Abstract
By developing a high-density murine immunophenotyping platform compatible with high-throughput genetic screening, we have established profound contributions of genetics and structure to immune variation (www.immunophenotype.org). Specifically, high-throughput phenotyping of 530 unique mouse gene knockouts identified 140 monogenic “hits”, of which most had no previous immunological association. Furthermore, hits were collectively enriched in genes for which humans show poor tolerance to loss-of-function. The immunophenotyping platform also exposed dense correlation networks linking immune parameters with one another and with specific physiologic traits. Such linkages limit freedom-of-movement for individual immune parameters, thereby imposing genetically regulated “immunological structures”, whose integrity was associated with immunocompetence. Hence, we provide an expanded genetic resource and structural perspective for understanding and monitoring immune variation in health and disease.
The increasing implication of immunology in myriad arenas of pathophysiology emphasises the importance of understanding and appropriately measuring inter-individual immune variation. Reflecting this are highly informative studies describing human immune system dynamics1–4, and investigations of the factors contributing to it5–8. Thus, SNP-based and deep sequencing-based Genome-Wide Association Studies (GWAS) and Twin-studies have associated defined genetic loci with autoimmunity and/or immunodeficiency9–14. However, it can be challenging to link discrete immunophenotypes to specific genes and/or genetic variants15. Conversely, Mendelian trait analysis, that is expanding through genome sequencing of “rare diseases”16,17, has established concrete links between specific genes and immune function. Nonetheless, this approach can be limited by the infrequency and uncertain clinical annotation of patients, and by practical limitations on phenotypic assays.
At the same time, sex, age, and environmental factors, including diet and the microbiome, make major contributions to human immune variation5,6,8, but assessing their full impacts is limited by appropriate constraints on interventions, and by human genetic diversity. In sum, immunoregulatory factors seem so numerous and diverse that resolving their individual contributions can seem intractable.
In this regard, animal model studies offer unique opportunities. Specifically, use of an inbred strain limits genetic variation; co-housing reduces microbiome and dietary variation; and age-matching limits physiologic variation. Thus, their study can establish a frame-of-reference for the nature and sources of variation in the “baseline immune system”. That frame-of-reference can aid myriad investigations of rodent immunology, and guide the design and interpretation of human immunological studies. Indeed, despite their limitations, gene knockout mouse studies have usefully modelled several human immunopathologies and the actions of many of the most widely prescribed medicines18. Superimposed upon this, the use of co-housed, age-matched mice for a genetic screen could offer insight into the fraction and nature of genes whose loss-of-function perturbs the immune system.
To achieve these goals at scale, we have developed a robust, broadly transferrable, high-density, high-throughput Infection and Immunity Immunophenotyping (“3i”) platform that has facilitated analysis of the baseline immune system and its response to challenge in aged-matched, co-housed, isogenic mouse strains collectively mutated in 530 genes (www.immunophenotype.org). Additionally, by integrating 3i into the International Mouse Phenotyping Consortium (IMPC) pipeline (www.mousephenotype.org), immune variation could be related to measures of general physiology. The expansive outputs (>1million data-points) have provided many discrete insights and data-rich resources that collectively build a revised frame-of-reference for viewing immune variation. Moreover, of 530 genes screened, the baseline immunophenotype and/or responses to challenge were affected by mutations in 140 genes (>25% hit-rate), most of which (57%) were never hitherto associated with immunobiology, but which were strikingly enriched in genes for which humans show little tolerance to loss-of-function.
Results
Immunophenotyping of mutant mice at scale
The IMPC aims to obtain and publicly disseminate phenotyping data for mice with targeted disruptions in each of ~18,000 annotated protein-coding genes, generated using either embryonic stem cells generated by the International Knockout Mouse Consortium19 or CRISPR technology. Although full-gene knockouts do not model most forms of human genetic variation, they can irrefutably implicate defined molecular processes in immunophenotypes, and those phenotypes may be directly related to biallelic human loss-of-function variants, revealed by studies of communities with parental relatedness20.
Contributing to IMPC, the Wellcome Trust Sanger Institute (WTSI) generated ~3 mutant lines per week. Given this scale, immunological assays were limited to peripheral blood lymphocytometry and responses to Salmonella and Citrobacter infection21 (Fig 1a; Supplementary Fig. 1a), potentially missing many immunoregulatory genes. We therefore developed a high-density infection and immunity immunophenotyping platform (3i) compatible with the IMPC high-throughput screen (HTS), that permitted us to assess the proportion and types of genes that may underpin immune variation. Moreover, by integration into IMPC, 3i could relate immunophenotypes to general physiological traits.
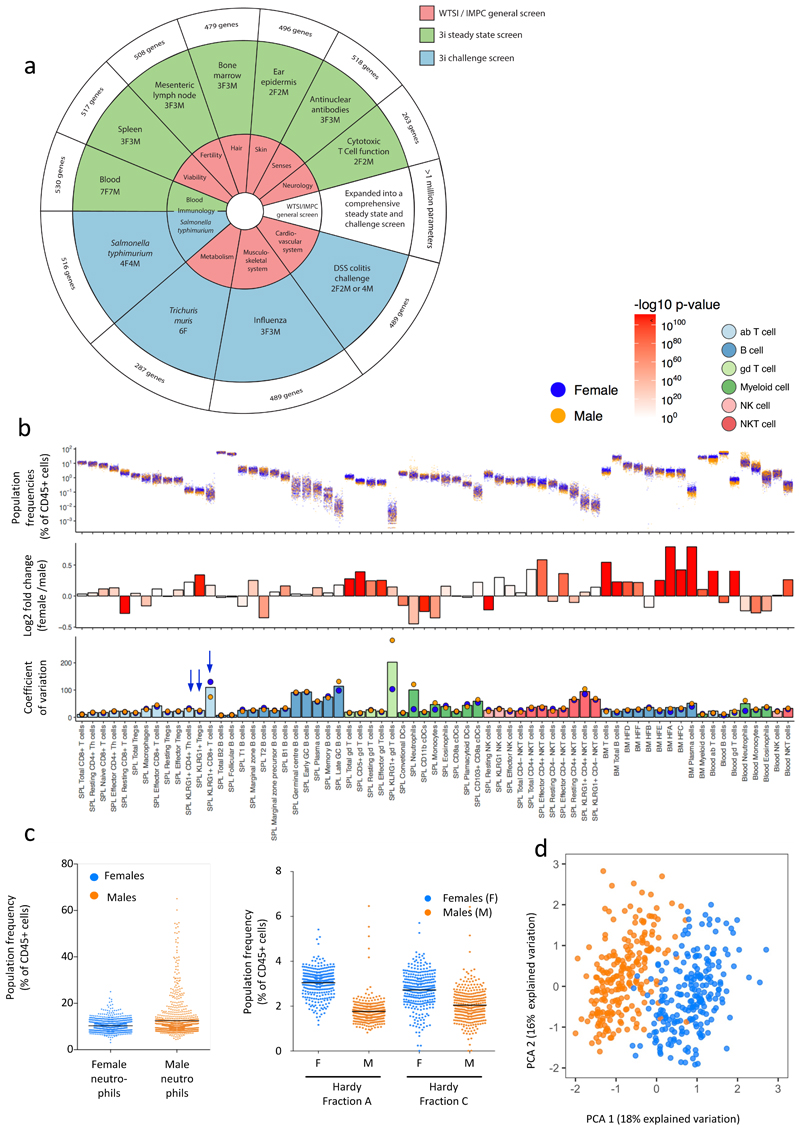
Figure 1. Variation in immune cell subset composition with sex as a contributory driver.
a. Overview of the tests performed by WTSI as part of IMPC (inner circle) and by 3i and WTSI as part of 3i (outer circle).
b. Sexual dimorphism in the immune system: Population sizes as % of CD45 cells (upper panel); sexual dimorphism of mean values of population sizes as log2 fold change (middle panel, female/male); and % coefficient of variation (lower panel) of immune populations in SPL, BM and PB from 16-week old male and female wt C57BL/6N mice (n>500). Blue arrows denote cell subsets mentioned in text. Blue and orange circles in bottom panel denote CV values for female and male mice, respectively. Non adjusted p-values from two-sided Wilcoxon test.
c. Left: Neutrophils from peripheral blood of 16-week old wt C57BL/6N mice (for females n=918 and for males n=913, bars represent means) Right: Hardy fractions A and C from the bone marrow of 16-week old WT C57BL/6N mice (for females n=308, for males n=315, bar represents mean)
d. PCA of cell type frequencies from four tissues (SPL, MLN, BM, PB), 60 subsets, and 451 mice; colour denotes sex.
At homeostasis, the baseline immune system is simultaneously poised to respond to infectious or toxic challenges and regulated to limit immunopathology. Hence, inter-individual baseline variation is likely manifest in differential immunocompetence and susceptibilities to autoimmune diseases. To capture this, 3i featured high-content flow cytometry analysis of lymphoid and myeloid cells and their activation states in spleen (SPL), mesenteric lymph nodes (MLN), bone marrow (BM) and peripheral blood (PB) at steady-state (Fig 1a; panels in Supplementary Table 1; populations quantitated in Supplementary Table 2; illustrative gating strategy for MLN T, NKT, and NK cell subsets shown in Supplementary Fig 1b; for other gating, see materials and methods). To sample an extra-lymphoid immune system, quantitative object-based imaging was applied to intraepidermal lymphoid and myeloid cells in situ (Supplementary Fig 1c). Anti-nuclear antibodies (ANA) were quantitated, since they commonly reflect impaired immunological tolerance (Supplementary Fig 1d), while effector potential was gauged by measuring SPL CD8 T cell-mediated cytolysis.
All observational assays requiring sacrifice were conducted at the IMPC termination-point of 16 weeks so that they neither diverted tissue from, nor operationally interfered with the basic phenotyping programme. Measuring absolute numbers of cells was largely precluded by IMPC requirements on organ usage, although the focus on percentage representation reflects common practice in immune monitoring2,3,5. In parallel, mice were assayed for responses to infection by a parasite (Trichuris muris), a virus (influenza), and a bacterium (Salmonella typhimurium), and to sodium dextran sulphate (DSS) that causes gut epithelial erosion and microbial translocation (Fig 1a; Supplementary Fig 1e). For each 3i component, experimental standard-operating procedures were established and stringently quality-controlled; instruments were well calibrated; and data reproducibility monitored longitudinally, with automated analysis minimizing any temporal variation (Supplementary Fig 1f). Additionally, and to comply with time and budgetary constraints typical of an HTS, we chose minimum numbers of data-points required to establish significance following application of bespoke statistical analyses (Fig 1a; Supplementary Table 3).
This study represents a five-year phase in which 3i phenotyped knockout strains for 530 genes (Supplementary Table 4). Most strains were nulls or severe hypomorphs of protein-coding genes, whereas 1.8% were lncRNA or miRNA mutants. For ~30% of genes, heterozygotes were screened because homozygotes were embryonically lethal or sub-viable. Of genes selected, 9% had been identified in GWAS screens for inflammatory bowel disease (IBD) or were linked to infection; some others had GWAS associations to non-immunological traits; and the majority were poorly understood genes, thereby maximising the potential for discovery. Panther Biological Process Gene Ontology showed that relative to the whole genome, the selected genes were neither enriched nor depleted in categories with immunological annotations, a point illustrated by a GO-Slim analysis in which the only categories with low but significant deviation from the whole genome were reproduction and protein modification (Supplementary Fig 1g – indicated by asterisks).
Overall, >1 million data-points were collected from 7 distinct steady-state assay systems applied to 2,100 -10,000 mice (Fig 1a), while additional cohorts of mice were subjected to challenges. Moreover, minimization of technical variation; fastidious control over batch variation, e.g. by spreading phenotyping of each strain over several separate experiments; optimization of data collection and analysis22; and innovative data management across heterologous platforms permitted 3i to make rigorous assessments of naturally-arising variation in the baseline immune system of many hundreds of genetically identical, age-matched, co-located, adult C57BL/6N mice, thereby creating a precise backdrop for analyzing mutants.
Immunophenotypic variation among controls
Most steady-state immune cell subsets in adult C57BL/6N controls showed low coefficients of variation (CV), which were further reduced by dynamic automated gating of flow cytometry data, particularly for numerically small cell subsets whose reproducible quantitation can be challenging (Supplementary Fig 1h). Hence, automated gating was adopted screen-wide to obtain the population sizes and CVs described in Fig 1b (see also Materials and Methods)22. The screen revealed greater variation for some cell types, including germinal centre (GC) B cells and various αβ and γδ T cell subsets expressing an activation marker, KLRG1 (Fig 1b; bottom-most panel). Activation-driven variation of adaptive subsets was anticipated since non-heritable, antigen receptor gene rearrangements dictate that syngeneic individuals respond differently to shared environments. Nonetheless, effects were highly selective, as evidenced by low CVs of KLRG1+ CD4 T helper cells and of KLRG1+ regulatory T cells (Treg cells) compared to a comparably-sized subset of KLRG1+ CD8 T cells (Fig 1b, compare top and bottom-most panels for subsets denoted with blue arrows).
Compared to most innate immune cell subsets, SPL and PB neutrophils showed relatively high CVs, particularly in males. Indeed, sex was a consistent source of variation for ~50% of PB, SPL, BM, and MLN cell subsets, reflected either in significantly different variance for female (F) and male (M) mice, e.g. for PB neutrophils (Fig 1c; left panel) and displayed broadly in Fig 1b (bottom panel - CVF, blue circles; CVM, orange circles), and/or in sexually dimorphic mean-values, e.g. for BM B cell progenitors (Fig 1c; right panel), and displayed broadly in Fig 1b (middle panel; meanF/meanM log2-transformed). Considered at scale, the impact of sex was such that principal component analysis (PCA) of 60 aggregated SPL, MLN, BM, and PBL flow cytometry parameters sex-segregated 451 mice with >99% accuracy, with the first two principal components explaining 34% of total variation (Fig 1d).
Several properties of Vγ5+ dendritic epidermal T cells (DETC) and Langerhans Cells (LC) were also sexually dimorphic, as were ANA outputs, evoking frequent gender imbalances in human autoimmunity (Supplementary Fig 1i)23. Conversely, DSS outputs did not segregate by sex, evoking gender-neutral incidences of IBD (Supplementary Fig 1i)24. In sum, widespread but highly selective sexual dimorphism characterizes the baseline immune system of adult C57BL/6N mice. As a practical response to this, all statistical tests of 3i data accounted for sex.
Significant correlations of discrete immune parameters
Although the immune system is multi-component, the inter-connectedness of its constituent cell populations is poorly understood. In this regard, the 3i analysis of >650 age-matched, co-located, genetically identical control mice identified significant positive (red) and negative (blue) correlations, as illustrated for 46 steady-state SPL parameters in male and female mice (Fig 2a; Supplementary Fig 2): note, all correlations shown exclude contingent relationships reflecting nested or directly paired technical measurements (see Supplementary Fig 1b; Materials and Methods). Conversely, the strong, negative, steady-state relationship of effector CD44+CD62L-CD8+ T cells to naive CD44-CD62L+CD8+ T cells is not contingent, since there is a variable population of resting CD44+CD62L+CD8+ T cells.
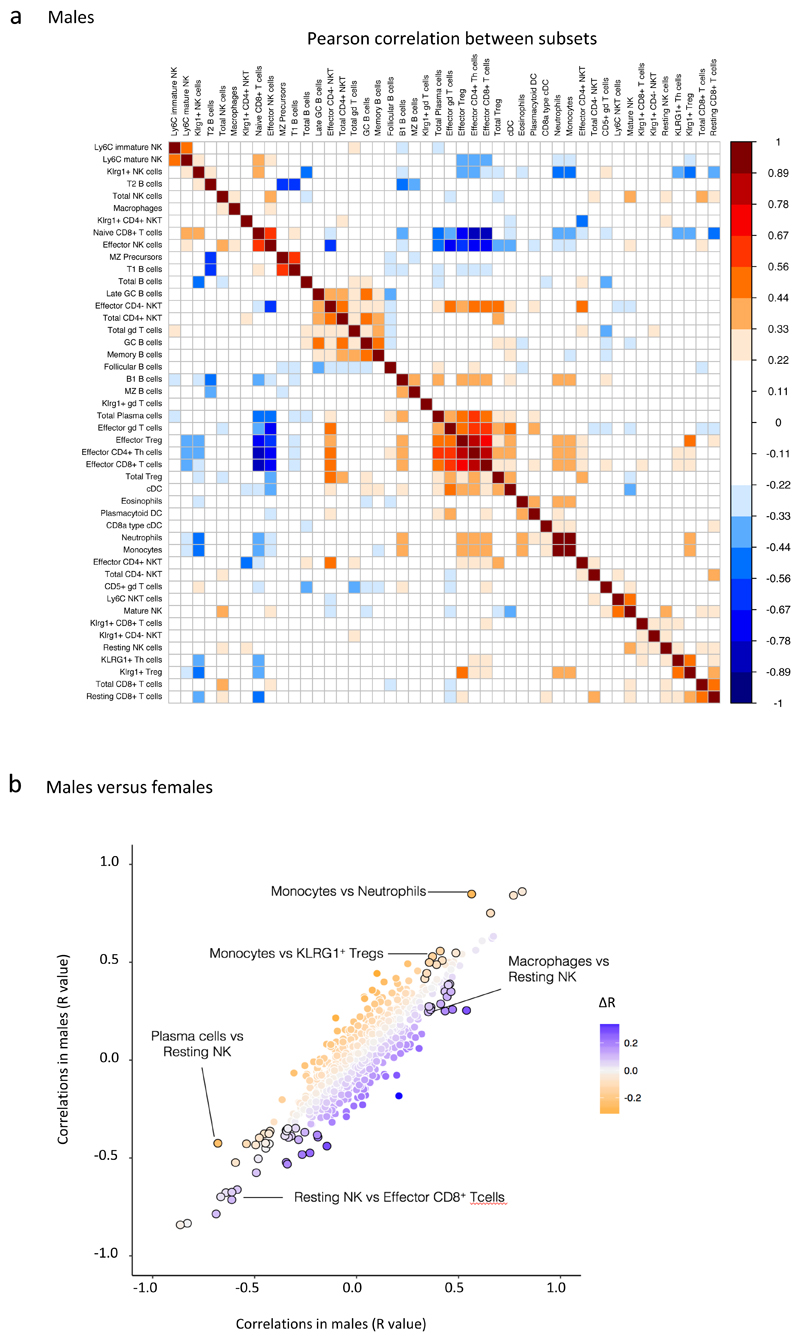
Figure 2. Correlations exist between immune parameters.
a. Heat map represents Pearson correlations of 46 splenic immune cell subsets with each other in wt males (n>230) as determined by flow cytometry. Dark red fields denote strong positive, dark blue fields strong negative correlations between frequencies of spleen immune cell subsets.
b. Correlation differences between males and females. Colour denotes ∆R, the difference between the correlation coefficient R for SPL subsets in male and female wt mice. Black circumferences mark parameter pairs that are significantly sexually dimorphic (see Materials and Methods). Correlation coefficients for male and female mice were derived from data depicted in Fig 2a and Fig S2, respectively.
As illustrated by male and female SPL data-sets (Fig 2a, Supplementary Fig 2), correlations included a “lymphoid activation-cluster” embracing effector CD4+, CD8+, NK and γδ T cells; Treg cells; conventional DC; and plasma cells (Fig 2a, Supplementary Fig 2; central red core). While some such correlations were anticipated, e.g. effector Th with effector Treg cells, others were not, e.g. effector CD8+T cells correlated positively with plasma cells (Fig 2a, 27 down, 23 across) but negatively with effector NK cells (Fig 2a, 27 down, 9 across). Such relationships may inform the design and monitoring of vaccines aimed at eliciting discrete effector responses.
Other correlations were related to immune homeostasis: thus, higher NK cell representation reflected increases in mature NK cells (Fig 2a, 5 down, 9 across), whereas higher CD8+ T cell representation reflected increases in resting but not effector CD8+ T cells (Fig 2a, 45 down versus 46 across or 27 across). Finally, some correlations seemed wholly unexplained; e.g. monocytes positively correlated with B1 B cells, and memory B cells positively correlated with γδ T cells (Fig 2a, 34 down, 20 across; 18 down, 16 across).
Many correlations were comparable in male and female mice (Fig 2a; Supplementary Fig 2; Fig 2b, colorless circles along the 45° axis), but some were sexually dimorphic, with R values and/or regression slopes being stronger in females (e.g. macrophages versus resting NK) or males (e.g. monocytes versus neutrophils) (Fig 2b, purple and orange circles, respectively). Whereas the spleen matrices shown are intra-organ, there were also many inter-organ correlations, collectively revealing that the baseline immune system of adult C57BL/6N mice is underpinned by dense, sexually dimorphic networks of >1000 correlations. These may reflect robust intercellular circuitry, such as exists for macrophages and fibroblasts25.
Integrating immunology with physiology
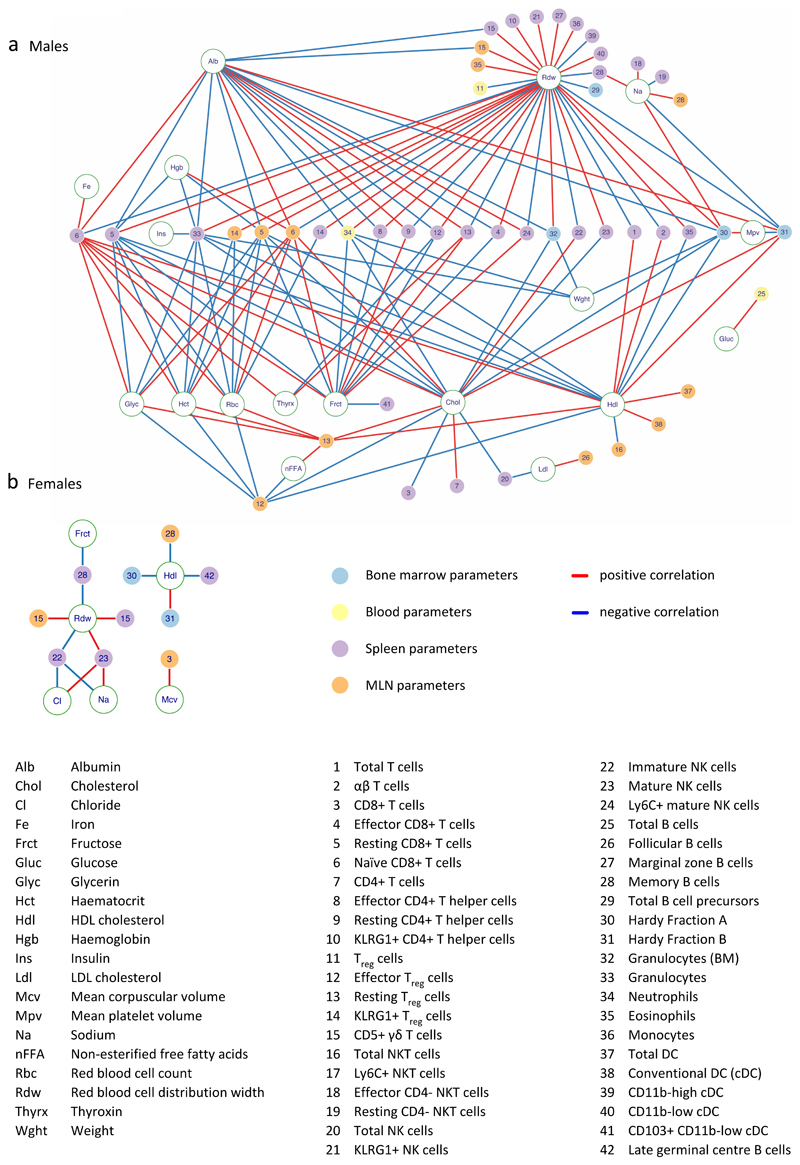
By coupling 3i to the IMPC, all animals in the observational screen were subject to measures of general physiology (see Fig 1a; Supplementary Fig 1a). As is well established, many physiologic traits are correlated with one another. By limiting the freedom-of-movement of individual parameters, those correlations impose phenotypic structures for female and male mice, reflecting sex-specific physiologies (Supplementary Fig 3a,b). Of note, the correlation network appeared much less dense in females, possibly because its full elucidation was masked by additional variation arising from sex-specific components, notably oestrous that cycles every 4-5 days26. This notwithstanding, core relationships were clearly conserved in females and males, e.g. positive correlations spanning cholesterol (Chol), HDL-cholesterol (HDL), insulin (Ins), and body-weight (Wght), and the relationship of total protein (Tp) to fructosamine (Fruct) which reflects blood protein glycation. Likewise, glucose (Gluc) correlated negatively with chloride (Cl) and sodium (Na). From a practical standpoint, the relatively low inter-individual variability of Na emphasised that prominent correlations did not merely reflect high dynamic ranges of specific parameters.
The more consistent variation of non-immunological parameters in males versus females allowed us to identify many highly significant correlations between those parameters and specific immunological parameters, with Chol, HDL, and Na being prominent (Fig 3). Although immunoregulatory roles for metabolic products and processes are well documented27,28, identified correlations were conspicuously selective, with, for example, no overt immunophenotypic correlations with triglycerides, creatinine, or calcium, but many with red blood cell distribution width (RDW), a common marker of anaemia29, frequently associated with chronic immune activation30, and recently shown to predict all-cause mortality in humans31.
Figure 3. Correlations between immune and non-immune parameters form a sex-specific network of interactions.
Correlations with a Pearson R-value >0.33 and p<0.001 between PBL, SPL, MLN, BM with haematological, clinical blood chemistry, and additional parameters (see Supplementary Fig 3) for males (a) and females (b). Circle colours denote organ assayed; red lines denote positive correlations; blue lines, negative correlations (n>180 per sex).
Clearly, significant correlations of immune parameters one with another and with discrete non-immunological traits will limit the freedom-to-vary of any single immune parameter, thereby imposing an immunological structure, as shown for males (Fig 3a). Immunological structure may reflect a balance of immune and non-immunological components that delivers immunocompetence while limiting immunopathology. For females, the sparse correlation network of non-immunological parameters (Supplementary Fig 3b) dictated that the immunological structure was also more sparse, although HDL, RDW and Na were again prominent within it (Fig 3b).
Many diverse genes affect the immunophenotype
The establishment of robust ranges for myriad immune parameters across age-matched, co-located, wild-type mice, coupled with the application of bespoke statistical tests (see Fig 1a; Supplementary Fig 1f; Supplementary Table 3; Materials and Methods) facilitated high-throughput screening of knockout lines collectively mutated in 530 genes. For steady-state immune parameters, “phenodeviants” (“hits”) which exerted monogenic dysregulation of the immunophenotype were called when ≥60% of samples from a strain located outside the 95th percentiles of the wild-type distribution for that parameter.
To further limit batch effects, each mutant strain was assayed on at least two separate occasions, with hits called in a supervised manner; i.e. no data-points were discarded from the dataset, but those possibly attributable to batch effects would not be scored as hits. Effects of temporal data drift in flow-cytometric data were avoided by comparing each knockout mouse to the 70 wild-type mice examined closest in time for any given parameter (Supplementary Table 3). Additionally, there was expert review of raw data for all hits and borderline candidates identified by the statistical pipeline.
False positive rates (FPRs) for each parameter were estimated by randomly sampling sets of wild-type controls in accordance with the real work flow so as to mimic 10,000 different mutant strains, and then assessing their phenodeviance (Supplementary Fig 4a-d). Resulting FPRs were far below observed hit-rates that collectively identified 140 phenodeviants, an overall hit-rate of >25% for genes that a priori showed no collective enrichment of immunological GO-annotation (Fig 4; Supplementary Fig 4; see Supplementary Fig 1g). Approximately ~20% of hits were heterozygotes, that collectively accounted for ~30% of the strains analyzed. The relevant code is available at: https://github.com/AnnaLorenc/3i_heatmapping; note that precise hit counts for genes with borderline read-outs may be subject to minor adjustment, based on ongoing analyses and expert review.
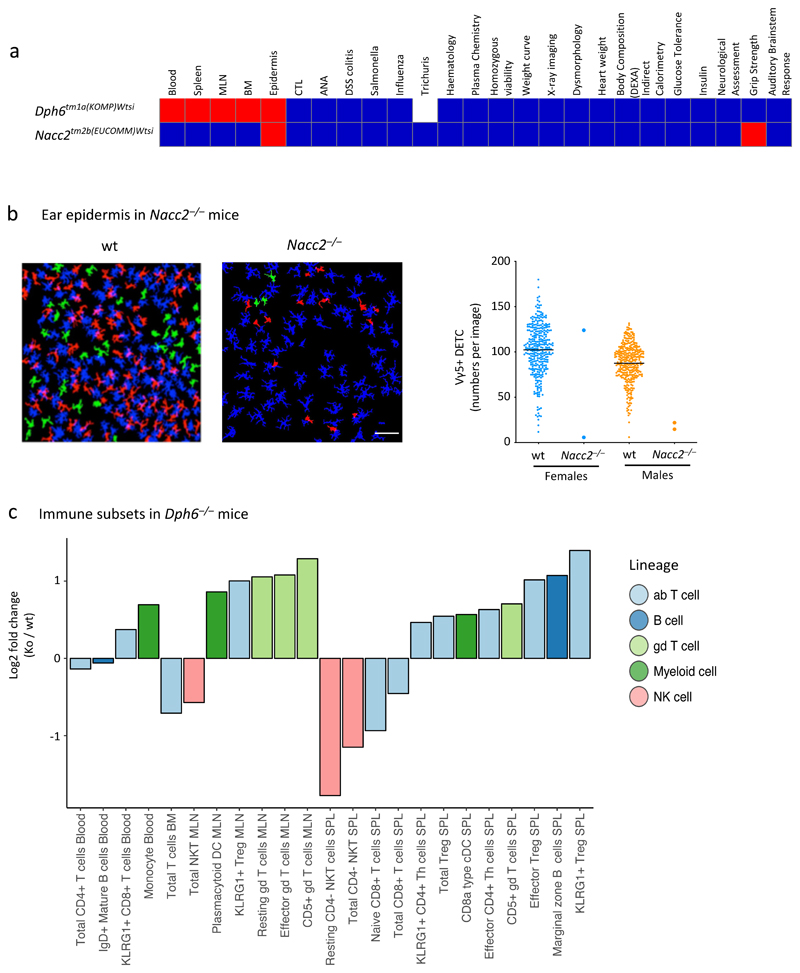
Figure 4. 140 out of 530 genes perturb the immunophenotype.
Red, significantly different from wt; blue, not significantly different from wt; white, not performed; grey, insufficient data to make a call. Each cell is coloured red when at least one of parameters within an assay is significant. Methods for determining significance of parameters are parameter-specific (see Materials and Methods and www.immunophenotyping.org).
Unsurprisingly, hit-rates varied among the assays (Supplementary Fig 5a), and the importance of assaying the same cell populations in different organs was emphasized by the fact that ~50% of flow cytometry hits showed no peripheral blood phenotype (hence would not have scored in IMPC), and of those 36% were unique to spleen, and 39% unique to MLN (Supplementary Fig 5b). Likewise, two thirds of challenge phenodeviants were unique to one pathogen (Supplementary Fig 5b).
Because most phenodeviants scored in only one or few assays, and because overall 3i hit-calling was conservative (Supplementary Table 3), a 25% hit-rate for monogenic effects may have under-estimated the contribution of genetics to immune variation. In short, 3i might have identified more hits if more immunological assays had been performed. For example, no direct assessments of immunological memory were made, and the steady-state gut was not analysed because of interference with IMPC phenotyping. Additionally, although sexually dimorphic trends were apparent for some hits, e.g. TCRαβ+ CD8 T cell percentages in Arpc1b–/– mice; TCRαβ+ Treg cell percentages in Setd5–/– mice; and low TCRαβ+CD44+CD62L– effector CD4 cell percentages in male IL-27p28 deficient mice (see www.immunophenotype.org), some sexually dimorphic phenodeviants might not have scored because, other than for PB cytometry and ANA, too few mutant mice were examined. Furthermore, no thorough analysis was made of the genetic impact of non-coding regions.
These considerations notwithstanding, it was conspicuous that the overall hit-rate was largely comparable irrespective of how well-studied genes were, as assessed by PubMed. Specifically, the hit-rate for genes in the lowest quartile of citations (from 0-26) was 23.3%, with the rates for the 3rd, 2nd, and 1st quartiles being 22.5%, 25% and 34%, respectively. This observation, combined with the fact that the genes screened collectively lacked a priori enrichment in any immunological annotations, permitted 3i to identify monogenic impacts on the C57BL/6N immune system of 80 genes never hitherto implicated in immunoregulation. Four case-studies, briefly considered below, illustrate the diversity of impacts of different phenodeviants (Figs 5, 6), while all phenodeviants are described at www.immunophenotype.org.
Figure 5. Examples of genes with specific impacts on the immune system.
a. Overview of two genes that display specific immunophenotypes. Colours as in Fig 4. Statistical methods and sample size differ between parameters – see Materials and Methods and www.immunophenotype.org for specific gene/parameter combination of n.
b. Vγ5 DETC in ear epidermis of wt and Nacc2–/–mice. The image represents a z-projection of cell outlines produced by image processing in Definiens Developer XD, which were used for quantitative object-based image analysis: blue, Langerhans cells (LC); red, Vγ5+ DETC contacting LC; green, Vγ5+ DETC not contacting LC. Bottom: cumulative data for Nacc2–/–mice (n=4) versus sex-matched wt controls (for females n=330, for males n=340, bar represents mean).
c. Phenotypic abnormalities in Dph6–/– mice. Fold change in immune cell subset proportions between Dph6–/– and wt mice across multiple tissues (Dph6–/–, n=6 for SPL, MLN, BM and n=14 for PBL; wt, n>500 for all parameters).
Figure 6. Examples of genes that impact upon the immune system and physiology.
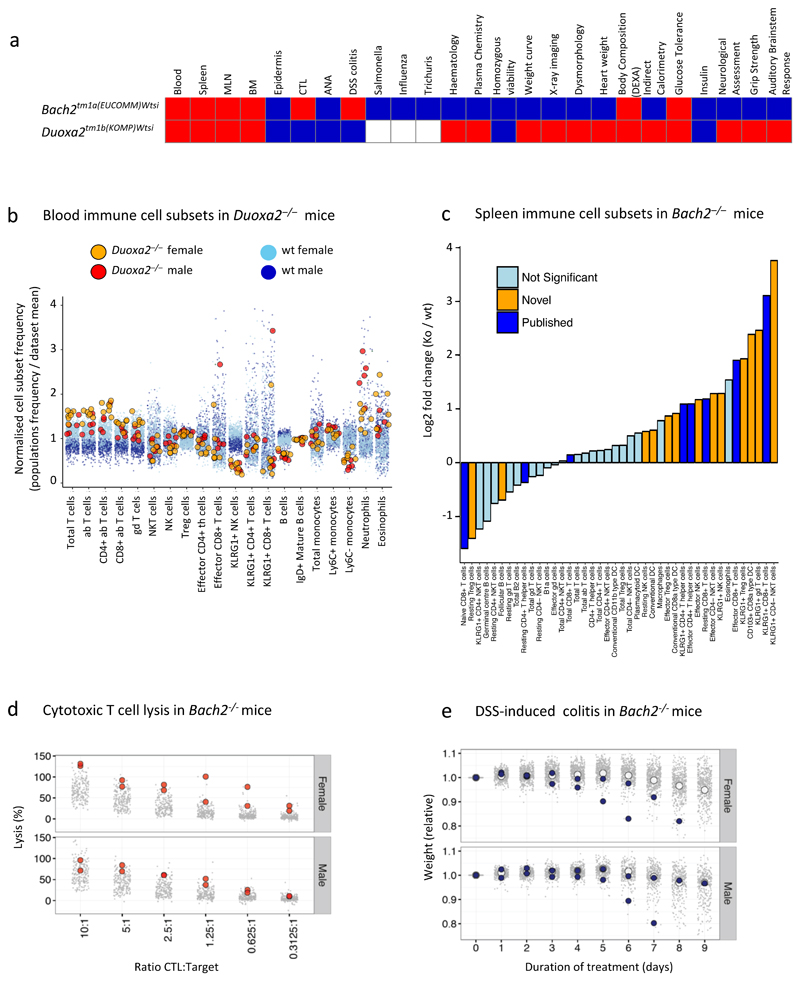
a. Overview of two genes that exhibit broad immunophenotypes. Colours as in Fig 4. Statistical methods and sample size differ between parameters – see Materials and Methods and www.immunophenotype.org for specific gene/parameter combination of n.
b. Phenotypic abnormalities PBL in Duoxa2–/– mice. Relative PBL cell subset frequencies for Duoxa2–/– (n=11) versus wt mice (n>450 per sex for all parameters). Dark and light blue denote wt males and females; red and orange denote Duoxa2–/– males and females.
c. Effector cell subsets in Bach2–/– mice. Fold-change in SPL cell subset composition in mutant mice (n=6) versus wt controls (n=76). Yellow represents significant cellular phenotypes not previously reported, as detected by reference range; dark blue represents significant cellular phenotypes previously reported; light blue represents non-significant differences. KLRG1+CD4- NK cells: 10-fold increase, p=1.1x10-9; reference range combined with Fisher’s exact test).
d. Cytotoxic T lymphocytes in Bach2–/– mice (n=4) compared to wt controls (212 female and 208 male): grey points represent individual wt mice; red circles represent individual Bach2–/– mice.
e. DSS colitis in Bach2–/– mice (n=4) compared to wt controls (n=481 female and n=315male): grey points represent individual wt mice; white circles represent wt mean values; blue circles represent individual Bach2–/– mice.
Different classes of genetically regulated immune variation
Nacc2 (Nucleus accumbens-associated protein 2) is a little-studied gene32 which specifically affected Vγ5+ DETC numbers (Fig 5a,b), a phenotype hitherto limited to mice mutant for Vγ5 or for Skint genes that encode poorly understood, epithelial DETC-selecting elements33,34. Thus, the 3i screen identified a novel immune regulator with potential to inform how a prototypic tissue-resident T cell compartment is selected and/or maintained in adulthood. In this regard, occasional mice scored as harboring wild-type DETC numbers, but those cells had low TCR expression, consistent with incomplete selection33.
Dph6–/– mice displayed an immune-specific but broad phenotype, with several innate and adaptive cell lineages differing significantly from wild-type (Fig 5a,c). Dph6 is ubiquitously expressed, encoding one of seven enzymes required for synthesizing diphthamide, a modified histidine residue incorporated into eukaryotic elongation factor 235. However, whereas Dph1, Dph3 or Dph4 deficiencies are embryonically lethal36,37, the 3i phenotype suggested an immune cell-specific, Dph6-dependent protein translation system of differential importance in different leukocyte subsets. This evokes growing evidence for immunoregulation by cell type-specific translation38.
In the third case, the broad phenotypic perturbation of several physiologic traits in Duoxa2–/– mice (Fig 6a) was unsurprising, since Duoxa2 contributes critically to iodine utilisation in thyroid hormone synthesis39. By contrast, the significant expansions of specific immune cell subsets, particularly CD4+ T cells, neutrophils and eosinophils, and the decreased representation of blood Ly6C–monocytes and KLRG1+ NK cells in Duoxa2–/– mice (Fig 6b) were unanticipated given little prior implication of the gene in immunobiology. Thus, 3i identified a potential immunoregulatory role of Duoxa2-dependent endocrine biology.
Finally, Bach2–/– mice illustrated how 3i could offer broader insights into a well-established, disease-related gene. BACH2 deficiencies have been associated with autoimmunity, inflammation, and allergy, commonly attributed to increased effector CD4+ and CD8+ T cells, as reported in Bach2–/– mice40–44. However, 3i also identified a greatly expanded range of innate and adaptive immune phenotypes of Bach2–/– mice (orange bars in Fig 6c), of which the most significant was a >10-fold increase in KLRG1+CD4–NK cells. Thus, some components of Bach2-dependent disease may reflect precocious innate immune cell activation. Moreover, Bach2–/– CD8+ T cells showed abnormally high cytolytic activity, and most Bach2–/– mice were high susceptibility to DSS-colitis (Fig 6d,e).
Genetic regulators of challenge responses
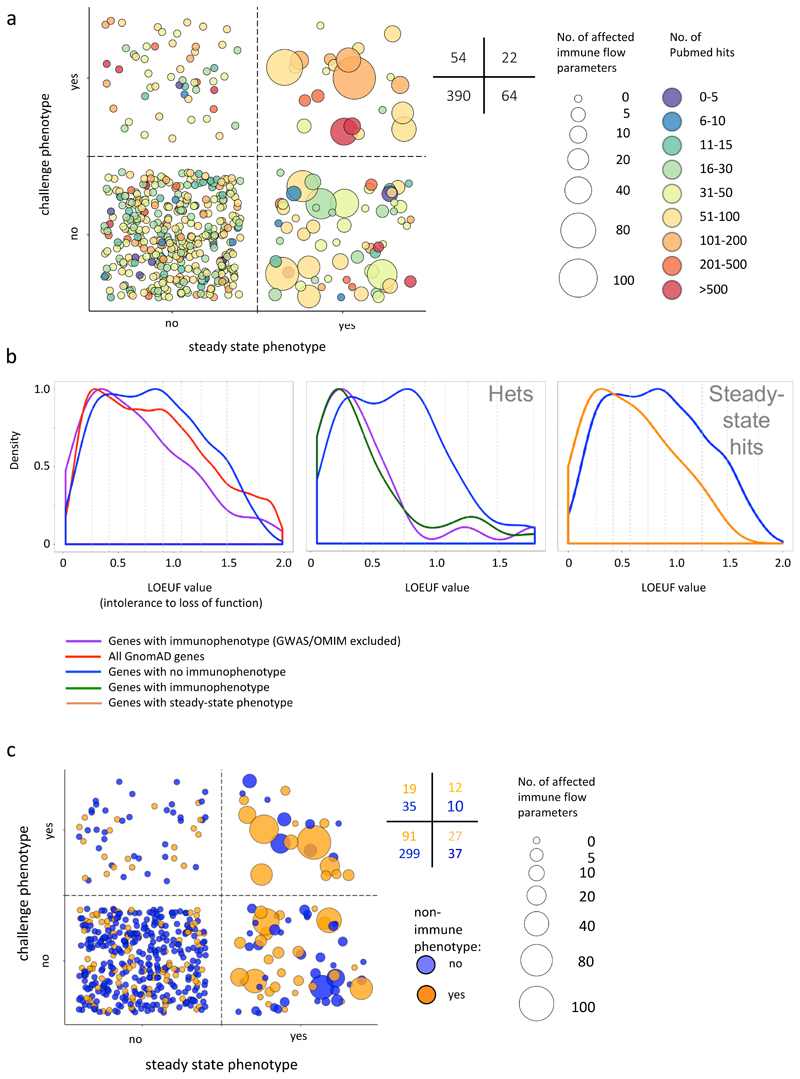
Of 86 phenodeviants affecting steady-state PB, SPL, MLN, BM, CTL, ANA or epidermal phenotypes, ~25% also affected responses to challenges, as judged against wild-type controls assayed contemporaneously (Fig 7a; Supplementary Fig 5b). Once more, those hits included several sparsely-studied genes (Fig 7a, green circles in upper-right quadrant). The fraction of phenodeviants affecting steady-state responses that also affected challenge responses might have been higher if 3i had employed additional challenges and/or assays of response. However, given the breadth of challenges utilized, and the biologically significant post-challenge outcome metrics, including survival and weight-loss, our findings most probably point to greater genetic redundancy in immune function than in baseline immunophenotypes. Indeed, genes affecting challenge outcomes were enriched in those perturbing more than one steady-state parameter (Fig 7a; larger circles are relatively enriched in upper-right vs lower-right quadrant). High functional redundancy might ensure that immunocompetence would be resilient to wide-ranging inter-individual variation in baseline immune cell subsets, as characterises humans1–3. Finally, some phenodeviants, including many sparsely-studied genes, affected functional responses but not steady-state immune parameters (Fig 7a, upper-left quadrant; Supplementary Fig 5b), possibly because they affect baseline immune parameter(s) not assayed by 3i and/or are expressed by non-immune cells which regulate challenge responses via non-immunological mechanisms such as barrier protection.
Figure 7. Genes affecting baseline immunophenotypes and challenge responses.
a. Effected parameters and Pubmed citations per gene. Each bubble represents one gene; size represents the number of immune flow phenotypes detected for that gene; colour represents the extent to which each gene has been reported on, determined by number of Pubmed citations, as of October 2018. Numbers denote the numbers of genes in quadrants of the figure.
b. Tolerance to LOF mutations in human orthologs of 3i genes. Left-hand panel: LOF tolerance scores derived from GnomAD for human orthologs of: 3i genes without hits (blue, n=334); 3i genes with hits OMIM/GWAS genes excluded (purple, n=77), and for all GnomAD genes (red, n=16037). A score of 0 denotes complete intolerance; a score of 2 complete tolerance to loss of function. Wilcoxon test: purple versus red p=0.010; purple versus blue p=0.009. Deciles of all genes marked with vertical grey broken lines. Middle panel: Genes tested in heterozygotes. 3i genes without hits (blue, n=109), 3i genes with hits (green, n=31), 3i genes with hits OMIM/GWAS genes excluded (purple, n=19). Wilcoxon test: purple versus blue p=0.0029; green versus blue p=0.003. Right-hand panel: Human orthologs of all 3i genes without hits (blue, n=334) versus genes with hits in steady state flow cytometry phenotypes but without challenge hits (orange, n=34); orange versus blue p=0.014, Wilcoxon test.
c. Relationship between immune phenotypes and non-immune phenotypes. Each bubble represents one gene; size represents the number of immune flow phenotypes detected for that gene; colour indicates non-immune phenotype. Genes with any immune hit and a non-immune phenotype (orange bubbles among bubbles in top and right bottom quadrants) versus not hit genes (orange bubbles among bubbles in left bottom quadrant); p=0.000072, Fisher exact test. Genes with baseline phenotype change and a non-immune phenotype (orange bubbles among bubbles in right bottom quadrant) versus not hit genes (orange bubbles among bubbles in left quadrants); p= p=0.00021, Fisher exact test.
Given that 57% of the 3i “hits” (80/140) had never hitherto been implicated in immunobiology, and that the others included many little-studied genes, it was striking that hits were collectively enriched in genes for which humans show poor tolerance to “loss-of-function” (LoF). For all human genes assessed by the Genome Aggregation Database (gnomAD) (n=16307), tolerance to LoF is illustrated by “LOEUF”: loss-of-function observed/expected upper bound fraction, for which 0 indicates no tolerance of loss-of-function and 2 indicates high tolerance to loss of function (Fig 7b; left panel, red line)45,46. We then compared this distribution with that for human orthologues of 3i hits, but first excluded genes with prior GWAS or OMIM classifications that might bias toward low LOEUF values. This notwithstanding, the residual 3i hits (n=77) (Fig 7b; left panel, purple line) showed a significantly different distribution, with conspicuous enrichment for genes with lower LOEUF. This outcome was even more striking when contrasted with human orthologues of 3i genes without immune phenotypes (n=334) which collectively showed no enrichment for LoF intolerance (Fig 7b; left panel, blue line).
It might be argued that immune hits were biased toward low LOEUF values by genes scored as heterozygotes because the strains were sub-viable. Indeed, LOEUF values were conspicuously low when only heterozygotes were scored for human orthologues of all 3i hits, or for 3i hits minus the OMIM/GWAS genes (as above) (Fig 7b, middle panel, green and purple lines, respectively). Nonetheless, those low-LOEUF-value distributions were significantly displaced from that of human orthologues of 3i heterozygotes that were not immune hits (Fig 7b; middle panel, blue line), strongly arguing that the low LOEUF scores for hits assessed as heterozygotes were not attributable solely to sub-viability but probably also to their immune phenodeviance.
Interestingly, enrichment for low LOEUF was also apparent for human orthologues of genes purely with steady-state flow cytometry phenotypes (p=0.014, Wilcoxon) (Fig 7b; right hand panel, orange versus blue lines). Seemingly consistent with this, the 140 immunological phenodeviants were strikingly enriched in genes that also had non-immunological phenotypes (58/140 [>41%]) relative to the genes that were not hits (91/390 [23%]), with almost identical enrichment seen for genes affecting only baseline immune variation (39/86 [>45%]) versus those that did not (110/444 [25%]) (Fig 7c).
Genetic regulation of correlation networks
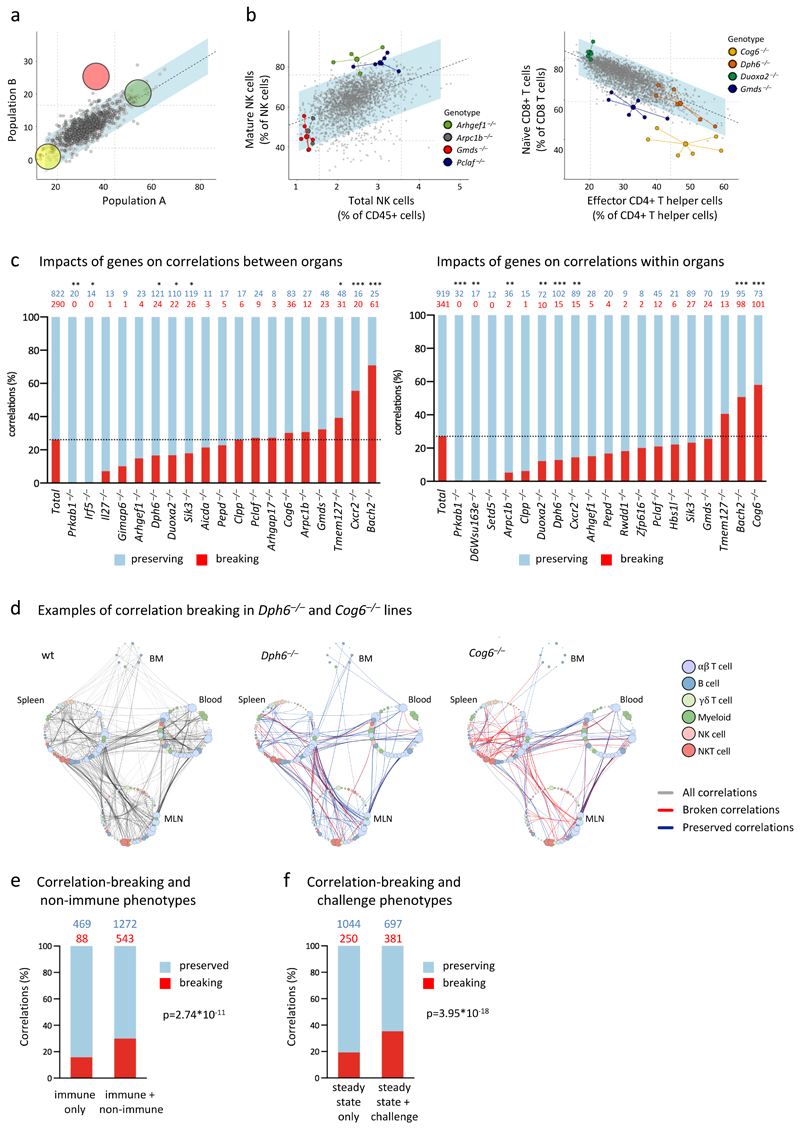
The 3i data-sets were next used to ask whether genes affecting one or more steady-state immune parameter(s) might segregate according to whether they also disrupted correlations connecting immune parameters: i.e. whether they perturbed immunological structure. Fig 8a (left panel) schematises hypothetical distribution clouds of data-points for three phenodeviants (yellow, pink, green) displaying extreme values for cell population B. Of note, whereas gene yellow and gene green maintained proportionality with population A, gene pink broke that correlation.
Figure 8. Phenodeviants preserve or break immunological structures.
a. Schematic illustrating the concept of structural perturbation. The yellow and green genes are theoretical hits in both correlated parameters (Pearson’s correlation), and exist as an exaggeration of the normal relationship that exists at steady state (grey dots represent wt mice). The pink gene is also a hit in both parameters, but breaks the correlation by falling outside the blue corridor that represents a 95% prediction confidence interval around the correlation line.
b. Examples of genes that preserve or disrupt immunological structure. Plotted are data frequencies of SPL subsets (indicated on x- and y-axes) as determined by flow cytometry. Data from mutant lines with phenotypes are highlighted in different colours; large dots represent the x/y centroid (mean) values, small dots represent data points for each mouse. Grey data points represent wt mice and mutant mice that are comparable to wt mice for both parameters shown. n=4 for Arhgef1–/–, n=5 for Arpc1b–/–, n=5 for Gmds–/–, n=4 for Pclaf–/–.
c. Genes differ in their capacity to break or preserve relationships. Stated in red font are the number of correlations that each cited gene disrupts and in blue font the residual number of correlations that each cited gene preserves, the total being all correlations contributed to by the steady-state parameters that the cited gene affects. These are represented as percentages in the left-hand graph (correlations across organs) and right-hand graph (correlations within the same organ). Only genes affecting parameters which contribute to >10 correlations are depicted. Statistical significance was determined by two-sided Fisher’s exact test in comparison to the data set average (dotted line). Number of correlations above bars, *p<0.05, **p<0.01, ***p<0.001.
d. Examples of genes that either preserve or disrupt many correlations. Plotted are parameters (colour-coded according to cell lineage) in BM, SPL, MLN, and PBL, and the correlations that link them in wt mice (left panel, grey lines) or in Dph6–/– mice (middle) or Cog6–/– mice (right), in which cases blue and red lines denote correlations that are preserved or broken, respectively.
e. Comparison of correlation breaking in genes that score in non-immune tests and immune tests versus genes that have hits only in immune parameters (Probit regression, p=2.74*10-11; p=7.45*10-7 when controlling for unequal numbers of correlations per gene; p=0.007 when allowing for any dependencies of correlations within a gene by using cluster-robust standard errors). Numbers of correlations above bars.
f. Comparison of correlation breaking in genes that score in challenge assays and steady-state immune tests versus genes that have hits only in steady-state immune parameters (Probit regression, p=3.95*10-18; p=9.10*10-16 when controlling for unequal numbers of correlations per gene; p=0.021 when allowing for any dependencies of correlations within a gene by using cluster-robust standard errors). Numbers of correlations above bars. Separate contributions of non-immune and challenge phenotypes: p= 1.56*10-6 for non-immune controlling for challenge; p=2.93*10-13 for challenge controlling for non-immune.
Real examples of this are shown in Fig 8b: Gmds and Arpc1b mutants displayed very few NK cells but maintained proportionality with mature NK cells, whereas most Arhgef1–/– and Pclaf1–/– mice sat outside the confidence limits of the relationship, displaying disproportionately high NK maturation. Likewise, Duxoa2–/– and Dph6–/– mice, whose effector CD4+ T cell numbers were atypically low and high, respectively, each preserved an inverse relationship of effector CD4+ Th cells and naïve CD8+ T cells, whereas this correlation was broken by Gmds and Cog6 deficiencies. Hence, correlations that confer structure on the immune system are differentially resilient to genetic perturbation.
Overall, we observed that genes with the potential to affect >10 correlates collectively preserved ~75% of intra-organ and inter-organ correlations (Fig 8c; left-hand column, ‘Total’). Thus, immunological structure was relatively resilient to genetic perturbation of discrete immune parameters. Nonetheless, mutations of some genes that affected larger numbers of immune parameters disrupted substantially more correlations, as was evident for Cog6 and Bach2 (Fig 8c). However, structural perturbation did not simply reflect the greater number of immunophenotypes affected by a gene, since Dph6–/– mice displayed a similar number of steady-state immunophenotypes (e.g. altered cell subsets) to Cog6–/– mice, but preserved most intra-organ and inter-organ correlations (Fig 8d; compare the density of red lines (broken correlations) versus blue lines (preserved correlations) in the spleens of the two strains). This argues that specific genes underpin the existence of certain correlations and hence shape immunological structure. Added to this, Arpc1b broke many inter-organ correlations while preserving most intra-organ correlations (Fig 8c), indicating that different genes regulate immune subset relationships in different ways.
Given that many immune parameters also correlated with specific physiologic measures, it was appropriate to ask whether genes with non-immunological phenotypes were more likely to disrupt immunological structure. Indeed, baseline immunological structure seemed much less resilient when discrete physiological traits were perturbed, with such genes collectively breaking significantly more correlations (30%) than did genes only affecting immune parameters (16%) (Fig 8e). Additionally, genes which affected challenge responses as well as the baseline immunophenotype broke significantly more correlations (35%) than genes affecting only the baseline immunophenotype (19%) (Fig 8f). By using cluster-robust standard errors, the results shown in Fig. 8e and 8f were robust vis-a-vis different genes contributing to different numbers of correlations, and correlations being underpinned by additional factors not tested for. Moreover, controlling for the influence of non-immune traits and challenge responses in the same regression analysis confirmed that non-immune and challenge phenotypes made separate contributions. Clearly, the association of defects in immunocompetence with compromises in baseline immunological structure makes a compelling case for assessing and better understanding immune subset correlations as a refined means for monitoring immune variation.
Discussion
3i has provided a community resource in the form of a broadly transferrable, high-throughput platform by which immunophenotypes can be measured in different settings; e.g. comparison of genotypic variants, or pre- and post-treatments. By combining genetic screening and immunophenotyping at scale, 3i has affirmed a substantial contribution of genetics to immunophenotypic variation, and has considerably expanded the number and diversity of genes known to have monogenic impacts on steady-state immune parameters and/or challenge responses. Those genes, most of which lacked prior implication in immunobiology, likely have disease-relevance given their wholly unanticipated enrichment in genes for which humans show poor tolerance to loss-of-function. Moreover, for well-studied genes such as Bach2, 3i identified hitherto unrealised endophenotypes that may contribute to disease mechanisms.
Of comparable value to 3i’s identification of immunoregulatory genes was the functional implication of pathways on which those genes lie. Complex, multigenic autoimmune pathologies such as systemic lupus erythematosus (SLE) and multiple sclerosis (MS) reflect combined dysfunctions in commonly unelucidated pathways. Germane to 3i, the recent demonstration that causative dysfunction in SLE and MS could be tracked via discrete endophenotyes15 emphasises the value of associating specific genes and pathways to specific endophenotypes. Likewise, relative proportions of murine splenic cell types have proved to be powerful indicators of autoimmune dysregulation47. Added to this, the web-based curation of 3i data-sets, comparing hundreds of single-gene ablations on the C57BL/6N background, one the most-widely used animal models, might help predict the outcomes of drug-mediated pathway inhibition18.
The scale, throughput, and robustness of 3i were also sufficient to provide insight into the nature and scope of immune variation in C57BL/6N mice. For example, whereas sexual dimorphism of immune parameters is well established48,49, 3i established that its impacts are highly selective. The analysis of immune variation at scale also revealed dense correlation networks of immune parameters with each other and with general physiologic traits. De facto, such correlations impose structures constraining immune variation. Although structure was more evident in males, it likely exists in females but in practical terms may be masked by higher variability in general physiology. This notwithstanding, correlations offer insights into physiologic and environmental factors, e.g. diet, that may regulate specific immunophenotypes, and may additionally provide practical surrogate measures of immune status.
While immune correlations and resulting structures will differ in detail across organs, strains and species, they provide a revised framework for viewing immune variation. For example, the co-ordination of immune cell subsets reflected by immunologic structure may confer on the immune system a critical agility to respond to diverse challenges. Thus, structure metrics (e.g. subset ratios versus consensus values) might usefully be included in immune-monitoring strategies. Supporting this, Bach2, Cog6 and Arpc1b, which were readily scored as disrupting correlations, are strongly associated with human disease40–42,50–52. Moreover, therapeutic responsiveness to checkpoint blockade was recently associated with a ratio of CD8+ T cell subtypes, rather than with individual subsets53. In parallel, the unanticipated density of correlation networks revealed by 3i makes the case for investigating their biological basis.
Materials and Methods
Contact for Reagent and Resource Sharing
All reagents used are listed in Supplementary Table 5. For additional information about reagents and resources, contact Adrian Hayday at adrian.hayday@kcl.ac.uk.
Experimental design
All assays relied on the fastidious application to cells and tissues of intensively piloted, robust, optimised Standard Operating Procedures (SOPs) employing high-resolution, quantitative protocols, whose high reproducibility was monitored over time (e.g. Supplementary Fig 1f).
Mice were randomly allocated to the experimental groups (wt versus ko) by Mendelian inheritance. The experimental unit in the study was the individual mouse. For the majority of tests, operators were blinded with regard to the genetic identities of mice. Further detailed experimental design information is captured by a standardized ontology as described19, and is available from the IMPC portal (www.mousephenotype.org). The steady-state screens integrated within the HTS followed a multi-batch design, in which a baseline set of control data was constantly collected by phenotyping wt mice of both sexes along with mutant mice at least once per week. As soon as mutant mice from the breeding colonies reached appropriate ages, they were issued to the pipeline until sufficient number of males and females of each genotype were assessed. In this way, animals from each mutant strain were tested on two to five days, interspersed throughout experiment duration, rather than within one batch. For advantages and robustness of such design in an HTS see 21,54–56. The challenge screens implemented a parallel group study design.
Compliant with HTS practice, the numbers of mice examined in each assay were dictated by costs, logistics, and power, with bespoke statistical tests (see below) applied to identify genes (so-called “hits”) perturbing defined components of the immune system. Two to seven homozygote mice of each sex per mutant line were assessed per test. If no homozygotes were obtained from >28 offspring of heterozygote intercrosses, the line was deemed homozygous lethal. Similarly, if <13% of the pups resulting from intercrossing were homozygous, the line was judged as being homozygous subviable. In either case, heterozygotes were phenotyped. The numbers and sex of animals tested per genotype and assay are summarized in Supplementary Table 3.
Ethical compliance
Mouse use in this study was justified based on their facilitating a large variety of phenotypic tests to be carried out on a sufficient number of individuals in a controlled environment. The care and use of mice in the study was conducted in accordance with UK Home Office regulations, UK Animals (Scientific Procedures) Act of 2013 under two UK Home Office licenses which approved this work (80/2076 and 80/2485) which were reviewed regularly by the WTSI Animal Welfare and Ethical Review Board.
All efforts were made to minimize suffering by considerate housing and husbandry, the details of which are available at the IMPC portal: http://www.mousephenotype.org/about-impc/arrive-guidelines. Animal welfare was assessed routinely for all mice involved. Adult mice were killed by terminal anaesthesia followed by exsanguination, and death was confirmed by either cervical dislocation or cessation of circulation.
Animals
Mice were maintained in a specific pathogen-free unit under a 12-hour light, 12-hour dark cycle with water and food ad libitum (Mouse Breeders Diet (LabDiets 5021-3, IPS, Richmond, USA), unless stated otherwise. Mice where housed in Tecniplast Sealsafe 1284L (overall dimensions of caging (L × W × H): 365 × 207 × 140 mm; floor area = 530 cm2) at a density of 3-5 animals per cage, and provided with a sterilized aspen bedding substrate and cardboard tubes and nestlets for environmental enrichment.
Mutant mouse production
Mice carrying knockout first conditional-ready alleles were generated on a C57BL/6N background using the EUCOMM/KOMP Embryonic Stem (ES) cell resource, with ES quality control and molecular characterization of mutant mouse strains performed as described previously 57. Upon completion of phenotyping, genotyping was repeated and data were only accepted from mice for which the second genotype was concordant with the original. The knock-in first strategy first generates mice that still possess the full sequence of the targeted gene, interrupted in a crucial exon by the inserted cassette. These tm1a alleles result in functional knockout lines in most cases, but can carry residual expression of the targeted gene. These tm1a alleles can be converted into unequivocal full knockout tm1b alleles by excising the inserted cassette with the targeted essential exon. Details of alleles used can be found in Skarnes et al19. All lines are available from www.knockoutmouse.org or mouseinterest@sanger.ac.uk.
Non-immune phenotyping
Non-immune phenotyping (summarized in Supplementary Fig 1a) was conducted as described 21. Screening for phenotypes in Citrobacter infection was replaced by the 3i challenges.
Immunological steady-state phenotyping
Tests conducted in the steady-state (PBL, SPL, MLN, BM, ear epidermis, antinuclear antibodies, cytotoxic T lymphocytes function) were conducted on the same 16-week-old mice that were subject to broad non-immunological phenotyping procedures21. Non-fasted mice were terminally anaesthetised using Ketamine (100 mg/kg)/Xylazine (10 mg/kg) injection. Organs were harvested and either analysed directly (PBL) or shipped in HBSS at 4°C for analysis on the same day off-site. Readouts of the respective tests and numbers of mice used are summarized in Supplementary Fig 1 and Supplementary Table 3.
Single cell preparation of immune cells from spleen
After removing the fat, spleens were transferred into Miltenyi C-tubes with 3 ml of enzyme buffer (PBS Ca2+/Mg2+, 2% FCS (v/v), 10 mM HEPES, Collagenase (1 mg/ml), and DNAse (0.1 mg/ml). Samples were then processed using a Miltenyi Gentle MACS dissociator (SPL program 01) and incubated at 37°C for 30 minutes. After the incubation, samples were processed again in the Miltenyi Gentle MACS dissociator (SPL program 02) and the enzyme reaction was stopped adding 300 μl of stop buffer (PBS, 0.1 M EDTA). Samples were filtered through 30 μm filters and centrifuged for 5 minutes at 400 x g at 8°C. The pellet was resuspended in FACS buffer (PBS, Ca2-/Mg2-, 0.5% FCS, EDTA 2mM), incubated for 60 seconds in RBC lysis buffer and then washed with FACS buffer. Samples were transferred to 96-well V bottom plates and incubated in 50 μl of RBC lysis buffer (eBioscience) for 1 minute prior to antibody staining.
Single cell preparation of immune cells from mesenteric lymph nodes (MLN)
After removing the fat, MLN were transferred into 1.7 ml microfuge tubes with 200 μl of buffer (PBS Ca2+/Mg2+, 2% FCS (v/v), 10 mM HEPES) and ruptured using small plastic pestles. 400 μl enzyme buffer (PBS Ca2+/Mg2+, 2% FCS (v/v), 10 mM HEPES), Collagenase (1 mg/ml), and DNAse (0.1 mg/ml) were and samples were incubated for 15 minutes at 37°C. The reaction was stopped by adding 60 μl stop buffer (PBS 0.1 M EDTA) and samples filtered through 50 μm filters and centrifuged for 5 minutes at 400 x g at 8°C. The cell pellet was resuspended in FACS buffer (PBS Ca2-/Mg2-, 0.5% FCS, EDTA 2 mM) and transferred to 96-well V bottom plates for antibody staining.
Single cell preparation of immune cells from bone marrow (BM)
The tibia was cleared of muscle tissue and cut below the knee and above the ankle. The open bone was placed into a cut pipette tip placed into a microfuge tube, thereby keeping the bone away from the bottom of the tube and allowing the bone marrow to be centrifuged out of the bone at 1000 × g for 1 minute. The bone was discarded and the pellet resuspended in 50 μl RBC lysis buffer at room temperature for 1 minute. Cells were washed in 200 μl FACS buffer and again centrifuged at 1000 × g for 1 minute. Cells were resuspended in 400 μl FACS buffer and transferred to 96-well V bottom plates for antibody staining.
Blood preparation
Blood was collected into EDTA coated tubes (peripheral blood leukocytes assay) via the retro-orbital sinus. Whole blood for peripheral blood leukocyte assays was stained with two titrated cocktails of antibodies (Supplementary Table 1). Using the white blood cell count obtained from the haematological analysis, absolute cell counts were derived for each population and reported as cells/μl.
Immunophenotyping by flow cytometry
Single cell suspensions were incubated with Fc-block for 10 minutes at room temperature, washed four times, first with FACS buffer and then with PBS, and then incubated with live/dead ZiR dye (BioLegend) for 10 minutes at room temperature. Samples were washed again with FACS buffer and incubated with antibody cocktails (see Supplementary Table 1) at 4°C for 20 minutes. Samples were washed twice with FACS buffer and measured on a BD Fortessa X-20 equipped with 405 nm, 488 nm 561 nm and 644 nm lasers (see Supplementary Table 1). Full details of instrument setup are available at www.immunophenotyping.org. Panels were modified slightly in summer 2014 in order to better correspond to the IMPC panels (T cell panel: 9th June 2014, B cell panel: 15th September 2014). Data before and after this data split were analysed separately. Data were analysed with FACSDiva and Flowjo software. FCS files are available from flowrepository.org.
Flow cytometry quantification (SPL, MLN and BM)
Additionally to the manual gating performed at the time of data acquisition, collected flow cytometry data for SPL, MLN and BM were gated computationally using flowClean, UFO, and flowDensity22,58. FlowClean was used to perform acquisition-based quality checking to remove anomalous events. Files with fewer than 20,000 events were then removed from further analysis. UFO was used to identify outlier samples (e.g., batch effects). FlowDensity was used to enumerate cell populations by automating a predefined gating approach using sequential, supervised bivariate gating analysis to set the best cut-off for an individual marker using characteristics of the density distribution. The parameters for each individual cell population were pre-defined once for all files. The automated analysis data was validated against matched manually analysed data. Gating strategies are outlined in Supplementary Fig 1b and in Supplementary Fig 6). Assessment of absolute cell counts were not compatible with the high-throughput workflow employed in the study for SPL, MLN and BM. All cell subset frequencies are presented as a percentages of a relevant parent populations.
Ear epidermis immunophenotyping
Epidermal sheets from mouse ears were treated with hair removal cream (Nair) for 4 minutes at room temperature. After removing the cream by extensive washing in PBS, ears were split into dorsal and ventral sides and incubated dermal side down for 35 minutes at 37°C in 0.5 M ammonium thiocyanate (Sigma-Aldrich). Epidermal sheets were gently peeled from the dermis in PBS, and fixed in cold acetone for 20 minutes at -20°C. After washing in PBS, epidermal sheets were incubated for 1 hour at room temperature with 5% (wt/vol) FCS in PBS and were stained for 1 hour at 37°C with Vγ5 TCR-FITC (clone 536; BD), MHCII-AF647 (I-A/IE; BioLegend) and CD45-eFluor450 (eBioscience). Epidermal sheets were washed extensively in PBS and mounted on slides with ProLong Gold antifade medium (Life Signalling Technologies). Leica SP2 or SP5 confocal microscopes equipped with 40 x 1.25 NA oil immersion lens and 405 nm, 488 nm and 633 nm lasers were used to record 1024 x 1024 pixel confocal z-stacks with Leica Acquisition Software. The confocal records were processed and quantified with Definiens Developer XDR software using a custom-made automated protocol where images were smoothed with a sliding window Gaussian pixel filter, segmented by an automated Otsu’s method and then filtered based on object size and morphology parameters to detect cells in each fluorescence channel. Further, in order to quantify the number and length of dendrites, the detected cells were skeletonised in 3D to determine the points where dendrites start to branch out of the cell body. Mean of 3 vision fields was used for quantification.
Antinuclear Antibody Immunophenotyping
Murine serum samples were obtained and stored at -20°C prior to analysis (after dilution 1:100 in PBS) by incubating on commercially-sourced substrate slides (A. Menarini Diagnostics Ltd.) coated with HEp-2 cells for 30 minutes at room temperature in a humidifying tray. Samples were removed and slides were washed twice with PBS for 5 minutes and once with water for 5 seconds. Slides were incubated with FITC-conjugated goat anti-mouse IgG, diluted 1:500 and incubated for 20 minutes at room temperature in a humidifying tray in the dark. The secondary antibody was removed and slides washed twice with PBS for 5 minutes and once with water for 5 seconds, both in the dark. Slides were mounted in medium and stored at 4°C prior to imaging for 400 ms at 20 x magnification in the GFP channel on a Nikon wide-field TE2000U Microscope or a Deltavision Elite widefield system based on an Olympus microscope. Images were subject to multi-parametric analysis in Fiji. Samples were scored from 0-4 according to intensity based on control samples and commercially sourced FITC QC beads. Samples scored ≥2 were marked as ANA positive. All sera flagged by automated image analysis as putative positives vis-à-vis a contemporaneous standard were manually cross-checked for bona fide nuclear localization before scoring.
Cytotoxic T Lymphocyte Immunophenotyping
Mouse splenocytes were isolated using 70 μM cell strainers (BD Plastipak) and cultured in T cell media (TCM: 500 ml RPMI, 500 μl B-ME, 5 ml NaPyr, 5 ml pen/strep, 5 ml L-glut, 50 ml 10% heat inactivated FCS and 100 μl IL-2) on 6-well plates pre-coated with 0.5 μg/ml anti-CD3ε antibody and 1 μg/ml anti-CD28 antibody (1.7×106cells/well) for 48 hours. Plates were washed and cells where cultured for a further 8 days with daily passage prior CTL assay.
Cytotoxicity assays were performed using a CytoTox96 Non-Radioactive Cytotoxicity Assay kit (Promega UK Ltd). Cells were washed and re-suspended in killing assay media (KAM: 500 ml RPMI-phenol red, 10 ml 10% heat inactivated FCS and 5ml pen/strep), with CTLs at a concentration of 0.1 × 106 and P815 target cells at a concentration of 0.1 × 106. Purified hamster anti-mouse CD3ε antibody was added to P815 target cells.
P815 cells were added to a serial dilution of CTL samples and incubated for 3 hours. Lysis buffer (Promega UK Ltd.) was added to control samples and incubated for 45 minutes. Supernatants were harvested and substrate mix (Promega UK Ltd) was added prior to a 30-minute incubation in the dark. Stop solution (Promega Corporation UK Ltd) was added to halt the reaction and results acquired using a spectrophotometer (VersaMax, molecular devices).
Flow cytometric analysis was performed to assess the percentage of CD4+ and CD8+ cells within the cell culture. The cell suspension was washed in FACS buffer and the cell pellet re-suspended in a staining master mix (FACS buffer solution + 1:200 anti-CD8α APC and 1:200 anti-CD4 PE). Tubes were then incubated in the dark for 7 minutes at room temperature before the antibody was washed off and cells resuspended in FACS buffer. Results were acquired on a FACS Calibur machine and analysed using FlowJo 10 software.
Challenge screens
Challenge screens were conducted on separate cohorts of mice from the same breeding colony used for the steady-state screens.
DSS colitis challenge
Colitis was induced by adding 1.5% (w/v) DSS (Affymetrix, Inc.) to drinking water for 7 days, followed by 3 days with regular drinking water, in animals aged between 5 and 18 weeks (mean age 9 weeks). Mice were weighed every day and culled if weight loss reached 20% of starting weight.
For histological assessment of intestinal inflammation, mice were sacrificed at day 10 by cervical dislocation, and samples from mid and distal colon taken. Tissue sections were fixed in buffered 10% formalin; paraffin-embedded; cut; and stained with haematoxylin and eosin. Colon histopathology was blind-graded semi-quantitatively on a scale from zero to three, for four criteria: (1) degree of epithelial hyperplasia/damage and goblet cell depletion, (2) leukocyte infiltration in lamina propria, (3) area of tissue affected, and (4) presence of markers of severe inflammation, including crypt abscesses, submucosal inflammation, and oedema. Scores for individual criteria were added for an overall inflammation score of between zero and twelve for each sample. Scores from mid and distal colon were then averaged to obtain inflammation scores for each mouse colon.
Salmonella typhimurium challenge
Groups of 8 mutant and 8 C57BL/6N wild type mice were challenged intravenously with 5 x 105 colony forming units (cfu) Salmonella typhimurium M525 :: TetC, (Fragment C of tetanus toxin, to act as an antigen for subsequent antibody quantification), and followed for 28 days. On day 14 post-infection (pi), four mutant and four wt mice were sacrificed by cervical dislocation and organs (spleen, liver and caecum) removed. A small piece of spleen and liver was fixed in 4% formalin and then later processed to paraffin blocks as an infected tissue biobank. The rest of the organs were weighed then homogenized, serially diluted and plated to determine viable bacterial load. At day 28 pi, the remaining four mice were culled by cardiac puncture under terminal anesthesia and organs removed and processed, as above. The blood was allowed to clot, then centrifuged, serum collected and used to detect TetC antigen specific antibodies by enzyme-linked immunosorbent assay (ELISA). Mice were weighed and monitored daily for signs of pathophysiology.
Influenza challenge
Mutant and wt mice (5-21 weeks of age) were lightly anesthetised and intranasally inoculated with 171 or 227 p.f.u. of A/X-31 (H3N2) influenza in 50 μl of sterile PBS. Mouse weight was recorded daily and the percent reduction was calculated from their weight on day 0. Mice were sacrificed by cervical dislocation on day 10 pi, and the area under the curve from day 0 to 9 pi was calculated. Mice exceeding 25% total weight loss were culled in accordance with UK Home Office guidelines.
Trichuris muris challenge
The Edinburgh (E) strain of Trichuris muris was used in all experiments. Female mice (6-12 weeks old) were orally infected with 400 embryonated eggs. Mice were culled by cervical dislocation at day 32 pi, blood was collected by cardiac puncture for serum recovery, and caecum/proximal colon was dissected to inspect for worm presence by stereomicroscope. Levels of parasite-specific serum IgG1 and IgG2a Ab were by ELISA: briefly, ELISA plates were coated with T. muris excretory/secretory (E/S) antigen at 5 μg/ml. Serum was diluted 1/40, and parasite-specific IgG1, IgG2a and IgE detected with biotinylated anti-mouse IgG1 (Biorad), biotinylated anti-mouse IgG2a (BD PharMingen), and anti-mouse IgE (BioLegend), respectively. To generate T. muris E/S antigen, live adult worms were incubated at 37°C for 24 hours in RPMI-1640 medium (Gibco, UK) supplemented with 500 U/ml penicillin, 500 μg/ml streptomycin and 2 mM L-glutamine (all Gibco, UK). Supernatants were removed, centrifuged at 2000 x g for 15 minutes, filtered through a 0.22 μm filter (Millipore, UK), concentrated using a 10 kD molecular weight cut off Centriprep concentrator (Amicon, UK) and dialysed against PBS over a 24-hour period. The supernatant was subsequently filtered again and protein concentration determined before use.
Statistical analysis
Sample sizes for each experimental group are included in the figure legends and Supplementary Table 3. Measures were taken from distinct animals in almost all experiments. Exceptions are body weight, where the weight of the same animal was recorded repeatedly during the course of the experiment and the CTL assay, in which T cells from the same culture were used for different effector : target ratios. Tests for all assays are listed in Supplementary Table 3. Reference-range tests which require no assumptions were used for most assays. Adjustments for multiple comparisons were not made unless stated in test description, as individual comparisons were not independent (e.g. percentages for different cell types determined by flow cytometry are often nested and dependent on each other). Low false positive rates were instead ensured by using conservative reference range cut-offs and monitoring false positive rates by simulation (Supplementary Fig 4). All tests used were two-sided. Co-variates were included in the analysis indirectly, by dividing animals into sex-specific groups or matching ko and wt by assay date/weight/age (MMR analysis described below) and in the analysis presented in figures 8e and 8f where covariates tested are described in the text. A description of statistical parameters is included into the figure legends. Effect sizes are given when fold change between sexes was analysed. All significant calls and borderline candidates were manually reviewed by biological experts.
Experimental batch effects were minimised by the experimental design, the statistical analysis and by expert review. Experimental set-up: Mice were phenotyped on at least two different days and typically three to four different days. Statistical analysis: Stringent reference range hit calling required >60% of samples to be in the upper 2.5% or lower 2.5% of the wt distribution. The influence of temporal drift in flowcytometric data was minimized by using as controls only the 70 wt mice that were closest in time for any given parameter. Reference ranges have been shown by the WTSI team to be stable for >60 mice. Expert review: Experts reviewed all hits and borderline candidates and excluded candidates that suffered from batch effects despite the measures taken above.
Coefficient of variation (CV) was estimated as a ratio of the standard deviation of the wt population to its mean (for both sexes together, unless indicated otherwise). Note that since a data-set is expressed as frequency of parent, it is bound and CV estimates are less accurate close to the boundary.
Sexual dimorphism
Significance of sex as a source of variation in wt animals was tested in a mixed model (sex as explanatory variable, assay date as a random effect) by examining the contribution of sex to the model and whether the variance was homogeneous between sexes59. The effect of multiple testing was managed with Bonferroni correction to control the family-wise error rate to 5%. For Principal Component Analysis and Linear discriminant analysis (LDA), only wt samples with a complete set of flow parameters were used. Data was scaled. Accuracy of classification by sex in LDA was checked with leave-one-out cross-validation.
Estimation of false positive rates
A mutant mouse line was mimicked by randomly selecting N wt animals (N depending on the screen) and assessed whether it would be called a hit using the RR approach. To reflect the data structure and address the potentially confounding batch effect of test days, the number of experimental days from which animals were drawn and their sex was set according to the distributions observed across all tested mutant lines. For example, in the lab, bone marrow was collected on five different days in 36% of mouse strains; in 38% on four different dates; in 11% from three different days, etc. When strains were tested on four assay days, in 78% of cases females were tested on two days, etc. The same data structure was imposed on the draws for the false positive rates. Thereupon, the same rubric used for calling hits from real data was applied, except that the expert data review step (see above) was replaced by filtering out samples from the days when the median of wt animals and non-significant mutant strain samples was further than two median absolute deviations (MADs) from the overall median in wt animals. 10,000 draws were performed for each parameter to determine the rate at which false positives would occur.
Correlation analysis
Pearson correlations between parameters in the flow cytometry screens as well as between the flow cytometry screens and non-immune screens were identified in wt mice for pairwise complete observations, accumulated throughout whole duration of the experiment. Correlations with parent and sister populations were excluded from the dataset. Correlations with R>0.33 and p<0.0001 were considered significant.
Comparison of correlations between male and female mice
R values of all significant correlations were compared with a t-test after Fisher z-transformation. Difference in dependence between parameters between sexes (slope of regression line) was tested by fitting a linear regression model to correlated parameters and assessing significance of interaction between sex and predictor variable. P-values were adjusted for multiple testing with Bonferroni correction to control the family wise error rate to 5%.
Immunological structure analysis
Starting from parameters that were affected in a mutant strain, a list was compiled of other parameters with which the affected parameter was ordinarily correlated. The wt C57BL/6N data were then used to predict a value for the correlated parameter in the mutant strain by sex-specific linear regression. If ≥60% of mice for a given mutant strain had predicted values outside the 95% confidence interval based on the wt distribution, the mutant line was defined as breaking this specific parameter correlation. Note that a single perturbed parameter could contribute more than one correlation to the dataset. For the purpose of this analysis, it was assumed that all correlations were equally likely to be broken. To assess if breaking of correlations was correlated with other characteristics, such as a non-immune phenotype or a challenge phenotype, a multivariate Probit regression analysis was undertaken. The same analysis was also used to control for unequal numbers of correlations per gene, allowing for dependencies of correlations within a gene by cluster-robust standard errors and for separate contributions of all characteristics tested. The analysis was carried out using R, RStudio and R packages ggplot2, data.table, dplyr and igraph, org.Mm.eg.db and org.Hs.eg.db.
GnomAD, OMIM and GWAS analyses
If homo-and heterozygotes were analysed from the same strain, only homozygotes were included in this analysis. Human orthologues were based on JAX definition (http://www.informatics.jax.org, accessed 05.05.2016) and only 1:1 human-mouse orthologues were used. GnomAD scores were extracted from release 2 (gs://gnomad-public/release/2.1/ht/constraint/constraint.ht)46.
OMIM annotations were obtained on 31 May 2017, with immune-related OMIM-listed genes considered as those implicated in phenotypes of immunodeficiency, recurrent infections, autoimmunity.
GWAS associations were obtained from the full NHGRI-EBI GWAS catalog database 78 on September 10, 2017 (file gwas_catalog_v1.0.1-associations_e89_r2017-07-31.tsv). In GWAS annotation, "immune-related" genes were considered as those mapping to susceptibility to autoimmune disease (systemic lupus erythematosus, psoriasis, rheumatoid arthritis, Sjögren syndrome, primary biliary cholangitis, ulcerative colitis, inflammatory bowel disease, Crohn's disease, multiple sclerosis, type 1 diabetes, Graves' disease, late-onset myasthenia gravis); immune response to virus (measured by secreted TNF-alpha); and functional units of gut microbiota.
Gene overrepresentation analysis
Enrichment analysis was performed with PANTHER Overrepresentation Test with the tool version 14.1 (Released 2019-03-12, www.pantherdb.org)60. We calculated significance of over- and underrepresentation of GO-slim Biological Process categories among 466/530 genes with available GO annotation, by Fisher's Exact test and with FDR multiple testing correction. Only categories with more than one gene expected in our dataset and less than 1000 genes in the mouse genome were included.
Supplementary Material
Acknowledgements
We thank many colleagues for advice, particularly Drs S. Heck of the Biomedical Research Centre of Guy’s and St Thomas’ Hospital and King’s College London; D. Davies of the Francis Crick Institute; F. Geissmann, J. Strid and O. Sobolev in the early stages of programme planning; and N. Karamanis and K. Hawkins for UX testing of the website. The project was funded by Wellcome Trust grants 100156/Z/12/Z. M.D-C received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 656347. K.O.B., J.W. and T.M., were supported by the NIH Common Fund [UM1 HG006370]. The development of automated flow analysis in R.B.’s group was supported by Genome BC (SOF152), NSERC, International Society for Advancement of Cytometry Genome Canada, Genome BC (252FLO), the National Institute of general medical sciences (R01GM118417) and CIHR. G.D. was supported by an NIHR grant to the Cambridge Biomedical Centre. W.J. was supported by NIH grant AI026170. C.M.L. is supported by a Wellcome Senior Fellowship in Basic Biomedical Science (107059/Z/15/Z). R.J.C. is a principal investigator of the MRC Human Immunology Unit. K.J.M. is funded by a Wellcome Trust Investigator Award (102972/Z/13/Z). R.K.G. holds a Wellcome Trust Investigator Award Z10661/Z/18/Z and is part of the Wellcome Trust Centre for Cell Matrix Research funded by award Z03128/Z/16/Z. In addition, the project was supported by grants and facilities provided to A.C.H. by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001093), the UK Medical Research Council (FC001093), and the Wellcome Trust (FC001093).
Footnotes
Data and Code Availability
The flow cytometry files that support the findings of this study are available from www.flowrepository.org with the identifiers listed below:
3i PBMC panel 1: FR-FCM-ZYPJ
3i PBMC panel 2: FR-FCM-ZYPK
3i T-cell Spleen IMPC: FR-FCM-ZYX9
3i T-cell MLN IMPC: FR-FCM-ZYXB
3i B-cell Spleen IMPC: FR-FCM-ZYXC
3i B-cell MLN IMPC: FR-FCM-ZYXE
3i M-cell Spleen IMPC: FR-FCM-ZYXF
3i M-cell MLN IMPC: FR-FCM-ZYXG
3i P2 SPL IMPC: FR-FCM-ZYXN
3i BM IMPC: FR-FCM-ZYXQ
Vignettes showing gating of affected cell populations in KO mice and controls are available for all phenotypes deemed significant in this study in flow cytometry assays through www.mousephenotype.org (access through Associated Images on a gene page).
Microscopy image files from ear epidermis assay, ANA assay and DSS histology are available from the corresponding author upon request and are submitted to www.mousephenotype.org (access through Associated Images on a gene page).
All assay results on mouse and strain level that support the findings of this study are available through http://www.immunophenotype.org (website entirely devoted to this project) and www.mousephenotype.org (access via gene name).
Figures with associated raw data are:
1bcd, 2AB,3ab, 5bc, 6bcde, 7abc, 8cef,S1eghi, S2, S3ab, S4abcd. Supporting data is in Source data files and also available from https://github.com/AnnaLorenc/3i_heatmapping.
Code used for initial hit calling and preprocessed per-mouse data for flow cytometry, ear epidermis and DSS assays are available from https://github.com/AnnaLorenc/3i_heatmapping.
The PhenStat R package used for Influenza analysis is available on Bioconductor (www.bioconductor.org). The ImageJ macro and the Python code used to score ANA positivity, the Definiens Developer code to assess the ear epidermis images and the R code used to assess the false positive rate are available on request.
Material Availability
All mouse lines analysed in this work are available from repositories linked to the International Mouse Phenotyping Consortium (www.mousephenotype.org) or from WTSI (mouseinterest@sanger.ac.uk). Cell lines are available upon request.
Author contributions (in order of appearance as authors):
Conceptualisation: J.K.W., F.P., G.D., W.J., C.M.L., R.J.C., K.J.M., G.M.G., R.K.G., D.J.A., A.C.H. Methodology: L.A.-D., A.G.L., A.L., D.S.U., S.C., A.O.S., N.Sa., M.D.-C., K.R.B., B.M., J.I. P.B., K.I.H., E.C., S.F., T.L.C., B.W., A.R., S.D., J.M., A.Y., M.L., G.X.S.-Z., A.C., R.B., G.D., W.J., C.M.L., R.J.C., K.J.M., G.M.G., R.K.G., D.J.A., A.C.H. Software: A.L., M.G., N.R., N.A.K., D.M., A.R., S.D., J.M., A.Y., M.L., J.A., R.B. Website: L.A.-D., A.L., K.O.B., J.W., J.M., A.T.M., T.M. Validation: L.A.-D., A.G.L., A.L., D.S.U., S.C., A.O.S., N.Sa., M.D., K.R.B., B.M., J.I., P.B., E.C. Formal analysis: L.A.-D., A.G.L., A.L., D.S.U., S.C., A.O.S., N.Sa., M.D.-C., K.R.B., B.M., J.I., P.B., E.C., A.C.H. Investigation: L.A.-D., A.G.L., D.S.U., S.C., A.O.S., N.Sa., M.D.-C., K.R.B., B.M., J.I., P.B., K.I.H., E.C., S.F., T.L.C., H.W., L.K., K.H., C.B., G.N., E.C., B.W., G.X.S.-Z., A.C., C.B.R., A.P-K., M.E., N.St., M.P. Data curation: L.A.-D, A.L. Writing – original draft: A.C.H. Writing – Review and editing: L.A.-D., A.G.L., A.L., D.S.U., A.O.S., N.Sa, N.A.K., J.A., G.D., R.J.C., G.K.G., R.K.G., D.J.A., A.C.H. Vizualisation: L.A.-D., A.G.L., A.L., A.C.H. Supervision: R.B., G.D., W.J., C.M.L., R.J.C., K.J.M., G.M.G., R.K.G., D.J.A., A.C.H. Project Administration: L.A.-D., J.K.W., S.C., A.O.S., R.R-S., A.C.H. Funding Acquisition: R.B., F.P., G.D., W.J., C.M.L., R.J.C., K.J.M., G.M.G., R.K.G., D.J.A., A.C.H.
References
- 1.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobolev O, et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat Immunol. 2016;17:204–213. doi: 10.1038/ni.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsang JS, et al. Global Analyses of Human Immune Variation Reveal Baseline Predictors of Postvaccination Responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei SC, et al. Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell. 2017:1–32. doi: 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr EJ, et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patin E, et al. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat Immunol. 2018;19:302–314. doi: 10.1038/s41590-018-0049-7. [DOI] [PubMed] [Google Scholar]
- 8.Alpert A, et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat Med. 2019:1–34. doi: 10.1038/s41591-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinghaus D, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orrù V, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelms KA, Goodnow CC. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity. 2001;15:409–418. doi: 10.1016/s1074-7613(01)00199-6. [DOI] [PubMed] [Google Scholar]
- 14.Wang T, et al. Real-time resolution of point mutations that cause phenovariance in mice. Proc Natl Acad Sci USA. 2015;112:E440–E449. doi: 10.1073/pnas.1423216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steri M, et al. Overexpression of the Cytokine BAFF and Autoimmunity Risk. N Engl J Med. 2017;376:1615–1626. doi: 10.1056/NEJMoa1610528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S-Y, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 17.Turnbull C, et al. The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687. doi: 10.1136/bmj.k1687. [DOI] [PubMed] [Google Scholar]
- 18.Zambrowicz BP, Sands AT, et al. Knockouts model the 100 best-selling drugs—will they model the next 100? Nat Rev Drug Discov. 2003;2:38–51. doi: 10.1038/nrd987. [DOI] [PubMed] [Google Scholar]
- 19.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saleheen D, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White JK, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahim A, et al. High throughput automated analysis of big flow cytometry data. Methods. 2018;134–135:164–176. doi: 10.1016/j.ymeth.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper GS, Stroehla BC. The epidemiology of autoimmune diseases. Autoimmunity Reviews. 2003;2:119–125. doi: 10.1016/s1568-9972(03)00006-5. [DOI] [PubMed] [Google Scholar]
- 24.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nature Publishing Group. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X, et al. Circuit Design Features of a Stable Two-Cell System. Cell. 2018;172:744–747.e17. doi: 10.1016/j.cell.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byers SL, Wiles MV, Dunn SL, Taft RA. Mouse estrous cycle identification tool and images. PLoS ONE. 2012;7:e35538. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang SC-C, et al. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity. 2016;45:817–830. doi: 10.1016/j.immuni.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolev M, et al. Complement Regulates Nutrient Influx and Metabolic Reprogramming during Th1 Cell Responses. Immunity. 2015;42:1033–1047. doi: 10.1016/j.immuni.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salvagno GL, Sanchis-Gomar F, Picanza A, Lippi G. Red blood cell distribution width: A simple parameter with multiple clinical applications. Crit Rev Clin Lab Sci. 2015;52:86–105. doi: 10.3109/10408363.2014.992064. [DOI] [PubMed] [Google Scholar]
- 30.Olumuyiwa-Akeredolu O-OO, Pretorius E. Platelet and red blood cell interactions and their role in rheumatoid arthritis. Rheumatol Int. 2015;35:1955–1964. doi: 10.1007/s00296-015-3300-7. [DOI] [PubMed] [Google Scholar]
- 31.Pilling LC, Atkins JL, Kuchel GA, Ferrucci L, Melzer D. Red cell distribution width and common disease onsets in 240,477 healthy volunteers followed for up to 9 years. PLoS ONE. 2018;13:e0203504–12. doi: 10.1371/journal.pone.0203504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xuan C, et al. RBB, a novel transcription repressor, represses the transcription of HDM2 oncogene. 2012;32:3711–3721. doi: 10.1038/onc.2012.386. [DOI] [PubMed] [Google Scholar]
- 33.Barbee SD, et al. Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc Natl Acad Sci USA. 2011;108:3330–3335. doi: 10.1073/pnas.1010890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narita T, Nitta T, Nitta S, Okamura T, Takayanagi H. Mice lacking all of the Skint family genes. International Immunology. 2018;30:301–309. doi: 10.1093/intimm/dxy030. [DOI] [PubMed] [Google Scholar]
- 35.Liu S, et al. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc Natl Acad Sci U.S.A. 2012;109:13817–13822. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C-M, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004;18:320–332. doi: 10.1101/gad.1162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, et al. Dph3, a small protein required for diphthamide biosynthesis, is essential in mouse development. Mol Cell Biol. 2006;26:3835–3841. doi: 10.1128/MCB.26.10.3835-3841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson R, et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature. 2018:1–20. doi: 10.1038/s41586-018-0794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grasberger H, Refetoff S. Identification of the maturation factor for dual oxidase. Evolution of an eukaryotic operon equivalent. J Biol Chem. 2006;281:18269–18272. doi: 10.1074/jbc.C600095200. [DOI] [PubMed] [Google Scholar]
- 40.Christodoulou K, et al. Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut. 2013;62:977–984. doi: 10.1136/gutjnl-2011-301833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper JD, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, et al. Identification of BACH2 as a susceptibility gene for Graves' disease in the Chinese Han population based on a three-stage genome-wide association study. Hum Genet. 2014;133:661–671. doi: 10.1007/s00439-013-1404-2. [DOI] [PubMed] [Google Scholar]
- 43.Roychoudhuri R, et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 2013;498:506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afzali B, et al. BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency. Nat Immunol. 2017;18:813–823. doi: 10.1038/ni.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karczewski KJ, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. 2019:1–39. Supplementary Information. [Google Scholar]
- 47.Goltsev Y, et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piasecka B, et al. Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges. Proc Natl Acad Sci USA. 2018;115:E488–E497. doi: 10.1073/pnas.1714765115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zalocusky KA, et al. The 10,000 Immunomes Project: Building a Resource for Human Immunology. Cell Rep. 2018;25:513–522.e3. doi: 10.1016/j.celrep.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Márquez A, et al. A combined large-scale meta-analysis identifies COG6 as a novel shared risk locus for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2017;76:286–294. doi: 10.1136/annrheumdis-2016-209436. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, et al. A Genome-Wide Association Study of Psoriasis and Psoriatic Arthritis Identifies New Disease Loci. PLoS Genet. 2008;4:e1000041–14. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuijpers TW, et al. Combined immunodeficiency with severe inflammation and allergy caused by ARPC1B deficiency. J Allergy Clin Immunol. 2017;140:273–277.e10. doi: 10.1016/j.jaci.2016.09.061. [DOI] [PubMed] [Google Scholar]
- 53.Sade-Feldman M, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 2018;175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karp NA, et al. Applying the ARRIVE Guidelines to an In Vivo Database. PLoS Biol. 2015;13:e1002151. doi: 10.1371/journal.pbio.1002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karp NA, Melvin D, Sanger Mouse Genetics Project. Mott RF. Robust and Sensitive Analysis of Mouse Knockout Phenotypes. PLoS ONE. 2012;7:e52410–11. doi: 10.1371/journal.pone.0052410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karp NA, et al. Impact of Temporal Variation on Design and Analysis of Mouse Knockout Phenotyping Studies. PLoS ONE. 2014;9:e111239–10. doi: 10.1371/journal.pone.0111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryder E, et al. Molecular characterization of mutant mouse strains generated from the EUCOMM/KOMP-CSD ES cell resource. Mamm Genome. 2013;24:286–294. doi: 10.1007/s00335-013-9467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malek M, et al. flowDensity: reproducing manual gating of flow cytometry data by automated density-based cell population identification. Bioinformatics. 2015;31:606–607. doi: 10.1093/bioinformatics/btu677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mason J, et al. Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat Commun. 2017;8:1–12. doi: 10.1038/ncomms15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurbatova N, Mason JC, Morgan H, Meehan TF, Karp NA. PhenStat: A Tool Kit for Standardized Analysis of High Throughput Phenotypic Data. PLoS ONE. 2015;10:e0131274. doi: 10.1371/journal.pone.0131274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osorio FG, et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002847. 106ra107–106ra107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.