Abstract
Background
Immune checkpoints target regulatory pathways in T cells that enhance antitumor immune responses and elicit durable clinical responses. As a novel immune checkpoint, CD96 is an attractive key target for cancer immunotherapy. However, there has been no integrative investigation of CD96 in glioma. Our study explored the relationship between CD96 expression and clinical prognosis in glioma.
Methods
RNA and clinical data for a total of 1,001 samples were included in this study, including 325 samples from the Chinese Glioma Genome Atlas (CGGA) database and 676 samples from The Cancer Genome Atlas (TCGA) dataset. The R programming language was employed to perform statistical analysis and draw figures.
Results
CD96 had a consistently positive relationship with glioblastoma and was highly enriched in IDH-wildtype and mesenchymal subtype glioma. Gene ontology enrichment and gene set variation analysis analyses suggested that CD96 was mostly involved in immune functions and was especially related to T cell-mediated immune response in glioma. Subsequent immune infiltration analysis showed that CD96 was positively correlated with infiltrating levels of CD4 + T and CD8 + T cells, macrophages, neutrophils, and DCs in glioblastoma multiforme and low-grade glioma. Additionally, CD96 was tightly associated with other immune checkpoints, including PD-1, CTLA-4, TIGIT, and TIM-3. Univariate and multivariate Cox analysis demonstrated that CD96 acts as an independent indicator of poor prognosis in glioma.
Conclusion
CD96 expression was increased in malignant phenotype and negatively associated with overall survival in glioma. CD96 also showed a positive correlation with other immune checkpoints, immune response, and inflammatory activity. Our findings indicate that CD96 is a promising clinical target for further immunotherapeutic use in glioma patients.
Keywords: CD96, glioma, immune checkpoint, prognosis, immunotherapy
Introduction
The most prevalent and devastating primary intracranial tumor, glioma, and especially glioblastoma multiforme (GBM, WHO grade IV), are characterized by heterogeneity and extensive invasion, with high recurrence and fatality rate (Jiang et al., 2016; Louis et al., 2016; Van Meir et al., 2010). Therefore, multiple attempts have been made to prolong life expectancies of glioma patients, comprising developing effective therapy, appropriate biomarkers, and molecular-targeted drugs. Numerous conventional treatment methods of central nervous system (CNS) tumors have emerged in recent decades, such as neurosurgical resection, radiotherapy, and chemotherapy (Stupp et al., 2009; Walker et al., 2017). Among them, immunotherapy is reckoned to be an encouraging treatment because it evokes an antitumor immune response to restrain immune evasion of the tumor (Louveau et al., 2015). In melanoma and non-small-cell lung cancer, immune checkpoint inhibitors like target programmed cell death protein 1 (PD-1)/programmed death ligand 1 (PD-L1) and cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) have been exploited and successfully applied in the clinic (Topalian et al., 2012; Wang et al., 2016; Huang et al., 2017), whereas many glioma patients are refractory to current immunotherapy, which arouses our interest in identifying additional immune checkpoints to enhance the therapeutic efficacy in glioma (Reardon et al., 2014; Huang et al., 2017).
A novel immune checkpoint receptor target, CD96, has recently entered the limelight in current cancer immunotherapies shown to inhibit natural killer (NK) cells (Georgiev et al., 2018). Compelling evidence confirmed that blocking CD96 enhanced the containment of primary tumor growth in murine model systems in a CD8 + T cell-dependent manner (Dougall et al., 2017; Mittal et al., 2019). Of note, the efficiency of the antitumor activity of anti-CD96 therapy improved in dual-combination with blockade of other immune checkpoints, like PD-1, PD-L1, TIGIT, and CTLA-4 (Mittal et al., 2019). Besides, CD96 represents several unique features that exhibit profound beneficial effects in the coming age of human cancer therapy (Georgiev et al., 2018; Deuss et al., 2019).
To make a systematic examination of CD96 in glioma, we gathered RNA-seq and clinical data for 325 glioma samples from the Chinese Glioma Genome Atlas (CGGA) project. Simultaneously, we obtained another data set containing 676 samples from The Cancer Genome Atlas (TCGA) cohort to further corroborate the findings. Overall, our comprehensive study of the molecular and clinicopathological features of CD96 through 1,001 samples will provide a better understanding of CD96 in glioma and pave the way for developing CD96-targeted cancer immunotherapies.
Materials and Methods
Data Collection From the CGGA and TCGA Projects
The CD96 genetic and clinical data of 325 glioma samples were downloaded from the CGGA cohort1; these ranged from WHO grade II–IV. The read counts for each GENCODE gene were calculated using RSEM and were transformed into a FPKM (fragments per kilobase transcriptome per million fragments) matrix. In this mRNA expression profile, an expressed gene was defined only if its expression level was larger than 0 in half of the samples.
Moreover, 161 GBM (glioblastoma multiforme) and 515 LGG (low-grade glioma) samples were obtained from the TCGA Pan-Cancer Atlas project through the cbioportal2 website3 including RNA-seq data and clinicopathological information. The read counts were also calculated using RSEM, and mRNA expression was batch corrected and normalized.
Gene Ontology and Gene Set Variation Analysis Analysis
After Spearman correlation analysis, gene ontology (GO) analysis of the most correlated genes was constructed by using the R package ‘clusterProfiler’ (Yu et al., 2012). Gene set variation analysis (GSVA) analysis was applied using standard settings, as implemented in the R package ‘GSVA’ (Hänzelmann et al., 2013). Additionally, inflammatory-related metagenes were described previously (Wang et al., 2016).
TIMER Database Analysis
The Tumor Immune Estimation Resource (TIMER) serves a comprehensive resource to analyze immune infiltrates among 10,897 samples across 32 cancer types (Li T. et al., 2017). The purity-corrected Spearman’s rho of CD96 expression with the abundance of immune infiltrates, including B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, dendritic cells, T cell NK, and NK cells were detected in GBM and LGG patients on TIMER4 and with TIMER 2.05 online tools.
Statistical Computations
Statistical analysis and figure construction were conducted using the R language, version 3.6.16. The t-test was performed to evaluate CD96 expression among different grades, pathological subsets, IDHWT/IDHMUT, and subtypes of glioma. Kaplan–Meier plots and Cox proportional hazard model analysis were generated using the R packages ‘survminer’ and ‘survival’ (Therneau and Lumley, 2015; Kassambara et al., 2018). Receiver operating characteristic (ROC) curves were calculated by the R package ‘pROC’ (Robin et al., 2013). Area under the curve (AUC) values were depicted from the ROC curves. Corrgram and corrplot plots were drawn using the R packages ‘corrgram’ and ‘corrplot’, separately (Friendly, 2002; Wei, 2017). All statistical tests were two-sided, and a p-value < 0.05 was regarded as indicating statistical significance.
Results
CD96 Was Enriched in Glioblastoma, IDH-Wildtype, and Mesenchymal Glioma
To clarify differences in the CD96 expression pattern in four grades of glioma malignancy, the mRNA level of CD96 was examined in the CGGA and TCGA databases separately. In the CGGA cohort, the CD96 transcript profiles in WHO grade III and IV displayed little difference (not significant in t-test) and had higher values than WHO II (Figure 1A). In the TCGA dataset, CD96 expression in glioblastoma (WHO IV) was higher than in WHO grade II and grade III glioma in TCGA (Figure 1B). The results revealed that CD96 expression was comparatively upregulated in higher-grade gliomas.
FIGURE 1.
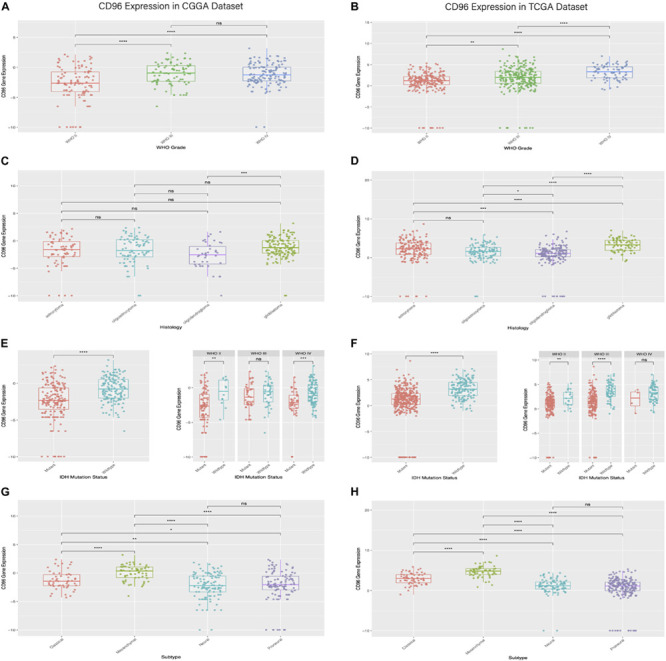
Relationship between CD96 expression and clinical glioma parameters in the CGGA and TCGA cohorts. (A,B) Correlation of CD96 transcript levels and WHO grade. (C,D) CD96 expression pattern in different WHO grades. (E,F) Association between CD96 expression and IDH-wildtype for all-grade glioma. (G,H) Correlation of CD96 transcript levels and glioma subtypes. Ns, **, ***, and **** represent no significant, p < 0.05, p < 0.01, and p < 0.0001, respectively.
The distribution of CD96 in different glioma subtypes was next analyzed. Compared to other pathological subsets (namely oligodendroglioma, oligoastrocytoma, and astrocytoma), CD96 expression was relatively upregulated in glioblastoma (Figures 1C,D). Isocitrate dehydrogenase (IDH) mutation is present in almost 40% of glioma, which has an outsized impact on glioma development and progression (Branzoli et al., 2018). To this end, the expression profile of CD96 in different IDH states was also explored. We found that CD96 was consistently enriched in IDH-wildtype (IDHWT) in both the CGGA and TCGA databases (Figures 1E,F). This result suggested that CD96 expression was more prevalent without IDH mutation (IDHWT) than with IDH mutation (IDHMUT) in glioma. Besides, CD96 was expressed at higher levels in the mesenchymal subtype compared with the other three subtypes (classical, neural, and proneural) in the CGGA cohort (Figure 1G). Highly consistent results were obtained using the TCGA dataset (Figure 1H).
CD96 Had Sufficient Sensitivity to Predict Mesenchymal Subtype Glioma in ROC Curve Analysis
We carried out ROC curves analysis of CD96 expression and mesenchymal subtype in glioma. Intriguingly, we observed that the AUC values of ROC curves were up to 78.6. and 92.8% in the CGGA and TCGA datasets, respectively (Figures 2A,C). These findings indicated that CD96 acted as a potential biomarker for mesenchymal subtype glioma. ROC curves analyses of CD96 in glioma of all WHO grades were next conducted. The AUC of WHO IV was 0.723 in the CGGA database, whereas the AUC of WHO IV was 0.591 in the TCGA database (Figures 2B,D).
FIGURE 2.
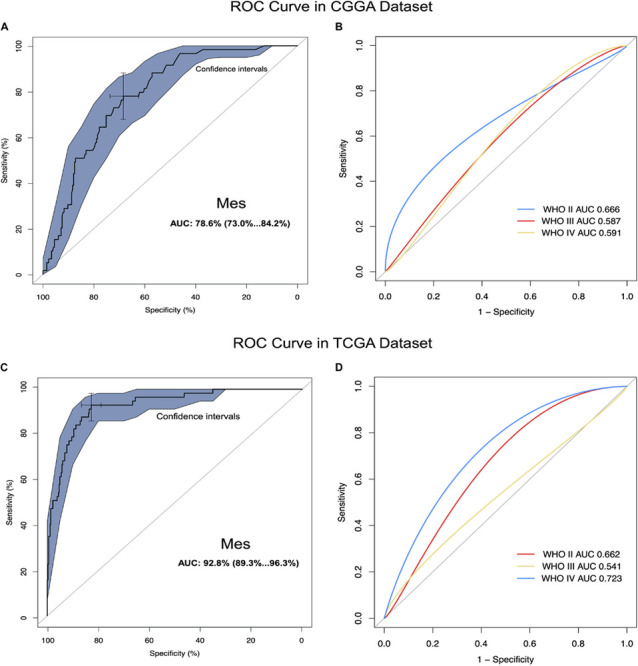
CD96 was highly enriched in mesenchymal molecular subtype glioma. (A,B) ROC curve analysis of CD96 in mesenchymal subtype and all-grade gliomas in the CGGA dataset. (C,D) ROC curve analysis of CD96 in mesenchymal subtype and all-grade gliomas in the TCGA dataset.
CD96 Was Significantly Associated With Immune Functions in Glioma
Considering that CD96 expression was closely tied to malignancy, we inferred that CD96 became an integral part of glioma progression and therefore performed GO analysis to uncover its role. Spearman correlation analysis (Spearman | rho| > 0.6) indicated that 54 and 176 genes were positively correlated with CD96 in the CGGA and TCGA datasets, respectively. According to GO biological process (BP) enrichment analysis for the top 50 related genes, we found that the genes most relevant to CD96 were involved in T-cell activation and regulation of lymphocyte proliferation in the CGGA and TCGA databases, respectively (Figures 3A,B; Supplementary Table S1). To further elucidate the immune function of CD96 in glioma, on the basis of the 1,540 genes reported to be associated with the immune response which were downloaded form the AmiGO 2 website7 (Li G. et al., 2017), we selected the 362 and 354 genes most relevant to CD96 (Spearman |rho| > 0.3) in the CCGA and TCGA cohorts (Supplementary Table S2) for heatmap drawing (Liu et al., 2019). Thereinto, 357 genes were strongly positively correlated with CD96 expression, while five genes had a significantly negative relationship with CD96 in the CGGA dataset. In the TCGA dataset, 347 and 7 genes were directly and inversely proportional to CD96 expression, respectively (Figure 4). To sum up, CD96 was directly correlated with most immune responses and negatively correlated with few immune responses in glioma.
FIGURE 3.
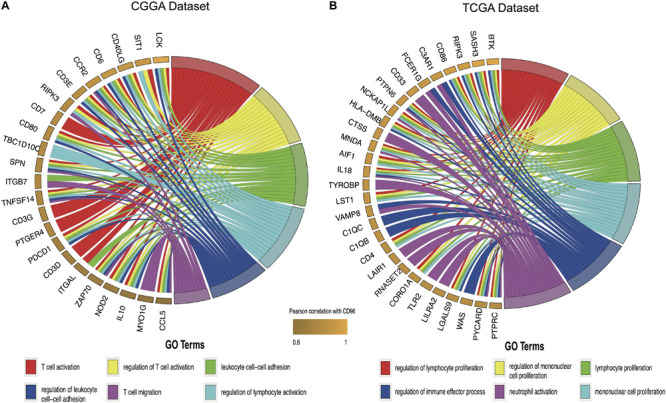
Gene ontology (GO) analysis for top 50 genes most relevant to CD96 in the CGGA database (A) and TCGA database (B).
FIGURE 4.
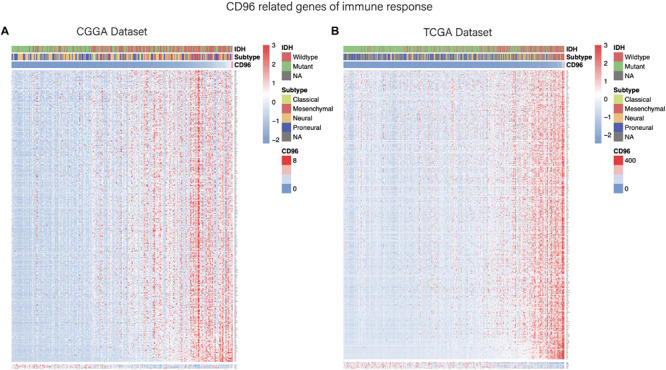
Heatmaps of CD96-related immune genes in glioma in the CGGA (A) and TCGA cohorts (B).
Correlation Between CD96 and T-Cell Mediated Immunity in Glioma
To fully understand the relationship between CD96 and T cell-related immunity in glioma, we performed GSVA to assess differential activities of pathways between sets of genes. As delineated in Figures 5A,B, these relationships were similar in both the CGGA and TCGA databases. Specifically, CD96 showed a positive correlation with T-helper 1/2 type immune response (GO:0042088 and GO:0042092), T-helper 1/2 cell cytokine production (GO:2000556 and GO:2000553), and NK cell-mediated cytotoxicity directed against tumor cell target (GO:0002860). Conversely, CD96 was correlated negatively with T cell-mediated immune response to tumor cell (GO:0002842) and T cell-mediated cytotoxicity directed against tumor cell target (GO:0002852). This result re-validated that the special immune function of CD96 is to act an inhibitory role in T cell-mediated immune response to tumor cells in glioma.
FIGURE 5.
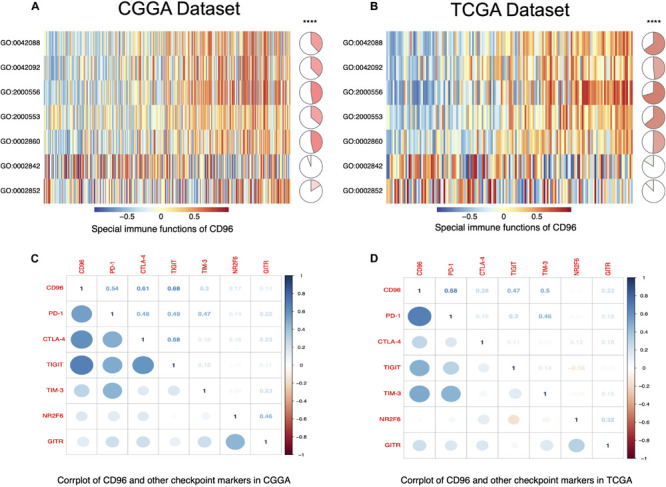
CD96-related T cell immunity and immune checkpoint markers in glioma. (A,B) The relationship between CD96 and T cell immunity in glioma in the CGGA and TCGA datasets. (C,D) Corrgram map of CD68 and immune checkpoint markers in glioma in the CGGA and TCGA databases.
Correlation Between CD96 and Other Immune Checkpoints
A growing number of immune checkpoints have been emerged as therapeutic targets and have been examined in clinical trials or clinical situations (Ribas and Wolchok, 2018; Feng et al., 2019). Therefore, we analyzed the relationship between CD96 and other immune checkpoints, such as PD-1, CTLA-4, TIGIT, TIM-3, NR2F6, and GITR. Pearson correlation analysis revealed that CD96 was tightly associated with PD-1, CTLA-4, TIGIT, and TIM-3. Co-expression of PD-1 with CD96 was consistent with the findings of previous research (Mittal et al., 2019). These results were validated in both the CGGA and TCGA datasets (Figures 5C,D), implying possible synergistic effects of CD96 with these checkpoint members. Accordingly, we postulated that CD96 may contribute significantly to the inflammatory response in glioma and used a previously described method to test this (Wang et al., 2016). As shown in Supplementary Figure S1, CD96 was positively associated with HCK, MHC-I, MHC-II, STAT1, STAT2, and FCGR2A, and especially with LCK. This finding additionally evidenced the vital immune function of CD96 in glioma.
Correlation Between CD96 and Immune Infiltration Level in GBM and LGG
Tumor-infiltrating lymphocytes are an independent predictor of sentinel lymph node status and survival in cancers. Hence, we investigated whether CD96 expression was connected with immune infiltration levels in GBM (glioblastoma multiforme) and LGG (Low-Grade Glioma) by using TIMER website tools. Our findings showed that CD96 expression related positively with infiltrating levels of dendritic cells (r = 0.509), neutrophils (r = 0.491), macrophages (r = 0.435), and CD8 + T cells (r = 0.437) in LGG. Simultaneously, CD96 had a marginal positively association with B cell (r = 0.355) and CD4 + T cell (r = 0.361) infiltration level in LGG. On the other hand, CD96 expression has no significant relationships with tumor purity (r = -0.117) or infiltrating levels of B cells (r = 0.101), CD8 + T cells (r = -0.157), CD4 + T cells (r = -0.1), and other cells in GBM (Figure 6). These differences implied that CD96 would make more of a contribution to immune infiltration in LGG than GBM, especially in dendritic cells. Using TIMER 2.0 tools, the associations between CD96 expression and T cell NK and NK cell immune infiltrates were explored. As shown in Supplementary Figure S2, the CD96 expression level showed a positive association with NK cell EPIC and a negative association with NK cell QUANTISEQ in both GBM and LGG. Additionally, CD96 was positively correlated with NK cell activated CIBERSORT-ABS, while it was negatively correlated with NK cell XCELL. The low purity-adjusted Spearman’s rho (all rho <0.5) implied a weak correlation of CD96 expression with the above immune infiltration levels (Figure 6 and Supplementary Figure S2). These findings could provide helpful information about the potential cellular targets of CD96 blockade.
FIGURE 6.
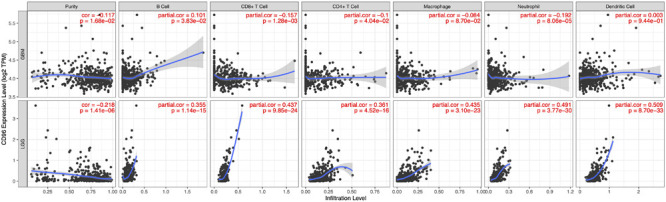
Correlation of CD96 expression with immune infiltration level in GBM and LGG.
CD96 Predicted Worse Survival in Glioma
Owing to the high relevance between CD96 and immune suppressor in glioma, the prognostic impact of CD96 was verified via the Kaplan–Meier method. Overall survival (OS) analysis in glioma patients demonstrated that high expression of CD96 predicted relatively poor survival in the CGGA and TCGA cohorts (Figures 7A,E). Higher CD96 expression is associated with worse OS in patients with WHO grades II, III, and IV glioma based on data from the CGGA dataset (Figures 7C,D,B, respectively). Similarly, as shown, strong associations were observed between higher expression of CD96 and shorter OS for all WHO grades patients in the TCGA dataset (Figures 7F–H). These findings suggested that CD96 is a negative prognostic indicator in glioma and GBM patients.
FIGURE 7.
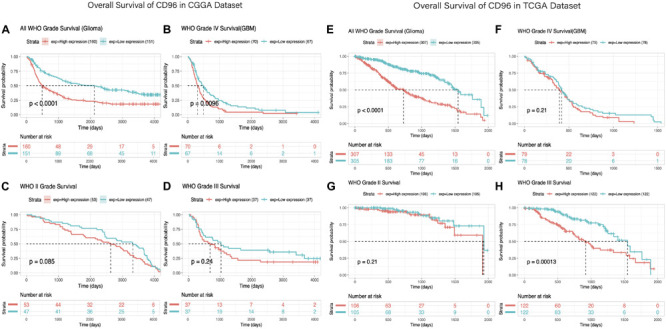
Survival analysis of glioma based on CD96 expression. (A–D) Overall survival analysis of CD96 in all-grade glioma and WHO IV, WHO II, and WHO III gliomas based on data from the CGGA cohort. (E–H) Overall survival analysis of CD96 in all-grade glioma and WHO IV, WHO II, and WHO III gliomas based on data from the TCGA cohort. High and low expressions were defined as the CD96 transcript level being more or less than the median level of all samples.
Eventually, we evaluated the independence of the clinicopathological significance of CD96 in glioma by univariate and multivariate Cox regression analyses. The results indicated that CD96, age, gender, WHO grade, IDH mutation, and h1p19q codeletion status were closely associated with OS and revealed that CD96 is an independent prognosticator for glioma patients (Supplementary Table S3).
Discussion
Recent studies on tumor immunotherapy continue to show strong results and bring hope to glioma patients. Among those immunotherapeutic strategies, immune checkpoint blockade offers remarkable benefits to the therapies of various tumor types by increasing antitumor immunity (Postow et al., 2018). However, research in the field of neural tumor immunotherapy currently mainly focuses on CTLA-4 and PD-1/PD-L1 blockade. Nonetheless, potential immune-related adverse events hinder the wide clinical application of immunotherapy (Postow et al., 2018). The identification of alternative checkpoint targets may facilitate the solution of this predicament and bring new therapeutic benefits for cancer treatment.
Increasingly, CD96 is emerging as a potent modulator of antitumor immune responses (Georgiev et al., 2018). CD96 has been shown to negatively regulate NK cell-mediated immune surveillance and to intervene in multidimensional adhesion, inhibition, and activation of participating cells (Fuchs et al., 2004; Chan et al., 2014; Georgiev et al., 2018; Roman Aguilera et al., 2018). The special effect of CD96 has also been reported in some tumors. Hepatocellular carcinoma (HCC) patients with high expression level of CD96 within tumor are strongly correlated with deteriorating disease situations and shorter disease-free survival and OS times (Sun et al., 2019). Targeting host CD96 appears to be an innovative strategy for clinical application in combination with current immunotherapies. Herein, we probed the biological functions of CD96 in glioma through large scale and in-depth analyses. CD96 expression was markedly enriched in higher-grade malignant pathological gliomas. Moreover, high expression of CD96 was observed in the malignant molecule phenotype, including IDH wildtype and mesenchymal subtype. Collectively, CD96 exhibited a malignant biological property in glioma. Meanwhile, through GO analysis of the relationship between CD96 and BPs, we noted that CD96 had a positive association with immune response and inflammatory activities.
A range of immunotherapies targeting checkpoint inhibitors and blocking monoclonal antibodies (mAbs) have been widely adopted, and several of them are ongoing in glioblastoma (Bouffet et al., 2016; Huang et al., 2017). Compared to monotherapy treatments, combination approaches were reported to be more effective and associated with substantially longer progression-free survival (Larkin et al., 2015; Postow et al., 2015). In our research, CD96 showed a high concordance with immune checkpoints PD-1, CTLA-4, TIGIT, TIM-3, NR2F6, and GITR, indicating the potential synergistic effects of these markers. We inferred that collaboration of CD96 with other checkpoint members, especially PD-1, may increase glioma immunotherapy effectiveness. Indeed, previous studies have demonstrated that concurrent blockade of CD96 and PD-1 increased antitumor immunity more than targeting PD-1 alone, potentially without inducing serious 908 immune-related toxicities (Blake et al., 2016; Harjunpää et al., 2018). This provided support to our research. We also discovered that higher CD96 expression predicted worse survival rates in glioma and GBM patients. This significant prognostic signature implied that CD96 blockade may significantly improve the prognosis of glioma patients, especially GBM patients.
Conclusion
To sum up, we initially explored the genetic and clinical characteristics of CD96 based on CGGA and TCGA datasets. Our results highlighted that CD96 may be a promising biomarker and therapeutic target for glioma that presents favorable application prospects.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://cancergenome.nih.gov/ and http://www.cgga.org.cn/.
Ethics Statement
All the procedures in this study were approved by the ethics committees of all hospitals, and written informed consent was obtained from all patients.
Author Contributions
SY and FC conceptualized and designed this study. HZ performed the data collection and analysis. QZ wrote the manuscript. YF, QL, and JS participated in constructing figures and revision. All authors gave final approval of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Tao Jiang and Dr. Zhao Zheng from the Beijing Neurosurgical Institute for assistance with data collection in the CGGA and helpful suggestions. This manuscript has been released as a pre-print at Research Square (Zhang et al.).
Abbreviations
- AUC
area under the curve
- CGGA
Chinese Glioma Genome Atlas
- GBM
Glioblastomas
- GO
gene ontology
- GSVA
gene set variation analysis
- IDH
Isocitrate dehydrogenase
- OS
Overall survival
- PD-L1
Programmed death-ligand 1
- TCGA
The Cancer Genome Atlas dataset
- TIM3
T cell immunoglobulin mucin-3
- WHO
World Health Organization.
Funding. This research was financially supported by the Natural Science Foundation of China (81660421) and the Young Talents Fund of Zunyi Medical University (18zy-005).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00592/full#supplementary-material
Relationship between CD96 and inflammatory activities in glioma.
Association between CD96 expression and T cell NK and NK cell immune infiltrates. Red and blue text represents positive and negative associations with statistical significance (p-value < 0.05), respectively, and gray text represents non-significant correlations.
GO biological process analysis results for top 50 genes most relevant to CD96 in the CGGA and TCGA databases.
The most relevant immune genes to CD96 in the CGGA and TCGA datasets. The red highlighted genes are involved in NK and T cell responses.
Univariate and multivariate analyses of clinical prognostic parameters in CGGA and TCGA datasets.
References
- Blake S. J., Stannard K., Liu J., Allen S., Yong M. C. R., Mittal D., et al. (2016). Suppression of metastases using a new lymphocyte checkpoint target for cancer immunotherapy. Cancer Discov. 6 446–459. 10.1158/2159-8290.CD-15-0944 [DOI] [PubMed] [Google Scholar]
- Bouffet E., Larouche V., Campbell B. B., Merico D., De Borja R., Aronson M., et al. (2016). Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 4 2206–2211. 10.1200/JCO.2016.66.6552 [DOI] [PubMed] [Google Scholar]
- Branzoli F., Di Stefano A. L., Capelle L., Ottolenghi C., Valabrègue R., Deelchand D. K., et al. (2018). Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro. Oncol. 20 907–916. 10.1093/neuonc/nox214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. J., Martinet L., Gilfillan S., Souza-Fonseca-Guimaraes F., Chow M. T., Town L., et al. (2014). The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 15 431–438. 10.1038/ni.2850 [DOI] [PubMed] [Google Scholar]
- Deuss F. A., Watson G. M., Fu Z., Rossjohn J., Berry R. (2019). Structural Basis for CD96 immune receptor recognition of nectin-like protein-5, CD155. Structure 27:219-228.e3. 10.1016/j.str.2018.10.023 [DOI] [PubMed] [Google Scholar]
- Dougall W. C., Kurtulus S., Smyth M. J., Anderson A. C. (2017). TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy. Immunol. Rev. 276 112–120. 10.1111/imr.12518 [DOI] [PubMed] [Google Scholar]
- Feng E., Liang T., Wang X., Du J., Tang K., Wang X., et al. (2019). Correlation of alteration of HLA-F expression and clinical characterization in 593 brain glioma samples. J. Neuroinflammation 16 1–8. 10.1186/s12974-019-1418-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friendly M. (2002). Corrgrams: exploratory displays for correlatigon matrices. Am. Stat. 56 316–324. 10.1198/000313002533 12611515 [DOI] [Google Scholar]
- Fuchs A., Cella M., Giurisato E., Shaw A. S., Colonna M. (2004). Cutting Edge: CD96 (Tactile) Promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J. Immunol. 172 3994–3998. 10.4049/jimmunol.172.7.3994 [DOI] [PubMed] [Google Scholar]
- Georgiev H., Ravens I., Papadogianni G., Bernhardt G. (2018). Coming of age: CD96 emerges as modulator of immune responses. Front. Immunol. 9:1072. 10.3389/fimmu.2018.01072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. (2013). GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjunpää H., Blake S. J., Ahern E., Allen S., Liu J., Yan J., et al. (2018). Deficiency of host CD96 and PD-1 or TIGIT enhances tumor immunity without significantly compromising immune homeostasis. Oncoimmunology 7 1–11. 10.1080/2162402X.2018.1445949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Liu F., Liu Z., Tang H., Wu H., Gong Q., et al. (2017). Immune checkpoint in glioblastoma: promising and challenging. Front. Pharmacol. 8:242. 10.3389/fphar.2017.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Mao Y., Ma W., Mao Q., You Y., Yang X., et al. (2016). CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 375 263–273. 10.1016/j.canlet.2016.01.024 [DOI] [PubMed] [Google Scholar]
- Kassambara A., Kosinski M., Biecek P., Fabian S. (2018). survminer: Drawing Survival Curves using “ggplot2” (R package). version 0.4.3. [Google Scholar]
- Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J. J., Cowey C. L., Lao C. D., et al. (2015). Combined nivolumab and ipilimumab or monotherapy in untreated Melanoma. N. Engl. J. Med. 373 23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang Z., Zhang C., Liu X., Cai J., Wang Z., et al. (2017). Molecular and clinical characterization of TIM-3 in glioma through 1,024 samples. Oncoimmunology 6:e1328339. 10.1080/2162402X.2017.1328339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J. S., et al. (2017). TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77 e108–e110. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Huang J., Xiong Y., Li S., Liu Z. (2019). Large-scale analysis reveals the specific clinical and immune features of CD155 in glioma. Aging 11 5463–5482. 10.18632/aging.102131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D. N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W. K., et al. (2016). The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131 803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- Louveau A., Smirnov I., Keyes T. J., Eccles J. D., Rouhani S. J., Peske J. D., et al. (2015). Structural and functional features of central nervous system lymphatic vessels. Nature 523 337–341. 10.1038/nature14432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D., Lepletier A., Madore J., Aguilera A. R., Stannard K., Blake S. J., et al. (2019). CD96 Is an Immune Checkpoint That Regulates CD8þ T-cell Antitumor Function. Cancer Immunol. Res. 7 559–571. 10.1158/2326-6066.CIR-18-0637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow M. A., Chesney J., Pavlick A. C., Robert C., Grossmann K., McDermott D., et al. (2015). Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 372 2006–2017. 10.1056/NEJMoa1414428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow M. A., Sidlow R., Hellmann M. D. (2018). Immune-related adverse events associated with immune checkpoint blockade. N. Engl. J. Med. 378 158–168. 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- Reardon D. A., Freeman G., Wu C., Chiocca E. A., Wucherpfennig K. W., Wen P. Y., et al. (2014). Immunotherapy advances for glioblastoma. Neuro. Oncol. 16 1441–1458. 10.1093/neuonc/nou212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A., Wolchok J. D. (2018). Cancer immunotherapy using checkpoint blockade. Science 359 1350–1355. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin A. X., Turck N., Hainard A., Lisacek F., Sanchez J., Müller M., et al. (2013). Package ‘ pROC.’ 2012-09-10 09:34:56. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman Aguilera A., Lutzky V. P., Mittal D., Li X. Y., Stannard K., Takeda K., et al. (2018). CD96 targeted antibodies need not block CD96-CD155 interactions to promote NK cell anti-metastatic activity. Oncoimmunology 7 1–10. 10.1080/2162402X.2018.1424677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R., Hegi M. E., Mason W. P., van den Bent M. J., Taphoorn M. J., Janzer R. C., et al. (2009). Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10 459–466. 10.1016/S1470-2045(09)70025-7 [DOI] [PubMed] [Google Scholar]
- Sun H., Huang Q., Huang M., Wen H., Lin R., Zheng M., et al. (2019). Human CD96 Correlates to Natural Killer Cell Exhaustion and Predicts the Prognosis of Human Hepatocellular Carcinoma. Hepatology 70 168–183. 10.1002/hep.30347 [DOI] [PubMed] [Google Scholar]
- Therneau T. M., Lumley T. (2015). Package ‘ survival.’ R Top. Doc. [Google Scholar]
- Topalian S. L., Hodi F. S., Brahmer J. R., Gettinger S. N., Smith D. C., McDermott D. F., et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366 2443–2454. 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meir E. G., Hadjipanayis C. G., Norden A. D., Shu H. K., Wen P. Y., Olson J. J. (2010). Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA. Cancer J. Clin. 60 166–193. 10.3322/caac.20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D., Bendel A., Stiller C., Indelicato D., Smith S., Murray M., et al. (2017). Central nervous system tumors. Pediatr. Oncol. 82 1271–1286. 10.1007/978-3-319-33679-4_14 [DOI] [Google Scholar]
- Wang Z., Zhang C., Liu X., Wang Z., Sun L., Li G., et al. (2016). Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology 5:e1196310. 10.1080/2162402X.2016.1196310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T. (2017). Package “corrplot” for R: Visualization of a Correlation Matrix. CRAN. [Google Scholar]
- Yu G., Wang L. G., Han Y., He Q. Y. (2012). ClusterProfiler: an R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 16 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relationship between CD96 and inflammatory activities in glioma.
Association between CD96 expression and T cell NK and NK cell immune infiltrates. Red and blue text represents positive and negative associations with statistical significance (p-value < 0.05), respectively, and gray text represents non-significant correlations.
GO biological process analysis results for top 50 genes most relevant to CD96 in the CGGA and TCGA databases.
The most relevant immune genes to CD96 in the CGGA and TCGA datasets. The red highlighted genes are involved in NK and T cell responses.
Univariate and multivariate analyses of clinical prognostic parameters in CGGA and TCGA datasets.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://cancergenome.nih.gov/ and http://www.cgga.org.cn/.


