Abstract
The analysis of CpG methylation in circulating tumor DNA fragments has emerged as a promising approach for the noninvasive early detection of solid tumors, including colorectal cancer (CRC). The most commonly employed assay involves bisulfite conversion of circulating tumor DNA, followed by targeted PCR, then real-time quantitative PCR (alias methylation-specific PCR). This report demonstrates the ability of a multiplex bisulfite PCR–ligase detection reaction–real-time quantitative PCR assay to detect seven methylated CpG markers (CRC or colon specific), in both simulated (approximately 30 copies of fragmented CRC cell line DNA mixed with approximately 3000 copies of fragmented peripheral blood DNA) and CRC patient–derived cell-free DNAs. This scalable assay is designed for multiplexing and incorporates steps for improved sensitivity and specificity, including the enrichment of methylated CpG fragments, ligase detection reaction, the incorporation of ribose bases in primers, and use of uracil DNA glycosylase. Six of the seven CpG markers (located in promoter regions of PPP1R16B, KCNA3, CLIP4, GDF6, SEPT9, and GSG1L) were identified through integrated analyses of genome-wide methylation data sets for 31 different types of cancer. These markers were mapped to CpG sites at the promoter region of VIM; VIM and SEPT9 are established epigenetic markers of CRC. Additional bioinformatics analyses show that the methylation at these CpG sites negatively correlates with the transcription of their corresponding genes.
Colorectal cancer (CRC) is the third most common cancer worldwide, with >1.8 million new cases in 2018.1 The global burden of CRC is projected to increase by 60% to >2.2 million new cases and 1.1 million deaths by 2030.1 The key to reducing mortality due to CRC is early detection, as it provides an opportunity to surgically remove premalignant lesions. Catching CRC at early stage saves lives, with 5-year survival rates (US data) for localized, regional, and distant CRC being 91%, 72%, and 13%, respectively.2
Colonoscopy still remains the standard screening tool for CRC, and is recommended at the age of 50 years (with 5- to 10-year screening intervals).3 Nevertheless, compliance rate for colonoscopy is only at 30% to 55%.4 What may hinder patients from visiting gastroenterologists is the uncomfortable cleansing preparation and the highly invasive nature of the procedure,4 prompting the pursuit of noninvasive and inexpensive alternatives. Such alternatives include procedures that detect heme (in case of fecal immunochemical test) or human globin (in case of fecal occult blood test) from blood released from advanced adenomas and adenocarcinomas into stool samples.5 Unfortunately, fecal tests also have low specificity and sensitivity, low compliance, and variability in the test results because of many confounding factors.6,7
The limitations of the current tests used to detect CRC encourage persistent efforts toward developing noninvasive screening approaches, such as liquid biopsies. Indeed, many assays to detect CRC biomarkers, based on circulating tumor cells, proteins, cell-free DNA (cfDNA), and RNA, have been identified and validated in various clinical settings.8 One of the underlying foundations of cancer liquid biopsy is the release of cfDNA from apoptotic cancer cells into the blood. The isolated cfDNA contains the same molecular aberrations as the solid tumors (mutations, hypermethylation or hypomethylation, copy number changes, or chromosomal rearrangements).9 Among the molecular markers present in cfDNA isolated from the blood of CRC patients are KRAS and BRAF mutations.10, 11, 12 In general, these mutation markers are found in late-stage primary cancers and metastases.10 Tests to detect these mutations from blood samples are limited because only 33% and 13% of all CRC tumors contain mutations for KRAS and BRAF, respectively.13 Furthermore, these mutations are commonly found in many different solid tumors, confounding the ability to determine where the cancer originated. According to the Catalogue of Somatic Mutations in Cancer,13 BRAF mutations frequently occur in melanoma (44%) and thyroid cancer (52%), whereas KRAS is also highly mutated in lung (15%) and pancreatic cancers (63%). In the most common type of pancreatic cancer (ductal adenocarcinoma), KRAS mutation occurs at a much higher rate (>90%).14
Tumor-specific CpG methylations have been detected in the plasma from patients from a variety of solid tumors,15,16 and detected by techniques involving bisulfite conversion of unmethylated cytosines, methylation-sensitive enzymes, or immunoprecipitation of 5-methylcytosines.17 Methylation signatures are promising as blood-based biomarkers as they demonstrate better specificity toward a particular cancer type because methylation patterns are highly tissue specific.18 The best-studied blood-based methylation markers for CRC detection are located in the promoter region of the SEPT9 gene.19, 20, 21, 22, 23, 24, 25
The clinical adaptation of any cancer diagnostic test is determined by several factors, foremost of which is whether it can perform within acceptable performance metrics, such as sensitivity, specificity, and utility, along with cost-effectiveness. For any CpG methylation-interrogating, noninvasive assay for early cancer detection, sound performance metrics may then largely depend on how it addresses biological realities regarding the CRC-specific methylation markers contained in cfDNAs isolated from patient plasma: that they are present in minuscule quantities (down to several copies), thus the assay should be highly sensitive, and that they can be identified in large excesses of genomic DNAs (gDNAs) from peripheral blood, as well as cfDNAs released by other tissues (including other types of cancer). The latter therefore requires the assay to be designed for methylation markers predicted to be negative in peripheral blood, as well as most other cancer types.
Approaches to address such concerns are presented in the current study. The described technique employs multiplexed amplification of the CpG cancer markers (relative to normal DNA), such that the marker can be confidently detected even at the single-molecule level. Moreover, the novel methylation markers described herein were identified through comprehensive bioinformatic approaches, integrating genome-wide methylation data sets for CRC, peripheral blood, immune infiltrating cells, and other cancer types.
Materials and Methods
Public Genomic Data Sets
The identification of candidate CRC-specific, plasma-based CpG methylation sites entailed integration of various publicly available genome-wide methylation data sets, including genome-wide methylation (Illumina 450K methylation array; Illumina, San Diego, CA) and expression (RNA-sequencing) data sets for colorectal adenocarcinoma (COADREAD), generated by The Cancer Genome Atlas (TCGA) project,26 and previously compiled (and processed) in the University of California, Santa Cruz, Cancer Genomics (now known as University of California, Santa Cruz, Xena) website (https://xena.ucsc.edu/welcome-to-ucsc-xena, last accessed January 23, 2020).27,28 Other TCGA-generated methylation data sets analyzed in this study include the following cohorts: adrenocortical carcinoma, bladder urothelial carcinoma, breast invasive adenocarcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, cholangiocarcinoma, lymphoid neoplasm diffuse large B-cell lymphoma, esophageal carcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, kidney chromophobe, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, brain lower-grade glioma, liver hepatocellular carcinoma, acute myeloid leukemia, lung adenocarcinoma, lung squamous cell carcinoma, mesothelioma, pancreatic adenocarcinoma, pheochromocytoma and paraganglioma, prostate adenocarcinoma, sarcoma, skin cutaneous melanoma, stomach adenocarcinoma, testicular germ cell tumors, thymoma, thyroid carcinoma, uterine corpus endometrial carcinoma, uterine carcinosarcoma, and uveal melanoma. Methylation data sets were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) for ovarian cancer primary tumors and matching normal tissues (GSE65820),29 normal tissues of the cervix (GSE46306),30 gastric mucosa (GSE99553), testis (GSE74104),31 adrenal tissues (GSE77871), dermis and epidermis (GSE51954),32 various brain tissues (GSE64509),33 peripheral blood (GSE42861),34 and various immune cells (GSE59250)35 from healthy individuals. The sources of methylation data for particular CRC lines were the data sets GSE5734236 and GSE68379.37
Bioinformatic and Statistical Approaches
Statistical and Bioinformatics Tools
All statistical analyses (ie, comparative statistics, normalization, correlation, and regression analyses, multivariate analyses, and hierarchical clustering) were performed using JMP Pro 13.2.1/JMP Genomics 9.0 software (SAS, Cary, NC), Gene-E (Broad Institute, Cambridge, MA), and Microsoft Excel 2016 Office Analysis ToolPak (Microsoft Corp., Redmond, WA). Genomic sequence extraction and alignment were performed through the University of California, Santa Cruz, Genome Browser (https://genome.ucsc.edu, last accessed January 23, 2020).38 The OligoAnalyzer Tool from Integrated DNA Technologies Inc. (Coralville, IA) aided primer designs.
Prediction of Blood-Based, Colorectal Cancer– and Colon-Specific Methylation Markers
The primary task was to identify CpG markers whose high degree of methylation in blood cfDNA may be indicative of primary CRC tumors, but not of peripheral blood, or other cancer types (overlap with methylation signals from normal colon was not preferred, but not as stringently selected against). Methylation marker prediction starts with defining the metric associated with every CpG marker P for a given cohort C (eg, TCGA–breast invasive adenocarcinoma, TCGA-COADREAD, GSE77871), and cohort subset S (eg, primary tumors and solid normal tissues). In the current analysis, this particular metric is the statistical value V, such as %UM, %IM, %LM, %HM, %UM + %IM, and %LM + %HM, where UM, IM, LM, and HM refer to UnMethylated (βP ≤ 0.15), Indeterminately Methylated (0.15 < βP ≤ 0.3), Lowly Methylated (0.3 < βP ≤ 0.6), and Highly Methylated (βP > 0.6), respectively. The candidate markers (Ps) are dynamically identified by isolating CpG sites that satisfy multiple criteria in the general form: VP(C,S) ≥ n; {0 ≤ n ≤ 100} (explained further in Results).
Assessment of Methylation Markers' Relationship with Various Clinicopathologic Data
Accompanying TCGA COADREAD data sets are clinicopathologic data, such as sample type (primary tumor or normal), primary tumor pathologic stage (I to III), microsatellite stability status [microsatellite (MS) stable, high MS instability (MSI-H), and low MSI], and anatomic origin (colon or rectal), sex, and age.
Cell Lines and Genomic DNAs
The colon adenocarcinoma cell lines HT29, LoVo, and SW1116 would serve as sources of cancer gDNAs. All cell lines were seeded in 60-cm2 culture dishes, kept in a humidified atmosphere containing 5% CO2, and grown in recommended media: HT29 cells in McCoy's 5a medium containing 4.5 g/L glucose, supplemented with 10% fetal bovine serum; SW1116 cells in Leibovitz's L-15 Medium containing 10% fetal bovine serum; and LoVo in ATCC (Manassas, VA)–formulated F-12K Medium, supplemented with 10% fetal bovine serum. Once cells reached 80% to 90% confluence, they were washed with phosphate-buffered saline (×3) and collected by centrifugation (500 × g). The gDNAs from these cell lines were isolated using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). Roche (Indianapolis, IN) DNA, which is pooled gDNA (>50-kb size) isolated from the blood (buffy coat) of approximately 80 healthy individuals was purchased from Roche. gDNAs were then fragmented (50 bp to 1 kb size) through nonrandom sonication method, using ultrasonicator from Covaris (Woburn, MA). The fragmentation size was assessed using Agilent Bioanalyzer system (Agilent Technologies, Santa Clara, CA). DNA concentrations were determined using Quant-iT Picogreen Assay (Life Technologies/Thermo Fisher Scientific, Waltham, MA).
Patient Plasma and Cell-Free DNA
The isolation of human plasma samples from both CRC and healthy patients was performed by the MT Group, Inc. (Van Nuys, CA). In the isolation procedure, 10 mL of blood was drawn into a Streck Cell-Free DNA BCT Tube (Streck, Omaha, NE) by venipuncture. The subsequent steps toward plasma isolation carefully adhered to the Streck Tube manufacturer's instructions to prevent the release of gDNA from nucleated blood cells. The cfDNAs were subsequently extracted from the plasma samples (5 mL) using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA), and quantified with the Quant-iT Picogreen Assay. Processing and handling of patient specimens were in accordance with the approved institutional review board protocols for our group at Weill Cornell Medicine (Cancer Serum Detection Project; institutional review board identifier 1308014272), as well as that of the MT Group, Inc. (institutional review board identifier 3764).
Enrichment of Methylated Genomic DNA and cfDNA
Two approaches were employed to enrich the population of genomic and cfDNA fragments containing the desired methylated CpG markers. In one approach, the gDNAs (500 ng) from the cell lines were digested with 10 units of the restriction enzyme Bsh1236I (BstUI) in 20 μL of reaction mixture containing 1× CutSmart buffer (50 mmol/L potassium acetate, 20 mmol/L Tris-acetate, 10 mmol/L magnesium acetate, and 100 μg/mL bovine serum albumin, pH 7.9 at 25°C). Bsh1236I is a methylation-sensitive restriction enzyme that specifically recognizes the sequence 5′CGCG if the Cs are unmethylated. The digestion reactions were performed at 37°C for 1 hour with subsequent enzyme inactivation by heating to 80°C for 20 minutes. An alternative enrichment strategy was the use of EpiMark Methylated DNA Enrichment Kit (New England BioLabs, Ipswich, MA). This approach uses selective binding of double-stranded methyl-CpG DNA to the methyl-CpG binding domain of human methyl-CpG-binding domain protein 2 (MBD2) fused to the Fc tail of human IgG1 (MBD2-Fc).39 The fused IgG1 (MBD2-Fc) antibody is coupled to paramagnetic hydrophilic protein A magnetic beads. Epimark enrichment was performed according to the manufacturer's instructions.
Bisulfite Conversion of Digested Genomic DNA and cfDNA
Bisulfite conversion of cytosine bases was accomplished using the EZ DNA Methylation-Lightning kit from Zymo Research Corp. (Irvine, CA). In brief, 130 μL of Lightning Conversion Reagent (Zymo Research Corp.) was added to 20 μL of previously enriched gDNA fragments (or cfDNA). Subsequent protocol steps (per manufacturer's instructions) led to elution of bisulfite converted DNA fragments in 10 μL of elution buffer.
PCR–Ligase Detection Reaction–Real-Time Quantitative PCR
The assay developed for detection of CRC-specific, plasma-based methylation markers is divided into several steps described in following subsections. All of the necessary primers (Table 1) were purchased from Integrated DNA Technologies Inc.
Table 1.
List of Primers Used for the Multiplex PCR-LDR-qPCR Assay
| Marker | Primer type | Sequence |
|---|---|---|
| m_SEPT9 | F (PCR) | 5′-TCCTCCGACGACTAACTCTACACrUACAG-C3-3′ |
| m_SEPT9 | R (PCR) | 5′-GGTGTCGTGAGGTAGCGGCGAGGAAGCrGTTTC-C3-3′ |
| m_SEPT9 | U (LDR) | 5′-TAGGAACACGGAGGACATCAACGACGACTAACTCTACACTACAAAAATGCrGAATA-C3-3′ |
| m_SEPT9 | D (LDR) | 5′-GAACGCGACGCCCCAACCAAC TTGTGGGTGGGTATAGGTCAGA-3′ |
| m_SEPT9 | P (qPCR) | 5′-FAM-TTAAAATGC-ZEN-GAACGCGACGCCC-IABkFQ-3′ |
| m_SEPT9 | F (qPCR) | 5′-TAGGAACACGGAGGACATCAA-3′ |
| m_SEPT9 | R (qPCR) | 5′-TCTGACCTATACCCACCCACAA-3′ |
| m_GSG1L | F (PCR) | 5′-AACCGAAACCGAACTAACCGCrCGCCT-C3-3′ |
| m_GSG1L | R (PCR) | 5′-GGTGTCGTGGGAATTTTTATATCGGTATTTGGTATTCGTGCrGAGGG-C3-3′ |
| m_GSG1L | U (LDR) | 5′-TTCGTCCCTGCACGCTAACCGAACTAACCGCCGTCCrGCGTA-C3-3′ |
| m_GSG1L | D (LDR) | 5′-GCGCGCACTCACCAAACCCGGTTCCATCACCGTTAGGCCA-3′ |
| m_GSG1L | P (qPCR) | 5′-FAM-TTCCGTCCG-ZEN-CGCGCA-IABkFQ-3′ |
| m_GSG1L | F (qPCR) | 5′-TTCGTCCCTGCACGCTAAC-3′ |
| m_GSG1L | R (qPCR) | 5′-TGGCCTAACGGTGATGGAAC-3′ |
| m_PP1R16B | F (PCR) | 5′-CTATTCCGAAACCTAACCACGTCCrCAACT-C3-3′ |
| m_PP1R16B | R (PCR) | 5′-GGTGTCGTGGAGGTGGGCGCGTTTAATTTTATTCrGGTTC-C3-3′ |
| m_PP1R16B | U (LDR) | 5′-TTCAGCAGCCTGGCATCACGAAACCTAACCACGTCCCAGCCrGATCC-C3-3′ |
| m_PP1R16B | D (LDR) | 5′-GATTTCAACTTCCTACAACTCAAAAAAAAAATCCCCACCGTGGAGCGCTAAGGTTGCA-3′ |
| m_PP1R16B | P (qPCR) | 5′-FAM-TTCGTCCCA-ZEN-GCCGATTTCAACTTCCTACAA-IABkFQ-3′ |
| m_PP1R16B | F (qPCR) | 5′-TTCAGCAGCCTGGCATCAC-3′ |
| m_PP1R16B | R (qPCR) | 5′-TGCAACCTTAGCGCTCCAC-3′ |
| m_KCNA3 | F (PCR) | 5′-GACTCGTAACGATCGCAACCGrCCGCT-C3-3′ |
| m_KCNA3 | R (PCR) | 5′-GGTGTCGTGGCGGTTACGCGGAGTTCGTCrGTAGA-C3-3′ |
| m_KCNA3 | U (LDR) | 5′-TCACAGAGACTTGCCGATCACGATCGCAACCGCCACCrGCCGT-C3-3′ |
| m_KCNA3 | D (LDR) | 5′-GCCACAACCGCCTTAAAACGAAACCCGTGTGTAGCTTAGACATGGCCA-3′ |
| m_KCNA3 | P (qPCR) | 5′-FAM-TTCGCCACC-ZEN-GCCACAACC-IABkFQ-3′ |
| m_KCNA3 | F (qPCR) | 5′-TCACAGAGACTTGCCGATCAC-3′ |
| m_KCNA3 | R (qPCR) | 5′-TGGCCATGTCTAAGCTACACAC-3′ |
| m_CLIP4 | F (PCR) | 5′-CGCGAGGTTGAGGGTTGTGrAAGGT-C3-3′ |
| m_CLIP4 | R (PCR) | 5′-GGTGTCGTGGGTCTACGAAATATCGCAATATTACCTCCrCCCGT-C3-3′ |
| m_CLIP4 | U (LDR) | 5′-TTCAGAGCACCTGCGTACCGAGGTTGAGGGTTGTGAAAGCrGGTAA-C3-3′ |
| m_CLIP4 | D (LDR) | 5′-GGTGGGTACGTACGGCGTGTCGGGTTCTTCGGCTGGCTCAA-3′ |
| m_CLIP4 | P (qPCR) | 5′-FAM-CCTTGTGAA-ZEN-AGCGGTGGGTACGTAC-IABkFQ-3′ |
| m_CLIP4 | F (qPCR) | 5′-TTCAGAGCACCTGCGTACC-3′ |
| m_CLIP4 | R (qPCR) | 5′-TTGAGCCAGCCGAAGAACC-3′ |
| m_GDF6 | F (PCR) | 5′-AACGCAAAAACCAACGAAAAACCrCGCGT-C3-3′ |
| m_GDF6 | R (PCR) | 5′-GGTGTCGTGGTGGAAAGTTTTGGGTAAAGTCGGTArUTAGA-C3-3′ |
| m_GDF6 | U (LDR) | 5′-TCTTACGCCCAGGGAATGTAACCGCCAAAACCAAAAAACTACCCAACrGCCAT-C3-3′ |
| m_GDF6 | D (LDR) | 5′-GCCGCTCGCGAACTAATTCCTCAAACTATAAAACGTTGTCCGGCTGTGGTTACA-3′ |
| m_GDF6 | P (qPCR) | 5′-FAM-TGTACCCAA-ZEN-CGCCGCTCGC-IABkFQ-3′ |
| m_GDF6 | F (qPCR) | 5′-TCTTACGCCCAGGGAATGTAAC-3′ |
| m_GDF6 | R (qPCR) | 5′-TGTAACCACAGCCGGACAAC-3′ |
| m_VIM | F (PCR) | 5′-GAACTCCAACCGAAACTACGTAArCTACA-C3-3′ |
| m_VIM | R (PCR) | 5′-GGTGTCGTGGACGAGGCGTAGAGGTTGCrGGTTA-C3-3′ |
| m_VIM | U (LDR) | 5′-TAGACACGAGCGAGGTCACAACTCCAACCGAAACTACGTAACTGCGrUCCGT-C3-3′ |
| m_VIM | D (LDR) | 5′-TCCACCCGCACCTACAACCTAAACAACGCGTGCAAAATTCAGGCTGTGCA-3′ |
| m_VIM | P (qPCR) | 5′-HEX-TAACTGCGT-ZEN-CCACCCGCACCTAC-IABkFQ-3′ |
| m_VIM | F (qPCR) | 5′-TAGACACGAGCGAGGTCAC-3′ |
| m_VIM | R (qPCR) | 5′-TGCACAGCCTGAATTTTGCAC-3′ |
F (PCR) and R (PCR) refer to forward and reverse primers for the PCR amplification step, respectively; U (LDR) and D (LDR) refer to upstream and downstream oligonucleotides used in the LDR step, respectively; P (qPCR), F (qPCR), and R (qPCR) refer to probes, forward primers, and reverse primers used in the qPCR detection step, respectively.
C3, C3 spacer; FAM, FAM fluorescent dye; HEX, HEX fluorescent dye; IABkFQ, Iowa Black fluorescent quencher; LDR, ligase detection reaction; qPCR, real-time quantitative PCR; r, ribonucleotide base; ZEN, ZEN fluorescent quencher.
Linear Amplification Step
In 25 μL of reaction volume, the linear amplification step was performed by mixing the following: 5 μL of 5× GoTaq Flexi buffer (no magnesium; Promega, Madison, WI), 2.5 μL of 25 mmol/L MgCl2 (Promega), 0.5 μL of 10 mmol/L dNTPs (dATP, dCTP, dGTP, and dTTP; Promega), 2.5 μL of the reverse primer (or primers in case of multiplex reaction; 1 μmol/L), 0.625 μL of 20 mU/μL RNAseH2 (diluted in RNAseH2 dilution buffer from Integrated DNA Technologies Inc.), 0.55 μL of KlenTaql polymerase (DNA Polymerase Technology, St. Louis, MO) mixed with Platinum Taq Antibody (Invitrogen/Thermo Fisher Scientific, Waltham, MA), and 5.0 μL of corresponding bisulfite converted DNA template (of 50 μL of eluted DNA after bisulfite conversion). The reactions were run in a ProFlex PCR system thermocycler (Applied Biosystems/Thermo Fisher Scientific, Waltham, MA) using the following program: 2 minutes at 94°C, 40 cycles of 20 seconds at 94°C, 40 seconds at 60°C, and 30 seconds at 72°C, and a final hold at 4°C. After the reaction, Platinum Taq antibodies were added in the reaction mixture to inhibit the KlenTaq DNA polymerase. The KlenTaql/Platinum Taq Antibody mixture was prepared by adding 0.02 μL of KlenTaql polymerase at 50 U/μL to 0.2 μL of Platinum Taq Antibody at 5 U/μL. In the initial assay development, uniplex formats were used, in which a single biomarker was detected, using 35 ng of bisulfite-converted gDNAs isolated from the CRC lines HT29, LoVo, and SW1417, or Roche DNA. To determine the limitation of the assay sensitivity, the templates consisted of 0.069, 0.034, or 0 ng of HT29 gDNA fragments (bisulfite converted) mixed with 8.6 ng of fragmented Roche DNA. In the testing of assays in a multiplex format, the templates are either the simulated plasma cfDNA (which is the mixture of 0.10 ng HT29 and 1.0 ng Roche bisulfite-converted DNA fragments) or the bisulfite-converted cfDNA isolated from the plasma of CRC patients or healthy individuals.
PCR Data
For the PCR, the 10 μL of linear amplification product (previous step) was mixed with 2 μL of 5× GoTaq Flexi buffer without magnesium, 1 μL of 25 mmol/L MgCl2, 0.4 μL of dNTPs (10 mmol/L each of dATP, dCTP, dGTP, and dUTP), 2 μL of 0.5 μmol/L forward primer (or primers in case of multiplex reaction), 0.4 μL of Antarctic Thermolabile uracil-N-DNA glycosylase (UDG; 1 U/μL; New England Biolabs), 0.25 μL of 20 mU/μL RNAseH2, and 0.44 μL of KlenTaql polymerase mixed with Platinum Taq Antibody. The KlenTaql/Platinum Taq Antibody mixture was prepared by adding 0.02 μL of 50 U/μL KlenTaql polymerase to 0.2 μL of 5 U/μL Platinum Taq Antibody. The 20-μL volume reactions were run in a ProFlex PCR system thermocycler, using the following program: 10 minutes at 37°C, 40 cycles of 20 seconds at 94°C, 40 seconds at 60°C, and 30 seconds at 72°C, 10 minutes at 99.5°C, and a final hold at 4°C.
LDR Step
The ligase detection reaction (LDR) step40, 41, 42, 43, 44, 45, 46, 47 was performed in a 20-μL reaction prepared by combining 5.82 μL of nuclease-free water (Integrated DNA Technologies Inc.), 2 μL of 10× AK16D ligase reaction buffer 0.5 μL of 40 mmol/L dithiothreitol (Sigma-Aldrich, St. Louis, MO), 0.25 μL of 40 mmol/L NAD+ (Sigma-Aldrich), 0.5 μL of 20 mU/μL RNAseH2, 0.4 μL of 500 nmol/L LDR upstream probe (or probes for multiplex reactions), 0.4 μL of 500 nmol/L LDR downstream probe (or probes for multiplex reactions), 0.57 μL of purified AK16D ligase (at 0.88 μmol/L), and 4 μL of PCR products from the previous step. The AK16D ligase reaction buffer (at 1×) contains the following: 20 mmol/L Tris-HCI at pH 8.5, 5 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L dithiothreitol, and 20 μg/mL of bovine serum albumin (all components purchased from Sigma-Aldrich). LDR reactions were run in a ProFlex PCR system thermocycler using the following program: 20 cycles of 10 seconds at 94°C and 4 minutes at 60°C, followed by a final hold at 4°C.
qPCR Step
The real-time quantitative PCR (qPCR) was accomplished in 10 μL of reaction mixture prepared by mixing 1.5 μL of nuclease-free water (Integrated DNA Technologies Inc.), 5 μL of 2× TaqMan Fast Universal PCR Master Mix (Fast AmpliTaq; UDG and dUTP; Applied Biosystems/Thermo Fisher Scientific), 1 μL 2.5 μmol/L forward primer, 1 μL of 2.5 μmol/L reverse primer, 0.5 μL of 5 μmol/L probe, and 1 μL of LDR reaction products from the previous step. All qPCRs were run in a ViiA7 real-time thermocycler from Applied Biosystems/Thermo Fisher Scientific, using MicroAmp Fast-96-Well Reaction 0.1-mL plates sealed with MicroAmp Optical adhesive film (Applied Biosystems/Thermo Fisher Scientific). The run settings were as follows: fast block, standard curve as experiment type, ROX (carboxyrhodamine) as passive reference, Ct as quantification method, TAMRA (tetramethylrhodamine) as reporter, and NFQ-MGB (nonfluorescent quencher, minor groove binder) as quencher; program at 2 minutes at 50°C and 40 cycles of 1 second at 95°C and 20 seconds at 60°C.
Results
Bioinformatic Identification of Colon Cancer–Specific CpG Markers
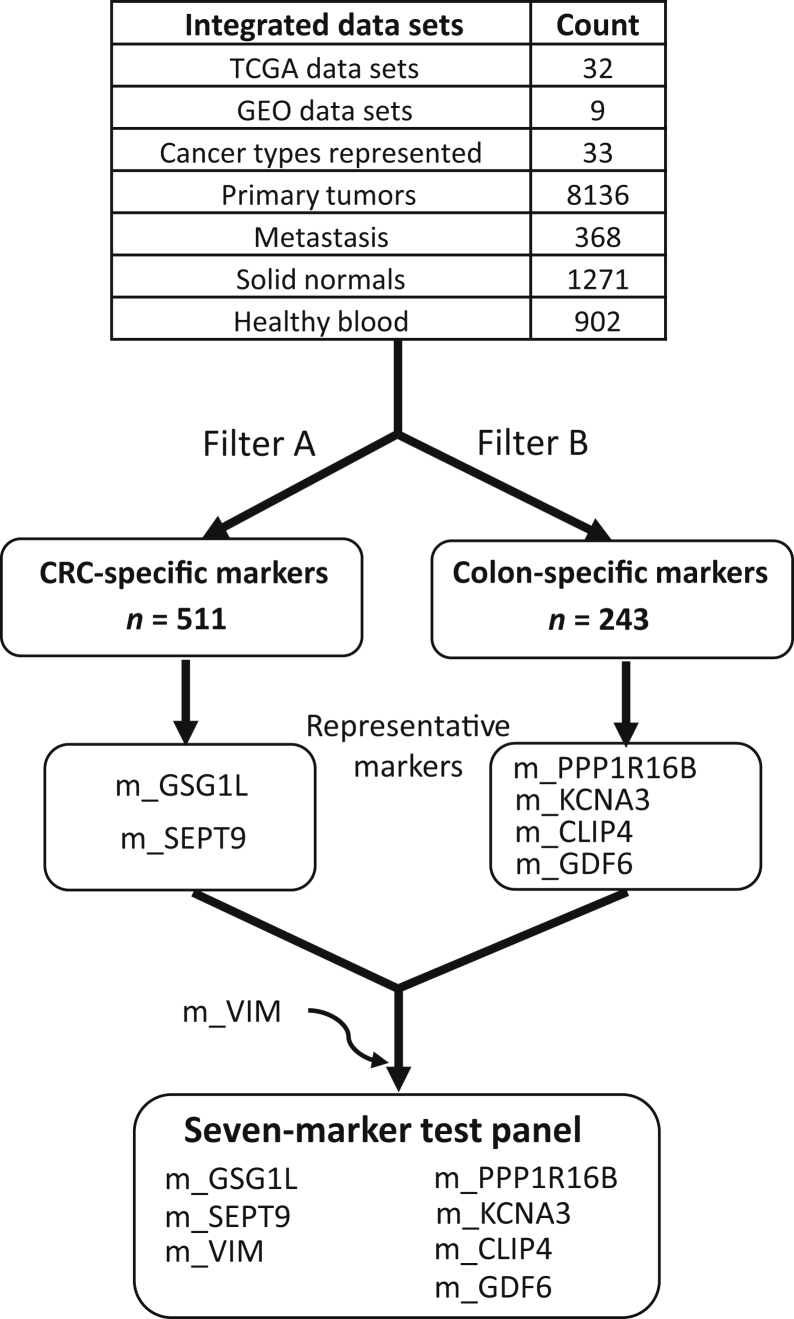
The identification of the methylation markers for assay design (Figure 1) was accomplished by applying the following filters (collectively referred to as filter A) on the integrated methylation data sets: i) (%LM + %HM)(COADREAD, PT) ≥ 85; PT = primary tumor; ii) (%UM + %IM)(COADREAD, SN) ≥ 85; SN = solid normal colon tissues; iii) average (%UM + %IM)(C, S) ≥ 70; C = any cancer cohort ≠ COADREAD, S = primary tumors; iv) average (%UM + %IM)(C, S) ≥ 85; C = any cancer cohort ≠ COADREAD; S = solid normal; and v) % UM(GSE42861,blood) ≥ 98. A total of 511 CpG markers passed the filters. On the other hand, the CRC-specific methylation markers were isolated through the following criteria (collectively referred to as filter B): i) (%LM + %HM)(COADREAD, PT) ≥ 85; ii) (%LM + %HM)(COADREAD, SN) ≥ 70; iii) average (%UM + %IM)(C, S) ≥ 45; C = any cancer cohort ≠ COADREAD; S = primary tumors; iv) average (%UM + %IM)(C, S) ≥ 75; C = any cancer cohort ≠ COADREAD; S = solid normal; and v) % UM(GSE42861,blood) ≥ 98. A total of 243 markers passed these filters. Among the CpG markers selected through filter A are the CpG sites within the promoter regions of SEPT9 (m_SEPT9) and GSG1L (m_GSG1L) (Supplemental Table S1). This list also included CpG sites in the promoter regions of THBD,48 C9orf50,48 ZNF154,49 and AGBL4, FLI1, and TWIST1,50 all previously reported as potential blood-based markers for CRC. The application of filter B led to the identification of CpG sites within the promoter regions of PPP1R16B (m_ PPP1R16B), KCNA3 (m_KCNA3), CLIP4 (m_CLIP4), and GDF6 (m_GDF6). The six aforementioned CpG sites, along with VIM promoter region CpG site (m_VIM), composed the panel of methylation markers against which assays were designed (and described further in subsequent sections). m_VIM was included in this panel because promoter methylation in the VIM locus has been previously used as a marker for CRC from fecal samples.51 The m_VIM marker passed all of the criteria for filter A, except (%LM + %HM)(COADREAD, PT), which equals 62.
Figure 1.
The scheme for identification and selection of CpG methylation markers tested for bisulfite PCR–ligase detection reaction–real-time quantitative PCR assay development. CRC, colorectal cancer; GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas.
CpG Markers' Association with Other Clinicopathologic Data
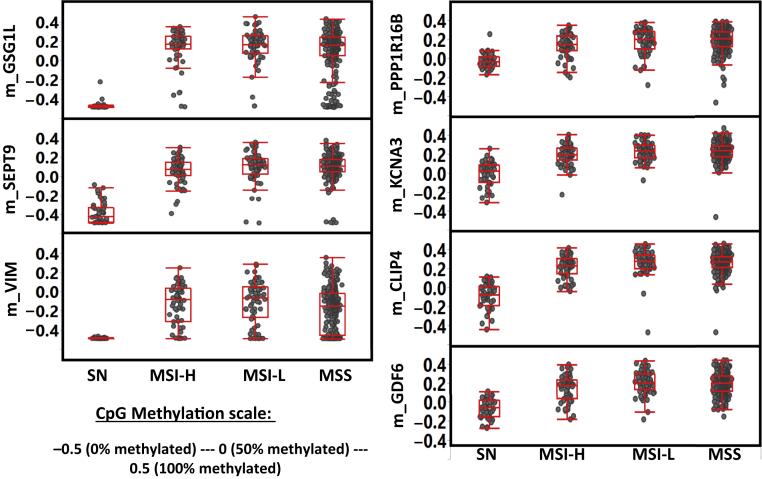
All seven CpG markers are highly methylated in primary CRC tumors (Figure 2 and Supplemental Table S2). However, the biggest difference between CRC-specific markers (m_SEPT9, m_GSG1L, and m_VIM) and colon-specific markers (m_ PPP1R16B, m_KCNA3, m_CLIP4, and m_GDF6) is that the former are virtually unmethylated in normal colon tissues. On the other hand, the latter group of markers still maintained a fairly high level of methylation in normal tissues, albeit at a relatively lower level of methylation compared with that of the primary tumors. If the primary tumors are subdivided according to MS status (MS stable, MSI-H, and low MSI), it is clear that each MS subgroup has significantly higher methylation level (at each of the seven CpG sites) when compared with normal colonic tissue (P < 0.0001; t-test) (Supplemental Table S3). However, among the primary tumors, the difference between each of the MS classes is generally less statistically significant.
Figure 2.
Comparative methylation of the seven CpG markers (which the multiplex assay was designed for) in normal colon tissues [solid normal (SN)] and each of the three microsatellite (MS) subtype colorectal cancer primary tumors: high MS instability (MSI-H), low MSI (MSI-L), and MS stable (MSS).
It is important to assess the influence of patient age on methylation, as previous studies have confirmed that, in general, the level of CpG methylation increases with age.52 Patient age does significantly contribute to the methylation status of each of the seven CpG markers, as far as primary tumors are concerned (R2 ranges from 0.01 to 0.05) (Supplemental Figure S1). However, for some of the markers (m_PPP1R16B, m_KCNA3, m_CLIP4, and m_GDF6 in particular), the differential methylation between primary tumors and normal tissues is not as pronounced in patients of more advanced age. This is expected for these markers in which the correlation between methylation and patient age is substantially positive among normal colon tissues. Furthermore, when the patients were divided into two groups (aged ≤65 and ≥66 years), it seems that the average CpG methylation (for each of seven markers) is statistically higher in primary tumors compared with normal colonic tissue regardless of the age (Supplemental Table S4). As our multivariate analysis indicated, age may be a factor that influences the methylation level at each of the CpG sites (Supplemental Table S5). However, the contribution of age is negligible compared with how methylation levels are influenced by sample type (Supplemental Table S5). In general, sex does not factor in the methylation of each marker. Some of the markers also appear to be influenced by anatomic origin (colon or rectal), but this factor is also negligible compared with the sample type.
Uniplex Assays Employed to Validate the Methylation Markers in Colon Cancer Cell Line Genomic DNA
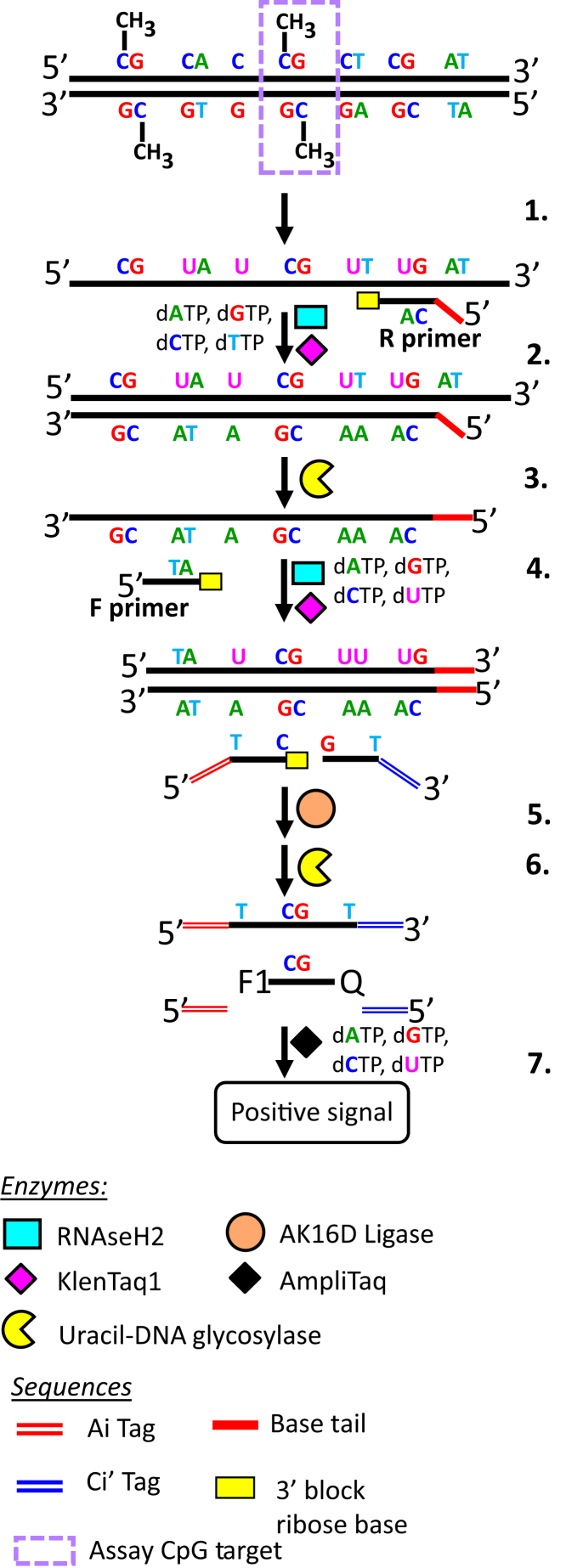
The eventual aim is to develop an assay that can detect early-stage CRC through the identification of methylation in the CpG markers described above, using plasma-derived cfDNA as templates. First, there was a need to ascertain whether this assay (Figure 3) could identify such markers at the low concentrations of DNA typically found in cfDNA samples. This was accomplished by using fragmented gDNAs derived from CRC cell lines as templates. Fortunately, it is now possible to assess the actual methylation levels at a particular CpG site in many of the widely used cancer cell lines through analysis of publicly available genome-wide methylation data sets. A closer look at the Gene Expression Omnibus data sets GSE5734236 and GSE683937 (generated by Illumina 450K methylation array) indicated that m_GDF6, m_GSG1L, m_SEPT9, m_KCNA3, and m_CLIP4 are all at least 92% methylated in CRC lines HT29, LoVo, and SW1417 (Supplemental Table S6). The m_VIM site is 46% methylated in LoVo, but 93% and 95% methylated in HT29 and SW1417, respectively. The marker m_PPP1R16B is approximately 67% methylated in HT29, 88% methylated in Lovo, and 74% methylated in SW11417. We were confident that the initial template of 35 ng of gDNAs isolated from the three CRC lines would include approximately 4000 to 9000 methylated copies of each of the seven CpG markers (on the assumption that average molar mass of a base pair is 650 g/mol, and the human genome size of 3.2 × 109 bp). Two possible methods were employed to enrich for the markers before bisulfite conversion. One possible enrichment step is the use of a fusion protein with a methyl-CpG binding domain, which can select for DNA fragments containing highly methylated CpG sites. Another approach is pretreatment of gDNA with the restriction enzyme Bsh1236I (BstUI), which cuts through the sequence 5′CGCG, which is unmethylated in Cs. This is particularly useful if the CpG marker being interrogated is located in the promoter region, where the BstUI recognition sites can be found at a much higher frequency compared with other parts of the genome. Using the BstUI method, the resulting (enriched) DNA fragments containing the target CpG site were bisulfite treated (Figure 3). This was followed by one- or two-step PCR amplification (separate reaction for each marker). The one-step PCR amplification simultaneously uses both the forward and reverse primers, whereas, in the two-step PCR, linear amplification was accomplished through the addition of solely the reverse primer followed by the forward primer in a second reaction. The resulting PCR amplicons then served as a template for LDR. The final step was the detection of the desired CpG site through TaqMan-based real-time PCR, using the LDR products as templates. Results showed that the average cell line Ct values (ie, average for the three cell lines) for m_GDF6, m_GSG1L, m_PPP1R16B, m_SEPT9, m_VIM, and m_KCNA3 were 16.6, 4.4, 9.8, 9.2, 12.2, and 7.5, respectively (Supplemental Table S6). For the peripheral blood DNA, these values were 37.6, 35.8, 32.6, >40, >40, and 31.5, respectively. The Ct value for m_CLIP4 was 21.3 for LoVo and >40 for normal peripheral blood. These results indicated that these seven CpG sites are highly methylated in CRC lines, whereas they are unmethylated in peripheral blood. However, our assay development included several features designed to minimize non-specific amplification and thus reduce false-positive signals. The reverse PCR primers contained identical 8 to 11 base tails designed to minimize primer dimer formation. The presence of ribose bases at the 3′ end of the PCR and LDR primers (which were removed by RNaseH2 only when the primers were bound to their targets) ensured highly specific extension (for PCR) and ligation (for LDR). UDG and the timely addition of either dUTP or dTTP in the dNTP mixture was also employed for carryover prevention.53 The tags in LDR primers (Ai and Ci’) allowed for uniformity (in terms of melting temperature) in our qPCRs.
Figure 3.
Schematic of bisulfite PCR–ligase detection reaction–real-time quantitative PCR assay for detection of CpG methylation. 1. Bisulfite conversion. 2. Linear amplification (reverse primer only). 3. Digestion by uracil-N-DNA glycosylase (UDG). 4. PCR (add forward primer). 5. Ligase detection reaction. 6. Digestion by UDG. 7. ViiA7 real-time PCR.
Single-Molecule Detection of VIM Methylation by Pixel PCR-LDR-qPCR
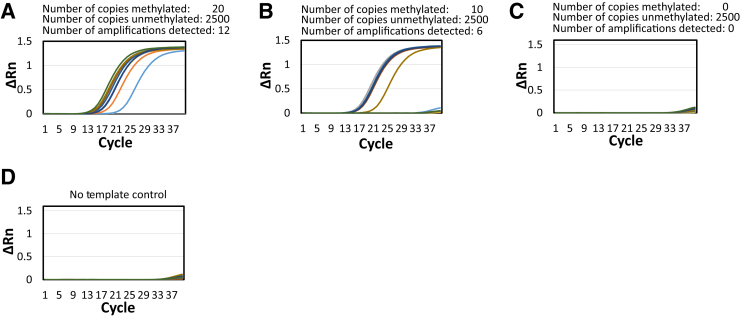
The sensitivity of the bisulfite PCR-LDR-qPCR assay and the limit of detection were next determined. Considering that a 10-mL blood sample from a patient may only yield a few copies of a CpG marker released by an early-stage CRC, the ability to detect a few methylation molecules is of utmost importance. Therefore, a series of experiments was conducted in which the initial DNA templates consisted of 20, 10, and 0 copies of the bisulfite-converted m_VIM CpG site, each of which was mixed with 2500 copies of normal peripheral blood gDNA. After PCR and LDR, the resulting LDR products were divided into 12 wells for subsequent qPCR. The LDR reactions with 20, 10, and 0 bisulfite-converted markers resulted in 12, 6, and 0 distinctly amplified wells, respectively (Figure 4). These results suggest that the current assay design is sensitive enough to detect a small number of copies of methylated CpG markers. In a separate article, a similar approach was employed to demonstrate the sensitivity of our PCR-LDR-qPCR assay, designed to interrogate mutations in cfDNA samples.54
Figure 4.
Single-molecule detection of methylation of m_VIM by pixel PCR–ligase detection reaction–real-time quantitative PCR. A–C: Detection of 12 (A), 6 (B), and 0 (C) methylated molecules in presence of 2500 nonmethylated molecules. D: No template control. Rn, normalized reporter value.
Multiplexed Assay Employed in Detection of the Methylation Markers
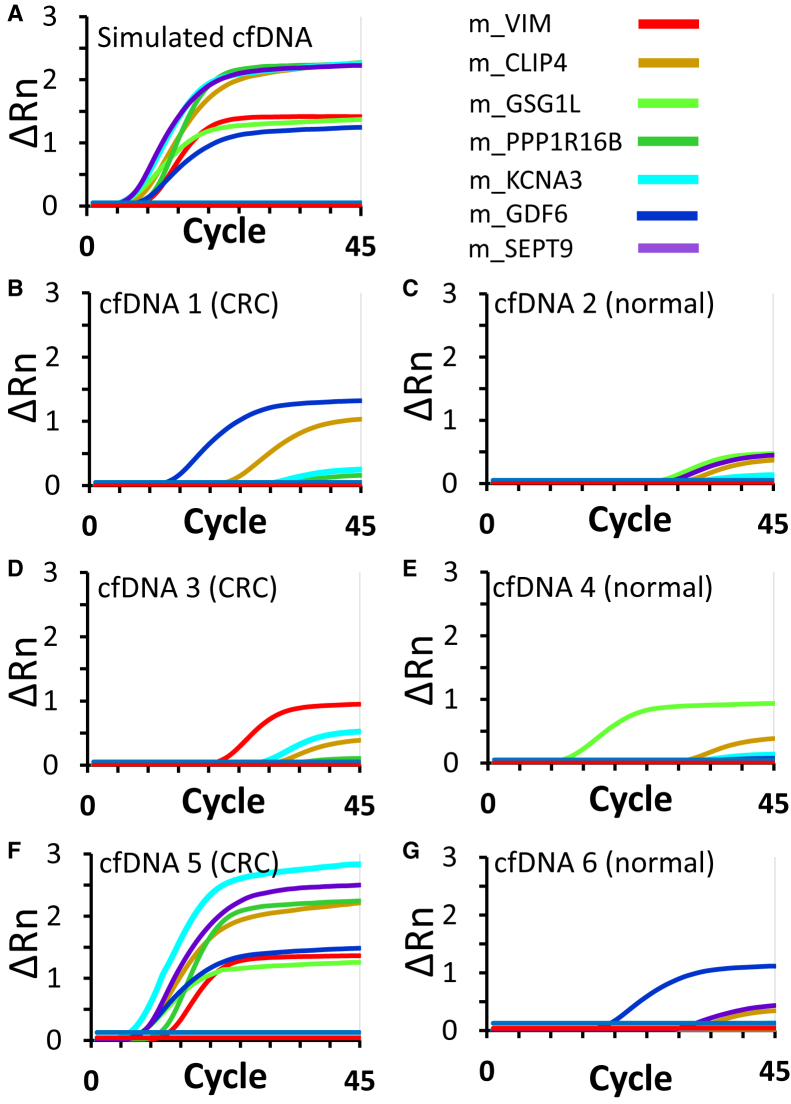
The next step was to combine the seven individual CpG assays into a multiplex format, which was preferred given the limited number of copies of each methylated CpG marker in a clinical plasma sample. In the multiplex format, all the marker-specific PCR amplification and LDR reaction primers were combined in a single reaction well. However, the qPCR for each marker was performed separately. As shown in the results (Figure 5A), all of the seven CpG markers tested positive in the simulated plasma cfDNA sample (which contained approximately 30 copies of fragmented gDNA from HT29, mixed with approximately 3000 copies of fragmented normal blood gDNA). The Ct values for the markers ranged from 5.5 (m_SEPT9 and m_KCNA3) to 9.1 (m_VIM), with an average of 7.1 for all of the seven markers. The multiplex assay was then tested in six plasma-derived CRC (cfDNAs 1, 3, and 5) and healthy control (cfDNAs 2, 4, and 6) samples (Supplemental Tables S7 and S8 and Figure 5, B–G). In the CRC sample cfDNA5, all the seven markers registered strong positive signals (average Ct value of 7.8). However, not every marker exhibited low Ct values for the two other CRC patient-derived cfDNAs. The plot for cfDNA 1 displayed relatively strong signals for m_GDF6 (Ct = 12.7) and m_CLIP4 (Ct = 22.7). For cfDNA 3, the signal for m_VIM (Ct = 21.4) was likely positive, whereas that of m_KCNA3 (Ct = 28.9) was weakly positive. The curves in the three healthy control cfDNA samples (normal) were mostly flat curves, indicative of background signals. In cfDNA 2, none of the Ct values was <27. However, each of the two other cfDNA samples from healthy controls included a marker with substantially low Ct values: m_GSG1L for cfDNA 4 (Ct = 11.7) and m_GDF6 for cfDNA 6 (Ct = 17.6). In summary, for the CRC patients, one sample had all seven markers positive, whereas the other two samples had two markers positive. For the normal patients, two samples had one strong positive marker, whereas the other did not show a positive marker.
Figure 5.
Multiplexed detection of seven colorectal cancer (CRC) methylation markers by bisulfite PCR–ligase detection reaction–real-time quantitative PCR in simulated plasma sample (ie, average of 30 molecules of each methylated CpG marker mixed in 3000 unmethylated CpG markers from normal DNA; A), as well as plasma samples from CRC patients (B, D, and F) and healthy individuals (C, E, and G). cfDNA, cell-free DNA; Rn, normalized reporter value.
Discussion
The highly invasive nature of colonoscopy and the limited success of stool-based tests (ie, fecal occult blood and immunochemical tests)5 have prompted the exploration of blood-based, CRC screening technologies. These noninvasive or minimally invasive approaches are predicted to be effective diagnostic tools as tumor cells can escape into the patient bloodstream, and may eventually release certain biomolecules through secretion (proteins and metabolites) or undergo apoptosis (cfDNA and RNA molecules contained in exosomes).8
In recent years, there has been a particular focus on the characterization of cfDNA from cancer cells [alias circulating tumor DNA (ctDNA)] as a surrogate marker for early cancer detection. This is primarily because ctDNAs naturally possess the same molecular attributes (ie, mutation, CpG methylation, and copy number variations)55, 56, 57 found in gDNAs extracted from tumor cells. In theory, therefore, analytical tools used for routine analysis of gDNAs (eg, sequencing, PCR, methyl-specific PCR, and microarrays) may also be employed to characterize ctDNAs. However, one important consideration is the fact that ctDNA fragments constitute just a small percentage of the total cfDNAs that can be isolated from patient blood. A recent report by Phallen et al58 has shown that the mutant allele fraction (based on sequencing 58 cancer-related genes) in cfDNAs isolated from stage I cancer (colorectal, breast, lung, and ovarian) patients is <1%, despite the fact that a higher amount of cfDNAs is released in the blood of cancer patients (29 ng/mL) compared with healthy individuals (7 ng/mL). This implies that most of the cfDNAs originate from noncancer cells. By examining the methylation signatures in pooled cfDNA samples from healthy individuals, Moss et al59 concluded that most cfDNAs are hematopoietic in origin (granulocytes, erythrocyte progenitors, monocytes, and lymphocytes). Hence, early CRC detection assay based on cfDNA analysis should be sensitive enough to detect limited amount of the cancer-specific markers, and be highly capable of distinguishing ctDNAs from the rest of cfDNAs. The current report explains that this goal can be attained using two complementary approaches: the identification of the most appropriate molecular biomarkers and the development of a highly sensitive assay to detect such markers. We decided to focus on methylation markers over mutation and copy number aberration for several reasons. Previous studies have indicated (and as our bioinformatics analysis has shown) that methylation markers can be tissue specific,18 thus highly useful in distinguishing one cancer type from another. A closer look at the data set GSE4868460 (which was integrated in our analyses) indicated that methylation markers positive in primary CRC tumors may already be positive in colon adenomas CRC.61,62 Also, CpG sites located in the promoter regions are often methylated in tandem. This attribute can be exploited by procedures meant to enrich DNA fragments containing the desired methylation markers (ie, the methylation-dependent capture and restriction techniques employed in this report).
The first aspect of this report is how bioinformatic predictive approaches were employed to select the site-specific CpG markers (m_CLIP4, m_PP1R16B, m_GDF6, m_KCNA3, m_GSG1L, m_VIM, and m_SEPT9) that would become the assay targets. Additional bioinformatic analyses would then indicate the potential association of the markers to processes that are related to cancer progression. Given that all the seven CpG sites are located in the promoter regions, it can be inferred that these sites are hypermethylated because they are part of genes that have tumor-suppressive properties. The expression levels of most of the genes in the panel (CLIP4, PP1R16B, KCNA3, and GSG1L; minimal/or not are VIM, SEPT9, and GDF6) are, in fact, down-regulated in primary colon tumors relative to normal colon tissues (Supplemental Figure S2). However, the integration of transcription and methylation data of the COADREAD cohort would reveal that of the seven CpG markers, only m_CLIP4, m_PP1R16B, m_GDF6, and m_KCNA3 have methylation levels that negatively correlate (R is −0.4 or lower) with the corresponding transcript levels (Supplemental Figure S3). For m_GSG1L, the correlation is negative but moderate (R = −0.3). However, there are many factors (apart from promoter methylation) that can determine the extent of measurable transcripts for any given gene. Histone modification,63 regulation by post-transcriptional controls (eg, miRNAs),64 and the competing rates of mRNA transcription and mRNA degradation65 influence the mRNA level of a gene at any given time.
Our bioinformatic approach was designed to identify blood-based markers for CRC detection, irrespective of the tumor's MS status. Nonetheless, it is also possible to identify such methylation markers more specific for identification of MSI-H CRCs (which constitutes approximately 15% of spontaneous CRCs).66 This can be accomplished if filter A (Figure 1) (explained in Results) is modified such that the first marker criterion [(%LM + %HM)(COADREAD, PT) ≥ 85)] is replaced with: (%LM + %HM)(COADREAD, PT-MSI-H) ≥ 65), wherein the COADREAD primary tumor (PT) subset is replaced with the smaller COADREAD MSI-H PT subset (Supplemental Figure S4A). This new filter (filter A′) has identified a total of 4088 candidate CpG markers, 5 of which are located in the promoter region of MLH1. Further analysis would reveal that the methylation level at each of these 5 CpG sites negatively correlates with MLH1 transcript count (Supplemental Figure S4B).
For some of the seven genes interrogated in our assays, experimental evidence concerning promoter hypermethylation, but not necessarily transcriptional down-regulation nor tumor-suppressive properties, has already been reported in the literature. In a 147 case-cohort study, CpG methylation at KCNA3 (potassium voltage-gated channel subfamily A member 3) promoter region has been observed in 76% of CRC cases.67 KCNA3 promoter methylation negatively correlated with its expression,67 and is associated with poor prognosis.67,68 Found to be hypermethylated among sessile serrated colon polyps (which may progress to CRC) are CpG sites in the promoter region of GSG1L, a gene that codes for a membrane protein previously shown to regulate α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor-mediated synaptic transmission,69 and was identified by genome-wide association study (mice study) as a CRC predisposition locus.70 Also reported to be highly methylated71 in CRC (relative to normal) is the promoter region of PP1R16B (protein phosphatase 1 regulatory subunit 16B), which codes for a membrane protein that regulates protein phosphatase 1 in endothelial cells.72 The locus of GDF6 (growth differentiation factor 6), a gene that has a role in vascular stabilization,73 is also highly methylated74 in CRC. CLIP4 (CAP-Gly domain-containing linker protein family member 4), whose function is largely unknown, has been reported to be hypermethylated in gastric cancer.75 The promoter region of VIM (vimentin), a gene coding for a cytoskeletal protein,76 is found to be highly methylated in colon adenoma and carcinoma.77 VIM promoter methylation is also the basis for a fecal-based test for early detection of CRC.78 The hypermethylation of the promoter region of SEPT9 (septin 9), belonging to a family of genes involved in tumor cell migration and invasion,79 has been observed in CRC tissues.80
The second part of the current report is the development of a multiplex assay designed to interrogate the methylation status of select CpG markers in cfDNAs isolated from CRC patient serum (a similar study concerning breast cancer was recently published81). The assay is designed to address the limitation of the most commonly used techniques in interrogating CpG methylation markers in cfDNA, methylation-specific PCR. In CRC, most of the studies that employed methylation-specific PCR (which usually involves bisulfite conversion of unmethylated cytosines, PCR, and qPCR) have been employed to detect methylation in the SEPT9 promoter region,19, 20, 21, 22, 23, 24, 25 which is the basis for Epi proColon test, a blood-based CRC detection assay being marketed by Epigenomics (Berlin, Germany).20 Other CpG sites that have been reported to be interrogated for CRC detection through methylation-specific PCR approach are located in the loci of ALX4,82,83 SRF2,84 BCAT1 and IKZP1,85 TMEFF2 and NGFR,20 RUNX3,86 NEUROG1,87 THBD and C9orf50,48 WIF1 and NPY,88 APC and RASSF1A,89 and SFRP1 SDC2 and PRIMA1.84 The assay described in this report aims to improve the widely used methylation-specific PCR by incorporating the following features: i) selective enrichment of methylated DNA, ii) signal amplification of the targeted CpG site through the use of two rounds of PCR and LDR, iii) multiple primer binding regions for orthogonal amplification and signal detection, iv) prevention of non-specific primer extension through the incorporation of RNaseH2-targeted ribose bases in the 3′ end of PCR and LDR, v) prevention of carryover contamination by PCR products originating from previous positive samples (through the use of UDG), and vi) capacity to multiplex to interrogate multiple CpG sites.
The enrichment of fragments containing methylated CpG sites was accomplished using two methods: digestion by BstUI restriction enzyme and the use of the MBD2-Fc enrichment kit. The BstUI digestion approach was successfully incorporated in our initial assay designs, which generated results using 35-ng cell line gDNA (thousands of copies of targeted methylated markers) as template (Supplemental Table S6). A caveat of the BstUI approach, if not properly optimized, is that non-specific nucleases may further nick the already miniscule amount of cfDNAs isolated from patient plasma. Moreover, BstUI recognition sequence (5′CGCG) is located in or directly adjacent to many, but not all, of the CpG markers identified in our initial analysis. Alternatively, the MBD2_Fc fusion protein can bind to and select for DNA fragments containing any methylated CpG sites. The MBD2_Fc approach, while enriching for the desired methylated DNA, may also have lower recovery yields, because methylated DNA represents only a fraction of total DNA (7% to 14.9% with a modified protocol).90
Our group and collaborators have incorporated PCR and LDR steps in developing tools for detection of low abundant mutations.44,54,91, 92, 93, 94 In the current report, signal amplification involved 40 cycles of linear amplification, 40 cycles of PCR, and 20 cycles of LDR. One crucial feature is the orthogonality (ie, noninteraction) of the LDR and PCR primers, ensuring that only the target 5-methylcytosines (originally methylated in interrogated DNA fragments) will be amplified before the qPCR detection step. Another important component of our assay is use of a blocked 3′ end and a ribose base near the 3′ end of both PCR primers, as well as the upstream LDR primer. This is another mechanism that can improve the assay's specificity, because RNaseH2 liberates the active 3′ hydroxyl only when the primers are bound to their respective targets. The addition of UDG, along with addition of dUTP during PCR, prevents carryover contamination. This feature ensures the removal of excess templates (from bisulfite conversion and PCR) that, if carried through the assay, can contribute to false-positive calls. This assay also allows for a flexible detection method. This report and a recent article81 demonstrated the reliability of TaqMan qPCR as the final detection step. In the future, a detection method, such as microfluidic devices,42,92,95, 96, 97, 98, 99, 100 may also be employed. Another important feature of our assay is its multiplex capability. The simultaneous interrogation of multiple markers (in our case, seven CpG sites, representing distinct genes) adds to the robustness of this assay. With multiplexing, it is possible to develop a diagnostic metric based on the number of markers that test positive in the assay (instead of just relying on the positive/negative call of just a single marker).
Despite the incorporation of the features listed above, there are still technical and biological limitations that may hinder the assay's translation to clinical use. Foremost, the bisulfite conversion process can degrade the already limited number of ctDNA fragments, resulting in false-negative calls for certain methylation markers. Because of bisulfite conversion, approximately 84% to 96% of the input cfDNA can be degraded.90,101 In two of the three cfDNA samples derived from CRC patients, several CpG methylation markers were undetectable, which may represent either absence of methylation at such positions in the primary tumor (true negative) or loss of marker DNA during purification of cfDNA, methylation capture, and bisulfite conversion, and/or purification of bisulfite-converted single-stranded DNA product (resulting in a false-negative call). Another important concern is a positive signal that may unexpectedly show up in cfDNAs from healthy individuals. Despite the seven CpG methylation markers being carefully selected (bioinformatically) to be negative in peripheral blood, it was possible to detect one positive signal in each of two (of three) cfDNA samples extracted from plasma of healthy individuals. It is now generally believed that such a positive signal may be attributed to a phenomenon known as clonal hematopoiesis, wherein hematopoietic cells can acquire genetic mutations and clonally expand, likely factoring in various diseases during the aging process.102 Recent studies have concluded that some mutations found in cfDNAs of cancer patients actually emanate from cfDNAs contributed by these hematopoietic neoclones, thus resulting in false-positive calls.103 It will not be surprising if these hematopoietic neoclones also possess epigenetic signatures (ie, methylation) that are distinct from peripheral blood. Among the most commonly acquired mutations in these hematopoietic neoclones are genes involved in epigenetic regulation: DNMT3A (DNA methyltransferase 3 α), TET2 (tet methylcytosine dioxygenase 2), and ASXL1 (ASXL transcriptional regulator 1).104 Mutations in TET2 can result in hypermethylated CpG sites.104,105 Thus, either de novo age-related methylation or clonal hematopoiesis mutations driving altered epigenetic signatures may lead to cfDNA methylated at specific sites (as a reflection of aging), even in patients who are purportedly cancer free. The noncancer peripheral blood cell data set was employed as a negative control (GSE42861),34 and revealed that a handful of samples exhibited β values that are significantly higher than the rest of the cohort (Supplemental Figure S5A). Of 689 samples, 3 (0.4%) registered m_GDF6 β values of 0.25, 0.31, and 0.46. There is a possibility that these elevated β values are indicative of the presence of neoclones in peripheral blood or, alternatively, a random methylation event occurred in one of these three individuals during the earliest stages of development, and is manifest in the peripheral blood. Our presumption is that the observed β value for any given marker (eg, m_GDF6) is simply the weighted average for all clones present in peripheral blood (Supplemental Figure S5B). On the basis of this assumption, the fractions of neoclones for the three outlier peripheral blood samples are 19%, 25%, and 43%. An earlier study by Jaiswal et al102 (using exome sequencing) concluded that with clonal hematopoiesis, neoclones can constitute approximately 20% of cells in the hematopoietic system.
Our working assumption is that in cfDNA- and methylation-based early cancer diagnostics, multiplexing (if approached correctly) can reduce both false-positive and false-negative calls arising from the assays’ technical limitations (eg, bisulfite conversion), as well as biological realities (low ctDNA count and clonal hematopoiesis). The pilot experiments described herein were limited to seven markers, which may be insufficient to distinguish between individuals with early CRC from those with age-related clonal hematopoiesis. We can only speculate how results from a multiplex assay interrogating 20 to 24 markers would be used to complement colonoscopy. Such a higher-order multiplexed assay may be interpreted on the basis of a total marker score (ie, count of markers registering positive signals). A total marker score of: i) 0 to 2 may be interpreted as negative in cancer; ii) 3 to 4 as intermediate call, which suggests retesting in 3 to 6 months; and iii) ≥5 as call for colonoscopy. In essence, this approach is based on the hypothesis that the clinical decision will be dependent on the overall cancer marker load and not specific markers. Accumulation of data on a substantial number of clinical samples using 20 to 24 markers would allow for refinement of this simple algorithm to provide a more accurate prediction of which patients would most likely benefit from follow-up colonoscopy.
Undeniably, bisulfite sequencing (whether targeting several loci or the entire genome) of blood-derived cfDNA is a promising tool for early cancer detection.106, 107, 108, 109, 110, 111 Nevertheless, this approach is also limited by the degradation of cfDNA input during bisulfite conversion employed in PCR-based methods, such as in this report.
In summary, this report demonstrates how careful biomarker prediction (using publicly available genomic data sets) can be integrated in the process of improving noninvasive tests for early CRC detection. Using highly sensitive PCR-LDR-qPCR assays, CRC-specific methylation markers could be detected in a multiplex manner in excess of normal genomic DNA and in clinical samples. Whether these CRC-specific markers combined with the assays developed herein will find broad use for early-stage cancer diagnostics awaits large-scale testing to establish clinical validity and clinical utility.
Acknowledgments
We thank David Gelfand, Bill Efcavitch, Cristian Ruiz Rueda, Olivier Elemento, Bert Gold, Steve Lipkin, Chris Mason, Ken Offit, Pat Paty, Bernard Peperstraete, Mark Pochapin, and Eugene Spier for helpful discussions.
Footnotes
Supported by The Biotechnology Resource Center of Biomodular Multi Scale Systems CBM2 for Precision Molecular Diagnostics NIH grant P41 EB020594, Weill Cornell Medicine funding through distribution of royalties from intellectual property generated by the Barany laboratory, a Sponsored Research Agreement between AcuamarkDx and Weill Cornell Medicine, an Earlier.org Friends for an Earlier Breast Cancer Test research grant (M.D.B.), and a Laura Crandall Brown Foundation and The Foundation for Women's Cancer research grant (S.F.G.).
Disclosures: The authors have submitted a patent on use of bisulfite PCR–ligase detection reaction–real-time quantitative PCR for detection of methylation markers of early colorectal cancer. M.D.B., A.H.M., J.H., S.F.G., P.B.F., and F.B. are shareholders in AcuamarkDx.
Current address of A.H.M., Department of Pharmacology, Weill Cornell Medicine, New York, NY.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.03.009.
Supplemental Data
The relationship between patient age (in years) and methylation level at each of the seven CpG sites, for both primary tumor (PT; red solid circles) and solid normal colon tissues (SN; blue solid circles) included in The Cancer Genome Atlas colorectal cancer (CRC; colorectal adenocarcinoma) cohort. The indicated methylation levels range from −0.5 (0% methylated), to 0 (50% methylated), to 0.5 (100% methylated). Insets: The squares of correlation coefficient (R2) between methylation level and age of diagnosis, for PT and SN tissues. The gray shade that covers each line (solid line for PT, and dashed line for SN) represents the confidence region for that particular linear fit.
Comparative heat map (primary tumor and normals) depicting the expression level of each gene whose corresponding CpG sites were interrogated in the multiplex assay. CRC, colorectal cancer; FC, fold change; Max, maximum; Min, minimum; SN, solid normal colon tissues.
The relationship between patient gene transcription and methylation at each of the seven CpG sites, in colon cancer cohort. The relationship between CpG methylation and the transcription level of its corresponding gene, among the primary tumor (red solid circles) and solid normal colon tissues (blue solid circles) included in The Cancer Genome Atlas colorectal cancer (CRC; colorectal adenocarcinoma) cohort. The indicated methylation levels range from −0.5 (0% methylated), to 0 (50% methylated), to 0.5 (100% methylated). Insets: The Pearson correlation coefficient (R) values between the two variables. The gray shade that covers each line represents the confidence region for that particular linear fit. Expr., expression.
A: An alternative scheme to identify potential blood-based methylation markers for high microsatellite instability (MSI-H) colorectal cancers. Filter A': similar to filter A (see Results), except the modification of requirement (i) to the following: (%LM + %HM)(COADREAD, PT-MSI) ≥ 65. B: Heat map depicting the methylation levels at MLH1 locus CpG sites (mostly within promoter region), across 394 The Cancer Genome Atlas (TCGA) colorectal adenocarcinoma (COADREAD) samples. Indicated (with solid, differently colored circles) are the five CpG sites identified through the scheme shown in A. The correlation value (R) between MLH1 expression (expr.) and CpG methylation level at each of these sites is highly negative. GEO, Gene Expression Omnibus; utr, untranslated region.
A: The methylation levels (β values) of the seven CpG markers, for peripheral blood (PB)–derived genomic DNAs (GSE42861). Also indicated are the average β values (βPB) for each of the healthy and rheumatoid arthritis subsets. B: The hypothetical presence of neoclones in certain PB samples (labeled 1, 2, and 3 in A) is illustrated. The main presumption in this model is that the measured β for a PB sample (for a specific marker, such as m_GDF6) is the weighted average of the β values for old clones (βOC) and neoclones (βNC; equation 1). If it is assumed that βOC is the average for samples minus the three outliers (referred to as base cluster), and that βNC for a given outlier sample is almost fully methylated (0.95), then the hypothetical fraction of neoclones (x) can be calculated using equation 1. n = 335 (healthy subset); n = 354 (rheumatoid arthritis subset).
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fedewa S.A., Ahnen D.J., Meester R.G.S., Barzi A., Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Rex D.K., Boland C.R., Dominitz J.A., Giardiello F.M., Johnson D.A., Kaltenbach T., Levin T.R., Lieberman D., Robertson D.J. Colorectal cancer screening: recommendations for physicians and patients from the U.S. Multi-Society task force on colorectal cancer. Gastroenterology. 2017;153:307–323. doi: 10.1053/j.gastro.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Beydoun H.A., Beydoun M.A. Predictors of colorectal cancer screening behaviors among average-risk older adults in the United States. Cancer Causes Control. 2008;19:339–359. doi: 10.1007/s10552-007-9100-y. [DOI] [PubMed] [Google Scholar]
- 5.Das V., Kalita J., Pal M. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed Pharmacother. 2016;87:8–19. doi: 10.1016/j.biopha.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 6.Morikawa T., Kato J., Yamaji Y., Wada R., Mitsushima T., Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129:422–428. doi: 10.1016/j.gastro.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 7.Oono Y., Iriguchi Y., Doi Y., Tomino Y., Kishi D., Oda J., Takayanagi S., Mizutani M., Fujisaki T., Yamamura A., Hosoi T., Taguchi H., Kosaka M., Delgado P. A retrospective study of immunochemical fecal occult blood testing for colorectal cancer detection. Clin Chim Acta. 2010;411:802–805. doi: 10.1016/j.cca.2010.02.057. [DOI] [PubMed] [Google Scholar]
- 8.Yoruker E.E., Holdenrieder S., Gezer U. Blood-based biomarkers for diagnosis, prognosis and treatment of colorectal cancer. Clin Chim Acta. 2016;455:26–32. doi: 10.1016/j.cca.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Ignatiadis M., Dawson S.J. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25:2304–2313. doi: 10.1093/annonc/mdu480. [DOI] [PubMed] [Google Scholar]
- 10.Spindler K.L., Pallisgaard N., Andersen R.F., Brandslund I., Jakobsen A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One. 2015;10:e0108247. doi: 10.1371/journal.pone.0108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Cao M., Mayo-de-Las-Casas C., Molina-Vila M.A., De Mattos-Arruda L., Munoz-Couselo E., Manzano J.L., Cortes J., Berros J.P., Drozdowskyj A., Sanmamed M., Gonzalez A., Alvarez C., Viteri S., Karachaliou N., Martin Algarra S., Bertran-Alamillo J., Jordana-Ariza N., Rosell R. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res. 2015;25:486–495. doi: 10.1097/CMR.0000000000000187. [DOI] [PubMed] [Google Scholar]
- 12.Sakai K., Tsurutani J., Yamanaka T., Yoneshige A., Ito A., Togashi Y., De Velasco M.A., Terashima M., Fujita Y., Tomida S., Tamura T., Nakagawa K., Nishio K. Extended RAS and BRAF mutation analysis using next-generation sequencing. PLoS One. 2015;10:e0121891. doi: 10.1371/journal.pone.0121891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., Kok C.Y., Jia M., De T., Teague J.W., Stratton M.R., McDermott U., Campbell P.J. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood L.D., Hruban R.H. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 2012;18:492–501. doi: 10.1097/PPO.0b013e31827459b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratt V.M. Are we ready for a blood-based test to detect colon cancer? Clin Chem. 2014;60:1141–1142. doi: 10.1373/clinchem.2014.227132. [DOI] [PubMed] [Google Scholar]
- 16.Warton K., Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci. 2015;2:13. doi: 10.3389/fmolb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorda M., Peinado M.A. Methods for DNA methylation analysis and applications in colon cancer. Mutat Res. 2010;693:84–93. doi: 10.1016/j.mrfmmm.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Issa J.P. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 19.Church T.R., Wandell M., Lofton-Day C., Mongin S.J., Burger M., Payne S.R., Castanos-Velez E., Blumenstein B.A., Rosch T., Osborn N., Snover D., Day R.W., Ransohoff D.F. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lofton-Day C., Model F., Devos T., Tetzner R., Distler J., Schuster M., Song X., Lesche R., Liebenberg V., Ebert M., Molnar B., Grutzmann R., Pilarsky C., Sledziewski A. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 21.Potter N.T., Hurban P., White M.N., Whitlock K.D., Lofton-Day C.E., Tetzner R., Koenig T., Quigley N.B., Weiss G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183–1191. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 22.Ravegnini G., Zolezzi Moraga J.M., Maffei F., Musti M., Zenesini C., Simeon V., Sammarini G., Festi D., Hrelia P., Angelini S. Simultaneous analysis of SEPT9 promoter methylation status, micronuclei frequency, and folate-related gene polymorphisms: the potential for a novel blood-based colorectal cancer biomarker. Int J Mol Sci. 2015;16:28486–28497. doi: 10.3390/ijms161226113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toth K., Sipos F., Kalmar A., Patai A.V., Wichmann B., Stoehr R., Golcher H., Schellerer V., Tulassay Z., Molnar B. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One. 2012;7:e46000. doi: 10.1371/journal.pone.0046000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toth K., Wasserkort R., Sipos F., Kalmar A., Wichmann B., Leiszter K., Valcz G., Juhasz M., Miheller P., Patai A.V., Tulassay Z., Molnar B. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One. 2014;9:e115415. doi: 10.1371/journal.pone.0115415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren J.D., Xiong W., Bunker A.M., Vaughn C.P., Furtado L.V., Roberts W.L., Fang J.C., Samowitz W.S., Heichman K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser J. National Institutes of Health: NCI gears up for cancer genome project. Science. 2005;307:1182. doi: 10.1126/science.307.5713.1182a. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J., Sanborn J.Z., Benz S., Szeto C., Hsu F., Kuhn R.M., Karolchik D., Archie J., Lenburg M.E., Esserman L.J., Kent W.J., Haussler D., Wang T. The UCSC cancer genomics browser. Nat Methods. 2009;6:239–240. doi: 10.1038/nmeth0409-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldman M., Craft B., Swatloski T., Ellrott K., Cline M., Diekhans M., Ma S., Wilks C., Stuart J., Haussler D., Zhu J. The UCSC cancer genomics browser: update 2013. Nucleic Acids Res. 2013;41:D949–954. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patch A.M., Christie E.L., Etemadmoghadam D., Garsed D.W., George J., Fereday S. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 30.Farkas S.A., Milutin-Gasperov N., Grce M., Nilsson T.K. Genome-wide DNA methylation assay reveals novel candidate biomarker genes in cervical cancer. Epigenetics. 2013;8:1213–1225. doi: 10.4161/epi.26346. [DOI] [PubMed] [Google Scholar]
- 31.Killian J.K., Dorssers L.C., Trabert B., Gillis A.J., Cook M.B., Wang Y., Waterfall J.J., Stevenson H., Smith W.I., Jr., Noyes N., Retnakumar P., Stoop J.H., Oosterhuis J.W., Meltzer P.S., McGlynn K.A., Looijenga L.H. Imprints and DPPA3 are bypassed during pluripotency- and differentiation-coupled methylation reprogramming in testicular germ cell tumors. Genome Res. 2016;26:1490–1504. doi: 10.1101/gr.201293.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandiver A.R., Irizarry R.A., Hansen K.D., Garza L.A., Runarsson A., Li X., Chien A.L., Wang T.S., Leung S.G., Kang S., Feinberg A.P. Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin. Genome Biol. 2015;16:80. doi: 10.1186/s13059-015-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horvath S., Mah V., Lu A.T., Woo J.S., Choi O.W., Jasinska A.J., Riancho J.A., Tung S., Coles N.S., Braun J., Vinters H.V., Coles L.S. The cerebellum ages slowly according to the epigenetic clock. Aging (Albany NY) 2015;7:294–306. doi: 10.18632/aging.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Aryee M.J., Padyukov L., Fallin M.D., Hesselberg E., Runarsson A., Reinius L., Acevedo N., Taub M., Ronninger M., Shchetynsky K., Scheynius A., Kere J., Alfredsson L., Klareskog L., Ekstrom T.J., Feinberg A.P. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Absher D.M., Li X., Waite L.L., Gibson A., Roberts K., Edberg J., Chatham W.W., Kimberly R.P. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Chiappinelli K.B., Guzzetta A.A., Easwaran H., Yen R.W., Vatapalli R., Topper M.J., Luo J., Connolly R.M., Azad N.S., Stearns V., Pardoll D.M., Davidson N., Jones P.A., Slamon D.J., Baylin S.B., Zahnow C.A., Ahuja N. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–598. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gebhard C., Schwarzfischer L., Pham T.H., Andreesen R., Mackensen A., Rehli M. Rapid and sensitive detection of CpG-methylation using methyl-binding (MB)-PCR. Nucleic Acids Res. 2006;34:e82. doi: 10.1093/nar/gkl437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khanna M., Park P., Zirvi M., Cao W., Picon A., Day J., Paty P., Barany F. Multiplex PCR/LDR for detection of K-ras mutations in primary colon tumors. Oncogene. 1999;18:27–38. doi: 10.1038/sj.onc.1202291. [DOI] [PubMed] [Google Scholar]
- 41.Tong J., Cao W., Barany F. Biochemical properties of a high fidelity DNA ligase from thermus species AK16D. Nucleic Acids Res. 1999;27:788–794. doi: 10.1093/nar/27.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinville R., Coyne J., Meagher R.J., Cheng Y.W., Barany F., Barron A., Soper S.A. Ligase detection reaction for the analysis of point mutations using free-solution conjugate electrophoresis in a polymer microfluidic device. Electrophoresis. 2008;29:4751–4760. doi: 10.1002/elps.200800197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Y.W., Pincas H., Bacolod M.D., Schemmann G., Giardina S.F., Huang J., Barral S., Idrees K., Khan S.A., Zeng Z., Rosenberg S., Notterman D.A., Ott J., Paty P., Barany F. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–6013. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci U S A. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barany F. The ligase chain reaction in a PCR world. PCR Methods Appl. 1991;1:5–16. doi: 10.1101/gr.1.1.5. [DOI] [PubMed] [Google Scholar]
- 46.Wiedmann M., Wilson W.J., Czajka J., Luo J., Barany F., Batt C.A. Ligase chain reaction (LCR)--overview and applications. PCR Methods Appl. 1994;3:S51–S64. doi: 10.1101/gr.3.4.s51. [DOI] [PubMed] [Google Scholar]
- 47.Khanna M., Cao W., Zirvi M., Paty P., Barany F. Ligase detection reaction for identification of low abundance mutations. Clin Biochem. 1999;32:287–290. doi: 10.1016/s0009-9120(99)00020-x. [DOI] [PubMed] [Google Scholar]
- 48.Lange C.P., Campan M., Hinoue T., Schmitz R.F., van der Meulen-de Jong A.E., Slingerland H., Kok P.J., van Dijk C.M., Weisenberger D.J., Shen H., Tollenaar R.A., Laird P.W. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One. 2012;7:e50266. doi: 10.1371/journal.pone.0050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margolin G., Petrykowska H.M., Jameel N., Bell D.W., Young A.C., Elnitski L. Robust detection of DNA hypermethylation of ZNF154 as a pan-cancer locus with in silico modeling for blood-based diagnostic development. J Mol Diagn. 2016;18:283–298. doi: 10.1016/j.jmoldx.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin P.C., Lin J.K., Lin C.H., Lin H.H., Yang S.H., Jiang J.K., Chen W.S., Chou C.C., Tsai S.F., Chang S.C. Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-wide high-resolution array. Ann Surg Oncol. 2015;22 Suppl 3:S1419–S1427. doi: 10.1245/s10434-014-4277-2. [DOI] [PubMed] [Google Scholar]
- 51.Chen W.D., Han Z.J., Skoletsky J., Olson J., Sah J., Myeroff L., Platzer P., Lu S., Dawson D., Willis J., Pretlow T.P., Lutterbaugh J., Kasturi L., Willson J.K., Rao J.S., Shuber A., Markowitz S.D. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 52.Lim U., Song M.A. DNA methylation as a biomarker of aging in epidemiologic studies. Methods Mol Biol. 2018;1856:219–231. doi: 10.1007/978-1-4939-8751-1_12. [DOI] [PubMed] [Google Scholar]
- 53.Longo M.C., Berninger M.S., Hartley J.L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz C., Huang J., Giardina S.F., Feinberg P.B., Mirza A.H., Bacolod M.D., Soper S.A., Barany F. Single-molecule detection of cancer mutations using a novel PCR-LDR-qPCR assay. Hum Mutat. 2020;41:1051–1068. doi: 10.1002/humu.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bacolod M.D., Barany F. Gene dysregulations driven by somatic copy number aberrations-biological and clinical implications in colon tumors: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2010;12:552–561. doi: 10.2353/jmoldx.2010.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bacolod M.D., Barany F. Molecular profiling of colon tumors: the search for clinically relevant biomarkers of progression, prognosis, therapeutics, and predisposition. Ann Surg Oncol. 2011;18:3694–3700. doi: 10.1245/s10434-011-1615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheffer M., Bacolod M.D., Zuk O., Giardina S.F., Pincas H., Barany F., Paty P.B., Gerald W.L., Notterman D.A., Domany E. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9:eaan2415. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., Samet Y., Maoz M., Druid H., Arner P., Fu K.Y., Kiss E., Spalding K.L., Landesberg G., Zick A., Grinshpun A., Shapiro A.M.J., Grompe M., Wittenberg A.D., Glaser B., Shemer R., Kaplan T., Dor Y. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9:5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo Y., Wong C.J., Kaz A.M., Dzieciatkowski S., Carter K.T., Morris S.M., Wang J., Willis J.E., Makar K.W., Ulrich C.M., Lutterbaugh J.D., Shrubsole M.J., Zheng W., Markowitz S.D., Grady W.M. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014;147:418–429.e418. doi: 10.1053/j.gastro.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.King W.D., Ashbury J.E., Taylor S.A., Tse M.Y., Pang S.C., Louw J.A., Vanner S.J. A cross-sectional study of global DNA methylation and risk of colorectal adenoma. BMC Cancer. 2014;14:488. doi: 10.1186/1471-2407-14-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cassinotti E., Melson J., Liggett T., Melnikov A., Yi Q., Replogle C., Mobarhan S., Boni L., Segato S., Levenson V. DNA methylation patterns in blood of patients with colorectal cancer and adenomatous colorectal polyps. Int J Cancer. 2012;131:1153–1157. doi: 10.1002/ijc.26484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swierczynski S., Klieser E., Illig R., Alinger-Scharinger B., Kiesslich T., Neureiter D. Histone deacetylation meets miRNA: epigenetics and post-transcriptional regulation in cancer and chronic diseases. Expert Opin Biol Ther. 2015;15:651–664. doi: 10.1517/14712598.2015.1025047. [DOI] [PubMed] [Google Scholar]
- 64.Wang S., Wu W., Claret F.X. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics. 2017;12:187–197. doi: 10.1080/15592294.2016.1273308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haimovich G., Medina D.A., Causse S.Z., Garber M., Millan-Zambrano G., Barkai O., Chavez S., Perez-Ortin J.E., Darzacq X., Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 66.Boland C.R., Thibodeau S.N., Hamilton S.R., Sidransky D., Eshleman J.R., Burt R.W., Meltzer S.J., Rodriguez-Bigas M.A., Fodde R., Ranzani G.N., Srivastava S. A National Cancer Institute Workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 67.He T., Wang C., Zhang M., Zhang X., Zheng S., Linghu E., Guo M. Epigenetic regulation of voltage-gated potassium ion channel molecule Kv1.3 in mechanisms of colorectal cancer. Discov Med. 2017;23:155–162. [PubMed] [Google Scholar]
- 68.Ouadid-Ahidouch H., Rodat-Despoix L., Matifat F., Morin G., Ahidouch A. DNA methylation of channel-related genes in cancers. Biochim Biophys Acta. 2015;1848:2621–2628. doi: 10.1016/j.bbamem.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Gu X., Mao X., Lussier M.P., Hutchison M.A., Zhou L., Hamra F.K., Roche K.W., Lu W. GSG1L suppresses AMPA receptor-mediated synaptic transmission and uniquely modulates AMPA receptor kinetics in hippocampal neurons. Nat Commun. 2016;7:10873. doi: 10.1038/ncomms10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu P., Lu Y., Liu H., Wen W., Jia D., Wang Y., You M. Genome-wide association and fine mapping of genetic loci predisposing to colon carcinogenesis in mice. Mol Cancer Res. 2012;10:66–74. doi: 10.1158/1541-7786.MCR-10-0540. [DOI] [PubMed] [Google Scholar]
- 71.Galamb O., Kalmar A., Bartak B.K., Patai A.V., Leiszter K., Peterfia B., Wichmann B., Valcz G., Veres G., Tulassay Z., Molnar B. Aging related methylation influences the gene expression of key control genes in colorectal cancer and adenoma. World J Gastroenterol. 2016;22:10325–10340. doi: 10.3748/wjg.v22.i47.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boratko A., Csortos C. TIMAP, the versatile protein phosphatase 1 regulator in endothelial cells. IUBMB Life. 2017;69:918–928. doi: 10.1002/iub.1695. [DOI] [PubMed] [Google Scholar]
- 73.Krispin S., Stratman A.N., Melick C.H., Stan R.V., Malinverno M., Gleklen J., Castranova D., Dejana E., Weinstein B.M. Growth differentiation factor 6 promotes vascular stability by restraining vascular endothelial growth factor signaling. Arterioscler Thromb Vasc Biol. 2018;38:353–362. doi: 10.1161/ATVBAHA.117.309571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X., Ji P., Zhang Y., LaComb J.F., Tian X., Li E., Williams J.L. Aberrant DNA methylation: implications in racial health disparity. PLoS One. 2016;11:e0153125. doi: 10.1371/journal.pone.0153125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pirini F., Noazin S., Jahuira-Arias M.H., Rodriguez-Torres S., Friess L., Michailidi C., Cok J., Combe J., Vargas G., Prado W., Soudry E., Perez J., Yudin T., Mancinelli A., Unger H., Ili-Gangas C., Brebi-Mieville P., Berg D.E., Hayashi M., Sidransky D., Gilman R.H., Guerrero-Preston R. Early detection of gastric cancer using global, genome-wide and IRF4, ELMO1, CLIP4 and MSC DNA methylation in endoscopic biopsies. Oncotarget. 2017;8:38501–38516. doi: 10.18632/oncotarget.16258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Battaglia R.A., Delic S., Herrmann H., Snider N.T. Vimentin on the move: new developments in cell migration. F1000Res. 2018;7 doi: 10.12688/f1000research.15967.1. F1000 Faculty Rev-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Q., Dong Y., Wu W., Zhu C., Chong H., Lu J., Yu D., Liu L., Lv F., Wang S. Detection and differential diagnosis of colon cancer by a cumulative analysis of promoter methylation. Nat Commun. 2012;3:1206. doi: 10.1038/ncomms2209. [DOI] [PubMed] [Google Scholar]
- 78.Ned R.M., Melillo S., Marrone M. Fecal DNA testing for colorectal cancer screening: the ColoSure test. PLoS Curr. 2011;3:RRN1220. doi: 10.1371/currents.RRN1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shankar J., Messenberg A., Chan J., Underhill T.M., Foster L.J., Nabi I.R. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 2010;70:3780–3790. doi: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 80.Toth K., Galamb O., Spisak S., Wichmann B., Sipos F., Valcz G., Leiszter K., Molnar B., Tulassay Z. The influence of methylated septin 9 gene on RNA and protein level in colorectal cancer. Pathol Oncol Res. 2011;17:503–509. doi: 10.1007/s12253-010-9338-7. [DOI] [PubMed] [Google Scholar]
- 81.Bacolod M.D., Huang J., Giardina S.F., Feinberg P.B., Mirza A.H., Swistel A., Soper S.A., Barany F. Prediction of blood-based biomarkers and subsequent design of bisulfite PCR-LDR-qPCR assay for breast cancer detection. BMC Cancer. 2020;20:85. doi: 10.1186/s12885-020-6574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanzer M., Balluff B., Distler J., Hale K., Leodolter A., Rocken C., Molnar B., Schmid R., Lofton-Day C., Schuster T., Ebert M.P. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5:e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ebert M.P., Model F., Mooney S., Hale K., Lograsso J., Tonnes-Priddy L., Hoffmann J., Csepregi A., Rocken C., Molnar B., Schulz H.U., Malfertheiner P., Lofton-Day C. Aristaless-like homeobox-4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology. 2006;131:1418–1430. doi: 10.1053/j.gastro.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 84.Bartak B.K., Kalmar A., Galamb O., Wichmann B., Nagy Z.B., Tulassay Z., Dank M., Igaz P., Molnar B. Blood collection and cell-free DNA isolation methods influence the sensitivity of liquid biopsy analysis for colorectal cancer detection. Pathol Oncol Res. 2018;25:915–923. doi: 10.1007/s12253-018-0382-z. [DOI] [PubMed] [Google Scholar]
- 85.Pedersen S.K., Symonds E.L., Baker R.T., Murray D.H., McEvoy A., Van Doorn S.C., Mundt M.W., Cole S.R., Gopalsamy G., Mangira D., LaPointe L.C., Dekker E., Young G.P. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer. 2015;15:654. doi: 10.1186/s12885-015-1674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishio M., Sakakura C., Nagata T., Komiyama S., Miyashita A., Hamada T., Kuryu Y., Ikoma H., Kubota T., Kimura A., Nakanishi M., Ichikawa D., Fujiwara H., Okamoto K., Ochiai T., Kokuba Y., Sonoyama T., Ida H., Ito K., Chiba T., Ito Y., Otsuji E. RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res. 2010;30:2673–2682. [PubMed] [Google Scholar]
- 87.Herbst A., Rahmig K., Stieber P., Philipp A., Jung A., Ofner A., Crispin A., Neumann J., Lamerz R., Kolligs F.T. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106:1110–1118. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 88.Garrigou S., Perkins G., Garlan F., Normand C., Didelot A., Le Corre D., Peyvandi S., Mulot C., Niarra R., Aucouturier P., Chatellier G., Nizard P., Perez-Toralla K., Zonta E., Charpy C., Pujals A., Barau C., Bouche O., Emile J.F., Pezet D., Bibeau F., Hutchison J.B., Link D.R., Zaanan A., Laurent-Puig P., Sobhani I., Taly V. A study of hypermethylated circulating tumor DNA as a universal colorectal cancer biomarker. Clin Chem. 2016;62:1129–1139. doi: 10.1373/clinchem.2015.253609. [DOI] [PubMed] [Google Scholar]
- 89.Matthaios D., Balgkouranidou I., Karayiannakis A., Bolanaki H., Xenidis N., Amarantidis K., Chelis L., Romanidis K., Chatzaki A., Lianidou E., Trypsianis G., Kakolyris S. Methylation status of the APC and RASSF1A promoter in cell-free circulating DNA and its prognostic role in patients with colorectal cancer. Oncol Lett. 2016;12:748–756. doi: 10.3892/ol.2016.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lissa D., Robles A.I. Methylation analyses in liquid biopsy. Transl Lung Cancer Res. 2016;5:492–504. doi: 10.21037/tlcr.2016.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han K., Lee T.Y., Nikitopoulos D.E., Soper S.A., Murphy M.C. A vertically stacked, polymer, microfluidic point mutation analyzer: rapid high accuracy detection of low-abundance K-ras mutations. Anal Biochem. 2011;417:211–219. doi: 10.1016/j.ab.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hashimoto M., Barany F., Soper S.A. Polymerase chain reaction/ligase detection reaction/hybridization assays using flow-through microfluidic devices for the detection of low-abundant DNA point mutations. Biosens Bioelectron. 2006;21:1915–1923. doi: 10.1016/j.bios.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 93.Albrecht J.C., Kotani A., Lin J.S., Soper S.A., Barron A.E. Simultaneous detection of 19 K-ras mutations by free-solution conjugate electrophoresis of ligase detection reaction products on glass microchips. Electrophoresis. 2013;34:590–597. doi: 10.1002/elps.201200462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gerry N.P., Witowski N.E., Day J., Hammer R.P., Barany G., Barany F. Universal DNA microarray method for multiplex detection of low abundance point mutations. J Mol Biol. 1999;292:251–262. doi: 10.1006/jmbi.1999.3063. [DOI] [PubMed] [Google Scholar]
- 95.Wang Y., Vaidya B., Farquar H.D., Stryjewski W., Hammer R.P., McCarley R.L., Soper S.A., Cheng Y.W., Barany F. Microarrays assembled in microfluidic chips fabricated from poly(methyl methacrylate) for the detection of low-abundant DNA mutations. Anal Chem. 2003;75:1130–1140. doi: 10.1021/ac020683w. [DOI] [PubMed] [Google Scholar]
- 96.Situma C., Wang Y., Hupert M., Barany F., McCarley R.L., Soper S.A. Fabrication of DNA microarrays onto poly(methyl methacrylate) with ultraviolet patterning and microfluidics for the detection of low-abundant point mutations. Anal Biochem. 2005;340:123–135. doi: 10.1016/j.ab.2005.01.044. [DOI] [PubMed] [Google Scholar]
- 97.Hashimoto M., Hupert M.L., Murphy M.C., Soper S.A., Cheng Y.W., Barany F. Ligase detection reaction/hybridization assays using three-dimensional microfluidic networks for the detection of low-abundant DNA point mutations. Anal Chem. 2005;77:3243–3255. doi: 10.1021/ac048184d. [DOI] [PubMed] [Google Scholar]
- 98.Soper S.A., Hashimoto M., Situma C., Murphy M.C., McCarley R.L., Cheng Y.W., Barany F. Fabrication of DNA microarrays onto polymer substrates using UV modification protocols with integration into microfluidic platforms for the sensing of low-abundant DNA point mutations. Methods. 2005;37:103–113. doi: 10.1016/j.ymeth.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 99.Hashimoto M., Barany F., Xu F., Soper S.A. Serial processing of biological reactions using flow-through microfluidic devices: coupled PCR/LDR for the detection of low-abundant DNA point mutations. Analyst. 2007;132:913–921. doi: 10.1039/b700071e. [DOI] [PubMed] [Google Scholar]
- 100.Dharmasiri U., Njoroge S.K., Witek M.A., Adebiyi M.G., Kamande J.W., Hupert M.L., Barany F., Soper S.A. High-throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grunau C., Clark S.J., Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., Higgins J.M., Moltchanov V., Kuo F.C., Kluk M.J., Henderson B., Kinnunen L., Koistinen H.A., Ladenvall C., Getz G., Correa A., Banahan B.F., Gabriel S., Kathiresan S., Stringham H.M., McCarthy M.I., Boehnke M., Tuomilehto J., Haiman C., Groop L., Atzmon G., Wilson J.G., Neuberg D., Altshuler D., Ebert B.L. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu Y., Ulrich B.C., Supplee J., Kuang Y., Lizotte P.H., Feeney N.B., Guibert N.M., Awad M.M., Wong K.K., Janne P.A., Paweletz C.P., Oxnard G.R. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24:4437–4443. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 104.Langstein J., Milsom M.D., Lipka D.B. Impact of DNA methylation programming on normal and pre-leukemic hematopoiesis. Semin Cancer Biol. 2018;51:89–100. doi: 10.1016/j.semcancer.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 105.Jaiswal S., Libby P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol. 2020;17:137–144. doi: 10.1038/s41569-019-0247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen S.Y., Singhania R., Fehringer G., Chakravarthy A., Roehrl M.H.A., Chadwick D. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–583. doi: 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 107.Maggi E.C., Gravina S., Cheng H., Piperdi B., Yuan Z., Dong X., Libutti S.K., Vijg J., Montagna C. Development of a method to implement whole-genome bisulfite sequencing of cfDNA from cancer patients and a mouse tumor model. Front Genet. 2018;9:6. doi: 10.3389/fgene.2018.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu L., Toung J.M., Jassowicz A.F., Vijayaraghavan R., Kang H., Zhang R., Kruglyak K.M., Huang H.J., Hinoue T., Shen H., Salathia N.S., Hong D.S., Naing A., Subbiah V., Piha-Paul S.A., Bibikova M., Granger G., Barnes B., Shen R., Gutekunst K., Fu S., Tsimberidou A.M., Lu C., Eng C., Moulder S.L., Kopetz E.S., Amaria R.N., Meric-Bernstam F., Laird P.W., Fan J.B., Janku F. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann Oncol. 2018;29:1445–1453. doi: 10.1093/annonc/mdy119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Holmila R., Sklias A., Muller D.C., Degli Esposti D., Guilloreau P., McKay J., Sangrajrang S., Srivatanakul P., Hainaut P., Merle P., Herceg Z., Nogueira da Costa A. Targeted deep sequencing of plasma circulating cell-free DNA reveals vimentin and fibulin 1 as potential epigenetic biomarkers for hepatocellular carcinoma. PLoS One. 2017;12:e0174265. doi: 10.1371/journal.pone.0174265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaca-Paniagua F., Oliver J., Nogueira da Costa A., Merle P., McKay J., Herceg Z., Holmila R. Targeted deep DNA methylation analysis of circulating cell-free DNA in plasma using massively parallel semiconductor sequencing. Epigenomics. 2015;7:353–362. doi: 10.2217/epi.14.94. [DOI] [PubMed] [Google Scholar]
- 111.Sun K., Jiang P., Chan K.C., Wong J., Cheng Y.K., Liang R.H., Chan W.K., Ma E.S., Chan S.L., Cheng S.H., Chan R.W., Tong Y.K., Ng S.S., Wong R.S., Hui D.S., Leung T.N., Leung T.Y., Lai P.B., Chiu R.W., Lo Y.M. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112:E5503–E5512. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relationship between patient age (in years) and methylation level at each of the seven CpG sites, for both primary tumor (PT; red solid circles) and solid normal colon tissues (SN; blue solid circles) included in The Cancer Genome Atlas colorectal cancer (CRC; colorectal adenocarcinoma) cohort. The indicated methylation levels range from −0.5 (0% methylated), to 0 (50% methylated), to 0.5 (100% methylated). Insets: The squares of correlation coefficient (R2) between methylation level and age of diagnosis, for PT and SN tissues. The gray shade that covers each line (solid line for PT, and dashed line for SN) represents the confidence region for that particular linear fit.
Comparative heat map (primary tumor and normals) depicting the expression level of each gene whose corresponding CpG sites were interrogated in the multiplex assay. CRC, colorectal cancer; FC, fold change; Max, maximum; Min, minimum; SN, solid normal colon tissues.
The relationship between patient gene transcription and methylation at each of the seven CpG sites, in colon cancer cohort. The relationship between CpG methylation and the transcription level of its corresponding gene, among the primary tumor (red solid circles) and solid normal colon tissues (blue solid circles) included in The Cancer Genome Atlas colorectal cancer (CRC; colorectal adenocarcinoma) cohort. The indicated methylation levels range from −0.5 (0% methylated), to 0 (50% methylated), to 0.5 (100% methylated). Insets: The Pearson correlation coefficient (R) values between the two variables. The gray shade that covers each line represents the confidence region for that particular linear fit. Expr., expression.
A: An alternative scheme to identify potential blood-based methylation markers for high microsatellite instability (MSI-H) colorectal cancers. Filter A': similar to filter A (see Results), except the modification of requirement (i) to the following: (%LM + %HM)(COADREAD, PT-MSI) ≥ 65. B: Heat map depicting the methylation levels at MLH1 locus CpG sites (mostly within promoter region), across 394 The Cancer Genome Atlas (TCGA) colorectal adenocarcinoma (COADREAD) samples. Indicated (with solid, differently colored circles) are the five CpG sites identified through the scheme shown in A. The correlation value (R) between MLH1 expression (expr.) and CpG methylation level at each of these sites is highly negative. GEO, Gene Expression Omnibus; utr, untranslated region.
A: The methylation levels (β values) of the seven CpG markers, for peripheral blood (PB)–derived genomic DNAs (GSE42861). Also indicated are the average β values (βPB) for each of the healthy and rheumatoid arthritis subsets. B: The hypothetical presence of neoclones in certain PB samples (labeled 1, 2, and 3 in A) is illustrated. The main presumption in this model is that the measured β for a PB sample (for a specific marker, such as m_GDF6) is the weighted average of the β values for old clones (βOC) and neoclones (βNC; equation 1). If it is assumed that βOC is the average for samples minus the three outliers (referred to as base cluster), and that βNC for a given outlier sample is almost fully methylated (0.95), then the hypothetical fraction of neoclones (x) can be calculated using equation 1. n = 335 (healthy subset); n = 354 (rheumatoid arthritis subset).