Abstract
In the current study, ramp-PCR fragments from improved RAPD (random amplified polymorphic DNA) amplification of Lycium (Goji) species or cultivars were cut and cloned into the vector of pGEM-T. A positive clone 10–5 was screened by PCR amplification, enzymatic digestion, and Sanger sequencing. A SCAR (sequence-characterized amplified region) marker, named Goji 10–5, with 949 nucleotides in length, was identified. Goji 10–5 is specific to Goji species Lycium chinense Miller from Jiangxi in China and Texas in the USA. A BLAST search of this nucleotide sequence in the GenBank database indicated that it shows no identity with any other species, including no any other Lycium species. As a new sequence, we have deposited it in the GenBank database with accession No. MN862323. PCR assays were developed and converted the nucleotide sequence to become a novel molecular marker for Lycium chinense Miller, named Goji 10–5. This marker may be used for the genetic identification of other samples. This study has successfully developed Goji 10–5, a specific SCAR marker to identify L. chinense and distinguish it from other species, including other Lycium species from different locations.
Keywords: Authentication, Lycium species, Molecular marker, Random amplified polymorphic DNA, Sequence-characterized amplified region
Introduction
The Lycium plants, called Gouji (pinyin: gǒuqǐ) or Goji, are well known and include species such as Lycium barbarum, L. chinense, L. europeaum (Jin et al. 2013). The Lycium fruit is called “Gou qi zi”, “Gojizi”, “Wolfberry”, or “Goji berry”. It is a red berry obtained from two closely related boxthorn species, L. chinense Miller and L. barbarum L., of the family Solanaceae. These species originated from northwest China (Ulbricht et al. 2015; Potterat 2010). Nowadays, the Lycium plant is extensively cultivated throughout China and around the world (Dong et al. 2008; Jin et al. 2013; Chang and So 2008). L. barbarum is called Ningxia Goji, and its products have important medicinal properties in Traditional Chinese Medicine (TCM). In China, their use in TCM can be traced back to over two millennia ago. Interestingly, the use of Lycium fruits for anti-aging, improving eyesight, and nourishment was documented already in the “Mingyi Bielu” in 500 C.E. (Yao et al. 2018; Liu et al. 2020). Identification and authentication of Lycium species are very important but challenging, as the morphology is very similar. The development of specific molecular markers is, therefore, essential to overcome this identification problem.
Sevaral DNA techniques have been developed in the era of modern biotechnological sciences over the last 3–4 decades (Williams et al. 1990; Agarwal et al. 2008; Fu et al. 2013). These techniques are generally being used for genetic characterization and authentication of organisms. Sequence characterized amplified region (SCAR) marker is a DNA fragment amplified by PCR using specific 18–26 bp primers, these primers link to traits of interest (Bortoloto et al. 2020; Baite et al. 2020) and designed from nucleotide sequences cloned from random amplified polymorphic DNA (RAPD) fragments (Yang et al. 2014; Lee et al. 2019; Noh et al. 2020; Mei et al. 2020), or other DNA marker derived fragments such as inter simple sequence repeat (ISSR) (Yao et al. 2019), simple sequence repeat (SSR) (Baite et al. 2020), and internal transcribed spacer (ITS) (Hiremani and Dubey 2019). Using longer PCR primers compared to RAPD primers, SCAR markers do not face the low reproducibility problem that is generally encountered with RAPD markers. Obtaining codominant markers might be an added bonus of converting RAPD markers into SCAR markers (Xin et al. 2014; Zhang et al. 2015). Thus, using the SCAR markers, molecular analysis is reduced to a simple PCR evaluation using PCR primers designed from the sequences of the ISSR/SSR/RAPD amplicons (Yang et al. 2014; Huang et al. 2018; Das et al. 2019; Sairkar et al. 2016). The development of SCAR markers from the RAPD fragments is a beneficial molecular approach for genetic identification of different species, substitutions, or adulterants (Yang et al. 2013). Discrimination and molecular genetic diversity in Lycium species were reported by taste pattern and betaine analysis (between Lycium chinense and Lycium barbarum) (Lee et al. 2014), RAPD (Zhang et al. 2001) or SSR markers (Zhao et al. 2010). Although the application of SCAR marker to authenticate wolfberry (L. barbarum) and discriminate it from its adulterants was reported (Sze et al. 2008), a specific SCAR marker to identify and discriminate the medicinal L. chinense Miller from other Lycium species was not yet developed. Here, we have successfully developed a novel SCAR marker after cloning a ramp-PCR RAPD fragment which is specific to L. chinense Miller.
Materials and methods
Extraction of DNA from Lycium species samples
A Nitraria sibirica Pall. sample was collected from Bohu County, Bayingolin Mongol Autonomous Prefecture, Xinjiang Uyghur Autonomous Region in China. The DNA was extracted from samples of different Lycium species or cultivars (Table 1), as previously described (Fu et al. 2013; Liu et al. 2020). DNA samples were then diluted to a final concentration of 10 ng/μL and stored at − 20 °C till used. The RAPD primer sequence for SBS-Q16 was 5′-AAGCGACCTG-3’.
Table 1.
The Goji species sources for RAPD-SCAR analysis
| No | Species or cultivars | Sources | Locality abbreviation |
|---|---|---|---|
| 1 | Lycium chinense Miller | Zhongning, Lingxia | ZN |
| 2 | Lycium chinense Miller | Gongzhou, Jiangxi | GZ |
| 3 | Lycium chinense Miller | Panzhihua, Sichuan | PZH |
| 4 | Lycium chinense Miller (207) | Haidong, Qinghai | HD |
| 5 | Lycium chinense Miller | Haidong, Qinghai | HD |
| 6 | Lycium barbarum 'Ningqi-1' | Haidong, Qinghai | HD |
| 7 | Lycium barbarum 'Ningqi-1' | NAAFS, Yinchuan, Lingxia | YC |
| 8 | Lycium barbarum 'Ningqi-2' | NAAFS, Yinchuan, Lingxia | YC |
| 9 | Lycium barbarum 'Ningqi-3' | NAAFS, Yinchuan, Lingxia | YC |
| 10 | Lycium barbarum 'Ningqi-4' | NAAFS, Yinchuan, Lingxia | YC |
| 11 | Lycium barbarum 'Ningqi-5' | NAAFS, Yinchuan, Lingxia | YC |
| 12 | Lycium barbarum 'Ningqi-6' | NAAFS, Yinchuan, Lingxia | YC |
| 13 | Lycium barbarum L.cv.'Ningqi-7' | NAAFS, Yinchuan, Lingxia | YC |
| 14 | Lycium chinense Mill..'Cai-1' | NAAFS, Yinchuan, Lingxia | YC |
| 15 | Lycium barbarum 'Ningqi-9' | NAAFS, Yinchuan, Lingxia | YC |
| 16 | Lycium chinense Miller | Houston, TX in the USA | TX |
NAAFS Ningxia Academy of Agriculture and Forestry Science
Ramp-PCR amplification and improved RAPD fragments isolation
The ramp-PCR was amplified with the random primer, SBS-Q16, using DNA from 16 Goji species (Table 1). The PCR reaction system (10 μL) consisted of 5 μL 2 × Taq PCR MasterMix (Tiangen Biotech, Beijing, China), 1 μL 2.5 μM SBS-Q16 RAPD primer, 1 μL genomic DNA, and 3 μL ddH2O. The amplification reactions were performed on a PCR machine, with the following steps: initial denaturation at 95 °C for 90 s, 40 cycles of denaturation at 94 °C for 40 s, annealing at 36 °C for 60 s, ramp rate from annealing to extension set to 0.125 °C/s (5% ramp rate), and extension at 72 °C for 90 s. The final extension step was at 72 °C for 5 min. The PCR products were detected by 1.8% agarose gel electrophoresis for 45 min. The bright band was excised from the gel, and purified using TIANgel Mini Purification kit (DP209, Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol (Fu et al. 2017).
DNA cloning
The purified DNA fragments were ligated into pGEM-T vectors (T vector, No. VT202, Tiangen, Beijing, China) at 16 °C for 6–8 h, and transformed into DH5α E. coli complement cells, and cultured at 37 °C overnight (Fu et al. 2017, 2015).
Screening for positive clones
The recombinant clones were sprayed on luria broth (LB) agar plates containing 100 μg/μL ampicillin, 40 mg X-gal and 160 μg isopropyl β-d-1-thiogalactopyranoside (IPTG). The white colonies were screened out by the blue/white screen method. Clones with the presence of correct inserts were verified by PCR amplification using T7/SP6 promoter primer pair (T7 primer: 5′-TAATACGACTCACTATAGGG-3′, SP6 primer: 5′-ATTTAGGTGACACTATAGAA-3′), specific to the T vector near to the ligation ends and the EcoR I digestion site (Yang et al. 2014).
Sequencing and homolog analysis
Cloned DNA fragments at the correct size were sequenced by the Sanger method, using the T7 and SP6 promoter primers for the T vector. Homology of the sequenced DNA to different species was searched and examined by bioinformatics using the online program BLAST (https://www.ncbi.nlm.nih.gov/BLAST/) (Cheng et al. 2020; Zhang et al. 2020).
SCAR primer design
The DNA sequences of the cloned ramp-PCR amplification RAPD fragment of were used to design pairs of SCAR primers utilizing the Primer 3 program (https://bioinfo.ut.ee/primer3-0.4.0/primer3/) (Fu et al. 2017). These designed primers are as follows: Goji10-5L,5′-gcctatcccttttccccata-3′; Goji10-5R,5′-tggtttgtgagtctgcttgg-3’.
Development of SCAR markers
To develop SCAR markers, PCR amplification was executed on the following 10 μL PCR reaction system: 5 μL 2 × Taq PCR Master Mix (Tiangen Biotech, Beijing, China), 1 μL of 2.5 μM SCAR primer pair (Goji10-5L and Goji10-5R), 1 μL genomic DNA (10 ng), and 3 μL ddH2O. PCR was performed on an “Applied Biosystems Veriti® 96-Well Thermal Cycler” (Life Technology, USA), with an initial pre-denaturation for 90 s at 95 °C, followed by 35 cycles of denaturation at 94 °C for 40 s, annealing at 60 °C for 30 s, and extension at 72 °C for 40 s. The final extension step was performed at 72 °C for 5 min. The SCAR analysis was performed on 36 DNA samples as templates to distinguish between different goji species and cultivars and other medicinal plant species. These samples included 16 samples of Lycium species (Table 1), 6 samples of Ganoderma species that were previously described (Fu et al. 2015), 2 specimens of Lonicera japonica collected from Guangdong and Hubei Provinces were previously described (Fu et al. 2013), two specimens of Dimocarpus longan collected from Fujian and Hainan Provinces were previously described (Yang et al. 2013), a specimen of Litchi chinensis from Guangdong, a specimen of Dimocarpus confinis from Guangxi Province, a specimen of Gardenia jasminoides, a specimen Chenopodium album from Luzhou City in Sichuan Province, a specimen of Gastrodia elata from Liangshan City in Sichuan Province, a specimen of Penthorum sedoides, and a specimen of Penthorum Chinense collected from Gulin County in Sichuan Province, a specimen of Angelica sinensis from Sichuan Province as previously described (Zhang et al. 2015), and a specimen of Viola philippica as previously described (Yang et al. 2013). A Nitraria sibirica Pall. sample was also used for PCR amplification by above primers specific for the SCAR marker Goji 10–5.
The products from the PCR amplification were then separated on a 1.5% agarose gel in 1 × TAE buffer with 140 V for 25 min. The DNA electrophoresis gels were stained by 0.5 μg/mL ethidium bromide, and images were acquired by ChemiDoc XRS (Bio-Rad, USA).
Results
Isolation of RAPD fragments from ramp-PCR amplification
The SBC-Q16 primer was used to improve RAPD amplification from the sixteen Goji species samples (Table 1). The ramp-PCR results are shown in Fig. 1a. The very strong band indicated by the blue arrow in the figure was specific to L. chinense Miller. The band, labeled with Goji 10, was cut from the agarose gel (we cut different PCR bands and named the indicated band in Fig. 1a as “10”). The DNA in the agarose gel was purified and eluted with 20 μL of ddH2O. To check DNA quality and measure the quantity used for ligation, the purified DNA samples were measured on an agarose gel by loading 2 μL of purified PCR products into each well and 0.5 μL of T-vector into another well. The results are shown in Fig. 1b.
Fig. 1.
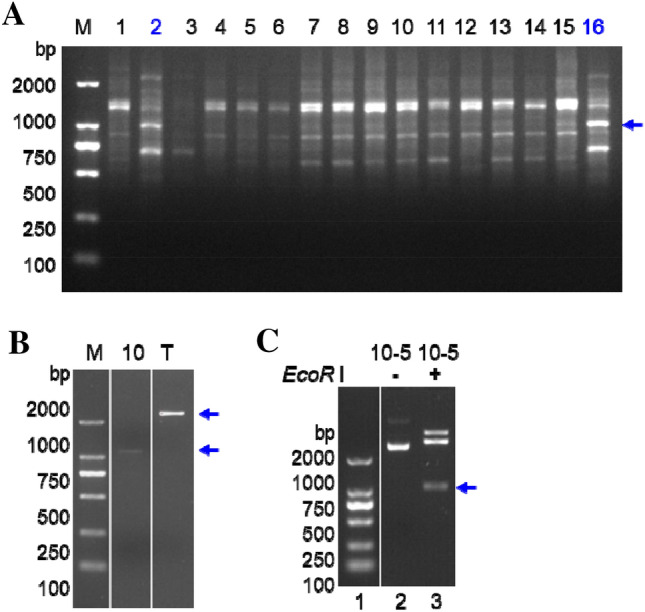
Amplification, DNA cloning and positive clone identification from ramp-PCR amplified fragments. a The ramp-PCR amplification from DNA samples of Lycium species. Lanes 1–16 are different Lycium species samples listed in Table 1 (Liu et al. 2020). The blue arrow in lanes 2 and 16 is indicated bands for cutting. b The quality and quantity checking of ramp-PCR fragments purified from an agarose gel. The blue arrows indicate a fragment named “10” which we cut different PCR bands and a band of T vector, respectively. c Identification of positive clone by plasmid DNA digestion. Lane 1 indicates the DNA molecular weight marker DL2000 with the fragment sizes (bp) 2000, 1000, 750, 500, 250, 100. Lanes 2 and 3 indicate clone 10–5 plasmid DNA without (−) or with (+) EcoR I digestion. The blue arrow indicates expected insert of RAPD DNA fragment
DNA cloning and sequencing information of the Goji 10–5 clone
According to the DNA quantity (Fig. 1b), an appropriate amount of PCR product and T vector were ligated and screened by the blue/white screen method. Positive clones were screened by PCR (data not shown) and enzymatic digestion by EcoR I from extracted plasmids (Fig. 1c). The clone in lane 3 of Fig. 1c, named Goji 10–5 (we screened different clones and clone number 5 was called Goji 10–5), showed an inserting DNA-fragment of ~ 1000 bp. We, therefore, sequenced it by the Sanger method.
Sequencing of the Goji 10–5 clone of the ramp-PCR RAPD fragment indicated that Goji 10–5 consists 949 nucleotides (Fig. 2). A BLAST search for this DNA sequence in the GenBank database indicated that it does not show identity to Goji species or other species (data not shown). This new sequence was deposited into the GenBank database with accession No. MN862323 (Fig. 2).
Fig. 2.
Cloned sequences information by Sanger-sequencing. a The DNA sequences of clone 10–5 with 949 bp in length. This sequences were GenBank database with accession No. MN862323
Developing a specific SCAR marker
SCAR primers were designed to establish stably diagnostic Goji species-specific SCAR markers from the ramp-PCR RAPD fragments. The results of the PCR reaction are shown in Fig. 3a. The SCAR marker Goji 10–5 showed the expected size, with very strong signals only in the two L. chinense Miller samples from Jiangxi in China and Houston, Texas, in the USA (lanes 2 and 16 in Fig. 3a). There was no strong amplification in any of the other 14 Lycium species (Fig. 3a) or the other 19 medicinal species tested (data not shown). These findings indicate that the SCAR marker Goji 10–5 is sample-specific. Lack of specific amplicons in the other Goji and other species indicates the Goji 10–5 marker’s efficacy in identifying L. chinense. Note that we did observe very weak bands with the same or larger size in the other lanes, but these should probably be non-specific binding.
Fig. 3.
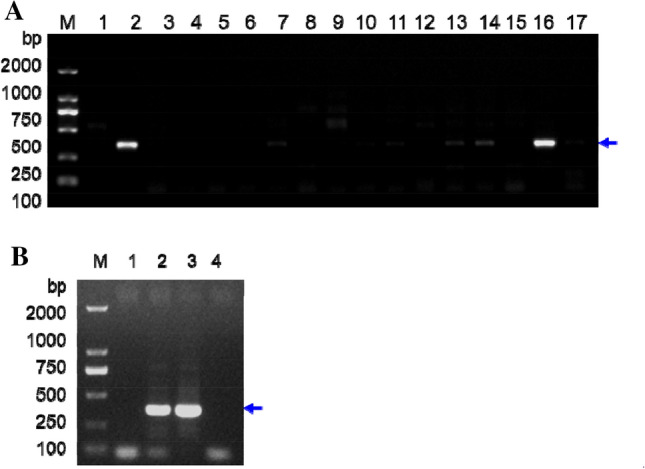
Development of RAPD-SCAR marker Goji 10–5. a Development of RAPD-SCAR marker Goji 10–5. Lanes 1–16 indicate the different Lycium species samples listed in Table 1 described in Material and Method and the previously published paper (Liu et al. 2020). Lane 17 is PCR amplification without any DNA samples as a negative control. b Discriminating Nitraria sibirica Pall. from Goji samples by Goji 10–5 marker. Lanes 1–4 indicate a negative control without any DNA samples, two samples from Jiangxi in China and Houston, Texas, in the USA, and a N. sibirica Pall. sample, respectively (b). No any specific PCR band is found in the lane 4 of N. sibirica Pall. sample DNA. Lane M indicates the DNA molecular weight marker DL2000 with the indicated fragment sizes
Discriminating Nitraria sibirica Pall. from Goji samples by Goji 10–5 marker
A Nitraria sibirica Pall. sample was collected to demonstrate efficacy for the discrimination of Nitraria sibirica Pall. from Goji samples by Goji 10–5 marker. Indeed, by PCR amplification we have successfully distinguished N. sibirica Pall. from Goji species, two samples from Jiangxi in China, and Houston, Texas, in the USA, respectively (Fig. 3b).
Discussion
The plant genus Lycium (Goji) in the Solanaceae family is known as boxthorn. It has a notable place in TCM thanks to its fruits. Substitutions and adulterants of Goji might be compromise the values of the TCM treatment or even jeopardize the safety of the consumers. Nevertheless, fruits of all the Lycium species are red and very similar in appearance and tissue structure (Konarska 2018). Since the Lycium genus is an umbrella term for different plant species that are closely related, adulterations or mislabeling are possible (Wetters et al. 2018). However, it is difficult to differentiate among these species using traditional morphological and histological analyses. Nitraria is a genus of flowering plants in the family Nitrariaceae, native to Africa, Asia, Europe, and Australia. Nitraria sibirica Pall., one of nine species of Nitraria (https://en.wikipedia.org/wiki/Nitraria), is distributed in the desert and coastal sand areas of north and northeast China. Even though N. sibirica Pall. is traditionally exercised as a folk medicine in China, its actively effective gradients are different from those in L. barbarum. The fruit from N. sibirica Pall. is smooth and fat and is used as a fake L. barbarum in China’s markets. With this regard, DNA markers, such as SCAR markers (Baite et al. 2020; Mei et al. 2020), would be useful for differentiation between cultivars, species, substitutions, or adulterants (Sze et al. 2008; Zhao et al. 2010; Zhang et al. 2001; Noh et al. 2020). In our previous study, sixteen Lycium samples were collected from different locations, and genetically characterized them by ramp-PCR RAPD and ISSR markers. This characterization provided valuable insights into the genetic and biological diversity of these medicinal plants (Liu et al. 2020). Nevertheless, the specific SCAR markers for L. chinense Miller have not been developed yet.
In the current study, the RAPD fragments from ramp-PCR amplification of the Lycium species or cultivars were cut and cloned into pGEM-T vectors. A positive clone Goji 10–5 was identified and sequenced successfully. One SCAR marker, named Goji 10–5, with 949 nucleotides, was developed. This marker is specific to Goji species L. chinense Miller, being positive with samples from Jiangxi in China and Houston, Texas, in the USA. BLAST search for this DNA sequence in the GenBank database found no identity to any other species. We deposited the sequence into the GenBank database with accession No. MN862323. PCR assays with primers for the designed marker confirmed that it as a novel molecular marker for L. chinense Miller. As such, it might be used for DNA authentication of other samples. Indeed, we have successfully distinguished N. sibirica Pall. from Goji species (Fig. 3b). Of course, it is better if more SCAR markers specific to L.chinense Miller are developed.
Conclusion
By cloning the ramp-PCR fragments, we have successfully developed the Goji 10–5 SCAR marker, a specific marker for L. chinense Miller. This novel specific SCAR marker could be useful for specific authentication and identification of the popular medicinal L. chinense and for differentiating it from other Goji samples, or adulterant samples, such as N. sibirica Pall.
Acknowledgements
This research was supported in part by the Science and Technology Innovation Team of Colleges and Universities in Sichuan Province (13TD0032), and the Joint Research Foundation of Luzhou City and Southwest Medical University (2018LZXNYD-YL01). We truly thank people for help in collecting samples.
Author contributions
JF and JP were in charge of the idea, project design. XL, JC, and CW performed DNA extraction, PCR amplification, sequencing and data analysis. ZM and XL collected N. sibirica Pall. sample and performed DNA extraction. JF wrote the manuscript, MK edited the manuscript draft and JF revised the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Agarwal M, Shrivastava N, Padh H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008;27(4):617–631. doi: 10.1007/s00299-008-0507-z. [DOI] [PubMed] [Google Scholar]
- Baite MS, Upadhyay BK, Dubey SC. Development of a sequence-characterized amplified region marker for detection of Ascochyta rabiei causing Ascochyta blight in chickpea. Folia Microbiol. 2020;65(1):103–108. doi: 10.1007/s12223-019-00711-5. [DOI] [PubMed] [Google Scholar]
- Bortoloto TM, Fuchs-Ferraz MCP, Kettener K, Martins Rubio L, Gonzalez ER, de Souza ICG, Oda S, Rossini BC, Marino CL. Identification of a molecular marker associated with lignotuber in Eucalyptus ssp. Sci Rep. 2020;10(1):3608. doi: 10.1038/s41598-020-60308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RC, So KF. Use of anti-aging herbal medicine, Lycium barbarum, against aging-associated diseases. What do we know so far? Cell Mol Neurobiol. 2008;28(5):643–652. doi: 10.1007/s10571-007-9181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Peng J, Fu J, Khan MA, Tan P, Wei C, Deng X, Chen H, Fu J. Identification of a novel germline BRCA2 variant in a Chinese breast cancer family. J Cell Mol Med. 2020;24(2):1676–1683. doi: 10.1111/jcmm.14861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AK, Nerkar S, Gawande N, Thakre N, Kumar A. SCAR marker for Phytophthora nicotianae and a multiplex PCR assay for simultaneous detection of P nicotianae and Candidatus Liberibacter asiaticus in citrus. J Appl Microbiol. 2019;127(4):1172–1183. doi: 10.1111/jam.14392. [DOI] [PubMed] [Google Scholar]
- Dong JZ, Yang JJ, Wang Y. Resources of Lycium species and related research progress. Zhongguo Zhong yao za zhi Zhongguo zhongyao zazhi China journal of Chinese materia medica. 2008;33(18):2020–2027. [PubMed] [Google Scholar]
- Fu J, Yang L, Khan MA, Mei Z. Genetic characterization and authentication of Lonicera japonica Thunb by using improved RAPD analysis. Mol Biol Rep. 2013;40(10):5993–5999. doi: 10.1007/s11033-013-2703-3. [DOI] [PubMed] [Google Scholar]
- Fu JJ, Mei ZQ, Tania M, Yang LQ, Cheng JL, Khan MA. Development of RAPD-SCAR markers for different Ganoderma species authentication by improved RAPD amplification and molecular cloning. Genet Mol Res. 2015;14(2):5667–5676. doi: 10.4238/2015.May.25.19. [DOI] [PubMed] [Google Scholar]
- Fu S, Cheng J, Wei C, Yang L, Xiao X, Zhang D, Stewart MD, Fu J. Development of diagnostic SCAR markers for genomic DNA amplifications in breast carcinoma by DNA cloning of high-GC RAMP-PCR fragments. Oncotarget. 2017;8(27):43866–43877. doi: 10.18632/oncotarget.16704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremani NS, Dubey SC. Phylogenetic relationship among Indian population of Fusarium oxysporum f. sp. lentis infecting lentil and development of specific SCAR markers for detection. 3 Biotech. 2019;9(5):196. doi: 10.1007/s13205-019-1734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Liou JS, Huang L, Watanabe K. Developing novel species-specific DNA markers for PCR-based species identification of the Lactobacillus sakei group. Lett Appl Microbiol. 2018;66(2):138–144. doi: 10.1111/lam.12825. [DOI] [PubMed] [Google Scholar]
- Jin M, Huang Q, Zhao K, Shang P. Biological activities and potential health benefit effects of polysaccharides isolated from Lycium barbarum L. Int J Biol Macromol. 2013;54:16–23. doi: 10.1016/j.ijbiomac.2012.11.023. [DOI] [PubMed] [Google Scholar]
- Konarska A. Microstructural and histochemical characteristics of Lycium barbarum L. fruits used in folk herbal medicine and as functional food. Protoplasma. 2018;255(6):1839–1854. doi: 10.1007/s00709-018-1277-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim YH, Kim YH, Lee GH, Lee MY. Discrimination of Lycium chinense and Lycium barbarum by taste pattern and betaine analysis. Int J Clin Exp Med. 2014;7(8):2053–2059. [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ali A, Ha B, Kim MK, Kong WS, Ryu JS. Development of a molecular marker linked to the A4 locus and the structure of HD genes in Pleurotus eryngii. Mycobiology. 2019;47(2):200–206. doi: 10.1080/12298093.2019.1619989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Du J, Khan MA, Cheng J, Wei C, Mei Z, Chen H, He T, Fu J. Analysis of genetic diversity and similarities between different Lycium varieties based on ISSR analysis and RAMP-PCR markers. World Acad Sci J. 2020;2(2):83–90. doi: 10.3892/wasj.2020.39. [DOI] [Google Scholar]
- Mei Z, Khan MA. Genetic authentication of Eclipta prostrate (Asteraceae) from Penthorum chinense (Penthoraceae) by sequence characterized amplified region (SCAR) markers. Rev Biol Trop. 2020;68(1):180–188. doi: 10.15517/RBT.V68I1.37046. [DOI] [Google Scholar]
- Noh P, Kim WJ, Song JH, Park I, Choi G, Moon BC. Rapid and simple species identification of cicada exuviae using COI-based SCAR assay. Insects. 2020 doi: 10.3390/insects11030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterat O. Goji (Lycium barbarum and L. chinense): phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med. 2010;76(1):7–19. doi: 10.1055/s-0029-1186218. [DOI] [PubMed] [Google Scholar]
- Sairkar PK, Sharma A, Shukla NP. SCAR marker for identification and discrimination of Commiphora wightii and C. myrrha. Mol Biol Int. 2016 doi: 10.1155/2016/1482796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze SC, Song JX, Wong RN, Feng YB, Ng TB, Tong Y, Zhang KY. Application of SCAR (sequence characterized amplified region) analysis to authenticate Lycium barbarum (wolfberry) and its adulterants. Biotechnol Appl Biochem. 2008;51(Pt 1):15–21. doi: 10.1042/BA20070096. [DOI] [PubMed] [Google Scholar]
- Ulbricht C, Bryan JK, Costa D, Culwell S, Giese N, Isaac R, Nummy K, Pham T, Rapp C, Rusie E, Weissner W, Windsor RC, Woods J, Zhou S. An evidence-based systematic review of goji (Lycium spp.) by the natural standard research collaboration. J Diet Suppl. 2015;12(2):184–240. doi: 10.3109/19390211.2014.904128. [DOI] [PubMed] [Google Scholar]
- Wetters S, Horn T, Nick P. Goji who? Morphological and DNA based authentication of a “Superfood”. Front Plant Sci. 2018;9:1859. doi: 10.3389/fpls.2018.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin GZ, Lam YC, Maiwulanjiang M, Chan GK, Zhu KY, Tang WL, Dong TT, Shi ZQ, Li P, Tsim KW. Authentication of Bulbus Fritillariae Cirrhosae by RAPD-derived DNA markers. Molecules. 2014;19(3):3450–3459. doi: 10.3390/molecules19033450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Fu S, Khan MA, Zeng W, Fu J. Molecular cloning and development of RAPD-SCAR markers for Dimocarpus longan variety authentication. SpringerPlus. 2013;2:501. doi: 10.1186/2193-1801-2-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Khan MA, Mei Z, Yang M, Zhang T, Wei C, Yang W, Zhu L, Long Y, Fu J. Development of RAPD-SCAR markers for Lonicera japonica (Caprifolicaceae) variety authentication by improved RAPD and DNA cloning. Rev Biol Trop. 2014;62(4):1649–1657. doi: 10.15517/rbt.v62i4.13493. [DOI] [PubMed] [Google Scholar]
- Yao R, Heinrich M, Weckerle CS. The genus Lycium as food and medicine: a botanical, ethnobotanical and historical review. J Ethnopharmacol. 2018;212:50–66. doi: 10.1016/j.jep.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Yao Z, Qin D, Chen D, Liu C, Chen W, Liu T, Liu B, Gao L. Development of ISSR-derived SCAR marker and SYBR green I real-time PCR method for detection of teliospores of Tilletia laevis Kuhn. Sci Rep. 2019;9(1):17651. doi: 10.1038/s41598-019-54163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KY, Leung HW, Yeung HW, Wong RN. Differentiation of Lycium barbarum from its related Lycium species using random amplified polymorphic DNA. Planta Med. 2001;67(4):379–381. doi: 10.1055/s-2001-14310. [DOI] [PubMed] [Google Scholar]
- Zhang C, Mei Z, Cheng J, He Y, Khan MA, Luo P, Imani S, Fu J. Development of SCAR markers based on improved RAPD amplification fragments and molecular cloning for authentication of herbal medicines Angelica sinensis, Angelica acutiloba and Levisticum officinale. Nat Prod Commun. 2015;10(10):1743–1747. [PubMed] [Google Scholar]
- Zhang L, Cheng J, Zhou Q, Khan MA, Fu J, Duan C, Sun S, Lv H, Fu J. Targeted next-generation sequencing identified novel compound heterozygous variants in the CDH23 gene causing usher syndrome type ID in a chinese patient. Front Genet. 2020;11:422. doi: 10.3389/fgene.2020.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WG, Chung JW, Cho YI, Rha WH, Lee GA, Ma KH, Han SH, Bang KH, Park CB, Kim SM, Park YJ. Molecular genetic diversity and population structure in Lycium accessions using SSR markers. CR Biol. 2010;333(11–12):793–800. doi: 10.1016/j.crvi.2010.10.002. [DOI] [PubMed] [Google Scholar]



