Abstract
Two laxaphycin type-B cyclic dodecapeptides, laxaphycins B5 and B6, were obtained from UIC 10484, a freshwater cf. Phormidium sp. Analysis using the 16S rRNA sequence found UIC 10484 to clade with UIC 10045, a known laxaphycin type-A and -B producer, and MS/MS analysis revealed the presence of two novel laxaphycin type-B compounds. The structures of the metabolites were elucidated using 2D NMR and MS/MS. The absolute configurations of the amino acids were determined by advanced Marfey’s analysis. Both metabolites were evaluated against the same three cancer cell lines. The IC50 of both laxaphycins B5 and B6 was near 1 μM against breast cancer MDA-MB-231, melanoma MDA-MB-435, and ovarian cancer OVCAR3 cell lines.
Keywords: cyanobacteria, cyclic lipopeptide, cytotoxic, laxaphycin, phylogenetics
Introduction
Secondary metabolites isolated from natural sources are integral to modern drug discovery.1 Cyanobacteria, a phylum of photosynthetic prokaryotic organisms, is a group found to possess considerable secondary metabolite producing capacity that has resulted in a range of biomedically relevant active molecules.2 In particular, strains within the phylum are known to produce anticancer drug leads, including the linear peptide Dolastatin 10, a natural product that inspired the development of the synthetic analog monomethyl auristatin E.3 The synthetic analog was approved as an antibody-drug conjugate to treat relapsed Hodgkin Lymphoma in addition to subtypes of T-cell lymphoma,4 highlighting the overall biomedical potential of the phylum.
In our ongoing investigation to identify antiproliferative drug leads produced by freshwater cyanobacteria, a fraction from UIC 10484 was found to be active against melanoma (MDA-MB-435), breast cancer (MDA-MB-231), and ovarian cancer (OVCAR3) cell lines. Utilizing both phylogenetic and MS/MS analyses, the presence of novel laxaphycin type-B metabolites was identified in the UIC 10484 active fractions. Isolation and structure elucidation led to the identification of cytotoxic laxaphycins B5 (1) and B6 (2) (Fig. 1).
Fig. 1:
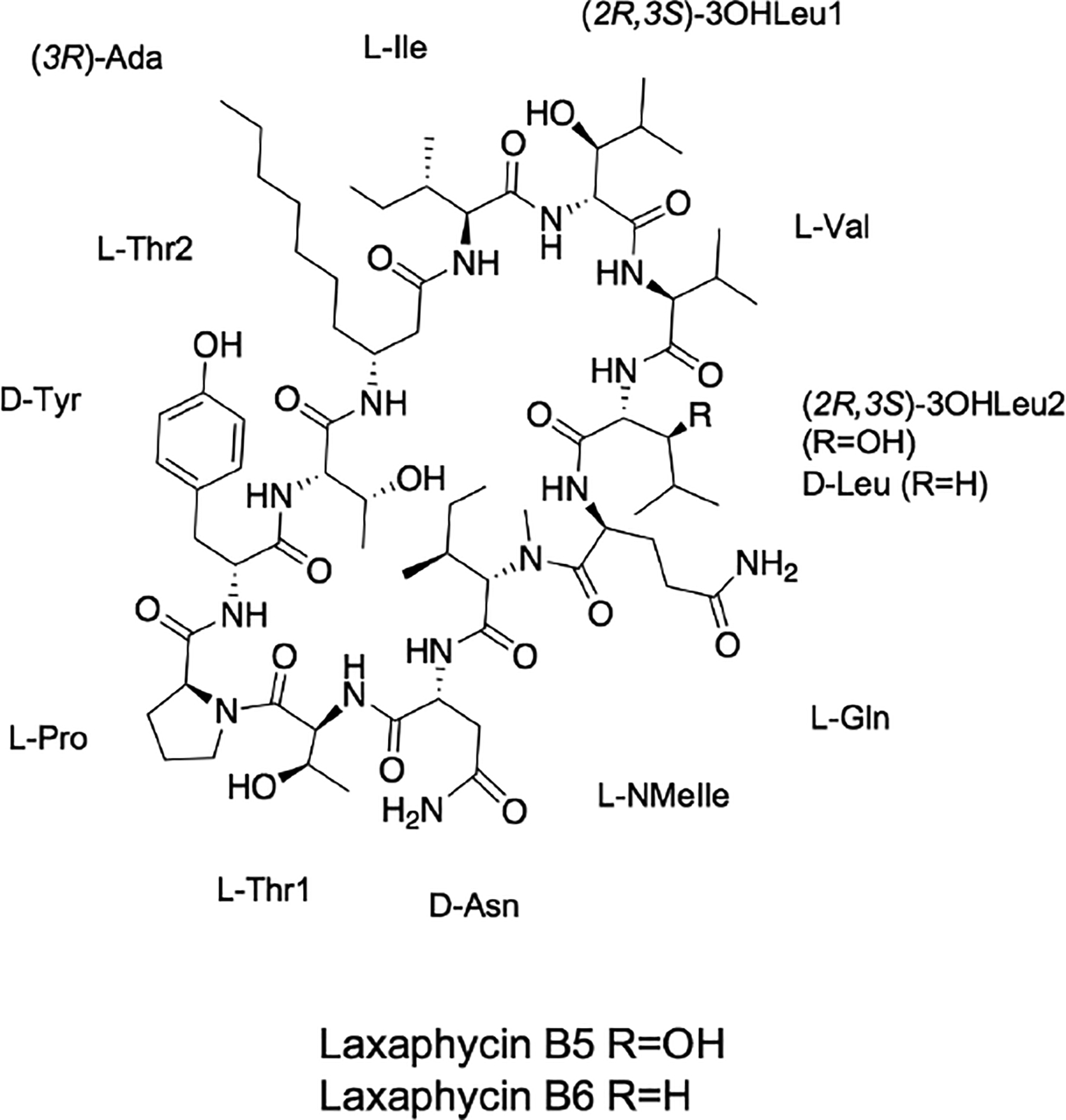
Structure of laxaphycins B5 (1) and B6 (2).
RESULTS
Laxaphycins B5 and B6 were obtained following a bioassay-guided fractionation strategy. After fractionating the cellular extract of UIC 10484, fraction 3 (F3, 40% IPA) demonstrated cell growth inhibition against the three cancer cell lines. A phylogenetic analysis using the 16S rRNA sequences of known secondary metabolite producers from the Orjala lab strain library revealed that UIC 10045 and 10484 are closely related (Fig. 2). UIC 10045, a cf. Oscillatoria sp., has previously been identified to produce laxaphycin-type compounds (trichormamides C and D).5 MS/MS fragmentation data were obtained from two active 10484 fraction components and the laxaphycin-type compounds (trichormamides A-D) in our compound library. Comparison of the fragmentation data (Supplementary fig. S2) in addition to evaluating the [M+H]+ values of the UIC 10484 constituents against all known laxaphycins indicated that the secondary metabolites were novel.
Fig. 2:
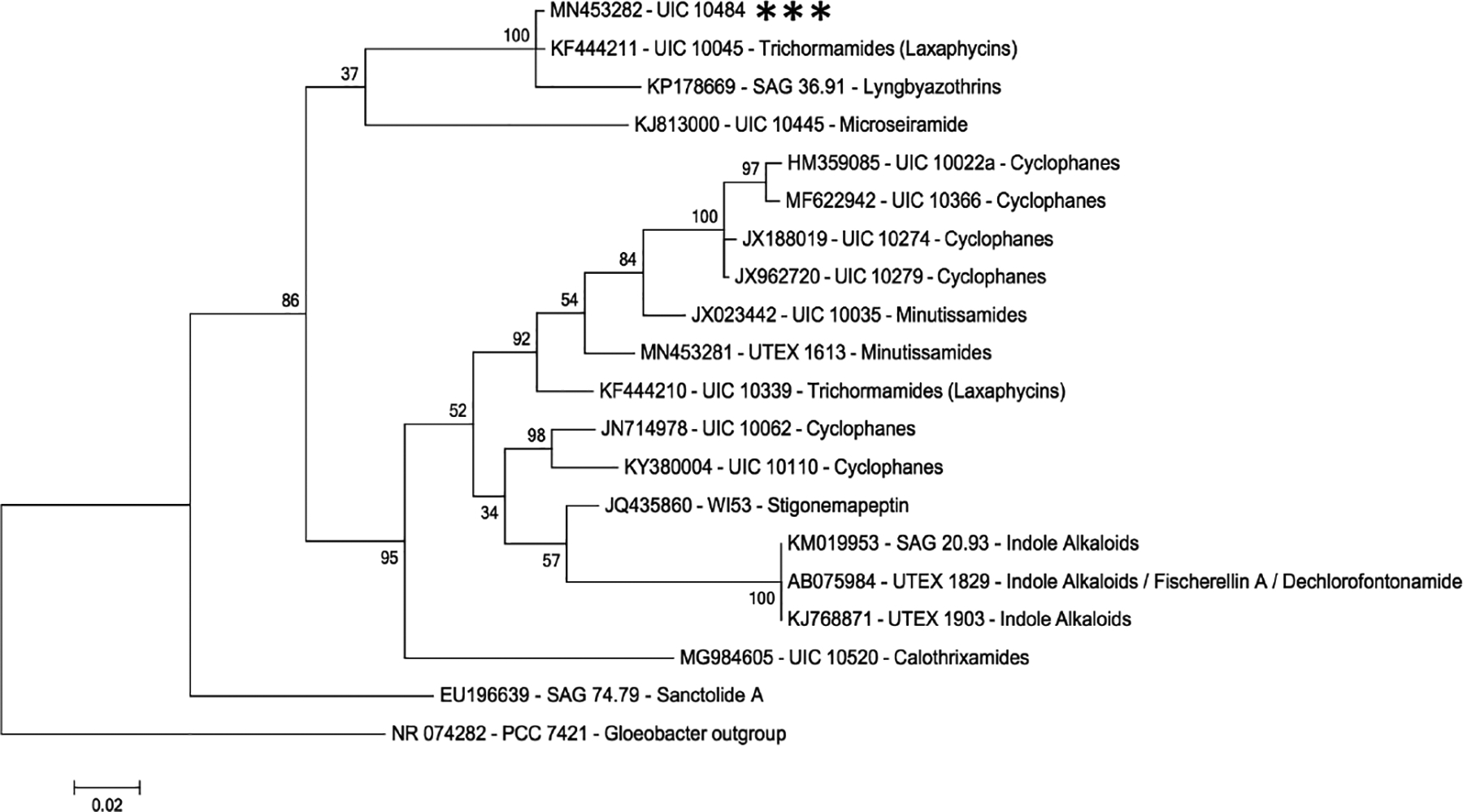
Phylogenetic tree using the SSU (16S) rRNA sequence. The evolutionary relatedness of 19 freshwater cyanobacterial strains from the Orjala library with published secondary metabolite data. PCC 7421 Gloeobacter strain served as the outgroup for the analysis. UIC 10484 (highlighted with ***) clades with UIC 10045, a known laxaphycin producer.
Laxaphycin B5 (1; 9.0 mg), a white, amorphous powder, was the major secondary metabolite isolated from UIC 10484. The molecular formula (C71H118N14O19) was determined based on its HRMS [M+H]+ 1471.8765 (−0.3 ppm error). Analysis of the NMR data (Supplementary figs. S3–S12) found 1 to be a dodecapeptide, consistent with other laxaphycin type-B secondary metabolites.5–11 Eight of the twelve residues are standard amino acids while four are nonstandard. Combining DEPTQ, HSQC, and band-selective HMBC NMR data confirmed that 1 was composed of 71 carbons and 118 hydrogens. COSY, HMBC, and TOCSY data analysis resulted in the elucidation of 11 amino acids; isoleucine (Ile), 3-hydroxylecuine (3-OHLeu1), valine (Val), a second 3-hydroxyleucine (3OHLeu2), glutamine (Gln), N-methylisoleucine (NMeIle), asparagine (Asn), threonine (Thr1), proline (Pro), tyrosine (Tyr), and a second threonine (Thr2) (Fig. 3). The long aliphatic chain of the aminodecanoic acid (Ada) resulted in substantial signal overlap in both 13C and 1H spectra (δc 27–34; δH 1.28). Substructures of Ada, H2-2 to H2-4 and H2-9 to H3-10, were elucidated using COSY. The full elucidation of the ten-carbon lipophilic β-amino acid required the combination of COSY, MS/MS fragmentation, molecular formula, and advanced Marfey’s data. This non-canonical amino acid is present in several laxaphycin type-B metabolites.5,9,11
Fig. 3:
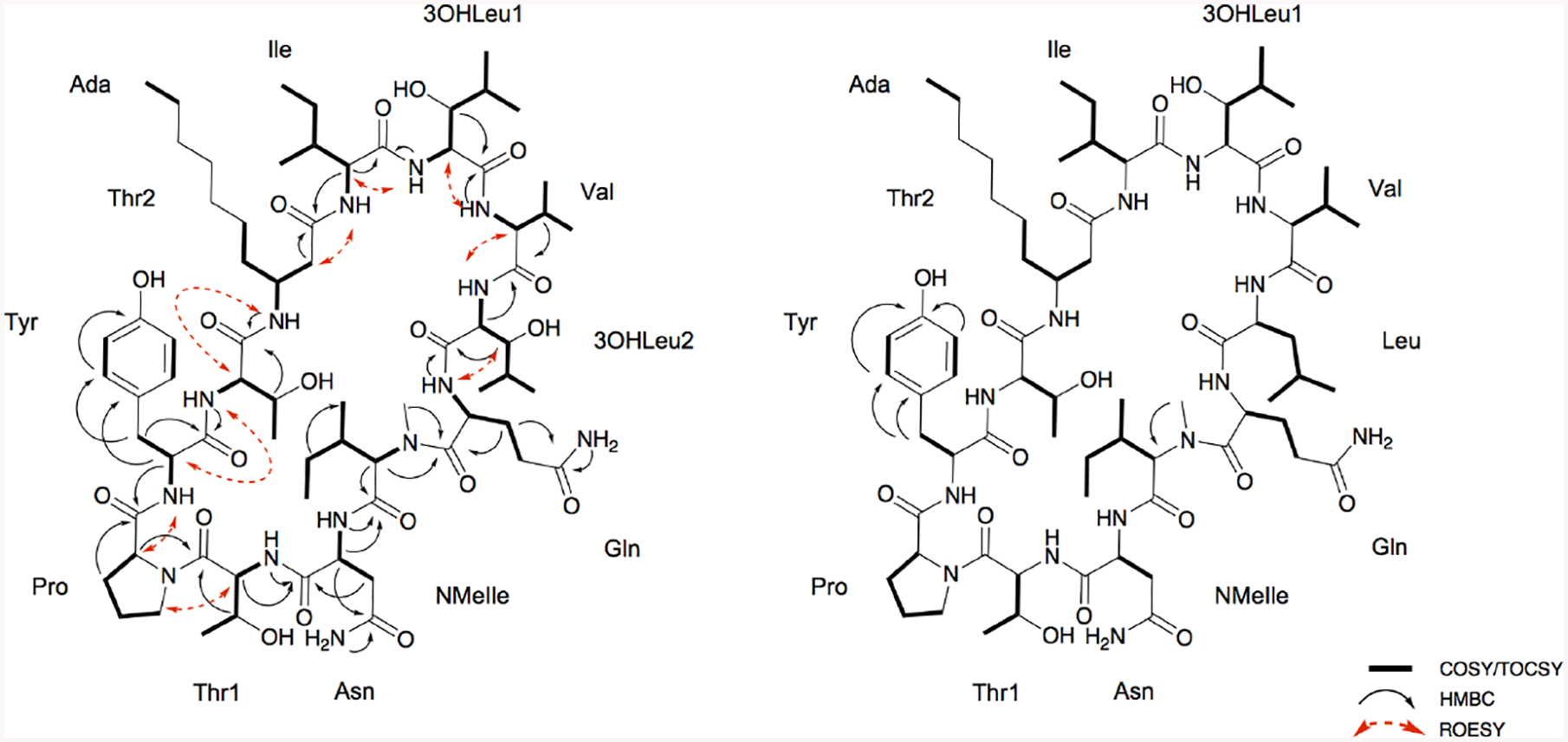
2D NMR correlations resulting in the planar structures laxaphycins B5 (1) and B6 (2).
The COSY spectrum acquired in CD3OH resulted in the correlation between each α-carbon proton and its corresponding amino acid NH. A band-selective HMBC experiment acquired in CD3OH was used to resolve substantial carbonyl overlap (δc 171–179) and resulted in distinct correlations between the amide carbonyls with the NHs, α-protons, and/or β-protons. Correlations between the β-protons and amide carbonyls established corresponding carbonyls for ten of the amino acids (Fig. 3). The other two residues’ (Ile and NMeIle) amide carbons were deduced based on correlations between the α-proton of each side chain and two carbonyl groups of the amide backbone.
Band-selective HMBC, ROESY, and MS/MS fragmentation data were also used to determine amino acid sequence of 1. Connectivity was primarily assessed using band-selective HMBC data (in CD3OH). Ada C-1 (δc 173.8) correlated with Ile H-2 (δH 4.31) and NH (δH 8.23); Ile C-1 (δc 174.3) with 3OHLeu1 H-2 (δH 4.58) and NH (δH 7.99); 3OHLeu1 C-1 (δc 173.8) with Val H-2 (δH 4.18) and NH (δH 7.90); Val C-1 (δc 174.1) with 3OHLeu2 H-2 (δH 4.52) and NH (δH 8.12); 3OHLeu2 C-1 (δc 173.7) with Gln H-2 (δH 4.76) and NH (δH 8.05); Gln C-1 (δc 174.7) with NMeIle H-2 (δH 4.71) and NMe (δH 3.17); NMeIle C-1 (δc 172.2) with Asn H-2 (δH 4.82) and NH (δH 8.24); Asn C-1 (δc 172.8) with Thr1 H-2 (δH 4.59) and NH (δH 7.56); Thr1 C-1 (δc 170.9) with Pro H-2 (δH 4.37); Pro C-1 (δc 174.4) with Tyr H-2 (δH 4.47); Tyr C-1 (δc 174.1) with Thr2 NH (δH 7.68); and Thr2 C-1 (δc 172.0) with Ada NH (δH 7.86). ROESY correlations confirmed the connectivity of the amino acid residues, although the Gln-NMeIle-Asn-Thr1 sequence had limited through space correlation data. However, this partial sequence was validated by MS/MS data of 1 (Fig. 4; Supplementary figs. S19–21). The degrees of unsaturation, calculated from the molecular formula, confirmed 1 is cyclized.
Fig. 4:
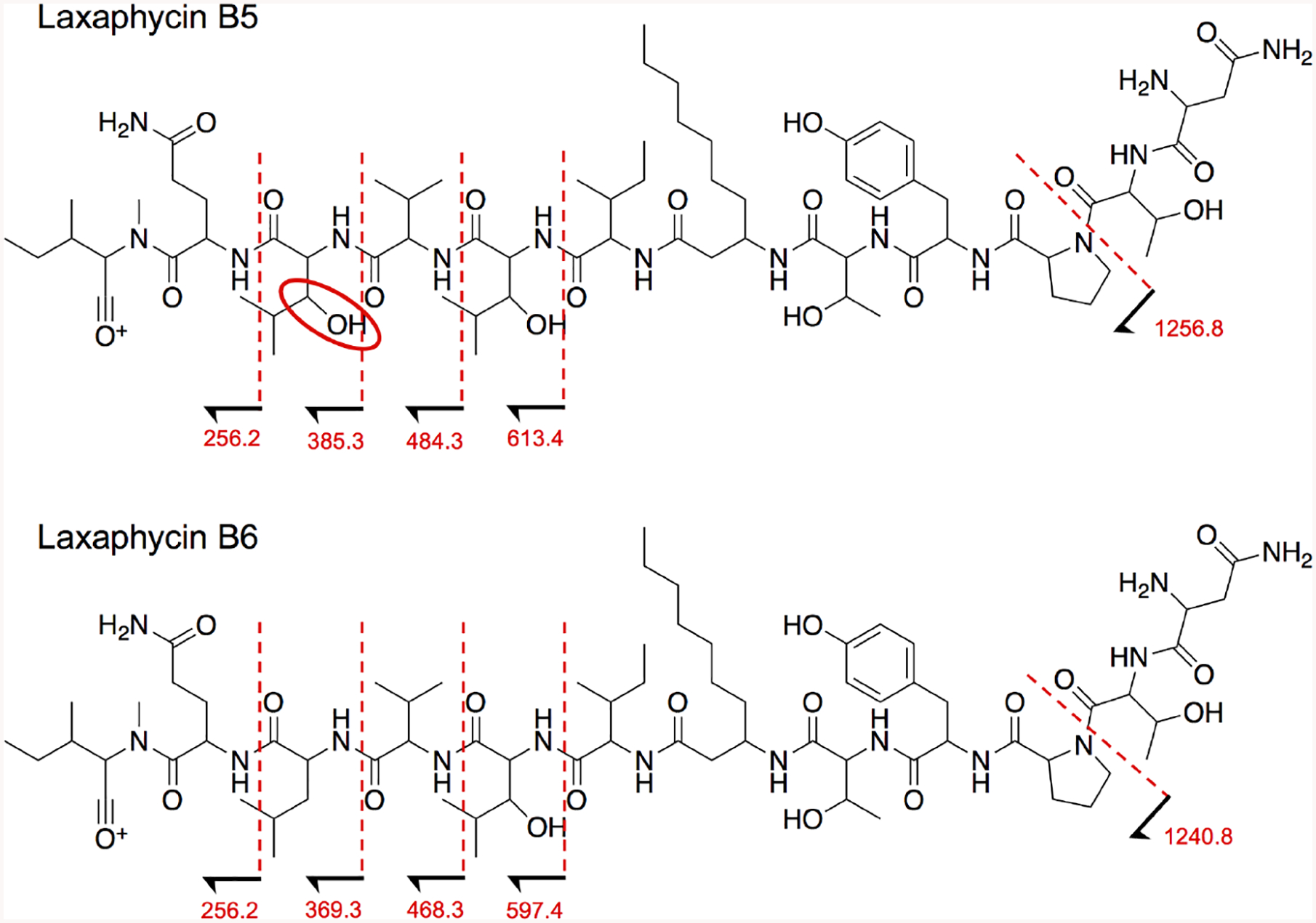
Laxaphycins B5 and B6 are structurally related and, thus, have similar fragmentation patterns. Comparing the MS/MS patterns of the two compounds, both metabolites have the same peptide sequence, however, B5 has an extra 16 Da until the fragments in which B5 lacks 3OHLeu2 and B6 lacks Leu. Fragmentation peaks that lack the differing amino acids share the same mass. For the mass spectrum of this fragmentation pattern, see supplementary fig. S19.
Absolute configuration was determined by Advanced Marfey’s analysis after acid hydrolysis of 1.12–14 The configuration of 11 of the 12 amino acids was established comparing the derivatized hydrolysate retention times to standard retention times. Val, Gln, and Pro were determined to be L while Tyr and Asn were identified as D. Thr1 and Thr2 were both found to be L (2S,3R) while NMeIle and Ile were established as L (2S,3S), and 3OHLeu1 and 3OHLeu2 were assigned as 2R,3S. Ada was assigned as R based on elution order.7 The complete structure was thus determined as cyclo[(3R)-Ada—L-Ile—(2R,3S)-3OHLeu—L-Val—(2R,3S)-3OHLeu—L-Gln—L-NMeIle—D-Asn—L-Thr—L-Pro—D-Tyr—L-Thr] (Fig. 1).
Laxaphycin B6 (2; 2.5 mg), a white amorphous powder, was found to have the molecular formula C71H118N14O18, deduced from the HRESIMS [M+H]+ 1455.8804 (−1.2 ppm), which is 16 Da less than 1. MS and NMR analysis confirmed that compound 2 is a laxaphycin type-B metabolite, with a leucine (Leu) rather than 3OHLeu2 compared to 1 (Fig. 1). 2D NMR experiments (Supplementary figs. S13–18) were performed using CD3OD to elucidate the side chains of each amino acid. HSQC data resulted in the assignment of protons to their carbons and COSY, TOCSY, and HMBC elucidated the planar structures of the side chains. The amino acid residue sequence of the cyclic peptide was determined by comparing MS/MS data between 1 and 2 (Fig. 4; Supplementary figs. S19–21). That of 3OHLeu2 from 1 when fragmented corresponds to the loss of Leu from 2. The advanced Marfey’s method was carried out to determine the absolute configuration of the structure, which was determined as cyclo[(3R)-Ada—L-Ile—(2R,3S)-3OHLeu—L-Val—D-Leu—L-Gln—L-NMeIle—D-Asn—L-Thr—L-Pro—D-Tyr—L-Thr] (Fig. 1).
DISCUSSION
Generally, cyanobacteria from marine and freshwater environments have been found to produce distinct chemistry.15 However, there is some overlap in secondary metabolite production among strains from the two different ecological niches.16,17,18 The class of cyclic lipopeptides characterized by either a β-aminodecanoic or β-aminooctanoic acid residue, comprised of hormothamnin A, the trichormamides, the laxaphycins, the lobocyclamides, the scytocyclamides, in addition to the lyngbyacyclamides, has been found from both marine as well as freshwater cyanobacteria and has been collected from several regions of the world (Supplementary fig. S22).5–11,19–21 This secondary metabolite class is sorted into two groups – the undecapeptide laxaphycin type-A and dodecapeptide laxaphycin type-B compounds (Supplementary fig. S23). All laxaphycin type secondary metabolites are cyclic with the exceptions proposed to be biosynthetic precursors to the cyclic structures or the result of altered pathways in which thioesterase-catalyzed hydrolysis linearizes the metabolites.20,22 Of the nine strains with reported laxaphycins prior to this publication, seven have been reported to produce both type-A and type-B.5–10,21 However, extensive MS/MS analysis found no laxaphycin type-A metabolites to be produced by UIC 10484.
While compounds within each type are structural analogs to each other, laxaphycin type-A and -B secondary metabolites share some of the same amino acids but have demonstrably different amino acid sequences. Both compound classes have shown cytotoxic activity against cancer cells, although previous research has suggested the modes of action are distinct from each other.23 Additionally, several studies have demonstrated a synergistic effect in both antifungal and anticancer bioassays when combining doses of laxaphycin type-A and type-B compounds.6–8,10,23
Using the partial 16S rRNA sequence, it was predicted that UIC 10484 would produce laxaphycin type compounds given its 99.4% sequence similarity to and phylogenetic grouping with laxaphycin producer UIC 10045 (Fig. 2). It is important to note, production of a particular secondary metabolite class often is not monophyletic.24–28 This holds true for the laxaphycin compound classes as laxaphycins are produced by at least three distinct phylogenetic clades from two different orders of cyanobacteria – Oscillatoriales and Nostocales.5,9,21 UIC 10339, a known laxaphycin producer collected from the Midwestern United States, clades distinctly from UIC 10484 and UIC 10045, which were also collected in the Midwest. This highlights the complexity of cyanobacterial secondary metabolite production and the evolutionary relatedness of the producers.
Both laxaphycins B5 (1) and B6 (2) displayed moderate activity against MDA-MB-231, OVCAR3, and MDA-MB-435 with IC50 of 2.2, 1.6, and 1.2 μM for 1 and 0.81, 0.92, and 0.58 μM for 2. These values are consistent with other laxaphycin type-B metabolites evaluated for cytotoxicity (IC50 range of 0.19 – 6.0 μM) against a variety of cancer cell lines.5,8,9,11 While 1 and 2 share the same 11 of 12 amino acids, only five of the 12 amino acid residues are conserved throughout all cyclic dodecapeptide laxaphycin type-B structures with either established antifungal or anticancer activity (Supplementary fig. 24). Of the seven variable residues, the differences are generally minor (i.e., Val rather than Ile). Thus, at least some structural variance is tolerated in order to maintain biomedical relevance.
EXPERIMENTAL SECTION
General experimental procedures
Optical rotations for 1 and 2 were evaluated on a PerkinElmer 241 polarimeter. UV spectra were obtained using a Varian Cary 5000 spectrophotometer. IR spectra were acquired on a Thermo-Nicolet 6700 FT-IR equipped with a SMART iTR sampling accessory. 1D and 2D NMR spectra were acquired on a Bruker AVII 900 MHz NMR spectrometer fitted with a 5 mm CPTCI cryoprobe. The CD3OH/CD3OD solvent chemical shifts were used as the reference signals for both 1H and 13C spectra (δH 3.31 and δC 49.0). MS/MS spectra and Marfey’s data were acquired using a Bruker Impact II UHR Qq-TOF coupled to a Shimadzu Nexera X2 UHPLC system using a reversed-phase C18 UPLC column (Phenomenex Kinetex, 50×2.1mm). Three collision energies were set to obtain MS/MS data on 1, 2, and trichormamides A-D – 65.0, 72.0, 80.0 eV. Additionally, Marfey’s analysis was performed on an Agilent 1100 along with an Agilent 6545 LC/Q-TOF instrument using an analytical HPLC column (Phenomenex, 250×4.6mm). For all LC-MS experiments, the solvent system consisted of H2O and CH3CN, each with 0.1% v/v formic acid.
Biological material
Freshwater sample UIC 2014–072-T3–1 was collected in August 2014 from Indiana, United States near the border of Michigan (latitude: 41.72572; longitude: −85.20015), roughly 250 km from laxaphycin producer UIC 10045. From that sample, strain UIC 10484 was isolated applying a micropipette technique29 and initially cultured in 150 mL BG-12 medium30 in which it was found to grow best. UIC 10484 was cultured in BG-12 in large scale; both in 2.8 L Fernbach flasks and 13 L flasks with 2 L and 10 L of medium, respectively, with a constant flow of 1.7 L/min of filtered air pumped into the flasks. The culture room temperature was set to 22 °C with cultures grown under 1.03 klx fluorescent lighting directly illuminating the flasks from either the side (13 L culture flasks) or directly above (2.8 L Fernbach flasks) with an 18/6h light/dark cycle. Each batch was grown for 6 weeks, harvested and lyophilized prior to extraction. The strain is maintained in the Orjala lab culture library.
Morphological and Phylogenetic Assessment
The morphology of UIC 10484 was established using a Zeiss Axiostar Plus light microscope coupled to a Canon PowerShot A620 camera. The strain was observed at 400x under brightfield illumination. Filaments were isopolar, unbranched, and no heterocysts were detected. The trichomes were cylindrical with rounded ends. Based on this phenotypic assessment, the strain was identified as a member of the freshwater Oscillatoriaceae family (Supplementary fig. S1).
To evaluate the evolutionary position of UIC 10484, the 16S rRNA sequence of the strain was used. Fifty mg of wet cell mass was initially exposed to a 1 mg/mL lysozyme treatment and 0.1 mg/mL proteinase K in 0.5 mL cell lysis solution (Promega) to reduce epiphytic bacterial contamination and digest the bacterial contaminant proteome, in particular contaminant DNAse. Following this, the DNA was extracted from the cell mass using the Nucleospin Soil Kit (Macherey-Nagel). To amplify the partial 16S sequence, cyanobacteria-specific primers 109F and 1509R were used and PCR was carried out using a previously described method.31,32
UIC 10484’s (16S acc. no. MN453282) phylogenetic position was evaluated amongst strains from the Orjala library with both published 16S and secondary metabolite data to gain insight into the potential secondary metabolite production. The secondary metabolite tree was comprised of 18 known freshwater cyanobacteria secondary metabolite producers (Fig. 2). Additionally, its taxonomic position was assessed using 24 cyanobacterial strains identified as type strains of the Oscillatoriaceae family from the database CyanoDB (Hauer T and Komárek J, 2019, database) provisional Oscillatoriaceae type strains from the Cyanotype database33, established reference strains from Bergey’s Manual,34 and strains with high sequence similarity (≥98%) with respect to UIC 10484 established using the National Center for Biotechnology Information’s (NCBI) Blast algorithm (Supplementary fig. S25).35 Taxonomic classification as cf. Phormidium sp. was established using CyanoDB as the authoritative source and CyanoType as an orthogonal source. For both trees, a Gloeobacter outgroup was used. The sequences in the secondary metabolite tree were trimmed to 950 bp and the taxonomy tree trimmed to 1150 bp.
MEGA 7.0 was used to align and build both phylogenetic trees to analyze how UIC 10484 clades amongst other cyanobacterial strains.36 For both the taxonomic and secondary metabolite trees, the respective sequences were aligned using the MUSCLE algorithm with a −400 gap penalty and no gap extend penalty. The trees were constructed using the maximum likelihood method with the GTR+I+G model. One thousand replicates were run for each tree.
Extraction and isolation
After lyophilization of all batches of grown cell mass (33.5 g total from 136 L of total medium), the dry cell mass was soaked in 500 mL of 1:1 MeOH:CH2Cl2 for 24 hours with the extract filtered. The remaining cell material was extracted an additional two times with the same solvent mixture. The extract (8.81 g total) was dried down and then separated into six fractions using Diaion HP-20SS resin with a step gradient of 0, 20, 40, 70, 90, and 100% IPA (referred to as fractions 1 to 6, in the same order). Fraction 3 (F3, eluting with 40% IPA) demonstrated significant growth inhibition against human melanoma cell line MDA-MB-435, breast adenocarcinoma cell line MDA-MB-231, and ovarian adenocarcinoma cell line OVCAR3. F3 was further separated using a Monolithic C18 HPLC column (Phenomenex, 100×4.6 mm) with the resulting subfractions assessed for activity. The active constituents were purified by means of a two-step HPLC process. The first step used a C18 semipreparative reversed-phase column (Agilent, 250×10.0mm) with an initial five minute 20% CH3CN isocratic 4.0 mL/min flow followed by a linear gradient of 20% to 40% CH3CN over ten minutes and a subsequent 100% CH3CN wash for six minutes at 6 mL/min. The cluster of laxaphycin-type compounds eluted between 17 and 19 minutes with 1 and 2 co-eluting at 18.5 minutes. The resulting impure sample containing 1 and 2 was then separated and purified using an analytical C18 column (Phenomenex, 250×4.6mm) with a preliminary five minute 35% CH3CN isocratic 1.0 mL/min flow followed by a linear gradient of 35–55.4% CH3CN over 17 minutes and a 100% CH3CN wash for five minutes. Compound 1 (9.0 mg; 0.03% of dry weight) eluted at 17.6 minutes while compound 2 (2.5 mg; 0.01% of dry weight) eluted at 19.3 minutes.
Biological Activity
Human melanoma MDA-MB-435, breast adenocarcinoma MDA-MB-231, and ovarian adenocarcinoma OVCAR3 cell lines were purchased from American Type Culture Collection. Cell growth inhibition was assessed using an established protocol.37 Vinblastine was used as the positive control. To establish the IC50 for B5 and B6, all three bioassays were run in triplicate.
Advanced Marfey’s analysis
To determine the absolute configuration of both 1 and 2, 0.3 mg of each compound was initially hydrolyzed in 1 mL of 6N hydrochloric acid (HCl) for 24 hours at 110 °C. The hydrolysate was dried and dissolved in 500 μL of water twice to remove residual HCl. Each hydrolysate sample was then split into two equal amounts and derivatized with stereoisomers of 1-fluoro-2,4-dinitrophenyl-5-leucinamide (L-FDLA or D/L-FDLA) as previously described.9,12–14 Standards for 3OHLeu and NMeIle had been previously synthesized.9 The amino acid standards were derivatized following the same protocol used for 1 and 2. Derivatized hydrolysate in addition to the standards was analyzed using a Bruker Impact II Q-TOF LC/MS with three different gradients. An additional fourth gradient was used on an Agilent 1100 Series HPLC system with an Agilent 6545 LC/Q-TOF to confirm the derivatized hydrolysate masses. The retention times of the compounds’ derivatized amino acids were compared to the standard retention times to establish absolute configuration (Supplementary tables S26–28). Gln and Asn configurations were determined based on the retention times of glutamic and aspartic acids.
Under the hydrolytic conditions stated above, two Val enantiomers were observed in the laxaphycin B5 sample and two Ile diastereomers were observed in both the laxaphycin B5 and B6 samples. Previous reports have noted that acid-induced manipulations in configuration occur while amino acids remain part of the intact structure.38 To reduce racemization, the same conditions as previously described were applied but the hydrolysis was reduced from 24 hours to 4 hours resulting in a single laxaphycin B5 Val enantiomer and reduced Ile racemization in both laxaphycins B5 and B6.
The laxaphycin B6 Tyr residue was not initially detected after hydrolysis and Marfey’s derivatization because of Tyr reaction with trace radicals in the HCl solution during hydrolysis. Phenol (0.1% w/v) was added to compete with Tyr for this reaction during hydrolysis to prevent unintended Tyr reactivity.39
Laxaphycin B5 (1)
White, amorphous powder; [a]22D −22.1 (c 0.3, MeOH); UV (MeOH) λmax (log ε) 222 (4.14), 277 (3.11); IR (neat) νmax 3309 (br), 2962, 2931, 1637, 1516, 1457, 1417 cm−1; 1H (CD3OD, CD3OH) and 13C (CD3OD) see table 1; HRESIMS m/z 1471.8765 [M+H]+ (calculated for C71H119N14O19).
Table 1:
Laxaphycin B5 (1) and B6 (2) NMR data in CD3OD and/or CD3OH(900 MHz)
| Laxaphycin B5 (1) | Laxaphycin B6 (2) | ||||
|---|---|---|---|---|---|
| Position | δC | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | |
| Ada | 1 | 173.8 | |||
| 2 | 41.4 | 2.48, m (7.1, 14.4) | 41.2 | 2.5, dd (6.6, 14.2) | |
| 2.58, dd (5.6, 14.4) | 2.57 (4.1, 14.2) | ||||
| 3 | 48.5 | 4.19, m | 48.5 | 4.18, m | |
| 4 | 34.8 | 1.54, q (7.4) | 34.7 | 1.56, m | |
| 5 | 27.3 | 1.31, m | 27.4 | 1.34 | |
| 6 | 30.5 | 1.28, m | 30.8 | 1.29 | |
| 7 | 30.5 | 1.28, m | 30.5 | 1.29 | |
| 8 | 32.2 | 1.27, m | 33.1 | 1.28 | |
| 9 | 23.8 | 1.31, m | 23.7 | 1.31, m | |
| 10 | 14.5 | 0.88a | 14.5 | 0.9a | |
| NH | 7.86, d (8.0) | nd | |||
| Ile | 1 | 174.3 | |||
| 2 | 59.9 | 4.31, t (7.8) | 59.8 | 4.34, m | |
| 3 | 37.9 | 1.89, m | 38.0 | 1.9, m | |
| 3-Me | 16.0 | 0.95, d (7.0) | 16.0 | 0.96a | |
| 4 | 26.3 | 1.21, m | 26.3 | 1.2, m | |
| 1.58, m | 1.58, m | ||||
| 5 | 11.6 | 0.91a | 11.6 | 0.91a | |
| NH | 8.23a | nd | |||
| 3-OHLeu1 | 1 | 173.8 | |||
| 2 | 57.0 | 4.58, m | 57.1 | 4.56, d (3.0) | |
| 3 | 77.5 | 3.68, dd (2.4, 8.2) | 77.3 | 3.68, m | |
| 4 | 32.1 | 1.66, m | 32.3 | 1.67, m | |
| 4-Me | 19.7 | 0.9a | 19.2 | 1.02 (6.5) | |
| 5 | 19.7 | 1.01a | 19.8 | 0.91a | |
| 3-OH | nd | nd | |||
| NH | 7.99, d (6.4) | nd | |||
| Val | 1 | 174.1 | |||
| 2 | 61.4 | 4.18, m | 61.4 | 4.07, m | |
| 3 | 31.5 | 2.14, m | 31.6 | 2.07, m | |
| 3-Me | 19.7 | 0.99a | 19.6 | 0.97a | |
| 4 | 19.3 | 1.00a | 19.6 | 0.97a | |
| NH | 7.90, br | nd | |||
| 3-OHLeu2 (1)/Leu (2) | 1 | 173.7 | |||
| 2 | 57.5 | 4.52, d (9.1) | 53.4 | 4.36, m | |
| 3 | 78.0 | 3.62, dd (1.8, 9.1) | 41.6 | 1.6, m | |
| 4 | 32.3 | 1.66, m | 25.9 | 1.68, m | |
| 4-Me | 19.4 | 0.87a | 23.7 | 0.95a | |
| 5 | 19.7 | 1.01a | 21.3 | 0.90a | |
| 3-OH | nd | x | |||
| NH | 8.12, d (8.4) | nd | |||
| Gln | 1 | 174.7 | |||
| 2 | 51.5 | 4.76, dd (5.2, 7.9) | 50.9 | 4.74, t (6.9) | |
| 3 | 27.6 | 1.98, m | 27.8 | 2.04, m | |
| 2.08, m | 2.06, m | ||||
| 4 | 32.2 | 2.27, q (7.8) | 32.3 | 2.25 | |
| 5 | 178.1 | 178.0 | |||
| NH2 | 6.82, b | nd | |||
| 7.47, b | nd | ||||
| NH | 8.05, d (5.2) | nd | |||
| NMeIle | 1 | 172.2 | |||
| 2 | 62.8 | 4.70, d (10.7) | 62.9 | 4.67, m | |
| 3 | 33.4 | 2.08, m | 33.1 | 2.1, m | |
| 3-Me | 16.1 | 0.91a | 16.0 | 0.92a | |
| 4 | 25.5 | 1.00a | 25.6 | 1.01, m | |
| 1.42, m | 1.41, m | ||||
| 5 | 11.0 | 0.87a | 11.0 | 0.88a | |
| N-Me | 31.9 | 3.16, s | 32.3 | 3.2, s | |
| Asn | 1 | 172.8 | |||
| 2 | 51.5 | 4.82, dd (5.7, 7.2) | 51.4 | 4.84, t (6.4) | |
| 3 | 37.1 | 2.7 (7.2, 15.8) | 37.2 | 2.7, dd (7.3, 15.7) | |
| 2.75 (5.7, 15.8) | 2.75 dd (5.5, 15.7) | ||||
| 4 | 175.0 | ||||
| NH2 | 6.9, b | nd | |||
| 7.58, b | nd | ||||
| NH | 8.24a | nd | |||
| Thr1 | 1 | 170.9 | |||
| 2 | 57.5 | 4.59a | 57.5 | 4.63, d (3.5) | |
| 3 | 68.1 | 4.16, m | 68.1 | 4.19, m | |
| 4 | 19.8 | 1.21 | 19.7 | 1.23, d (6.4) | |
| 3-OH | nd | nd | |||
| NH | 7.56, d (6.6) | nd | |||
| Pro | 1 | 174.4 | |||
| 2 | 62.0 | 4.37, dd (6.3, 7.5) | 62.0 | 4.4, t (6.6) | |
| 3 | 30.5 | 1.71, m | 30.5 | 1.76, m | |
| 2.10, m | 2.12, m | ||||
| 4 | 25.8 | 1.90, m | 25.8 | 1.93, m | |
| 1.92, m | |||||
| 5 | 49.2 | 3.72, m | 49.2 | 3.75, m | |
| 3.78, m | 3.79, m | ||||
| Tyr | 1 | 174.1 | |||
| 2 | 57.0 | 4.47, t (7.8) | 57.1 | 4.46, t (8.2) | |
| 3 | 37.5 | 2.88, dd (7.8,14.1) | 37.3 | 2.89, m | |
| 3.05, dd (7.8, 14.1) | 3.05, m | ||||
| 4 | 128.7 | 129.0 | |||
| 5/9 | 131.3 | 7.03, d (8.4) | 131.3 | 7.03, d (8.0) | |
| 6/8 | 116.4 | 6.69, d (8.4) | 116.4 | 6.69, d (8.0) | |
| 7 | 157.7 | 157.5 | |||
| OH | nd | nd | |||
| NH | 8.15, b | nd | |||
| Thr2 | 1 | 172.0 | |||
| 2 | 60.6 | 4.13, m | 60.7 | 4.12, m | |
| 3 | 67.9 | 4.21, m | 67.8 | 4.23, m | |
| 4 | 20.2 | 0.89a | 14.5 | 0.88a | |
| 3-OH | nd | nd | |||
| NH | 7.68, d (8.2) | nd | |||
Laxaphycin B6 carbonyl groups not included due to signal overlap
overlapping signal
not in compound
no detected signal
Laxaphycin B6 (2)
White, amorphous powder; [a]22D −18.1 (c 0.08, MeOH); UV (MeOH) λmax (log ε) 222 (4.17), 277 (3.20); IR (neat) νmax 3327 (br), 2961, 2927, 1654, 1517, 1450 cm−1; 1H (CD3OD) and 13C (CD3OD) see table 1; HRESIMS m/z 1455.8804 [M+H]+ (calculated for C71H119N14O18).
Supplementary Material
Acknowledgements
This research was supported by the NCI/NIH P01 CA12506 grant and The Office of the Director, NIH National Center for Complementary and Integrative Health (NCCIH) T32AT007533-05 training grant (PS). We thank Dr. Jonathan Bisson and Dr. Charlotte Simmer for their assistance using the Q-TOF mass spectrometer, Rojin Ahadi and Angel Antunez for isolating and culturing strain UIC 10484, Dr. Ben Ramirez for his guidance using the UIC Center for Structure Biology NMR instrumentation, Dr. Chen of the Research Resources Center’s Mass Spectrometry Core for use of the Agilent 6454 LC/Q-TOF, and Dr. Young Jeong for access to their HPLC system. We’d also like to acknowledge the James R. Fuchs lab of Ohio State University for synthesizing the NMeIle and 3OHLeu standards.
Footnotes
Dedicated to Professor William Fenical in recognition of his contributions to marine derived secondary metabolites.
Supplementary information is available at the Journal of Antibiotics website.
REFERENCES
- 1.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. 2016;79(3):629–661. [DOI] [PubMed] [Google Scholar]
- 2.Singh RK, Tiwari SP, Rai AK, Mohapatra TM. Cyanobacteria: An emerging source for drug discovery. J Antibiot (Tokyo). 2011;64(6):401–412. [DOI] [PubMed] [Google Scholar]
- 3.Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. Isolation of Dolastatin 10 from the Marine Cyanobacterium Symploca Species VP642 and Total Stereochemistry and Biological Evaluation of Its Analogue Symplostatin 1. J Nat Prod. 2001;64(7):907–910. [DOI] [PubMed] [Google Scholar]
- 4.Senter PD, Sievers EL. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat Biotechnol. 2012;30(7):631–637. [DOI] [PubMed] [Google Scholar]
- 5.Luo S, et al. Trichormamides C and D, antiproliferative cyclic lipopeptides from the cultured freshwater cyanobacterium cf. Oscillatoria sp. UIC 10045. Bioorganic Med Chem. 2015;23(13):3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frankmölle WP, Knübel G, Moore RE, Patterson GM. Antifungal cyclic peptides from the terrestrial blue-green alga Anabaena laxa. II: Structures of laxaphycins A, B, D and E. J Antibiot (Tokyo). 1992;45(9):1458–1466. [DOI] [PubMed] [Google Scholar]
- 7.MacMillan JB, Ernst-Russell MA, De Ropp JS, Molinski TF. Lobocyclamides A-C, lipopeptides from a cryptic cyanobacterial mat containing Lyngbya confervoides. J Org Chem. 2002;67(23):8210–8215. [DOI] [PubMed] [Google Scholar]
- 8.Bonnard I, Rolland M, Salmon JM, Debiton E, Barthomeuf C, Banaigs B. Total structure and inhibition of tumor cell proliferation of laxaphycins. J Med Chem. 2007;50(6):1266–1279. [DOI] [PubMed] [Google Scholar]
- 9.Luo S, et al. Trichormamides A and B with antiproliferative activity from the cultured freshwater cyanobacterium Trichormus sp. UIC 10339. J Nat Prod. 2014;77(8):1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai W, Matthew S, Chen QY, Paul VJ, Luesch H. Discovery of new A- and B-type laxaphycins with synergistic anticancer activity. Bioorganic Med Chem. 2018;26(9):2310–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maru N, Ohno O, Uemura D. Lyngbyacyclamides A and B, novel cytotoxic peptides from marine cyanobacteria Lyngbya sp. Tetrahedron Lett. 2010;51(49):6384–6387. [Google Scholar]
- 12.Harada K, et al. A Method Using LC/MS for Determination of Absolute Configuration of Constituent Amino Acids in Peptide --- Advanced Marfey’s Method. Tetrahedron Lett. 1995;36(9):1515–1518. [Google Scholar]
- 13.Fujii K, Harada KI. A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: Combination of Marfey’s method with mass spectrometry and its practical application. Anal Chem. 1997;69(24):5146–5151. [Google Scholar]
- 14.Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada KI. A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: Elucidation of limitations of Marfey’s method and of its separation mechanism. Anal Chem. 1997;69(16):3346–3352. [Google Scholar]
- 15.Luzzatto-Knaan T, et al. Digitizing mass spectrometry data to explore the chemical diversity and distribution of marine cyanobacteria and algae. Elife. 2017;6:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp K, et al. Phylogenetic and chemical diversity of three chemotypes of bloom-forming Lyngbya species (cyanobacteria: Oscillatoriales) from reefs of southeastern Florida. Appl Environ Microbiol. 2009;75(9):2879–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami M, Suzuki S, Itou Y, Kodani S, Ishida K. New anabaenopeptins, carboxypeptidaze-A inhibitors from the cyanobacterium Aphanizomenon flos-aquae. J Nat Prod. 2000;63(9):1280–1282. [DOI] [PubMed] [Google Scholar]
- 18.Matthew S, Ross C, Paul VJ, Luesch H. Pompanopeptins A and B, new cyclic peptides from the marine cyanobacterium Lyngbya confervoides. Tetrahedron. 2008;64(18):4081–4089. [Google Scholar]
- 19.Gerwick WH, Jiang ZD, Agarwal SK, Farmer BT. Total structure of hormothamnin A, A toxic cyclic undecapeptide from the tropical marine cyanobacterium Hormothamnion enteromorphoides. Tetrahedron. 1992;48(12):2313–2324. [Google Scholar]
- 20.Bornancin L, et al. Structure and biological evaluation of new cyclic and acyclic laxaphycin-A type peptides. Bioorg Med Chem. 2019;27(10):1966–1980. [DOI] [PubMed] [Google Scholar]
- 21.Grewe CJ. Cyanopeptoline und Scytocyclamide: Zyklische Peptide aus Scytonema hofmanni PCC 7110 Struktur und biologische Aktivität. 2005. [Google Scholar]
- 22.Bornancin L, et al. Isolation and synthesis of laxaphycin b-type peptides: A case study and clues to their biosynthesis. Mar Drugs. 2015;13(12):7285–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gbanktoto A, Vigo J, Dramane K, Banaigs B, Aina E, Salmon JM. Cytotoxic Effect of Laxaphycins A and B on Human Lymphoblastic Cells (CCRF-CEM) Using Digitised Videomicrofluorometry. In Vivo. 2005;19:577–582. [PubMed] [Google Scholar]
- 24.Fujii K, Sivonen K, Nakana T, Harada K. Structural elucidation of cyanobacterial peptides encoded by peptide synthetase gene in Anabaena species. Tetrahedron. 2002;58:6863–6871. [Google Scholar]
- 25.Bister B, et al. Cyanopeptolin 963A, a chymotrypsin inhibitor of Microcystis PCC 7806. J Nat Prod. 2004;67(10):1755–1757. [DOI] [PubMed] [Google Scholar]
- 26.Itou Y, Ishida K, Shin HJ, Murakami M. Oscillapeptins A to F, Serine Protease Inhibitors from the Three Strains of Oscilltoria agardhii. Tetrahedron. 1999;55:6871–6882. [Google Scholar]
- 27.Okumura HS, Philmus B, Portmann C, Hemscheidt TK. Homotyrosine-containing cyanopeptolins 880 and 960 and anabaenopeptins 908 and 915 from Planktothrix agardhii CYA 126/8. J Nat Prod. 2009;72(1):172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guljamow A, et al. High-density cultivation of terrestrial Nostoc strains leads to reprogramming of secondary metabolome. Appl Environ Microbiol. 2017;83(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein JR, ed. Handbook of Phycological Methods. Cambridge: Cambridge University Press; 1973. [Google Scholar]
- 30.Chlipala G, Mo S, Carcache De Blanco EJ, Ito A, Bazarek S, Orjala J. Investigation of antimicrobial and protease-inhibitory activity from cultured cyanobacteria. Pharm Biol. 2009;47(1):53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.May DS, et al. Merocyclophanes C and D from the Cultured Freshwater Cyanobacterium Nostoc sp. (UIC 10110). J Nat Prod. 2017;80(4):1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63(8):3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos V, Morais J, Vasconcelos VM. A curated database of cyanobacterial strains relevant for modern taxonomy and phylogenetic studies. Sci Data. 2017;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrity George M.; Boone David R.; Castenholz RW Bergey’s Manual of Systematic Bacteriology. 2nd ed. New York: Springer; 2001. [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren Y, et al. Cardiac Glycoside Constituents of Streblus asper with Potential Antineoplastic Activity. J Nat Prod. 2017;80(3):648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser K, Benner R. Hydrolysis-induced racemization of amino acids. Limnol Oceanogr Methods. 2005;3:318–325. [Google Scholar]
- 39.Davidson I Hydrolysis of Samples for Amino Analysis In: Smith BJ, ed. Methods in Molecular Biology. 2nd ed. Humana Press Inc.; 2003:111–122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


