Abstract
To learn more about the mechanisms of human dietary fat perception, we asked 398 human twins to rate the fattiness and how much they liked 6 types of potato chips that differed in triglyceride content (2.5%, 5%, 10%, and 15% corn oil); reliability estimates were obtained from a subset (n = 50) who did the task twice. Some chips also had a saturated long-chain fatty acid (FA; hexadecanoic acid, 16:0) added (0.2%) to evaluate its effect on fattiness and liking. We computed the heritability of these measures and conducted a genome-wide association study (GWAS) to identify regions of the genome that co-segregate with fattiness and liking. Perceived fattiness of and liking for the potato chips were reliable (r = 0.31–0.62, P < 0.05) and heritable (up to h2 = 0.29, P < 0.001, for liking). Adding hexadecanoic acid to the potato chips significantly increased ratings of fattiness but decreased liking. Twins with the G allele of rs263429 near GATA3-AS1 or the G allele of rs8103990 within ZNF729 reported more liking for potato chips than did twins with the other allele (multivariate GWAS, P < 1 × 10–5), with results reaching genome-wide suggestive but not significance criteria. Person-to-person variation in the perception and liking of dietary fat was 1) negatively affected by the addition of a saturated FA and 2) related to inborn genetic variants. These data suggest that liking for dietary fat is not due solely to FA content and highlight new candidate genes and proteins within this sensory pathway.
Keywords: fat perception, genetics, oleogustus, sensory, taste, taste receptors
Introduction
Sensory nutrition is a research area that investigates how the taste, smell, and flavor of food and drink affect food choices and diet quality, and how food choice, in turn, affects human health and disease (Hayes 2015; Forde 2018). While food is essential to our survival and eating may be pleasant, it can also be dangerous, especially for those who “dig their grave with a spoon” (Card 2013) and die from heart disease or diabetes, health conditions that arise in whole or in part from dietary choices (Reed and Knaapila 2010). Some of the pleasure of food arises from its dietary fat and sugar content. The sweetness of sugar is well understood from a sensory perspective (Nelson et al. 2001), with direct links between taste cells and brain areas of reward, for example, (Veldhuizen et al. 2017). In contrast, the initial sensory cues responsible for the perception of dietary fat are less well understood, and what is known is contentious: whether there is a distinct taste quality for fat or fatty acids (FA) and which of the chemical and texture components of fat are responsible for the sensations it evokes (Reed and Xia 2015; Running et al. 2015; Running and Mattes 2016).
One unresolved conundrum is mounting evidence that, while triglycerides and FA both impart fatty sensations in foods, triglycerides tend to have a positive hedonic valance (e.g., Bakke et al. 2016), whereas FA typically have a negative hedonic valence, for example, scratchy (Voigt et al. 2014) or otherwise “bad” (Running and Mattes 2016). These data suggest that multiple sensory pathways are involved in the perception of fats in foods (Drewnowski 1992). One method to learn more about these multiple pathways is to evaluate origins of person-to-person or animal-to-animal differences—this type of genetics-driven approach helped identify the bitter and sweet receptors (Reed and Knaapila 2010; Reed et al. 2006). Here, we reasoned that people differ in their response to fat in food, that these differences are heritable, and that genome-wide methods are likely useful to identify the relevant genes.
To establish heritability, we selected a classic twin design, comparing monozygotic (MZ) and dizygotic (DZ) twins for their response to fat in foods. We also had to choose appropriate test stimuli that would generalize to real foods (vs. model systems) and appropriate behavioral methods. No one standard method has been adopted, with investigators in this area using many different stimuli to measure fat perception, including oil-and-water mixtures (Heinze et al. 2017); oil in salad dressing (Keller et al. 2012); fat in puddings (Mennella et al. 2012), in scrambled eggs or mashed potatoes (Mela and Sacchetti 1991), or in ice cream (Rolon et al. 2017) or added FA in chocolate (Running et al. 2017). Here, we used potato chips that varied in amounts of corn oil and an added FA, capitalizing on our technical expertise in their production and practical constraints of our testing environment (an annual convention of twins; see below). We also tested the twins’ ability to discriminate high- and low-fat milk samples.
Materials and methods
Participants
We tested adult MZ and DZ twins who attended an annual convention of twins, the Twin Days Festival in Twinsburg, OH. This event is held each August, and all data reported here were collected during the 2018 convention. The exclusion criteria for participation were age less than 18 years, pregnancy, or an allergy or sensitivity to milk. All data were collected under protocols approved by the University of Pennsylvania Institutional Review Board (#701426).
Stimuli
Three types of stimuli were used: potato chips that differed in triglyceride and FA content, multiple prototypical tastants, and milk that was either high (18.00%) or low (2.35%) in fat. Six types of potato chips were prepared, following standard methods at PepsiCo research laboratories: chips that contained 2.5%, 5.0%, 10%, or 15% corn oil and chips with 2.5% or 5.0% corn oil with added 0.2% (w/w) hexadecanoic acid, a saturated long-chain FA (16:0). Time constraints prevented us from testing all combinations of triglycerides and FA. Ascending amounts of corn oil were chosen to minimize carryover effects across samples; the FA was added to gauge its impact on ratings of fattiness and liking.
We chose 0.2% hexadecanoic acid as the add-in to the potato chips based on a pilot test we conducted. In this pilot test, 17 adults compared potato chips that had 0.12% or 0.2% FA (oleic, linoleic, or hexadecanoic) added versus plain potato chips. We found the largest differences in fattiness compared with the plain potato chips occurred with the 0.2% hexadecanoic acid. This concentration is similar to those used in other human sensory studies using FA (e.g., Heinze et al. 2017) and is within the receptive range of the known mammalian FA receptors, GPR40, GPR120, and CD36 (Sclafani et al. 2007a; Dramane et al. 2012; Galindo et al. 2012; Voigt et al. 2014).
As an aside, we noted that, for potato chips made with oleic acid, 13 of 17 subjects used descriptors like “odd,” “aftertaste,” “funny,” or “cheesy” for chips with either 0.12% or 0.20% oleic acid, which discouraged us from choosing this commonly used FA.
The second type of stimulus comprised standard solutions (5 mL) used in taste psychophysics: plain deionized water, sucrose (12% w/v, 350 mM), NaCl (1.5% w/v, 256 mM), and the bitter compound phenylthiocarbamide (PTC; 1.8 × 10–4 M), all purchased from Sigma (as the last 2 stimuli, we also tested menthol [1 mM] and capsaicin [3 μM] for an unrelated project; those results are not reported here). The third type of stimuli was milk with 18.00% or 2.35% fat mixed at the Monell Chemical Senses Center using Shop Rite brand instant nonfat dry milk (SKU/UPC 041190010189) purchased at a local grocery store and anhydrous dairy fat (Table 1). All ingredients were combined in a homogenizer (GEA) and processed with 5 passes at 250 bars of pressure; resulting particle sizes were within the expected range.
Table 1.
High- and low-fat milk ingredients
| Milk type | Fat content (%) | Water (mL) | Dry milk (g) | Dairy fat (g) | Casein (g) |
|---|---|---|---|---|---|
| Low fat | 2.35 | 890 | 90.7 | 23.7 | 10.09 |
| High fat | 18.00 | 890 | 90.7 | 216.8 | 12.04 |
Sample presentation
Single potato chips of roughly equivalent size and weight were placed in clear 3–5-oz plastic souffle cups with plastic lids (Universal Product Code [UPC] #742010492467). Participants were given potato chips in a fixed, predetermined order (as follows: 2.5% corn oil, 15% corn oil, 5% corn oil, 10% corn oil, 5% corn oil with 0.2% hexadecanoic acid, and 2.5% corn oil with 0.2% hexadecanoic acid), and asked to rate the potato chips for “fattiness” and “liking” on visual analog scales presented on an Apple iPad Air (9.7-inch display; Apple Inc.). The fixed and predetermined order made the testing procedure easier logistically and reduced carryover effects but was not ideal because there may be order effects of testing. Liking scales were anchored with “do not like at all” on the left and “like extremely” on the right. Similarly, the fattiness scale was anchored on the left with “not fatty at all” and on the right with “extremely fatty.” We also asked about “crispiness” and “saltiness” to prevent halo-dumping effects, a bias in sensory ratings, which can occur when subjects are provided too few salient rating options (Clark and Lawless 1994). Participants were instructed to rinse their mouths with water (Nestle Pure Life, UPC 068274934711) after each sample. For logistical reasons and to enhance ecological validity, participants did not wear nose clips but rather chewed (and swallowed) all potato chip samples.
For the taste solutions, participants rated each for the qualities of “liking,” “saltiness,” “sweetness,” “sourness,” “bitterness,” and “burn” on visual analog scales, with the left side anchored with “no [quality] at all” and the right side anchored with “extreme [quality]” as previously described (Knaapila et al. 2012). To focus on taste and reduce odor cues, participants wore nose clips (GENEXA LLC, UPC 708981350007). Participants were asked to hold each solution in their mouth for 5 s, rate it on the scale provided, spit out the solution, and rinse their mouth with water afterward.
For the milk fat discrimination test, a 2-alternative forced-choice task was used. Before testing began, each participant was given 2 references as warm-up samples; these were verbally identified to participants as “low-fat” and “high-fat” samples, respectively. Participants were then given 10 pairs of opaque bottles (EP-34434, Berry Global Group, Inc.). Each pair contained one low-fat and one high-fat sample (each 5 mL) presented in a fixed order. Participants wore nose clips; they were instructed to hold each sample in their mouth for 5 s, spit out the sample, and rinse their mouth with water afterward. For each pair, participants were asked, “Which solution tastes fattier?” If they were unsure, they were instructed to guess. Discrimination ability was defined as the number correct across all 10 trials (i.e., perfect discrimination would be 10 out of 10 trials correct). On the day of testing, the milk discrimination was first, followed by taste solutions and potato chip testing; this order was driven by practical concerns to maintain participant engagement because most participants enjoyed the most.
Saliva collection and DNA extraction
We obtained saliva samples from all participants by asking them to expectorate into collection tubes; DNA was extracted from the saliva using procedures recommended by Oragene (DNA Genotek). We measured and recorded DNA concentration and quality scores using a Nanodrop 1000 Spectrophotometer (Thermo Fisher Scientific).
Genotyping
We conducted both single-marker and high-throughput-based genotyping. Using the single-marker method, we typed 3 variant sites in the TAS2R38 gene in all twins as a quality-control step 1) to ensure that the DNA extracted from saliva could be genotyped, 2) to confirm that the genotype matched the psychophysical ratings of PTC bitterness, and 3) to get preliminary confirmation of twin zygosity (each pair of MZ twins is expected to have the same genotype). For these assays, DNA samples were diluted to a concentration of 10 ng/μL and used as templates in Taqman assays (rs713598, C___8876467_10; rs1726866, C___9506827_10; and rs10246939, C___9506826_10; Applied Biosystems) using previously established methods.
For the DNA high-throughput genotyping, we sent the DNA samples to the Center for Inherited Disease Research (CIDR), which typed them for the Illumina OmniExpress panel (Infinium OmniExpressExome-8, v1.6; Illumina) following the manufacturer’s procedures and the CIDR’s standard quality-control methods. For 176 MZ twin pairs, we used high-throughput genotyping for only one twin of each pair and imputed the genotype of the other member of the pair because of their presumed identical genomes.
Twin zygosity
Twin zygosity was measured in 3 ways. Twins self-reported their zygosity status as 1) MZ (identical), 2) DZ (fraternal), or 3) uncertain; photographs were taken of each twin and rated for physical similarity by a research assistant blind to self-reported zygosity, and all twins were genotyped for the 3 markers described above. In rare cases where zygosity status was still uncertain, both members of the pair were genotyped using the high-throughput-based genotyping method (see above).
Data analysis
We conducted 4 types of statistical analysis: 1) descriptive statistics of the psychophysical data, 2) calculation of heritability, 3) tests of genome-wide association between genetic variants and the measures of fat perception, and 4) gene expression (RNASeq) and bioinformatics (enrichment) analyses. All descriptive statistics, such as means, standard deviations (SDs), and correlations among variables, were computed using R (v. 3.53) and R-Studio (v. 1.1.456).
Sensory analyses
For descriptive analyses, we plotted the probability density of the data (smoothed by a kernel density estimator) as a violin plot, calculated mean and SD, and checked for sex, race, and age effects on the sensory measures in a general linear model (GLM) using race and sex as fixed effects and age as a covariate. For all GLM analyses, individual group means were evaluated for difference using Tukey post hoc tests (honestly significant difference [HSD]). If race and sex had a significant effect in the GLM analysis, to better understand their effects on psychophysical outcomes, we grouped participants by these factors and compared the mean ratings. For age and its relationship to the psychophysical measures, we computed Pearson correlations.
To evaluate whether there were consistent person-to-person differences in the rating of the potato chips overall, Pearson correlations of intensity and liking measures among the 6 types of potato chip were calculated. In addition, we calculated Cronbach’s alpha for psychophysical measures across all 6 types of potato chips. To understand the reliability of the measures, we assessed test–retest correlations among the same measures taken twice in a subset of participants (n = 50).
To gauge the effect of corn oil concentrations and hexadecanoic acid on the sensory measures, we reconducted a linear mixed-model analysis with corn oil concentration (2.5% and 5.0%) and hexadecanoic acid (added or not) as 2 separate factors and treated the psychophysical data as repeated measurements, with race and age as covariates in the model (we did not include sex in this model because results indicated that male and females were similar in their ratings). In a complementary analysis, we reconducted the analysis using potato chip type as a single factor (with 6 levels, one for each type of potato chip). These complementary analyses were included because of the unbalanced design: not all concentrations of corn oil were presented with and without the added 0.2% hexadecanoic acid.
Heritability
For the heritability analysis, the Cholesky model was used to evaluate the magnitude of genetic and environmental influences on the traits, and the phenotypic variance was decomposed into an additive genetic component (a2), shared environmental factors (c2), and nonshared environmental or individual-specific factors (e2) as described previously (Wise et al. 2007). Variance accounted for by each of these components was calculated by comparing MZ twin correlations to DZ twin correlations. The computation of the heritability was conducted using R package OpenMx (v. 2.13) (Boker et al. 2011).
Genome-wide association studies
For GWAS, we expanded variants from ~720,000 to 11,315,231 by imputation using the Michigan Imputation Server (Das et al. 2016) with the reference genome HRCr1.1 (McCarthy et al. 2016). We filtered out markers with a low minor allele frequency (<5%) and removed markers that had P-values associated with Hardy–Weinberg disequilibrium <1e-6, genotype call rate <0.9, and imputation score <0.3. The remaining 4 234 798 variants on the 22 autosomes were used for GWAS for each trait (univariate GWAS [uvGWAS]), with genetic relatedness matrix (20 eigenvalues) calculated by principal components analysis, and sex and age used as covariates (Liu et al. 2018; Wu et al. 2018; Hwang et al. 2019). The genome-wide significance threshold was P = 5.0e-8 and, for suggestive associations, it was P = 1e-5 (International HapMap 2005; Pe’er et al. 2008).
We reasoned that there would be more statistical power to detect associations if we considered the liking and fattiness ratings from all potato chips simultaneously, especially because, as the results indicated, these measures were correlated (e.g., people with high liking ratings for the 5% corn oil chip also liked the 10% chip more). Thus, we conducted multivariate GWAS (mvGWAS) using the correlated ratings for all the potato chips. The covariates are the same as uvGWAS procedure; the computation was done using GEMMA (Zhou and Stephens 2012), and regional associational plots were created using LocusZoom (Pruim et al. 2010). For the mvGWAS, GEMMA adjusted for testing multiple phenotypes and applied a correction for multiple phenotypes (Fatumo et al. 2019). For the milk discrimination task, the trait was not heritable (see Results), so we did not conduct a GWAS.
Candidate gene analyses
We extracted variants from the candidate genes that were previously implicated in the sensory signaling of fat taste from either animal models (mouse and rat) or human studies: CD36 (Abumrad 2005; Gaillard et al. 2007; Sclafani et al. 2007a ; Laugerette et al. 2005; Keller et al. 2012; Pepino et al. 2012), GNAT3 (Sclafani et al. 2007b), GPR120 (Matsumura et al. 2007; Tsuzuki 2007; Cartoni et al. 2010), GPR40 (Cartoni et al. 2007, 2010; Matsumura et al. 2007), TRPM5 (Sclafani et al. 2007b; Liu et al. 2011), GPR41 and GPR43 (Brown et al. 2003), GPR84 (Wang et al. 2006), and KCNA2 (Gilbertson et al. 1998; Liu et al. 2005). In addition, we looked at genes for salivary enzymes (lipase, lysozyme, and amylase) and protein (lipocalin, mucin, and protein rich in proline) because these proteins change in response to dietary fat consumption (Feron and Poette 2013; Mounayar et al. 2014).
To extract the results of genotype–phenotype association for these candidate genes, we conducted analyses using 2 methods. In method 1, we identified the most significant variant within each candidate genes for each trait and extracted the relevant P-value and other test statistics. In method 2, we chose the most significant variant for traits of the potato chip with 5% corn oil (with no added FA) and examined all the sensory measures for the same variant; that is, we chose the 5% corn oil chip as the baseline from which to compare the other associations. These methods are complementary because method 1 detects associations that are specific to a particular concentration of triglyceride and FA combination, while method 2 detects common variants affecting the intensity and liking measures across the potato chip types. We also examined the effect of the variant rs1761667 within CD36 because it was previously associated with fat sensory perception in humans (Keller et al. 2012; Pepino et al. 2012; Mrizak et al. 2015; Sayed et al. 2015).
Gene expression in human taste tissue using the RNASeq method
To understand whether the genes identified by GWAS might be acting at the level of the receptors in taste tissue (as opposed to in the brain or in other tongue tissue, e.g., the filiform papillae), we compared the mRNA expression of these genes to those previously implicated in the peripheral aspects of fat taste perception (e.g., the candidate gene CD36) in human taste tissue. To do so, we collected fungiform papillae from subjects recruited for our previous study (Douglas et al. 2019) using published procedures (Spielman et al. 2010) and isolated the RNA following the manufacturer’s directions, processing the taste tissue with Quick-RNA MiniPrep R1054 (Zymo Research). We evaluated RNA quality expressed as an RNA integrity number (RIN) using the Agilent 2200 TapeStation system (Agilent Technologies). The 6 samples with sufficient RNA quality as determined by the Next-Generation Sequencing Core of the University of Pennsylvania (RIN >7; 5 males and 1 female) were used to perform library preparation and sequencing (100 bp single-end) on the HiSeq 4000 sequencer (Illumina) following the manufacturer’s sequencing protocols. We mapped reads to the reference genome (GRCh38.p10) after the raw sequence data in fastq format passed standard quality filters equipped in Trimmomatic (Bolger et al. 2014) and then normalized the counts using the R package Ballgown (Frazee et al. 2014). The expression level in reads per kilobase per million mapped reads (RPKM) of each gene for each sample was used to compare their expression level.
Pathway and gene set enrichment analysis
We reasoned that genes identified through GWAS may be partners with other genes that code for proteins in related sensory pathways. Thus, we conducted pathway analyses of the genes identified by uvGWAS and mvGWAS. Using the background of the genes from the database of Gene Ontology annotations (Thomas et al. 2003) and Reactome annotations (Fabregat et al. 2017; Fabregat et al. 2018), we used Fisher’s exact test to examine whether there was enrichment of these pathways versus all annotated human genes using GENEVESTIGATOR (Hruz et al. 2008).
Results
Participant characteristics
The twins (n = 398) were predominantly female (72%, n = 285; and 28% male, n = 113), middle aged (38.6 ± 16.7, mean ± SD), and members of MZ twin pairs (n = 360 twins, 90.4%). Most were of European descent (n = 331, 83.2%), but some participants were of African descent (n = 50, 12.6%). The remaining racial groups (e.g., Asian) were grouped into an “other” category for the analyses described below (n = 19, 4.8%). A total of 213 individual subjects were genotyped using the chip-based platform (MZ, n = 184; DZ, n = 29), and 176 MZ twins had their genotypes imputed.
Liking and intensity measures
Liking and fattiness ratings differed across potato chips with variable fat content
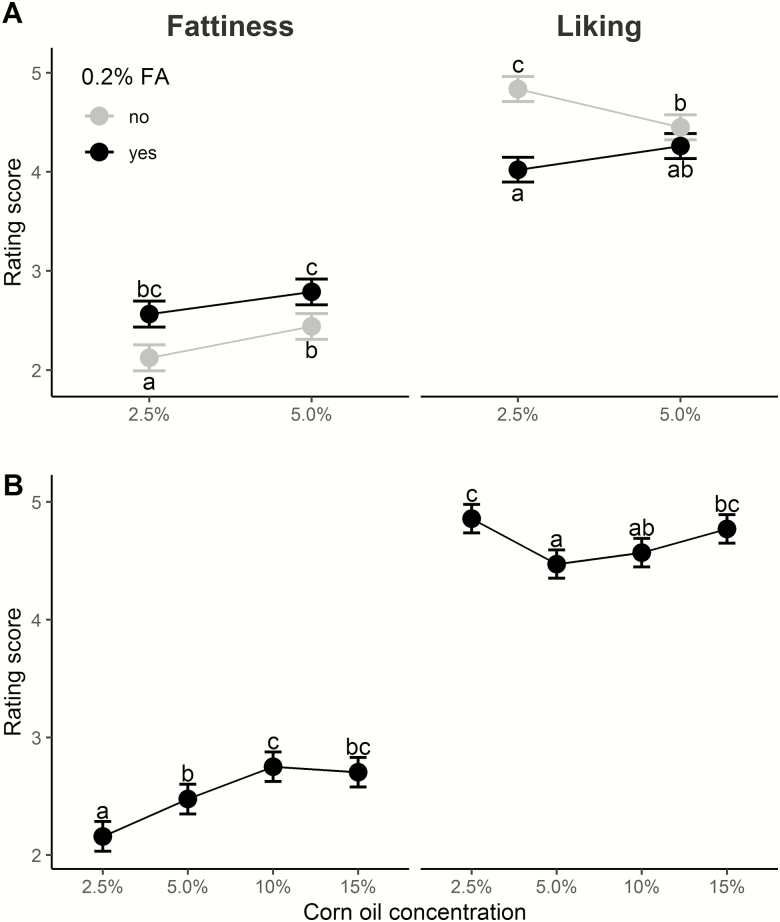
Overall, participants liked the potato chips and were able to accurately rate them for fattiness. Adding 0.2% hexadecanoic acid to the potato chips increased fattiness at both corn oil concentrations tested (Figure 1A). The effect of added hexadecanoic acid on liking was less straightforward: for the 5% corn oil chips, adding a 16:0 FA did not alter liking, while, for the 2.5% corn oil chips, adding the FA decreased liking (Figure 1A). For chips with no added FA, there was a mostly linear increase in ratings of fattiness as corn oil concentration increased, although a plateau was reached above 10% oil (Figure 1B). For liking, there was a J-shaped curve: participants liked the 2.5% and 15% corn oil potato chips best (Figure 1B). See Supplementary Figures 1 and 2 and Supplementary Table 1 for additional details.
Figure 1.
Corn oil and corn oil spiked with 0.2% hexadecanoic acid (FA) modify ratings of fattiness and liking of potato chips. (A) Potato chips with more corn oil plus added FA increased fattiness and decreased liking. (B) As corn oil concentration (2.5%, 5.0%, 10%, and 15% without added FA) increased, fattiness ratings increased linearly but liking changed in a J-curve: participants liked potato chips more with corn oil at the lowest and highest concentrations (2.5% and 15%). The points and bars show least square mean (LSM) and standard error of rating scores, and different letters (a, b, and c) indicate a significant LSM difference between groups.
Relationship between liking and fattiness relative to benchmarks
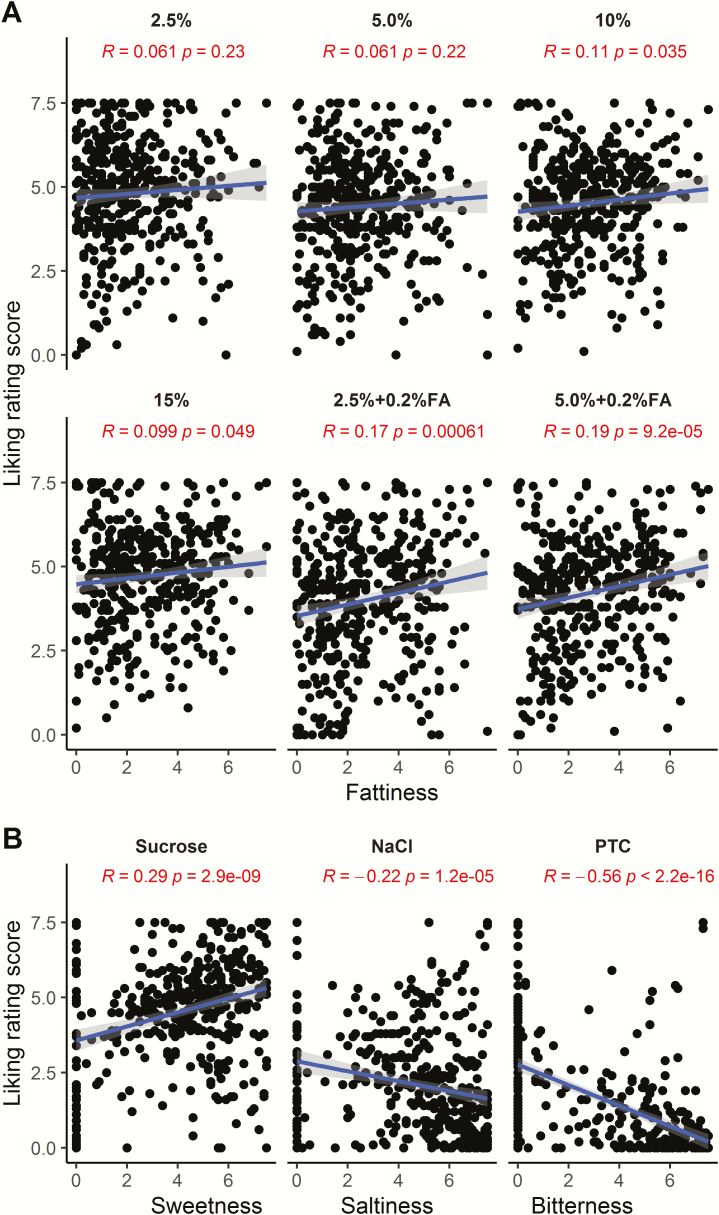
Within each type of potato chip, the ratings of liking and fattiness were only slightly or not at all related (Figure 2A). This relationship between liking and sensory quality differed from those for the prototypical taste solutions; for example, participants liked sucrose better if they rated it as sweeter (Figure 2B). For potato chip liking, even at the low corn oil concentrations, ratings were high on average, so there may have been a ceiling effect that restricts the range of responses and accounted for the low correlation between fattiness and liking.
Figure 2.
Pearson correlations between sensory measures indicate multiple mechanisms underlying dietary fat perception (n = 398). (A) No or weak correlations between ratings of liking and fattiness depending on the type of potato chip. (B) Strong correlations between liking and other taste ratings (sweetness, saltiness, and bitterness) for the standard taste solutions sucrose, NaCl, and PTC.
Reliability of liking and fattiness relative to benchmarks
The ratings of both fattiness and liking for the potato chips were reliable (r = 0.31–0.62, P < 0.05; Supplementary Figure 3), slightly lower than (but mostly similar to) those for the prototypical taste solutions (sucrose, NaCl, and PTC; r = 0.54–0.74, P < 0.0001, except for NaCl saltiness; Supplementary Figure 3).
Age, race, and sex effects on fattiness and liking
Men and women were similar in their ratings of all sensory stimuli (Supplementary Table 2). Race and age had significant effects on some sensory ratings (P < 0.01; Supplementary Table 2). Younger participants liked some of the potato chip types more than did the older participants (r = −0.17 to −0.14, P < 0.001; Supplementary Figure 4). People of European ancestry rated some potato chips as less fatty than did people of African ancestry (5.0% corn oil without added FA; P < 0.05, GLM analysis followed by post hoc Tukey HSD tests; Supplementary Figure 5). There were also race effects for the other sensory stimuli, for example, for the liking of sucrose and PTC. Supplementary Figure 5 summarizes all sensory results that differed by race.
Relationships of ratings across potato chip type
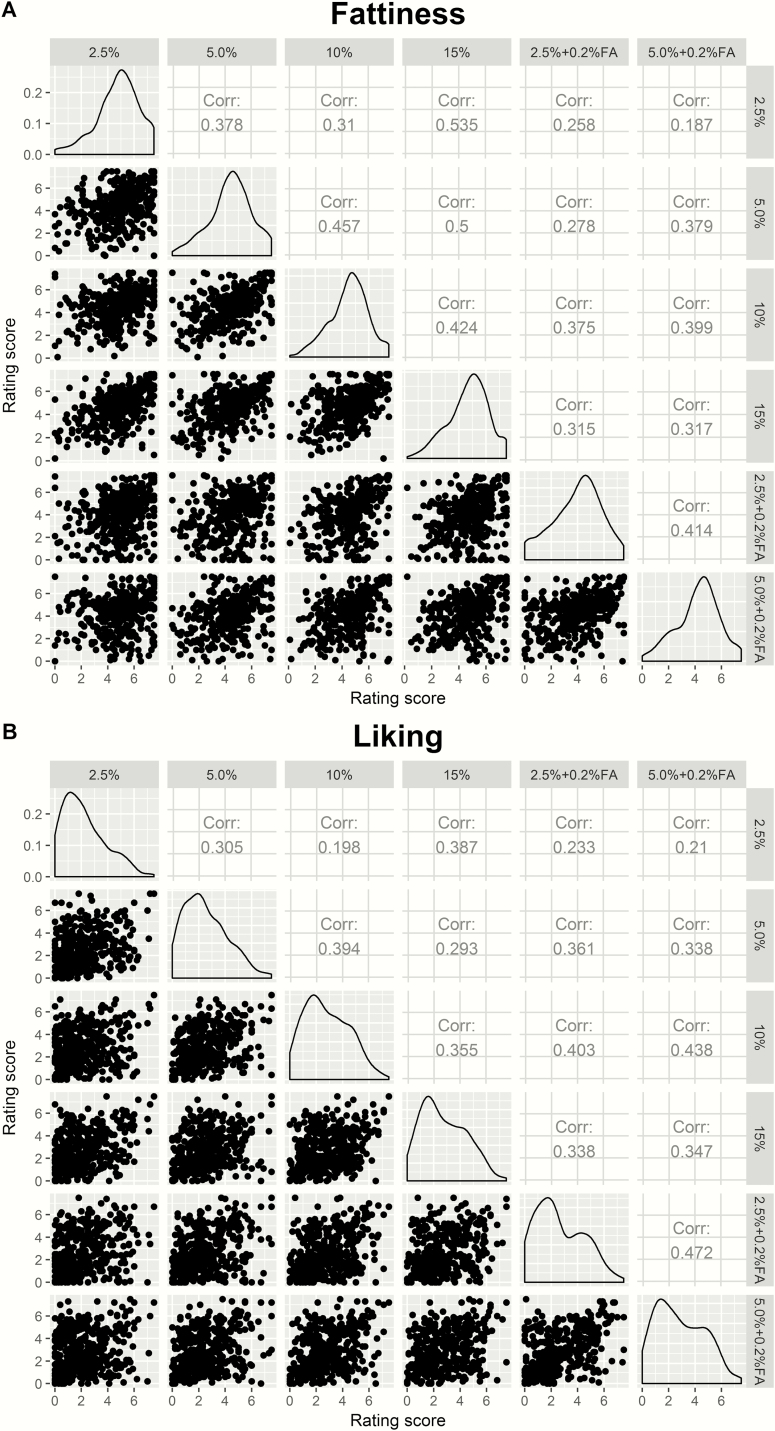
Each participant tasted and rated 6 potato chips, and there were correlations among each participant’s ratings of fattiness (Cronbach’s alpha = 0.75, 95% confidence boundaries = 0.72–0.79) and liking (Cronbach’s alpha = 0.77, 95% confidence boundaries = 0.74–0.81). Fattiness correlations tended to be higher among the chips without added FA than with the chips with added FA. A scatter matrix of pairwise correlations between potato chips types is shown in Figure 3.
Figure 3.
Strong and positive interrelated correlations of ratings of fattiness (A) and liking (B) across the 6 types of potato chips: scatter plots (lower left), density distributions (diagonal line), and correlations (upper right).
Discrimination of milk fattiness
On average, participants could discriminate the high- and low-fat milk samples (exact binomial test, one-tailed, P < 0.0001), but only slightly above chance (probability of success = 0.53; Supplementary Figure 6A). This ability to discriminate was only somewhat reliable when testing the same participant twice (retest correlation, r = 0.36; P > 0.05; Supplementary Figure 3). We had expected based on our pilot data collected in our sensory laboratory that about 30% of participants would perform this discrimination perfectly every time, with 10 out of 10 samples correctly identified, but our results showed that only 3% of subjects could do so.
Heritability
Between about 10% and 30% of the variation in potato chip liking arose from genetics (h2), but only accounted for about 5–15% for ratings of fattiness arose from genetics (Table 2). The heritability confidence intervals are large, which is due in part to the small number of DZ relative to MZ twins. For comparison, for the bitter compound PTC, the most heritable taste trait currently known, liking heritability was 53% and, for sucrose, which has a midrange heritability, it was 46%. The pattern of heritability for NaCl was similar to that for potato chips as rating of NaCl liking has more genetic variation than does rating of NaCl saltiness. We did not calculate heritability for the milk fat discrimination because there was no similarity in milk discrimination scores between the twins (Supplementary Figure 6B).
Table 2.
Heritability (h2) of fat sensory traits, with NaCl, sucrose, and PTC as a benchmarks (n = 199 twin pairs)
| Stimulus | h 2 | CI |
|---|---|---|
| Liking (chips) | ||
| 2.5% corn oil | 0.21* | 0.07–0.34 |
| 5.0% corn oil | 0.10 | 0.00–0.24 |
| 10% corn oil | 0.10 | 0.00–0.24 |
| 5% corn oil | 0.29* | 0.15–0.41 |
| 2.5% corn oil with 0.2% hexadecanoic acid | 0.10 | 0.00–0.24 |
| 5.0% corn oil with 0.2% hexadecanoic acid | 0.10 | 0.00–0.24 |
| Fattiness | ||
| 2.5% corn oil | 0.05 | 0.00–0.20 |
| 5.0% corn oil | 0.11 | 0.00–0.25 |
| 10% corn oil | 0.12 | 0.00–0.27 |
| 15% corn oil | 0.07 | 0.00–0.22 |
| 2.5% corn oil with 0.2% hexadecanoic acid | 0.03 | 0.00–0.17 |
| 5.0% corn oil with 0.2% hexadecanoic acid | 0.15 | 0.00–0.29 |
| Other solutions | ||
| Sucrose sweetness | 0.11 | 0.00–0.25 |
| Sucrose liking | 0.46* | 0.33–0.56 |
| NaCl saltiness | 0.19* | 0.05–0.32 |
| NaCl liking | 0.38* | 0.25–0.49 |
| PTC bitterness | 0.49* | 0.38–0.59 |
| PTC liking | 0.53* | 0.42–0.62 |
CI, confidence interval.
*Different from zero.
Genome-wide association
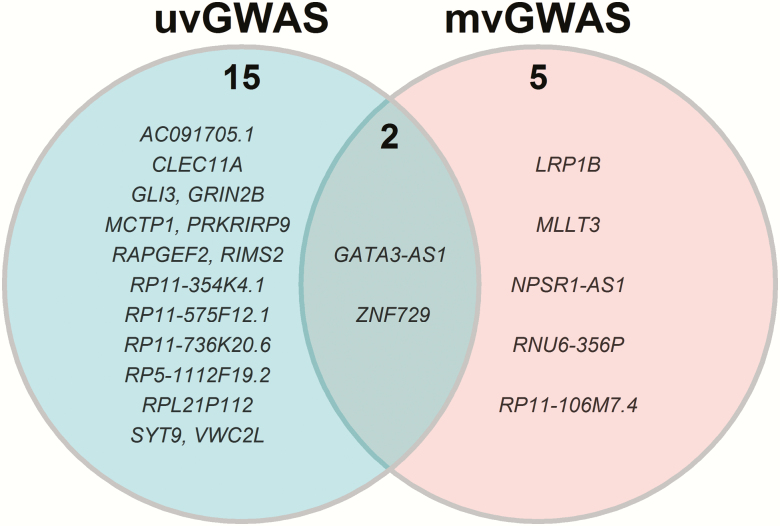
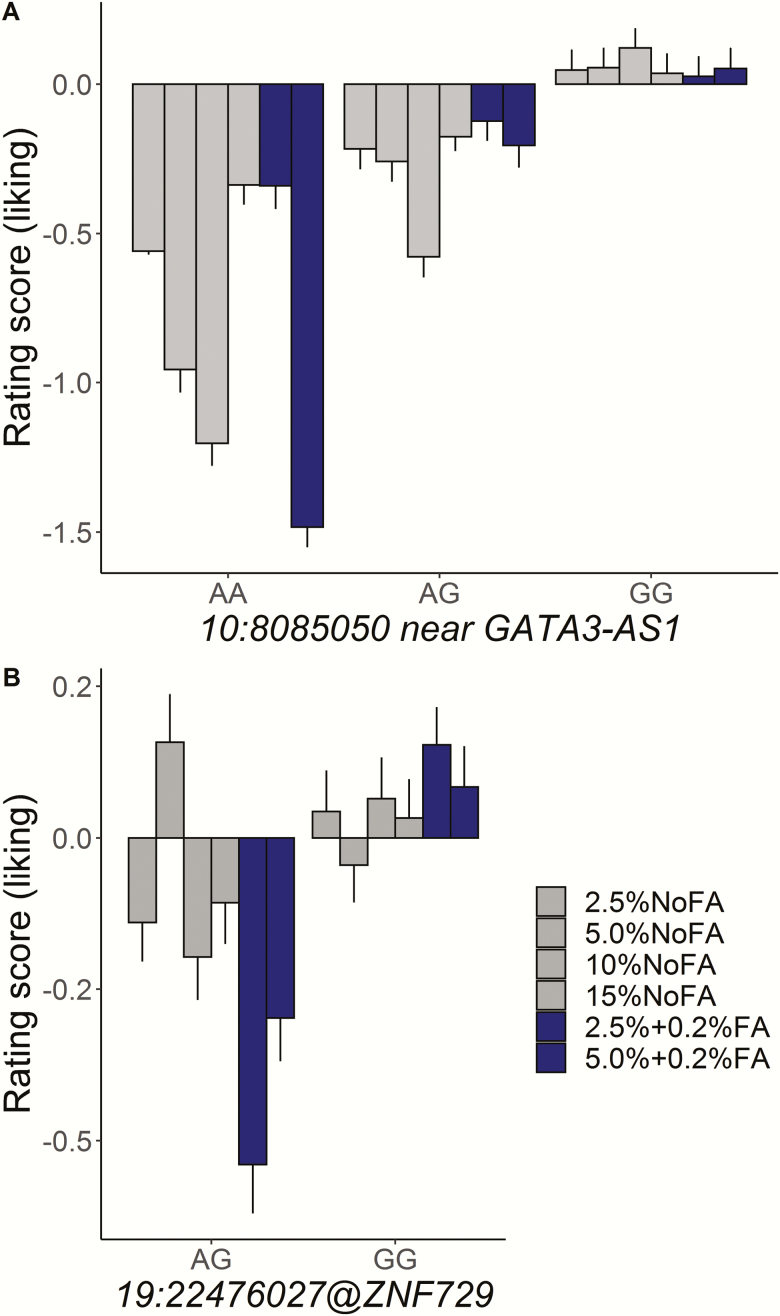
No associations met the commonly accepted genome-wide significance threshold, but we did identify suggestive variants using the univariate and multivariate methods. uvGWAS identified 9 associations for fattiness and 8 for liking (Table 3). All these associations were specific for potato chip type. The mvGWAS detected 2 variants for chip fattiness and 5 variants for chip liking (Table 4). We reasoned that associations detected with both uvGWAS and mvGWAS would be most valid. Of the 7 genotype associations detected by mvGWAS, 2 (GATA3-AS1 and ZNF729) were also detected by uvGWAS (Figure 4): twins with the G allele of rs263429 (10:8085050, near GATA3-AS1) reported more liking for the potato chips than did twins with the other allele and the same was true for the G allele of rs8103990 (19:22476027, within ZNF729; mvGWAS, P < 1 × 10–5; Table 4). We show the allelic effects for these 2 variants in Figure 5. The effects of the novel variants were larger than those for CD36, the candidate gene previously associated with fat perception (Supplementary Figure 7).
Table 3.
Suggestive associations for ratings of potato chip fattiness and liking identified by uvGWAS
| Stimuli | CHR | SNP (CHR:BP) | ALT | REF | MAF | Beta | SE | P | Gene | HIT_TYPE | SNP_TYPE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fattiness | |||||||||||
| 5.0% NoFA | 4 | 4:45361270 | G | C | 0.12 | 0.92 | 0.20 | 6.14E-06 | PRKRIRP9 | Nearest | Imputed |
| 5.0% NoFA | 7 | 7:42032565 | T | C | 0.26 | 0.62 | 0.14 | 8.25E-06 | GLI3 | Within | Genotyped |
| 5.0% NoFA | 8 | 8:105135473 | A | T | 0.15 | 0.86 | 0.18 | 1.92E-06 | RIMS2 | Within | Imputed |
| 5.0% NoFA | 11 | 11:7476601 | A | G | 0.21 | 0.74 | 0.15 | 9.51E-07 | SYT9 | Within | Imputed |
| 10% NoFA | 7 | 7:25514020 | C | G | 0.18 | 0.74 | 0.16 | 4.91E-06 | AC091705.1 | Nearest | Imputed |
| 15% NoFA | 5 | 5:94138997 | C | T | 0.17 | 0.76 | 0.17 | 4.37E-06 | MCTP1 | Within | Genotyped |
| 15% NoFA | 6 | 6:77345798 | C | T | 0.10 | 0.87 | 0.20 | 9.64E-06 | RP11-354K4.1 | Nearest | Imputed |
| 15% NoFA | 11 | 11:86707909 | G | C | 0.09 | 0.88 | 0.20 | 9.45E-06 | RP11-736K20.6 | Within | Imputed |
| 15% NoFA | 19 | 19:51226244 | T | C | 0.38 | 0.59 | 0.13 | 9.19E-06 | CLEC11A | Nearest | Imputed |
| Liking | |||||||||||
| 5.0% NoFA | 2 | 2:215358759 | C | T | 0.06 | −1.12 | 0.25 | 8.47E-06 | VWC2L | Within | Imputed |
| 5.0% NoFA | 20 | 20:50443885 | T | C | 0.25 | 0.63 | 0.13 | 1.84E-06 | RP5-1112F19.2 | Nearest | Imputed |
| 10% NoFA | 10 | 10:8085050 | A | G | 0.08 | −0.92 | 0.20 | 5.27E-06 | GATA3-AS1 | Nearest | Imputed |
| 10% NoFA | 12 | 12:127482374 | C | G | 0.07 | −0.95 | 0.21 | 9.08E-06 | RP11-575F12.1 | Within | Imputed |
| 2.5% with 0.2% FA | 4 | 4:160267304 | A | G | 0.06 | −1.32 | 0.30 | 8.02E-06 | RAPGEF2 | Within | Imputed |
| 2.5% with 0.2% FA | 12 | 12:13948270 | A | G | 0.10 | −1.13 | 0.24 | 3.58E-06 | GRIN2B | Within | Imputed |
| 2.5% with 0.2% FA | 19 | 19:22476027 | A | G | 0.11 | −1.07 | 0.23 | 4.92E-06 | ZNF729 | Within | Imputed |
| 5.0% with 0.2% FA | 13 | 13:95498608 | T | C | 0.09 | −0.98 | 0.22 | 5.94E-06 | RPL21P112 | Nearest | Imputed |
Two genes (PRKRIRP9 and RP11-270L13.1) on chr 4 for ratings of potato chip fattiness reached genome-wide suggestive threshold (1e-5), but only one with a relatively lower P-value is reported in this table. Variants for potato chip liking shown in boldface were also detected by the mvGWAS across all corn oil concentrations; see Table 4.
ALT, alternative allele; BP, base pair; CHR, chromosome; MAF, minor allele frequency; NoFA, no added hexadecanoic acid; REF, reference allele; SNP, single-nucleotide polymorphism; SNP_TYPE, individual’s genotype was genotyped or imputed.
Table 4.
Suggestive genes for ratings of potato chip fattiness and liking identified by mvGWAS
| Trait | CHR | SNP (CHR:BP) | ALT | REF | MAF | P | Gene | HIT_TYPE | SNP_TYPE |
|---|---|---|---|---|---|---|---|---|---|
| Fattiness | 2 | 2:141755773 | A | T | 0.32 | 2.96E-06 | LRP1B | Within | Imputed |
| 10 | 10:116662107 | C | T | 0.05 | 8.97E-06 | RP11-106M7.4 | Nearest | Imputed | |
| Liking | 7 | 7:34676953 | T | A | 0.06 | 3.05E-07 | NPSR1-AS1 | Within | Imputed |
| 8 | 8:40998854 | A | T | 0.04 | 5.14E-07 | RNU6-356P | Nearest | Imputed | |
| 9 | 9:20636973 | T | C | 0.48 | 1.15E-06 | MLLT3 | Nearest | Genotyped | |
| 10 | 10:8085050 | G | A | 0.08 | 8.19E-07 | GATA3-AS1 | Nearest | Imputed | |
| 19 | 19:22476027 | A | G | 0.11 | 4.71E-06 | ZNF729 | Within | Imputed |
Two genes on chr 2 (LINC00486 and LRP1B) and two genes on chr 10 (FAM160B1 and BP11-106M7.4) for potato chip fattiness, and 2 genes on chr 7 (EEPD1 and NPSR1-AS1) and 2 genes on chr 8 (RNU6-356P and SULf1) for potato chip liking reached genome-wide suggestive threshold (1e-5), but only one gene with a relatively lower P-value on each chromosome is reported in this table. Variants for potato chip liking shown in boldface were also detected by the uvGWAS across all corn oil concentrations. See Table 3 for abbreviations and other details.
Figure 4.
Venn diagram comparing loci identified by uvGWAS and mvGWAS (see Methods for details). Two variants were detected by both methods: 10:8085050 near the gene GATA3-AS1 and 19:22476027 within ZNF729.
Figure 5.
Allele effect of variants 10:8085050 near gene GATA3-AS1 (A) and 19:22476027 within the gene ZNF729 (B) on ratings of liking across types of potato chips. For both variants, participants with G allele rated higher liking for all potato chip types than did those with other allele. The standard residual scores for liking were calculated in the general linear model with covariates of sex, age, and 20 eigenvalues.
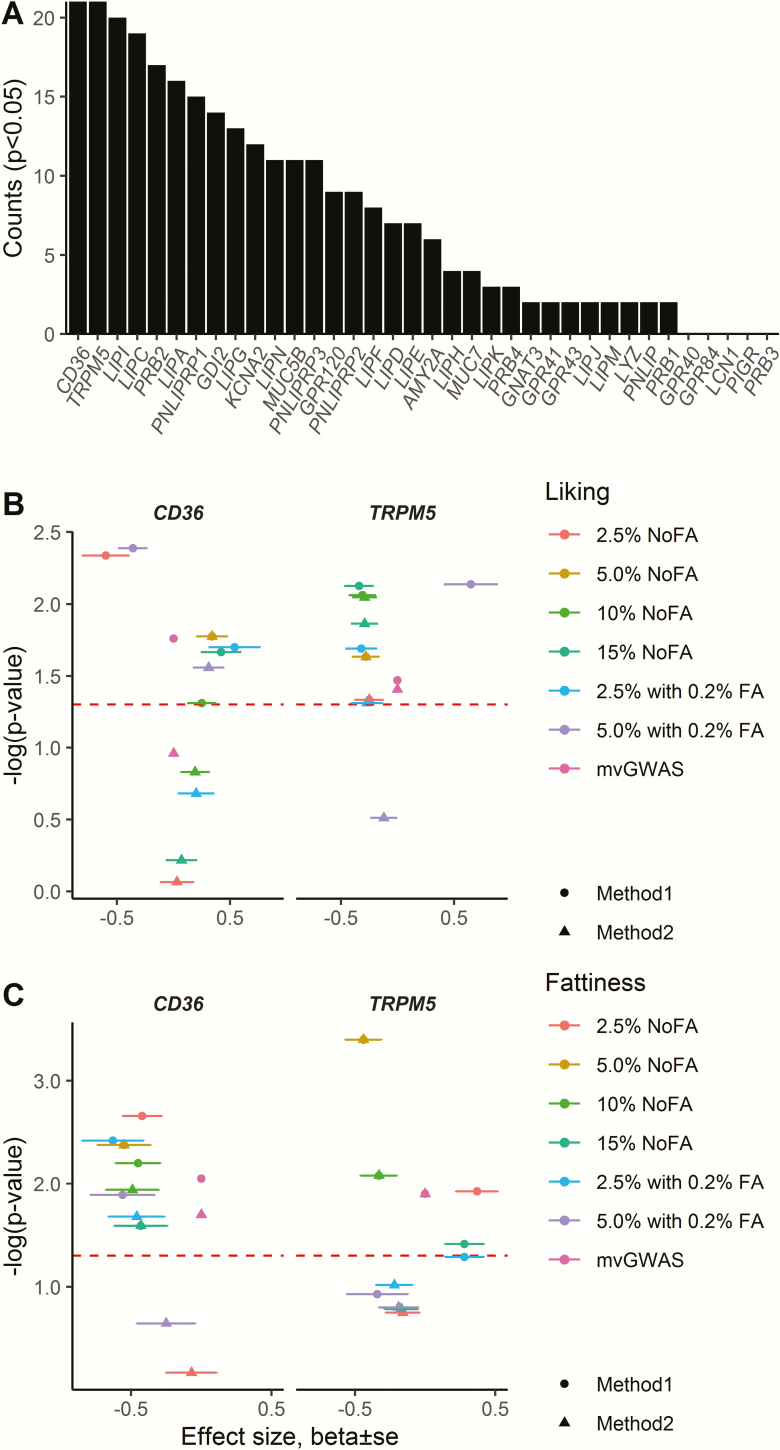
Candidate genes
None of the candidate genes consistently met a genome-wide statistical threshold, but some candidate genes were more often associated with potato chip fattiness or liking than others at a nominal significance threshold (P < 0.05; Figure 6A). The most notable results were significant variants within CD36 and TRPM5 associated with potato chip liking and fattiness (Figure 6B,C; Supplementary Figure 8, Supplementary Tables 3 and 4). For CD36, the variant rs1761667 (which was associated with fat perception in previous studies) did not pass the quality-control filters, but we examined a nearby variant, rs1722501, that was in nearly perfect linkage disequilibrium (R2 > 0.99) with rs1761667. However, participants did not differ in ratings of potato chip fattiness or liking for this proxy marker (Supplementary Table 5), although there were many associations for other variants within CD36 as noted above (see Figure 6).
Figure 6.
Candidate gene effect on fat perception for potato chips. (A) Total counts of nominal P < 0.05 out of 28 tests for each candidate gene for the 2 methods of candidate gene analysis (methods 1 and 2; see Materials and Methods) in the outputs from uvGWAS and mvGWAS. (B, C) Associations of top variants within candidate genes CD36 and TRPM5 with ratings of liking (B) and fattiness (C) for each type of potato chip. The x axis shows effect size (β ± SE), obtained from uvGWAS, and y axes show –log(P-value), obtained from uvGWAS and mvGWAS, for the top variants within CD36 and TRPM5 (no β ± standard error [SE] data were available from mvGWAS; i.e., β ± SE = 0 is not true). Red dashed lines indicate P = 0.05; the points above this line indicate a nominal significant effect on the trait. For other details of the data, see Supplementary Tables 3 and 4.
Gene expression, pathway, and gene enrichment analysis
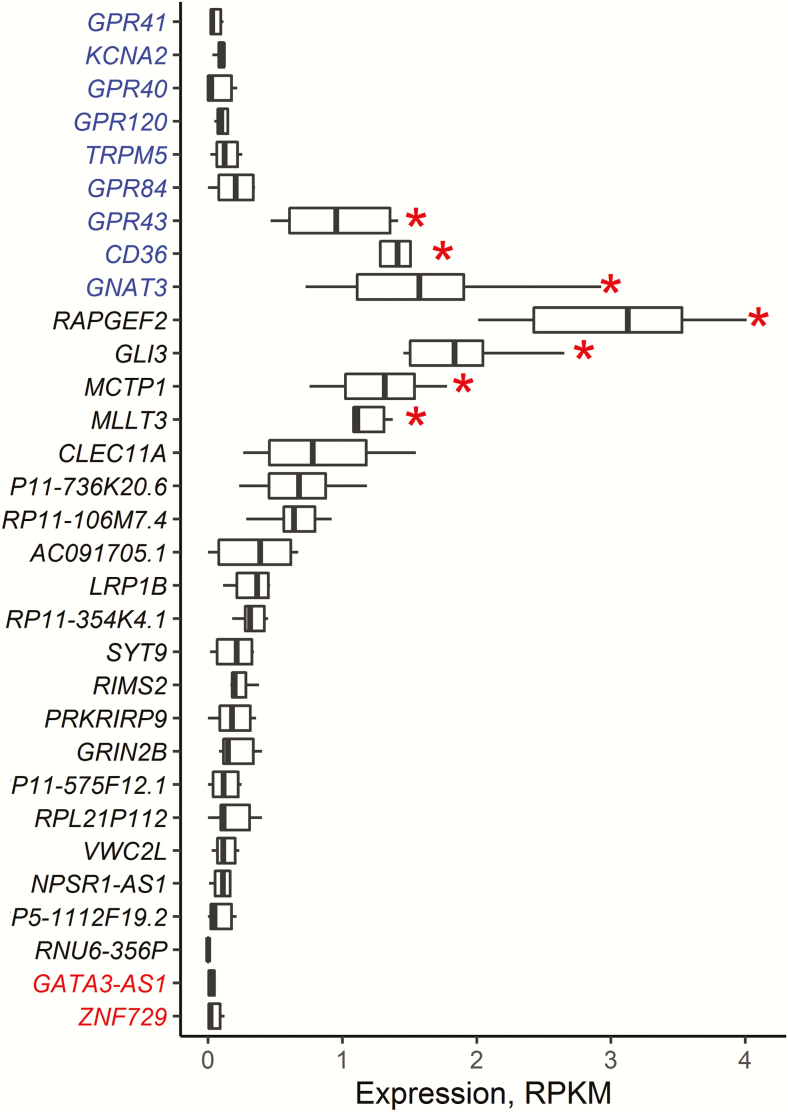
We reasoned that the expression of fat candidate genes (those that have a proposed role in peripheral fat or FA signaling) would be a benchmark to compare the taste-tissue expression of the novel genes identified from the GWAS results. Compared with receptor and other signaling candidate genes (GPR40, GPR41, GRP43, GPR84, GPR120, TRPM5, CD36, KCNA2, and GNAT3), the novel genes have relatively higher expression levels in fungiform papillae, especially for RAPGEF2, GLI3, MCTP1, and MLLT3 (Figure 7). ZNF729 and GATA3-AS1 had a similar expression abundance as the candidate genes GPR40, GPR41, GPR84, GPR120, KCNA2, and TRPM5 but much lower than the candidate genes GRP43, CD36, and GNAT3. The presence of many of the novel genes in taste tissue is consistent with a role in peripheral perception, but some candidate genes had a very low abundance. This subset of low-abundance novel genes may be nearly undetectable in the taste tissue sampled because only a few of the relevant cells may have been present in the tissue sample or because the genes may act at different times (e.g., early development) or in different tissues (e.g., the filiform papillae or the brain).
Figure 7.
Box plots of taste tissue expression abundance of genes near the peak statistical associations from the GWAS (novel hits) and for candidate genes (shown in blue) known from prior studies to contribute to fat perception. Two genes, ZNF729 and GATA3-AS1 (shown in red), were commonly detected by both uvGWAS and mvGWAS in the present study. RNU6-356P had no expression in any sample. Outliers are not shown. Red asterisks indicate genes with statistically higher expression level compared with other genes in taste tissue (P < 0.05/351 = 0.000142, Bonferroni corrections for multiple tests).
We conducted pathway analysis to understand the function of as many of the novel genes identified as possible. In the GENEVESTIGATOR analysis, 21 of the 22 associated genes identified by GWAS (RP11-575F12.1 is not found in the database) were tested against the 74,727 background genes. Three gene sets were enriched using the associated genes as bait (P < 0.001, Fisher’s exact test; Supplementary Figure 9; Supplementary Table 6) from the Gene Ontology categories synapse GO:0045202, cell–cell signaling GO:0007267, and positive regulation of neurogenesis GO:0050769. Overall, these results point to a role of these genes and their protein products in sensory signaling and perhaps regulation of sensory cell types.
Discussion
Dietary fat is added to food to increase its flavor and palatability, but whether fat is sensed by chemical cues (e.g., from FA), textural cues, or both is contentious. The results from this study support the hypothesis that fattiness has both a positive effect (more is better) and a negative effect (more is worse) and that other sensory cues drive the positive fattiness, whereas FA contribute to the negative fattiness. Texture or chemical cues from triglycerides may provide this positive effect because previous studies show that texture is not the cue associated with the ability to detect FA (Laugerette et al. 2005; Chale-Rush et al. 2007a, 2007b; Mattes 2009; Cartoni et al. 2010; Dramane et al. 2012; Voigt et al. 2014). Overall, these data are consistent with previous observations that FA provide a chemical cue for fattiness and reduce detection thresholds when added to oil (Heinze et al. 2017) but that this component of fattiness is not desirable (Running et al. 2017). Parenthetically, subjects did not wear nose clips for the potato chip testing; thus, the chemical cue for fattiness could either be from the taste or smell of the FA. When hexadecanoic acid (a saturated 16-carbon FA) was added to the potato chip lowest in fat, it was rated as fattier but was less liked than a potato chip with a comparable amount of fat but without the added FA. Thus, presumably, taking a broader view and generalizing, this result suggests that increasing “fattiness” by adding FA to foods would not make them better liked and raises the possibility that recently discovered antagonists to the FA receptors (Milligan et al. 2017) might improve fat flavor.
In addition to studying the relationship between fattiness and liking, we also attempted to study fat discrimination, asking participants to choose the fattier milk solution from a pair of high- and low-fat samples. This task was difficult for the participants, and almost no one correctly identified the high-fat sample 10 times out of the 10 trials. This result came as a surprise because our preliminary testing suggested that this task was easy; however, most preliminary testing was conducted with commercially available low- and high-fat milk samples and in a quiet sensory laboratory, making discrimination easier. The prepared milk samples used for testing here were the same in all aspects except for the amount of dietary fat added and, for many people, the oral cues alone (as opposed to visual or olfactory cues) are insufficient to discriminate low-fat from high-fat samples. One additional concern was the effect of transportation on the milk which was prepared and then driven by truck several hundred miles to the test location—conceivably, vibration may have caused coalescence of the fat globules that altered the ability to discriminate between samples.
The main focus of this study was to examine whether person-to-person differences in the liking or perception of fattiness are due in part to individual genetic variation. To establish the heritability of a trait, it is essential to have a reliable measurement, that is, a trait that can be measured reproducibly; accordingly, demonstrating that the measures used were reliable was an essential precondition for the heritability calculations. We learned from the reliability and heritability analyses that liking for this solid food matrix, potato chips, with differing fat concentrations was more similar among genetically identical (MZ) twins than among nonidentical (DZ) twins. Ratings of fattiness were also heritable but less so, aligning with results from our studies of other taste modalities, which, for example, demonstrated that liking for a concentrated salt solution is more heritable than are salty intensity ratings (Knaapila et al. 2012). Our results contradict a prior twin study on the effect of diet on FA perception, which found few or no genetic effects (Costanzo et al. 2018); however, these 2 studies differed in methods, as did the number of twins investigated, 88 (Costanzo et al. 2018) versus 398 here.
Thus, despite the logistical challenges posed by measuring percepts from dietary fat, there is evidence for a genetic determinant on par with other traits that have been studied using GWAS methods (Clarke et al. 2017). Building on the heritability analysis, we also performed 2 types of GWAS, which are agnostic to prior information about which genes and variants might be previously known or suspected to contribute to the perception of dietary fat. This part of the study was underpowered and returned no results that met the classic statistical threshold for GWAS results but did provide, in tandem with the bioinformatic analysis, clues about which genes and pathways might be worth pursuing in future work, specifically cell-to-cell communication and perhaps cell type. For the lead genetic variants, those with the largest effects, the effect of the major allele was to reduce liking, suggesting that they may increase the ability to taste the “bad” or negative aspects of the fat or FA.
Of particular interest is the association between fat liking and variants in a transcription factor (Gli3) that contribute to the development of taste cells (Ermilov et al. 2016; Qin et al. 2018). We could detect this gene in the whole-tissue transcriptome of human fungiform papillae, making it an attractive candidate gene for further study. However, the study of bulk tissue is a crude approach and single-cell studies from all regions of the oral cavity would be a step forward to understand the cell type expressing this and other candidate genes. This experimental step is increasingly feasible as methods of isolating single cells from human tissue improve, although the most complete experimental paradigm would also include the sensory pathways, including brain regions that process the sensory properties of dietary fat information (Grabenhorst and Rolls 2014).
The results of the candidate gene analyses were more compelling in the sense that, although none of the results were individually very striking, multiple methods of analysis indicated a role for CD36 and TRPM5 in the perception of dietary fat in the current study. As an aside, we did not see associations with the proxy marker we used to try to replicate the previous studies exactly (Keller et al. 2012; Pepino et al. 2012; Mrizak et al. 2015; Sayed et al. 2015), but CD36 is a large gene with many potentially functional variants and, therefore, a fine-mapping study in multiple populations is warranted because there may be multiple variants that cause a spectrum of effects that differ by ancestral population, for example, (Gurdasani et al. 2019). We failed to observe associations for other candidate genes previously reported, which could be due to differences in the FA or triglycerides tested, small effect sizes relative to the sample size, methodological differences or the absence of influential genetic variants in our study population.
We speculate that sensory nutrition and taste perception offer a way to reduce nutrition-related human diseases by studying the nuanced and often misunderstood relationship between liking and intake (Hayes 2020). GWAS allows us to screen and identify common genetic variants associated with fat consumption (Tanaka et al. 2013) and, our findings, combined with future functional genomic analyses, especially single-cell profiling, will delineate the causal genetic variants and biological mechanisms underlying the observed statistical associations (Gallagher and Chen-Plotkin 2018). These approaches can help us to understand personalized nutrition in the realm of fat perception, liking, and intake.
Supplementary material
Supplementary material can be found at Chemical Senses online.
Supplemental Figure 1. Changes in ratings of fattiness and liking by corn oil concentration across the six types of potato chips. FA=fatty acid (hexadecanoic acid). For other details, see Figure 1.
Supplemental Figure 2. Violin plots for ratings of the sensory traits. The violin area shows the estimated density of each rating score point. The dots and bars show means and SDs.
Supplemental Figure 3. Pearson correlations between test and retest of each rating (n = 50).
Supplemental Figure 4. Pearson correlations between age and sensory measures for potato chips and other taste stimuli. Young participants were more sensitive to taste stimuli than were older participants.
Supplemental Figure 5. Least square mean (LSM) and standard error of sensory measures by race. EA=European Americans, AA=African Americans, Oth=others (Asian, Hispanic, Native American, mixed). Different letters (a, b) show a significant LSM difference.
Supplemental Figure 6. Most participants had difficultly discriminating milk fat content, with near chance levels overall. (A) Histogram of milk fat discrimination scores. The dashed white line shows probability of success, which is near the chance level of 5, but it is significantly different from the chance level, p < 0.001. (B) No significant correlations were observed between twin 1 and twin 2 for milk discrimination for either DZ or MZ twins; thus, no heritability for milk fat discrimination scores was calculated.
Supplemental Figure 7. Regional associational plots, based on mvGWAS results, for single-nucleotide polymorphisms in linkage disequilibrium (r2) with the peak variants 10:8085050 near the gene GATA3-AS1 (A) and 19:22476027 within the gene ZNF729 (B) for ratings of liking, and for the fat perception candidate gene CD36 for ratings of liking (C) and fattiness (D) for potato chips. The highlighted chromosome regions show the target genes.
Supplemental Figure 8. Associations of top variants within each candidate gene with ratings of liking (A) and fattiness (B) for each type of potato chip. For details see Figure 6.
Supplemental Figure 9. Gene set enrichment analyses. Venn diagram visualizes overlapping genes among the top three gene sets and the target genes (21 out of 22 GWAS hits; RP11-575F12.1 is not found the database). All genes (n = 74,727 total genes) were selected from Reactome annotations and Gene Ontology annotations as background collection. The top three gene sets identified are synapse GO:0045202, cell-cell signaling GO:0007267, and positive regulation of neurogenesis GO:0050769 (see Supplemental Table 6).
Supplemental Table 1. Summary statistics for linear mixed model analyses
FA, fatty acid; ICC, intraclass correlation. Boldface indicates the test statistic meets a significance threshold of p < 0.01.
Supplemental Table 2. The effect of sex, race, and age on sensory measures for potato chips, taste stimuli, and milk discrimination
PTC, phenylthiocarbamide. Highlighting indicates suggestive effects with a p-value < 0.05.
Supplemental Table 3. The effect of the top variant within each candidate gene on ratings of potato chip fattiness and liking. FA, fatty acid; mvGWAS, multivariate genome-wide association study. Highlighting indicates suggestive effects with a p-value < 0.05. *For GPR41 and GPR84, no variant within the genes was available from the association data, so we expanded the region to 500 bp up- and downstream for each site when extracting the variant to examine for association. For other details see Supplementary Tables 5 and 6.
Supplemental Table 4. The top variant within each candidate gene with effects on ratings of potato chips with 5% corn oil (without added fatty acid) had effects on fattiness and liking for other types of potato chips. For details, see Supplemental Table 3.
Supplemental Table 5. The variant rs1722501 (chr7:80244694) as proxy for rs1761667 within CD36 has no significant effect on ratings of potato chip fattiness and liking.
Supplemental Table 6. Gene set enrichment analysis
Acknowledgments
We thank the following people for assistance with data collection, listed in alphabetical order: Charles J. Arayata, Nuala Bobowski, Fujiko Duke, Hillary Ellis, Brad Fesi, Nicole Greenbaum, Aurora Hannikainen, Desmond Johnson, Katherine Leung, Durpri Lin, Alex Mangroo, Corrine Mansfield, Michael Marquis, Elliott McDowell, Tiffany Murray, Lauren Shaw, Lindsey Snyder, Molly Spencer, Amber Suk, Alyssa Treff, and Casey Trimmer. We thank the twins for their participation and the administration of TwinsDays, including Sandy Miller and Janine Bregitzer for their assistance during data collection. We thank two anonymous reviewers for their constructive comments, which improved the quality of this manuscript.
Funding
This work was supported in part by PepsiCo R&D, Diageo and Monell Institutional Funds. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of PepsiCo, Inc., or Diageo. Some genotyping was performed at the Monell DNA and RNA Analysis Core, which is supported, in part, by funding from the National Institutes of Health–National Institute on Deafness and Other Communication Disorders Core Grant 1P30DC011735 using an instrument purchased using National Institutes of Health funds (S10 OD018125).
Conflict of interest
This study was partially funded by PepsiCo, Inc. PepsiCo, Inc., had no role in the design, execution, interpretation, or writing of the study. L.F. discloses her conflict of interest as she is an employee of PepsiCo, Inc., and claims no other conflicts of interest. This study was also partially funded by Diageo. Diageo had no role in the design, execution, interpretation, or writing of the study. A.S. and D.B. disclose their conflict of interest as they are employees of Diageo and claim no other conflicts of interest.
Author Contributions
D.R.R. designed the study. L.C., F.G., I.M., and P.J. collected data. P.W., P.A.S.B., N.E.R., L.S., D.B., and J.E.H. contributed to data interpretation. L.F. contributed reagents and also contributed to data interpretation, and A.S. and A.M. assisted in stimuli preparation and data collection. All authors were involved in drafting the article or revising it for intellectual content and have read and approved the final version of the manuscript.
References
- Abumrad NA. 2005. CD36 may determine our desire for dietary fats. J Clin Invest. 115(11):2965–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke AJ, Shehan CV, Hayes JE. 2016. Type of milk typically consumed, and stated preference, but not health consciousness affect revealed preferences for fat in milk. Food Qual Prefer. 49:92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Spies J, Estabrook R, Kenny S, Bates T, et al. 2011. OpenMx: an open source extended structural equation modeling framework. Psychometrika. 76(2):306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. 2003. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 278(13):11312–11319. [DOI] [PubMed] [Google Scholar]
- Card MM. 2013. America, you are digging your grave with your spoon–should the FDA tell you that on food labels? Food Drug Law J. 68(3):309–327, ii. [PubMed] [Google Scholar]
- Cartoni C, Yasumatsu K, le Coutre J, Ninomiya Y, Damak S. 2007. Diminished taste responses to fatty acids and oils in GPR40 knockout mice. 5th International Symposium on Molecular and Neural Mechanisms of Taste and Olfactory Perception. 2007. Nov 2; Fukuoka (Japan). [Google Scholar]
- Cartoni C, Yasumatsu K, Ohkuri T, Shigemura N, Yoshida R, Godinot N, le Coutre J, Ninomiya Y, Damak S. 2010. Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci. 30(25):8376–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalé-Rush A, Burgess JR, Mattes RD. 2007a. Evidence for human orosensory (taste?) sensitivity to free fatty acids. Chem Senses. 32(5):423–431. [DOI] [PubMed] [Google Scholar]
- Chalé-Rush A, Burgess JR, Mattes RD. 2007b. Multiple routes of chemosensitivity to free fatty acids in humans. Am J Physiol Gastrointest Liver Physiol. 292(5):G1206–G1212. [DOI] [PubMed] [Google Scholar]
- Clark CC, Lawless HT. 1994. Limiting response alternatives in time-intensity scaling: an examination of the halo-dumping effect. Chem Senses. 19(6):583–594. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, et al. 2017. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry. 22(10):1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A, Nowson C, Orellana L, Bolhuis D, Duesing K, Keast R. 2018. Effect of dietary fat intake and genetics on fat taste sensitivity: a co-twin randomized controlled trial. Am J Clin Nutr. 107(5):683–694. [DOI] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, et al. 2016. Next-generation genotype imputation service and methods. Nat Genet. 48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JE, Lin C, Mansfield CJ, Arayata CJ, Cowart BJ, Spielman AI, Adappa ND, Palmer JN, Cohen NA, Reed DR. 2019. Tissue-dependent expression of bitter receptor TAS2R38 mRNA. Chem Senses. 44(1):33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramane G, Abdoul-Azize S, Hichami A, Vögtle T, Akpona S, Chouabe C, Sadou H, Nieswandt B, Besnard P, Khan NA. 2012. STIM1 regulates calcium signaling in taste bud cells and preference for fat in mice. J Clin Invest. 122(6):2267–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A. 1992. Sensory properties of fats and fat replacements. Nutr Rev. 50(4 (Pt 2)):17–20. [DOI] [PubMed] [Google Scholar]
- Ermilov AN, Kumari A, Li L, Joiner AM, Grachtchouk MA, Allen BL, Dlugosz AA, Mistretta CM. 2016. Maintenance of taste organs is strictly dependent on epithelial hedgehog/GLI signaling. PLoS Genet. 12(11):e1006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. 2018. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46(D1):D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D’Eustachio P, Stein L, Hermjakob H. 2017. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinf. 18(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatumo S, Carstensen T, Nashiru O, Gurdasani D, Sandhu M, Kaleebu P. 2019. Complimentary methods for multivariate genome-wide association study identify new susceptibility genes for blood cell traits. Front Genet. 10:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feron G, Poette J. 2013. In-mouth mechanism leading to the perception of fat in humans: from detection to preferences. The particular role of saliva. OCL. 20: 102–107. [Google Scholar]
- Forde CG. 2018. From perception to ingestion; the role of sensory properties in energy selection, eating behaviour and food intake. Food Qual Prefer. 66: 171–177. [Google Scholar]
- Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. 2014. Flexible analysis of transcriptome assemblies with Ballgown. bioRxiv. doi: 10.1101/003665 [DOI] [Google Scholar]
- Gaillard D, Laugerette F, Darcel N, El-Yassimi A, Passilly-Degrace P, Hichami A, Khan NA, Montmayeur JP, Besnard P. 2008. The gustatory pathway is involved in CD36-mediated orosensory perception of long-chain fatty acids in the mouse. FASEB J. 22(5):1458–1468. [DOI] [PubMed] [Google Scholar]
- Galindo MM, Voigt N, Stein J, van Lengerich J, Raguse JD, Hofmann T, Meyerhof W, Behrens M. 2012. G protein-coupled receptors in human fat taste perception. Chem Senses. 37(2):123–139. [DOI] [PubMed] [Google Scholar]
- Gallagher MD, Chen-Plotkin AS. 2018. The post-GWAS Era: from association to function. Am J Hum Genet. 102(5):717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson TA, Liu L, York DA, Bray GA. 1998. Dietary fat preferences are inversely correlated with peripheral gustatory fatty acid sensitivity. Ann NY Acad Sci. 855:165–168. [DOI] [PubMed] [Google Scholar]
- Grabenhorst F, Rolls ET. 2014. The representation of oral fat texture in the human somatosensory cortex. Hum Brain Mapp. 35(6):2521–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdasani D, Barroso I, Zeggini E, Sandhu MS. 2019. Author correction: genomics of disease risk in globally diverse populations. Nat Rev Genet. 20(9):562. [DOI] [PubMed] [Google Scholar]
- Hayes J. 2015. Measuring sensory perception in relation to consumer behavior. In: Delarue J, Lawlor J, Rogeaux M, editors. Rapid sensory profiling techniques. Cambridge UK: Woodhead Publishing; p. 53–69. [Google Scholar]
- Hayes JE. 2020. Influence of sensation and liking on eating and drinking. In: Meiselman HL, editor. Handbook of eating and drinking. Cham: Springer; p. 131–155. [Google Scholar]
- Heinze JM, Costanzo A, Baselier I, Fritsche A, Lidolt M, Hinrichs J, Frank-Podlech S, Keast R. 2017. Oil perception-detection thresholds for varying fatty stimuli and inter-individual differences. Chem Senses. 42(7):585–592. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008:420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LD, Lin C, Gharahkhani P, Cuellar-Partida G, Ong JS, An J, Gordon SD, Zhu G, MacGregor S, Lawlor DA, et al. 2019. New insight into human sweet taste: a genome-wide association study of the perception and intake of sweet substances. Am J Clin Nutr. 109(6):1724–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap C 2005. A haplotype map of the human genome. Nature. 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller KL, Liang LC, Sakimura J, May D, van Belle C, Breen C, Driggin E, Tepper BJ, Lanzano PC, Deng L, et al. 2012. Common variants in the CD36 gene are associated with oral fat perception, fat preferences, and obesity in African Americans. Obesity (Silver Spring). 20(5):1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaapila A, Hwang LD, Lysenko A, Duke FF, Fesi B, Khoshnevisan A, James RS, Wysocki CJ, Rhyu M, Tordoff MG, et al. 2012. Genetic analysis of chemosensory traits in human twins. Chem Senses. 37(9):869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugerette F, Passilly-Degrace P, Patris B, Niot I, Febbraio M, Montmayeur JP, Besnard P. 2005. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest. 115(11):3177–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Hansen DR, Kim I, Gilbertson TA. 2005. Expression and characterization of delayed rectifying K+ channels in anterior rat taste buds. Am J Physiol Cell Physiol. 289(4):C868–C880. [DOI] [PubMed] [Google Scholar]
- Liu P, Shah BP, Croasdell S, Gilbertson TA. 2011. Transient receptor potential channel type M5 is essential for fat taste. J Neurosci. 31(23):8634–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang W, Zhang C, Xu C, Duan H, Tian X, Zhang D. 2018. Heritability and genome-wide association study of plasma cholesterol in Chinese adult twins. Front Endocrinol (Lausanne). 9:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S, Mizushige T, Yoneda T, Iwanaga T, Tsuzuki S, Inoue K, Fushiki T. 2007. GPR expression in the rat taste bud relating to fatty acid sensing. Biomed Res. 28(1):49–55. [DOI] [PubMed] [Google Scholar]
- Mattes RD. 2009. Oral detection of short-, medium-, and long-chain free fatty acids in humans. Chem Senses. 34(2):145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. ; Haplotype Reference Consortium 2016. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mela DJ, Sacchetti DA. 1991. Sensory preferences for fats: relationships with diet and body composition. Am J Clin Nutr. 53(4):908–915. [DOI] [PubMed] [Google Scholar]
- Mennella JA, Finkbeiner S, Reed DR. 2012. The proof is in the pudding: children prefer lower fat but higher sugar than do mothers. Int J Obes (Lond). 36(10):1285–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan G, Alvarez-Curto E, Hudson BD, Prihandoko R, Tobin AB. 2017. FFA4/GPR120: pharmacology and therapeutic opportunities. Trends Pharmacol Sci. 38(9):809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounayar R, Morzel M, Brignot H, Tremblay-Franco M, Canlet C, Lucchi G, Ducoroy P, Feron G, Neyraud E. 2014. Nutri-metabolomics applied to taste perception phenotype: human subjects with high and low sensitivity to taste of fat differ in salivary response to oleic acid. OMICS. 18(11):666–672. [DOI] [PubMed] [Google Scholar]
- Mrizak I, Šerý O, Plesnik J, Arfa A, Fekih M, Bouslema A, Zaouali M, Tabka Z, Khan NA. 2015. The A allele of cluster of differentiation 36 (CD36) SNP 1761667 associates with decreased lipid taste perception in obese Tunisian women. Br J Nutr. 113(8):1330–1337. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. 2001. Mammalian sweet taste receptors. Cell. 106(3):381–390. [DOI] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 32(4):381–385. [DOI] [PubMed] [Google Scholar]
- Pepino MY, Love-Gregory L, Klein S, Abumrad NA. 2012. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J Lipid Res. 53(3):561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. 2010. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Sukumaran SK, Jyotaki M, Redding K, Jiang P, Margolskee RF. 2018. Gli3 is a negative regulator of Tas1r3-expressing taste cells. PLoS Genet. 14(2):e1007058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, Knaapila A. 2010. Genetics of taste and smell: poisons and pleasures. Prog Mol Biol Transl Sci. 94:213–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, Tanaka T, McDaniel AH. 2006. Diverse tastes: genetics of sweet and bitter perception. Physiol Behav. 88(3):215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, Xia MB. 2015. Recent advances in fatty acid perception and genetics. Adv Nutr. 6(3):353S–360S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolon ML, Bakke AJ, Coupland JN, Hayes JE, Roberts RF. 2017. Effect of fat content on the physical properties and consumer acceptability of vanilla ice cream. J Dairy Sci. 100(7):5217–5227. [DOI] [PubMed] [Google Scholar]
- Running CA, Craig BA, Mattes RD. 2015. Oleogustus: the unique taste of fat. Chem Senses. 40(7):507–516. [DOI] [PubMed] [Google Scholar]
- Running CA, Hayes JE, Ziegler GR. 2017. Degree of free fatty acid saturation influences chocolate rejection in human assessors. Chem Senses. 42(2):161–166. [DOI] [PubMed] [Google Scholar]
- Running CA, Mattes RD. 2016. A review of the evidence supporting the taste of non-esterified fatty acids in humans. J Am Oil Chem Soc. 93: 1325–1336. [Google Scholar]
- Sayed A, Šerý O, Plesnik J, Daoudi H, Rouabah A, Rouabah L, Khan NA. 2015. CD36 AA genotype is associated with decreased lipid taste perception in young obese, but not lean, children. Int J Obes (Lond). 39(6):920–924. [DOI] [PubMed] [Google Scholar]
- Sclafani A, Ackroff K, Abumrad NA. 2007a. CD36 gene deletion reduces fat preference and intake but not post-oral fat conditioning in mice. Am J Physiol Regul Integr Comp Physiol. 293(5):R1823–R1832. [DOI] [PubMed] [Google Scholar]
- Sclafani A, Zukerman S, Glendinning JI, Margolskee RF. 2007b. Fat and carbohydrate preferences in mice: the contribution of alpha-gustducin and Trpm5 taste-signaling proteins. Am J Physiol Regul Integr Comp Physiol. 293(4):R1504–R1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman AI, Pepino MY, Feldman R, Brand JG. 2010. Technique to collect fungiform (taste) papillae from human tongue. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ngwa JS, van Rooij FJ, Zillikens MC, Wojczynski MK, Frazier-Wood AC, Houston DK, Kanoni S, Lemaitre RN, Luan J, et al. 2013. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 97(6):1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13(9):2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki S. 2007. Mechanisms on the oral chemoreception of fats: the possible participation of FAT/CD36 and GPR120. 5th International Symposium on Molecular and Neural Mechanisms of Taste and Olfactory Perception. 2007 Nov 2; Fukuoka (Japan). [Google Scholar]
- Veldhuizen MG, Babbs RK, Patel B, Fobbs W, Kroemer NB, Garcia E, Yeomans MR, Small DM. 2017. Integration of sweet taste and metabolism determines carbohydrate reward. Curr Biol. 27(16):2476–2485.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt N, Stein J, Galindo MM, Dunkel A, Raguse JD, Meyerhof W, Hofmann T, Behrens M. 2014. The role of lipolysis in human orosensory fat perception. J Lipid Res. 55(5):870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu X, Simonavicius N, Tian H, Ling L. 2006. Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J Biol Chem. 281(45):34457–34464. [DOI] [PubMed] [Google Scholar]
- Wise PM, Hansen JL, Reed DR, Breslin PA. 2007. Twin study of the heritability of recognition thresholds for sour and salty taste. Chem Senses. 32(8):749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Duan H, Tian X, Xu C, Wang W, Jiang W, Pang Z, Zhang D, Tan Q. 2018. Genetics of obesity traits: a bivariate genome-wide association analysis. Front Genet. 9:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. 2012. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 44(7):821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.