A predominance of HLA-I bound Asn deamidated peptides have been found to be generated through the ERAD pathway by quantitative MS-based proteomics. The Asn deamidated peptides in the peptidome of HLA-I, but not in HLA-II peptidomes or shotgun proteolysis, bear a concensus the N-glycosylation motif. The results reported provide new insights into the generation deamidated peptide antigens and provides a strategy for predicting deamidated T cell epitopes derived from glycoproteins.
Keywords: Immunology, peptidomics, post-translational modifications, N-glycosylation, peptides, de-glycosylation, deamidation, HLA, immunopeptidomics
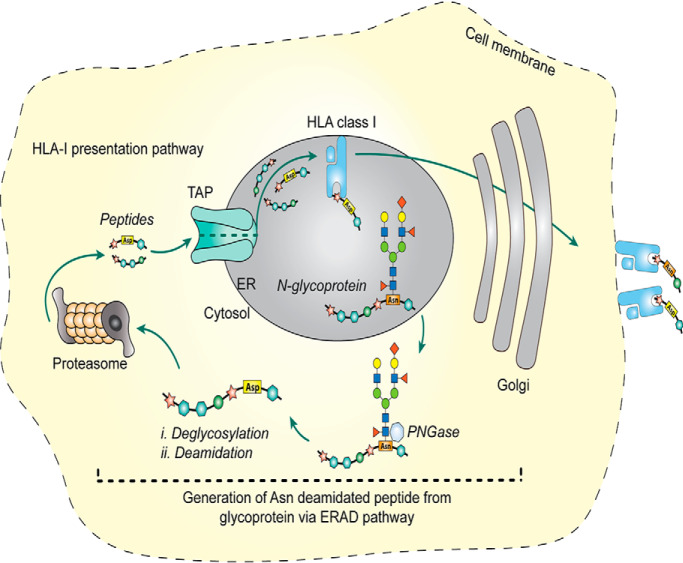
Graphical Abstract
Highlights
A predominance of HLA-I bound deamidated peptides are generated through ERAD pathway.
Deamidation of peptides with N-glycosylation motifs not in peptidome of HLA-II or proteolysis.
Many precursors of ERAD generated deamidated peptides are glycoproteins.
Abstract
The presentation of post-translationally modified (PTM) peptides by cell surface HLA molecules has the potential to increase the diversity of targets for surveilling T cells. Although immunopeptidomics studies routinely identify thousands of HLA-bound peptides from cell lines and tissue samples, in-depth analyses of the proportion and nature of peptides bearing one or more PTMs remains challenging. Here we have analyzed HLA-bound peptides from a variety of allotypes and assessed the distribution of mass spectrometry-detected PTMs, finding deamidation of asparagine or glutamine to be highly prevalent. Given that asparagine deamidation may arise either spontaneously or through enzymatic reaction, we assessed allele-specific and global motifs flanking the modified residues. Notably, we found that the N-linked glycosylation motif NX(S/T) was highly abundant across asparagine-deamidated HLA-bound peptides. This finding, demonstrated previously for a handful of deamidated T cell epitopes, implicates a more global role for the retrograde transport of nascently N-glycosylated polypeptides from the ER and their subsequent degradation within the cytosol to form HLA-ligand precursors. Chemical inhibition of Peptide:N-Glycanase (PNGase), the endoglycosidase responsible for the removal of glycans from misfolded and retrotranslocated glycoproteins, greatly reduced presentation of this subset of deamidated HLA-bound peptides. Importantly, there was no impact of PNGase inhibition on peptides not containing a consensus NX(S/T) motif. This indicates that a large proportion of HLA-I bound asparagine deamidated peptides are generated from formerly glycosylated proteins that have undergone deglycosylation via the ER-associated protein degradation (ERAD) pathway. The information herein will help train deamidation prediction models for HLA-peptide repertoires and aid in the design of novel T cell therapeutic targets derived from glycoprotein antigens.
Post-translational modification (PTM) including phosphorylation (1), ubiquitinylation (2), acylation (3), deamidation (4) and other structural modifications (5) has the potential to vastly expand the ligand repertoire presented by cell surface human leukocyte antigen (HLA) class I molecules for recognition by T cells. The alteration of physiochemical properties following PTM may modulate binding affinity to a given HLA allotype (5) as well as eliciting or abrogating T cell responses compared with the native sequence (4, 6–10). For example, several citrullinated or phosphorylated peptides have higher binding affinity to HLA-A2 and can upregulate the response of CD8+ T cells compared with their unmodified forms in subjects with type 1 diabetes (4), and phosphorylation of tumor epitopes has been show to stabilize the peptide-HLA complex (11). In addition, selective deamidation of HLA class II-restricted gliadin peptides enhances HLA-DQ2 and HLA-DQ8 binding with concomitant increases in immunogenicity (12). In contrast, the CD8+ T cell response to A-gliadin123–132 is abolished when the position 123 glutamine is deamidated, likely because of reduced HLA binding (10).
Our understanding of the human immunopeptidome—that is, the repertoire of peptides bound to and presented by HLA molecules—has grown rapidly because of increases in the speed and sensitivity of mass spectrometers and improvements in the specific isolation of peptide-HLA (pHLA) complexes from cells and tissues (13–19). These studies rely on computational algorithms to infer the peptide sequence from a reference protein database. Despite PTMs not being explicitly encoded in these databases, such algorithms have the capacity to search for and detect the appropriate mass shift associated with a large array of PTMs (20–22). However, except for S/T phosphorylation (23–25), there have been few systematic studies of PTM peptides in the immunopeptidome. Herein we describe the analysis of several comprehensive immunopeptidomics data sets (our data and those of others), with a focus on assessing the distribution of PTMs across different HLA allotypes. Other than oxidation of methionine deamidation of asparagine and glutamine was the next most prevalent type of modification across all HLA-I allotypes. Detailed analysis of the amino acid residues flanking the site of peptide ligand deamidation revealed a strong prevalence of the known N-linked glycosylation motif NX(S/T), where N is the deamidated asparagine residue and X is any amino acid except proline (26). Notably no motif was observed flanking the site of glutamine deamidation or for asparagine deamidation in peptides isolated from HLA class II molecules. Subsequent blocking of PNGase activity confirmed the prevalent role of deglycosylation of NX(S/T)-bearing antigenic precursors in the generation of asparagine-deamidated HLA ligands. Although this mechanism is known for a handful of deamidated T cell epitopes (6, 7, 27–31), these data indicate that the immunopeptidome is substantially enriched for peptides derived from formerly glycosylated proteins and specifically those that have been retro-translocated from the ER and targeted for deglycosylation and degradation in the cytoplasm. These large data sets will help to train algorithms for the prediction of deamidated peptide ligands and the discovery of novel T cell targets. Moreover, our data highlights a relatively unappreciated surveillance mechanism for glycoprotein antigens via the HLA-I antigen presentation pathway.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
In this study, peptides from three sources were analyzed: (1) HLA-I immunopeptidome; (2) HLA-II immunopeptidome; (3) shot gun proteomics. Each source contains nine data sets. For data sets from HLA-I immunopeptidome, three of them were identified from in-house experiments that involved biological duplicates of 5 × 109 cells which were used for the isolation of each HLA-I allotype (three in total) and analysis of their bound peptides. The remaining data sets from HLA-I immunopeptidome and other two sources come from publicly available data sets. For PNGase inhibition experiments, quadruplicate biological replicates of 1 × 108 cells were tested, with equivalent numbers of control samples (adding equivalent of DMSO instead of PNGase). t Test (32), one-way ANOVA (33), and two-way ANOVA (19) were used in this study.
Cell Lines
The Epstein-Barr virus-transformed B-lymphoblastoid cell line C1R, which expresses very low levels of endogenous HLAB*35:03 and low levels HLA-C*04:01 (34, 35), was stably transfected with either HLA-A*01:01, HLA-A*02:01 or HLA-A*24:02 by electroporation as previously described (36). Transfected cells were grown in RPMI 1640 (Invitrogen) supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, 7.5 mm HEPES (Sigma, St Louis, MO), 2 mm l-glutamine (MP Biomedical), 75 μm β-mercaptoethanolamine (Sigma), 0.1 mm non-essential amino acids (Invitrogen, Carlsbad, CA) and 10% fetal calf serum (FCS) (RF-10). 0.3 mg/ml hygromycin (Invitrogen) was added to select for stable expression of the transfected HLA-I allotypes, which was confirmed by flow cytometry. Cells were harvested by centrifugation (3724 × g at 4 °C for 15 min), washed with PBS, pelleted by centrifugation (524 × g at 4 °C for 10 min), and snap frozen in liquid nitrogen. Pellets were stored at −80 °C until processing.
PNGase Inhibition
The pan-caspase inhibitor Z-VAD-FMK (ab120487, Abcam, Cambridge, UK) was used to block PNGase activity, as described by Altrich-VanLith et al. (28). Briefly, 1×108 cells were pelleted by centrifugation and resuspended in 10 ml of RF-10 containing either 50 μm Z-VAD-FMK or vehicle control (2.5 μl/ml DMSO) and incubated for 30 min at 37 °C, 5% CO2. The cells were then treated with an isotonic acid stripping buffer (0.066 m Na2HPO4 and 0.131 m citric acid, pH 3.3) to remove existing cell surface HLA class I complexes (37, 38), resuspended in 10 ml of fresh RF-10 containing the 50 μm Z-VAD-FMK or vehicle alone and incubated at 37 °C, 5% CO2 for 5 h to allow re-expression of HLA class I molecules. Cells were harvested, washed with PBS, snap frozen in liquid nitrogen, and stored at −80 °C until processed.
Isolation of HLA-I Bound Peptides
was performed as previously described (18, 39). Briefly, cell pellets were resuspended in lysis buffer (0.5% IGEPAL (Sigma), 50 mm Tris, pH 8 (Sigma), 150 mm NaCl (Merck-Millipore, Darmstadt, Germany) and protease inhibitors (Complete Protease Inhibitor Mixture Tablet [1 tablet per 50 ml solution]; Roche Molecular Biochemicals, Basel, Switzerland)) and incubated for 45 min at 4 °C. Lysates were cleared by centrifugation at 16,000 × g for 40 min at 4 °C. pHLA complexes were immunoaffinity purified from lysates using W6/32 monoclonal antibody (10 mg/109 cells) crosslinked to protein A Sepharose (40). Bound complexes were eluted with 10% acetic acid and fractionated by RP-HPLC as previously described (18). Briefly, the mixture of eluted peptides, class I heavy chain and β2-microglobulin (β2m) was fractionated by RP-HPLC using a 4.6 × 100 mm monolithic C18 column (Chromolith Speed Rod, Merck-Millipore), an AK̈TAmicro™ HPLC system (GE Healthcare, UK) and mobile phases consisting of buffer A (0.1% trifluoroacetic acid (TFA) [Thermo Scientific, San Jose, CA]) and buffer B (80% acetonitrile (ACN) [Fisher Scientific, Waltham, MA] and 0.1% TFA). Fractions were combined, concentrated by vacuum centrifugation, and reconstituted in 2% v/v acetonitrile in 0.1% v/v aqueous formic acid. Each pool contained 25 fmol/μl iRT peptides (Biognosys, Schlieren, Switzerland (41)) as an internal retention time standard.
LC-MS/MS Acquisition
LC-MS/MS of HLA-I bound peptides was carried out using a SCIEX TripleTOF® 6600 equipped with an on-line Eksigent Ekspert nanoLC 415 (SCIEX, Toronto, Canada). 10 μl of each sample was directly loaded onto a trap column (ChromXP C18, 3 μm 120 Å, 350 μm × 0.5 mm [SCIEX]) maintained at an isocratic flow of buffer A (2% v/v acetonitrile in water supplemented with 0.1% v/v formic acid) at 5 μl/min for 10 min and then separated using an analytical column (ChromXP C18, 3 μm 120 Å, 75 μm × 15 cm [SCIEX]) by increasing linear concentrations of buffer B (0.1% v/v formic acid, 80% v/v acetonitrile) at a flow rate of 300 nL/min for 75 min. Up to 20 MS/MS spectra were acquired per cycle using an IDA strategy with accumulation times of 200 ms and 150 ms for MS1 and MS2, respectively. The MS1 scan range was set to 300–1800 m/z and MS2 set to 80–2000 m/z. To prevent multiple sequencing of the same peptide, MS1 masses were excluded for sequencing after two occurrences for 30 s.
LC-Multiple Reaction Monitoring (MRM)
HLA bound peptides purified from Z-VAD-FMK inhibition experiments (four biological replicates for each condition; Tier 3 analysis) were analyzed using a QTRAP® 6500+ (SCIEX) mass spectrometer equipped with an on-line Eksigent Ekspert nanoLC 415 (SCIEX) using an identical trap-elute schema to the LC-MS/MS experiments above. Data were acquired in positive ion MRM mode (refer to supplemental Table S1 for full transition list parameters) at unit resolution, with a triggered Enhanced Product Ion scan (80–1000 m/z; dynamic fill time; rolling collision energy) for any MRM transition exceeding 1000 counts per second. Data were analyzed in Skyline (v19.1.0.193) and manually assessed for any background or nonspecific interference, as well as ensuring all dot-products were ≥0.8 when comparing against the spectral library of pHLA derived from C1R-A1 cells. Peak areas for transitions were summed and values normalized to levels of iRT peptides present in each sample.
Peptide Sequence Identification
MS/MS data were searched against the human proteome [UniProt v2018_11] by PEAKS Studio 8.5 (Bioinformatics Solutions, Toronto, Canada) using the Homo sapiens Uniprot database (71975 entries, dated 2018–11). Both in-house generated MS data files and public MS data files from PRIDE repository (42) (PRIDE accession: (HLA-I): PXD004894, PXD000394, PXD005084, PXD004023, PXD008570, PXD008571, PXD008572; (HLA-II): PXD006939, PXD001205; (Proteolysis): PXD000394, PXD009797, PXD004447, PXD003977) were imported into PEAKS Studio 8.5 and subjected to default data refinement. For in-house generated MS data, the parent mass error tolerance was set to 50 ppm and the fragment mass error tolerance to 0.02 Da. For public collected MS data, the parameters were set as based on the default settings for the MS instrument used. For HLA and elastase data sets, enzyme specificity was turned off. For tryptic data sets, enzyme was set to trypsin and 3 missed cleavages were allowed. Oxidation of Methionine, deamidation of Asn or Gln, phosphorylation of Ser, Thr or Tyr and citrullination of Arg were included as variable modifications in the database peptide searches. After this analysis, a PEAKS PTM search was carried out for each data set incorporating all classes of PTMs (in total 313 types). A 1% false discovery rate (FDR, calculated by PEAKS using a target-decoy approach) threshold was applied to allow selection of high-confidence peptides.
Statistics
The differences in flanking residues surrounding the sites of Asn deamidation from different samples (Fig. 4D) and the comparison of the subcellular location of source antigens (Fig. 5B/C) were evaluated using two-way analysis of variance (ANOVA) multiple-comparison Bonferroni test. Differences in the proportion of known N-linked glycosylation sites among the peptides with NX(S/T) motif (Fig. 5A) was evaluated using a multiple-sample t test. PNGase inhibition experiments (Fig. 6C) were assessed by a one-way ANOVA multiple-comparison test. All analyses were calculated using GraphPad Prism 7, and a p value ≤ 0.05 was considered statistically significant.
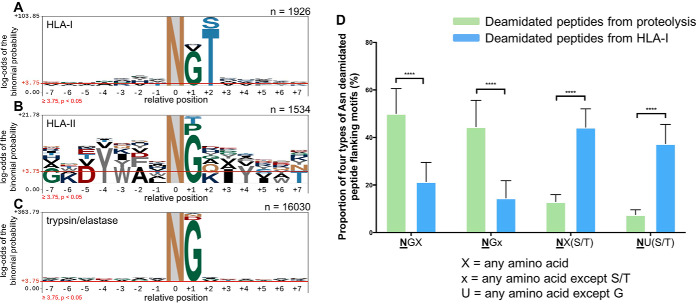
Fig. 4.
Motif analysis of asparagine deamidated peptides and proportion of four motif types across proteome and immunopeptidomics data sets. A, Asparagine-centralized motif of deamidated peptides presented from a mixture of HLA-I allotypes. B, Asparagine-centralized motif of deamidated peptides presented from a mixture of HLA-II allotypes. C, Asparagine-centralized motif of deamidated peptides from a mixture of trypsin and elastase whole cell digests. D, Proportion of four types of asparagine deamidated peptide flanking motifs from proteome digests (trypsin and elastase) and HLA-I immunopeptidome data sets. NGX, where X can be any amino acid but proline; NGx, where x can be any amino acid except for and serine and threonine; NX(S/T), where X can be any amino acid; NU(S/T), where U can be any amino acid except for glycine. In each case, N denotes a deamidated asparagine residue. The significant difference level was determined by the two-way analysis of variance (ANOVA) performed by using GraphPad Prism 7 (****, p < 0.0001). The motif analysis was generated by using pLogo (81). Amino acid above the red threshold line indicate the log-odds of the binomial probability is significant (p < 0.05).
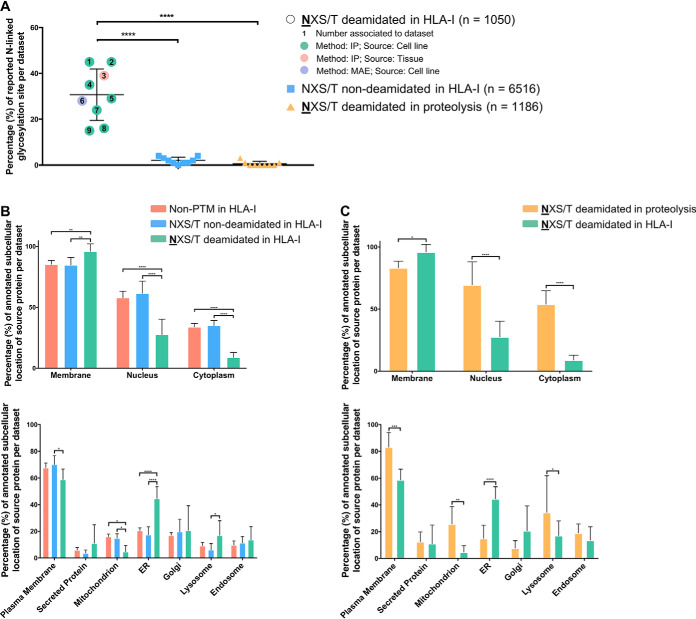
Fig. 5.
Subcellular location of source proteins and the relationship between deamidation of peptides containing the NX(S/T) motif. A, A high proportion of deamidated HLA-I bound peptides contain a consensus N-linked glycosylation motif which is not reflected in peptides derived from tryptic/elastase digestion of cellular material. B, A comparison of the subcellular location of the source antigens for HLA-I bound peptides among unmodified and peptides with native or deamidated Asparagine residues that harbor the NX(S/T) glycosylation motif. C, A comparison of the subcellular location of the source proteins of deamidated peptides bearing the NX(S/T) motif shows significant differences between tryptic/elastase-digested proteomes and HLA-I bound peptides. Significance was determined by two-way analysis of variance (ANOVA) performed using Prism 7 (***, p = 0.0002, **, p = 0.0021).
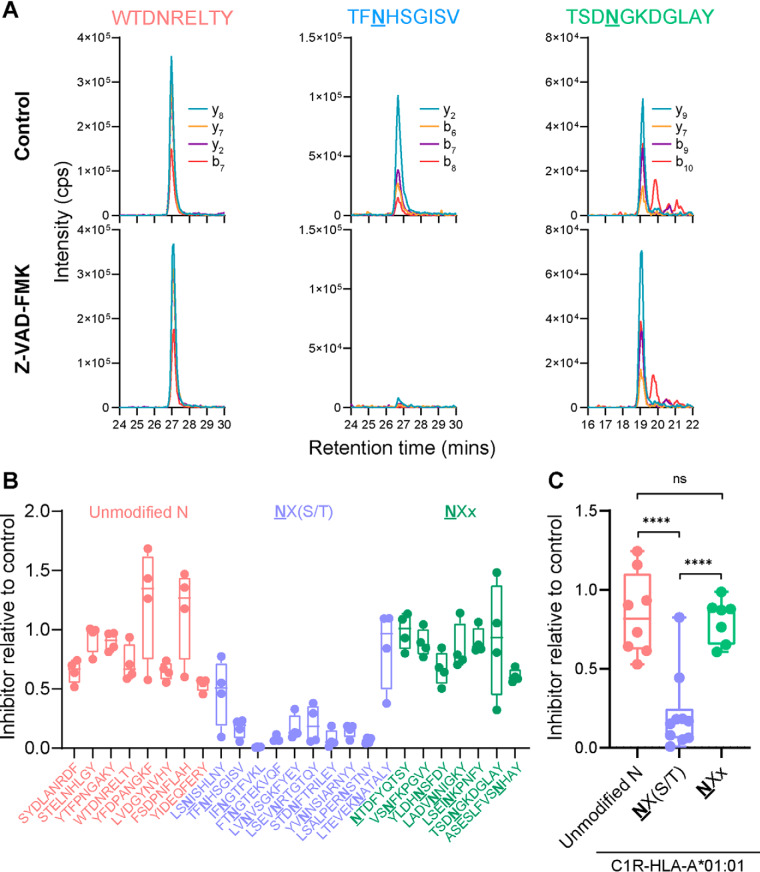
Fig. 6.
NX(S/T)-bearing pHLA are dependent on PNGase processing prior to presentation. LC-MRM was used to quantify the relative presentation of a set of HLA-bound peptides on C1R-A*01:01 cells after incubation for 5 h in the presence or absence of the PNGase inhibitor Z-VAD-FMK. A, Representative LC-MRM traces from control (upper) or inhibitor-treated (lower) cells showing specific detection of peptides from three categories: (left) unmodified asparagine, or deamidated asparagine containing peptides with (middle) or without the NX(S/T) motif (right). Each LC-MRM trace consists of four transitions, with specific ions as indicated. B–C, LC-MRM quantitation of peptide detection in inhibitor-treated relative to control-treated cells (quantitation change on individual peptide (B), quantitation change on overall peptides from three categories (C)). Datapoints represent individual peptides from four biological replicates in (B) and the average of replicates in (C). Significance was determined using an ordinary one-way ANOVA multiple comparison test (****, p < 0.0001; ***, p = 0.0002).
RESULTS
PTM Profile of Peptides Presented by Various HLA Allotypes
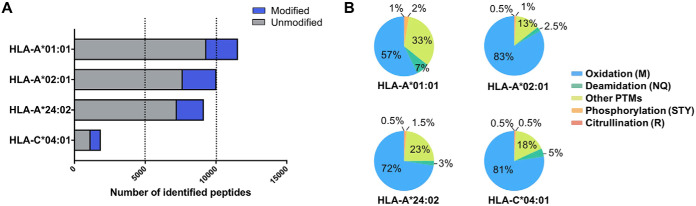
We investigated the prevalence of PTMs presented by three common HLA-A allotypes using C1R cell lines expressing HLA-A*01:01, -A*02:01 or -A*24:02. We exploited the common expression of low levels of endogenous HLA-C*04:01 as an additional control. HLA-peptide complexes were purified by immunoaffinity capture, bound peptides eluted by mild acid treatment and sequenced by liquid chromatography coupled to high-resolution mass spectrometry. As part of this process, we used the PEAKS PTM algorithm (43) to detect the diversity of PTMs across the immunopeptidomes of these allotypes (Fig. 1). On average, 10,000 peptides were detected for each of the transfected alleles (range: 9000–11,000), with around 1800 peptides identified from the parental C1R cells, derived from HLA-C*04:01. This unbiased PTM search revealed ∼21%, 22%, 17 and 38% of these peptides were post-translationally modified within the HLA-A*01:01, -A*02:01, -A*24:02 and -C*04:01 data sets, respectively (Fig. 1A). On assessment of the distribution of modification types, oxidation of Met accounted for the largest proportion (57–83%) of PTMs across all four allotypes (Fig. 1B), followed by deamidation of Asn or Gln (2.5–7%) (Fig. 1B). Phosphorylation (Ser, Thr, or Tyr) and citrullination of Arg accounted for less than 3% of the detected PTM repertoire—all other modifications were individually lower in frequency, yet in total they represented up to 33% of the detectable PTM space.
Fig. 1.
Immunopeptidome profile and PTM prevalence of peptides identified from four HLA-I alleles. A, The number of modified and unmodified peptides sequenced from each HLA allotype. B, The proportion of different PTMs identified on bound peptides isolated from each HLA-I allotype (data sets are from two biological replicates).
Properties of Deamidated Peptides Presented by HLA Allotypes
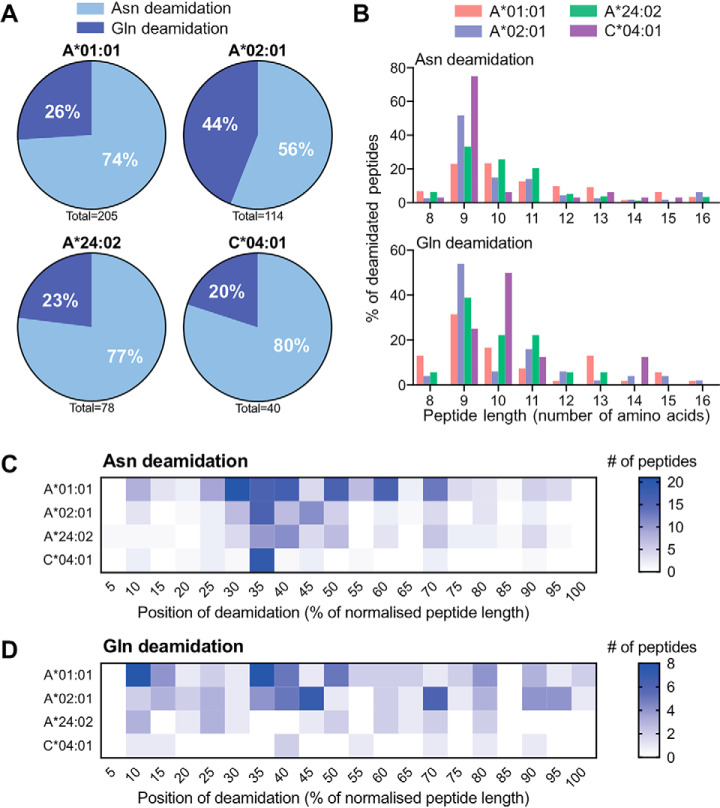
Next, we investigated the properties of peptides bearing modified residues. Despite the high prevalence of oxidized Met, it is difficult to attribute this modification to bona fide biological origin over sample preparation artifacts (44, 45); as such, we chose to focus on deamidation, the next most prevalent PTM (Fig. 2). Both Asn and Gln deamidation were observed across each of the transfected alleles (Fig. 1B, Fig. 2A) and the endogenous HLA-C*04:01 allele expressed by the parental C1R cell line (46). Deamidation of Asn was observed to be more prevalent than for Gln across all allotypes (Fig. 2A). The length of deamidated peptides was predominantly 9–13 amino acids (Fig. 2B), consistent with the total peptide repertoire of the corresponding HLA-I allotypes (supplemental Fig. S1). To assess any positional bias, we normalized the position of the deamidated residue as a proportion of peptide length and expressed the results as a heatmap (Fig. 2C and 2D). This analysis showed some degree of positional bias for asparagine deamidation (Fig. 2C), with a higher proportion of peptides bearing a deamidated residue in the N-terminal half of the ligand. These biases reflected the individual HLA allotype preferences for aspartic acid residues at P3 for HLA-C*04:01 (46), P3 for HLA-A*01:01 (36) and to a lesser extent P4 for HLA-A*02:01 (for all binding motifs, see supplemental Fig. S2). For Gln deamidation, the spread of the modified residue position appears more random (Fig. 2D), with only a moderate change in density for HLA-A*01:01 and -A*02:01 consistent with the peptide binding motifs of these alleles favoring Glu residues at position 3 or 4 (supplemental Fig. S2).
Fig. 2.
Immunopeptidome profile of deamidated peptides presented by four HLA allotypes. A, The proportion of asparagine and glutamine deamidation in the total deamidation set of each allotype. B, The length distribution of asparagine and glutamine deamidation from each allotype. C and D, Heatmap of the normalized position of deamidated asparagine or glutamine across each allotype (data sets are from two biological replicates).
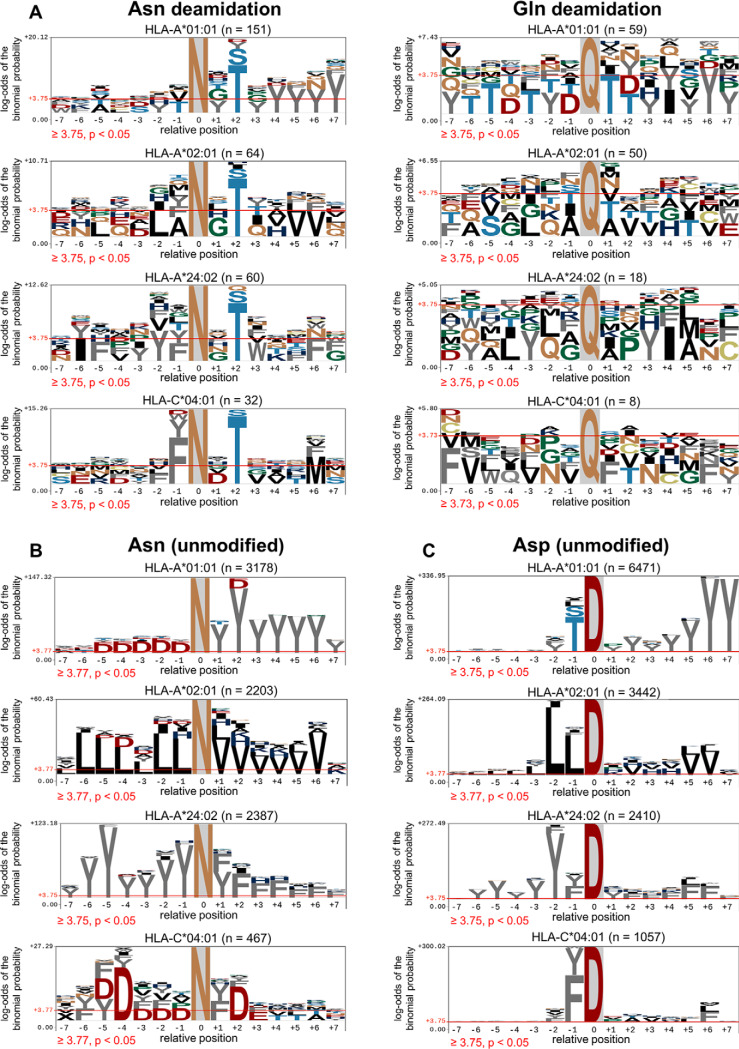
Next, we assessed the amino acids flanking the deamidated Asn or Gln residues (Fig. 3). A motif analysis was performed for deamidated peptides by centralizing the deamidated residue and expanding the sequence to the seven naturally occurring amino acids either side of the modification (as a means to extend beyond either termini of the final bound HLA ligand). As shown in Fig. 3A, a preference of Thr or Ser at the +2 site was observed in Asn deamidation across each HLA allotype, whereas no obvious bias was observed for Gln deamidation. To verify that the +2 Ser/Thr bias was a feature of Asn deamidated sequences, we carried out the same type of centralized motif analysis on peptides bearing unmodified Asn residues (Fig. 3B). The results for these unmodified peptides do not demonstrate +2 Ser/Thr preference, but show clear dominance of the allele-specific anchor residues. Finally, to demonstrate the specific enrichment of the NX(S/T) motif in the deamidated peptide ligands, we carried out the same type of centralized motif analysis for Asp residues found across the peptide data sets (Fig. 3C) and once again no enrichment of a +2 Ser/Thr bias was observed.
Fig. 3.
Enrichment of amino acid residues flanking the NX(S/T) motif in Asparagine deamidated peptides. A, Flanking residue analysis on asparagine or glutamine deamidated peptides. B, Flanking residue motif analysis of peptides containing unmodified asparagine. C, Flanking residue analysis of peptides containing native aspartic acid. The motif analysis was generated using pLogo (81). Amino acids above the red threshold line indicate the log-odds of the binomial probability is significant (p < 0.05).
The NX(S/T) motif is the canonical motif for N-linked glycosylation (47). The data from Fig. 3A show a clear enrichment of NX(S/T) for the four HLA class I data sets analyzed (on average, 59% of Asn deamidated peptides contain the NX(S/T) motif). We further extended our analysis to public immunopeptidomics data sets (1, 16, 39, 48, 49). In total, this comprised 42 HLA-I allotypes and 153,006 peptides of length 8–20 amino acids, of which 2206 (∼1.4%) contained one or more deamidated Asn residues (supplemental Table S2). As shown in Fig. 4A, this combined data revealed and strengthened the same +2 Ser/Thr motif observed in our data. An additional significant preference for a Gly residue at the +1 position was observed (Fig. 4A), which was apparent to a lesser extent in our own data sets (see Fig. 3A). The presence of Gly at position +1 is of note because this motif is known to drive spontaneous deamidation of Asn (50, 51).
In order to investigate the division between +1 Gly and +2 Ser/Thr motifs, we initially analyzed data sets from either HLA-II immunopeptidomes (52, 53) (supplemental Table S3) or a selection of conventional shotgun proteomics data sets (19, 49, 54–57) (supplemental Table S4). In the former, HLA-II bound peptides will not derive from proteins degraded and processed via the ERAD pathway (58), whereas the latter will snapshot the intact protein content of a cell through specific enzymatic digestion (in this case, trypsin (49, 54–57) or elastase (19, 57)). Fig. 4B displays the motif flanking centralized deamidated Asn residues from pHLA-II, showing a clear enrichment of Gly at the +1 position. The global proteome analysis (Fig. 4C) shows an even stronger enrichment for deamidated Asn residues to bear a glycine at the +1 position. Neither data sets, however, revealed a bias for a +2 Ser/Thr motif.
Next, we segregated the deamidated Asn sequences from the HLA-I and global proteome analysis based on their motif types, separating into four categories: NGX, where X denotes any amino acid; NGx, where x can be any amino acid except S/T; NX(S/T); NU(S/T) where U can be any amino acid except Gly (Fig. 4D). This analysis shows that the proportion of deamidated Asn peptides with an NGX or NGx motif from the shotgun proteomics data sets are significantly higher than for peptides derived from the HLA-I presentation pathway (p < 0.0001). Conversely, we observe that ∼48% of HLA-I bound deamidated peptides bear the NX(S/T) motif and only a small fraction (∼7%) of these comprise the motif where X can be glycine (Fig. 4D). The results obtained here further indicate the enrichment of the NX(S/T) motif in the deamidated pHLA-I repertoire.
Analysis of N-linked Glycosylation Sites Across Deamidated Asparagine Peptides
We next compared the concordance of annotated N-linked glycosylation sites with peptides bearing this motif within immunopeptidome and proteome data sets (Fig. 5A). These data show a 30% concordance of asparagine-deamidated pHLA-I with known N-linked glycosylation sites, whereas only ∼2% of non-deamidated peptides with the motif (number = 6516) overlapped with known glycosylation sites. It should be noted that around 130 known N-linked glycosylation sites are still found in non-deamidated peptides with the motif. In contrast only ∼0.5% of NX(S/T)-bearing peptides (number = 1186) map to known glycosylation sites in the proteomics data sets (Fig. 5A).
Subcellular Distribution of Source Proteins of Peptides Bearing the NX(S/T) Motif
We next investigated the subcellular distribution of the source protein of each NX(S/T)-containing HLA-I bound peptide. As shown in Fig. 5B–5C, the proportion of these source proteins that mapped to membrane or ER categories is significantly higher than sources of either non-deamidated NX(S/T) pHLA-I or trypsin/elastase derived NX(S/T) peptides. In contrast, the proportion of NX(S/T) pHLA-I source proteins mapped to the nucleus or cytoplasm categories are significantly lower compared with the corresponding protein source in the other peptide categories. As a comparison, we analyzed subcellular distribution from a comprehensive N-linked glycosylation site data set that has been recently curated (4008 experimentally verified N-linked glycosylation sites) (59). The result shows that most of the N-linked glycosylation sites are derived from proteins that can be mapped in the membrane category and/or ER ((59); data not shown). This is consistent with the subcellular location and further corroborates the glycoprotein origins of the of NX(S/T) deamidated peptides bound to HLA-I.
The Presentation of NX(S/T)-Containing pHLA is Dependent On Prior Enzymatic Removal of N-linked Glycosylation
It is well established that deamidation of the Asn residue of N-linked glycosylation sites occurs as a by-product of peptide:N-glycanase (PNGase)-mediated cleavage between the Asn and the innermost N-acetylglucosamine (GlcNAc) monosaccharide of glycosylated proteins (60–63). In addition, PNGase is primarily responsible for the removal of N-linked glycans following the dislocation of misfolded glycoproteins from the ER into the cytosol (64). Therefore, peptides bound to HLA-I with an NX(S/T) motif are expected to be dependent on this enzyme for their genesis, and indeed this has been shown for a HLA-I restricted epitope derived from tyrosinase (28). We used LC-MRM-MS to quantify the relative levels of a subset of peptides originally observed in our C1R-HLA-A*01:01 data set when purified from these cells cultured in the presence or absence of the potent PNGase inhibitor carbobenzyloxy-Val-Ala-Asp-α-fluoromethylketone (Z-VAD-FMK) (61) (Fig. 6). Peptides were selected from the HLA-A*01:01/C*04:01 repertoire, consisting of eight control unmodified peptides (seven with unmodified and one without any asparagine residue), ten deamidated peptides with the NX(S/T) motif, and seven deamidated peptides with a NXx motif (X is any amino acid; x is any amino acid except Ser or Thr). Fig. 6A shows raw MRM traces of one representative peptide from each category, with each robustly detected in control-treated cells. However, PNGase inhibition showed marked reduction of the signal from peptide TFNHSGISV yet had no impact on the unmodified sequence WTDNRELTY nor the NGx motif deamidated peptide TSDNGKDGLAY. Fig. 6B shows levels of each peptide across the three categories from four biological replicates comparing inhibitor-treated versus control cells, with Fig. 6C showing that levels of NX(S/T)-containing peptides were significantly (p < 0.0001, one-way ANOVA multiple-comparison test) decreased compared with unmodified or NXx motif-bearing peptides.
DISCUSSION
Despite the potential importance of PTM of peptide antigens in the development of a range of human diseases (4, 65–77), few systematic studies of the nature and properties of PTM peptides in the immunopeptidome have been performed. In this study we have focused on deamidation of HLA-I bound peptides. Although prior studies have identified several deamidated asparagine-containing T cell epitopes generated following retrograde transport of glycoproteins from the ER to the cytosol and subsequent removal of the N-linked glycans, here we extend these studies to show that a large proportion of HLA-I bound Asparagine deamidated peptides are generated through this mechanism. Quantitative measurements of peptide presentation confirmed the selective reduction of deamidated sequences bearing the N-linked glycosylation motif when the enzyme responsible for catalyzing glycan removal and generation of neo-aspartic acid residues (PNGase) was inhibited. To our knowledge, this is the most detailed study of asparagine deamidation to date, including the analysis of deamidated peptides from the proteome as well as HLA-II immunopeptidomes for comparison. The significant lack of deamidated peptides with the N-linked glycosylation motif in related proteome and HLA-II immunopeptidome data sets further reinforces the enrichment of these peptides by the class I pathway. In contrast a much smaller (15%) fraction of asparagine deamidation in the HLA-I immunopeptidome could be attributed to spontaneous conversion of asparagine to aspartic acid. Spontaneous deamidation is associated with a NG motif allowing these peptides to be distinguished as potential sample preparation artifacts. As anticipated, the proportion of asparagine deamidated peptides with NG motif in proteomics data sets was significantly higher than those in the HLA-I immunopeptidome.
Protein glycosylation plays important roles in cancers (78) and critical cellular pathways (79). We suggest that the enrichment of HLA-I bound asparagine deamidated peptides that are derived from glycosylated precursors provides a means for the immune system to survey the fidelity of N-glycosylation. This mechanism may allow CD8+ killer T cells to eradicate cells with perturbed glycosylation resulting from metabolic abnormalities infection or malignancy. Finally, we anticipate the NX(S/T) motif of asparagine deamidation in the HLA-I immunopeptidome as well as other features of deamidated peptides can be used to build models to predict HLA-I bound asparagine deamidated peptides for inclusion in vaccines and other immunotherapies that target glycosylated antigens.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (80) partner repository with the data set identifier PXD014754, including raw .wiff files and peptide csv files exported by PEAKS Studio 8.5, as well as mzIdentML files. MRM data is available at http://www.peptideatlas.org/PASS/PASS01473.
Supplementary Material
Acknowledgments
We thank staff at the Monash Proteomics and Metabolomics Facility and Monash Bioinformatics Facility for technical assistance and Nicola Ternette (University of Oxford) for sharing elastase digestion data.
This article contains supplemental data.
Funding and additional information—This project was funded in part by Australian National Health and Medical Research Council (NHMRC) Project Grants 1165490 (to A.W.P.) and 1084283 (to A.W.P. and N.P.C.). AWP is supported by a NHMRC Principal Research Fellowship (1137739). Computational resources were supported by the R@CMon/Monash Node of the NeCTAR Research Cloud, an initiative of the Australian Government's Super Science Scheme and the Education Investment Fund.
Author contributions—S.M., A.W.P., and N.P.C. designed research; S.M. and R.A. performed research; S.M., S.H.R., P.F., J.S., and N.P.C. analyzed data; S.M., A.W.P., and N.P.C. wrote the paper; R.A., S.H.R., P.T.I., and P.F. contributed new reagents/analytic tools.
Conflict of interest—Authors declare no competing interests.
Abbreviations—The abbreviations used are:
- PTM
- Post-translational modification
- HLA
- human leukocyte antigen
- MRM
- Multiple Reaction Monitoring
- FDR
- false discovery rate.
REFERENCES
- 1. Alpízar A., Marino F., Ramos-Fernández A., Lombardía M., Jeko A., Pazos F., Paradela A., Santiago C., Heck A. J., and Marcilla M. (2017) A molecular basis for the presentation of phosphorylated peptides by HLA-B antigens. Mol. Cell. Proteomics 16, 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wei J., Zanker D., Di Carluccio A. R., Smelkinson M. G., Takeda K., Seedhom M. O., Dersh D., Gibbs J. S., Yang N., and Jadhav A. (2017) Varied role of ubiquitylation in generating MHC class I peptide ligands. J. Immunol. 198, 3835–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beresford G. W., and Boss J. M. (2001) CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat. Immunol. 2, 652. [DOI] [PubMed] [Google Scholar]
- 4. McGinty J. W., Marré M. L., Bajzik V., Piganelli J. D., and James E. A. (2015) T cell epitopes and post-translationally modified epitopes in type 1 diabetes. Current Diabetes Reports 15, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sidney J., Vela J. L., Friedrich D., Kolla R., von Herrath M., Wesley J. D., and Sette A. (2018) Low HLA binding of diabetes-associated CD8+ T-cell epitopes is increased by post translational modifications. BMC Immunol. 19, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skipper J., Hendrickson R. C., Gulden P. H., Brichard V., Van Pel A., Chen Y., Shabanowitz J., Wolfel T., Slingluff C. L., and Boon T. (1996) An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 183, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferris R. L., Hall C., Sipsas N. V., Safrit J. T., Trocha A., Koup R. A., Johnson R. P., and Siliciano R. F. (1999) Processing of HIV-1 envelope glycoprotein for class I-restricted recognition: dependence on TAP1/2 and mechanisms for cytosolic localization. J. Immunol. 162, 1324–1332 [PubMed] [Google Scholar]
- 8. Yagüe J., Vázquez J., and De Castro J. A. L. (2000) A post-translational modification of nuclear proteins, N G, N G-dimethyl-Arg, found in a natural HLA class I peptide ligand. Protein Sci. 9, 2210–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meadows L., Wang W., den Haan J. M., Blokland E., Reinhardus C., Drijfhout J. W., Shabanowitz J., Pierce R., Agulnik A. I., and Bishop C. E. (1997) The HLA-A* 0201-restricted HY antigen contains a posttranslationally modified cysteine that significantly affects T cell recognition. Immunity 6, 273–281 [DOI] [PubMed] [Google Scholar]
- 10. Gianfrani C., Troncone R., Mugione P., Cosentini E., De Pascale M., Faruolo C., Senger S., Terrazzano G., Southwood S., and Auricchio S. (2003) Celiac disease association with CD8+ T cell responses: identification of a novel gliadin-derived HLA-A2-restricted epitope. J. Immunol. 170, 2719–2726 [DOI] [PubMed] [Google Scholar]
- 11. Petersen J., Wurzbacher S. J., Williamson N. A., Ramarathinam S. H., Reid H. H., Nair A. K., Zhao A. Y., Nastovska R., Rudge G., and Rossjohn J. (2009) Phosphorylated self-peptides alter human leukocyte antigen class I-restricted antigen presentation and generate tumor-specific epitopes. Pro, Natl. Acad. Sci. U.S.A. 106, 2776–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arentz-Hansen H., Körner R., Molberg Ø Quarsten. H, Vader W., Kooy Y. M., Lundin K. E., Koning F., Roepstorff P., and Sollid L. M. (2000) The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J. Exp. Med. 191, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Purcell A. W., and Gorman J. J. (2004) Immunoproteomics mass spectrometry-based methods to study the targets of the immune response. Mol. Cell. Proteomics 3, 193–208 [DOI] [PubMed] [Google Scholar]
- 14. Cravatt B. F., Simon G. M., and Yates Iii J. R. (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991. [DOI] [PubMed] [Google Scholar]
- 15. Mommen G. P., Frese C. K., Meiring H. D., van Gaans-van den Brink J., de Jong A. P., van Els C. A., and Heck A. J. (2014) Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Pro, Natl. Acad. Sci. U.S.A. 111, 4507–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bassani-Sternberg M., Bräunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., and Specht K. (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Communications 7, 13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caron E., Aebersold R., Banaei-Esfahani A., Chong C., and Bassani-Sternberg M. (2017) A case for a human immuno-peptidome project consortium. Immunity 47, 203–208 [DOI] [PubMed] [Google Scholar]
- 18. Purcell A. W., Ramarathinam S. H., and Ternette N. (2019) Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nat. Protocols 14, 1687. [DOI] [PubMed] [Google Scholar]
- 19. Faridi P., Li C., Ramarathinam S. H., Vivian J. P., Illing P. T., Mifsud N. A., Ayala R., Song J., Gearing L. J., Hertzog P. J., Ternette N., Rossjohn J., Croft N. P., and Purcell A. W. (2018) A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 3, eaar3947. [DOI] [PubMed] [Google Scholar]
- 20. Witze E. S., Old W. M., Resing K. A., and Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798. [DOI] [PubMed] [Google Scholar]
- 21. Mann M., and Jensen O. N. (2003) Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255. [DOI] [PubMed] [Google Scholar]
- 22. Olsen J. V., and Mann M. (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zarling A. L., Polefrone J. M., Evans A. M., Mikesh L. M., Shabanowitz J., Lewis S. T., Engelhard V. H., and Hunt D. F. (2006) Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Pro, Natl. Acad. Sci. U.S.A. 103, 14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abelin J. G., Trantham P. D., Penny S. A., Patterson A. M., Ward S. T., Hildebrand W. H., Cobbold M., Bai D. L., Shabanowitz J., and Hunt D. F. (2015) Complementary IMAC enrichment methods for HLA-associated phosphopeptide identification by mass spectrometry. Nat. Protocols 10, 1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cobbold M., De La Peña H., Norris A., Polefrone J. M., Qian J., English A. M., Cummings K. L., Penny S., Turner J. E., and Cottine J. (2013) MHC class I–associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci. Translational Med. 5, 203ra125–203ra125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao L., Diedrich J. K., Kulp D. W., Pauthner M., He L., Park S.-K. R., Sok D., Su C. Y., Delahunty C. M., and Menis S. (2017) Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Communications 8, 14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ostankovitch M., Altrich-VanLith M., Robila V., and Engelhard V. H. (2009) N-glycosylation enhances presentation of a MHC class I-restricted epitope from tyrosinase. J. Immunol. 182, 4830–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Altrich-VanLith M. L., Ostankovitch M., Polefrone J. M., Mosse C. A., Shabanowitz J., Hunt D. F., and Engelhard V. H. (2006) Processing of a class I-restricted epitope from tyrosinase requires peptide N-glycanase and the cooperative action of endoplasmic reticulum aminopeptidase 1 and cytosolic proteases. J. Immunol. 177, 5440–5450 [DOI] [PubMed] [Google Scholar]
- 29. Mosse C. A., Meadows L., Luckey C. J., Kittlesen D. J., Huczko E. L., Slingluff C. L., Shabanowitz J., Hunt D. F., and Engelhard V. H. (1998) The class I antigen-processing pathway for the membrane protein tyrosinase involves translation in the endoplasmic reticulum and processing in the cytosol. J. Exp. Med. 187, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dalet A., Robbins P. F., Stroobant V., Vigneron N., Li Y. F., El-Gamil M., Hanada K.-i., Yang J. C., Rosenberg S. A., and Van den Eynde B. J. (2011) An antigenic peptide produced by reverse splicing and double asparagine deamidation. Pro, Natl. Acad. Sci. U.S.A. 108, E323–E331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Selby M., Erickson A., Dong C., Cooper S., Parham P., Houghton M., and Walker C. M. (1999) Hepatitis C virus envelope glycoprotein E1 originates in the endoplasmic reticulum and requires cytoplasmic processing for presentation by class I MHC molecules. J. Immunol. 162, 669–676 [PubMed] [Google Scholar]
- 32. Abelin J. G., Keskin D. B., Sarkizova S., Hartigan C. R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G. L., Eisenhaure T. M., Clauser K. R., Hacohen N., Rooney M. S., Carr S. A., and Wu C. J. (2017) Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weinzierl A. O., Rudolf D., Hillen N., Tenzer S., van Endert P., Schild H., Rammensee H.-G., and Stevanović S. (2008) Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur. J. Immunol. 38, 1503–1510 [DOI] [PubMed] [Google Scholar]
- 34. Storkus W., Howell D., Salter R., Dawson J., and Cresswell P. (1987) NK susceptibility varies inversely with target cell class I HLA antigen expression. J. Immunol. 138, 1657–1659 [PubMed] [Google Scholar]
- 35. Zemmour J., Little A., Schendel D., and Parham P. (1992) The HLA-A, B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148, 1941–1948 [PubMed] [Google Scholar]
- 36. Giam K., Ayala-Perez R., Illing P. T., Schittenhelm R. B., Croft N. P., Purcell A. W., and Dudek N. L. (2015) A comprehensive analysis of peptides presented by HLA-A1. HLA 85, 492–496 [DOI] [PubMed] [Google Scholar]
- 37. Wang W., Man S., Gulden P. H., Hunt D. F., and Engelhard V. H. (1998) Class I-restricted alloreactive cytotoxic T lymphocytes recognize a complex array of specific MHC-associated peptides. J. Immunol. 160, 1091–1097 [PubMed] [Google Scholar]
- 38. Luckey C. J., Marto J. A., Partridge M., Hall E., White F. M., Lippolis J. D., Shabanowitz J., Hunt D. F., and Engelhard V. H. (2001) Differences in the expression of human class I MHC alleles and their associated peptides in the presence of proteasome inhibitors. J. Immunol. 167, 1212–1221 [DOI] [PubMed] [Google Scholar]
- 39. Illing P. T., Pymm P., Croft N. P., Hilton H. G., Jojic V., Han A. S., Mendoza J. L., Mifsud N. A., Dudek N. L., and McCluskey J. (2018) HLA-B57 micropolymorphism defines the sequence and conformational breadth of the immunopeptidome. Nat. Communications 9, 4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Purcell A. W., Gorman J. J., Garcia-Peydró M., Paradela A., Burrows S. R., Talbo G. H., Laham N., Peh C. A., Reynolds E. C., and de Castro J. A. L. (2001) Quantitative and qualitative influences of tapasin on the class I peptide repertoire. J. Immunol. 166, 1016–1027 [DOI] [PubMed] [Google Scholar]
- 41. Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., MacCoss M. J., and Rinner O. (2012) Using i RT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., and Ternent T. (2015) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han X., He L., Xin L., Shan B., and Ma B. (2011) PeaksPTM: mass spectrometry-based identification of peptides with unspecified modifications. J. Proteome Res. 10, 2930–2936 [DOI] [PubMed] [Google Scholar]
- 44. Liu H., Ponniah G., Neill A., Patel R., and Andrien B. (2013) Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis. Anal. Chem. 85, 11705–11709 [DOI] [PubMed] [Google Scholar]
- 45. Morand K., Talbo G., and Mann M. (1993) Oxidation of peptides during electrospray ionization. Rapid Commun. Mass Spectrometry 7, 738–743 [DOI] [PubMed] [Google Scholar]
- 46. Schittenhelm R., Dudek N., Croft N., Ramarathinam S., and Purcell A. (2013) A comprehensive analysis of constitutive naturally processed and presented HLA-C* 04: 01 (Cw4)–specific peptides. HLA 83, 174–179 [DOI] [PubMed] [Google Scholar]
- 47. Li F., Li C., Revote J., Zhang Y., Webb G. I., Li J., Song J., and Lithgow T. (2016) GlycoMine struct: a new bioinformatics tool for highly accurate mapping of the human N-linked and O-linked glycoproteomes by incorporating structural features. Sci. Reports 6, 34595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pearson H., Daouda T., Granados D. P., Durette C., Bonneil E., Courcelles M., Rodenbrock A., Laverdure J.-P., Côté C., and Mader S. (2016) MHC class I–associated peptides derive from selective regions of the human genome. J. Clin. Investig. 126, 4690–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bassani-Sternberg M., Pletscher-Frankild S., Jensen L. J., and Mann M. (2015) Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell. Proteomics 14, 658–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wright H. T. (1991) Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit. Rev. Biochem. Mol. Biol. 26, 1–52 [DOI] [PubMed] [Google Scholar]
- 51. Wright H. T. (1991) Sequence and structure determinants of the nonenzymatic deamidation of asparagine and glutamine residues in proteins. Protein Engineering, Design Selection 4, 283–294 [DOI] [PubMed] [Google Scholar]
- 52. Chong C., Marino F., Pak H., Racle J., Daniel R. T., Müller M., Gfeller D., Coukos G., and Bassani-Sternberg M. (2018) High-throughput and sensitive immunopeptidomics platform reveals profound interferonγ-mediated remodeling of the Human Leukocyte Antigen (HLA) Ligandome. Mol. Cell. Proteomics 17, 533–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bergseng E., Dørum S., Arntzen M. Ø., Nielsen M., Nygård S., Buus S., de Souza G. A., and Sollid L. M. (2015) Different binding motifs of the celiac disease-associated HLA molecules DQ2. 5, DQ2. 2, and DQ7. 5 revealed by relative quantitative proteomics of endogenous peptide repertoires. Immunogenetics 67, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liepe J., Marino F., Sidney J., Jeko A., Bunting D. E., Sette A., Kloetzel P. M., Stumpf M. P., Heck A. J., and Mishto M. (2016) A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 [DOI] [PubMed] [Google Scholar]
- 55. Wu Z., Huang J., Huang J., Li Q., and Zhang X. (2018) Lys-C/Arg-C, a More Specific and Efficient Digestion Approach for Proteomics Studies. Anal. Chem. 90, 9700–9707 [DOI] [PubMed] [Google Scholar]
- 56. Tsiatsiani L., Giansanti P., Scheltema R. A., van den Toorn H., Overall C. M., Altelaar A. M., and Heck A. J. (2016) Opposite electron-transfer dissociation and higher-energy collisional dissociation fragmentation characteristics of proteolytic K/R (X) n and (X) n K/R Peptides Provide Benefits for Peptide Sequencing in Proteomics and Phosphoproteomics. J. Proteome Res. 16, 852–861 [DOI] [PubMed] [Google Scholar]
- 57. Davis S., Charles P. D., He L., Mowlds P., Kessler B. M., and Fischer R. (2017) Expanding proteome coverage with CHarge Ordered Parallel Ion aNalysis (CHOPIN) combined with broad specificity proteolysis. J. Proteome Res. 16, 1288–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Neefjes J., Jongsma M. L., Paul P., and Bakke O. (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823. [DOI] [PubMed] [Google Scholar]
- 59. Li F., Zhang Y., Purcell A. W., Webb G. I., Chou K.-C., Lithgow T., Li C., and Song J. (2019) Positive-unlabelled learning of glycosylation sites in the human proteome. BMC Bioinformatics 20, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Plummer T. H., Elder J. H., Alexander S., Phelan A., and Tarentino A. L. (1984) Demonstration of peptide: N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. J. Biol. Chem. 259, 10700–10704 [PubMed] [Google Scholar]
- 61. Misaghi S., Pacold M. E., Blom D., Ploegh H. L., and Korbel G. A. (2004) Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turnover. Chem. Biol. 11, 1677–1687 [DOI] [PubMed] [Google Scholar]
- 62. Palmisano G., Melo-Braga M. N., Engholm-Keller K., Parker B. L., and Larsen M. R. (2012) Chemical deamidation: a common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 11, 1949–1957 [DOI] [PubMed] [Google Scholar]
- 63. Hao P., Ren Y., Alpert A. J., and Sze S. K. (2011) Detection, evaluation and minimization of nonenzymatic deamidation in proteomic sample preparation. Mol. Cell. Proteomics 10, O111.009381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hirsch C., Blom D., and Ploegh H. L. (2003) A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22, 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gudmann N. S., Hansen N. U., Jensen A. C., Karsdal M. A., and Siebuhr A. S. (2015) Biological relevance of citrullinations: diagnostic, prognostic and therapeutic options. Autoimmunity 48, 73–79 [DOI] [PubMed] [Google Scholar]
- 66. Koning F., Thomas R., Rossjohn J., and Toes R. E. (2015) Coeliac disease and rheumatoid arthritis: similar mechanisms, different antigens. Nat. Rev. Rheumatol. 11, 450. [DOI] [PubMed] [Google Scholar]
- 67. Scally S. W., Petersen J., Law S. C., Dudek N. L., Nel H. J., Loh K. L., Wijeyewickrema L. C., Eckle S. B., van Heemst J., and Pike R. N. (2013) A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 210, 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sollid L. M., and Jabri B. (2011) Celiac disease and transglutaminase 2: a model for posttranslational modification of antigens and HLA association in the pathogenesis of autoimmune disorders. Curr. Opinion Immunol. 23, 732–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Babon J. A. B., DeNicola M. E., Blodgett D. M., Crèvecoeur I., Buttrick T. S., Maehr R., Bottino R., Naji A., Kaddis J., and Elyaman W. (2016) Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat. Med. 22, 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rondas D., Crèvecoeur I., D'Hertog W., Ferreira G. B., Staes A., Garg A. D., Eizirik D. L., Agostinis P., Gevaert K., and Overbergh L. (2015) Citrullinated glucose-regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes 64, 573–586 [DOI] [PubMed] [Google Scholar]
- 71. Strollo R., Vinci C., Arshad M. H., Perrett D., Tiberti C., Chiarelli F., Napoli N., Pozzilli P., and Nissim A. (2015) Antibodies to post-translationally modified insulin in type 1 diabetes. Diabetologia 58, 2851–2860 [DOI] [PubMed] [Google Scholar]
- 72. Schloot N. C., Willemen S., Duinkerken G., de Vries R. R., and Roep B. O. (1998) Cloned T cells from a recent onset IDDM patient reactive with insulin B-chain. J. Autoimmunity 11, 169–175 [DOI] [PubMed] [Google Scholar]
- 73. Yang J., Chow I. T., Sosinowski T., Torres-Chinn N., Greenbaum C. J., James E. A., Kappler J. W., Davidson H. W., and Kwok W. W. (2014) Autoreactive T cells specific for insulin B:11–23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc. Natl. Acad. Sci. U.S.A. 111, 14840–14845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Durinovic-Bello I., Schlosser M., Riedl M., Maisel N., Rosinger S., Kalbacher H., Deeg M., Ziegler M., Elliott J., and Roep B. (2004) Pro-and anti-inflammatory cytokine production by autoimmune T cells against preproinsulin in HLA-DRB1* 04, DQ8 Type 1 diabetes. Diabetologia 47, 439–450 [DOI] [PubMed] [Google Scholar]
- 75. McGinty J. W., Chow I.-T., Greenbaum C., Odegard J., Kwok W. W., and James E. A. (2014) Recognition of post-translationally modified glutamic acid decarboxylase 65 epitopes in subjects with type 1 diabetes. Diabetes 63, 3033–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Scotto M., Afonso G., Larger E., Raverdy C., Lemonnier F., Carel J., Dubois-Laforgue D., Baz B., Levy D., and Gautier J. (2012) Zinc transporter (ZnT) 8 186–194 is an immunodominant CD8+ T cell epitope in HLA-A2+ type 1 diabetic patients. Diabetologia 55, 2026–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dang M., Rockell J., Wagner R., Wenzlau J. M., Yu L., Hutton J. C., Gottlieb P. A., and Davidson H. W. (2011) Human type 1 diabetes is associated with T cell autoimmunity to zinc transporter 8. J. Immunol. 186, 6056–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pinho S. S., and Reis C. A. (2015) Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540. [DOI] [PubMed] [Google Scholar]
- 79. Ohtsubo K., and Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 80. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., and Eisenacher M. (2018) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. O'shea J. P., Chou M. F., Quader S. A., Ryan J. K., Church G. M., and Schwartz D. (2013) pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods 10, 1211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (80) partner repository with the data set identifier PXD014754, including raw .wiff files and peptide csv files exported by PEAKS Studio 8.5, as well as mzIdentML files. MRM data is available at http://www.peptideatlas.org/PASS/PASS01473.