Abstract
Background
Aberrant fucosylation plays a critical role in lung cancer progression. Nevertheless, the key fucosyltransferase with prognostic significance in lung cancer patients, the enzyme's intracellular targets, and complex molecular mechanisms underlying lung cancer metastasis remain incompletely understood.
Methods
We performed a large-scale transcriptome-clinical correlation to identify major fucosyltransferases with significant prognostic values. Invasion, migration, cell adhesion assays were performed using lung cancer cells subject to genetic manipulation of FUT4 levels. Genome-wide RNA-seq and immunoprecipitation-mass spectrometry were used to characterize major cellular processes driven by FUT4, as well as profiling its intracellular protein targets. We also performed lung homing and metastasis assays in mouse xenograft models to determine in vivo phenotypes of high FUT4-expressing cancer cells.
Findings
We show that FUT4 is associated with poor overall survival in lung adenocarcinoma patients. High FUT4 expression promotes lung cancer invasion, migration, epithelial-to-mesenchymal transition, and cell adhesion. FUT4-mediated aberrant fucosylation markedly activates multiple cellular processes, including membrane trafficking, cell cycle, and major oncogenic signaling pathways. The effects are independent of receptor tyrosine kinase mutations. Notably, genetic depletion of FUT4 or targeting FUT4-driven pathways diminishes lung colonization and distant metastases of lung cancer cells in mouse xenograft models.
Interpretation
We propose that FUT4 can be a prognostic predictor and therapeutic target in lung cancer metastasis. Our data provide a scientific basis for a potential therapeutic strategy using targeted therapy in a subset of patients with high FUT4-expressing tumors with no targetable mutations.
Keywords: Fucosyltransferase, Lung cancer, Cancer metastasis, Terminal fucosylation, Glycoproteomics
Research in context.
Evidence before this study
Aberrant expressions of terminal fucosylated epitopes such as Lewis antigens have been reported in non-small cell lung cancer tissues. Several fucosyltransferases such as FUT4, FUT7, among others, have been shown to involve in cancer progression. Nevertheless, the key enzymes with clinical prognostic values have been controversial. While some groups found that terminal fucosyltransferases such as FUT4 or FUT7 may promote lung cancer progression via epithelial-mesenchymal transition (EMT) and epidermal growth factor receptor (EGFR) activation, others demonstrated that FUT4- or FUT6-mediated fucosylation could suppress EGFR activation thereby hindering cancer invasiveness. Moreover, previous mechanistic studies of fucosyltransferases in cancer progression lacked a global view from genome-wide approaches and were limited in potential therapeutic implications.
Added value of this study
Our study took a genome-wide approach in identifying FUT4 as the key fucosyltransferase with significant prognostic value in lung cancer and is the first report in delineating the comprehensive cellular and molecular processes on the omics level that promote the FUT4-mediated malignant phenotype. Furthermore, we provide a valuable data resource of global intracellular targets fucosylated by FUT4 via a glycoproteomic approach. Most importantly, our data demonstrated that FUT4 can be a promising therapeutic target through in vitro and in vivo experiments.
Implication of all the available evidence
Our report bridges clinical significance and mechanistic insights, not only identifying the key prognostic enzyme catalyzing aberrant terminal fucosylation in lung cancer patients, but also elucidating molecular networks altered by FUT4 and its intracellular targets through combined transcriptomic and proteomic analyses. Our data open a new possibility in targeting FUT4 or FUT4-mediated networks such as vesicular transport or oncogenic signaling to curtail cancer metastasis and highlight the potential for integration of glycomics into precision medicine-based therapeutics.
Alt-text: Unlabelled box
1. Introduction
Non-small cell lung cancer is a disease with genetic and gene regulatory complexities. Molecular analysis of individual tumors is essential for the clinical choice of therapeutic strategies in the era of precision medicine. Despite the growing dimensions of lung cancer-associated molecular abnormalities, current practices for lung cancer therapy rely primarily on pathological, mutational, and immunological characterizations. Abundant evidence indicates that aberrant glycosylation plays critical roles in fundamental steps of tumor development and progression, including cell-cell/cell-matrix interactions, metastasis, cancer metabolism as well as immune surveillance [[1], [2], [3]]. In particular, patterns of fucosylation, which adds a fucose (6-deoxy-l-galactose) residue to surface oligosaccharides/proteins catalyzed by the fucosyltransferase (FUT) gene family, are frequently altered in various types of cancer [4, 5]. Depending on the site of the oligosaccharide chain to which the fucose is added, two types of fucosylation— core and terminal fucosylation—were defined [4]. So far, there are 13 FUTs identified in humans, each of which catalyzes the synthesis of fucosylated glycans with designated glycosidic linkages and targets different substrate proteins/glycans in a tissue-specific manner.
In search of the key fucosyltransferases underlying aberrant fucosylation patterns specific for lung cancer progression, we and others have previously discovered that fucosyltransferase 8 (FUT8), the only enzyme responsible for the core fucosylation with α1,6-linkage, mediated the malignant phenotypes of non-small cell lung cancer [6, 7]. On the other hand, the clinical significance of fucosyltransferases involved in terminal fucosylation (α1,3- or α1,4- linkage) in lung cancer remains controversial. Past studies have demonstrated aberrant expression of terminal fucosylated epitopes such as Lewis antigens in non-small cell lung cancer tissues [[8], [9], [10]]. Nevertheless, the prognostic values of various Lewis antigens appeared to differ. Expressions of sialyl Lewis x (sLex) and Lewis x (Lex) were associated with shortened survival times [10]. In contrast, Lewis y (Ley) seemed to predict better survival or confer limited clinical significance [8, 9]. Furthermore, while some groups found that terminal fucosyltransferases such as FUT4 or FUT7 may promote lung cancer progression [[11], [12], [13]], another group demonstrated that FUT4- or FUT6-mediated fucosylation of epidermal growth factor receptor (EGFR) could suppress EGFR dimerization and activation [14]. As multiple fucosyltransferases (FUT3–7, 9–11) are involved in terminal fucosylation and Lewis antigen synthesis, a systemic approach on all α1,3- or α1,4- fucosyltransferases with large scale clinical correlation in individual subtypes of non-small cell lung cancers, followed by in-depth mechanistic and molecular studies, is needed to identify the key enzyme that accounts for malignant phenotype of lung cancer and decipher the complex molecular networks involved in cancer progression.
In the present study, our group examined 81 surgically-resected tumor tissues from patients with non-small cell lung cancer for molecular and prognostic correlations on all terminal α1,3- or α1,4- fucosyltransferases, and independently validated our results with the TCGA lung cancer cohorts for cross-ethnic generalization. We identified fucosyltransferase 4 (FUT4) as the main indicator for poor clinical outcome in lung adenocarcinoma patients. We conducted mechanistic and functional studies in vitro and in vivo by altering FUT4 expression levels in lung cancer cells and deciphered the molecular networks affected by FUT4 through integrated transcriptomic and glycoproteomic analyses. We found that FUT4 activates intracellular transport machinery and enhances oncogenic signaling via aberrant fucosylation of membrane trafficking components as well as signaling cascade proteins. Finally, we demonstrated the therapeutic implications of our findings by directly targeting FUT4 or by FUT4-related pathway inhibitors. Our study pinpointed a clinically-significant fucosylation enzyme for the malignant phenotype of lung adenocarcinoma and delineated its complex glycoproteomic networks for developing therapeutics for cancer progression in the future.
2. Materials and methods
2.1. TCGA data acquisition and analysis
Normalized RNA-seq expression datasets and EGFR mutation status of The Cancer Genome Atlas (TCGA) lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) cohorts were acquired from Firehose data repository using the R/Bioconductor package “RTCGAToolbox” (version 2.10.0, run data 2016–01–28) [15]. We used the log-rank test to compare between-group survival differences. Patients were censored at the last follow-up or death, with post-surgery recurrence (>3 months) classified as an event for relapse-free survival. As for overall survival, patients were censored at the last follow-up, and death was classified as an event. Heatmaps of gene expression profiles were created by R package “pheatmap” version 1.0.10.
2.2. Human tissues and subject follow-up data
Surgically-resected lung tumors and adjacent uninvolved lung tissues were obtained from patients with non-small cell lung cancer from National Taiwan University Hospital (NTUH, Taipei, Taiwan) from 1994 to 2010. Clinical information, including gender, histological type, stage, and overall survival, was collected. Median follow-up time was 51 months (range 2–164 months). The staging of lung cancer was based on the American Joint Committee on Cancer (AJCC) TNM system, a standard classification system of the anatomical extent of cancers describing the size of the tumor (T), the extent of spread to the lymph nodes (N), and the presence of metastasis (M). This study has been approved by the Institutional Review Board (IRB) of National Taiwan University Hospital (Approval No.: 201701010RINB).
2.3. Quantitative PCR analysis
PCR was carried out on a CFX96 TouchTM Real-Time PCR Detection System using Sybr Green I detection (BIO-RAD) according to the manufacturer's instructions. Relative expression levels were normalized to TBP (TATA-box-binding protein) and calculated using the 2−ΔΔCt method as described. Primers were purchased from IDT (Integrated DNA Technologies), and the sequences are listed in Supplemental Table S6. mRNA expression levels of FUT4 were measured in 75 human lung adenocarcinoma tissues obtained from NTUH and matched with their clinical data.
2.4. Human lung cancer cell lines
CL1–0 and CL1–5 human lung adenocarcinoma cell lines were kindly provided by Prof. Pan-Chyr Yang (Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan) [16]. All other human lung cancer cell lines were obtained from ATCC. Cell line authentication was performed by STR analysis for all cell lines used in this study. Cells were maintained in complete RPMI-1640 media with 10% (v/v) FBS, 1% (v/v) l-glutamine, and 1% (v/v) Penicillin-Streptomycin (Gibco™) at 37 °C and 5% CO2. Detection of mycoplasma in cell culture was performed before a new experiment starts and every three months for ongoing experiments. Additional testing may be done as needed. For signaling transduction assays, cells were cultured in medium containing 0.5% (v/v) FBS for 16 h for cell cycle synchronization. Then cells were treated with EGF (10 ng/mL; AF-100–15, PeproTech) or TGF-β1 (5 ng/mL; 100–21, PeproTech) for 0.5, 1, 2, 6 h, respectively.
2.5. RNA interference
FUT4 expression was knocked down using shERWOOD UltramiR lentiviral inducible shFUT4 system. pZIP-Mcmv-shFUT4 plasmids (15 μg) were transfected into cells (5 × 106 cells) using Lipofectamine™ 3000 transfection reagents (45 μL). Cells were incubated for 16 h and selected by fresh RPMI medium with 2 μg/mL puromycin for 24 h. The effects of shRNA knockdown will be evaluated using proliferation assays, migration assays, and invasion assays.
2.6. Single-cell tracking
Cells were seeded on the 6-well plate overnight. On the next day, time-lapse imaging of the cells was performed using an automated inverted microscope (Leica DMI 6000B) for 24 h. Data were analyzed by MetaMorph® software. For each sample, at least 20 cells were selected and tracked for cell motility calculation.
2.7. Invasion/migration assay
Cell invasion assays were performed using 24-well transwell units with permeable polycarbonate filter with pore sizes of 8 μm. 24-well transwell units were pre-coated with 2 mg/mL BD Matrigel® (BD Biosciences), and incubated at 37 °C for 1 h. Each lower compartment of the transwell unit contained a complete medium with 10% FBS. 5 × 104 cells in 0.5 ml RPMI with 0.5% BSA were added into the upper compartment of the transwell unit and were incubated at 37 °C overnight. On the next day, the Matrigel® coated on the filter was removed with cotton swabs, and the filter was gently separated from the transwell unit. The cells attached to the lower surface of the filter membrane were fixed with 4% paraformaldehyde in PBS and stained with 300 nM DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride). The number of cells was counted at 400X magnification under a fluorescent microscope. For each sample, the average number of cells from eight high-power fields (HPF) was recorded. Cell migration assays were performed using the same 24-well transwell units without the Matrigel® coating.
2.8. Mouse experiments
4–6 weeks old male C57BL/6JNarl, NOD.CB17-Prkdcscid/JNarl (NOD SCID) and BALB/cAnN.Cg-Foxn1nu/CrlNarl (NUDE) mice were purchased from the National Laboratory Animal Center (Taiwan) and maintained under standard pathogen-free conditions.
In vivo adhesion/extravasation assays: A549 (FUT4high and vector control), CL1–0 (FUT4 and vector control) and CL1–5 (shFUT4 and scramble control) human lung cancer cells were pre-stained with Cyto-ID™ long-term dyes (Enzo Life Sciences), and intracardially injected into the right ventricle of 6-week-old C57BL/6JNarl mice (5 mice per group; 5 × 105 cells/mouse). Mice were kept warm with electric heating blankets for 20 min. Mice were then sacrificed and intravenously perfused with phosphate-buffered saline to remove blood cells and unattached human cancer cells. The lungs were removed and fixed with 1% agarose on a 35 mm imaging μ-Dish with a high glass bottom (ibidi, 81158). The lungs were examined by the Axio Observer Inverted Microscope System (Zeiss). Tail vein assay for cancer metastases: A549 (FUT4high and vector control) and CL1–5 (shFUT4 and scramble control) human lung cancer cells were pre-stained with Cyto-ID™ long-term dyes (Enzo Life Sciences) and were injected into the tail vein of 6-week-old nude mice with 5 × 105 cells per mouse. In vivo bioluminescent imaging to monitor cancer cell metastasis was performed using the IVIS® Spectrum in vivo imaging system at day 1 and day 7 after tail vein injection. Mice were euthanized with carbon dioxide at 28 days. All organs were removed and fixed in 10% formalin. The lung nodules were counted through gross inspection and under microscopic examination. The number of mice used in each group (n = 6) was based on the goal of having 98% power to detect a 2-fold difference in nodule numbers between groups at p < 0.05. Spontaneous metastasis of subcutaneous tumor xenografts: Tumor xenografts were established in 6-week-old NOD SCID mice via subcutaneous injection into the dorsal region of each mouse with 1 × 107 cells/0.15 ml Hank's balanced salt solution containing Matrigel® (BD Biosciences). Experiments were performed at five mice in each group (A549_Vector, A549_FUT4med, and A549_FUT4high). Mice were then sacrificed at 8 weeks. Distant metastases in the lungs were evaluated by gross visualization of internal organs and by microscopic imaging of lung tissue sections. In vivo drug treatment: A549 (FUT4high and vector control) human lung cancer cells were injected into the tail vein of 6-week-old nude mice with 5 × 105 cells per mouse to establish the lung cancer metastatic model. Mice were intraperitoneally injected with DMSO (mock control), afatinib (0.25 mg/Kg BW) (TargetMol TM-T2510), and LY2157299 (15 mg/Kg BW) (TargetMol TM-T2303) three times per week from week 2 to week 7 post-injection to inhibit tumor metastasis. Mice were euthanized with carbon dioxide at 49 days. The lungs were removed and fixed in 10% formalin. The lung nodules were counted through gross inspection and under microscopic examination. The number of mice used in each group (n = 6) was based on the goal of having 98% power to detect a 2-fold difference in nodule numbers between groups at p < 0.05.
2.9. Adhesion assay
The uncharged polystyrene 96-well plates were coated with different recombinant proteins- Collagen IV (Merck Millipore), P-Selectin, E-Selectin, or L-selectin (PeproTech), and incubated at room temperature for 2 h. The coated 96-well plates were blocked with 5% BSA/PBS at 37 °C for 1 h. Subsequently, 5 × 104 cells in serum-free medium were added onto the coated 96-well plate. After incubation at 37 °C for 1 h, non-adherent cells were gently washed away twice with 1x PBS. Adherent cells were fixed with 4% paraformaldehyde/PBS for 20 min and stained with 300 nM DAPI. Cell images were obtained with MD ImageXpress Micro XL High-Content Analysis System (Molecular Devices). The number of adherent cells was calculated with the MetaMorph software (Molecular Devices).
2.10. RNA-sequencing and data analysis
Total RNAs were isolated from human lung cancer tissues from National Taiwan University Hospital and lung cancer cell lines. RNA integrity was evaluated with an Agilent 2100 Bioanalyzer using an RNA 6000 nano kit (Agilent Technologies). RNA sequencing was performed using Illumina HiSeq 4000® (for lung cancer tissue RNAs) and Illumina NextSeq 500® systems (for cell line RNAs). Raw reads in FASTAQ format were processed using Trimmomatic software version 0.33 for adapter trimming and quality filtering [17]. The processed sequencing reads were mapped to the human reference genome (hg19) using bowtie v2.2.6 with parameters: –forward-prob 0 –output- genome-bam for cell lines and clinical samples [18, 19]. The mapped reads in BAM files were annotated with GENCODE Release 25 (GRCh37), using GenomicFeatures (version 1.32.2) and GenomicAlignments (version 1.16.0) packages. Finally, DESeq2 package (version 1.20.0) was used to generate a raw-counts matrix followed by FPKM normalization and differential expression analysis [20]. Gene Set Enrichment Analysis (GSEA) was performed using the official software javaGSEA version 3.0 with the Molecular Signatures Database (MSigDB, version 6.1) containing BioCarta, Reactome and KEGG gene sets [21, 22]. The signal-to-noise metric was used to rank differentially-expressed genes in GSEA. The number of permutations was set at 1000, and the enrichment statistic was set as weighted. The resulting gene sets with a nominal p value < 0.05 were considered as enriched. All RNA-seq data have been deposited in GEO: GSE120622.
2.11. Immunofluorescence staining
For immunolocalization, samples were blocked with 1% (wt/vol) BSA in PBS for 1 h, followed by incubation at 4 °C with primary antibodies for 16 h and with secondary antibodies (donkey anti-mouse IgG Alexa 488 or donkey anti-goat IgG Alexa 488, 1:250 dilution) for 1 h. Cells were counterstained with 300 nM DAPI for 10 min before mounted with ProLong Gold Antifade Mountant (ThermoFisher Scientific). Images were obtained with the Zeiss LSM 880 confocal microscope and analyzed with ZEN microscope software. Antibodies used in this study were listed in Supplemental Table S5.
2.12. Immunoprecipitation
Lewis x and TβRI immunoprecipitations were performed using PierceTM Protein L Magnetic Beads (ThermoFisher Scientific, #88849) and PierceTM Protein A/G Magnetic Beads (ThermoFisher Scientific, #88802) respectively, according to the manufacturer's protocol. Cells were lysed with RIPA buffer containing a protease inhibitor cocktail (GOAL Bio) and incubated on ice for 20 min. The cell lysate was centrifuged for 15 min at 4 °C, and an aliquot of the supernatant was kept aside on ice as input. 1 mg of cell lysate was incubated with 40 μg of primary antibody per IP at 4 °C on a shaker overnight. The following antibodies were used for IP: TGF beta receptor I (Abcam, ab31013) and Lewis x (Abcam, ab3358). The next day, the magnetic beads were loaded in the cell lysate for 4 h at room temperature and washed according to the manufacturer's protocol. For Western blotting and mass spectrometry, beads were resuspended in the SDS sample buffer, boiled for 5 min, and loaded onto an 8% acrylamide gel.
2.13. Mass spectrometry
The post-IP samples were subjected to SDS-PAGE and the subsequent in-gel trypsin digestion. The peptides were dried using the CentriVap Benchtop Vacuum Concentrator (Labconco Corp.), dissolved in 0.1% trifluoroacetic acid (TFA), and desalted with C18 Ziptips (Millipore) according to the manufacturer's protocol. NanoLC−nanoESI-MS/MS identification was performed on an Ultimate system 3000 nanoLC system (ThermoFisher Scientific) connected to the Orbitrap Fusion Lumos mass spectrometer equipped with NanoSpray Flex ion source (ThermoFisher Scientific). Briefly, peptide mixtures conditioned in 0.1% formic acid were injected into the HPLC instrument and enriched on a C18 Acclaim PepMap NanoLC reverse phase column of 25 cm length, 75 μm internal diameter and 2 μm particle size with a pore of 100 Å (ThermoFisher Scientific). The mobile phase consisted of aqueous solvent A--0.1% formic acid, and organic solvent B--0.1% formic acid in acetonitrile. Peptides were separated by 90 min of the segmented gradient from 2% to 40% solvent B at a flow rate of 300 nL/min. The mass spectra were acquired using one full MS scan followed by data-dependent MS/MS of the most intense ions in 3 s. Full MS scans were recorded with an automatic gain control (AGC) target set at 500,000 ions and a resolution of 120,000 at m/z = 200. For MS/MS spectra, selected parent ions within a 1.4 Da isolation window were fragmented by high-energy collision activated dissociation (HCD) with normalized collision energies of 32 eV. HCD-MS/MS with a resolution of 15,000, AGC at 50,000, and potential charge status of 2+ to 7+ were performed. A dynamic exclusion duration for the selection of parent ions was set at 180 s with a repeat count. Spectra were collected by Xcalibur tool version 4.1 (ThermoFisher Scientific). The resulting RAW files were directly processed and analyzed by using MaxQuant software version 1.6.0.16. MS/MS peaks were searched in the Andromeda peptide search engine against the forward and reverse (as a decoy database) of reviewed UniProtKB/Swiss-Prot human proteome database. The common contaminants list attached in MaxQuant was also provided during the search (Tyanova et al., 2016). Peptides with a minimum of 6 amino acids and maximum trypsin missed cleavages of 3 were considered. Variable modifications including methionine oxidation, N-terminal acetylation, Serine/Threonine/Tyrosine phosphorylation, and Asparagine/Glutamine deamidation, and the fixed modification of carbamidomethyl cysteine were applied. Initial parent peptide and fragment mass tolerance were set to 4.5 and 20 ppm, respectively. False discovery rate (FDR) filtration of the peptide-spectrum match and protein levels were applied at 0.05 and 0.01, respectively. Finally, proteins identified as reverse, common contaminations, or matched with only one unique peptide were excluded.
2.14. Western blotting
Cells were lysed in cold RIPA buffer supplemented with protease inhibitors (Merck, 539,137) and phosphatase inhibitors (GoalBio, HC100–008). Equal amounts of proteins were resolved by 8% and 10% SDS-PAGE, transferred onto PVDF membranes, and blocked in a blocking buffer (Visual protein, BP01–1 L) for 1 h at room temperature. Primary antibodies were diluted in the blocking buffer at 1:1000. Secondary antibodies—anti-mouse or anti-rabbit-HRP (Bio-Rad)—were diluted at 1:5000. Antibodies used in this study were listed in Supplemental Table S5. Blots were developed by using LuminataTM Crescendo Western HRP Substrate (Millipore, WBLUR0500), and images were taken in ChemiDoc® Touch imaging system. Raw densitometry for each antibody was normalized to GAPDH using Image Studio Lite. All quantification was performed in at least triplicate on original unmodified blots. One-way ANOVA with a Dunnett multiple comparison test was performed to compare the normalized signal for each antibody using GraphPad Prism 7. Error bars represent mean normalized signals ± SEM.
2.15. Data availability
All RNA-seq data are available at the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) under the accession number GSE120622. All proteomics data are available at the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under the accession number: MSV000083028
2.16. MS-based glycomic analyses
Proteins were extracted from cell lysates, reduced, alkylated, and digested by trypsin/chymotrypsin before subject to glycan release. The glycans were subsequently recovered, permethylated, and cleaned-up for both MALDI-MS and nanoLC-MS2-product dependent MS3, exactly as described before [23]. MALDI-MS was performed on a TOF/TOF 5800 system (AB Sciex) and data manually assigned. NanoLC-MS/MS was carried out on an Orbitrap Fusion Tribrid system (ThermoFisher Scientific), using the same C18 nanoLC conditions, instrument settings, data acquisition, and GlyPick-based data processing and analysis methods described in Hsiao et al., 2017.
3. Statistical analysis
Statistical analysis of expression data was performed by R for Mac (Version 3.5.0) and GraphPad Prism 7 for Mac software. Patient demographics and survival information were gathered from the database registry, electronic medical record, and Taiwan Cancer Registry (for NTUH cohort). Cox proportional hazard modeling was used to examine the effect of expression levels of various fucosyltransferases on overall survival (time until death). The correlation between all members in the FUTs family was analyzed using Pearson correlation coefficients. Statistical analysis of the experimental data, including migration/invasion assays, drug treatments, etc., were performed using the Mann-Whitney test or one-way ANOVA with Dunnett's test. Data are presented as mean ± SEM. p value < 0.05 is considered statistically significant.
4. Ethics statement
The study on primary tumor tissues from lung cancer patients was approved by the Institutional Review Board (IRB) of National Taiwan University Hospital (Approval No.: 201701010RINB). Written informed consent was obtained from all participants prior to inclusion in the study. Every participant was assigned a number, and the data were analyzed at the group level for patient de-identification. All mice experiments were performed in compliance with an experimental protocol (20160126) approved by the NTU College of Medicine Institutional Animal Care and Use Committee (IACUC).
5. Results
5.1. Higher levels of FUT4 is associated with poor prognosis in lung adenocarcinoma
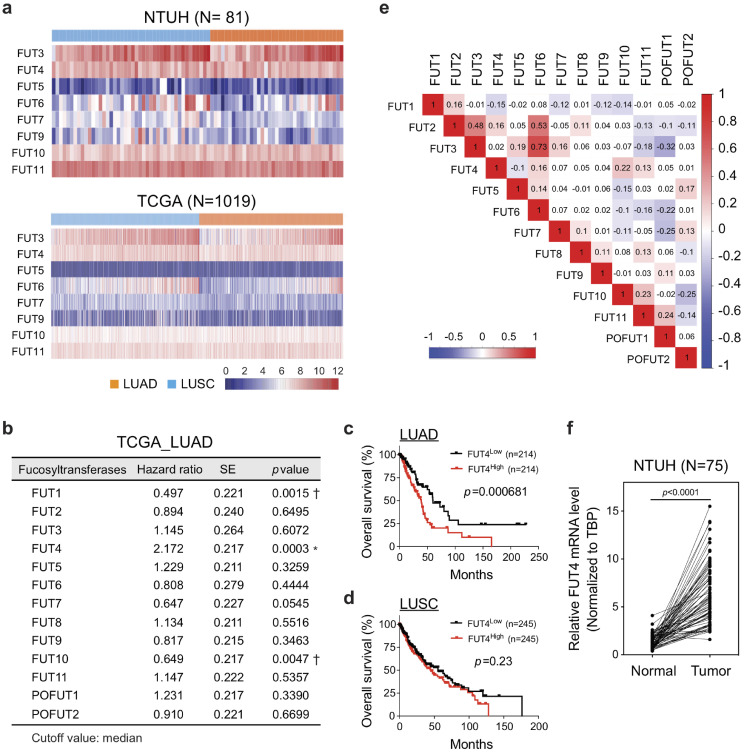
Terminal fucosyltransferases, such as FUT 3–7 and 9–11, are known to catalyze fucosylation with α1,3- and α1,4-linkage. In order to decipher the clinical impact of these fucosyltransferases, we performed transcriptome-clinical correlation on FUT 3–7 and 9–11 using the genome-wide RNA-seq data of 81 primary lung cancer tissues (adenocarcinoma, N = 44; squamous cell carcinoma, N = 37) from National Taiwan University Hospital (NTUH) (GEO: GSE120622). We observed relatively high expression of FUT3, 4, 6, 10, 11 in lung cancer tissues, while little expressions of FUT5, 7, and 9 were detected (Fig 1a). Interestingly, similar patterns were observed in another cohort of 517 lung adenocarcinoma and 502 squamous cell carcinoma patients from The Cancer Genome Atlas (TCGA) despite the ethnic differences and distinct mutational profiles between the two cohorts (Fig. 1a) [24], [25], [26], [27]. To determine the clinical significance of individual fucosyltransferases in terms of overall survival, we divided the patient cohorts into high-expressing and low-expressing groups by the median value of individual fucosyltransferases and performed survival analysis on the two groups using Cox proportional hazard model. The analysis in lung adenocarcinoma patients demonstrated a negative prognostic effect of FUT4 (*, hazard ratio 2.172 ± 0.217; p = 0.0003, Cox regression model), and protective effects of FUT1 (†, hazard ratio = 0.497 ± 0.220, p = 0.0015) and FUT10 (†, hazard ratio = 0.649 ± 0.217, p = 0.0047) on overall survivals (Fig. 1b). Kaplan-Meier curves also showed a significantly worse overall survival of the FUT4High group (p = 0.00068; Fig. 1c). Notably, we did not observe a significant association between FUT4 expression and overall survival in lung squamous cell carcinoma (p = 0.23; Fig. 1d and Table S1), indicating a distinct glycobiological mechanism may be involved in the progression of this cancer subtype. Moreover, to investigate potential functional coordination between different FUTs, we performed correlation analysis based on mRNA levels of FUTs in 517 lung adenocarcinoma tissues. We found little or modest concordance between the expressions of FUT4 and other α1,3- or α1,4-fucosyltransferases (Pearson correlation coefficient between −0.15 to 0.22, Fig. 1e). This suggests the unique role of FUT4 plays in promoting the aggressive phenotypes of lung adenocarcinoma. In contrast, while not having prognostic values, expressions of FUT3 and FUT6 showed a strong correlation (Pearson correlation coefficient, R = 0.73; Fig. 1e), consistent with a previous study showing functional coordination between these two enzymes in colorectal cancers [28]. Moreover, expression levels of FUT4 in 75 surgically-removed lung adenocarcinoma tissues at NTUH were significantly upregulated compared to the adjacent unaffected lung tissues measured by quantitative real-time PCR (p = 0.001) (Fig. 1f), indicating that elevated FUT4 expression may contribute to the tumorigenesis of lung adenocarcinoma.
Fig. 1.
Higher levels of FUT4 is associated with poor prognosis in lung adenocarcinoma. (a) Heatmap of RNA-seq transcriptomic analysis for eight α-(1,3)-fucosyltransferases in surgically-resected lung cancer tissues at National Taiwan University Hospital (LUAD, N = 44, and LUSC, N = 37) and from TCGA database (LUAD, N = 517, and LUSC, N = 502), respectively. LUAD: lung adenocarcinoma. LUSC: lung squamous cell carcinoma. (b) Relationships between expression levels of individual fucosyltransferases and overall survival in patients with lung adenocarcinoma. (N = 428, High vs. Low expressions, cutoff=median; Cox proportional hazard models) (c,d) Kaplan-Meier curves of overall survival for patients with lung adenocarcinoma (LUAD) (c) or squamous cell carcinoma (LUSC) (d). Patients were divided into two groups based on the median value of FUT4 expressions. (e) The correlation matrix of all members in the FUT family based on the RNA-seq data of the TCGA lung adenocarcinoma samples. Numbers in the colored boxes represent Pearson correlation coefficients. Red and blue colors denote positive and negative correlations with statistical significance (p < 0.05), respectively. (f) mRNA levels of FUT4 in lung adenocarcinoma tissues compared to those of paired adjacent unaffected tissues from National Taiwan University Hospital (N = 75). Data information: p value in (b–d) was calculated by the log-rank test, and in (f) was calculated by Wilcoxon matched-pairs signed-rank test.
5.2. FUT4 promotes aggressive phenotypes of human lung cancer cells
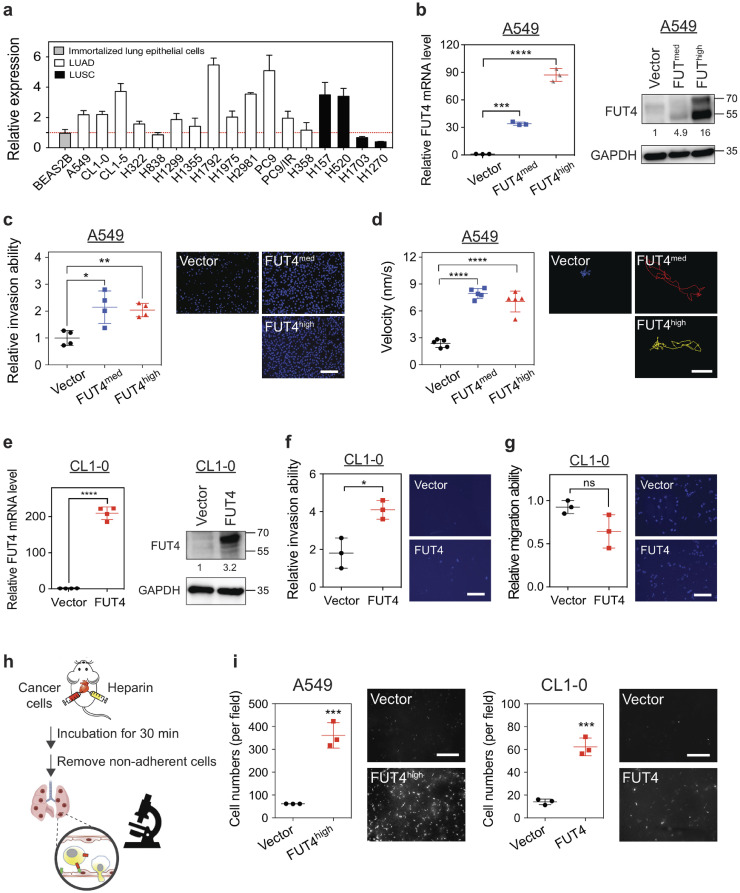
To decipher the mechanisms of FUT4 that account for poor prognosis in lung cancer patients, we established FUT4-overexpressing lung adenocarcinoma cell lines using the tetracycline-controlled transcriptional regulatory system for a series of in vitro and in vivo mechanistic studies. First, we examined expression levels of FUT4 in a panel of 17 human non-small cell lung cancer lines. Higher expressions of FUT4 were observed in 13 of the 17 cell lines as compared with immortalized BEAS-2B lung epithelial cells (Fig. 2a). Subsequently, we stably transfected two human lung adenocarcinoma cell lines, A549 (Fig. 2b) and CL1–0 cells (Fig. 2e), with FUT4. For A549 cells, we selected two clones with different levels of FUT4 expression, A549_FUT4high, and A549_FUT4med, to investigate the dose-responsive relationship of FUT4 protein. We found that A549_FUT4highand A549_FUT4med showed markedly increased invasion abilities through the Matrigel®-coated Boyden chamber (Fig. 2c). Moreover, single-cell migration assays in a live-cell imaging and tracking system revealed increased migration abilities of A549_FUT4high and A549_FUT4med cells. (Fig. 2d). Interestingly, A549_FUT4med cells showed similar invasion and migration abilities with A549_FUT4high cells, which indicates that moderate elevation of FUT4 protein level was sufficient to cause significant functional changes. Similarly, CL1–0_FUT4 cells showed enhanced invasion ability (Fig. 2f), although there was no significant change in cell migration velocity (Fig. 2g).
Fig. 2.
FUT4 promotes aggressive phenotypes of human lung cancer cells. (a) FUT4 mRNA expressions in various human lung cancer cell lines analyzed by quantitative real-time PCR. All experiments were performed in triplicates and presented as mean ± SEM. LUAD: adenocarcinoma. LUSC: squamous cell carcinoma. (b) FUT4 mRNA (left panel) and protein levels (right panel) in A549 human lung cancer cells transduced with FUT4 (A549_vector, A549_FUT4med, A549_FUT4high) measured by quantitative real-time PCR and western blots. (c) Dot plots showing relative invasion ability of A549 cells measured by matrigel-based transwell invasion assays. Representative images of cells with DAPI nuclear stains in the lower chambers are shown in the right panels. Relative invasion ability was calculated using the cell number in the lower chamber of the transwell system for each clone compared to that of the vector control. (d) Dot plots showing migration velocity of A549 cells measured by single-cell tracking assays under a fluorescence microscope. Data were analyzed using Metamorph® software. The paths of cell migration were delineated in the right panels using pseudo-colors. (e) FUT4 mRNA (left panel) and protein levels (right panel) in CL1–0 human lung cancer cells overexpressed with FUT4 (CL1–0_vector, CL1–0_FUT4) measured by quantitative real-time PCR and western blots. (f) Dot plots showing relative invasion ability of CL1–0 cells measured by matrigel-based transwell invasion assays. Representative images of cells with DAPI nuclear stains in the lower chambers are shown in the right panels. Relative invasion ability was calculated using the cell number in the lower chamber of the transwell system for each clone compared to that of the vector control. (g) Dot plots showing the migration ability of CL1–0 cells measured by transwell migration assays. Relative migration ability was calculated using the cell number in the lower chamber of the transwell system for each sample compared to that of the vector control. (h) Diagram of in vivo extravasation and lung colonization assay following injection of lung cancer cells into the right ventricle of C57BL/6 mice. Mice were sacrificed 30 mins after intracardial injection. Whole lung perfusion with normal saline was performed to remove blood and cells not adhered to the pulmonary vasculature. (i) Numbers of lung cancer cells with FUT4 over-expression (left panel, A549_FUT4high; right panel, CL1–0_FUT4) adhered to the vascular walls or retained in the lung tissues following intracardial injection were visualized under Zeiss Axio Observer microscope.
Scale bar in c, d, f, and g; 100 μm. Scale bar in i ; 1 mm. p value in (b-d) was calculated by one-way ANOVA with Dunnett's test, and in (e, f, g, and i) was by Mann-Whitney test. All experiments were performed in three biological replicates and presented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. ns: not significant.
Furthermore, to confirm whether FUT4-mediated aberrant fucosylation may lead to a stronger cell adhesion—an early step in the metastatic cascade, we performed in vitro adhesion assays with several common adhesion molecules within human tissues and blood vessels. Consistent with recent reports [12, 29, 30], our data showed that FUT4 facilitated binding to E-selectin (on endothelial cells) and L-selectin (on lymphocytes), but not P-selectin (on platelets) or collagen IV (Supplemental Fig. 1). In light of this, we further performed an in vivo assay to investigate the effects of FUT4 on organotropic extravasation ability to model the metastatic process in which the cells travel and lodge themselves at distant sites. We pre-labeled lung cancer cells with CYTO-ID® red tracer dye and injected the cells into the right ventricle of C57BL6 mice. Thirty minutes later, mice were sacrificed and perfused with normal saline intracardially to remove cells not adhered to the pulmonary vasculature (Fig. 2h). Marked retention of A549_FUT4high and CL1–0_FUT4 cells in the lungs was observed under a microscope as opposed to their respective vector controls (Fig. 2i). These data suggest that FUT4 is a key player in promoting invasion and colonization of lung adenocarcinoma cells.
5.3. FUT4 drives metastasis of lung cancer cells in mouse xenograft models
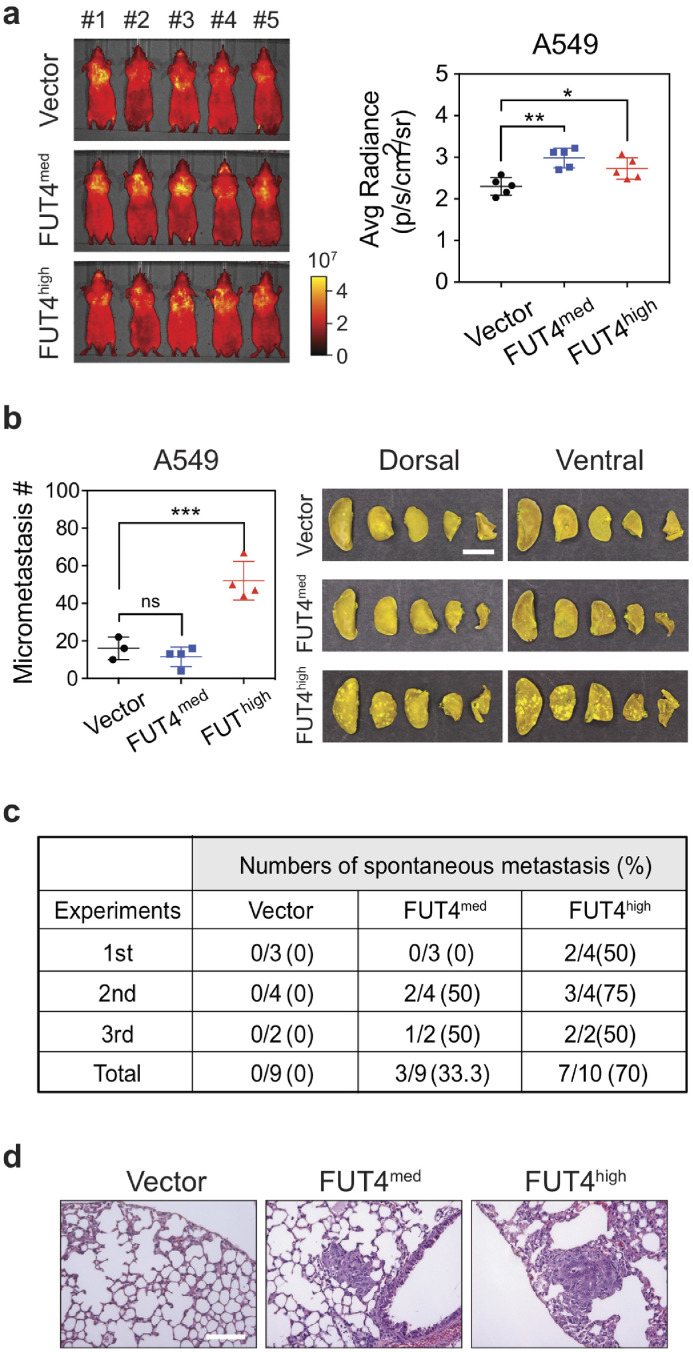
To substantiate the role of FUT4 on lung cancer metastasis, we investigated whether aberrant FUT4 expression led to spontaneous distant metastasis using two mouse xenograft models. In the tail vein assay for cancer metastasis in nude mice, we observed a rapid lung homing phenomenon enhanced by FUT4. As short as one day post tail vein injection, more A549_FUT4med and A549_FUT4high tumor cells were trapped in the lung areas shown by the IVIS® Spectrum imaging system compared to A549_vector cells (Fig. 3a). When we sacrificed mice 28 days post-injections, we found that mice in the FUT4high group had a much greater number of metastatic lung nodules compared to those in the FUT4med and vector groups (p < 0.001) (Fig. 3b). In the second xenograft model, we subcutaneously injected A549_FUT4med, A549_FUT4high cells, and A549_vector into NOD/SCID immunocompromised mice and evaluated spontaneous metastasis of cancer cells. When mice were sacrificed 56 days post-injection, we observed that mice carrying subcutaneous tumors of A549_FUT4high cells and A549_FUT4med cells had higher frequencies of metastatic lung nodules (Fig. 3c and d). Moreover, higher FUT4 levels were correlated with higher metastatic potentials (Fig. 3c and d). These data support the significant role of FUT4 in driving metastasis of lung adenocarcinoma cells in vivo.
Fig. 3.
FUT4 potentiates lung homing ability and drives in vivo metastasis of lung cancer cells. (a) Whole animal imaging of in vivo tumor metastasis in nude mice with the IVIS® Spectrum imaging system at 24 hours following tail vein injections of A549 lung cancer cells pre-stained with Cyto-IDTM long-term dyes. Quantification of signal radiances of metastatic foci in the dorsal and ventral sides of animals was graphed on the right panel. (b) Numbers of metastatic foci in the lungs of nude mice at 28 days after tail vein injection of lung cancer cells with FUT4 overexpression (A549_FUT4med, A549_FUT4high). Pictures of lung nodules from one representative mouse in each group are also shown on the right. (c) Numbers (percentages) of mice with spontaneous lung metastases in NOD/SCID mice bearing subcutaneous tumors of A549_vector, A549_FUT4med, and A549_FUT4high lung cancer cells. Data are summarized from three independent mouse experiments. (d) Representative hematoxylin & eosin (H&E) stained images of lung tissues from mice bearing subcutaneous xenograft lung tumors from (c). Scale bar in (b): 5 mm. Scale bar in (d): 0.1 mm. Data information: p value in (a, b) was calculated by one-way ANOVA with Dunnett's test. * p < 0.05, ** p < 0.01, *** p < 0.001. ns: not significant. All experiments were performed in three biological replicates and presented as mean ± SEM.
5.4. FUT4 mediates activation of membrane trafficking and oncogenic pathways
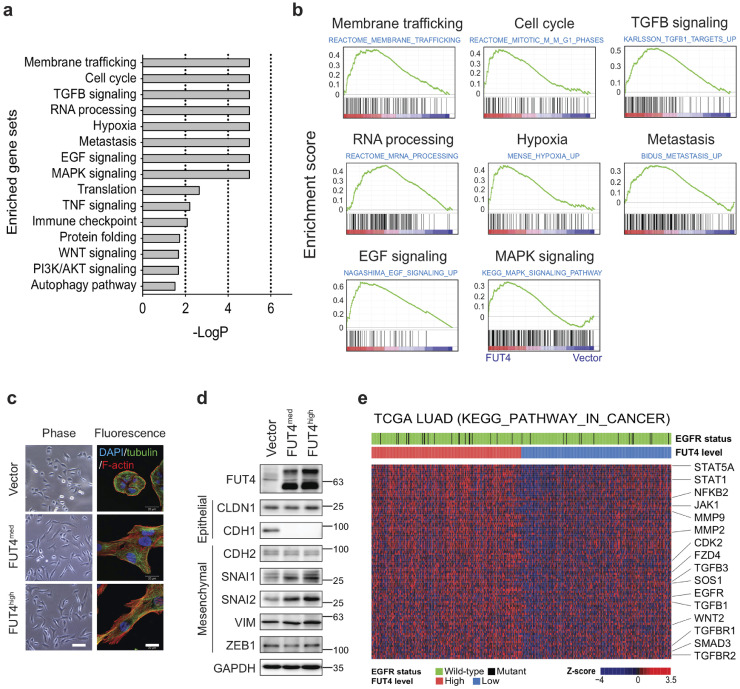
Fucosylation has been shown to play an important regulatory role in protein structure and functions. An in-depth understanding of global transcriptomic profiling on FUT4-associated cellular processes and pathway alterations may facilitate the development of therapeutic strategies to reverse malignant phenotypes. Thus, we performed a transcriptomic analysis on A549_FUT4high and CL1–0_FUT4 cells using genome-wide RNA-seq technologies (GEO: GSE120622). We observed that there were 193 and 291 genes significantly upregulated, and 183 and 330 genes significantly downregulated by more than 1.4-fold (p < 0.05) between FUT4-overexpressing cells and parental cells in A549 and CL1–0, respectively. For genes commonly upregulated in both cell lines, Gene Set Enrichment Analysis [22] revealed activation of multiple pathways related to membrane trafficking, cell cycle, RNA processing, as well as EGF and TGFβ signaling pathways, among others (Fig. 4a and b). The membrane trafficking system mediates intracellular membrane transport of proteins, carbohydrates, lipids between organelles such as endoplasmic reticulum (ER), Golgi complex, and plasma membrane [31]. For instance, SEC31A, SEC13, SEC23A, SEC24B, C, D, components of the coat protein complex II (COPII), which facilitates transport vesicle formation from ER, are upregulated in FUT4-overexpressing cells. Similarly, transcriptomic activation of COPA, COPB, COPE, ARCN1, components of another coatomer complex, which mediates Golgi outbound cargos, are also noted (Table S2). The data indicate that FUT4-overexpressing cells are in a highly active state of biosynthesis and biomolecule trafficking.
Fig. 4.
FUT4 mediates the activation of membrane trafficking and oncogenic pathways. (a) Top 15 positivelyenriched gene sets in genome-wide RNA-seq data of A549_FUT4high and CL1–0_FUT4 cells versus respective vector controls. Enriched gene set data from Gene Set Enrichment Analysis (GSEA) with a nominal p value less than 0.05 and false discovery rate (FDR) less than 0.25 are presented. (b) GSEA enrichment plots of top 8 positively enriched gene sets in A549_FUT4high and CL1–0_FUT4 cells, including membrane trafficking, cell cycle, TGFβ signaling, RNA processing, hypoxia, metastasis, EGF, and MAPK signaling pathways. (c) Immunofluorescent imaging analysis of cytoskeleton and cell morphology of A549 lung cancer cells with various levels of FUT4 over-expression (A549_vector, A549_FUT4med, and A549_FUT4high). Scale bar: 50 μm (left panel) and 20 μm (right panel). (d) Western blot analyses of EMT marker proteins in A549 lung cancer cells with various levels of FUT4 over-expression. (e) Heatmap of RNA-seq transcriptomes for the leading-edge genes of KEGG_pathway_in_cancer from GSEA analyses of TCGA lung adenocarcinoma. Top sidebars denote the mutation status of EGFR and expression levels of FUT4 grouped with the median value cut-off. p value in (a, b) was calculated by one-way ANOVA with Dunnett's test. * p < 0.05, ** p < 0.01, *** p < 0.001. ns: not significant. Experiments were performed in three biological replicates and presented as mean ± SEM.
In addition, FUT4 appears to modulate pathways related to cell cycle (e.g., CDC6, CDC23, etc.), oncogenes (e.g., NRAS, CBFB, FOS), cancer metastasis (e.g., AURKA, CDK1, PLK4), as well as major oncogenic signaling pathways – TGFβ, EGF, MAPK, and WNT signaling (Fig. 4a, b, and Table S2). As a functional validation to activating TGFβ and EGF signaling in FUT4-overexpressing cells, we demonstrated that FUT4 leads to morphological changes characteristic of EMT, a phenomenon mediated by the two signaling pathways, with a prominent reorganization of the cytoskeleton toward a more mesenchymal phenotype in A549_FUT4med, A549_FUT4high and CL1–0_FUT4 cells (Fig. 4c and Supplemental Fig. 2). Consistently, these changes are associated with the down-regulation of CDH1 (E-cadherin) and up-regulation of SNAI1 (a.k.a SNAIL) and SNAI2 (a.k.a SLUG), two EMT-inducing transcription factors (Fig. 4d). Interestingly, minimal increases of mesenchymal proteins—VIM (vimentin) and CDH2 (N-cadherin) are noted, suggesting FUT4 induces an intermediate EMT phenotype in A549 lung cancer cells (Fig. 4d).
To investigate whether these FUT4-mediated pathway alterations were also shown in primary tumor tissues, we examined transcriptomic data in the TCGA cohort of 517 lung adenocarcinomas. Consistent with cell line data, we found that high FUT4-expressing tumors displayed significantly enhanced activities in oncogenic signaling networks, including TGFβ, EGF, MAPK, and WNT as opposed to low FUT4-expressing tumors (Fig. 4e). Notably, this FUT4-mediated pathway activation appears to be independent of EGFR mutation status. Collectively, our analysis suggests that FUT4 induces global reprogramming of the transcriptomic program towards a more malignant phenotype via concurrent alterations of multiple cellular processes and signaling pathways.
5.5. FUT4 induces aberrant fucosylation of intracellular transport and signaling proteins
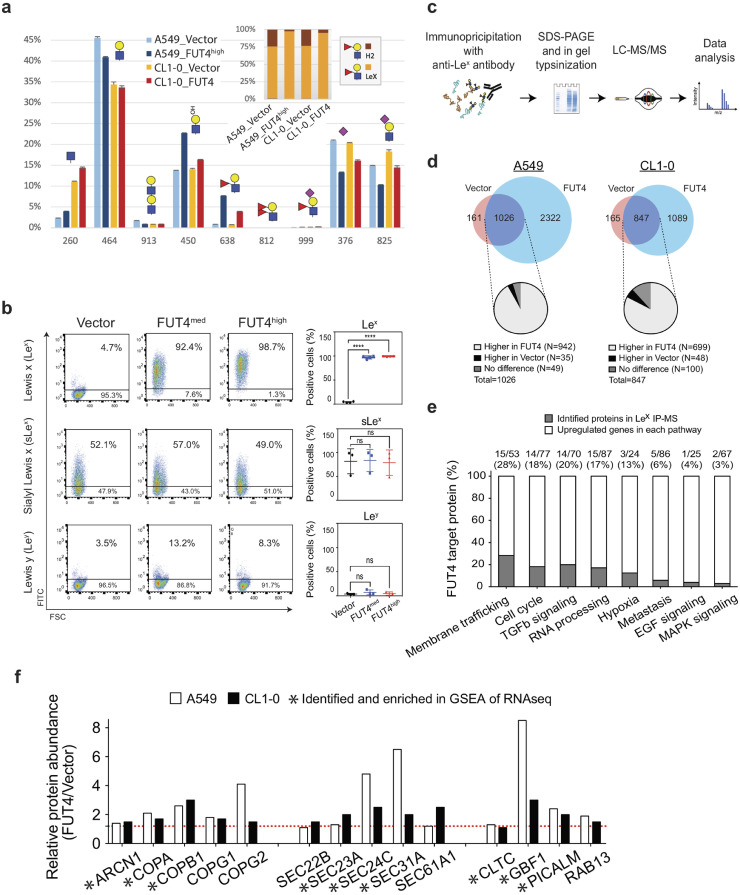
To gain further insights into FUT4-mediated glycomic changes, we performed MALDI-MS mapping followed by LC-MS2/MS3 analysis of N-glycans in A549 and CL1–0 cells upon FUT4 overexpression. Comparing the profiles between that afforded by A549 cells with and without overexpressing FUT4 (Supplemental Fig. 3a), it is clear that there is a significant increase in molecular ion signals assigned as complex type N-glycans carrying two or more fucoses in the latter. A less obvious but distinct increase was likewise found in CL1–0 cells (Supplemental Fig. 3b). To ascertain if the additional fucoses indeed contributed to an increase in Lex, the permethylated N-glycans were further subject to LC-MS2/MS3 analysis. In this approach, all signals detected above a predefined intensity threshold were comprehensively selected for MS2. The presence and total amount of terminal LacNAc (Hex-HexNAc), sialyl LacNAc, and mono-fucosylated LacNAc, can each be inferred from the sum intensity of the MS2 oxonium ions at m/z 464, 825, and 638, respectively, extracted and computed by the GlyPick data processing tool from all productive MS2 spectra contributed by the entire N-glycome. Moreover, the data acquisition was programmed to further trigger MS3 on the MS2 ion at m/z 638 in order to define the relative amount of each of the possible isomeric constituents, namely Lex, Lea, H type 1 and 2, by their respective diagnostic ions [23]. It is clear from the resulting plots (Fig. 5a) that there is a significant increase in fucosyl LacNAc in both A549 and CL1–0 overexpressing FUT4, concomitant with an overall decrease in terminal sialylation (m/z 376) and sialyl LacNAc. Notably, the increase in fucosylation did not contribute to any appreciable amount of Ley/b (difucosyl LacNAc, m/z 812) or sialyl Lex/a (m/z 999). Upon further MS3, the majority of fucoses were shown to be added on to LacNAc as Lex, to the extent that it reduced the relative amount of H type 2 in total fucosyl LacNAc glycotope (Fig 5a, inset), with hardly any detectable Lea. The finding was also validated using flow cytometric analysis, which showed that Lex-expressing cell populations were markedly increased in A549_FUT4med, A549_FUT4high , and CL1–0_FUT4 cells. In contrast, Ley and sLex were not altered (Fig. 5a and Supplemental Fig. 4). Moreover, immunoblotting with anti-Lex showed a global increase of glycans that carry Lex antigen in the whole cell lysate of FUT4-overexpressing A549 and CL1–0 cells. After treatment with PNGase F, an amidase that removes almost all N-linked oligosaccharides, the FUT4-mediated increases of glycans were no longer observed (Supplemental Fig. 4). In addition to glycoproteins, a moderate increase of Lex on glycolipids was also observed in FUT4-overexpressing cells (Supplemental Fig. 5).
Fig. 5.
FUT4 induces aberrant fucosylation of intracellular transport and signaling proteins. (a) LC-MS2/MS3 analysis of N-glycomic profiles of A549 and CL1–0 lung cancer cells with FUT4 overexpression. A significant increase in fucosyl LacNAc (m/z 638) contributed mostly by Lex, concomitant with a decrease in sialyl LacNAc (m/z 825) was registered in both cell lines. (b) Flow cytometric analysis of cell surface glycans, Lewis x (Lex), sialyl Lewis x (sLex), and Lewis y (Ley) in FUT4-overexpressing A549 lung cancer cells. Left panels, representative flow cytometric dot plots of Lex, sLex, and Ley expressions are shown. Right panels, dot plots showing percentages of cells expressing individual surface glycans in three biological replicates. (c) Experimental diagram of immunoprecipitation (IP) with anti-Lex antibody followed by liquid chromatography-mass spectrometry (LC/MS-MS). (d) Venn diagram comparing the numbers of identified proteins in FUT4-overexpressing cells vs. vector controls in A549 (left panel) and CL1–0 (right panel) cells. Pie charts show percentages of increased or decreased fucosylated proteins in FUT4-overexpressing cells. (e) Bar graphs reveal the percentages of FUT4 acceptor proteins (gray bars) in the core enriched genes of top 8 positively enriched gene sets upon FUT4 overexpression. The numbers above the bars denote the numbers (numerator) of identified proteins by anti-Lex IP-MS and the numbers (denominator) of core enriched genes in individual gene sets. (f) Bar graphs show the relative abundance of fucosylated membrane trafficking-related proteins in A549_FUT4high and CL1–0_FUT4 cells compared to their respective vector controls. The proteins also identified in RNA-seq GSEA analysis are marked with asterisks below.
To delineate the cellular effects of FUT4-mediated fucosylation, we went on to probe acceptor proteins fucosylated by FUT4 using immunoprecipitation (IP) with anti-Lex antibody followed by tandem mass spectrometry (LC-MS/MS)(Fig. 5c). We identified 3348 and 1936 proteins bearing Lex antigen in A549_FUT4high and CL1–0_FUT4 cells, and 1187 and 1012 proteins in their respective vector controls (Fig. 5c and d) (MassIVE: MSV000083028). Among the Lex-bearing proteins upregulated in A549_FUT4high and CL1–0_FUT4 cells, many of them participate in the activated cellular processes and signaling pathways revealed by RNA-seq, including membrane trafficking, cell cycle, TGFβ signaling, etc. (Fig. 5e). The data suggest a strong link between FUT4-mediated aberrant fucosylation and heightened activities of cellular processes and signaling. In particular, 28% of upregulated genes that drive the enrichment of the membrane trafficking process are aberrantly fucosylated in FUT4-overexpressing cells (Fig. 5e). In particular, components of a coatomer complex—ARCN1, COPA1, COPB1, as well as SEC family proteins that form the coat protein complex II (COPII)—SEC23, 24, 31A, 61A1, appear to be enriched in the lysates of FUT4-overexpressing cells after pull down by anti-Lex antibody (Fig. 5f). Moreover, aberrant fucosylation of RAB13, a small GTPase regulating vesicular trafficking between trans-Golgi network and recycling endosomes [32], is also noted (Fig. 5f). These data strongly suggest that FUT4 high-expressing cells have a disturbed intracellular transport state.
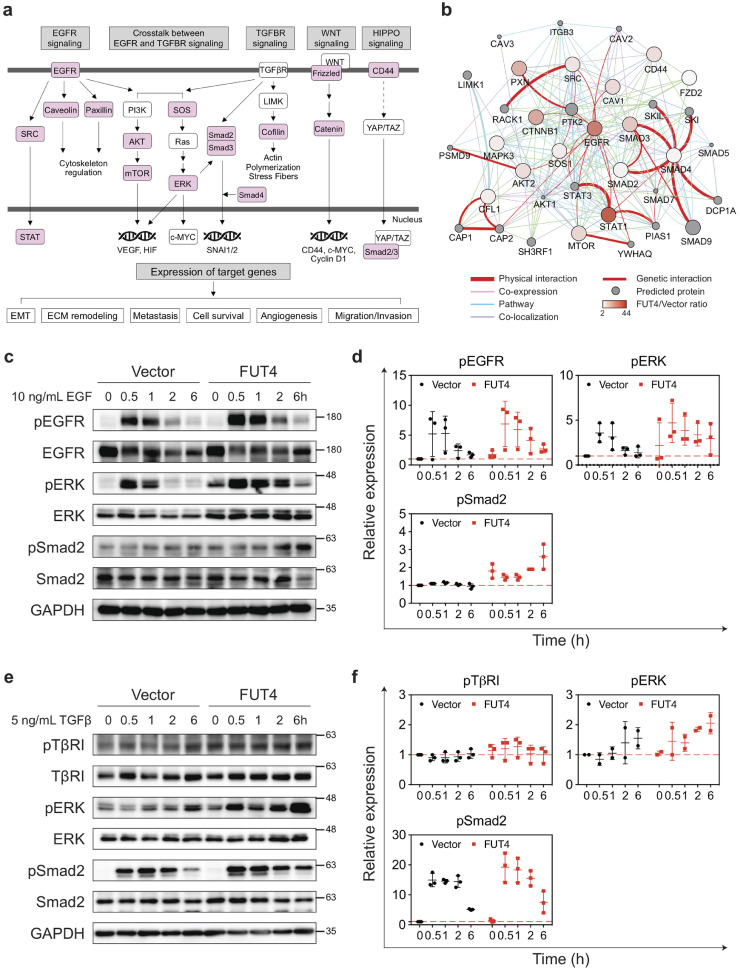
5.6. FUT4 enhances metastasis-related signaling via fucosylation of cascade proteins
In addition to facilitating intracellular transport, FUT4-fucosylated proteins are simultaneously involved in several major signaling pathways known for metastasis, angiogenesis, and EMT, including EGF, TGFβ, WNT and HIPPO pathways (Fig. 6a). With immunoprecipitation-western blot analysis, we observed increased expression of Lex antigen on TGFβ and EGF receptors (Supplemental Fig. 6). Furthermore, putative FUT4 acceptor proteins revealed by anti-Lex immunoprecipitation-mass spectrometry interact physically and form an intricate network of key mediator proteins at multiple levels downstream of these pathways (Fig. 6b). To further elucidate the potentiating effect of FUT4-mediated aberrant fucosylation on signal transduction, we found that FUT4 high-expressing cells exhibit enhanced signaling activity as demonstrated by an increase of phospho-ERK (pERK) and phosphor-Smad2 (pSmad2) at 0.5 to 2 h post EGF (Fig. 6c and d) or TGFβ (Fig. 6e and f) pathway activation in A549_FUT4high compared to the vector control. A similar phenomenon was also observed in CL1–0_FUT4 cells (Supplemental Fig. 7). This suggests that FUT4-mediated fucosylation augments signal transduction upon pathway activation.
Fig. 6.
FUT4 enhances metastasis-related signaling via the fucosylation of cascade proteins. (a) Immunoprecipitation (IP) with anti-Lex antibody followed by tandem mass spectrometry (LC-MS/MS) in A549_FUT4high and CL1–0_FUT4 cells reveals key mediator proteins (in pink) in oncogenic signaling cascades including EGF, TGFβ, WNT and HIPPO pathways. (b) Protein networks showing protein-protein interactions between FUT4 acceptor proteins bearing higher levels of Lex antigens in A549_FUT4high cells relative to A549_vector cells. The data were analyzed by Cytoscape (ver. 3.6.1) using the GeneMania application (ver. 3.4.1). (c) Western blot analyses of signaling cascade proteins in the EGFR pathway in A549_FUT4highversus A549_vector cells at 0, 0.5, 1, 2, and 6 hrs following the addition of 10 ng/mL EGF. (d) Quantifications of signal intensities in (c) for phospho-EGFR, phospho-ERK, and phospho-Smad2 from three biological replicates. (e) Western blot analyses of signaling cascade proteins in the TGFβ signaling pathway in A549_FUT4highversus A549_vector cells at 0, 0.5, 1, 2, and 6 hrs following the addition of 5 ng/mL TGFβ. (f) Quantifications of signal intensities in (e) for phospho-TGFβ receptor I, phospho-ERK, and phospho-Smad2 from three biological replicates. p value in (d, f) was calculated by one-way ANOVA with Dunnett's test. * p < 0.05, ** p < 0.01, *** p < 0.001. ns: not significant. All experiments were performed in three biological replicates and presented as mean ± SEM.
5.7. Genetic depletion of FUT4 diminishes the aggressive phenotypes of lung cancer cells
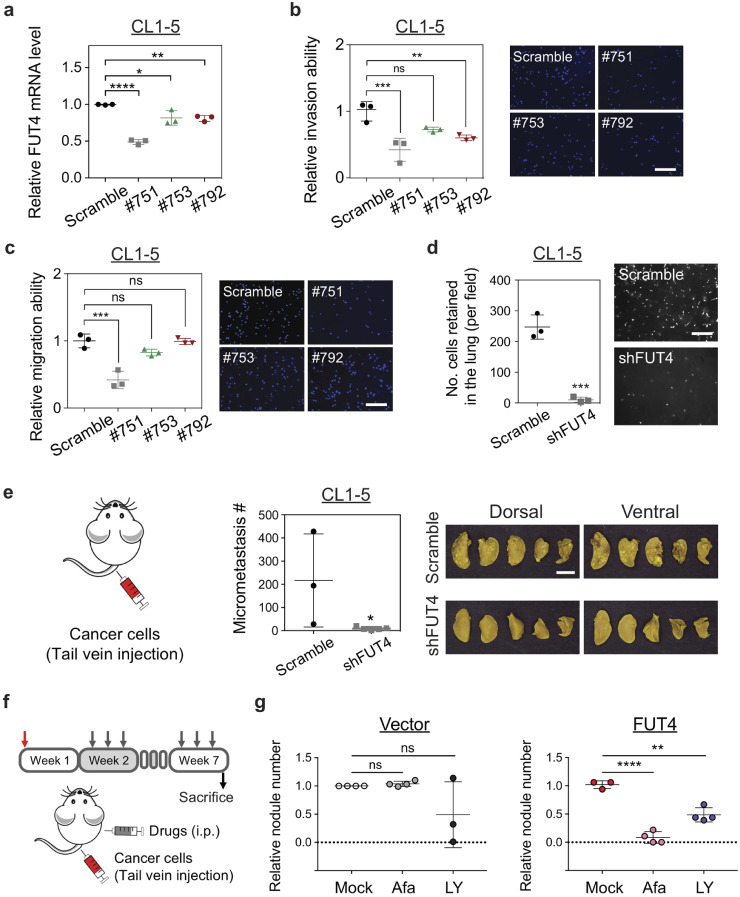
To investigate the potentials of FUT4 as a therapeutic target, we knocked down FUT4 using an shRNA approach in CL1–5 lung adenocarcinoma cells (Fig 7a), which have aggressive migration/invasion behaviors characterized in the previous study [16], and relatively high expression of FUT4 (Fig 2a). Migration and invasion abilities of cancer cells appeared to be reduced in CL1–5_shFUT4#751 cells, which contain the lowest expression level of FUT4 among all shRNA clones (#751, #753 and #792) compared to the vector control (Fig 7b and c). The other two knockdown clones (#753 and #792) also showed a consistent statistical trend towards significance. Moreover, knock-down of FUT4 significantly decreased the extravasation and retention ability of CL1–5 cells in the pulmonary vasculature when we injected the cells into the right ventricle of C57BL6 mice to evaluate their organotropic extravasation ability (Fig 7d). Furthermore, when we silenced FUT4 using shRNA in CL1–5 lung cancer cells, which have spontaneous in vivo metastatic ability [16], we found that FUT4 silencing markedly abolished metastasis of lung cancer cells (Fig. 7e).
Fig. 7.
Genetic depletion of FUT4 diminishes the aggressive phenotypes of human lung cancer cells. (a) FUT4 mRNA levels in CL1–5 lung cancer cells transfected with FUT4 shRNA (#751, #753, #792) measured by quantitative real-time PCR. (b) Dot plots showing relative invasion ability of CL1–5 cells with FUT4 knockdowns measured by Matrigel-based transwell invasion assays. Representative images of cells with DAPI nuclear stains in the lower chambers are shown on the right. (c) Dot plots showing relative migration ability of CL15 cells with FUT4 knockdowns. Relative migration ability was calculated using the cell number in the lower chamber of the transwell system for each clone compared to that of the scramble control. Representative images of cells with DAPI nuclear stains in the lower chambers are shown on the right. (d) Numbers of lung cancer cells with FUT4 knockdowns (CL1–5_shFUT4) adhered to the vascular walls or retained in the lung tissues following intracardial injection visualized under Zeiss Axio Observer microscope. Scale bar: 5 mm. p value was calculated by the Mann-Whitney test. *** p < 0.001. (e) Numbers of metastatic foci in the lungs of nude mice at 28 days after tail vein injection of lung cancer cells with FUT4 knockdown (CL1–5_shFUT4). Pictures of lung nodules from one representative mouse in each group are shown in the right panels. (f) Diagram of in vivo lung cancer metastasis assay via tail vein injection in nude mice subject to treatments with EGFR or TGF-β inhibitors. (g) Dot plots showing relative metastatic abilities of A549_vector or A549_FUT4high lung cancer cells in nude mice receiving 0.25 mg/kg Afatinib or 15 mg/kg LY2157299 treatment. Numbers of metastatic foci in the lungs were counted and normalized to the numbers in the mock (normal saline)-treated groups. p value in (a, b, c, g) was calculated by one-way ANOVA with Dunnett's test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 and in (d, e) was calculated by Mann-Whitney test. ns: not significant. Data in (a–f) were performed in three biological replicates and presented as mean ± SEM.
In addition, to test whether FUT4-mediated malignant phenotype could be pharmacologically diminished by targeting signaling networks provoked by FUT4, we performed in vivo lung cancer metastasis assay via tail vein injection of A549_vector or A549_FUT4high cells into nude mice, which subsequently received intraperitoneal treatment of either 0.25 mg/kg afatinib (an EGFR inhibitor) or 15 mg/kg LY2157299 (a TGF-β inhibitor) three times a week (Fig. 7f). We found that both drugs partially diminished lung metastasis in A549_FUT4high cells (Fig. 7g). Interestingly, afatinib demonstrated differential inhibitory effects between A549_FUT4high and A549_vector cells (Fig. 7g), which implies A459_FUT4high cells have a higher dependency on EGF signaling for distant metastasis. This proof of principle data indicates that targeting FUT4-potentiated pathways may provide a therapeutic opportunity against metastasis of FUT4 high-expressing tumors and that combination therapy against multiple FUT4-medicated cellular processes may be required to achieve better efficacy in patients with FUT4-high-expressing tumors.
6. Discussion
Aberrant expression of fucosylated epitopes such as Lewis antigens has been widely reported in various types of cancer [4]. These Lewis antigens are mainly synthesized by α1–3/ α1–4 fucosyltransferases in a tissue-specific manner. In the current study, we holistically evaluated all α1–3/ α1–4 fucosyltransferases and identified FUT4 as a prognostic biomarker in Asian and Western cohorts of lung cancer patients through large scale transcriptomic analysis. Asian lung cancer is known to be a molecularly and etiologically distinct identity composed of a much higher percentage of never smokers and EGFR mutations [26, 27] as compared to the western counterpart [24, 25]. Yet, comparative transcriptomic analyses of Eastern and Western cohorts revealed striking similarities in the expression patterns of α1–3 terminal fucosyltransferases in non-small cell lung cancers despite differences in the mutational status and ethnicities. Among all α1–3 FUTs, FUT3, 4, 6, 10, and 11 appeared to be highly expressed in both lung adenocarcinoma and squamous cell carcinoma. Moreover, despite the general conception that FUTs may facilitate cell proliferation or invasion in cancers [28, [33], [34], [35]], our genome-wide analysis in large patient cohorts revealed that FUTs may also have protective prognostic effects, which is generally underappreciated. In our data, while FUT4 is correlated with poor survival, FUT1 and FUT10 appeared to confer a survival benefit in lung cancer. Interestingly, FUT1 has been shown to promote proliferation and spreading potential of ovarian carcinoma [36, 37]. This suggests that the same fucosyltransferase may act on different sets of acceptor proteins in different tumor types. While we delineated the role of FUT4 in lung adenocarcinoma, detailed mechanistic and clinical studies may be required to elucidate the precise roles of other FUTs in individual tumor types.
Furthermore, our combined transcriptomic and glycoproteomic approach provides a comprehensive view of cellular processes and intracellular signaling altered by FUT4. This glycosylation-related reprogramming of cellular transcriptome collectively contributes to the malignant phenotype of cancer cells, instead of one or two pathways being held accountable as shown in previous reports [11, 28, 38, 39]. In addition to major oncogenic pathways such as EGF/MAPK, NF-κB/p65, and TGF-beta pathways, our data reveal that FUT4 induces an active intracellular trafficking state as the top enriched cellular process. Notably, the alterations of intracellular vesicle trafficking have recently been shown to drive oncogenesis and regulate cancer behavior [40, 41]. FUT4 not only upregulates but also appears to aberrantly fucosylate many components of vesicle transport proteins including SEC protein family in COPII complex, and regulators of vesicle trafficking such as small GTPases (Fig. 5f). The hyperactive intracellular transport system may underly the neoplastic phenotypes, including proliferation, migration, and invasion [40, 41]. Moreover, it is increasingly acknowledged that intracellular transport and signaling pathways (e.g., PI3K-Akt, EFGR, and FGFR signaling) are interconnected [42]. COPII trafficking was shown to involve in regulating surface levels of receptor tyrosine kinases [43]. In our data, overexpression of FUT4 markedly reinforces the crosstalks between membrane trafficking and signal transduction. This provides a molecular basis for targeting these pathways as novel therapeutic strategies in treating high FUT4-expressing tumors.
In addition, based on the glycomic profiles in the FUT4 overexpressing cells, we observed a preference for the formation of fucosyl LacNAc (m/z 638) over Sialyl LacNAc (m/z 999), with Lex being a main glycan synthesized by FUT4. This can be explained by the presumed biosynthetic pathways where FUT4 is primarily responsible for the addition of a fucose residue on the LacNAc (Gal β−1,4 GlcNAc-R) via α−1,3 linkage to form Lex (Galβ−1,4(Fucα−1,3) GlcNAc-R). This marks the end of glycan synthesis as fucose residues are usually involved in the synthesis of the terminal structure of glycans. On the other hand, the formation of sialyl Lex, for example, requires the addition of a sialyl group by ST3GAL3/4/6 first, followed by terminal fucosylation. The biosynthetic pathway of sialylation and its key enzymes do not appear to be enhanced upon FUT4 overexpression [44, 45].
Most importantly, our in vitro and in vivo studies demonstrated a prime role of FUT4 during the process of lung cancer metastasis, which highlights the potential of FUT4 as an attractive therapeutic target. Indeed, knockdown of FUT4 in our study significantly diminished lung metastases in mouse xenograft models without apparent toxicities. Currently, glycosylation-related therapeutics is under active development, ranging from modulating the stability of therapeutic proteins via glycosylation to glycoprotein-based biodrugs [46]. Multiple attempts have been taken to identify inhibitors of sialyl- and fucosyl-transferases via high-throughput screening [[47], [48], [49]]. Non-selective metabolic inhibitors such as synthetically-derived fucose-related molecules have been shown to inhibit global fucosylation, thereby diminishing neutrophil extravasation or xenograft tumor growth in immunocompromised mice [50]. As our data showed that individual fucosyltransferases may have opposite effects on patient prognosis, we think that the development of FUT4-specific inhibitors may be necessary for preventing lung cancer metastases, and at the same time minimize potential off-target effects due to non-selective ablation of global fucosylation.
On the other hand, for a broader clinical implication, we show that FUT4-mediated EGFR or TGFβ activation can occur in lung tumors that do not carry pathway-specific driver mutations. As shown in the current study, EGFR- or TGFβ- targeted therapy had therapeutic efficacy to a certain extent in FUT4 high-expressing cells. This indicates that pharmacologically targeting FUT4-mediated pathways, such as membrane trafficking, EGF, TGFβ, WNT signaling, may be a reasonable therapeutic option alternative to FUT4 depletion therapy for treating a subset of EGFR-wild type lung cancers with medium-to-high FUT4 expressions. As inhibitors targeting these FUT4-mediated oncogenic signaling pathways are either ready for trials or FDA-approved for clinical use, it would be rather straightforward to design future clinical trials and test the use of these targeted therapies in patients with wild-type EGFR lung cancers expressing higher levels of FUT4.
In summary, our results suggest that FUT4, a terminal α1,3-fucosyltransferase, is an important prognostic indicator in lung adenocarcinoma patients. FUT4 promotes lung cancer progression at multiple steps of the metastatic process — increased migration/invasion, vascular adhesion, extravasation, and tumor metastasis at distant organs. These profound cellular and functional effects can be attributable to the augmentation of global transcriptomic alterations such as intracellular membrane trafficking, oncogenic signaling pathways, which may be reversed by targeting FUT4 directly or pathway-specific inhibitors. Our study may pave the way to the development of novel glycol-based therapeutic strategies for clinical management in lung cancer metastases.
Funding sources
This study was supported by the Ministry of Science and Technology (MOST 107-2314-B-002-241, MOST 105-2628-B-002-040), and National Health Research Institutes (NHRI-EX106-10610BC) in Taiwan. K.-H. K. is supported by an Academia Sinica Investigator Award grant AS-IA-105-L02. The funding agencies did not have any role in study design, data collection, data analysis, interpretation, and writing of the manuscript.
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Acknowledgments
We acknowledge the service provided by the Flow Cytometric Analyzing and Sorting Core of the First Core Laboratory and Laboratory Animal Center of National Taiwan University College of Medicine, Instrumentation Center, National Taiwan University, as well as Confocal Microscopy Core Facility, Institute of Biomedical Sciences, Academia Sinica. The glycomics data by LC-MS/MS were acquired at the Academia Sinica Common Mass Spectrometry Facilities for Proteomics and Protein Modification Analysis located at the Institute of Biological Chemistry, Academia Sinica, supported by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-107). We sincerely thank Dr. Min-Chuan Huang at National Taiwan University for his valuable feedback on the manuscript. We also thank Ms. Min-Chuan Hsu and Ms. Pei-Chun Tsai for their administrative assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102846.
Contributor Information
Chong-Jen Yu, Email: jefferycjyu@ntu.edu.tw.
Hsing-Chen Tsai, Email: htsai@ntu.edu.tw.
Appendix. Supplementary materials
References
- 1.Ohtsubo K., Marth J.D. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Pinho S.S., Reis C.A. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira-Ferrer L., Legler K., Milde-Langosch K. Role of protein glycosylation in cancer metastasis. Semin Cancer Biol. 2017;44:141–152. doi: 10.1016/j.semcancer.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Blanas A., Sahasrabudhe N.M., Rodriguez E., van Kooyk Y., van Vliet S.J. Fucosylated antigens in cancer: an alliance toward tumor progression, metastasis, and resistance to chemotherapy. Front Oncol. 2018;8:39. doi: 10.3389/fonc.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyoshi E., Moriwaki K., Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143(6):725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 6.Chen C.Y., Jan Y.H., Juan Y.H., Yang C.J., Huang M.S., Yu C.J. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc Natl Acad Sci USA. 2013;110(2):630–635. doi: 10.1073/pnas.1220425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honma R., Kinoshita I., Miyoshi E., Tomaru U., Matsuno Y., Shimizu Y. Expression of fucosyltransferase 8 is associated with an unfavorable clinical outcome in non-small cell lung cancers. Oncology. 2015;88(5):298–308. doi: 10.1159/000369495. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa J., Sano A., Inoue H., Koide S. Expression of Lewis-related antigen and prognosis in stage I non-small cell lung cancer. Ann Thorac Surg. 1995;59(2):412–415. doi: 10.1016/0003-4975(94)00866-6. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka F., Miyahara R., Ohtake Y., Yanagihara K., Fukuse T., Hitomi S. Lewis Y antigen expression and postoperative survival in non-small cell lung cancer. Ann Thorac Surg. 1998;66(5):1745–1750. doi: 10.1016/s0003-4975(98)00937-0. [DOI] [PubMed] [Google Scholar]
- 10.Kadota A., Masutani M., Takei M., Horie T. Evaluation of expression of CD15 and sCD15 in non-small cell lung cancer. Int J Oncol. 1999;15(6):1081–1089. doi: 10.3892/ijo.15.6.1081. [DOI] [PubMed] [Google Scholar]
- 11.Tian L., Shen D., Li X., Shan X., Wang X., Yan Q. Ginsenoside Rg3 inhibits epithelial-mesenchymal transition (EMT) and invasion of lung cancer by down-regulating FUT4. Oncotarget. 2016;7(2):1619–1632. doi: 10.18632/oncotarget.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jassam S.A., Maherally Z., Ashkan K., Pilkington G.J., Fillmore H.L. Fucosyltransferase 4 and 7 mediates adhesion of non-small cell lung cancer cells to brain-derived endothelial cells and results in modification of the blood-brain-barrier: in vitro investigation of CD15 and CD15s in lung-to-brain metastasis. J Neurooncol. 2019;143(3):405–415. doi: 10.1007/s11060-019-03188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang J.X., Gao W., Cai L. Fucosyltransferase VII promotes proliferation via the EGFR/AKT/mTOR pathway in A549 cells. Onco Targets Ther. 2017;10:3971–3978. doi: 10.2147/OTT.S140940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y.C., Yen H.Y., Chen C.Y., Chen C.H., Cheng P.F., Juan Y.H. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA. 2011;108(28):11332–11337. doi: 10.1073/pnas.1107385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samur M.K. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu Y.W., Yang P.C., Yang S.C., Shyu Y.C., Hendrix M.J., Wu R. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 1997;17(3):353–360. doi: 10.1165/ajrcmb.17.3.2837. [DOI] [PubMed] [Google Scholar]
- 17.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao C.T., Wang P.W., Chang H.C., Chen Y.Y., Wang S.H., Chern Y. Advancing a High Throughput Glycotope-centric Glycomics Workflow Based on nanoLC-MS(2)-product Dependent-MS(3) Analysis of Permethylated Glycans. Mol Cell Proteomics. 2017;16(12):2268–2280. doi: 10.1074/mcp.TIR117.000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan Q., Hsiung C.A., Matsuo K., Hong Y.C., Seow A., Wang Z. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44(12):1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y., Au J.S., Thongprasert S., Srinivasan S., Tsai C.M., Khoa M.T. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9(2):154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirakawa M., Takimoto R., Tamura F., Yoshida M., Ono M., Murase K. Fucosylated TGF-beta receptors transduces a signal for epithelial-mesenchymal transition in colorectal cancer cells. Br J Cancer. 2014;110(1):156–163. doi: 10.1038/bjc.2013.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jassam S.A., Maherally Z., Smith J.R., Ashkan K., Roncaroli F., Fillmore H.L. TNF-alpha enhancement of CD62E mediates adhesion of non-small cell lung cancer cells to brain endothelium via CD15 in lung-brain metastasis. Neuro Oncol. 2016;18(5):679–690. doi: 10.1093/neuonc/nov248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jassam S.A., Maherally Z., Smith J.R., Ashkan K., Roncaroli F., Fillmore H.L. CD15s/CD62E interaction mediates the adhesion of non-small cell lung cancer cells on brain endothelial cells: implications for cerebral metastasis. Int J Mol Sci. 2017;18(7):1474–1489. doi: 10.3390/ijms18071474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lippincott-Schwartz J., Roberts T.H., Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nokes R.L., Fields I.C., Collins R.N., Folsch H. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J Cell Biol. 2008;182(5):845–853. doi: 10.1083/jcb.200802176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin X., Rana K., Ponmudi V., King M.R. Knockdown of fucosyltransferase III disrupts the adhesion of circulating cancer cells to E-selectin without affecting hematopoietic cell adhesion. Carbohydr Res. 2010;345(16):2334–2342. doi: 10.1016/j.carres.2010.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang L., Gao C., Li Y., Sun M., Xu J., Li H. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8(8):e2968. doi: 10.1038/cddis.2017.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X.S., Liu S., Liu Y.J., Liu J.W., Liu T.J., Wang X.Q. Overexpression of fucosyltransferase IV promotes A431 cell proliferation through activating MAPK and PI3K/Akt signaling pathways. J Cell Physiol. 2010;225(2):612–619. doi: 10.1002/jcp.22250. [DOI] [PubMed] [Google Scholar]
- 36.Yan L.M., Lin B., Zhu L.C., Hao Y.Y., Qi Y., Wang C.Z. Enhancement of the adhesive and spreading potentials of ovarian carcinoma RMG-1 cells due to increased expression of integrin alpha5beta1 with the Lewis Y-structure on transfection of the alpha1,2-fucosyltransferase gene. Biochimie. 2010;92(7):852–857. doi: 10.1016/j.biochi.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Liu J.J., Lin B., Hao Y.Y., Li F.F., Liu D.W., Qi Y. Lewis(y) antigen stimulates the growth of ovarian cancer cells via regulation of the epidermal growth factor receptor pathway. Oncol Rep. 2010;23(3):833–841. [PubMed] [Google Scholar]
- 38.Giordano G., Febbraro A., Tomaselli E., Sarnicola M.L., Parcesepe P., Parente D. Cancer-related CD15/FUT4 overexpression decreases benefit to agents targeting EGFR or VEGF acting as a novel RAF-MEK-ERK kinase downstream regulator in metastatic colorectal cancer. J Exp Clin Cancer Res. 2015;34:108–118. doi: 10.1186/s13046-015-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shan X., Tian L.L., Zhang Y.M., Wang X.Q., Yan Q., Liu J.W. Ginsenoside Rg3 suppresses FUT4 expression through inhibiting NF-kappaB/p65 signaling pathway to promote melanoma cell death. Int J Oncol. 2015;47(2):701–709. doi: 10.3892/ijo.2015.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ioannou M.S., McPherson P.S. Regulation of Cancer Cell Behavior by the Small GTPase Rab13. J Biol Chem. 2016;291(19):9929–9937. doi: 10.1074/jbc.R116.715193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldenring J.R. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat Rev Cancer. 2013;13(11):813–820. doi: 10.1038/nrc3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tripathi K.P., Piccirillo M., Guarracino M.R. An integrated approach to infer cross-talks between intracellular protein transport and signaling pathways. BMC Bioinformatics. 2018;19(Suppl 2):58. doi: 10.1186/s12859-018-2036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farhan H. Regulation of EGFR surface levels by COPII-dependent trafficking. J Cell Biol. 2016;215(4):441–443. doi: 10.1083/jcb.201611014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trinchera M., Aronica A., Dall’Olio F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers. Biology. 2017;6(1):16–33. doi: 10.3390/biology6010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diswall M. Biochemical studies of carbohydrate blood group antigens: carbohydrate phenotype in relation to cellular glycosyltransferases2009.
- 46.Sola R.J., Griebenow K. Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs. 2010;24(1):9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rillahan C.D., Brown S.J., Register A.C., Rosen H., Paulson J.C. High-throughput screening for inhibitors of sialyl- and fucosyltransferases. Angew Chem Int Ed Engl. 2011;50(52):12534–12537. doi: 10.1002/anie.201105065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahsen O., Voigtmann U., Klotz M., Nifantiev N., Schottelius A., Ernst A. A miniaturized high-throughput screening assay for fucosyltransferase VII. Anal Biochem. 2008;372(1):96–105. doi: 10.1016/j.ab.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 49.Rillahan C.D., Antonopoulos A., Lefort C.T., Sonon R., Azadi P., Ley K. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8(7):661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okeley N.M., Alley S.C., Anderson M.E., Boursalian T.E., Burke P.J., Emmerton K.M. Development of orally active inhibitors of protein and cellular fucosylation. Proc Natl Acad Sci USA. 2013;110(14):5404–5409. doi: 10.1073/pnas.1222263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data are available at the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) under the accession number GSE120622. All proteomics data are available at the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under the accession number: MSV000083028