Abstract
Many highly ordered complex systems form by the spontaneous self-assembly of simpler subunits. An important biophysical tool that relies on self-assembly are Nanodiscs, which find extensive use as native-like environments for studying membrane proteins. Nanodiscs are self-assembled from detergent-solubilized mixtures of phospholipids and engineered helical proteins called membrane scaffold proteins (MSPs)1. Detergent removal results in the formation of nanoscale bilayers stabilized by two MSP “belts.” Despite their numerous applications in biology, and contributions from many laboratories world-wide, little is known about the self-assembly process such as when the bilayer forms or when the MSP associates with lipids. We use fluorescence and optical spectroscopy to probe self-assembly at various equilibria defined by the detergent concentration. We show that the bilayer begins forming below the critical micellar concentration of the detergent (10 mM), and the association of MSP and lipids begins soon at lower detergent levels, showing a dependence on the concentrations of MSP and lipids. Following the dissolution process by adding detergent to purified Nanodiscs demonstrates that the self-assembly is reversible. Our data demonstrate that Nanodisc self-assembly is experimentally accessible, and that controlling the detergent concentration allows exquisite control over the self-assembly reaction. This improved understanding of self-assembly could lead to better functional incorporation of hitherto intractable membrane protein target proteins.
Keywords: Nanodiscs, self-assembly, fluorescence, optical spectroscopy
Graphical Abstract
Fluorescence and optical spectroscopy reveals that Nanodisc assembly is thermodynamically reversible and controllable.
Introduction
Self-assembly occurs widely in physical, chemical, and biological systems, from the formation of crystals to the assembly of the lipid bilayer2. The formation of ordered structures from weakly interacting components plays a key role in the chemistry of life. The base pairing of DNA, folding of proteins, and crystal formation all rely on the thermodynamic assembly of biochemical components giving rise to complex structures and new functions. Aside from its ubiquity in natural processes, one can apply the principles of self-assembly to the fabrication of new materials, circuits, and biological structures2. Important biochemical applications of self-assembly are self-assembled phospholipid systems such as self-assembled monolayers (SAMs)3, supported lipid bilayers (SLBs)4, liposomes5, and Nanodiscs1. While each can be used to study biophysical and biochemical processes that occur at or near the membrane, the widely used Nanodisc system provides precise control over both lipid content and protein stoichiometry.
Nanodisc technology entails the self-assembly of phospholipids and membrane scaffold proteins (MSPs) into a nanometer-scale bilayer which is stabilized by two encircling MSP “belts.” Reconstitution of membrane protein targets into the Nanodisc requires only the addition of the detergent-solubilized target to the self-assembly reaction. Once all components are detergent-solubilized, the reaction proceeds spontaneously upon the removal of detergent6. Purification of the reaction by HPLC results in a population of Nanodiscs with the target protein reconstituted into a native-like environment. This technique has been applied to study many different classes of membrane proteins7–11 and the assembly of oligomeric peptides at the membrane surface12 as has been recently reviewed13,14. Other kinds of nanodiscs include those that are polymer-based, which can also be assembled from mixtures of liposomes and amphiphilic polymers15,16, and peptide-based nanodiscs, which are formed from the self-assembly of peptide and lipids without detergent17. We reserve the capital ‘Nanodisc’ term for engineered MSP-based discs for clarity. Peptide-based nanodiscs have been shown to undergo lipid exchange over the course of several hours after preparation18, indicating that these nanodiscs are highly dynamic following lipid/peptide mixing. Polymer based nanodiscs have been shown to be devoid of a phase transition19, and the polymers can directly solubilize phospholipid vesicles20 unlike MSP. On the other hand, extensive studies on MSP-based Nanodiscs document a clear phase transition as observed by x-ray scattering, fluorescence, and calorimetry21,22.
Despite the wide applications of Nanodiscs to membrane biochemistry, little is known about the process of Nanodisc assembly, and protein targets sometimes aggregate irreversibly after detergent removal, leading to prohibitively low yield of functional protein in Nanodiscs. Published guidelines and protocols are mostly empirical, with each target requiring a trial-and-error approach to obtain maximum yield of protein in Nanodiscs23, and some work has been done to optimize the yield of certain CYPs10 and bacteriorhodopsin7. Binary systems of phospholipids and detergents have been investigated in depth both experimentally and theoretically24,25, and several models for bilayer solubilization have been proposed26. More complex systems that include human HDL27 or cyclodextrin-induced protein reconstitution28 have also been studied. While much is known empirically about how one can reconstitute Nanodiscs and incorporate targets, the actual process of assembly remains experimentally unexplored. Detailed knowledge of this process, such as when the lipid bilayer forms, or when the MSP first associates with the nascent bilayer, could lead to better reconstitution guidelines and a better understanding of the “branch point” where target proteins either aggregate or assemble into Nanodiscs. This in turn might open new protein targets to structural and functional characterization.
In this work, we apply fluorescence and optical spectroscopy to probe the self-assembly process of simple Nanodiscs composed of phospholipid and membrane scaffold protein (MSP) as detergent is removed. Using the hydration-sensitive dye laurdan, we demonstrate that the lipid bilayer begins forming below the critical micelle concentration (CMC) of the detergent used (sodium cholate). We also apply fluorescence anisotropy of a long-lifetime dye covalently attached to the MSP to show that the MSP and lipids form a larger complex near 5 mM cholate concentration. Second derivative optical spectroscopy shows that the hydration of MSP tyrosine residues decreases as detergent is removed. Our data collectively show that we can observe the self-assembly process at various equilibria defined by the detergent concentration, and that we can control the self-assembly reaction by precisely controlling the amount of detergent.
Experimental
Materials
All buffers were prepared with MilliQ H2O (MilliPoreSigma, Burlington, MA) with 18.2 MOhm resistance/cm. Sodium cholate was purchased from Anatrace (Anatrace, Maumee, OH). The fluorescent dye ([Ru(bpy)2(5-iodoacetamido-1,10-phenanthroline)](PF6)2), herein RuBPY, was synthesized according to published methods29. Laurdan was purchased in powder form from Molecular Probes (Molecular Probes, Eugene, OR). The dye was dissolved in methanol before use and stored at room temperature protected from light. Dimyristoyl phosphatidyl choline (DMPC) was purchased in powder form from NOF Corporation (NOF Corporation, Tokyo, Japan). Lipids were dissolved in chloroform and concentration was measured using a phosphate analysis30. Lipid concentrations were determined by the following procedure: separate aliquots of both 1 μL and 2 μL lipid stock were drawn using a glass syringe. 225 μL of 8.9N sulfuric acid was added and the tubes were heated at 225°C for 25 minutes, followed by cooling and addition of 75 μL 30% hydrogen peroxide, followed by another round of heating for 30 minutes. After cooling to room temperature, 1.95 mL of deionized water was added, then 250 μL of 2.5% ammonium molybdate (VI). Tubes were then mixed by vortexing, followed by the addition of 250 μL of 10% ascorbic acid and another round of vortexing. After a final round of heating at 100°C for 7 minutes, the tubes are allowed to cool to room temperature, and samples are drawn for reading at 820 nm on a microplate reader. A standard curve is measured along with the lipid stock to determine the concentration of the lipid stock. MSP1D1 was expressed and purified according to published protocols23. The 7X His-tag was cleaved using tobacco etch virus (TEV) protease at 35°C for 4 hours in a water bath while agitating. The reaction proceeded overnight at 4°C, after which the cleaved protein was purified via Ni-NTA affinity chromatography. Protein concentration was measured via spectrophotometry using the published extinction coefficient at 280 nm (18435 M−1cm−1)23. To produce fluorescent MSP, a cysteine mutant of MSP1D1 (D73C) was purified using the same protocol discussed above. The 7X His-Tag was cleaved before labeling. The protein sulfhydryls were reduced with a fourfold molar excess of tris(2-carboxyethyl)phosphine (TCEP) for 15 minutes while stirring at room temperature. A tenfold molar excess of 30mM solution of RuBPY dye dissolved in dry DMSO was added drop by drop. The reaction continued for 4 hours at room temperature and was moved to 4°C overnight on a stir plate. The reaction was purified the next day on a G-25 gel filtration column into 20 mM HEPES, 150 mM NaCl, pH 7.3. Labeling efficiency and protein concentration were measured using spectrophotometry and a published correction factor of 3.8829. According to the Beer-Lambert law, the total absorbance of the labeled protein at 280 nm is
Where and are the molar extinction coefficients of the protein and dye at 280 nm and Cprotein and CDye are the molar concentrations of protein and dye. The reported correction factor is equal to the ratio ε280/ε450 for the ruthenium dye. If one measures the absorbance at 450 nm (the absorbance peak of the dye, which is not confounded by protein absorbance), then the concentration of the dye can be measured independently, since the extinction coefficient of the dye is known (16,600 M−1cm−1):
Multiplying the above by the above equation by the correction factor CF results in
and substituting this into the equation for total absorbance results in
where A280 is the total absorbance recorded at 280 nm. The final concentration of MSP after labeling was measured as 65 μM. We use the same method to measure the concentration of labeled Nanodiscs after elution from HPLC, and dilutions are prepared of either 500 nM or 5 μM MSP concentration.
Nanodisc Reconstitution
To make Nanodiscs for the disassembly experiments, we followed standard published protocols for the production and HPLC purification of Nanodiscs1. Briefly, lipids were drawn from chloroform stocks in the desired amount into a glass tube and dried briefly under nitrogen and for at least 4 hours under vacuum. For experiments requiring laurdan-labeled Nanodiscs, laurdan dissolved in methanol was added along with the lipids, and the mixture was dried together. Laurdan was added in a 1:2 ratio with the MSP. Buffer with detergent (0.1 M potassium phosphate, 50 mM NaCl, 50 mM Na cholate, pH 7.4) was then added and the lipid or lipid/dye mixture was solubilized by repeated vortexing and sonication. MSP was added in a 1:85 molar ratio with lipids for all experiments. For all experiments, we used MSP1D1(−) which corresponds to MSP1D1 with the 7X-His tag removed. For the anisotropy experiments, we used a fluorescent MSP1D1(−) labeled with RuBPY. Amberlite XAD-2 beads were added in a ratio of 0.5g beads per mL of total volume. Reactions were then left shaking at room temperature for at least 4 hours. Nanodiscs were then purified via HPLC into 20 mM HEPES, 150 mM NaCl, pH 7.3. Peak fractions were stored at 4°C until needed. For experiments requiring higher concentration of Nanodiscs, peak fractions from separate injections were pooled and concentrated using 10K MWCO filters. For all experiments in this work, error bars are reported. These error bars refer to independent samples, not repeated measurements of the same sample.
Nanodisc Disassembly Experiments
To disassemble Nanodiscs, we titrated sodium cholate into purified Nanodiscs. Samples were left stirring for 5 minutes (laurdan and anisotropy experiments) or were mixed using several rounds of pipetting (tyrosine absorbance experiments). Each aliquot was added sequentially. We repeated each experiment three times; each replicate was an independently prepared sample drawn from the HPLC fractions.
Nanodisc Self-Assembly Experiments
To perform the self-assembly experiments, we created reconstitution mixtures as outlined in the “Nanodisc reconstitution” section (see above) but stopped before adding the detergent-removing beads. We drew small aliquots of this mixture and diluted it into buffer to decrease the cholate concentration. We mixed one buffer with detergent (0.1 M potassium phosphate, 50 mM NaCl, 50 mM Na cholate) and the same buffer without detergent to achieve intermediate values of cholate. Each experimental data point therefore represents a separate aliquot of self-assembly reconstitution mixture. This allowed us to vary the cholate concentration while maintaining the same concentration of MSP, lipids, and dye in each sample.
Laurdan Generalized Polarization
Laurdan generalized polarization (GP) was measured using the K2 fluorometer from ISS (ISS, Champaign, IL) using a 376 nm LED with a 376 nm interference filter. Each individual sample was measured at least 5 times at each wavelength (440 nm and 490 nm) for each concentration of cholate. Temperature was maintained at 20°C with a water bath. Data were analyzed using MATLAB scripts.
MSP Lifetime and Anisotropy Measurements
Time-domain lifetimes were measured with a 20 μs window and fitted to a single exponential. Anisotropy measurements were made using the K520 digital frequency domain system on our K2 fluorometer. A fundamental frequency of 50 kHz was selected, and 30 harmonics were generated up to 1.5 MHz. Screens and a neutral density filter were used to reduce intensity incident on the photomultiplier tubes. The phase component of the signal was fit to a single rotational correlation time according to standard fitting methods31. Data analysis and fitting were performed using VINCI software supplied with the K2 fluorometer. Temperature was maintained at 20°C with a water bath for all fluorescence experiments.
Tyrosine 2nd Derivative Absorbance Spectroscopy
Absorbance of the MSP during self-assembly and disassembly experiments was measured using a Cary UV/Vis spectrophotometer. The scan was performed from 270 nm – 300 nm with a 10 s averaging time, 1 nm slit width, and 0.5 nm resolution. Spectra were measured at ambient temperature (22° C). Each sample was scanned three times and averaged to produce a single data point. This procedure was repeated three times for each experiment (assembly and disassembly), and the standard deviations reported in this work refer to the fitting results of different samples, not the different scans of individual samples. To fit the data, we used published equations to calculate the degree of tyrosine hydration using the 2nd derivative of the absorbance signal using peaks at 287 nm, 283 nm, 295 nm, and 290.5 nm32. The 2nd derivative spectra were interpolated to 0.25 nm resolution and smoothed before fitting the peak ratios to the model. Fitting was performed using a MATLAB script.
Results
The Lipid Bilayer Begins Forming Immediately Below the CMC of Cholate
To probe the formation of the bilayer during Nanodisc assembly, we chose the lipophilic dye laurdan for its hydration-sensitive emission spectrum. The emission shifts from 440 nm to 490 nm upon solvent exposure, allowing one to measure the extent of hydration in the microenvironment of the dye as given by the generalized polarization (GP)33:
We measured the steady-state GP of laurdan as a function of detergent concentration (sodium cholate in this work). By diluting the self-assembly reconstitution reaction directly in a fluorescence cuvette, we lower the concentration of detergent while holding the amount of lipids and MSP constant. The GP changes from −1.5 to 0.4 as cholate is diluted (Fig. 1). This indicates that the hydration near the lipids decreases as cholate is removed and signifies the formation of the lipid bilayer. This change in hydration apparently begins once the concentration of cholate decreases below its critical micellar concentration (CMC) value. Once the detergent concentration reaches approximately 4 mM, the GP stabilizes near 0.4, indicating bilayer formation is mostly finished. This data demonstrates that by manipulating the amount of cholate, one can exercise precise control over Nanodisc self-assembly, and that one can observe the process using steady-state spectroscopic measurements.
Figure 1:
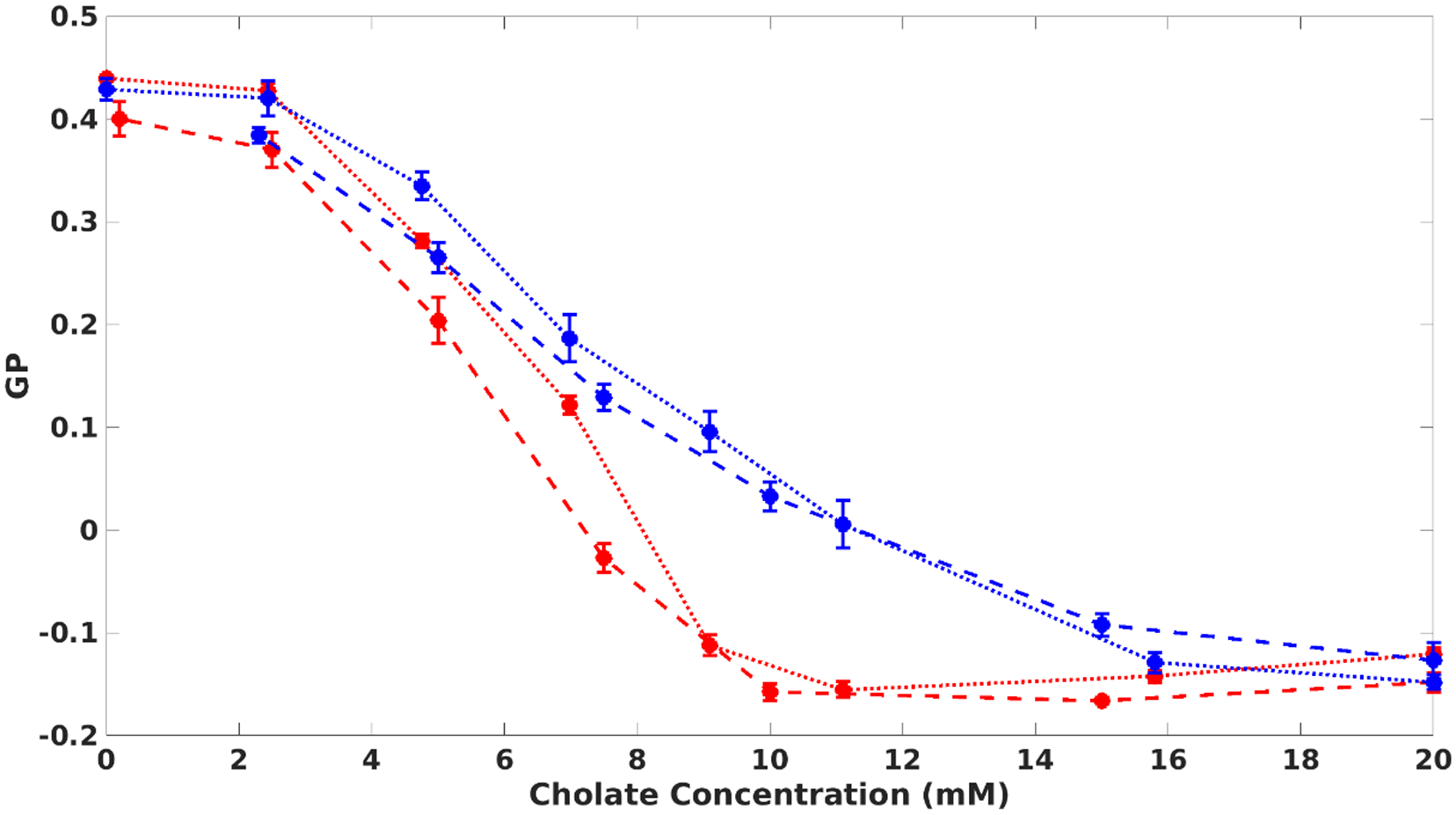
The generalized polarization (GP) of laurdan probes the formation of the lipid bilayer during Nanodisc assembly and disassembly. As cholate is removed, the GP increases from approximately −1.5 to 0.4. As cholate is added to purified Nanodiscs, the GP decreases. Dashed lines represent self-assembly experiments; dotted lines are disassembly. The concentration of MSP was either 500 nM (red) or 5 μM (blue).
We hypothesized that Nanodisc self-assembly is a thermodynamic process, and that it should be possible to observe the GP of laurdan decrease when Nanodiscs are “disassembled” by adding detergent to preformed Nanodiscs. We titrated small aliquots of sodium cholate into purified Nanodiscs and indeed observed decreasing laurdan GP with increasing cholate (Fig. 1, red dotted line). In the disassembly experiment, the concentration of MSP and lipids changes little over the course of the experiment. The GP measurements for these self-assembly and disassembly experiments (Fig. 2) show that each data point obtained during assembly and disassembly experiments represents a point of thermodynamic equilibrium, with cholate concentration acting as the thermodynamic coordinate. To our knowledge, this represents the first direct evidence that Nanodisc formation is a thermodynamic process. Additionally, experiments performed with different amounts of MSP and lipids show that the process is concentration-dependent, with bilayer formation apparently starting at higher cholate concentrations when more MSP and lipids are present (Fig. 1, blue and red). This is expected since at higher concentrations of MSP and lipids, the ratio of detergent to Nanodisc materials is lower, which means that more detergent must be present to achieve the same degree of solubilization. These results provide evidence that Nanodisc formation is thermodynamically reversible, that the bilayer formation occurs below the CMC of the detergent, and that the detergent to Nanodisc material ratio influences the course of assembly.
Figure 2:
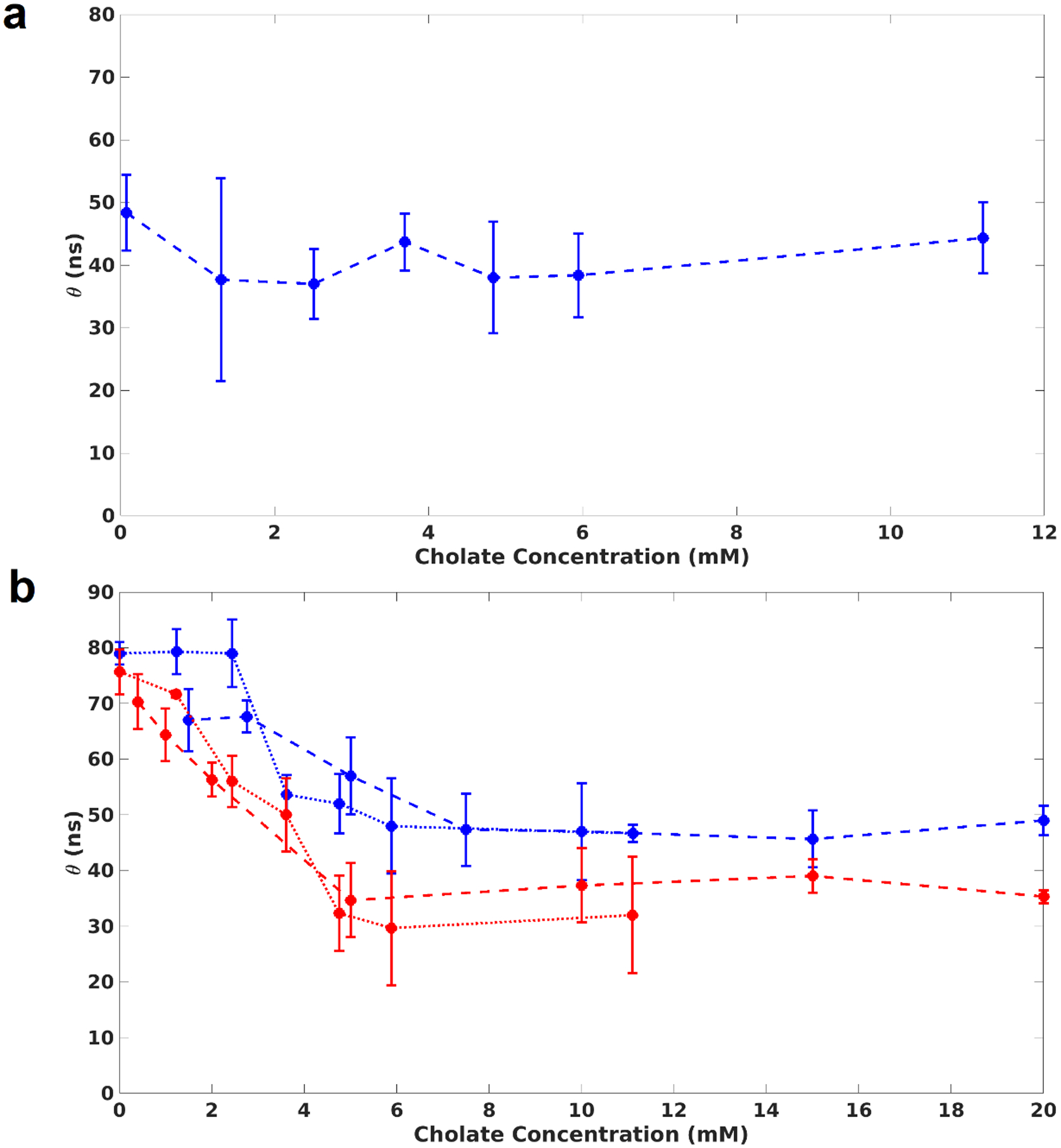
Rotational correlation time of MSP during assembly and disassembly. (a): MSP1D1(−), the MSP used in all experiments in this work, does not aggregate when cholate is removed. (b): Overlaid rotational correlation times measured during self-assembly and disassembly of Nanodiscs. The rotational correlation time changes significantly when the cholate concentration is lower than 5 mM. Dashed lines represent self-assembly experiments; dotted lines are disassembly. The concentration of MSP was either 500 nM (red) or 5 μM (blue).
The MSP and Lipids Form A Larger Complex Below 5 mM Cholate
Besides bilayer formation, we also sought to investigate how the MSP and lipids interact during self-assembly. We have previously demonstrated the use of a covalently attached long-lifetime dye for measuring rotational correlation times of protein-Nanodisc complexes11. Here, we apply the same technique to quantify the rotational dynamics of the nascent MSP-lipid complex as the detergent is removed. We use frequency-domain fluorescence anisotropy to measure the rotational correlation time during both self-assembly and disassembly, and we fit a single correlation time to capture the overall rotation of the complex. We first measured the correlation time of MSP alone as cholate is removed, and we found that there is no significant change in the fitted rotation (Fig. 2A), indicating that the MSP used does not appreciably aggregate with itself during these experiments. Next, we performed the self-assembly and disassembly experiments with a full reconstitution reaction where the concentration of MSP and lipids were held constant. In contrast to the bilayer formation, the association of MSP with lipids does not begin at the CMC of cholate. As shown by Fig. 2B, the rotational correlation time does not change significantly until the cholate concentration is below ~5 mM. When the concentrations of MSP and lipids are higher, this change in rotational correlation time occurs at a higher concentration of cholate. This indicates that bilayer formation is partially complete before there is a measurable change in the rotational mobility of the nascent Nanodisc complex. The disassembly data overlap closely and recapitulate the trend observed for the laurdan experiments (see Fig. 1). We rationalize the fact that the bilayer formation begins before the MSP rotational correlation time changes by noting that the lipid-MSP interactions consist of weak hydrophobic interactions between the lipid acyl chains and the aliphatic portions of the MSP helices. Because these interactions are weak, many lipids must present their acyl chains to the MSP for the interaction to be stable which avoids demanding a high curvature of the amphipathic MSP helices. When there is a higher concentration of detergent, the lipid particles are small, and the MSP does not encircle them. However, as detergent is removed, the particles become bigger and the MSP begins to bury its hydrophobic face into the larger bilayer. This ultimately leads to the change in rotational correlation time that is seen in Figure 2 – which happens at a lower detergent concentration relative to the changes observed with the laurdan GP shown in Figure 1. While we do not know the precise structure of the detergent/lipid particles (see the Discussion), they must be large and numerous enough to form stable interactions with the MSP.
Tyrosine Hydration Decreases as Self-Assembly Proceeds
Second derivative optical spectroscopy can be used to measure the degree to which tyrosine residues are exposed to solvent. This technique has revealed the extent of tyrosine hydration in several soluble proteins, and it has also sensitively measured the conformational changes in a membrane protein32,34. We chose this method as another probe of Nanodisc assembly based on the solid state NMR structure of MSP1D1ΔH535, which showed that some of the tyrosines apparently buried in the bilayer in Nanodiscs. We hypothesized that during self-assembly, the microenvironment of the tyrosines on MSP1D1(−) would change from mostly solvent-exposed when the MSP is fully solubilized by cholate to partially buried after cholate is removed. The degree of hydration of tyrosines in the presence of confounding absorbance by tryptophan residues is given by32:
where ΔA1 and ΔA2 are the differential absorbances at two pairs of wavelengths (287 – 283 nm and 295 – 290.5 nm), and the double prime indicates the values should come from the second derivative spectra. In order to interpret our experimental results, we calculated the expected rp for free tryptophan and tyrosine amino acids in the same molar ratio as occur in MSP1D1(−), based on published experimental measurements32. Using a Trp/Tyr ratio of 2.5, we find that the “buried” state, corresponding to measurements made in ethylene glycol, a reasonable analog of a hydrophobic core-like environment, yields a rp of 0.21, whereas the “exposed” state, corresponding to measurements in water, results in 1.39. Second derivative spectra of purified Nanodiscs before and after cholate addition are shown in Fig. 3A. The calculated peak ratios for self-assembly and disassembly with MSP concentration at 5 μM are shown in Fig. 3B. The hydration of tyrosine residues decreases from 1.1 to 0.7 during self-assembly. The disassembly experiments show that the tyrosines transition from mostly buried (0.6) to mostly exposed (1.1) as cholate is added. During self-assembly, the peak ratio began to change as soon as the cholate concentration dropped below 10 mM, indicating that tyrosines begin to bury just after the lipid hydration begins to decrease (Fig. 1). This might indicate that some lipids associate with the hydrophobic portion of the MSP helices as soon as the detergent concentration drops below the CMC. The self-assembly and disassembly data overlay closely. The combined data show that second derivative optical spectroscopy serves as a sensitive probe of Nanodisc self-assembly.
Figure 3:
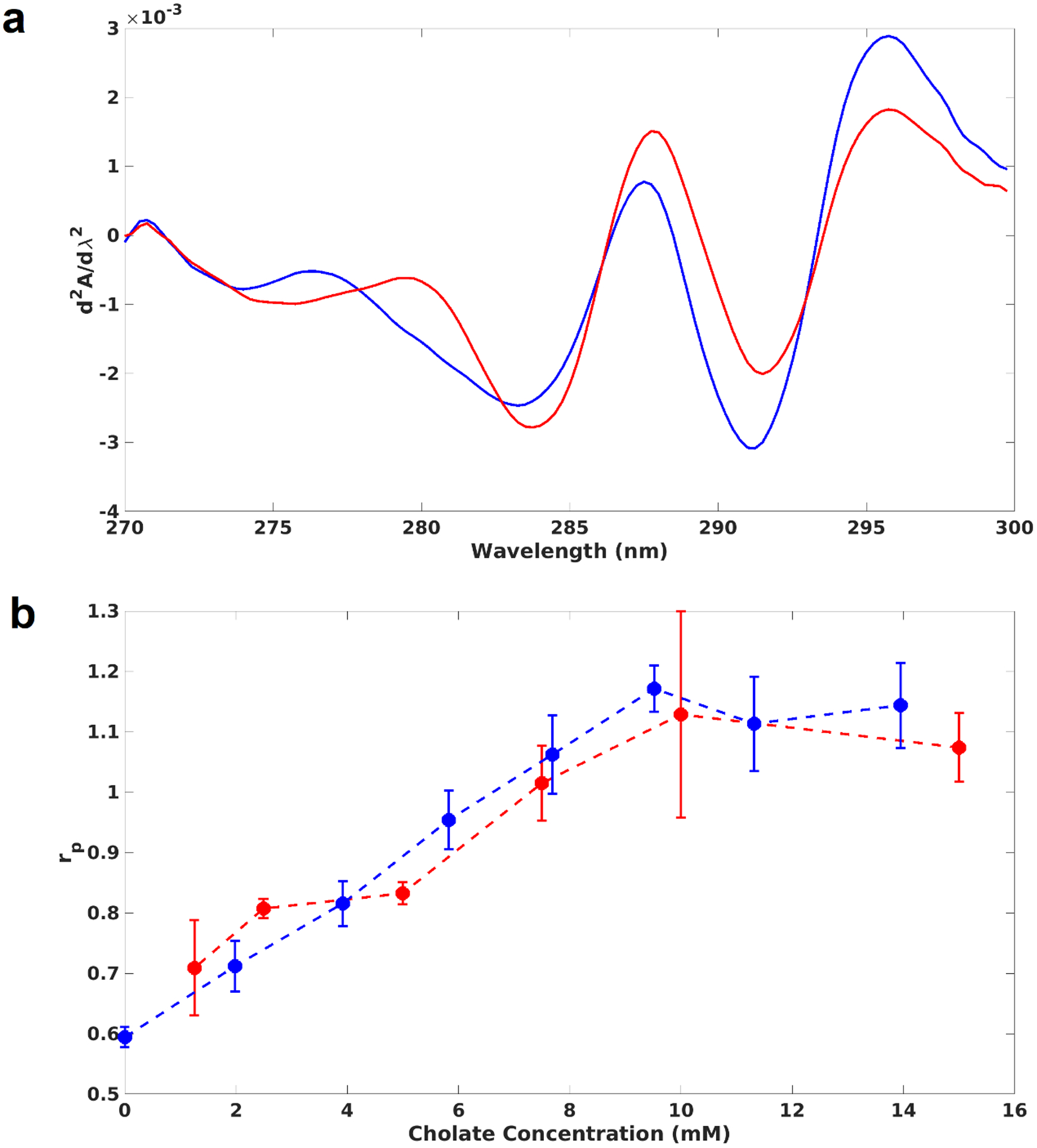
Tyrosine hydration changes during Nanodisc assembly and disassembly. (a): Interpolated and smoothed second derivative absorbance spectra of Nanodiscs. Purified Nanodiscs were measured (blue) and then remeasured after a single large cholate addition (red). The shift in peak position and intensity indicates a shift in the degree of solvent exposure of tyrosines. (b): Peak ratios calculated from the second derivative spectra of Nanodisc self-assembly (red) and disassembly (blue) reactions. Higher peak ratios indicate a more solvated microenvironment; low ratios represent a more buried environment.
Discussion
We have shown by fluorescence methods that the lipid bilayer begins to form below the CMC of cholate, while the MSP and lipids coalesce and form larger complexes at a lower detergent concentration. The second derivative optical spectra also show that tyrosine residues become less hydrated once the cholate concentration drops below the CMC. The concentrations of cholate at which these events occur depends on the total amount of MSP and lipids present during self-assembly. The data show that Nanodisc assembly is thermodynamically reversible in that, as detergent is either added or removed, the reaction proceeds along the same “pathway” (i.e. through the same points of thermodynamic equilibrium as defined by the cholate concentration). Apart from being theoretically interesting, the discovery that Nanodisc assembly is truly thermodynamic opens the door to more precise control over the reaction by controlling the detergent concentration.
Because assembly is a function of the total amount of MSP and lipids present relative to detergent, we can focus on experimental data where the concentration of MSP was 5 μM to interpret the data in a self-consistent manner. First, as the cholate concentration drops to 10 mM, the nascent bilayer begins to form as lipid molecules begin to find each other amongst the mixed micelles of detergent, lipids, and MSP. Then, as the concentration of cholate drops further, the tyrosine residues on the MSP become less hydrated, most likely by accumulating lipids near the hydrophobic portions of the helices. Then, as cholate is diluted further, the rotational correlation time of the MSP increases, indicating that the MSP and lipids have begun to coalesce. This process leads to the formation of larger, slowly rotating complexes as detergent removal continues. A diagram of this process is shown in Fig. 4 above the dotted line.
Figure 4:
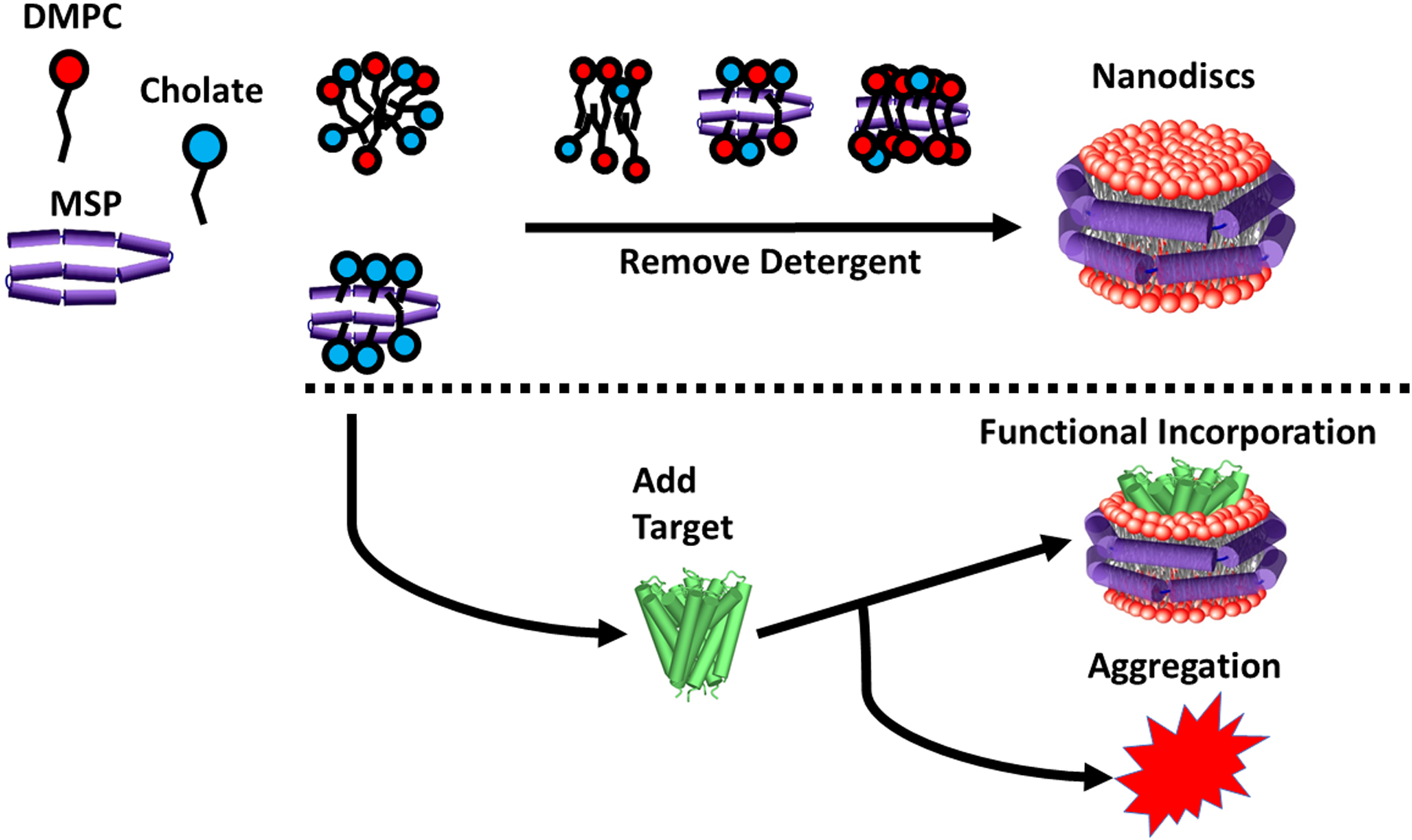
Diagram of the Nanodisc self-assembly process. The reaction begins with MSP (purple), lipids (red), and detergent (blue) all together in micelles. As detergent is removed, bilayer patches begin forming, tyrosines become more buried, and eventually lipids and MSP associate significantly and form a larger complex. Below the dotted line, the diagram is extended to include target protein, which results in two divergent pathways, with one resulting in functional incorporation of target into Nanodiscs, and another resulting in target protein aggregation.
This interpretation represents a major improvement in our understanding of Nanodisc self-assembly, and it builds on previous SAXS measurements and computational modeling of Nanodisc disassembly36. While most previous experimental efforts have focused on empirical methods to optimizing Nanodisc yield or the effect of reconstitution conditions on monodispersity37, this work represents an experimental investigation of Nanodisc self-assembly. We expect that this improved understanding of how the MSP, lipids, and cholate thermodynamically assemble into Nanodiscs will contribute to understanding the complex process of Nanodisc self-assembly, although we recognize that our description is but one model. We note that while methods such as electron microscopy or nuclear magnetic resonance (NMR) would help to provide a molecular picture of assembly to refine our model, the presence of multiple species in solution (lipid/MSP aggregates, mixed micelles of varying composition) would likely result in severe line broadening of NMR signals and would render data nearly impossible to interpret.
We have shown (Figures 1 and 2) that the lipid bilayer begins forming before the MSP rotational correlation time changes. We consider a model wherein lipid/detergent particles, termed “transitional bilayers”, form as detergent is removed. As evidenced by the GP data, these particles become less hydrated as detergent is removed, indicating partial bilayer character. The MSP-lipid interface consists of many hydrophobic interactions between lipid acyl chains and the aliphatic faces of the MSP helices. This means that, if the lipid particles are too small, or if there are very few of them, they would not be expected to interact stably with the semi-rigid helical MSP. However, if there were a preponderance of these particles presenting acyl chains to the MSP, and if they were large enough, then the MSP could stably wrap around the newly forming bilayer, and the rotational correlation time would increase. The GP of laurdan indicates that, by the time the rotation time of the MSP increases, the lipids are already partially ordered into these “transitional bilayer” patches. Thus, both our GP and anisotropy data support this model, although we do not know the exact structure and stoichiometry of these particles.
Our work could also lead to improved reconstitution protocols for a variety of membrane protein targets which are difficult to express, purify, and isolate. The “branch point” wherein addition of a target protein results in either functional incorporation into the Nanodisc or aggregation is depicted below the dotted line in Fig. 4. We expect further studies of Nanodisc assembly that include target proteins will shed light on the molecular events leading to these outcomes. We expect the presence of a target protein might cause Nanodisc assembly to finish at relatively higher detergent concentrations. Large membrane proteins with several membrane-spanning helices, for example, take up significant space in the bilayer, and thus fewer lipids must be recruited to the Nanodisc. Thus, one might expect that less cholate must be removed before Nanodisc formation is finished. We also note that our experiments used only one of several possible detergents compatible with Nanodisc assembly. While we have not here reported experiments with other detergents such as octyl glucoside, CHAPS, or Triton, we speculate that due to their different CMCs, these detergents might result in bilayer formation occurring within a different range of detergent concentrations. They may also modify the point at which the Nanodisc complex forms as measured by the rotational correlation time of the MSP. We also have not reported or discussed the behavior of circularized MSP in this work. Since we have less experience with circularized MSP-based Nanodiscs, we have opted not to include a discussion about these covalently closed MSPs (either by sortase or disulfide) as they have significantly different behavior.
Recent coarse-grained and all-atom molecular dynamics simulations of the self-assembly of MSP-based Nanodiscs38 and polymer discs39 have also shed light on the molecular details of self-assembly. In MSP-based Nanodiscs, water contacts from MD simulations as well as experimental measurements of the lifetime of a lipid-linked AS probe40 indicate that water penetrates significantly more near the edge of the MSP-lipid interface in fully assembled Nanodiscs. While our measurements do not have the resolution to differentiate “annular” from “bulk” lipids, it is interesting to note that during Nanodisc disassembly, the hydration of MSP tyrosine residues begins to increase immediately upon even small additions of cholate (Fig. 4B). This suggests that cholate may initially partition near the MSP-lipid interface, partially solvating tyrosine residues and annular lipids before eventually breaking apart the entire Nanodisc complex. Indeed we see that when the MSP concentration is 5 μM, both the laurdan GP and MSP rotational correlation times do not change much until the concentration of cholate reaches ~4–5mM, well after the tyrosine hydration has already increased.
Conclusions
We have described the application of fluorescence and optical spectroscopy to the study of Nanodisc self-assembly and disassembly. Our data collectively show not only that the self-assembly reaction is experimentally accessible, but that the process is thermodynamically reversible, and various points of thermodynamic equilibrium can be accessed by strictly controlling the amount of detergent. The formation of the lipid bilayer, decreasing hydration of the MSP tyrosines, and association of MSP with lipids have been monitored as cholate is removed (leading to self-assembly) or added to purified Nanodiscs (disassembly). The data in turn have informed a structural model of Nanodisc assembly which may lead to improved reconstitution protocols. This work significantly advances our understanding of the self-assembly of a widely used biophysical tool. Aside from increasing the yield of currently unwieldy targets, an improved understanding of Nanodisc assembly could lead to the isolation and characterization of new and exciting membrane proteins that play hitherto unknown roles in the chemistry of life.
Acknowledgements
This work was supported by NIH GM118145 to S.G.S. We thank Drs. Denisov and McLean for many useful discussions.
References
- 1.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J Am Chem Soc. 2004;126(11):3477–3487. doi: 10.1021/ja0393574 [DOI] [PubMed] [Google Scholar]
- 2.Whitesides GM, Grzybowski B. Self-Assembly at All Scales. Science (80- ). 2002;295(5564):2418–2421. http://science.sciencemag.org/. Accessed November 3, 2018. [DOI] [PubMed] [Google Scholar]
- 3.Möhwald H Surfactant layers at water surfaces. Reports Prog Phys. 1993;56(5):653–685. doi: 10.1088/0034-4885/56/5/002 [DOI] [Google Scholar]
- 4.Sackmann E Supported membranes: Scientific and practical applications. Science (80- ). 1996;271(5245):43–48. doi: 10.1126/science.271.5245.43 [DOI] [PubMed] [Google Scholar]
- 5.Pautot S, Frisken BJ, Weitz DA. Production of unilamellar vesicles using an inverted emulsion. Langmuir. 2003;19(7):2870–2879. doi: 10.1021/la026100v [DOI] [Google Scholar]
- 6.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002;2(8):853–856. doi: 10.1021/nl025623k [DOI] [Google Scholar]
- 7.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12(11):2476–2481. doi: 10.1110/ps.03267503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282(9):6556–6563. doi: 10.1074/jbc.M607973200 [DOI] [PubMed] [Google Scholar]
- 9.Ye X, McLean MA, Sligar SG. Conformational equilibrium of talin is regulated by anionic lipids. Biochim Biophys Acta -Biomembr. 2016;1858(8):1833–1840. doi: 10.1016/j.bbamem.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc Natl Acad Sci. 2002;99(10):6725–6730. doi: 10.1073/pnas.062565599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory MC, McLean MA, Sligar SG. Interaction of KRas4b with anionic membranes: A special role for PIP2. Biochem Biophys Res Commun. 2017;487(2):351–355. doi: 10.1016/j.bbrc.2017.04.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez Camargo DC, Korshavn KJ, Jussupow A, et al. Stabilization and structural analysis of a membrane-associated hIAPP aggregation intermediate. Elife. 2017;6:1–22. doi: 10.7554/eLife.31226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denisov IG, Sligar SG. Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev. 2017;117(6):4669–4713. doi: 10.1021/acs.chemrev.6b00690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLean MA, Gregory MC, Sligar SG. Nanodiscs: A Controlled Bilayer Surface for the Study of Membrane Proteins. Annu Rev Biophys. 2018;47:107–124. doi: 10.1146/annurev-biophys [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuhara K, Arakida J, Ravula T, et al. Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers. J Am Chem Soc. 2017;139(51):18657–18663. doi: 10.1021/jacs.7b10591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A. Bioinspired, Size-Tunable Self-Assembly of Polymer–Lipid Bilayer Nanodiscs. Angew Chemie - Int Ed. 2017;56(38):11466–11470. doi: 10.1002/anie.201705569 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 17.Zhang M, Huang R, Ackermann R, et al. Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy. Angew Chemie - Int Ed. 2016;55(14):4497–4499. doi: 10.1002/anie.201600073 [DOI] [PubMed] [Google Scholar]
- 18.Ravula T, Ishikuro D, Kodera N, Ando T, Anantharamaiah GM, Ramamoorthy A. Real-Time Monitoring of Lipid Exchange via Fusion of Peptide Based Lipid-Nanodiscs. Chem Mater. 2018;30(10):3204–3207. doi: 10.1021/acs.chemmater.8b00946 [DOI] [Google Scholar]
- 19.Dominguez Pardo JJ, Dörr JM, Renne MF, et al. Thermotropic properties of phosphatidylcholine nanodiscs bounded by styrene-maleic acid copolymers. Chem Phys Lipids. 2017;208(September):58–64. doi: 10.1016/j.chemphyslip.2017.08.010 [DOI] [PubMed] [Google Scholar]
- 20.Grethen A, Oluwole AO, Danielczak B, Vargas C, Keller S. Thermodynamics of nanodisc formation mediated by styrene/maleic acid (2:1) copolymer. Sci Rep. 2017;7(1):1–14. doi: 10.1038/s41598-017-11616-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denisov IG, McLean MA, Shaw AW, Grinkova YV., Sligar SG. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109(32):15580–15588. doi: 10.1021/jp051385g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2004;556(1–3):260–264. doi: 10.1016/S0014-5793(03)01400-5 [DOI] [PubMed] [Google Scholar]
- 23.Ritchie TK, Grinkova YV., Bayburt TH, et al. Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs In: Methods in Enzymology. Vol 464 1st ed Elsevier Inc.; 2009:211–231. doi: 10.1016/S0076-6879(09)64011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand A, Neubert R, Garidel P, Blume A. Bile salt induced solubilization of synthetic phosphatidylcholine vesicles studied by isothermal titration calorimetry. Langmuir. 2002;18(7):2836–2847. doi: 10.1021/la011421c [DOI] [Google Scholar]
- 25.Garidel P, Hildebrand A. Thermodynamic properties of association colloids. J Therm Anal Calorim. 2005;82(2):483–489. doi: 10.1007/s10973-005-0921-1 [DOI] [Google Scholar]
- 26.Lichtenberg D, Ahyayauch H, Goñi FM. The mechanism of detergent solubilization of lipid bilayers. Biophys J. 2013;105(2):289–299. doi: 10.1016/j.bpj.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonas A Reconstitution of High-Density Lipoproteins. In: Methods in Enzymology. Vol 128 ; 1986:553–582. doi: 10.1016/0076-6879(86)28092-1 [DOI] [PubMed] [Google Scholar]
- 28.Textor M, Vargas C, Keller S. Calorimetric quantification of linked equilibria in cyclodextrin/lipid/detergent mixtures for membrane-protein reconstitution. Methods. 2015;76(January):183–193. doi: 10.1016/j.ymeth.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 29.Castellano FN, Dattelbaum JD, Lakowicz JR. Long-Lifetime Ru(II) Complexes as Labeling Reagents for Sulfhydryl Groups. Anal Biochem. 1998;255:165–170. https://ac.els-cdn.com/S0003269797924684/1-s2.0-S0003269797924684-main.pdf?_tid=ff725b9f-d434-43b5-951b-bc5f71effc83&acdnat=1540927791_b980b8ee6d6b130a1907694aa42730f4. Accessed October 30, 2018. [DOI] [PubMed] [Google Scholar]
- 30.Chen PS, Toribara TY, Warner H. Microdetermination of phosphorus. Anal Chem. 1956;28(11):1756–1758. https://pubs.acs.org/sharingguidelines. Accessed October 31, 2018. [Google Scholar]
- 31.Lakowicz JR. Time-Dependent Anisotropy Decays In: Principles of Fluorescence Spectroscopy. 3rd ed Springer; 2006:383. doi: 10.1007/978-0-387-46312-4_11 [DOI] [Google Scholar]
- 32.Ragone SR, Colonna G, Balestrieri C, Servillo L, Irace G. Determination of Tyrosine Exposure in Proteins by Second-Derivative. Vol 23; 1984. https://pubs.acs.org/sharingguidelines. [DOI] [PubMed] [Google Scholar]
- 33.Parasassi T, De Stasio G, d’Ubaldo A, Gratton E. Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophys J. 1990;57(6):1179–1186. doi: 10.1016/S0006-3495(90)82637-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher MT, Sligar SG. Tyrosine Motions in Relation to the Ferric Spin Equilibrium of Cytochrome P-450cam. Biochemistry. 1985;24(23):6696–6701. https://pubs.acs.org/sharingguidelines. [DOI] [PubMed] [Google Scholar]
- 35.Bibow S, Polyhach Y, Eichmann C, et al. Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nat Struct Mol Biol. 2017;24(2):187–193. doi: 10.1038/nsmb.3345 [DOI] [PubMed] [Google Scholar]
- 36.Shih AY, Freddolino PL, Sligar SG, Schulten K. Disassembly of nanodiscs with cholate. Nano Lett. 2007;7(6):1692–1696. doi: 10.1021/nl0706906 [DOI] [PubMed] [Google Scholar]
- 37.Skar-Gislinge N, Johansen NT, Hoiberg-Nielsen R, Arleth L. Comprehensive Study of the Self-Assembly of Phospholipid Nanodiscs: What Determines Their Shape and Stoichiometry? Langmuir. 2018;34(42):12569–12582. doi: 10.1021/acs.langmuir.8b01503 [DOI] [PubMed] [Google Scholar]
- 38.Debnath A, Schäfer LV. Structure and Dynamics of Phospholipid Nanodiscs from All-Atom and Coarse-Grained Simulations. J Phys Chem B. 2015;119(23):6991–7002. doi: 10.1021/acs.jpcb.5b02101 [DOI] [PubMed] [Google Scholar]
- 39.Sahoo BR, Genjo T, Moharana KC, Ramamoorthy A. Self-Assembly of Polymer-Encased Lipid Nanodiscs and Membrane Protein Reconstitution. J Phys Chem B. 2019;123(21):4562–4570. doi: 10.1021/acs.jpcb.9b03681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano M, Fukuda M, Kudo T, et al. Static and dynamic properties of phospholipid bilayer nanodiscs. J Am Chem Soc. 2009;131(23):8308–8312. doi: 10.1021/ja9017013 [DOI] [PubMed] [Google Scholar]


