with additional sterile sticks to generate four quadrants of decreasing bacterial concentration with the goal of isolating single colonies.
-
Invert the plate and incubate it upside down in a 28°C incubator overnight.
Other temperatures, such as 24°C or room temperature will also permit growth.
Store plate at 4°C for up to one week.
BASIC PROTOCOL 2: GROWTH OF V. fischeri IN COMPLEX LIQUID MEDIUM
V. fischeri is routinely grown in complex media like LBS or SWT (a seawater-based medium). Depending on the assay, the choice of medium may result in significantly different phenotypic outcomes (Marsden, Grudzinski, Ondrey, DeLoney-Marino, & Visick, 2017).
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1) or from frozen stock (see Basic Protocol 3)
LBS or SWT medium (see recipes)
Sterile applicator stick
Sterile capped glass culture tube (routinely 18 × 150-mm)
28°C shaking incubator
1a. From agar plates: Pick a single colony of V. fischeri from an LBS streak plate using a sterile applicator stick.
1b. From frozen stocks: Scrape some V. fischeri cells from the frozen culture stock with an applicator stick.
-
2. Inoculate a liquid culture by suspending the cells from the stick in 5 mL of media in a sterile 18 × 150-mm, capped glass culture tube. Remove the stick and cap the test tube.
V. fischeri grows better with good aeration. Therefore, smaller tubes can be used with a proportionally smaller volume.
3. Grow the culture overnight at 28°C with aeration by shaking at 220 rpm.
ALTERNATE PROTOCOL 1: GROWTH OF V. fischeri IN MINIMAL MEDIUM
Nutritional requirements or auxotrophies of wild-type and mutant strains of V. fischeri can be assessed by growth in a defined minimal medium. Similarly, minimal medium allows one to control the exact composition of metals, nitrogen sources, and carbon sources provided to the culture.
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1) or from frozen stock (see Basic Protocol 3)
Minimal medium (TMM or HMM) (see recipes)
Sterile applicator stick
Sterile capped glass culture tube (routinely 18 × 150-mm)
28°C shaking incubator
1a. From agar plates: Pick a single colony of V. fischeri from an LBS streak plate using a sterile applicator stick.
1b. From frozen stocks: Scrape some V. fischeri cells from the frozen culture stock with an applicator stick.
2. Inoculate a liquid culture by suspending the cells from the stick in 5 mL of TMM (or HMM) in a sterile 18 × 150-mm, capped glass culture tube. Remove the stick and cap the test tube.
-
3. Grow the culture overnight at 28°C with aeration by shaking at 220 rpm.
The cells may not recover well from the frozen stock. Addition of casamino acids to 0.1% can help cells to recover.
BASIC PROTOCOL 3: STORAGE OF V. fischeri IN FROZEN STOCKS
For long-term storage of V. fischeri, the bacteria can be stored in a 20% glycerol stock at −80°C and subsequently recovered using Basic Protocol 1.
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1)
LBS medium (see recipe)
Autoclaved plastic or glass cryovial containing 335 μL of 80% glycerol
Sterile applicator stick
Sterile 18 × 150-mm, capped glass culture tube
28°C shaking incubator
Using a sterile applicator stick, pick a single V. fischeri colony from an LBS agar plate.
Inoculate an 18 × 150-mm glass culture tube containing 5 mL LBS by suspending the cells from the applicator stick in the medium. Remove the applicator stick and cap the test tube.
-
Aerate the inoculated tube at 28°C at 220 rpm overnight.
To maintain the health of the culture, incubate for only 14 – 18 hours.
The following day, harvest 1 mL of the overnight culture and add it to the sterile cryovial with glycerol. Pipette up and down or invert the tube a few times to ensure complete mixing of glycerol and culture.
Store this cryovial containing the strain stock in a −80°C freezer.
Check viability of the frozen stock the next day by streaking onto LBS agar and incubating the streak at 28°C overnight. This is particularly important if LBS broth containing glycerol was used for growth of the culture.
BASIC PROTOCOL 4: BIOFILM ASSAY – SOLID AGAR
Biofilm formation by V. fischeri is a critical step in establishing symbiosis with its host, and a correlation has been drawn between host colonization and biofilm formation under laboratory conditions (Shibata, Yip, Quirke, Ondrey, & Visick, 2012; Yip, Geszvain, DeLoney-Marino, & Visick, 2006). Wild-type V. fischeri strain ES114 does not form robust biofilms under these common laboratory conditions. However, certain genetically-altered ES114 derivatives as well as some other squid isolates are biofilm-competent and exhibit colony morphologies such as colony cohesion, adherence to the plate, and/or wrinkles/corrugation on solid agar plates (Figure 2). For further reading on this technique, see (Ray, Morris, & Visick, 2012).
Figure 2. Examples of different biofilm phenotypes on a solid medium.
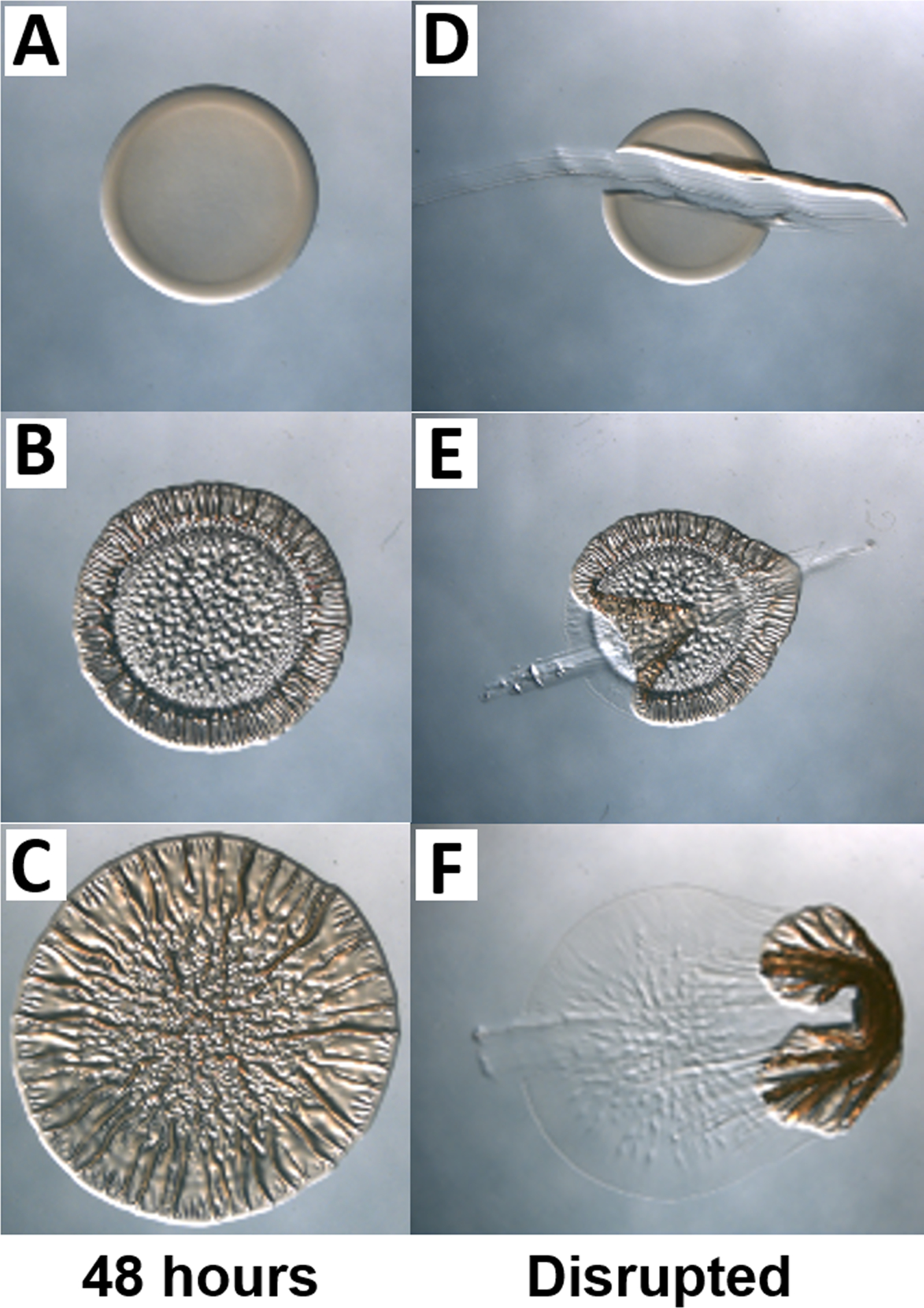
Images were taken after growth for 48 hours and before (A-C) and after (D-F) toothpick disruption. (A) Smooth colony; (B and C) Wrinkled colonies; (D) No biofilm, as evidenced by lack of adherence and coherence; (E) Colony adherent to plate; and (F) Coherent colony. Images (A-C) are magnified relative to images (D-F).
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1)
LBS medium (see recipe)
LBS agar (or LBS agar containing calcium chloride) plate(s) (see recipe)
Sterile applicator stick
Sterile 18 × 150-mm, capped glass culture tube
28°C shaking incubator
Spectrophotometer
24°C incubator
Dissecting (stereoscopic) microscope
Toothpick
Using a sterile applicator stick, pick a single V. fischeri colony from an LBS agar plate.
Inoculate an 18 × 150-mm glass culture tube containing 5 mL LBS by suspending cells from the applicator stick in the medium. Remove the applicator stick and cap the test tube.
-
Aerate the inoculated tube at 28°C at 220 rpm overnight.
Try not to exceed 16 hours to maintain the health of the culture.
Subculture the overnight culture by pipetting 50 μL from the overnight culture into an 18 × 150-mm glass culture tube containing 5 mL LBS (this results in a 1:100 dilution of the overnight culture).
-
Aerate the cultures until they reach log phase, determine the OD600 using a spectrophotometer, and normalize the cultures to an OD600 of 0.2 using LBS.
For example, if a culture reaches an OD600 of 0.3, add 67 μL culture to 33 μL LBS. If another culture reaches an OD600 of 0.4, add 50 μL culture to 50 μL LBS. When this is done, mix the cell suspensions by vortexing.
V. fischeri grows rapidly and should only take about 1 – 2 hours to achieve the desired OD600. However, time to achieve log phase should be independently determined for each strain via growth curve analysis.
-
Divide an LBS plate into four to eight sections. Evenly spot 10 μL of each normalized culture onto the LBS agar plate.
For consistent results within an experiment that requires more than one plate and between experiments, the agar plates should be made with a standard volume such as 25 mL.
LBS medium can be supplemented with a nutrient/compound of interest (such as 10 mM calcium chloride, which is known to promote V. fischeri biofilm formation) prior to autoclaving or prior to dispensing the autoclaved and cooled agar into petri dishes or it can be substituted with a distinct medium of choice.
Samples can be spotted in duplicate or other multiples on the same plate, both to evaluate reproducibility and to permit multiple “endpoint” analyses as described in step 8 below.
-
Leave the plate upright until the spot soaks in, and then invert the plates and incubate in a 24°C incubator. Observe the spots at various times after spotting using a dissecting microscope.
A biofilm-competent strain may produce spots with visible wrinkles that continue to develop over time.
Some strains will form biofilms at higher temperatures, such as 28°C, so this condition can be modified as needed.
The timing of biofilm formation varies depending on the choice of strain, genetic manipulation, and media/growth conditions. If the timing is unknown, a reasonable time course would be to evaluate colony morphology every 24 hours for three to five days.
-
After the last time point, disrupt the colony with a toothpick by gently dragging it across the agar surface and through the colony from one side to the other.
This “toothpick assay” can sometimes reveal colony cohesiveness (the cells in the colony stick together) or adhesiveness (the cells in the colony stick to the plate) that cannot be visually observed by eye or by using the dissecting microscope. Alternatively, in some cases, architecture is observed without a corresponding “stickiness” (cohesiveness or adhesiveness). The “toothpick assay” thus permits an assessment of colony stickiness, permitting a determination of whether or not the colony has formed a biofilm (is cohesive or adhesive).
The “toothpick assay” can be used at multiple timepoints if replicate samples were spotted.
ALTERNATE PROTOCOL 2: BIOFILM ASSAY – SHAKING LIQUID CULTURE
In addition to forming biofilms on solid agar, V. fischeri can produce biofilms in a shaking liquid culture in a test tube (Tischler, Lie, Thompson, & Visick, 2018). While the wild type V. fischeri strain ES114 does not form robust biofilms, genetically-altered, biofilm-competent strains form white/opaque rings at the air-liquid interface of a shaking culture and/or clumps of cells at the bottom of the test tube, which may be cohesive (Figure 3). Calcium chloride (final concentration of 10 mM) is commonly used as an inducing signal for certain ES114 mutants (e.g., ΔbinK) (Tischler et al., 2018).
Figure 3. Biofilm formation during growth with shaking in liquid medium.
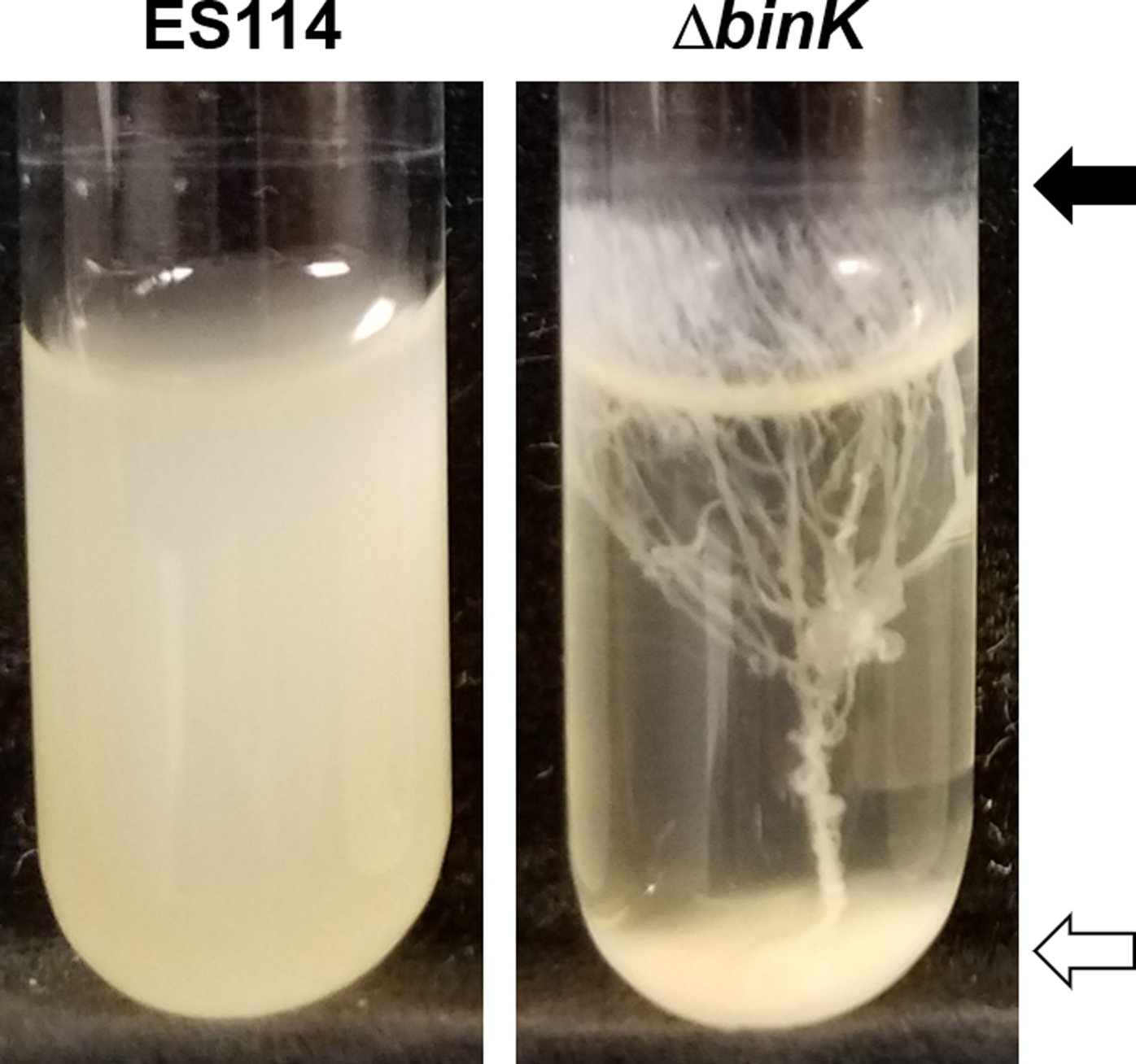
Following growth with shaking in LBS medium supplemented with 10 mM CaCl2, wild-type strain ES114 (left) presents a very faint ring and no clump, while the ΔbinK mutant (right) exhibits clumps (white arrow) and rings (black arrow) connected by “trees”. These cultures were imaged after 24 hours of shaking at 24°C.
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1)
LBS medium (see recipe)
1 M CaCl2 stock
Sterile applicator stick
Sterile 18 × 150-mm, capped glass culture tube
Sterile 13 × 100-mm, capped glass culture tube
28°C shaking incubator
Spectrophotometer
24°C shaking incubator
Using a sterile applicator stick, pick a single V. fischeri colony from an LBS agar plate.
Inoculate an 18 × 150-mm glass culture tube containing 5 mL LBS by suspending the cells from the applicator stick in the medium. Remove the applicator stick and cap the test tube.
Aerate the inoculated tube at 28°C at 220 rpm overnight.
-
The following day, prepare a volume of LBS supplemented with 10 mM CaCl2 sufficient for the needs of the experiment (e.g., add 1 mL 1 M CaCl2 stock to 100 mL LBS).
Making this medium fresh helps to prevent precipitation of calcium that can be observed during extended storage of the medium.
Pipette 2 mL of the LBS supplemented with 10 mM CaCl2 into a 13 × 100-mm, capped glass culture tube.
-
Determine the optical density of the overnight culture using the spectrophotometer and pipette an appropriate volume of the overnight culture to achieve a final optical density of 0.05 OD600 in the 2 mL LBS supplemented with 10 mM CaCl2.
For example, if an overnight culture reaches an OD600 of 5, add 20 μL of that culture into a tube containing 2 mL LBS. If another culture reaches an OD600 of 6, add 16.7 μL of that culture into another tube containing 2 mL LBS.
-
Aerate the tube with shaking at 220 rpm at 24°C for 24 hours.
Timing can be important for observing this phenotype. Earlier time points may reveal deficiencies for certain mutants that may be missed at later time points.
-
Visually assess and/or photo-document the biofilms formed. The optical density of the planktonic cells in the culture medium can be measured with a spectrophotometer, if desired, for a quantitative measure of biofilm formation; the lower the optical density of the planktonic cells, the greater the biofilm formation.
If measuring optical density, a no biofilm control should be included for comparison.
If a biofilm has formed, do not disturb the biofilm when taking optical density readings of the culture medium.
ALTERNATE PROTOCOL 3: BIOFILM ASSAY – STATIC LIQUID CULTURE
A third method to assess biofilms formed by V. fischeri is by observing biofilms formed during static liquid culture – i.e., pellicles (Figure 4). Wild type V. fischeri strain ES114 does not form a robust pellicle, but genetically-altered biofilm-competent ES114 or strains that are naturally biofilm competent will form a film at the air-liquid interface of a bacterial culture that may develop wrinkles. Calcium chloride is commonly required to induce robust pellicles for certain ES114 mutants (e.g., ΔbinK) (Tischler et al., 2018), though it may not always be required.
Figure 4. Biofilm formation during growth in static culture.
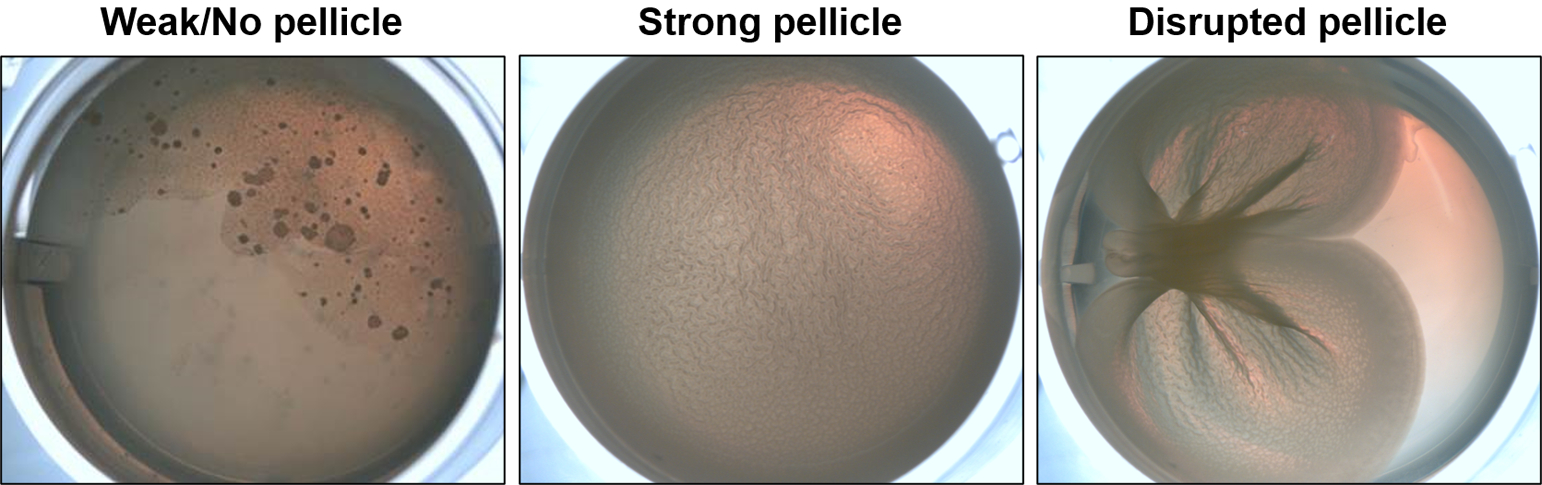
Pellicles of V. fischeri in static cultures grown at 24°C. (Left) Image of a poor pellicle producing strain with a brittle pellicle and dark microcolonies, following disruption with a toothpick. (Middle) Image of a strain with a pellicle with visible architecture (wrinkles). (Right) Image of the pellicle in the middle panel following disruption by a toothpick to assist visualization. This “toothpick test” permits an assessment of pellicle robustness/strength that is not possible to determine by a visual examination alone.
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1)
LBS medium (see recipe)
1 M CaCl2
Sterile applicator stick
Sterile 18 × 150-mm, capped glass culture tube
28°C shaking incubator
Spectrophotometer
24-well microwell plate
24°C incubator
Dissecting (stereoscopic) microscope
Resealable plastic bag
Toothpick
Using a sterile applicator stick, pick a single V. fischeri colony from an LBS agar plate.
Inoculate an 18 × 150-mm glass culture tube containing 5 mL LBS by suspending the cells from the applicator stick in the medium. Remove the applicator stick and cap the test tube.
Aerate the inoculated tube at 28°C by shaking at 220 rpm overnight.
-
If using LBS supplemented with 10 mM CaCl2, add a volume of sterile 1 M CaCl2 to LBS to achieve a final concentration of 10 mM CaCl2.
Making this medium fresh helps to prevent precipitation of calcium that can be observed during extended storage of the medium.
-
Pipette 2 mL LBS or LBS supplemented with 10 mM CaCl2 into a well of the 24-well plate.
To avoid differences in aeration between the outer wells and inner wells, only the inner 8 wells should be used.
Determine the optical density of the overnight culture using the spectrophotometer and pipette an appropriate volume of the overnight culture into the well to achieve a final optical density of 0.2 OD600.
-
Place the plate in a resealable plastic bag. Incubate the 24-well plate at 24°C and observe the pellicles under a dissecting microscope at intervals, such as 24 and 48 hours.
The plate can be incubated for shorter or longer times, although longer periods may result in evaporation of the medium.
Care should be taken not to disturb the pellicles when observing pellicle growth at the 24-hour time point.
-
After the last time point, drag a toothpick or plastic pipette tip across the pellicle from one side to the other to disrupt the pellicle.
Disruption of the pellicle helps to visualize the biofilm atop the turbidity of the culture and can impart information about the stickiness, cohesion, and robustness of the biofilm.
BASIC PROTOCOL 5: MOTILITY ASSAY
Migration of V. fischeri must occur for the bacteria to travel into the light organ of the squid (Graf et al., 1994; Nyholm et al., 2000). To study the migration of V. fischeri in the lab, low percentage agar plates are used to observe flagella-based motility of the bacteria on a macroscopic scale (Figure 5).
Figure 5. Motility in semi-solid agar medium.
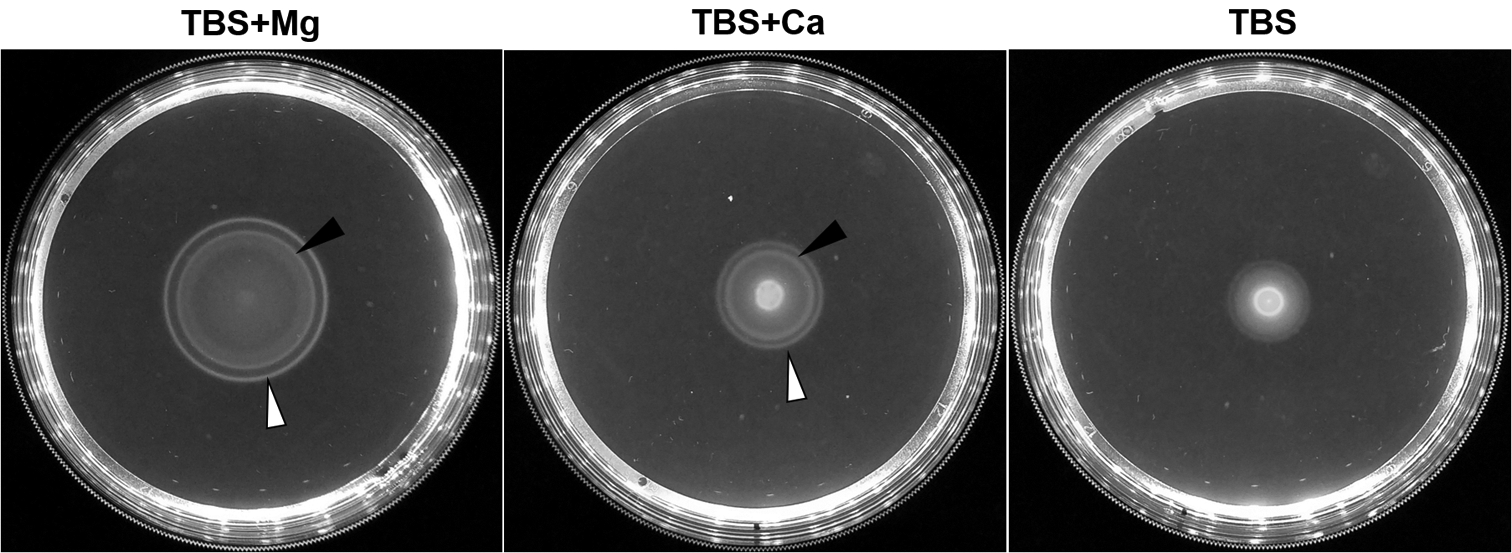
Migration of V. fischeri across TBS soft agar plates supplemented with (Left) 35 mM MgSO4 or (Middle) 10 mM CaCl2, or (Right) without supplement. The inner ring (black arrow) and outer ring (white arrow) can be readily seen for magnesium and calcium supplementation, and represent cells migrating to thymidine and serine, respectively. The rings take longer to form when cells are inoculated onto plates containing TBS without supplementation (Right).
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1)
TBS medium (see recipe)
Low percentage (0.25%) TBS-Mg or TBSW agar plate (see recipes)
Sterile applicator stick
Sterile 18 × 150-mm, capped glass culture tube
28°C shaking incubator
Spectrophotometer
24°C incubator
Ruler
Using a sterile applicator stick, pick a single V. fischeri colony from an LBS agar plate.
Inoculate an 18 × 150-mm glass culture tube containing 5 mL LBS by suspending cells from the applicator stick in the medium. Remove the applicator stick and cap the test tube.
Aerate the inoculated tube at 28°C by shaking at 220 rpm overnight.
Subculture 50 μL of the overnight culture in an 18 × 150-mm glass culture tube containing 5 mL TBS (for a 1:100 dilution of the overnight culture).
Aerate the culture by shaking at 220 rpm at 28°C until the cells have reached log phase.
Determine the optical density of the overnight culture using the spectrophotometer and normalize the culture to an OD600 of 0.2 using TBS as the diluent.
-
Spot 10 μL of the normalized culture onto either a low percentage TBS-Mg or TBSW plate.
The plate can be divided into three or four parts to spot three or four strains on a single plate.
A control strain should be included on each plate to take into account plate-to-plate variation.
-
Incubate the plate right side up at 24°C or 28°C.
Be careful and gentle with the low percentage plates as the agar gel can break easily. They should NOT be inverted during incubation.
-
At a fixed time point or hourly, measure the zone of migration using a ruler and/or image plates with a camera.
ES114 migrates rapidly through TBS-Mg (or TBSW) motility agar, visibly moving within 1 or 2 hours and reaching the edges of the petri plate within 8 or 9 hours. Two major chemotaxis rings are formed that are comprised of cells sensing thymidine (outer ring) or serine (inner ring) (DeLoney-Marino, Wolfe, & Visick, 2003).
BASIC PROTOCOL 6: LUMINESCENCE ASSAY
In the symbiosis between V. fischeri and its squid host, the squid uses the bacterially-produced light to avoid detection by predators (Jones & Nishiguchi, 2004). As a result, bacteria that cannot produce light appear to be “sanctioned” by the host, as they fail to form long-term associations with the host (Bose, Rosenberg, & Stabb, 2008; Koch, Miyashiro, McFall-Ngai, & Ruby, 2014; Visick, Foster, Doino, McFall-Ngai, & Ruby, 2000). In culture, wild type V. fischeri ES114 does not produce visibly-detectable levels of luminescence, but within the squid, the bacteria induce the production of light that is about 1000-fold higher than in laboratory culture (K. J. Boettcher & Ruby, 1990; Bose et al., 2007). This phenomenon suggests a lack of understanding of signals/conditions present in the squid (or lacking in standard laboratory culture conditions). This experimental design can be used to assess the bioluminescence produced by various strains.
Materials
V. fischeri on LBS agar plate (see Basic Protocol 1)
SWTO medium (see recipe)
Sterile applicator stick
Sterile 18 × 150-mm, capped glass culture tube
28°C shaking incubator
Sterile 250 mL baffled flask
Spectrophotometer
1.5 mL cuvette (10 mm path length)
24°C shaking incubator
Luminometer
20 mL glass (scintillation) vial or appropriately sized container for luminometer
Using a sterile applicator stick, pick a single V. fischeri colony from an LBS agar plate.
Inoculate an 18 × 150-mm glass culture tube containing 5 mL SWTO by suspending cells from the applicator stick in the medium. Remove the applicator stick and cap the test tube.
Aerate the inoculated tube at 28°C by shaking at 220 rpm overnight.
Determine the optical density of the overnight culture using the spectrophotometer. Pipette a volume of the overnight culture into 30 mL SWTO in a 250 mL baffled flask to achieve a final starting optical density of 0.005 OD600.
Aerate the flask at 24°C by shaking at 220 rpm.
-
At desired time points, take an OD600 reading and a luminescence reading. Transfer 1 mL of the culture to a 1.5 mL cuvette and measure the OD600. To take a luminescence reading, transfer the 1 mL culture to a scintillation vial or an appropriate container suitable for the luminometer. Shake the container for 5 seconds to aerate the sample, and then measure the luminescence using a luminometer.
The luminescence reaction requires oxygen. If the sample is not adequately aerated, the luminescence readings will be inaccurate.
Hourly time points can provide initial details about the dynamics of growth and luminescence. In future experiments, time points can be taken more or less frequently.
REAGENTS AND SOLUTIONS
Artificial seawater stock, 2× (ASW)
| 197.6 g | MgSO4 * 7 H4O |
| 23.2 g | CaCl2 * 2 H2O |
| 280.8 g | NaCl |
| 12 g | KCl |
Add 4 L dH2O into an 8 L carboy. Dissolve magnesium sulfate heptahydrate in 1 L dH2O and add to carboy. Dissolve calcium chloride dihydrate into 1 L dH2O and add to carboy. Dissolve sodium chloride and potassium chloride directly into the carboy. Bring the final volume in the carboy to 8 L.
The final concentrations of each salt in this stock are 100 mM MgSO4, 19.7 mM CaCl2, 600 mM NaCl, and 20.1 mM KCl.
Low percentage (0.25%) agar plates for motility
TBS-Mg
| 10 g | Tryptone |
| 20 g | NaCl |
| 2.5 g | Agar |
| 8.6 g | MgSO4 * 7 H2O |
Add the above reagents and dH2O to 1000 mL in a 2 L flask. Autoclave and allow to cool until the flask can be comfortably held. Pipette 25 mL of medium into sterile, plastic, 100 mm petri dishes. Store plates at room temperature and use the plates within two days to retain proper moisture.
These plates are very fragile and should never be inverted.
Pipetting exactly 25 mL of agar medium into each petri dish helps ensure reproducibility from plate to plate and from experiment to experiment.
V. fischeri requires magnesium for optimal motility, and its omission from the medium results in a severely decreased rate of migration through the soft agar (O’Shea et al., 2005).
TBSW
| 10 g | Tryptone |
| 350 mL | 2× ASW (see recipe) |
| 2.5 g | Agar |
Add the above reagents and add dH2O to a final volume of 1000 mL in a 2 L flask. Autoclave and allow to cool until the flask can be comfortably held. Pipette 25 mL of medium into sterile petri dishes. Store plates at room temperature and use the plates within two days to retain proper moisture.
Pipetting exactly 25 mL of agar medium into each petri dish helps ensure reproducibility from plate to plate and from experiment to experiment.
Note that the ASW is used at less than 1X in this recipe.
Calcium chloride, 1M
| 14.7 g | Calcium Chloride Dihydrate |
Add the calcium chloride dihydrate powder and dH2O to a final volume of 100 mL. Autoclave the solution and store at room temperature.
LB broth
| 10 g | Tryptone |
| 5 g | Yeast Extract |
| 10 g | NaCl |
Add the above reagents and dH2O to a final volume of 1000 mL in a 2 L flask or beaker and stir to completely dissolve solids. Distribute into clean bottles and autoclave. Store at room temperature.
LB was originally abbreviated from lysogeny broth but can be referred to in the literature as Luria-Bertani medium, as well (Bertani, 2004).
LB agar plates
| 10 g | Tryptone |
| 5 g | Yeast Extract |
| 10 g | NaCl |
| 15 g | Agar |
Add the above reagents and dH2O to a final volume of 1000 mL in a 2 L flask and autoclave the mixture. Allow to cool until the flask can be comfortably held. As desired, add an antibiotic to the cooled medium and mix well. Pour approximately 30 mL into sterile, plastic, 100 mm petri dishes and allow agar to solidify. Let the plates sit for one to two days at room temperature until sufficiently dry. Then, store the plates at 4°C for up to 3 months. Antibiotic-containing media may have a shorter shelf-life and may be light sensitive.
LB salt (LBS) broth
| 10 g | Tryptone |
| 5 g | Yeast Extract |
| 20 g | NaCl |
Add the above reagents, 975 mL dH2O, and 25 mL of 2 M Tris pH 7.5 to a 2 L flask or beaker. Stir the mixture until solids are completely dissolved. Distribute the medium into clean bottles at the desired volume. Autoclave the bottles and store at room temperature.
Some researchers include glycerol in this recipe to a final concentration of 0.3%. If added, V. fischeri will metabolize the glycerol and secrete acid into the medium. With prolonged growth under these conditions, V. fischeri will begin to die, and thus glycerol addition should be carefully considered and strain growth carefully monitored, as the Tris buffer won’t sufficiently prevent acidification produced by V. fishceri.
When performing a shaking biofilm assay, 10 mL of sterile 1 M calcium chloride can be added after autoclaving, for a final concentration of 10 mM.
LB salt (LBS) agar plates
| 10 g | Tryptone |
| 5 g | Yeast Extract |
| 20 g | NaCl |
| 15 g | Agar |
Add the above reagents, 975 mL dH2O, and 25 mL of 2 M Tris pH 7.5 to a 2 L flask. Autoclave the mixture. Allow to cool until the flask can be comfortably held. As desired, add an antibiotic to the cooled medium and mix well. Pour approximately 30 mL into sterile, plastic, 100 mm petri dishes and allow agar to solidify. Let the plates sit for one to two days at room temperature until sufficiently dry. Then, store the plates at 4°C for up to 3 months.
For solid agar-based biofilm experiments, pipette 25 mL of medium into sterile petri dishes. For calcium-induced biofilms, add 10 mL of sterile 1 M CaCl2 liquid stock to the autoclaved medium and mix well. Store plates at room temperature and use the plates within two days to retain proper moisture.
Pipetting exactly 25 mL of agar medium into each petri dish helps ensure reproducibility of biofilm assays from plate to plate and from experiment to experiment.
Minimal medium (HEPES, HMM or Tris, TMM)
| 75.3 mL | dH2O |
| 10 mL | 1 M HEPES, pH 7.5 or 1 M Tris, pH 7.5 |
| 6 mL | 5 M NaCl |
| 5 mL | 1 M MgSO4 heptahydrate |
| 100 μL | 33 mM K2HPO4 |
| 100 μL | 10 mM Ferrous Ammonium Sulfate |
| 500 μL | 20% NH4Cl |
| 1 mL | 20% D-glucose |
| 1 mL | 1 M KCl |
| 1 mL | 1 M CaCl2 dihydrate |
Filter sterilize the 1M HEPES (or 1 M Tris) stock. Prepare the water and other component stocks in individual bottles and autoclave each. Combine the appropriate volumes of sterile water, Tris, and component reagents. Use the medium immediately, or store the medium at 4°C for up to two days.
Substitute glucose for any carbon source desired.
The individual reagents can be made up separately, as listed here, so that the concentrations of each component may be adjusted depending on the experimental design. This strategy also prevents precipitation of calcium and phosphate. Alternatively, the sodium chloride, magnesium sulfate, potassium chloride, and calcium chloride solutions can be replaced with 35 ml of 2X ASW stock per 100 mL, with the amount of water adjusted accordingly.
If carbon sources are not being studied, 1 mL of a 10% casamino acids stock can be added to enhance growth in this medium.
The ferrous ammonium sulfate stock will settle to the bottom of the bottle following autoclaving and must be resuspended prior to addition to the TMM mixture.
Seawater tryptone (SWT)
| 5 g | Tryptone |
| 350 mL | 2× ASW |
| 3 g | Yeast extract |
Add the above reagents and dH2O to a final volume of 1000 mL in a 2 L bottle. Autoclave the mixture. This should be made the day before use for best effect.
Some researchers include glycerol in this recipe to a final concentration of 0.3%. If added, V. fischeri will metabolize the glycerol and secrete acid into the medium. With prolonged growth under these conditions, V. fischeri will begin to die. Thus, glycerol addition should be carefully considered, and strain growth carefully monitored.
Note that the ASW is used at less than 1X in this recipe.
Seawater tryptone (SWTO)
| 5 g | Tryptone |
| 350 mL | 2× ASW |
| 3 g | Yeast extract |
| 20 g | NaCl |
Add the above reagents and dH2O to a final volume of 1000 mL in a 2 L bottle. Autoclave the mixture. This should be made the day before use for best effect.
The additional NaCl brings the solution near to the osmolarity of seawater.
Some researchers use Instant Ocean instead of 2X ASW.
As with SWT, some researchers include glycerol in this recipe to a final concentration of 0.3%. V. fischeri will metabolize the glycerol and secrete acid into the medium, which causes cell death with prolonged exposure. However, glycerol addition may also increase luminescence production by V. fischeri.
Note that the ASW is used at less than 1X in this recipe.
Tris pH 7.5, 2 M
| 270 g | Tris HCl |
| 46 g | Tris base |
Dissolve the Tris-HCl and Tris base into 800 mL dH2O. Adjust the pH to 7.5 with HCl. Bring the final volume to 1000 mL, and store at 4°C.
TBS broth
| 10 g | Tryptone |
| 20 g | NaCl |
Dissolve the above reagents in 1000 mL dH2O in a 2 L flask or beaker. Stir the mixture until solids are completely dissolved. Distribute the medium into clean bottles at the desired volume. Autoclave the bottles and store at room temperature.
COMMENTARY
Background Information
Biofilm assays
Efficient colonization by V. fischeri depends on the ability of this microbe to form a biofilm on the surface of the squid’s symbiotic light organ and to subsequently disperse from the biofilm to migrate inside where colonization occurs (Nyholm et al., 2000; Yip et al., 2006). The ability to form a biofilm in laboratory culture strongly correlates with colonization competence, with mutants that fail to form biofilms in vitro exhibiting colonization defects and strains with enhanced biofilm formation exhibiting increased colonization proficiency (e.g., (Shibata et al., 2012; Yip et al., 2006)). Studies with genetically altered strains of ES114 with increased biofilm capability in vitro and the assays described here have permitted a mechanistic understanding of the requirement for symbiotic biofilm formation and colonization as well as the identification of the syp (symbiosis polysaccharide) genes as required for biofilm formation and symbiotic colonization.
There are numerous assays of biofilm formation. Strains of ES114 with increased biofilm capability (e.g., overexpressing rscS) form wrinkled colonies on solid agar and strong pellicles in static liquid culture (Yip et al., 2006). Null mutants defective for the syp genes form smooth colonies and weak pellicles, and they colonize squid poorly. Thus, these two assays are highly predictive of symbiotic competence. In contrast, the crystal violet assay (O’Toole, 2011) is not a good predictor of symbiotic competence (Shibata et al., 2012) and is not included here. Finally, certain strains can form biofilms upon exposure to calcium under shaking liquid culture conditions (Tischler et al., 2018). Recent work has shown that this assay also can provide insights into the requirements for symbiotic colonization (Thompson, Tischler, Tarnowski, Mandel, & Visick, 2019).
Motility assay
To date, motility is the one phenotype that appears to be absolutely critical for colonization by V. fischeri: non-motile strains fail to colonize (Graf et al., 1994). Other strains with altered motility or with chemotactic defects also have shown altered colonization dynamics (e.g., (Mandel et al., 2012; Millikan & Ruby, 2002)). While other assays exist, we present here the simplest assay for motility, tryptone-based soft agar plates. This simple assay can be performed by the most junior researcher and will yield results within a short time frame (less than six hours). By measuring the diameter of the outer migrating band of cells at specific intervals (with a low-tech instrument, a ruler), subtle defects in the motility of a given strain relative to another can be uncovered. Furthermore, this assay can reveal differences in behavior, such as differences in the response or chemotaxis to nutrients in the medium. This assay permitted both the discovery that magnesium promotes migration by ES114 and the identification of two chemoattractants, serine and thymidine, sensed by V. fischeri (DeLoney-Marino et al., 2003; O’Shea et al., 2005). This assay can be adapted by using different media such as a minimal medium to test specific nutrients as chemoattractants.
Luminescence assay
Luminescence is the key “product” of the symbiosis between V. fischeri and its squid host. Mutants of V. fischeri that are defective for light production can initially colonize, but fail to sustain a long-term colonization (Koch et al., 2014; Visick et al., 2000). Examination of lux gene expression in the light organ has revealed distinct patterns in the different crypts, a result that provides insight into the environments found in the three crypts (Dunn, Millikan, Adin, Bose, & Stabb, 2006). Finally, symbiotic ES114 produces about 1000× the amount of light relative to that which it produces in laboratory culture (K. J. Boettcher & Ruby, 1990). This phenomenon permitted the discovery of ArcA as one factor that inhibits bioluminescence in culture (Bose et al., 2007). In addition to control by ArcA, light production depends on stimulation by multiple autoinducers that are produced when the cells reach high cell density (Verma & Miyashiro, 2013). Thus, understanding regulation of light production provides insight into the behaviors of V. fischeri—and the underlying mechanism—during colonization of the light organ. The protocol included here uses a complex medium that contains high levels of sodium chloride and other salts present in seawater, as this combination of salts was shown to result in increased light production by ES114 (Bose et al., 2007).
Critical Parameters and Troubleshooting
Biofilm assay – solid agar
To obtain reproducible results within and across experiments, the agar plates should be made with a standard volume, such as 25 mL. Additionally, the plates should be fresh but dry. Differences in moisture of the agar plates can affect the ability of V. fischeri to form biofilms. Thus, the duration between pouring and use should be noted for each experiment. Finally, it is possible that different batches of tryptone or yeast extract from different lots or companies may result in differences in the exact timing in the development of the wrinkled colony phenotypes. In our biofilm work, we use Gibco™ Bacto™ Yeast Extract and Gibco™ Bacto™ Tryptone. The consistent use of the same reagents with positive and negative controls should permit reproducible data collection.
If two or more nutrients (e.g., CaCl2 and MgSO4) are to be compared for their effects on biofilm formation, one single flask of medium should be used. Following autoclaving, the medium can be split into sterile flasks, followed by delivery and mixing of specific additives. Alternatively, additives may be added to each plate individually from liquid stocks. This These approaches avoid inconsistencies that can arise from preparing independent flasks of media that may have slight differences in composition due to measurement errors or liquid loss from autoclaving. If the volume of additives is different or a no addition condition is compared, a corresponding volume of sterile water should be added to each flask or plate to ensure equivalent final volumes. These additives can be added to sterile flasks or petri dishes while the medium is autoclaving and cooling. It is critical to ensure the additives are fully integrated into the medium in the plate either by swirling or pipetting. Otherwise, uneven pockets of the nutrient may be found across the plate.
Samples should be spotted as equidistantly as possible. Proximity of the spots to one another and to the edge of the plate may affect their phenotypes. Do not puncture the agar or introduce air bubbles when spotting.
Biofilm assay – shaking liquid culture
The size of tube can affect results of this assay. A biofilm-competent V. fischeri strain grown in a 13 × 100-mm glass tube with 2 mL medium often results in a strong biofilm with a clear culture. However, the same strain in an 18 × 150-mm glass tube with 5 mL medium will often show reduced biofilm phenotypes, with the culture retaining some turbidity. A positive control with a known behavior should be used whenever altering conditions.
Biofilm assay – static liquid culture
Cultures grown in the outer wells in a 24-well plate may not produce the same pellicle phenotypes as the same strains grown in the inner wells, and thus the outer wells should not be used for bacterial culture. The differences in aeration and/or evaporation rates between these inner and outer wells may account for the observed differences in pellicle formation. Placing the plate in a sealed plastic bag or plastic box helps to mitigate these differences. In two to four outer wells, uninoculated medium controls should be added that flank the samples. These negative controls help to ensure that there was no contamination during pipetting into the wells.
Take care when moving the plate once pellicles have formed. Jostling of the plate may cause the pellicles to detach from the sides of the well.
When disrupting pellicles with a toothpick, the pellicle may stick to the toothpick. To overcome this issue, pull the adherent part of the pellicle to the edge of the well in the direction of disruption. Then, gently wipe the pellicle off of the toothpick on the edge. Repeat until the pellicle has dislodged from the toothpick.
Motility assay
To obtain consistent results, exactly 25 mL of agar should be dispensed into each petri plate. These plates must be made fresh and used within 1 – 2 days after pouring. Differences in moisture can have significant effects on reproducibility between experiments. Once solidified, care must be taken with motility plates as the low percentage agar is very easily broken. Do not puncture the agar when spotting. Ideally, if multiple plates are used in a single experiment, each plate should have a control spot, such as a wild-type strain, as a standard to permit comparisons across plates. The experiment thus typically ends when a wild-type control spreads to or into the growth of a less motile strain. If it is necessary to verify that a strain is fully non-motile, then that strain may also be placed on its own plate to permit extended incubation.
V. fischeri requires magnesium for optimal motility. Tryptone powder from different companies or lots from the same company may have differing amounts of magnesium, which may influence migration. Growth of the cells in LBS instead of TBS prior to spotting may affect the migration rates and/or patterns of migration.
Luminescence assay
It is essential that the samples are aerated for 5 seconds prior to reading luminescence (by vigorous pipetting or shaking the vial). The aeration is required for an accurate measurement of light produced.
Understanding Results
Biofilm assay – solid agar
Biofilm competent colonies may be adherent to the plate, cohesive, or produce architecture/wrinkles (Figure 2). Strains that cannot form biofilms will be smooth and lack adherence, cohesion, or architecture (Figure 2A,D). The degree of wrinkling can vary from a minor phenotype (Figure 2B) to a strong phenotype (Figure 2C) for different strains at the same time point. Disrupting a spotted colony with a toothpick is a critical tool for assessing biofilm formation, as some smooth-appearing colonies are cohesive (and thus categorized as biofilm-forming) and some colonies exhibit a non-smooth architecture but remain non-cohesive (and thus categorized as non-biofilm forming). Furthermore, a time course provides valuable information about the dynamics of biofilm formation.
Biofilm assay – shaking liquid culture
A strain of V. fischeri that cannot form a biofilm in this assay will remain as a turbid culture. If a strain can form a biofilm (Figure 3), the culture will be mostly clear with a ring at the air-liquid interface, a clump at the bottom of the test tube, or both. When both are present, a stringy “tree” can connect from the ring to the clump. For biofilm-competent derivatives of ES114, the ring at the top of the tube depends primarily on the polysaccharide cellulose, while the clump at the bottom of the tube depends primarily on the Syp polysaccharide (Tischler et al., 2018).
Biofilm assay – static liquid culture
The expectation is that a biofilm-forming strain will form a pellicle that extends across the air-liquid interface of the well. Like in the solid agar biofilm assay, these pellicles may be smooth or have wrinkles. After disruption with a toothpick, a pellicle may tear or remain coherent. In some cases, the pellicle may stick to the toothpick. The stickiness and cohesion of the pellicle is primarily imparted by the polysaccharide Syp (Shibata et al., 2012).
Motility assay
Upon spotting normalized cultures onto motility agar plates, the bacteria will swim outward from that spot. Hourly measurement of the diameter of the spot quantifiably measures migration by the bacteria. The distance that the bacteria migrate from the initial spot depends on when the cells synthesize flagella as well as the frequency of the bacteria swimming in a single direction. Monitoring the distance moved over time can help to inform whether the cells start swimming earlier or whether they swim faster.
Luminescence assay
Strains may either produce luminescence or they may not. For the strains that do luminesce, luminescence should increase with optical density. At some point, the luminescence may plateau and/or may decrease.
Time Considerations
Growth of V. fischeri from frozen stocks
The time to grow V. fischeri from frozen stocks is very short at permissive conditions: after streaking and placing the plate at 28°C, a healthy V. fischeri strain will produce large colonies after an average overnight of 16 hours, though colonies may arise in even less time. If the cells are incubated at 24°C or room temperature, it will take more than 16 hours for the development of large colonies. On antibiotic-containing media, resistant colonies will take longer to achieve the same size as their counterparts on LBS alone.
Growth of V. fischeri in rich, undefined liquid medium
V. fischeri cultures started from frozen stock or taken from a single colony on a plate will take very little time (~8 hours) to achieve high optical densities. Routinely, cultures incubated at 24°C or 28°C should be grown overnight for no longer than 16–18 hours to preserve the health of the culture.
Growth of V. fischeri in minimal medium
As with many other bacteria, V. fischeri grows more slowly in minimal medium than it does in rich medium. The transition from a frozen stock or a rich, plate-bound lifestyle may be stressful for the bacteria and the growth may be poor or variable. However, overnight growth at 24°C or 28°C in this medium for 14–18 hours often will yield dense cultures, with an OD of about 1.0–1.2, that are readily subcultured the next day. A shorter overnight growth period of around 14 hours results in “young” cells that have not yet reached late stationary phase and will re-grow more quickly and reliably in the subculture, an approach that can substantially reduce the lag phase of growth. When V. fischeri is growing well in minimal medium, the culture will reach an OD of about 0.5 in about 5 – 6 hours following a 1:50 dilution from an overnight culture; higher inocula will decrease the time to reach that optical density.
Storage of V. fischeri in frozen stocks
Frozen stocks are typically generated with fully turbid cultures (i.e., OD600 > 3) of V. fischeri, which can take as little as 8 hours or as many as 16–18 hours to obtain. Thus, the time required to take a colony from a plate and prepare a frozen stock depends on how long it takes to obtain a turbid culture. Glycerol cryovials should be prepared in advance by pipetting the 80% glycerol solution into plastic or glass cryovials and sterilizing by autoclaving. Alternatively, the 80% glycerol and cryovials can be sterilized separately, and then the 80% glycerol can be transferred into the sterile cryovials.
Biofilm assay – solid agar
The time required to perform a biofilm assay is between 2 and 3 days of setup with 3 to 5 days of analysis. For a quicker exploratory experiment, an overnight culture can be started directly from frozen cells on the first day. On the second day, the overnight culture is subcultured 1:100 in fresh medium for about 1 to 2 hours. Depending on the number of strains being spotted, the optical density readings, normalization, and spotting could take minutes or up to an hour. To achieve accuracy in assessments of the timing of biofilm formation, experiments should be designed to have sufficiently few strains/conditions such that it takes less than one hour to spot the cultures. To speed up this process, tubes and plates can be labeled before or during growth of the subculture. For a more precise experiment, the first day will instead require streaking the cells from frozen stocks onto LBS plates and then the work can proceed as described for the subsequent two days.
After spotting onto the plates, analysis can be performed at a single time point each day for up to 5 days. The amount of time each day for analysis and imaging will be proportional to the number of spots. In some cases, differences will be observed in a time frame less than 24 hours, and thus subsequent assessment could be hourly from, for example, 16 to 24 hours.
Biofilm assay – shaking liquid culture
The shaking liquid culture biofilm assay will take 3 to 4 days. Like the solid agar biofilm assay, this timing depends on whether frozen cells are directly inoculated to make an overnight culture (1 day) or instead are streaked onto a plate for single colonies and the following day inoculated into a liquid medium (2 days). On the following day, the overnight culture will be normalized and subcultured. After a specified (consistent) period of incubation time (e.g., 16 or 24 hours), the tubes can be assessed for biofilm formation.
Biofilm assay – static liquid culture
Starting with single colonies on a plate, a static liquid culture assay will take 2 days with 1 to 3 days for analysis. The day following growth of the overnight culture, optical densities of each strain are determined and subcultured into a 24-well plate containing fresh medium. The pellicles may develop after only 24 hours, or they may take up to three days. The length of time of analysis is dependent on the number of wells used.
Motility assay
The motility assay is more time-intensive than the biofilm assays if performing a time course. If starting with single colonies on a plate, the assay will take only take 2 days. On the first day, a single colony will be used to start an overnight culture in TBS. The low percentage agar plates should also be made on this day. On the second day after subculturing from the overnight culture, the culture will be grown for 1 to 3 hours to achieve an OD600 of 0.2. Depending on the number of strains or conditions, normalization of the cultures and spotting may only take minutes and should take less than an hour. Once the spotted plates are incubating, the experiment may continue for as many hours as desired. Every time point taken requires measurement of the diameter of each spot, which takes an amount of time proportional to the number of spots. The duration of migration will depend on how closely strains were inoculated onto a single plate, as the migrating cells will run into each other, preventing further accurate measurement.
Luminescence assay
Like the motility assay, the assay will only take 2 days when starting from single colonies on a plate. The first day requires starting an overnight culture. The subsequent day, optical densities are taken to permit standardization of the subculture. The subcultured cells are grown throughout the day, with time points taken each hour or potentially more frequently. The duration of each time point will depend on the number of samples, as each sample requires time for harvesting, an optical density reading, and a luminescence reading.
Acknowledgements.
We thank members of the lab for their comments on these protocols. We thank Kevin Grudzinski for the pictures of the smooth and wrinkled colony spots. This work was supported by NIH grant 1R35 GM130355.
Literature Cited
- Aschtgen M-S, Brennan CA, Nikolakakis K, Cohen S, McFall-Ngai M, & Ruby EG (2019). Insights into flagellar function and mechanism from the squid–vibrio symbiosis. npj Biofilms and Microbiomes, 5(1), 32. doi: 10.1038/s41522-019-0106-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G (2004). Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol, 186(3), 595–600. doi: 10.1128/jb.186.3.595-600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, & Ruby EG (1990). Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol, 172(7), 3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, & Ruby EG (1994). Occurrence of plasmid DNA in the sepiolid squid symbiont Vibrio fischeri. Current Microbiology, 29(5), 279–286. doi: 10.1007/BF01577441 [DOI] [Google Scholar]
- Bongrand C, Koch EJ, Moriano-Gutierrez S, Cordero OX, McFall-Ngai M, Polz MF, & Ruby EG (2016). A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J, 10(12), 2907–2917. doi: 10.1038/ismej.2016.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongrand C, & Ruby EG (2019). Achieving a multi-strain symbiosis: strain behavior and infection dynamics. ISME J, 13(3), 698–706. doi: 10.1038/s41396-018-0305-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, …Stabb EV, (2007). Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol, 65(2), 538–553. doi: 10.1111/j.1365-2958.2007.05809.x [DOI] [PubMed] [Google Scholar]
- Bose JL, Rosenberg CS, & Stabb EV (2008). Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol, 190(2), 169–183. doi: 10.1007/s00203-008-0387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman KM, Cecere AG, Miyashiro T, Septer AN, & Mandel MJ (2019). Draft genome sequences of type VI secretion system-encoding Vibrio fischeri strains FQ-A001 and ES401. Microbiol Resour Announc, 8(20). doi: 10.1128/MRA.00385-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ML, Mashanova EV, Rosen NM, & Soto W (2019). Adaptation to temperature stress by Vibrio fischeri facilitates this microbe’s symbiosis with the Hawaiian bobtail squid (Euprymna scolopes). Evolution, 73(9), 1885–1897. doi: 10.1111/evo.13819 [DOI] [PubMed] [Google Scholar]
- DeLoney-Marino CR, Wolfe AJ, & Visick KL (2003). Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl Environ Microbiol, 69(12), 7527–7530. doi: 10.1128/aem.69.12.7527-7530.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, & Stabb EV (2006). New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol, 72(1), 802–810. doi: 10.1128/AEM.72.1.802-810.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, & Ruby EG (1994). Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol, 176(22), 6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, & Ruby EG (1998). Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci U S A, 95(4), 1818–1822. doi: 10.1073/pnas.95.4.1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, & Nishiguchi MK (2004). Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Marine Biology, 144(6), 1151–1155. doi: 10.1007/s00227-003-1285-3 [DOI] [Google Scholar]
- Koch EJ, Miyashiro T, McFall-Ngai MJ, & Ruby EG (2014). Features governing symbiont persistence in the squid-vibrio association. Mol Ecol, 23(6), 1624–1634. doi: 10.1111/mec.12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler S, Gaedeke R, Thompson C, Bongrand C, Visick KL, Ruby E, & McFall-Ngai M (2018). The model squid-vibrio symbiosis provides a window into the impact of strain- and species-level differences during the initial stages of symbiont engagement. Environ Microbiol. doi: 10.1111/1462-2920.14392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, & Ruby EG (1994). Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol, 60(5), 1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EA, Deloney-Marino CR, McFall-Ngai MJ, & Ruby EG (2012). Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol, 78(13), 4620–4626. doi: 10.1128/AEM.00377-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden AE, Grudzinski K, Ondrey JM, DeLoney-Marino CR, & Visick KL (2017). Impact of salt and nutrient content on biofilm formation by Vibrio fischeri. PLoS One, 12(1), e0169521. doi: 10.1371/journal.pone.0169521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ, & Ruby EG (1991). Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science, 254(5037), 1491–1494. doi: 10.1126/science.1962208 [DOI] [PubMed] [Google Scholar]
- Millikan DS, & Ruby EG (2002). Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl Environ Microbiol, 68(5), 2519–2528. doi: 10.1128/aem.68.5.2519-2528.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth JC, Guo H, Koch E, Heath-Heckman EAC, Hermanson JC, Ruby EG, … McFall-Ngai M (2017). Motile cilia create fluid-mechanical microhabitats for the active recruitment of the host microbiome. Proc Natl Acad Sci U S A, 114(36), 9510–9516. doi: 10.1073/pnas.1706926114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi M (2001). Cospeciation between hosts and symbionts: The sepiolid squid-Vibrio mutualism In Seckbach(Ed.), Symbiosis (pp. 757–774): Kluwer Academic Publishers. [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, & McFall-Ngai MJ (2000). Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A, 97(18), 10231–10235. doi: 10.1073/pnas.97.18.10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea TM, Deloney-Marino CR, Shibata S, Aizawa S, Wolfe AJ, & Visick KL (2005). Magnesium promotes flagellation of Vibrio fischeri. J Bacteriol, 187(6), 2058–2065. doi: 10.1128/JB.187.6.2058-2065.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA (2011). Microtiter dish biofilm formation assay. J Vis Exp(47). doi: 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray VA, Morris AR, & Visick KL (2012). A semi-quantitative approach to assess biofilm formation using wrinkled colony development. J Vis Exp(64), e4035. doi: 10.3791/4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, & Nealson KH (1976). Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica; a model of symbiosis based on bacterial studies. Biol Bull, 151(3), 574–586. doi: 10.2307/1540507 [DOI] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, … Greenberg EP (2005). Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A, 102(8), 3004–3009. doi: 10.1073/pnas.0409900102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Yip ES, Quirke KP, Ondrey JM, & Visick KL (2012). Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol, 194(24), 6736–6747. doi: 10.1128/JB.00707-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, … Septer AN (2018). Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci U S A, 115(36), E8528–E8537. doi: 10.1073/pnas.1808302115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, & Visick KL (2013). Vibrio fischeri: A bioluminescent light-organ symbiont of the bobtail squid Euprymna scolopes In Rosenberg EDEF., Lory S, Stackebrandt E, Thompson F (Ed.), The Prokaryotes (pp. 497–532). Berlin, Heidelberg: Springer. [Google Scholar]
- Sun Y, LaSota ED, Cecere AG, LaPenna KB, Larios-Valencia J, Wollenberg MS, & Miyashiro T (2016). Intraspecific competition impacts Vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl Environ Microbiol, 82(10), 3082–3091. doi: 10.1128/AEM.04143-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Tischler AH, Tarnowski DA, Mandel MJ, & Visick KL (2019). Nitric oxide inhibits biofilm formation by Vibrio fischeri via the nitric oxide sensor HnoX. Mol Microbiol, 111(1), 187–203. doi: 10.1111/mmi.14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AH, Lie L, Thompson CM, & Visick KL (2018). Discovery of calcium as a biofilm-promoting signal for Vibrio fischeri reveals new phenotypes and underlying regulatory complexity. J Bacteriol, 200(15). doi: 10.1128/JB.00016-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanczyk H, Ast JC, Higgins MJ, Carson J, & Dunlap PV (2007). Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int J Syst Evol Microbiol, 57(Pt 12), 2823–2829. doi: 10.1099/ijs.0.65081-0 [DOI] [PubMed] [Google Scholar]
- Verma SC, & Miyashiro T (2013). Quorum sensing in the squid-Vibrio symbiosis. Int J Mol Sci, 14(8), 16386–16401. doi: 10.3390/ijms140816386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Foster J, Doino J, McFall-Ngai M, & Ruby EG (2000). Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol, 182(16), 4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Hodge-Hanson KM, Tischler AH, Bennett AK, & Mastrodomenico V (2018). Tools for rapid genetic engineering of Vibrio fischeri. Appl Environ Microbiol, 84(14). doi: 10.1128/AEM.00850-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé RP, Schaefer AL, Koroleva I, …McFall-Ngai MJ(2010). Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A, 107(5), 2259–2264. doi: 10.1073/pnas.0909712107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Geszvain K, DeLoney-Marino CR, & Visick KL (2006). The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol, 62(6), 1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x [DOI] [PMC free article] [PubMed] [Google Scholar]


