Abstract
Seaweeds are an important component of human diets, especially in Asia and the Pacific islands, and have shown chemopreventive as well as anti-inflammatory properties. However, structural characterization and mechanistic insight of seaweed components responsible for their biological activities are lacking. We isolated cymopol and related natural products from the marine green alga Cymopolia barbata and demonstrated their function as activators of transcription factor Nrf2-mediated antioxidant response to increase the cellular antioxidant status. We probed the reactivity of the bioactivation product of cymopol, cymopol quinone, which was able to modify various cysteine residues of Nrf2’s cytoplasmic repressor protein Keap1. The observed adducts are reflective of the polypharmacology at the level of natural product, due to multiple electrophilic centers, and at the amino acid level of the cysteine-rich target protein Keap1. The non-polar C. barbata extract and its major active component cymopol, reduced inflammatory gene transcription in vitro in macrophages and mouse embryonic fibroblasts in an Nrf2-dependent manner. Cymopol-containing extracts attenuated neutrophil migration in a zebrafish tail wound model. RNA-seq analysis of colonic tissues of mice exposed to non-polar extract or cymopol showed an antioxidant and anti-inflammatory response, with more pronounced effects exhibited by the extract. Cymopolia extract reduced DSS-induced colitis as measured by fecal lipocalin concentration. RNA-seq showed that mucosal-associated bacterial composition and transcriptional profile in large intestines were beneficially altered to varying degrees in mice treated with either the extract or cymopol. We conclude that seaweed-derived compounds, especially cymopol, alter Nrf2-mediated host and microbial gene expression, thereby providing polypharmacological effects.
Keywords: Seaweed, natural products, cancer prevention, Nrf2, anti-inflammatory agents, zebrafish, microbiome
1. Introduction
Marine algae (seaweeds), including green algae (Chlorophyta), red algae (Rhodophyta) and brown algae (Ochrophyta), have been used as food source and medicine since ancient history [1], particularly in certain Asian countries and mainly Japan. In addition to the high nutritional value of edible seaweeds, providing minerals, proteins, vitamins, polysaccharides and antioxidants [2], seaweed consumption is suggested to be inversely related to various cancers, including colon, rectal and stomach cancers [3, 4]. Seaweeds and their constituents have been linked to beneficial activities, including recent reports that demonstrated anti-oxidant and life expansion activities of algal extracts [5–7].
Eukaryotes are equipped with an endogenous defense system comprised of a series of signaling cascades that protect them against various insults and maintain cellular redox homeostasis. One oxidative stress response is activation of the nuclear factor erythroid 2-related factor 2 (Nrf2)-driven antioxidant response element (ARE), which leads to the induction of numerous cytoprotective phase II enzymes [8, 9]. The Kelch ECH associating protein 1 (Keap1)-dependent regulation of Nrf2 is the most characterized mechanism of ARE activation. The key signaling protein, Nrf2 is an inducible cap ‘n’ collar type transcription factor and, under normal conditions, its repressor Keap1 binds to Nrf2 through Cul3-based E3 ubiquitin ligase complex and promotes its degradation by the ubiquitin proteasome pathway. Keap1 is a cysteine rich protein when exposed to stressors undergoes modifications leading to the dissociation of Keap1-Cul3 complex resulting in discontinuation of Nrf2 ubiquitination [10–12] This results in the accumulation of Nrf2, its nuclear translocation, and the subsequent binding to the ARE that drives the expression of Nrf2 target genes, such as NAD(P)H:quinone oxidoreductase 1 (NQO1), heme oxygenase (HMOX1), glutamate cysteine ligase (GCL) and glutathione S-transferase (GST) [13].
Nrf2 has six functional domains (Nrf2-ECH homology 1–6, or Neh1–6). While the C-terminal Neh1, Neh4 and Neh5 domains are critical for ARE binding and transactivation, the N-terminal Neh2 domain is responsible for binding with Keap1 through its DLG and ETGE motifs [14] Keap1 contains three functional domains, including the N-terminal Broad complex, Tramtrack, and Bric-a-Brac (BTB) domain, the intervening region (IVR), and the C-terminal DC domain that comprises the double glycine repeat or Kelch repeat (DGR) and the C-terminal region (CTR). The N-terminal BTB domains of two molecules of Keap1 form a homodimer with the two C-terminal ‘arms’ (the DC domains) that ‘hold’ each of the DLG (lower affinity) and ETGE (high affinity) motifs in the Neh2 domain of Nrf2. This arrangement allows the Cul3-E3 ubiquitin ligase (bound to the BTB region) to ubiquitinate the seven lysine residues between the DLG and ETGE [15]
Keap1 is a redox-sensor and a negative regulator of Nrf2 [8, 16–19]. Modifications of a number of conserved cysteine residues of Keap1 result in de-repression of Nrf2 [19–22]. Amino acid Cys151, found in the heterodimerization (BTB) domain of Keap1, was the most heavily targeted while two other residues, Cys273 and Cys288, have also been shown to be extremely important targets for Nrf2 activation [21] Targeting of cysteine residues of Keap1 is the antioxidant mechanism of popular dietary components, such as sulforaphane found in terrestrial vegetables like broccoli [23]. Phase II enzymes are conjugating enzymes that function to make potentially harmful endogenous and exogenous compounds more water soluble, and thus more easily excreted. Some endogenous ligands include glutathione, glucuronic acid, and sulfate for which conjugation can be catalyzed by glutathione S-transferases (GST), UDP-glucuronosyl transferases (UGTs), and sulfotransferases, respectively. Collectively, detoxification enzymes function by metabolizing and excreting harmful agents and by-products of oxidative stress. Interestingly, there is a substantial amount of crosstalk between the Nrf2/ARE and TNF/NFκB pathways [24, 25]. One of the key players in the crosstalk between these pathways is heme oxygenase 1 (HMOX1), which has been reported to inhibit the pro-inflammatory signals of NFκB [26], making Nrf2 an attractive target for chemopreventive agents to combat both oxidative and inflammatory stresses.
The promise exhibited by our studies on green algae [5, 7], prompted us to analyze Cymopolia barbata for the presence of small molecule ARE inducers. C. barbata is a green marine alga commonly found in the shallow coastal waters near the Florida Keys. Molecules collectively referred to as cymopols were among the first halogenated natural products derived from green algae [27]. These molecules, containing a bromohydroquinone motif, have since been associated with various bioactivities: antimutagenic [28], phospholipase A2 inhibition [29], inhibition of LFA-1/ICAM-1 mediated cell adhesion [30], antifungal and antimicrobial [31], as well as antioxidative properties by way of free-radical sequestration, as determined by the 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay [32]. Another important component of host response to dietary exposure is the microbiota [33]. Microbial composition and activity are influenced by various conditions such as inflammation, infection, antibiotic treatment and diet [34]. Interestingly, some algae, including Cymopolia barbata, have antimicrobial activity [31]. However, limited understanding is available on the effect of seaweed extracts on microbial activities and how this integrates with host responses. Herein we characterize cymopol and a crude cymopol-containing extract in the Nrf2 pathway, inflammatory and microbial genomic responses.
2. Materials and methods
2.1. General procedures
Nuclear magnetic resonance (NMR) spectra were recorded on an Agilent VNMR 400 and 600 MHz spectrometer as indicated in the data list. Chemical shifts for proton nuclear magnetic resonance (1H NMR) spectra are reported in parts per million (ppm) relative to the residual signals for CDCl3 at 7.26 ppm and d6-DMSO at 2.50; Chemicals shifts for carbon nuclear magnetic resonance (13C NMR) spectra are reported in ppm relative to the residual CDCl3 triplet at 77.16 ppm and d6-DMSO multiplet at 39.5 ppm; The abbreviations s, d, t, stand for the resonance multiplicity singlet, doublet, triplet, respectively. High resolution mass spectral (HRMS) data was obtained using an Agilent-LC-TOF mass spectrometer with DART/ESI multimode ion source detectors.
2.2. Chemicals and reagents
HPLC grade solvents were from Fisher Scientific and all other chemicals were purchased from Sigma, unless indicated otherwise. All commercial reagents were used without further purification. CH2Cl2 for quinone synthesis was distilled from CaH2. Thin layer chromatography was performed on EMD silica gel 60Å F254 glass plates and preparative thin layer chromatography was performed on Whatman silica gel 60Å F254 glass plates (layer thick 1000 μm). Anti-NQO1 (mouse) and anti-Nrf2 (rabbit) antibodies were from Abcam, anti-Oct-1 (C-21), anti-Gal4 (DBD), and anti-Keap1 (E-20) from Santa Cruz Biotechnology, anti-HA (mouse) from Covance, anti-goat-HRP from Chemicon (Millipore) and anti-β-actin (rabbit), anti-α-tubulin (rabbit), anti-mouse-HRP, and anti-rabbit-HRP from Cell Signaling Technology. Protein A/G-agarose beads were purchased from Santa Cruz Biotechnology and chitin magnetic beads from New England Biolabs.
2.3. Plasmid constructs
The ARE-luciferase reporter construct (ARE-luc) contains the core sequence of human NQO1-ARE [35]. The mock plasmid (pcDNA3-mRFP) was purchased from Addgene (plasmid 13032). The following plasmids were previously reported. The Keap1-CBD plasmid carries the entire open reading frame (ORF) of human Keap1 fused to the chitin binding domain (CBD) of the Bacillus circulans chitinase A1 gene upstream of the Keap1 stop codon [36]. Oligonucleotide-directed mutagenesis was used to generate the Keap1-C151S-CBD plasmids. The HA-Cul3 plasmid was constructed by fusing an N-terminal hemagglutinin (HA) tag with a cDNA sequence coding for amino acids 1‒380 of human Cul3 gene. The Gal4-Neh2 expression vector contains the codons for the first 97 amino acids (Neh2 domain) of human Nrf2 fused to the ORF of the Gal4 DNA-binding domain.
2.4. ARE-luciferase reporter assay
An ARE-luciferase reporter plasmid (1) and CMV-GFP plasmid (to monitor transfection efficiency) were co-transfected into a human androgen-sensitive prostate cancer cell line, LNCaP (6 × 104 cells/well), and a human neuroblastoma cell line, IMR-32 (3 × 104 cells/well), using Fugene® HD (Roche Diagnostics) in 96-well format. LNCaP and IMR-32 cells have been previously used as cellular models of oxidative stress [5, 37–40]. After 24 hours of incubation, cells were treated with extracts from Cymopolia barbata, a negative control (DMSO, 1%, v/v), and a positive control (10 μM sulforaphane, SF, and tert-butylhydroquinone, tBHQ, respectively). 24 hours post-treatment, luciferase activity was detected using BriteLite detection reagent (PerkinElmer). ARE-luc activity was used to guide fractionation of the extracts.
2.5. Collection, extraction, isolation and structure determination
The Cymopolia barbata was collected in Boca Grande Key Florida in May 2009 and was immediately placed into −20 °C freezer. The algae was later lyophilized and stored at −20 °C before being extracted with solvents of varying polarities. The non-polar (NP) extract was generated with 1 L EtOAc added to 214.10 g of freeze dried Cymopolia barbata on a stir plate at room temperature for two consecutive nights, filtering and adding fresh solvent the second day (2 L total). The solvent was evaporated, yielding 4.09 g of the non-polar (NP) extract. A 2 g sample of the NP extract was further partitioned between hexane and 80% MeOH:H2O v/v. The 80% solution was brought to a 50% MeOH:H2O v/v and partitioned against equal amounts of DCM. The DCM fraction (921.66 mg total) was fractionated using size exclusion chromatography (Sephadex LH20, 40 cm in length × 2 cm in diameter column bed) in 50% DCM:MeOH v/v. Nine fractions were collected. The 6th fraction, a yellow band, was collected between 42–50 mL elution (597.5 mg). The fraction was subjected to a C18 column (20 cm height × 3 cm diameter) with 2 column volumes of 10% MeOH:H2O v/v, 2 column volumes of 50% MeOH:H2O v/v, 4 column volumes of MeOH, and 4 column volumes of iPrOH. The MeOH fraction (557.0 mg) was purified using reversed-phase HPLC (Phenomenex Luna 5u C18(2); 250 × 10 mm; 5 micron) in a 70–100% MeOH:H2O gradient over 32 min (2 mL/min). Four major compounds, collectively referred to as cymopols, were eluted from the non-polar extract. These molecules include cymopol (1, 16 min, 170.1 mg, 8.50% yield), 7-hydroxy cymopol (2, 12 min, 12.1 mg, 0.61% yield), cymobarbatol (3, 15 min, 100.6 mg, 5.03% yield), and cyclocymopol monomethyl ether (4, 22 min, 38.0 mg, 1.90% yield). The compound yields are calculated against the NP extract that was used for oral gavage experiments in mice to highlight the bioavailabilty of the active cymopols in the feeding experiments. Recollections at the same collection site were carried out in May 2011 and extract composition verified by LC-MS and functional equivalence ensured through cell-based bioactivity assays.
The structures of the four pure compounds were determined using 400 MHz (1D) and 500 MHz NMR (2D) and high-resolution mass spectrometry and confirmed by comparison to data reported in literature [28, 32, 41] (SI NMR Spectra).
Cymopol (1), 170.1 mg, 1H NMR (400 MHz, CDCl3): δ ppm 1.60 (3H, s), 1.69 (3H, s), 1.73 (3H, s), 5.0–5.2 (1H, m) 2.04–2.20 (2H, m), 2.04–2.20 (2H, m), 2.28 (2H, d, J = 7.4), 5.27 (1H, t, J = 7.4), 6.79 (1H, S), 6.92 (1H, S). 13C NMR (100 MHz, CDCl3) δ ppm 16.2, 17.9, 25.9, 26.5, 29.6, 39.8, 107.1, 116.8, 118.8, 121, 123.9, 128.9, 132.2,139.2, 146.3, 148.7. ESIMS m/z 323.07/325.06 (1:1) [M-H]−; HRESI(−)MS m/z 323.0652/325.0640 ([M-H]−, C16H2079/81BrO2 calcd. 323.0647/325.0626).
7-Hydroxycymopol (2), 12.1 mg, 1H NMR (400 MHz, CDCl3): δ ppm 1.21 (6H, s), 1.45 (2H, m), 1.49 (2H, m), 1.73 (3H s), 2.05 (2H m), 3.27 (2H, d, J = 7.3), 5.28 (1H, tq, J = 7.3, 1.0), 6.81 (1H, s), 6.92 (1H, s). 13C NMR (100 MHz, CDCl3) δ ppm 16.3, 22.5, 29.3, 29.4, 29.4, 39.9, 43.2, 71.4, 106.9, 116.8, 121.3, 128.8, 139.0, 146.5, 148.4, 188.8. ESIMS m/z 341.1/343.2 (1:1) [M-H]−; HRESI(+)MS m/z 365.0714/367.0698 ([M+Na]+, C16H2279/81BrNaO3 calcd. 365.0728/367.0708).
Cymobarbatol (3), 100.6 mg, 1H NMR (400 MHz, CDCl3): δ ppm 0.85 (s), 1.22 (s), 1.12 (s), 1.90 (m), 1.90 (d, J = 8.21), 2.23 (m), 2.43 (dq, J = 13.9, 2.8), 2.66 (d, J = 18.0), 3.00 (dd, J = 18.0, 8.0), 4.31(s), 5.04 (s), 6.74 (s), 6.86 (s). 13C NMR (100 MHz, CDCl3) δ ppm 22.11, 22.92, 26.5, 27.5, 32.04, 33.81, 38.69, 38.81, 69.7, 75.17, 107.67, 115.23, 119.96, 123.1, 145.83, 148.36. ESIMS m/z 400.7/402.8/404.8 (1:2:1) [M-H]−; DART-HRMS m/z 401.9805/403.9789/405.9770 ([M]+; C16H2179/81Br2O2 calcd. /401.9830/403.9810/405.9789).
Cyclocymopol monomethyl ether (4), 38.0 mg, 1H NMR (400 MHz, CDCl3): δ ppm 1.08 (3H, s), 1.23 (3H, s), 2.02–2.13 (1H, m), 2.17–2.27 (2H, m), 2.39 (1H, m), 2.45 (1H, dd, J =3.5, 11.2 Hz). 2.62 (1H, dd, J = 11.6. 12.8 Hz), 2.92 (1H, dd, J = 3.5, 13.3 HZ), 3.80 (3H, s), 4.31 (1H. br s), 4.44 (1H, dd, J = 4.4, 11.2Hz), 4.63 (1H, s, OH), 4.63 (1H. brs), 6.53 (1H, s), 6.93 (1H, s); 13C NMR (100 MHz, CDCl3) δ ppm 23.75, 27.84, 28.05, 31.5, 34.7, 40.09, 55.45, 57.14, 63.18, 108.63, 112.64, 115.08, 120.1, 127.98, 145.33, 147.88, 149.99. ESIMS m/z 414.9/416.9/418.6 (1:2:1) [M-H]−; DART-HRMS m/z 434.0307/436.0298/438.0283 ([M+NH4]+, C17H2679/81Br2NO2 calcd. 434.0330/436.0310/438.0290).
2.6. Oxidation of cymopol (1) to cymopol quinone (CymQ, 5)
Silver oxide (14.2 mg) was added to a solution of cymopol (6.6 mg) in dichloromethane (2.5 mL). The reaction mixture was stirred at room temperature for 15 min before being evaporated in vacuum. The crude product was purified by preparative TLC plate (ethyl acetate/hexane 1:4, v/v, Rf 0.8) to give 4.92 mg product (75% yield). It should be noted that the product, cymopol quinone, is not air stable, so should be handled quickly and stored in inert gas at low temperature. 1H NMR (600 MHz, d6-DMSO): δ ppm 1.52 (s, 3H), 1.55 (s, 3H), 1.61 (s, 3H), 2.02‒1.98 (m, 2H), 2.06‒2.02 (m, 2H), 3.03 (d, J = 12.0 Hz, 2H), 5.02 (t, J =12.0 Hz, 1H), 5.10 (t, J = 6.0 Hz, 1H), 6.64 (t, J = 1.65 Hz, 1H), 7.50 (s, 1H). 13C NMR (150 MHz, d6-DMSO) δ ppm 15.8, 17.6, 25.5, 25.9, 27.0, 39.1, 118.3, 124.0, 131.0, 131.1, 136.4, 138.3, 138.9, 148.6, 179.8, 184.8. DART-HRMS m/z 323.0634/325.0667 ([M+H]+, C16H2079/81BrO2 calcd. 323.0641/325.0626).
2.7. Immunoblot analysis
IMR-32 cells were seeded into 6-well plates (7 × 105 cells/well) and incubated at 37 °C for 24 h. The cells were then treated with a solvent control (DMSO, 1%, v/v), compounds, or extracts. After 24 h, cells were lysed in 200 μL of PhosphoSafe buffer (Novagen). Protein concentrations were measured by using a BCA assay kit (Pierce). Equal amounts of total protein were separated using SDS-PAGE (NuPAGE® Novex® 4–12% Bis-Tris Mini gels, Invitrogen) and transferred onto a PVDF membrane. Membranes were blocked overnight with 5% BSA at 4 °C and incubated with indicated primary antibodies for 2 h at room temperature. After washing, the membranes were then incubated with the corresponding secondary antibodies (HRP-linked) for 1 h at room temperature and detected with Supersignal Femto Western Blotting kit (Pierce). For assays in the presence of N-acetylcysteine (NAC), the cells were pretreated with the antioxidant (NAC at 1 mM) for 2 h before being treated with the solvent control, compounds, or extracts.
2.8. RNA extraction, cDNA synthesis, and quantitative PCR
The cells were seeded in 6-well format (see immunoblot) and treated with a solvent control (DMSO, 1%, v/v), compounds, or extracts 12 h. The RNeasy Mini Kit (Qiagen) was used to collect total RNA from cells, while for mouse tissue, TRIzol reagent (Invitrogen) was used. In each case, total RNAs (2 μg) were reverse-transcribed into cDNAs, which were used as templates for TaqMan gene expression assay (Applied Biosystems) and detected using the 7300 Real-Time PCR System (Applied Biosystems). Each qPCR sample was tested in triplicate as a 25 μL total reaction volume (12.5 μL of TaqMan 2× universal master mix, 1.25 μL of a 20× TaqMan gene expression assay probe, 1 μL of the above cDNA, and 10.25 μL of RNase-free sterile water). The qPCR method was designed as following: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Endogenous controls used included GAPDH for IMR-32 cells and β-actin to normalize for RAW264.7 cells and mouse tissues.
2.9. RNA interference assay
IMR-32 cells were seeded into 6-well plates (6 × 105 cells/well) and incubated for 24 h at 37 °C. The media was then carefully aspirated and replaced with a transfection mixture composed of siRNAs (50 nM) and siLentFectTM lipid transfection reagent (Bio-Rad Laboratories) in fresh medium. After another 48 h of incubation, the cells were treated with a solvent control (DMSO, 1%, v/v) or serial dilutions of compounds or extracts for an additional 24 h. Protein was isolated using Phosphosafe (see immunoblot). The siRNAs, siGENOME Non-Targeting siRNA Pools and siGENOME SMARTpool (human NFE2L2), were purchased from Dharmacon.
2.10. Glutathione assay
IMR-32 cells were seeded in 6-well format (8 × 105 cells/well) and incubated at 37 °C overnight. The glutathione assay was performed using the standard manufacturer’s protocol (Sigma). Briefly, the cells were treated with a solvent control (DMSO, 1%, v/v) or serial dilutions of compounds or extracts for the indicated periods of time. Following treatment, cells were very gently washed twice with 100 μL Dulbecco’s phosphate buffered saline (DPBS). The cells were then re-suspended in 200 μL DPBS and pelleted at 600 × g for 10 min at 4 °C. The supernatant was aspirated and the pellet was deproteinized and re-suspended in 3 volumes (approximately 30 μL) of 5% sulfosalicylic acid solution (v/v). Two freeze-thaw cycles were performed between liquid N2 and a water bath at 37 °C. The suspension was then incubated for 5 min at 4°C and the cellular debris was pelleted at 10,000 × g for 10 min at 4 °C. The supernatant was transferred to a new tube and used as glutathione stock. A kinetic assay was performed measuring the absorbance of 5-thio-2-nitrobenzoic acid (TNB) spectrophotometrically at 412 nm over 10 minutes. A standard curve of reduced GSH was used to determine the amount of GSH in the biological samples. All calculations were performed according to the manufacturer’s protocol.
2.11. Preparation of cytosolic and nuclear extracts
IMR-32 cells were seeded in 6-well format at 2 × 106 cells/well and incubated at 37 °C overnight. The cells were treated with a solvent control (DMSO, 1%, v/v) or serial dilutions of compounds or extracts for the indicated periods of time. The cytosolic and nuclear extracts were separated and prepared using a commercial kit, NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). Briefly, cells were gently washed once with 200 μL DPBS before being resuspended in 1 mL DPBS, transferred to a pre-chilled tube, and pelleted at 500 × g for 3 min at 4 °C. The supernatant was removed and cells were resuspended in ice-cold CER I containing 1% cOmplete, EDTA-free Protease Inhibitor Cocktail (v/v, Roche Diagnostics) by vigorous vortexing. After addition of ice-cold buffer CER II, the samples were vortexed vigorously for 5 s and incubated on ice for 1 min twice before being pelleted at 16,000 × g for 5 min at 4 °C. The supernatant was transferred to a fresh tube and used as cytosolic stock. The pellet was washed twice by adding 75 μL ice-cold DPBS, flicking, centrifuging, and removing the supernatant before resuspension in ice-cold buffer NER supplemented with 1% cOmplete, EDTA-free Protease Inhibitor Cocktail (v/v). Samples underwent rounds of vigorous vortexing (15 s) followed by incubation on ice for 10 min over a period of 40 min. Samples were centrifuged at 16,000 × g for 10 min at 4 °C. The supernatant was used as nuclear extract stocks. Subsequently, the cellular and nuclear extracts were analyzed by immunoblot.
2.12. Reaction of CymQ (5) with NAC
To test the hypothesis that Keap1 undergoes alkylation at Cys thiol position in the presence of CymQ (5), an in vitro alkylation experiment was conducted in which 5 was incubated with N-acetyl cysteine (NAC) [22]. The reaction products were analyzed by LC-HRMS. Briefly, 10 μM NAC dissolved in milliQ water was reacted with 2- and 10-fold excess of CymQ (5) and cymopol (1) prepared in DMSO. The reaction was incubated for 2 h at room temperature. The reaction was incubated for 2 hours at room temperature and a 10 μL volume re-suspended in 1 mL MeOH/H2O (1:1) before subjecting to LC-HR(+)ESIMS analysis. The resulting adducts were separated on a Phenomenex Synergi Hydro-RP analytical column (150 × 4.6 mm; 4 micron) using a linear gradient from 90% to 100% B over 5 min and 100% B over 10 min (Solvent B: acetonitrile with 0.1% formic acid). The mass spectrometer was set to acquire the total ion chromatogram (TIC) and corresponding ions (m/z) for [M+H]+ adducts were filtered by ion extraction (EIC). The potential adducts formed as identified for the corresponding [M+H]+ ions for the CymQ-NAC reaction are shown in SI, Fig. S1B.
2.13. Reactions of CymQ (5) with Keap1
Recombinant Keap1 protein (Keap1-DDK) (4.2 μM, Origene) dissolved in 20 mM TRIS-HCl (adjusted pH 8.0) was incubated with a solvent control (final 5% DMSO) or 1- and 10-fold excess of CymQ (5) (Experiment-1) and solvent control plus 5- and 10-fold excess of CymQ (5) (Experiments-2 and −3) for 30 min at room temperature with each experiment being carried out in sets of two (6 samples each). Each reaction (20 μL) was prepared for LC/MS/MS analysis as described below.
2.13.1. Sample preparation for mass spectrometry (experiment 1)
All 6 samples (DMSO ×2; 1-fold ×2; 10-fold ×2) were treated with 1mM DTT (15 min) to quench excess CymQ. The 6 samples were solubilized in 50 mM ammonium bicarbonate, pH 8.5, and then reduced by 10 mM tris(2-carboxyethyl) phosphine at 37 ˚C for 1h, followed by addition of 1mM Dithiothreitol to samples (15 min). Next, alkylation by addition of 30 mM iodoacetamide (final IAA concentration of 3 mM) for 3 samples (DMSO; 1-fold and 10-fold) was completed in the dark for 45 min at room temperature. 45 mM Dithiothreitol (final DTT concentration 5mM) was added to remove excess iodoacetamide in the samples. All 6 samples were treated with Chymotrypsin (Promega) for digestion (w/w for enzyme: sample = 1:50), and the samples were incubated at 25 °C overnight. The digested peptides were desalted using micro ZipTip mini-reverse phase (Millipore), and then lyophilized to dryness. Only IAA modified samples showed cysteine alkylation by CymQ in this experiment.
2.13.2. Sample preparation for mass spectrometry (experiments 2 and 3)
A set of 6 samples was first treated with DTT to quench excess CymQ and all 12 samples subjected for proteomics analysis as follows. Samples were first solubilized in 50 mM ammonium bicarbonate, pH 8.5, and then reduced by 10 mM tris(2-carboxyethyl) phosphine at 37 ˚C for 1h. Alkylation by addition of 30 mM iodoacetamide (final IAA concentration of 3 mM) to samples 1–12 was carried out in the dark for 45 min at room temperature. 45 mM Dithiothreitol (final DTT concentration 5mM) was added to remove excess iodoacetamide in all samples. Trypsin (Promega, Fitchburg, WI) was added for digestion (w/w for enzyme: sample = 1:50) for 1.5 hours at 37 °C for to 6 samples. Chymotrypsin was added for digestion (w/w for enzyme: sample = 1:50) for 1.5 hours at 37 °C to the remaining 6 samples. The digested peptides were desalted using micro ZipTip mini-reverse phase (Millipore), and then lyophilized to dryness.
2.13.3. Nano-LC-MS
Peptides derived from the protein samples and resuspended in 0.1% formic acid for mass spectrometric analysis. The bottom-up proteomics data acquisition was performed on an EASY-nLC 1200 ultraperformance liquid chromatography system (Thermo Scientific) connected to an Orbitrap Fusion Tribrid instrument equipped with a nano-electrospray source (Thermo Scientific, San Jose, CA). The peptide samples were loaded to a C18 trapping column (75 μm i.d. × 2 cm, Acclaim PepMap® 100 particles with 3 μm size and 100 Å pores) and then eluted using a C18 analytical column (75 μm i.d. × 25 cm, 2 μm particles with 100 Å pore size). The flow rate was set at 300 nL/minute with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid and 99.9% acetonitrile) as the mobile phases. Separation was conducted using the following gradient: 2% of B over 0 – 3 min; 2 – 40 % of B over 3 – 150 min, 40 – 98 % of B over 150 – 151 min, and isocratic at 98% of B over 151–166 min, and then from 98 – 2% of B from 166 – 167 min. The equilibration at 2% B is from 167 to 180 min.
The full MS1 scan (m/z 350 – 2000) was performed on the Orbitrap with a resolution of 120,000 at m/z 400. The automatic gain control (AGC) target is 2e5 with 50 ms as the maximum injection time. Monoisotopic precursor selection (MIPS) was enforced to filter for peptides. Peptides bearing +2 – 6 charges were selected with an intensity threshold of 1e4. Dynamic exclusion of 15 s was used to prevent resampling the high abundance peptides. Top speed method was used for data dependent acquisition within a cycle of 3 s. The MS/MS was carried out in the ion trap, with a quadrupole isolation window of 1.3 Da. Fragmentation of the selected peptides by collision induced dissociation (CID) was done at 35% of normalized collision energy. The MS2 spectra were detected in the linear ion trap with the AGC target as 1e4 and the maximum injection time as 35 ms.
Database searching: All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.4.1). Mascot was set up to search the IPI_human database (entries) assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 10.0 PPM. Carbamidomethyl of cysteine was specified in Mascot as a fixed modification. Gln->pyro-Glu of the n-terminus, deamidated of asparagine and glutamine and oxidation of methionine were specified in Mascot as variable modifications.
Scaffold (version Scaffold_4.2.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm with Scaffold delta-mass correction [42]. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm [43]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
2.14. NO assay
Wild-type (8 × 103 cells/well), Nrf2−/− (7 × 103 cells/well), and Keap1−/− (5 × 103 cells/well) mouse embryonic fibroblasts (MEF) cells were graciously provided by Dr. Dinkova-Kostova. RAW264.7 cells (2 × 104 cells/well), wild-type (8 × 103 cells/well), Nrf2−/− (7 × 103 cells/well), and Keap1−/− (5 × 103 cells/well) MEF were seeded into 96-well format. Cells were pretreated for 1 h with a solvent control (DMSO, 1%, v/v) or serial dilutions of compounds or extracts before addition of LPS (1 μg/mL) or IFN-γ (10 ng/mL). Nitric oxide (NO) production levels were determined in cell culture supernatant after 24 h for RAW264.7 cells or 20 h for MEF cells. In each case, 50 μL of the media was combined with 50 μL Griess Reagent (Promega). Absorbance was measured at 540 nm and compared with a calibration curve generated using the provided sodium nitrate standard.
2.15. Nqo1 activity assay
Wild-type (8 × 103 cells/well), Nrf2−/− (8 × 103 cells/well), and Keap1−/− (4 × 103 cells/well) MEFs were seeded into 96-well format and incubated overnight at 37 °C. Cells were treated with a solvent control (DMSO, 1%, v/v) or serial dilutions of compounds or extracts for 40 h and the Nqo1 activity was measured using a previously described method [44]. Briefly, solvent or drug treated cells were lysed and quinone reductase activity measured by the addition of a reaction mixture generating NADPH, menadione (2-methyl-1,4-naphthoquinone) and MTT. The MTT is non-enzymatically reduced by menadiol and the formation of the purple color is quantified by UV absorbance. Activities are relative to the vehicle control for the given cell line.
2.16. PGE2 assay
RAW 264.7 cells were seeded into 96-well format at 4 × 104 cells/well and incubated overnight at 37 °C. Cells were treated with a solvent control (DMSO, 1%, v/v) or serial dilutions of compounds or extracts for 1 h before the addition of IFN-γ (10 ng/mL). Cells were incubated for an additional 24 h and the supernatant was analyzed for PGE2 production using the Amersham Prostaglandin E2 Biotrak Enzyme immunoassay (EIA) system (GE Healthcare). The experiment was performed using the standard manufacturer’s protocol.
2.17. Induction of iNOS and Cox2 transcript levels in macrophage cells
RAW264.7 cells were seeded in 6-well plates (8 × 105 cells/well) and incubated at 37 °C overnight. Cells were treated with a solvent control (DMSO, 1%, v/v) or serial dilutions of compounds or extracts for 1 h before the addition of 1 μg/mL lipopolysaccharide (LPS) and incubated at 37 °C for 12 h before extraction of total RNA (see RNA extraction, cDNA synthesis, and quantitative PCR). qPCR analyses used to detect transcript levels of iNOS, Cox2, Nqo1, and β-actin (endogenous control).
2.18. Zebrafish neutrophil migration assays
All zebrafish procedures in this study complied with the National Advisory Committee For Laboratory Animal Research (NACLAR) Guidelines set out by the Agri-Food and Veterinary Authority (AVA) of Singapore and was overseen by the Institutional Animal Care and Use Committee (IACUC) of the Biological Resource Centre BRC (IACUC Protocol Number: 140924). Larvae were obtained through natural crosses of Tg(mpx:egfp)i114Tg adults [45], kept in the IMCB zebrafish facility and staged as per Kimmel et al. 1995 [46]. Tg(mpx:egfp)i114Tg transgenic zebrafish larvae (6 dpf) were pre-treated in duplicate with a solvent control ((DMSO, 1%, v/v) or serial dilutions of the compound or extract in 12-well plate format with five larvae per well containing 1 mL E2 medium containing 1 mM Tris pH 7.4. After 3 hours, larvae were anesthetized with Tricaine and tailfins were amputated using sharp blades to induce the migration of neutrophils. The larvae were then placed into fresh wells to be post-treated for an additional 3 hours. A fluorescence stereomicroscope was used to count the number of neutrophils that migrated to the site of injury. The assay was performed in triplicate to determine statistical significance. This neutrophil migration assay was previously reported [47].
We first performed several dose-response analyses on zebrafish wild-type AB embryos in order to determine the concentration at which to treat the reporter line. A concentration was selected in which no phenotypic toxicity was noted in AB embryos treated 1.5 hpf through embryonic development. Additionally, due to the nature of the fin clip assay, any slight phenotypic toxicity will generally cause the tail to slightly degrade. A concentration was chosen at which both embryos and full adults demonstrated no phenotypic toxicity. Cymopol (1) and the NP extract demonstrated toxicity at fairly low concentrations, leaving the therapeutic window in fish smaller rather small. Regardless, there was a therapeutic window similar to what was seen in the cellular studies. 4dpf fish were pre-treated for 9 h with either a vehicle control, 1, or the NP extract. The fish were anesthetized in tricaine before their fins were cut with a sharp razor blade. The fish were then quickly transferred into fresh media containing the treatments for an additional 3 h before the GFP-tagged neutrophils were counted under the fluorescent microscope.
2.19. Mouse experiments for tissue distribution and functional response
The experiments were approved by the Institutional Animal Care and Use Committee at the University of Florida (IACUC Protocol Number: 201101386). Groups of five 4-week old wild type male mice (C57BL/6J) were used for each treatment type (n = 5). The mice were maintained under approved standard conditions. Cymopol (low dose: 0.1 g/kg bw, high dose: 0.4 g/kg bw) and the non-polar extract (low dose: 0.3 g/kg bw, high dose: 1.0 g/kg bw) were dissolved in Cremophor EL (Sigma) containing 10% DMSO and administered via oral gavage for 3 consecutive days, with 24 h between each treatment. No apparent toxicity was observed for the animals. 12 h following the final treatment, the mice were euthanized in 100% CO2. The tissues were harvested immediately, frozen on dry ice, and kept at −80 °C until analyzed. RNA was isolated for RT-qPCR and RNA-seq using the TRIzol method as described above.
2.20. RNA-seq libraries preparation using CloneTech RiboGone, SMARTer Universal Low Input RNA kit for sequencing combined with Illumina Nextera DNA sample preparation kit
Illumina RNA library construction was performed at the Interdisciplinary Center for Biotechnology Research (ICBR) Gene Expression Core, University of Florida (UF). Quantitation was done on a NanoDrop Spectrophotometer (NanoDrop Technologies, Inc.), and sample quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc). rRNAs first were removed started with 90 ng of total RNA by ClonTech RiboGone –Mammalian-Low input ribosomal RNA removal kit for Human, Mouse and Rat Samples (cat#: 634848) following the manufacturer’s protocol, then the depleted RNA were used for library construction with SMARTer Universal Low input RNA kit for sequencing (cat#: 634940) combined with Illumina Nextera DNA Library Preparation Kit (cat#: FC-121–1030) according to the user guide.
Briefly, 1st strand cDNA is primed by a modified N6 primer (the SMART N6 CDS primer), then base-pairs with these additional nucleotides, creating an extended template. The reverse transcriptase then switches templates and continues transcribing to the end of the oligonucleotide, resulting single-stranded cDNA contains sequences that are complementary to the SMARTer oligonucleotide. The SMARTer anchor sequence and the N6 sequence serve as universal priming sites for DNA amplification by PCR for 10 cycles. Then Illumina sequencing libraries were generated with 125 pg of cDNA using Illumina Nextera DNA Sample Preparation Kit (Cat#: FC-131–1024) according to manufacturer’s instructions. Briefly, 125 pg of cDNA was fragmented by tagmentation reaction and then adapter sequences added onto template cDNA by PCR amplification. Libraries were quantitated by Bioanalyzer and qPCR (Kapa Biosystems, catalog number: KK4824). Finally, the libraries were pooled equal molar concentration and sequenced by Illumina 2X75 NextSeq 500. Data are deposited in GenBank, accession number GSE107623.
2.21. NextSeq500 sequencing: RNA-seq
In preparation for sequencing, barcoded libraries were sized on the bioanalyzer, quantitated by QUBIT and qPCR (Kapa Biosystems, catalog number: KK4824). Individual samples were pooled equimolarly at 4 nM. This “working pool” was used as input in the NextSeq500 instrument sample preparation protocol (Illumina, Part # 15048776, Rev A). Typically, a 1.3 pM library concentration resulted in optimum clustering density in our instrument (i.e., ~200,000 clusters per mm2). Samples were sequenced on a single flowcell, using a 2×75 cycles (paired-end) configuration. A typical sequencing run in the NextSeq500 produced 750–800 million paired-end read with a Q30>=85%. For RNA seq, 50–100 million reads provided sufficient depth for transcriptome analysis.
2.22. DSS-induced inflammation
All animal procedures were performed according to the guidelines of the University of Florida Institutional Animal Care and Use Committee (IACUC Protocol Number: 201608025). The DSS model was performed with aged matched WT C57Bl/6 mice (6–8 weeks old). Dose optimization experiments were carried out with three concentrations (1.0, 1.5 and 2.0 mg/kg) using the formulation described above and otherwise identical conditions (daily treatment for 3 days followed by harvesting large intestines and cecum 12 h after the last dose). Nqo1 and Hmox1 mRNA levels were analyzed by RT-qPCR as described above. Based on this data, mice were then administered NP extract (2.0 g/kg body weight, daily) or vehicle (10% DMSO, 10% Cremophor) via oral gavage. After 3 days of pre-treatment, mice were given 3% DSS-drinking water ad libitum for 7 days along with daily extract administration. Mice were sacrificed and colon snips and stools were collected and snap frozen for further processing.
2.23. Lipocalin immunoassay
Lipocalin-2 protein levels were detected using the DuoSet Mouse lipocalin-2 ELISA kit (R&D Systems). Briefly, frozen fecal pellets were weighed and re-suspended in sterile PBS. The supernatant was collected and used for ELISA per application instructions. Data were normalized by stool weight.
2.24. RNA-seq based intestinal microbiome analysis
Reads were quality filtered at Q20 and trimmed to remove remaining adaptors using Trimmomatic version 0.36 [48]. Quality filtered and trimmed reads were aligned to iGenome Mus musculus GRCm38 reference genome using BWA version 0.7.16a [49] and reads with alignments were excluded from further analysis. Microbial classifications were assigned to the unaligned reads from the step above using centrifuge version 1.0.3 [50] (in pair-end mode utilizing both the forward and reverse reads together) and the bacteria and archaea database provided by the software developers (ftp://ftp.ccb.jhu.edu/pub/infphilo/centrifuge/data/p_compressed.tar.gz). Reads with archaeal taxonomy assignment were removed from subsequent analysis.
Moreover, the unaligned reads from above were used to generated closed-reference OTUs at 97% similarity using Quantitative Insights into Microbial Ecology (QIIME) version 1.9.1 [51] and the Green Gene reference dataset version 13.8. We generated OTUs from the forward reads and the reverse reads independently. Taxonomy assignment for the resulting OTUs was done in QIIME through the ribosomal database project (RDP) classifier after training on Green Gene reference dataset with confidence set to 50% [52]. We filtered out singleton OTUs from the resulting OTU tables. The final OTU tables contained a minimum of 2,448,533 and 2,702,466 reads per sample for the forward and reverse ends, respectively. Those counts were then normalized and log10 transformed using the following formula [53]:
Principle Coordinate Analysis (PCoA) was generated from Bray-Curtis dissimilarity matrix obtained from the normalized and log10 transformed OTU counts using phyloseq R package [54, 55].
Significant differences between the groups (Control, High and Low) were detected using the lm function in the R, using a linear model of the form: variable ~ group + ɛ; where variable indicates either the PCoA axis or taxa (OTU, genus, family, order, class or phylum) normalized count (we only considered taxa present in at least 25% of the samples). We then ran an ANOVA analysis on the above model to generate a P-value for the group. All comparisons were done in pair-wise fashion (group: control vs. low, group: control vs. high and group: high vs. low). All p-values were adjusted for multiple hypothesis testing in R using the p.adjust function employing the method of Benjamini & Hochberg [56].
2.25. Metatranscriptome analysis
Quality filtered and trimmed reads from above were then filtered from rRNA and tRNA sequences by aligning (using BWA) to a collection of NCBI rRNA and tRNA sequences and SLIVA database sequences. The resulting reads were used for de novo assembly using Trinity version 2.4.0 [57]. The resulting assembly was annotated using Trinotate version 3.0.1 (http://trinotate.github.io) [58] with the following databases: uniprot_uniref90, uniprot_sprot [59], Pfam [60] and Virulence Factor Database (VFDB) [61]. The resulting annotations were examined and sequences annotated as non-bacterial were removed. Transcripts abundance was determined using RNA-Seq by Expectation Maximization (RSEM) [62] through Trinity’s align_and_estimate_abundance.pl script and the counts were imported to edgeR [63] version 3.16.5 for differential expression analysis. A gene was considered for differential expression test if it was present in at least two samples. We considered a transcript DE if its edgeR FDR adjusted P-value < 0.05.
3. Results and Discussion
3.1. Discovery of Nrf2 Activators from Cymopolia barbata
An ARE-luciferase reporter assay [35] using two different cell lines was used to guide fractionation of a subtropical marine alga, Cymopolia barbata, for activators of the Nrf2/ARE pathway. Cymopolia extract, resulting fractions and pure compounds were analyzed in IMR-32 neuroblastoma cells, a model system that is highly responsive to ARE induction, and in the androgen-sensitive LNCaP prostate cancer cells, a model for early stage prostate cancer, where Nrf2 activation is postulated to be beneficial [5, 37–40]. The dichloromethane (DCM) fraction resulting from a liquid-liquid partitioning of the non-polar ethylacetate (EtOAc) extract of Cymopolia retained a majority of the mass and ARE-luc activity. Additional fractionation using size exclusion chromatography and subsequent reversed-phase chromatography yielded cymopol (1, 8.50% yield), 7-hydroxycymopol (2, 0.61% yield), cymobarbatol (3, 5.03% yield), and cyclocymopol monomethyl ether (4, 1.90% yield), collectively referred to as cymopols (Fig. 1A). Purified compounds 1 and 2 were able to activate ARE-luc in a dose-response manner in both LNCaP (Fig. 1B) and IMR-32 (Fig. 1C) cells, with cymopol (1) being the most active, rivaling the activity of sulforaphane. The dual activity excluded cell type specificity.
Fig. 1.
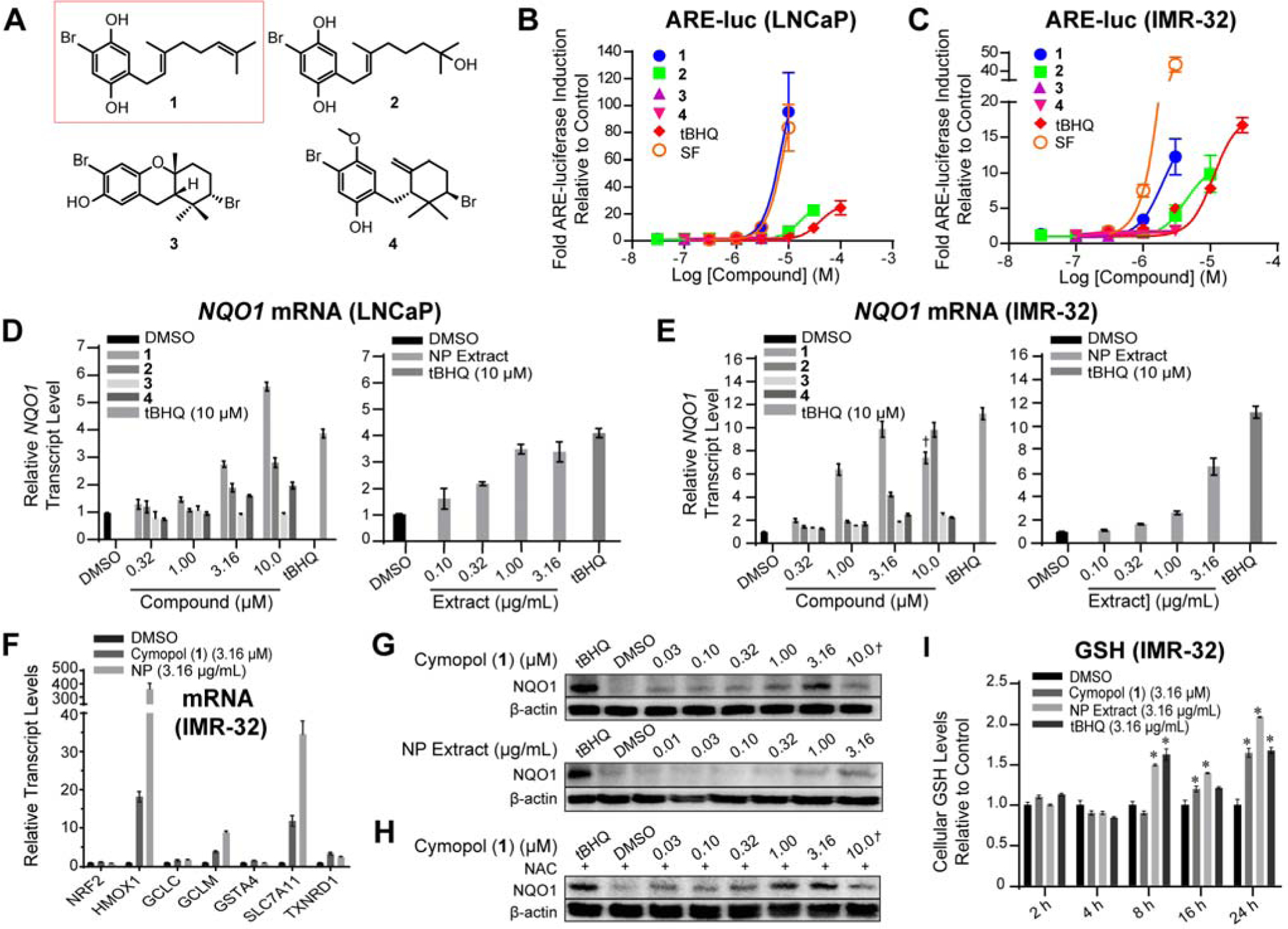
Structures and cellular activities of compounds isolated from Cymopolia barbata. (A) Structures of cymopol (1), 7-hydroxycymopol (2), cymobarbatol (3), cyclocymopol monomethyl ether (4). (B,C) Cymopols activated the ARE-luc reporter (24 h) in a dose-dependent manner in (B) LNCaP and (C) IMR-32 cells (n = 4). (D-F) Cells were treated with cymopols for 12 h and transcript levels were analyzed using qPCR analysis (n = 3). Cymopols were able to induce relative transcript levels of ARE-driven NQO1 in (D) IMR-32 and (E) LNCaP cells. (F) Cymopols and the NP extract increased transcript levels of various other ARE-regulated genes, without increasing levels of Nrf2. Symbol (✝) indicates cytotoxicity. (G) This induction of ARE-driven genes correlates with an induction of NQO1 protein levels after 24 h treatment in IMR-32 cells. (H) The induction of NQO1 by cymopol was not inhibited by pre-treatment with NAC. (I) Cymopol (1) and the NP extract were able to increase cellular GSH levels (n = 4, p < 0.05, t-test). tBHQ and sulforaphane (SF) served as positive controls.
3.2. Validation of antioxidant properties
Compounds 1–4 and their parent fraction, the non-polar (NP) extract, were analyzed in IMR-32 and LNCaP cells for effects on endogenous gene transcription. Compounds 1 and 2 as well as the NP extract all induced the transcription of the Nrf2/ARE regulated gene NQO1 in a dose dependent manner in both LNCaP and IMR-32 cells (Fig. 1D,E). Further evaluation of transcript regulation in IMR-32 cells by cymopol (1) and the NP extract revealed that these samples induce the transcription of a series of ARE-driven genes, demonstrating bioactivity in the Nrf2/ARE pathway (Fig. 1F). Nrf2 target genes, including GCLC (glutamate-cysteine ligase, catalytic subunit), GCLM (glutamate-cysteine ligase, modifier subunit), TXNRD1 (thioredoxin reductase 1), and GSTA4 (glutathione S-transferase alpha-4), were induced 3- to 34-fold in IMR-32 cells. Of particular interest was an extreme transcript induction of HMOX1 (heme oxygenase 1), by the NP extract by 360-fold in IMR-32 cells. The induction of NQO1 was further confirmed at the protein level using Western blot analysis (Fig. 1G). The in vitro activity of the NP extract was functionally equivalent to the active concentration of cymopol (1) (NP extract 3.16 μg/mL vs. cymopol 1 μM/0.32 μg/mL) and could be accounted for by the presence of ARE-active cymopols (~10 wt% of extract). In order to evaluate whether this rise in cytoprotective signaling products was due to inducing oxidative stress, cells were pre-treated with the antioxidant N-acetylcysteine (NAC) which is able to sequester free radicals, minimizing toxic effects of applied compounds. Even in the presence of NAC, 1 was able to induce the translation of NQO1, which suggests that the stress response pathway was activated independently of oxidative stress (Fig. 1H). The natural products were able to increase cellular glutathione levels at 16 and 24 h post-treatment (Fig. 1I), validating that the increase in transcript levels of the GSH synthetic machinery, GCLC and GCLM, leads to generation of a cellular product with cytoprotective activity.
3.3. Mechanism of action
Due to the hydroquinone structure of the compounds, it was hypothesized that the cymopols may act via alkylation of cysteine residues on Keap1 to induce the nuclear translocation of Nrf2. It is likely that these compounds would undergo oxidation, forming the presumed bioactive quinone with a Michael acceptor capable of alkylating Keap1 cysteines as is known for the tert-butylhydroquinone (tBHQ)/tert-butylquinone (tBQ) (SI, Fig. S1A) redox cycling pair [64]. This hypothesis is supported by reduced antioxidant efficacy of compounds 3 and 4 relative to compounds 1 and 2, which are more prone to redox cycling with the presence of a para-OH substitution pattern. The most active compound, which was also the major cymopol (1), was subjected to further analysis. In order to confirm the active species, 1 was oxidized chemically to produce the corresponding cymopol quinone, (5, CymQ, Fig. 2A). The hydroquinone/quinone pair 1/5 was examined besides model compound tBHQ and its corresponding quinone tBQ for the ability to induce NQO1 transcript levels in IMR-32 cells. In both cases, the quinones were able to retain the bioactivity in a dose-dependent manner similar to that of the hydroquinone counterpart, supporting the hypothesis that cymopols function via an α,β-unsaturated ketone (quinone) moiety (Fig. 2B).
Fig. 2.
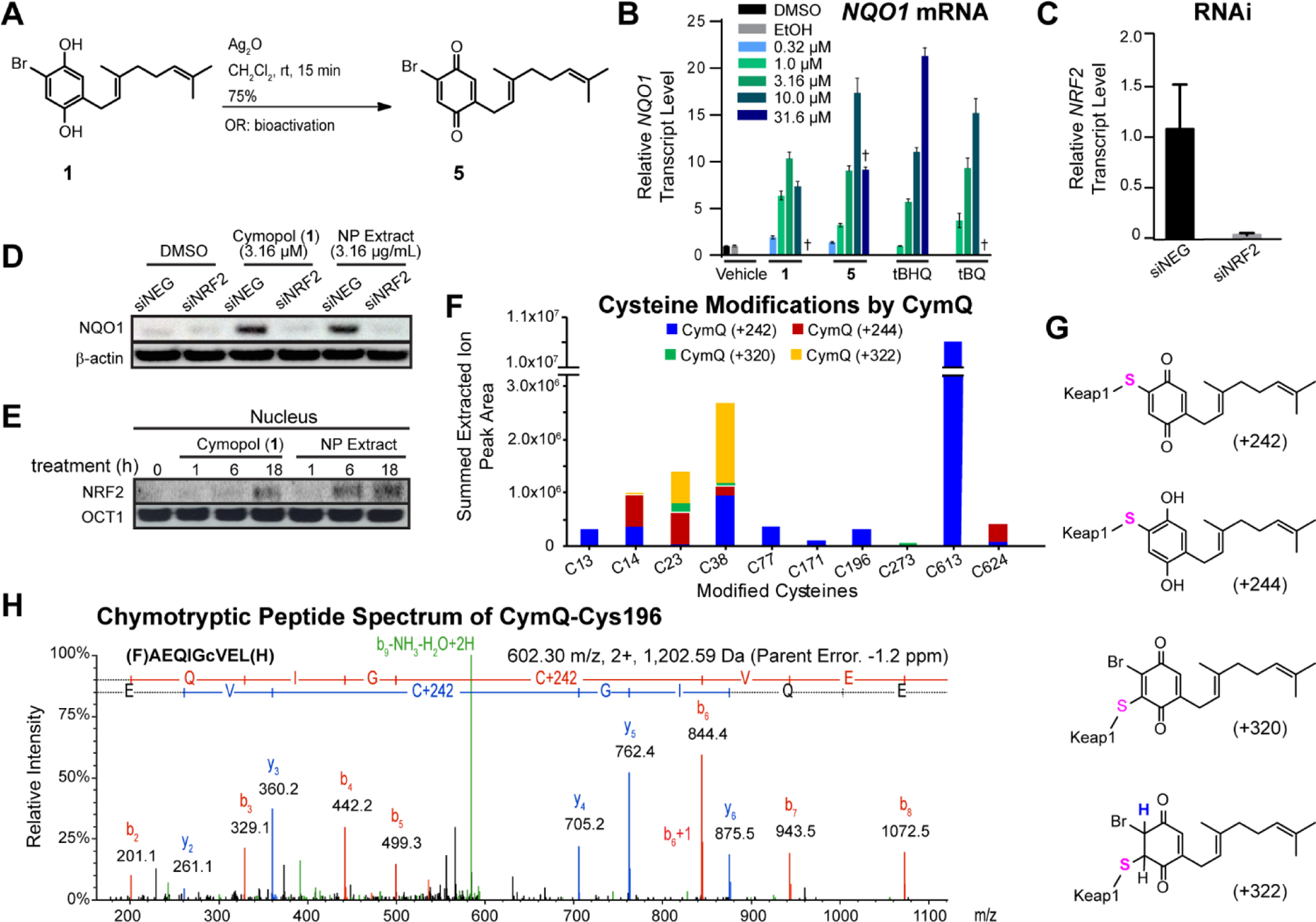
Mechanism of action of Nrf2 activation by cymopol. (A) Silver catalyzed oxidation reaction of cymopol (1) to form the corresponding cymopol quinone (5). (B) IMR-32 cells were treated for 12 h with cymopol (hydroquinone) and its corresponding quinone, alongside structurally related positive control tBHQ and its active quinone metabolite, tBQ, and mRNA levels of the Nrf2 target gene NQO1 assessed (n = 3). Cymopol quinone and tBQ are able to induce NQO1 similar to that of their hydroquinone counterparts in a dose-dependent manner. (C) Knockdown efficiency of the siNRF2 in IMR-32 cells after 48 h (n = 3). (D) Nrf2 is required for the induction of NQO1 protein levels for cymopol and the NP extract. (E) Cymopol and the NP extract induce Nrf2 nuclear translocation and possibly also stabilization of nuclear Nrf2 in IMR-32 cells. (F, G) Summed CymQ modifications for different cysteines of Keap1 protein from three separate experiments using five different sample preparation conditions. The total ion current (TIC) peak area (>95% confidence level) for each CymQ adduct (242, 244, 320 and 322) from all experiments were summed for individual cysteine residues where modification occurred. The most abundant modification across multiple cysteine residues was shown to be with CymQ(+242) adduction. (H) Representative chymotryptic peptide spectrum containing CymQ(+242) alkylated Cys196.
In order to determine whether NQO1 induction is Nrf2 dependent, IMR-32 cells were transfected with siRNA targeting Nrf2. After 48 h, NRF2 transcript levels were reduced by 96% compared to control (Fig. 2C) at which point cells were treated with the samples or the vehicle control for 24 h. The induction of NQO1 transcript levels was prevented in siNRF2 transfected cells (Fig. 2D), indicating that Nrf2 is required for the induction of ARE-driven genes by cymopol and the NP extract. Nuclear proteins were isolated from IMR-32 cells following treatment with cymopol (1) or NP extract for 0, 1, 6, or 18 h to determine whether cymopols cause an increase in nuclear NRF2. Both 1 and the NP extract induced nuclear translocation of NRF2 (and possibly the stability of nuclear NRF2), with NP extract showing increased levels after 6 h, and 1 only after 18 h (Fig. 2E). This further confirms the mechanism of action of cymopols in the Nrf2/ARE pathway, which is negatively regulated by the cytoplasmic repressor protein Keap1 to be the predicted target of these compounds.
The CymQ structure has several potential reactive sites that could lead to conjugate (Michael) addition or addition-elimination since bromide could act as a leaving group. To probe the chemical reactivity of CymQ (5), we conducted in vitro experiments with excess of N-acetylcysteine (NAC) for 2 h at room temperature and analyzed the reaction mixtures by LCMS. CymQ-NAC adducts were observed by ion extraction for relevant mass tags [M+H]+. We detected mass tags that corresponded to m/z 485 and 483 for a conjugate addition reaction and m/z 405 and 407 for addition-elimination reaction of NAC with CymQ (5). Additional adducts with two NAC molecules observed for mass tags corresponding to m/z 566 and 568 confirmed multiple reactive sites for 5 (SI, Fig. S1B).
We next investigated the reactivity at the amino acid level of Keap1. We first tested cymopol’s cellular effect on the functionality of Keap1 as an adaptor for the Cul3-ubiquitin ligase complex, similarly as we have described for synthetic Nrf2 activators AI-1 and AI-3 (7, 35). Using a candidate approach to evaluate specific cysteine residues which may be alkylated in the presence of cymopols, we have initially focused on Cys-151 and co-transfected HEK293 cells with either a mock plasmid, a wild-type Keap1-CBD, or a Keap1-C151S-CBD and HA-Cul3 (to evaluate Cul3 binding inhibition upon cymopol addition), HA-Nrf2 (to evaluate Nrf2-Keap1 interactions), or Gal4-Neh2 and HA-Ub (to evaluate ubiquitination patterns of Nrf2). Our data suggested that cymopol (1) and the NP extract function at least in part through modification at Cys-151 of Keap1 as reduced Neh2 ubiquitination, higher levels of Neh2 and increased Keap1 dimerization were observed in the wild-type Keap1 upon addition of 1 (SI, Fig. S2).
To obtain a global picture of cysteine modification of Keap1, rather than focusing on known functionally relevant residues, we then utilized a proteomics approach, monitoring the effect of activated cymopol (CymQ, 5) on cysteine residues of full-length human recombinant KEAP1 in vitro, similarly as described [22, 65]. Briefly, KEAP1 was treated with excess CymQ (5) and adducts were mapped by LC-MS/MS in three separate experiments (five different conditions) using DTT, chymotrypsin and trypsin as the variables for different sample preparations (see Material and Methods), in order to capture more potential adducts. The most abundant modification observed was a mass increase of 242 resulting from an addition-elimination reaction of 5 (Fig. 2F,G,H and SI, Fig. S3) with loss of bromine, which is unique to CymQ compared with tBQ. Additional representative spectra observed for potential CymQ-Keap1 adduct formation represented by different mass tags can be found in SI, Fig. S4. Among the ten cysteines detected to be modified by 5, only two cysteine residues (Cys23 and Cys38) showed common adducts (244 and 322 mass tags) under at least two different experimental conditions (Fig. 2F). These and other cysteine modifications were predominantly scattered across the N-terminal, BTB and C-terminal domains of the Keap1 protein. Interestingly, none of the previously reported reactive cysteines including Cys151 [21], which is most consistently detected as being of high reactivity with many ARE activators failed to alkylate under the conditions we employed, which could be likely due to reversibility of the reaction. Critical cysteines in Keap1 and their function have been categorized into six classes using a “cysteine code” [66]. Most ARE activators belong to class 1 of this cysteine code where Cys151 is readily modified, although contradictory results are reported for these compounds, depending on the experimental conditions used [23]. Since iodoacetamide, used for free cysteine modification during sample preparation, could cause a reversible effect of alkylated cysteines as demonstrated previously for sulforaphane [23], it is possible to speculate that either CymQ-Cys151 is a reversible adduct or 5 is more favoured towards alkylating different cysteines similar to other ARE activators [67, 68]. Although we have identified that CymQ (5) has the capability to modify Keap1 and activate Nrf2, exactly which cysteines could act as the critical sensors is yet to be confirmed. Overall, it is clear the compound possesses polypharmacology at the cysteine level of Keap1 (and probably additional proteins), potentially reversible in nature, which will produce a net effect that translates into the observed functional consequences.
3.4. Validation of anti-inflammatory properties
Due to the strong induction of HMOX1 transcript levels and the aforementioned crosstalk between antioxidant (Nrf2) and anti-inflammatory (NFκB) pathways [24, 25], we sought to further evaluate the anti-inflammatory properties of cymopol (1) and the NP extract. RAW264.7 macrophage cells activated by IFN-γ were treated with 1 or the NP extract. IFN-γ induced transcript levels of the pro-inflammatory genes iNOS, leading to the downstream reduction in levels of NO. NO production was reduced by >96% in the presence of 10 μM cymopol (1) and >75% in the presence of 10 μg/mL NP extract (Fig. 3A,B). Additionally, transcript levels of pro-inflammatory Cox2 were reduced by 90% in the presence of both 10 μM cymopol (1) and 10 μg/mL NP extract, with concomitant ablation of PGE2 production (Fig. 3C,D) and increase in Nqo1 transcript levels by >96%. (Fig. 3E). When comparing anti-inflammatory activity at 3.16 μM concentration of pure cymopol (1) with 10 μg/mL concentration of NP extract containing an approximate chemical equivalence of 1, both samples show similar activity against cytokine induced iNOS and Cox2 transcript levels.
Fig. 3.
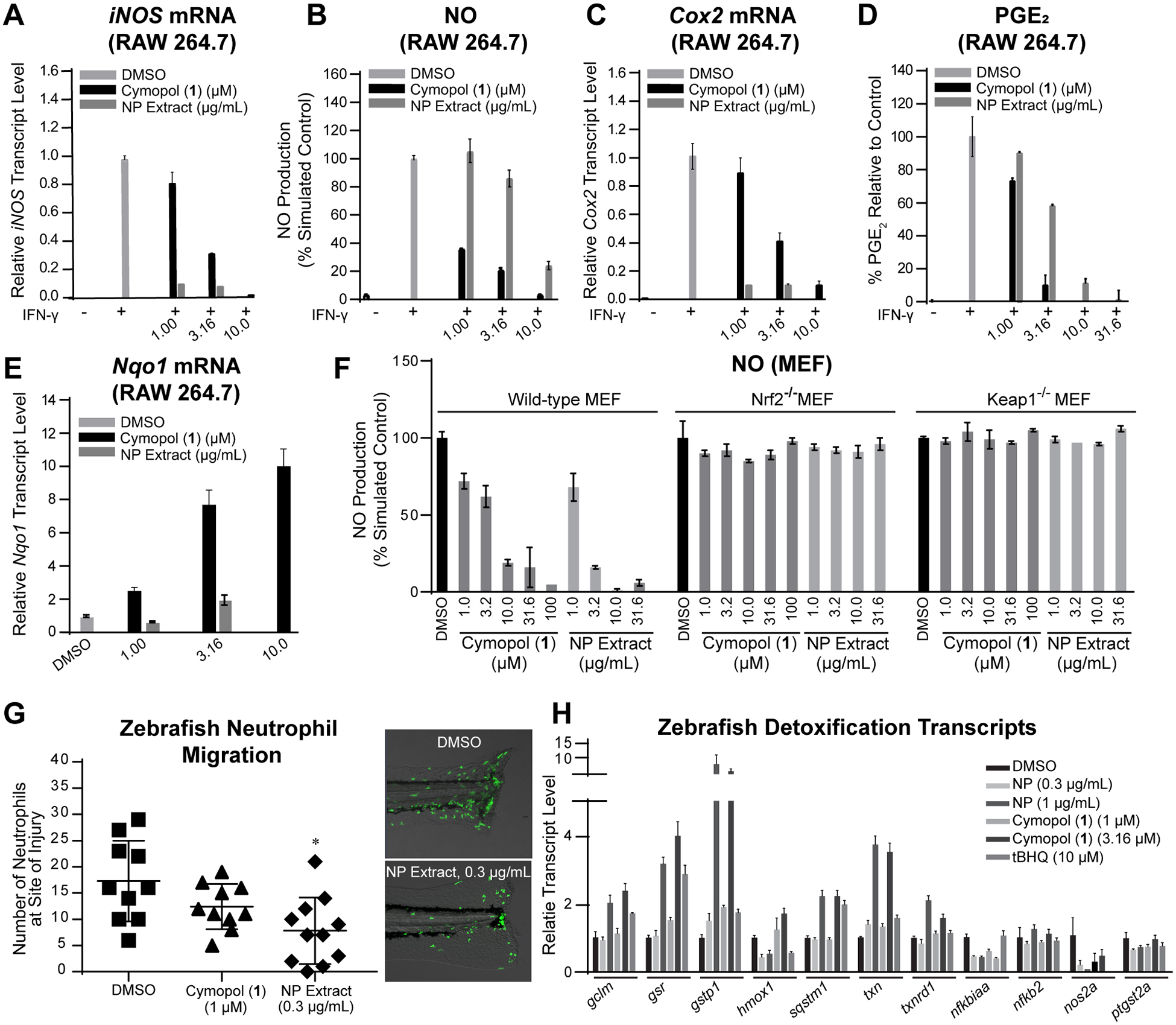
Anti-inflammatory activity in various cellular models in vitro and in vivo (zebrafish). (A-E) Cymopol (1) and the NP extract can dose-dependently ameliorate the pro-inflammatory effects of IFN-γ in macrophage cells, by reducing pro-inflammatory gene transcription and induce Nrf2 target gene transcription. RAW264.7 macrophage cells were pretreated with various doses of 1 or NP extract prior to activation with inflammatory activating IFN-γ. (A) iNOS transcript levels (n = 3) and consequently (B) NO production (n = 6); (C) Cox2 transcript levels (n = 3) and consequently (D) PGE2 production (n = 3) were decreased in a dose dependent manner, while (E) increasing levels of Nqo1 mRNA (n = 3). (F) The reduction of pro-inflammatory NO levels are dependent on Nrf2/Keap1 reactivity. Cymopol (1) and NP extract were able to restore basal level of NO in wildtype MEFs, while Nrf2−/− and Keap1−/− MEF cells were resistant towards the anti-inflammatory properties of the cymopols (n = 6). (G,H) Anti-inflammatory activity in zebrafish. (G) 9 h pre-treatment with the NP extract significantly (n = 10, p < 0.05) decreased the number of neutrophils to the cut site of the Tg(mpx:egfp)i114Tg zebrafish line. (I) Treatment of wild type zebrafish embryos of the same age with equal and greater concentrations of 1 and NP extract resulted in an increase in ARE-driven gene transcription and a decrease in select NFκB regulated genes, although these effects are mostly seen in the higher (and slightly toxic) doses (n = 3).
We next wanted to determine whether cymopols exert their anti-inflammatory function through the Nrf2/Keap1 pathway in biologically relevant murine embryonic fibroblast (MEF) lines and whether this correlated to Nqo1 activity. The Nqo1 activity and NO synthesis inhibition properties of the NP extract and cymopol (1) were evaluated in previously described manner [69]. At high concentrations, 1 and the NP extract are able to induce relative Nqo1 activity more so in wild-type than in Nrf2 and Keap1 knockout MEFs at a given concentration (SI, Fig. S5). Cymopol (1) and the NP extract minimized NO production in wild-type MEFs stimulated with IFN-γ and TNF-α (Fig. 3F). However, these anti-inflammatory effects were lost in Nrf2−/− and Keap1−/− MEFs, suggesting that cymopol’s anti-inflammatory effects function through an Nrf2/Keap1 mechanism of action.
For initial in vivo evaluation, we used a previously reported neutrophil migration assay in the model organism, Danio rerio [45]. In this assay, neutrophils migrate rapidly to a site of injury in response to a variety of chemo-attractants, including N-formyl methionine-leucine-phenylalanine (fMLF), interleukin-8, and ROS, providing an additional link between the ARE/Nrf2 pathway and the inflammatory pathway [70–73]. A transgenic zebrafish neutrophil-specific reporter line, Tg(mpx:egfp)i114Tg, containing neutrophils labelled with an enhanced green fluorescent protein (egfp), was used to test whether these natural products minimized neutrophil recruitment to an injury (45, 47). We performed several dose-response analyses on zebrafish wild-type AB embryos in order to determine the concentration at which to treat the reporter line. A concentration was chosen at which larvae demonstrated no phenotypic toxicity. The NP extract demonstrated anti-inflammatory properties relative to a DMSO vehicle control with a statistically significant decrease in the number of neutrophils at the cut site (p < 0.05, Fig. 3G). The overall number of neutrophils which flood into the tail post-injury and those which make it to the cut site in fish pre-treated with the NP extract were lesser than those in the DMSO treated embryos (SI, Movies S1: DMSO control vs. Movie S2: treatment with NP extract). Both samples demonstrated the ability to induce detoxification enzymes in zebrafish (Fig. 3H).
3.5. In vivo validation of antioxidant and anti-inflammatory activity in mice
Two doses each of purified cymopol (1) or the NP extract were given to 4-week old male mice via oral gavage. As dose starting points, we used molar equivalents for amounts used in animal studies for the chemically and functionally similar tBHQ, which was reported to be non-toxic via i.p. at 0.2 g/kg bw (74, 75), corresponding to ~0.39 g/kg bw (1.2 mmol/kg bw) for cymopol (1) (high dose). We also wanted to evaluate if a 3-fold lower dose ~0.13 g/kg bw (0.4 mmol kg bw) of 1 (low dose) can induce a functional response. For the NP extract, we used doses of 1.0 g/kg bw (high dose) and 0.3 g/kg bw (low dose). The high dose of NP extract contained roughly equal amounts of cymopol (1) as the low-dose treatment with purified cymopol 1 and both concentrations were also functionally equivalent in IMR-32 cells (SI, Fig. S6). Our dose selection was designed to allow us to evaluate potential cooperative effects in vivo for the NP extract (see below). Mice were treated by oral gavage for three consecutive days every 24 h and euthanized 12 h following the last treatment. Nine organs were isolated and evaluated for transcript levels of Nqo1 (Fig. 4A and SI, Fig. S7) and Hmox1 (Fig. 4B and SI, Fig. S8). Of particular interest was a large increase in the anti-inflammatory and antioxidant genes within the digestive tract, with the large intestine and cecum showing the highest induction levels of Nqo1 and Hmox1 (Fig. 4C,D), which was roughly equivalent for high-dose NP extract and high-dose cymopol treatment.
Fig. 4.
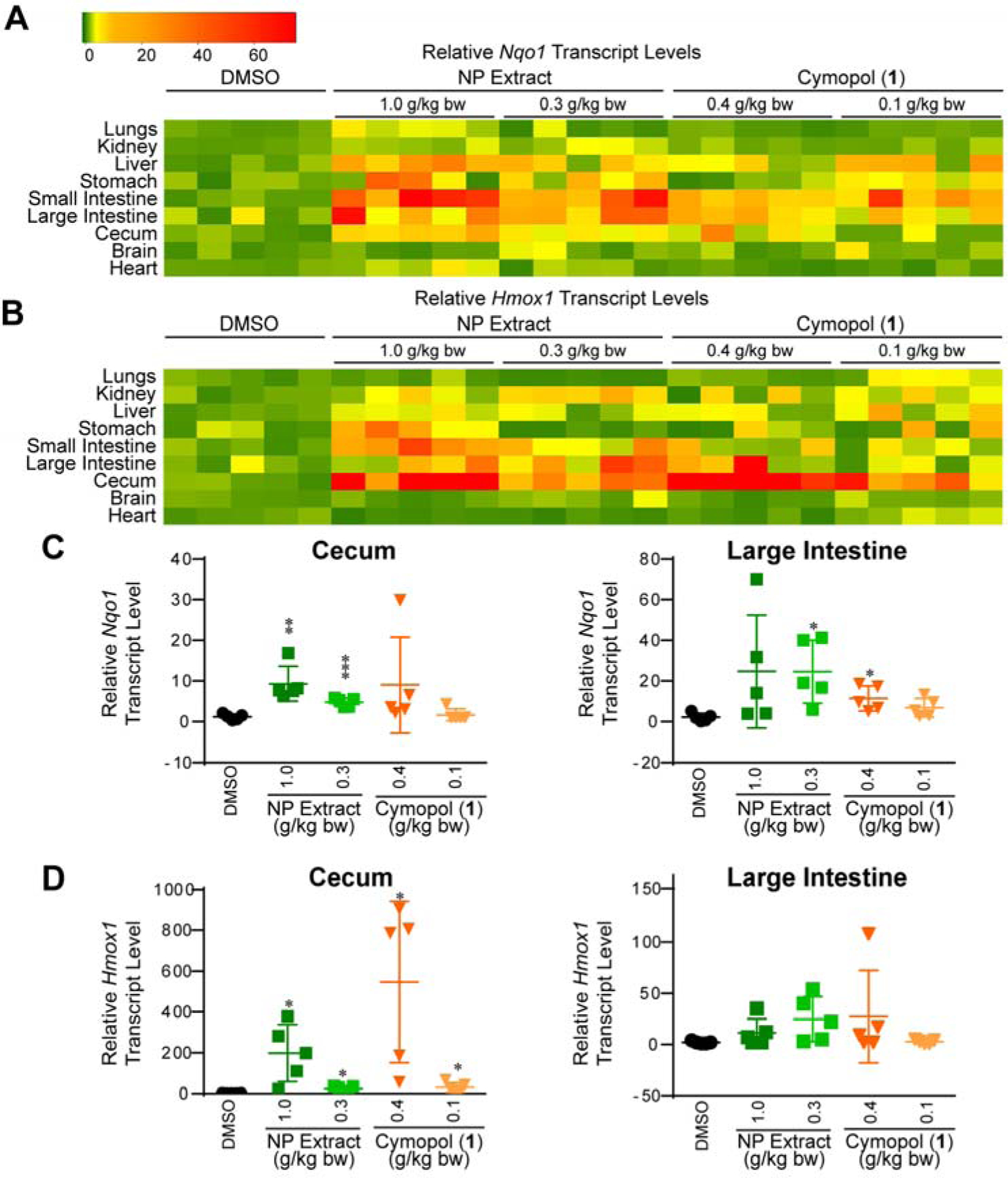
Functional response to cymopol and NP extract in mouse tissue. Mice (n = 5) were treated for 3 consecutive days (every 24 h) via oral gavage. Organs were extracted for analysis 12 h following the final treatment and evaluated using RT-qPCR. (A,B) Heatmap summary of mice given cymopol or NP extract via oral gavage, resulting in elevated transcript levels of Nqo1 (A) and Hmox1 (B) in various organs, particularly in the digestive tract. (C,D) Induction of (C) Nqo1 and (D) Hmox1 in cecum and large intestines (* indicates p-value < 0.05; ** indicates p-value < 0.005; *** indicates p-values < 0.0005).
To further evaluate the effect of cymopol (1) and the NP extract, the large intestines were examined by RNA-seq experiments using three out of the five mice tissue samples, excluding the samples showing the highest and lowest Nqo1 and Hmox1 transcriptional expression in each set. Genes were prioritized based on those with a 1.5-fold change in transcript level with p < 0.05. Ingenuity Pathway Analysis (IPA) was used to further investigate the effect of cymopols on the large intestine. Consistent with our in vitro and in vivo model systems, we identified multiple canonical pathways associated with antioxidant and anti-inflammatory activities (Fig. 5A). In support of our experimental design targeting activators of the Nrf2/ARE pathway, the canonical pathway entitled ‘NRF2-mediated Oxidative Stress Response’ appeared within our top 20 canonical pathways in an IPA comparison analysis. Furthermore, the ERK/MAPK signaling pathway was downregulated, which indirectly stabilizes Nrf2 in mammalian systems [76]. Additionally, the canonical pathway, ‘Production of Nitric Oxide and Reactive Oxygen Species in Macrophages’ was reduced, which supports our earlier findings that 1 and the NP extract have anti-inflammatory and antioxidant properties. Furthermore, the overall decrease in ‘Colorectal Cancer Metastasis Signaling’ is consistent with a potential anticancer effect. A decrease in the PI3K/AKT signaling pathway coupled with an increase in PTEN signaling (which negatively regulates the AKT pathway) also point to an anticancer effect of these compounds. These findings suggest that the cymopols could show potential in the treatment or prevention of colorectal cancers. Purified cymopol (1) (0.1 g/kg = low dose) elicited a weaker host response than the NP extract (1.0 g/kg = high dose) containing similar amounts of cymopol (0.1 g/kg). Instead, the functional response in vivo on the host was more similar across these signaling pathways for both high-dose treatments (see activation z-scores/heatmap in Fig. 5A), with the NP extract still having a slightly higher effect on Nrf2 and most other signaling. It is possible that other inactive cymopols lacking the hydroquinone moiety, including compounds 3 and 4 (Fig. 1A), might also be metabolically bio-activated via oxidative dealkylation to the hydroquinone that then can undergo the same reactions as cymopol 1, and the less active compound 2 might get converted into 1 via dehydration. The presence of additional functionally dormant (“protected”) cymopols cannot be excluded. The active cymopol content would then increase to >16%, which could then at least partially account for the superior in vivo activity of the NP extract, although additional factors might play a role.
Fig. 5.
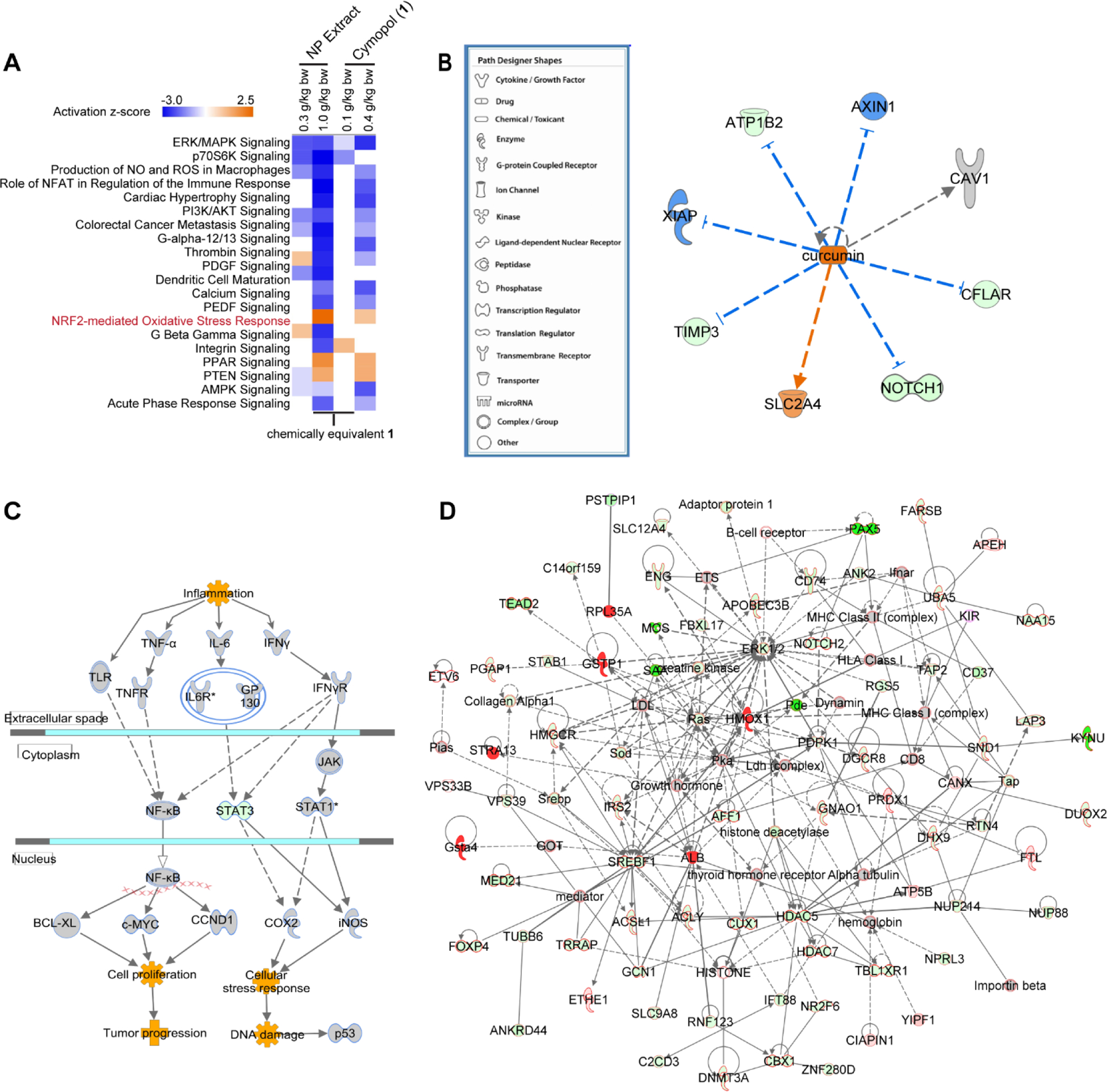
RNA-seq data analysis of mouse large intestines. (A-D) IPA analysis on RNA-seq data (n = 3) from mice large intestines. (A) Top canonical pathways derived from comparison analysis of all doses/treatments (B) Similar directional changes of cymopol (1) to curcumin. The blue lines indicate genes which are downregulated by curcumin, whereas the orange lines are for genes which are up-regulated. (C) Colorectal Cancer Metastasis Signaling pathway indicates that proinflammatory and oncogenic genes such as Tnf-α, Il6r, iNOS, and c-Myc are all downregulated, while cytoprotective Stat1 is up-regulated. (D) Top cancer relevant pathways regulated by NP extract in mice large intestine, green fill = downregulated; red fill = upregulated; outline of black = cancer functionality, outline of blue = abdominal cancer functionality, outline of red = both general cancer and abdominal cancer functionalities. Nrf2 target genes, Gsta4, Hmox1, and Gstp1, are all notably upregulated.
One interesting finding is that cymopol (1) and the NP extract function quite similarly to curcumin, a natural product well characterized for its function in the prevention of inflammation, cancer, and neurological diseases (Fig. 5B) [77]. A recent review has highlighted the benefits of curcumin in the targeting of colorectal cancer stem cells, reducing tumor occurrence via pathways such as the Wnt/β-catenin, Sonic Hedgehog, Notch and PI3K/AKT/mTOR signaling pathways [78]. While no notable difference in the Wnt/β-catenin or Sonic Hedgehog signalling was seen, the PI3K/AKT/mTOR pathway was greatly affected. Elevated levels of TNF at sites of inflammation have been associated with the pathogenesis of these chronic inflammatory diseases. Transcript levels of TNF-α are down-regulated at low dose of 1 and the NP extract (Fig. 5C). A decrease in interleukin-6 receptor transcription supports the overall decrease in the pro-inflammatory response, and results in a decrease in transcript levels of oncogenic c-Myc (Fig. 5C). Furthermore, a decrease in the oncogenic Stat3 as well as reduced Cox2 levels result in a decrease in DNA damage associated with cellular stress responses. STAT3 is important for cellular responses to stimuli such as TNF-α and is associated with an increase in ROS formation. A decrease in STAT3 is also important in reducing inflammation, as it has been shown to facilitate the nuclear accumulation of NFκB [79]. A slight induction of STAT1 is also seen in the ‘Colorectal Cancer Metastasis Signaling’. This may contain chemotherapeutic properties, as STAT1 activation has been associated with antitumor properties including suppression of tumor proliferation, induction of apoptosis, and inhibition of angiogenesis [80–84]. To evaluate the most active treatment in cancer pathways, a network was generated by overlaying top networks associated with cancer (Fig. 5D). Genes associated with ‘cancer’ are outlined in black, while those specifically associated with ‘abdominal cancer’ are shown in blue. Those associated with both general ‘cancer’ and ‘abdominal cancer’ are outlined in red and make up a majority of the network. We observed strong up-regulation of ARE-driven cytoprotective enzymes Hmox1, Gsta4 and Gstp1 on the transcript level, which support the earlier findings of their induction in cell culture and zebrafish (Fig. 3). Collectively, these results indicate the promising properties of cymopols and cymopol-containing extracts for the prevention of cancer caused by oxidative stress-mediated chronic inflammatory disease in the digestive tract.
We next explored if the transcriptional response leads to potentially disease-modifying readout. There is evidence that bioavailable Nrf2 activators can attenuate dextran sodium sulfate (DSS)-induced colitis [85–87]. In addition, Nrf2−/− mice are more susceptible to colitis-associated colorectal cancer [88]. Thus, we tested whether the Cymopolia extract was able to reduce inflammation in our mouse model of DSS-induced colitis by measuring level of the inflammatory marker Lipocalin 2 (Lcn-2) [89]. Mice (6–8 weeks old) were older than the mice used for the tissue distribution and transcriptional assays, so we initially performed dose optimization studies using otherwise the same condition as above (3 days daily treatment), indicating that a slightly higher dose (2.0 g/kg) was required in these older mice to induce a similar robust response in the large intestines and cecum (SI, Fig. S9). Mice were pretreated with extract (2.0 g/kg) or vehicle for 3 days prior to and during DSS administration for a total of 10 days. Lcn-2 level was significantly reduced (p = 0.0012) in extract-gavaged, DSS-exposed mice compared to vehicle treated mice, while there was a trend that baseline Lcn-2 is reduced as well (Fig. 6).
Fig. 6.
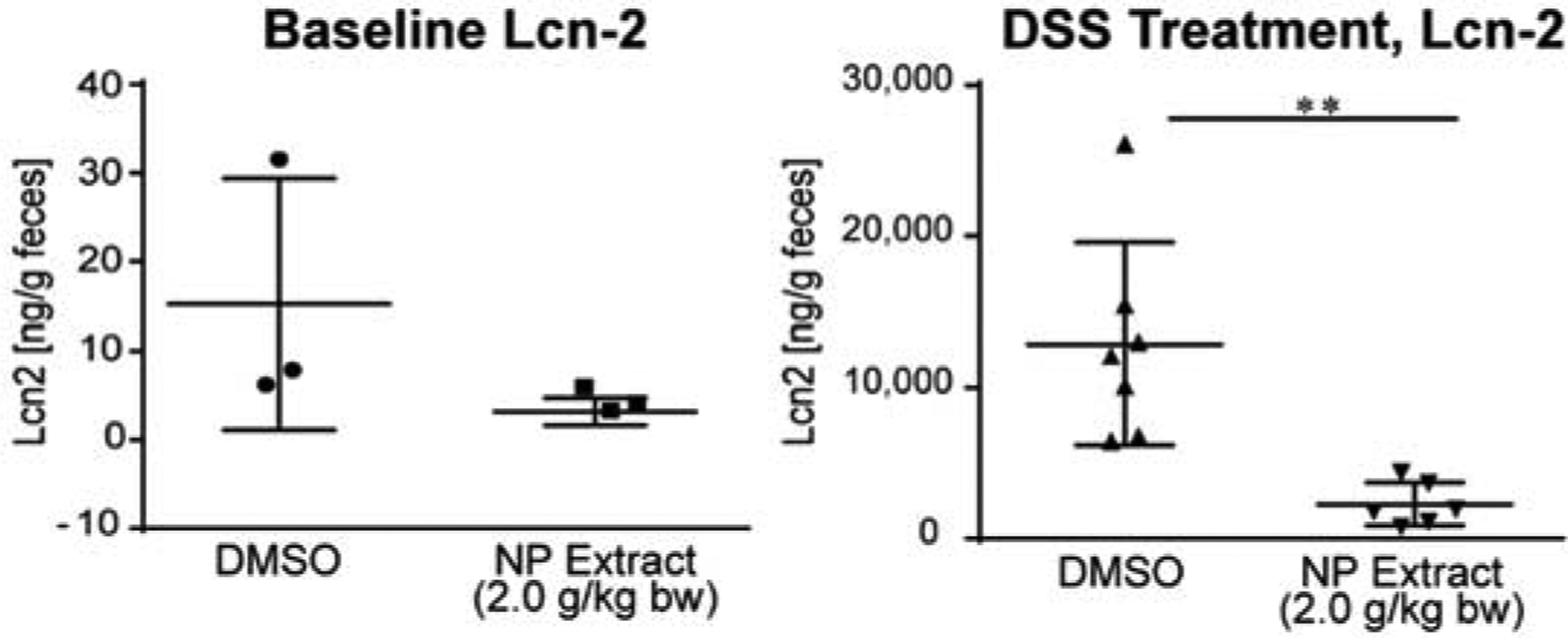
Activity of cymopol-containing NP extract in the DSS model. NP extract (2.0 g/kg) significantly reduced DSS-induced inflammatory marker Lipocalin 2 (Lcn-2) (right, n = 7, DMSO and n = 6 for extract), while there was a trend that NP extract also reduced the baseline level (left, n = 3 for both groups). For DSS treatment ** indicates p-value < 0.0012 using the Mann-Whitney, unpaired t-test.
3.6. RNA-seq based intestinal microbiome analysis
Because microbiota is an important component of intestinal health, we studied the effect of cymopol (1) and NP extract (NP) on mucosal associated bacterial gene expression using RNA-seq. Sequences generated from mucosal tissues obtained from cymopol (1) and NP extract (NP) exposed mice were classified using two independent pipelines: centrifuge [50] (which considered all the non-mouse reads) and QIIME closed-reference (which considered only 16S bacterial sequences present in the dataset) [51]. Interestingly, mice treated with either the extract or cymopol show different gut microbial composition than the control group (Fig. 7,B). We also detected a significant shift in the microbiota composition between the low and the high groups (Fig. 7C). These shifts were detected in the dataset regardless of the pipeline or the read group (forward or reverse) used (SI, Fig. S10). No significant differences were detected between the groups that induced most similar functional responses in the host (see Fig. 4 and Fig. 5A and above; 1 low vs. NP low or 1 high vs NP high) but we detected significant difference in the microbiota composition between control and NP high but not between control and 1 high (SI, Fig. S10). We did not detect any significant differences between the groups in terms of their alpha diversity (data not shown).
Fig. 7.
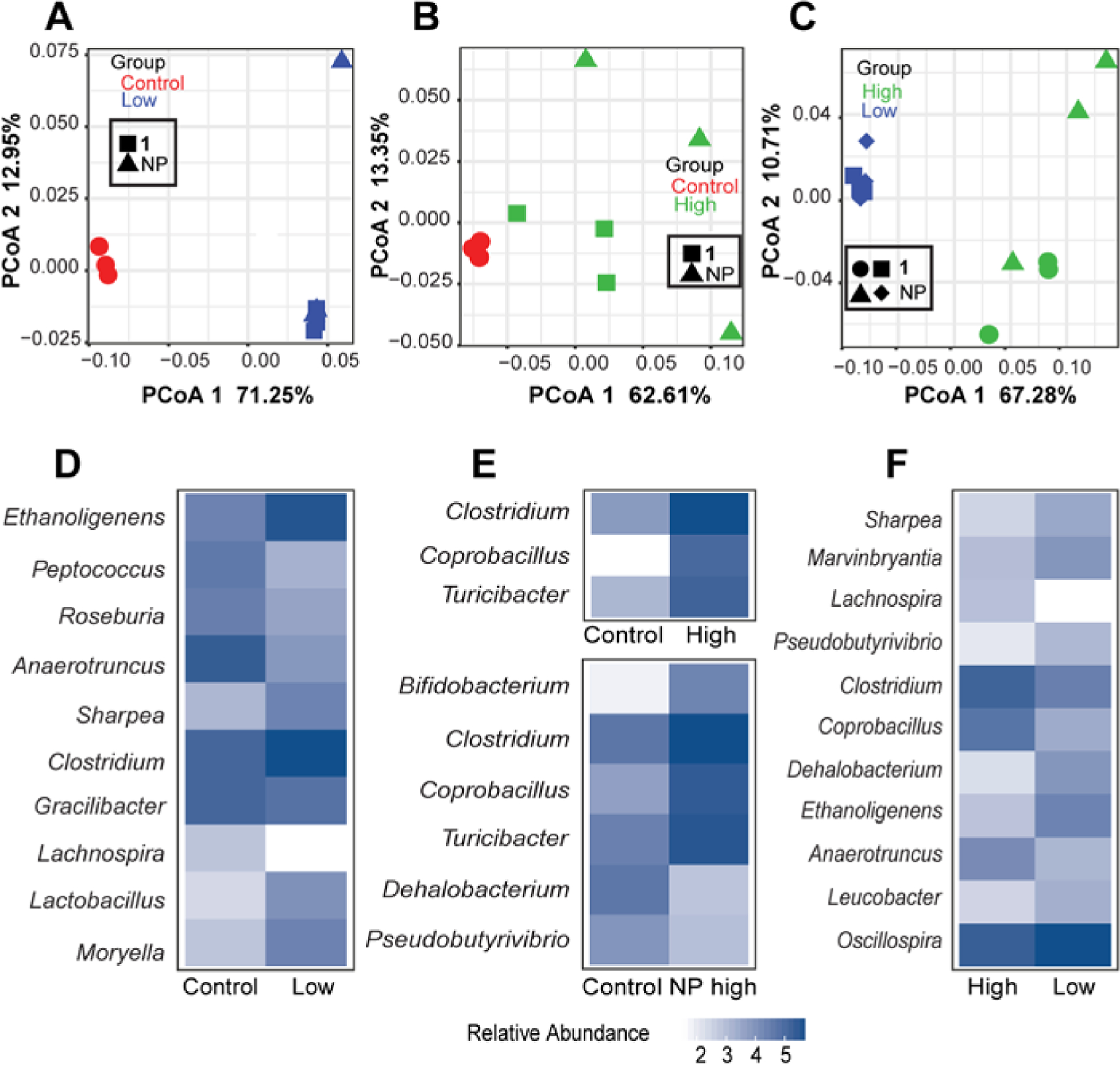
Analysis of the microbiome in the large intestines using 16S sequences extracted from RNA-seq data. The extract and cymopol administration changed the composition of mouse gut microbiota. PCoA analysis using QIIME close-reference OTUs generated from the forward reads shows significant shift in microbial composition between (A) control and low groups PCoA1 FDR p =1.0E-08, (B) control and high groups, PCoA1 FDR p = 0.04 and (C) high and low groups, PCoA1 FDR p = 2.8e-06. Heatmaps showing the mean log10 normalized relative abundances of genera that were significantly different (FDR p < 0.05) between (D) control and low groups, (E) control and high groups and (F) high and low groups.
To characterize the shift in the microbiota, we examined the genera that show significant difference between the three groups and we saw the largest shift in the control versus the low groups (ten genera, Fig. 7D and SI, Table S1) and the high versus the low groups (eleven genera, Fig. 7F, SI, Table S1). The control and the high groups showed a more modest differential abundance with only three genera significantly different (Fig. 7E and SI, Table S1). When we examined each of the high components (NP high and 1 high) separately, we did not detect any significantly different genera between control and 1 high but found six genera that are significantly different between control and NP high (Fig. 7E), including Clostridium and Bifidobacterium which are increased in abundance. The Nrf2 activator sulforaphane was previously shown to increase Clostridium cluster I and Bacteroides fragilis to normalize the composition of intestinal microbiota based on genomic DNA analysis [90], indicating that cymopol and cymopol-containing NP extracts have some overlapping effects with compounds that possess similar effects on the host system [90, 91]. Importantly, Bifidobacterium is considered one of the beneficial gut bacteria and is included as live-component of functional foods [92] and has recently been linked to positive response to PD-1 immunotherapy in cancer patients [93]. Further, we examined the bacterial families that show significant (FDR p < 0.05) differences between the three groups. We found that low versus control comparison showed the highest number of changes. A total of seven bacterial families show changes in their relative abundance. Interestingly, all except one family (Peptococcaceae), increased in abundance in response to treatment with the low group (SI, Fig. S11A and Table S2) compared to the control group. In the high versus control group, only four families significantly changed and all increased in response to treatment with the high group (SI, Fig. S11B and Table S2). In the third comparison between high and low groups, we detected four families (SI, Fig. S11C and Table S2), half of them increased in the low, while the other half increased in the high group. The anti-inflammatory natural product curcumin, which showed overlapping functional responses based on IPA of host RNA-seq data (Fig. 5B), also modulated the gut microbiota, including Bacteroidaceae [91] which is affected by cymopol as well (Table S2).
Our microbiota community abundance survey was conducted using RNA-seq, which we fully acknowledge it has its limitation for this kind of analysis. However, we ran two parallel analyses utilizing two different approaches, one using 16S rRNA gene (Qiime closed-reference) and the other using unique k-mers (centrifuge) and both showed similar results (SI, Fig. S10). Moreover, our analysis that utilized centrifuge output also showed that both Bifidobacterium pseudolongum and Bifidobacterium adolescentis were significantly higher (FDR p < 0.05) in the high-dose NP compared to control.
To gain a better understanding of the microbial response to 1 and NP exposure, we next analyzed its gene expression and pathways. Our transcriptomic analysis identified 141 genes differentially expressed between 1 and NP administered in high concentration and control mice (96 genes up-regulated in the high group and 45 up-regulated in the control group). We detected 605 genes differentially expressed between 1 and NP administered in low concentration and control mice (239 genes upregulated and 366 downregulated in the low group). Regarding the genes differentially expressed between the low and high groups, we found 10,107 genes differentially expressed with 4,573 genes up-regulated in the low group and 5,534 up-regulated in the high group. Principal component analysis (PCA) revealed that microbial transcriptomes of 1 and NP administered in low concentration were different from those of control mice (FDR p = 0.01) (Fig. 8A), 1 and NP administered in high concentration were different from those of control mice (FDR p = 0.001) (Fig. 8B) and 1 and NP administered in high concentration were different from those of 1 and NP administered in low concentration (FDR p = 9.1E-06) (Fig. 8C). We also detected significant difference in the microbial transcriptomes between control and NP high, control and 1 high and NP high and 1 high (SI, Fig. S12). Interestingly, a number of genes downregulated in the treated mice are well known bacterial virulence factors, including Enolase, Internalin (Fig. 8D and SI, Fig. S12), Flagellar basal–body rod protein FlgG and Putative flagellin YvzB (Fig. 8E and SI, Fig. S12). Enolase is a known immunogenic protein contributing to bacterial virulence in many infectious diseases [94], and Internalins help pathogenic bacteria adhere and invade mammalian cells through E-cadherin [95]. Flagellar genes are among the well-known virulence factors and play important role in bacterial motility, adherence and biofilm formation [96, 97].
Fig. 8.
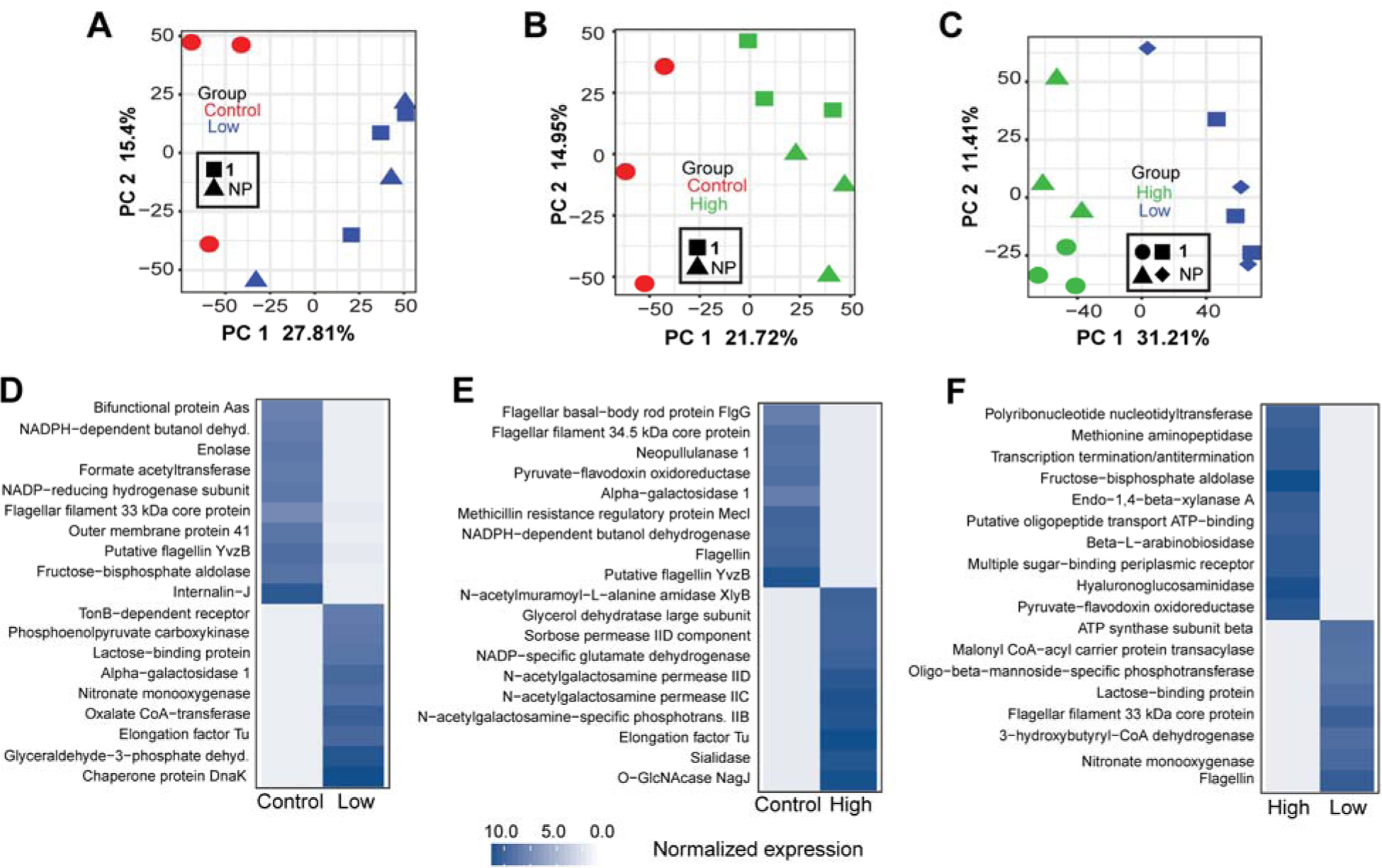
Analysis of the microbial gene expression in the large intestines using microbial transcripts extracted from RNA-seq data. The extract and cymopol administration changed gene expression profile for mouse gut microbiota. PCA analysis of microbial gene expression shows significant changes in microbial gene expression between (A) control and low groups PC1 FDR p = 0.01, (B) control and high groups, PC1 FDR p = 0.001 and (C) high and low groups, PC1 FDR p = 9.1E-06. Heatmaps showing the mean log2 edgeR normalized gene expression of representative significantly differentially (FDR p < 0.05) expressed genes between (D) control and low groups, (E) control and high groups, control and NP high groups and (F) high and low groups.
4. Conclusion
Cymopols isolated from the subtropical marine algae Cymopolia barbata were shown to have both antioxidant and anti-inflammatory activities in a functionally dependent manner. These cytoprotective properties have been validated in various cell culture assays as well as in zebrafish (D. rerio) and mice. Most of the biological activity in orally-gavaged mice was targeted to the digestive tract, namely the small intestine, large intestine, and cecum, increasing the transcript levels of cytoprotective genes, Nqo1 and Hmox1. In addition, using RNA-seq technology, we determined that several pathways are affected by cymopol and the NP extract which are related to oxidative stress, inflammation, and cancer. Most notably, we have seen a decrease in pro-inflammatory transcripts such as that of Tnfα and Cox2 with an increase in ARE-driven genes, such as Gsta4. Such compounds could be useful in the prevention of diseases of the digestive system, which often stem from oxidative stress and chronic inflammation, such as Crohn’s disease that is associated with elevated levels of TNF-α [98]. As with any stress response pathway, the regulation needs to be tightly controlled. Considerations for doses and off-target effects would be critical in determining clinical application of cymopols for the prevention of inflammatory mediated disease. However, based on our analyses in vitro and in vivo, cymopols offer great potential as attenuators of chronic inflammation and oxidative stress associated with the development of diseases such as colorectal carcinoma. The contribution of microbiota compositional changes and altered microbial gene expression to modulation of Nrf2 signaling is still unclear and will require further investigation. However, given the known beneficial role of probiotic cocktails containing Bifidobacterium to reduce symptoms of inflammatory bowel disease and ulcerative colitis and improvements in colon health in general [92], our studies suggest that the positive regulation of Bifidobacterium (and possibly modulation of the abundance of other bacteria) by the seaweed extract could provide additional health benefits. The extract-induced microbiome shift might potentially lead to an increased response to cancer immunotherapy [93], which might be explored in the future. On the functional level, we observed pronounced downregulation of microbial virulence factors, suggesting an additional health benefit. In general, consumption of dietary seaweeds may have functional consequences by modulating host signaling and the microbiome, and the net effect will determine the overall health benefit or potentially adverse effects. Even though many algae, including Cymopolia, are not necessarily expected to be a main human dietary, they might be prepared in various ways for ingestion.
In perspective, we demonstrated the similarities of cymopol (1) and its corresponding NP extract to other well-established natural products including sulforaphane from broccoli and curcumin, a bioactive compound of turmeric. While well characterized for the antioxidant and anti-inflammatory effects, the chemical stability and oral bioavailability of curcumin has been reputably low in both animal studies and humans, and thus several experiments have been conducted to explore benefits and stabilities of various analogues, metabolites, and oral delivery systems [99, 100] Curcumin has been shown to significantly affect the abundance of several families of gut microbial communities, including Prevotellaceae, Bacteroidaceae, and Rikenellaceae (91), some of which are also affected by cymopol Although the oral bioavailability and thus serum levels of curcumin is undetectable in high oral doses up to 8g/day [101], it has been hypothesized that many of the benefits may be attributed to the effect on the gut microbiome [91, 102]. Sulforaphane has been shown to normalize the composition of intestinal microbiota and repair physiological destruction of gut barriers, decrease, inflammation, and immune response in BBN-induced bladder cancer murine models [90]. Of note, one of the major microbiota associated with this benefit on bladder cancer is increased Clostridium, which is known for its involvement in butyric acid production that provides multiple beneficial effects [90]. Similarly, our RNA-seq analysis of colonic tissues demonstrated that the number of Clostridium increased upon treatment with extracts from Cymopolia barbata. Recent studies have shown the benefits of glucoraphanin (precursor for sulforaphane) on breast cancer. Although, similarly, more work is required to evaluate dose-response and ways to improve bioavailability of sulforaphane [103, 104]. In contrast, our functional tissue distribution analysis indirectly indicated sufficient bioavailability of cymopol, especially in the intestines. Additionally, the increase of Bifidobacterium in response to cymopol-containing extracts might be distinct to other compounds that modulate host Nrf2 signaling. The availability of host and bacterial RNA-seq data sets enabled us to extend our analysis to evaluate a functional impact on the intestinal microbiome and revealed a dual function effect through host and bacterial gene expression modulation. Further studies will address the contributions of host and bacterial gene expression changes to the in vivo activity and overall health benefits.
Supplementary Material
Acknowledgements
This research was supported by the National Institutes of Health, NCI grants R01CA172310 (H.L.), R21CA133681 (H.L.), R50CA211487 (R.R.), the Debbie and Sylvia DeSantis Chair Professorship (H.L.), R01DK73338 (C.J.) the University of Florida, Department of Medicine Gatorade Fund (C.J.) and the UF Health Cancer Center funds (R.Z.G. and R.R.). We are grateful to the Agency for Science, Technology and Research (A*STAR) Singapore for funding through the Research Attachment Programme (M.S.B.) and to the University of Florida (UF) College of Pharmacy and UF Health (D. Guzick) for supporting the sabbatical visit at the IMCB (H.L.). We thank the Florida Institute of Oceanography for supporting use of the R/V Bellows and its crew for enabling the collection, J. Johnson for the ARE-luc plasmid, A. Dinkova-Kostova for the isogenic MEFs, D. Zhang for tagged Keap1 and Keap1 mutant plasmids, A. Lewin and P. Y. Han for assistance with oral gavage, P. Havre for mouse tissue processing and L. Imperial for assistance with proteomics and DSS studies. We also thank Y. Zhang from the Gene Expression and Genotyping Core, W. Farmerie and T. Gu from the Bioinformatics Core, and J. Koh from the Proteomics Core of the Interdisciplinary Center for Biotechnology Research at the University of Florida.
Footnotes
Appendix A
Supporting Information: NMR spectra; Figures S1-S12; Tables S1 and S2
Appendix B
Movies S1 and S2
Conflict of Interest
M.S.B. and H.L are listed as inventors on a patent application filed by the University of Florida.
References
- [1].Chapman VJ & Chapman DJ (1980) Seaweeds and Their Uses (Chapman and Hall, New York: ) 3 Ed. [Google Scholar]
- [2].Arasaki S & Arasaki T (1983) Low calorie, high nutrition vegetables from the sea To help you look and feel better. (Japan Publications, Inc., Tokyo: ). [Google Scholar]
- [3].Hoshiyama Y & Sasaba T (1992) A case-control study of single and multiple stomach cancers in Saitama Prefecture, Japan. Jpn J Cancer Res 83(9):937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hoshiyama Y, Sekine T, & Sasaba T (1993) A case-control study of colorectal cancer and its relation to diet, cigarettes, and alcohol consumption in Saitama Prefecture, Japan. Tohoku J Exp Med 171(2):153–165. [DOI] [PubMed] [Google Scholar]
- [5].Ratnayake R, Liu Y, Paul VJ, & Luesch H (2013) Cultivated sea lettuce is a multiorgan protector from oxidative and inflammatory stress by enhancing the endogenous antioxidant defense system. Cancer Prev Res (Phila) 6(9):989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Snare DJ, Fields AM, Snell TW, & Kubanek J (2013) Lifespan extension of rotifers by treatment with red algal extracts. Exp Gerontol 48(12):1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang R, Paul VJ, & Luesch H (2013) Seaweed extracts and unsaturated fatty acid constituents from the green alga Ulva lactuca as activators of the cytoprotective Nrf2-ARE pathway. Free Radic Biol Med 57:141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, & Nabeshima Y (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236(2):313–322. [DOI] [PubMed] [Google Scholar]
- [9].Miao W, Hu L, Scrivens PJ, & Batist G (2005) Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem 280(21):20340–20348. [DOI] [PubMed] [Google Scholar]
- [10].Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, & Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol 24:7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cullinan SB, Gordan JD, Jin J, Harper JW, & Diehl JA (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol 24:8477–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang DD, Lo SC, Cross JV, Templeton DJ, & Hannink M (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol 24:10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dinkova-Kostova AT, Holtzclaw WD, & Kensler TW (2005) The role of Keap1 in cellular protective responses. Chem Res Toxicol 18:1779–1791. [DOI] [PubMed] [Google Scholar]
- [14].Zhang DD (2006) Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev 38:769–789. [DOI] [PubMed] [Google Scholar]
- [15].McMahon M, Thomas N, Itoh K, Yamamoto M, & Hayes JD (2006) Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem 281, 24756–24768. [DOI] [PubMed] [Google Scholar]
- [16].Zipper LM & Mulcahy RT (2002) The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem 277(39):36544–36552. [DOI] [PubMed] [Google Scholar]
- [17].Tong KI, Kobayashi A, Katsuoka F, & Yamamoto M (2006) Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem 387(10–11):1311–1320. [DOI] [PubMed] [Google Scholar]
- [18].McMahon M, Itoh K, Yamamoto M, & Hayes JD (2003) Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem 278(24):21592–21600. [DOI] [PubMed] [Google Scholar]
- [19].Bryan HK, Olayanju A, Goldring CE, & Park BK (2013) The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85(6):705–717. [DOI] [PubMed] [Google Scholar]
- [20].Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, & Talalay P (2004) Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A 101(7):2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Holland R & Fishbein JC (2010) Chemistry of the cysteine sensors in Kelch-like ECH-associated protein 1. Antioxid Redox Signal 13(11):1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang R, Mason DE, Choe KP, Lewin AS, Peters EC, & Luesch H (2013) In vitro and in vivo characterization of a tunable dual-reactivity probe of the Nrf2-ARE pathway. ACS Chem Biol 8(8):1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hu C, Eggler AL, Mesecar AD, & van Breemen RB (2011) Modification of Keap1 cysteine residues by sulforaphane. Chem Res Toxicol 24(4):515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nair S, Doh ST, Chan JY, Kong AN, & Cai L (2008) Regulatory potential for concerted modulation of Nrf2- and Nfkb1-mediated gene expression in inflammation and carcinogenesis. Br J Cancer 99(12):2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, Li CW, Ding Q, Liao TL, Lai CC, Lin AC, Chang YH, Tsai SF, Li LY, & Hung MC (2009) KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell 36(1):131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Alcaraz MJ, Vicente AM, Araico A, Dominguez JN, Terencio MC, & Ferrándiz ML (2004) Role of nuclear factor-kappaB and heme oxygenase-1 in the mechanism of action of an anti-inflammatory chalcone derivative in RAW 264.7 cells. Br J Pharmacol 142(7):1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Högberg HE, J. TR, & King JT (1976) The cymopols, a group of prenylated bromohydroquinones from the green calcareous alga Cymopolia barbata. J Chem Soc 1:1696–1701. [Google Scholar]
- [28].Wall ME, Wani MC, Manikumar G, Taylor H, Hughes TJ, Gaetano K, Gerwick HW, McPhail AT, & McPhail DR (1989) Plant antimutagenic agents, 7. Structure and antimutagenic properties of cymobarbatol and 4-isocymobarbatol, new cymopols from green alga (Cymopolia barbata). J Nat Prod 52(5):1092–1099. [DOI] [PubMed] [Google Scholar]
- [29].Mayer RJ & Marshall LA (1993) New insights on mammalian phospholipase A2(s); comparison of arachidonoyl-selective and -nonselective enzymes. FASEB J 7(2):339–348. [DOI] [PubMed] [Google Scholar]
- [30].Takamatsu S, Nagle DG, & Gerwick WH (2004) Secondary metabolites from marine cyanobacteria and algae inhibit LFA-1/ICAM-1 mediated cell adhesion. Planta Med 70(2):127–131. [DOI] [PubMed] [Google Scholar]
- [31].Martínez Nadal N, Casillas Chapel C, Rodríguez L, Rodriguez Perazza J, & Torres Vera L (1964) Antibiotic properties of marine algae. III. Cymopolia barbata. Botanica Marina 9(1–2):21–26. [Google Scholar]
- [32].Takamatsu S, Hodges TW, Rajbhandari I, Gerwick WH, Hamann MT, & Nagle DG (2002) Marine natural products as novel antioxidant prototypes. J Nat Prod 66:605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gentile CL, Weir TL (2018) The gut microbiota at the intersection of diet and human health. Science 362(6416):776–780. [DOI] [PubMed] [Google Scholar]
- [34].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, & Turnbaugh PJ (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505(7484):559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Moehlenkamp JD & Johnson JA (1999) Activation of antioxidant/electrophile-responsive elements in IMR-32 human neuroblastoma cells. Arch Biochem Biophys 363(1):98–106. [DOI] [PubMed] [Google Scholar]
- [36].Zhang DD & Hannink M (2003) Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23(22):8137–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Brooks JD, Paton VG, & Vidanes G (2001) Potent induction of phase 2 enzymes in human prostate cells by sulforaphane. Cancer Epidemiol Biomarkers Prev 10(9):949–954. [PubMed] [Google Scholar]
- [38].Lee JM, Hanson JM, Chu WA, & Johnson JA (2001) Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J Biol Chem 276(23):20011–20016. [DOI] [PubMed] [Google Scholar]
- [39].Lee JM, Moehlenkamp JD, Hanson JM, & Johnson JA (2001) Nrf2-dependent activation of the antioxidant responsive element by tert-butylhydroquinone is independent of oxidative stress in IMR-32 human neuroblastoma cells. Biochem Biophys Res Commun 280(1):286–292. [DOI] [PubMed] [Google Scholar]
- [40].Li J, Lee JM, & Johnson JA (2002) Microarray analysis reveals an antioxidant responsive element-driven gene set involved in conferring protection from an oxidative stress-induced apoptosis in IMR-32 cells. J Biol Chem 277(1):388–394. [DOI] [PubMed] [Google Scholar]
- [41].McConnell OJ, Hughes PA, & Targett NM (1982) Diastereomers of cyclocymopol and cyclocymopol monomethyl ether from Cymopolia barbata. Phytochemistry 21(8):2139–2141. [Google Scholar]
- [42].Keller A, Nesvizhskii AI, Kolker E, & Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74(20):5383–5392. [DOI] [PubMed] [Google Scholar]
- [43].Nesvizhskii AI, Keller A, Kolker E, & Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75(17):4646–4658. [DOI] [PubMed] [Google Scholar]
- [44].Prochaska HJ & Santamaria AB (1988) Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal Biochem 169(2):328–336. [DOI] [PubMed] [Google Scholar]
- [45].Renshaw SA, Loynes CA, Trushell DM., Elworthy S, Ingham PW, & Whyte MK (2006) A transgenic zebrafish model of neutrophilic inflammation. Blood 108:3976–3978. [DOI] [PubMed] [Google Scholar]
- [46].Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, & Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. [DOI] [PubMed] [Google Scholar]
- [47].Wang X, Robertson AL, Li J, Chai RJ, Haishan W, Sadiku P, Ogryzko NV, Everett M, Yoganathan K, Luo HR, Renshaw SA, & Ingham PW (2014) Inhibitors of neutrophil recruitment identified using transgenic zebrafish to screen a natural product library. Dis Model Mech 7(1):163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bolger AM, Lohse M, & Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li H & Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kim D, Song L, Breitwieser FP, & Salzberg SL (2016) Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26(12):1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Caporaso JG, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang Q, Garrity GM, Tiedje JM, & Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McCafferty J, Mühlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, & Fodor AA (2013) Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J 7(11):2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].McMurdie PJ & Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Team RC (2015) R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing; ). [Google Scholar]
- [56].Benjamini Y & Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- [57].Grabherr MG, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Haas BJ, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].The UniProt C (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45(D1):D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, & Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen L, Zheng D, Liu B, Yang J, & Jin Q (2016) VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res 44(D1):D694–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li B & Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Robinson MD, McCarthy DJ, & Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dinkova-Kostova AT & Wang XJ (2011) Induction of the Keap1/Nrf2/ARE pathway by oxidizable diphenols. Chem Biol Interact 192(1–2):101–106. [DOI] [PubMed] [Google Scholar]
- [65].Hur W, Sun Z, Jiang T, Mason DE, Peters EC, Zhang DD, Luesch H, Schultz PG, & Gray NS (2010) A small-molecule inducer of the antioxidant response element. Chem Biol 17(5):537–547. [DOI] [PubMed] [Google Scholar]
- [66].Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, & Yamamoto M (2009) The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol 29(2):493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, & van Breemen RB (2007) Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom 18(12):2226–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chen H, Fu J, Chen H, Hu Y, Soroka DN, Prigge JR, Schmidt EE, Yan F, Major MB, Chen X, & Sang S (2014) Ginger compound [6]-shogaol and its cysteine-conjugated metabolite (M2) activate Nrf2 in colon epithelial cells in vitro and in vivo. Chem Res Toxicol 27(9):1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dinkova-Kostova AT, Holtzclaw WD, & Kensler TW (2005) The role of Keap1 in cellular protective responses. Chem Res Toxicol 18(12):1779–1791. [DOI] [PubMed] [Google Scholar]
- [70].Clark RA & Klebanoff SJ (1979) Chemotactic factor inactivation by the myeloperoxidase-hydrogen peroxide-halide system. J Clin Invest 64(4):913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ellett F, Pase L, Hayman JW, Andrianopoulos A, & Lieschke GJ (2011) mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117(4):e49–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hattori H, Subramanian KK, Sakai J, Jia Y, Li Y, Porter TF, Loison F, Sarraj B, Kasorn A, Jo H, Blanchard C, Zirkle D, McDonald D, Pai SY, Serhan CN, & Luo HR (2010) Small-molecule screen identifies reactive oxygen species as key regulators of neutrophil chemotaxis. Proc Natl Acad Sci U S A 107(8):3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lekstrom-Himes JA, Kuhns DB, Alvord WG, & Gallin JI (2005) Inhibition of human neutrophil IL-8 production by hydrogen peroxide and dysregulation in chronic granulomatous disease. J Immunol 174(1):411–417. [DOI] [PubMed] [Google Scholar]
- [74].The forty-ninth meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA) Anonymous (1998) Safety evaluation of certain food additives and contaminants. WHO Food Additives Series, No. 40. IPCS INCHEM. (http://www.inchem.org/documents/jecfa/jecmono/v040je01.htm) (accessed 9 September 2019). [Google Scholar]
- [75].Peters MM, Lau SS, Dulik D, Murphy D, van Ommen B, van Bladeren PJ, & Monks TJ (1996) Metabolism of tert-butylhydroquinone to S-substituted conjugates in the male Fischer 344 rat. Chem Res Toxicol 9(1):133–139. [DOI] [PubMed] [Google Scholar]
- [76].Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, Han J, Agarwal A, & Kong AN (2006) Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res 66(17):8804–8813. [DOI] [PubMed] [Google Scholar]
- [77].Mahmood K, Zia KM, Zuber M, Salman M, & Anjum MN (2015) Recent developments in curcumin and curcumin based polymeric materials for biomedical applications: A review. Int J Biol Macromol 81:877–890. [DOI] [PubMed] [Google Scholar]
- [78].Ramasamy TS, Ayob AZ, Myint HH, Thiagarajah S, & Amini F (2015) Targeting colorectal cancer stem cells using curcumin and curcumin analogues: insights into the mechanism of the therapeutic efficacy. Cancer Cell Int 15:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zouein FA, Duhé RJ, Arany I, Shirey K, Hosler JP, Liu H, Saad I, Kurdi M, & Booz GW (2014) Loss of STAT3 in mouse embryonic fibroblasts reveals its Janus-like actions on mitochondrial function and cell viability. Cytokine 66(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Battle TE, Lynch RA, & Frank DA (2006) Signal transducer and activator of transcription 1 activation in endothelial cells is a negative regulator of angiogenesis. Cancer Res 66(7):3649–3657. [DOI] [PubMed] [Google Scholar]
- [81].Bromberg JF, Horvath CM, Wen Z, Schreiber RD, & Darnell JE Jr., (1996) Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A 93(15):7673–7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hosui A, Klover P, Tatsumi T, Uemura A, Nagano H, Doki Y, Mori M, Hiramatsu N, Kanto T, Hennighausen L, Hayashi N, & Takehara T (2012) Suppression of signal transducers and activators of transcription 1 in hepatocellular carcinoma is associated with tumor progression. Int J Cancer 131(12):2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Huang S, Bucana CD, Van Arsdall M, & Fidler IJ (2002) Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene 21(16):2504–2512. [DOI] [PubMed] [Google Scholar]
- [84].Stephanou A & Latchman DS (2003) STAT-1: a novel regulator of apoptosis. Int J Exp Pathol 84(6):239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Liu X, Zhou W, Zhang X, Lu P, Du Q, Tao L, Ding Y, Wang Y, & Hu R (2016) Dimethyl fumarate ameliorates dextran sulfate sodium-induced murine experimental colitis by activating Nrf2 and suppressing NLRP3 inflammasome activation. Biochem Pharmacol 112:37–49. [DOI] [PubMed] [Google Scholar]
- [86].Wang Y, Wang H, Qian C, Tang J, Zhou W, Liu X, You Q, & Hu R (2016) 3-(2-oxo-2-phenylethylidene)-2,3,6,7-tetrahydro-1H-pyrazino[2,1-a]isoquinolin-4(1 1bH)-one (compound 1), a novel potent Nrf2/ARE inducer, protects against DSS-induced colitis via inhibiting NLRP3 inflammasome. Biochem Pharmacol 101:71–86. [DOI] [PubMed] [Google Scholar]
- [87].Xi MY, et al. (2013) 3-aroylmethylene-2,3,6,7-tetrahydro-1H-pyrazino[2,1-a]isoquinolin-4(11bH)-ones as potent Nrf2/ARE inducers in human cancer cells and AOM-DSS treated mice. J Med Chem 56(20):7925–7938. [DOI] [PubMed] [Google Scholar]
- [88].Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS, & Kong AN (2008) Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res (Phila) 1(3):187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, & Vijay-Kumar M (2012) Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One 7(9):e44328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].He C, Huang L, Lei P, Liu X, Li B, & Shan Y (2018) Sulforaphane normalizes intestinal flora and enhances gut barrier in mice with BBN-induced bladder cancer. Mol Nutr Food Res. 62(24):e1800427. (doi: 10.1002/mnfr.201800427). [DOI] [PubMed] [Google Scholar]
- [91].Shen L, Liu L, & Ji HF (2017) Regulative effects of curcumin spice administration on gut microbiota and its pharmacological implications. Food Nutr Res. 61(1):1361780. (doi: 10.1080/16546628.2017.1361780). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].O’Callaghan A, & van Sinderen D (2016) Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol 7:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, & Gajewski TF (2018) The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359(6371):104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Henderson B & Martin A (2011) Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79(9):3476–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bonazzi M, Lecuit M, & Cossart P (2009) Listeria monocytogenes internalin and E-cadherin: from bench to bedside. Cold Spring Harb Perspect Biol 1(4):a003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Saeb AT, David SK, & Al-Brahim H (2014) In silico detection of virulence gene homologues in the human pathogen sphingomonas spp. Evol Bioinform Online 10:229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Elhadad D, Desai P, Rahav G, McClelland M, & Gal-Mor O (2015) Flagellin is required for host cell invasion and normal Salmonella pathogenicity island 1 expression by Salmonella enterica serovar Paratyphi A. Infect Immun 83(9):3355–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Reimund JM, Ratajczyk J, Sola B, Justum AM, & Muller CD (2007) Anti-tumor necrosis factor-alpha (TNF-alpha) treatment strategies in Crohn’s disease. Recent Pat Inflamm Allergy Drug Discov 1(1):21–34. [DOI] [PubMed] [Google Scholar]
- [99].Anand P, Kunnumakkara AB, Newman RA, & Aggarwal BB (2007) Bioavailability of curcumin: problems and promises. Mol Pharm 4(6):807–818. [DOI] [PubMed] [Google Scholar]
- [100].Xu XY, Meng X, Li S, Gan RY, Li Y, & Li HB (2018) Bioactivity, Health Benefits, and Related Molecular Mechanisms of Curcumin: Current Progress, Challenges, and Perspectives. Nutrients 10(10):E1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lao CD, Ruffin MT 4th, Normolle D, Heath DD, Murray SI, Bailey JM, Boggs ME, Crowell J, Rock CL, & Brenner DE (2006) Dose escalation of a curcuminoid formulation. BMC Complement Altern Med 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Jin T, Song Z, Weng J, & Fantus IG (2018) Curcumin and other dietary polyphenols: potential mechanisms of metabolic actions and therapy for diabetes and obesity. Am J Physiol Endocrinol Metab 314(3):E201–E205. [DOI] [PubMed] [Google Scholar]
- [103].Atwell LL, Zhang Z, Mori M, Farris P, Vetto JT, Naik AM, Oh KY, Thuillier P, Ho E, & Shannon J (2015) Sulforaphane bioavailability and chemopreventive activity in women scheduled for breast biopsy. Cancer Prev Res (Phila) 8(12):1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Atwell LL, Hsu A, Wong CP, Stevens JF, Bella D, Yu TW, Pereira CB, Löhr CV, Christensen JM, Dashwood RH, Williams DE, Shannon J, & Ho E (2015) Absorption and chemopreventive targets of sulforaphane in humans following consumption of broccoli sprouts or a myrosinase-treated broccoli sprout extract. Mol Nutr Food Res 59(3):424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


