Abstract
The last two decades saw the establishment of three-dimensional (3D) cell cultures as an acknowledged tool to investigate cell behaviour in a tissue-like environment. Cells growing in spheroids differentiate and develop different characteristics in comparison to their two-dimensionally grown counterparts and are hence seen to exhibit a more in vivo-like phenotype. However, generating, treating and analysing spheroids in high quantities remains labour intensive and therefore limits its applicability in drugs and compound research. Here we present a fully automated pipetting robot that is able to (a) seed hanging drops from single cell suspensions, (b) treat the spheroids formed in these hanging drops with drugs and (c) analyse the viability of the spheroids by an image-based deep learning based convolutional neuronal network (CNN). The model is trained to classify between ‘unaffected’, ‘mildly affected’ and ‘affected’ spheroids after drug exposure. All corresponding spheroids are initially analysed by viability flow cytometry analysis to build a labelled training set for the CNN to subsequently reduce the number of misclassifications. Hence, this approach allows to efficiently examine the efficacy of drug combinatorics or new compounds in 3D cell culture. Additionally, it may provide a valuable instrument to screen for new and individualized systemic therapeutic strategies in second and third line treatment of solid malignancies using patient derived primary cells.
Subject terms: Biological techniques, Cytological techniques, Experimental organisms, High-throughput screening, Software
Introduction
Systemic therapies, i.e. chemotherapies and targeted-therapies, constitute one of the major pillars of modern cancer therapies and are of particular importance for the treatment of advanced and metastasized diseases. Today, the vast majority of all cancer patients dies due to metastases and their associated complications1. Despite the availability of evidence-based sequential or parallel therapeutic regimens for the most of all known cancer entities, physicians and patients alike regularly face the challenge of evolving resistance mechanisms that demand a robust fallback level. Once patients experience a disease progression under the respective first line treatment, they need to be administered a promising second or third line regimen. As guidelines tend to present treatments beyond the first or second line as equal options within the respective group, individual or institutional experiences drive the decision, if the enrolment in a suitable clinical trial is not an option. The search for a solution of this issue has contributed to pave the way for the field of personalized medicine over the past years, which focuses on the individual characteristics of a patient’s disease and custom tailors the best treatment option, respectively. The paramount interest in this field has just recently been underscored by the NIH’s Cancer Moonshot Blue Ribbon Panel that particularly emphasized the need for novel technologies to facilitate comprehensive cancer research with so-called human-derived next-generation cancer models2. The work presented in this article aims to contribute precisely to this proposed goal.
Major progress in the field of personalized medicine in cancer therapies was achieved through a better and more profound understanding of cancer itself: solid tumours are no longer seen as a collection of homogeneous malignant cells, but are rather considered a micro-evolutionary system that undergoes a constant transformation in response to endogenous and exogenous stimuli3,4. Several so-called lead mutations have been known for many years and are, in some cases, already important predictive indicators for a specific treatment. Nonetheless, these mutations only initiate the genomic dysregulation of malignant cells and are typically followed by further mutations. These so-called follow mutations contribute further to the genetic instability and diversity of a tumorous mass and eventually lead to the evolvement of numerous, genetically heterogeneous sub-entities5. Under a standard therapy, some of these sub-entities either develop mechanisms of resistance towards the applied agents or persevere better when nutrients and oxygen are at scarce—and hence gain dominance. Numerous approaches have been undertaken to approximate tumour response by modelling6–8. However, predictions about which sub-entities can persevere and the mechanisms they apply are, up to date, hard to make and can therefore not yet be taken into consideration when choosing the right regimen for an advanced cancer disease9,10. Just recently, Tuveson and Clevers presented a comprehensive review on cancer modelling by the means of organoid technologies and emphasized a number of key characteristics of three dimensionally grown cell cultures, namely the more accurate representation of interpatient variations, the ability to employ complex patient-derived cell sources and their capacity to report drug responses more reliably than conventional flat dish culture systems11.
In line with their promotion of sophisticated organoid models and a versatile usability thereof, several promising attempts on the generation of high-throughput screening assays for organoid cultures have been published. Yet, only few managed to integrate automation, size and spatial control, high numbers of replicates and a suitable interface for follow-up investigations so far. For example, Amann et al. demonstrated the generation of complex mono- or co-cultures. However, due to the complex generation mechanism discussed below, only low numbers of replicates were thoroughly examined by flow cytometry or microscopy12. Similar limitations apply to the work of Anastasov et al., who generated organoids similarly and focused on the detection of radiation resistant organoids and the effects of a simultaneous chemotherapy13. Other groups recognize scaffold-free 3D cultures as most suitable for mimicking in vivo phenotypes, but fall short of evaluating the tumour evolution14. Finally, some groups demonstrate a true high-throughput approach, but lack size and shape control to generate homogeneous organoids for follow-up investigations15. Although the results of this promising research as well as its limitations are clear indicators for the high complexity in this field, none of the approaches above incorporated a genuine computer-based readout for their experiments. From our perspective, this allows, with a reasonable effort, to reduce the complexity of a molecular or histochemical readout with comparable results.
Previous research in our group proposes an up-scale printing device that, in a next step, allows the simulation of the aforementioned micro-evolutionary process by generating a high number of homogenous cancer cell organoids from a tissue sample: organoids are generated by a printing platform employing a drop-on-demand (DOD) print head and are cultivated via the hanging-droplet method16. As using an automated drop-on-demand printing procedure increased the efficiency of spheroid production significantly, we now aim to apply evolutionary pressure, i.e. a specific cytostatic agent, a decrease in nutrients or pO2, to the spheroids and detect specific reactions of the individual spheroids. However, biochemical analysis remains labour intensive, as standard biochemical analysis methods demand a large quantity of source material. For instance, several hundred spheroids need to be pooled in order to yield a sufficient protein concentration to perform Western blot or qPCR analysis. Working on this problem, it became evident that spheroids undergo characteristic morphological changes when they are stressed, e.g. by cytotoxic agents. Images of individual spheroids can hence be analysed and classified by deep learning models17. Convolutional Neuronal Networks (CNN), if trained accordingly, allow to classify images in an efficient manner and are therefore predestined to assist—and eventually replace—costly and complex biochemical analysis. First introduced by LeCun and Bengio18, the application of CNN has enjoyed a rise of reputation in the past few years. In comparison to other neural networks, a CNN takes advantage of the spatial localisation inherent in images and hence identifies and extracts features.
The precise detection of morphologically aberrant spheroids and their subsequent analysis regarding cell viability and increase in cell death after exposure to cytotoxic agents by deep learning are at the focus of this paper. The project aims to provide a methodological feasibility study and therefore employs established and well characterized cell lines.
Material and methods
Cells and cell culture
All experiments were performed using immortalized adherent cell lines. HCT116, HEp2, HEK293T or mIMCD3 cells were maintained in standard cell culture vessels in DMEM medium supplemented with 10% FCS and 1% Pen/Strep at 37 °C in a 5% CO2 atmosphere. To maintain exponential growth, cells were split once they reached confluence, typically after 3 days. In order to grow the cells in spheroids, cells were washed with PBS, trypsinised with Trypsin/EDTA for 5–10 min and after all cells lost contact to the culture vessel, the reaction was stopped with the addition of an excess of DMEM. Cells were counted using a Neubauer counting chamber and diluted to a density of 1 × 106 cells per ml.
Automated spheroid production
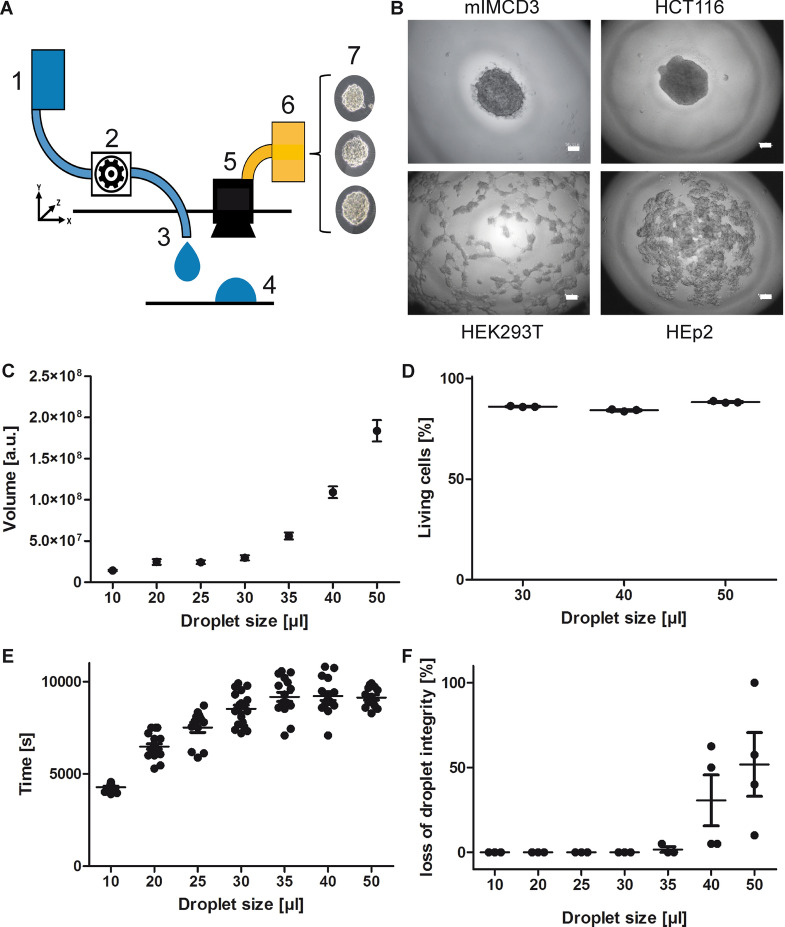
The DOD printing platform was developed on the basis of a computerized numerical control (CNC) milling machine (Sainsmart, Lexana, Kansas, USA), supplemented with an additive printer control chip (Anycubic, Frankfurt, Germany). The nozzle (200 µm size) was mounted to the x–y–z arm and connected to an infusion pump operated by a stepper motor, adjustable to the desired volumes after gravimetrical calibration. The nozzle aims at an inverted cell culture plate lid of 10 or 15 cm diameter that is placed 7 mm below the nozzle. A CCD camera (KKmoon, Shenzhen, China) is mounted next to the nozzle on the x–y–z arm. At 20 mm operation distance the camera captures images of individual spheroids and saves it to a database (Fig. 1a).
Figure 1.
Spheroids form spontaneously in hanging drops. (A) Schematic representation of the spheroid seeding tool used throughout this work. Spheroids in hanging drops or cytotoxic drugs are seeded onto an inverted cell culture plate at various volumes using a peristaltic pump and a high precision x–y–z robot arm. A CCD camera monitors the spheroid quality and saves the images to a database. (1) Reservoir for cell suspension, (2) Peristaltic pump, (3) Nozzle for DOD droplet generation, (4) Droplet of homogenous shape and volume, (5) CCD camera, (6) Database, (7) Detection of shape and conformational alterations. (B) Representative images of spheroids of mIMCD3, HCT116, HEK293T or HEp2 cell lines forming spontaneously in 30 µl medium droplets containing 1 × 106 cells/ml of trypsinised cells after 24 h. Size marker = 50 µm. (C) The volume of the spheroids consisting of mIMCD3 cells increases logarithmically depending on the droplet size/cell number. The volume was calculated measuring pixels of the captured images and is therefore set to arbitrary units [a.u.]. Cell concentration = 1 × 106 cells/ml. Shown are means (horizontal line) ± s.d., n > 20. (D) Cell viability of mIMCD3 cells inside the spheroid after 48 h of incubation in spheroids of 30, 40 and 50 µl of volume. Cell viability was assessed by flow cytometry using Annexin-V counterstain. Cell concentration = 1 × 106 cells/ml. Shown are individual survival rates (dots), means (horizontal line) ± s.d., n = 3. (E) Evaporation time of hanging drops (DMEM containing 10% FCS and 1% Pen/Strep) in a non-humid environment. Droplet sizes larger than 30 µl plateau at 8,500 s = 140 min. Shown are individual times (dots), means (horizontal line) ± s.d., n > 10. (F) The formation of hanging drops depends on an inversion of the carrier plate that holds the droplets. The larger the droplets are, the higher the probability that the droplets spontaneously trickle away during this process. Shown are percentages of spontaneously lost droplets on a plate bearing 80 droplets in three different experiments (dots), means (horizontal line) ± s.d., n = 3.
Setup and process of spheroid production
Trypsinised floating cells at a concentration of 1 × 106 cells per ml were loaded into the printing device. Droplets in the size of 30 µl were printed onto the inverted lid of a 15 cm cell culture plate in an array of 10 × 10 droplets. After seeding, the plate was carefully removed from the seeding device and quickly inverted. To avoid evaporation, the bottom of the 10 cm cell culture plate was filled with 10 ml PBS and the droplets bearing lid was put on the bottom plate. The spheroids were allowed to form in the droplets for 24 h in an incubator at 37 °C and 5% CO2. After 24 h the lid containing the spheroids was again loaded onto the seeding machine and 5 µl liquid containing 7 × concentrated drugs were applied into the droplets to yield a final 1 × concentration of drugs in the droplet containing the spheroid (35 µl final volume). Drugs used were Etoposide (ETO, final concentration: 20 and 100 mM) and Staurosporine (STS, final concentration: 2 and 20 µM). The lid bearing the droplets was again inverted and incubated for 24 h. To analyse the effect of the drugs on the spheroids, droplets were either scanned by the AI-connected camera or analysed by flow cytometry.
Flow cytometry analysis
After imaging the spheroids, the lid containing the droplets was inverted, rinsed with 20 ml of PBS and all the spheroids were collected in a 50 ml tube. After centrifugation (5 min, 400 g), spheroids were trypsinised to obtain a single cell solution. After 5 min, DMEM + 10%FCS was added and cells were sedimented again. The pellet was resuspended in Annexin-V binding Buffer (10 mM Hepes, 140 mM NaCl, 2.5 mM CaCl2 in ddH2O) containing Annexin-V-APC (1:500). Cells were measured using a LSRII flow cytometer (Beckton Dickinson).
Statistical analysis
Statistical analysis was performed where appropriate. Statistical significance was assessed by a two-way ANOVA and a Bonferroni post-test.
Deep learning
The implementation is based on the python neural networks library Keras19. This library offers an ImageDataGenerator class that allows to generate batches of images with data augmentation. The following parameters for real-time data augmentation are used: rotation_range, rescale, shear_range, zoom_range, horizontal_flip. Designing the model, we focused on using a small convolutional network with few layers and a small amount of filters (neurons) per layer. The number of filters corresponds to the number of feature maps. A high dropout factor will regularize the model by randomly selecting neurons and ignore them during the training. This results in various independent representations and the model will incorporate these into its learning process to avoid overfitting20. Specifically, we trained a convolutional network with 4 convolutional layers. Each layer has a rectified linear unit (ReLU) activation function, which projects negative weights to zero, and is followed by max-pooling layers with a pool size of 2 × 2. Pooling is needed to eliminate redundant information and speed up training in addition to ReLU and dropout. After the fourth pooling layer, activations were flattened and two subsequent layers with 128 and 3 features were fully connected. The model ends with a softmax activation to produce a probability distribution over given classes. The categorical model summary can be seen in Table 1 and has overall 6,913,091 trainable parameters.
Table 1.
Categorical model summary.
| Layer (type) | Output shape | Parameter # |
|---|---|---|
| conv2d_1 (Conv2D) | (None, 398, 318, 32) | 320 |
| activation_1 (Activation) | (None, 398, 318, 32) | 0 |
| max_pooling2d_1 (MaxPooling2D) | (None, 199, 159, 32) | 0 |
| conv2d_2 (Conv2D) | (None, 197, 157, 64) | 18,496 |
| activation_2 (Activation) | (None, 197, 157, 64) | 0 |
| max_pooling2d_2 (MaxPooling2D) | (None, 98, 78, 64) | 0 |
| conv2d_3 (Conv2D) | (None, 96, 76, 64) | 36,928 |
| activation_3 (Activation) | (None, 96, 76, 64) | 0 |
| max_pooling2d_3 (MaxPooling2D) | (None, 48, 38, 64) | 0 |
| conv2d_4 (Conv2D) | (None, 46, 36, 128) | 73,856 |
| activation_4 (Activation) | (None, 46, 36, 128) | 0 |
| max_pooling2d_4 (MaxPooling2D) | (None, 23, 18, 128) | 0 |
| flatten_1 (Flatten) | (None, 52,992) | 0 |
| dense_1 (Dense) | (None, 128) | 6,783,104 |
| activation_4 (Activation) | (None, 128) | 0 |
| dropout_1 (Dropout) | (None, 128) | 0 |
| dense_2 (Dense) | (None, 3) | 387 |
| activation_5 (Activation) | (None, 3) | 0 |
The complete source code is uploaded to GitHub and can be accessed upon reasonable request.
Results
We aimed to develop a tool that allows the generation of spheroids in high quantities and in a constant quality with regard to spatial control and size distribution. Furthermore, it needed to be able to treat the generated spheroids with preselected drugs and should be able to capture each individual spheroid in a hanging drop with a CCD camera. The resulting tool consists of an x–y–z platform that holds a peristaltic pump, which applies defined amounts of liquid containing trypsinised epithelial cells in a defined pattern on the lid of a cell culture plate (Fig. 1a). The droplet sizes generated by the infusion pump remained constant with only very small standard deviations (Supplementary Fig. 1). After applying the cells in a specific grid-pattern, the cell culture plate is carefully inverted and incubated for 24 h in a standard cell culture incubator. Next, the plate is inverted again and placed into the printing device. Once the pre-formed pattern is automatically identified, therapeutics in different concentrations are infused into the existing droplets via DOD printing, as outlined above. After a 24-h incubation period, the effect of the drugs is assessed by a CCD camera that identifies the individual spheroids and compares the condition of each separate spheroid to a database of previously diagnosed, i.e. labelled, spheroids.
Tumour spheroids form spontaneously in hanging drops
As shown in various previous studies, cells from diverse sources will form spheroids spontaneously when grown in an environment where cells can only attach to each other, as it happens in hanging drops21,22. However, depending on the cell line or differentiation status, cells can aggregate and form a ball shaped single spheroid (Fig. 1b, mIMCD3 and HCT116), or form several interconnected small spheroids on the bottom of the droplet (Fig. 1b, HKT293T and HEp2).
The volume of the spheroids grows logarithmically in relation to a linear increase in the cell number contributing to the spheroid (Fig. 1c, mIMCD3 cells). The formation of a spheroid rapidly establishes a supply gradient that exposes the inner cell mass to a gradually increasing scarcity in nutrients and oxygen. However, the increase in cell number included into the spheroid does not lead to an increase in spontaneous apoptosis after 48 h in culture, averaging around 80% living cells in spheroids larger than 60.000 cells (Fig. 1d). Nonetheless, too small droplets bear the risk of rapid evaporation of the medium before the spheroids can be placed in the incubator (Fig. 1e) and too large droplets, containing more than 35 µl (or 35.000 cells), cannot be inverted securely without the danger of the droplets of flowing uncontrollably on the carrier material and losing its pre-formed grid-pattern (Fig. 1f).
Based on this finding we chose a concentration of 1 × 106 cells/ml as an optimal medium-to-cell ratio when growing spheroids in further experiments. Droplet sizes of 30 µl proved to be the ideal volume to form spheroids since handling and efficacy demonstrated to be optimal in this setting (Fig. 1c, e, f and Supplementary Fig. 1).
Spheroids react to drug exposure
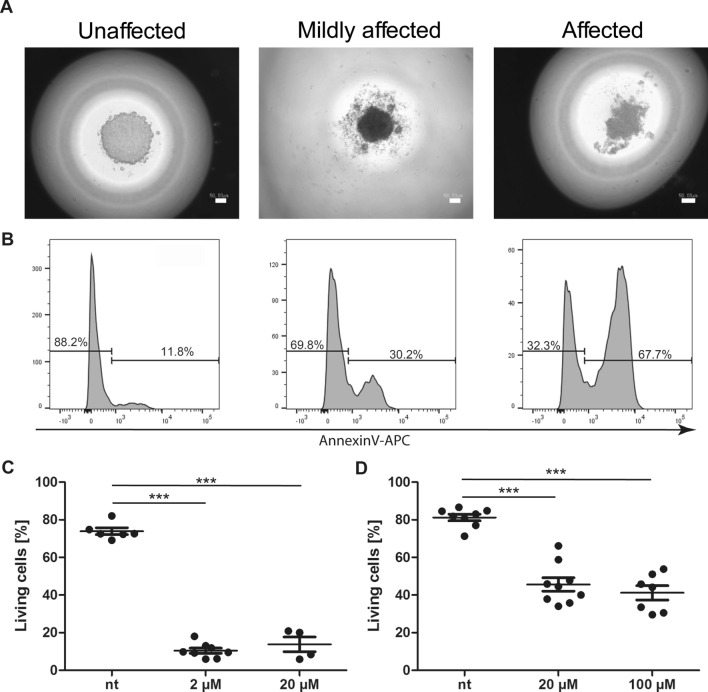
Cells in hanging drops start to form spheroids within minutes after seeding. After 24 h under suitable incubation conditions, the spheroids of mIMCD3 or HCT116 cells condense to a stable cell mass (Fig. 1b). These matured spheroids can then be treated with cytotoxic agents. Directly applying the agent into the liquid of the hanging drop enables a penetration of the agent into the spheroid and exposes the cells to its biological effect. Depending on the sensitivity of the cells towards the cytotoxic agent, cells exhibit clear signs of cytotoxicity in a time and dose dependent manner: apoptotic cells within the spheroid disturb the cohesion of the cells and cause a shift from high contrast edges towards an irregular appearance. This finding was particularly prominent in mIMCD3 spheroids after 24 h of incubation (Fig. 2a), but was also seen in other spheroid-forming epithelial cell lines. The association of the altered morphology and the viability of the spheroids was evaluated via flow cytometry (Fig. 2b). Applying the pan kinase inhibitor Staurosporin to the spheroids induces massive cell death within the spheroid after 24 h of incubation (Fig. 2c). The topoisomerase inhibitor Etoposide, however, only induces mild cell death in spheroids, even despite high concentrations (Fig. 2d), probably because cells in spheroids do not actively enter the cell cycle anymore and therefore are not specifically sensitive to DNA damage23. The same cells grown in standard 2D cell culture conditions still show high sensitivity towards Etoposide exposure (Supplementary Fig. 2).
Figure 2.
Spheroid integrity is affected by the addition of cytotoxic drugs. (A) Representative images of spheroids consisting of mIMCD3 cells that are either labelled ‘unaffected’ (left image, > 80% living cells), ‘mildly affected’ (middle image, 40–60% viability) or ‘affected’ (right image, < 40% viability. Size marker = 50 µm. (B) Corresponding flow cytometry analysis to images presented in (A). Cell viability was assessed by flow cytometry using Annexin-V counterstain. (C) Viability of spheroids consisting of mIMCD3 cells exposed for 24 h to the pan kinase inhibitor Staurosporine in the indicated concentrations. N = 4–9, p < 0.001 for nt against both treated conditions. (D) Viability of spheroids consisting of mIMCD3 cells exposed for 24 h to the topoisomerase inhibitor Etoposide in the indicated concentrations. N = 7–9, p < 0.001 for nt against both treated conditions.
Neural networks are able to discriminate between drug sensitive and non-sensitive spheroids
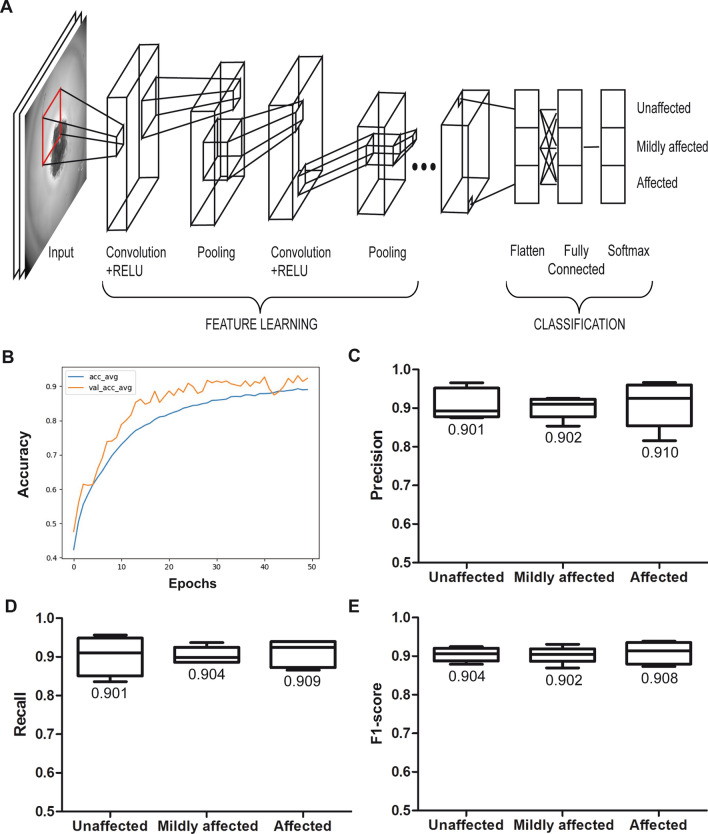
The analysis of spheroid sensitivity towards cytotoxic agents, as presented in Fig. 2, is very cost and work intensive and a pool of > 100 equally treated spheroids had to be measured by flow cytometry to yield reasonable cell numbers. To circumvent this detriment, we trained a deep learning model to estimate the sensitivity of spheroids toward a given drug. All spheroids that were analysed in Fig. 2 were individually micrographed before disassembling the spheroids for flow cytometry analysis. After determining the viability of the spheroids by flow cytometry, the corresponding images were grouped into three categories: ‘unaffected’ (> 80% viability), ‘mildly affected’ (40–60% viability) and ‘affected’ spheroids (< 40% viability). This set of categorised images was then used to train a deep learning model based on an AlexNet CNN architecture (Fig. 3a). For our model different CNN architectures (DenseNet, VGG, ResNet24–26) were tested, but did not delivered satisfactory results. AlexNet performed best, marked a starting point for our architecture and resulted in the described model27. 400 images per category were generated for the training and test phase, leading to a total of 1,200 images. To evaluate the model five-fold cross validation was used. We split our data into 80% training and 20% test set first. Regarding to fivefold cross validation, this training set was then used to create 5 splits of training and validation sets. Averaged training accuracy and validation accuracy over all folds are presented in Fig. 3b. After 50 epochs, our model reached in average a training accuracy of 0.89 and a validation accuracy of 0.92 (Fig. 3b). The best performing model was then used on the separate test set and predicted all three categories with a precision of about 0.9 (Fig. 3c), where 1.0 could be considered as the perfect result with 100% correct classifications. Relevant designations were identified by the recall measure and yielded values for all three categories higher than 0.9 (Fig. 3d), which further proves the efficacy of the model. A harmonic mean between precision and recall results into the F1-score (Fig. 3e) and further supports the findings above: for all three categories the mean of the F1-score is around 0.9. However, the standard deviation seems to increase for spheroids classified as ‘affected’. The cross validation demonstrates that the CNN is able to discriminate visual images between untreated spheroids and spheroids that are exposed to chemical compounds that induce cell death.
Figure 3.
Neural Networks are able to discriminate between drug sensitive and non-sensitive spheroids. (A) Graphical representation of the convolutional neural network (CNN). Images are convoluted with kernel matrices in order to reduce complexity but to keep the image information. Fully connected layers are then used to annotate an input image to a given classification. (B) Averaged training accuracy (‘acc_avg’, blue line) and validation accuracy (‘val_acc_avg’, orange line) of the CNN in a five-fold cross validation for 50 epochs. (C) The level of precision of the image classification by the CNN as calculated by a five-fold cross validation for the three individual categories “unaffected”, “mildly affected” and “affected. Numerical values are the means. (D) The level of recall of the image classification by the CNN as calculated by a five-fold cross validation for the three individual categories “unaffected”, “mildly affected” and “affected. Numerical values are the means. (E) The F1-score as a function from precision (C) and recall (D) for the three individual categories of spheroid viability: F1 = 2 × (precision × recall)/(precision + recall). Numerical values are the means.
Discussion
The last two decades brought the insight, that many cells show entirely different physiological behaviours when they grow in a 3D culture environment28,29. This effect may constitute one of the reasons, why so many promising new drug candidates fail in the clinical application30. To tackle this issue, testing new compounds on three dimensionally grown spheroids or organoids is more and more turning into the new standard of drug research30,31. However, efficient production and handling is a major issue of spheroid research and numbers for statistical analysis remain notoriously low32. Several commercial applications offer tailored kits that aim to address this issue, but fail to fully overcome the current limitations, as discussed in the introductory paragraph. Particularly, the reliance on multi-component commercial kits that require a repeated transfer of cells and spheroids12, the limited variation of the experimental setup and a low spatial control of spontaneously aggregated spheroids are of major concern. Furthermore, we consider the frequent use of Matrigel as a structural supplement as noteworthy15, since it is derived from the murine Engelbreth-Holm-Swarm Sarkoma. Although it provides a widely accepted and particularly in vivo-like model for extracellular matrix (ECM), it has ever since lacked a precise classification of its components and characteristics33,34. It therefore accounts for a high level of heterogeneity in any model for 3D cell culture. We are well aware that heterogeneity is a crucial characteristic of any tumour ECM—which is not yet sufficiently reflected by our model -, but consider this as a particular challenge for deep learning based classifications that needs to be addressed by further research. Here, we focus on a new approach on generating, treating and analysing spheroids with the perspective of providing supplementary information for patient specific therapy decisions. The design of an automated x–y–z pipetting device allows (a) efficient mass production of hanging drops on the lids of 15 cm cell culture dishes, (b) efficient treatment of hanging drops with drugs and (c) rapid analysis of spheroid viability by an image-based deep learning algorithm. While the automated seeding of spheroids did not yield significant time savings compared to manual spheroid production, the precise application of drugs into the existing hanging drops was much faster and more accurate than the manual approach. Generating vast quantities of spheroids and treating them with drugs for further biochemical analysis becomes hence much more time and resource efficient.
Feeding the CCD-generated close caption images to a deep learning model enabled a classification of the spheroids. Treatment of spheroids with cytotoxic agents activates cell stress responses and ultimately the cells in the spheroid enter apoptosis. Up to date, the gold standard methods to measure cell death are usually either protein based or single cell based, but these methods demand a high number of cells that cannot be provided by a single spheroid. We therefore used the flow cytometry based Annexin V staining on phosphatidylserine as a criterion standard35 on a pool of 80 identically treated spheroids. Before the biochemical analysis was performed, all 80 spheroids were imaged and later tagged according to the survival rate measured by flow cytometry. These photos and the respective tags were then employed to train the deep learning model based on a CNN, as it has been presented for other diagnostic issues with comparable results36,37. We are aware that our set of labelled images is relatively small in comparison to other datasets used to train neural networks. We hence aim to further underline the capacity of our model with independent training and validation sets. Yet, for the here presented proof of concept, we consider a cross validation a suitable and reasonable approach to demonstrate the feasibility of our setup. Furthermore, the limited number of images and the requirement of our setup to work robustly on these data sets represents the general setting we expect for the envisaged clinical application of the presented approach. It is, nonetheless, noteworthy that small datasets consequently lead to the problem of overfitting. An overfitted model is not able to generalize, i.e. yielding a high accuracy at classification with the data used in the training period, but with a significantly lower accuracy when new test data is used. To avoid overfitting, we employed measures that can help to improve a model, like data augmentation or the entropic capacity of a model. Entropic capacity represents the amount of information a model will store. The less features a model can store, the more it will focus on the most significant features. To address these issues, we designed a model with few layers and filters. Furthermore, images were manipulated via data augmentation. However, as outlined above, we aim to increase the number of labelled images used for training to improve the overall generalization capabilities of our model. Additionally, the application of more complex neural networks designs, like the competition-winning architecture ResNet24 or VGG25, would be facilitated through more available training data. Besides increasing the learning data set, we also aim to use a pretrained model, which already has learned features that are relevant for our specific classification problem of spheroids or organoids. Another promising approach would be the use of a generative adversarial network (GAN). This system consists of two networks that challenge each other. The generative network (generator) generates image candidates and tries to trick the discriminative network (discriminator), which aims to differ between real and generated images38. Hence, our model could serve as an initial discriminator in a GAN and is at the focus of a future project.
Although methodologically different, we consider our project as a complement to the previous work of other groups, which have achieved significant progress in the field of high throughput 3D culture screening. Particularly, we aim to discuss the compatibility of the different approaches to define further fields of study. Celli et al. presented an interesting work flow for the evaluation of 3D tumour models, but relied on pre-defined parameters for their quantitative readout39. From our experience, the incorporation of calcein and ethidium bromide as well as trypan blue is difficult to establish on 3D cultures, since all agents tend to distribute unevenly. This phenomenon has already been described before40 and is even more relevant when Matrigel-embedded heterogeneous 3D cultures are employed, as this group reports. Although the latter aspect mimics an in vivo situation better than homogeneously distributed 3D cultures, as highlighted above, it complicates the characterization of the organoids on the basis of a deep learning classification enormously. Furthermore, our setup is by now not able to detect fluorescent signals and can hence not apply a respective algorithm to detect the distribution of fluorescent dyes39. For future works, we will evaluate the validity of fluorescent markers in 3D cultures, but tend to incorporate metabolic sensors for a more specific analysis. Another promising approach has been presented by Zanoni et al., who put a particular focus on the spherical configuration and volume of the 3D cultures. However, our findings do not support their proposed theory that more irregularly shaped spheroids yield a higher viability than spherical ones41. Yet, we consider larger 3D cultures with diameters of 800–900 µm a suitable option to increase the in vivo similarity of our approach, but failed to generate spheroids of this size by the hanging droplet method, as outlined above. Again, our experience is that trypan blue exclusion is not a reliable parameter for 3D cultures and, although thoroughly investigated by this group, bioluminescence based assays neither seem to penetrate complex large 3D culture systems sufficiently. Other groups propose intracellular stains, i.e. calcein AM and ethidium bromide or familiar substances, as a suitable qualitative marker, but do not extend their use to quantitative comparisons42,43. In general, we question that simple morphological parameters, i.e. diameter, perimeter, volume, surface area and sphericity are suited as individual markers for 3D culture screening, as proposed by various groups44–46. However, an integrated deep learning approach, as presented in this article, allows the incorporation of these and many more parameters into the classification process. A very sophisticated approach towards the screening of 3D cultures was presented by Bulin et al., which included a stromal component into the setup. In order to mimic in vivo-like circumstances, we consider this an essential step and aim to incorporate stromal cells into our screening approach in future works, as our homogeneous 3D cultures without a suitable ECM do not yet mimic an in vivo environment to a sufficient degree. However, although complex, we consider the presented multi-step workflow as error-prone, since (a) conventional fluorescent stains are once again used as a quantitative readout and (b) the analysis is essentially based on two pre-defined parameters47. Nonetheless, we understand the presented platform as a robust tool for the screening of complex and structurally heterogeneous 3D cultures and see an opportunity to increase the versatility of our deep learning classification to more complex organoids.
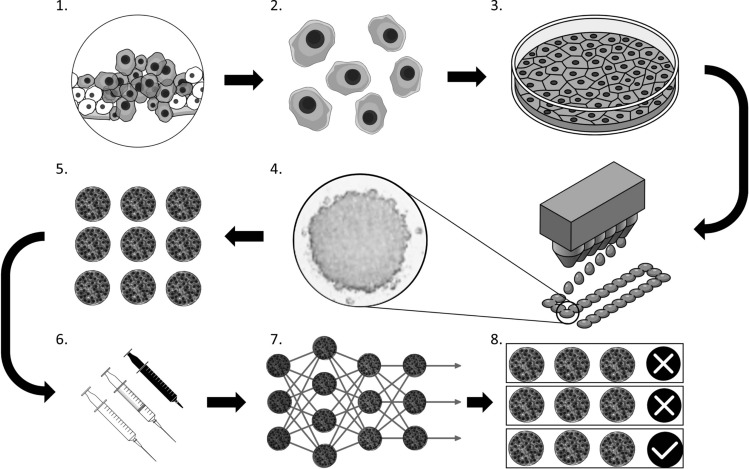
The promise of personalized medicine rests on the tailored treatment of a patient with a therapeutic regime that demands efficient determination of drug-disease-interaction for each individual patient. Our approach promises an easy to operate all-in-one tool that combines the strength of two innovative concepts—in vivo-like 3D cell culture and a deep learning environment, to enable personalized medicine in clinical applications (Fig. 4). By establishing a reliable automated screening approach for spheroids and their respective responses to the administered therapeutic agents or environmental parameters, we aim to detect mutation tendencies within the primary tissue samples that, as a long term goal, could support the decision making process for specific and personalized treatment regimens.
Figure 4.
Proposed workflow to optimize drug screening in solid tumours. (1) A malignant lesion grows in vivo. (2) Primary cells are secured by a biopsy and (3) expanded to yield a sufficient number of cells in vitro. (4) Homogeneous spheroids are generated via the hanging drop method and (5) are incubated until spheroids are fully matured. (6) Combinations of different drugs and concentrations are applied and the response of each spheroid is tracked via imaging. (7) Images are analysed by a pre-trained CNN and (8) allow a classification regarding the sensitivity towards a specific drug or concentration.
Supplementary information
Acknowledgements
LP was funded by the DFG SFB 1140. We thank the Bayer Foundation for enabling this work by a StartUp Grant to LP. LB was a participant of the MOTIVATE programme at the University of Freiburg. The article processing charge was funded by the Baden-Wuerttemberg Ministry of Science, Research and Art and the University of Freiburg in the funding programme Open Access Publishing.
Abbreviations
- CNN
Convolutional neural network
- DOD
Drop on demand
- ECM
Extracellular matrix
- GAN
Generative adversarial network
- 2D
Standard two-dimensional cell culture
- 3D
Three-dimensionally grown spheroids in cell culture
Author contributions
LP, GF and LB conceived and designed the project; LB and LP edited the figures; LB, AP, GF and LP wrote the manuscript; LB and LP performed the experiments, AP conceived the deep learning model and trained it upon images made by LP.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Leo Benning and Andreas Peintner.
Supplementary information
is available for this paper at 10.1038/s41598-020-67960-0.
References
- 1.Anderson RL, et al. A framework for the development of effective anti-metastatic agents. Nat. Rev. Clin. Oncol. 2019;16:185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Cancer Institute. Cancer Moonshot, https://www.cancer.gov/research/key-initiatives/moonshot-cancer-initiative/
- 3.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 4.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 5.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability: an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 6.Garland J. Unravelling the complexity of signalling networks in cancer: a review of the increasing role for computational modelling. Crit. Rev. Oncol. Hematol. 2017;117:73–113. doi: 10.1016/j.critrevonc.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Rudrapatna VA, Cagan RL, Das TK. Drosophila cancer models. Dev. Dyn. 2012;241:107–118. doi: 10.1002/dvdy.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balani S, Nguyen LV, Eaves CJ. Modeling the process of human tumorigenesis. Nat. Commun. 2017;8:15422. doi: 10.1038/ncomms15422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh R, et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer Res. 2011;71:1871–1882. doi: 10.1158/0008-5472.CAN-10-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stintzing S, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol. 2016;17:1426–1434. doi: 10.1016/S1470-2045(16)30269-8. [DOI] [PubMed] [Google Scholar]
- 11.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 12.Amann A, et al. Development of an innovative 3D cell culture system to study tumour–stroma interactions in non-small cell lung cancer cells. PLoS ONE. 2014;9:e92511. doi: 10.1371/journal.pone.0092511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastasov N, et al. A 3D-microtissue-based phenotypic screening of radiation resistant tumor cells with synchronized chemotherapeutic treatment. BMC Cancer. 2015;15:466. doi: 10.1186/s12885-015-1481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkenberg N, et al. Three-dimensional microtissues essentially contribute to preclinical validations of therapeutic targets in breast cancer. Cancer Med. 2016;5:703–710. doi: 10.1002/cam4.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehnke K, et al. Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. J. Biomol. Screen. 2016;21:931–941. doi: 10.1177/1087057116650965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutzweiler L, et al. Large scale production and controlled deposition of single HUVEC spheroids for bioprinting applications. Biofabrication. 2017;9:025027. doi: 10.1088/1758-5090/aa7218. [DOI] [PubMed] [Google Scholar]
- 17.Marx V. Machine learning, practically speaking. Nat. Methods. 2019;16:463–467. doi: 10.1038/s41592-019-0432-9. [DOI] [PubMed] [Google Scholar]
- 18.Lecun, Y. & Bengio, Y. In The Handbook of Brain Theory and Neural Networks (MIT Press, Cambridge, 1995).
- 19.Keras. The phyton deep learning library.
- 20.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- 21.Muller I, Kulms D. A 3D organotypic melanoma spheroid skin model. J. Vis. Exp.: JoVE. 2018 doi: 10.3791/57500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peintner L. 3D printed scaffold to monitor hanging drops on inverted microscopes. J. Sci. Humanit. Arts. 2019 doi: 10.17160/josha.6.2.531. [DOI] [Google Scholar]
- 23.Lagies S, et al. Cells grown in three-dimensional spheroids mirror in vivo metabolic response of epithelial cells. Commun. Biol. 2020;3:246. doi: 10.1038/s42003-020-0973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He, K., Zhang, X., Ren, S. & Sun, J. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 770–778.
- 25.Simonyan, K. & Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. http://arxiv.org/abs/1409.1556 (2015).
- 26.Huang, G., Liu, Z., van der Maaten, L. & Weinberger, K. Q. Densely Connected Convolutional Networks. arXiv e-prints, arXiv:1608.06993 (2016). https://ui.adsabs.harvard.edu/abs/2016arXiv160806993H.
- 27.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2017;60:84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 28.Fitzgerald KA, Malhotra M, Curtin CM, O'Brien FJ, O'Driscoll CM. Life in 3D is never flat: 3D models to optimise drug delivery. J. Control Release. 2015;215:39–54. doi: 10.1016/j.jconrel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 29.Abbott A. Biology's new dimension. Nature. 2003;424:870. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 30.Breslin S, O'Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov. Today. 2013;18:240–249. doi: 10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Laschke MW, Menger MD. Life is 3D: boosting spheroid function for tissue engineering. Trends Biotechnol. 2017;35:133–144. doi: 10.1016/j.tibtech.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Dumas-Mallet E, Button KS, Boraud T, Gonon F, Munafo MR. Low statistical power in biomedical science: a review of three human research domains. R. Soc. Open Sci. 2017;4:160254. doi: 10.1098/rsos.160254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Benton G, Arnaoutova I, George J, Kleinman HK, Koblinski J. Matrigel: from discovery and ECM mimicry to assays and models for cancer research. Adv. Drug Deliv. Rev. 2014;79–80:3–18. doi: 10.1016/j.addr.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Peintner L, Borner C. Role of apoptosis in the development of autosomal dominant polycystic kidney disease (ADPKD) Cell Tissue Res. 2017;369:27–39. doi: 10.1007/s00441-017-2628-6. [DOI] [PubMed] [Google Scholar]
- 36.Haenssle HA, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018;29:1836–1842. doi: 10.1093/annonc/mdy166. [DOI] [PubMed] [Google Scholar]
- 37.Garnavi R, Aldeen M, Bailey J. Computer-aided diagnosis of melanoma using border and wavelet-based texture analysis. IEEE Trans. Inf. Technol. Biomed. 2012;16:1239–1252. doi: 10.1109/TITB.2012.2212282. [DOI] [PubMed] [Google Scholar]
- 38.Goodfellow, I. J. et al. In Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2 2672–2680 (MIT Press, Montreal, 2014).
- 39.Celli JP, et al. An imaging-based platform for high-content, quantitative evaluation of therapeutic response in 3D tumour models. Sci. Rep. 2014;4:3751. doi: 10.1038/srep03751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizvi I, et al. Synergistic enhancement of carboplatin efficacy with photodynamic therapy in a three-dimensional model for micrometastatic ovarian cancer. Cancer Res. 2010;70:9319–9328. doi: 10.1158/0008-5472.CAN-10-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanoni M, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016;6:19103. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smalley KS, et al. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol. Cancer Ther. 2006;5:1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 43.Stehn JR, et al. A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Cancer Res. 2013;73:5169–5182. doi: 10.1158/0008-5472.CAN-12-4501. [DOI] [PubMed] [Google Scholar]
- 44.Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003;83:173–180. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]
- 45.Dufau I, et al. Multicellular tumor spheroid model to evaluate spatio-temporal dynamics effect of chemotherapeutics: application to the gemcitabine/CHK1 inhibitor combination in pancreatic cancer. BMC Cancer. 2012;12:15. doi: 10.1186/1471-2407-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen AG, et al. Comparison of diameter and perimeter methods for tumor volume calculation. J. Clin. Oncol. 2001;19:551–557. doi: 10.1200/JCO.2001.19.2.551. [DOI] [PubMed] [Google Scholar]
- 47.Bulin AL, Broekgaarden M, Hasan T. Comprehensive high-throughput image analysis for therapeutic efficacy of architecturally complex heterotypic organoids. Sci. Rep. 2017;7:16645. doi: 10.1038/s41598-017-16622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.