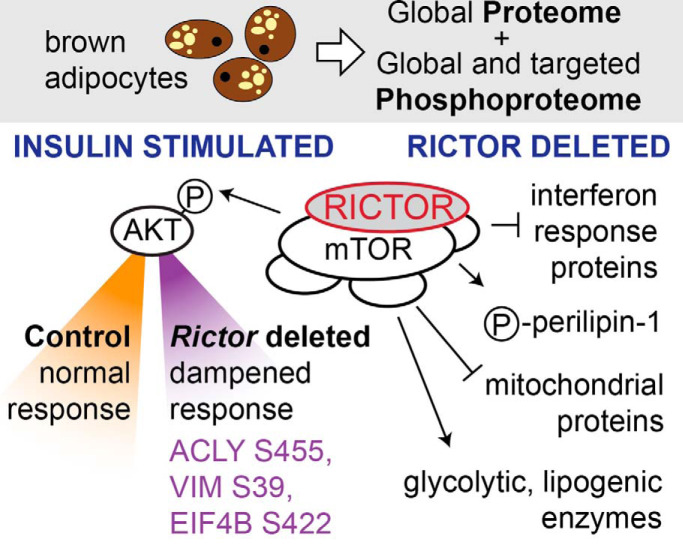
We applied DDA proteomics along with DDA and PRM phosphoproteomics to study brown adipocytes lacking RICTOR, an essential subunit of mTOR complex 2, over a time series of insulin stimulation. RICTOR loss caused many protein abundance changes, including increased levels of interferon response proteins. The insulin phosphoproteome response in RICTOR-deficient cells was dampened, including known functional sites ACLY S455, VIM S39, and EIF4B S422, potentially helping to bridge the gap between the acute and chronic effects of RICTOR loss.
Keywords: Phosphoproteome, phosphorylation, targeted mass spectrometry, signal transduction, signaling circuits, adipocytes, insulin signaling, mTOR
Graphical Abstract
Highlights
Global and targeted phosphoproteomics in RICTOR-deficient brown adipocytes.
RICTOR loss leads to higher levels of many interferon response-associated proteins.
RICTOR loss dampens the dynamic insulin-dependent phosphoproteome response.
ACLY S455, VIM S39, and EIF4B S422 are among the most dampened phosphosites.
Abstract
Stimulating brown adipose tissue (BAT) activity represents a promising therapy for overcoming metabolic diseases. mTORC2 is important for regulating BAT metabolism, but its downstream targets have not been fully characterized. In this study, we apply proteomics and phosphoproteomics to investigate the downstream effectors of mTORC2 in brown adipocytes. We compare wild-type controls to isogenic cells with an induced knockout of the mTORC2 subunit RICTOR (Rictor-iKO) by stimulating each with insulin for a 30-min time course. In Rictor-iKO cells, we identify decreases to the abundance of glycolytic and de novo lipogenesis enzymes, and increases to mitochondrial proteins as well as a set of proteins known to increase upon interferon stimulation. We also observe significant differences to basal phosphorylation because of chronic RICTOR loss including decreased phosphorylation of the lipid droplet protein perilipin-1 in Rictor-iKO cells, suggesting that RICTOR could be involved with regulating basal lipolysis or droplet dynamics. Finally, we observe mild dampening of acute insulin signaling response in Rictor-iKO cells, and a subset of AKT substrates exhibiting statistically significant dependence on RICTOR.
The mammalian target of rapamycin (mTOR)1 is a serine/threonine kinase and a critical regulator of metabolism and cell growth. mTOR exists in two protein complexes called mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which are commonly defined by their regulatory subunits, RAPTOR and RICTOR, respectively. Of the two complexes, mTORC2 is less well understood, though it is essential for development, the metabolic functions of many tissues, and the progression of certain tumors (1, 2). In brown adipose tissue (BAT), mTORC2 is a key regulator of lipid metabolism. Tissue-specific knockout of Rictor in mouse BAT inhibits de novo lipogenesis (DNL), and increases lipid uptake, catabolic gene and UCP1 expression (3, 4). Most notably, Rictor knockout in BAT protects against diet-induced obesity and hepatic steatosis in mice living at thermoneutrality. Although the underlying molecular and cellular mechanisms of Rictor/mTORC2 loss in BAT are beginning to be uncovered (3, 5), the full extent of biological processes and functions that are affected is not known.
mTORC2 phosphorylates the hydrophobic motif of AGC-family kinases AKT, SGK1, and PKCα. In 3T3L1 adipocytes, AKT is rapidly activated upon stimulation by insulin and IGF-1 and exhibits precise temporal regulation where AKT hydrophobic motif phosphorylation (S473 in AKT1, S474 in AKT2) by mTORC2 occurs after phosphorylation of the T-loop (T308 in AKT1, T309 in AKT2) by PDK1 (6, 7). Mutagenesis studies suggest S473 phosphorylation is required for maximal in vitro AKT activity (8). However, in vivo studies where essential mTORC2 components have been deleted reported minimal effects on the phosphorylation of AKT substrates, with some exceptions (4, 5, 9–11). By applying targeted phosphoproteomics, we recently identified some AKT substrates with partial sensitivity to mTORC2 loss in brown preadipocytes, including ACLY S455, whose phosphorylation is important for brown adipocyte differentiation, histone acetylation, and expression of de novo lipogenesis genes and GLUT4 in differentiated brown adipocytes (5). Collectively, these studies suggest that loss of mTORC2-dependent AKT hydrophobic motif phosphorylation may differentially affect AKT substrate phosphorylation yet leave many open questions regarding the role of mTORC2 in regulating the global proteome and the dynamic insulin-AKT signaling response.
While our prior work in brown preadipocytes focused on the targeted phosphoproteomic analysis of AKT substrates at a single time point of insulin stimulation (5), we reasoned that a global, systems-level, time course analysis of protein abundance and phosphorylation might better elucidate downstream mTORC2 actions in brown adipocytes. Here, we apply mass spectrometry (MS)-based proteomics to profile protein and dynamic, insulin-stimulated phosphorylation in wild-type and mTORC2-deficient differentiated, mature brown adipocytes in which Rictor is deleted using an inducible knockout system. Applying both discovery-driven and targeted proteomic approaches, we assessed changes to baseline protein abundance, insulin response amplitude, and temporal dynamics. We find that RICTOR loss mildly, yet broadly, dampens insulin-stimulated phosphorylation of AKT substrates in brown adipocytes. This dampening varies in magnitude depending on the phosphosite and is most strongly observed on a subset of AKT substrates that includes ATP citrate lyase (ACLY), vimentin (VIM), and eukaryotic translation initiation factor 4B (EIF4B). RICTOR loss also increases abundance of several proteins that are consistent with an interferon-like response, decreases carbohydrate and lipid metabolism enzymes, and down-regulates phosphorylation of multiple sites on perilipin-1, independently of acute insulin stimulation. This study expands on our prior work (5) by offering a global view of how the brown adipocyte proteome is remodeled in the absence of functional mTORC2, and provides the most comprehensive proteomic analysis to date of the effect of mTORC2 loss on insulin/AKT signaling in mature brown adipocytes.
EXPERIMENTAL PROCEDURES
Brown Adipocyte Cell Culture
To obtain inducible-knockout cell lines, the interscapular BAT was removed from Ubc-CreERT2;Rictorl/l mice at postnatal day 1, dissected and minced, and added 500 μl of collagenase solution. The tubes were vortexed for 10 s and placed into a thermomixer at 800 rpm at 37 °C for 40 min with a mechanical disaggregation at 20 min. Afterward the digested tissue was filtered through a 100 μm filter into a fresh microcentrifuge tube, and centrifuged at room temperature at 1500 rpm for 5 min. The supernatant was removed, and the pellet resuspended in culture medium (DMEM-high glucose, 20% FBS, 20 mm HEPES) and transferred into a 6-well plate. The cells were left overnight and given fresh medium the next day. On day 3 the cells were immortalized by adding a virus solution that consisted of 50% primary culture media, 50% viral stock (pBABE Zeocin-SV40 Large T) and polybrene (8 mg/ml). On day 4 the media was changed to DMEM high-glucose media (supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin) and on day 5, Zeocin selection (500 μg/ml) began and remained for 1 week.
The viral stock was generated as follows: On the day of the transfection a 100 μl serum-free DMEM-high glucose media aliquot that contained 8 μl of FuGENE, 1 μg of the transfer vector (pBABE zeocin-SV40 Large T) and 1 μg of the pCL vector (Ampho) was prepared in an microcentrifuge tube and incubated at room temperature for 20 min. The FuGENE + DNA mixture was dripped onto a 10-cm dish of 293T cells at 70% confluence that were growing in DMEM-high glucose culture media and incubated overnight. On the following day, transfection the media was replaced with fresh DMEM-high glucose culture media and the viral supernatant was harvested and filtered through a 45-μm syringe filter at 48 and 72 h post transfection.
To differentiate brown adipocytes, cell lines were split into differentiation media (high-glucose DMEM including 10% FBS, 1% antibiotics, 20 nm insulin and 1 nm T3) at day 1. Differentiation was induced with induction media (high-glucose DMEM including 10% FBS, 1% antibiotics, insulin 20 nm, 1 nm T3, 0.125 mm indomethacin, 2 μg/ml dexamethasone and 0.5 mm 3-isobutyl-1-methylxanthine (IBMX)) after they reached confluence at day 4. On day 6, cells were put back into differentiation media. The media was changed one more time on day 8. To generate Rictor-iKO cells, 4-hydroxytamoxifen was added into the differentiation media on day 8. At the end of day 12 the control and Rictor-iKO cells were treated in DMEM high-glucose without FBS for 12 h and stimulated with 150 nm of insulin for 1, 2, 5, 10, 15, or 30 min. Cells were harvested by washing with ice-cold PBS, and flash freezing of the plates in liquid nitrogen.
Mass Spectrometry Sample Preparation
Snap-frozen cells were lysed using ice-cold urea buffer (8 m urea, 50 mm Tris pH 8.2, 75 mm NaCl, 50 mm β-glycerophosphate, 1 mm sodium orthovanadate, 10 mm sodium pyrophosphate, 50 mm sodium fluoride, and 1x EDTA-free protease inhibitors (Pierce, Rockford, IL). Two wells of a six-well plate were pooled for each replicate. Cells from each well were lysed in 125 μl of urea lysis buffer, and subsequently vortexed and homogenized using a probe sonicator 2 × each while kept ice-cold. Lysates were then centrifuged at 16,000 × g to remove solid debris and lipids, and proteins amounts were quantified with a BCA assay (Pierce). Protein cysteine residues were then reduced with dithiothreitol (DTT) for 45 min at 55 °C, alkylated with 15 mm iodoacetamide for 30 min at room temperature in the dark, and quenched with an additional 5 mm DTT for 15 min at room temperature. Samples were then diluted one-fifth with 50 mm Tris pH 8.2, and digested with trypsin for 16 h at 37 °C. Digestions were quenched with trifluoroacetic acid (TFA) to pH < 2, and centrifuged for 10 min at 16,000 × g to remove precipitated debris. Clarified peptide samples were desalted on 50 mg Sep-Pak columns (Waters, Milford, MA), using 0.1% TFA to wash and 70% acetonitrile with 0.25% acetic acid to elute. De-salted samples dried using vacuum centrifugation, and a portion of these dried peptides were used for whole proteome analysis.
Phosphopeptides were enriched using iron(III) immobilized metal-ion affinity chromatography (IMAC) using a Kingfisher Flex (Thermo Scientific, San Jose, CA). 250 μg of peptides were used with 70 μl of 5% iron(III)-loaded IMAC beads (Cube Biotech, Monheim, Germany). Peptide binding to beads was done for 30 min in 80% acetonitrile and 0.1% TFA. Beads were then washed with 3 × 80% acetonitrile and 0.1% TFA, eluted with 2.5% NH4OH and 50% acetonitrile, immediately acidified with 70% acetonitrile and 10% formic acid. Eluates were then filtered with 2-layer StageTips (12) to remove any residual beads, and dried using vacuum centrifugation.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Data Acquisition
Whole proteome analysis was done using data-dependent acquisition (DDA) on a EASY-nLC II (Thermo Scientific) coupled to a QExactive mass spectrometer (Thermo Scientific). LC was performed with a 40 cm long, 100 μm inner diameter column packed in-house with Reprosil C18 3 μm beads (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany), with a column oven set to 50 °C. Prior to LC-MS/MS analysis, peptide samples were dissolved in a solution with 4% formic acid and 3% acetonitrile. Samples were separated with a linear gradient of 9–32% acetonitrile in 0.15% formic acid over 66 min. MS acquisition was performed using data-dependent acquisition with a “Top-20” method and 40 s dynamic exclusion. Precursor fragmentation was performed using higher-energy collisional dissociation with a 26% normalized collision energy. MS/MS acquisition was performed as follows: full-scan MS was acquired between 300–1500 m/z at 70,000 resolution, with a 3 × 106 automatic gain control (AGC) target and 100 ms maximum injection time. Precursor fragmentation was performed with a 2 m/z isolation window, and MS/MS acquisition was performed at 17,500 resolution with a 50 ms maximum injection time. All MS data was collected in centroid mode.
Global phosphoproteomics was performed using DDA on a nanoACQUITY (Waters) coupled to a Orbitrap Fusion mass spectrometer (Thermo Scientific). Column length and bead specifications were the same as for the whole proteome analysis. Prior to LC-MS/MS analysis, peptide samples were dissolved in a solution with 4% formic acid and 3% acetonitrile. Chromatographic separation was performed with a linear 5–23% acetonitrile over 90 min. Precursor fragmentation was performed using higher-energy collisional dissociation with a 29% normalized collision energy. MS/MS acquisition was performed as follows: full-scan MS was acquired between 375–1575 m/z at 120,000 resolution, with a 7 × 105 AGC target and 50 ms maximum injection time. Precursor fragmentation was performed with a 1.6 m/z isolation window with 30 s dynamic exclusion. MS/MS acquisition was performed at 15,000 resolution with a 50 ms maximum injection time and a flexible AGC target to maximize parallelizable time. The number of precursor fragmentations was tuned to achieve a 3 s duty cycle. All MS data was collected in centroid mode.
Targeted phosphoproteomics measurements were performed using parallel reaction monitoring (PRM) on a EASY-nLC II (Thermo Scientific) coupled to a QExactive mass spectrometer (Thermo Scientific). LC was performed with a 20 cm long, 100 μm inner diameter column packed in-house with Reprosil C18 1.9 μm beads (Dr. Maisch GmbH), with a column oven set to 50 °C. Chromatographic separation was performed with a linear 6–25% acetonitrile over 30 min at 500 nl/min. Precursor fragmentation was performed using higher-energy collisional dissociation with a 27% normalized collision energy. Full-scan MS was acquired between 300–1500 m/z at 70,000 resolution, with a 3 × 106 AGC target and 240 ms maximum injection time. 20 targeted precursor fragmentations were performed per duty cycle. Isolation was performed with a 2 m/z isolation window, and MS/MS acquisition was performed at 17,500 resolution with a 50 ms maximum injection time and a 2 × 105 AGC target. All MS data was collected in centroid mode. For the initial “calibration assay,” retention time prediction windows were ± 4 min, and for the final “quantification assay,” retention time prediction windows were ± 1.5 min.
Experimental Design and Statistical Rationale
All proteomics experiments were collected in six biological replicates, in two batches of three. Each batch was performed on cells derived from different mice, thawed on separate days, and subject to independent differentiation and knockout induction. We previously determined that six replicates is sufficient for uncovering meaningful significant changes, corrected for multiple hypotheses, in global, unbiased analysis of phosphopeptides. For all proteomics and phosphoproteomics data, batch effects caused by sample preparation batches and chromatographic instability were removed using the removeBatchEffect function from the R package “limma.” All subsequent data analysis was performed on the batch effect-corrected samples. For the targeted phosphoproteome analysis, only phosphopeptides that were quantified in at least two replicates per condition were included, and the remaining values were imputed to the lowest measurement for that phosphopeptide across all replicates. We observed that quantification values of each protein and phosphorylation site roughly followed a normal distribution in each experimental condition. Because of the multiple variables in our time course analysis, statistical significance of phosphorylation sites or proteins that respond to insulin, Rictor-iKO, or an interaction between the two was determined by ANOVA corrected by the Benjamini-Hochberg method. To determine phosphorylation sites that significantly increase or decrease in response to insulin in control or Rictor-iKO cells specifically, we implemented a natural cubic spline model that has been described previously (13). Specifically, for each protein the model tries to fit a flexible natural cubic spline function to the log2 change in expression over time and assesses its goodness-of-fit relative to a flat horizontal line. Prior to fitting the model, we imputed missing values to the lowest measurement for each site across all replicates, only keeping sites that were measured in at least three replicates per condition. Whenever single time points were compared between control and Rictor-iKO samples, a Student's t test was used, corrected by the Benjamini-Hochberg method when appropriate. Statistical significance was defined as q < 0.05. In rare cases, biological replicates were removed from analysis if their protein or phosphorylation quantifications showed low Pearson's r correlations with all other samples. Vehicle-treated Ubc-CreERT2;Rictorl/l brown adipocytes were used as a control to establish baseline serum-starved and insulin-stimulated protein and phosphorylation levels. No technical or injection replicates were analyzed for the samples. Samples were analyzed by LC-MS/MS in an unblinded fashion, in blocks containing one replicate of each condition, and the order within each block was randomized using the sample() function in R.
Peptide Identification, Pooling, and Quantification for DDA Experiments
Raw files were converted to mzXML format and searched using Comet (14) (version 2015.02 rev.2) against the mouse SwissProt database including isoforms (downloaded May 10, 2015, 24,750 protein sequences). Methionine oxidation and N-terminal protein acetylation were treated as variable modifications, and carbamidomethylation of cysteine residues was treated as a constant modification. In the case of phosphoproteome analysis, phosphorylated serine/threonine/tyrosine residues were also treated as variable modifications. Trypsin digestion was defined with cut-sites at lysine/arginine except when followed by proline, and peptides were allowed to have up to two missed cleavages. Precursor mass tolerance was set to 50 ppm, and fragment mass tolerance was 0.02 Daltons. Because of its consideration of many score features, Percolator (15) (version 3.1.2) was used to filter the search results to obtain a list of peptide-spectrum matches at a <1% false discovery rate. Phosphorylation site localization was performed using an in-house implementation of Ascore (16) and sites with an Ascore > 13 (p < 0.05) were considered localized, consistent with prior work (17). Quantification was performed using ThunderQuant, an in-house developed software for peptide quantification that relies on MS1 peak area integration. Peptide quantifications were pooled into protein quantifications, where the mean peptide quantification for each protein was taken to represent the protein. Protein quantifications were normalized to the total signal from each run. Proteins, occasionally including multiple isoforms per sample, were inferred from identified peptides using ProteinProphet (18) with a probability threshold of > 0.95. Phosphorylated peptides were pooled into phosphorylation site quantifications using the mean quantification of peptides that represented each site. Phosphorylation site localization was performed using an in-house implementation of Ascore that admits any post-translational modification setting. We set an Ascore threshold of >13, which corresponds to a >95% probability of correct assignment. When uploading annotated spectra to a public repository (see Data Availability), raw MS/MS data was converted to mzML using msconvert from ProteoWizard release 3.0.20002.
Development and Analytical Validation of Targeted MS Measurements
We performed Tier 3 targeted PRM analysis in this study. The list of targets for targeted phosphoproteomics was assembled by manual curation of phosphorylation sites from the literature and referencing an in-house database of mouse phosphopeptides detected using DDA to determine the best precursor m/z and an indexed retention time for each phosphorylation site of interest. In some cases where one peptide was not the obvious choice for a phosphorylation site, multiple peptides were chosen. The initial list consisted of nearly 400 phosphopeptides. To narrow down the list into 100–200 that can fit in a single one-hour targeted MS method, the 400 peptides were split up into six separate targeted methods and measured with wide retention time windows in a pooled brown adipocyte phosphopeptide sample. This allowed us to prioritize peptides that were abundant in our brown adipocyte sample, and ensure that only one peptide per site, or combination of sites, was included in the final assay. A response-to-concentration curve was not generated for this analysis. An additional 12 phosphopeptides were used for run-to-run normalization, as they did not change across our time series conditions in our DDA data.
Raw files were analyzed directly in Skyline Daily (19) version 19.0.9.149, where the data was manually analyzed using a custom transition list generated for small-molecule mode. Peak picking was performed by Skyline and manually inspected. Interfering fragment ions were removed by manual inspection in Skyline based on co-elution, mass error, and relative abundance compared with other fragment ions. Quantification was performed by summing the fragment ion intensities. Identification of peptides in PRM was determined using a database of DDA data generated in-house, matched by m/z and retention time prediction, an approach that was described previously (20). Raw files were also searched using the same pipeline that was used for the DDA data to identify peptides and phosphorylation site localizations from each PRM MS/MS spectrum. Manual annotation of PRM targets, informed by the search results, allowed for confident identification of peptides and assignment of their respective chromatographic peaks.
Bioinformatic Analysis
Normalization of phosphorylation sites to their respective proteins was done in two ways, dependending on the analysis. ANOVA statistics for protein-normalized phosphorylation were calculated by normalizing phosphorylation site quantifications to their respective protein in each replicate, and calculating statistics based on these new quantitative values. When only average fold-changes were required for an analysis, the protein normalization was performed at the level of averages per condition with a threshold of ≥ 3 replicates observed per condition. Hierarchical clustering heatmaps of phosphorylation sites were performed, using the R package “gplots,” on any phosphorylation site that significantly responds to insulin, as defined by its spline fit, in either the control or Rictor-iKO cells. To focus the clustering analysis on the insulin response, each condition was normalized to the median condition in each time course. Principal component analysis was performed on the 500 phosphorylation sites with the highest coefficient of variation across all samples. Gene ontology enrichment analysis on proteomics data was performed using the 1D enrichment tool in Perseus (21).
Analysis of kinase substrate sets was performed by downloading kinase-substrate annotations for the human and mouse proteomes from the PhosphositePlus database (22), and mapping them to our data via the PhosphositePlus group ID. Enrichment of kinase substrates in conjunction with clustering was performed using a Fisher's Exact test in Perseus, corrected using the Benjamini-Hochberg method. For analysis of RKO response ratios of kinase substrate sets, significance was assessed using one-sample Student's t-tests corrected by the Benjamini-Hochberg method. Analysis of subcellular localizations was performed similar to kinase substrate sets, except the localization terms were selected GO cellular components, namely “mitochondrion,” “nucleus,” “Golgi apparatus,” “endoplasmic reticulum,” “lysosome,” “spliceosomal complex,” “ribosome,” “cytosol,” “plasma membrane,” “cytoskeleton,” and “extracellular matrix.”
In plots showing individual data points, bars or lines represent the median. For volcano plots, the mean log2 fold-change ratio is shown on the x axis. Fold-changes and the RKO response ratio were calculated based on the means at each time point condition. When a fold-change is averaged across time points, it is calculated as the mean of time point means.
Mice
C57BL/6J mice (JAX stock 000664) were obtained from Jackson Laboratory (Bar Harbor, ME). Ucp1-Cre (JAX stock 024670) was kindly provided by Dr. Evan Rosen. Ucp1-Cre;Rictorl/l mice was generated by crossing Ucp1-Cre with Rictorl/l. Mice were housed in the UMMS Animal Medicine facilities in a room set at 22 °C in 45% humidity under daily 12 light/dark cycles. For all in vivo studies (Western blot analysis and RT-PCR analysis) we used 14 week-old Rictorl/l (as control) or Ucp1-Cre;Rictorl/l mice (as iBAT specific Rictor KO). All mouse experiments were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Western Blot Analysis
Cells were homogenized in a Triton-X lysis buffer containing 150 mm NaCl, 50 mm Tris pH 7.5, 0.5% deoxycholate, 1% Triton X-100 and 0.1% SDS, a protease inhibitor mixture (Biotool Cat. Number B14011; Houston, TX), and a phosphatase inhibitor mixture (Sigma Cat. Number P5726; St. Louis, MO). For in vivo analysis, tissue was homogenized in RIPA lysis buffer containing 150 mm NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris, pH 7.5 including a protease (Biotool Cat. Number B14011) and phosphatase inhibitor mixture (Sigma Cat. Number P5726). Samples were run on 10% SDS-Polyacrylamide gels for general protein analysis. Equal sample loading was verified by Ponceau S staining after transfer to the blotting membrane. The antibodies used for Western blot analysis were: Rictor (#9476), pan-AKT (#4691), phospho-AKT (S473) (#4058), phospho-AKT (T308) (#4056), β-tubulin (#2146), and PKA substrate motif (#9624) from Cell Signaling Technologies (Danvers, MA).
Gene Expression Analysis
Cells or tissues were lysed with Qiazol (Invitrogen, Carlsbad, CA) and total RNA was isolated with the RNeasy kit (Invitrogen). Equal amounts of RNA (2 μg) were retro-transcribed to cDNA using a high-capacity cDNA reverse transcription kit (#4368813, Applied Biosystems, Foster City, CA). Quantitative real-time PCR was performed in 10 μl reactions using a StepOnePlus real-time PCR machine from Applied Biosystems using SYBR Green PCR master mix (#4309156, Applied Biosystems) according to manufacturer's instructions. Relative mRNA expression was determined by the ΔΔCt method and Tbp expression was used as a normalization gene in all conventional PCR with reverse transcription experiments. Primer information is listed in supplemental Table S1.
RESULTS
Time Course Analysis of Insulin Signaling in Control and Rictor-iKO Brown Adipocytes
To identify mTORC2 effectors in brown adipocytes, we employed a recently-developed model whereby mTORC2 can be inducibly inhibited in differentiated brown adipocytes (3). Briefly, preadipocytes isolated from Ubc-CreERT2;Rictorl/l mice were immortalized, differentiated, and treated with 4-hydroxytamoxifen late during differentiation to induce Rictor deletion after PPARγ induction (hereafter Rictor-iKO), or vehicle to generate isogenic controls (Fig. 1A). Western blots confirm loss of RICTOR protein and attenuation of insulin-stimulated AKT hydrophobic motif phosphorylation (supplemental Fig. S1A).
Fig. 1.
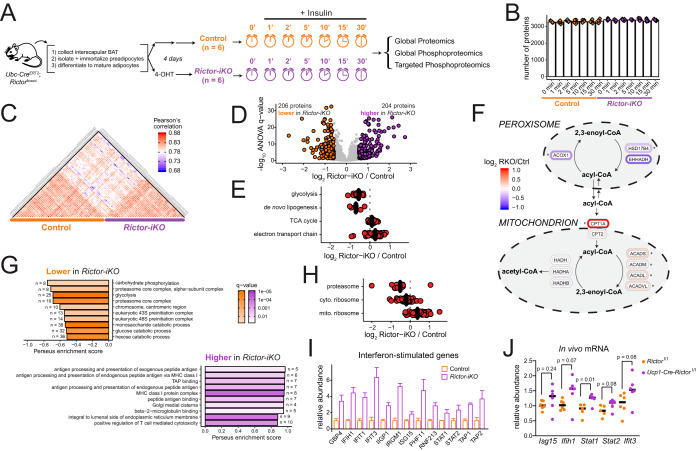
Rictor knockout reprograms metabolism and up-regulates an interferon-like response in brown adipocytes. A, A description of the inducible knockout system (left) and a schematic overview of proteome and phosphoproteome analysis over an insulin time series of brown adipocytes. B, The number of quantified proteins in each sample, organized by experimental condition. Bar height represents the median of all replicates for that condition. C, Heatmap showing Pearson's r correlation of protein abundance measurements between samples. D, Volcano plot showing the Rictor-iKO to control log2 fold-change averaged across all time points (x axis) and the -log10 ANOVA q-value (y axis) for proteins. Proteins significantly decreased in Rictor-iKO cells are shown in orange, and those significantly increased in Rictor-iKO cells are shown in purple. E, Protein abundance fold-change between Rictor-iKO and control (the mean of all time points) are shown for enzymes in the indicated metabolic pathways. F, Rictor-iKO versus control fold-change protein abundance data (the mean of all time points) mapped to a pathway model of fatty acid oxidation in the mitochondria and peroxisome. Asterisk (*) next to proteins indicates ANOVA q < 0.005. G, Top ten GO enrichment results of proteins that are higher in the control (top) or higher in the Rictor-iKO (bottom). H, Protein abundance fold-change between Rictor-iKO and control (mean of all time points) are shown for proteins in the proteasome, and the cytosolic and mitochondrial ribosomes. I, Bar plots for protein abundance of selected proteins previously reported as “interferon-stimulated genes.” Bar height is the mean fold-change across all time points, normalized to the control, and the error bars are standard deviation. J, RT-PCR analysis of selected interferon-stimulated genes from interscapular brown fat from Rictorl/l (control) and Ucp1-Cre;Rictorl/l mice.
Phosphorylation downstream of insulin stimulation in the 3T3-L1 adipocyte model is highly dynamic during the first 10 min following stimulation (6). To probe the temporal dynamics of insulin-stimulated mTORC2 signaling in brown adipocytes, we performed a similar time-course analysis. Control and Rictor-iKO brown adipocytes were serum-deprived for 12 h, then stimulated with 150 nm insulin for 0, 1, 2, 5, 10, 15, and 30 min, where “0 min” is the unstimulated control (Fig 1A). To control reproducibility, in vitro differentiation was performed in two batches on separate days, and three replicates were harvested per time point per batch. Each biological replicate was analyzed in a 'single-shot' injection with label-free quantification based on MS1 peak area. Quantification values were corrected for batch effects because of sample processing and chromatographic drift as described in the Methods. In rare cases, specific replicates were removed from analysis if their quantifications had low Pearson r correlations with all other samples. Subsequent proteome and phosphoproteome analysis were integrated to identify protein level and protein phosphorylation differences between control and Rictor-iKO cells.
Proteome Analysis Reveals Changes to Diverse Cellular Processes in Rictor-iKO Brown Adipocytes
First we quantified protein abundance to identify changes in the proteome induced within 30-min insulin stimulation and protein abundance changes between control and Rictor-iKO cell (supplemental Data set S1). Each sample was analyzed in a single run on a QExactive mass spectrometer, and each identified over 3000 proteins, including isomers (Fig. 1B). Global comparison of protein abundances between samples revealed higher correlation between sample replicates or different timepoints in the same group (control or Rictor-iKO) than when comparing the control and Rictor-iKO groups (Fig. 1C).
To assess the insulin responsiveness of the control and Rictor-iKO proteomes, we implemented a non-linear model (13) independently for the control and Rictor-iKO groups, and found no statistically significant differences in protein abundance as a result of acute insulin stimulation.
To determine the effect of RICTOR loss on the proteome independently of acute insulin stimulation, protein abundance measurements from each insulin time point were averaged and compared between control and Rictor-iKO cells. At a 5% false-discovery rate (FDR) threshold, we identified 206 proteins that significantly decrease in abundance ≥ 1.5-fold in RICTOR deficient cells, and 204 proteins that increase ≥ 1.5-fold (Fig. 1D), in total amounting to 15% of all proteins quantified in at least 3 replicates per condition. Thus, the effect of RICTOR loss on the brown adipocyte proteome was substantial.
mTORC2 is known to regulate the abundance of de novo lipogenesis (DNL) enzymes in BAT (4, 5). To further investigate the role of mTORC2 in brown adipocyte metabolism, we analyzed protein abundance levels in a set of core metabolic pathways. This analysis confirms that, in addition to DNL enzymes, all 21 glycolytic enzymes we measured are decreased in abundance by Rictor-iKO (Fig. 1E, supplemental Data set S1). Expression of the glycolytic genes Hk2, AldoA, and Pfkp is also decreased in isolated BAT from Ucp1-Cre;Rictorl/l relative to control Rictorl/l mice (supplemental Fig. S1B), suggesting that this response is relevant in vivo. This is consistent with previously-reported mTORC2-dependent glycolytic gene expression in glioblastoma cells (23). In contrast to the aforementioned cytosolic pathways, we find that mitochondrial tricarboxylic acid (TCA) cycle enzymes (43% with q < 0.05) and electron transport chain components (58% with q < 0.05) are on average more abundant in Rictor-iKO cells (Fig. 1E, supplemental Data set S1). Fatty acid oxidation (FAO) enzymes follow a similar pattern, as peroxisomal FAO enzymes are less abundant in Rictor-iKO cells whereas mitochondrial FAO enzymes are unchanged or more abundant (Fig. 1F, supplemental Data set S1). This is consistent with previous findings that expression of mitochondrial genes, mitochondrial size, and oxygen consumption rate, are slightly higher in Rictor-deficient BAT (4). In summary, core cytosolic and peroxisomal metabolic enzymes are less abundant in Rictor-iKO, whereas mitochondrial enzymes are increased.
We performed gene ontology (GO) enrichment analysis to identify additional molecular functions, biological processes, and cellular compartments that are overrepresented among the RICTOR-regulated proteins. Terms related to the proteasome and ribosome were enriched among proteins that are lower in the Rictor-iKO cells (Figs. 1G, supplemental Fig. S1C). This is also apparent by the overall decrease in core proteasome and cytosolic ribosome proteins in Rictor-iKO cells (Fig. 1H) and suggests that protein turnover may be slower in Rictor-iKO cells. In contrast, mitochondrial ribosomal proteins are on average more abundant in Rictor-iKO cells (Fig. 1H), consistent with differential regulation of cytosolic versus ribosomal proteins. Among proteins that increase in Rictor-iKO brown adipocytes, several GO terms related to antigen presentation, as well as the term “response to interferon-beta”, are significantly enriched (Figs 1G, supplemental Fig. S1D). We assessed known interferon-regulated proteins in our data, and several significantly increased as a result of RICTOR loss (Fig. 1I). We measured expression levels of a set of interferon-stimulated genes in isolated BAT from Ucp1-Cre;Rictorl/l and control Rictorl/l mice using RT-PCR, and found that the transcript abundance of Stat1 significantly increases in Rictor-deficient BAT with p = 0.05, whereas Ifih1, Ifit3, and Stat2 are increased with p ≤ 0.08 (Fig. 1J). Although the in vivo results are preliminary, they underscore a potential physiological role for mTORC2 in suppressing interferon signaling in brown adipocytes. Collectively, our results suggest that beyond regulating DNL and glycolysis, mTORC2, directly or indirectly, may regulate mitochondrial metabolism, protein turnover and immune response.
Rictor-iKO Decreases Phosphorylation on Basophilic Motif Sites on Perilipin-1
We Next Assessed Global Differences in protein phosphorylation between control and Rictor-iKO cells during the 30 min insulin time course (supplemental Data set S2). Phosphopeptide samples were obtained by immobilized metal ion affinity chromatography enrichment and subjected to single shot LC-MS/MS analysis on an Orbitrap Fusion mass spectrometer. This analysis identified over 5,000 unique phosphorylation sites or protein regions per sample (Fig. 2A). Phosphorylation site localization was performed using an in-house implementation of Ascore (16). Pairwise Pearson's correlation r values between replicates shows a clear separation between control and Rictor-iKO samples (Fig. 2B). Principal component analysis (PCA) distinguishes control and Rictor-iKO cells by principal component 1, whereas duration of insulin stimulation separates samples along principal component 2 (Fig. 2C). To distinguish whether this PC1 separation between control and Rictor-iKO is caused by differences in total protein levels or phosphorylation site occupancy, we compared fold-changes for phosphorylation sites and their respective total protein levels between control and Rictor-iKO cells for each time point. Correlations between phosphorylation and total protein levels were poor, with r2 ≤ 0.062 for each time point (supplemental Fig. S2A). This indicates that control and Rictor-iKO brown adipocytes have markedly different phosphorylation patterns that are likely because of site occupancy rather than protein abundance.
Fig. 2.
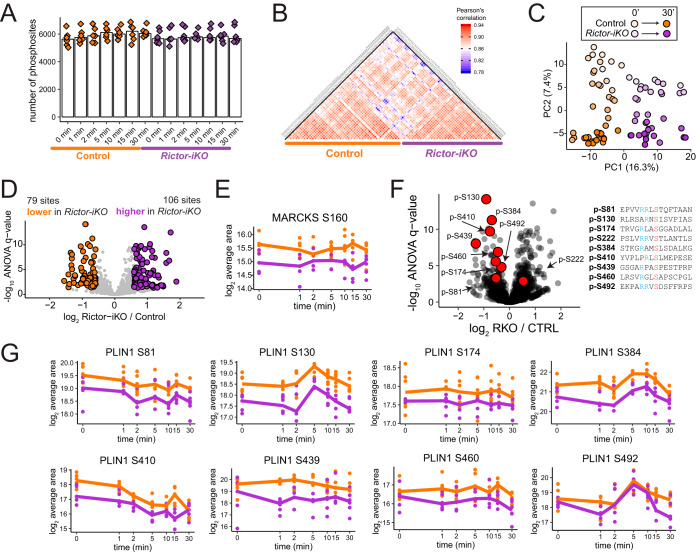
Basal phosphorylation levels in brown adipocytes are significantly altered by Rictor knockout. A, The number of quantified phosphorylation sites in all samples, organized by experimental condition. B, Heatmap showing Pearson's r correlation of phosphorylation site abundance measurements between samples. C, Principal component analysis showing the effect of both Rictor-iKO and insulin on brown adipocyte phosphorylation levels. D, Volcano plot showing the Rictor-iKO to control log2 fold-change averaged across all time points (x axis) and the -log10 ANOVA q-value (y axis) for protein-normalized phosphorylation sites. Sites significantly decreased in Rictor-iKO cells are shown in orange, and those significantly increased in Rictor-iKO cells are shown in purple. E, Protein-normalized quantification of MARCKS S160 phosphorylation. F, Identical volcano plot to 2D, except that sites on perilipin-1 are highlighted in red. The amino acid sequence windows for these sites are shown to the right with the phosphorylated residue in red and upstream arginine residues in blue. G, Protein-normalized quantification across the time series for phosphosites on perilipin-1.
To identify phosphorylation differences arising from RICTOR loss regardless of insulin sensitivity we normalized the phosphorylation measurements to total protein levels. After normalization, the distributions of log2 fold-changes between time points and between genetic backgrounds have similar spread, meaning that acute insulin stimulation and chronic RICTOR loss cause phosphorylation changes of similar magnitude (supplemental Fig. S2B). We next applied ANOVA to phosphorylation measurements that were protein-normalized on a replicate-to-replicate basis, and found that at a 5% FDR threshold, 79 phosphorylation sites are significantly lower in Rictor-iKO cells and 106 sites are significantly higher by 1.5 fold averaged across all time points (Fig. 2D). Among the phosphosites significantly decreased in Rictor-iKO cells, we found S160 on the myristoylated alanine-rich protein kinase C substrate (MARCKS) (Fig. 2E), consistent with decreased activity of PKC, a known mTORC2 substrate. In addition, eight basophilic residues on perilipin-1 (PLIN1), which coats lipid droplets and regulates lipolysis, have lower phosphorylation levels in Rictor-iKO cells (Figs. 2F, 2G). Some of these PLIN1 sites increase in response to insulin, whereas others do not (Fig. 2G). Phosphosite S81 is within a protein kinase A (PKA) consensus motif (RRx[S/T]) that decreases across the insulin time series (Fig. 2G), and is one of three residues whose phosphorylation together enable maximum cAMP-driven lipolysis (24). Phosphorylation at S410 (an Rxx[S/T] motif) also decreases in response to insulin and is similarly RICTOR-dependent (Fig. 2G). Phosphorylation at PLIN1 S492 (RRx[S/T] motif) is reported to control cAMP-induced lipid droplet fragmentation (25). This phosphosite shows an increase with insulin that peaks at 5 min, suggesting that AKT or a different insulin-activated basophilic kinase, but not PKA, may phosphorylate this site (Fig. 2G). Rictor-iKO cells also have increased phosphorylation on diverse proteins, including the proteasome core subunit PSMA5 S56, and filamin-A S2152. In sum, the phosphoproteome is altered in Rictor-iKO cells, with changes including several PLIN1 phosphosites that are dependent on RICTOR, suggesting the potential to regulate lipolysis and lipid droplet dynamics.
RICTOR-dependence of Inferred Kinase Activities and Subcellular Localizations in Serum-starved Brown Adipocytes
We next sought to characterize RICTOR-dependent phosphorylation differences which occur in the serum-starved (insulin-unstimulated, time 0), referred to hereafter as “basal” state. We annotated phosphoproteins according to a custom list of GO cellular compartments and globally assessed phosphosite abundance ratios between Rictor-iKO and control cells on each compartment. This analysis showed that the mTORC2-regulated phosphoproteome in basal state is not specific to particular subcellular localizations (supplemental Fig. S2C). Next, we assessed basal differences in kinase activities using annotations of kinase-substrate relationships from PhosphositePlus (22). Despite identifying RICTOR-dependent PLIN1 phosphosites that match a PKA motif, we did not observe a significant difference of PKA substrate phosphorylation in the basal state (supplemental Fig. S2D). Consistently, we did not observe significant differences in global PKA motif phosphorylation between BAT from control and Ucp1-Cre;Rictorl/l mice (supplemental Fig. S2E). By expanding this analysis to other kinases, however, we found that AKT1 has significantly higher basal substrate phosphorylation, by ∼1.4-fold, in Rictor-iKO cells relative to controls (supplemental Figs. S2D, S2F). Although interesting, we note that these results should be interpreted with caution. First, because substrate specificity of AKT isoforms is poorly understood, annotations of AKT1 substrates may also include targets of other AKT isoforms. Second, it is also possible that the basal phosphorylation levels of AKT substrates in the Rictor-iKO could be set by a kinase different than AKT or a phosphatase.
The Brown Adipocyte Insulin Response Follows Temporal Progression from the Plasma Membrane to the Nucleus
To characterize dynamic insulin signaling in brown adipocytes, we first identified distinct temporal patterns of phosphorylation in the control by performing hierarchical clustering on the phosphorylation sites according to their abundance over the time course. This analysis reveals 11 clusters of phosphosites that distinguish earlier versus later responders, acute versus sustained responders, and increasing versus decreasing phosphosite levels over time (Fig. 3A). Cluster 1 contains sites that reach near to their maximum at 1 min (Figs 3A, 3B). Clusters 3, 4, and 5 contain later responders, and cluster 3 sites additionally exhibit a drop in their levels by 30 min, suggesting negative feedback or increased susceptibility to phosphatase activity (Figs. 3A, 3B, supplemental Fig. S3B). Cluster 6, 7, 9, and 11 comprise sites that decrease in response to insulin, although cluster 9 also exhibits a slight increase at 30 min (Figs. 3A, 3B, supplemental Fig. S3B, S3C). We next benchmarked our data against a previous dynamic insulin phosphoproteome study in 3T3-L1 adipocytes (6). We classified data from Humphrey et al. according to our clusters, and found that median temporal profiles for the phosphosites in each cluster are largely similar in both data sets (supplemental Fig. S3A). Thus, our time series analysis recapitulates previously described patterns of insulin-responsive phosphorylation in 3T3-L1 adipocytes.
Fig. 3.
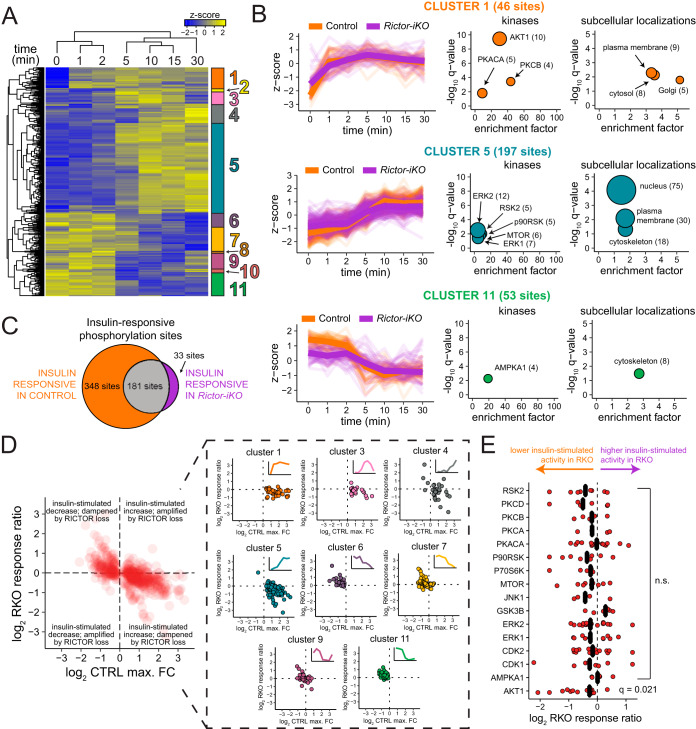
Rictor knockout dampens early and late insulin response. A, Hierarchical clustering of phosphorylation site abundances in the control reveals distinct patterns of increased and decreased phosphorylation over the insulin time series. Heat map shows the z-score of the mean phosphorylation site abundance for each condition. B, Z-scores (left) for each cluster defined in (A), with the median for each time point shown in bold. Each selected cluster was analyzed for enrichment of kinase substrates (middle) and subcellular localizations (right), where the fold-enrichment is plotted on the x axis, the y axis contains the -log10 q-value from a Fisher's exact test corrected by the Benjamini-Hochberg method, and the size of the circles corresponds to the number of sites represented by each term. C, Euler diagram showing the number of phosphorylation sites, in the control and Rictor-iKO, that fit a natural cubic spline function (q ≤ 0.05). D, Scatter plots assessing the RICTOR-dependence of the insulin response. The maximum insulin log2 fold-change in the control cells is shown on the x axis, and the log2 RKO response ratio is shown on the y axis. All insulin responsive sites are shown to the left in red; on the right they are organized by cluster. E, Analysis of kinase substrates from PhosphositePlus. The log2 RKO response ratio is shown on the x axis, q-values correspond to one-sample Student's t test followed by Benjamini-Hochberg correction.
To functionally characterize the temporal clusters, we annotated kinase-substrate relationships (22) and GO cellular components in each cluster to calculate enrichments for specific kinase substrates and subcellular localizations, respectively. This revealed that Cluster 1 (early insulin responders) is significantly enriched for AKT substrates (Fig. 3B). PKA catalytic subunit A (PKACA) and protein kinase C beta substrates are also enriched in cluster 1 (Fig. 3B), but many of these sites are also annotated as AKT1 substrates; thus, their enrichments are likely secondary to AKT1. Cluster 1 is also enriched for proteins annotated with the GO cellular component term “plasma membrane,” including AKT2, SHC1, and CAV1 (Fig. 3B, supplemental Data set S2). Cluster 5 is enriched for substrates of mTOR, ERK, p90RSK, and RSK2, consistent with cluster 5 sites being relatively distal to insulin receptor activation at the plasma membrane. Likewise, clusters 3, 4, and 5 show enrichment for nuclear proteins (Figs 3B, (supplemental Fig. S3B)). Cluster 11 is enriched for AMPK substrates and cytoskeletal proteins (Fig. 3B), consistent with insulin promoting anabolism and suppressing catabolism. In agreement, a reported inhibitory AKT phosphorylation site on AMPK (S496) (26) increases after 1 min insulin stimulation (supplemental Data set S2), preceding the average decrease in cluster 11 AMPK substrate sites (Fig. 3B). In sum, clustering the temporal response to insulin in the control brown adipocytes reveals distinct waves of kinase activity with an early (∼1 min) enrichment of plasma membrane and Golgi sites followed by later (∼5 min) enrichment of nuclear sites.
RICTOR Loss Mildly but Broadly Dampens Insulin Signaling
To assess insulin responsiveness in control and Rictor-iKO cells, we applied the natural cubic spline model described above to each insulin time series. We identified 528 phosphosites that significantly change in response to insulin in control cells, and 214 that significantly change in the Rictor-iKO cells (Fig. 3C). This difference suggests that the insulin response is weaker and/or less robust in Rictor-iKO brown adipocytes relative to control. We next compared the temporal profiles of the control and Rictor-iKO insulin responses by mapping the z-score-normalized temporal profiles of Rictor-iKO cells to the clusters defined above from the control cells. This revealed that the profiles are largely the same (Figs 3B, supplemental Fig. S3B, S3C), meaning that RICTOR loss does not cause a temporal shift in the insulin response.
To quantify the effect of RICTOR loss on the magnitude of the insulin response, we calculated a Rictor-iKO versus control insulin response ratio (“RKO response ratio” hereafter) that reflects differences in the maximum insulin response because of RICTOR loss. The RKO response ratio was calculated in three steps by first determining the time point of highest absolute response to insulin (either positive or negative) relative to the unstimulated condition, then calculating the fold-change relative to unstimulated for that time point (i.e. maximum insulin response), and finally determining the ratio of the Rictor-iKO fold-change to the control fold-change. Plotting the RKO response ratios against the maximum insulin response in the control shows that RICTOR loss dampens, to some degree, both insulin-stimulated increases and decreases in phosphorylation (Fig. 3D). The most RICTOR-dependent clusters were clusters 1, 4, 5, and 6. Of those, clusters of increasing sites—clusters 1, 4, and 5—all had an average of ∼1.2-fold dampening in Rictor-iKO cells, whereas decreasing sites in cluster 6 had an average of ∼1.3-fold dampening. RKO response ratio did not appear to correlate with subcellular localization (supplemental Fig. S3D). We also surveyed annotated kinase substrate sets to identify putative RICTOR/mTORC2-dependent, insulin-responsive kinases. Although almost all sets surveyed had an average insulin response dampening of 1.1-fold or greater in Rictor-iKO cells, only AKT1 substrates were significantly (q < 0.05) dampened, by a median of 1.2-fold (Fig. 3E). In sum, when assessing the global phosphoproteome, RICTOR loss broadly, yet mildly, dampens insulin-sensitive phosphorylation in brown adipocytes.
To identify individual phosphorylation sites with RICTOR-dependent insulin responses, we compared the insulin temporal profiles of Rictor-iKO and control cells using 2-way ANOVA. This approach revealed AMFR S542 to have a 1.5-fold, significantly decreased insulin response in RICTOR-deficient brown adipocytes (q = 7 × 10−4) (supplemental Fig. S3E, supplemental Data set S2). AMFR is an E3 ligase, member of the VCP/p97-AMFR/gp78 complex that participates in the final step of endoplasmic reticulum-associated degradation, and phosphorylation of S542 on AMFR by p38 has been shown to regulate mitochondrial-endoplasmic reticulum interactions and mitochondrial motility (27). In addition, we observed a 2.1-fold decreased insulin response of ACLY S455 at q = 0.07 (supplemental Fig. S3E, supplemental Data set S2). ACLY is a key enzyme in de novo lipogenesis, its protein levels are decreased in Rictor-iKO BAT (4). ACLY S455 is an AKT substrate and we recently showed that its phosphorylation is RICTOR-dependent in brown preadipocytes and is important for differentiation into mature adipocytes (5). As a third example, TPR S2223 is a cluster 3 site which has a markedly different temporal profile characterized by dampening of both its maximum insulin response (1.2-fold), and its decrease at 30 min (1.7-fold). TPR is a nuclear pore complex protein and TPR S2223 is an ERK substrate of unknown function (28). In sum, our global phosphoproteomic analysis of the insulin response in brown adipocytes revealed temporally distinct classes of phosphorylation sites and a general dampening of insulin action in Rictor-iKO cells, most significantly in phosphorylation of sites on ACLY, AMFR, and TPR.
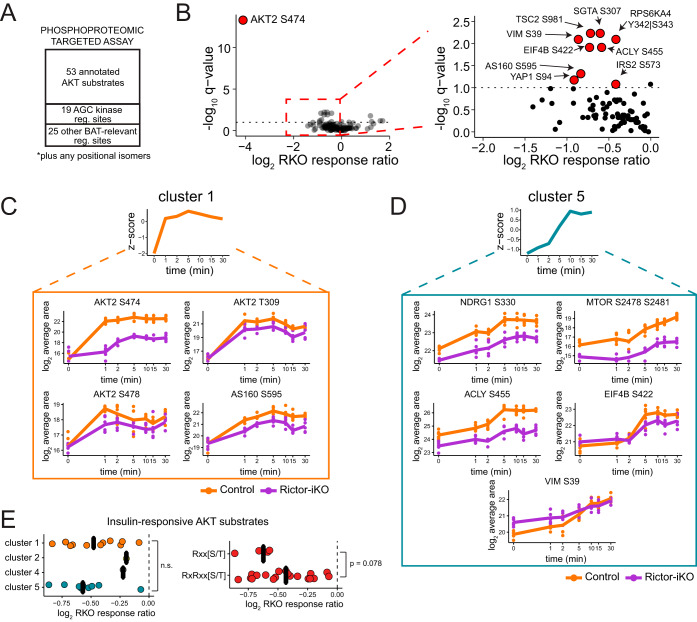
Targeted Analysis of a Panel of Phosphorylation Sites in the Insulin/AKT Pathway
To improve sampling and quantitative accuracy of the insulin/AKT pathway and more specifically identify Rictor-dependent AKT substrates in mature brown adipocytes, we complemented our global phosphoproteomics with a targeted MS assay that systematically measures phosphopeptides of choice. This targeted analysis is a more refined version of the assay performed previously on brown preadipocytes (5), with a few new phosphosites added to the assay to more broadly cover known AKT substrates and other signaling and BAT-associated phosphorylation sites (Fig. 4A). To configure the targeted assay, we used Phosphopedia (20), a web-portal that provides chromatographic and mass spectrometric properties informed by the aggregation of hundreds of phosphoproteomic experiments, along with an in-house mouse phosphoproteome database. The prior phosphoproteomic experiments served as evidence for identification of the phosphopeptides, along with manual inspection of fragment ions in Skyline (19). After optimization (see Methods), the targeted MS assay contained 166 phosphopeptides, which we applied to the two insulin time series (Rictor-iKO and control).
Fig. 4.
Targeted phosphoproteomics reveals that RICTOR loss dampens AKT- and ERK-dependent signaling downstream of insulin. A, Overview of targeted MS strategy for systematically measuring phosphopeptides corresponding to AKT substrates, AGC kinase phosphorylation sites, and other functional phosphorylation sites, across the control and Rictor-iKO insulin time series samples. B, Volcano plot showing the log2 RKO response ratio on the x axis, and the -log10 ANOVA interaction q-value on the y axis. Entire plot is shown to the left, and a detailed view is shown to the right. Phosphopeptides representing sites with the most significantly different insulin response are highlighted. C–D, Example phosphopeptides corresponding to clusters 1 (C) and 5 (D) in Fig 3A. E, The insulin response ratio is shown for all measured AKT substrates, organized by cluster (left, see Fig 3A) and phosphorylation motif (right). p values are from 1-way ANOVA.
Of all measured phosphopeptides, 110 yielded high-quality quantifications -based on the summed MS2 fragment intensities and assessed by their chromatographic peak shape and relative fragment ion intensity- in at least two replicates per condition (supplemental Data set S3). Quantification values were corrected for batch effects because of sample processing and chromatographic drift as described in the Methods. Because of its higher sensitivity, the targeted analysis allowed us to measure many phosphorylation sites that were absent or incompletely measured across the time series in the global phosphoproteomic analysis (supplemental Fig. S4A). Of the 110 quantified phosphopeptides, 52 were insulin-responsive in control cells, and 32 were insulin-responsive in Rictor-iKO cells as determined by the cubic spline model described above (supplemental Data set S3). Insulin-responsive phosphopeptides in either the control or the Rictor-iKO cells were assigned to one of the clusters defined in Fig. 3A by calculating the Euclidean distance to the median temporal response from each cluster (supplemental Data set S3).
Activation loop, turn motif, and hydrophobic motif phosphorylation sites for all three AKT isoforms were targeted in our assay; however, only AKT2 sites were consistently measured across all conditions. Phosphosites on the AKT2 activation loop (T309) and hydrophobic motif (S474 and S478) are early insulin responders and were assigned to cluster 1. As expected, the insulin response of AKT2 S474 is significantly diminished (17-fold) in the Rictor-iKO (Figs. 4B, 4C, supplemental Data set S3). The insulin response of AKT2 S478 and T309 is also diminished in the Rictor-iKO, yet to a lesser extent (1.7-fold and 2.3-fold, respectively) (Fig. 4C, supplemental Data set S3) and not significantly (at 10% FDR). The phosphopeptide spanning the turn-motif region of AKT2 does not have a significant insulin response, but is significantly decreased in the knockout (supplemental Fig. S4B, supplemental Data set S3) relative to AKT2 protein levels, which do not change (supplemental Data set S1). This is consistent with the known mTORC2 dependence of AKT turn motif phosphorylation (29, 30). Of the other AGC kinase phosphorylation sites measured in our assay, the hydrophobic motif site S369 on RPS6KA1 (a.k.a. p90RSK1) had a dampened insulin-dependent increase response, albeit not statistically significant (supplemental Fig. S4C). In addition, a phosphopeptide from RPS6KA4 (a.k.a. MSK2) spanning the turn motif site, has statistically significant dampening (Figs. 4B, supplemental Fig. S4C). Beyond AGC kinases, previously-reported mTORC2-dependent phosphorylation sites at mTOR S2481 and NDRG1 S330, both classified as later insulin responders (cluster 5), are lower in Rictor-iKO cells (Fig. 4D) whereas their protein levels do not change (supplemental Data set S1), also consistent with loss of functional mTORC2.
We find that seven previously-characterized AKT substrate sites have significant RICTOR-dependent insulin responses at 10% FDR (Fig. 4B, supplemental Data set S3): TSC2 S981 and AS160 S595 (early insulin responding, cluster 1) (Figs 4B, 4C, supplemental Fig. S4D), VIM S39, EIF4B S422, SGTA S307, and as we described earlier, ACLY S455 (late insulin-responding, cluster 5) (Figs 4B, 4D). AKT substrates where the control insulin response is similar (i.e. within 1.2-fold) to the Rictor-iKO insulin response include known AKT substrates AS160 T649, FLNC S2234, and SLC9A1 S707 (supplemental Figs. S4B, S4E, supplemental Data set S3). We did not find any relationship between the temporal dynamics of insulin-responsive AKT substrates (clusters 1, 3, 4, and 5) and their RKO response ratio (Fig. 4E). However, among all annotated AKT substrates we targeted whose phosphorylation increased in response to insulin, sites matching a partial AKT motif (Rxx[S/T]) have more dampening in Rictor-iKO cells, on average, than sites matching the full AKT consensus motif (RxRxx[S/T]) (Fig. 4E).
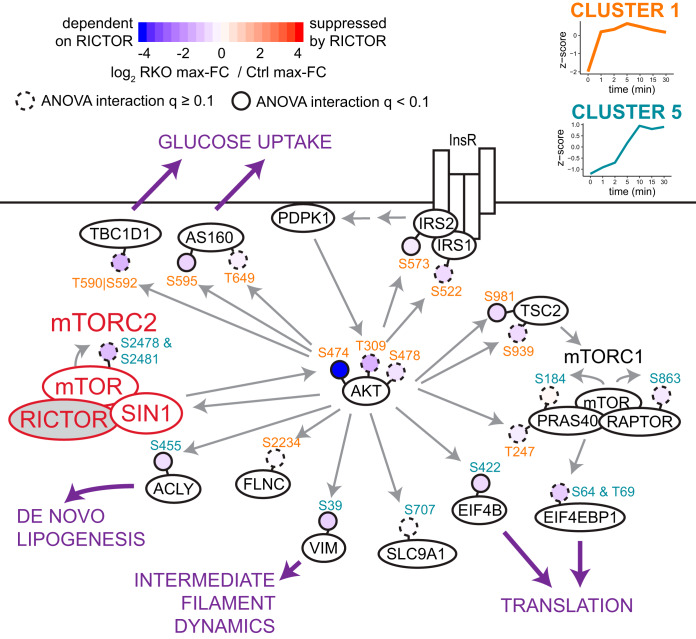
Fig. 5 shows the variety of AKT substrates whose insulin response is dampened in Rictor-iKO cells to different extents, along with their associated downstream functions. In sum, targeted phosphoproteomics confirms that RICTOR loss mildly dampens the insulin response of many phosphosites, with the most significant suppression observed for AKT2 S474 and set of AKT substrates that includes ACLY S455, EIF4B S422, and VIM S39.
Fig. 5.
Diverse impacts of RICTOR on the insulin signaling response. Pathway map showing the difference in insulin response between control and Rictor-iKO for selected phosphorylation sites (shaded circles) that increase in response to insulin in the control. Membership of phosphosites to clusters from Fig. 3A are color-coded in the site name. Solid gray arrows indicate direct regulation, typically driven by phosphorylation as indicated when the arrow points to a phosphorylation site. Purple arrows highlight downstream functional consequences that could be regulated by RICTOR-dependent insulin signaling.
DISCUSSION
mTORC2 is the least understood mTOR complex in terms of its downstream effectors and associated functions, despite its clear importance in regulating glucose and lipid metabolism. mTORC2 has been shown to act as an AKT kinase in response to insulin and other growth factors, yet the nature of its regulation of AKT activity is unclear. To address this, we analyzed the effects of mTORC2 loss on dynamic insulin-dependent phosphorylation and global protein abundance. Our results show that RICTOR deletion mildly dampens insulin-dependent phosphorylation; AKT substrate sites more strongly affected by RICTOR include ACLY S455, EIF4B S422, and VIM S39. In addition, we find that RICTOR loss decreases phosphorylation levels of several sites on perilipin-1, including some insulin-sensitive and some insulin-insensitive sites. Finally, we observe that independently of acute insulin stimulation, chronic RICTOR loss induces an interferon-like response in brown adipocytes.
How might RICTOR loss broadly dampen insulin signaling? The dampening of AKT substrate phosphorylation in response to insulin in Rictor-iKO cells is comparable to that of AKT2 T309 phosphorylation. The essential role of activation loop phosphorylation for AKT catalytic activity suggests that the dampened insulin/AKT response in Rictor-iKO cells could be explained by the decrease in AKT2 T309 phosphorylation. mTORC2 has also been shown to act in a positive feedback loop with AKT involving phosphorylation of the mTORC2 subunit SIN1 at T86 by AKT (31), but the effect of this feedback mechanism on broad downstream signaling has not been described. Although we were not able to measure SIN1 T86 phosphorylation with our phosphoproteomics approach, nor did we obtain good measurement of SIN1 protein levels, the global dampening of AKT signaling that we observed would be consistent with the loss of a positive feedback loop. Alternatively, because AKT activation loop phosphorylation is sensitive to acute mTOR inhibition, it is possible that prolonged mTORC2 loss triggers a compensatory mechanism for AKT activation loop phosphorylation, with perhaps altered regulation. Interestingly, basal phosphorylation of known AKT substrates is higher in Rictor-iKO cells than control (supplemental Figs. S2D, S2F), suggesting that RICTOR loss might inhibit a phosphatase or activate a kinase that target known AKT substrates under basal (i.e. serum-starved) condition. Regardless of the mechanism, increased basal phosphorylation rather than decreased insulin-dependent phosphorylation contributes to insulin response dampening in Rictor-iKO cells in some cases, e.g. VIM S39 and TSC2 S981 (Fig. 4D, (supplemental Fig. S4B)). Additionally, a recent study showed a dampening of insulin-dependent cellular responses, as well as decreased insulin-dependent phosphorylation of four AKT substrates in 3T3-L1 adipocytes expressing inhibitor-resistant AKT2 S474A (32), suggesting that regulation of AKT hydrophobic motif phosphorylation could explain the dampening of insulin-dependent phospho-signaling we observe in Rictor-null cells. It should also be noted that, whereas the degree of dampening followed a distribution, the average dampening of insulin-sensitive sites was quite mild at an average of ∼1.2 fold for both increasing and decreasing sites. It is possible that the average insulin response dampening because of RICTOR loss could increase in magnitude if certain conditions were changed, e.g. using a lower concentration of insulin, or using a different stimulus such as a β-adrenergic receptor agonist.
In our previous targeted phosphoproteomic analysis of brown preadipocytes, we found that mTORC2 loss reduces insulin-dependent phosphorylation of some AKT substrates more than others (5). Here, we expand on these findings with both targeted and global phosphoproteomics on an insulin time course in a post-differentiation brown adipocyte model, and show that the effect of induced Rictor loss varies across the 348 phosphosites that respond to insulin in the control cells, and that on average insulin/AKT-dependent phosphorylation is dampened. Three of the most dampened AKT substrates, EIF4B, VIM, and ACLY, have reported functional consequences. Phosphorylation of EIF4B may alter translation rate (33), whereas phosphorylation of VIM is known to regulate the polymerization of intermediate filaments (34), suggesting novel roles for mTORC2 in these processes. Phosphorylation of S455 on ACLY promotes its enzymatic activity (35). We recently showed evidence supporting a model in which mTORC2-AKT-ACLY signaling is important for acetyl-CoA production and brown adipocyte differentiation in vitro. In this work, we found that ACLY S455 phosphorylation depends on RICTOR/mTORC2 in brown preadipocytes. Moreover, through rescue experiments overexpressing a ACLY S455D phospho-mimetic in Rictor-deficient brown preadipocytes undergoing differentiation, we provided additional evidence that mTORC2 promotes DNL enzyme and transcript abundance, histone acetylation, and differentiation of brown adipocytes through AKT-mediated ACLY phosphorylation (5).
Our current work reveals the full extent of the gradient of mTORC2 sensitivity that exists in the insulin response of mature brown adipocytes. Why the insulin response for some AKT substrates is more dampened than for others with RICTOR loss is not yet clear, but may reflect differences in the abundance or linear range of phosphorylation of the substrate (36), or perhaps AKT selectivity is altered upon phosphorylation of its hydrophobic motif. Consistent with our previous study (5), AKT substrates that match the minimal AKT consensus motif (Rxx[S/T]), exemplified by VIM S39 and ACLY S455 are more sensitive to mTORC2 loss than those matching the full motif (RxRxx[S/T]). Although the number of AKT substrates measured herein is too small to make broad conclusions, this hypothesis could be tested in future work.
In addition to assessing the effect of Rictor loss on acute insulin signaling, we also sought to identify proteome and phosphoproteome changes that likely occur as the result of chronic Rictor loss. In our analysis we found that under basal (serum-starved) conditions Rictor loss altered the abundance of some phosphosites. Of particular interest, we measured eight basophilic phosphorylation sites on perilipin-1 whose abundance is lower in Rictor-iKO cells, two of which (S81 and S492) are understood to be PKA substrates and important for regulating cAMP-induced lipolysis (24) and lipid droplet fragmentation (25), respectively. Of these eight sites, we find that PLIN1 S130, S384, and S492 increase in response to insulin. This is inconsistent with them being a PKA substrate in our brown adipocyte model, as insulin is known to inhibit PKA activity through stimulation of phosphodiesterase activity (37). Other RICTOR-dependent PLIN1 sites that do not increase in response to insulin could be candidate PKA substrates; however, we find no evidence overall of altered PKA activity in RICTOR-null cells. Thus, it is possible that mTORC2 controls a localized pool of PKA activity toward perilipin-1, perhaps via a specific protein kinase A anchoring protein, while having a minimal effect on total cellular PKA activity. Future studies should aim to elucidate the mechanism by which RICTOR/mTORC2 regulates perilipin-1 phosphorylation.
We find that across all insulin time points, Rictor-iKO cells had widespread differences in protein abundance compared with control cells. Consistent with previous results in various cell and tissue types (4, 23, 38), we observed an overall decrease of glycolysis and DNL enzymes in Rictor-iKO cells. Conversely, TCA cycle, electron transport, and mitochondrial fatty acid oxidation enzymes are, on average, increased. This is in line with prior findings that mitochondrial gene expression and thermogenesis are heightened in Rictor-null BAT (4). Another striking finding from our proteome analysis is the apparently coordinated up-regulation of antigen presentation proteins and immunogenic signaling in Rictor-iKO cells. This includes several MHC class I antigens, as well as TAP1/2, and the transcriptional regulators STAT1 and STAT2. Stat1, Stat2, Ifih1, and Ifit3 are higher in isolated BAT from Ucp1-Cre;Rictorl/l mice at q ≤ 0.1, providing preliminary evidence that RICTOR loss also induces an interferon-like response in vivo. Kissig et al. showed that suppression of type-I interferon signaling by the transcription factor PRDM16 is important for the thermogenic program in brown adipocytes (39). Rictor loss does not phenocopy type-1 interferon stimulation in brown adipocytes, as Kissig et al. observed mitochondrial dysfunction in response to interferon. Nevertheless, the connection we find between RICTOR/mTORC2 and interferon responses in vivo remains an interesting area of future investigation.
To conclude, this work offers the broadest analysis to date of the role of RICTOR in the context of dynamic cell signaling. Our proteomic analysis reveals that globally insulin-dependent phosphorylation events are dampened in cells without functional mTORC2, including particularly prominent dampening of AKT substrate sites on ACLY, EIF4B, and VIM. RICTOR loss also alters insulin-independent phosphorylation, including sites on perilipin-1 that may have relevance in brown adipocyte lipid droplet dynamics. And finally, chronic RICTOR loss causes substantial protein-level changes consistent with more respiration and less glycolysis and DNL, as well as an increased interferon-like response. The latter suggests a potentially novel function of mTORC2, and future work should seek to validate this finding and understand the mechanism by which RICTOR, or mTORC2-mediated signaling, may regulate the expression of interferon-stimulated genes in BAT, and its impact is on metabolic regulation in vivo. In sum, we have uncovered diverse cellular roles of mTORC2 in brown adipocytes, suggesting a path forward to better understand its role in promoting healthy metabolism in BAT.
DATA AVAILABILITY
All mass spectrometry files presented here, along with detailed tables reporting m/z, charge, identification and phosphosite scores, and other details for all peptide identifications, have been deposited to the MassIVE proteomics repository with project identifier MSV000084310.
Supplementary Material
Acknowledgments
We thank members of the Villén and Guertin laboratories for useful discussions. We also thank William Doxsey for assisting C.M.C. with RT-PCR experiments.
Footnotes
* S.W.E. and R.T.L. were supported by NIH T32 HG00035 Interdisciplinary Training grant in Genome Sciences and Samuel and Althea Stroum Endowed Graduate Fellowships. A.V. was supported by NIH T32 LM012419 Big Data in Genomics and Neurosciences training grant. C.M.C was supported by American Diabetes Association Fellowship 1-16-PMF-008. This work is supported by NIH grants R35 GM119536, R01 AG056359, and R01 NS098329, and a Research Grant from the W.M. Keck Foundation to J.V.; and NIH grants R01 CA196986 and R01 DK094004, and a Career Development Award from the Leukemia and Lymphoma Society to D.A.G. The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- mTORC2
- mTOR complex 2
- BAT
- brown adipose tissue
- DNL
- de novo lipogenesis
- ACLY
- ATP citrate lyase
- VIM
- vimentin
- EIF4B
- eukaryotic translation initiation factor 4B.
REFERENCES
- 1. Saxton R. A., and Sabatini D. M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee P. L., Jung S. M., and Guertin D. A. (2017) The complex roles of mechanistic target of rapamycin in adipocytes and beyond. Trends Endocrinol. Metab. 28, 319–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung S. M., Hung C.-M., Hildebrand S. R., Sanchez-Gurmaches J., Martinez-Pastor B., Gengatharan J. M., Wallace M., Mukhopadhyay D., Martinez Calejman C., Luciano A. K., Hsiao W.-Y., Tang Y., Li H., Daniels D. L., Mostoslavsky R., Metallo C. M., and Guertin D. A. (2019) Non-canonical mTORC2 signaling regulates brown adipocyte lipid catabolism through SIRT6-FoxO1. Mol. Cell 75, 807–822.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hung C.-M., Calejman C. M., Sanchez-Gurmaches J., Li H., Clish C. B., Hettmer S., Wagers A. J., and Guertin D. A. (2014) Rictor/mTORC2 loss in the Myf5 lineage reprograms brown fat metabolism and protects mice against obesity and metabolic disease. Cell Rep. 8, 256–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martinez Calejman C., Trefely S., Entwisle S. W., Luciano A., Jung S. M., Hsiao W., Torres A., Hung C. M., Li H., Snyder N. W., Villén J., Wellen K. E., and Guertin D. A. (2020) mTORC2-AKT signaling to ATP-citrate lyase drives brown adipogenesis and de novo lipogenesis. Nat. Commun. 11, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Humphrey S. J., Azimifar S. B., and Mann M. (2015) High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 [DOI] [PubMed] [Google Scholar]
- 8. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., and Hemmings B. A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 9. Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., and Su B. (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 10. Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., and Sabatini D. M. (2006) Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell 11, 859–871 [DOI] [PubMed] [Google Scholar]
- 11. Shiota C., Woo J.-T., Lindner J., Shelton K. D., and Magnuson M. A. (2006) Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell 11, 583–589 [DOI] [PubMed] [Google Scholar]
- 12. Rappsilber J., Ishihama Y. and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 13. Storey J. D., Xiao W., Leek J. T., Tompkins R. G., and Davis R. W. (2005) Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. U.S.A. 102, 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eng J. K., Jahan T. A. and Hoopmann M. R. (2013) Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 [DOI] [PubMed] [Google Scholar]
- 15. Käll L., Canterbury J. D., Weston J., Noble W. S. and MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 16. Beausoleil S. A., Villén J., Gerber S. A., Rush J., and Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 17. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nesvizhskii A. I., Keller A., Kolker E. and Aebersold R. A. (2003) Statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 19. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinforma. Oxf. Engl. 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lawrence R. T., Searle B. C., Llovet A., and Villén J. (2016) Plug-and-play analysis of the human phosphoproteome by targeted high-resolution mass spectrometry. Nat. Methods 13, 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M. and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- 22. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Masui K., Tanaka K., Akhavan D., Babic I., Gini B., Matsutani T., Iwanami A., Liu F., Villa G. R., Gu Y., Campos C., Zhu S., Yang H., Yong W. H., Cloughesy T. F., Mellinghoff I. K., Cavenee W. K., Shaw R. J., and Mischel P. S. (2013) mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 18, 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Souza S. C., Muliro K. V., Liscum L., Lien P., Yamamoto M. T., Schaffer J. E., Dallal G. E., Wang X., Kraemer F. B., Obin M., and Greenberg A. S. (2002) Modulation of hormone-sensitive lipase and protein kinase A-mediated lipolysis by perilipin A in an adenoviral reconstituted system. J. Biol. Chem. 277, 8267–8272 [DOI] [PubMed] [Google Scholar]
- 25. Marcinkiewicz A., Gauthier D., Garcia A., and Brasaemle D. L. (2006) The phosphorylation of serine 492 of perilipin A directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 281, 11901–11909 [DOI] [PubMed] [Google Scholar]
- 26. Valentine R. J., Coughlan K. A., Ruderman N. B., and Saha A. K. (2014) Insulin inhibits AMPK activity and phosphorylates AMPK Ser485/491 through Akt in hepatocytes, myotubes and incubated rat skeletal muscle. Arch. Biochem. Biophys. 562, 62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li L., Gao G., Shankar J., Joshi B., Foster L. J., and Nabi I. R. (2015) p38 MAP kinase-dependent phosphorylation of the Gp78 E3 ubiquitin ligase controls ER-mitochondria association and mitochondria motility. Mol. Biol. Cell 26, 3828–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carlson S. M., Chouinard C. R., Labadorf A., Lam C. J., Schmelzle K., Fraenkel E., and White F. M. (2011) Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Sci. Signal. 4, rs11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ikenoue T., Inoki K., Yang Q., Zhou X., and Guan K.-L. (2008) Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bellacosa A., Chan T. O., Ahmed N. N., Datta K., Malstrom S., Stokoe D., McCormick F., Feng J., and Tsichlis P. (1998) Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene 17, 313–325 [DOI] [PubMed] [Google Scholar]
- 31. Yang G., Murashige D. S., Humphrey S. J., and James D. E. (2015) A positive feedback loop between Akt and mTORC2 via SIN1 phosphorylation. Cell Rep. 12, 937–943 [DOI] [PubMed] [Google Scholar]
- 32. Kearney A. L., et al. (2019) Serine 474 phosphorylation is essential for maximal Akt2 kinase activity in adipocytes. J. Biol. Chem. 294, 16729–16739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raught B., Peiretti F., Gingras A.-C., Livingstone M., Shahbazian D., Mayeur G. L., Polakiewicz R. D., Sonenberg N., and Hershey J. W. B. (2004) Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 23, 1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eriksson J. E., He T., Trejo-Skalli A. V., Härmälä-Braskén A.-S., Hellman J., Chou Y.-H., and Goldman R. D. (2004) Specific in vivo phosphorylation sites determine the assembly dynamics of vimentin intermediate filaments. J. Cell Sci. 117, 919–932 [DOI] [PubMed] [Google Scholar]
- 35. Potapova I. A., El-Maghrabi M. R., Doronin S. V., and Benjamin W. B. (2000) Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry 39, 1169–1179 [DOI] [PubMed] [Google Scholar]
- 36. Hoehn K. L., Hohnen-Behrens C., Cederberg A., Wu L. E., Turner N., Yuasa T., Ebina Y., and James D. E. (2008) IRS1-independent defects define major nodes of insulin resistance. Cell Metab. 7, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Solomon S. S., Palazzolo M., and King L. E. (1977) Cyclic nucleotide phosphodiesterase. Insulin activation detected in adipose tissue by gel electrophoresis. Diabetes 26, 967–972 [DOI] [PubMed] [Google Scholar]
- 38. Hagiwara A., Cornu M., Cybulski N., Polak P., Betz C., Trapani F., Terracciano L., Heim M. H., Rüegg M. A., and Hall M. N. (2012) Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 15, 725–738 [DOI] [PubMed] [Google Scholar]
- 39. Kissig M., Ishibashi J., Harms M. J., Lim H.-W., Stine R. R., Won K.-J., and Seale P. (2017) PRDM16 represses the type I interferon response in adipocytes to promote mitochondrial and thermogenic programing. EMBO J. 36, 1528–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mass spectrometry files presented here, along with detailed tables reporting m/z, charge, identification and phosphosite scores, and other details for all peptide identifications, have been deposited to the MassIVE proteomics repository with project identifier MSV000084310.