Abstract
Aims
Lipotoxic cardiomyopathy in diabetic and obese patients typically encompasses increased cardiac fatty acid (FA) uptake eventually surpassing the mitochondrial oxidative capacity. Lowering FA utilization via inhibition of lipolysis represents a strategy to counteract the development of lipotoxic heart dysfunction. However, defective cardiac triacylglycerol (TAG) catabolism and FA oxidation in humans (and mice) carrying mutated ATGL alleles provokes lipotoxic heart dysfunction questioning a therapeutic approach to decrease cardiac lipolysis. Interestingly, decreased lipolysis via cardiac overexpression of Perilipin 5 (Plin5), a binding partner of ATGL, is compatible with normal heart function and lifespan despite massive cardiac lipid accumulation. Herein, we decipher mechanisms that protect Plin5 transgenic mice from the development of heart dysfunction.
Methods and results
We generated mice with cardiac-specific overexpression of Plin5 encoding a serine-155 to alanine exchange (Plin5-S155A) of the protein kinase A phosphorylation site, which has been suggested as a prerequisite to stimulate lipolysis and may play a crucial role in the preservation of heart function. Plin5-S155A mice showed a substantial increase in cardiac TAG and ceramide levels, which was comparable to mice overexpressing non-mutated Plin5. Lipid accumulation was compatible with normal heart function even under mild stress. Plin5-S155A mice showed reduced cardiac FA oxidation but normal ATP production and changes in the Plin5-S155A phosphoproteome compared to Plin5 transgenic mice. Interestingly, mitochondrial recruitment of dynamin-related protein 1 (Drp1) was markedly reduced in cardiac muscle of Plin5-S155A and Plin5 transgenic mice accompanied by decreased phosphorylation of mitochondrial fission factor, a mitochondrial receptor of Drp1.
Conclusions
This study suggests that low cardiac lipolysis is associated with reduced mitochondrial fission and may represent a strategy to combat the development of lipotoxic heart dysfunction.
Keywords: Cardiac lipolysis, Lipotoxicity, Perilipin 5, Mitochondrial dynamics, Heart dysfunction
1. Introduction
Fatty acids (FAs) are the predominant energy fuel of the adult heart. Cardiomyocytes obtain FAs from the hydrolysis of triacylglycerol (TAG)-rich lipoproteins or from albumin-bound non-esterified FAs.1 The healthy heart exhibits very low numbers of small lipid droplets (LDs). Increased cardiac muscle (CM) TAG deposition is often associated with elevated ceramide and diacylglycerol (DAG) levels, which are potentially harmful lipids linked to lipotoxic heart dysfunction.2,3 Moreover, increased cardiac FA uptake in obesity and diabetes has been linked to lipotoxic heart dysfunction. This clinical picture is phenocopied in mice with cardiac overexpression of PPARα showing increased FA catabolism leading to pathologic cardiac hypertrophy and left ventricular (LV) dysfunction.4 Additionally, reperfusion injury has been attributed to increased fatty acid oxidation (FAO) and pharmacological inhibition of mitochondrial FA uptake has improved mechanical recovery during reperfusion of the ischaemic heart.5 Together, these findings suggest that lowering FA utilization is a promising therapeutic strategy to combat the development of lipotoxic heart dysfunction. However, defective lipolysis in humans and mice encoding adipose triglyceride lipase (ATGL) loss of function mutations provokes massive cardiac fat accumulation and lethal heart dysfunction.6 ATGL-mediated cardiac lipolysis has been demonstrated as a prerequisite for PPARα- and PGC-1α-induced expression of FA oxidizing genes.7 Findings in ATGL-deficient mice revealed that the cardiac TAG pool is dynamic and that cardiac lipolysis plays a significant role in the supply of FAs as oxidative fuel. The catabolism of cardiac TAG is under the regulation of Perilipin 5 (Plin5), a member of the PAT protein family of LD-associated proteins.8 Current evidence suggests that Plin5 interacts with ATGL and/or its co-activator CGI-58 thereby blocking ATGL-mediated lipolysis at the LD. This interaction can be resolved upon protein kinase A (PKA)-mediated phosphorylation of Plin5.9–11 In strong contrast to ATGL-deficiency, reduced lipolysis caused by CM-specific Plin5 overexpression does not negatively affect heart function and life span despite massive cardiac fat accumulation.12,13
In the present study, we decipher mechanisms protecting Plin5 transgenic mice from lipid-induced cardiac dysfunction, which may have important implications for preservation of heart function in metabolic disease. Cardiac overexpression of either wild-type (wt) or mutant Plin5 encoding a serine-155 to alanine exchange (Plin5-S155A) is compatible with normal heart function despite a distinct lipotoxic lipid profile. Low cardiac lipolysis in the transgenic mice is associated with reduced mitochondrial fission, which may protect from lipotoxicity-induced heart dysfunction.
2. Methods
2.1. Animals
Mice with CM-specific overexpression of Plin5-S155A were generated as previously described.12 See Supplementary material online, for details. Animals were housed in a specific pathogen-free facility and maintained on a regular light-dark cycle (14 h/10 h) with ad libitum access to water and a standard laboratory chow diet. Unless otherwise indicated, non-fasted 12–14 weeks old mice were used. Mice were anaesthetized with isoflurane and euthanized by cervical dislocation. Maintenance, handling, experiments, and tissue collection of mice were approved by the Austrian Federal Ministry for Science, Research and Economy and by the Ethics Committee of the University of Graz and the University of Veterinary Medicine Vienna (protocol number BMWFW-66.007/0006-WF/V/3b/2014) according to the EU (Directive 2010/63/EU) ethical guidelines.
2.2. Magnetic resonance imaging
Magnetic resonance imaging (MRI) was performed on a 7 T small animal MRI platform (Bruker BioSpec). See Supplementary material online for details.
2.3. Cell culture
COS-7 cells (ATCC CRL-1651) and H9c2 myoblasts (ATCC CRL-1446) were cultured according to the manufactures instructions. H9c2 cells were differentiated in medium containing 1% FCS and 10 nM retinoic acid for 7 days. Cell culture experiments are described in Supplementary material online.
2.4. Cardiac lipid analyses and lipidomics
Methods are described in Supplementary material online.
2.5. Lipase activity assays
TAG hydrolase activity and LD-associated lipolysis were determined as described in Supplementary material online.
2.6. (Phospho)proteomics and NMR metabolic profiling
Procedures are described in Supplementary material online.
2.7. Analyses of cardiac ROS levels
Procedures are described in Supplementary material online.
2.8. Cell fractionation
Cardiac mitochondria were isolated as previously described.14 Briefly, CM samples were homogenized in buffer A (0.25 M sucrose pH 7, 1 mM EDTA, 1 mM DTT) using UltraTurrax homogenizer and filtered through a 100 μm cell strainer. Mitochondria were isolated by differential centrifugation.
LDs surrounded by peridroplet mitochondria were prepared as previously described15 with slight modifications as detailed in Supplementary material online.
2.9. FAO in cardiac mitochondria
Palmitic acid oxidation was measured in cardiac mitochondria from fasted mice as previously described16 using [1-14C]-palmitic acid as tracer. Details are described in Supplementary material online.
2.10. RT-qPCR
RT-qPCR was performed as previously described.12 Relative mRNA levels were quantified according to the ΔΔCT method with 36b4 as reference gene. Primer sequences are listed in the Supplementary material online.
2.11. Western blot analyses
Western blots were performed according to standard protocols. Antibodies are listed in the Supplementary material online.
2.12. Histology, TEM, and confocal laser scanning microscopy
Methods are described in the Supplementary material online.
2.13. Statistical analyses
Data are presented as means ± SEM. Unless otherwise indicated, statistical significance was determined by unpaired Student’s two-tailed t-test. Differences between two groups were considered statistically significant for *P < 0.05, **P < 0.01, and ***P < 0.001.
3. Results
3.1. Plin5 serine 155 to alanine substitution lowers lipolysis and FA oxidation in H9c2 cardiac cells
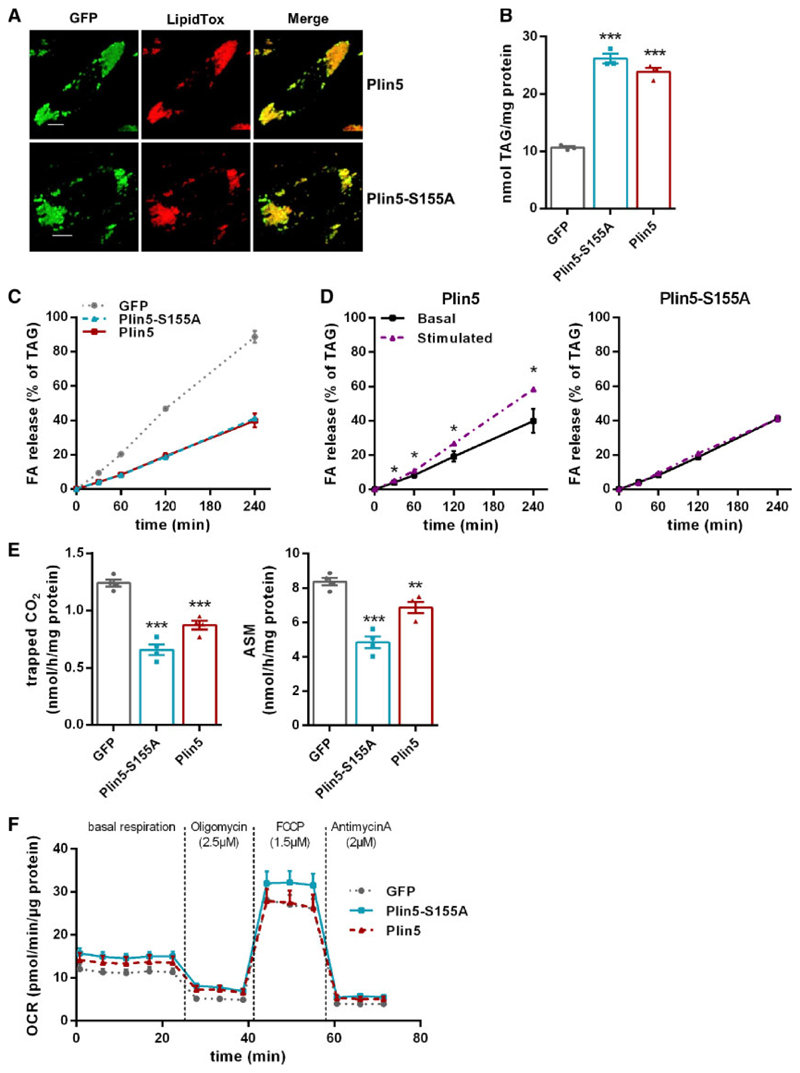
To explore whether the Plin5-S155A mutation of the PKA phosphorylation site interferes with TAG homeostasis of cardiac cells, we generated H9c2 cardiomyocytes stably overexpressing GFP, GFP-tagged murine Plin5, or Plin5-S155A. Recombinant Plin5 as well as Plin5-S155A localized to the surface of cellular LDs (Figure 1A) and were found at the interface of LDs and mitochondria (Supplementary material online, Figure S1A). Overexpression of Plin5 or Plin5-S155A markedly increased cellular TAG content (Figure 1B) during an oleic acid pulse labelling experiment, implicating that overexpression of non-mutant and mutant Plin5 similarly counteracts cellular TAG catabolism. In accordance with reduced TAG turnover, FA release into the medium (as a measure of lipolysis) was significantly reduced upon Plin5 or Plin5-S155A overexpression under basal conditions (Figure 1C). ß-Adrenergic stimulation via forskolin and IBMX incubation increased FA release in cells overexpressing non-mutant Plin5 (Figure 1D), but not in cells overexpressing Plin5-S155A. In line, FAO was significantly reduced in both, Plin5 and Plin5-S155A overexpressing cells (Figure 1E) albeit the reduction was more pronounced in cells expressing Plin5-S155A. Moreover, oxygen consumption rate (OCR) was moderately but non-significantly increased in Plin5-S155A H9c2 cells (Figure 1F). These findings suggest that serine-155 of Plin5 impacts TAG catabolism in cardiomyocytes.
Figure 1. Overexpression of Plin5-S155A reduces lipolysis in H9c2 cardiac cells.
(A) Confocal life cell imaging in cells overexpressing GFP-tagged Plin5 or Plin5-S155A upon incubation with 400 μM oleic acid (OA) and lipid staining using LipidTox™. Bar: 10 μm. (B) Incorporation of radioactivity into TAG upon incubation with OA and 3H-labelled OA for 20 h (n = 3). (C) Measurement of (FA) release into the medium after incubation with cold and 3H-labelled OA. (D) FA release upon incubation with 20 μM forskolin and 0.5 mM IBMX. (E) Released CO2 and production of acid soluble metabolites (ASM) after incubation with 14C-palmitic acid for 90 min (n = 4). (F) Oxygen consumption rate (OCR) during sequential addition of oligomycin, FCCP, and antimycin A, as indicated. Data are shown as means ± SEM. Statistical significance determined by unpaired Student’s t-test (*P < 0.05; **P > 0.01; ***P < 0.001).
3.2. 3.2 Cardiomyocyte-specific overexpression of Plin5-S155A provokes cardiac steatosis and increases cardiac ceramide and DAG levels
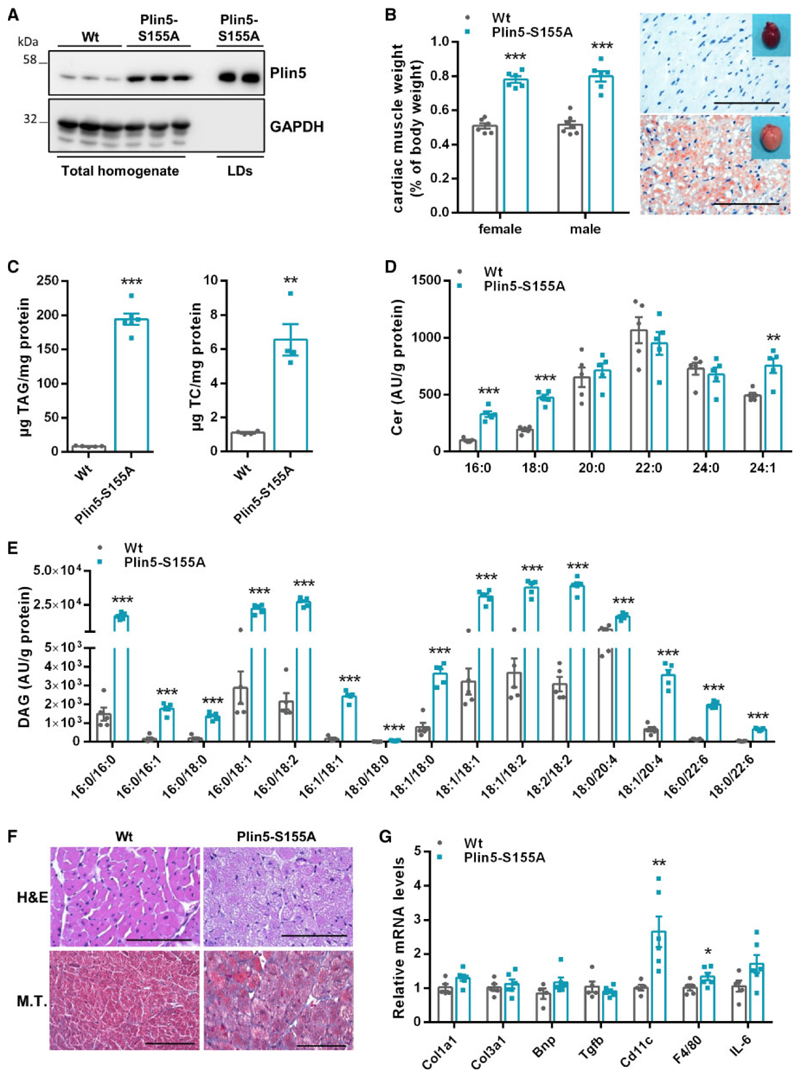
To examine the in vivo role of Plin5-S155 phosphorylation in cardiac TAG catabolism, we generated transgenic mice overexpressing murine Plin5-S155A (Supplementary material online, Figure S2A) exclusively in cardiomyocytes. We have previously shown that cardiac-specific over-expression of non-mutant Plin5 provokes cardiac steatosis via decelerated lipolysis.12 If applicable, we included mice overexpressing non-mutant Plin5 in our analyses to emphasize the specific impact of Plin5-S155 in cardiac lipid and energy metabolism. Cardiac mRNA expression of the Plin5-S155A transgene was markedly increased, whereas endogenous Plin5 expression was significantly reduced (-37%) in Plin5-S155A transgenic compared to Wt mice (Supplementary material online, Figure S2B). Total Plin5 protein levels were markedly increased in CM preparations of Plin5-S155A mice (Figure 2A) and highly abundant in the LD fraction. Plin5 protein levels were similarly increased in Plin5-S155A and Plin5 transgenic mice (five- and six-fold, respectively) (Supplementary material online, Figure S2C). Body weight (Supplementary material online, Figure S2D) and plasma parameters (Supplementary material online, Table S1) were comparable among Plin5-S155A and Wt mice. Heart weight was significantly increased in Plin5-S155A mice and hearts showed stark neutral lipid staining (Figure 2B) due to a drastic increase in TAG (22-fold) and total cholesterol (TC) (six-fold) content (Figure 2C). Interestingly, ceramide levels, especially C16:0 and C18:0 ceramides, were strongly increased (up to eight-fold) in CM of Plin5-S155A and Plin5 transgenic mice (Figure 2D and Supplementary material online, Figure S2E) compared to Wt. Moreover, Plin5-S155A and Plin5 overexpression increased cardiac DAG levels (Figure 2E and Supplementary material online, Figure S2F). Notably, DAG species in Plin5-S155A mice reflect the FA composition of TAGs (Supplementary material online, Figure S2G). The increase in these lipid species can lead to cardiac stress and lipotoxicity. Unexpectedly, histological analyses revealed only mild fibrosis in Plin5-S155A CM (Figure 2F) and absence of periodic acid schiff (PAS)-staining (not shown). Furthermore, mRNA levels of genes involved in extracellular matrix remodelling as well as expression of proinflammatory marker genes (except for Cd11c) were unchanged (Figure 2G).
Figure 2. Cardiomyocyte-specific overexpression of Plin5-S155A provokes cardiac steatosis and increases ceramide and DAG levels.
(A) Plin5 protein levels in cardiac homogenates of Wt and Plin5-S155A mice and in LD fractions of Plin5-S155A mice. (B) Heart to body weight ratio (n = 6–7) and heart images. (C) Oil red O staining (ORO) of cardiac tissue sections (Scale bar: 200 μm) and cardiac tissue TAG and TC levels (n = 4–5). Targeted lipidomic analyses of ceramide (D) and DAG species (E) in CM of Wt and Plin5-S155A mice (n = 5). (F) Representative histological images of heart sections stained with Haematoxylin and eosin (H&E) and Masson’s trichrome (M.T.) (Scale bar: H&E 200 μm; M.T. 100 μm). (G) Cardiac mRNA expression levels of genes associated with cardiac fibrosis and hypertrophy (Col1a1, Col3a1, Bnp, Tgfb) and inflammation (Cd11c, F4/80, IL-6) determined by RT-qPCR with 36b4 as reference gene (n = 5–6). Data are presented as means ± SEM. Statistical significance was tested using unpaired Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
3.3. Plin5-S155A divergently affects cardiac lipolysis compared to Plin5
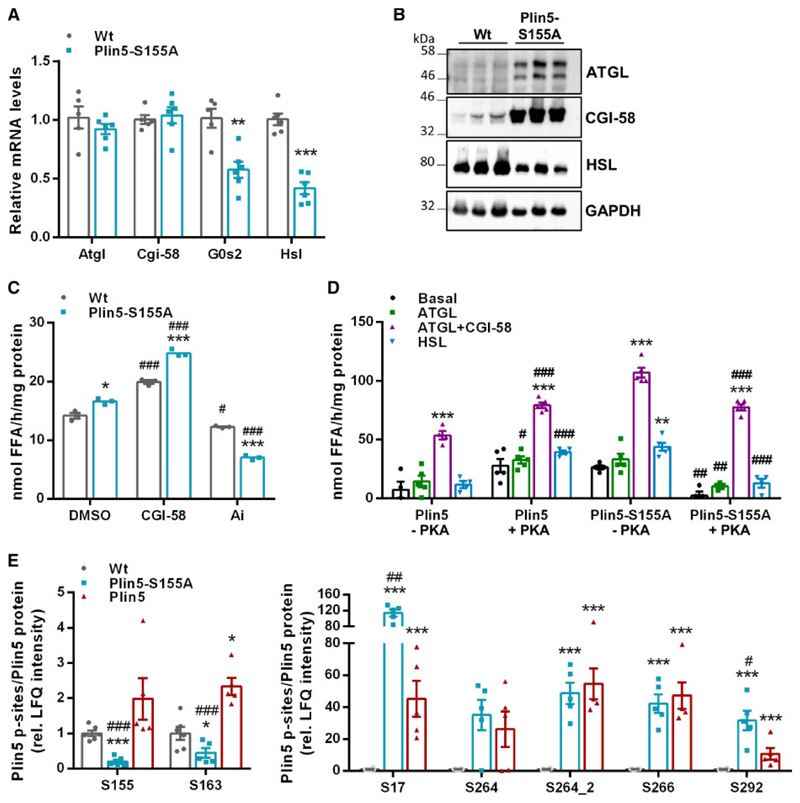
TAG accumulation in CM of Plin5-S155A mice implicates alterations in cardiac lipolysis, which prompted us to measure expression levels of lipases and co-regulators. As shown in Figure 3A, mRNA levels of Atgl and its co-activator Cgi-58 were comparable between Plin5-S155A and Wt mice, whereas mRNA expression of the ATGL inhibitor G0s2 and hormone-sensitive lipase (Hsl) were significantly reduced (-42% and -58%). In contrast to mRNA expression, and in line with previous studies investigating Plin5 transgenic mice,12,13 cardiac protein levels of ATGL and CGI-58 (Figure 3B) were substantially increased in Plin5-S155A mice compared to Wt. Interestingly, cardiac protein content of HSL, which is limiting for DAG hydrolysis,17 was markedly reduced in Plin5-S155A mice. We have previously shown that conventional measurement of TAG hydrolytic activities in cardiac lysates devoid of LDs applying a synthetic micellar phospholipid/TAG substrate mainly reflects the tissue protein levels of ATGL and its co-activator CGI-58.12 In contrast, in vivo TAG catabolism of LDs is under tight regulation mediated by Plin5 and other LD proteins, which recruit or release ATGL and its co-activator CGI-58.8 In accordance with increased cardiac CGI-58 and ATGL protein content, TAG hydrolase activity applying a micellar TAG substrate was increased (1.2-fold) in cardiac tissue preparations of Plin5-S155A mice compared to Wt (Figure 3C). The marked reduction of TAG hydrolase activities in Plin5-S155A and Wt cardiac lysates upon incubation with the ATGL-specific inhibitor Atglistatin18 demonstrated that increased lipolysis is due to elevated ATGL protein content. We previously showed that Plin5 acts as a lipolytic barrier at the LD surface and that incubation with the PKA catalytic subunit restores lipolysis.12 These findings prompted us to measure PKA-mediated FA release from cardiac LDs prepared from Plin5 compared to Plin5-S155A transgenic mice incubated with cell lysates enriched with ATGL, ATGL/CGI-58 or HSL. The extremely low abundance of LDs in CM of Wt mice hinders the isolation of LDs in sufficient quantities for measuring FA release. As previously shown, PKA incubation markedly stimulated FA release from Plin5-enriched LDs (up to 6.8-fold) (Figure 3D).10 Unexpectedly, FA release from LDs prepared from Plin5-S155A mice was even reduced upon PKA incubation compared to preparations without PKA. However, FA release was elevated from LDs of PlinS155A compared to Plin5 transgenic mice in the absence of PKA. In line, FA liberation from freshly isolated Plin5-S155A LDs was already elevated (in the absence of lipase-containing cell lysates) compared to Plin5 LDs (Supplementary material online, Figure S3A). Elevated basal LD-associated lipolysis in Plin5-S155A hearts could arise from differences in cardiac phospho-PKA (p-PKA) levels compared to mice with cardiac overexpression of non-mutant Plin5. However, cardiac p-PKA levels were unchanged between Wt, Plin5, and Plin5-S155A mice (Supplementary material online, Figure S3B), which contradicts previous findings.10
Figure 3. Plin5-S155A divergently affects cardiac lipolysis compared to Plin5 and displays changes in the Plin5 phosphorylation pattern.
(A) Cardiac mRNA expression levels of Atgl, Cgi-58, G0s2, and Hsl determined by RT-qPCR with 36b4 as reference gene. Data are presented as means ± SEM (n = 5–6). Statistical significance determined by unpaired Student’s t-test (**P < 0.01; ***P < 0.001). (B) Protein levels of ATGL, CGI-58, and HSL in cardiac homogenates. (C) TAG hydrolase activities in cardiac lysates from Wt and Plin5-S155A mice (n = 5 per genotype) applying a micellar TAG substrate. Recombinant CGI-58 or Atglistatin (Ai) was added for stimulation or inhibition of ATGL activity (*P < 0.05; ***P < 0.001 vs. Wt; #P < 0.05; ###P < 0.001 vs. DMSO conditions). (D) Cardiac LDs incubated with COS-7 cell lysates enriched with LacZ (basal), ATGL, ATGLþCGI-58, or HSL in the absence or presence of recombinant PKA (-PKA/þPKA) (n = 5). Data are shown as means ± SEM. Statistical significance was tested using unpaired Student’s t-test (**P < 0.01; ***P < 0.001 vs. basal levels; #P < 0.05; ##P < 0.01; ###P < 0.001 vs. LDs without PKA). (E) Cardiac Plin5-phosphosites determined by Q-TOF MS. Values were normalized to total Plin5 levels determined by western blot analyses (n = 5–6). Data are presented as means ± SEM. ANOVA with subsequent multiple testing correction by Permutation-based FDR method was used to identify altered protein groups (*P < 0.05; ***P < 0.001 transgenic vs. Wt; #P < 0.05; ##P < 0.01; ###P < 0.001 Plin5 vs. Plin5-S155A).
Alterations in lipolysis of Plin5 compared to Plin5-S155A transgenic mice prompted us to analyse the cardiac phosphoproteome applying Q-TOF mass spectrometry. We detected phosphorylated Plin5 peptides in Wt and transgenic mice of the phosphopeptide (p-peptide) isolations. In contrast, we hardly detected non-phosphorylated Plin5 peptides in the flow-troughs from Wt mice, which is most likely explained by the much lower abundance of Plin5 protein. This prompted us to normalize p-peptide levels to the relative Plin5 protein intensities (Supplementary material online, Figure S2C). Notably, we detected several currently unknown p-sites of Plin5 that were increased in CM of the transgenic mice (Figure 3E). In line with the proposed role of Plin5 S155 as a PKA phosphorylation site, levels of p-S155 peptides were strongly reduced in Plin5-S155A mice, whereas p-peptide levels were increased in mice overexpressing non-mutant Plin5. Together, these findings suggest that Plin5-mediated regulation of cardiac lipolysis involves multiple Plin5 phosphorylation sites.
3.4. Cardiac steatosis in Plin5-S155A mice is compatible with normal heart function
Cardiac steatosis and a rise in ceramide levels is a common characteristic of lipotoxic heart disease,2,4 which prompted us to explore cardiac parameters and heart function under normal conditions or during mild cardiac stress (Table 1). Cardiac MRI19 revealed a significant increase in LV mass of Plin5-S155A and Plin5 transgenic mice (1.6- and 1.9-fold, respectively). In line, interventricular septum and LV posterior wall thickness were markedly enlarged (up to 1.7-fold) in Plin5-S155A and Plin5 transgenic mice. Notably, LV internal diameter was increased in transgenic mice, whereas relative wall thickness was comparable among Wt and transgenic mice. These morphological changes in the hearts of transgenic mice are indicative for physiologic eccentric cardiac growth.20 Next, we examined heart function under normal conditions or under dobutamine-induced mild stress. Heart rates were significantly reduced in Plin5-S155A and Plin5 transgenic mice compared to Wt under normal conditions (-32% and -27%) and upon dobutamine application (-21% and -22%). In contrast, LV stroke volumes were increased in Plin5-S155A and Plin5 transgenic mice in the absence (1.4- and 1.5-fold, respectively) and presence of dobutamine (1.2-fold). Ejection fraction and cardiac output were essentially unchanged in transgenic mice, further indicating physiologic cardiac growth in Plin5-S155A and Plin5 mice.
Table 1. Cardiac lipid accumulation is compatible with adequate heart function in Plin5-S155A mice.
| Parameters | Wt | Plin5-S155A | Plin5 | |
|---|---|---|---|---|
| LV mass (mg) | 97.37 ± 3.48 | 152.07 ± 8.84*** | 184.97 ± 11.59***,$ | |
| Interventricular septum (mm) | 0.95 ± 0.03 | 1.16 ± 0.06* | 1.16 ± 0.07* | |
| LV posterior wall thickness (mm) | 0.91 ± 0.03 | 1.19 ± 0.08** | 1.29 ± 0.05*** | |
| LV internal diameter (mm) | 3.49 ± 0.09 | 3.96 ± 0.11* | 4.21 ± 0.11*** | |
| Relative wall thickness | 0.54 ± 0.02 | 0.59 ± 0.02 | 0.57 ± 0.01 | |
| Ejection fraction (%) | Basal | 76.10 ± 1.52 | 84.76 ± 1.41 | 74.83 ± 3.98 |
| Dobutamine | 85.72 ± 1.32 | 86.30 ± 2.00 | 74.51 ± 6.000.058 | |
| LV stroke volume (μl) | Basal | 34.83 ± 1.43 | 50.03 ± 1.86*** | 50.36 ± 2.68*** |
| Dobutamine | 35.48 ± 1.39 | 43.99 ± 1.84* | 43.32 ± 2.110.059 | |
| Heart rate (b.p.m.) | Basal | 448.0 ± 19.7 | 303.0 ± 18.6*** | 331.2 ± 15.5*** |
| Dobutamine | 490.2 ± 6.6 | 384.2 ± 8.3***,# | 381.4 ± 15.7*** | |
| Cardiac output (mL/min) | Basal | 15.63 ± 0.98 | 15.11 ± 0.91 | 16.62 ± 1.01 |
| Dobutamine | 17.42 ± 0.80 | 16.85 ± 0.54 | 16.56 ± 1.22 |
Cardiac morphological and functional parameters determined by MRI in 14 weeks old Wt, Plin5-S155A, and Plin5 mice. Moderate cardiac stress was induced by i.p. injection of the β1-adrenergic agonist dobutamine (1.5 mg/kg). Data are presented as means ± SEM (n = 6–9). Statistical significance was determined by one-way ANOVA and Bonferroni post hoc tests as appropriate (SPSS Statistics 24, IBM, Armonk, NY, USA)
P < 0.05
P < 0.01
P < 0.001 vs. Wt
P < 0.05 vs. Plin5
P < 0.05 vs. basal conditions.)
LV, left ventricular.
3.5. Plin5-S155A hearts exhibit decreased cardiac FA oxidation and changes in amino acid homeostasis, but normal energy production
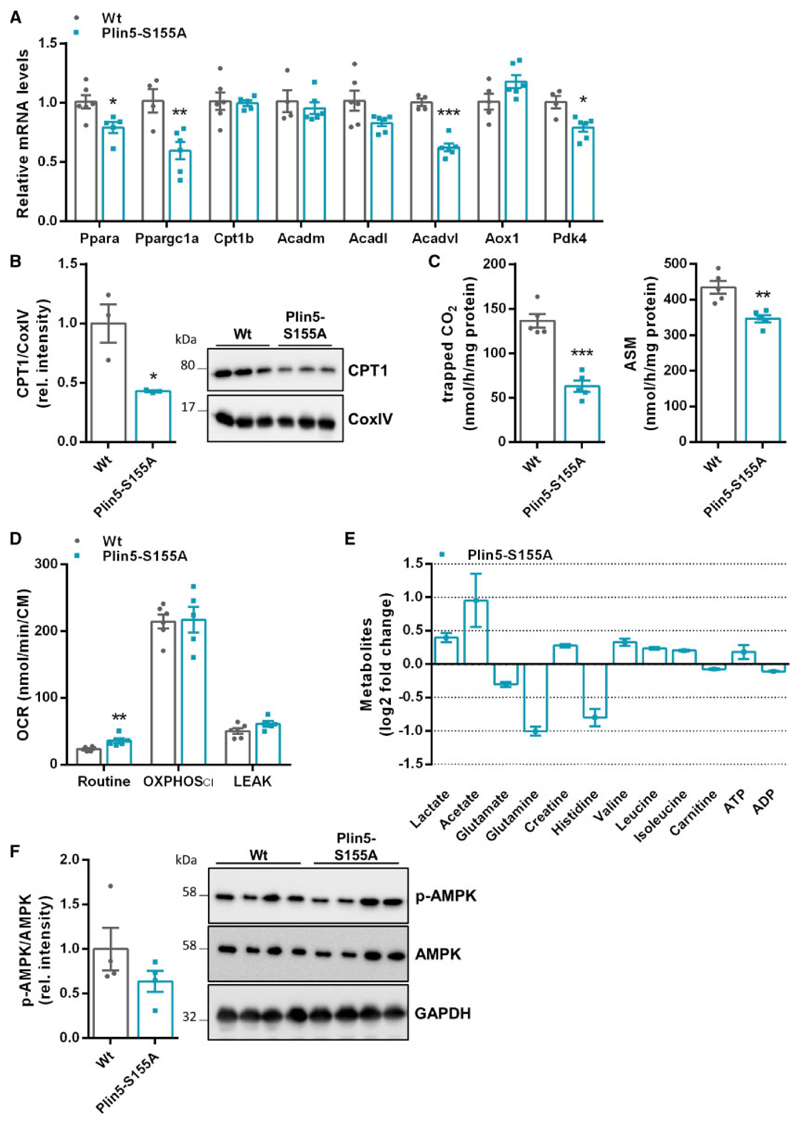
Metabolic flexibility is a characteristic of the healthy heart. We suggest that low cardiac lipolysis in Plin5-S155A cardiomyocytes induces changes in cardiac energy metabolism to deliver adequate ATP levels. Cardiac Plin5-S155A overexpression lowered Pparα and Pgc1α mRNA levels and partially reduced mRNA expression of FAO genes in CM (Figure 4A). mRNA expression of genes involved in FA uptake including Cd36 and Lpl were markedly decreased in CM of Plin5-S155A mice (Supplementary material online, Figure S4), whereas lipogenic gene expression was comparable to Wt. The decrease in mitochondrial CPT1 protein expression (-57%) (Figure 4B) indicated reduced FAO in Plin5-S155A CM. In line, palmitic acid oxidation, determined by the release of 14CO2 and the generation of acid soluble metabolites (ASM) (Figure 4C), were significantly reduced in cardiac mitochondria of Plin5-S155A compared to Wt mice. As shown in Figure 4D, basal OCR (Routine) was 1.5-fold increased in Plin5-S155A cardiac homogenates compared to Wt, whereas the addition of pyruvate and ADP (OXPHOS CI) similarly increased OCR in both genotypes. The decline in OCR upon addition of the ATPase-inhibitor oligomycin indicates low proton leakage. To further investigate energy metabolism in Plin5-S155A mice, we applied an untargeted NMR-based metabolic phenotyping approach to analyse cardiac metabolites. Figure 4E depicts relative changes in cardiac metabolite levels from Plin5-S155A mice in relation to Wt. Cardiac ATP levels were even moderately increased in Plin5-S155A mice indicating normal energy homeostasis. Elevated lactate and acetate levels are in line with reduced utilization of FAs as energy substrate. The significant drop in glutamate, glutamine, and histidine levels may reflect an increased anaplerotic carbon flux into the TCA via α-ketoglutarate. Finally, cardiac levels of phospho-AMPK, a major sensor of the cellular energy status, were maintained indicating robust metabolic control in Plin5-S155A hearts (Figure 4F).
Figure 4. Cardiac Plin5-S155A overexpression decreases FAO and affects amino acid homeostasis without changes in overall energy production.
(A) Cardiac mRNA levels of Ppara and target genes of fasted mice determined by RT-qPCR using 36b4 as reference gene (n = 4 –6). (B) Mitochondrial CPT1 protein levels in CM of Wt and Plin5-S155A mice using CoxIV as loading control (n = 3). (C) Measurement of 14CO2 release and the production of ASM in mitochondria preparations from CM of Wt and Plin5-S155A mice (n = 5). (D) Cardiac metabolites in fasted Wt and Plin5-S155A mice quantified by NMR spectroscopy (n = 5). (E) pAMPK and total AMPK protein levels in cardiac homogenates from fasted mice. (F) Oxygen consumption rates (OCR) in cardiac homogenates from fasted mice measured with Oroboros under basal conditions (Routine), after addition of pyruvate and ADP (OXPHOS CI), and after addition of oligomycin (LEAK). Data are presented as means ± SEM. Statistical significance was determined by unpaired Student’s t-test. (*P <0.05; **P <0.01; ***P <0.001).
3.6. Cardiac overexpression of Plin5-S155A or Plin5 reduces mitochondrial fission
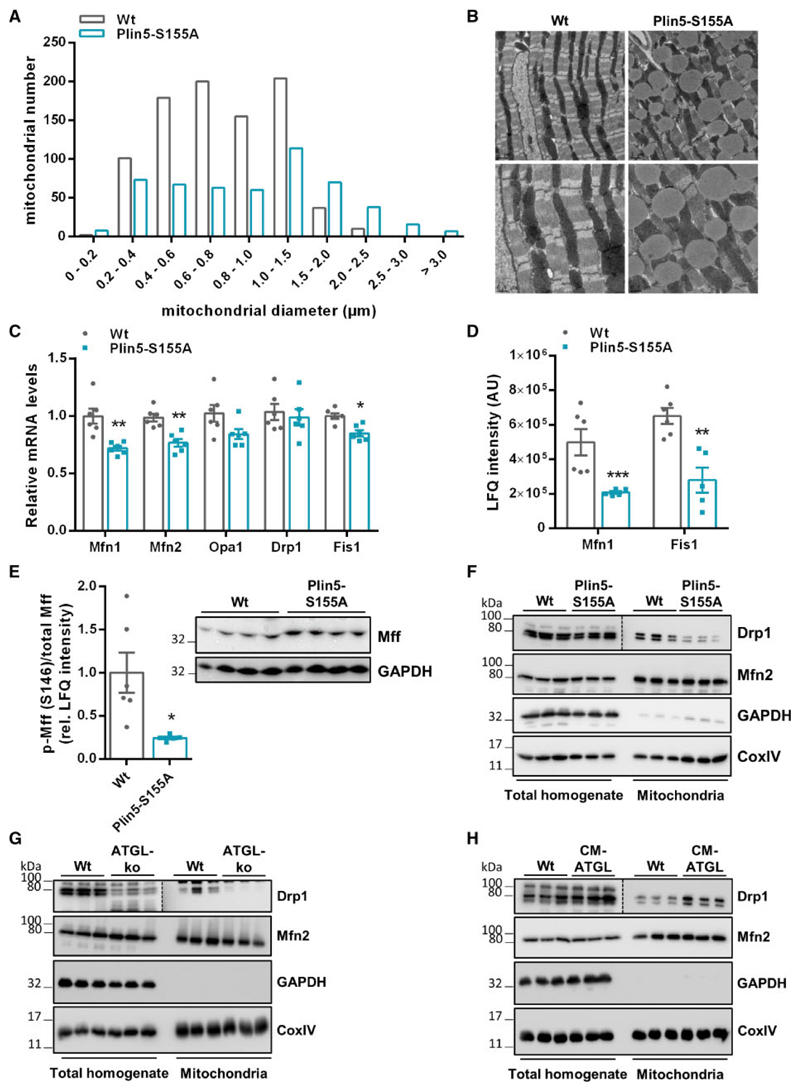
Cardiac Plin5 overexpression markedly increased mitochondria size and led to tight association of mitochondria and LDs.12,13 Mitochondria of Plin5-S155A mice were also increased in size (Figure 5A) and in close proximity to LDs (Figure 5B). Moreover, total mitochondrial protein levels were significantly increased in Plin5-S155A hearts compared to Wt (Supplementary material online, Figure S5A). Recently, a method for the preparation of LD-associated mitochondria (designated as peridroplet mitochondria) from tissues has been established.15 Applying this protocol, we observed the presence of mitochondria on nearly all LDs prepared from Plin5-S155A mice demonstrating the tight association of LDs and mitochondria in CM of Plin5-S155A mice ex vivo (Supplementary material online, Figure S5B). Starvation and a metabolic undersupply have been linked to mitochondrial elongation,21,22 which prompted us to examine expression levels of genes that regulate mitochondrial dynamics. mRNA expression of genes mediating mitochondrial fission or fusion were moderately decreased or unchanged in Plin5-S155A (Figure 5C) or Plin5 (Supplementary material online, Figure S5C) transgenic mice. Protein abundance of Mitofusin 1 (Mfn1) and Fission 1 (Fis1), two proteins that inversely affect mitochondrial dynamics, were reduced in Plin5-S155A mice compared to Wt (Figure 5D). Phosphorylation of mitochondrial fission factor (Mff) recruits dynamin-related protein 1 (Drp1) to mitochondria thereby inducing mitochondrial fission.23 Levels of phosphorylated Mff (p-S146) were markedly reduced in CM of Plin5-S155A (Figure 5E) and Plin5 (Supplementary material online, Figure S5D) transgenic mice suggesting reduced mitochondrial Drp1 recruitment and fission. Notably, cardiac mitochondria prepared from Plin5-S155A and Plin5 transgenic mice showed markedly reduced Drp1 protein levels compared to Wt (Figure 5F and Supplementary material online, Figure S5E). Protein levels of Mitofusin 2 (Mfn2) were unchanged among Plin5-S155A, Plin5 and Wt mice. Decreased mitochondrial Drp1-localization was also present in CM of global ATGL-deficient mice harbouring a defect in cardiac lipolysis (Figure 5G). In contrast, increased cardiac lipolysis via CM-specific ATGL overexpression (CM-ATGL)24 enhanced mitochondrial Drp1 shuttling (Figure 5H) suggesting that cardiac lipolysis may regulate mitochondrial dynamics in vivo.
Figure 5. Cardiac Plin5-S155A overexpression reduces mitochondrial fission.
(A) Mitochondria size range analysed from electron microscopy images of heart sections (Analysis of 41 and 48 images from Wt and Plin5-S155A mice, respectively). (B) Representative transmission electron microscopy images of CM slides from Wt and Plin5-S155A mice. (C) Cardiac mRNA levels of genes involved in mitochondrial fusion (Mfn1, Mfn2, Opa1) and fission (Drp1, Fis1) analysed by RT-qPCR using 36b4 as reference gene (n = 6). (D) Mfn1 and Fis1 corresponding peptide levels in CM from fasted mice quantified by MS proteomics analysis (n = 5–6). (E) Cardiac phospho-Mff (S146) levels quantified by phosphoproteome analyses applying Q-TOF MS (n = 5–6) and normalized to total Mff levels determined by western blot analyses. Protein levels of Drp1 and Mfn2 in total homogenates and mitochondrial fractions of Wt, Plin5-S155A (F), ATGL-ko (G), and CM-ATGL mice (H) using GAPDH (homogenate) and CoxIV (mitochondria) as loading controls. Data are presented as means ± SEM. Statistical significance was determined by unpaired Student’s t-test. For (phospho)proteomics, ANOVA with subsequent multiple testing correction by Permutation-based FDR method was used to identify altered protein groups (*P < 0.05; **P < 0.01; ***P < 0.001).
3.7. Mild oxidative stress in Plin5-S155A mice is linked to increased cardiac expression of ROS defense genes
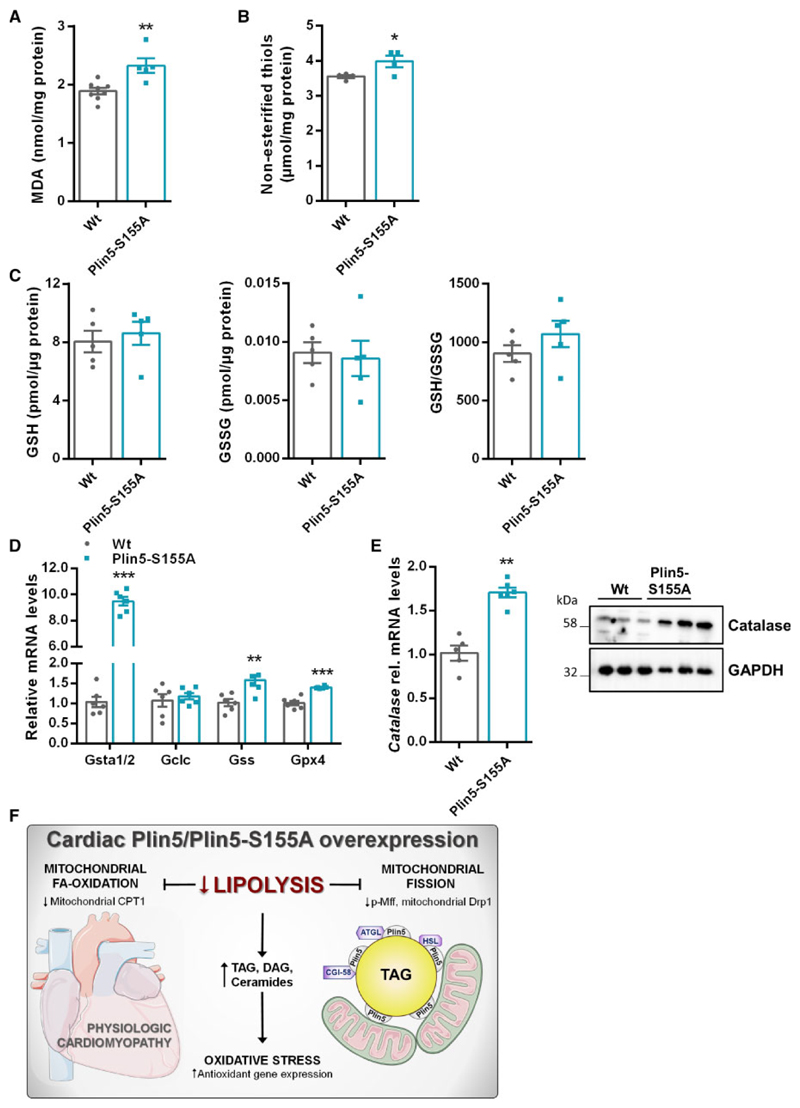
Lipotoxicity is linked to increased ceramide levels, which trigger ROS generation and oxidative damage.2 Preserved heart function despite augmented ceramide levels prompted us to examine cardiac ROS levels in Plin5-S155A mice. Malondialdehyde (MDA) concentration, a marker for lipid peroxidation, was moderately increased in Plin5-S155A CM compared to Wt (Figure 6A). Oxidative stress promotes the oxidation of thiol groups. Cardiac levels of non-esterified thiols were slightly increased in Plin5-S155A hearts compared to Wt (Figure 6B). Reduced glutathione (GSH) is an important antioxidant and the ratio with its oxidized form (GSSG) can be a marker for oxidative stress. The GSH:GSSG ratio was comparable between Wt and Plin5-S155A mice (Figure 6C) indicating that upregulation of the ROS-defense gene machinery may protect Plin5-S155A mice from oxidative stress which has been shown in Plin5 transgenic mice.13 Cardiac mRNA levels of Glutathione-S-transferase α1/2 (Gsta1/2), Glutathione synthetase (Gss), and Glutathione peroxidase 4 (Gpx4) were increased in both, Plin5-S155A and Plin5 transgenic mice, whereas glutamate-cysteine ligase catalytic subunit (Gclc) mRNA was unchanged (Figure 6D and Supplementary material online, Figure S6A). Expression of catalase, a peroxisomal enzyme that eliminates hydrogen peroxide, was markedly increased in Plin5-S155A (Figure 6E) and Plin5 (Supplementary material online, Figure S6B) transgenic mice compared to Wt. Moreover, mRNA levels of Nfr2, which promotes expression of ROS defense genes, were moderately increased in CM of Plin5-S155A and Plin5 transgenic mice (Supplementary material online, Figure S6C). Proteomic analyses revealed a marked increase in the protein content of several other enzymes from the antioxidant pathway including glutathione S-transferase Mu1 and 2 (Gstm1/2), glutathione S-transferase A4 (Gsta4), Gpx4, and NAD(P)H:quinone oxidoreductase (Nqo1) (Supplementary material online, Table S2).
Figure 6. Increased cardiac expression of ROS defense genes in Plin5-S155A mice.
(A) Cardiac MDA levels were quantified in fasted mice using a colorimetric kit (Wt: n = 8, Plin5-S155A: n = 5). (B) Non-esterified thiol groups determined by the Ellman’s reaction in CM of fasted mice (n = 4). (C) CM glutathione levels determined by MS analysis in fasted mice (n = 5). (D) Cardiac mRNA expression of Gsta1/2, Gclc, Gss, and Gpx4 using 36b4 as reference gene (n = 6). (E) Catalase expression in CM of Wt and Plin5-S155A mice determined by western blot and RT-qPCR (n = 6). Data are presented as means ± SEM. Statistical significance was determined by unpaired Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). (F) Scheme depicting the impact of Plin5-S155A or Plin5 overexpression on lipolysis, mitochondrial fission, and ROS defense.
4. Discussion
Obesity and diabetes are often associated with cardiac lipid overload due to increased cardiomyocyte FA uptake and oxidation2,3 eventually surpassing the mitochondrial FAO capacity. Conversely, TAG accumulation protects from FA-induced lipotoxicity by channelling palmitate into the cellular TAG pool thereby counteracting the production of potentially harmful lipids like ceramides.25 Defective cardiac lipolysis caused by ATGL loss of function provokes severe heart dysfunction in mice and humans26 linked to massive cardiac steatosis, impaired FAO and ROS-induced mitochondrial damage. In contrast, substantial cardiac TAG accumulation and reduced mitochondrial FAO is compatible with normal heart function and life span in mice overexpressing Plin5 in CM12,13 asking for the underlying mechanisms. We hypothesized that PKA-mediated phosphorylation of Plin5 at S155 partially recovers lipolysis thereby promoting FAO and counteracting the production of deleterious lipids. To address this hypothesis, we generated and characterized a cardiac cell line and transgenic mice overexpressing Plin5 carrying the S155A mutation.
FA release from H9c2 cardiac cells overexpressing Plin5-S155A was not inducible upon ß-adrenergic stimulation compared to Plin5 overexpression demonstrating that S155 phosphorylation increases lipolysis. Moreover, the reduction in FAO was even more pronounced in Plin5-S155A compared to Plin5 overexpressing H9c2 cells indicating that lipolysis is coupled to FAO. These findings prompted us to study the impact of CM-specific Plin5-S155A overexpression on cardiac energy metabolism in vivo in mice. Plin5-S155A transgenic mice showed massive TAG accumulation in CM accompanied by a substantial increase in C16:0 and C18:0 ceramide and DAG levels. A similar lipid profile was also present in mice overexpressing non-mutated Plin5. Considering that mRNA levels of genes involved in cardiac FA uptake (Lpl and Cd36) are substantially reduced, it is unlikely that the increase in ceramides and DAG originates from augmented cardiac FA influx as shown in mice overexpressing PPARα.4 Reduced FAO in H9c2 cells or in CM of Plin5-S155A transgenic mice suggests that the increase in ceramide levels is due to impaired FAO and augmented palmitic acid channelling into ceramides. Moreover, cardiac DAG species reflect the FA profile of the TAG pool. Considering that lipolysis is reduced in Plin5-S155A/Plin5 mice, DAGs likely originate from lipogenesis to protect from the accumulation of non-esterified FAs. Interestingly, expression of HSL, which efficiently hydrolyzes cholesteryl esters,27 is decreased in CM of Plin5-S155A mice. Hence, it is conceivable that TC accumulation originates from low HSL levels. Unexpectedly, cardiac lipid accumulation in Plin5-S155A (and Plin5) transgenic mice is compatible with normal heart function even under mild stress. Analysis of heart parameters revealed a significant increase in LV internal diameter, whereas relative wall thickness was comparable to Wt mice indicating physiologic eccentric cardiac growth20 in Plin5-S155A and Plin5 transgenic mice.
Which mechanisms protect Plin5-S155A (and Plin5) transgenic mice from lipotoxic-induced heart dysfunction? The study by Wang et al. and our data suggest that induction of the ROS defense gene machinery counteract mitochondrial damage and consequently the development of heart failure. Recently, Plin5 has been demonstrated to shuttle into the nucleus in a PKA-dependent manner, which may be linked to the increase in antioxidant gene expression.11 Nutrient overflow strongly determines mitochondrial dynamics28 and increased levels of saturated FAs have been demonstrated to trigger mitochondrial division. Only recently, the study by Tsushima et al.29 demonstrated that increased myocardial FA uptake increases mitochondrial FAO and ROS production, and triggers Drp1-induced mitochondrial fission. Mitochondrial fission requires the translocation of cytosolic Drp1 to mitochondria.28 Most strikingly, mitochondria prepared from Plin5-S155A (and Plin5) transgenic mice exhibited low Drp1 protein levels compared to Wt. Moreover, ATGL-deficiency also reduced mitochondrial Drp1 localization suggesting that reduced cardiac lipolysis may lower mitochondrial fission. In line, increased lipolysis upon cardiac overexpression of ATGL24 promotes mitochondrial Drp1 recruitment. Notably, levels of phosphorylated Mff,23 a mitochondrial outer-membrane receptor of Drp1, were markedly decreased in CM of Plin5-S155A and Plin5 mice further suggesting that low cardiac lipolysis reduces mitochondrial fission. Notably, triple knock-out mice lacking Drp1, Mfn1, and Mfn2 show life span extension compared to mice solely lacking Drp1 or Mfn1/Mfn2,30 indicating that an imbalance in fission/fusion is more detrimental than the simultaneous shut down of both processes. Interestingly, Mfn1 protein levels were significantly reduced in Plin5-S155A mice likely representing a compensatory adaptation to reduced fission.
A recent study showed that a subfraction of mitochondria, designated as peridroplet mitochondria, is increased in size and tightly tethered to LDs in brown adipocytes.15 These mitochondria showed reduced DrpI recruitment and augmented respiration associated with low FAO. Moreover, elongated mitochondria are considered to be less vulnerable to oxidative stress and are metabolically more efficient.21 Plin5 overexpression has been demonstrated to increase mitochondrial size.13 In accordance, mitochondria were also enlarged in CM of Plin5-S155A mice. Virtually, the vast majority of mitochondria in CM of Plin5-S155A transgenic mice can be regarded as peridroplet mitochondria considering the strong association with LDs. In accordance, we show that LDs prepared from Plin5-S155A CM are enriched with mitochondria. These changes in mitochondrial dynamics and morphology may contribute to adequate ATP production.
We hypothesized that S155 of Plin5 is a critical regulator of cardiac lipolysis and predicted impaired heart function in Plin5-S155A compared to Plin5 transgenic mice. However, the very similar impact on cardiac lipid accumulation, mitochondrial FAO, and PPARα target gene expression suggests that cardiac lipolysis is similarly restricted in Plin5-S155A compared to Plin5 transgenic mice. Interestingly, comparative analysis of the cardiac phosphoproteome of Plin5-S155A and Plin5 transgenic mice revealed a different phosphorylation pattern. As expected, relative phosphorylation of S155 was substantially blunted in Plin5-S155A CM. Reduced phosphorylation of adjacent S163 indicates coordinated regulation of both sites. Moreover, the significant increase in phosphorylation of S17 and S292 in Plin5-S155A CM implicates and adaptation to the lack of S155, which requires further investigation. Interestingly, except for S266, all discovered phosphosites are potential PKA motives (Supplementary material online, Table S3) suggesting that cardiac lipolysis is under regulation of PKA-mediated Plin5 phosphorylation. We previously showed that p-PKA levels are significantly increased in CM of Plin5 transgenic mice compared to Wt. However, this finding could not be reproduced and p-PKA levels of Plin5-S155A mice were comparable to Wt applying p-PKA specific antibodies. An explanation for this discrepancy may be the cross reaction of the p-PKA specific antibody from a former batch with an additional cAMP-dependent protein kinase,31 which is expressed in the heart and shows high homology to PKA. Nonetheless, the increase in several potential PKA phosphorylation sites in the transgenic mice suggests that levels of p-PKA are adequate.
Normal heart function implicates sufficient ATP generation despite reduced FAO in Plin5-S155A mice. We have previously shown that glucose uptake is markedly increased in Plin5 transgenic mice10 and the study by Wang et al.13 suggest a switch to glucose as energy substrate. OCR measurements upon glucose incubation of H9c2 cells overexpressing Plin5-S155A together with an increase in CM lactate levels as a measure of increased glycolysis argument for a switch towards glucose oxidation. In line, basal OCR in CM of Plin5-S155A was increased and overall OCR was similar compared to Wt upon addition of pyruvate. Finally, low levels of glutamate, glutamine, and histidine may indicate increased utilization of amino acids as energy fuel.
Together, we show that low cardiac lipolysis in Plin5-S155A and Plin5 transgenic mice is associated with reduced mitochondrial fission (Figure 6F), which may protect from the development of lipotoxic heart dysfunction. Low lipolysis induces the expression of ROS defense genes and a switch to alternative energy fuels. Accordingly, the transient inhibition of cardiac lipolysis might serve as a therapeutic strategy to combat mitochondrial dysfunction and cardiac lipotoxicity.
Supplementary Material
Supplementary material is available at Cardiovascular Research online.
Translational perspective.
Obesity and type 2 diabetes typically lead to increased fatty acid (FA) utilizationin the heart. However, cardiac FA uptake may surpass the oxidativecapacity causing cellular stress and mitochondrial damage designatedas lipotoxicity. Recently, augmented mitochondrial fission has been linkedto lipotoxic heart dysfunction. In this study, we show that decelerated30 cardiac lipolysis upon cardiac-specific overexpression of the lipid dropletbinding protein Perilipin 5 reduces mitochondrial FA oxidation and fissionin cardiomyocytes, which is compatible with normal heart functiondespite cardiac steatosis. Thus, lowering cardiac lipolysis and consequentlymitochondrial fission may protect from lipotoxicity-induced35 heart dysfunction in metabolic disease.
Acknowledgements
We thank B. Juritsch, A. Steiner, and K. Zierler for assistance with animal breeding, housing, and genotyping. We thank L. Liesinger for helping with (Phospho)proteomics analysis and data processing. We thank S. Schauer for histological analyses and D. Pernitsch for TEM analyses.
Funding
This study was supported by the grants P29253 (G.H.), P28882-B21 (G.S.), SFB-Lipotox F30-B05, and the DK Metabolic and Cardiovascular Disease W1226 (G.H.), which are all funded by the Austrian Science Fund (FWF).
Footnotes
Author contributions
G.H. designed the study and prepared the manuscript. S.K. performed experiments and prepared the manuscript. B.K. isolated peridroplet mitochondria. T.R. produced transgenic mice. C.D. performed MRI measurements. T.O.E. performed Lipidomics. S.S. and T.M. performed NMR analyses. P.K., T.T., and R.B.G. performed (Phospho)proteomics. D.K. performed TEM. G.H. performed histological analyses. C.H. and H.W. performed fluorescence microscopy. G.S., R.S., L.K.M., and M.S. assisted with experiments and data interpretation.
Conflict of interest: none declared.
References
- 1.Goldberg IJ, Trent CM, Schulze PC. Lipid metabolism and toxicity in the heart. Cell Metab. 2012;15:805–812. doi: 10.1016/j.cmet.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaffer JE. Lipotoxicity: when tissues overeat. Curr Opin Lipidol. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Szczepaniak LS, Victor RG, Orci L, Unger RH. Forgotten but not gone: the rediscovery of fatty heart, the most common unrecognized disease in America. Circ Res. 2007;101:759–767. doi: 10.1161/CIRCRESAHA.107.160457. [DOI] [PubMed] [Google Scholar]
- 4.Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, Han X, Gross RW, Kozak R, Lopaschuk GD, Kelly DP. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopaschuk GD, Spafford MA, Davies NJ, Wall SR. Glucose and palmitate oxidation in isolated working rat hearts reperfused after a period of transient global ischemia. Circ Res. 1990;66:546–553. doi: 10.1161/01.res.66.2.546. [DOI] [PubMed] [Google Scholar]
- 6.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 7.Haemmerle G, Moustafa T, Woelkart G, Büttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, Schreiber R, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimmel AR, Sztalryd C. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr Opin Lipidol. 2014;25:110–117. doi: 10.1097/MOL.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Bell M, Sreenevasan U, Hu H, Liu J, Dalen K, Londos C, Yamaguchi T, Rizzo MA, Coleman R, Gong D, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. 2011;286:15707–15715. doi: 10.1074/jbc.M110.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollak NM, Jaeger D, Kolleritsch S, Zimmermann R, Zechner R, Lass A, Haemmerle G. The interplay of protein kinase A and Perilipin 5 regulates cardiac lipolysis. J Biol Chem. 2015;290:1295–1306. doi: 10.1074/jbc.M114.604744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallardo-Montejano VI, Saxena G, Kusminski CM, Yang C, McAfee JL, Hahner L, Hoch K, Dubinsky W, Narkar VA, Bickel PE. Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollak NM, Schweiger M, Jaeger D, Kolb D, Kumari M, Schreiber R, Kolleritsch S, Markolin P, Grabner GF, Heier C, Zierler KA, et al. Cardiac-specific overexpression of perilipin 5 provokes severe cardiac steatosis via the formation of a lipolytic barrier. J Lipid Res. 2013;54:1092–1102. doi: 10.1194/jlr.M034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Sreenivasan U, Gong D-W, O’Connell KA, Dabkowski ER, Hecker PA, Ionica N, Konig M, Mahurkar A, Sun Y, Stanley WC, et al. Cardiomyocyte-specific perilipin 5 overexpression leads to myocardial steatosis and modest cardiac dysfunction. J Lipid Res. 2013;54:953–965. doi: 10.1194/jlr.M032466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber R, Diwoky C, Schoiswohl G, Feiler U, Wongsiriroj N, Abdellatif M, Kolb D, Hoeks J, Kershaw EE, Sedej S, Schrauwen P, et al. Cold-induced thermogenesis depends on ATGL-mediated lipolysis in cardiac muscle, but not brown adipose tissue. Cell Metab. 2017;26:753–763.e7. doi: 10.1016/j.cmet.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.zBenador IY, Veliova M, Mahdaviani K, Petcherski A, Wikstrom JD, Assali EA, Acín-Pérez R, Shum MMM, Oliveira MF, Cinti S, Sztalryd C, et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 2018;27:869–885. doi: 10.1016/j.cmet.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zierler KA, Jaeger D, Pollak NM, Eder S, Rechberger GN, Radner FPW, Woelkart G, Kolb D, Schmidt A, Kumari M, Preiss-Landl K, et al. Functional cardiac lipolysis in mice critically depends on comparative gene identification-58. J Biol Chem. 2013;288:9892–9904. doi: 10.1074/jbc.M112.420620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haemmerle G, Zimmermann R, Hayn M, Theussl C, Waeg G, Wagner E, Sattler W, Magin TM, Wagner EF, Zechner R. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 18.Mayer N, Schweiger M, Romauch M, Grabner GF, Eichmann TO, Fuchs E, Ivkovic J, Heier C, Mrak I, Lass A, Höfler G, et al. Development of small-molecule inhibitors targeting adipose triglyceride lipase. Nat Chem Biol. 2013;9:785–787. doi: 10.1038/nchembio.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Shafei AIM, Wise RG, Gresham GA, Bronns G, Carpenter TA, Hall LD, Huang C-H. Non-invasive magnetic resonance imaging assessment of myocardial changes and the effects of angiotensin-converting enzyme inhibition in diabetic rats. J Physiol. 2002;538:541–553. doi: 10.1113/jphysiol.2001.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega RB, Konhilas JP, Kelly DP, Leinwand LA. Molecular mechanisms underlying cardiac adaptation to exercise. Cell Metab. 2017;25:1012–1026. doi: 10.1016/j.cmet.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes LC, Benedetto G, Di Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyama EQ, Herzig S, Courchet J, Lewis TL, Losón OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreiber R, Hofer P, Taschler U, Voshol PJ, Rechberger GN, Kotzbeck P, Jaeger D, Preiss-Landl K, Lord CC, Brown JM, Haemmerle G, et al. Hypophagia and metabolic adaptations in mice with defective ATGL-mediated lipolysis cause resistance to HFD-induced obesity. Proc Natl Acad Sci USA. 2015;112:13850–13855. doi: 10.1073/pnas.1516004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coassin S, Schweiger M, Kloss-Brandstätter A, Lamina C, Haun M, Erhart G, Paulweber B, Rahman Y, Olpin S, Wolinski H, Cornaciu I, et al. Investigation and functional characterization of rare genetic variants in the adipose triglyceride lipase in a large healthy working population. PLoS Genet. 2010;6:e1001239. doi: 10.1371/journal.pgen.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeaman SJ. Hormone-sensitive lipase–a multipurpose enzyme in lipid metabolism. Biochim Biophys Acta. 1990;1052:128–132. doi: 10.1016/0167-4889(90)90067-n. [DOI] [PubMed] [Google Scholar]
- 28.Mishra P, Chan DC. Metabolic regulation of mitochondrial dynamics. J Cell Biol. 2016;212:379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsushima K, Bugger H, Wende AR, Soto J, Jenson GA, Tor AR, McGlauflin R, Kenny HC, Zhang Y, Souvenir R, Hu XX, et al. Mitochondrial reactive oxygen species in lipotoxic hearts induce post-translational modifications of AKAP121, DRP1, and OPA1 that promote mitochondrial fission. Circ Res. 2018;122:58–73. doi: 10.1161/CIRCRESAHA.117.311307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song M, Mihara K, Chen Y, Scorrano L, Dorn GW. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–285. doi: 10.1016/j.cmet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S, Li Q, Alberts I, Li X. PRKX, a novel cAMP-dependent protein kinase member, plays an important role in development. J Cell Biochem. 2016;117:566–573. doi: 10.1002/jcb.25304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.