Abstract
The investigation of the causative agents for foodborne diseases and subsequent development of preventive steps to control the outbreak and related economic loss is the basic goal and priority of a rational food safety program. The entero-pathogenic Vibrio spp., which are Gram negative bacteria inhabiting estuarine ecosystems, are the major cause of foodborne illness associated with the consumption of raw or undercooked contaminated seafood or shellfish. To survey the Vibrio contamination in sea snails (Neptunea cumingi Crosse and Busycon canaliculatu), a total of 20 samples were collected from traditional market, at Qingdao city in Shandong province, China and analyzed for Vibrio species contamination. Presumptive-positive colonies grown on a specific Vibrio agar-based medium were picked and identified by the VITEKTM. Vibrio alginolyticus, V. parahaemolyticus, and V. vulgaris were isolated and identified in 11, 8, and 2 seafood samples, respectively. Among the 8 isolates of V. parahaemolyticus. The V. parahaemolyticus isolates were further tested for the tdh, trh, and tlh virulence factors. All the V. parahaemolyticus isolates were tlh-positive, however, all of them were tdh-negative. Interestingly 2 V. parahaemolyticus isolates were positive for trh virulence factor. These results indicated that there is a high incidence of V. alginolyticus and V. parahaemolyticus in sea snails. Therefore, food safety regulations for fishery auction markets should be established to control these species in addition to other Vibrio pathogenic contaminants. Our study provides the first evidence for the prevalence of Vibrio spp. in sea snail samples from traditional market in the Qingdao province of China.
Keywords: Foodborne disease surveillance, Vibrio alginolyticus, V. parahaemolyticus, V. vulgaris, sea snails
Introduction
Sea snail is a nutritious food that contains various desirable nutritional components of a healthy diet. Nevertheless, there are many health risks associated with the careless consumption of seafood products (Zhang et al., 2018). One of the major risks, seen in a few Asian countries, including China, Korea, and Japan, involves the consumption of raw or undercooked seafood that may be naturally contaminated by foodborne pathogens present in the marine environment (Weissfeld, 2014). The risk of marine contamination can be increased if the food is mishandled during production and processing, allowing microorganisms to multiply exponentially in favorable environments (Amagliani et al., 2012). Vibrio spp. are Gram negative, facultatively anaerobic, rod shaped, non-spore-forming bacteria that are ubiquitously and naturally present in estuarine waters throughout the world’s aquatic environments and are especially resistant to high salt concentrations (Su and Liu, 2007). Vibrio was first identified as a foodborne pathogen in Japan in the 1950s (Fujino et al., 1953; Letchumanan et al., 2019). By the early 1970s, Vibrio parahaemolyticus was recognized as a cause of diarrheal disease worldwide, most commonly in Asia and the United States. Vibrio parahaemolyticus associated with sea foods is one of the leading agent for food borne illness in China and across the world (Chen et al., 2017; Yang et al., 2017). Although cooking destroys this organism, sea snails are sometime eaten raw and will lead to infections associated with V. parahaemolyticus. Therefore, factors contributing to Vibrio contamination, including improperly handled seafood products, could pose a risk of exposure to these infectious agents that are transmissible to humans. Several species of the genus Vibrio are associated with foodborne spoilage and contamination of seafood products (Heng et al., 2017; Lee et al., 2018). The route of Vibrio entry into the biliary system remains unknown, and it was assumed to be via cutaneous lesions or due to gastrointestinal translocation upon consumption of contaminated seafood, as in other Vibrio infections (Liu et al., 2011). A few other species are more hazardous to humans, including V. parahaemolyticus and Vibrio cholerae, which are known to cause severe intestinal diseases (Kass and Riemann, 2006). The majority of V. parahaemolyticus isolates are avirulent, but they are still the leading cause of gastroenteritis linked to seafood consumption in several countries, including the United States (Iwamoto et al., 2010). Some Vibrio spp. pose a significant health risk to the immunocompromised elderly people and children. Vibrio may enter human hosts via wounds or direct consumption of raw seafood, including shellfish (primarily oysters), and frequently lead to septicemia and, ultimately, death in susceptible individuals (Harwood et al., 2004).
Researchers have shown a significant association between rising seawater temperatures and an increase in the number of Vibrio infections, suggesting that global warming might also be a factor affecting the increasing incidents of Vibrio contamination in seafood (Huehn et al., 2014). Over the past few years, V. parahaemolyticus strains have often been isolated from fish, shrimp, and other types of seafood. Additionally, Cho et al. (2016) and Mala et al. (2016) isolated V. parahaemolyticus in ready-to-eat food. In the Zhejiang province of China alone, there were 71 outbreaks caused by V. parahaemolyticus, resulting in 933 illnesses and 117 hospitalizations from 2010 to 2014. Recently, a multistate outbreak of gastrointestinal illnesses linked to oysters imported from Mexico occurred in June 2019; 16 people became severely ill, and almost all patients had Vibrio infections, especially those involving V. parahaemolyticus and Vibrio albensis (CDC, 2019).
Reports of outbreaks causing life threatening infections with Vibrio species associated with the consumption of a variety of seafoods are increasing. Therefore, to understand the risk of acquiring Vibrio infections through the consumption of traditional market-based seafood, it is important to have data on the prevalence of the virulent strains in traditional market-based seafood. This study aimed to examine the distribution of Vibrio species in sea snail (Neptunea cumingi Crosse and Busycon canaliculatu), Chinese traditional market-based seafood in the Qingdao province of China and determine the prevalence of the virulence genes of thermostable direct hemolysin (tdh), thermostable direct hemolysin-related hemolysin (trh), and thermolabile hemolysin (tlh) in V. parahaemolyticus isolates for food safety purposes.
Materials and Methods
Sample Collection
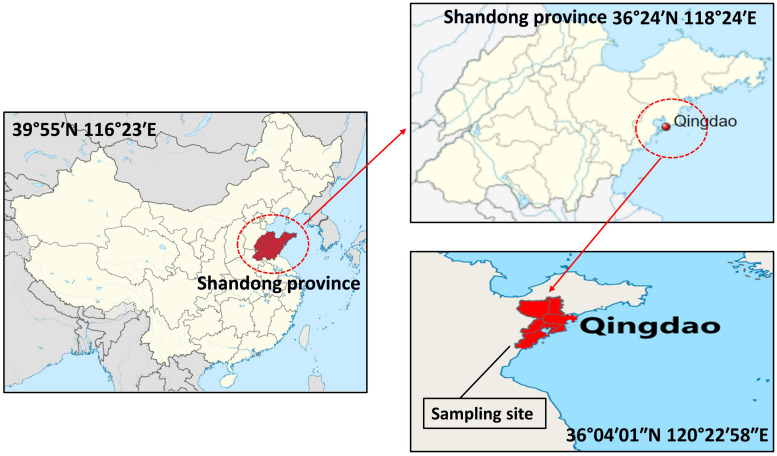
In total, 20 samples of sea snail were randomly collected from four traditional markets in the Chinese city of Qingdao (36°04′01″N 120°22′58″E) during March–September, 2018 (Figure 1). Approximately 1 kg of each sample was collected. All samples were packed in sterile plastic zipper bags, stored in an airtight box containing crushed ice, and transferred to the laboratory in 4 h for further processing.
FIGURE 1.
Map of samples (sea snail) collection site in Qingdao city (36°04′01″N 120°22′58″E) of Shandong province, China.
Sample Processing
All samples were thawed and pre-incubated in buffered peptone water for 16–18 h. Thereafter the samples were serially diluted, cultured and the bacteria were counted according to standard microbiological methods. Briefly, 25 g of each seafood sample was transferred into a sterile stomacher bag under aseptic conditions and blended with 225 mL of sterile buffered peptone water using a homogenizer set at 200 rpm for 15–20 min. Further, serial dilutions were made using sterile 0.1% peptone water to allow for culturing and bacterial enumeration.
Microbiological Analysis
Vibrio spp. were isolated and enumerated from all samples. The bacterial counts were determined using Vibrio spp. selective thiosulfate citrate bile salt (TCBS) agar plates. TCBS agar plates were inoculated by spreading 0.1 mL of the serial dilutions of samples and incubated at 37°C for 48 h. TCBS plates were examined for the presence of either yellow, round, 2–3 mm diameter colonies (suspect: V. alginolyticus) or green, round, 2–3 mm diameter colonies (suspect: V. parahaemolyticus). Each isolated suspected Vibrio colony was streaked onto another TCBS agar plate and incubated overnight at 37°C to obtain pure colonies.
Biochemical Characteristic Analysis
VITEK 2TM (BioMérieux, Marcy-l’Étoile, France) automated system was used according to the manufacturer’s instructions to identify the Vibrio spp. The method is a combination of photoelectric technology, computer technology, and bacterial octet digital identification technology. VITEK is frequently used for identification owing to its simplicity, accuracy and prompt outcome (Funke et al., 1998). In VITEK 2TM system, each test card contained 64 wells to carry out biochemical reactions (Funke et al., 1998). Each group was divided into three terms, and each positive reaction in the group was evaluated as 1, 2, 4, and then the values of each group were calculated. Based on the 64 biochemical reactions, biological identification numbers were obtained. With the help of the computer, the test cards were read after every 15 min and the dynamic reaction changes could be observed via optical scanning of the substrate of each reaction hole.
The specific operation procedure was as follows. The isolated bacteria were streaked on TCBS plates and incubated for 24 h at 35°C. A homogeneous suspension was prepared with an optical density (600 nm) between 0.55 and 0.63, measured with a DensiCHEKTM VITEK Caliper. Sixty-four biochemical tests were performed using the gram-negative bacteria identification VITEK cards. The VITEK biochemical tests for Vibrio parahaemolyticus (yellow color colony in TCBS medium) are described positive for indole, nitrate reduction, citrate utilization, and growth at 7% NaCl, while negative for urease, arginine, sucrose fermentation, swarming, and growth at 10% NaCl. Whereas Vibrio alginolyticus (appeared as green color colony in TCBS medium) are positive for indole, Voges–Proskauer (VP), nitrate reduction, citrate utilization, sucrose fermentation, swarming, and growth at 7 and 10% NaCl, while negative for urease, arginine.
Virulence Gene Detection
The virulence genes, namely tlh (450 bp) (Gutierrez West et al., 2013), tdh (251 bp) (Tada et al., 1992), and trh (250 bp) (Tada et al., 1992), were tested by polymerase chain reaction (PCR) amplification. The sequences of primers were as follows:
tlh-F: 5′-AAAGCGGATTATGCAGAAGCACTG-3′; tlh-R: 5′-GCTACTTTCTAGCATTTTCTCTGC-3′; tdh-F: 5′-CCACTACCACTC TCATATGC-3′;
tdh-R: 5′-GGTACTAAATGGCTGACATC-3′; trh-F: 5′-GGCTCAAAATGGTTAAGCG-3′; trh-R: 5′-CATTTCCGCTCTCATATGC-3′.
The PCR was carried out in a final volume of 20 μL containing 1 × PCR buffer (containing MgCl2) (Takara, Dalian, China), 0.5 mM dNTPs (Takara), 0.25 μM forward primer and reverse primer (Songon, Shanghai, China), 1.0 U Taq polymerase (Takara), and 50 ng genomic DNA. The PCR amplification was conducted under the following conditions: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 45 s, and extension at 72°C for 45 s, and an additional 7-min extension at 72°C. The amplicons were analyzed by 1.2% agarose gel electrophoresis and photographed in the gel imaging system (Vilber Lourmat, France). The amplicons were gel purified by TIANgel Midi Purification Kit (Tiangen) and send to Sangon Biotech Co., Ltd. for sequencing to confirm the identities. The sequence data were compared with those in the NCBI nucleotide sequence database by means of Basic Local Alignment Search Tool.
Results and Discussion
Total 35 Vibrio spp. were isolated from the tested 20 seafood samples using selective (TCBS) agar medium. All the isolates were tested for three major Vibrio species: V. parahaemolyticus, V. alginolyticus, and V. vulgaris using VITEK test kits, and tested for biochemical activity, serotype, and the presence of known virulence markers (tlh, tdh, and trh genes). The tdh and trh genes are strongly correlated with the virulence of pathogenic V. parahaemolyticus (Gutierrez West et al., 2013; Raghunath, 2015). To our knowledge, very few studies have characterized such a diverse panel of V. parahaemolyticus isolates in seafood samples from traditional markets at this level. Table 1 shows detailed information on the Vibrio species identified in all samples. The reliability and reproducibility of the VITEK system for Vibrio strain identification was tested every other day. Out of the 20 seafood samples, V. alginolyticus was identified in 55% (n = 11), while V. parahaemolyticus was identified in 40% (n = 8) samples. Interestingly, V. parahaemolyticus is highly co-occurred with V. alginolyticus (8/11). Besides these two Vibrio spp., V. vulgaris was also present in 10% samples (n = 2), and it always co-occurred with V. alginolyticus and V. parahaemolyticus (Table 1). These results indicate that there is a high incidence of V. alginolyticus and V. parahaemolyticus in seafood distribution channels. The V. parahaemolyticus isolates were further tested for the tdh, trh, and tlh virulence factors. Of these, tlh is suggested to be an important molecular marker for the identification of V. parahaemolyticus (Gutierrez West et al., 2013). Results showed that all the VITEK-identified V. parahaemolyticus isolates (n = 8) were positive for tlh, thus confirming the success of the VITEK identification method. Results are good agreement with the previous published reports showing the presence of tlh positive V. parahaemolyticus in many sea food samples (Letchumanan et al., 2014). Consistent with this Terzi Gulel and Martinez-Urtaza (2016) identified and confirmed many strains of V. parahaemolyticus using tlh virulence marker. We further noticed that among all the identified V. parahaemolyticus isolates, only 25.0% (n = 2) were positive for trh. Moreover, all the identified V. parahaemolyticus isolates were tdh-negative. These results are consistent with the findings of Jones et al. (2012) who obtained tdh-negative and trh-positive isolates of Vibrio species from Canada, Maine, and Washington during the early summer, indicating a potential preferential distribution of these strains in northern areas. A study carried out by Tran et al. (2018) on 385 sea food samples demonstrated the presence of tdh and trh positive V. parahaemolyticus strains in 25 samples. Out of theses 25 samples, only 2 samples have V. parahaemolyticus strains with both tdh and trh gene and rest 23 samples have V. parahaemolyticus strains either with tdh and trh virulence gene. In another similar study done by Yang et al. (2017) examined the prevalence of V. parahaemolyticus in various oyster, fish and shrimp samples collected form the South China domestic market and revealed 8.16 and 12.24% tdh and trh positive isolates, respectively. The difference in the distribution and evolution of tdh and trh in V. parahaemolyticus in different samples are highly associated with geographical location, sample source and environmental conditions (Raghunath, 2015). This was also reflected in a previous report that trh-positive V. parahaemolyticus strains constitute a higher proportion of the total V. parahaemolyticus population in the Mid-Atlantic region than in other areas during the summer (DePaola et al., 2010).
TABLE 1.
Positive samples of Vibrio species in sea snail available in a traditional market in Qingdao city, Shandong province, China.
| Positive sea snail samples | TCCB Agar plate* | VITEK Identification | Presence of virulence gene |
||
| tlh | tdh | trh | |||
| 1 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| 2 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| 3 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | −ve | +ve | ||
| 4 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | −ve | −ve | ||
| 5 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | -ve | -ve | ||
| 6 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| 7 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | −ve | −ve | ||
| 8 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | −ve | −ve | ||
| V. vulgaris | −ve | −ve | −ve | ||
| 9 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | −ve | −ve | ||
| V. vulgaris | |||||
| 10 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | −ve | −ve | ||
| 11 | +ve for Vibrio spp. | V. alginolyticus | −ve | −ve | −ve |
| V. parahaemolyticus | +ve | −ve | +ve | ||
*TCBS, thiosulfate citrate bile salt (Vibrio spp. Selective medium); −ve: negative; +ve: positive. Bold value is highlight of the positive result.
Similar to our study, V. alginolyticus was also found to be the most frequently isolated species in two culture-based studies from Italy and Malaysia (Elhadi et al., 2004). In 2011, an interesting study on the prevalence of V. parahaemolyticus in retail oysters sold at a local market in São Paulo State (Brazil) reported that 100% of the samples was contaminated with V. parahaemolyticus. In addition, none of the 1,943 tested isolates in that study harbored tdh and/or trh gene(s) (Sobrinho et al., 2011). Our data provides the first information on exposure to Vibrio spp. in traditional market-based seafood in China. However, it must be recognized that using only culture-based methods to detect Vibrio spp. could be a limitation. In our study, the VITEK along with tdh, trh, and tlh virulence factors identification system was used to identify the presence of potentially pathogenic Vibrio spp. in traditional market-based seafood.
Conclusion
This study identified several Vibrio species in sea snail samples locally available in the traditional Chinese market of Qingdao city, Shandong province, China through culturing on selective media and VITEK biochemical techniques. V. alginolyticus was found in 55% (n = 11) of selected sea snails while V. parahaemolyticus was found in 40% (n = 8) of the samples. Besides, V. vulgaris was also present in 10% samples (n = 2), and it always co-occurred with V. alginolyticus and V. parahaemolyticus in samples. All V. parahaemolyticus isolates were positive for tlh virulence factor. In addition 2 V. parahaemolyticus isolates found positive for trh virulence factor. Eating raw seafood in Asian countries, including China, Korea, and Japan is the main cause of emerging gastrointestinal diseases. Therefore, based on the results obtained in this study, further efforts are needed for rapid pathogen identification and for controlling Vibrio contamination in sea snails. Additionally, meta-analyses or systematic studies on the worldwide prevalence of antimicrobial-resistant V. parahaemolyticus in seafood are needed. Furthermore, to minimize the risk of V. parahaemolyticus infection and ensure seafood safety, consultation and collaboration between researchers and governments are urgently needed. We further suggest avoiding the consumption the raw sea snails owing to their high chance of contamination with Vibrio species.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
XSo wrote the manuscript. XSo, JZ, WY, and YW performed the experiments and analyzed the data. XSh evaluated the drafts of manuscript and approved the manuscript for submission. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was part of the national risk surveillance program in China.
References
- Amagliani G., Brandi G., Schiavano G. F. (2012). Incidence and role of Salmonella in seafood safety. Food Res. Int. 45 780–788. 10.1016/j.foodres.2011.06.022 [DOI] [Google Scholar]
- CDC (2019). Multistate Outbreak of Gastrointestinal Illnesses Linked to Oysters Imported from Mexico. Available online at: https://www.cdc.gov/vibrio/investigations/rawoysters-05-19/index.html (accessed June 21, 2019). [Google Scholar]
- Chen A. J., Hasan N. A., Haley B. J., Taviani E., Tarnowski M., Brohawn K., et al. (2017). Characterization of pathogenic Vibrio parahaemolyticus from the Chesapeake Bay, Maryland. Front. Microbiol. 8:2460. 10.3389/fmicb.2017.02460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho T., Kim N., Kim S., Song J., Rhee M.-S. (2016). Survival of foodborne pathogens (Escherichia coli O157: H7, Salmonella Typhimurium, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus) in raw ready-to-eat crab marinated in soy sauce. Int. J. Food Microbiol. 238 50–55. 10.1016/j.ijfoodmicro.2016.08.041 [DOI] [PubMed] [Google Scholar]
- DePaola A., Jones J. L., Woods J., Burkhardt W., Calci K. R., Krantz J. A., et al. (2010). Bacterial and viral pathogens in live oysters: 2007 United States market survey. Appl. Environ. Microbiol. 76 2754–2768. 10.1128/aem.02590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhadi N., Radu S., Chen C., Nishibuchi M. (2004). Prevalence of potentially pathogenic Vibrio species in the seafood marketed in Malaysia. J. Food Protect. 67 1469–1475. 10.4315/0362-028x-67.7.1469 [DOI] [PubMed] [Google Scholar]
- Fujino T., Okuno Y., Nakada D., Aoyoma A., Fukai K., Mukai T., et al. (1953). On the bacteriological examination of Shirasu food poisoning. Med. J. Osaka Univ. 4 299–304. [Google Scholar]
- Funke G., Monnet D., deBernardis C., von Graevenitz A., Freney J. (1998). Evaluation of the VITEK 2TM system for rapid identification of medically relevant gram-negative rods. J. Clin. Microbiol. 36 1948–1952. 10.1128/jcm.36.7.1948-1952.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez West C. K., Klein S. L., Lovell C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79 2247–2252. 10.1128/aem.03792-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood V. J., Gandhi J. P., Wright A. C. (2004). Methods for isolation and confirmation of Vibrio vulnificus from oysters and environmental sources: a review. J. Microbiol. Methods 59 301–316. 10.1016/j.mimet.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Heng S.-P., Letchumanan V., Deng C.-Y., Ab Mutalib N.-S., Khan T. M., Chuah L.-H., et al. (2017). Vibrio vulnificus: an environmental and clinical burden. Front. Microbiol. 8:997. 10.3389/fmicb.2017.00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn S., Eichhorn C., Urmersbach S., Breidenbach J., Bechlars S., Bier N., et al. (2014). Pathogenic vibrios in environmental, seafood and clinical sources in Germany. Int. J. Med. Microbiol. 304 842–850. [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Ayers T., Mahon B. E., Swerdlow D. L. (2010). Epidemiology of seafood associated infections in the United States. Clin. Microbiol. Rev. 23 399–411. 10.1128/cmr.00059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. L., Lüdeke C. H., Bowers J. C., Garrett N., Fischer M., Parsons M. B., et al. (2012). Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 50 2343–2352. 10.1128/jcm.00196-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass P. H., Riemann H. P. (2006). Foodborne Infections and Intoxications, 3rd Edn Cambridge, MA: Academic Press, 3–26. [Google Scholar]
- Lee L.-H., Ab Mutalib N.-S., Law J. W.-F., Wong S. H., Letchumanan V. (2018). Discovery on antibiotic resistance Patterns of Vibrio parahaemolyticus in selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front. Microbiol. 9:2513. 10.3389/fmicb.2018.02513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchumanan V., Chan K.-G., Lee L.-H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. 10.3389/fmicb.2014.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchumanan V., Loo K. Y., Law J. W., Wong S. H., Goh B., Ab Mutalib N., et al. (2019). Vibrio parahaemolyticus: the protagonist of foodborne diseases. Prog. Microbes Mol. Biol. 2:a0000029 10.36877/pmmb.a0000029 [DOI] [Google Scholar]
- Liu W. L., Chiu Y. H., Chao C. M., Hou C. C., Lai C. C. (2011). Biliary tract infection caused by Vibrio fluvialis in an immunocompromised patient. Infection 39 495–496. 10.1007/s15010-011-0146-0 [DOI] [PubMed] [Google Scholar]
- Mala W., Alam M., Angkititrakul S., Wongwajana S., Lulitanond V., Huttayananont S., et al. (2016). Serogroup, virulence, and molecular traits of Vibrio parahaemolyticus isolated from clinical and cockle sources in northeastern Thailand. Infection Genet. Evol. 39 212–218. 10.1016/j.meegid.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Raghunath P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. 10.3389/fmicb.2014.00805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrinho P. S. C., Destro M. T., Franco B. D., Landgraf M. (2011). Occurrence and distribution of Vibrio parahaemolyticus in retail oysters in São Paulo State, Brazil. Food Microbiol. 28 137–140. 10.1016/j.fm.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Su Y. C., Liu C. C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24 549–558. 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Tada J., Ohashi T., Nishimura N., Shirasaki Y., Ozaki H., Fukushima S., et al. (1992). Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol. Cell. Probes 6 477–487. 10.1016/0890-8508(92)90044-x [DOI] [PubMed] [Google Scholar]
- Terzi Gulel G., Martinez-Urtaza J. (2016). Molecular characterizations of Vibrio parahaemolyticus in seafood from the Black Sea, Turkey. Lett. Appl. Microbiol. 62 494–500. 10.1111/lam.12579 [DOI] [PubMed] [Google Scholar]
- Tran T. H. T., Yanagawa H., Nguyen K. T., Hara-Kudo Y., Taniguchi T., Hayashidani H. (2018). Prevalence of Vibrio parahaemolyticus in seafood and water environment in the Mekong Delta, Vietnam. J. Veter. Med. Sci. 80 1737–1742. 10.1292/jvms.18-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissfeld A. S. (2014). Infections from eating raw or undercooked seafood. Clin. Microbiol. Newslett. 36:3. [Google Scholar]
- Yang Y., Xie J., Li H., Tan S., Chen Y., Yu H. (2017). Prevalence, antibiotic susceptibility and diversity of Vibrio parahaemolyticus isolates in seafood from South China. Front. Microbiol. 8:2566. 10.3389/fmicb.2017.02566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. S., Han L. W., Shi Y. P., Li X. B., Zhang X. M., Hou H. R., et al. (2018). Two novel multi-functional peptides from meat and visceral mass of marine sanil Neptunea arthritica cumingii and their activities in vitro and in vivo. Mar. Drugs 16:473. 10.3390/md16120473 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.