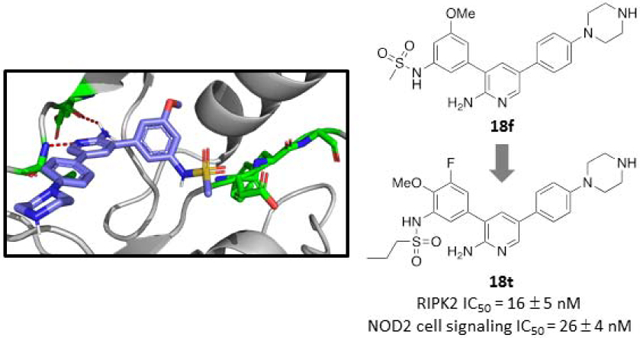
Abstract
Receptor-interacting protein kinase 2 (RIPK2) is a key mediator of nucleotide-binding oligomerization domain (NOD) cell signaling that has been implicated in various chronic inflammatory conditions. A new class of RIPK2 kinase/NOD signaling inhibitors based on a 3,5-diphenyl-2-aminopyridine scaffold was developed. Several co-crystal structures of RIPK2•inhibitor complexes were analyzed to provide insights into inhibitor selectivity versus the structurally related activin receptor-like kinase 2 (ALK2) demonstrating that the inhibitor sits deeper in the hydrophobic binding pocket of RIPK2 perturbing the orientation of the DFG motif. In addition, the structure-activity relationship study revealed that in addition to anchoring to the hinge and DFG via the 2-aminopyridine and 3-phenylsulfonamide, respectively, appropriate occupancy of the region between the gatekeeper and the αC-helix provided by substituents in the 4- and 5-positions of the 3-phenylsulfonamide were necessary to achieve potent NOD cell signaling inhibition. For example, compound 18t (e.g. CSLP37) displayed potent biochemical RIPK2 kinase inhibition (IC50 = 16 ± 5 nM), > 20-fold selectivity versus ALK2 and potent NOD cell signaling inhibition (IC50 = 26 ± 4 nM) in the HEKBlue assay. Finally, in vitro ADME and pharmacokinetic characterization of 18t further supports the prospects of the 3,5-diphenyl-2-aminopyridine scaffold for the generation of in vivo pharmacology probes of RIPK2 kinase and NOD cell signaling functions.
Keywords: inhibitor, kinase, NOD, nucleotide-binding oligomerization domain, receptor-interacting protein kinase 2, RIPK2
Graphical Abstract
1. Introduction
The nucleotide-binding oligomerization domain (NOD)-containing proteins 1 and 2 are members of the NOD-like receptor (NLR) family that are involved in the innate immune system’s detection of bacterial peptidoglycan (PG) derivatives. NOD1 is stimulated by bacterial PG fragments containing diaminopimelic acid (DAP), while NOD2 senses muramyl dipeptide (MDP). NOD1/2 then initiates assembly of signaling complexes by oligomerization through the nucleotide-binding oligomerization domains (NBD), which triggers the recruitment of interacting proteins through homotypic caspase-activated recruitment domain (CARD)-mediated interactions [1-4]. Receptor-interacting protein kinase 2 (RIPK2) is one of the key molecules in NOD-dependent signaling as it plays an essential role in the activation of NF-κB pathway and mitogen-activated protein kinase (MAPK) pathways that ultimately lead to synthesis of pro-inflammatory cytokines and antimicrobial molecules [2, 5].
RIPK2, which has dual serine/threonine and tyrosine kinase function, was identified as an essential mediator of signaling in both innate and adaptive immune systems [6]. In addition, nonkinase activity, particularly X-linked inhibitor of apoptosis (XIAP)-mediated ubiquitination of RIPK2, significantly contributes to the functions of this protein [7-14]. Aberrant RIPK2-NOD signaling pathways appear to play key roles in various inflammatory diseases. Most importantly, positive or negative dysregulation of the NOD2-dependent signaling pathway has been shown to facilitate several chronic inflammatory disorders such as Crohn’s disease [15-17], Blau syndrome [18-19], early-onset sarcoidosis [19] and multiple sclerosis [20]. Therefore, RIPK2–NOD has emerged as a crucial mediator of inflammatory pathways and thus a potential therapeutic target for treating an array of inflammatory and autoimmune diseases.
With growing appreciation of RIPK2–NOD involvement in inflammatory and autoimmune cellular pathways and diseases, interest in RIPK2 small molecule ligands has accelerated. Several compound classes have been reported to inhibit RIPK2’s kinase activity (Figure 1), including SB203580 (1) [21], gefitinib (2) [22], OD36 (3a) and OD38 (3b) [23], WEHI-345 (4) [24], ponatinib (5) [25], GSK583 (6a) [26], 6b and its prodrug 6c [27], 7a and 7b [28], and 8 [29]. Several of these compounds (e.g. 1, 2, 4, 6a, 6b and 7b) have been shown to bind the active (DFG-in/αC-helix-in) conformation of RIPK2 via a type I manner, while 5 demonstrated binding to the inactive (DFG-out/αC-helix-in) conformation via a type II binding mode. These molecules have been shown to inhibit various downstream processes in cells associated with NOD signaling, such as NF-κB activation and MDP stimulated IL-8 or TNFα release. In addition, 3a and 3b [23] have demonstrated in vivo efficacy in an MDP-induced peritonitis model, 4 [24] and 8 [29] have shown activity in the experimental autoimmune encephalomyelitis model of multiple sclerosis and intestinal and lung inflammation models, respectively, while 7a [28] inhibited inflammatory cytokine release in the ex vivo experiments using human inflammatory bowel disease biopsy samples. Herein we report the structure-activity relationship (SAR) of a new structure class of RIPK2 kinase and NOD cell signaling inhibitors that will be useful as probes for further understanding this protein, its role in NOD cell signaling and potentially as lead compounds for therapeutic development.
Figure 1.
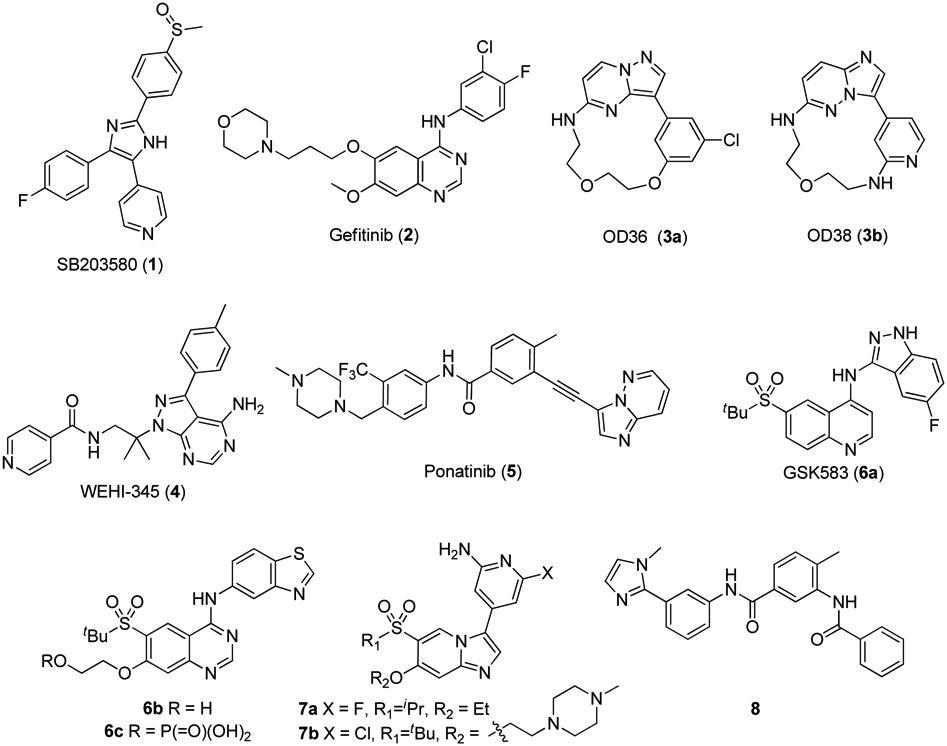
Reported RIPK2 kinase inhibitors.
2. Design and synthesis
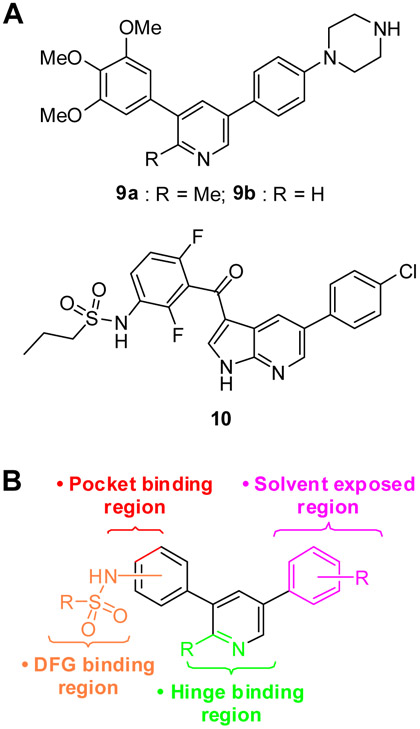
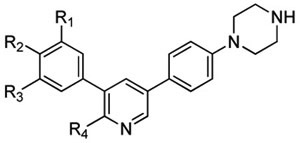
In contemplating a new class of RIPK2 kinase inhibitors we considered two scaffolds (9a and 9b) that had previously been used for activin receptor-like kinase 2 (ALK2) inhibitors. In both cases, RIPK2 was a prevailing off-target revealed by kinome-wide selectivity profiling [30-31]. The 3,5-diphenyl-2-aminopyridine scaffold, exemplified by LDN-214117 (9a) [30] that displayed a modest IC50 of 100 nM in the RIPK2 ADPGlo kinase assay, was chosen to pursue (Figure 2A, Table I).
Figure 2.
A) Structures of ALK2 inhibitors 9a–b and B-RafV600E inhibitor 10, all of which block RIPK2 kinase activity. B) RIPK2 inhibitor design incorporating the sulfonamide DFG binding fragment of 10 into scaffold 9.
Table 1.
RIPK2 and ALK2 kinases, and NOD2 cell signaling inhibitory activities of 9a and 18a–k.
 |
|||||||
|---|---|---|---|---|---|---|---|
| Compounds | R1 | R2 | R3 | R4 | Kinase IC50 (nM) |
RIPK2/NOD2 Cell Assay IC50 (nM) |
|
| RIPK2 | ALK2 | ||||||
| 9a | OMe | OMe | OMe | Me | 100 ± 23 | 24 [30] | NI |
| 18a | OMe | NHSO2nPr | H | Me | NI | NI | 2944 ± 420 |
| 18b | OMe | H | NHSO2Me | Me | 296 ± 10 | 31 ± 34 | 3705 ± 85 |
| 18c | OMe | H | NHSO2Et | Me | 127 ± 18 | 86 ± 45 | 5239 ± 861 |
| 18d | OMe | H | NHSO2nPr | Me | 164 ± 7 | 108 ± 92 | 2669 ± 138 |
| 18e | OMe | H | NHSO2iPr | Me | 76 ± 12 | 239 ± 170 | 523 ± 18 |
| 18f | OMe | H | NHSO2Me | NH2 | 51 ± 26 | 5 ± 6 | 390 ± 22 |
| 18g | OMe | H | NHSO2nPr | NH2 | 14 ± 0.6 | 9 ± 3 | 243 ± 14 |
| 18h | OMe | H | NHSO2iPr | NH2 | 13 ± 0.04 | 11 ± 10 | 121 ± 27 |
| 18i | OMe | H | NHSO2Ph | NH2 | 23 ± 0.5 | 5 ± 7 | 53 ± 36 |
| 18j | OEt | H | NHSO2iPr | NH2 | 21 ± 2 | 299 ± 180 | 46 ± 0.5 |
| 18k | OiPr | H | NHSO2iPr | NH2 | 18 ± 8 | NI | 323 ± 12 |
Values are shown as the mean of two or three determinations ± standard deviation; NI: No inhibition up to 10 μM
To improve potency, we desired to introduce a functional group into the 3,5-diaryl-2-aminopyridine scaffold that would ensure an interaction with the DFG segment of RIPK2. In order to identify a candidate moiety, the Harvard Medical School Library of Integrated Network-based Cellular Signatures (http://lincs.hms.harvard.edu) database was reviewed seeking to find inhibitors with non-overlapping kinase profiles except for RIPK2. Over 160 compounds in the database displayed RIPK2 inhibition and among them PLX4032 (10, vemurafenib, Figure 2A), a B-RafV600E kinase inhibitor, showed ~96% inhibition at 10 μM. A B-RafV600E•10 co-crystal structure reveals a DFG-in type I½ binding mode, a hydrogen bond between the sulfonamide and the backbone NH of Phe595 and Gly596 in the DFG motif and displacement of the αC-helix [32-34].
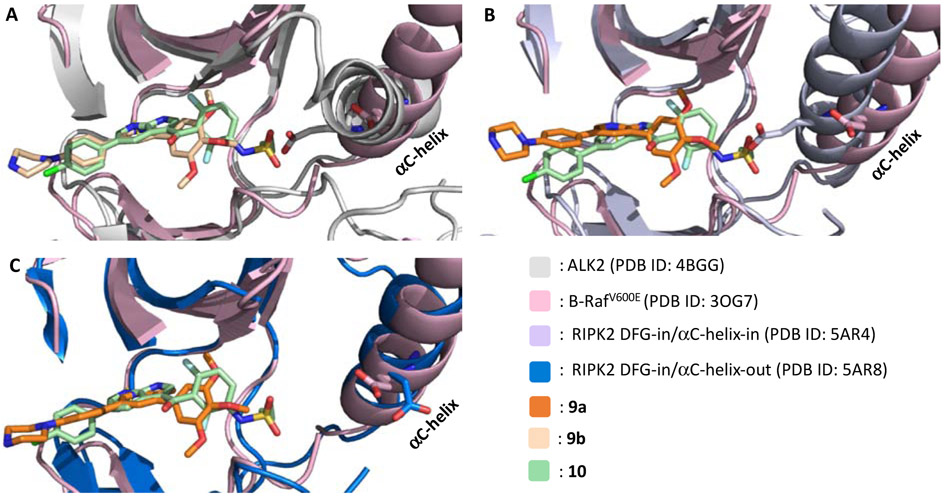
Next, 9a was docked into the DFG-in/αC-helix-in (PDB 5AR4) and DFG-in/αC-helix-out (PDB 5AR8) structures of RIPK2 using AutodockTool-1.5.6. The co-crystal structures of ALK2•9b (PDB 4BGG) and B-RafV600E•10 (PDB 3OG7) as well as the RIPK2•9a docked structures were compared to identify potential positions on 9a for installation of the sulfonamide from 10 (Figure 3) in order to engage the DFG of RIPK2. The overlay results, with either the DFG-in/αC-helix-in or DFG-in/αC-helix-out conformations, suggested that the sulfonamide should be installed into the 4-position of the 3-phenyl to generate 9a/10-hybrids as a potential new class of RIPK2 inhibitors (Figure 2B).
Figure 3.
Overlaid structures of 10 vs 9b and 10 vs docked 9a suggested the position to install the sulfonamide moiety on the 3,5-diphenyl-2-aminopyridine scaffold. A) Superimposed crystal structures of 10 in B-RafV600E and 9b in ALK2; B) and C) Docking result of 9a in RIPK2 DFG-in/αC-helix-in and DFG-in/αC-helix-out structures, respectively, overlaid with co-crystal structure of 10 in B-RafV600E.
Various 1-(4-(5-phenylpyridin-3-yl)phenyl)piperazines utilized in this study were prepared using modifications of the reported syntheses of 9a and 9b [30]. Primary or secondary bromoanilines 11 and 12, many of which were commercially available, were used as starting materials. However, several non-commercial anilines were prepared by either nucleophilic aromatic substitution or nitration, followed by iron-mediated nitro reduction. The bromoanilines 11 and 12 were allowed to react with sulfonyl chlorides in the presence of pyridine using slightly modified conditions to provide 13 (Scheme 1). Miyaura borylation reaction of 13 with bis(pinacolato)diboron using either PdCl2(dppf) in THF or PdCl2(dppf) in 1,4-dioxane furnished arylboronic esters 14. Palladium-catalyzed Suzuki–Miyaura coupling of 14 with 3-bromo-5-iodopyridines 15a–c delivered 3-bromo-5-(phenylalkylsulfonamide)pyridines 16.
Scheme 1. Synthesis of 3-bromo-5-arylpyridine intermediates 16.*.
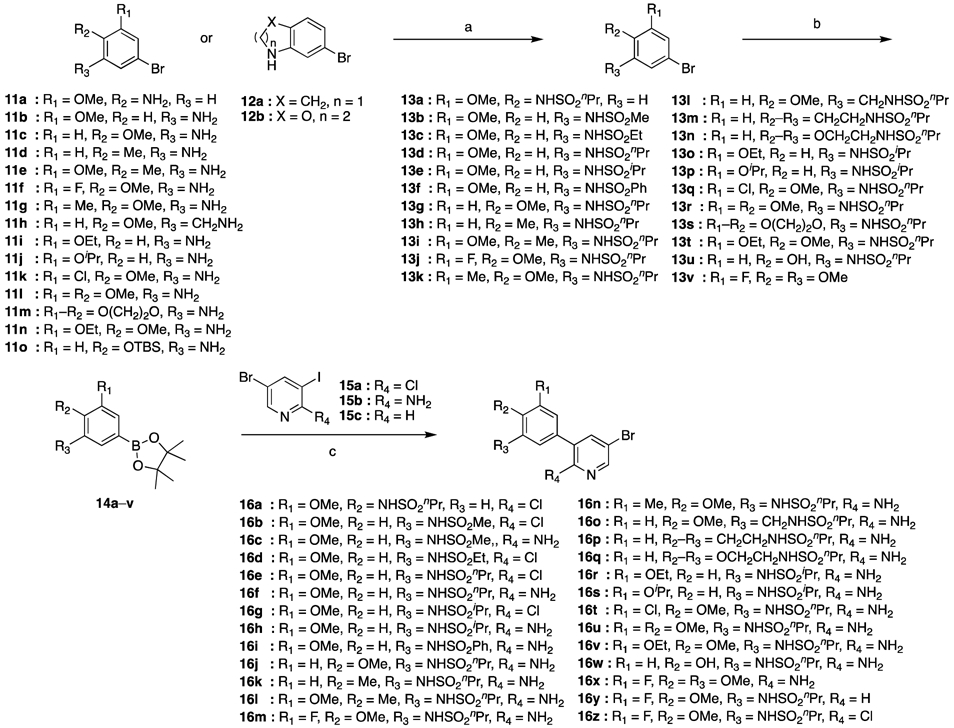
*Reagents and Conditions: (a) alkyl or phenyl sulfonyl chloride, Method A: pyridine, CH2Cl2, rt 16–48 h, Method B: pyridine, cat. DMAP, CH2Cl2, rt, 16–48 h, Method C: cat. DMAP, pyridine, 50 °C, 3 h, Method D: pyridine, 50 °C, 2.5 h or Method E: pyridine, rt, 16–48 h (30–99%); (b) bis(pinacolato)diboron, Method F: 3 mol% PdCl2(dppf), KOAc, THF, reflux, 16 h or Method G: 10 mol% PdCl2(dppf), KOAc, 1,4-dioxane, 80 °C, 40 h (24–90%); (c) 15a, 15b or 15c, 10 mol% Pd(PPh3)4, 1 M Na2CO3, MeCN, DMF, 90 °C, 16 h (38–96%).
2-Chloropyridine intermediates 16 (R4 = Cl) were subjected to a second Suzuki reaction with 4-(4-tert-butoxycarbonylpiperazinyl)phenylboronic acid pinacol ester to deliver 17, which were treated with trimethylboroxine in the presence of a palladium catalyst followed by Boc deprotection using trifluoroacetic acid (TFA) in dichloromethane at room temperature to furnish 18a–e and 18aa (Scheme 2). Similarly, 2-aminopyridine or pyridine intermediates 16 (R4 = NH2 or H) were coupled with 4-(4-tert-butoxycarbonylpiperazinyl)phenylboronic acid pinacol ester and deprotected to give 18f-z.
Scheme 2. Synthesis of 1-(4-(5-phenylpyridin-3-yl)phenyl)piperazine derivatives 18a–z and 18aa.*.
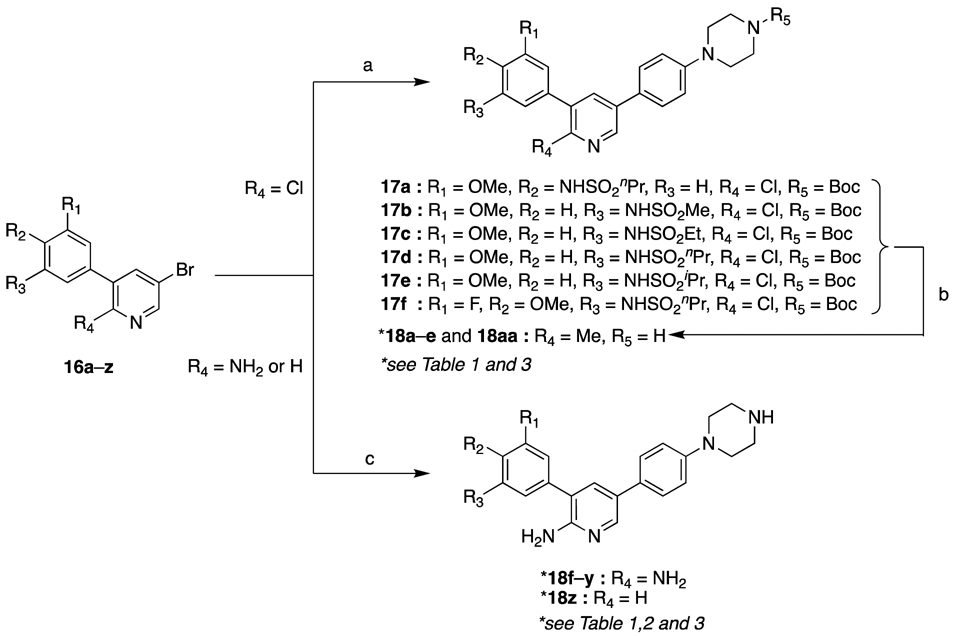
*Reagents and Conditions: (a) 4-(4-tert-butoxycarbonylpiperazinyl)phenylboronic acid pinacol ester, 15 mol% Pd(PPh3)4, 1 M Na2CO3, DME, 90 °C, 16 h (79–99%); (b) 1) trimethylboroxine, 20 mol% Pd(PPh3)4, K2CO3, 1,4-dioxane, 110 °C, 16 h; 2) 10% TFA in CH2Cl2, rt, 16 h (62–96% over two steps); (c) 1) 4-tert-butoxycarbonylpiperazinyl)phenylboronic acid pinacol ester, 15 mol% Pd(PPh3)4, 1 M Na2CO3, DME, 90 °C, 16 h; 2) 10% TFA in CH2Cl2, rt, 16 h (18–62% over two steps).
Finally, a set of compounds was prepared with modifications to the solvent exposed region. In this case, intermediate 16j was coupled with 2-, 3-, and 4-(methanesulfonyl)phenylboronic acid to give 18ab, 18ac and 18ad, respectively (Scheme 3). Similarly, 16m was coupled with (3-(4-tert-butoxycarbonyl)piperazin-1-yl)phenyl)boronic acid pinacol ester and deprotected to provide 18ae.
Scheme 3. Synthesis of 18ab–18ae.*.
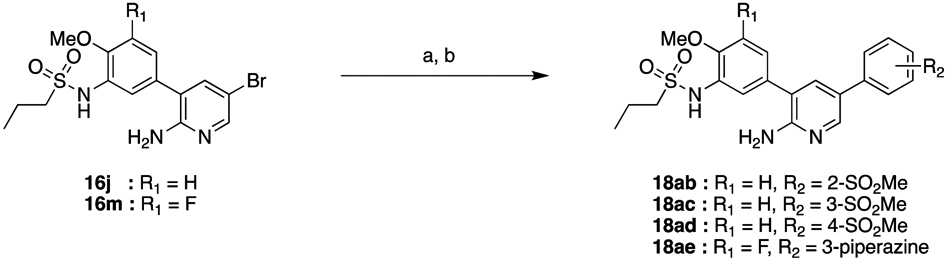
*Reagents and Conditions: (a) 2-, 3-, or 4-(methanesulfonyl)phenylboronic acid or (3-(4-(tert-butoxycarbonyl)piperazin-1-yl)phenyl)boronic acid pinacol ester, 10 mol% Pd(PPh3)4, 1 M Na2CO3, MeCN, DMF, 90 °C, 16 h (22–83%); (b) In the case of 3-(4-(tert-butoxycarbonyl)piperazin-1-yl)phenyl: 10% TFA in CH2Cl2, rt, 16 h (48% over two steps).
3. Results and discussion
As previously mentioned, structure analyses using either the DFG-in/αC-helix-in or DFG-in/αC-helix-out conformations suggested that the sulfonamide should be installed into the para-position of the 3-phenyl group occupying the binding pocket region. To test this initial model, a derivative of 9a containing a 1-propanesulfonamide in this position (e.g. 18a) was evaluated for RIPK2 inhibitory activity. However, it was not active (Table 1). Therefore, the 1-propanesulfonamide was changed to a methylsulfonamide and moved to the meta-position (18b) to further test the strategy. Gratifyingly, this compound proved to be a moderately potent inhibitor of RIPK2 (IC50 = 296 nM). Since the primary component of these compounds was 9a, ALK2 kinase activity was chosen as the counter-screen for assessing selectivity [30]. Indeed, 18b proved to be more potent against ALK2 (IC50 = 31 nM). However, these preliminary results indicated that placement of the sulfonamide in the meta-position was preferred. Several additional derivatives (18c–e) examined homologation and branching of the sulfonamide. In general, RIPK2 kinase inhibitory activity was modestly improved, but selectivity versus ALK2 was not significantly altered. Next, an amino group was introduced in place of the methyl at the 2-position of the pyridine to provide better hinge binding interactions. As expected, this set of compounds (18f–h) was more potent against both RIPK2 and ALK2. In addition, phenylsulfonamide 18i was found to also be tolerated. Interestingly, increasing the size of the ether to an ethyl or isopropyl (18j or 18k) was allowed by RIPK2, but less so for ALK2.
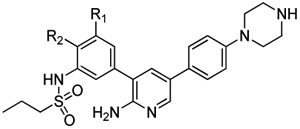
Given the previously demonstrated importance of methoxy groups on the 3-phenyl ring of 3,5-diphenyl-2-aminopyridine ALK2 inhibitors, a series of compounds was evaluated that offered changes to this region, while maintaining a 1-propanesulfonamide in the meta-position (Table 2) [30]. Installing a methyl in the para-position (181) was detrimental for both RIPK2 and ALK2 inhibitory activities. However, moving the methoxy to the para-position (18m) proved advantageous for improving selectivity (to 86-fold) for RIPK2 versus ALK2. Replacing this group with hydroxyl (18n) or methyl (18o) resulted in reduced RIPK2 inhibitory activity and selectivity over ALK2.
Table 2.
RIPK2 and ALK2 kinase, and NOD2 cell signaling inhibitory activities of 18g and 18l–o.
 |
|||||
|---|---|---|---|---|---|
| Compounds | R1 | R2 | Kinase IC50 (nM) |
RIPK2/NOD2 Cell Assay IC50 (nM) |
|
| RIPK2 | ALK2 | ||||
| 18g | OMe | H | 14 ± 0.8 | 9 ± 4 | 243 ± 14 |
| 18l | OMe | Me | NI | 355 ± 318 | 3347 ± 495 |
| 18m | H | OMe | 41 ± 19 | 3545 ± 2114 | 420 ± 34 |
| 18n | H | OH | 54 ± 6 | 387 ± 42 | > 5000 |
| 18o | H | Me | 102 ± 22 | 2120 ± 509 | 1064 ± 109 |
Values are shown as the mean of two or three determinations ± standard deviation; NI: No inhibition up to 10 μM
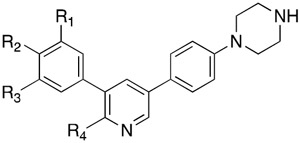
Based on the encouraging results with a methoxy group in the ortho-position relative to the 3-phenylsulfonamide, additional changes were examined in this region (Table 3). Reintroducing a meta-methoxy (18p) retained potent RIPK2 inhibitory activity, but predictably resulted in reduced selectivity versus ALK2. Increasing the size to ethoxy (18q) or tethering the ethers in an ethylenedioxy (18r) resulted in loss of inhibition for both RIPK2 and ALK2. However, replacement with methyl (18s), fluoro (18t) or chloro (18u) was well tolerated with 10 to 20-fold selectivity versus ALK2. Not surprisingly, replacement of the sulfonamide with a methoxy (18v) completely abolished selectivity. Installing a methylene between the sulfonamide and phenyl (18w) eroded both enzyme inhibitory activity and selectivity. In addition, incorporation of the sulfonamide in a ring (18x–y) was also not well tolerated. Interestingly, removal of the 2-amino group on the pyridine of 18t (e.g. 18z) did not erode kinase inhibitory activity. However, replacing the amino group with methyl (18aa) decreased both RIPK2 and ALK2 inhibitory activities, as expected.
Table 3.
RIPK2 and ALK2 kinase, and NOD2 cell signaling inhibitory activities of 18m, 18p–z and 18aa.
 |
|||||||
|---|---|---|---|---|---|---|---|
| Compounds | R1 | R2 | R3 | R4 | Kinase IC50 (nM) |
RIPK2/NOD 2 Cell Assay IC50 (nM) |
|
| RIPK2 | ALK2 | ||||||
| 18m | H | OMe | NHSO2nPr | NH2 | 32 ± 9 | 3545 ± 2114 |
476 ± 97 |
| 18p | OMe | OMe | NHSO2nPr | NH2 | 20 ± 0.8 | 57 ± 13 | 1 ± 0.4 |
| 18q | OEt | OMe | NHSO2nPr | NH2 | NI | NI | NI |
| 18r | OCH2CH2O | NHSO2nPr | NH2 | 520 ± 13 | NI | 2458 ± 309 | |
| 18s | Me | OMe | NHSO2nPr | NH2 | 41 ± 1 | 395 ± 245 | 129 ± 38 |
| 18t | F | OMe | NHSO2nPr | NH2 | 16 ± 5 | 348 ± 107 | 26 ± 4 |
| 18u | Cl | OMe | NHSO2nPr | NH2 | 21 ± 4 | 202 ± 88 | 46 ± 0.4 |
| 18v | F | OMe | OMe | NH2 | 39 ± 4 | 29 ± 14 | 595 ± 70 |
| 18w | H | OMe | CH2NHSO2nPr | NH2 | 99 ± 20 | 145 ± 57 | 1290 ± 94 |
| 18x | H | CH2CH2N(SO2nPr) | NH2 | 185 ± 16 | NI | 2426 ± 116 | |
| 18y | H | OCH2CH2N(SO2nPr) | NH2 | 78 ± 20 | NI | 1011 ± 27 | |
| 18z | F | OMe | NHSO2nPr | H | 27 ± 2 | NT | 352 ± 83 |
| 18aa | F | OMe | NHSO2nPr | Me | 1414 ± 312 | NI | 2556 ± 253 |
Values are shown as the mean of two or three determinations ± standard deviation; NI: No inhibition up to 10 μM; NT: Not tested
Finally, the solvent exposed region of the inhibitor was briefly examined. Previous studies have demonstrated that selectivity for RIPK2 can be improved by introducing functional groups in this area that engage Ser25 in the glycine-rich loop, which is fairly unique to this particular kinase [26]. For example, a co-crystal structure of 6 with RIPK2 reported by Haile and co-worker showed a hydrogen bonding interaction of a methylsulfone with this residue [26]. The potency of the methylsulfone derivative was > 200-fold greater compared with the unsubstituted analog. Based on these observations, (methylsulfonyl)phenyl moieties were introduced to the solvent exposed region of the hybrid series (Table 4). However, the 2-, 3-, and 4-(methylsulfonyl) derivatives (18ab, 18ac and 18ad) demonstrated significantly less potency. On the other hand, moving the piperazine from the para- to meta-position (18ae) retained potent RIPK2 kinase inhibition and improved selectivity versus ALK2 raising the possible that this group provides a similar interaction with Ser25.
Table 4.
RIPK2 and ALK2 kinase, and NOD2 cell signaling inhibitory activities of 18t and 18ab–18ae
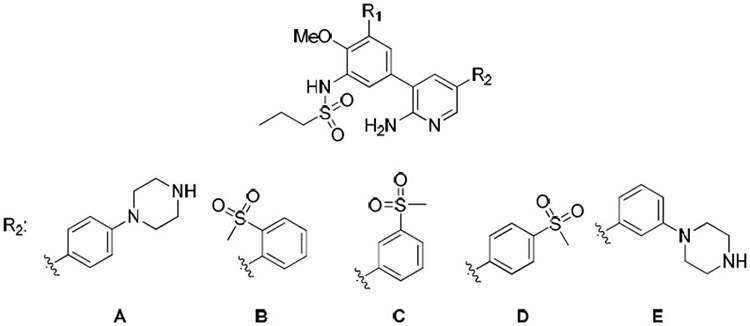 | |||||
|---|---|---|---|---|---|
| Compounds | R1 | R2 | Kinase IC50 (nM) |
RIPK2/NOD2 Cell Assay IC50 (nM) |
|
| RIPK2 | ALK2 | ||||
| 18t | F | A | 16 ± 5 | 348 ± 76 | 26 ± 4 |
| 18ab | H | B | NI | NI | NI |
| 18ac | H | C | 103 ± 16 | NI | 2655 ± 49 |
| 18ad | H | D | 264 ± 38 | NI | NI |
| 18ae | F | E | 11 ± 0.6 | 522 ± 317 | 16 ± 0.6 |
Values is are shown as the mean of two or three determinations ± standard deviation; NI: No inhibition up to 10 μM
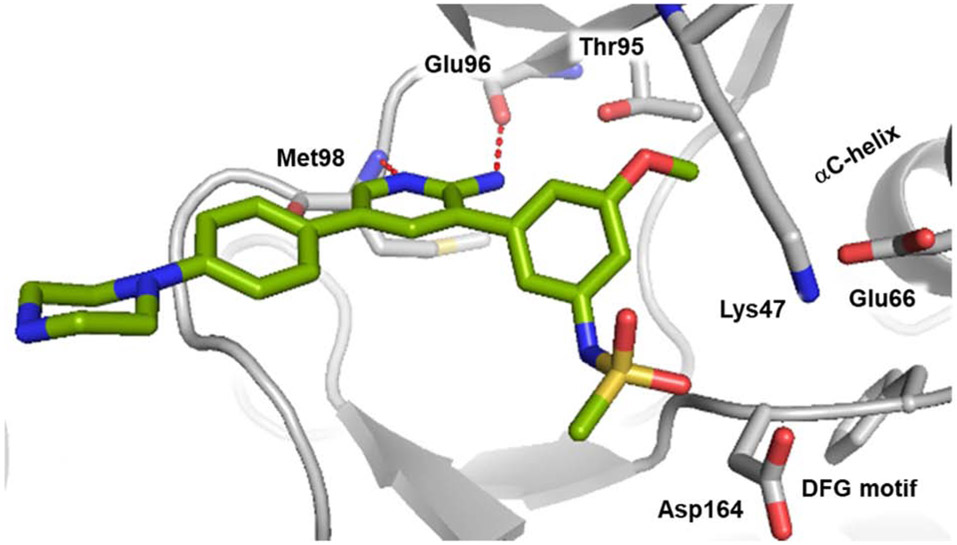
In order to understand inhibitor interactions with RIPK2 and to gain insights for preferential binding for a subset of compounds versus ALK2, compound 18f was co-crystallized with RIPK2 (PDB 6S1F). The 18f•RIPK2 structure was resolved at a resolution of 3.1 Å (see Table S1 for data collection and refinement statistics) and displayed RIPK2 in an inactive DFG-in/Glu-in conformation with several critical residues misplaced from their active positions, including misorientation of the catalytic residues, perturbation of the R-spine and P-loop, as well as a disordered activation segment (Figure 4) similar to the 18m•RIPK2 complex [7]. The 2-aminopyridine formed hydrogen bonds with the backbone NH and carbonyl of hinge residues Met98 and Glu96, respectively. In addition, the 3-phenylsulfonamide of 18f projected towards the hydrophobic pocket comprised of Val32, Leu70, Leu79, Ile93, Leu153 and Ala163, with the sulfonamide oriented towards Asp164 in the DFG motif. Finally, an ionic-ionic interaction was evident between Lys47 and Glu66 maintains the αC-helix in the Glu-in conformation.
Figure 4.
Co-crystal structure of 18f•RIPK2 (PDB 6S1F). The inhibitor (green) forms hydrogen bonds to backbone NH and carbonyl of Met98 and Glu96, respectively, in the hinge region, while the methylsulfonamide is oriented towards the Asp164 in the DFG motif. Lys47 forms an ionic-ionic interaction with Glu66, in the αC-helix, stabilizing the DFG-in/Glu-in conformation.
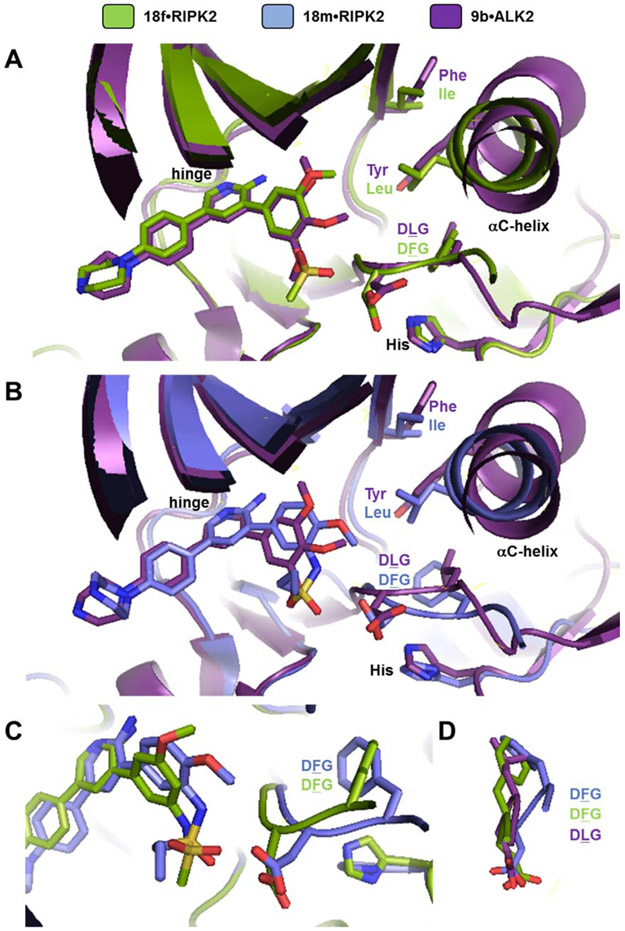
A number of interactions observed in the 18f•RIPK2 co-crystal structure were seen for inhibitor 9b bound to ALK2 with the two inhibitors occupying similar space in their respective kinases (Figure 5A) [30]. Also the DFG (in RIPK2) and DLG (in ALK2) motifs, as well as the αC-helix, were comparable. However, juxtaposition of the 9b•ALK2 and 18m•RIPK2 co-crystal structures (Figure 5B) demonstrated that the latter inhibitor, which has the methoxy at the 4-position on the 3-phenylsulfonamide and is more selective for RIPK2 versus ALK2, sits deeper in the hydrophobic pocket. In addition, it perturbs the orientation of the DFG relative to the DLG in ALK2. Interestingly, the DLG in ALK2 appears to be less flexible as evidenced by a lack of disclosed DLG-out ALK2 inhibitors, whereas adoption of a DFG-out conformation in RIPK2 has been previously observed with 5 [25]. The ability of the DFG in RIPK2 to move, in the case of 18m relative to 18f in RIPK2 and 9b in ALK2 (Figures 5C and D), may play a role in its enhanced selectivity for RIPK2 versus ALK2. One additional difference noted for 18m versus 18f is rotation of the sulfonamide (Figure 5C) potentially induced by the presence of the methoxy in the 4-position.
Figure 5.
(A) Alignment of 18f•RIPK2 (green; PDB 6S1F) and 9b•ALK2 (light purple; PDB 4BGG) co-crystal structures. Several residues, the DFG and DLG motifs, as well as the αC-helices are labeled. (B) Alignment of 18m•RIPK2 (dark purple; PDB 6FU5) and 9b•ALK2 (light purple; PDB 4BGG) co-crystal structures. Several residues, the DFG and DLG motifs, as well as the αC-helices are labeled. (C) Alignment of 18f•RIPK2 (green) and 18m•RIPK2 (dark purple) co-crystal structures. The DFG motifs are labeled. (D) Comparison of the DFG and DLG of 18f•RIPK2 (green) and 9b•ALK2 (light purple) with the DFG of 18m•RIPK2 (dark purple).
Although kinase activity is a function of RIPK2, this protein is also an ubiquitination substrate in response to NOD1/2 stimulation [8-9]. Several ubiquitin (Ub) ligases participate in the conjugation of Ub chains to RIPK2. Particularly critical is XIAP-mediated ubiquitination of RIPK2, which results in recruitment [10] and linear ubiquitin chain assembly complex (LUBAC) mediated Met1-Ub conjugation of RIPK2 [11]. These latter processes are intimately involved in the downstream signaling step leading to the assembly of the NF-κB-activating IKK kinase complex [12]. Disruption of XIAP-RIPK2 interactions through genetic mutations in XIAP [14] or RIPK2 inhibitors mitigates NOD2 inflammatory signaling [7, 13]. Given this array of functional roles of RIPK2, the newly developed kinase inhibitors were assessed in a HEKBlue assay that measures NF-κB activation, downstream of XIAP-mediated RIPK2 ubiquitination, in response to L18-MDP stimulation.
A significant portion of the compounds, despite demonstrating potent RIPK2 kinase inhibition (IC50 < 100 nM), either modestly blocked NOD2 signaling in the HEKBlue assay (100 nM < IC50 < 1 μM) or were comparatively inactive (IC50 > 1 μM), suggesting that inhibition of RIPK2 catalytic activity was not sufficient for inhibition of NOD signaling in cells. In a recently published report, Hrdinka, et al. demonstrated that a subset of these 3,5-diaryl-2-aminopyridine based RIPK2 inhibitors (e.g. 18p and 18t) engaged RIPK2 to impede XIAP-mediated RIPK2 ubiquitination, leading to efficient inhibition of L18-MDP-induced NOD-dependent signaling responses, e.g. CXCL8 production in U2OS/NOD2 cells and TNFα release from RAW264.7 macrophages. Both 18p and 18t interfered with the interaction between GST-BIR2-cIAP1 and RIPK2 blocked XIAP-mediated RIPK2 ubiquitination resulting in disruption of NOD2 signaling similar to ponatinib and GSK583 [7]. Furthermore, 18t did not block lipopolysaccharide and pan-caspase inhibitor induced RIPK1 and RIPK3 dependent necroptosis [35] in RAW264.7 macrophages at 1 μM (data not shown) demonstrating the compound’s selectivity for RIPK2/NOD signaling. The mechanism of how allosteric binding of compounds to the ATP pocket disrupts XIAP-mediated RIPK2 ubiquitination is currently not known. However, the structural features that appear to achieve NOD cell signaling inhibitory function for a subset of compounds in Tables 1-4 as assessed in the NF-κB activation HEKBlue assay were the 2-aminopyridine and 3-phenylsulfonamide anchoring to the hinge and DFG, respectively, as well as appropriate occupancy of the region between the gatekeeper and the αC-helix provided by substituents (e.g. H or OMe) in the 4-position and relatively small substituent (e.g. F, Cl, or OMe) in the 5-positions of the 3-phenylsulfonamide. Absence of this last structural feature was sufficient to eliminate NOD cell signaling, but not kinase inhibitory activity. Further interrogation of this portion of the molecule will likely provide additional insights into the SAR for disruption of NOD cell signaling.
Since 18t was among a set of compounds that displayed potent biochemical RIPK2 kinase and NOD cell signaling inhibitory activities, it was selected as a representative compound for further in vitro characterization and pharmacokinetic study (Table 5). Its solubility in pH 7.4 PBS buffer was ≥100 μM. The compound also demonstrated both moderate permeability of 2.9 × 10−6 cm/s in the PAMPA and mouse liver microsome stability (CLint = 27 μL/min/mg; t1/2 = 25.6 min) assays. After a single 10 mg/kg intraperitoneal administration [6% Captisol® in water formulation] to 8-week old female C57BL/6 mice (n = 5 mice/per group), the compound reached a maximum plasma concentration (Cmax) of 0.55 ± 0.2 μM in 1.2 h (Tmax) with a plasma elimination half-life (t1/2) of 4.7 ± 0.2 h, an AUC of 175.2 ± 59.9 min•μg/mL, a volume of distribution of 24.0 ± 6.2 L/kg and clearance (CL) of 59.7 ± 15.1 mL/min/kg, well below mouse hepatic blood flow. Overall, these properties support 18t as a potential useful in vivo probe for RIPK2 kinase and NOD cell signaling inhibition.
Table 5.
Physicochemical properties and pharmacokinetic parameters of 18t.
| Aqueous Solubility at pH 7.4 (μM) |
Permeability (×10−6 cm/s) |
Mouse Microsome Stability | |||
|---|---|---|---|---|---|
| t1/2 (min) | CLint (μL/min/mg) | ||||
| ≥100 | 2.9 | 25.6 | 27 | ||
| In vivo Pharmacokinetics* | |||||
| Cmax (μM) | Tmax (h) | t1/2 (h) | AUC (min•μg/mL) |
Vd (L/kg) | CL (mL/min/kg) |
| 0.55 ± 0.2 | 1.2 | 4.7 ± 0.2 | 175.2 ± 59.9 | 24.0 ± 6.2 | 59.7 ± 15.1 |
Single 10 mg/kg intraperitoneal administration of 18t to female C57BL/6 mice.
4. Conclusion
A new class of RIPK2 kinase/NOD cell signaling inhibitors based on a 3,5-diphenyl-2-aminopyridine scaffold was developed. Several co-crystal structures of RIPK2•inhibitor complexes were analyzed to provide insights into inhibitor selectivity versus a related kinase (e.g. ALK2). The inhibitors occupy space further into the hydrophobic binding pocket of RIPK2 altering the position of the DFG motif. The SAR studies also revealed that achieving potent NOD cell signaling inhibition depends on three structural features of the inhibitors: anchoring to the hinge and DFG via the 2-aminopyridine and 3-phenylsulfonamide, respectively, as well as appropriate occupancy of the region between the gatekeeper and the αC-helix provided by substituents in the 4- and 5-positions of the 3-phenylsulfonamide. For example, a representative compound from the series 18t demonstrated both potent inhibition of recombinant RIPK2 kinase (IC50 = 16 ± 5 nM), > 20-fold selectivity versus ALK2 and NOD cell signaling inhibition (IC50 = 26 ± 4 nM). In addition, this compound was found to have in vitro ADME and pharmacokinetic characteristics further supporting the use of 3,5-diphenyl-2-aminopyridines (e.g. 18t – CSLP37 and 18ae – CSLP58) as the basis for generating in vivo pharmacology probes of RIPK2 kinase and NOD cell signaling functions.
5. Experimental section
Unless otherwise stated, all reagents and solvents were obtained from commercial sources and used directly without further purification. All reactions involving air-sensitive reagents were carried out with magnetic stirring and in oven-dried glassware with rubber septa under argon unless otherwise noted. Reactions were monitored by thin-layer chromatography on Baker-flex® silica gel plates (IB2-F) using UV-light (254 and 365 nm) detection or visualizing agent (phosphomolybdic acid stain). Flash chromatography was performed on silica gel (230–400 mesh) utilizing Teledyne ISCO CombiFlash® Rf. The NMR spectra were recorded at 25 °C using a JEOL ECA (1H NMR at 400, 500, or 600 MHz, and 13C NMR at 100, 125, or 150 MHz). Chemical shifts (δ) are given in parts per million (ppm) with reference to solvent signals [1H-NMR: CDCl3 (7.26 ppm), CD3OD (3.30 ppm), DMSO-d6 (2.49 ppm); 13C-NMR: CDCl3 (77.0 ppm), CD3OD (49.0 ppm), DMSO-d6 (39.5 ppm)]. Coupling constants (J) are given in Hz. The C-F coupling patterns labeled in 13C-NMR indicate visible patterns in spectra. High resolution mass spectra (HRMS) were carried out using AccuTOF by the Department of Chemistry, The University of Texas at Austin. The spectra were measured using TOF-MS with an ESI ionization source and reported as m/z (relative intensity) for the molecular ion [M]. All tested compounds had a purity ≥95% as determined by high-performance liquid chromatography (HPLC) analyses using a Waters 1525 instrument equipped with a quaternary pump and a Proteo-C12 column (250 mm × 1 mm, 4 μm). UV absorption was monitored at λ = 220 nm. HPLC gradient went from 0% (method A) or 2% (method B) MeCN in H2O to 90% MeCN in H2O (both solvents contain 0.1% trifluoroacetic acid) with a total run time of 30 min and a flow rate of 0.5 mL/min.
General Procedure for the Preparation of N-Bromophenyl)sulfonamides (Method A); N-(4-bromo-2-methoxyphenyl)propane-1-sulfonamide (13a).
To a solution of 4-bromo-2-methoxyaniline (11a) (300 mg, 1.48 mmol) in anhydrous CH2Cl2 (15 mL) was added pyridine (0.24 mL, 2.97 mmol) and 1-propanesulfonyl chloride (0.18 mL, 1.63 mmol) under an argon atmosphere. The mixture was stirred at room temperature for 16 h. After being quenched with 1 N HCl(aq) (1.0 mL), water and CH2Cl2 were added and then the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13a (490.1 mg, 99%) as a light yellow oil; 1H NMR (CDCl3, 500 MHz) 7.41 (1 H, d, J = 8.6 Hz), 7.08 (1 H, dd, J = 8.3, 2.3 Hz), 7.03 (1 H, d, J = 2.5 Hz), 6.74 (1 H, br), 3.88 (3 H, s), 3.02–2.98 (2 H, m), 1.83–1.79 (2 H, m), 1.00 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 149.5, 125.5, 124.2, 121.0, 117.5, 114.3, 56.1, 53.1, 17.1, 12.8.
N-(3-Bromo-5-methoxyphenyl)methanesulfonamide (13b).
Prepared from 11b and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 13b (82%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 6.96–6.95 (2 H, m), 6.86 (1 H, s), 6.75–6.74 (1 H, m), 3.79 (3 H, s), 3.05 (3 H, s); 13C NMR (CDCl3, 100 MHz) 161.1, 137.8, 123.5, 115.1, 113.8, 105.0, 55.7, 39.5.
N-(3-Bromo-5-methoxyphenyl)ethanesulfonamide (13c).
Prepared from 11b and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 13c (88%) as a yellow oil; 1H NMR (CDCl3, 400 MHz) 7.34 (1 H, br), 6.96 (1 H, t, J = 1.8 Hz), 6.82 (1 H, t, J = 1.8 Hz), 6.75 (1 H, t, J = 1.8 Hz), 3.78 (3 H, s), 3.18 (2 H, q, J = 7.3 Hz), 1.36 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 161.0, 139.0, 123.4, 114.7, 113.5, 104.5, 55.6, 46.0, 8.1.
N-(3-Bromo-5-methoxyphenyl)propane-1-sulfonamide (13d).
Prepared from 11b and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 13d (86%) as a yellow oil; 1H NMR (CDCl3, 400 MHz) 7.34 (1 H, br), 6.96 (1 H, t, J = 1.8 Hz), 6.82 (1 H, t, J = 1.8 Hz), 6.75 (1 H, d, J = 2.3 Hz), 3.78 (3 H, s), 3.13–3.09 (2 H, m), 1.90–1.79 (2 H, m), 1.02 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 161.0, 139.0, 123.4, 114.6, 113.4, 104.5, 55.6, 53.3, 17.1, 12.8.
N-(5-Bromo-3-fluoro-2-methoxyphenyl)propane-1-sulfonamide (13j).
Prepared from 11e and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13j (76%) as a pale yellow solid; 1H NMR (CDCl3, 400 MHz) 7.49 (1 H, s), 7.04–7.00 (2 H, m), 3.99 (3 H, d, J = 2.3 Hz), 3.12–3.08 (2 H, m), 1.89–1.80 (2 H, m), 1.04 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 154.6 (d, JCF = 251.4 Hz), 135.7 (d, JCF = 12.7 Hz), 132.1 (d, JCF = 5.9 Hz), 116.7 (d, JCF = 2.9 Hz), 115.7 (d, JCF = 22.5 Hz), 115.4 (d, JCF = 10.8 Hz), 61.6 (d, JCF = 6.8 Hz), 53.6, 17.2, 12.8.
N-(5-Bromo-2-methoxy-3-methylphenyl)propane-1-sulfonamide (13k).
Prepared from 11g and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13k (75%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 7.51 (1 H, d, J = 2.3 Hz), 7.13 (1 H, br), 7.04 (1 H, d, J = 1.7 Hz), 3.74 (3 H, s), 3.12–3.09 (2 H, m), 2.26 (3 H, s), 1.86–1.81 (2 H, m), 1.02 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 146.2, 133.1, 131.8, 128.9, 118.4, 117.2, 60.6, 53.5, 17.1, 15.9, 12.7.
General Procedure for the Preparation of N-(Bromophenyl)sulfonamides (Method B); N-(3-bromo-5-methoxyphenyl)propane-2-sulfonamide (13e).
To a solution of 3-bromo-5-methoxyaniline (11b) (150.0 mg, 0.74 mmol) and a catalytic amount of DMAP in anhydrous CH2Cl2 (8 mL) was added pyridine (0.08 mL, 0.55 mmol) and 2-propanesulfonyl chloride (0.09 mL, 0.82 mmol) under an argon atmosphere. The mixture was stirred at room temperature for 40 h. After being quenched with 1 N HCl(aq) (0.5 mL) and water, CH2Cl2 were added, and the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 13e (67.8 mg, 30%) as a yellow oil; 1H NMR (CDCl3, 600 MHz) 7.12 (1 H, br), 6.96 (1 H, t, J = 2.1 Hz), 6.81 (1 H, t, J = 2.1 Hz), 6.76 (1 H, t, J = 2.1 Hz), 3.78 (3 H, s), 3.35 (1 H, sep, J = 6.9 Hz), 1.40 (6 H, d, J = 6.9 Hz); 13C NMR (CDCl3, 150 MHz) 161.0, 139.4, 123.4, 114.6, 113.3, 104.5, 55.6, 52.8, 16.4.
N-(3-Bromo-5-methoxyphenyl)benzenesulfonamide (13f).
Prepared from 11b and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13f (79%) as a light brown solid; 1H NMR (CDCl3, 400 MHz) 7.83 (2 H, d, J = 7.3 Hz), 7.57 (1 H, t, J = 7.8 Hz), 7.48 (2 H, t, J = 7.8 Hz), 7.14 (1 H, br), 6.81 (1 H, s), 6.77 (1 H, s), 6.64 (1 H, t, J = 1.8 Hz), 3.71 (3 H, s); 13C NMR (CDCl3, 100 MHz) 160.7, 138.5, 138.5, 133.4, 129.2, 127.2, 123.0, 115.9, 113.9, 105.5, 55.6.
N-(5-Bromo-2-methoxyphenyl)propane-1-sulfonamide (13g).
Prepared from 11c and the mixture was stirred at room temperature for 48 h. The reaction was purified by column chromatography on silica gel (EtOAc/hexane, 20:80 to 30:70) to give 13g (85%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 7.65 (1 H, d, J = 2.3 Hz), 7.18 (1 H, dd, J = 8.6, 2.3 Hz), 6.86 (1 H, br), 6.76 (1 H, d, J = 8.6 Hz), 3.86 (3 H, s), 3.05–3.02 (2 H, m), 1.83–1.79 (2 H, m), 1.00 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 147.7, 127.6, 127.3, 122.0, 113.3, 112.0, 56.0, 53.2, 17.1, 12.7.
N-(5-Bromo-2-methylphenyl)propane-1-sulfonamide (13h).
Prepared from 11d and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13h (96%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.64 (1 H, d, J = 2.3 Hz), 7.22 (1 H, dd, J = 8.0, 1.7 Hz), 7.07 (1 H, d, J = 8.0 Hz), 6.42 (1 H, br), 3.13–3.10 (2 H, m), 2.25 (3 H, s), 1.90–1.82 (2 H, m), 1.05 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 136.3, 132.3, 128.2, 127.7, 123.8, 120.3, 54.0, 17.6, 17.2, 12.9.
N-(5-Bromo-3-methoxy-2-methylphenyl)propane-1-sulfonamide (13i).
Prepared from 11e and purified by column chromatography on silica gel (EtOAc/hexane, 10:90) to give 13i (78%) as a yellow oil; 1H NMR (CDCl3, 400 MHz) 7.05 (1 H, d, J = 2.8 Hz), 6.97 (1 H, d, J = 2.8 Hz), 6.71 (1 H, br), 3.76 (3 H, s), 3.09–3.05 (2 H, m), 2.33 (3 H, s), 1.86–1.81 (2 H, m), 1.02 (3 H, t, J = 7.8 Hz); 13C NMR (CDCl3, 100 MHz) 158.2, 136.2, 125.9, 121.6, 115.2, 107.9, 55.6, 53.8, 17.2, 17.1, 12.8.
N-(5-Bromo-2-methoxybenzyl)propane-1-sulfonamide (13l).
Prepared from 11h and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13l (57%) as a clear oil; 1H NMR (CDCl3, 500 MHz) 7.39–7.38 (2 H, m), 6.77–675 (1 H, m), 5.06 (1 H, br), 4.22 (2 H, d, J = 6.3 Hz), 3.83 (3 H, s), 2.84–2.81 (2 H, m), 1.72–1.65 (2 H, m), 0.92 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 156.4, 132.1, 132.0, 127.4, 112.7, 112.0, 55.6, 54.8, 43.0, 17.2, 12.8.
N-(3-Bromo-5-ethoxyphenyl)propane-2-sulfonamide (13o).
Prepared from 11i and purified by column chromatography on silica gel (EtOAc/hexane, 5:95 to 10:90) to give 13o (58%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 7.21 (1 H, br), 6.95 (1 H, s), 6.79 (1 H, s), 6.76 (1 H, s), 3.98 (2 H, q, J = 7.4 Hz), 3.35 (1 H, sep, J = 7.4 Hz), 1.40–1.37 (9 H, m); 13C NMR (CDCl3, 125 MHz) 160.4, 139.3, 123.3, 114.4, 113.7, 104.9, 64.0, 52.7, 16.4, 14.6.
Preparation of 6-Bromo-1-(propylsulfonyl)indoline (13m) (Method C).
To a solution of 6-bromoindoline (12a) (100.0 mg, 0.51 mmol) and a catalytic amount of DMAP in pyridine (2 mL) was added 1-propanesulfonyl chloride (0.09 mL, 0.76 mmol) under an argon atmosphere. The mixture was stirred at 50 °C for 3 h. After being quenched with 1 N HCl(aq) (1.0 mL) and water, EtOAc were added, and the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13m (102.1 mg, 66%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.50 (1 H, d, J = 1.7 Hz), 7.10 (1 H, dd, J = 8.0, 1.7 Hz), 7.03 (1 H, d, J = 8.0 Hz), 4.02 (2 H, t, J = 8.6 Hz), 3.08 (2 H, t, J = 8.0 Hz), 3.04–3.00 (2 H, m), 1.92–1.84 (2 H, m), 1.05 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 143.4, 130.0, 126.4, 126.1, 121.2, 116.4, 51.0, 50.6, 27.5, 16.7, 13.0.
Preparation of 6-Bromo-4-(propylsulfonyl)-3,4-dihydro-2H-benzo[b][1,4]oxazine (13n) (Method D).
To a solution of 6-bromo-3,4-dihydro-2H-benzo[b][1,4]oxazine (12b) (44.0 mg, 0.22 mmol) in pyridine (1 mL) was added 1-propanesulfonyl chloride (0.04 mL, 0.33 mmol) under an argon atmosphere. The mixture was stirred at 50 °C for 2.5 h. After being quenched with 1 N HCl(aq) (1.0 mL) and water, EtOAc were added, and the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 13n (49.6 mg, 70%) as a brown solid; 1H NMR (CDCl3, 500 MHz) 7.76 (1 H, d, J = 2.3 Hz), 7.13 (1 H, dd, J = 8.6, 2.3 Hz), 6.79 (1 H, d, J = 8.6 Hz), 4.26 (2 H, t, J = 4.6 Hz), 3.85 (2 H, t, J = 4.6 Hz), 3.09–3.06 (2 H, m), 1.92–1.85 (2 H, m), 1.06 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 145.0, 128.1, 125.6, 124.3, 119.2, 113.0, 64.7, 54.0, 44.0, 17.0, 12.9.
General Procedure for the Preparation of N-(Bromophenyl)sulfonamides (Method E); N-(3-bromo-5-isopropoxyphenyl)propane-2-sulfonamide (13p).
To a solution of 3-bromo-5-isopropoxyaniline (11j) (108.5 mg, 0.47 mmol) in pyridine (2.5 mL) was added 2-propanesulfonyl chloride (0.08 mL, 0.71 mmol) under an argon atmosphere. The mixture was stirred at room temperature for 24 h. After being quenched with 1 N HCl(aq) (1.0 mL), water and EtOAc were added, the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13p (73.1 mg, 46%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 7.31 (1 H, s), 6.94 (1 H, t, J = 1.7 Hz), 6.78 (1 H, t, J = 1.7 Hz), 6.75 (1 H, t, J = 1.7 Hz), 4.49 (1 H, sep, J = 5.7 Hz), 3.35 (1 H, sep, J = 6.9 Hz), 1.39 (6 H, d, J = 6.9 Hz), 1.31 (6 H, d, J = 5.7 Hz); 13C NMR (CDCl3, 125 MHz) 159.4, 139.4, 123.3, 114.9, 114.3, 105.9, 70.5, 52.7, 21.8, 16.4.
N-(5-Bromo-3-chloro-2-methoxyphenyl)propane-1-sulfonamide (13q).
Prepared from 11k and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13q (58%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 7.61 (1 H, d, J = 2.3 Hz), 7.25 (1 H, d, J = 2.3 Hz), 7.10 (1 H, br), 3.90 (3 H, s), 3.14–3.11 (2 H, m), 1.88–1.83 (2 H, m), 1.04 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 144.1, 133.2, 128.4, 127.8, 119.4, 117.4, 61.2, 53.9, 17.2, 12.8.
N-(5-Bromo-2,3-dimethoxyphenyl)propane-1-su1fonamide (13r).
Prepared from 11l and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 13r (85%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 7.33 (1 H, d, J = 1.8 Hz), 7.01 (1 H, br), 6.80 (1 H, d, J =2.3 Hz), 3.86 (6 H, s), 3.10–3.06 (2 H, m), 1.86–1.80 (2 H, m), 1.02 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 152.8, 136.6, 131.9, 116.8, 113.3, 111.3, 60.9, 56.0, 53.3, 17.1, 12.7.
N-(7-Bromo-2,3-dihydrobenzo[b][1,4]dioxin-5-yl)propane-1-sulfonamide (13s).
Prepared from 11m and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 13s (80%) as a yellow oil; 1H NMR (CDCl3, 400 MHz) 7.25 (1 H, d, J = 2.3 Hz), 6.82 (1 H, d, J = 1.8 Hz), 6.67 (1 H br), 4.31–4.29 (2 H , m), 4.28–4.26 (2 H, m), 3.10–3.06 (2 H, m), 1.88–1.79 (2 H, m), 1.03 (3 H, t, J = 7.8 Hz); 13C NMR (CDCl3, 100 MHz) 144.2, 132.3, 127.3, 116.0, 114.3, 113.2, 64.6, 64.2, 53.4, 17.2, 12.8.
N-(5-Bromo-3-ethoxy-2-methoxyphenyl)propane-1-su1fonamide (13t).
Prepared from 11n and purified by column chromatography on silica gel (EtOAc/hexane, 15:85) to give 13t (87%) as a pale yellow solid; 1H NMR (CDCl3, 400 MHz) 7.22 (1 H, s), 6.98 (1 H, s), 6.51 (1 H, s), 4.04 (2 H, q, J = 7.3 Hz), 3.84 (3 H, s), 3.01–2.97 (2 H, m), 1.88–1.78 (2 H, m), 1.44 (3 H, t, J = 6.9 Hz), 0.99 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 149.3, 146.8, 127.7, 115.8, 107.6, 105.9, 64.8, 56.1, 53.6, 17.1, 14.6, 12.9.
N-(5-Bromo-2-hydroxyphenyl)propane-1-su1fonamide (13u).
N-(5-Bromo-2-((tert-butyldimethylsilyl)oxy)phenyl)propane-1-sulfonamide was prepared from 11o and purified by column chromatography on silica gel (EtOAc/hexane, 5:95 to 10:90) as a brown oil (86%); 1H NMR (CDCl3, 400 MHz) 7.65 (1 H, d, J = 2.3 Hz), 7.08 (1 H, dd, J = 8.5, 2.3 Hz), 6.72 (1 H, d, J = 8.2 Hz), 6.75 (1 H, br), 3.08–3.04 (2 H, m), 1.86–1.77 (2 H, m), 1.03–0.99 (12 H, m), 0.27 (6 H, s); 13C NMR (CDCl3, 100 MHz) 143.5, 129.8, 127.0, 121.4, 119.1, 114.0, 53.3, 25.6, 18.1, 17.2, 12.7, −4.3. To a solution of N-(5-bromo-2-((tert-butyldimethylsilyl)oxy)phenyl)propane-1-sulfonamide (35.0 mg, 0.09 mmol) in anhydrous THF (1 mL) were added 1 M tetrabutylammonium fluoride in THF (0.17 mL, 0.17 mmol) at 0 °C. The mixture was stirred at 0 °C for 1 h. After being quenched with saturated aqueous NH4Cl (5 mL), EtOAc were added, and the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 30:70) to give 13u (23.6 mg, 93%) as a brown oil; 1H NMR (CDCl3, 500 MHz) 7.46 (1 H, d, J = 2.3 Hz), 7.21 (1 H, dd, J = 8.6, 2.3 Hz), 6.82 (1 H, d, J = 8.6 Hz), 6.47 (1 H, br), 3.09–3.06 (2 H, m), 1.90–1.86 (2 H, m), 1.05 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 100 MHz) 147.1, 129.3, 125.3, 125.2, 117.6, 112.6, 53.1, 17.0, 12.8.
General Procedure for the Preparation of N-((4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)sulfonamides (Method F); N-(2-methoxy-4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14a).
To a mixture of 13a (50.0 mg, 0.16 mmol), bis(pinacolato)diboron (49.4 mg, 0.20 mmol), KOAc (47.7 mg, 0.49 mmol) and PdCl2(dppf) (3.6 mg, 0.005 mmol) was added anhydrous THF (1.5 mL) under argon. The reaction was stirred at room temperature for 10 min then was refluxed for 16 h. After being quenched by the addition of water, the aqueous layer was extracted with EtOAc (2 × 15 mL). The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 20:80) to afford 14a (46.6 mg, 81%) as a white solid; ; 1H NMR (CDCl3, 400 MHz) 7.53 (1 H, d, J = 7.8 Hz), 7.41 (1 H, d, J = 7.8 Hz), 7.29 (1 H, s), 6.99 (1 H, br), 3.91 (3 H, s), 3.04–3.00 (2 H, m), 1.82–1.74 (2 H, m), 1.33 (12 H, s), 0.97 (1 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 147.6, 129.2, 128.3, 117.8, 116.0, 83.9, 55.8, 52.9, 24.8, 17.1, 12.8.
N-(3-Methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)methane sulfonamide (14b).
Prepared from 13b and purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 50:50) to give 14b (90%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.37 (1 H, br), 7.14 (1 H, d, J = 1.7 Hz), 7.10 (1 H, d, J = 2.9 Hz), 6.99, (1 H, t, J = 2.3 Hz), 3.78 (3 H, s), 2.98 (3 H, s), 1.30 (12 H, s); 13C NMR (CDCl3, 125 MHz) 159.9, 137.6, 118.8, 115.8, 110.0, 84.0, 55.3, 39.0, 24.6.
N-(3-Methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)ethanesulfonamide (14c).
Prepared from 13c and purified by column chromatography on silica gel (EtOAc/hexane, 15:85 to 20:80) to give 14c (74%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 7.10–7.09 (2 H, m), 7.01 (1 H, t, J = 2.3 Hz), 6.93 (1 H, br), 7.02 (1 H, s), 3.81 (3 H, s), 3.13 (2 H, q, J = 7.4 Hz), 1.36–1.32 (15 H, m); 13C NMR (CDCl3, 125 MHz) 160.1, 137.6, 118.5, 115.5, 109.6, 84.1, 55.4, 45.8, 24.8, 8.2.
N-(3-Methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14d).
Prepared from 13d and purified by column chromatography on silica gel (EtOAc/hexane, 15:85) to give 14d (78%) as a pale yellow solid; 1H NMR (CDCl3, 400 MHz) 7.11–7.10 (1 H, m), 7.06 (1 H, s), 7.02 (1 H, s), 6.68 (1 H, br), 3.82 (3 H, s), 3.09–3.05 (2 H, m), 1.89–1.79 (2 H, m), 1.33 (12 H, s), 1.00 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 160.1, 137.6, 118.4, 115.5, 109.5, 84.1, 55.5, 53.2, 24.8, 17.2, 12.8.
N-(3-Methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-2-sulfonamide (14e).
Prepared from 13e and purified by column chromatography on silica gel (EtOAc/hexane, 15:85 to 20:80) to give 14e (69%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 7.09–7.07 (2 H, m), 7.06–7.05 (1 H, m), 6.76 (1 H, br), 3.81 (3 H, s), 3.32 (1 H, sep, J = 6.9 Hz), 1.38 (6 H, d, J = 6.9 Hz), 1.33 (12 H, s); 13C NMR (CDCl3, 125 MHz) 160.1, 137.9, 118.2, 115.2, 109.4, 84.1, 55.4, 52.4, 24.8, 16.5.
N-(3-Methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)benzenesulfonamide (14f).
Prepared from 13f and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 14f (86%) as a white solid; 1H NMR (CDCl3, 400 MHz) 7.79 (2 H, d, J = 7.3 Hz), 7.51 (1 H, t, J = 7.3 Hz), 7.42 (2 H, t, J = 7.3 Hz), 7.04 (1 H, s), 6.93–6.92 (3 H, m), 3.75 (3 H, s), 1.29 (12 H, s); 13C NMR (CDCl3, 100 MHz) 159.8, 138.9, 137.1, 133.0, 129.0, 127.2, 119.5, 115.8, 110.2, 84.0, 55.4, 24.8.
N-(2-Methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14g).
Prepared from 13g and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 14g (82%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.90 (1 H, s), 7.58 (1 H, d, J = 8.0 Hz), 6.89 (1 H, d, J = 8.0 Hz), 6.72 (1 H, br), 3.90 (3 H, s), 3.04–3.03 (2 H, t, J = 8.0 Hz), 1.82 (2 H, sex, J = 7.4 Hz), 1.32 (12 H, s), 0.99 (3 H, t, J = 6.7 Hz); 13C NMR (CDCl3, 125 MHz) 151.6, 132.4, 126.4, 125.7, 109.9, 83.8, 55.8, 53.0, 24.8, 17.2, 12.8.
N-(2-Methyl-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14h).
Prepared from 13h and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 14h (78%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.77 (1 H, s), 7.56 (1 H, d, J = 7.4 Hz), 7.23 (1 H, d, J = 7.4 Hz), 6.43 (1 H, br), 3.13–3.10 (2 H, m), 2.36 (3 H, s), 1.88 (2 H, sex, J = 8.0 Hz), 1.32 (12 H, s), 1.04 (3 H, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 135.0, 134.4, 132.6, 130.7, 129.4, 83.9, 53.9, 24.8, 18.5, 17.3, 12.9.
N-(3-Methoxy-2-methyl-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14i).
Prepared from 13i and purified by column chromatography on silica gel (EtOAc/hexane, 10:90) to give 14i (54%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.20 (1 d, J = 2.7 Hz), 7.14 (1 H, d, J = 2.7 Hz), 6.31 (1 H, br), 3.80 (3 H, s), 3.06–3.03 (2 H, m), 2.42 (3 H, s), 1.87–1.79 (2 H, m), 1.34 (12 H, s), 1.01 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 157.7, 135.9, 126.4, 117.5, 110.1, 83.9, 55.4, 53.6, 24.8, 17.2, 15.5, 12.9.
N-(3-Fluoro-2-methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14j).
Prepared from 13j and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 14j (68%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.67 (1 H, s), 7.29 (1 H, dd, J = 12.0, 1.2 Hz), 6.90 (1 H, s), 4.03 (3 H, d, J = 2.9 Hz), 3.10–3.07 (2 H, m), 1.87–1.80 (2 H, m), 1.31 (12 H, s), 1.02 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 153.8 (d, JCF = 248.6 Hz), 139.2 (d, JCF = 12.3 Hz), 130.4 (d, JCF = 3.7 Hz), 120.6 (d, JCF = 2.5 Hz), 118.7 (d, JCF = 17.2 Hz), 84.2, 61.4 (d, JCF = 8.6 Hz), 53.3, 24.8, 17.2, 12.8.
N-(2-Methoxy-3-methyl-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14k).
Prepared from 13k and purified by column chromatography on silica gel (EtOAc/hexane, 15:85 to 20:80) to give 14k (24%) as a pale yellow solid; 1H NMR (CDCl3, 400 MHz) 7.73 (1 H, s), 7.39 (1 H, s), 6.89 (1 H, s), 3.77 (3 H, s), 3.15–3.11 (2 H, m), 2.30 (3 H, s), 1.90–1.80 (2 H, m), 1.00 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 150.0, 133.5, 130.4, 130.1, 122.2, 83.9, 60.5, 53.4, 24.8, 17.2, 16.0, 12.8.
N-(2-Methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)benzyl)propane-1-sulfonamide (14l).
Prepared from 13l and purified by column chromatography on silica gel (EtOAc/hexane, 2.5:97.5) to give 14l (71%) as a clear oil; 1H NMR (CDCl3, 500 MHz) 7.77 (1 H, dd, J = 8.0, 1.7 Hz), 7.68 (1 H, d, J = 1.7 Hz), 6.89 (1 H, d, J = 8.6 Hz), 4.28 (2 H, d, J = 6.9 Hz), 3.88 (3 H, s), 2.82–2.79 (2 H, m), 1.68–1.60 (2 H, m), 1.33 (12 H, s), 0.87 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 160.0, 136.8, 136.2, 124.4, 109.8, 83.7, 55.4, 54.6, 43.7, 24.8, 12.8.
1-(Propylsulfonyl)-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)indoline (14m).
Prepared from 13m and purified by column chromatography on silica gel (EtOAc/hexane, 10:90) to give 14m (70%) as a yellow oil; 1H NMR (CDCl3, 500 MHz) 7.74 (1 H, s), 7.46 (1 H, d, J = 7.4 Hz), 7.20 (1 H, d, J = 7.4 Hz), 4.02 (2 H, t, J = 8.6 Hz), 3.14 (2 H, t, J = 8.6 Hz), 3.05–3.02 (2 H, m), 1.92–1.84 (2 H, m), 1.32 (12 H, s), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 141.7, 134.5, 130.2, 124.8, 118.9, 83.8, 50.9, 50.2, 28.2, 24.8, 16.7, 13.1.
4-(Propylsulfonyl)-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-3,4-dihydro-2H-benzo[b][1,4]oxazine (14n).
Prepared from 13n and purified by column chromatography on silica gel (EtOAc/hexane, 10:90) to give 14n (74%) as a yellow oil; 1H NMR (CDCl3, 500 MHz) 7.95 (1 H, d, J = 1.2 Hz), 7.48 (1 H, dd, J = 8.3, 1.2 Hz), 6.90 (1 H, d, J = 8.6 Hz), 4.30 (2 H, t, J = 4.6 Hz), 3.86 (2 H, t, J = 4.6 Hz), 3.13–3.09 (2 H, m), 1.93–1.85 (2 H, m), 1.31 (12 H, s), 1.05 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 148.7, 132.2, 129.0, 124.0, 117.2, 83.7, 65.1, 54.4, 44.0, 24.8, 17.0, 13.0.
N-(3-Ethoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-2-sulfonamide (14o).
Prepared from 13o and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to afford 14o (63%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.09–7.08 (1 H, m), 7.05–7.04 (2 H, m), 6.63 (1 H, br), 4.04 (2 H, q, J = 6.9 Hz), 3.32 (1 H, sep, J = 6.9 Hz), 1.40–1.37 (9 H, m), 1.32 (12 H, s); 13C NMR (CDCl3, 125 MHz) 159.5, 137.8, 118.2, 116.0, 109.9, 84.0, 63.6, 52.3, 24.8, 16.5, 14.7.
N-(3-Isopropoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-2-sulfonamide (14p).
Prepared from 13p and purified by column chromatography on silica gel (EtOAc/hexane, 2.5:97.5) to give 14p (34%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 7.09 (1 H, d, J = 2.3 Hz), 7.04 (1 H, t, J = 2.3 Hz), 7.00 (1 H, d, J = 1.7 Hz), 6.04 (1 H, s), 4.59 (1 H, sep, J = 5.7 Hz), 3.32 (1 H, sep, J = 6.9 Hz), 1.38 (6 H, d, J = 6.9 Hz), 1.32–1.31 (18 H, m); 13C NMR (CDCl3, 125 MHz) 158.4, 137.8, 118.0, 117.7, 111.0, 84.0, 70.0, 52.3, 24.8, 22.0, 16.5.
N-(3-Chloro-2-methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14q).
Prepared from 13q and purified by column chromatography on silica gel (EtOAc/hexane, 2.5:97.5) to give 14q (46%) as a white solid; 1H NMR (CDCl3, 500 MHz) 7.79 (1 H, d, J = 1.2 Hz), 7.56 (1 H, d, J = 1.2 Hz), 6.96 (1 H, br), 3.92 (3 H, s), 3.15–3.12 (2 H, m), 1.88–1.83 (2 H, m), 1.32 (12 H, s), 1.03 (3 H, t, J = 8.0 Hz); 13C NMR (CDCl3, 125 MHz) 147.6, 132.1, 131.8, 127.1, 122.9, 84.3, 61.1, 53.7, 24.8, 17.2, 12.8
N-(2,3-Dimethoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14r).
Prepared from 13r and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 14r (25%) as a clear oil; 1H NMR (CDCl3, 400 MHz) 7.56 (1 H, d, J = 0.9 Hz), 7.12 (1 H, d, J = 1.4 Hz), 6.92 (1 H, br), 3.91 (3 H, s), 3.90 (3 H, s), 3.11–3.07 (2 H, m), 1.88–1.78 (2 H, m), 1.32 (12 H, s), 1.00 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 151.6, 140.3, 130.5, 117.3, 113.9, 84.0, 60.9, 55.9, 53.2, 24.8, 17.2, 12.8.
N-(7-(4,4,5,5-Tetramethyl-1,3,2-dioxaborolan-2-yl)-2,3-dihydrobenzo[b][1,4]dioxin-5-yl)propane-1-sulfonamide (14s).
Prepared from 13s and purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 25:75) to give 14s (56%) as a white solid; 1H NMR (CDCl3, 600 MHz) 7.48 (1 H, s), 7.13 (1 H, s), 6.57 (1 H, s), 4.33–4.32 (2 H, m), 4.26–4.24 (2 H, m), 3.08–3.06 (2 H, m), 1.84 (2 H, sex, J = 8.2 Hz), 1.30 (12 H, s), 1.01 (3 H, t, J = 7.6 Hz); 13C NMR (CDCl3, 150 MHz) 143.1, 136.3, 125.8, 119.9, 118.7, 83.8, 64.9, 63.9, 53.2, 24.8, 17.2, 12.8.
N-(2-Hydroxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14u).
Prepared from 13u and purified by column chromatography on silica gel (EtOAc/hexane, 20:80) to give 14u (73%) as a clear oil; 1H NMR (CDCl3, 500 MHz) 7.67 (1 H, d, J = 1.2 Hz), 7.56 (1 H, dd, J = 8.0, 1.2 Hz), 6.94 (1 H, d, J = 8.0 Hz), 6.60 (1 H, s), 3.06–3.03 (2 H, m), 1.90–1.83 (2 H, m), 1.32 (12 H, s), 1.00 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 152.6, 134.6, 131.0, 123.0, 116.4, 83.9, 52.5, 24.8, 17.0, 12.8.
2-(3-Fluoro-4,5-dimethoxyphenyl)-4,4,5,5-tetramethyl-1,3,2-dioxaborolane (14v).
Prepared from 5-bromo-1-fluoro-2,3-dimethoxybenzene (13v) and purified by column chromatography on silica gel (EtOAc/hexane, 5:95) to give 14v (66%) as a clear oil; 1H NMR (CDCl3, 400 MHz) 7.16 (1 H, dd, J = 10.8, 1.4 Hz), 7.09 (1 H, s), 3.94 (3 H, d, J = 1.4 Hz), 3.90 (3 H, s), 1.32 (12 H, s); 13C NMR (CDCl3, 100 MHz) 155.4 (d, JCF = 245.5 Hz), 153.0 (d, JCF = 4.9 Hz), 139.6 (d, JCF = 12.7 Hz), 115.4 (d, JCF = 18.6 Hz), 113.2 (d, JCF = 2.0 Hz), 84.0, 61.3 (d, JCF = 4.9 Hz), 56.3, 24.8.
Preparation of N-(3-Ethoxy-2-methoxy-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)propane-1-sulfonamide (14t) (Method G).
To a mixture of 13t (60.0 mg, 0.17 mmol), bis(pinacolato)diboron (64.9 mg, 0.26 mmol), KOAc (50.0 mg, 0.51 mmol) and PdCl2(dppf) (12.4 mg, 0.017 mmol) was added anhydrous 1,4-dioxane (0.9 mL) under argon. The reaction was put into a preheated oil bath (80 °C) and stirred for 40 h. After being quenched by the addition of water, the aqueous layer was extracted with EtOAc (2 × 15 mL). The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 10:90) to afford 14t (19.8 mg, 29%) as a yellow oil; 1H NMR (CDCl3, 500 MHz) 8.24 (1 H, s), 7.26 (1 H, s), 7.18 (1 H, s), 4.10 (2 H, q, J = 6.9 Hz), 3.89 (3 H, s), 2.98–2.95 (2 H, m), 1.82-1.74 (2 H, m), 1.45 (3 H, t, J = 6.9 Hz), 1.34 (12 H, s), 0.95 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 153.0, 144.7, 138.7, 119.0, 103.4, 84.4, 64.5, 55.9, 52.5, 24.8. 17.0, 14.8, 12.9.
General Procedure for the Preparation of 3-Bromo-5-phenylpyridines; N-(4-(5-bromo-2-chloropyridin-3-yl)-2-methoxyphenyl)propane-1-sulfonamide (16a).
To a mixture of 14a (46.6 mg, 0.13 mmol), 15a (41.7 mg, 0.13 mmol) and Pd(PPh3)4 (3.1 mg, 0.002 mmol) was added anhydrous acetonitrile (1 mL) and anhydrous DMF (0.5 mL) under argon. The reaction was stirred at room temperature for 10 min then 1 M Na2CO3 (0.3 mL, 0.30 mmol) was added. The mixture was put into a preheated oil bath (90 °C) and stirred for 16 h. After being quenched by the addition of water, the aqueous layer was extracted with EtOAc (2 × 10 mL). The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to afford 16a (31.5 mg, 57%) as a white solid; 1H NMR (CDCl3, 500 MHz) 8.44 (1 H, d, J = 2.3 Hz), 7.81 (1 H, d, J = 2.3 Hz), 7.61 (1 H, d, J = 8.0 Hz), 7.02–6.99 (2 H, m), 6.94 (1 H, br), 3.92 (3 H, s), 3.11–3.08 (2 H, m), 1.89–1.84 (2 H, m), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 149.1, 148.2, 148.1, 141.7, 137.6, 132.4, 127.1, 122.3, 119.0, 118.8, 111.6, 56.0, 53.5, 17.2, 12.8.
N-(3-(5-Bromo-2-chloropyridin-3-yl)-5-methoxyphenyl)methanesulfonamide (16b).
Prepared from 14b and 15a, and then purified by column chromatography on silica gel (EtOAc/hexane, 20:80 to 30:70) to give 16b (88%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 8.45 (1 H, s), 7.80 (1 H, s), 7.45 (1 H, s), 6.88 (2 H, s), 6.74 (1 H, s), 3.83 (3 H, s), 3.08 (3 H, s); 13C NMR (CDCl3, 100 MHz) 165.0, 149.5, 147.9, 141.7, 138.4, 138.2, 137.4, 119.1, 112.9, 111.6, 106.2, 55.6, 39.4.
N-(3-(2-Amino-5-bromopyridin-3-yl)-5-methoxyphenyl)methanesulfonamide (16c).
Prepared from 14b and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 20:80 to 60:40) to give 16c (38%) as a yellow oil; 1H NMR (CDCl3, 400 MHz) 8.08 (1 H, s), 7.44 (1 H, d, J = 1.8 Hz), 6.88–6.87 (1 H, m), 6.85 (1 H, s), 6.72 (1 H, s), 4.79 (2 H, br), 3.81 (3 H, s), 3.05 (3 H, s); 13C NMR (CDCl3, 100 MHz) 161.1, 154.4, 147.8, 139.7, 139.2, 139.0, 122.6, 112.3, 110.5, 108.2, 105.7, 55.6, 39.7.
N-(3-(5-Bromo-2-chloropyridin-3-yl)-5-methoxyphenyl)ethanesulfonamide (16d).
Prepared from 14c and 15a, and then purified by column chromatography on silica gel (EtOAc/hexane, 15:85 to 25:75) to give 16d (81%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 8.47 (1 H, d, J = 2.3 Hz), 7.80 (1 H, d, J = 2.9 Hz), 7.00 (1 H, br), 6.87 (1 H, t, J = 1.7 Hz), 6.86 (1 H, t, J = 1.7 Hz), 6.72 (1 H, t, J = 2.3 Hz), 3.84 (3 H, s), 3.20 (2 H, q, J = 7.4 Hz), 1.40 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 160.6, 149.5, 148.0, 141.7, 138.5, 138.3, 137.4, 119.1, 112.6, 111.3, 105.9, 55.6, 46.1, 8.3.
N-(3-(5-Bromo-2-chloropyridin-3-yl)-5-methoxyphenyl)propane-1-sulfonamide (16e).
Prepared from 14d and 15a, and then purified by column chromatography on silica gel (EtOAc/hexane, 20:80) to give 16e (96%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.46 (1 H, d, J = 2.3 Hz), 7.80 (1 H, d, J = 2.9 Hz), 7.28 (1 H, s), 6.87 (1 H, t, J = 1.7 Hz), 6.86 (1 H, t, J = 1.7 Hz), 6.72 (1 H, t, J = 1.7 Hz), 3.84 (3 H, s), 3.16–3.13 (2 H, m), 1.91–1.82 (2 H, m), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 160.5, 149.5, 148.0, 141.7, 138.4, 138.3, 137.4, 119.1, 112.6, 111.3, 105.8, 55.6, 53.4, 17.2, 12.8.
N-(3-(2-Amino-5-bromopyridin-3-yl)-5-methoxyphenyl)propane-1-sulfonamide (16f).
Prepared from 14d and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 45:55) to give 16f (65%) as a pale yellow solid; 1H NMR (CD3OD, 500 MHz) 7.98 (1 H, d, J = 2.3 Hz), 7.50 (1 H, d, J = 2.3 Hz), 6.87 (1 H, t, J = 1.7 Hz), 6.84 (1 H, t, J = 1.7 Hz), 6.74 (1 H, t, J = 1.7 Hz), 3.82 (3 H, s), 3.13–3.10 (2 H, m), 1.81 (2 H, sex, J = 7.4 Hz), 1.02 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 162.6, 148.2, 141.6, 141.0, 140.2, 124.7, 112.9, 110.6, 108.1, 107.9, 106.4, 56.0, 54.0, 18.4, 13.1.
N-(3-(5-Bromo-2-chloropyridin-3-yl)-5-methoxyphenyl)propane-2-sulfonamide (16g).
Prepared from 14e and 15a, and then purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 20:80) to give 16g (92%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.46 (1 H, d, J = 2.9 Hz), 7.80 (1 H, d, J = 2.3 Hz), 7.19 (1 H, br), 6.90 (1 H, t, J = 2.3 Hz), 6.88 (1 H, t, J = 1.7 Hz), 6.70 (1 H, t, J = 1.7 Hz), 3.84 (3 H, s), 3.38 (1 H, sep, J = 6.9 Hz), 1.42 (6 H, d, J = 6.9 Hz); 13C NMR (CDCl3, 125 MHz) 160.5, 149.5, 148.0, 141.7, 138.6, 138.4, 137.5, 119.0, 112.5, 111.1, 105.8, 55.6, 52.8, 16.5.
N-(3-(2-Amino-5-bromopyridin-3-yl)-5-methoxyphenyl)propane-2-sulfonamide (16h).
Prepared from 14e and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 45:55) to give 16h (88%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 8.08 (1 H, d, J = 1.7 Hz), 7.76 (1 H, br), 7.45 (1 H, d, J = 2.3 Hz), 6.85 (1 H, t, J = 1.7 Hz), 6.84 (1 H, t, J = 1.7 Hz), 6.69 (1 H, t, J = 2.3 Hz), 4.82 (2 H, br), 3.82 (3 H, s), 3.36 (1 H, sep, J = 6.9 Hz), 1.41 (6 H, d, J = 6.9 Hz); 13C NMR (CDCl3, 125 MHz) 161.2, 154.5, 147.8, 139.7, 139.5, 138.9, 122.7, 111.9, 110.1, 108.1, 105.5, 55.6, 53.0, 16.5.
N-(3-(2-Amino-5-bromopyridin-3-yl)-5-methoxyphenyl)benzenesulfonamide (16i).
Prepared from 14f and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 40:60) to give 16i (85%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 7.98 (1 H, s), 7.79 (2 H, d, J = 8.0 Hz), 7.53 (1 H, t, J = 7.4 Hz), 7.43 (2 H, t, J = 7.4 Hz), 7.30 (1 H, d, J = 2.3 Hz), 6.72 (1 H, s), 6.62 (1 H, s), 6.58 (1 H, s), 3.73 (3 H, s); 13C NMR (CDCl3, 125 MHz) 160.8, 154.3, 147.3, 139.8, 139.0, 139.0, 138.2, 133.0, 129.0, 127.1, 122.8, 112.5, 110.4, 107.9, 106.2, 55.4.
N-(5-(2-Amino-5-bromopyridin-3-yl)-2-methoxyphenyl)propane-1-sulfonamide (16j).
Prepared from 14g and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 50:50) to give 16j (92%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 8.08 (1 H, s), 7.59 (1 H, d, J = 1.7 Hz), 7.44 (1 H, d, J = 2.3 Hz), 7.17 (2 H, dd, J = 8.6, 2.3 Hz), 6.99 (1 H, d, J = 8.6 Hz), 6.89 (1 H, s), 4.65 (2 H, br), 3.94 (3 H, s), 3.08–3.04 (2 H, m), 1.90–1.83 (2 H, m), 1.03 (3 H, t, J = 6.9 Hz); 13C NMR (CDCl3, 125 MHz) 154.6, 148.8, 147.6, 139.7, 132.0, 129.8, 128.4, 127.0, 125.1, 119.9, 111.3, 56.0, 53.8, 17.2, 12.9.
N-(5-(2-Amino-5-bromopyridin-3-yl)-2-methylphenyl)propane-1-sulfonamide (16k).
Prepared from 14h and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 50:50) to give 16k (82%) as a pale yellow solid; 1H NMR (CDCl3, 400 MHz) 8.06 (1 H, d, J = 2.3 Hz), 7.51 (1 H, d, J = 1.4 Hz), 7.46 (1 H, d, J = 2.3 Hz), 7.31 (1 H, d, J = 7.8 Hz), 7.17 (1 H, dd, J = 7.8, 1.4 Hz), 6.80 (1 H, br), 4.75 (2 H, br), 3.15–3.11 (2 H, m), 2.36 (3 H, s), 1.90 (2 H, sex, J = 7.8 Hz), 1.06 (3 H, t, J = 7.8 Hz); 13C NMR (CDCl3, 100 MHz) 154.6, 147.7, 139.7, 135.7, 135.7, 132.0, 129.9, 125.6, 122.4, 122.1, 108.3, 54.7, 17.8, 17.3, 12.9.
N-(5-(2-Amino-5-bromopyridin-3-yl)-3-methoxy-2-methylphenyl)propane-1-sulfonamide (16l).
Prepared from 14i and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 40:60) to give 16l (81%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.10 (1 H, d, J = 2.3 Hz), 7.37 (1 H, d, J = 2.8 Hz), 7.13 (1 H, d, J = 2.8 Hz), 6.80 (1 H, br), 6.57 (1 H, d, J = 2.3 Hz), 4.48 (2 H, br), 3.79 (3 H, s), 3.16–3.12 (2 H, m), 2.02 (3 H, s), 1.93–1.84 (2 H, m), 1.06 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 125 MHz) 158.5, 154.5, 148.0, 139.7, 137.7, 137.0, 122.6, 119.8, 112.0, 107.9, 107.8, 55.5, 54.1, 17.3, 13.8, 12.9.
N-(5-(2-Amino-5-bromopyridin-3-yl)-3-fluoro-2-methoxyphenyl)propane-1-sulfonamide (16m).
Prepared from 14j and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 50:50) to give 16m (83%) as a pale yellow solid; 1H NMR (CDCl3, 600 MHz) 8.09 (1 H, d, J = 1.4 Hz), 7.43 (1 H, d, J = 1.4 Hz), 7.37 (1 H, s), 7.24 (1 H, s), 6.96 (1 H, d, J = 11.7 Hz), 4.75 (2 H, s), 4.06 (3 H, d, J = 2.1 Hz), 3.12–3.10 (2 H, m), 1.87 (2 H, sex, J = 7.6 Hz), 1.05 (3 H, t, J = 6.9 Hz); 13C NMR (CDCl3, 150 MHz) 154.5 (d, JCF = 248.4 Hz), 154.4, 148.2, 139.7, 136.6 (d, JCF = 11.8 Hz), 132.2 (d, JCF = 8.9 Hz), 131.7 (d, JCF = 5.9 Hz), 121.2, 114.6, 112.7 (d, JCF = 20.7 Hz), 108.3, 61.6 (d, JCF = 7.4 Hz), 54.2, 17.2, 12.8.
N-(5-(2-Amino-5-bromopyridin-3-yl)-2-methoxy-3-methylphenyl)propane-1-sulfonamide (16n).
Prepared from 14k and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 40:60) to give 16n (87%) as a light brown solid; 1H NMR (CDCl3, 500 MHz) 8.08 (1 H, d, J = 2.3 Hz), 7.44 (1 H, d, J = 2.3 Hz), 7.37 (1 H, d, J = 1.7 Hz), 7.04 (1 H, br), 6.97 (1 H, d, J = 1.2 Hz), 4.72 (2 H, br), 3.82 (3 H, s), 3.15–3.12 (2 H, m), 2.35 (3 H, s), 1.92–1.86 (2 H, m), 1.05 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 154.5, 147.7, 147.2, 139.7, 133.1, 132.3, 131.3, 126.6, 122.5, 116.0, 108.3, 60.7, 54.2, 17.3, 16.3, 12.9.
N-(5-(2-Amino-5-bromopyridin-3-yl)-2-methoxybenzyl)propane-1-sulfonamide (16o).
Prepared from 14l and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 50:50) to give 16o (78%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 7.98 (1 H, s), 7.39–7.33 (3 H, m), 6.96 (1 H, d, J = 8.6 Hz), 5.45 (1 H, t, J = 6.3 Hz), 4.72 (2 H, br), 4.29 (2 H, d, J = 6.3 Hz), 3.90 (3 H, s), 2.91–2.88 (2 H, m), 1.79–1.72 (2 H, m), 0.96 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 157.2, 154.8, 147.2, 139.6, 129.7, 129.6, 128.9, 126.3, 122.7, 111.0, 108.2, 55.6, 54.8, 43.2, 17.3, 12.9.
5-Bromo-3-(1-(propylsulfonyl)indolin-6-yl)pyridin-2-amine (16p).
Prepared from 14m and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 40:60) to give 16p (70%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 8.07 (1 H, d, J = 2.3 Hz), 7.43 (1 H, d, J = 2.3 Hz), 7.39 (1 H, d, J = 1.2 Hz), 7.27 (1 H, d, J = 8.6 Hz), 7.04 (1 H, dd, J = 7.4, 1.2 Hz), 4.67 (2 H, br), 4.08 (2 H, t, J = 8.6 Hz), 3.18 (2 H, t, J = 8.6 Hz), 3.06–3.03 (2 H, m), 1.93–1.85 (2 H, m), 1.04 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 154.5, 147.8, 143.0, 139.7, 136.6, 131.2, 126.0, 123.4, 123.0, 113.2, 108.2, 51.4, 50.4, 27.8, 16.7, 13.0.
5-Bromo-3-(4-(propylsulfonyl)-3,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)pyridin-2-amine (16q).
Prepared from 14n and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 40:60) to give 16q (85%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 8.07 (1 H, s), 7.60 (1 H, d, J = 1.8 Hz), 7.45 (1 H, d, J = 1.4 Hz), 7.08 (1 H, d, J = 8.7, 1.8 Hz), 7.00 (1 H, d, J = 8.7 Hz), 7.24 (1 H, s), 4.72 (2 H, br), 4.33 (2 H, t, J = 4.1 Hz), 3.87 (2 H, t, J = 4.6 Hz), 3.15–3.11 (2 H, m), 1.91 (2 H, sex, J = 7.8 Hz), 1.07 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 154.7, 147.5, 146.0, 139.6, 129.2, 125.6, 124.6, 122.3, 118.6, 65.0, 54.8, 44.0, 17.1, 12.9.
N-(3-(2-Amino-5-bromopyridin-3-yl)-5-ethoxyphenyl)propane-2-sulfonamide (16r).
Prepared from 14o and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 35:65) to afford 16r (85%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 8.07 (1 H, d, J = 2.3 Hz), 7.80 (1 H, br), 7.45 (1 H, d, J = 2.3 Hz), 6.85 (1 H, s), 6.82 (1 H, br), 6.68(1 H, s), 4.84 (2 H, br), 4.03 (2 H, q, J = 6.9 Hz), 3.36 (1 H, sep, J = 6.9 Hz), 1.43–1.40 (9 H, m); 13C NMR (CDCl3, 100 MHz) 160.5, 154.5, 147.6, 139.7, 139.4, 138.8, 122.8, 111.7, 110.6, 108.1, 105.9, 63.9, 52.9, 24.8, 16.5, 14.6.
N-(3-(2-Amino-5-bromopyridin-3-yl)-5-isopropoxyphenyl)propane-2-sulfonamide (16s).
Prepared from 14p and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 50:50) to give 16s (81%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.08 (1 H, s), 7.68 (1 H, s), 7.45 (1 H, d, J = 2.3 Hz), 6.84 (1 H, t, J = 2.3 Hz), 6.80 (1 H, s), 6.68 (1 H, s), 4.84 (2 H, br), 4.56 (1 H, sep, J = 5.7 Hz), 3.36 (1 H, sep, J = 6.9 Hz), 1.41 (6 H, d, J = 6.9 Hz), 1.34 (6 H, d, J = 6.3 Hz); 13C NMR (CDCl3, 125 MHz) 159.5, 154.5, 147.6, 139.7, 139.5, 138.9, 122.8, 111.7, 111.6, 108.1, 107.0, 70.3, 52.9, 21.9, 16.5.
N-(5-(2-Amino-5-bromopyridin-3-yl)-3-chloro-2-methoxyphenyl)propane-1-sulfonamide (16t).
Prepared from 14q and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70) to give 16t (78%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.11 (1 H, d, J = 2.3 Hz), 7.50 (1 H, d, J = 1.7 Hz), 7.44 (1 H, d, J = 2.3 Hz), 7.19 (1 H, d, J = 2.3 Hz), 7.13 (1 H, br), 4.68 (2 H, br), 3.97 (3 H, s), 3.16–3.13 (2 H, m), 1.93–1.86 (2 H, m), 1.06 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 154.3, 148.4, 144.9, 130.7, 134.0, 132.9, 128.3, 125.5, 121.0, 116.9, 108.4, 61.3, 54.5, 17.3, 12.8.
N-(5-(2-Amino-5-bromopyridin-3-yl)-2,3-dimethoxyphenyl)propane-1-sulfonamide (16u).
Prepared from 14r and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70) to give 16u (69%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.09 (1 H, d, J = 2.3 Hz), 7.46 (1 H, d, J = 2.3 Hz), 7.20 (1 H, d, J = 1.7 Hz), 7.12 (1 H, br), 6.72 (1 H, d, J = 2.3 Hz), 4.72 (2 H, br), 3.93 (3 H, s), 3.90 (3 H, s), 3.12–3.09 (2 H, m), 1.90–1.83 (2 H, m), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 154.5, 152.8, 147.8, 139.6, 137.5, 132.9, 131.6, 122.6, 110.8, 108.2, 61.0, 56.0, 54.0, 17.2, 12.8.
N-(5-(2-Amino-5-bromopyridin-3-yl)-3-ethoxy-2-methoxyphenyl)propane-1-sulfonamide (16v).
Prepared from 14t and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 40:60) to give 16v (89%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 8.19 (1 H, d, J = 2.3 Hz), 7.50 (1 H, d, J = 2.3 Hz), 7.21 (1 H, s), 6.88 (1 H, s), 6.70 (1 H, s), 4.52 (2 H, br), 4.14–4.02 (2 H, m), 3.92 (3 H, s), 2.76–2.69 (1 H, m), 2.60–2.53 (1 H, m), 1.60–1.51 (1 H, m), 1.47 (3 H, t, J = 6.9 Hz), 1.29–1.20 (1 H, m), 0.84 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 154.2, 150.2, 148.8, 147.3, 140.8, 127.2, 121.5, 121.0, 113.8, 110.0, 109.5, 64.8, 56.2, 53.8, 17.1, 14.7, 12.9.
N-(5-(2-Amino-5-bromopyridin-3-yl)-2-hydroxyphenyl)propane-1-sulfonamide (16w).
Prepared from 14u and 15b, and then purified by column chromatography on silica gel (MeOH/CH2Cl2, 1:99 to 3:97) to give 16w (61%) as a brown solid; 1H NMR (CD3OD, 500 MHz) 7.93 (1 H, s), 7.46 (1 H, d, J = 2.3 Hz), 7.43 (1 H, d, J = 1.7 Hz), 7.11 (1 H, dd, J = 7.7, 2.9 Hz), 6.97 (1 H, d, J = 8.6 Hz), 3.07–3.04 (2 H, m), 1.86 (2 H, sex, J = 8.0 Hz), 1.00 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 156.9, 151.4, 147.4, 141.0, 129.2, 127.8, 126.6, 126.2, 124.8, 117.1, 108.2, 25.0, 18.3, 13.2.
5-Bromo-3-(3-fluoro-4,5-dimethoxyphenyl)pyridin-2-amine (16x).
Prepared from 14v and 15b, and then purified by column chromatography on silica gel (EtOAc/hexane, 30:70) to give 16x (39%) as a white solid; 1H NMR (CDCl3, 500 MHz) 8.09 (1 H, d, J = 2.3 Hz), 7.45 (1 H, d, J = 2.3 Hz), 6.81 (1 H, dd, J = 10.9, 2.3 Hz), 6.73 (1 H, s), 4.68 (2 H, br), 3.97 (3 H, s), 3.90 (3 H, s); 13C NMR (CDCl3, 125 MHz) 156.0 (d, JCF = 246.1 Hz), 154.4, 154.1 (d, JCF = 6.2 Hz), 147.9, 139.6, 137.0 (d, JCF = 13.5 Hz), 131.7 (d, JCF = 9.8 Hz), 122.2, 109.3 (d, JCF = 20.9 Hz), 108.3, 108.0 (d, JCF = 2.5 Hz), 61.5 (d, JCF = 3.7 Hz), 56.4.
N-(5-(5-Bromopyridin-3-yl)-3-fluoro-2-methoxyphenyl)propane-1-sulfonamide (16y).
Prepared from 14j and 15c, and then purified by column chromatography on silica gel (EtOAc/hexane, 0:100 to 25:75) to give 16y (89%) as a pale yellow solid; 1H NMR (CDCl3, 600 MHz) 8.69 (1 H, s), 8.67 (1 H, s), 7.96 (1 H, s), 7.54 (1 H, s), 7.09–7.05 (2 H, m), 4.08 (3 H, d, J = 2.4 Hz), 3.11 (2 H, t, J = 8.1 Hz), 1.95–1.84 (2 H, sex, J = 7.8 Hz), 1.06–1.03 (3 H, t, J = 7.5 Hz); 13C NMR (CDCl3, 150 MHz) 153.7 (d, JCF = 247.1 Hz), 149.9, 146.1, 136.9 (d, JCF = 19.5 Hz), 136.7, 136.3, 132.0 (d, JCF = 9.0 Hz), 131.7 (d, JCF = 6.0 Hz), 121.0, 112.8, 111.2 (d, JCF = 19.5 Hz), 61.6 (d, JCF = 7.2 Hz), 53.7, 17.3, 12.9.
N-(5-(5-Bromo-2-chloropyridin-3-yl)-3-fluoro-2-methoxyphenyl)propane-1-sulfonamide (16z).
Prepared from 14j and 15a, and then purified by column chromatography on silica gel (EtOAc/hexane, 10:90 to 15:85) to give 16z (85%) as a yellow oil; 1H NMR (CDCl3, 600 MHz) 8.46 (1 H, d, J = 2.1 Hz), 7.78 (1 H, d, J = 2.1 Hz), 7.41 (1 H, s), 7.06 (1 H, s), 6.96 (1 H, dd, J = 12.4, 1.4 Hz), 4.09 (3 H, d, J = 2.1 Hz), 3.13 (2 H, t, J = 7.6 Hz), 1.87 (2 H, sex, J = 7.6 Hz), 1.04 (3 H, t, J = 7.6 Hz); 13C NMR (CDCl3, 150 MHz) 153.8 (d, JCF = 246.9 Hz), 149.6, 148.0, 141.6, 136.8 (d, JCF = 11.8 Hz), 136.4, 131.4 (d, JCF = 8.9 Hz), 131.1 (d, JCF = 4.4 Hz), 119.1, 114.8, 113.3 (d, JCF = 22.2 Hz), 108.3, 61.6 (d, JCF = 7.4 Hz), 53.7, 17.2, 12.8.
General Procedure for the Preparation of tert-Butyl 4-(4-(6-chloro-5-phenylpyridin-3-yl)phenyl)piperazine-1-carboxylate; tert-butyl 4-(4-(6-chloro-5-(3-methoxy-4-(propylsulfonamido)phenyl)pyridin-3-yl)phenyl)piperazine-1-carboxylate (17a).
To a mixture of 16a (31.0 mg, 0.07 mmol), 4-(4-tert-butoxycarbonylpiperazinyl)phenylboronic acid pinacol ester (31.6 mg, 0.08 mmol) and Pd(PPh3)4 (12.8 mg, 0.01 mmol) was added anhydrous DME (2 mL) under argon. The reaction was stirred at room temperature for 10 min then 1 M Na2CO3 (0.15 mL, 0.15 mmol) was added. The mixture was refluxed for 16 h. After being quenched by the addition of water, the aqueous layer was extracted with EtOAc (2 × 15 mL). The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 20:80 to 30:70) to afford 17a (35.6 mg, 80%) as a white solid; 1H NMR (CDCl3, 500 MHz) 8.56 (1 H, d, J = 2.3 Hz), 7.80 (1 H, d, J = 2.3 Hz), 7.62 (1 H, d, J = 8.6 Hz), 7.51 (2 d, J = 8.6 Hz), 7.07–7.05 (2 H, m), 7.00 (2 H, d, J = 9.2 Hz), 6.93 (1 H, s), 3.92 (3 H, s), 3.61–3.59 (4 H, m), 3.22–3.21 (4 H, m), 3.12–3.08 (2 H, m), 1.92–1.84 (2 H, m), 1.48 (9 H, s), 1.04 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 154.6, 151.3, 148.2, 147.2, 145.9, 137.1, 135.8, 135.5, 134.0, 127.7, 127.0, 126.5, 122.3, 119.1, 116.2, 111.9, 80.0, 56.0, 53.4, 48.6, 28.3, 17.2, 12.8.
tert-Butyl 4-(4-(6-chloro-5-(3-methoxy-5-(methylsulfonamido)phenyl)pyridin-3-yl)phenyl)piperazine-1-carboxylate (17b).
Prepared from 16b and purified by column chromatography on silica gel (EtOAc/hexane, 20:80 to 60:40) to give 17b (83%) as a white solid; 1H NMR (CDCl3, 400 MHz) 8.56 (1 H, d, J = 2.3 Hz), 7.79 (1 H, d, J = 2.3 Hz), 7.53 (1 H, br), 7.49 (2 H, d, J = 8.7 Hz), 6.98 (2 H, d, J = 8.7 Hz), 6.95–6.94 (1 H, m), 6.91 (1 H, t, J = 2.3 Hz), 6.81–6.80 (1 H, m), 3.84 (3 H, s), 3.60–3.58 (4 H, m), 3.21–3.19 (4 H, m), 3.08 (3 H, s), 1.48 (9 H, s); 13C NMR (CDCl3, 100 MHz) 160.4, 154.7, 151.3, 147.0, 146.2, 139.8, 138.2, 135.7, 135.6, 127.8, 126.8, 116.5, 113.2, 111.7, 105.8, 80.1, 55.6, 48.6, 39.4, 28.4.
tert-Butyl 4-(4-(6-chloro-5-(3-(ethylsulfonamido)-5-methoxyphenyl)pyridin-3-yl)phenyl)piperazine-1-carboxylate (17c).
Prepared from 16d and purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 45:55) to give 17c (99%) as a white solid; 1H NMR (CDCl3, 600 MHz) 8.57 (1 H, d, J = 2.1 Hz), 7.79 (1 H, d, J = 2.1 Hz), 7.49 (2 H, d, J = 8.9 Hz), 7.47 (1 H, s), 6.99 (2 H, d, J = 8.9 Hz), 6.94 (1 H, s), 6.91 (1 H, s), 6.78 (1 H, s), 3.84 (3 H, s), 3.59–3.59 (4 H, m), 3.21–3.19 (6 H, m), 1.48 (9 H, s), 1.39 (3 H, t, J = 7.6 Hz); 13C NMR (CDCl3, 150 MHz) 160.4, 154.6, 151.3, 147.0, 146.2, 139.8, 138.3, 137.0, 135.7, 135.5, 127.8, 126.9, 116.5, 113.0, 111.4, 105.5, 80.0, 55.6, 55.5, 48.6, 45.9, 28.4, 8.22.
tert-Butyl 4-(4-(6-chloro-5-(3-methoxy-5-(propylsulfonamido)phenyl)pyridin-3-yl)phenyl)piperazine-1-carboxylate (17d).
Prepared from 16e and purified by column chromatography on silica gel (EtOAc/hexane, 20:80 to 40:60) to give 17d (95%) as a pale yellow solid; 1H NMR (CDCl3, 500 MHz) 8.57 (1 H, d, J = 2.9 Hz), 7.79 (1 H, d, J = 2.9 Hz), 7.49 (2 H, d, J = 8.6 Hz), 7.47 (1 H, br), 6.99 (2 H, d, J = 8.6 Hz), 6.93 (1 H, s), 6.90 (1 H, t, J = 1.7 Hz), 6.78–6.77 (1 H, m), 3.84 (3 H, s), 3.60–3.58 (4 H, m), 3.21–3.19 (4 H, m), 3.16–3.13 (2 H, m), 1.91–1.84 (2 H, m), 1.48 (9 H, s), 1.02 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 160.4, 154.7, 151.3, 147.0, 146.2, 139.8, 138.3, 137.0, 135.7, 135.5, 127.8, 126.9, 116.5, 112.9, 111.3, 105.5, 80.1, 55.5, 53.3, 48.6, 28.4, 24.8, 17.2, 12.8.
tert-Butyl 4-(4-(6-ch\oro-5-(3-methoxy-5-(1-methylethylsulfonamido)phenyl)pyridin-3-yl)phenyl)piperazine-1-carboxylate (17e).
Prepared from 16g and purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 50:50) to give 17e (79%) as a white solid; 1H NMR (CDCl3, 500 MHz) 8.57 (1 H, d, J = 2.7 Hz), 7.79 (1 H, d, J = 2.3 Hz), 7.49 (2 H, d, J = 8.6 Hz), 7.37 (1 H, s), 6.99 (2 H, d, J = 8.6 Hz), 6.95 (1 H, s), 6.92 (1 H, t, J = 2.3 Hz), 6.76–6.75 (1 H, m), 3.83 (3 H, s), 3.60–3.58 (4 H, m), 3.39 (1 H, sep, J = 6.9 Hz), 3.21–3.20 (4 H, m), 1.48 (9 H, s), 1.42 (6 H, d, J = 6.9 Hz); 13C NMR (CDCl3, 125 MHz) 160.4, 154.6, 151.3, 147.0, 146.2, 139.7, 138.6, 137.0, 135.8, 135.5, 127.8, 126.9, 116.5, 112.9, 111.2, 105.4, 80.0, 55.5, 52.6, 48.6, 30.9, 28.4, 16.5.
tert-Butyl 4-(4-(6-chloro-5-(3-fluoro-4-methoxy-5-(propylsulfonamido)phenyl)pyridin-3-yl)phenyl)piperazine-1-carboxylate (17f).
Prepared from 16z and purified by column chromatography on silica gel (EtOAc/hexane, 20:80) to give 17f (69%) as a white solid; 1H NMR (CDCl3, 500 MHz) 8.57 (1 H, d, J = 2.3 Hz), 7.82 (1 H, d, J = 2.3 Hz), 7.51 (2 H, d, J = 8.6 Hz), 7.39 (1 H, t, J = 8.0 Hz), 7.07 (1 H, dd, J = 6.6, 1.7 Hz), 7.03, 7.02 (1 H, d, J = 2.3, 1.7 Hz), 7.01–6.99 (3 H, m), 3.85 (3 H, s), 3.61–3.59 (4 H, m), 3.21 (4 H, br), 1.49 (9 H, s); 13C NMR (CDCl3, 125 MHz) 154.6, 153.9 (d, JCF = 248.6 Hz), 151.4, 147.0, 146.4, 137.0, 136.4 (d, JCF = 12.3 Hz), 135.7, 134.6, 133.0 (d, JCF = 8.6 Hz), 131.0 (d, JCF = 4.9 Hz), 127.8, 126.9, 116.5, 114.8 (d, JCF = 2.5 Hz), 113.4 (d, JCF = 20.9 Hz), 80.0, 61.6 (d, JCF = 7.4 Hz), 53.6, 48.6, 28.4, 17.2, 12.9.
General Procedure for the Preparation of 1-(4-(6-methyl-5-phenylpyridin-3-yl)phenyl)piperazines; N-(2-methoxy-4-(2-methyl-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)phenyl)propane-1-sulfonamide (18a).
To a mixture of 17a (35.0 mg, 0.06 mmol), K2CO3 (16.0 mg, 0.12 mmol) and Pd(PPh3)4 (13.4 mg, 0.01 mmol) was added anhydrous 1,4-dioxane (1.5 mL) and trimethylboroxine (0.03 mL, 0.23 mmol) under argon. The reaction was stirred at room temperature for 10 min then the mixture was refluxed for 16 h. After being quenched by the addition of water, the aqueous layer was extracted with EtOAc (2 × 15 mL). The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 20:80 to 70:30) to afford the methylated pyridine product then anhydrous CH2Cl2 (3 mL) and trifluoroacetic acid (0.3 mL) were added. The mixture was stirred at room temperature for 16 h. After being quenched with saturated aqueous NaHCO3 (5 mL), CH2CL2 were added, and the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18a (24.7 mg, 89%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 8.70 (1 H, d, J = 1.8 Hz), 7.67 (1 H, d, J = 1.8 Hz), 7.60 (1 H, d, J = 8.2 Hz), 7.52 (2 H, d, J = 8.7 Hz), 7.00 (2 H, d, J = 8.7 Hz), 6.96 (2 H, dd, J = 8.2, 1.8 Hz), 6.88 (1 H, d, J = 1.4 Hz), 3.90 (3 H, s), 3.24–3.22 (4 H, m), 3.11–3.07 (5 H, m), 2.53 (3 H, s), 1.88 (2 H, sex, J = 7.8 Hz), 1.04 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 153.5, 151.3, 148.6, 145.8, 136.8, 136.0, 134.8, 133.7, 128.3, 127.6, 125.7, 122.0, 119.6, 116.2, 111.5 55.9, 53.3, 49.6, 45.7, 23.0, 17.2, 12.9; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H33N4O3S 481.2268; found 481.2274; Purity (method A) >99%, tR = 14.8 min.
N-(3-Methoxy-5-(2-methyl-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)phenyl)methanesulfonamide (18b).
Prepared from 17b and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 20:80) to give 18b (81%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.71 (1 H, d, J = 2.3 Hz), 7.65 (1 H, s), 7.50 (2 H, d, J = 8.6 Hz), 6.99 (2 H, d, J = 8.6 Hz), 6.86 (1 H, s), 6.77 (1 H, s), 6.70 (1 H, s), 3.84 (3 H, s), 3.24–3.22 (4 H, m), 3.09–3.07 (7 H, m), 2.52 (3 H, s); 13C NMR (CDCl3, 125 MHz) 160.6, 153.3, 151.4, 146.0, 142.6, 138.2, 135.9, 134.6, 133.7, 128.2, 127.6, 116.2, 113.0, 111.5, 105.1, 55.6, 49.6, 45.8, 39.6, 22.9; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C24H29N4O2S 453.1955; found 453.1963; Purity (method A) 98%, tR = 13.2 min.
N-(3-Methoxy-5-(2-methyl-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)phenyl)ethanesulfonamide (18c).
Prepared from 17c and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 20:80) to give 18c (62%) as a yellow solid; 1H NMR (CDCl3, 400 MHz) 8.70 (1 H, d, J = 1.8 Hz), 7.64 (1 H, d, J = 1.8 Hz), 7.51 (2 H, d, J = 8.7 Hz), 7.00 (2 H, d, J = 8.7 Hz), 6.86 (1 H, s), 6.77 (1 H, s), 6.68 (1 H, s), 3.84 (3 H, s), 3.31–3.28 (4 H, m), 3.22–3.16 (6 H, m), 2.52 (3 H, s), 1.40 (3 H, t, J = 7.3 Hz); 13C NMR (CDCl3, 100 MHz) 160.6, 153.4, 151.0, 146.1, 142.5, 138.2, 135.9, 134.7, 133.7, 128.7, 127.7, 116.5, 112.8, 111.2, 104.8, 55.6, 49.0, 46.1, 45.3, 22.9, 8.3; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H31N4O3S 467.2111; found 467.2116; Purity (method A) 98%, tR = 13.9 min.
N-(3-Methoxy-5-(2-methyl-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)phenyl)propane-1-sulfonamide (18d).
Prepared from 17d and purified by column chromatography on silica gel (MeOH/CH2Cl2, 10:90 to 20:80) to give 18d (65%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.71 (1 H, d, J = 2.3 Hz), 7.65 (1 H, d, J = 2.3 Hz), 7.50 (2 H, d, J = 8.6 Hz), 7.00 (2 H, d, J = 8.6 Hz), 6.85 (1 H, t, J = 2.3 Hz), 6.75 (1 H, t, J = 1.7 Hz), 6.68 (1 H, t, J = 2.3 Hz), 3.84 (3 H, s), 3.23–3.21 (4 H, m), 3.15–3.12 (2 H, m), 3.08–3.06 (4 H, m), 2.52 (3 H, s), 1.92–1.84 (2 H, m), 1.04 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 160.6, 153.3, 151.4, 146.0, 142.6, 138.2, 135.9, 134.6, 133.8, 128.2, 127.6, 116.2, 112.8, 111.2, 105.8, 55.5, 53.5, 49.8, 45.9, 22.9, 17.3, 12.9; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H33N4O3S 481.2268; found 481.2274; Purity (method A) 98%, tR = 15.0 min.
N-(3-Methoxy-5-(2-methyl-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)phenyl)propane-2-sulfonamide (18e).
Prepared from 17e and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18e (96%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.62 (1 H, d, J = 2.3 Hz), 7.81 (1 H, d, J = 2.3 Hz), 7.58 (2 H, d, J = 8.6 Hz), 7.08 (2 H, d, J = 9.2 Hz), 6.91 (1 H, t, J = 1.7 Hz), 6.84, (1 H, t, J = 1.7 Hz), 6.70 (1 H, t, J = 1.2 Hz), 3.82 (3 H, s), 3.35–3.29 (1 H, m), 3.23–3.21 (4 H, m), 3.02–3.00 (4 H, m), 2.48 (3 H, s), 1.35 (6 H, d, J = 6.9 Hz); 13C NMR (CD3OD, 125 MHz) 162.2, 154.1, 153.0, 145.9, 143.0, 141.2, 138.5, 136.3, 135.9, 129.1, 128.6, 117.6, 113.4, 111.2, 105.6, 56.0, 53.5, 50.3, 46.3, 22.4, 16.7; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H33N4O3S 481.2268; found 481.2273; Purity (method B) 98%, tR = 15.8 min.
N-(3-Fluoro-2-methoxy-5-(2-methyl-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)phenyl)propane-1-sulfonamide (18aa).
Prepared from 17f and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 10:90) to give 18aa (90%) as a yellow solid; 1H NMR (CDCl3, 600 MHz) 8.71 (1 H, d, J = 2.1 Hz), 7.63 (1 H, d, J = 2.1 Hz), 7.50 (2 H, d, J = 8.9 Hz), 7.34 (1 H, s), 7.01 (2 H, d, J = 8.2 Hz), 6.88 (1 H, dd, J = 11.7, 1.4 Hz), 4.08 (3 H, d, J = 1.4 Hz), 3.23–3.22 (4 H, m), 3.12–3.10 (2 H, m), 3.08–3.06 (4 H, m), 2.53 (3 H, s), 1.87 (2 H, sex, J = 7.6 Hz), 1.04 (3 H, t, J = 7.6 Hz); 13C NMR (CDCl3, 150 MHz) 154.1 (d, JCF = 246.9 Hz), 153.3, 151.5, 146.2 (d, JCF = 3.0 Hz), 135.9 (d, JCF = 10.4 Hz), 135.8 (d, JCF = 8.9 Hz), 134.8, 134.6, 133.9, 131.0 (d, JCF = 5.9 Hz), 128.1, 127.6, 116.2, 114.8, 113.1 (d, JCF = 20.7 Hz), 61.6, 53.8, 49.8, 45.9, 23.0, 17.2, 12.9; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H32FN4O3S 499.2174; found 499.2181, Purity (method A) 97%, tR = 15.4 min.
General Procedure for the Preparation of 3-phenyl-5-(4-(piperazin-1-yl)phenyl)pyridin-2-amines; N-(3-(2-amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-5-methoxyphenyl)methanesulfonamide (18f).
To a mixture of 16c (26.0 mg, 0.07 mmol), 4-(4-tert-butoxycarbonylpiperazinyl)phenylboronic acid pinacol ester (29.8 mg, 0.08 mmol) and Pd(PPh3)4 (13.4 mg, 0.01 mmol) was added anhydrous DME (1.5 mL) under argon. The reaction was stirred at room temperature for 10 min then 1 M Na2CO3 (0.14 mL, 0.14 mmol) was added. The mixture was refluxed for 16 h. After being quenched by the addition of water, the aqueous layer was extracted with EtOAc (2 × 15 mL). The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered and concentrated. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 70:30) to afford the coupling product then anhydrous CH2Cl2 (3 mL), and trifluoroacetic acid (0.3 mL) were added. The mixture was stirred at room temperature for 16 h. After being quenched with saturated aqueous NaHCO3 (5 mL), CH2Cl2 were added, and the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (MeOH/CH2Cl2, 10:90 to 30:70) to give 18f (9.9 mg, 31%) as a yellow solid; m.p. 155 - 157°C; 1H NMR (CD3OD, 500 MHz) 8.15 (1 H, d, J = 2.3 Hz), 7.62 (1 H, d, J = 2.3 Hz), 7.48 (2 H, d, J = 8.6 Hz), 7.06 (2 H, d, J = 8.6 Hz), 6.92 (1 H, s), 6.89 (1 H, t, J = 1.7 Hz), 6.83 (1 H, s), 3.84 (3 H, s), 3.30–3.27 (4 H, m), 3.17–3.15 (4 H, m), 3.02 (3 H, s); 13C NMR (CD3OD, 125 MHz) 162.6, 156.4, 151.6, 144.9, 141.5, 141.4, 137.4, 131.2, 128.3, 127.8, 123.1, 118.1, 113.5, 111.2, 106.3, 66.9, 56.0, 45.7, 39.4; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C23H28N5O3S 454.1907; found 454.1913; Purity (method A) 98%, tR = 13.2 min.
N-(3-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-5-methoxyphenyl)propane-1-sulfonamide (18g).
Prepared from 16f and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18g (49%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.15 (1 H, d, J = 2.3 Hz), 7.62 (1 H, d, J = 2.3 Hz), 7.48 (2 H, d, J = 8.6 Hz), 7.06 (2 H, d, J = 8.6 Hz), 6.92 (1 H, t, J = 1.7 Hz), 6.87 (1 H, t, J = 1.7 Hz), 6.81 (1 H, t, J = 1.7 Hz), 3.83 (3 H, s), 3.33–3.29 (4 H, m), 3.22–3.20 (4 H, m), 3.14–3.10 (2 H, m), 1.82 (2 H, sex, J = 7.4 Hz), 1.02 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 161.2, 154.2, 150.8, 145.1, 140.5, 139.1, 136.0, 129.1, 127.7, 126.9, 120.9, 116.4, 112.3, 110.1, 105.1, 55.6, 53.7, 50.0, 45.9, 17.3, 12.9; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H32N5O3S 482.2220; found 482.2225; Purity (method A) 99%, tR = 14.6 min.
N-(3-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-5-methoxyphenyl)propane-2-sulfonamide (18h).
Prepared from 16g and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18h (31%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.14 (1 H, d, J = 1.7 Hz), 7.60 (1 H, d, J = 1.7 Hz), 7.45 (2 H, d, J = 8.6 Hz), 7.02 (2 H, d, J = 8.6 Hz), 6.93 (1 H, s), 6.89 (1 H, s), 6.78 (1 H, s), 3.82 (3 H, s), 3.37–3.30 (1 H, m), 3.21–3.19 (4 H, m), 3.06–3.04 (4 H, m), 1.35 (6 H, d, J = 6.9 Hz); 13C NMR (CD3OD, 125 MHz) 162.5, 156.3, 152.0, 144.9, 141.7, 141.4, 137.4, 130.8, 128.4, 127.7, 123.1, 118.0, 113.0, 110.7, 105.9, 55.9, 53.4, 50.3, 46.2, 16.7; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H32N5O3S 482.2220; found 482.2225; Purity (method A) >99%, tR = 14.4 min.
N-(3-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-5 methoxyphenyl)benzenesulfonamide (18i).
Prepared from 16i and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18i (24%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.23 (1 H, d, J = 1.7 Hz), 7.85 (2 H, d, J = 7.4 Hz), 7.55 (1 H, t, J = 7.4 Hz), 7.47 (2 H, t, J = 7.4 Hz), 7.43 (1 H, d, J = 2.3 Hz), 7.38 (2 H, d, J = 8.6 Hz), 6.95 (2 H, d, J = 8.6 Hz), 6.78 (1 H, t, J = 1.7 Hz), 6.74 (1 H, s), 6.69 (1 H, s), 4.55 (2 H, br), 3.78 (3 H, s), 3.19–3.17 (4 H, m), 3.08–3.06 (4 H, m); 13C NMR (CDCl3, 125 MHz) 160.9, 154.2, 150.8, 144.9, 140.0, 139.1, 138.6, 135.9, 133.1, 129.1, 129.0, 127.6, 127.3, 126.9, 120.8, 116.4, 113.4, 111.2, 106.4, 55.5, 50.0, 45.9; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C28H30N5O3S 516.2064; found 516.2069; Purity (method A) 97%, tR = 15.7 min.
N-(3-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-5-ethoxyphenyl)propane-2-sulfonamide (18j).
Prepared from 16r and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18j (37%) as a yellow solid; 1H NMR (CD3OD, 600 MHz) 8.14 (1 H, d, J = 2.1 Hz), 7.59 (1 H, d, J = 2.1 Hz), 7.44 (2 H, d, J = 8.9 Hz), 7.02 (2 H, d, J = 8.9 Hz), 6.92 (1 H, s), 6.88 (1 H, t, J = 2.1 Hz), 6.76 (1 H, s), 4.05 (2 H, q, J = 6.9 Hz), 3.35–3.32 (1 H, m), 3.24–3.22 (4 H, m), 3.11–3.09 (4 H, m), 1.39 (3 H, t, J = 6.9 Hz), 1.34 (6 H, d, J = 6.9 Hz); 13C NMR (CD3OD, 125 MHz) 161.8, 156.3, 151.8, 144.9, 141.7, 141.3, 137.4, 130.9, 128.3, 127.8, 123.2, 118.0, 112.9, 111.2, 106.4, 64.8, 53.4, 49.3, 45.9, 16.7, 15.1; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H34N5O3S 496.2377; found 496.2381; Purity (method B) >99%, tR = 16.5 min.
N-(3-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-5-isopropoxyphenyl)propane-2-sulfonamide (18k).
Prepared from 16s and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18k (37%) as a yellow solid; 1H NMR (CD3OD, 400 MHz) 8.21 (1 H, d, J = 2.3 Hz), 7.50–7.47 (3 H, m), 6.97 (2 H, d, J = 9.2 Hz), 6.88 (1 H, s), 6.77 (1 H, s), 6.71 (1 H, s), 5.64 (2 H, br), 4.61 (1 H, sep, J = 6.3 Hz), 3.39–3.33 (1 H, m), 3.12–3.10 (4 H, m), 2.93–2.91 (4 H, m), 1.28–1.24 (12 H, m); 13C NMR (CDCl3, 100 MHz) 159.3, 150.2, 144.5, 139.9, 139.6, 136.1, 129.6, 127.4, 126.9, 121.3, 116.7, 111.7, 111.6, 106.5, 70.2, 52.6, 49.1, 45.0, 21.9, 16.4; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C27H36N5O3S 510.2533; found 510.2538; Purity (method B) >99%, tR = 17.0 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-3-methoxy-2-methylphenyl)propane-1-sulfonamide (18l).
Prepared from 16l and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18l (30%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.19 (1 H, d, J = 2.3 Hz), 7.54 (1 H, d, J = 2.3 Hz), 7.50 (2 H, d, J = 8.6 Hz), 7.08 (2 H, d, J = 9.2 Hz), 7.04 (1 H, d, J = 2.9 Hz), 6.73 (1 H, d, J = 2.9 Hz), 3.80 (3 H, s), 3.41–3.39 (4 H, m), 3.35–3.32 (4 H, m), 3.16–3.13 (2 H, m), 2.09 (3 H, s), 1.88 (2 H, sex, J = 8.0 Hz), 1.07 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 159.7, 156.6, 150.8, 145.0, 140.3, 138.4, 137.6, 131.8, 127.9, 127.6, 125.4, 123.4, 118.4, 114.5, 112.6, 55.9, 55.2, 48.1, 45.0, 18.4, 14.7, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H34N5O3S 496.2377; found 496.2377; Purity (method B) 99%, tR = 15.9 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-2-methoxyphenyl)propane-1-sulfonamide (18m).
Prepared from 16j and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18m (39%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.12 (1 H, s), 7.60 (1 H, d, J = 1.7 Hz), 7.56 (1 H, d, J = 1.7 Hz), 7.46 (2 H, d, J = 8.6 Hz), 7.32 (1 H, dd, J = 8.6, 1.7 Hz), 7.16 (1 H, d, J = 8.6 Hz), 7.03 (2 H, d, J = 8.6 Hz), 3.94 (3 H, s), 3.21–3.19 (4 H, m), 3.06–3.03 (6 H, m), 1.84 (2 H, sex, J = 8.0 Hz), 1.01 (3 H, t, J = 7.5 Hz); 13C NMR (CD3OD, 125 MHz) 156.5, 152.7, 152.0, 144.4, 137.5, 131.6, 130.9, 128.6, 127.9, 127.8, 127.7, 125.7, 122.9, 118.0, 113.0, 56.5, 54.6, 50.4, 46.2, 18.3, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H32N5O3S 482.2220; found 482.2223; Purity (method A) 98%, tR = 14.1 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-2-hydroxyphenyl)propane-1-sulfonamide (18n).
Prepared from 16w and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18n (18%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.11 (1 H, s), 7.59 (1 H, d, J = 2.3 Hz), 7.50–7.48 (3 H, m), 7.20 (1 H, dd, J = 8.6, 2.3 Hz), 7.07 (2 H, d, J = 9.2 Hz), 7.00 (1 H ,d , J = 8.0 Hz), 3.39–3.37 (4 H, m), 3.30–3.29 (4 H, m), 3.08–3.05 (2 H, m), 1.91–1.84 (2 H, m), 1.02 (3 H, t, J = 7.4 Hz); HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C24H29N5O3S 468.2064; found 468.2071; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C24H30N5O3S 468.2064; found 468.2071; Purity (method A) >95%, tR = 13.2 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-2-methylphenyl)propane-1-sulfonamide (18o).
Prepared from 16k and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18o (39%) as a pale yellow solid; 1H NMR (CD3OD, 500 MHz) 8.13 (1 H, d, J = 2.3 Hz), 7.60 (1 H, d, J = 2.3 Hz), 7.48 (1 H, d, J = 1.7 Hz), 7.44 (2 H, d, J = 8.6 Hz), 7.36 (1 H, d, J = 8.0 Hz), 7.27 (1 H, dd, J = 7.7, 1.7 Hz), 7.01 (2 H, d, J = 8.6 Hz), 3.19–3.17 (4 H, m), 3.13–3.10 (2 H, m), 3.21–3.18 (4 H, m), 3.04–3.02 (4 H, m), 2.40 (3 H, s), 1.89–1.81 (2 H, m), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 156.4, 152.0, 144.7, 137.6, 137.5, 137.3, 134.5, 133.0, 128.5, 127.7, 126.9, 122.8, 117.9, 55.4, 50.4, 46.2, 18.4, 18.3, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H32N5O2S 466.2271; found 466.2275; Purity (method A) 99%, tR = 14.2 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-2,3-dimethoxyphenyl)propane-1-sulfonamide (18p).
Prepared from 16u and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18p (49%) as a yellow solid; 1H NMR (CD3OD, 600 MHz) 8.14 (1 H, d, J = 2.1 Hz), 7.62 (1 H, d, J = 2.8 Hz), 7.47 (2 H, d, J = 8.9 Hz), 7.20 (1 H, d, J = 1.4 Hz), 7.04 (2 H, d, J = 8.9 Hz), 6.94 (1 H, d, J = 2.1 Hz), 3.01 (3 H, s), 3.90 (3 H, s), 3.26–3.24 (4 H, m), 3.13–3.10 (6 H, m), 1.84 (2 H, sex, J = 7.6 Hz), 1.02 (3 H, t, J = 7.6 Hz); 13C NMR (CD3OD, 150 MHz) 156.4, 154.6, 151.7, 144.7, 141.2, 137.5, 134.8, 132.8, 131.1, 128.4, 127.8, 123.1, 118.1, 115.8, 110.6, 61.4, 56.6, 54.8, 49.7, 45.8, 18.4, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H34N5O4S 512.2326; found 512.2332; Purity (method A) 98%, tR = 14.9 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-3-ethoxy-2-methoxyphenyl)propane-1-sulfonamide (18q).
Prepared from 16v and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18q (52%) as a yellow solid; 1H NMR (CD3OD, 600 MHz) 8.24 (1 H, s), 7.70 (1 H, d, J = 2.1 Hz), 7.52 (2 H, d, J = 8.9 Hz), 7.10 (1 H, s), 7.06 (2 H, d, J = 8.9 Hz), 6.91 (1 H, s), 4.08 (2 H, q, J = 6.9 Hz), 3.88 (3 H, s), 3.30–3.29 (4 H, m), 3.18–3.17 (4 H, m), 2.74–2.69 (1 H, m), 2.67–2.62 (1 H, m), 1.46–1.42 (1 H, m), 1.41–1.36 (4 H, m), 0.77 (3 H, t, J = 7.6 Hz); 13C NMR* (CD3OD, 150 MHz) 156.7, 151.6, 151.0, 149.2, 145.2, 139.2, 131.0, 128.7, 128.6, 127.9, 127.8, 121.3, 118.1, 116.0, 113.7, 65.9, 56.6, 55.4, 45.6, 18.2, 15.1, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C27H36N5O4S 526.2483; found 526.2493; Purity (method A) 95%, tR = 14.5 min. *One of 13C NMR peak was hidden in CD3OD peaks due to low sample concentration.
N-(7-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-2,3-dihydrobenzo[b][1,4]dioxin-5-yl)propane-1-sulfonamide (18r).
Prepared from 5-bromo-3-(8-((propylsulfonyl)methyl)-2,3-dihydrobenzo[b][1,4]dioxin-6-yl)pyridin-2-amine (16 where R1–R2 = OCH2CH2O, R3 = NHSO2nPr and R4 = NH2), which was generated from 14s, but was not characterized. The product was purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18r (37% over three steps) as a light brown solid; 1H NMR (CD3OD, 600 MHz) 8.12 (1 H, s), 7.58 (1 H, d, J = 1.4 Hz), 7.48 (2 H, d, J = 8.9 Hz), 7.12 (1 H, s), 7.06 (2 H, d, J = 8.2 Hz), 6.85 (1 H, s), 4.38–4.37 (2 H, m), 4.32–4.31 (2 H, m), 3.36–3.34 (4 H, m), 3.27–3.25 (4 H, m), 3.09 (2 H, t, J = 7.6 Hz), 1.86 (2 H, sex, J = 7.6 Hz), 1.03 (3 H, t, J = 7.6 Hz); 13C NMR (CD3OD, 150 MHz) 156.5, 151.1, 145.8, 144.6, 137.4, 137.3, 131.6, 131.5, 128.2, 128.0, 127.9, 122.8, 118.3, 117.7, 115.5, 66.0, 65.6, 54.7, 45.3, 30.7, 18.4, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H32N5O4S 510.2170; found 510.2173; Purity (method A) 98%, tR = 14.2 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-2-methoxy-3-methylphenyl)propane-1-sulfonamide (18s).
Prepared from 16n and purified by column chromatography on silica gel (MeOH/CH2Cl2, 10:90 to 15:85) to give 18s (53%) as a yellow solid; 1H NMR (CD3OD, 600 MHz) 8.13 (1 H, d, J = 2.1 Hz), 7.59 (1 H, d, J = 2.1 Hz), 7.46 (1 H, d, J = 8.9 Hz), 7.43 (1 H, d, J = 1.4 Hz), 7.11 (1 H, s), 7.03 (2 H, d, J = 8.9 Hz), 3.82 (3 H, s), 3.24–3.23 (4 H, m), 3.18–3.16 (2 H, m), 3.11–3.09 (4 H, m), 2.36 (3 H, s), 1.86 (2 H, sex, J = 7.6 Hz), 1.03 (3 H, t, J = 7.6 Hz); 13C NMR (CD3OD, 150 MHz) 156.4, 151.8, 150.8, 144.6, 137.5, 135.1, 133.9, 132.7, 131.0, 128.7, 128.4, 127.8, 123.0, 121.2, 118.0, 61.2, 55.2, 49.9, 45.9, 18.4, 16.4, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H34N5O3S 496.2377; found 496.2383; Purity (method A) 97%, tR = 15.1 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-3-fluoro-2-methoxyphenyl)propane-1-sulfonamide (18t).
Prepared from 16m and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18t (52%) as a yellow solid; m.p. 142 – 144 °C; 1H NMR (CD3OD, 500 MHz) 8.13 (1 H, d, J = 2.3 Hz), 7.58 (1 H, d, J = 2.3 Hz), 7.44–7.40 (3 H, m), 7.10 (1 H, dd, J = 12.0, 2.3 Hz), 7.00 (2 H, d, J = 8.6 Hz), 4.01 (3 H, d, J = 1.7 Hz), 3.19–3.17 (4 H, m), 3.14–3.11 (2 H, m), 3.04–3.02 (4 H, m), 1.88–1.81 (2 H, m), 1.02 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 156.8 (d, JCF = 246.1 Hz), 156.2, 152.0, 145.1, 140.1 (d, JCF = 13.5 Hz), 137.5, 134.6 (d, JCF = 9.8 Hz), 133.6 (d, JCF = 4.9 Hz), 130.6, 128.4, 127.7, 121.8, 119.6, 117.9, 114.4, 62.1 (d, JCF = 7.4 Hz), 55.0, 50.3, 46.1, 18.4, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H31FN5O3S 500.2126; found 500.2137; Purity (method B) 98%, tR = 15.9 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-3-chloro-2-methoxyphenyl)propane-1-sulfonamide (18u).
Prepared from 16t and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18u (62%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.16 (1 H, d, J = 2.3 Hz), 7.60 (1 H, d, J = 2.3 Hz), 7.57 (1 H, d, J = 1.7 Hz), 7.46 (2 H, d, J = 9.2 Hz), 7.31 (1 H, d, J = 1.7 Hz), 7.03 (2 H, d, J = 8.6 Hz), 3.93 (3 H, s), 3.27–3.25 (4 H, m), 3.20–3.12 (6 H, m), 1.86 (2 H, sex, J = 8.0 Hz), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 156.4, 151.6, 148.2, 145.3, 137.6, 136.2, 134.7, 130.9, 129.7, 128.4, 127.8, 127.2, 122.0, 121.6, 118.1, 61.7, 55.4, 49.6, 45.8, 18.4, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H31ClN5O3S 516.1831; found 516.1836; Purity (method B) 96%, tR = 16.4 min.
3-(3-Fluoro-4,5-dimethoxyphenyl)-5-(4-(piperazin-1-yl)phenyl)pyridin-2-amine (18v).
Prepared from 16x and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18v (47%) as a pale yellow solid; 1H NMR (CDCl3, 400 MHz) 8.273 (1 H, d, J = 1.8 Hz), 7.55 (1 H, d, J = 2.3 Hz), 7.46 (2 H, d, J = 8.2 Hz), 6.99 (2 H, d, J = 8.7 Hz), 6.88 (1 H, dd, J = 11.0, 1.8 Hz), 6.81 (1 H, s), 4.72 (2 H, br), 3.98 (3 H, s), 3.90 (3 H, s), 3.42 (4 H, br), 3.29 (4 H, br); 13C NMR (CDCl3, 100 MHz) 156.1 (d, JCF = 246.5 Hz), 154.4, 154.1 (d, JCF = 5.9 Hz), 149.6, 144.9, 136.1, 132.9, 130.5, 127.6 (d, JCF = 35.2 Hz), 127.1, 120.7, 117.3, 113.8, 109.5, (d, JCF = 20.5 Hz), 108.2, 61.6 (d, JCF = 3.9 Hz), 56.4, 47.9, 44.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C23H26FN4O2 409.2034; found 409.2038; Purity (method A) 99%, tR = 14.6 min.
N-(5-(2-Amino-5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)-2-methoxybenzyl)propane-1-sulfonamide (18w).
Prepared from 16o and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18w (39%) as a yellow solid; 1H NMR (CD3OD, 500 MHz) 8.10 (1 H, d, J = 2.3 Hz), 7.57 (1 H, d, J = 2.3 Hz), 7.48 (1 H, d, J = 2.3 Hz), 7.45–7.40 (3 H, m), 7.10 (1 H, d, J = 8.6 Hz), 7.02 (2 H, d, J = 9.2 Hz), 4.28 (2 H, s), 3.91 (3 H, s), 3.22–3.20 (4 H, m), 3.08–3.06 (4 H, m), 2.94–2.90 (2 H, m), 1.77–1.69 (2 H, m), 0.97 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 158.4, 156.6, 151.8, 144.2, 137.5, 131.1, 131.0, 130.8, 130.5, 128.5, 128.0, 127.7, 123.3, 118.0, 112.2, 56.1, 55.2, 50.1, 46.0, 42.8, 18.4, 13.3; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H34N5O3S 496.2377; found 496.2386; Purity (method B) 98%, tR = 15.8 min.
5-(4-(Piperazin-1-yl)phenyl)-3-(1-(propylsulfonyl)indolin-6-yl)pyridin-2-amine (18x).
Prepared from 16p and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 10:90) to give 18x (38%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.27 (1 H, d, J = 2.3 Hz), 7.55 (1 H, d, J = 2.3 Hz), 7.47–7.43 (3 H, m), 7.29 (1 H, d, J = 8.0 Hz), 7.13 (1 H, dd, J = 7.7, 1.2 Hz), 6.98 (2 H, d, J = 8.6 Hz), 4.66 (2 H, br), 4.09 (2 H, t, J = 8.6 Hz), 3.22–3.18 (6 H, m), 3.09–3.04 (6 H, m), 1.94–1.86 (2 H, m), 1.04 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 154.4, 150.7, 144.9, 143.0, 138.0, 136.1, 130.7, 129.4, 127.7, 127.0, 125.9, 123.6, 121.4, 116.5, 113.5, 51.3, 50.5, 49.9, 45.8, 27.8, 16.7, 13.1; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H32N5O2S 478.2271; found 478.2277; Purity (method A) 98%, tR = 15.4 min.
5-(4-(Piperazin-1-yl)phenyl)-3-(4-(propylsulfonyl)-3,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)pyridin-2-amine (18y).
Prepared from 16q and purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18y (56%) as a pale yellow solid; 1H NMR (CD3OD, 500 MHz) 8.13 (1 H, d, J = 2.3 Hz), 7.71 (1 H, d, J = 2.3 Hz), 7.60 (1 H, d, J = 2.3 Hz), 7.48 (2 d, J = 9.2 Hz), 7.19 (1 H, dd, J = 8.3, 2.3 Hz), 7.07–7.03 (3 H, m), 4.34 (2 H, t, J = 4.6 Hz), 3.88 (2 H, t, J = 4.6 Hz), 3.30–3.24 (6 H, m), 3.17–3.15 (4 H, m), 1.85 (2 H, sex, J = 8.0 Hz), 1.04 (3 H, t, J = 7.4 Hz); 13C NMR (CD3OD, 125 MHz) 156.5, 152.0, 147.6, 144.4, 137.4, 131.4, 130.8, 128.5, 127.7, 126.9, 126.0, 124.1, 123.0, 119.5, 117.9, 66.2, 54.9, 50.4, 46.2, 45.2, 18.2, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C26H32N5O3S 494.2220; found 494.2226; Purity (method A) >99%, tR = 15.5 min.
N-(3-Fluoro-2-methoxy-5-(5-(4-(piperazin-1-yl)phenyl)pyridin-3-yl)phenyl)propane-1-sulfonamide (18z).
Prepared from 16y and purified by column chromatography on silica gel (EtOAc/hexane, 0:100 to 30:70) to give 18z (30%) as a pale yellow solid; 1H NMR (CDCl3, 600 MHz) 8.80 (1 H, s), 8.68 (1 H, s), 7.93 (1 H, s), 7.61 (1 H, s), 7.55 (2 H, d, J = 7.8 Hz), 7.14 (1 H, d, J = 12.6 Hz), 7.05 (2 H, d, J = 9.0 Hz), 4.08 (1 H, s), 3.24 (4 H, t, J = 4.8 Hz), 3.12 (2 H, t, J = 8.1 Hz), 3.07 (4 H, t, J = 4.2 Hz), 1.91–1.84 (2 H, sex, J = 7.8 Hz), 1.04 (3 H, t, J = 7.5 Hz); 13C NMR (CDCl3, 150 MHz) 153.8 (d, JCF = 245.7 Hz), 151.8, 147.1, 145.70, 136.6 (d, JCF = 11.6 Hz), 136.4, 134.6, 133.9 (d, JCF = 9.0 Hz), 131.8, 131.6 (d, JCF = 23.4 Hz), 127.9, 116.1, 112.9, 111.2 (d, JCF = 21.6 Hz), 61.6, 53.6, 49.8, 46.0, 17.3, 12.9; HRMS (ESI-QTOF) m/z: [M + Na]+ calculated for C25H29FN4O3S 507.1837; found 507.1848; Purity (method A) 100%, tR = 16.9 min.
Preparation of 3-phenyl-5-(methylsulfonyl)phenyl)pyridin-2-amines.
Intermediate 16j was coupled to either 2-, 3-, or 4-(methanesulfonyl)phenylboronic acid using the conditions described for the general procedure for the preparation of 3-bromo-5-phenylpyridines.
N-(5-(2-Amino-5-(2-(methylsulfonyl)phenyl)pyridin-3-yl)-2-methoxyphenyl)propane-1-sulfonamide (18ab).
Purified by column chromatography on silica gel (EtOAc/hexane, 50:50 to 80:20) to give 18ab (22%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.24 (1 H, dd, J = 7.7, 1.7 Hz), 8.05 (1 H, d, J = 2.3 Hz), 7.69–7.66 (3 H, m), 7.57 (1 H, td, J = 7.4, 1.2 Hz), 7.42 (1 H, dd, J = 7.4, 1.2 Hz), 7.29 (1 H, dd, J = 8.2, 2.3 Hz), 6.99 (1 H, d, J = 8.6 Hz), 6.83 (1 H, s), 4.84 (2 H, br), 3.93 (3 H, s), 3.11–3.08 (2 H, m), 2.79 (3 H, s), 1.87 (2 H, sex, J = 8.0 Hz), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 155.7, 148.6, 146.8, 140.4, 139.6, 138.3, 133.4, 133.2, 130.3, 129.0, 128.1, 126.8, 125.5, 125.0, 119.9, 119.3, 111.4, 56.0, 53.9, 43.5, 17.2, 12.8; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C22H26N3O5S2 476.1308; found 476.1313; Purity (method B) 95%, tR = 18.0 min.
N-(5-(2-Amino-5-(3-(methylsulfonyl)phenyl)pyridin-3-yl)-2-methoxyphenyl)propane-1-sulfonamide (18ac).
Purified by column chromatography on silica gel (MeOH/CH2Cl2, 2.5:97.5 to 5:95) to give 18ac (60%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.32 (1 H, s), 8.08 (1 H, s), 7.88 (1 H, d, J = 7.4 Hz), 7.82 (1 H, d, J = 8.0 Hz), 7.66–7.60 (3 H, m), 7.24 (1 H, dd, J = 8.3, 2.9 Hz), 7.03 (1 H, d, J = 8.6 Hz), 6.95 (1 H, s), 4.83 (2 H, br), 3.95 (3 H, s), 3.10–3.06 (5 H, m), 1.91–1.84 (2 H, m), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 155.9, 148.8, 145.6, 141.2, 139.8, 136.4, 131.2, 130.4, 130.0, 126.9, 125.6, 125.4, 124.8, 121.0, 120.3, 120.3, 111.4 50.0, 53.7, 44.5, 17.2, 12.9; HRMS (ESI-QTOF) m/z: [M + Na]+ calculated for C22H25N3NaO5S2 498.1128; found 498.1132; Purity (method B) 96%, tR = 17.7 min.
N-(5-(2-Amino-5-(4-(methylsulfonyl)phenyl)pyridin-3-yl)-2-methoxyphenyl)propane-1-sulfonamide (18ad).
Purified by column chromatography on silica gel (MeOH/CH2Cl2, 2.5:97.5 to 5:95) to give 18ad (83%) as a yellow solid; 1H NMR (CDCl3, 500 MHz) 8.35 (1 H, d, J = 1.7 Hz), 7.98 (2 H, d, J = 8.6 Hz), 7.72 (2 H, d, J = 8.0 Hz), 7.66 (1 H, d, J = 1.7 Hz), 7.60 (1 H, d, J = 2.3 Hz), 7.20–7.24 (1 H, m), 7.03 (1 H, d, J = 8.6 Hz), 6.90 (1 H, s), 4.84 (2 H, br), 3.95 (3 H, s), 3.09–3.06 (5 H, m), 1.88 (2 H, sex, J = 8.0 Hz), 1.03 (3 H, t, J = 7.4 Hz); 13C NMR (CDCl3, 125 MHz) 156.1, 148.8, 145.9, 143.7, 138.5, 136.4, 130.4, 128.1, 127.0, 126.8, 125.6, 125.3, 120.9, 120.2, 111.3, 56.0, 53.8, 44.6, 17.2, 12.9; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C22H26N3O5S2 476.1308; found 476.1318; Purity (method B) 97%, tR = 17.5 min.
Preparation of N-(5-(2-Amino-5-(3-(piperazin-1-yl)phenyl)pyridin-3-yl)-3-fluoro-2-methoxyphenyl)propane-1-sulfonamide (18ae).
Prepared from 16m and 3-(4-tert-butoxycarbonylpiperazinyl)phenylboronic acid pinacol ester using the conditions described for the general procedure for the preparation of 3-bromo-5-phenylpyridines. The residue was purified by column chromatography on silica gel (EtOAc/hexane, 30:70 to 70:30) to afford the coupling product then anhydrous CH2Cl2 (3 mL), and trifluoroacetic acid (0.3 mL) were added. The mixture was stirred at room temperature for 16 h. After being quenched with saturated aqueous NaHCO3 (5 mL), CH2Cl2 were added, and the layers were separated. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (MeOH/CH2Cl2, 5:95 to 15:85) to give 18ae (48%) as a pale yellow solid; 1H NMR (CD3OD, 500 MHz) 8.18 (1 H, d, J = 2.1 Hz), 7.64 (1 H, d, J = 2.1 Hz), 7.42 (1 H, d, J = 1.4 Hz), 7.30 (1 H, t, J = 7.6 Hz), 7.15–7.13 (2 H, m), 7.07 (1 H, d, J = 7.7 Hz), 6.94 (1 H, dd, J = 8.2, 2.1 Hz), 4.01 (3 H, d, J = 2.1 Hz), 3.28–3.27 (4 H, m), 3.15–3.11 (6 H, m), 1.89–1.82 (2 H, m), 1.04 (3 H, t, J = 7.6 Hz); 13C NMR (CD3OD, 150 MHz) 156.9, 156.8 (d, JCF = 246.9 Hz), 153.3, 145.8, 140.2 (d, JCF = 11.8 Hz), 140.0, 138.1, 134.5 (d, JCF = 8.9 Hz), 133.6 (d, JCF = 5.9 Hz), 130.8, 128.9, 121.7, 119.6, 119.5, 116.5, 115.6, 114.5 (d, JCF = 19.2 Hz), 62.1 (d, JCF = 7.4 Hz), 55.0, 50.1, 46.0, 18.4, 13.2; HRMS (ESI-QTOF) m/z: [M + H]+ calculated for C25H31FN5O3S 500.2126; found 500.2132; Purity (method B) >99%, tR = 16.5 min.
RIPK2 enzyme assay.
Recombinant RIPK2 protein (20 ng per reaction) is diluted in the reaction buffer consisting of 40 mM Tris (pH 7.5); 20 mM MgCl2; 0.1 mg/ml BSA; 50 μM DTT. Diluted protein is added to low volume white 384 well plates (2 μL/well). Inhibitors are diluted in reaction buffer (final 25% DMSO), 1 μL is added to each well and incubated 5 min at room temperature. Reactions are initiated by the addition of 2 μL of 100 μM ATP and 1 mg/ml RS repeat peptide (SignalChem) in the reaction buffer. Plates are sealed with plastic coverslips and incubated at room temperature for 2 h. Reactions are stopped by the addition of 5 μL of ADPGlo reagent (Promega) and ADP generation reaction is performed for 40 min at room temperature. Luminescence signal is generated by the addition of 10 μL of Kinase detection reagent (Promega) for 30 min at room temperature. Luminescence signals are determined using appropriate luminescence plate-reader (typical integration time 0.3-1 sec). To calculate percent inhibition, average background signal is subtracted from test well and maximal signal wells. Inhibition, % = (1- (test signal/maximal signal))*100. The percent inhibition at a specified concentration is determined or IC50 values are calculated based on a dose range of inhibitor concentrations using non-linear regression in GraphPad Prism software.
ALK2 enzyme assay.
Enzyme inhibitory activity was evaluated in a standard kinase enzyme assay by incubating human ALK2 with the protein substrate casein (1 mg/mL) and γ-33ATP (10 μM) in the presence of various concentrations of test compounds (10 nM – 100 μM). After 30 min the amount of 33P-casein was determined. A plot of inhibitor concentration versus % activity was constructed and from this plot an IC50 value was determined.
Human NOD2 signaling using a HEKBlue reporter assay.
HEK293 cell line expressing hNOD2 and NFκB-SEAP reporter cassette was obtained from Invivogen. Cells were maintained in Dulbecco's Modified Eagle Medium /10% fetal bovine serum media supplemented with zeocin, blasticidin and normocin as suggested by Invivogen. For the experiments, cells were seeded at 7.5-15x103 cells/well in clear 96-well plates. After 24–48 h, cells were changed into HEKBlue detection media (Invivogen) and treated with 11-point dose ranges for each inhibitor along with 1 ng/mL L18-MDP (Invivogen). After 7–9 h, absorbance at 620 nm was measured using a Victor3V plate reader (Perkin Elmer, Waltham, MA). Values of media-only wells were subtracted and %inhibition for each compound concentration relative to the DMSO/L18-MDP-treated controls was calculated. Inhibition values were fitted by non- linear regression using Prism software (GraphPad Software, La Jolla, CA) to calculate IC50 values.
Data collection and Structure determination.
The kinase domain of human RIPK2 (Uniprot: O43353, residues 3-317) was recombinantly expressed in Sf9 insect cells and purified by nickel-affinity and size exclusion chromatography as previously described [7]. Mass spectrometry revealed the phosphorylation state to be a mixture of between 2 to 5 phosphorylations. Protein buffered in 50 mM HEPES pH 7.5, 300 mM NaCl, 5% glycerol, and 1 mM tris(2-carboxyethyl)phosphine was concentrated to 10 mg/mL and 2 mM 18f added. This sample was then incubated for 10 minutes, spun down, and filtered to 0.22 μm. Crystals were grown using the vapor-diffusion technique at 4 °C in 150 nL sitting drops containing 100 nL protein and 50 nL of a reservoir solution containing 20% High PEG Smear, 0.1M citrate pH 5.5 [36]. Crystals were cryo-protected by addition of 25% ethylene glycol before being vitrified in liquid nitrogen. Diffraction data were collected at 100 K on Diamond Light Source beamline I24 using 0.9785 Å light. Data were indexed and integrated using XDS [37-38] and scaled using AIMLESS [39]. Phases were identified using molecular replacement in PHASER [40] and the PDB 6FU5 as a search model. The structure was refined and modified using PHENIX.REFINE [41] and COOT [42]. The refined structure was validated with MolProbity [43] and the atomic coordinate files deposited in the Protein Data Bank with Autodep [44].
In Vivo Pharmacokinetics.
The pharmacokinetic study was performed in accordance with an approved protocol by the Institutional Care and Use Committee (IACUC) at the University of Houston (PROTO201800072: PK and Perfusion in Mice). For in vivo pharmacokinetic experiments, 18t was administered to 8 week old female C57BL/6 mice (N = 5) intraperitoneally at a dose of 10 mg/kg with vehicle (6% Captisol® in water). Blood samples were collected per mouse under light anesthesia at 0.25, 0.5, 1, 2, 4, 6, and 24 h after dosing. An LC–MS/MS bioanalysis method was developed. A plasma concentration-time profile was obtained and the data subjected to non-compartmental analysis using Phoenix WinNonlin (version 8.1) to assess pharmacokinetic parameters.
Supplementary Material
Highlights.
Demonstration of 3,5-diphenyl-2-aminopyridines as potent inhibitors of receptor-interacting protein kinase 2 (RIPK2).
SAR insights into achieving RIPK2 dependent nucleotide-binding oligomerization domain (NOD) cell signaling inhibition.
SAR insights into selectivity versus structurally related activin receptor-like kinase 2 (ALK2).
In vitro ADME and in vivo pharmacokinetic characterization of a representative RIPK2 kinase/NOD cell signaling inhibitor illustated.
Acknowledgments
This work was supported in whole or in part with funds from the National Institutes of Health, Department of Health and Human Services (CA190542, AI124049, AG058642, AI144400 and NS111395). The authors would like to thank Diamond Light Source for beamtime (proposal mx15433), as well as the staff of beamlines I04-1 and I24 for assistance with crystal testing and data collection. The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome [106169/ZZ14/Z]. M.G-H is supported by the Ludwig Institute for Cancer Research Ltd and a Wellcome Trust Fellowship (102894/Z/13/Z).
Abbreviations used
- ADME
adsorption, distribution, metabolism and excretion
- ALK2
activin receptor-like kinase 2
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- AUC
area under the curve
- BRET
bioluminescence resonance energy transfer
- CARD
caspase-activated recruitment domain
- CL
clearance
- Cmax
concentration maximum
- CXCL8
C-X-C motif chemokine ligand 8
- DAP
diaminopimelic acid
- DFG
aspartic acid-phenyl alanine-glycine
- DTT
dithiothreitol
- HEPES
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid)
- HPLC
high-performance liquid chromatography
- HRMS
high resolution mass spectra
- IL
interleukin
- ip
intraperitoneal
- LUBAC
linear ubiquitin chain assembly complex
- MAPK
mitogen-activated protein kinase
- NBD
nucleotide-binding oligomerization domains
- NLR
NOD-like receptor
- NOD
nucleotide-binding oligomerization domain
- PAMPA
parallel artificial membrane permeability assay
- PBS
phosphate-buffered saline
- PDB
protein data bank
- PG
peptidoglycan
- PPI
protein-protein interaction
- RIPK2
receptor-interacting protein kinase 2
- SAR
structure-activity relationship
- SEAP
secreted embryonic alkaline phosphatase
- TNF
tumor necrosis factor
- Tris
tris(hydroxymethyl)aminomethane
- Ub
ubiquitin
- XIAP
X-linked inhibitor of apoptosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
C.S., A.D. and G.D.C. declare the following financial interests/personal relationships that may be considered as potential competing interests: PCT patent application (Publication Number 20200030303) assigned to the University of Houston System and Trustees of Tufts College. All other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Accession codes
Atomic coordinates for the x-ray structures of 18f•RIPK2 (PDB 6S1F), 18m•RIPK2 (PDB 6FU5) and 9b•ALK2 (PDB 4BGG) are available from the RCSB Protein Data Bank (www.rscb.org).
Appendix A. Supporting data
Synthesis scheme and procedures of starting materials 11i-o, 12b, 13v and intermediate 19, the pharmacokinetic procedure, copies of NMR spectra, and data collection and refinement statistics for the RIPK2•18f crystal structure are provided. The supporting data to this article can be found online at
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Inohara N; Koseki T; del Peso L; Hu Y; Yee C; Chen S; Carrio R; Merino J; Liu D; Ni J; Núñez G Nodl, an Apaf-1-like Activator of Caspase-9 and Nuclear Factor-κB. J. Biol. Chem 1999, 274 (21), 14560 10.1074/jbc.274.21.14560 [DOI] [PubMed] [Google Scholar]
- 2.Inohara N; Koseki T; Lin J; del Peso L; Lucas PC; Chen FF; Ogura Y; Núñez G An Induced Proximity Model for NF-κB Activation in the Nod1/RICK and RIP Signaling Pathways. J. Biol. Chem 2000, 275 (36), 27823 10.1074/jbc.M003415200 [DOI] [PubMed] [Google Scholar]
- 3.Ogura Y; Inohara N; Benito A; Chen FF; Yamaoka S; Núñez G Nod2, a Nod1/Apaf-1 Family Member That Is Restricted to Monocytes and Activates NF-κB. J. Biol. Chem 2001, 276 (7), 4812 10.1074/jbc.M008072200 [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa M; Fujimoto Y; Lucas PC; Nakano H; Fukase K; Núñez G; Inohara N A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 2008, 27 (2), 373 10.1038/sj.emboj.7601962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Windheim M; Lang C; Peggie M; Plater Lorna A.; Cohen P Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem. J 2007, 404 (Pt 2), 179 10.1042/BJ20061704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi K; Inohara N; Hernandez LD; Galan JE; Nunez G; Janeway CA; Medzhitov R; Flavell RA RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 2002, 416 (6877), 194 10.1038/416194a [DOI] [PubMed] [Google Scholar]
- 7.Hrdinka M; Schlicher L; Dai B; Pinkas DM; Bufton JC; Picaud S; Ward JA; Rogers C; Suebsuwong C; Nikhar S; Cuny GD; Huber KVM; Filippakopoulos P; Bullock AN; Degterev A; Gyrd-Hansen M Small molecule inhibitors reveal an indispensable scaffolding role of RIPK2 in NOD2 signaling. EMBO J. 2018, 37 (17), e99372 10.15252/embj.201899372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girardin SE; Boneca IG; Carneiro LAM; Antignac A; Jéhanno M; Viala J; Tedin K; Taha M-K; Labigne A; Zäthringer U; Coyle AJ; DiStefano PS; Bertin J; Sansonetti PJ; Philpott DJ Nod1 Detects a Unique Muropeptide from Gram-Negative Bacterial Peptidoglycan. Science 2003, 300 (5625), 1584 10.1126/science.1084677 [DOI] [PubMed] [Google Scholar]
- 9.Girardin SE; Boneca IG; Viala J; Chamaillard M; Labigne A; Thomas G; Philpott DJ; Sansonetti PJ Nod2 Is a General Sensor of Peptidoglycan through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem 2003, 278 (11), 8869 10.1074/jbc.C200651200 [DOI] [PubMed] [Google Scholar]
- 10.Damgaard Rune B.; Nachbur U; Yabal M; Wong Wendy W.-L.; Fiil Berthe K.; Kastirr M; Rieser E; Rickard James A.; Bankovacki A; Peschel C; Ruland J; Bekker-Jensen S; Mailand N; Kaufmann T; Strasser A; Walczak H; Silke J; Jost Philipp J.; Gyrd-Hansen M The Ubiquitin Ligase XIAP Recruits LUBAC for NOD2 Signaling in Inflammation and Innate Immunity. Mol. Cell 2012, 46 (6), 746 10.1016/j.molcel.2012.04.014 [DOI] [PubMed] [Google Scholar]
- 11.Fiil Berthe K.; Damgaard Rune B.; Wagner Sebastian A.; Keusekotten K; Fritsch M; Bekker-Jensen S; Mailand N; Choudhary C; Komander D; Gyrd-Hansen M OTULIN Restricts Met1-Linked Ubiquitination to Control Innate Immune Signaling. Mol. Cell 2013, 50 (6), 818 10.1016/j.molcel.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrdinka M; Gyrd-Hansen M The Met1-Linked Ubiquitin Machinery: Emerging Themes of (De)regulation. Mol. Cell 2017, 68 (2), 265 10.1016/j.molcel.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 13.Goncharov T; Hedayati S; Mulvihill MM; Izrael-Tomasevic A; Zobel K; Jeet S; Fedorova AV; Eidenschenk C; deVoss J; Yu K; Shaw AS; Kirkpatrick DS; Fairbrother WJ; Deshayes K; Vucic D Disruption of XIAP-RIP2 Association Blocks NOD2-Mediated Inflammatory Signaling. Mol. Cell 2018, 69 (4), 551 10.1016/j.molcel.2018.01.016 [DOI] [PubMed] [Google Scholar]
- 14.Damgaard RB; Fiil BK; Speckmann C; Yabal M; Stadt U. z.; Bekker-Jensen S; Jost PJ; Ehl S; Mailand N; Gyrd-Hansen M Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Molecular Medicine 2013, 5 (8), 1278 10.1002/emmm.201303090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugot J-P; Chamaillard M; Zouali H; Lesage S; Cezard J-P; Belaiche J; Almer S; Tysk C; O'Morain CA; Gassull M; Binder V; Finkel Y; Cortot A; Modigliani R; Laurent-Puig P; Gower-Rousseau C; Macry J; Colombel J-F; Sahbatou M; Thomas G Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411 (6837), 599 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- 16.Caruso R; Warner N; Inohara N; Núñez G NOD1 and NOD2: Signaling, Host Defense, and Inflammatory Disease. Immunity 2014, 41 (6), 898 10.1016/j.immuni.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin MC; Jirholt P; Wramstedt A; Johansson ME; Lundberg AM; Trajkovska MG; Ståhlman M; Fogelstrand P; Brisslert M; Fogelstrand L; Yan ZQ; Hansson GK; Björkbacka H; Olofsson SO; Borén J Rip2 deficiency leads to increased atherosclerosis despite decreased inflammation. Circ. Res 2011, 109 (11), 1210 10.1161/CIRCRESAHA.111.246702 [DOI] [PubMed] [Google Scholar]
- 18.Miceli-Richard C; Lesage S; Rybojad M; Prieur A-M; Manouvrier-Hanu S; Hafner R; Chamaillard M; Zouali H; Thomas G; Hugot J-P CARD15 mutations in Blau syndrome. Nat. Genet 2001, 29 (1), 19 10.1038/ng720 [DOI] [PubMed] [Google Scholar]
- 19.Caso F; Costa L; Rigante D; Vitale A; Cimaz R; Lucherini OM; Sfriso P; Verrecchia E; Tognon S; Bascherini V; Galeazzi M; Punzi L; Cantarini L Caveats and truths in genetic, clinical, autoimmune and autoinflammatory issues in Blau syndrome and early onset sarcoidosis. Autoimmun. Rev 2014, 13 (12), 1220 10.1016/j.autrev.2014.08.010 [DOI] [PubMed] [Google Scholar]
- 20.Shaw PJ; Barr MJ; Lukens JR; McGargill MA; Chi H; Mak TW; Kanneganti T-D Signaling via the RIP2 Adaptor Protein in Central Nervous System-Infiltrating Dendritic Cells Promotes Inflammation and Autoimmunity. Immunity 2011, 34 (1), 75 10.1016/j.immuni.2010.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Argast GM; Fausto N; Campbell JS Inhibition of RIP2/RICK/CARDIAK activity by pyridinyl imidazole inhibitors of p38 MAPK. Mol. Cell. Biochem 2005, 268 (1), 129 10.1007/s11010-005-3701-0 [DOI] [PubMed] [Google Scholar]
- 22.Tigno-Aranjuez JT; Asara JM; Abbott DW Inhibition of RIP2's tyrosine kinase activity limits NOD2-driven cytokine responses. Genes Dev. 2010, 24 (23), 2666 10.1101/gad.1964410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tigno-Aranjuez JT; Benderitter P; Rombouts F; Deroose F; Bai X; Mattioli B; Cominelli F; Pizarro TT; Hoflack J; Abbott DW In Vivo Inhibition of RIPK2 Kinase Alleviates Inflammatory Disease. J. Biol. Chem 2014, 289 (43), 29651 10.1074/jbc.M114.591388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nachbur U; Stafford CA; Bankovacki A; Zhan Y; Lindqvist LM; Fiil BK; Khakham Y; Ko HJ; Sandow JJ; Falk H; Holien JK; Chau D; Hildebrand J; Vince JE; Sharp PP; Webb AI; Jackman KA; Muhlen S; Kennedy CL; Lowes KN; Murphy JM; Gyrd-Hansen M; Parker MW; Hartland EL; Lew AM; Huang DC; Lessene G; Silke J A RIPK2 inhibitor delays NOD signalling events yet prevents inflammatory cytokine production. Nat. Commun 2015, 6 6442 10.1038/ncomms7442 [DOI] [PubMed] [Google Scholar]
- 25.Canning P; Ruan Q; Schwerd T; Hrdinka M; Maki JL; Saleh D; Suebsuwong C; Ray S; Brennan PE; Cuny GD; Uhlig HH; Gyrd-Hansen M; Degterev A; Bullock N Inflammatory Signaling by NOD-RIPK2 Is Inhibited by Clinically Relevant Type II Kinase Inhibitors. Chem. Biol 2015, 22 (9), 1174 10.1016/j.chembiol.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haile PA; Votta BJ; Marquis RW; Bury MJ; Mehlmann JF; Singhaus R; Charnley AK; Lakdawala AS; Convery MA; Lipshutz DB; Desai BM; Swift B; Capriotti CA; Berger SB; Mahajan MK; Reilly MA; Rivera EJ; Sun HH; Nagilla R; Beal AM; Finger JN; Cook MN; King BW; Ouellette MT; Totoritis RD; Pierdomenico M; Negroni A; Stronati L; Cucchiara S; Ziołkowski B; Vossenkämper A; MacDonald TT; Gough PJ; Bertin J; Casillas LN The Identification and Pharmacological Characterization of 6-(tert-Butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a Highly Potent and Selective Inhibitor of RIP2 Kinase. J. Med. Chem 2016, 59 (10), 4867 10.1021/acs.jmedchem.6b00211 [DOI] [PubMed] [Google Scholar]
- 27.Haile PA; Casillas LN; Votta BJ; Wang GZ; Charnley AK; Dong X; Bury MJ; Romano JJ; Mehlmann JF; King BW; Erhard KF; Hanning CR; Lipshutz DB; Desai BM; Capriotti CA; Schaeffer MC; Berger SB; Mahajan MK; Reilly MA; Nagilla R; Rivera EJ; Sun HH; Kenna JK; Beal AM; Ouellette MT; Kelly M; Stemp G; Convery MA; Vossenkämper A; MacDonald TT; Gough PJ; Bertin J; Marquis RW Discovery of a First-in-Class Receptor Interacting Protein 2 (RIP2) Kinase Specific Clinical Candidate, 2-((4-(Benzo[d]thiazol-5-ylamino)-6-(tert-butylsulfonyl)quinazolin-7-yl)oxy)ethyl Dihydrogen Phosphate, for the Treatment of Inflammatory Diseases. J. Med. Chem 2019, 62 (14), 6482 10.1021/acs.jmedchem.9b00575 [DOI] [PubMed] [Google Scholar]
- 28.He X; Da Ros S; Nelson J; Zhu X; Jiang T; Okram B; Jiang S; Michellys P-Y; Iskandar M; Espinola S; Jia Y; Bursulaya B; Kreusch A; Gao M-Y; Spraggon G; Baaten J; Clemmer L; Meeusen S; Huang D; Hill R; Nguyen-Tran V; Fathman J; Liu B; Tuntland T; Gordon P; Hollenbeck T; Ng K; Shi J; Bordone L; Liu H Identification of Potent and Selective RIPK2 Inhibitors for the Treatment of Inflammatory Diseases. ACS Medicinal Chemistry Letters 2017, 8 (10), 1048 10.1021/acsmedchemlett.7b00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salla M; Aguayo-Ortiz R; Danmaliki GI; Zare A; Said A; Moore J; Pandya V; Manaloor R; Fong S; Blankstein AR; Gibson SB; Garcia LR; Meier P; Bhullar KS; Hubbard BP; Fiteh Y; Vliagoftis H; Goping IS; Brocks D; Hwang P; Velázquez-Martínez CA; Baksh S Identification and Characterization of Novel Receptor-Interacting Serine/Threonine-Protein Kinase 2 Inhibitors Using Structural Similarity Analysis. J. Pharmacol. Exp. Ther 2018, 365 (2), 354 10.1124/jpet.117.247163 [DOI] [PubMed] [Google Scholar]
- 30.Mohedas AH; Wang Y; Sanvitale CE; Canning P; Choi S; Xing X; Bullock AN; Cuny GD; Yu PB Structure–Activity Relationship of 3,5-Diaryl-2-aminopyridine ALK2 Inhibitors Reveals Unaltered Binding Affinity for Fibrodysplasia Ossificans Progressiva Causing Mutants. J. Med. Chem 2014, 57 (19), 7900 10.1021/jm501177w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanvitale CE; Kerr G; Chaikuad A; Ramel M-C; Mohedas AH; Reichert S; Wang Y; Triffitt JT; Cuny GD; Yu PB; Hill CS; Bullock AN A New Class of Small Molecule Inhibitor of BMP Signaling. PLoS One 2013, 8 (4), e62721 10.1371/journal.pone.0062721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bollag G; Hirth P; Tsai J; Zhang J; Ibrahim PN; Cho H; Spevak W; Chao Z; Ying Z; Habets G; Burton EA; Bernice W; Garson T; West BL; Powell B; Shellooe R; Marimuthu A; Nguyen H; Zhang KYJ; Artis DR Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 2010, 467 (7315), 596 10.1038/nature09454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai J; Lee JT; Wang W; Zhang J; Cho H; Mamo S; Bremer R; Gillette S; Kong J; Haass NK; Sproesser K; Li L; Smalley KSM; Fong D; Zhu Y-L; Marimuthu A; Nguyen H; Lam B; Liu J; Cheung I; Rice J; Suzuki Y; Luu C; Settachatgul C; Shellooe R; Cantwell J; Kim S-H; Schlessinger J; Zhang KYJ; West BL; Powell B; Habets G; Zhang C; Ibrahim PN; Hirth P; Artis DR; Herlyn M; Bollag G Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (8), 3041 10.1073/pnas.0711741105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bollag G; Tsai J; Zhang J; Zhang C; Ibrahim P; Nolop K; Hirth P Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat. Rev. Drug Discovery 2012, 11 (11), 873 10.1038/nrd3847 [DOI] [PubMed] [Google Scholar]
- 35.Schworer SA; Smirnova II; Kurbatova I; Bagina U; Churova M; Fowler T; Roy AL; Degterev A; Poltorak A Toll-like Receptor-mediated Down-regulation of the Deubiquitinase Cylindromatosis (CYLD) Protects Macrophages from Necroptosis in Wild-derived Mice. J. Biol. Chem 2014, 289 (20), 14422 10.1074/jbc.M114.547547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaikuad A; Knapp S; von Delft F Defined PEG smears as an alternative approach to enhance the search for crystallization conditions and crystal-quality improvement in reduced screens. Acta Crystallogr., Sect. D: Biol. Crystallogr 2015, 71 (Pt 8), 1627 10.1107/S1399004715007968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W XDS. Acta Crystallogr. D Biol. Crystallogr 2010, 66 (Pt 2), 125 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leslie AGW; Powell HR In Processing diffraction data with mosflm, Evolving Methods for Macromolecular Crystallography, Dordrecht, Read RJ; Sussman JL, Eds. Springer Netherlands: Dordrecht, 2007, pp 41 10.1007/978-1-4020-6316-9_4 [DOI] [Google Scholar]
- 39.Evans PR; Murshudov GN How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr 2013, 69 (Pt 7), 1204 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCoy AJ; Grosse-Kunstleve RW; Adams PD; Winn MD; Storoni LC; Read RJ Phaser crystallographic software. J. Appl. Crystallogr 2007, 40 (4), 658 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung L-W; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 2010, 66 (2), 213 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P; Lohkamp B; Scott WG; Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 2010, 66 (Pt 4), 486 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen VB; Arendall WB 3rd; Headd JJ; Keedy DA; Immormino RM; Kapral GJ; Murray LW; Richardson JS; Richardson DC MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 2010, 66 (Pt 1), 12 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H; Guranovic V; Dutta S; Feng Z; Berman HM; Westbrook JD Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr 2004, 60 (10), 1833 10.1107/S0907444904019419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.