Abstract
Drivers of RNA interference are potent for manipulating gene and protein levels, which enable the restoration of dysregulated mRNA expression that is commonly associated with injuries and diseases. This review summarizes the potential of targeting neuroglial cells, using RNA interference, to treat nerve injuries sustained in the central nervous system. In addition, the various methods of delivering these RNA interference effectors will be discussed.
Keywords: RNA interference, central nervous system, nerve injury, drug delivery
Introduction
Elucidated by Fire et al. in 19981 and hailed as the ‘Breakthrough of the year’ in 2002,2 RNA interference (RNAi) can be employed as a potent mechanism for overwriting DNA-based regulation in cells, thereby enabling manipulation of cellular functions and behaviour. This was a huge revelation at that time because RNAs have always been thought to be inferior to DNAs and only function to convert genetic information into proteins.2 It turns out that RNAs are more than that and are capable of executing RNAi, which is a natural phenomenon that exists even across various kingdoms (in plants, fungi and animals)3 for regulating gene transcription and protein translation. In addition, the mechanism of RNAi is well-established and more information regarding this process can be found in these excellent reviews.3–6
Although its role has not been fully explored to date, it is clear that RNAi is rightfully an evolutionarily conserved defence mechanism against molecular invaders, such as viruses and transposable elements (TEs), where foreign double stranded RNA (dsRNA) were often introduced.3,7 More recently, it has also been recognized that RNAi plays an important role in development,8 such as neurogenesis,9,10 axonal outgrowth11–13 and myelination14 in the CNS. Given these huge responsibilities in ensuring the smooth progress of such crucial cellular processes, it is almost certain that RNAi is involved in maintaining cell/tissue homeostasis as well. Indeed, using RNAi microarray analysis, studies are revealing that microRNAs (miRNAs), naturally occurring components for regulating RNAi, are commonly dysregulated following nerve injuries6,15,16 and during disease progression.14 Therefore, with the goal of either enhancing tissue regeneration or reverting the dysregulated nucleic acid levels to normalcy, effectors of RNAi such as small interfering RNAs (siRNAs), miRNAs, short hairpin RNAs (shRNAs) and antisense oligonucleotides (ASOs) are useful as therapeutics for treating deficiencies in the CNS.
Traumatic nerve injuries often lead to prolonged disruption of the functional coherence of the tissue. This phenomenon, besides partly attributed to the lack of regeneration in neurons, also stems from the inhibitive status of the seemingly inconspicuous glial cells.17 Being the most abundant but underappreciated cell type in the CNS, these supportive cells are starting to rack up interests from researchers in recent years given their crucial roles in maintaining an optimal environment for neurons to function. Correspondingly, there are also increasing evidences that prudent regulation of their gene and protein expressions following traumatic injuries can promote functional recovery. This suggests that glial cells should also be considered as potential treatment candidates.
Here, we review currently known nucleic acid materials that are capable of eliciting RNAi, as well as methods for efficient therapeutic delivery. In addition, the possibility of targeting glial cells for treating CNS nerve injuries will also be discussed.
Drivers of RNAi
RNAi is fundamentally triggered by the presence of short sequences of RNA strands, which may be single or double stranded and contains around 19 to 29 nucleotides, within the cell. Currently, the predominantly known drivers of RNAi are siRNAs, miRNAs, shRNAs and ASOs. These nucleic acid sequences in their mature form can be easily obtained through commercial means and despite being so similar in their size, each of them have their own uniqueness and efficiency in achieving the intended outcome.
SiRNA
SiRNAs form the main arm of RNAi and are, in design, the most potent molecules to elicit a transient single gene target knockdown once it interacts with the cell’s machinery that is responsible for eliciting RNAi.3–6 Functional siRNAs were observed to be within 21 to 23 base pairs in length and can easily be synthesized in vitro by cleaving long dsRNAs into a pool of siRNAs using Dicer.3 The main mechanism of siRNA in leading to RNAi is gene regulation, where siRNA binds to perfectly complementary segments of messenger RNA (mRNA) to initiate degradation and prevent transcription from occurring.18
MiRNA
MiRNAs, unlike siRNAs, exist endogenously. They are transcribed by RNA polymerase II and usually consist of 19 to 25 nucleotides. Contrary to siRNAs, miRNAs induce RNAi by mainly repressing translation,4 although occasional degradation of mRNA to prevent transcription can occur as well.19 The modulation of protein at the translational level in animals usually stems from their imperfect base-pairing with target mRNAs.20,21 However in plants, their miRNAs generally bind to mRNAs with near-perfect complementarity, which leads to target mRNA degradation instead,22,23 a mechanism that is similar to that displayed by siRNAs. In a sense, the difference in RNAi regulatory mechanisms between siRNAs and miRNAs is determined by the degree of complementarity during the binding to mRNAs. Yet, due to the slight mismatch of miRNAs towards mRNAs, miRNAs have no true target mRNAs and can, therefore, bind to hundreds or even thousands of them,24 resulting in massive amounts of translational regulation. Since their discovery,25 we now know that miRNAs govern physiology at an organism level, especially since they are involved in the entire range of cellular processes such as cell proliferation,26,27 differentiation,10,26,28,29 senescence30–32 and even apoptosis.33,34
Plasmids encoding shRNA
The shRNAs are 19–22 base pairs in length, linked by a short loop of 4–11 nucleotides and drive RNAi similarly as siRNAs.35 A major difference between shRNAs and siRNAs is that shRNAs are encoded by plasmids, and long-term expression can be achieved if the plasmids are introduced into the nucleus through viral means. By incorporating inducible promotors in the transcription vector, the expression of shRNAs can also be turned on and off, allowing RNAi to be regulated as desired.36,37
ASOs
ASOs are synthetic, single-stranded DNAs that are 8–50 nucleotides in length. They can bind to target mRNAs through complementary base-pairing to induce endonuclease-mediated transcript knockdown and subsequently, protein downregulation.38,39 The concept of ASOs was first described in 1978 by Stephenson and Zamecnik,40 and ASOs were the first generation of biomolecules that were used for disrupting protein expression, albeit in viruses. Further refinement of the synthesis procedures have since allowed potent modifications to be made on the nucleotides, conferring ASOs enhanced pharmacological properties, such as altering pre-mRNA splicing (gene regulation) and blocking mRNA translation (protein regulation).38,41 Certain modifications, such as the use of a phosphorothioate backbone as well as 2′-O-methyl (2′-OMe) and 2′-O-methoxy-ethyl (2′-MOE) oligonucleotides, can also extend their potency beyond functional effects to increase their circulation duration in serum,42,43 increase their resistance to nuclease degradation,44 increase their hybridization affinity to their target RNA44–46 and even reduce immunostimulatory activities.47
RNAi mechanism
The mechanism for eliciting RNAi is generally similar for these four classes of small nucleic acids. One notable difference between them, however, is the degree of modification required to be functional.5,48 In their mature form, these nucleic acids will associate with the RNA-induced silencing complex (RISC), which comprises of Argonaute-2 (Ago-2) and Dicer, before binding to their complementary sequences present in the target mRNAs (Figure 1).5 Through this binding, the mRNAs will not be able to undergo RNA translation as it will thereafter be either degraded or physically hindered from associating with ribosomes.
Figure 1.
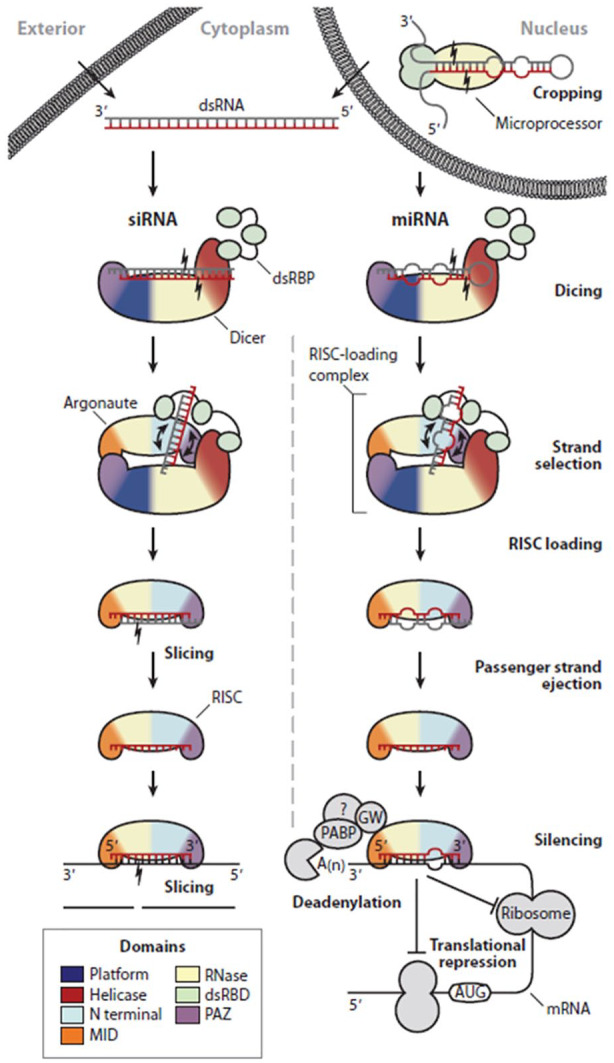
Mechanism of RNAi. RNAi is a general mechanism that siRNAs, miRNAs, shRNAs and ASOs can adopt. The main difference between them lies in the extent of modifications before being functional. Figure depicts a well-established RNAi pathway by siRNAs and miRNAs. Adapted with permission from RC Wilson and Doudna in Annu Rev Biophys, 2013.5
Delivery methods
While these nucleic acids are known to induce therapeutic effects, the benefits in using these molecules for treating pathologies in the CNS would not be attainable if these biomolecules are unable to reach their intended target environment, that is, the CNS and cytoplasm of cells. Fundamentally, as these nucleic acids possess numerous phosphodiester linkages between each nucleotides, the entire structure tends to be anionic.49 This indicates that electrostatic attraction is their main form of interaction with other biological materials. In addition, they are also hydrophilic and unable to penetrate effectively into tissue structures within the body. Delivery approaches can, therefore, be designed to either cater to or capitalize on their inherent properties for achieving a therapeutic outcome.
Systemic delivery
There are many strategies for delivering nucleic acids into the body. However, if the delivery region of interest is the CNS and the therapeutics are nucleic acids, then options are limited since the presence of the blood–brain barrier (BBB) and the blood–spinal cord barrier (BSCB) would reduce the efficacy of many conventional (intravenous, intramuscular, oral, transdermal) delivery methods substantially.
Surprisingly, gene/RNAi therapy is currently still in its infant stage of application in the clinics. After conducting a comprehensive search on the list of approved clinical trials related to RNAi within the CNS, it was found that there is currently no approved RNAi therapy for treating CNS-related injuries or diseases. Plasmid deliveries are mostly enabled through viral vectors and are only applicable for neurodegenerative diseases or tumours in the CNS. However, none of them have, to date, reached Phase 3 clinical trial (clinicaltrials.gov, identifier: NCT00876863, NCT00985517, NCT00004080, NCT00004041). To clarify, the physical limitations posed by the BBB and BSCB remain true only under physiologically healthy conditions. In the event of an injury in the CNS, depending on the injury intensity and site, their functional integrity may be compromised. Under these circumstances, there may exist a window for therapeutic interventions and the widely used intravenous delivery route may be the preferred method for delivering nucleic acids before the barriers regenerate. Alternatively, if surgery is unavoidable, then the nucleic acids can also be loaded into a biomaterial, most commonly hydrogels,50–54 and implanted into or placed near the injury site for localized and sustained delivery.
In the event when bypassing the BBB or BSCB is unfavourable, then nucleic acids can also be delivered directly into the CNS through intrathecal infusion. Although not for nerve injuries, this was the strategy employed to deliver ASOs in the first-ever approved treatment of spinal muscular atrophy (SMA).55 This method of delivery was also used previously in another completed Phase 1 trial for amyotrophic lateral sclerosis (ALS).56
Besides intrathecal injection, another notable method for the delivery of therapeutics into the CNS is microbubble-assisted focused-ultrasound blood–brain barrier disruption (MB + FUS BBBD).57 By focusing ultrasonic waves onto acoustically responsive microbubbles injected into the blood vessels, acoustic cavitations are induced. This in turn leads to the transient disruption of the BBB, thereby, allowing intravenously administered drugs to enter the brain parenchyma efficiently.57 However, it should be noted that optimization of the parameters (microbubble composition, size distribution and concentration as well as ultrasound settings) is required to prevent thermomechanical-induced haemorrhaging58 and immunoactivation.59
Tissue-localized delivery
Technically, nucleic acids are usually delivered non-virally in either an encapsulated, complexed or naked form. Encapsulation of these labile nucleic acids protects them from free nucleases and ensures that these drugs are not degraded before they can even be taken up by cells. On the contrary, complexed and naked delivery of nucleic acids44 will require chemical modifications in order to enhance their resistance to biodegradation. This is highlighted by a previous study which demonstrated that direct administration of both naked siRNAs and ASOs into the brain through intracerebroventricular application led only to the detection of ASOs but not siRNAs.60
As nucleic acids complexed with a delivery vector are not spared from the harshness of the in vivo milieu, chemical modifications of the former are recommended as well to prevent its degradation. Based on current knowledge, the single most important consideration for the most basic cellular delivery is electrostatic interactions. As both nucleic acids and cellular membrane are similarly charged (anionic), it is not efficient for naked, unmodified nucleic acids to be taken up by cells naturally.61 Therefore, vectors that are suitable for non-viral delivery are usually cationic (Lipofectamine 2000 reagent,61,62 TransIT-TKO,63,64 polyethylenimine (PEI),65 etc.). Due to their high net positive charge, these delivery vectors can form complexes with nucleic acids through electrostatic attraction and still remain cationic, which facilitates cellular uptake by enabling the nucleic acid-vehicle complex to attach to the surface of cells for various vesicular transports (endocytosis and/or macropinocytosis) to occur.66
In addition, the delivery of such complexes can be further augmented through scaffold-mediated approaches. In essence, a scaffolding system can be designed to function as a depot to house the nucleic acids, either naked or complexed with a cationic carrier, thereby ensuring their localization. By concentrating the nucleic acids in a confined region, cells that are present will have an increased chance of encountering them, which promotes their uptake and silencing efficiency. Accordingly, common strategies for retaining the naked and complexed nucleic acids within current conceivable scaffolds include electrostatic interaction,67,68 surface adsorption69–71 and also encapsulation.51,72,73
Besides providing localized delivery, scaffolds can also be tuned to manipulate the extent and even the location of uptake within cells.74 This can be achieved by altering the physico-chemical properties (elasticity, topology, composition, etc.) of the substrate, which the cells are attached to, thereby regulating cytoskeletal remodelling75 and subsequently internalization pathways74 (Figure 2 and Table 1).
Figure 2.
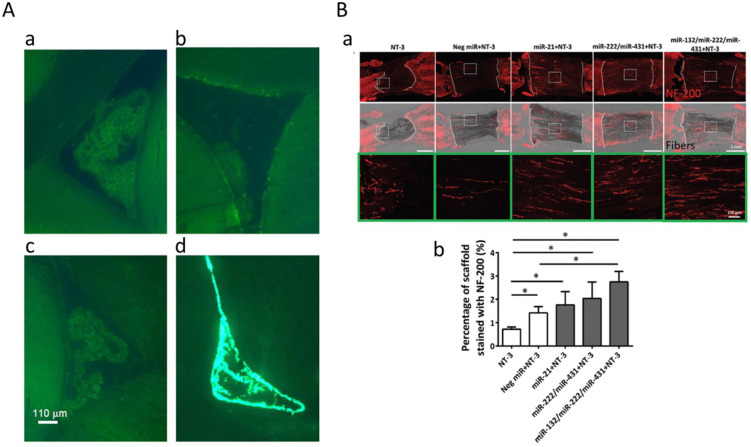
Tissue-localized delivery of RNAi. (A) The area around the lateral ventricle of the rat brain 3 h after intracerebroventricular (i.c.v.) injection of fluorescence-labelled siRNA. The i.c.v. injection of 2 μg siRNA and cell detergent i-Fect with 1:4 w/v ratio (d) was only detected but others not (a: 50 μg siRNA dissolved in siRNA suspension buffer without detergent, b: 2 μg siRNA and the other cell detergent DOTAP (1:4 w/v), c: i-Fect only). Adapted with permission from C Senn et al.60 (B) MiRNAs stimulated extensive neurite ingrowth into fibre-hydrogel scaffolds 2 weeks after spinal cord injury. Neurotrophin-3 (NT-3) and/or miRNAs were treated with PCL fibre-collagen scaffold (a). Neurofilament protein-200 (NF-200) was significantly higher in miRNA-treated groups compared to control group (NT only) (b). Adapted with permission from N Zhang et al.54
Table 1.
Table comparing the advantages and disadvantages of various nucleic acid delivery strategies. These comparisons provide different perspectives to the choice of delivery method.
| Format of nucleic acid delivery | Advantages | Disadvantages |
|---|---|---|
| Encapsulated | 1. Nucleic acids are protected from degradation by nucleases.76
2. Delivery can be relatively localized if nucleic acids are confined within a scaffold prior to their release.77 3. Delivery through nanoparticles can potentially allow nucleic acids to reach specific cell/tissue targets. |
1. May not be uptaken into cells efficiently without a
cationic delivery vehicle. 2. Require proper context for implementation, e.g. not ideal to inject scaffold encapsulating nucleic acids for intravenous delivery. 3. Nanoparticles encapsulating nucleic acids are not able to penetrate deep into tissue structures as compared to complexed/naked nucleic acids. |
| Complexed | 1. Can be taken up into cells much more efficiently due to the cationic vehicle.54 | 1. May get degraded easily without modifications to its
nucleic acid strands. 2. Due to the electrostatic interactions between the anionic nucleic acids and the cationic vehicle, aggregation may occur, which impedes delivery and uptake. 3. Delivery unlikely to be localized. |
| Naked | 1. Due to its extremely small size, nucleic acids can
potentially penetrate deep into tissues.78
2. Nucleic acid strands can be modified such that administration of large doses will not elicit an immune response.47 |
1. Requires modification to the nucleic acid strands to
enhance its resistance towards nucleases and uptake by cells.44
2. Delivery unlikely to be localized. |
CNS and neuroglial cells
The CNS is the part of the nervous system that consists of the brain and spinal cord. The CNS tissue is comprised of two main cell types, namely, the neurons and the neuroglia cells and is enclosed by connective tissue membranes of meninges.79 The neuroglial cells further consist of astrocytes, microglial and oligodendrocytes (OL). Neurons are the most studied cell type in the nervous system since they have been discovered, in part due to the importance of their role and also probably due to the lack of appreciation of the other neuroglia cells. This notion is rapidly changing recently as more studies are revealing that the neuroglial cells are, in fact, equally as crucial as neurons in maintaining the functionality of the CNS.80 The highly branched neuroglial cells that are located between neurons have intimate functional relationships with the neurons, providing both mechanical and physiological support.81,82
CNS diseases are a type of neurological disorders caused by various factors including trauma, infections, degeneration, autoimmune diseases and stroke, and can alter and degrade the function or structure of cells and tissues in the CNS.83,84 In particular, the age-dependent neurodegenerative diseases represent a major disease in human because the elderly population has increased in recent years.85 The major types of neurodegenerative diseases include Alzheimer’s (AD), Parkinson’s (PD), Huntington’s (HD), ALS, multiple sclerosis (MS), Creutzfeldt–Jakob disease (CJD) and others.86 The pathological characteristics of these neurodegenerative diseases include aggregation and accumulation of specific proteins in the CNS that are associated with neuronal defect in the CNS. In addition, the gliosis that proliferates and activates neuroglial cells is a major hallmark of neurodegenerative diseases.87,88
Among the neuroglial cells, astrocytes are morphologically heterogeneous cells that provide physical support for neurons and mediate the metabolite exchange between the neurons and the blood vessels, including glutamate re-uptake, ionic buffering and water balance.89,90 In addition, astrocytes play an important role in BBB maintenance, neuroinflammatory regulation and the repair of the CNS.91,92 In the case of AD, astrocytes become reactive as a result of generation and deposition of beta-amyloid (Aβ), which in turn leads to decreased glutamate uptake due to reduced expression of uptake transporters, altered energy metabolism and ion homeostasis, increased release of cytokines and inflammatory mediators and disruption of astrocytic networks.91,93 Recently, it was reported that neuroinflammation and ischaemia induced two different types of reactive astrocytes, A1 and A2. A1 astrocytes highly upregulate many classical inflammatory cascade genes to be destructive to synapses, whereas A2 astrocytes upregulate many neurotrophic factors. The A1 astrocyte that is induced by activated inflammatory microglia via the secretory cytokines, including IL-1a, TNF (tumour necrosis factor) and C1q, induces the death of neurons and OLs.94
Microglia, the resident immune cells of the CNS, have immunological surveillance95 and defence functions against invading micro-organisms, damaged cells and the debris of apoptotic cells.96–98 Microglia are derived from monocyte progenitor cells and their function is tightly regulated by the CNS microenvironment.99 Microglia express many pattern-recognition receptors (PRRs) and detect pathogen-associated molecular patterns (PAMPs) or damage–associated molecular patterns (DAMPs), including Toll-like receptors (TLRs) 4 and TLR1/2; their coreceptors; CD14; NOD-like receptors (NLRs); receptors for nucleic acids; C-type lectin receptors (CLRs), such as CLEC7A; and chemokine receptors, such as CX3CR1 and CXCR4. In addition, microglia express immune receptors such as TREM2 (triggering receptor expressed on myeloid cells 2) and multiple receptors for neurotransmitters and neuropeptides released by neurons.97 Therefore, the microglia are highly responsive to injury or neuroinflammatory disease and are activated under the chronic neurodegenerative diseases of the CNS such as AD, PD, MS and ALS.100,101 For example, proliferation and activation of microglia is a hallmark of AD and the impaired activities and altered microglial responses to Aβ increase the AD pathogenesis and can be harmful to neurons.102 The over-activated microglia can release cytotoxic factor and inflammatory cytokines. For this reason, inhibiting the activity of microglia appropriately may be an effective way for the treatment of neurodegenerative diseases.103
OLs are responsible for the formation and maintenance of myelin sheath by neurotrophic support in the CNS.104–106 In order to myelinate properly, OLs have high metabolic activities with oxygen and adenosine triphosphate (ATP) consumption and Reactive oxygen species (ROS) or hydrogen peroxide formation as toxic by-products. Hence, oxidative damage is a common cause to OL disorder or loss under many neuropathological diseases like MS and ischaemia.107 The damage of OL is observed in neurodegenerative disorders of the CNS, especially chronic demyelinating diseases such as MS, spinal cord injury, AD, PD and ALS.108 For example, the toxic effects of Aβ accumulation, a major pathological process in AD, induced OL dysfunction and demyelination.109 In the case of MS, autoimmune system attacks mainly the myelin and OLs. The consequence of such an autoimmune attack is a local demyelination and subsequent axonal loss that is one of the pathological hallmarks of MS.110
Consequently, the deprivation or dysfunction of these various neuroglial cell populations following traumatic nerve injuries and neurodegenerative diseases hinders recovery given the lack of support, as well as inhibitory effects posed by necrotic cellular debris.111 As such, the maintenance of spared glial cells followed by their quantitative and steady-state functional restoration is a potential area of focus for facilitating treatment.
Neuroglial cells as potential targets of RNAi for nerve injury treatment
RNAi in astrocytes
Astrocytes constitute the largest proportion of all cells in the mammalian CNS (around 20%–40%).112 They control neuronal activity and well-being through neurometabolic coupling113 and help to remove excess neurotransmitters, potassium114 and glutamate115 from the extracellular space. Following a traumatic nerve insult, astrocytes become activated or reactive, displaying functional and morphological changes that constitute astrogliosis.116 Over time, they may also migrate and align to form a glial scar that surrounds the injury site,117 physically and chemically inhibiting local axonal growth.118
Studies involving RNAi, conducted in both in vitro and in vivo settings, are thus commonly aimed at circumventing the repercussions caused by this barrier of astrocytes. However, in order to achieve effective gene silencing, it is first imperative to establish a suitable working concentration of RNAi effectors; excessive amounts may be cytotoxic while the opposite may lack potency for an outcome.
Ki et al. conducted a comprehensive testing of the optimal concentration of siRNA for gene silencing in primary cultured rat astrocytes in vitro. Using Lipofectamine 2000 as a transfection reagent, they showed that a siRNA concentration of 20 nM is sufficient to induce efficient uptake (~80%) and RNAi (silencing efficiency ~95%), while preserving the cells’ viability.119
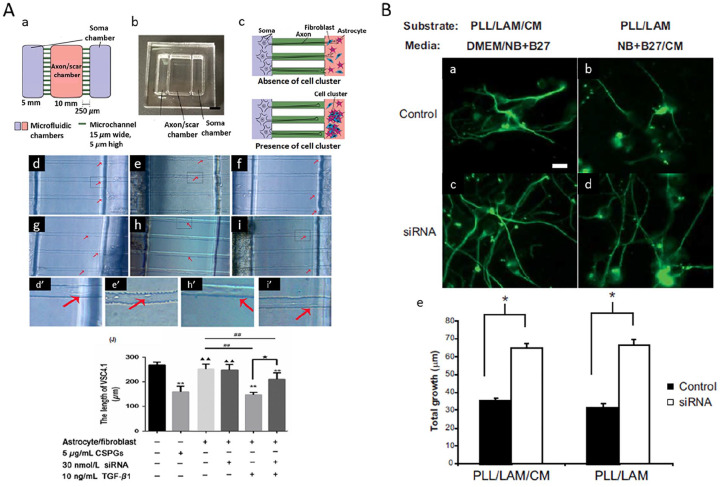
A similar concentration range was also used in another in vitro study by Li et al., where they investigated the relationship between ephrin-B2 and glial scar formation. Using a custom-made microfluidic platform containing chimeric ventral spinal cord 4.1 (VSC4.1) motoneurons, primary rat astrocytes and rat meningeal fibroblasts (MFb), they demonstrated that the administration of 30 nM of siRNA against ephrin-B2 complexed with SuperFectinTMII reagent (silencing efficiency ~84%) was able to ameliorate the effects of transforming growth factor-β1 (TGF-β1)-induced astroglial-fibrotic scar formation and enhance axonal growth of VSC4.1 motoneurons (Figure 3).120
Figure 3.
RNAi targeting in astrocyte. Silencing of inhibiting factors in astrocytes enhanced axonal growth. (A) Ephrin B2-targeting siRNA restored the motor axon outgrown from VSC4.1 culture towards the axon/scar chamber on the microfluidic platform. (Aa) Diagram depicting the design of the microfluidic platform. (Ab) Photograph of the actual device. (Ac) Schemes depicting motor axon growth towards the axon/scar chamber. In the absence of TGF-β1, astrocytes/MFb coculture allows ingrowth of motor axons into the axon/scar chamber; in the presence of TGF-β1, the coculture forms cell clusters, which resemble astrocyte/fibrotic scar, and inhibits the ingrowth of motor axons. (d, d′) The growth of motoneuron axons slowed down as they approached the axon/scar chamber when CSPGs was present in the axon/scar chamber (e, e′), or when the astrocytes/MFb coculture in the axon/scar chamber was treated with TGF-β1 (h, h′). When the axon/scar chamber was added with astrocytes/MFb coculture (f), or astrocytes/MFb coculture with siRNA (g), VSC4.1 motoneurons extended fine and long axons from the soma chamber and entered the microchannels. (i, i′) Addition of TGF-β1 and ephrin B2-targeting siRNA enhanced length of VSC4.1 axons as compared to TGF-β1 alone. Adapted with permission from Y Li et al.120(B) Increased neurite outgrowth on/in conditioned media (CM) from chondroitin polymerizing factor (ChPF)-targeting siRNA-treated Neu7 astrocyte cell line. Cerebellar granule neurons were either grown on a combination of poly-L-lysine (PLL), laminin (LAM) and immobilized CM (a, c) in half DMEM and half neurobasal (NB) + B27 or on a combination of PLL and LAM in half NB + B27 and half CM from the Neu7 cells (b, d). ChPF siRNA attenuated the detrimental effect of CM to neurite outgrowth (e). Adapted with permission from TL Laabs et al.121
Laabs et al. targeted chondroitin polymerizing factor (ChPF), a key enzyme in the chondroitin sulphate proteoglycan (CSPG) biosynthetic pathway, in Neu7 cells (astrocytic cell line) using plasmids encoding shRNA through nucleofection (a method of transfecting nucleic acids into the cellular nucleus). Although the concentration of plasmids used was not stated, they were able to achieve up to 75% uptake efficiency and ~50% silencing efficiency, resulting in reduced CSPG glycosaminoglycans (GAGs) expression by Neu7 cells. The depletion of these inhibitive polysaccharides subsequently promoted axonal growth (Figure 3).121
In another recent work, Smith et al. delivered a modified form of siRNA, siRNA three-way junction (siRNA-3WJ), against astrocyte reactivity effectors, namely lipocalin 2 (Lcn2), glial fibrillary acidic protein (GFAP) and vimentin (Vim) into mice astrocytes using Lipofectamine RNAiMAX. Due to the enhanced potency of siRNA-3WJ, they reported ~87%, ~60% and ~65% gene knockdown of Lcn2, GFAP and Vim, respectively, in activated astrocytes using a dosage of only 5 nM. In addition, the uptake efficiency of the astrocytes, both quiescent and activated, was consistently more than 70%.122 Following this, they proceeded to administer 10 µg of siRNA-3WJ against Lcn2 in mice with a contused spinal cord intralesionally and were able to obtain ~55% reduction in Lcn2 gene expression. Consequently, the contused region showed mitigated levels of Lcn2 protein and was relatively void of cyst formation as compared to the sham control.122 More examples that are relevant to this review have been compiled in Table 2 and readers are advised to refer to it for completeness. Collectively, all these studies suggest that the modulation of astrogliosis is indeed a potential strategy for enhancing nerve and functional recovery.
Table 2.
A summary of the RNAi-induced gene targets of various glial cells for treating spinal cord injury. Commercially available Lipofectamine is the preferred method for transfecting these cells.
| Glial cell type | RNAi effector | Target | Intent | Transfection vehicle and delivery method | Dosage of effectors | Setting | Uptake efficiency | Silencing efficiency | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Astrocytes | siRNA | GFP | Evaluate optimal transfection dosage | Lipofectamine 2000, bolus | 5–80 nM | In vitro | ~80% | ~95% | Ki et al.119 |
| Ephrin-B2 | Attenuate astroglial-fibrotic scar | SuperFectinTMII, bolus | 30 nM | In vitro | – | ~84% | Li et al.120 | ||
| OASIS | Inhibit astrogliosis | PTD-DRBD, intralesion injection | 2 µL from 12.5 µM siRNA stock | In vivo | – | ~50% | Takazawa et al.141 | ||
| GFAP, Vim | Reducing reactivity of astrocytes | Atelocollagen, intralesion injection, photomechanical waves | 25 µL from 10 µM siRNA stock | In vivo | – | – | Ando et al.142 | ||
| siRNA-3WJ | Lcn2, GFAP, Vim | Ameliorate astroglial reactivity | Lipofectamine RNAiMAX, bolus | 5 nM | In vitro | >70% | ~87%, ~60%, ~65%, respectively | Smith et al.122 | |
| Lcn2 | Ameliorate astroglial reactivity | Lipofectamine RNAiMAX, intralesion injection | 10 µg | In vivo | – | ~55% | Smith et al.122 | ||
| Plasmid encoding shRNA | ChPF | Inhibit GAG chain polymerization | Nucleofection | – | In vitro | ~75% | ~50% | Laabs et al.121 | |
| miRNA | miR-494 (CDK6) | Promote astrocyte proliferation | Naked, intrathecal infusion | 20 nM | In vivo | – | ~50% | Wang et al.143 | |
| miR-125b (Sema4D) | Reduce glial scar | Pluronic gel, intralesion application | 10 µM | In vivo | – | ~50% | Diaz Quiroz et al.144 | ||
| ASO | CX43 | Reduce glial scar activation | Pluronic gel, intralesion application | 1–15 µM | In vivo | – | – | Cronin et al.145 | |
| Microglial | siRNA | CD73 | Attenuate inflammation | Lipofectamine 2000, bolus | – | In vitro | – | ~50% | Xu et al.146 |
| GFP | Evaluate optimal transfection dosage | Lipofectamine 2000, bolus | 5–80 nM | In vitro | ~80% | ~40% | Ki et al.119 | ||
| MALAT1 | Reduce inflammatory response | Lipofectamine 2000, bolus | 1 µg | In vitro | – | ~69% | Zhou et al.147 | ||
| Prdx1 | Promote microglial proliferation | Lipofectamine 2000, bolus | – | In vitro | – | ~66% | Huang et al.148 | ||
| RGMa | Reduce microglial inhibition of axonal growth | Lipofectamine 2000, bolus | – | In vitro | – | ~60% | Kitayama et al.149 | ||
| USP4 | Attenuate microglial activation | Lipofectamine 2000, bolus | 100 nM | In vitro | – | ~50% | Jiang et al.150 | ||
| IRF5 | Facilitate macrophage M1 to M2 polarization | Lipofectamine 2000, bolus | 400 nM | In vitro | – | ~50% | Li et al.129 | ||
| IRF5 | Facilitate macrophage M1 to M2 polarization | Cationic liposome, intravenous injection | 0.5 mg siRNA/kg of weight | In vivo | – | – | Li et al.129 | ||
| STAT1 | Reduce inflammatory response | In vivo-jetPEI, intraperitoneal injection | 1 mg siRNA/kg of weight | In vivo | – | >80% | Wu et al.151 | ||
| miRNA | miR-199b (IKKβ) | Ameliorate inflammatory response | Lipofectamine 2000, bolus | – | In vitro | – | ~60% | Zhou et al.130 | |
| miR-124 (TNF-α) | Reduce microglial activation | Chitosan nanoparticles, bolus | 5–50 nM | In vitro | – | ~31% | Louw et al.152 | ||
| miR-124 | Reduce microglial activation | Chitosan nanoparticles, intraspinal injection | 35 ng/site, 3 sites in total | In vivo | – | – | Louw et al.152 | ||
| miR-30a-5p (Neurod1) | Attenuate inflammatory response | Lipofectamine 2000, bolus | – | In vitro | – | ~60% | Fu et al.153 | ||
| miR-199b (IKKβ) | Reduce inflammatory response | Naked, intrathecal injection | 100 nM | In vivo | – | >50% | Zhou et al.130 | ||
| ASO | TNF-α | Reduce pro-inflammatory cytokines | Lipofectamine, GENEfector, NOVAfector, polyet–hylenimine, bolus | 5–200 nM | In vitro | 79%–100% | ~33% to ~78% | Pearse et al.154 | |
| CX43 | Reduce inflammation | Pluronic gel, intralesion application | 1–15 µM | In vivo | – | – | Cronin et al.145 | ||
| Oligodendrocytes/Oligodendrocyte progenitor cells | siRNA | Nogo-A | Promote OLs’ process branching | Lipofectamine 2000, bolus | 50 nM | In vitro | – | ~60% | Zhao et al.139 |
| miRNA | miR-219/miR-338 | Enhance OPCs’ differentiation and maturation | TransIT-TKO, bolus and scaffold-mediated | 50 nM and 4 µg | In vitro | – | ~40% to ~70% | Diao et al.140 | |
| miR-219/miR-338 | Control OPCs’ development | TransIT-TKO, scaffold-mediated | 4 µg | In vitro | – | ~40% to ~70% | Diao et al.71 | ||
| miR-219/miR-338 | Enhance OPCs’ differentiation and myelination | TransIT-TKO, scaffold-mediated | 2 µg | In vitro | – | – | Ong et al.64 | ||
| miR-219/miR-338 | Enhance OPCs’ differentiation and myelination | TransIT-TKO, scaffold-mediated | 2 µg | In vivo | – | – | Milbreta et al.50 |
RNAi: RNA interference; GFAP: glial fibrillary acidic protein; GAG: glycosaminoglycans; ASO: antisense oligonucleotide; IRF5: interferon regulatory factor 5; TNF-α: tumour necrosis factor.
RNAi in microglia
Microglia are the resident innate immune cells within the CNS.123 Following a traumatic injury, microglia become activated and along with monocyte-derived macrophages found in blood, begin releasing ROS and pro-inflammatory cytokines to attract more immune cells to the injured region.124 Once there, these immune sentinels will mediate the confinement of the lesion,125 as well as facilitate debris clearance.126 Studies have also demonstrated that these activities are actually neuroprotective.124,127 While these events appeared to depict rosiness, it is, in reality, very volatile. Specifically, neuroprotective outcomes can easily be replaced with neurotoxic endings if chronic inflammation persists since microglial cells are phenotypically plastic128 and may react negatively when they are continually exposed to inflammatory signals. Yet, precisely because they are extremely plastic, efforts to encourage nerve repair are possible and usually consist of factors that are capable of skewing them towards a restorative phenotype.
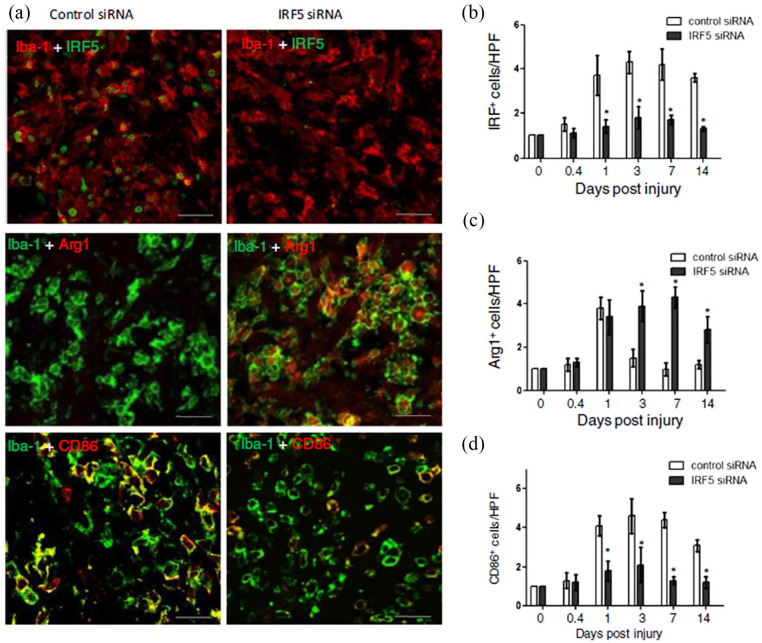
For instance, Li et al. utilized siRNAs to target a transcription factor, interferon regulatory factor 5 (IRF5), known to upregulate genes produced by classically activated macrophages. Using 400 nM of siRNAs against IRF5, they demonstrated that transfected microglial and macrophages isolated from the injured spinal cord had significantly decreased expression of IRF5 (silencing efficiency ~50%) and corresponding amounts of pro-inflammatory cytokines, such as TNF-α, IL-6 and IL-12. Conversely, the anti-inflammatory cytokine IL-10 was enhanced.129 When these siRNAs against IRF5 were intravenously administered into mice with contusion injuries, similar trends with reduction and enhancement of pro-inflammatory (CD86) and anti-inflammatory (Arg1) microglial, respectively, were observed. Importantly, these observations were associative with limited spinal cord tissue damage and improved locomotor functional recovery (Figure 4).129
Figure 4.
RNAi targeting in microglial. Modulation of macrophage phenotype provided a more conducive environment for tissue regeneration. IRF5 siRNA treatment reduced IRF+ cells (a, b) and CD86 presenting M1 macrophage (a, d). On the other hand, it increased Arginase 1 (Arg1) presenting M2 macrophages (a, c) in the wound of SCI animals. Adapted with permission from J Li et al.129
In another study, Zhou et al. utilized miR-199b, which they established as a negative regulator of IKKβ, to modulate production of inflammatory cytokines. They demonstrated in vitro in primary rat microglial that overexpression of miR-199b through transfection of miR-199b mimic led to a corresponding suppression of IKKβ gene expression (silencing efficiency ~60%). As IKKβ positively regulates inflammatory cytokines such as TNF-α and IL-1β, the downregulation of IKKβ also resulted in the downregulation of these cytokines.130 Further administration of 100 nM of miR-199b through intrathecal injection in rats with a compression-induced spinal cord injury similarly attenuated the levels of IKKβ (silencing efficiency > 50%) as well as TNF-α and IL-1β. Subsequent behaviour tests were also performed better by miR-199b-treated rats.130
While it appears that, in these examples mentioned, along with many more in Table 2, the reduction of pro-inflammatory signals is correlated with enhanced functional outcomes, it is advisable to be most prudent when attempting to manipulate the immune cells’ secretome as a treatment option. Besides the contrasting observations in a very recent work suggesting that prescribed inflammation could promote recovery,131 it should be recognized that the response of the immune system is far more complex to influence than it seems given its spatiotemporal sensitivity.
RNAi in OLs
OLs are responsible for forming myelin sheath around the axons in the CNS,132 consequently enabling essential saltatory signal conduction.133 Following a traumatic nerve injury, OLs become necrotic and disintegrate, leaving behind highly inhibiting myelin components that hinder new myelin formation. Without these sheaths, energy-efficient conduction in axons are impeded, resulting in their eventual degeneration and functional impairment.134 By targeting OLs and their immature progenitor cells, oligodendrocyte progenitor cells (OPCs), the intended outcome is straightforward: restore the formation of myelin sheaths in the region. Interestingly, while searching the literature, it was observed that viral-based approaches appeared to be the preferred choice135–138 for enacting RNAi in OLs to enhance their myelinating capacity and subsequent extent of recovery from nerve injuries. Despite this trend, it does not imply that non-viral delivery methods are ineffective for transfecting OLs.
Zhao et al. demonstrated that Lipofectamine 2000 could be used to transfect primary OL culture with siRNAs against Nogo-A (50 nM) and achieve a ~60% gene knockdown efficiency. Besides that, they showed that the suppression of Nogo-A in OLs enhanced its process branching, which is a crucial event for initiating myelination.139
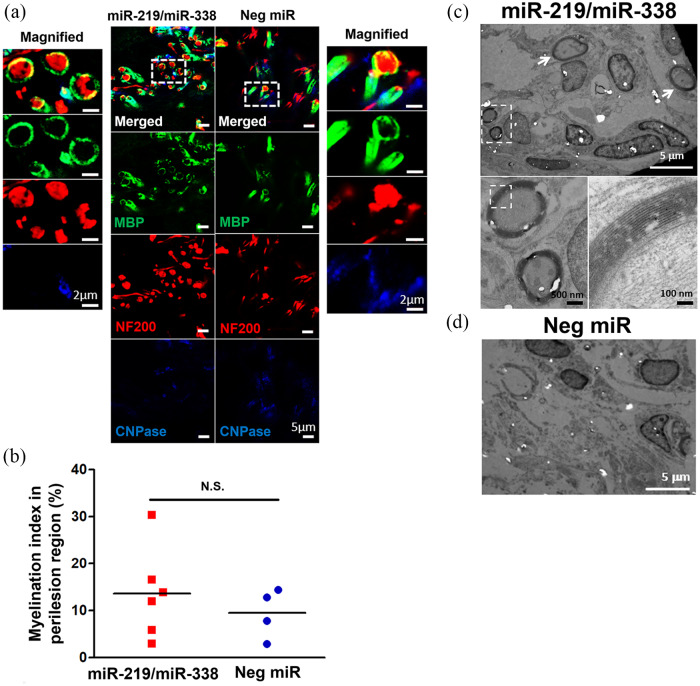
In our lab, we employed a non-viral, scaffold-mediated strategy to transfect OLs. Specifically, we complexed a cocktail of miR-219 and miR-338 (2–4 µg or ~200–400 nM of miRs in total) with a commercial transfection vehicle, TransIT-TKO, and adsorbed these complexes onto polydopamine-coated electrospun fibrous scaffolds before allowing primary rat OPCs to be reverse transfected after they were seeded onto these scaffolds. Through this method of delivery, we were able to obtain ~40% to ~70% of gene silencing efficiency71,140 as well as observe the biological effects of miR-219/miR-338 in enhancing OPC differentiation64,140 and OL myelination.50,64 Furthermore, when we loaded the miR-TKO complex into a three-dimensional (3D) hybrid scaffold containing electrospun fibres and collagen and implanted it into a spinally injured rat, we found that despite the inhibitive microenvironment, miR-219/miR-338 was able to preserve the viability of oligodendroglial lineage cells, promote the extent and rate of their differentiation, and also most importantly, augment their myelinating capacity (Figure 5 and Table 2).50
Figure 5.
RNAi targeting in oligodendrocytes. Promoting oligodendrocyte remyelination will facilitate the functionalization of regenerated axons. (a) Representative images obtained at Week 4 depicting MBP+ tubular structures surrounding NF+ axons. Scale bar represents 5 µm in the normal images and 2 µm in the magnified images. (b) Myelination index obtained at host-implant interface. (c, d) Representative transmission electron microscopy images showing the (c) presence and (d) lack of myelinated axon formation in (c) miR-219/miR-338 and (d) Neg miR groups, respectively. N.S.: not significant (Student’s T-test). Scale bars for the TEM images are labelled with their respective scale bar size. Adapted with permission from U Milbreta et al.50
Authors’ perspectives and conclusion
In our opinion, the discovery of RNAi and the effectors driving this process is both ground-breaking and exciting. Specifically, the effector nucleic acids are able to provide more utility as compared to normal conventional drugs. For instance, siRNAs can be used to target a single specific gene while miRNAs are often used for targeting multiple genes. On the contrary, shRNAs can be employed to prolong gene regulation with the added benefit of being switchable (on or off). Finally, ASOs can be modified to be used for naked and efficient delivery. However, there are also some downsides pertaining to the use of these nucleic acids for RNAi such as competition with endogenous RNAs,155 possible activation of innate immune responses156 and also accidental suppression of off-target genes.157 In addition, there is always a need to consider the method of intracellular delivery, which usually involves their complexation with a cationic delivery vehicle or encapsulation into nanoparticles.
In the context of spinal cord injury, we feel that miRNAs have the greatest potential for treatment purposes. Recent RNA sequencing of the rodent spinal cord158,159 revealed thousands of dysregulated genes post traumatic injury. Given the ability of miRNAs to regulate numerous genes concurrently,24 future studies can be conducted to identify the miRNAs required to re-regulate these dysregulated genes back to normalcy and perhaps to map the regulated pathways to the respective cell type.
RNAi holds great potential for treating injuries and diseases, which are frequently associated with dysregulated cellular gene and protein expressions. A major concern, as discussed in this review, is the method to efficiently deliver RNAi drivers into the traumatically injured CNS in order to regulate cellular gene and protein expressions towards an advantageous state, where functional recovery may be attained. Glial cells, being more abundant than neurons, are ideal targets for RNAi therapy. Besides being more amenable than neurons in accepting cationic carrier-complexed small nucleic acids, glial cells are also more proficient in shaping the injured/diseased milieu, through signalling molecules to enhance tissue regrowth. More importantly, besides the previously approved ASO-based therapeutics, with the recent (2018) FDA approval of Onpattro (patisiran), a siRNA-based therapeutic for the treatment of peripheral nerve disease (polyneuropathy) in adult patients, RNAi has become a clinically viable treatment option. Following this landmark acceptance, we can now anticipate that other potent and safe RNAi therapeutics will soon be approved as well. A greater implication of this approval, however, is the motivation for researchers to look towards RNAi as a preferred treatment choice. Specifically, as described, using RNAi to elicit changes in glial cells may provide an avenue to attain a more permissive environment for neuronal regeneration and/or locomotor recovery.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: SY Chew and J Lin would like to acknowledge partial funding support from the Singapore National Research Foundation under its National Medical Research Council Cooperative Basic Research Grant (NMRC-CBRG, NMRC/CBRG/0096/2015) and administered by the Singapore Ministry of Health’s National Medical Research Council; Ministry of Education Tier 1 grant (RG38/19). This research was supported in part by the National Research Foundation (NRF), Republic of Korea (2015K1A1A2032163, 2018K1A4A3A01064257, 2018R1A2B3003446).
ORCID iDs: Hae-Won Kim  https://orcid.org/0000-0001-6400-6100
https://orcid.org/0000-0001-6400-6100
Sing Yian Chew  https://orcid.org/0000-0002-6084-5967
https://orcid.org/0000-0002-6084-5967
References
- 1. Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391: 806–811. [DOI] [PubMed] [Google Scholar]
- 2. Couzin J. Small RNAs make big splash. Science 2002; 298: 2296–2297. [DOI] [PubMed] [Google Scholar]
- 3. Farrell RE. Chapter 23 – RNAi: take a RISC – role the dicer. In: RE Farrell. (ed.) RNA methodologies. 4th ed. San Diego, CA: Academic Press, 2010, pp. 539–560. [Google Scholar]
- 4. Kim D, Rossi J. RNAi mechanisms and applications. Biotechniques 2008; 44: 613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Ann Rev Biophys 2013; 42: 217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nieto-Díaz M, Esteban F, Reigada D, et al. MicroRNA dysregulation in spinal cord injury: causes, consequences and therapeutics. Front Cell Neurosci 2014; 8: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Obbard DJ, Gordon KHJ, Buck AH, et al. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci 2009; 364: 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrington JC, Ambros V. Role of MicroRNAs in plant and animal development. Science 2003; 301: 336–338. [DOI] [PubMed] [Google Scholar]
- 9. Jiao S, Liu Y, Yao Y, et al. miR-124 promotes proliferation and differentiation of neuronal stem cells through inactivating Notch pathway. Cell Biosci 2017; 7: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng L-C, Pastrana E, Tavazoie M, et al. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 2009; 12: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dajas-Bailador F, Bonev B, Garcez P, et al. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci 2012; 15: 697–699. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y, Ueno Y, Liu XS, et al. The MicroRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J Neurosci 2013; 33: 6885–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vo N, Klein ME, Varlamova O, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A 2005; 102: 16426–16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao X, He X, Han X, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 2010; 65: 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tigchelaar S, Streijger F, Sinha S, et al. Serum microRNAs reflect injury severity in a large animal model of thoracic spinal cord injury. Sci Rep 2017; 7: 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madathil SK, Nelson PT, Saatman KE, et al. MicroRNAs in CNS injury: potential roles and therapeutic implications. Bioessays 2011; 33: 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci 2006; 7: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dana H, Chalbatani GM, Mahmoodzadeh H, et al. Molecular mechanisms and biological functions of siRNA. Int J Biomed Sci 2017; 13: 48–57. [PMC free article] [PubMed] [Google Scholar]
- 19. Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005; 122: 553–563. [DOI] [PubMed] [Google Scholar]
- 20. Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol 1999; 216: 671–680. [DOI] [PubMed] [Google Scholar]
- 21. Seggerson K, Tang L, Moss EG. Two genetic circuits repress the caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol 2002; 243: 215–225. [DOI] [PubMed] [Google Scholar]
- 22. Rhoades MW, Reinhart BJ, Lim LP, et al. Prediction of plant MicroRNA targets. Cell 2002; 110: 513–520. [DOI] [PubMed] [Google Scholar]
- 23. Llave C, Xie Z, Kasschau KD, et al. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002; 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- 24. Giza DE, Vasilescu C, Calin GA. Key principles of miRNA involvement in human diseases. Discoveries 2014; 2: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 26. Bian S, Xu T-l, Sun T. Tuning the cell fate of neurons and glia by microRNAs. Curr Opin Neurobiol 2013; 23: 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shatseva T, Lee DY, Deng Z, et al. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci 2011; 124: 2826–2836. [DOI] [PubMed] [Google Scholar]
- 28. Visvanathan J, Lee S, Lee B, et al. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 2007; 21: 744–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mondanizadeh M, Arefian E, Mosayebi G, et al. MicroRNA-124 regulates neuronal differentiation of mesenchymal stem cells by targeting Sp1 mRNA. J Cell Biochem 2015; 116: 943–953. [DOI] [PubMed] [Google Scholar]
- 30. Marasa BS, Srikantan S, Martindale JL, et al. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging 2010; 2: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bu H, Wedel S, Cavinato M, et al. MicroRNA regulation of oxidative stress-induced cellular senescence. Oxid Med Cell Longev 2017; 2017: 2398696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams J, Smith F, Kumar S, et al. Are microRNAs true sensors of ageing and cellular senescence? Ageing Res Rev 2017; 35: 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature 2005; 435: 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fawzy MS, Toraih EA, Ibrahiem A, et al. Evaluation of miRNA-196a2 and apoptosis-related target genes: ANXA1, DFFA and PDCD4 expression in gastrointestinal cancer patients: a pilot study. PLoS ONE 2017; 12: e0187310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore CB, Guthrie EH, Huang MT-H, Taxman DJ. Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol Biol 2010; 629: 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herold MJ, van den Brandt J, Seibler J, et al. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci U S A 2008; 105: 18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsushita N, Matsushita S, Hirakawa S, et al. Doxycycline-dependent inducible and reversible RNA interference mediated by a single lentivirus vector. Biosci Biotechnol Biochem 2013; 77: 776–781. [DOI] [PubMed] [Google Scholar]
- 38. Muntoni F, Wood MJA. Targeting RNA to treat neuromuscular disease. Nat Rev Drug Discov 2011; 10: 621. [DOI] [PubMed] [Google Scholar]
- 39. Opalinska JB, Gewirtz AM. Nucleic-acid therapeutics: basic principles and recent applications. Nat Rev Drug Discov 2002; 1: 503–514. [DOI] [PubMed] [Google Scholar]
- 40. Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci U S A 1978; 75: 285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rinaldi C, Wood MJA. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol 2017; 14: 9. [DOI] [PubMed] [Google Scholar]
- 42. Watanabe TA, Geary RS, Levin AA. Plasma protein binding of an antisense oligonucleotide targeting human ICAM-1 (ISIS 2302). Oligonucleotides 2006; 16: 169–180. [DOI] [PubMed] [Google Scholar]
- 43. Rifai A, Brysch W, Fadden K, et al. Clearance kinetics, biodistribution, and organ saturability of phosphorothioate oligodeoxynucleotides in mice. Am J Pathol 1996; 149: 717–725. [PMC free article] [PubMed] [Google Scholar]
- 44. McKay RA, Miraglia LJ, Cummins LL, et al. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-α expression. J Biol Chem 1999; 274: 1715–1722. [DOI] [PubMed] [Google Scholar]
- 45. Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res 1997; 25: 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lubini P, Zürcher W, Egli M. Stabilizing effects of the RNA 2′-substituent: crystal structure of an oligodeoxynucleotide duplex containing 2′-O-methylated adenosines. Chem Biol 1994; 1: 39–45. [DOI] [PubMed] [Google Scholar]
- 47. Hamm S, Latz E, Hangel D, et al. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology 2010; 215: 559–569. [DOI] [PubMed] [Google Scholar]
- 48. Chery J. RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc J 2016; 4: 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jacobson DR, Saleh OA. Counting the ions surrounding nucleic acids. Nucleic Acids Res 2016; 45: 1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Milbreta U, Lin J, Pinese C, et al. Scaffold-mediated sustained, non-viral delivery of miR-219/miR-338 promotes CNS remyelination. Mol Ther 2019; 27: 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Milbreta U, Nguyen LH, Diao H, et al. Three-dimensional nanofiber hybrid scaffold directs and enhances axonal regeneration after spinal cord injury. ACS Biomater Sci Eng 2016; 2: 1319–1329. [DOI] [PubMed] [Google Scholar]
- 52. Gao W, Li J. Targeted siRNA delivery reduces nitric oxide mediated cell death after spinal cord injury. J Nanobiotechnology 2017; 15: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moreno PMD, Ferreira AR, Salvador D, et al. Hydrogel-assisted antisense LNA gapmer delivery for in situ gene silencing in spinal cord injury. Mol Ther Nucleic Acids 2018; 11: 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang N, Milbreta U, Chin JS, et al. Biomimicking fiber scaffold as an effective in vitro and in vivo microRNA screening platform for directing tissue regeneration. Adv Sci 2019; 6: 1800808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Finkel RS, Chiriboga CA, Vajsar J, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 2016; 388: 3017–3026. [DOI] [PubMed] [Google Scholar]
- 56. Miller T, Smith R, Pestronk A, et al. Results of a phase 1, double-blind, placebo-controlled, dose-escalation study of the safety, tolerability, and pharmacokinetics of ISIS 333611 administered intrathecally to patients with familial ALS due to SOD1 gene mutations (S25.001). Neurology 2012; 78: S25.001. [Google Scholar]
- 57. Song K-H, Harvey BK, Borden MA. State-of-the-art of microbubble-assisted blood-brain barrier disruption. Theranostics 2018; 8: 4393–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hynynen K, McDannold N, Vykhodtseva N, et al. Noninvasive MR imaging-guided focal opening of the blood-brain barrier in rabbits. Radiology 2001; 220: 640–646. [DOI] [PubMed] [Google Scholar]
- 59. Kovacs ZI, Kim S, Jikaria N, et al. Disrupting the blood-brain barrier by focused ultrasound induces sterile inflammation. Proc Natl Acad Sci U S A 2017; 114: E75–e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Senn C, Hangartner C, Moes S, et al. Central administration of small interfering RNAs in rats: a comparison with antisense oligonucleotides. Eur J Pharmacol 2005; 522: 30–37. [DOI] [PubMed] [Google Scholar]
- 61. Dalby B, Cates S, Harris A, et al. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 2004; 33: 95–103. [DOI] [PubMed] [Google Scholar]
- 62. Lasiecka ZM, Winckler B. Chapter 19 – studying endosomes in cultured neurons by live-cell imaging. In: KK Pfister. (ed.) Methods in cell biology. San Diego, CA: Academic Press, 2016, pp. 389–408. [DOI] [PubMed] [Google Scholar]
- 63. Akhtar S, Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: potential impact on siRNA-mediated gene silencing activity and specificity. Adv Drug Deliv Rev 2007; 59: 164–182. [DOI] [PubMed] [Google Scholar]
- 64. Ong W, Lin J, Bechler ME, et al. Microfiber drug/gene delivery platform for study of myelination. Acta Biomater 2018; 75: 152–160. [DOI] [PubMed] [Google Scholar]
- 65. Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 1995; 92: 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest 2007; 117: 3623–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim HS, Yoo HS. Matrix metalloproteinase-inspired suicidal treatments of diabetic ulcers with siRNA-decorated nanofibrous meshes. Gene Ther 2013; 20: 378–385. [DOI] [PubMed] [Google Scholar]
- 68. Nabzdyk CS, Chun MC, Oliver-Allen HS, et al. Gene silencing in human aortic smooth muscle cells induced by PEI-siRNA complexes released from dip-coated electrospun poly(ethylene terephthalate) grafts. Biomaterials 2014; 35: 3071–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Low WC, Rujitanaroj P-O, Lee D-K, et al. Nanofibrous scaffold-mediated REST knockdown to enhance neuronal differentiation of stem cells. Biomaterials 2013; 34: 3581–3590. [DOI] [PubMed] [Google Scholar]
- 70. Chooi WH, Ong W, Murray A, et al. Scaffold mediated gene knockdown for neuronal differentiation of human neural progenitor cells. Biomater Sci 2018; 6: 3019–3029. [DOI] [PubMed] [Google Scholar]
- 71. Diao HJ, Low WC, Lu QR, et al. Topographical effects on fiber-mediated microRNA delivery to control oligodendroglial precursor cells development. Biomaterials 2015; 70: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rujitanaroj PO, Jao B, Yang J, et al. Controlling fibrous capsule formation through long-term down-regulation of collagen type I (COL1A1) expression by nanofiber-mediated siRNA gene silencing. Acta Biomater 2013; 9: 4513–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rujitanaroj PO, Wang YC, Wang J, et al. Nanofiber-mediated controlled release of siRNA complexes for long term gene-silencing applications. Biomaterials 2011; 32: 5915–5923. [DOI] [PubMed] [Google Scholar]
- 74. Modaresi S, Pacelli S, Whitlow J, et al. Deciphering the role of substrate stiffness in enhancing the internalization efficiency of plasmid DNA in stem cells using lipid-based nanocarriers. Nanoscale 2018; 10: 8947–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Missirlis D. The effect of substrate elasticity and actomyosin contractility on different forms of endocytosis. PLoS ONE 2014; 9: e96548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fliervoet LAL, Engbersen JFJ, Schiffelers RM, et al. Polymers and hydrogels for local nucleic acid delivery. J Mater Chem B 2018; 6: 5651–5670. [DOI] [PubMed] [Google Scholar]
- 77. Nguyen LH, Gao M, Lin J, et al. Three-dimensional aligned nanofibers-hydrogel scaffold for controlled non-viral drug/gene delivery to direct axon regeneration in spinal cord injury treatment. Scientific Reports 2017; 7: 42212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Catuogno S, Esposito CL, Condorelli G, et al. Nucleic acids delivering nucleic acids. Adv Drug Deliv Rev 2018; 134: 79–93. [DOI] [PubMed] [Google Scholar]
- 79. Valente LA, Begg LR, Filiano AJ. Updating neuroimmune targets in central nervous system dysfunction. Trends Pharmacol Sci 2019; 40: 482–494. [DOI] [PubMed] [Google Scholar]
- 80. Jäkel S, Dimou L. Glial cells and their function in the adult brain: a journey through the history of their ablation. Front Cell Neurosci 2017; 11: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Verkhratsky A, Ho MS, Zorec R, et al. The concept of neuroglia. Adv Exp Med Biol 2019; 1175: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Verkhratsky A. Physiology of neuronal-glial networking. Neurochem Int 2010; 57: 332–343. [DOI] [PubMed] [Google Scholar]
- 83. Stephenson J, Nutma E, van der Valk P, et al. Inflammation in CNS neurodegenerative diseases. Immunology 2018; 154: 204–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pardieck J, Sakiyama-Elbert S. Genome engineering for CNS injury and disease. Curr Opin Biotechnol 2018; 52: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gitler AD, Dhillon P, Shorter J. Neurodegenerative disease: models, mechanisms, and a new hope. Dis Model Mech 2017; 10: 499–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Prinz M, Priller J. The role of peripheral immune cells in the CNS in steady state and disease. Nat Neurosci 2017; 20: 136–144. [DOI] [PubMed] [Google Scholar]
- 87. Meyer-Luehmann M, Prinz M. Myeloid cells in Alzheimer’s disease: culprits, victims or innocent bystanders? Trends Neurosci 2015; 38: 659–668. [DOI] [PubMed] [Google Scholar]
- 88. Burda JE, Sofroniew MV. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 2014; 81: 229–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stobart JL, Anderson CM. Multifunctional role of astrocytes as gatekeepers of neuronal energy supply. Front Cell Neurosci 2013; 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science 2016; 353: 777–783. [DOI] [PubMed] [Google Scholar]
- 91. Assefa BT, Gebre AK, Altaye BM. Reactive astrocytes as drug target in Alzheimer’s disease. Biomed Res Int 2018; 2018: 4160247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sofroniew MV. Astrocyte barriers to neurotoxic inflammation. Nat Rev Neurosci 2015; 16: 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Acosta C, Anderson HD, Anderson CM. Astrocyte dysfunction in Alzheimer disease. J Neurosci Res 2017; 95: 2430–2447. [DOI] [PubMed] [Google Scholar]
- 94. Liddelow SA, Guttenplan KA, Clarke LE, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017; 541: 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Marin IA, Kipnis J. Central nervous system: (immunological) ivory tower or not? Neuropsychopharmacology 2017; 42: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wolf SA, Boddeke HW, Kettenmann H. Microglia in physiology and disease. Ann Rev Physiol 2017; 79: 619–643. [DOI] [PubMed] [Google Scholar]
- 97. Colonna M, Butovsky O. Microglia function in the central nervous system during health and neurodegeneration. Annu Rev Immunol 2017; 35: 441–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ma Y, Wang J, Wang Y, et al. The biphasic function of microglia in ischemic stroke. Prog Neurobiol 2017; 157: 247–272. [DOI] [PubMed] [Google Scholar]
- 99. Perry VH, Teeling J. Microglia and macrophages of the central nervous system: the contribution of microglia priming and systemic inflammation to chronic neurodegeneration. Semin Immunopathol 2013; 35: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cunningham C. Microglia and neurodegeneration: the role of systemic inflammation. Glia 2013; 61: 71–90. [DOI] [PubMed] [Google Scholar]
- 101. Deczkowska A, Keren-Shaul H, Weiner A, et al. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell 2018; 173: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 102. Hansen DV, Hanson JE, Sheng M. Microglia in Alzheimer’s disease. J Cell Biol 2018; 217: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xu L, He D, Bai Y. Microglia-mediated inflammation and neurodegenerative disease. Mol Neurobiol 2016; 53: 6709–6715. [DOI] [PubMed] [Google Scholar]
- 104. Barateiro A, Brites D, Fernandes A. Oligodendrocyte development and myelination in neurodevelopment: molecular mechanisms in health and disease. Curr Pharm Des 2016; 22: 656–679. [DOI] [PubMed] [Google Scholar]
- 105. Bergles DE, Richardson WD. Oligodendrocyte development and plasticity. Cold Spring Harb Perspect Biol 2015; 8: a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nave KA. Myelination and support of axonal integrity by glia. Nature 2010; 468: 244–252. [DOI] [PubMed] [Google Scholar]
- 107. Bradl M, Lassmann H. Oligodendrocytes: biology and pathology. Acta Neuropathol 2010; 119: 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Peferoen L, Kipp M, van der Valk P, et al. Oligodendrocyte-microglia cross-talk in the central nervous system. Immunology 2014; 141: 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cai Z, Xiao M. Oligodendrocytes and Alzheimer’s disease. Int J Neurosci 2016; 126: 97–104. [DOI] [PubMed] [Google Scholar]
- 110. Tognatta R, Miller RH. Contribution of the oligodendrocyte lineage to CNS repair and neurodegenerative pathologies. Neuropharmacology 2016; 110: 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wang H-f, Liu X-k, Li R, et al. Effect of glial cells on remyelination after spinal cord injury. Neural Regen Res 2017; 12: 1724–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Herculano-Houzel S. The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 2014; 62: 1377–1391. [DOI] [PubMed] [Google Scholar]
- 113. Nortley R, Attwell D. Control of brain energy supply by astrocytes. Curr Opin Neurobiol 2017; 47: 80–85. [DOI] [PubMed] [Google Scholar]
- 114. Filosa JA, Bonev AD, Straub SV, et al. Local potassium signaling couples neuronal activity to vasodilation in the brain. Nat Neurosci 2006; 9: 1397–1403. [DOI] [PubMed] [Google Scholar]
- 115. Petr GT, Sun Y, Frederick NM, et al. Conditional deletion of the Glutamate transporter GLT-1 reveals that astrocytic GLT-1 protects against fatal epilepsy while neuronal GLT-1 contributes significantly to glutamate uptake into synaptosomes. J Neurosci 2015; 35: 5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sofroniew MV. Astrogliosis. Cold Spring Harb Perspect Biol 2015; 7: a020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Cregg JM, DePaul MA, Filous AR, et al. Functional regeneration beyond the glial scar. Exp Neurol 2014; 253: 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. McKeon RJ, Schreiber RC, Rudge JS, et al. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J Neurosci 1991; 11: 3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ki KH, Park DY, Lee SH, et al. The optimal concentration of siRNA for gene silencing in primary cultured astrocytes and microglial cells of rats. Korean J Anesthesiol 2010; 59: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Li Y, Chen Y, Tan L, et al. RNAi-mediated ephrin-B2 silencing attenuates astroglial-fibrotic scar formation and improves spinal cord axon growth. CNS Neurosci Ther 2017; 23: 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Laabs TL, Wang H, Katagiri Y, et al. Inhibiting glycosaminoglycan chain polymerization decreases the inhibitory activity of astrocyte-derived chondroitin sulfate proteoglycans. J Neurosci 2007; 27: 14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Smith JA, Braga A, Verheyen J, et al. RNA nanotherapeutics for the amelioration of astroglial reactivity. Mol Ther Nucleic Acids 2018; 10: 103–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lehnardt S. Innate immunity and neuroinflammation in the CNS: the role of microglia in Toll-like receptor-mediated neuronal injury. Glia 2010; 58: 253–263. [DOI] [PubMed] [Google Scholar]
- 124. Kroner A, Rosas Almanza J. Role of microglia in spinal cord injury. Neurosci Lett 2019; 709: 134370. [DOI] [PubMed] [Google Scholar]
- 125. Hines DJ, Hines RM, Mulligan SJ, et al. Microglia processes block the spread of damage in the brain and require functional chloride channels. Glia 2009; 57: 1610–1618. [DOI] [PubMed] [Google Scholar]
- 126. Akhmetzyanova E, Kletenkov K, Mukhamedshina Y, et al. Different approaches to modulation of microglia phenotypes after spinal cord injury. Front Syst Neurosci 2019; 13: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bellver-Landete V, Bretheau F, Mailhot B, et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat Commun 2019; 10: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. David S, Kroner A. Repertoire of microglial and macrophage responses after spinal cord injury. Nat Rev Neurosci 2011; 12: 388–399. [DOI] [PubMed] [Google Scholar]
- 129. Li J, Liu Y, Xu H, et al. Nanoparticle-delivered IRF5 siRNA facilitates M1 to M2 transition, reduces demyelination and neurofilament loss, and promotes functional recovery after spinal cord injury in mice. Inflammation 2016; 39: 1704–1717. [DOI] [PubMed] [Google Scholar]
- 130. Zhou H-J, Wang L-Q, Xu Q-S, et al. Downregulation of miR-199b promotes the acute spinal cord injury through IKKβ-NF-κB signaling pathway activating microglial cells. Exp Cell Res 2016; 349: 60–67. [DOI] [PubMed] [Google Scholar]
- 131. Torres-Espín A, Forero J, Fenrich KK, et al. Eliciting inflammation enables successful rehabilitative training in chronic spinal cord injury. Brain 2018; 141: 1946–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Almad A, Sahinkaya FR, McTigue DM. Oligodendrocyte fate after spinal cord injury. Neurotherapeutics 2011; 8: 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Miron VE, Kuhlmann T, Antel JP. Cells of the oligodendroglial lineage, myelination, and remyelination. Biochim Biophys Acta 2011; 1812: 184–193. [DOI] [PubMed] [Google Scholar]
- 134. Franklin RJM, Ffrench-Constant C. Regenerating CNS myelin – from mechanisms to experimental medicines. Nat Rev Neurosci 2017; 18: 753–769. [DOI] [PubMed] [Google Scholar]
- 135. Wu H-F, Cen J-S, Zhong Q, et al. The promotion of functional recovery and nerve regeneration after spinal cord injury by lentiviral vectors encoding Lingo-1 shRNA delivered by Pluronic F-127. Biomaterials 2013; 34: 1686–1700. [DOI] [PubMed] [Google Scholar]
- 136. Zhang H-L, Wang J, Tang L. Sema4D knockdown in oligodendrocytes promotes functional recovery after spinal cord injury. Cell Biochem Biophys 2014; 68: 489–496. [DOI] [PubMed] [Google Scholar]
- 137. Mi S, Miller RH, Lee X, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci 2005; 8: 745–751. [DOI] [PubMed] [Google Scholar]
- 138. Yu CG, Li Y, Raza K, et al. Calpain 1 knockdown improves tissue sparing and functional outcomes after spinal cord injury in rats. J Neurotrauma 2012; 30: 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zhao X, Wu J, Kuang F, et al. Silencing of Nogo-A in rat oligodendrocyte cultures enhances process branching. Neurosci Lett 2011; 499: 32–36. [DOI] [PubMed] [Google Scholar]
- 140. Diao HJ, Low WC, Milbreta U, et al. Nanofiber-mediated microRNA delivery to enhance differentiation and maturation of oligodendroglial precursor cells. J Control Release 2015; 208: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Takazawa A, Kamei N, Adachi N, et al. Endoplasmic reticulum stress transducer old astrocyte specifically induced substance contributes to astrogliosis after spinal cord injury. Neural Regen Res 2018; 13: 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ando T, Sato S, Toyooka T, et al. Photomechanical wave-driven delivery of siRNAs targeting intermediate filament proteins promotes functional recovery after spinal cord injury in rats. PLoS ONE 2012; 7: e51744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang Y, Sun JC, Wang HB, et al. Effects of microRNA-494 on astrocyte proliferation and synaptic remodeling in the spinal cord of a rat model of chronic compressive spinal cord injury by regulating the Nogo/Ngr signaling pathway. Cell Physiol Biochem 2018; 48: 919–933. [DOI] [PubMed] [Google Scholar]
- 144. Diaz Quiroz JF, Tsai E, Coyle M, et al. Precise control of miR-125b levels is required to create a regeneration-permissive environment after spinal cord injury: a cross-species comparison between salamander and rat. Dis Model Mech 2014; 7: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Cronin M, Anderson PN, Cook JE, et al. Blocking connexin43 expression reduces inflammation and improves functional recovery after spinal cord injury. Mol Cell Neurosci 2008; 39: 152–160. [DOI] [PubMed] [Google Scholar]
- 146. Xu S, Zhu W, Shao M, et al. Ecto-5′-nucleotidase (CD73) attenuates inflammation after spinal cord injury by promoting macrophages/microglia M2 polarization in mice. J Neuroinflammation 2018; 15: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zhou H-J, Wang L-Q, Wang D-B, et al. Long noncoding RNA MALAT1 contributes to inflammatory response of microglia following spinal cord injury via the modulation of a miR-199b/IKKβ/NF-κB signaling pathway. Am J Physiol Cell Physiol 2018; 315: C52–C61. [DOI] [PubMed] [Google Scholar]
- 148. Huang S, Liu X, Zhang J, et al. Expression of peroxiredoxin 1 after traumatic spinal cord injury in rats. Cell Mol Neurobiol 2015; 35: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kitayama M, Ueno M, Itakura T, et al. Activated microglia inhibit axonal growth through RGMa. PLoS ONE 2011; 6: e25234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Jiang X, Yu M, Ou Y, et al. Downregulation of USP4 promotes activation of microglia and subsequent neuronal inflammation in rat spinal cord after injury. Neurochem Res 2017; 42: 3245–3253. [DOI] [PubMed] [Google Scholar]
- 151. Wu Y, Yang L, Mei X, et al. Selective inhibition of STAT1 reduces spinal cord injury in mice. Neurosci Lett 2014; 580: 7–11. [DOI] [PubMed] [Google Scholar]
- 152. Louw AM, Kolar MK, Novikova LN, et al. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomedicine 2016; 12: 643–653. [DOI] [PubMed] [Google Scholar]
- 153. Fu X, Shen Y, Wang W, et al. MiR-30a-5p ameliorates spinal cord injury-induced inflammatory responses and oxidative stress by targeting Neurod 1 through MAPK/ERK signalling. Clin Exp Pharmacol Physiol 2018; 45: 68–74. [DOI] [PubMed] [Google Scholar]
- 154. Pearse DD, Pereira FC, Stolyarova A, et al. Inhibition of tumour necrosis factor-α by antisense targeting produces immunophenotypical and morphological changes in injury-activated microglia and macrophages. Euro J Neurosci 2004; 20: 3387–3396. [DOI] [PubMed] [Google Scholar]
- 155. Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006; 441: 537–541. [DOI] [PubMed] [Google Scholar]
- 156. Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 2005; 11: 263–270. [DOI] [PubMed] [Google Scholar]
- 157. Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 2003; 21: 635–637. [DOI] [PubMed] [Google Scholar]
- 158. Li Y, Chen Y, Li X, et al. RNA sequencing screening of differentially expressed genes after spinal cord injury. Neural Regen Res 2019; 14: 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Poplawski GHD, Kawaguchi R, Van Niekerk E, et al. Injured adult neurons regress to an embryonic transcriptional growth state. Nature 2020; 581: 77–82. [DOI] [PubMed] [Google Scholar]