ABSTRACT
Asymmetric cell division (ACD) is an evolutionarily conserved mechanism used by prokaryotes and eukaryotes alike to control cell fate and generate cell diversity. A detailed mechanistic understanding of ACD is therefore necessary to understand cell fate decisions in health and disease. ACD can be manifested in the biased segregation of macromolecules, the differential partitioning of cell organelles, or differences in sibling cell size or shape. These events are usually preceded by and influenced by symmetry breaking events and cell polarization. In this Review, we focus predominantly on cell intrinsic mechanisms and their contribution to cell polarization, ACD and binary cell fate decisions. We discuss examples of polarized systems and detail how polarization is established and, whenever possible, how it contributes to ACD. Established and emerging model organisms will be considered alike, illuminating both well-documented and underexplored forms of polarization and ACD.
KEY WORDS: Asymmetric cell division, Cell fate decisions, Cell fate determinants, Non-random segregation, Organelle partitioning
Summary: This Review discusses recent insights into the mechanisms driving asymmetric cell division, and its contribution to binary cell fate decisions, using established and emergent model organisms.
Introduction
Asymmetric cell division (ACD) is a conserved mechanism evolved to generate cellular diversity. A key principle of ACD is the establishment of distinct sibling cell fates by mechanisms linked to mitosis. Asymmetric fate can be established through exposure to cell extrinsic signaling cues. Alternatively, asymmetric inheritance of cell intrinsic cell fate determinants such as proteins or RNA can induce asymmetric cell fate decisions. The latter was demonstrated more than 100 years ago by Edwin Conklin who showed that, during early divisions in ascidian embryos, yellow cytoplasm segregates asymmetrically to specify muscle cell fate (Conklin, 1905). In addition to macromolecules, cell organelles such as centrosomes, midbodies, mitochondria, endoplasmic reticulum (ER) or lysosomes have been described to segregate asymmetrically. Interestingly, the asymmetric inheritance of cellular organelles appears to be the norm rather than the exception, but its connection to differential cell fate establishment remains unclear in many instances (Fig. 1).
Fig. 1.
Basic concepts of asymmetry and biased segregation. (A) Asymmetric cell division can be induced by cell extrinsic cues that are provided by the local niche. (B) Asymmetric cell division can also be induced by cell intrinsic mechanisms. Four examples that have been shown to, or potentially can, result in binary cell fate decisions, entail: (i) polarized cortical distribution of proteins, such as cell fate determinants; (ii) biased localization and segregation of RNA species; (iii) biased non-random distribution of organelles; (iv) biased distribution of protein aggregates.
ACD is relevant during the development of both invertebrates and vertebrates, and our current knowledge about this process stems to a large extent from genetically tractable invertebrate systems such as Caenorhabditis elegans or Drosophila melanogaster. However, classic embryological studies on animals that are currently not amenable to precise genetic manipulation, such as the mollusc Ilyanassa obsoleta, have also contributed a wealth of information to our current understanding of ACD. Here, we review recent literature that has provided new insights into symmetry breaking events. We highlight studies that have revealed asymmetric segregation of macromolecules or organelles and review the documented or potential influence of these events on cell fate decisions in yeast and metazoans. We also discuss how asymmetries in the cytoskeleton, spindle, centromeres and histones can contribute to ACD.
Polarized localization and biased inheritance of RNAs and proteins
Cell fate decisions can be induced through asymmetric partitioning of molecular determinants such as RNA species or proteins. For example, mRNAs that are segregated into one sibling cell can quickly produce proteins to elicit a specific cell behavior. Alternatively, regulatory RNA species or proteins can affect gene expression, protein localization and function. The polarized distribution of molecules often precedes biased segregation. In this section, we discuss examples and mechanisms of how the localization and segregation of RNA species and proteins is controlled and regulated. We also discuss potential contributions of this biased segregation to binary cell fate decisions.
RNAs
Differential RNA localization has been documented in a number of different species and cell types. One of the first examples of asymmetric RNA localization was reported for Actin isoforms in the early ascidian embryo Styela plicata using in situ hybridization (Jeffery et al., 1983). A recent global analysis of mRNA localization dynamics using high-resolution fluorescent in situ hybridization (FISH) during early Drosophila melanogaster development revealed that 71% of genes expressed during this time window display specific subcellular localization patterns. Notably, a number of these genes display a polarized distribution, being preferentially localized to either the apical or basal cell cortex (Lécuyer et al., 2007). Subcellular localization of different RNA species, including mRNAs, long non-coding RNAs and circular RNAs, was also found in a recent study using a combination of cell fractionation combined with RNA sequencing of human and Drosophila cells (Bouvrette et al., 2017). Although this study did not assay polarized distribution of RNA species per se, it clearly demonstrated that the majority of RNAs are localized in specific cellular compartments.
A clear example of how biased localization of mRNAs results in their asymmetric segregation has been shown during spiralian development. For instance, in I. obsoleta, mRNAs of the developmental patterning genes Eve, DPP and Tld are localized to the centrosome during the early cleavage cycles and subsequently segregate into one daughter cell during cell division (Lambert and Nagy, 2002). Centrosomal localization of mRNAs appears to be a dominant mechanism for embryonic patterning in this system, as it has also been observed for several other mRNAs (Kingsley et al., 2007; Lambert and Nagy, 2002). Interestingly, all of the centrosome-localized mRNAs show two discrete types of intracellular movement: an initial recruitment to particular interphase centrosomes, presumably mediated by minus-end directed transport, followed by movement to a cortical region that is inherited exclusively by one daughter cell during cell division. An intact microtubule cytoskeleton is necessary for centrosomal accumulation of mRNA but not for the subsequent relocalization to the cell cortex, which instead depends on Actin filaments (Lambert and Nagy, 2002). The cytoskeleton is also important for asymmetric localization and segregation of ASH1 mRNA in yeast; in this context, ASH1 is asymmetrically localized on the cortex via a process that involves Actin and Type V myosin Myo4 (Long et al., 1997). Centrosomal localization of mRNAs also occurs in oocytes of the surf clam (Spisula solidissima) (Alliegro et al., 2006) and in D. melanogaster embryos (Lécuyer et al., 2007).
Additional mechanistic and functional insight into polarized RNA distribution and segregation has been learned from studying asymmetrically dividing Drosophila neural stem cells, called neuroblasts. Neuroblasts divide asymmetrically to give rise to a self-renewing neuroblast and a differentiating cell termed a ganglion mother (GMC) (Gallaud et al., 2017; Homem and Knoblich, 2012). mRNA of the transcription factor Prospero (Pros; Prox1 in vertebrates) is localized on the apical cortex of neuroblasts in interphase before switching to the basal cell cortex in mitosis (Li et al., 1997; Schuldt et al., 1998). Apical pros mRNA localization is dependent on Inscuteable (Insc) protein and the RNA-binding protein Staufen (STAU1/2 in vertebrates), which binds the Prospero 3′ untranslated region (Li et al., 1997; Schuldt et al., 1998). Miranda protein, another neuroblast cell fate determinant, colocalizes with Staufen protein and pros mRNA during neuroblast division, and both molecules depend on Miranda for their correct localization (Schuldt et al., 1998). Interestingly, inscuteable mRNA has also been observed to localize to the apical prophase neuroblast cortex independently of Staufen; Inscuteable and Staufen proteins are both components of an apical complex (Li et al., 1997). Recently, the MS2 system (Bertrand et al., 1998; Fusco et al., 2003) has made it possible to assay the dynamics of mRNA localization in neuroblasts via live cell imaging (Ramat et al., 2017). Using this system, it was found that Miranda is localized in two distinct pools: around the apical centrosome and on the basal neuroblast cortex. This study revealed that the apical localization of Miranda depends on centrosomes and Staufen, and that basal miranda mRNA is necessary to maintain basal Miranda protein localization (Ramat et al., 2017). Interestingly, STAU2 is associated with cell fate determinants and is distributed asymmetrically during progenitor divisions in the developing mouse cortex (Kusek et al., 2012).
Other RNA species have also been shown to exhibit specific subcellular localization patterns and functions. For example, the long non-coding RNA cherub was discovered to be localized on the basal cortex of mitotic larval neuroblasts and to segregate asymmetrically into the immature neural progenitor cell (Landskron et al., 2018). cherub depends on Staufen for its basal localization and establishes a molecular link between the RNA-binding proteins Staufen and Syncrip [Syp; heterogeneous nuclear ribonucleoprotein R (HNRNPR) in humans]. Interestingly, cherub is not required for normal ACD nor development but is necessary for tumor progression in brain tumor (brat; Trim2, Trim3, Trim32 in vertebrates) mutant brains; specifically, cherub compromises normal temporal neuroblast progression, thereby allowing tumor cells to divide indefinitely (Landskron et al., 2018). Long non-coding RNAs are also important during cell fate establishment in mammalian embryos. In early mouse embryos, the long non-coding RNA lincGET (Gm45011) was shown to be expressed asymmetrically and transiently at the two- to four-cell stage transition of preimplantation embryos (Wang et al., 2018). LincGET physically binds to coactivator-associated arginine methyltransferase 1 (CARM1), promoting its nuclear localization, and biases blastomeres towards an inner cell mass (ICM) fate by activating ICM genes (Wang et al., 2018). This report suggests that cell fate bias in mammalian embryos is already established at the two- to four-cell stage. Previously, it was thought that lineage separation does not occur until later and might be linked to expression of the transcription factor CDX2, transcripts of which are localized apically at the eight-cell stage and subsequently inherited asymmetrically to separate pluripotent from differentiating cells (Skamagki et al., 2013).
These examples illustrate how RNAs can be localized in a polarized fashion and, subsequently, how they can be segregated asymmetrically. However, additional mechanistic details of RNA localization dynamics as well as the cellular and developmental functions of localized RNA species remain to be defined.
Proteins
Polarized protein distribution and biased segregation has been investigated extensively in fly neuroblasts and C. elegans embryos. Recent reviews have covered this topic quite extensively (Gallaud et al., 2017; Loyer and Januschke, 2020) so, below, we only briefly introduce some key concepts and new findings.
One of the earliest examples of proteins that regulate asymmetric cell division are the Partitioning defective (PAR) polarity proteins. PAR proteins were initially discovered in C. elegans as proteins that polarize the zygote by forming two opposing protein domains (Kemphues et al., 1988), but are now known to be evolutionarily conserved, acting as universal regulators of apical-basal polarity (Boxem and Heuvel, 2019; Cravo and van den Heuvel, 2019). During asymmetric cell division of Drosophila neuroblasts, apical-basal polarity is determined when the PAR complex proteins Par-3 (BAZ), PAR-6 and atypical protein kinase C (aPKC) form an apical polarity cap. Recent data suggest that apical aPKC localization starts in early prophase with aPKC forming discontinuous patches on the apical neuroblast hemisphere (Oon and Prehoda, 2019). These patches increase in size and subsequently coalesce into an apical crescent by metaphase, which disassembles again into cortical patches in telophase. How the early aPKC patches are positioned in the apical hemisphere is unknown but the subsequent crescent formation depends on cortical flow. A similar mechanism has also been proposed for Par-3 (Oon and Prehoda, 2019).
Recently, a novel interaction between the second PDZ domain of Par-3 and a highly conserved PDZ-binding motif (PBM) in aPKC was discovered (Holly et al., 2020). Par-3 is phosphorylated by the full PAR complex and phosphorylation induces dissociation of the Par-3 phosphorylation site from the aPKC kinase domain but does not disrupt the Par-3 PDZ2-aPKC PBM interaction. This is the first Par-3-aPKC interaction shown to be required for the cortical recruitment and polarization of aPKC in neuroblasts (Holly et al., 2020).
The activity of the PAR complex is instrumental in establishing the localization of basal cell fate determinants such as Miranda and Numb, which segregate specifically into the GMC to induce a change in cell fate (Loyer and Januschke, 2020). Miranda is initially localized on the apical interphase centrosome in embryonic neuroblasts (Mollinari et al., 2002) but is uniformly cortical in interphase larval neuroblasts (Sousa-Nunes et al., 2009). In metaphase, Miranda starts forming a basal crescent (Matsuzaki et al., 1998). Basal localization is induced through aPKC, which excludes it from the apical cortex through direct phosphorylation (Atwood and Prehoda, 2009), and maintained through the actomyosin cytoskeleton (Hannaford et al., 2018). Miranda phosphorylation status and subcellular localization are also regulated through Protein phosphatase 4 and associated proteins such as the Phosphotyrosyl phosphatase activator (PTPA) (Sousa-Nunes et al., 2009; Zhang et al., 2015). Miranda itself acts as a cargo protein, transporting the translational inhibitor Brat and Pros (reviewed by Gallaud et al., 2017). In GMCs, Pros represses genes required for self-renewal, such as stem cell fate genes and cell cycle genes, but also activates genes for terminal differentiation (Choksi et al., 2006).
It is of interest to note that, in fly neuroblasts, several transcripts and their corresponding proteins co-localize. However, it is currently unclear whether polarized mRNA localization correlates with local protein translation, and the function of localized mRNA in this context thus needs to be further explored.
Asymmetry of the cytoskeleton
Centrosome asymmetry and biased segregation
Centrosomal mRNA localization is suggestive of molecular and/or structural differences between centrosomes within a given cell. A prime example of this is seen in Drosophila neuroblasts, which display differences in microtubule organizing center (MTOC) activity, specifically during interphase. Centrosomes contain two centrioles, surrounded by pericentriolar matrix (PCM) proteins, which are necessary to form an MTOC. Centrioles replicate only once during the cell cycle: a ‘daughter’ centriole forms at a right angle to the older ‘mother’ centriole. The two centrioles later disengage in the cell cycle, generating two mature MTOCs that form a bipolar spindle (Conduit et al., 2015). This replication cycle implies an intrinsic asymmetry in centriole age, which has also been recognized by molecular criteria (Jakobsen et al., 2011; Januschke et al., 2011). In neuroblasts, the daughter centriole-containing centrosome maintains the ability to nucleate microtubules throughout interphase, whereas the mother centriole-containing centrosome downregulates such MTOC activity as neuroblasts enter interphase. This biased MTOC activity helps orient the mitotic spindle along the neuroblast intrinsic apical-basal polarity axis, as the active MTOC remains tethered to the apical neuroblast cortex. The daughter centriole-containing centrosome downregulates MTOC activity through ‘shedding’ of its PCM content, thereby displacing the inactivated centrosome from the apical cortex. During prophase, both centrosomes accumulate PCM components and nucleate microtubules (Conduit and Raff, 2010; Januschke and Gonzalez, 2010; Lerit and Rusan, 2013; Rebollo et al., 2007; Rusan and Peifer, 2007; Singh et al., 2014). This stereotypical MTOC behavior results in biased centrosome segregation. The apical centrosome always contains the younger daughter centriole, which will remain in the self-renewed neuroblast after division. By contrast, the mother centriole-containing centrosome segregates into the differentiating GMC (Januschke et al., 2011).
Similar segregation bias has also been observed in Drosophila germline stem cells (Salzmann et al., 2014; Yamashita et al., 2007), mouse neural stem cells (Wang et al., 2009) and, originally, in yeast (Pereira et al., 2001). However, few examples clearly demonstrate the functional consequences of biased centrosome inheritance. Recently, this issue was specifically addressed with the help of a genetically reengineered yeast strain in which inheritance of the old spindle pole body (SPB; the yeast equivalent of the centrosome/MTOC) was biased towards the mother cell instead of the younger daughter cell (bud). This study showed that inverted MTOC fate compromises the proper distribution of protein aggregates and aged mitochondria, resulting in reduced replicative lifespan of the new daughter cell (Manzano-López et al., 2019). It has also been reported that centrosomal localization of the Notch regulator Mindbomb1 (Mib1) has functional consequences (Tozer et al., 2017). Mib1m, which is a mono-ubiquitin ligase that regulates the trafficking of Notch ligands and promotes their activity (Weinmaster and Fischer, 2011), localizes asymmetrically on daughter centriole satellites in interphase chick progenitors (Tozer et al., 2017). In this context, Mib1 either remains asymmetrically localized during mitosis or is distributed symmetrically on both centrosomes through a compensatory mechanism from the Golgi. In the case of asymmetric localization, Mib1 correlates with neurogenic divisions (i.e. giving rise to a progenitor and a neuron), with Mib1 being inherited by the prospective neuron. This suggests that centrosomal localization of Mib1 is essential for the asymmetric activation of Notch signaling in neurogenic divisions and regulates the balance between proliferation and differentiation. Furthermore, it provides evidence for a centrosomal fate determinant that is localized and segregated asymmetrically (Tozer et al., 2017).
Biased MTOC activity can also induce asymmetric nuclear envelope breakdown (Ranjan et al., 2019). Previously it was shown that, before nuclear envelope breakdown (NEBD), early spindle microtubules induce folds and invaginations in the nuclear envelope, creating mechanical tension that results in local nuclear envelope rupture (Beaudouin et al., 2002). Mechanistically, this could involve nuclear envelope localized Dynein/Dynactin, which pull on nuclear membranes poleward along astral microtubules (Salina et al., 2002). Such asymmetric NEBD can have consequences on chromosome attachment and segregation (see below).
What is the mechanism underlying biased MTOC activity and asymmetric centrosome segregation? In Drosophila male germline stem cells (GSCs), differential MTOC activity correlates with centrosome maturity; the older, mother centriole-containing centrosome retains PCM and MTOC activity, thereby retaining the centrosome near the stem cell niche (Yamashita et al., 2007). However, in Drosophila neuroblasts, cortical cues mediated through the polarity protein Partner of inscuteable [Pins; LGN (Gpsm2) and AGS3 (Gpsm1) in vertebrates] influence asymmetric MTOC activity (Rebollo et al., 2007). Similarly, in yeast it was found that Kar9 and spatial cues, rather than the kinetics of SPB maturation, control the asymmetry of astral microtubule organization between the preexisting and new SPBs (Lengefeld et al., 2017). How such spatial cues affect differential MTOC activity is currently unclear. From studies of Drosophila neuroblasts, we have learned that differential MTOC activity can be regulated by the mitotic kinase Polo (Plk1 in vertebrates). Polo phosphorylates a number of PCM proteins, and this is necessary for MTOC activity (Conduit et al., 2014, 2015; Feng et al., 2017; Novak et al., 2016). Maintaining Polo/Plk1 at the daughter centriole is necessary for PCM maintenance (Conduit and Raff, 2010; Januschke and Gonzalez, 2010). Although the apical daughter centriole maintains Polo/Plk1, and thus MTOC activity, the mother centriole downregulates Polo/Plk1 and its PCM content, thereby losing MTOC activity and the ability to remain tethered to the apical neuroblast cortex (Conduit and Raff, 2010; Januschke et al., 2013; Lerit and Rusan, 2013; Ramdas Nair et al., 2016; Rebollo et al., 2007; Rusan and Peifer, 2007; Singh et al., 2014). Recently, Polo-like kinase 4 (Plk4; SAK), an important regulator of centriole duplication, has also been implicated in establishing centriolar asymmetry and biased MTOC activity. Plk4 phosphorylates Spd-2, which triggers basal-like centriole behavior (Gambarotto et al., 2019). Interestingly, biased MTOC activity in neuroblasts disappears during mitosis when the mother centriole-containing centrosome initiates maturation, reforming a second active MTOC. In yeast, differential microtubule growth has been attributed to the kinesin Kip2, which is specifically recruited to the old SPB (Chen et al., 2019). Kip2 prevents microtubule catastrophe and promotes microtubule extension (Carvalho et al., 2004; Cottingham and Hoyt, 1997; Hibbel et al., 2015; Huyett et al., 1998). Phosphorylation of Kip2 prevents random microtubule binding, restricting it initially to the minus-end. Thus, Kip2 recruitment, which depends on Bub2 and Bfa (Bfa1), could account for the generation of longer astral microtubules emanating from the old SPB through the ability of Kip2 to prevent microtubule catastrophe and promote microtubule elongation (Chen et al., 2019).
Spindle and cell size asymmetry
An immediate consequence of centrosome asymmetry and biased MTOC activity is asymmetric centrosome segregation, as discussed above. However, as MTOCs are the main contributor to bipolar spindle formation, biased MTOC activity could also affect bipolar spindle asymmetry, with potential implications for cell shape and sibling cell size. Recently, it was found that the microtubule-depolymerizing kinesin Klp10A is specifically localized on the stem cell centrosome of Drosophila male GSCs. Loss of Klp10A results in an abnormal elongation of the GSC mother centrosome and a considerably larger MTOC and associated half spindle, giving rise to an asymmetric mitotic spindle. This ultimately results in the GSCs dividing asymmetrically by size (Chen et al., 2016). Thus, Klp10A specifically counteracts the formation of an asymmetric spindle to prevent unequal sized sibling cell formation. Other cell types, such as Drosophila neuroblasts, have implemented mechanisms to establish spindle asymmetry deliberately. The classic literature discussing spindle asymmetry in Drosophila neuroblasts has recently been reviewed (Gallaud et al., 2017). In brief, mutations in cell polarity proteins can affect spindle asymmetry (Cai et al., 2003; Yu et al., 2003) but the mechanisms remain unclear (see also below). Evidence for centrosomal proteins affecting spindle symmetry has also been found in human cells. For example, the centrosomal protein coiled-coil domain containing 61 (CCDC61) is required for spindle assembly and chromosome alignment in cultured human cells; depletion of CCDC61 reveals a clear loss of the intrinsic symmetry of microtubule tracks within the spindle (Bärenz et al., 2018).
Spindle morphology must also be controlled in cells containing acentrosomal spindles, such as oocytes. Acentrosomal Drosophila oocyte spindles are usually symmetric, but a lack of Kinesin-5 (Klp61F) results in asymmetric bipolar spindles, leading to one half of the spindle containing more microtubules (Radford et al., 2016). It is unclear how Kinesin-5, which is a microtubule bundling protein, prevents the formation of asymmetric acentrosomal spindles under normal conditions, but knocking out Kinesin-6 (Subito) together with Klp61F enhances the asymmetric spindle phenotype of Klp61F depletion. This suggests that Kinesin-5 and -6 promote spindle symmetry in Drosophila oocytes (Radford et al., 2016).
Spindle asymmetry has been shown to be an important regulator of Notch signaling in asymmetrically dividing Drosophila sensory organ precursor cells (SOPs). In this system, it was shown that Klp10A and its antagonist Patronin generate central spindle asymmetry, which in turn is used for polarized endosome motility, causing biased transport of Sara-containing endosomes into one sibling cell (Derivery et al., 2015). Biased dispatch of Sara endosomes is an important mechanism that mediates asymmetric Notch/Delta signaling during the asymmetric division of Drosophila SOPs (Coumailleau et al., 2009). Spindle asymmetry could also be a mechanism for biased chromosome segregation in meiosis, often referred to as ‘meiotic drive’ (Kursel and Malik, 2018). This process was recently shown in mouse oocytes, which rely on CDC42 signaling from the cell cortex to regulate microtubule tyrosination, inducing spindle asymmetry and non-Mendelian bivalent segregation (Akera et al., 2017; 2019) (see also below).
What is the relationship between spindle asymmetry and sibling cell size asymmetry? As studies of Klp10A mutant male GSCs have shown, increasing MTOC activity of the stem cell centrosome results in the formation of asymmetric spindles and, subsequently, physically asymmetrically dividing GSCs; note that GSCs usually divide symmetrically with regard to size (Chen et al., 2016). Similarly, loss of the Drosophila polarity protein Pins, which also affects biased MTOC activity in interphase (Rebollo et al., 2007; see also above) and spindle asymmetry during mitosis (Cai et al., 2003; Yu et al., 2000), causes neuroblasts to divide symmetrically by size (Cabernard et al., 2010). However, whether sibling cell size asymmetry is exclusively determined by spindle asymmetry is not entirely clear. Recently, it was shown that cortical cues can override intrinsic spindle asymmetry. For example, Drosophila neuroblasts lacking Protein kinase N (Pkn; Pkn1-3 in vertebrates) show transient defects in sibling cell size asymmetry during mitosis; in contrast to wild-type neuroblasts, which form a large apical neuroblast and a small GMC, these pkn mutant neuroblasts initially display a reduced apical and an expanded basal cortex in early anaphase (Tsankova et al., 2017). Although transient, this inverted asymmetry appears to be a consequence of altered Non-muscle myosin II (Myosin, hereafter) localization. Specifically, in contrast to wild-type neuroblasts, pkn mutant neuroblasts fail to initiate timely apical Myosin clearing, which could prevent expansion of the apical neuroblast cortex (Cabernard et al., 2010; Connell et al., 2011; Roubinet et al., 2017; Tsankova et al., 2017). Moreover, forced apical localization of activated Myosin can result in inverted ACDs (Roubinet et al., 2017; Tsankova et al., 2017). These and other observations suggest that spindle asymmetry is partially dependent on the stiffness of the apical cell cortex. Indeed, recent stiffness measurements have revealed a complex picture of stiffness changes during asymmetric neuroblast division (Pham et al., 2019). Before the establishment of an asymmetric anaphase spindle, activated Myosin localizes on the cortex and engages with filamentous Actin (F-Actin). Asymmetric Myosin distribution was observed some time ago (Barros et al., 2003), but more recent quantitative analyses have revealed that Myosin moves away from the apical cortex towards the cleavage furrow through a cortical flow mechanism in early anaphase (Barros et al., 2003; Cabernard et al., 2010; Connell et al., 2011; Roubinet et al., 2017). This ‘relief’ of Myosin-dependent cortical tension could allow microtubules to expand asymmetrically, thereby pushing out the apical cell cortex. However, in Drosophila neuroblasts, apical cortical expansion is spindle-independent (Connell et al., 2011). Instead, recent biophysical measurements suggest that intracellular pressure contributes to the initial apical expansion (Pham et al., 2019). Taken together, the combination of apical Myosin clearing, through cortical flow, and the localized dissipation of hydrostatic pressure contribute towards the generation of sibling cell size asymmetry (Pham et al., 2019).
Myosin localization is also important during the establishment of physical asymmetry in C. elegans neuroblasts (Ou et al., 2010). The molecular mechanisms triggering asymmetric Myosin establishment remain to be elucidated but insight has been obtained from studies in C. elegans embryos, which initiate polarization shortly after fertilization through reorganization of the actomyosin contractile network (Munro et al., 2004). Recently, it was found that the mitotic kinase Aurora-A accumulates around centrosomes to locally disrupt actomyosin contractile activity at the proximal cortex, thereby promoting a cortical flow towards the anterior pole (Klinkert et al., 2019; Zhao et al., 2019). Other cortical proteins, such as the ERM (Ezrin, Radixin, Moesin) family protein Moesin, have also been shown to be apically enriched during asymmetric neuroblast division and are required for the formation of sibling cell size asymmetry (Abeysundara et al., 2017).
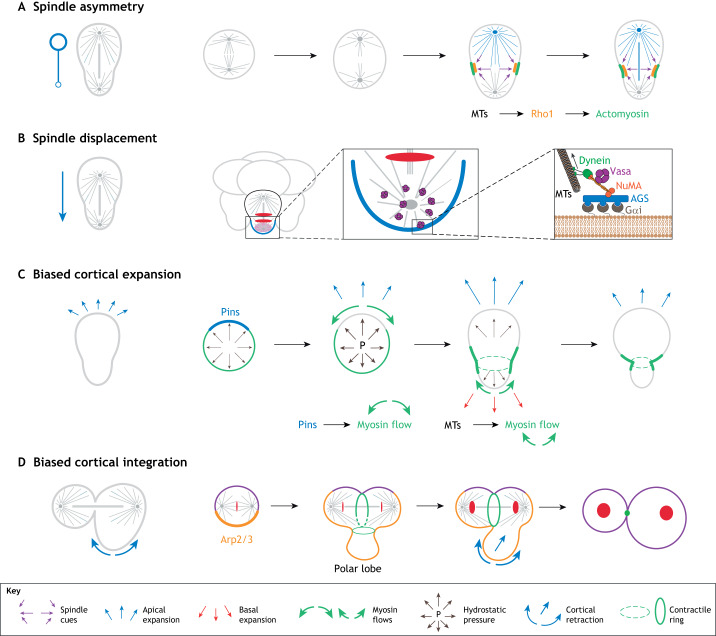
A key principle emerging from these studies is that cells utilize several different mechanisms to establish sibling cell size asymmetry (Roubinet and Cabernard, 2014). As outlined above, centrosome-dependent and centrosome-independent mechanisms can result in asymmetric mitotic spindles, which shift the cleavage furrow to one side of the cell cortex (Fig. 2A). Alternatively, cortical cues can pull the mitotic spindle towards one side of the cell cortex, thereby displacing furrow positioning cues off center (Fig. 2B). A recent demonstration of this concept was shown in symmetrically dividing sea urchin zygotes, by pulling the mitotic spindle towards one side of the cell cortex using injected magnetic beads and a local magnetic field. This revealed that displacing the mitotic spindle under these conditions induces physical ACD (Sallé et al., 2018). Similarly, sea urchin embryos undergo ACD to produce micromeres – small organizers that provide inducing cues to adjacent cells important for gastrulation – at the vegetal poles of embryos as they transition from the 8- to 16-cell stage. Micromeres are smaller than their macromere sibling cells and inherit the RNA helicase protein Vasa (Juliano et al., 2006). The polarity factor AGS is both necessary and sufficient to establish this physical and molecular ACD. Sea urchin AGS contains three GoLoco motifs, whereas sea star AGS lacks GoLoco motif #1, and it was recently shown that expressing sea urchin AGS in sea star embryos is sufficient to induce physical ACDs (Poon et al., 2019). It has also been shown that the core cortical force-generating machinery responsible for such cortical pulling forces is evolutionarily conserved and contains the Dynein-Dynactin complex, NuMA (Lin5 in C. elegans; Mud in Drosophila) and the Gαi complex (GOA-1, GPA-16 in C. elegans; Gαi in Drosophila) (Kiyomitsu, 2019; Poon et al., 2019) (Fig. 2B). Cell shape, adhesion geometry, cellular junctions and mechanical tension are other factors determining spindle orientation and positioning (Finegan and Bergstralh, 2019; van Leen et al., 2020; Li et al., 2019). Finally, as seen in Drosophila and C. elegans neuroblasts, measurable dynamic changes in the cell cortex during anaphase can induce sibling cell size asymmetry (Fig. 2C). Interestingly, a recent study of the developing notochord of the chordate Ciona revealed that different notochord blastomeres utilize a combination of polarized displacement of mitotic spindles, mother cell shape and post-anaphase mechanisms between different rounds of cell division to establish unequal sibling cell size (Winkley et al., 2019).
Fig. 2.
Mechanisms that can generate sibling cell size asymmetry. (A) Asymmetric spindles – due to biased MTOC activity generating a larger half-spindle – can push cleavage furrow-inducing cues towards the opposite cell cortex. Such spindle-dependent cues activate the RhoA pathway, which is necessary to assemble an actomyosin-containing contractile ring (Basant and Glotzer, 2018; D'Avino et al., 2015). (B) Cortical pulling forces can pull the mitotic spindle towards one cell pole, thereby shifting cleavage furrow positioning cues off-center. This mechanism largely depends on the conserved Gαi/LGN or AGS, NuMA, Dynein complex (shown in inset). In sea urchins, the cell fate determinant Vasa only forms a complex with Dynein and NuMA on the vegetal pole, ensuring its biased segregation into micromeres. (C) Biased cortical expansion, based on a build up of hydrostatic pressure and the timely relocalization of Myosin, can initiate the formation of sibling cell size asymmetry. Polarity- and spindle-dependent cues (e.g. Pins) control the relocalization of Myosin via cortical flow mechanisms. (D) Molluscs and some annelids form polar lobes, which reintegrate into one sibling cell only, thereby generating two cells differing in their size. The Arp2/3 complex, which nucleates the polymerization of branched Actin networks, has been implicated in defining the zone of cortex to be sequestered into the polar lobe. MTs, microtubules.
An additional, evolutionarily ancient but poorly described mechanism to generate unequal sized sibling cells is applied in many molluscs and some annelid species (Chen et al., 2006). These invertebrates generate two sibling cells differing in their size by forming and reintegrating a polar lobe (Fig. 2D). Polar lobes, also known as antipolar or yolk lobes, are transient vegetal protrusions, forming during the first and second embryonic divisions, which sequester vegetal cytoplasm that is inherited by the CD and D blastomeres, respectively (Conrad and Williams, 1974; Conrad et al., 1973, 1990; Morgan, 1933; Schmidt et al., 1980). Most studies on polar lobe formation and resorption have been conducted in the mud snail I. obsoleta, revealing that both Actin and Myosin are necessary for polar lobe formation and resorption (Hejnol and Pfannenstiel, 1998). The most recent study, using two closely related species of scallops, Chlamys hastada and C. rubida, revealed that the patch of cell cortex destined for polar lobe sequestration is marked by enrichment of the Arp2/3 complex. In addition, inhibition of Arp2/3 affects polar lobe formation and partitioning of cytoplasm into the two daughter cells, suggesting that Arp2/3 plays a functional role in defining the zone of cortex to be sequestered into the polar lobe (Toledo-Jacobo et al., 2019). The molecular mechanisms underlying polar lobe formation are currently unknown, but classic microsurgery experiments revealed that polar lobes are instrumental in cell fate decisions (Atkinson, 1971; Clement, 1952; Crampton and Wilson, 1896; Render, 1989).
Although much more could be learned about ACD by examining non-traditional or emerging model systems such as snails or scallops, the scarcity of molecular methods for studying these species is currently a confounding factor. Therefore, studies of both traditional and non-traditional model systems will be instrumental for investigating how sibling cell size asymmetry contributes to cell fate decisions. Moreover, although mutants that affect sibling cell size asymmetry can provide valuable mechanistic insight, they are often ill suited to address functional questions, owing to pleiotropic effects or early developmental requirements of the corresponding gene products. Thus, precise manipulations of sibling cell size asymmetry with high spatiotemporal control should be used to gain a better understanding of this process.
Centromere asymmetry, histone asymmetry and biased chromosome segregation
Epigenetic mechanisms play important roles in cell fate specification by altering chromatin structure and gene expression. How epigenetic mechanisms are linked to ACD is currently one area of active investigation (Fig. 3). For example, it has been shown that during asymmetric division of Drosophila male GSCs, the pre-existing canonical histones H3 and H4 are preferentially retained by the stem cell, whereas newly synthesized H3 and H4 segregate into the differentiating daughter cell, which is known as a gonialblast (Tran et al., 2012; Wooten et al., 2019a,b). In contrast, H2A and H2B segregate symmetrically (Wooten et al., 2019a). One proposed mechanism underlying this biased segregation is the phosphorylation of threonine 3 of H3 (H3T3P), which can distinguish old versus new H3 in the prophase of GSCs. Loss of H3T3P phosphorylation disrupts asymmetric H3 inheritance, resulting in stem cell loss and the formation of early-stage germline tumors (Xie et al., 2015). Such biased histone segregation could be explained by biased replication fork movement coupled with a strand preference in histone incorporation (Wooten et al., 2019a).
Fig. 3.
Spindle and centromere asymmetry bias sister chromatid segregation. In Drosophila male germline stem cells, mother centrosomes generate an active MTOC before the daughter centrosomes do. Asymmetry in nuclear envelope breakdown then allows microtubules from the mother centrosome to attach to sister chromatids containing larger kinetochores. Sister centromeres are differentially enriched with proteins involved in centromere specification and kinetochore function. This results in preferential recognition and attachment of microtubules to asymmetric sister kinetochores and sister centromeres. These mechanisms ensure that epigenetically distinct sister chromatids are partitioned asymmetrically in male germline stem cells. MTs, microtubules.
Another form of epigenetic modification occurs at centromeres, which together with kinetochore proteins form the microtubule attachment sites necessary for faithful chromosome segregation. Centromeric chromatin does not contain a specific DNA sequence but is epigenetically defined by the histone H3 variant CENP-A (CID in flies) (Allshire and Karpen, 2008). Drosophila intestinal stem cells predominantly retain previously synthesized CENP-A, whereas differentiating progenitor cells are enriched with newly formed CENP-A (García del Arco et al., 2018). The mechanisms and function of this biased CENP-A segregation remain to be further explored. Similarly, CENP-A has been found to be enriched on the sister chromatid, segregating into GSCs in the male Drosophila testis (Ranjan et al., 2019). How does this epigenetic modification influence chromatid segregation and, potentially, cell fate decisions? New evidence, mostly from studies of Drosophila GSCs, suggests that the kinetochore protein Ndc80 is also asymmetrically localized, correlating with CENP-A enrichment. As mentioned above, the nuclear envelope specifically ruptures first on the prospective GSC side, creating an opening for microtubules from the more active mother centrosome to penetrate the nuclear envelope and attach to chromatids exhibiting a higher Ndc80 concentration. This in turn can result in biased chromatid segregation (Ranjan et al., 2019) (Fig. 3). This mechanism is very similar to that observed in mouse oocytes, which also display asymmetric microtubules that preferentially attach to a set of kinetochore complexes to bias chromosome segregation (Akera et al., 2017, 2019; Wu et al., 2018). This ‘meiotic drive’ in oocytes is determined through differences in centromeres between homologous chromosomes, whereas ‘mitotic drive’ occurs between genetically identical sister chromatids. As sister centromeres are theoretically identical in their sequence, CENP-A must assemble asymmetrically through a currently unknown mechanism (Wooten et al., 2019b), and further studies are needed to reveal the mechanisms underlying this event.
Asymmetric segregation of protein aggregates and organelles
Protein aggregates
Protein aggregates form when hydrophobic patches of multiple unfolded proteins stick to each other. Small heat-shock proteins (Hsps) function as the first line of defense in preventing irreversible protein aggregation under stressful conditions or during cellular ageing. For example, in heat-shocked cells, unfolded proteins may expose ‘hydrophobic patches’ leading to improper protein-protein interactions and, often, to the formation of cytotoxic insoluble aggregated complexes. The association of small Hsps with unfolded or misfolded protein substrates prevents aggregation and accumulation of cytotoxic protein complexes and facilitates their active repair and refolding through ATP-dependent chaperones such as Hsp104p (Cashikar et al., 2005; Haslbeck et al., 2005; Liberek et al., 2008).
The accumulation of protein aggregates can be prevented through asymmetric partitioning. Dividing budding yeast cells, for example, confine protein aggregates to the aging mother cell in order to produce rejuvenated daughters. Hsp26p is a small heat-shock chaperone that is involved in protein homeostasis and associated with a variety of proteins that are prone to aggregation (Cashikar et al., 2005). Hsp26p is repressed under optimal growth conditions but highly upregulated upon heat-shock oxidative stress or nutrient depletion when cells enter the stationary phase (Franzmann et al., 2008). Hsp26p belongs to a group of long-lived asymmetrically retained proteins (LARPs) that assemble into distinct cytoplasmic, microscopically visible foci when cells enter the stationary phase or after a heat shock. These Hsp26p foci, which are potentially associated with protein aggregates, are retained almost exclusively in mother cells upon re-entry to proliferation or upon recovery to normal temperature (Thayer et al., 2014).
Confining protein aggregates to the aging mother cell requires asymmetry-generating genes (AGGs), which govern the asymmetrical inheritance of aggregated proteins. A recent genome-wide screen for candidate AGGs in yeast revealed a role for vesicle trafficking, membrane fusion and Myosin-dependent vacuole inheritance in this asymmetric inheritance process. For example, the Myosin-dependent vacuole inheritance adaptor protein Vac17 and the endocytic vesicle-associated protein dynamin-like protein Vps1 were shown to regulate asymmetry and replicative lifespan through Myo2-dependent effects on endocytosis and spatial protein quality (Hill et al., 2016). Asymmetric segregation of protein aggregates in yeast can also be achieved through an ER compartmentalization-based process. For example, protein deposit precursors, which are misfolded proteins and aggregation seeds, are captured by the ER membrane-bound chaperone Ydj1. Then, the ER lateral diffusion barrier – a distinct ER membrane domain at the bud neck, separating the ER in the mother from the bud (Clay et al., 2014; Luedeke et al., 2005) – facilitates the asymmetric segregation of aggregates (Fig. 4A) (Saarikangas et al., 2017).
Fig. 4.
Asymmetric segregation of protein aggregates and organelles. (A) In yeast cells, the ER membrane-bound chaperone Ydj1 captures protein aggregates in the mother cell. ER compartmentalization by diffusion barriers then facilitates the retention of protein aggregates in the mother cell. (B) In hematopoietic stem cells (HSCs), lysosomes, autophagosomes and mitophagosomes are partitioned non-randomly. The sibling cell that inherits more lysosomes, autophagosomes and mitophagosomes retains stem cell properties. (C) In epithelial cells within mitotic domain 1 of stage 8 Drosophila embryos, the ER (purple) is asymmetrically localized during mitosis, resulting in its biased partitioning. In mitotic domain 4, by contrast, the ER segregates symmetrically.
Asymmetric partitioning of damaged proteins is not confined to yeast and has been implicated in various important biological processes, including neurodegeneration protection in multicellular organisms and rejuvenation of newborn microbial cells upon succeeding cell divisions or during recovery from unfavorable environmental conditions (Hill et al., 2017; Moore and Jessberger, 2017). It has also been demonstrated that ER diffusion barriers help with asymmetric segregation of damaged proteins between daughter cells of mouse neural stem cells (Moore et al., 2015).
Mitochondria
Mitochondria – the power houses of cells – produce ATP, which is the standard currency for cellular energy. Accurate segregation of mitochondria is important for the health and fate of daughter cells (Mishra and Chan, 2014). In yeast, the quantity of mitochondria partitioned to the bud is precisely controlled, whereas the quantity of mitochondria retained in the mother cell declines with age (Rafelski et al., 2012). Furthermore, less functional and aged mitochondria are thought to be retained in mother cells, whereas buds receive highly functional organelles (Hughes and Gottschling, 2012; McFaline-Figueroa et al., 2011; Pernice et al., 2016). Anterograde (toward the bud) mitochondrial transport is mediated by Myo2 (Altmann et al., 2008; Chernyakov et al., 2013). A recent genetic screen revealed an unexpected interaction between myo2 and genes required for mitochondrial fusion; when Myo2 transport capacity is limited, mitochondria must be in a fused state to ensure that enough mitochondria are partitioned into the bud. Similarly, fused mitochondria ensure retention of a crucial mitochondrial quantity in the mother cell when bud-directed transport is enforced (Böckler et al., 2017). Interestingly, the machinery used for the retention of damaged cytosolic proteins and aggregates in the mother cell may also be involved in biased mitochondrial inheritance (Zhou et al., 2014). Based on these findings, it was proposed that minimal activity of Myo2 is required to distribute mitochondria in the mother cell and allow capture of protein aggregates, a prerequisite for aggregate retention. On the other hand, increased activity of Myo2 shifts mitochondria-associated aggregates into the bud and thereby perturbs aggregate retention. Thus, fine-tuned Myo2-dependent mitochondrial transport is essential for retention of cytosolic protein aggregates in the mother cell (Böckler et al., 2017).
Evidence for asymmetric partitioning of mitochondria has also been linked with stemness. Stem-like cells (SLCs) of immortalized human mammary epithelial cells inherit fewer old mitochondria and retain stem cell properties, manifested in their ability to form mammospheres (Katajisto et al., 2015). Stem cells normally confine mitochondria, containing old proteins, to distinct sub-cellular domains by a mechanism involving Dynamin related protein (Drp1), a protein that is essential for mitochondrial fission and autophagy (Mao et al., 2013; Mishra and Chan, 2014). Inhibition of mitochondrial fission disrupts both the age-dependent subcellular localization and segregation of mitochondria and causes loss of stem cell properties in the progeny cells (Katajisto et al., 2015). Notably, SLCs have a higher mitophagy/autophagy ratio, suggesting that a high quality of mitochondria might be relevant for the SLC state and for asymmetric segregation of mitochondria. These data suggest that any perturbation that challenges normal mitochondrial quality control mechanisms will either serve as a signal for an SLC to stop asymmetric segregation of mitochondria or, alternatively, overload the capacity of SLCs to effectively apportion old mitochondria asymmetrically (Katajisto et al., 2015).
A similar Drp1-dependent mechanism has been observed in activated lymphocytes, which use ACD to couple differentiation and self-renewal (Adams et al., 2016). In this context, it was shown that unequal elimination of aged mitochondria leads to different sibling cell fates: daughter cells clearing more mitochondria self-renew, whereas siblings retaining more mitochondria differentiate (Adams et al., 2016). Accordingly, genetically and pharmacologically inhibiting Drp1 leads to enhanced differentiation as well as elevated levels of mitochondrial and cellular reactive oxygen species (ROS) levels. Cells with higher levels of mitochondrial ROS have reduced aged mitochondria clearance. Conversely, scavenging ROS with N-acetyl-L-cysteine (NAC) stimulates the clearance of aged mitochondria and promotes self-renewal. Confocal microscopy revealed that the subcellular distribution of aged mitochondria is generally symmetric in metaphase and early telophase cells. In cytokinesis, however, the abundance of aged mitochondria between sibling pairs is frequently discordant. In summary, ROS mediates the effects of, and reinforces the clearing of, aged mitochondria in differentiated daughter cells.
Mitochondria also segregate asymmetrically during meiosis 1 in mice, such that the majority of mitochondria end up in the oocyte opposite to the polar body, which is eventually degraded (Brunet and Verlhac, 2010; Dalton and Carroll, 2013). Asymmetric segregation of mitochondria in this case is important because the oocyte does not multiply mitochondria until fertilization. Furthermore, a block in glycolysis until the blastocyst stage makes mitochondria the sole source of ATP during initial stages of development (Dumollard et al., 2007). Biased mitochondria segregation during meiosis involves the meiotic spindle and spindle displacement mechanisms. Initially, mitochondria accumulate around the spindle but, during anaphase, they are then transported towards the oocyte side on microtubules in a kinesin- and dynein-dependent manner (Dalton and Carroll, 2013). Subsequently, the meiotic spindle migrates to the cortex during polar body extrusion in meiosis 1 (Ledan et al., 2001). This process of spindle migration requires the actin cytoskeleton; in the absence of actin-dependent spindle migration, mitochondria remain symmetrically distributed (Mogessie et al., 2018).
Lysosomes
Lysosomes have been shown to segregate asymmetrically in dividing keratinocytes: they concentrate on the centrosome side of the nucleus immediately before mitosis, and then preferentially segregate into one daughter cell (Lång et al., 2018). Lysosome-rich keratinocytes have a higher two-cell colony rotation rate, an indicator of human keratinocyte stemness (Nanba et al., 2016), and also give rise to colonies that express the stem cell marker cytokeratin 15 (K15; KRT15) (Lång et al., 2018).
Another system showing biased lysosome segregation is that of hematopoietic stem cells (HSCs). HSCs undergo self-renewing divisions, reforming HSCs while also giving rise to sibling cells that can differentiate into all blood lineages. Using quantitative live cell imaging, a recent report revealed that the cellular degradative machinery – including lysosomes, autophagosomes and mitophagosomes – can be asymmetrically inherited by HSC daughter cells (Loeffler et al., 2019). Moreover, HSC daughter cells with low levels of lysosomes are induced to differentiate, without inducing specific lineage choices (Fig. 4B). This report also showed that Numb, an inhibitor of Notch signaling (Kopan and Ilagan, 2009; Kovall et al., 2017), can segregate asymmetrically in HSCs; daughter cells receiving less Numb are metabolically activated and induced to differentiate (Loeffler et al., 2019).
Endoplasmic reticulum
Experiments in yeast have revealed that, under conditions of high and low stress, the ER segregates asymmetrically. Induction of ER stress by pharmacological agents such as tunicamycin activates the ER stress surveillance (ERSU) mechanism (Babour et al., 2010). This ERSU pathway results in re-localization of the cytokinesis-associated septin complex away from the bud neck, thereby blocking cortical ER inheritance while allowing perinuclear ER segregation. The ERSU pathway also delays cytokinesis until a minimal threshold of ER functional capacity is reached, ensuring mother cell viability (Babour et al., 2010). The ERSU pathway may be activated by the accumulation of protein aggregates in the ER. For example, in yeast, the ERSU delays the passage of damaged ER along with the protein aggregates in it to daughter cells through the formation of a lipid barrier at the bud neck (Piña and Niwa, 2015). The ERSU is a non-transcriptional mechanism that operates through the MAP kinase Slt2. Removing the ERSU by knocking out Slt2 under stress conditions causes both mother and daughter cells to die, suggesting that it is an essential pathway for adaptation (Jonas et al., 2018; Piña and Niwa, 2015). The ERSU acts independently of the unfolded protein response (UPR), which is activated when unfolded or misfolded proteins accumulate in the lumen of the ER and maintains ER function by upregulating ER chaperones and protein-folding components (Walter and Ron, 2011). However, it was shown that both the UPR and ERSU mechanisms can operate under low stress, suggesting that both pathways are important for adaptation under physiologically relevant environmental changes. For example, under low stress, activation of the ERSU pathway leads to retention of ER by the mother cell and decreased ER segregation into the daughter cell. This asymmetric inheritance necessitates UPR activation and a delayed cell cycle in daughter cells so that they can recover functionality before commencing growth (Jonas et al., 2018).
Recent data from Drosophila has revealed that, in a subset of epithelial cells of mitotic domain 1 (which is located in two dorsolateral clusters at the anterior part of the Drosophila embryo), the ER segregates asymmetrically during normal embryonic development. Interestingly, this asymmetric ER partitioning occurs in mitotic domains that give rise to brain neuroblasts, although a functional connection between biased ER segregation and neuroblast fate has not been demonstrated. Biased ER segregation requires the highly conserved ER membrane protein Jagunal (Jagn; JAGN1 in humans). Jagn itself is localized asymmetrically from prometaphase onward and segregates asymmetrically in this population of proneural cells (Eritano et al., 2017) (Fig. 4C).
Concluding remarks
Since the original description of ACD in 1905 (Conklin, 1905), much has been learned about the molecular and cellular mechanisms underlying ACD. Established model systems such as yeast, C. elegans and D. melanogaster will continue to provide molecular insight into this evolutionarily conserved mechanism and inform studies in vertebrate systems. However, biology is incredibly diverse, and much can be discovered about ACD by further exploring emerging or non-model systems. Novel techniques in genome engineering such as CRISPR/Cas9, as well as the advent of less damaging imaging methods (e.g. light-sheet microscopy), could open the door for a new frontier in ACD research, revealing new concepts and mechanisms.
In light of this, it is likely that both established and emerging systems will be instrumental in addressing an array of open questions in the field. For example, many cellular structures have been shown to segregate in a biased, non-random fashion, but how these segregation patterns affect, or contribute to, cell fate decisions remains to be defined. Similarly, several mechanisms associated with the creation of physical asymmetry have been discovered, but precisely how these arise and function remains unclear. Whether and how sibling cell size asymmetry is instrumental in cell fate decisions also remains to be investigated. Moreover, how cell intrinsic and extrinsic mechanical forces contribute to cell polarization and biased segregation of macromolecules is an underexplored area in the field. Finally, we also know very little about how biased segregation of organelles impacts cell metabolism and physiology and whether such changes are implicated in binary cell fate decision.
As ACD is a dynamic process, many of these questions need be addressed by first performing a detailed phenomenological description that can capture the dynamic nature of the process. Similarly, as traditional loss-of-function experiments perturb multiple cellular functions, spatiotemporal perturbation approaches for acute protein mislocalization or knockdowns (e.g. using optogenetics or magnetogenetics) must be developed to gain mechanistic insight. Overall, a combination of these approaches will no doubt shed further light on ACD and aid our understanding of how this process influences cell fate decisions in healthy and diseased states.
Acknowledgements
We thank members of the Cabernard laboratory for helpful discussions. We apologize to all of the researchers and authors whose work we were unable to cite because of space constraints.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the National Institutes of Health (1R01GM126029-03) and a Research Scholar grant from the American Cancer Society (130285-RSG-16-253-01-CSM). Deposited in PMC for release after 12 months.
References
- Abeysundara N., Simmonds A. J. and Hughes S. C. (2017). Moesin is involved in polarity maintenance and cortical remodeling during asymmetric cell division. Mol. Biol. Cell 29, mbc.E17-05-0294 10.1091/mbc.E17-05-0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams W. C., Chen Y.-H., Kratchmarov R., Yen ., Nish S. A., Lin W.-H. W., Rothman N. J., Luchsinger L. L., Klein U., Busslinger M. et al. (2016). Anabolism-associated mitochondrial stasis driving lymphocyte differentiation over self-renewal. Cell Rep. 17, 3142-3152. 10.1016/j.celrep.2016.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akera T., Chmátal L., Trimm E., Yang K., Aonbangkhen C., Chenoweth D. M., Janke C., Schultz R. M. and Lampson M. A. (2017). Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358, 668-672. 10.1126/science.aan0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akera T., Trimm E. and Lampson M. A. (2019). Molecular strategies of meiotic cheating by selfish centromeres. Cell 178, 1132-1144.e10. 10.1016/j.cell.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliegro M. C., Alliegro M. A. and Palazzo R. E. (2006). Centrosome-associated RNA in surf clam oocytes. Proc. Natl. Acad. Sci. USA 103, 9034-9038. 10.1073/pnas.0602859103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C. and Karpen G. H. (2008). Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9, 923-937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann K., Frank M., Neumann D., Jakobs S. and Westermann B. (2008). The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J. Cell Biol. 181, 119-130. 10.1083/jcb.200709099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J. W. (1971). Organogenesis in normal and lobeless embryos of the marine prosobranch gastropod Ilyanassa obsoleta. J. Morphol. 133, 339-352. 10.1002/jmor.1051330307 [DOI] [PubMed] [Google Scholar]
- Atwood S. X. and Prehoda K. E. (2009). aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19, 723-729. 10.1016/j.cub.2009.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babour A., Bicknell A. A., Tourtellotte J. and Niwa M. (2010). A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell 142, 256-269. 10.1016/j.cell.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bärenz F., Kschonsak Y. T., Meyer A., Jafarpour A., Lorenz H. and Hoffmann I. (2018). Ccdc61 controls centrosomal localization of Cep170 and is required for spindle assembly and symmetry. Mol. Biol. Cell 29, mbcE18020115 10.1091/mbc.E18-02-0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros C. S., Phelps C. B. and Brand A. H. (2003). Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev. Cell 5, 829-840. 10.1016/S1534-5807(03)00359-9 [DOI] [PubMed] [Google Scholar]
- Basant A. and Glotzer M. (2018). Spatiotemporal regulation of RhoA during cytokinesis. Curr. Biol. 28, R570-R580. 10.1016/j.cub.2018.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J., Gerlich D., Daigle N., Eils R. and Ellenberg J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, 83-96. 10.1016/S0092-8674(01)00627-4 [DOI] [PubMed] [Google Scholar]
- Bertrand E., Chartrand P., Shenoy S. M., Singer R. H. and Long R. M. (1998). Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437-445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Böckler S., Chelius X., Hock N., Klecker T., Wolter M., Weiss M., Braun R. J. and Westermann B. (2017). Fusion, fission, and transport control asymmetric inheritance of mitochondria and protein aggregates. J. Cell Biol. 216, 2481-2498. 10.1083/jcb.201611197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvrette L. P. B., Cody N. A. L., Bergalet J., Lefebvre F. A., Diot C., Wang X., Blanchette M. and Lécuyer E. (2017). CeFra-seq reveals broad asymmetric mRNA and noncoding RNA distribution profiles in Drosophila and human cells. RNA 24, 98-113. 10.1261/rna.063172.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem M. and van del Heuvel S. (2019). Cell polarity: getting the PARty started. Curr. Biol. 29, R637-R639. 10.1016/j.cub.2019.05.032 [DOI] [PubMed] [Google Scholar]
- Brunet S. and Verlhac M. H. (2010). Positioning to get out of meiosis: the asymmetry of division. Hum. Reprod. Update 17, 68-75. 10.1093/humupd/dmq044 [DOI] [PubMed] [Google Scholar]
- Cabernard C., Prehoda K. E. and Doe C. Q. (2010). A spindle-independent cleavage furrow positioning pathway. Nature 467, 91-94. 10.1038/nature09334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Yu F., Lin S., Chia W. and Yang X. (2003). Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell 112, 51-62. 10.1016/S0092-8674(02)01170-4 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Gupta M. L., Hoyt M. A. and Pellman D. (2004). Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell 6, 815-829. 10.1016/j.devcel.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Cashikar A. G., Duennwald M. and Lindquist S. L. (2005). A chaperone pathway in protein disaggregation. J. Biol. Chem. 280, 23869-23875. 10.1074/jbc.M502854200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-Y., Bottjer D. J., Davidson E. H., Dornbos S. Q., Gao X., Yang Y.-H., Li C.-W., Li G., Wang X.-Q., Xian D.-C. et al. (2006). Phosphatized polar lobe-forming embryos from the Precambrian of Southwest China. Science 312, 1644-1646. 10.1126/science.1125964 [DOI] [PubMed] [Google Scholar]
- Chen C., Inaba M., Venkei Z. G. and Yamashita Y. M. (2016). Klp10A, a stem cell centrosome-enriched kinesin, balances asymmetries in Drosophila male germline stem cell division. eLife 5, e20977 10.7554/eLife.20977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Widmer L. A., Stangier M. M., Steinmetz M. O., Stelling J. and Barral Y. (2019). Remote control of microtubule plus-end dynamics and function from the minus-end. eLife 8, e48627 10.7554/eLife.48627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I., Santiago-Tirado F. and Bretscher A. (2013). Active segregation of yeast mitochondria by Myo2 is essential and mediated by Mmr1 and Ypt11. Curr. Biol. 23, 1818-1824. 10.1016/j.cub.2013.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S. P., Southall T. D., Bossing T., Edoff K., de Wit E., Fischer B. E., van Steensel B., Micklem G. and Brand A. H. (2006). Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775-789. 10.1016/j.devcel.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Clay L., Caudron F., Denoth-Lippuner A., Boettcher B., Frei S. B., Snapp E. L. and Barral Y. (2014). A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. eLife 3, e01883 10.7554/eLife.01883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement A. C. (1952). Experimental studies on germinal localization in Ilyanassa. I. The role of the polar lobe in determination of the cleavage pattern and its influence in later development. J. Exp. Zool. 121, 593-625. 10.1002/jez.1401210310 [DOI] [Google Scholar]
- Conduit P. T. and Raff J. W. (2010). Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20, 2187-2192. 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- Conduit P. T., Feng Z., Richens J. H., Baumbach J., Wainman A., Bakshi S. D., Dobbelaere J., Johnson S., Lea S. M. and Raff J. W. (2014). The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell 28, 659-669. 10.1016/j.devcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P. T., Wainman A. and Raff J. W. (2015). Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell. Biol. 16, 611-624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- Conklin E. G. (1905). Mosaic development in ascidian eggs. J. Exp. Zool. 2, 145-223. 10.1002/jez.1400020202 [DOI] [Google Scholar]
- Connell M., Cabernard C., Ricketson D., Doe C. Q. and Prehoda K. E. (2011). Asymmetric cortical extension shifts cleavage furrow position in Drosophila neuroblasts. Mol. Biol. Cell 22, 4205-4459. 10.1091/mbc.e11-02-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad G. W. and Williams D. C. (1974). Polar lobe formation and cytokinesis in fertilized eggs of Ilyanassa obsoleta: I. Ultrastructure and effects of cytochalasin B and colchicine. Dev. Biol. 36, 363-378. 10.1016/0012-1606(74)90058-X [DOI] [PubMed] [Google Scholar]
- Conrad G. W., Williams D. C., Turner F. R., Newrock K. M. and Raff R. A. (1973). Microfilaments in the polar lobe constriction of fertilized eggs of Ilyanassa obsoleta. J. Cell Biol. 59, 228-233. 10.1083/jcb.59.1.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad G. W., Schantz A. R. and Patron R. R. (1990). Mechanisms of polar lobe formation in fertilized eggs of molluscs. Ann. NY Acad. Sci. 582, 273-294. 10.1111/j.1749-6632.1990.tb21686.x [DOI] [PubMed] [Google Scholar]
- Cottingham F. R. and Hoyt M. A. (1997). Mitotic spindle positioning in saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138, 1041-1053. 10.1083/jcb.138.5.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumailleau F., Fürthauer M., Knoblich J. A. and González-Gaitán M. (2009). Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature 458, 1051-1055. 10.1038/nature07854 [DOI] [PubMed] [Google Scholar]
- Crampton H. E. and Wilson E. B. (1896). Experimental studies on gasteropod development. Arch. Entwicklungsmechan. Der Org. 3, 1-19. 10.1007/BF02156309 [DOI] [Google Scholar]
- Cravo J. and van den Heuvel S. (2019). Tissue polarity and PCP protein function: C. elegans as an emerging model. Curr. Opin. Cell Biol. 62, 159-167. 10.1016/j.ceb.2019.11.004 [DOI] [PubMed] [Google Scholar]
- Dalton C. M. and Carroll J. (2013). Biased inheritance of mitochondria during asymmetric cell division in the mouse oocyte. J. Cell Sci. 126, 2955-2964. 10.1242/jcs.128744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Avino P. P., Giansanti M. G. and Petronczki M. (2015). Cytokinesis in animal cells. Cold Spring Harb. Perspect Biol 7, a015834 10.1101/cshperspect.a015834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Seum C., Daeden A., Loubéry S., Holtzer L., Jülicher F. and Gonzalez-Gaitan M. (2015). Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature 528, 280-285. 10.1038/nature16443 [DOI] [PubMed] [Google Scholar]
- Dumollard R., Duchen M. and Carroll J. (2007). Current topics in developmental biology. Curr. Top. Dev. Biol. 77, 21-49. 10.1016/S0070-2153(06)77002-8 [DOI] [PubMed] [Google Scholar]
- Eritano A. S., Altamirano A., Beyeler S., Gaytan N., Velasquez M. and Riggs B. (2017). The endoplasmic reticulum is partitioned asymmetrically during mitosis before cell fate selection in proneuronal cells in the early Drosophila embryo. Mol. Biol. Cell 28, 1530-1538. 10.1091/mbc.e16-09-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Caballe A., Wainman A., Johnson S., Haensele A. F. M., Cottee M. A., Conduit P. T., Lea S. M. and Raff J. W. (2017). Structural basis for mitotic centrosome assembly in flies. Cell 169, 1078-1089.e13. 10.1016/j.cell.2017.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegan T. M. and Bergstralh D. T. (2019). Division orientation: disentangling shape and mechanical forces. Cell Cycle 18, 1187-1198. 10.1080/15384101.2019.1617006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann T. M., Menhorn P., Walter S. and Buchner J. (2008). Activation of the chaperone Hsp26 is controlled by the rearrangement of its thermosensor domain. Mol. Cell 29, 207-216. 10.1016/j.molcel.2007.11.025 [DOI] [PubMed] [Google Scholar]
- Fusco D., Accornero N., Lavoie B., Shenoy S. M., Blanchard J.-M., Singer R. H. and Bertrand E. (2003). Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13, 161-167. 10.1016/S0960-9822(02)01436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud E., Pham T. and Cabernard C. (2017). Drosophila melanogaster neuroblasts: a model for asymmetric stem cell divisions. Results Problems Cell Differ. 61, 183-210. 10.1007/978-3-319-53150-2_8 [DOI] [PubMed] [Google Scholar]
- Gambarotto D., Pennetier C., Ryniawec J. M., Buster D. W., Gogendeau D., Goupil A., Nano M., Simon A., Blanc D., Racine V. et al. (2019). Plk4 regulates centriole asymmetry and spindle orientation in neural stem cells. Dev. Cell 50, 11-24.e10. 10.1016/j.devcel.2019.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García del Arco A., Edgar B. A. and Erhardt S. (2018). In vivo analysis of centromeric proteins reveals a stem cell-specific asymmetry and an essential role in differentiated, non-proliferating cells. Cell Rep. 22, 1982-1993. 10.1016/j.celrep.2018.01.079 [DOI] [PubMed] [Google Scholar]
- Hannaford M. R., Ramat A., Loyer N. and Januschke J. (2018). aPKC-mediated displacement and actomyosin-mediated retention polarize Miranda in Drosophila neuroblasts. eLife 7, e29939 10.7554/eLife.29939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., Miess A., Stromer T., Walter S. and Buchner J. (2005). Disassembling protein aggregates in the yeast cytosol. J. Biol. Chem. 280, 23861-23868. 10.1074/jbc.M502697200 [DOI] [PubMed] [Google Scholar]
- Hejnol A. and Pfannenstiel H.-D. (1998). Myosin and actin are necessary for polar lobe formation and resorption in Ilyanassa obsoleta embryos. Dev. Genes Evol. 208, 229-233. 10.1007/s004270050177 [DOI] [PubMed] [Google Scholar]
- Hibbel A., Bogdanova A., Mahamdeh M., Jannasch A., Storch M., Schäffer E., Liakopoulos D. and Howard J. (2015). Kinesin Kip2 enhances microtubule growth in vitro through length-dependent feedback on polymerization and catastrophe. eLife 4, e10542 10.7554/eLife.10542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. M., Hao X., Grönvall J., Spikings-Nordby S., Widlund P. O., Amen T., Jörhov A., Josefson R., Kaganovich D., Liu B. et al. (2016). Asymmetric inheritance of aggregated proteins and age reset in yeast are regulated by Vac17-dependent vacuolar functions. Cell Rep. 16, 826-838. 10.1016/j.celrep.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. M., Hanzén S. and Nyström T. (2017). Restricted access: spatial sequestration of damaged proteins during stress and aging. EMBO Rep. 18, 377-391. 10.15252/embr.201643458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly R. W., Jones K. and Prehoda K. E. (2020). A conserved PDZ-binding motif in aPKC interacts with Par-3 and mediates cortical polarity. Curr. Biol. 30, 893-898.e5. 10.1016/j.cub.2019.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C. C. F. and Knoblich J. A. (2012). Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297-4310. 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Hughes A. L. and Gottschling D. E. (2012). An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261-265. 10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyett A., Kahana J., Silver P., Zeng X. and Saunders W. S. (1998). The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci. 111, 295-301. [DOI] [PubMed] [Google Scholar]
- Jakobsen L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L. G., Bennetzen M., Westendorf J., Nigg E. A. et al. (2011). Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 30, 1520-1535. 10.1038/emboj.2011.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J. and Gonzalez C. (2010). The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 188, 693-706. 10.1083/jcb.200905024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Llamazares S., Reina J. and Gonzalez C. (2011). Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2, 243 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J. and Gonzalez C. (2013). Centrobin controls mother–daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15, 241-248. 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Jeffery W. R., Tomlinson C. R. and Brodeur R. D. (1983). Localization of actin messenger RNA during early ascidian development. Dev. Biol. 99, 408-417. 10.1016/0012-1606(83)90290-7 [DOI] [PubMed] [Google Scholar]
- Jonas F. R. H., Royle K. E., Aw R., Stan G.-B. V. and Polizzi K. M. (2018). Investigating the consequences of asymmetric endoplasmic reticulum inheritance in Saccharomyces cerevisiae under stress using a combination of single cell measurements and mathematical modelling. Synthetic Syst. Biotechnol. 3, 64-75. 10.1016/j.synbio.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C. E., Voronina E., Stack C., Aldrich M., Cameron A. R. and Wessel G. M. (2006). Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev. Biol. 300, 406-415. 10.1016/j.ydbio.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Katajisto P., Dohla J., Chaffer C. L., Pentinmikko N., Marjanovic N., Iqbal S., Zoncu R., Chen W., Weinberg R. A. and Sabatini D. M. (2015). Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340-343. 10.1126/science.1260384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G. and Cheng N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311-320. 10.1016/s0092-8674(88)80024-2 [DOI] [PubMed] [Google Scholar]
- Kingsley E. P., Chan X. Y., Duan Y. and Lambert J. D. (2007). Widespread RNA segregation in a spiralian embryo. Evol. Dev. 9, 527-539. 10.1111/j.1525-142X.2007.00194.x [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T. (2019). The cortical force-generating machinery: how cortical spindle-pulling forces are generated. Curr. Opin. Cell Biol. 60, 1-8. 10.1016/j.ceb.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Klinkert K., Levernier N., Gross P., Gentili C., von Tobel L., Pierron M., Busso C., Herrman S., Grill S. W., Kruse K. et al. (2019). Aurora A depletion reveals centrosome-independent polarization mechanism in Caenorhabditis elegans. eLife 8, e44552 10.7554/eLife.44552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R. and Ilagan M. X. G. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216-233. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R. A., Gebelein B., Sprinzak D. and Kopan R. (2017). The canonical Notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell 41, 228-241. 10.1016/j.devcel.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel L. E. and Malik H. S. (2018). The cellular mechanisms and consequences of centromere drive. Curr. Opin. Cell Biol. 52, 58-65. 10.1016/j.ceb.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusek G., Campbell M., Doyle F., Tenenbaum S. A., Kiebler M. and Temple S. (2012). Asymmetric segregation of the double-stranded RNA binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression. Cell Stem Cell 11, 505-516. 10.1016/j.stem.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. D. and Nagy L. M. (2002). Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature 420, 682-686. 10.1038/nature01241 [DOI] [PubMed] [Google Scholar]
- Landskron L., Steinmann V., Bonnay F., Burkard T. R., Steinmann J., Reichardt I., Harzer H., Laurenson A.-S., Reichert H. and Knoblich J. A. (2018). The asymmetrically segregating lncRNA cherub is required for transforming stem cells into malignant cells. eLife 7, R106 10.7554/eLife.31347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lång E., Połeć A., Lång A., Valk M., Blicher P., Rowe A. D., Tønseth K. A., Jackson C. J., Utheim T. P., Janssen L. M. C. et al. (2018). Coordinated collective migration and asymmetric cell division in confluent human keratinocytes without wounding. Nat. Commun. 9, 3665 10.1038/s41467-018-05578-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T. R., Tomancak P. and Krause H. M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174-187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Ledan E., Polanski Z., Terret M.-E. and Maro B. (2001). Meiotic maturation of the mouse oocyte requires an equilibrium between Cyclin B synthesis and degradation. Dev. Biol. 232, 400-413. 10.1006/dbio.2001.0188 [DOI] [PubMed] [Google Scholar]
- Lengefeld J., Yen E., Chen X., Leary A., Vogel J. and Barral Y. (2017). Spatial cues and not spindle pole maturation drive the asymmetry of astral microtubules between new and preexisting spindle poles. Mol. Biol. Cell 29, 10-28. 10.1091/mbc.E16-10-0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit D. A. and Rusan N. M. (2013). PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202, 1013-1022. 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Yang X., Wasser M., Cai Y. and Chia W. (1997). Inscuteable and staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell 90, 437-447. 10.1016/S0092-8674(00)80504-8 [DOI] [PubMed] [Google Scholar]
- Li J., Cheng L. and Jiang H. (2019). Cell shape and intercellular adhesion regulate mitotic spindle orientation. Mol. Biol. Cell 30, 2458-2468. 10.1091/mbc.E19-04-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K., Lewandowska A. and Ziętkiewicz S. (2008). Chaperones in control of protein disaggregation. EMBO J. 27, 328-335. 10.1038/sj.emboj.7601970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler D., Wehling A., Schneiter F., Zhang Y., Müller-Bötticher N., Hoppe P. S., Hilsenbeck O., Kokkaliaris K. D., Endele M. and Schroeder T. (2019). Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature 573, 426-429. 10.1038/s41586-019-1531-6 [DOI] [PubMed] [Google Scholar]
- Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K. and Jansen R.-P. (1997). Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383-387. 10.1126/science.277.5324.383 [DOI] [PubMed] [Google Scholar]
- Loyer N. and Januschke J. (2020). Where does asymmetry come from? Illustrating principles of polarity and asymmetry establishment in Drosophila neuroblasts. Curr. Opin. Cell Biol. 62, 70-77. 10.1016/j.ceb.2019.07.018 [DOI] [PubMed] [Google Scholar]
- Luedeke C., Frei S. B., Sbalzarini I., Schwarz H., Spang A. and Barral Y. (2005). Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 169, 897-908. 10.1083/jcb.200412143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-López J., Matellán L., Álvarez-Llamas A., Blanco-Mira J. C. and Monje-Casas F. (2019). Asymmetric inheritance of spindle microtubule-organizing centres preserves replicative lifespan. Nat. Cell Biol. 21, 952-965. 10.1038/s41556-019-0364-8 [DOI] [PubMed] [Google Scholar]
- Mao K., Wang K., Liu X. and Klionsky D. J. (2013). The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell 26, 9-18. 10.1016/j.devcel.2013.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F., Ohshiro T., Ikeshima-Kataoka H. and Izumi H. (1998). miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development 125, 4089-4098. [DOI] [PubMed] [Google Scholar]
- McFaline-Figueroa J. R., Vevea J., Swayne T. C., Zhou C., Liu C., Leung G., Boldogh I. R. and Pon L. A. (2011). Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 10, 885-895. 10.1111/j.1474-9726.2011.00731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. and Chan D. C. (2014). Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell. Biol. 15, nrm3877 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogessie B., Scheffler K. and Schuh M. (2018). Assembly and positioning of the oocyte meiotic spindle. Annu. Rev. Cell Dev. Biol. 34, 381-403. 10.1146/annurev-cellbio-100616-060553 [DOI] [PubMed] [Google Scholar]
- Mollinari C., Lange B. and González C. (2002). Miranda, a protein involved in neuroblast asymmetric division, is associated with embryonic centrosomes of Drosophila melanogaster. Biol. Cell 94, 1-13. 10.1016/S0248-4900(02)01181-4 [DOI] [PubMed] [Google Scholar]
- Moore D. L. and Jessberger S. (2017). Creating age asymmetry: consequences of inheriting damaged goods in mammalian cells. Trends Cell Biol. 27, 82-92. 10.1016/j.tcb.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Moore D. L., Pilz G. A., Araúzo-Bravo M. J., Barral Y. and Jessberger S. (2015). A mechanism for the segregation of age in mammalian neural stem cells. Science 349, 1334-1338. 10.1126/science.aac9868 [DOI] [PubMed] [Google Scholar]
- Morgan T. H. (1933). The formation of the antipolar lobe in Ilyanassa. J. Exp. Zool. 64, 433-467. 10.1002/jez.1400640305 [DOI] [Google Scholar]
- Munro E., Nance J. and Priess J. R. (2004). Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7, 413-424. 10.1016/j.devcel.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Nanba D., Toki F., Tate S., Imai M., Matsushita N., Toki H., Higashiyama S. and Barrandon Y. (2016). Cell motion predicts human epidermal stemness. J. Dermatol. Sci. 84, e51 10.1016/j.jdermsci.2016.08.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak Z. A., Wainman A., Gartenmann L. and Raff J. W. (2016). Cdk1 phosphorylates Drosophila Sas-4 to recruit polo to daughter centrioles and convert them to centrosomes. Dev. Cell 37, 545-557. 10.1016/j.devcel.2016.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oon C. H. and Prehoda K. E. (2019). Asymmetric recruitment and actin-dependent cortical flows drive the neuroblast polarity cycle. eLife 8, e45815 10.7554/eLife.45815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Stuurman N., D'Ambrosio M. and Vale R. D. (2010). Polarized myosin produces unequal-size daughters during asymmetric cell division. Science 330, 677-680. 10.1126/science.1196112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Tanaka T. U., Nasmyth K. and Schiebel E. (2001). Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20, 6359-6370. 10.1093/emboj/20.22.6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernice W. M., Vevea J. D. and Pon L. A. (2016). A role for Mfb1p in region-specific anchorage of high-functioning mitochondria and lifespan in Saccharomyces cerevisiae. Nat. Commun. 7, 10595 10.1038/ncomms10595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T. T., Monnard A., Helenius J., Lund E., Lee N., Müller D. J. and Cabernard C. (2019). Spatiotemporally controlled Myosin relocalization and internal pressure generate sibling cell size asymmetry. Iscience 13, 9-19. 10.1016/j.isci.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña F. J. and Niwa M. (2015). The ER Stress Surveillance (ERSU) pathway regulates daughter cell ER protein aggregate inheritance. eLife 4, e06970 10.7554/eLife.06970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon J., Fries A., Wessel G. M. and Yajima M. (2019). Evolutionary modification of AGS protein contributes to formation of micromeres in sea urchins. Nat. Commun. 10, 3779 10.1038/s41467-019-11560-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., Go A. M. M. and McKim K. S. (2016). Cooperation between kinesin motors promotes spindle symmetry and chromosome organization in oocytes. Genetics 205, 517-527. 10.1534/genetics.116.194647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski S. M., Viana M. P., Zhang Y., Chan Y.-H. M., Thorn K. S., Yam P., Fung J. C., Li H., Costa L. D. F. and Marshall W. F. (2012). Mitochondrial network size scaling in budding yeast. Science 338, 822-824. 10.1126/science.1225720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramat A., Hannaford M. and Januschke J. (2017). Maintenance of Miranda localization in Drosophila neuroblasts involves interaction with the cognate mRNA. Curr. Biol. 27, 2101-2111.e5. 10.1016/j.cub.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas Nair A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B. and Cabernard C. (2016). The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 14, 1100-1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Ranjan R., Snedeker J. and Chen X. (2019). Asymmetric centromeres differentially coordinate with mitotic machinery to ensure biased sister chromatid segregation in germline stem cells. Cell Stem Cell 25, 666-681.e5. 10.1016/j.stem.2019.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H. and González C. (2007). Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell 12, 467-474. 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Render J. (1989). Development of Ilyanassa obsoleta embryos after equal distribution of polar lobe material at first cleavage. Dev. Biol. 132, 241-250. 10.1016/0012-1606(89)90220-0 [DOI] [PubMed] [Google Scholar]
- Roubinet C. and Cabernard C. (2014). Control of asymmetric cell division. Curr. Opin. Cell Biol. 31, 84-91. 10.1016/j.ceb.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Roubinet C., Tsankova A., Pham T. T., Monnard A., Caussinus E., Affolter M. and Cabernard C. (2017). Spatio-temporally separated cortical flows and spindle geometry establish physical asymmetry in fly neural stem cells. Nat. Commun. 8, 1383 10.1038/s41467-017-01391-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N. M. and Peifer M. (2007). A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177, 13-20. 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J., Caudron F., Prasad R., Moreno D. F., Bolognesi A., Aldea M. and Barral Y. (2017). Compartmentalization of ER-bound chaperone confines protein deposit formation to the aging yeast cell. Curr. Biol. 27, 773-783. 10.1016/j.cub.2017.01.069 [DOI] [PubMed] [Google Scholar]
- Salina D., Bodoor K., Eckley D. M., Schroer T. A., Rattner J. B. and Burke B. (2002). Cytoplasmic Dynein as a facilitator of nuclear envelope breakdown. Cell 108, 97-107. 10.1016/S0092-8674(01)00628-6 [DOI] [PubMed] [Google Scholar]
- Sallé J., Xie J., Ershov D., Lacassin M., Dmitrieff S. and Minc N. (2018). Asymmetric division through a reduction of microtubule centering forces. J. Cell Biol. 218, 771-782. 10.1083/jcb.201807102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann V., Chen C., Chiang C.-Y. A., Tiyaboonchai A., Mayer M. and Yamashita Y. M. (2014). Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol. Biol. Cell 25, 267-275. 10.1091/mbc.e13-09-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B. A., Kelly P. T., May M. C., Davis S. E. and Conrad G. W. (1980). Characterization of actin from fertilized eggs of Ilyanassa obsoleta during polar lobe formation and cytokinesis. Dev. Biol. 76, 126-140. 10.1016/0012-1606(80)90367-X [DOI] [PubMed] [Google Scholar]
- Schuldt A. J., Adams J. H. J., Davidson C. M., Micklem D. R., Haseloff J., Johnston D. S. and Brand A. H. (1998). Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Gene Dev. 12, 1847-1857. 10.1101/gad.12.12.1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Nair A. R. and Cabernard C. (2014). The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24, 1548-1555. 10.1016/j.cub.2014.05.050 [DOI] [PubMed] [Google Scholar]
- Skamagki M., Wicher K. B., Jedrusik A., Ganguly S. and Zernicka-Goetz M. (2013). Asymmetric localization of Cdx2 mRNA during the first cell-fate decision in early mouse development. Cell Rep. 3, 442-457. 10.1016/j.celrep.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R., Chia W. and Somers W. G. (2009). Protein Phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Gene Dev. 23, 359-372. 10.1101/gad.1723609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer N. H., Leverich C. K., Fitzgibbon M. P., Nelson Z. W., Henderson K. A., Gafken P. R., Hsu J. J. and Gottschling D. E. (2014). Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc. Natl. Acad. Sci. USA 111, 14019-14026. 10.1073/pnas.1416079111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Jacobo L., Henson J. H. and Shuster C. B. (2019). Cytoskeletal polarization and cytokinetic signaling drives polar lobe formation in spiralian embryos. Dev. Biol. 456, 201-211. 10.1016/j.ydbio.2019.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozer S., Baek C., Fischer E., Goiame R. and Morin X. (2017). Differential routing of Mindbomb1 via centriolar satellites regulates asymmetric divisions of neural progenitors. Neuron 93, 542-551.e4. 10.1016/j.neuron.2016.12.042 [DOI] [PubMed] [Google Scholar]
- Tran V., Lim C., Xie J. and Chen X. (2012). Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science 338, 679-682. 10.1126/science.1226028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova A., Pham T. T., Garcia D. S., Otte F. and Cabernard C. (2017). Cell polarity regulates biased myosin activity and dynamics during asymmetric cell division via drosophila rho kinase and protein kinase N. Dev. Cell 42, 143-155.e5. 10.1016/j.devcel.2017.06.012 [DOI] [PubMed] [Google Scholar]
- van Leen E. V., di Pietro F. , and Bellaïche Y. (2020). Oriented cell divisions in epithelia: from force generation to force anisotropy by tension, shape and vertices. Curr. Opin. Cell Biol. 62, 9-16. 10.1016/j.ceb.2019.07.013 [DOI] [PubMed] [Google Scholar]
- Walter P. and Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081-1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang X., Tsai J.-W., Imai J. H., Lian W.-N., Vallee R. B. and Shi S.-H. (2009). Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947-955. 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang L., Feng G., Wang Y., Li Y., Li X., Liu C., Jiao G., Huang C., Shi J. et al. (2018). Asymmetric expression of LincGET biases cell fate in two-cell mouse embryos. Cell 175, 1887-1901.e18. 10.1016/j.cell.2018.11.039 [DOI] [PubMed] [Google Scholar]
- Weinmaster G. and Fischer J. A. (2011). Notch ligand ubiquitylation: what is it good for? Dev. Cell 21, 134-144. 10.1016/j.devcel.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkley K., Ward S., Reeves W. and Veeman M. (2019). Iterative and complex asymmetric divisions control cell volume differences in Ciona notochord tapering. Curr. Biol. 29, 3466-3477.e4. 10.1016/j.cub.2019.08.056 [DOI] [PubMed] [Google Scholar]
- Wooten M., Snedeker J., Nizami Z. F., Yang X., Ranjan R., Urban E., Kim J. M., Gall J., Xiao J. and Chen X. (2019a). Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat. Struct. Mol. Biol. 26, 732-743. 10.1038/s41594-019-0269-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M., Ranjan R. and Chen X. (2019b). Asymmetric histone inheritance in asymmetrically dividing stem cells. Trends Genet. 36, 30-43. 10.1016/j.tig.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Lane S. I. R., Morgan S. L. and Jones K. T. (2018). Spindle tubulin and MTOC asymmetries may explain meiotic drive in oocytes. Nat. Commun. 9, 2952 10.1038/s41467-018-05338-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Wooten M., Tran V., Chen B.-C., Pozmanter C., Simbolon C., Betzig E. and Chen X. (2015). Histone H3 threonine phosphorylation regulates asymmetric histone inheritance in the Drosophila male germline. Cell 163, 920-933. 10.1016/j.cell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y. M., Mahowald A. P., Perlin J. R. and Fuller M. T. (2007). Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315, 518-521. 10.1126/science.1134910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X. and Chia W. (2000). Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399-409. 10.1016/S0092-8674(00)80676-5 [DOI] [PubMed] [Google Scholar]
- Yu F., Cai Y., Kaushik R., Yang X. and Chia W. (2003). Distinct roles of Gαi and Gβ13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 162, 623-633. 10.1083/jcb.200303174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Huang Z.-X., Bao H., Cong F., Wang H., Chai P. C., Xi Y., Ge W., Somers W. G., Yang Y. et al. (2015). Phosphotyrosyl phosphatase activator facilitates localization of Miranda through dephosphorylation in dividing neuroblasts. Development 143, dev.127233 10.1242/dev.127233 [DOI] [PubMed] [Google Scholar]
- Zhao P., Teng X., Tantirimudalige S. N., Nishikawa M., Wohland T., Toyama Y. and Motegi F. (2019). Aurora-A breaks symmetry in contractile actomyosin networks independently of its role in centrosome maturation. Dev. Cell 48, 631-645.e6. 10.1016/j.devcel.2019.02.012 [DOI] [PubMed] [Google Scholar]
- Zhou C., Slaughter B. D., Unruh J. R., Guo F., Yu Z., Mickey K., Narkar A., Ross R. T., McClain M. and Li R. (2014). Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell 159, 530-542. 10.1016/j.cell.2014.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]