Abstract
Background
Information regarding risk factors associated with severe coronavirus disease (COVID-19) is limited. This study aimed to develop a model for predicting COVID-19 severity.
Methods
Overall, 690 patients with confirmed COVID-19 were recruited between 1 January and 18 March 2020 from hospitals in Honghu and Nanchang; finally, 442 patients were assessed. Data were categorised into the training and test sets to develop and validate the model, respectively.
Findings
A predictive HNC-LL (Hypertension, Neutrophil count, C-reactive protein, Lymphocyte count, Lactate dehydrogenase) score was established using multivariate logistic regression analysis. The HNC-LL score accurately predicted disease severity in the Honghu training cohort (area under the curve [AUC]=0.861, 95% confidence interval [CI]: 0.800–0.922; P<0.001); Honghu internal validation cohort (AUC=0.871, 95% CI: 0.769–0.972; P<0.001); and Nanchang external validation cohort (AUC=0.826, 95% CI: 0.746–0.907; P<0.001) and outperformed other models, including CURB-65 (confusion, uraemia, respiratory rate, BP, age ≥65 years) score model, MuLBSTA (multilobular infiltration, hypo-lymphocytosis, bacterial coinfection, smoking history, hypertension, and age) score model, and neutrophil-to-lymphocyte ratio model. The clinical significance of HNC-LL in accurately predicting the risk of future development of severe COVID-19 was confirmed.
Interpretation
We developed an accurate tool for predicting disease severity among COVID-19 patients. This model can potentially be used to identify patients at risks of developing severe disease in the early stage and therefore guide treatment decisions.
Funding
This work was supported by the National Nature Science Foundation of China (grant no. 81972897) and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2015).
Keywords: COVID-19, SARS-COV-2, Severity, Prediction, HNC-LL
1. Introduction
Coronavirus disease (COVID-19) is a communicable disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first outbreak was reported in Wuhan,China in December 2019, and it was declared a pandemic on 11 March 2020 by the World Health Organization [1]. Measures aimed at improving patient care, increasing the efficiency of resource use, and decreasing the mortality risk are urgently needed.
Research in context.
Evidence before this study
Articles probing into risk factors of severe coronavirus disease (COVID-19) and early identification of COVID-19 patients with a high risk of disease progression were searched on PubMed on 25 May 2020 using the search terms (“SARS-Cov-2” OR “novel coronavirus” OR “COVID-19”) AND (“progression” OR “deterioration” OR “severe disease”) AND (“prediction” OR “predict” OR “early identification”) without time or language restrictions. A limited number of reports have identified age, Sequential Organ Failure Assessment score, D-dimer level, coagulation dysfunction, and high neutrophil count as risk factors for severe COVID-19. However, their application in predicting disease severity has not been completely delineated. One study proposed a clinical risk score, including 10 parameters, to predict critical-type COVID-19. However, concise and effective clinical models to predict disease progression with respect to severe COVID-19 (including severe-type and critical-type) have not been completely investigated and thus deserves intensive research.
Added value of this study
In this retrospective multicohort study, the clinical characteristics of 442 hospitalised COVID-19 patients were comprehensively assessed. A predictive model (named the HNC-LL [hypertension, neutrophil count, C-reactive protein, lymphocyte count, lactate dehydrogenase] score) was developed and validated to enable early and accurate identification of COVID-19 patients at a high risk of severe disease. Compared with the CURB-65 score model, MuLBSTA score model, and neutrophil-to-lymphocyte ratio model, the HNC-LL score performed better in predicting COVID-19 severity. More importantly, the clinical significance of the HNC-LL score in accurately predicting the risk of future development of severe COVID-19 was confirmed in subgroup analyses.
Implications of all the available evidence
There is a need to pay particular attention to the neutrophil count, lymphocyte count, lactate dehydrogenase level, hypertension, and C-reactive protein level when COVID-19 patients are admitted to the hospital. Risk assessment based on the HNC-LL score is recommended for all COVID-19 patients on admission. For those mild- and moderate-type (non-severe) patients who are classified as high-risk based on the HNC-LL score, active treatment and close monitoring, with the intensity higher than routine management for non-severe patients, should be provided, so as to prevent disease progression.
Alt-text: Unlabelled box
According to the reported data, the overall mortality rate was 2.3%, and no deaths were reported among non-severe cases [2]. An estimated 19% of patients with SARS-CoV-2 infection developed severe disease [2]. Furthermore, 61.5% of patients with the severe disease died within 28 days of admission [3]. In addition, treatment of a large number of patients with severe COVID-19 constitutes a huge strain on medical resources. Therefore, reducing the number of patients with severe COVID-19 would contribute to decreasing the associated mortality and the burden on medical resources. Although symptomatic and supportive therapies are the mainstays of treatment, these have limited efficacy since approximately 50% of patients do not attain significant clinical and imaging remission after 10 days in the hospital [4]. Therefore, early identification and triaging of patients with potential severe COVID-19 and a high risk of mortality are effective strategies for improving the cure rate and alleviating the burden on the healthcare system.
The risk factors for severe disease are not well-known. There is a paucity of reliable tools based on risk factors that enable the early identification of patients with severe COVID-19. A limited number of reports have identified age, Sequential Organ Failure Assessment score, D-dimer level, coagulation dysfunction, and high neutrophil count as risk factors for severe disease [5], [6], [7]. Moreover, severe COVID-19 may be associated with cytokine storm in some patients [8]. Despite these findings, their application in the early prediction of severity has not been investigated. In addition to the absence of effective and accurate predictive tools, few studies have focused on a specific patient population who had non-severe disease on admission but progressed to severe disease after admission. This subgroup of patients will benefit greatly from early identification and intervention. Therefore, in the present study, we developed a COVID-19-specific predictive HNC-LL (Hypertension, Neutrophil count, C-reactive protein, Lymphocyte count, and Lactate dehydrogenase) score that could accurately identify patients with a high risk of severe disease from this specific patient subgroup in the early stage.
2. Materials and methods
2.1. Patient cohort and study design
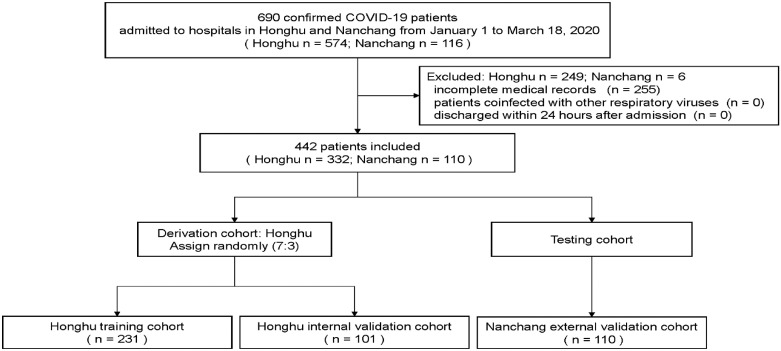
Data of a total of 690 patients with confirmed COVID-19 were retrospectively collected between 1 January 2020 and 18 March 2020 from hospitals in Honghu and Nanchang. Overall, 574 patients in Honghu were enrolled from the People's Hospital of Honghu and 116 patients in Nanchang were recruited from the First Affiliated Hospital of Nanchang University. All patients included in this study tested positive for SARS-CoV-2 nucleic acid and met the diagnostic criteria according to the guideline. A flowchart regarding the selection of the study participants for the training and validation cohorts is shown in Fig. 1. The protocol of this retrospective study was approved by the medical ethics committee of Nanfang Hospital of Southern Medical University and the institutional ethics review boards of participating hospitals. Written informed consent was obtained from all participants.
Fig. 1.
Study flowchart.
A total of 690 patients with confirmed COVID-19 between 1 January and 18 March 2020 were included in this study. After excluding patients who had incomplete clinical data, those who were coinfected with other respiratory viruses, and those who were discharged within 24 h after admission, 442 patients were retained in the final analysis. Of these, 333 were hospitalised in Honghu. Simple random sampling in a ratio of 7:3 was performed to assign 231 patients into a training cohort (the Honghu training cohort) and 101 patients into an internal validation cohort (the Honghu internal validation cohort). In addition, 110 patients hospitalised in Nanchang were used as an external validation cohort (the Nanchang external validation cohort).
2.2. Definition
The spectrum of the severity of COVID-19 ranges from mild to critical cases according to some recently reported literature and the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) [7,[9], [10], [11], [12]]. Patients with mild disease refer to those with mild clinical manifestations and no sign of pneumonia on imaging. Patients with moderate disease refer to those with fever and respiratory symptoms with radiological features of pneumonia. Patients with severe disease refer to those who meet any of the following criteria: respiratory distress (respiratory rate ≥30 breaths/min), oxygen saturation ≤93% at rest, arterial partial pressure of oxygen/fraction of inspired oxygen ≤300 mmHg, and >50% obvious lesion progression on chest imaging within 24–48 h. People with critical disease refer to those who meet any of the following criteria: respiratory failure and requiring mechanical ventilation, shock, and other forms of organ failure that require intensive care. In the present study, all patients were diagnosed and classified based on the aforementioned definition. Patients with mild and moderate pneumonia were included in the non-severe group, while those with severe and critical disease on admission and those who had mild and moderate pneumonia on admission but developed severe disease after admission were included in the severe group.
Using the above definition of severity as a foundation, severe illness-free survival was defined as the time from admission to disease progression to a severe state.
2.3. Data collection and outcome evaluation
Clinical electronic medical records, laboratory findings, nursing records, and radiological reports for all patients with confirmed COVID-19 were reviewed. Detailed admission data, including demographic information, comorbidities, signs and symptoms, laboratory test results, and imaging reports, of each patient were collected. After admission, treatment, disease severity, outcomes, and length of stay in hospital were also recorded. Two researchers independently reviewed the electronic medical records manually and recorded the disease severity on admission, daily assessment of disease severity afterwards, progression from non-severe to severe disease, and date of progression.
The clinical data will be made available to others on reasonable request after publication. A proposal along with a detailed description of the study objectives will be needed to evaluate the reasonability of requests. After approval from the corresponding authors, the First Affiliated Hospital of Nanchang University, and the People's Hospital of Honghu, de-identified patient data will be provided.
2.4. Statistical analysis
Continuous variables are presented as medians and interquartile ranges. Categorical variables are presented as frequencies and proportion. The comparison between the non-severe and severe groups were performed using the Mann–Whitney U test, Pearson's chi-squared test, or Fisher's exact test, as appropriate. To explore the risk factors associated with severity, univariable and multivariable logistic regression models were used. In the univariate logistic regression analyses, variables with P<0.05 were regarded as potential risk factors and included in the multivariate regression analysis with a backward elimination procedure (likelihood ratio test and elimination if P>0.1).
To assign patients into different cohorts, for 332 hospitalised patients in Honghu, simple random sampling in a ratio of 7:3 was performed using the “Select Cases” function in SPSS version 25.0 (IBM Corp., Armonk, New York, USA) under the parameter of “Random sample of cases”. The training cohort comprised 231 patients, and the internal validation cohort comprised 101 patients. In addition, 110 hospitalised patients from Nanchang were included as a testing cohort and further assessed as an external validation cohort.
Receiver operating characteristic (ROC) curve analysis and the Hosmer–Lemeshow test were performed to assess the discrimination and calibration of the model. The area under the ROC curve (AUC) was compared using DeLong's test. The cut-off value of the HNC-LL score was selected based on the maximum value of the Youden index. Net reclassification improvement analysis was also adopted to evaluate improvement to risk prediction.
Survival analysis was performed in the subgroup of patients who were classified as having non-severe disease on admission. The endpoint in this subgroup was defined as disease progression to a severe state after admission. The survival curves of the low and high risk groups were compared using the log-rank test, and hazard ratios (HRs) were calculated using the Cox proportional hazards model.
Statistical analyses and plots were performed using SPSS version 25.0 and R version 3.5.2 (The R Development Core Team, Vienna, Austria). The pROC package was used to plot the ROC curves and calculate the AUC. The survminer package was used to plot the severe illness-free survival curves. All statistical tests were two-sided, and analysis items with P values ≤0.05 were considered statistically significant.
3. Results
3.1. Patient characteristics and clinical differences between patients with severe and non-severe COVID-19
A total of 690 patients with confirmed COVID-19 from hospitals in Honghu and Nanchang were enrolled. According to the inclusion and exclusion criteria, 442 patients were finally included and divided into three cohorts (Fig. 1). The demographic and baseline clinical characteristics of the patients in the Honghu and Nanchang cohorts are shown in Table S1. The proportions of men were 47.0%, 60.0%, and 50.2% and the proportions of patients older than 60 years were 27.4%, 17.3%, and 24.9%, in the Honghu, Nanchang, and entire cohorts, respectively. Common comorbidities included hypertension and diabetes mellitus. Relevant signs and symptoms included bacterial coinfection, cough, and fever. In total, 96.4%, 100%, and 97.3% of patients were discharged from the hospital and 3.6%, 0.0%, and 2.7% of the patients died in the Honghu, Nanchang, and entire cohorts, respectively.
Overall, there were 339 and 103 patients in the non-severe and severe groups, respectively. Patients in the severe group were older, had lower lymphocyte counts, higher neutrophil counts, lower albumin levels, and higher urea nitrogen, lactate dehydrogenase (LDH), and C-reactive protein (CRP) levels than those in the non-severe group. The differences were significant in all three cohorts. In terms of comorbidities and pneumonia-related signs and symptoms, the proportions of patients with hypertension and bacterial coinfection were significantly higher in the severe group than in the non-severe group (Tables 1 and S2).
Table 1.
Demographics and Laboratory characteristics of patients with confirmed COVID-19.
| Characteristic | Training cohort |
Internal validation cohort |
External validation cohort |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-severe (n = 191) | Severe (n = 40) | P value | Non-severe (n = 81) | Severe (n = 20) | P value | Non-severe (n = 67) | Severe (n = 43) | P value | |
| Demographics | |||||||||
| Age (≥60 years) | 43 (22.51) | 22 (55.00) | <0.001 | 14 (17.28) | 12 (60.00) | <0.001 | 6 (8.96) | 13 (30.23) | 0.004 |
| Male | 84 (43.98) | 19 (47.50) | 0.684 | 41 (50.62) | 12 (60.00) | 0.452 | 32 (47.76) | 34 (79.07) | 0.001 |
| Hypertension | 20 (10.47) | 13 (32.50) | <0.001 | 6 (7.41) | 8 (40.00) | <0.001 | 7 (10.45) | 7 (16.28) | 0.371 |
| Diabetes mellitus | 6 (3.14) | 5 (12.50) | 0.011 | 4 (4.94) | 1 (5.00) | >0.999 | 4 (5.97) | 9 (20.93) | 0.018 |
| Highest temperature ( °C) | 36.90 (36.50–37.70) | 37.65 (36.55–38.45) | 0.016 | 37.95 (36.95–38.50) | 37.10 (36.50–38.00) | 0.015 | 37.90 (37.40–38.40) | 38.20 (37.80–39.00) | 0.009 |
| Laboratory findings | |||||||||
| White blood cell count <4 × 109/L | 29 (15.18) | 5 (12.50) | 0.026 | 13 (16.05) | 1 (5.00) | 0.022 | 25 (37.31) | 12 (27.91) | 0.419 |
| <10 × 109/L | 152 (79.58) | 28 (70.00) | 65 (80.25) | 15 (75.00) | 39 (58.21) | 27 (62.79) | |||
| ≥10 × 109/L | 10 (5.24) | 7 (17.50) | 3 (3.70) | 4 (20.00) | 3 (4.48) | 4 (9.30) | |||
| Lymphocyte count (<1.1 × 109/ L) | 42 (21.99) | 27 (67.50) | <0.001 | 15 (18.52) | 12 (60.00) | <0.001 | 28 (41.79) | 38 (88.37) | <0.001 |
| Neutrophil count(>6.3 × 109/L) | 17 (8.90) | 11 (27.50) | 0.001 | 6 (7.41) | 9 (45.00) | <0.001 | 4 (5.97) | 8 (18.60) | 0.038 |
| Platelet count (<150 × 109/L) | 19 (9.95) | 11 (27.50) | 0.003 | 8 (9.88) | 3 (15.00) | 0.510 | 19 (28.36) | 18 (41.86) | 0.144 |
| Alblumin (<34 g/L) | 15 (7.85) | 16 (40.00) | <0.001 | 8 (9.88) | 9 (45.00) | <0.001 | 0 (0.00) | 6 (13.95) | 0.002 |
| Direct bilirubin, μmol/L | 2.80 (2.10–3.70) | 4.00 (2.95–4.90) | <0.001 | 2.90 (2.40–4.00) | 4.05 (3.05–4.60) | 0.077 | 2.40 (1.80–3.60) | 3.60 (2.40–5.60) | 0.001 |
| Creatinine, μmol/L | 60.80 (50.30–73.20) | 74.20 (62.55–90.55) | <0.001 | 62.30 (51.80–72.30) | 63.65 (58.35–92.45) | 0.104 | 61.80 (49.20–69.90) | 72.80 (62.40–86.50) | 0.001 |
| Urea nitrogen, mmol/L | 4.07 (3.26–5.07) | 5.19 (3.61–8.33) | 0.002 | 3.89 (3.24–4.47) | 5.81 (4.21–7.23) | <0.001 | 3.90 (3.20–4.90) | 4.90 (3.90–6.30) | 0.001 |
| Lactate dehydrogenase (>245 U/L) | 44 (23.04) | 25 (62.50) | <0.001 | 17 (20.99) | 16 (80.00) | <0.001 | 17 (25.37) | 33 (76.74) | <0.001 |
| C-reactive protein (≥10 mg/L) | 46 (24.08) | 31 (77.50) | <0.001 | 17 (20.99) | 16 (80.00) | <0.001 | 26 (38.81) | 32 (74.42) | <0.001 |
Continuous variable data are presented as median (interquartile ranges, IQR).
Classified variable data are presented as n(%).
3.2. Development of the HNC-LL score for the prediction of disease severity
To effectively identify patients with potentially severe disease, a clinical characteristic-based predictive model was developed. Univariate logistic regression analysis revealed significant predictors (Table 2). The multivariate logistic regression analysis identified neutrophil count (odds ratio [OR] = 2.82, 95% confidence interval [CI]: 1.01–7.88; P = 0.049), lymphocyte count (OR = 0.29, 95% CI: 0.12–0.69; P = 0.005), CRP level (OR = 4.13, 95% CI: 4.13–10.32; P = 0.002), LDH level (OR = 2.90, 95% CI: 1.26–6.68; P = 0.012), and hypertension (OR = 2.64, 95% CI: 1.00–6.95; P = 0.050) as independent risk factors for disease severity, and the model was built using these five variables (Table 2). The HNC-LL score was calculated for each patient using a formula derived from values of these five clinical variables weighted by their regression coefficients, as follows:
HNC-LL score = 1.035 × neutrophil count (1: ≥6.3 × 109/L; 0: <6.3 × 109/L) − 1.237 × lymphocyte count (1: ≥1.1 × 109/L; 0: <1.1 × 109/L) + 1.419 × CRP level (1: ≥10 mg/L; 0: <10 mg/L) + 1.066 × LDH (1: >245 U/L; 0: ≤245 U/L) + 0.969 × hypertension (1: with; 0: without) − 2.425
Table 2.
Risk factors associated with severe illness among patients with confirmed COVID-19.
| Characteristic | Univariate |
Multivariate |
||
|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Age (≥60 years) | 4.207 (2.069–8.552) | <0.001 | ||
| Sex | 0.868 (0.438–1.718) | 0.684 | ||
| Current smokers | 2.423 (0.214–27.388) | 0.474 | ||
| Hypertension | 4.117 (1.836–9.232) | 0.001 | 2.635 (0.999–6.945) | 0.050 |
| Diabetes mellitus | 4.405 (1.274–15.230) | 0.019 | ||
| Highest temperature ( °C) | 1.697 (1.181–2.440) | 0.004 | ||
| White blood cell count, × 109/L | ||||
| 4–10 | ref | |||
| <4 | 0.983 (0.548–1.761) | 0.953 | ||
| >10 | 3.429 (1.616–7.276) | 0.043 | ||
| Lymphocyte count (<1.1 × 109/L) | 0.136 (0.064–0.286) | 0.002 | 0.290 (0.123–0.686) | 0.005 |
| Neutrophil count (>6.3 × 109/L) | 3.882 (1.652–9.122) | 0.002 | 2.815 (1.005–7.884) | 0.049 |
| eosinophilcount, × 109/l | 0.155 (0.011–2.128) | 0.163 | ||
| hemoglobin, g/dl | 0.996 (0.977–1.015) | 0.671 | ||
| Platelet count (<150 × 109/L) | 0.291 (0.126–0.675) | 0.004 | ||
| Albumin (<34 g/L) | 0.128 (0.056–0.291) | <0.001 | ||
| Total bilirubin | 1.036 (0.991–1.083) | 0.121 | ||
| Alanine aminotransferase (>40 U/L) | 0.595 (0.218–1.622) | 0.310 | ||
| Prothrombin time (≥16 s) | 2.423 (0.214–27.388) | 0.474 | ||
| Gamma-glutamyl transferase, U/L | 1.000 (0.998–1.002) | 0.974 | ||
| Direct bilirubin, μmol/L | 1.094 (1.004–1.193) | 0.041 | ||
| Lactate dehydrogenase (>245 U/L) | 5.568 (2.701–11.478) | <0.001 | 2.903 (1.262–6.679) | 0.012 |
| Creatinine, μmol/L | 1.021 (1.009–1.033) | <0.001 | ||
| Urea nitrogen, mmol/L | 1.203 (1.081–1.338) | 0.001 | ||
| Creatinine kinase (>120 U/L) | 1.498 (0.650–3.457) | 0.343 | ||
| C-reactive protein (≥10 mg/L) | 10.857 (4.816–24.477) | <0.001 | 4.133 (1.655–10.320) | 0.002 |
| Ground-glass opacity | 0.929 (0.470–1.837) | 0.833 | ||
| Local patchy shadowing | 0.705 (0.232–2.145) | 0.538 | ||
| Bilateral patchy shadowing | 1.517 (0.753–3.055) | 0.244 | ||
| Interstitial abnormalities | 0.791 (0.093–6.753) | 0.83 | ||
| Multi-lobular infiltration | 1.655 (0.743–3.690) | 0.218 | ||
OR, odds ratio; CI, confidence interval.
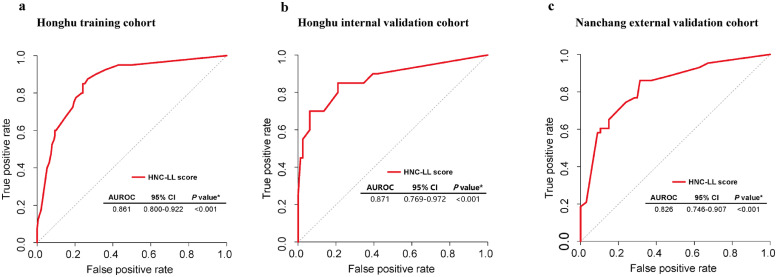
The HNC-LL score had a P value of 0.882 in the Hosmer–Lemeshow test. According to the ROC analysis, the HNC-LL score performed exceptionally in categorizing patients into the severe and non-severe groups (AUC = 0.861, 95% CI: 0.800–0.922; P < 0.001) (Fig. 2a).
Fig. 2.
Receiver operating characteristic (ROC) curves for evaluating the predictive ability of the HNC-LL score for disease severity in different cohorts.
ROC curve of the HNC-LL score in (a) the Honghu training cohort, (b) the Honghu internal validation cohort, and (c) the Nanchang external validation cohort.
AUC, area under the ROC curve; CI, confidence interval.
Based on the largest Youden's index, we determined an optimal cut-off value (–1.508) to stratify hospitalised patients into the HNC-LLhigh (≥–1.508) and HNC-LLlow (<–1.508) groups in the Honghu training cohort. The sensitivity and specificity of the model were 0.85 and 0.76, respectively. Patients in the HNC-LLhigh group were estimated to have a high risk of severe disease, while those in the HNC-LLlow group were estimated to have a low risk. The HNC-LL score classified 34 (85.0%) of 40 patients with severe COVID-19 into the HNC-LLhigh group and 6 (15.0%) of 40 patients with severe COVID-19 into the HNC-LLlow group in the Honghu training cohort. The estimated risk of severe disease, specificity, and sensitivity of the corresponding HNC-LL score are presented in Table S3.
3.3. Validation of the HNC-LL score for predicting disease severity among COVID-19 patients
To examine the generalisability of the HNC-LL score in predicting disease severity, validation and verification of the model were necessary. The HNC-LL score was validated in the Honghu internal validation cohort, and it maintained its good predictive performance (AUC = 0.871, 95% CI: 0.769–0.972; P < 0.001) (Fig. 2b). To further confirm the predictive efficacy, the score was validated in the Nanchang external validation cohort, in which it performed well consistently (AUC = 0.826, 95% CI: 0.746–0.907; P < 0.001) (Fig. 2c).
3.4. Comparison of different predictive models
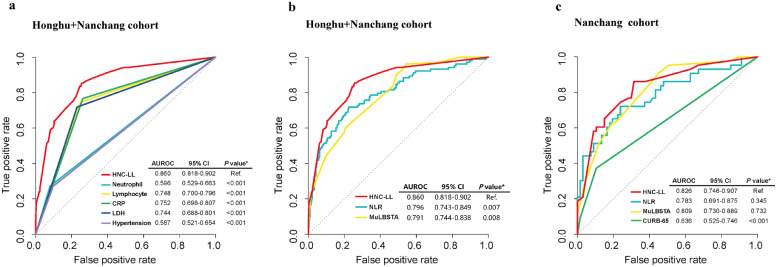
The effectiveness and efficiency of the HNC-LL score were further assessed by comparing different predictive models. When compared with each independent risk factor within the entire cohort, the HNC-LL score demonstrated a superior predictive capability with the highest AUC, which was statistically significant (Fig. 3a). The CURB-65 (confusion, uraemia, respiratory rate, blood pressure, age ≥65 years) score is a predictive model used to stratify patients with community-acquired pneumonia (CAP) into different management groups [13]. The neutrophil-to-lymphocyte ratio (NLR) is reported to be an independent risk factor for patients with severe COVID-19 [14]. The MuLBSTA (multilobular infiltration, hypo-lymphocytosis, bacterial coinfection, smoking history, hypertension, and age) score is used to stratify patients with viral pneumonia into relevant risk groups [15]. The HNC-LL score was compared with these models. Only patients in the Nanchang cohort had complete clinical information that was required to calculate the CURB-65 score. The HNC-LL and CURB-65 score models were thus compared using the Nanchang cohort. The HNC-LL score outperformed the NLR and MuLBSTA score models in both the Nanchang and entire cohorts (Fig. 3b, c) and demonstrated better predictive performance than the CURB-65 score in the Nanchang cohort (Fig. 3c).
Fig. 3.
Comparison of predictive performance for disease severity among the HNC-LL score and independent risk factors, MuLBSTA score, CURB-65, and NLR using ROC curves.
(a) ROC curves of the HNC-LL score and independent risk factors in the entire cohort. (b) ROC curves of the HNC-LL, NLR, and MuLBSTA scores in the entire cohort. (c) ROC curves of the HNC-LL and CURB-65 scores in the Nanchang external validation cohort.
Neutrophil, neutrophil count; Lymphocyte, lymphocyte count; CRP, C-reactive protein; LDH, lactate dehydrogenase; NLR, neutrophil-to-lymphocyte ratio model; ROC, receiver operating characteristic curve; MuLBSTA score, a model based on multilobular infiltrates, lymphocyte ≤0.8 × 109/L, bacterial coinfection, acute smoking, smoking cessation, hypertension, and age ≥60 years; CURB-65, a model based on age, confusion, urea, respiratory rate, and blood pressure. P values were calculated using DeLong's test.
Net reclassification improvement (NRI) analysis was also adopted for comparison. Results showed that HNC-LL can provide >12% net reclassification improvement (P = 0.019) when compared to NLR and >17% net reclassification improvement when compared to MuLBSTA (P = 0.001) (Table S4). It revealed that HNC-LL outperformed the other two models in predicting COVID-19 severity, appreciating the incremental predictive value of the HNC-LL furtherly.
In addition, we explored the performance of the HNC-LL score in predicting mortality. Compared with independent risk factors constituting the HNC-LL score (Figure S1a), the HNC-LL score had the highest AUC value, significant for all factors except the CRP level. When compared with the NLR and MuLBSTA score models (Figure S1b), the HNC-LL score consistently yielded the highest AUC, although the differences in AUC were not significant.
3.5. Clinical significance of the HNC-LL score in predicting the risk of future development of severe COVID-19 among patients
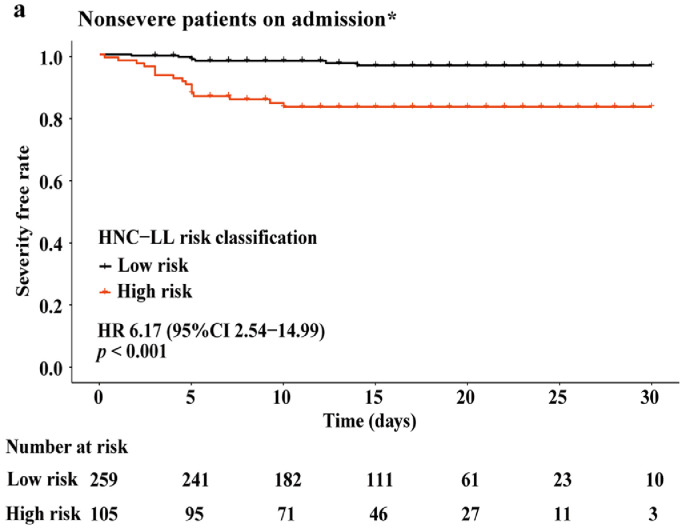
We further investigated whether the HNC-LL score could predict the risk of future development of severe COVID-19. Subgroup analysis was performed among patients diagnosed with non-severe disease on admission, including 285 and 80 patients from the Honghu and Nanchang cohorts, respectively. Moreover, 339 patients maintained their non-severe disease status and 26 progressed to severe disease after admission. The clinical characteristics of this specific patient population and the differences between the non-severe and severe groups are shown in Table S5. The five variables constituting the HNC-LL score remained significantly different between the groups even in this population. The HNC-LL score classified 259 (71.2%) and 105 (28.8%) patients into the high severe risk and low severe risk groups, respectively. The severe illness-free survival curves showed that patients in the high risk group had an unfavorable severe illness-free survival compared with those in the low risk group (HR = 6.17, 95% CI: 2.54–14.99, P < 0.001), thereby confirming the excellent discrimination ability of the HNC-LL score and suggesting that the HNC-LL score can identify patients with potentially severe COVID-19 in the early stage (Fig. 4).
Fig. 4.
Severe illness-free survival curves for high and low severity risk groups.
Patients in three cohorts diagnosed with non-severe disease on admission were included in this analysis. Based on their HNC-LL scores, they were stratified into the high and low severe risk groups according to the cut-off value of –1.508.
HR, hazard ratio; CI, confidence interval. P value was calculated using the log-rank test.
*A patient was excluded from this analysis for lack of follow-up information.
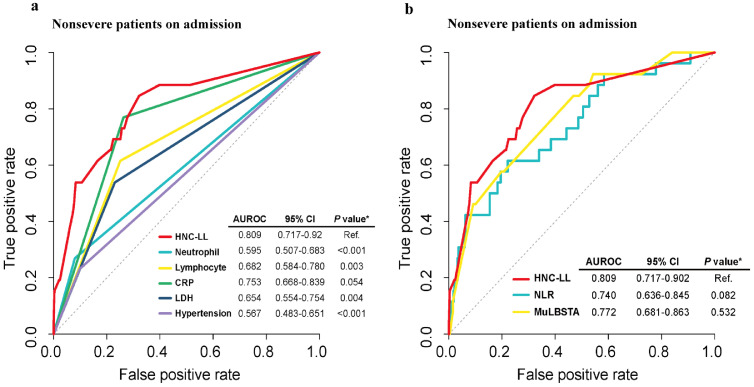
Furthermore, the predictive power was compared between the HNC-LL score and other models to verify its clinical utility. ROC curve analysis revealed that the HNC-LL score outperformed independent risk factors that constituted the HNC-LL score (Fig. 5a) as well as the MuLBSTA score model and NLR model (Fig. 5b) in distinguishing patients with a high risk of disease progression with the highest AUC. Compared with the CRP level, NLR model and the MuLBSTA score, the performance of the HNC-LL score was preferable, although it yielded non-significant results. These results may indicate the clinical significance of HNC-LL from another perspective.
Fig. 5.
Comparison of the ability to predict severe disease between the HNC-LL score and independent risk factors, MuLBSTA score, and NLR using ROC curves.
Patients in the entire cohort diagnosed as non-severe on admission were included in this analysis. (a) Comparison of clinical utility between the HNC-LL score and independent risk factors. (b) Comparison of clinical utility among the HNC-LL, NLR, and MuLBSTA scores. P values were calculated using DeLong's test.
4. Discussion
In this retrospective multicohort study, the clinical characteristics of 442 hospitalised patients with COVID-19 were comprehensively assessed. A predictive model (named the HNC-LL score) was developed and validated to enable early and accurate identification of COVID-19 patients at a high risk of severe disease.
To our knowledge, this is the first study to develop a predictive model for the assessment of COVID-19 severity using a three-step approach (training, internal validation, and external validation) and to focus on a specific population of patients who presented with non-severe disease on admission but progressed to severe disease after admission. The HNC-LL score was capable of accurately stratifying COVID-19 patients diagnosed with non-severe disease on admission into the high severe risk and low severe risk groups from a mixed cohort of 365 patients. This model can facilitate the triage of hospitalised COVID-19 patients on admission and help identify patients who require observation in the intensive care unit and special care to prevent disease progression. Therefore, this model may reduce the mortality owing to severe COVID-19. In addition, the predictors in the HNC-LL score are easily available in clinical practice and can be readily obtained; thus, it holds promise for large-scale use.
Risk factors for patients with severe COVID-19 were identified. A low lymphocyte count is very common among patients with severe COVID-19 [3,16,17]. Increased neutrophil count was observed among patients with severe COVID-19 in our study and a previous study [3]. However, low neutrophil counts were reported to be correlated with initial SARS-COV-2 infection [18]. Since bacterial coinfection was more prevalent among patients with severe disease than among patients with non-severe disease in our study and a previous study [3], it may be reasonable to speculate that bacterial coinfection would aggravate the condition and increase the neutrophil count. It was hypothesised that patients with hypertension have a high risk of developing severe and fatal COVID-19 because those patients tend to have a high level of angiotensin-converting enzyme 2, which would facilitate the infection with SARS-COV-2 [19]. The CRP level is associated with the state of infection and inflammation among patients with severe COVID-19 [20]. Elevated LDH levels in patients with severe COVID-19 suggested that SARS-COV-2 can damage organs such as cardiac muscles and the liver. The predictive role of CRP and LDH levels for severe COVID-19 has been previously reported [21,22,7]. Overall, these studies revealed that the five indicators included in our model had critical clinical significance, and they further affirm the practical value of the HNC-LL score.
To prove the accuracy of prediction, we compared the HNC-LL score with other potentially useful predictive scores. The CURB-65 score is an accurate predictor of disease severity and mortality among patients with CAP. The HNC-LL score performed better than the CURB-65 score in predicting disease severity among COVID-19 patients. This may be due to the fact that the CURB-65 score was initially designed for CAP and not for COVID-19. In addition, we compared the HNC-LL score to other recently developed score systems. The MuLBSTA score can be used as an early predictor of mortality among patients with viral pneumonia. It was proposed that the MuLBSTA score could play a role in the early prediction of mortality owing to COVID-19 [15]. The NLR model [14] was able to determine severe illness among COVID-19 patients in the early stage according to age and NLR. However, the HNC-LL score outperformed both models in predicting disease severity. One possible reason is that the HNC-LL score was derived from a multicentre and multicohort study, had undergone training and validation, and considered the heterogeneity of COVID-19 to a large extent, while the NLR model and MuLBSTA scoring model were based on single-center cohort studies. These results indicated that the HNC-LL score may be an accurate and reliable predictive tool that can be widely used. Furthermore, we compared the HNC-LL score to a recently proposed risk score (COVID-GRAM) based on a nationwide cohort to predict critical COVID-19 disease at admission by Liang et al. On one hand, the target population for the HNC-LL score was patients with severe COVID-19, including those with severe and critical disease, unlike the COVID-GRAM, which merely targeted critically ill patients. Thus, we believe that the HNC-LL and COVID-GRAM models targeted different clinical settings and yielded significantly different values. On the other hand, compared with some relatively subjective markers such as haemoptysis and dyspnea in COVID-GRAM, the five predictors assessed in the HNC-LL score (hypertension, CRP level, neutrophil count, lymphocyte count, and LDH level) were more objective. Subjective markers were highly practice based, and we did not identify markers with sufficient statistical power to be included. Objective markers were the major focus of the present study. The fewer markers assessed in the HNC-LL score also make it easier to apply in clinical practice.
We also compared HNC-LL to a recently proposed risk score [23] (COVID-GRAM) which was designed to predict critical type of COVID-19. It had to be pointed out that the two studies predict outcomes of two different target groups and are useful in different clinical settings. Our study aimed to predict disease severity, in an attempt to estimate future risk of developing of severe COVID-19 (including severe and critical type of COVID-19). Of course, we tried to evaluate the power of COVID-GRAM to predict risk of progression from non-severe type to severe type within our study population. The assessment did not pass the feasibility analysis phase for following reasons: first, hemoptysis, a predictor in COVID-GRAM, was rare within our cohort; second, all the patients included in our study underwent an initial CT scan, and no abnormal X-ray data was available. It is a pity that we were unable to conduct this performance assessment. However, considering that the two models are useful within distinct clinical settings, we do not feel that this detracts from findings of either study. Use of a combination of the tools at different decision-making checkpoints throughout the clinical management of disease will benefit patients with COVID-19, especially since so much remains unknown regarding disease progression. Further, we would like to address the strength of the HNC-LL model in clinical practice, since factors incorporated are objective, easy to acquire, time-saving and cost-effective. Evaluation of risk is based on objective markers, which decreases variation between individuals, increases reliability and improves quality control. The ease in which factors may be determined in routine clinical practice and the low cost of the assessment will facilitate performing risk predictions in daily practice, which is extremely important for COVID-19 management as a result of the severe limitations that occur with regard to healthcare resources.
Clinical significance is of profound importance in model assessment. Early identification of patients with severe COVID-19 and early intervention by clinicians are important for saving lives. Therefore, the initial purpose of the HNC-LL score was to distinguish patients with potentially severe COVID-19 at admission. The HNC-LL score has a good discrimination power for classifying patients into the high severe risk and low severe risk groups and has significantly better predictive performance than the NLR model in this setting. The NLR model utilised age and the ratio of neutrophil count to lymphocyte count to predict severe illness. The neutrophil and lymphocyte counts were also included in the HNC-LL score. Age is a “general” factor, which involves the effect of many risk factors such as hypertension and diabetes mellitus. Age was excluded from the final model based on multivariate logistic regression analysis in this study, suggesting that it was not an independent risk predictor. The HNC-LL score includes more independent risk factors than the NLR model; thus, it may better reflect the microenvironment in patients with severe COVID-19 than the NLR model and may facilitate early and accurate identification. These results further support the clinical utility of the HNC-LL score.
The present study had several limitations. First, the number of patients included in the analysis was small, and this may limit the interpretation of our model. Additional data from different regions should be collected to expand the sample size and further validate the predictive value of the HNC-LL score. Second, some clinical indicators such as D-dimer and cytokine levels were not included owing to the unavailability of data or a large amount of missing data (>15%). The predictive effect of these indicators may be underestimated. Third, it is true that we included individuals who progressed to severe COVID-19 after admission, and also some patients who were diagnosed with severe COVID-19 upon admission, within the severe group. But the results of our subgroup analyses still firmly demonstrated the predictive power of HNC-LL in predicting disease progression. Additional comprehensive research is needed to clarify the mechanism underlying severe COVID-19, and studies with large cohorts are needed to improve this model and increase its interpretability from both biological and population perspectives.
In conclusion, we developed an effective predictive model (called the HNC-LL score) to stratify hospitalised patients into high severe risk and low severe risk groups. We suggest that when patients with COVID-19 are admitted to the hospital, it is important to pay attention to the five indicators in the HNC-LL model (neutrophil count, lymphocyte count, LDH level, CRP level, and hypertension). Risk assessment based on the HNC-LL score is recommended for all COVID-19 patients on admission. For those mild- and moderate-type (non-severe) patients who are classified as high-risk based on the HNC-LL score, active treatment and close monitoring, with the intensity higher than routine management for non-severe patients, should be provided, so as to prevent disease progression. We hope that the HNC-LL score can provide clinical guidance to doctors in controlling the pandemic.
Author contributions
Lu-shan Xiao and Wen-Feng Zhang designed the study, analysed the data, and wrote the paper. Lu-shan Xiao, Hongbo Zhu and Wen-Feng Zhang performed the literature search and constructed the figures. Mengchun Gong and Yanpei Zhang performed the data analysis and wrote the paper. Liya Chen and Pei Kang performed the statistical analyses. Li Liu and Hong Zhu designed the study, analysed the clinical data, and wrote the paper. Li Liu, Hong Zhu, and Mengchun Gong collected the data in Honghu. Wen-Feng Zhang undertook data collection in Nanchang.
Declaration of Competing Interest
The authors declare that they do not have any conflicts of interest.
Acknowledgments
Funding
This work was supported by the National Nature Science Foundation of China (grant no. 81972897) and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2015). The funders had no role in the study design, data collection, data analysis, interpretation, and writing of the report. This study was supported by National Key Research & Development Program of China (Project No. 2018YFC0116901). We have one co-first author, Dr. Mengchun Gong, whose research and resources used for this article has been supported by the fund listed above.
Acknowledgements
We would like to thank all study participants, their families, medical staff, and participating hospitals for their involvement and support in the present study. We thank Yao-Hong Tong, the chief of IT department, and Qing-Lang Zeng, the chief of infectious disease department at the People's Hospital of Honghu, and Yu-lin He, a physician at the First Affiliated Hospital of Nanchang University, for their assistance in data collection in Honghu and Nanchang, respectively. We would like to thank Editage (www.editage.cn) for English language editing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102880.
Contributor Information
Li Liu, Email: liuli@i.smu.edu.cn.
Hong Zhu, Email: zhnfyy@yeah.net.
Appendix. Supplementary materials
References
- 1.Sohrabi C., Alsafi Z., O'Neill N. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mo P., Xing Y., Xiao Y. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa270. ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss P., Murdoch D.R. Clinical course and mortality risk of severe COVID-19. Lancet. 2020;395:1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C., Chen X., Cai Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Health Commission & State Administration of Traditional Chinese Medicine Diagnosis and treatment protocol for novel coronavirus pneumonia (Trial Version 7) Chin Med J. 2020;133:E027. doi: 10.1097/CM9.0000000000000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J., Li W., Shi X. Early antiviral treatment contributes to alleviate the severity and improve the prognosis of patients with novel coronavirus disease (COVID-19) J Intern Med. 2020 doi: 10.1111/joim.13063. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 11.Chen G., Wu D., Guo W. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin X., Lian J.S., Hu J.H. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim W.S., van der Eerden M.M., Laing R. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377–382. doi: 10.1136/thorax.58.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Liu Y., Xiang P. Neutrophil-to-lymphocyte ratio predicts severe illness patients with 2019 novel coronavirus in the early stage. medRxiv. 2020 doi: 10.1186/s12967-020-02374-0. 2020.02.10.20021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo L., Wei D., Zhang X. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol. 2019;10:2752. doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian S., Hu W., Niu L., Liu H., Xu H., Xiao S.Y. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020;15:700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y., Koh V., Marimuthu K. Epidemiological and clinical predictors of COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa322. ciaa322. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepys M.B., Hirschfield G.M. C-reactive protein: a critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling Y., Ling Y., Qian Z. Clinical analysis of risk factors for severe illness in COVID-19 patients. Chin J Infect Dis. 2020;(00):E023. [Google Scholar]
- 22.Yan L., Zhang H.-.T., Goncalves J. A machine learning-based model for survival prediction in patients with severe COVID-19 infection. medRxiv. 2020 2020.02.27.20028027. [Google Scholar]
- 23.Liang W., Liang H., Ou L. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.