Abstract
Background:
Low success rates in oncology drug development are prompting re-evaluation of preclinical models, including orthotopic tumor engraftment. In breast cancer models, tumor cells are typically injected into mouse mammary fat pads (MFP). However, this approach bypasses the epithelial microenvironment, potentially altering tumor properties in ways that affect translational application.
Materials and Methods:
Tumors were generated by mammary intraductal (MIND) engraftment of 4T1 carcinoma cells. Growth, histopathology, and molecular features were quantified.
Results:
Despite growth similar to that of 4T1 MFP tumors, 4T1 MIND tumors exhibit distinct histopathology and increased metastasis. Furthermore, >6,000 transcripts were found to be uniquely up-regulated in 4T1 MIND tumor cells, including genes that drive several cancer hallmarks, in addition to two known therapeutic targets that were not up-regulated in 4T1 MFP tumor cells.
Conclusion:
Engraftment into the epithelial microenvironment generates tumors that more closely recapitulate the complexity of malignancy, suggesting that intraductal adaptation of orthotopic mammary models may be an important step towards improving outcomes in preclinical drug screening and development.
Keywords: Mammary carcinoma, mammary epithelium, microenvironment, orthotopic tumor model, triple-negative breast cancer
A major challenge in oncology drug development is that more than 90% of new drugs fail to show efficacy in clinical trials (1). At the heart of this low success rate is the inability of conventional models, such as tumor cell lines and genetically engineered mice, to reliably predict patient response to drug candidates (2). To overcome these limitations, orthotopic tumor engraftment, including patient-derived xenograft, has gained traction as an alternative strategy for modeling solid tumor malignancies (3). Because orthotopic mouse models entail inoculating tumor cells into the same type of organ in which the cancer was initiated, the engrafted cells are exposed to microenvironmental conditions that simulate those which supported the original tumor. Given the powerful influence of the microenvironment in modulating cancer initiation, progression, and metastasis (4, 5), recapitulation of organ-specific microenvironmental conditions is a highly advantageous feature of orthotopic tumor models.
For breast cancer models, the standard practice is to inject tumor cells into the mouse mammary fat pad (MFP) (6). While this approach ensures generation of tumors within the mammary gland, there are potential drawbacks. Firstly, MFP engraftment bypasses tumor cell interaction with the mammary ductal epithelium, an omission that may be significant because breast carcinomas arise within the ductal tissue rather than the stromal fat pad. Indeed, the mammary ductal epithelium has a well-established role in regulating not only normal mammary development (7–9), but also tumor cell behavior (10), suggesting that contact with the epithelial microenvironment may alter orthotopic tumor properties. Secondly, given the relatively large area and diverse histology of the fat pad, engrafted tumor cells can be exposed to different microenvironmental milieux accordingto injection site, e.g. variable proximity to blood vessels, lymph nodes, or gradients of extracellular matrix density (11, 12), which in turn, may lead to inconsistencies in tumor progression or therapeutic response.
One approach to circumvent these limitations is to inoculate tumor cells by injection into the teat, which enables standardized delivery and facilitates tumor cell interaction with the epithelial microenvironment (13). Initially developed for studies of ductal carcinoma in situ, an early-stage neoplasia that is confined within the ductal epithelial lumen (13), the mammary intraductal (MIND) injection technique has recently been adapted for metastatic tumor studies, including the syngeneic 4T1 triple-negative breast cancer (TNBC) mouse model (14–16). MIND delivery of 4T1 mammary carcinoma cells has shown that the epithelial microenvironment effectively supports tumors that progress from localized ductal lesions to lung metastases (17). However, detailed analysis of molecular, histopathological, and other features of 4T1 MIND tumors has yet to be reported, leaving it unclear whether exposure to the ductal epithelial microenvironment produces tumors that differ from those generated via MFP engraftment. A recent study has made comparison between 4T1 MIND and MFP tumors generated in mice immediately following weaning (15), but changes in the ductal epithelial microenvironment that occur with pregnancy, lactation, and post-lactational involution confound interpretation and may be of limited translational relevance given that most drug development studies are conducted in non-lactating mice.
To determine the effects of the naive mammary epithelium on 4T1 tumor development, we examined growth, histopathology, and transcriptional signature of 4T1 MIND tumors generated in nulliparous BALB/c mice.
Materials and Methods
4T1-Luc2-RFP cell line.
The parental 4T1-Luc2 cell line was purchased from the American Type Culture Collection (Manassas, VA, USA). Red fluorescent protein (RFP) was inserted into the 4T1-Luc2 cell line using a PWPT-RFP lentiviral transduction. The PWPT-RFP vector was generated by replacing GFP sequence with RFP sequence in PWPT-GFP vector. 4T1-Luc2-RFP cells were maintained in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin (Sigma-Aldrich St. Louis, MO, USA) and 100 μg/ml streptomycin (Sigma-Aldrich) at 37°C in a humidified CO2 incubator. The 4T1 cell line was authenticated in 2016 by IDEXX Laboratories (Case #7479–2016; IDEXX BioResearch. Westbrook, ME, USA) (18).
Mice.
Sixteen-week-old female BALB/c mice (22–25 g) were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Mice were housed in the University of South Carolina Animal Research Facility. All experimental methods were conducted under a protocol (2267-101030-082615) approved by the Institutional Animal Care and Use Committee of the University of South Carolina according to National Institutes of Health guidelines.
4T1 MIND tumor engraftment. Before implantation, 4T1-Luc2-RFP cells were grown for 2 days.
Cells from the second passage were collected, counted, and re-suspended in sterile phosphate-buffered saline (PBS) at 106 cells/ml. Mice were anesthetized using isoflurane and 5 μl of cell suspension (i.e. 5,000 4T1-Luc2-RFP cells) were injected intraductally with a 30-gauge blunt-ended 0.5-inch needle into the #8 right thoracic mammary gland (19) as previously described (13), but without surgically opening the mouse. Prior to injection, the tip of the teat was snipped and the needle was inserted directly into the mammary duct through the teat opening.
Tumor growth quantification.
After 4T1 MIND tumors become palpable, tumors were measured semi-weekly using digital calipers to calculate their volume [(length×width2)/2]. Tumors were also monitored by weekly bioluminescence imaging using the IVIS Lumina system (PerkinElmer, Waltham, MA, USA) as described elsewhere (20). D-Luciferase (PerkinElmer) (10 μg/g of body weight) was injected intraperitoneally, and images were acquired within 10 min following D-Luciferase injection. Live imaging software (PerkinElmer) was used to quantify photons emitted by the tumor cells.
Histopathology.
Mice were sacrificed at day 35 post tumor cells injection. Tumors were fixed in 4% paraformaldehyde and processed for paraffin sectioning. Tumor sections of 7 μm were stained with trichrome staining and used for histological examination of regional invasion, necrosis, collagen deposition, and fibrosis of primary tumors. In order to quantify metastasis in lungs, liver, and spleen, organs were processed for paraffin sectioning of 7 μm followed by hematoxylin and eosin staining. A total of 25 sections per organ (spanning the full depth of the organ) were examined and photographed using a Nikon Eclipse Ni microscope (Melville, NY, USA).
Immunohistochemistry.
Tissue sections of 4 μm were deparaffinized in xylene and rehydrated through a graded ethanol series. Antigen retrieval was performed on slides heated in 10 mM citrate buffer (pH=6.0) for 30 min at 95°C. After blocking with 5% donkey serum, primary antibody to RFP (Novus Biologicals, Centennial, CO, USA) was added at a concentration of 3 μg/ml. Slides were incubated for 1 h at room temperature followed by overnight incubation at 4°C. After washing, slides were incubated with secondary antibody (1 μg/ml - Alexa Fluor 594) for 2 h at room temperature. 4’,6-Diamidino-2-phenylindole (DAPI) staining was added for nuclear labeling followed by mounting media and coverslips. Slides were photographed using a Nikon Eclipse Ni camera.
Tumor sphere assay.
Five weeks post tumor cell injection, 4T1 MIND tumors were mechanically dissociated into <1 mm3 pieces using sterile scalpels, then enzymatically dissociated in Leibovitz’s L-15 medium without phenol red, 1.5 mg/ml trypsin, and 3 mg/ml collagenase I (all from Invitrogen Carlsbad, CA, USA) at 37°C for 60 min to generate a single-cell suspension as described (21) and plated on ultra-low-adherence 96-well plates (Corning Corning NY,USA) at 300 cells/well in 300 μl tumor sphere medium (serumfree and phenol red-free Eagle’s minimal essential medium (DMEM) (Gibco Gaithersburg, MD, USA) supplemented with 1X B-27 (Gibco), 20 ng/ml basic fibroblast growth factor (Invitrogen), and 20 ng/ml epidermal growth factor (EGF; Invitrogen). Tumor spheres were counted and imaged 10 days after plating. Sphere-forming efficiency (SFE) was calculated by dividing the number of spheres by the number of plated cells per well times 100.
Lapatinib (Santa Cruz Dallas, TX, USA) and SB-505124 (Sigma) were diluted in dimethyl sulfoxide (DMSO). Vehicle controls were treated with 0.1% DMSO. Cells were maintained in drug or vehicle for the duration of the experiment. Vehicle controls and drug treatments were run in technical and biological triplicates. The data are presented as the mean±standard error of the mean (SEM). One-way ANOVA followed by Tukey multiple comparison test were used to test for significant differences among treatment groups. Statistical analyses were performed using GraphPad Prism software (San Diego, CA, USA).
4T1-Luc2-RFP cell isolation and RNA extraction.
Tumors were harvested at day 24 post tumor cell engraftment and dissociated into single-cell suspensions as described above. RFP-positive 4T1 cells were isolated by fluorescence-activated cell sorting (FACS) (FACSAria San Francisco, CA, USA) and collected in tubes containing QIAzol lysis reagent (Qiagen Germantown, MD, USA). RNA was extracted using the RNeasy Mini Kit (Qiagen).
RNA-Seq analysis.
Three independent RNA preparations from cultured 4T1-Luc2-RFP cells and FACS-isolated 4T1-Luc2-RFP cells were submitted for RNA sequencing analysis. The RNA-Seq libraries were created using TruSeq RNA Sample Prep kit (Illumina, San Diego, CA, USA) Libraries were sequenced on HiSeq 2500 (Illumina), following the manufacturer’s protocols. The mapping of the processed reads was performed using HISAT (22) with mm10 reference genome; reads were counted using HTSeq (23). Genes showing less than 10 counts on average across all samples were removed, resulting in 14,027 genes. The remaining values were normalized with trimmed mean of M values normalization method (24) and then transformed with voom transformation (25).
Statistical analysis.
Limma package (26) was used to examine differential expression between the groups of interest; obtained p-values were corrected with Benjamini-Hochberg correction for multiple testing (27), and genes exhibiting corrected p-values of less than 0.05 were considered to be significant and were used in further analysis. BINGO plugin (28) in Cytoscape environment (29) was used to examine Gene Ontology (GO) Biological processes. Hierarchical clustering was performed by using gplots package [https://cran.r-project.org/web/packages/gplots/index.html]. The 395 genes reported by Tabaries et al. (30) were selected to compare between those found to be differentially expressed between the FACS-isolated 4T1 MIND tumor cells and non-injected 4T1-Luc2-RFP cells versus those differentially expressed between 4T1 MFP tumor cells and non-injected 4T1 cells (30). A total of 384 out of 395 probes were annotated with Gene Symbols. These 384 probes represented 323 unique genes, which were used for the comparison.
Results
Following MIND engraftment of 4T1-Luc2-RFP cells into the #8 thoracic gland (19) of adult BALB/c mice, tumors formed with 100% tumor take and were palpable by day 10 post-injection. In vivo monitoring of 4T1 MIND tumors by bioluminescent imaging showed steadily increased growth over a 4-week period (Figure 1A and B). Tumor volume was also quantified bi-weekly using a digital caliper, which showed that 4T1 MIND tumors had reached an average of 1.8 cm3 in volume (Figure 1C). By 5 weeks post-injection, tumors had reached an average mass of 1.9 g (Figure 1D). Altogether, these results are comparable to the reported growth characteristics of 4T1 MFP tumors (15).
Figure 1.
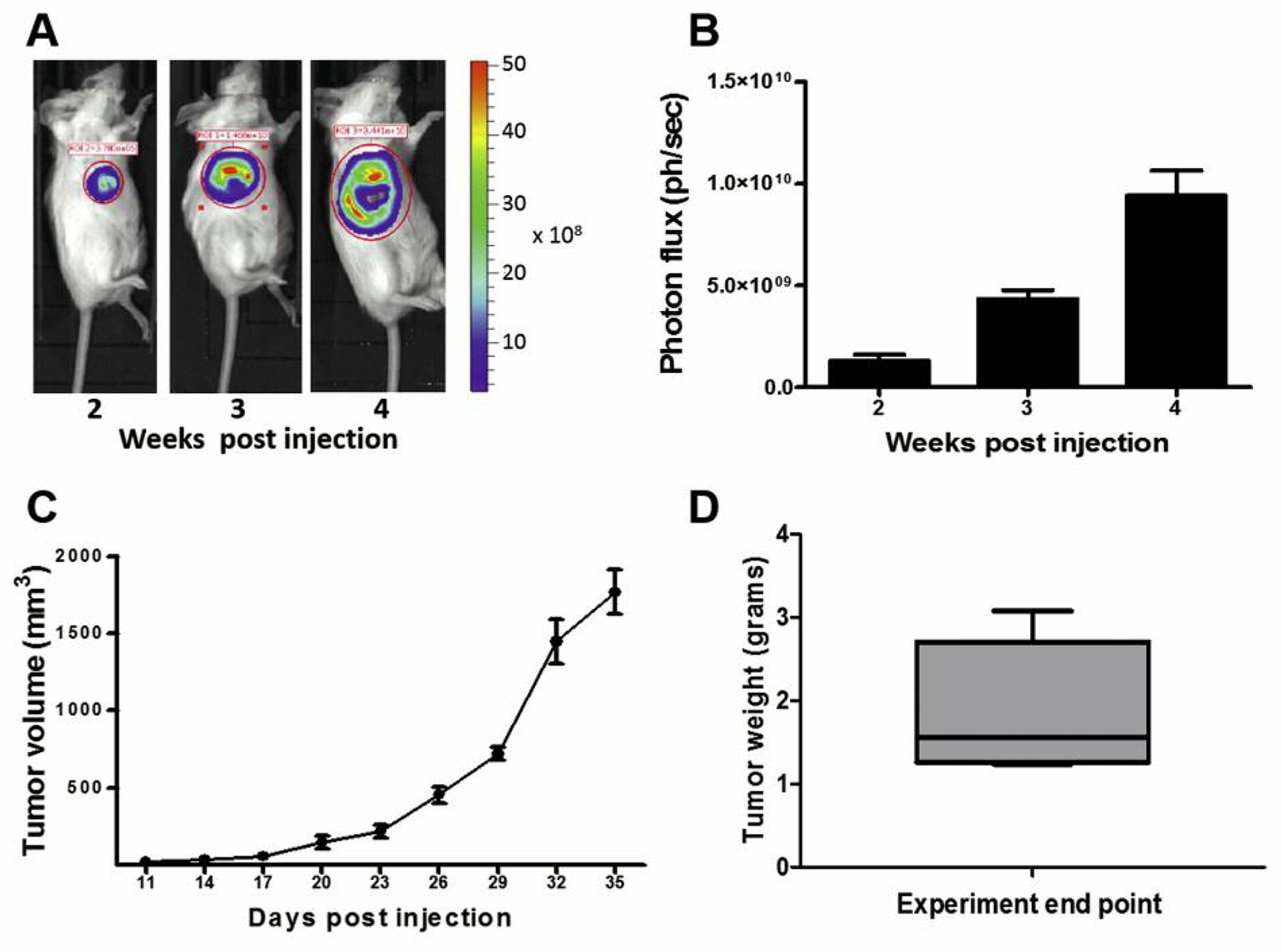
4T1 tumor growth. 4T1-RFP-luc cells were intraductally implanted into thoracic mammary glands of wild-type female BALB/C mice (n=6). A: Tumor growth was measured at the indicated time points by whole animal bioluminescence imaging as shown in a representative mouse. B: Bioluminescence expressed as photon flux per second (ph/s). C: Tumor growth was also measured semi-weekly using digital calipers. D: Tumor mass was quantified upon sacrifice. Data are presented as mean±standard error.
Upon the experimental end-point, histopathology of 4T1 MIND tumors was examined to determine if their features differed from what has been reported for 4T1 MFP tumors. Trichrome staining revealed adjacent muscle invasion (Figure 2A) and regions of necrosis with extensive immune cell infiltration (Figure 2B), features that have also been identified in 4T1 MFP tumors (31, 32). In addition, 4T1 MIND tumors also showed abundant collagen deposition (Figure 2C) and regions of fibrosis (Figure 2C), two indicators of malignant progression that have not been reported for 4T1 MFP tumors as far as we are aware (33, 34).
Figure 2.
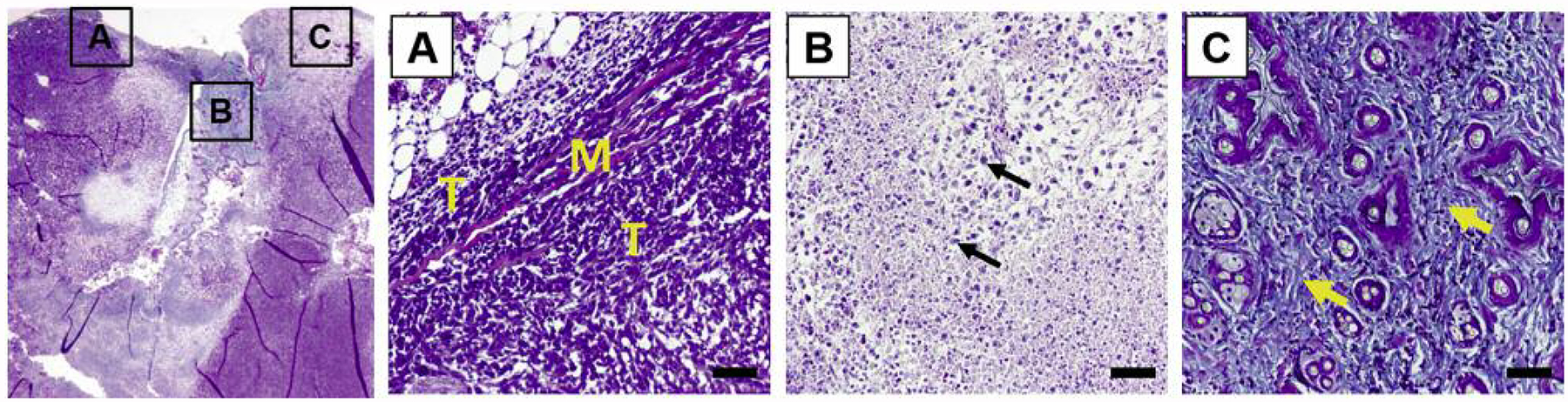
4T1 tumor histopathology. A: Trichrome-stained 4T1 tumor sections showing chest wall muscle (M) invasion by tumor cells (T). B: Necrosis and leukocyte (arrows) infiltration. C: Fibrosis and collagen deposition (arrows) (scale bar=50 μm).
To assess metastatic activity of 4T1 MIND tumors, lung sections were examined for the presence of macrometastatic nodules. At 3 weeks post-engraftment, 87% of mice had detectable lung lesions (Figure 3A and B). This high incidence is a striking difference from the metastatic activity of 4T1 MFP tumors which, despite similar growth rates, do not give rise to lung macrometastases at this early time point (35). By week 5, lung lesions were found in all 4T1 MIND tumor-bearing mice (Figure 3B), which is also higher compared to the 60–80% incidence reported for 4T1 MFP tumors at 5 weeks post-engraftment (35). Consistent with increased incidence at both time points, the metastatic burden in 4T1 MIND tumor-bearing mice also increased between weeks 3 and 5, as indicated by the greater number of lesions per lung (Figure 3C). Although the 4T1 line used in this study metastasizes only to the lungs when engrafted into the MFP (36), we nevertheless examined the liver, spleen, bones, and brain to determine if MIND delivery alters tropism. Macrometastatic lesions were not detected in these organs, although extensive extramedullary hematopoiesis was present in the liver (Figure 3D) and spleen (Figure 3E) at week 5 post engraftment.
Figure 3.
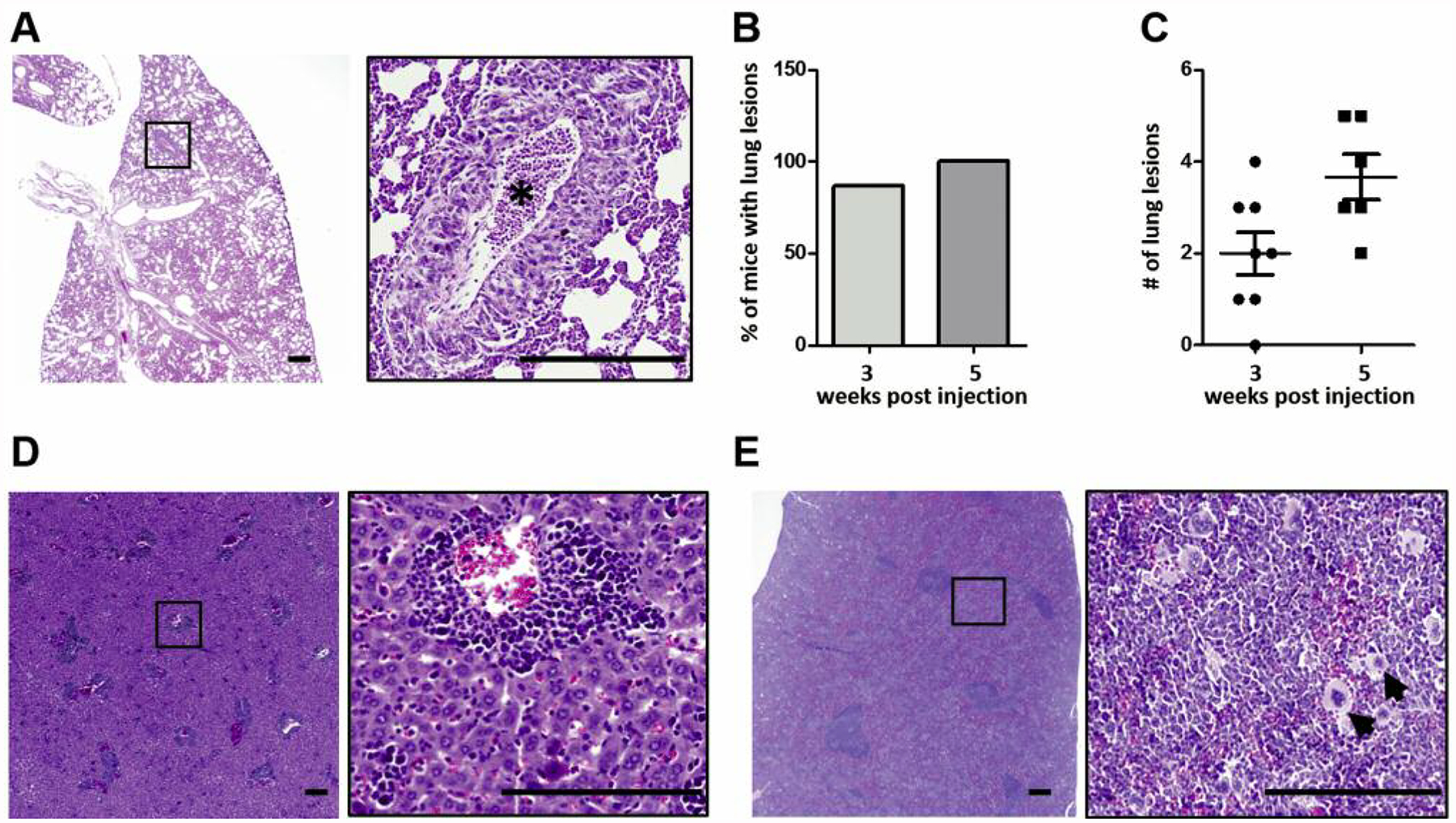
4T1 tumor metastasis. A: Hematoxylin and eosin staining of tissue sections showing macro-metastases in lung (asterisk). B: Incidence of lung metastasis. C: Number of lung lesions at 3- and 5-week time points. Extramedullary hematopoiesis in liver (D) and spleen (E) as evidenced by the presence of megakaryocytes (scale bar=50 μm). Data are presented as the mean±standard error.
The changes in tumor histopathology and metastatic activity in 4T1 MIND tumors prompted us to probe for accompanying changes in gene expression in 4T1 cells following MIND engraftment. In order to isolate 4T1 cells from stroma and other cell types within the 4T1 MIND tumors, we utilized their RFP expression, which was first confirmed by immunofluorescent staining of tumor sections. As shown in Figure 4, approximately 28% of 4T1 MIND tumor cells were RFP+.
Figure 4.
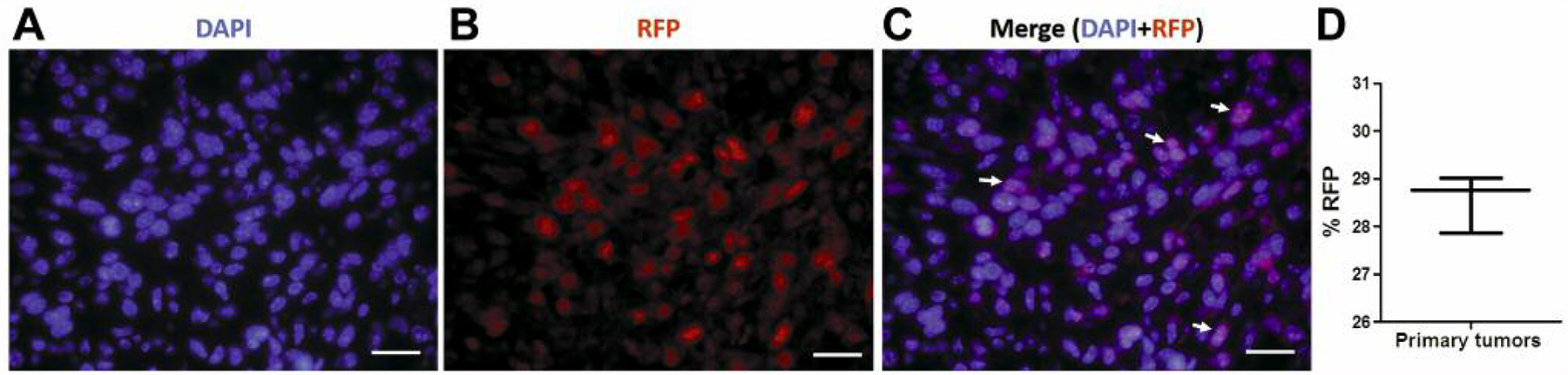
RFP expression in 4T1-Luc2-RFP tumor cells. A: Immunofluorescence staining of a 4T1 tumor section showing the RFP expression in tumor cells (arrows) (scale bar=25 μm). D: The percentage of RFP-positive cells in primary 4T1 tumors. Data are presented as the mean±standard error. DAPI: 4’,6-Diamidino-2-phenylindole.
Next, 4T1 MIND tumor cells were isolated by FACS for RFP+ cells. RNA was extracted from RFP+ 4T1 MIND tumor cells, as well as the cultured 4T1-Luc2-RFP cells used in inoculations, and samples were submitted for RNA sequencing. Differential expression analysis revealed 6,297 genes that were differentially expressed in the isolated 4T1-Luc2-RFP cells compared to non-injected 4T1-Luc2-RFP cells (Figure 5A, GSE129106). This robust difference in gene expression significantly differs from what has been reported for the molecular analysis of 4T1 tumor cells isolated from MFP, which exhibit only 395 differentially expressed genes compared to non-injected 4T1 cells (30). Of the 395 genes reported by Tabaries et al., 384 out of 395 probes had been annotated with Gene Symbols. These 384 probes represented 323 unique genes, which we compared with the differentially genes found in the present study (Figure 5B). Remarkably, some of the genes that become up-regulated in 4T1-Luc2-RFP cells after MIND implantation, but not MFP implantation, include known therapeutic targets such as cytotoxic T-lymphocyte antigen-4 (CTLA4) (37, 38) and androgen receptor (AR) (39).
Figure 5.
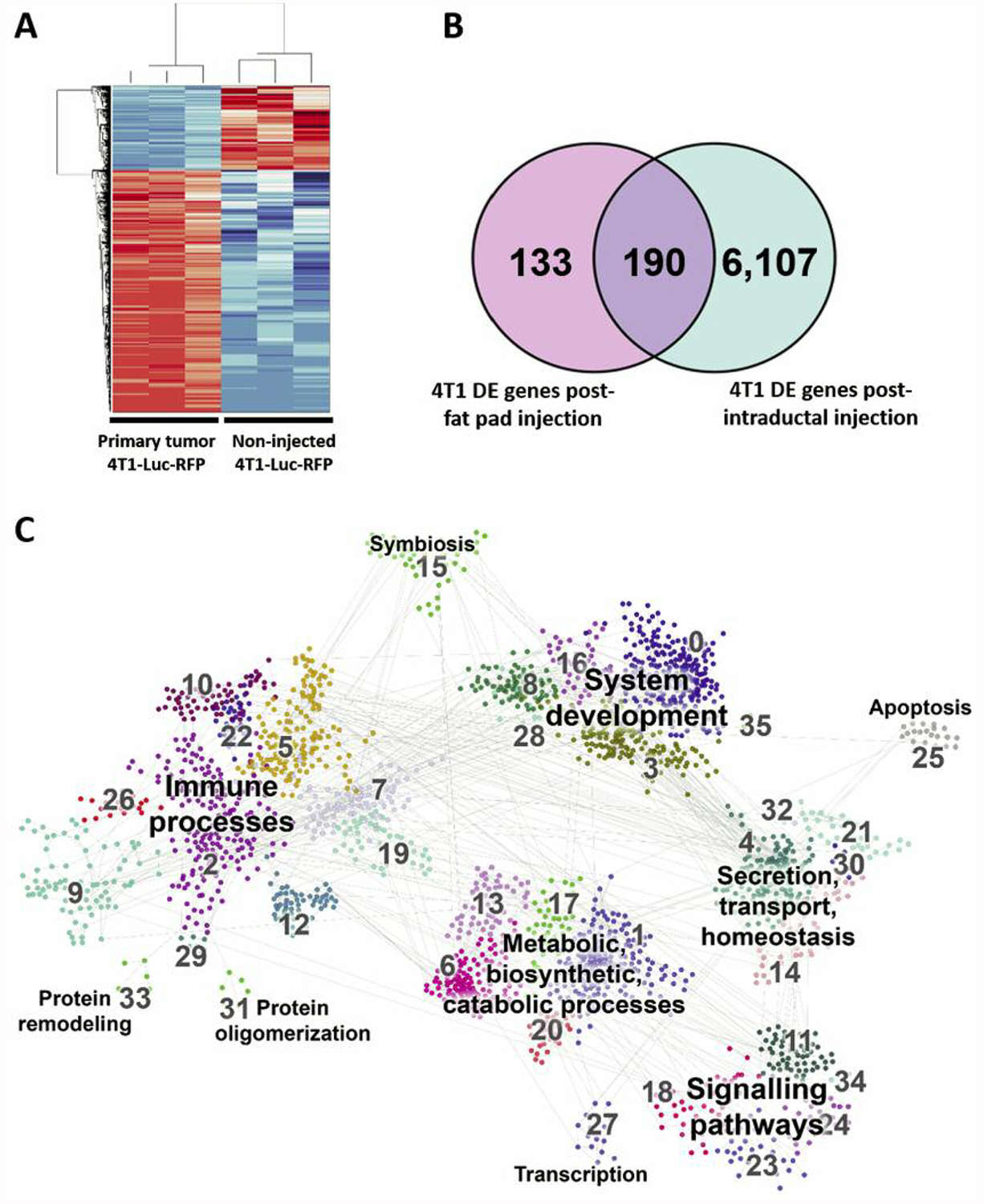
Distinct gene-expression profiling associated with the intraductal implantation of 4T1-Luc2-RFP cells. A: A heatmap displaying 6,375 genes that were differentially expressed after mammary intraductal (MIND) injection of 4T1-Luc2-RFP cells compared to non-injected 4T1-Luc2-RFP cells. B: Identification of differentially expressed genes in 4T1-Luc2-RFP post MIND injection compared to data of Tabaries et al. (30) of differentially expressed genes in 4T1 cells post mammary fat pad (MFD) injection (Venn diagram). C: Pathway analysis showing the number of differentially expressed genes in 4T1-Luc2-RFP post MIND injection.
We next selected genes that were up-regulated with a fold change of at least 2.0 in samples from the intraductal implantation relative to the non-injected cells. These 3,356 genes were subjected to GO analysis, focusing on the Biological Processes component, which showed significantly affected processes associated with intraductal engrafted 4T1-Luc2-RFP cells. When comparing our results to the pathways that are affected in MFP-engrafted cells, we found that many of the MIND-affected pathways were not reported to be affected by the MFP implantation of the 4T1 cells. Pathways affected only in MIND-implanted cells included angiogenesis, Wingless/Integrated signaling (Wnt), interleukin 10 (IL10) signaling, Janus kinases-signal transducer and activator of transcription proteins JAK-STAT signaling, and RAS signaling, all of which have been shown to be directly involved in supporting tumor progression and metastasis, and which represent putative targets for development of biologically-based therapies (40, 41). Pathway analysis also showed several regulators of the tumor immune microenvironment, including interferon gamma and toll-like receptor pathways, in addition to regulators of macrophage recruitment and polarization, i.e. IL10, IL4, and IL6 signaling.
Lastly, because tumor sphere assays are often used as an in vitro platform for high-throughput drug screening (42), we also examined the ability of 4T1 MIND tumors to produce tumor spheres following dissociation and placement into non-adherent culture. Tumor spheres formed within 1 week of culture, and upon passage formed secondary spheres, indicative of self-renewal capacity (Figure 6A and B). Consistent with up-regulation of EGF receptor expression in 4T1 MIND tumor cells, 4T1 MIND tumor sphere-forming efficiency was attenuated by lapatinib, a small-molecule inhibitor of EGF receptor/erb-b2 receptor tyrosine kinase (ERBB2) (43) (Figure 6C), but not by SB-505124, a small molecule inhibitor of activin-like kinase 4/5/7 receptor signaling (44) (Figure 6D).
Figure 6.
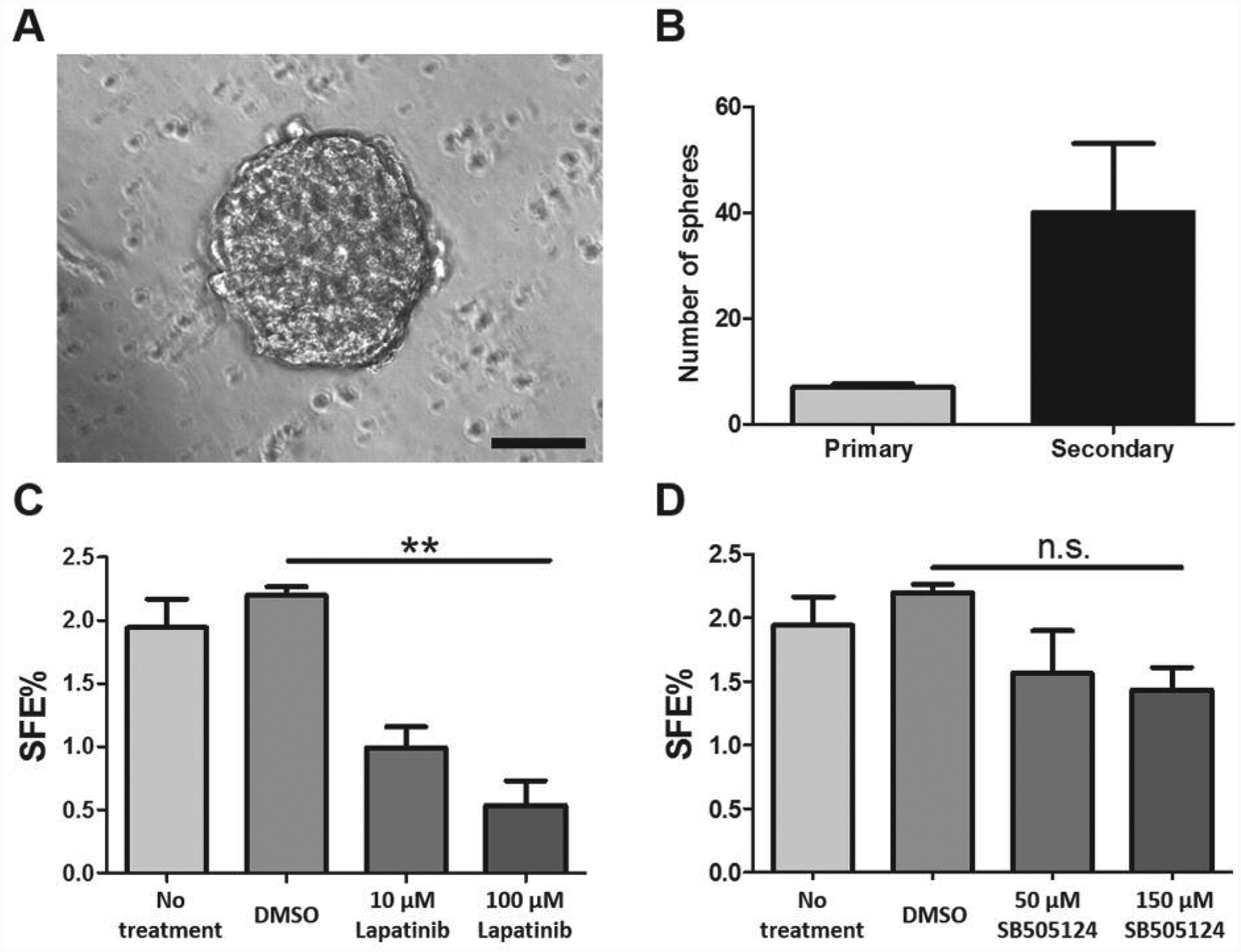
Tumor sphere formation and drug sensitivity. A: Representative image of 4T1 tumor sphere (scale bar=100 μm). B: Numbers of primary and secondary 4T1 tumor spheres (n=3). C: Lapatinib reduced 4T1 tumor sphere formation. D: Inhibitor of activin-like kinase 4/5/7 receptor signaling SB-505124 had no effect on 4T1 tumor sphere formation. Data are presented as the mean±standard error. DMSO: Dimethyl sulfoxide; SFE: Sphere-forming efficiency.
Discussion
The growth, histopathological, metastatic, and molecular properties of 4T1 MFP tumors are well documented (30, 35, 45); however, very little characterization has been reported for 4T1 MIND tumors. Our results demonstrate that when 4T1 cells are MIND-engrafted, tumors acquire a more aggressive malignant phenotype, as reflected by differences in histopathology, metastatic activity, and transcriptional signature. Quantification of macrometastatic lesions revealed increased metastatic activity of 4T1 MIND tumors, both in the number of mice with distant metastases and the number of nodules within lungs of mice with metastatic burden. Heightened metastatic activity was preceded by enhanced collagen deposition and fibrosis in primary tumors in addition to local invasion of tumor cells into adjacent muscle tissue, indicating that exposure to the ductal epithelial microenvironment promotes a more malignant phenotype.
The differences in 4T1 MIND tumor progression occurred despite similarity in tumor volume and endpoint mass compared with 4T1 MFP, indicating that the changes in gene expression and metastatic progression are unrelated to tumor size. The similarity in growth rate of 4T1 MIND and MFP tumors appears to be restricted to those generated in nulliparous mice; when engraftments are performed in lactating mice, 4T1 MIND tumor growth is delayed compared to MFP tumors (15). Although it remains to be determined how the differential physiological status of mammary epithelium promotes differential tumor growth, one likely possibility, as suggested by Steenbrugge et al. (16), is that the immunological compartment of the mammary microenvironment plays a contributory role. Consistent with this idea, the stroma of the weaning gland undergoes a marked shift to a pro-inflammatory signature, resembling a wound-healing phenotype that would be expected to delay tumor growth (46, 47). By contrast, the transcriptional signature derived from 4T1 MIND tumors generated in the naive mammary epithelium reflects a mixed inflammatory/immunosuppressive immune environment. Indeed, in recognition of the ability of the immune microenvironment to control tumor progression and metastasis in breast and other cancer types, we chose to inoculate 4T1 cells without surgical opening of the gland in order to mitigate inflammation induced by experimental manipulation (48). In addition to immunological influence, there may also be a hormonal contribution; differential MIND tumor growth has been reported for estrogen receptor positive (ER+) xenografts, which showed delayed growth compared to ER+ MFP tumors when engrafted into immunodeficient mice (14).
Results obtained from RNA sequencing offer insight into the molecular processes underlying the more aggressive metastatic progression of 4T1 MIND tumors. With respect to the parental 4T1 cell line, 4T1 tumor cells exhibit a striking differential expression of 6,297 genes following intraductal engraftment, many of which represent pathways that are directly associated with progression and metastasis of TNBC, e.g. homobox (Hoxb) actin-alpha 2 (Acta2), actin-gamma2 (Actg2), and Wnt (49, 50). Pathway analysis of differentially expressed genes yielded over 90 distinct pathways that were affected by intraductal implantation, uncovering those which are not affected by MFP implantation and that represent putative candidates for biologically based TNBC therapies, e.g. angiogenesis, WNT signaling, IL10 signaling, JAK-STAT signaling, EGF receptor signaling, and RAS signaling (30). Moreover, the 4T1 MIND transcriptional signature also includes two definitive drug targets, cytotoxic Ctla4, which was up-regulated 89-fold, and AR, up-regulated 67-fold. Ctla4 encodes an immunosuppressive protein that transmits an inhibitory signal to T-cells (51). Tremelimumab, a monoclonal antibody to CTLA4, has recently been used in clinical trials alone or in combination with other therapeutics to successfully treat metastatic cancer, including TNBC (38). Yet despite this clinical advancement, preclinical studies using 4T1 MFP tumors failed to detect efficacy of this drug (52), presumably due to lack of CTL4 expression following fat pad engraftment.
A similar discrepancy also appears to be emerging for AR, a steroid hormone receptor whose expression is absent from 4T1 MFP tumors, yet robust in 4T1 MIND tumors. The lack of AR expression in 4T1 MFP tumors would predict inefficacy of targeting this pathway in TNBC. However, there is a subset of triple-negative breast tumors that express high levels of AR that are showing promising results in clinical trials with enzalutamide, an inhibitor of AR signaling (53). Given that patients with the TNBC subtype exhibit a particularly poor response to chemotherapy (54), utilization of appropriate preclinical animal models will accelerate progress in this new therapeutic direction. Along this line, it is notable that differences in AR expression have also been observed in ER+ MCF7 xenografts: MCF7-MFP tumors lack AR expression, but intraductal engraftment causes markedly elevated AR expression (14), again providing evidence that tumor cell exposure to the ductal epithelial microenvironment alters tumor properties in ways that are translationally relevant. Thus, in light of these and the many other molecular differences that arise when 4T1 cells are intraductally engrafted, we propose that selection of the inoculation site is a parameter that warrants careful consideration when using orthotopic mammary tumor models, a decision that may significantly affect outcomes in preclinical drug development and testing.
Acknowledgements
This work was supported in part by P20GM103499 (SCINBRE) from the National Institute of General Medical Sciences, NIH, a USC School of Medicine Research and Development Fund, and R01CA218578 and P01AT003961-8455. H.A. was supported by MOHESR- Iraqi scholarship and SPARC funding. A.D.-G. was supported by Canadian Institutes of Health operating grant (142353). S.S.P. was supported by a USC Science Undergraduate Research Fellowship and a USC Magellan Scholar Award. R.L.P was supported by the USC School of Medicine Research Program for Medical Students.
Footnotes
Conflicts of Interest
The Authors have no conflicts of interest.
References
- 1.Hait WN: Anticancer drug development: the grand challenges. Nat Rev Drug Discov 9(4): 253–254, 2010. DOI: 10.1038/nrd3144 [DOI] [PubMed] [Google Scholar]
- 2.Mak IW, Evaniew N and Ghert M: Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res 6(2): 114–118, 2014. [PMC free article] [PubMed] [Google Scholar]
- 3.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, Factor R, Matsen C, Milash BA, Nelson E, Neumayer L, Randall RL, Stijleman IJ, Welm BE and Welm AL: Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med 17(11): 1514–1520, 2011. DOI: 10.1038/nm.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polyak K and Kalluri R: The role of the microenvironment in mammary gland development and cancer. Cold Spring Harb Perspect Biol 2(11): a003244, 2010. DOI: 10.1101/cshperspect.a003244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Place AE, Jin Huh S and Polyak K: The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res 13(6): 227, 2011. DOI: 10.1186/bcr2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kocaturk B and Versteeg HH: Orthotopic injection of breast cancer cells into the mammary fat pad of mice to study tumor growth. J Vis Exp 96, 2015 DOI: 10.3791/51967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bussard KM and Smith GH: The mammary gland microenvironment directs progenitor cell fate in vivo. Int J Cell Biol, 2011. DOI: 10.1155/2011/451676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Booth BW, Mack DL, Androutsellis-Theotokis A, McKay RD, Boulanger CA and Smith GH: The mammary microenvironment alters the differentiation repertoire of neural stem cells. Proc Natl Acad Sci USA 105(39): 14891–14896, 2008. DOI: 10.1073/pnas.0803214105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulanger CA, Mack DL, Booth BW and Smith GH: Interaction with the mammary microenvironment redirects spermatogenic cell fate in vivo. Proc Natl Acad Sci USA 104(10): 3871–3876, 2007. DOI: 10.1073/pnas.0611637104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booth BW, Boulanger CA, Anderson LH and Smith GH: The normal mammary microenvironment suppresses the tumorigenic phenotype of mouse mammary tumor virus-neu-transformed mammary tumor cells. Oncogene 30(6): 679–689, 2011. DOI: 10.1038/onc.2010.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fata JE, Werb Z and Bissell MJ: Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res 6(1): 1–11, 2004. DOI: 10.1186/bcr634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schedin P and Keely PJ: Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression. Cold Spring Harb Perspect Biol 3(1): a003228, 2011. DOI: 10.1101/cshperspect.a003228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, Young E, Mukhopadhyay P, Yeh HW, Allred DC, Hu M, Polyak K, Rosen JM and Medina D: An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res 11(5): R66, 2009. DOI: 10.1186/bcr2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sflomos G, Dormoy V, Metsalu T, Jeitziner R, Battista L, Scabia V, Raffoul W, Delaloye JF, Treboux A, Fiche M, Vilo J, Ayyanan A and Brisken C: A preclinical model for ERalpha-positive breast cancer points to the epithelial microenvironment as determinant of luminal phenotype and hormone response. Cancer Cell 29(3): 407–422, 2016. DOI: 10.1016/j.ccell.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 15.Steenbrugge J, Breyne K, Denies S, Dekimpe M, Demeyere K, De Wever O, Vermeulen P, Van Laere S, Sanders NN and Meyer E: Comparison of the adipose and luminal mammary gland compartment as orthotopic inoculation sites in a 4t1-based immunocompetent preclinical model for triple-negative breast cancer. J Mammary Gland Biol Neoplasia 21(3–4): 113–122, 2016. DOI: 10.1007/s10911-016-9362-7 [DOI] [PubMed] [Google Scholar]
- 16.Steenbrugge J, Breyne K, Demeyere K, De Wever O, Sanders NN, Van Den Broeck W, Colpaert C, Vermeulen P, Van Laere S and Meyer E: Anti-inflammatory signaling by mammary tumor cells mediates prometastatic macrophage polarization in an innovative intraductal mouse model for triple-negative breast cancer. J Exp Clin Cancer Res 37(1): 191, 2018. DOI: 10.1186/s13046-018-0860-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh A, Sarkar S, Banerjee S, Behbod F, Tawfik O, McGregor D, Graff S and Banerjee SK: MIND model for triple-negative breast cancer in syngeneic mice for quick and sequential progression analysis of lung metastasis. PLoS One 13(5): e0198143, 2018. DOI: 10.1371/journal.pone.0198143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwanowycz S, Wang J, Hodge J, Wang Y, Yu F and Fan D: Emodin inhibits breast cancer growth by blocking the tumor-promoting feedforward loop between cancer cells and macrophages. Mol Cancer Ther 15(8): 1931–1942, 2016. DOI: 10.1158/1535-7163.MCT-15-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veltmaat JM, Ramsdell AF and Sterneck E: Positional variations in mammary gland development and cancer. J Mammary Gland Biol Neoplasia 18(2): 179–188, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao K, Fang M, Alroy J and Sahagian GG: Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer 8: 228, 2008. DOI: 10.1186/1471-2407-8-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smalley MJ: Isolation, culture and analysis of mouse mammary epithelial cells. Methods Mol Biol 633: 139–170, 2010. DOI: 10.1007/978-1-59745-019-5_11 [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Langmead B and Salzberg SL: HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12(4): 357–360, 2015. DOI: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT and Huber W: HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2): 166–169, 2015. DOI: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD and Oshlack A: A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11(3): R25, 2010. DOI: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law CW, Chen Y, Shi W and Smyth GK: Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15(2): R29, 2014. DOI: 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W and Smyth GK: Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7): e47, 2015. DOI: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Shi Y and Toga AW: Controlling false discovery rate in signal space for transformation-invariant thresholding of statistical maps. Inf Process Med Imaging 24: 125–136, 2015. DOI: 10.1007/978-3-319-19992-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maere S, Heymans K and Kuiper M: BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21(16): 3448–3449, 2005. DOI: 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11): 2498–2504, 2013. DOI: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabaries S, Ouellet V, Hsu BE, Annis MG, Rose AA, Meunier L, Carmona E, Tam CE, Mes-Masson AM and Siegel PM: Granulocytic immune infiltrates are essential for the efficient formation of breast cancer liver metastases. Breast Cancer Res 17: 45, 2015. DOI: 10.1186/s13058-015-0558-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peixoto RC, Miranda-Vilela AL, de Souza Filho J, Carneiro ML, Oliveira RG, da Silva MO, de Souza AR and Bao SN: Antitumor effect of free rhodium (II) citrate and rhodium (II) citrate-loaded maghemite nanoparticles on mice bearing breast cancer: a systemic toxicity assay. Tumour Biol 36(5): 3325–3336, 2015. DOI: 10.1007/s13277-014-2966-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeap SK, Mohd Yusof H, Mohamad NE, Beh BK, Ho WY, Ali NM, Alitheen NB, Koh SP and Long K: In Vivo immunomodulation and lipid peroxidation activities contributed to chemoprevention effects of fermented mung bean against breast cancer. Evid Based Complement Alternat Med 2013: 708464, 2013. DOI: 10.1155/2013/708464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris BA, Burkel B, Ponik SM, Fan J, Condeelis JS, Aguirre-Ghiso JA, Castracane J, Denu JM and Keely PJ: Collagen matrix density drives the metabolic shift in breast cancer cells. EBioMedicine 13: 146–156, 2016. DOI: 10.1016/j.ebiom.2016.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barcus CE, Keely PJ, Eliceiri KW and Schuler LA: Stiff collagen matrices increase tumorigenic prolactin signaling in breast cancer cells. J Biol Chem 288(18): 12722–12732, 2013. DOI: 10.1074/jbc.M112.447631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey-Downs LC, Thorpe JE, Disch BC, Bastian A, Hauser PJ, Farasyn T, Berry WL, Hurst RE and Ihnat MA: Development and characterization of a preclinical model of breast cancer lung micrometastatic to macrometastatic progression. PLoS One 9(5): e98624, 2014. DOI: 10.1371/journal.pone.0098624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia X, Yu F, Wang J, Iwanowycz S, Saaoud F, Wang Y, Hu J, Wang Q and Fan D: Emodin suppresses pulmonary metastasis of breast cancer accompanied with decreased macrophage recruitment and M2 polarization in the lungs. Breast Cancer Res Treat 148(2): 291–302, 2014. DOI: 10.1007/s10549-014-3164-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan MK, Wolchok JD and Allison JP: Anti-CTLA-4 antibody therapy: immune monitoring during clinical development of a novel immunotherapy. Semin Oncol 37(5): 473–484, 2013. DOI: 10.1053/j.seminoncol.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comin-Anduix B, Escuin-Ordinas H and Ibarrondo FJ: Tremelimumab: Research and clinical development. Onco Targets Ther 9: 1767–1776, 2016. DOI: 10.2147/OTT.S65802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietri E, Conteduca V, Andreis D, Massa I, Melegari E, Sarti S, Cecconetto L, Schirone A, Bravaccini S, Serra P, Fedeli A, Maltoni R, Amadori D, De Giorgi U and Rocca A: Androgen receptor signaling pathways as a target for breast cancer treatment. Endocr Relat Cancer 23(10): R485–98, 2016. DOI: 10.1530/ERC-16-0190 [DOI] [PubMed] [Google Scholar]
- 40.Dey N, Barwick BG, Moreno CS, Ordanic-Kodani M, Chen Z, Oprea-Ilies G, Tang W, Catzavelos C, Kerstann KF, Sledge GW Jr., Abramovitz M, Bouzyk M, De P and Leyland-Jones BR: Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer 13: 537, 2013. DOI: 10.1186/1471-2407-13-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamidullah, Changkija B and Konwar R: Role of interleukin-10 in breast cancer. Breast Cancer Res Treat 133(1): 11–21, 2012. DOI: 10.1007/s10549-011-1855-x [DOI] [PubMed] [Google Scholar]
- 42.Lee CH, Yu CC, Wang BY and Chang WW: Tumorsphere as an effective in vitro platform for screening anti-cancer stem cell drugs. Oncotarget 7(2): 1215–1226, 2016. DOI: 10.18632/oncotarget.6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eccles SA: The epidermal growth factor receptor/Erb-B/HER family in normal and malignant breast biology. Int J Dev Biol 55(7–9): 685–696, 2011. DOI: 10.1387/ijdb.113396se [DOI] [PubMed] [Google Scholar]
- 44.DaCosta Byfield S, Major C, Laping NJ and Roberts AB: SB-505124 is a selective inhibitor of transforming growth factorbeta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol 65(3): 744–752, 2004. DOI: 10.1124/mol.65.3.744 [DOI] [PubMed] [Google Scholar]
- 45.DuPre SA, Redelman D and Hunter KW Jr: The mouse mammary carcinoma 4T1: characterization of the cellular landscape of primary tumours and metastatic tumour foci. Int J Exp Pathol 88(5): 351–360, 2007. DOI: 10.1111/j.1365-2613.2007.00539.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schedin P: Pregnancy-associated breast cancer and metastasis. Nat Rev Cancer 6(4): 281–291, 2006. DOI: 10.1038/nrc1839 [DOI] [PubMed] [Google Scholar]
- 47.Schedin P, O’Brien J, Rudolph M, Stein T and Borges V: Microenvironment of the involuting mammary gland mediates mammary cancer progression. J Mammary Gland Biol Neoplasia 12(1): 71–82, 2007. DOI: 10.1007/s10911-007-9039-3 [DOI] [PubMed] [Google Scholar]
- 48.Russell TD, Jindal S, Agunbiade S, Gao D, Troxell M, Borges VF and Schedin P: Myoepithelial cell differentiation markers in ductal carcinoma in situ progression. Am J Pathol 185(11): 3076–3089, 2015. DOI: 10.1016/j.ajpath.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pohl SG, Brook N, Agostino M, Arfuso F, Kumar AP and Dharmarajan A: Wnt signaling in triple-negative breast cancer. Oncogenesis 6(4): e310, 2017. DOI: 10.1038/oncsis.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah N and Sukumar S: The Hox genes and their roles in oncogenesis. Nat Rev Cancer 10(5): 361–371, 2010. DOI: 10.1038/nrc2826 [DOI] [PubMed] [Google Scholar]
- 51.Contardi E, Palmisano GL, Tazzari PL, Martelli AM, Fala F, Fabbi M, Kato T, Lucarelli E, Donati D, Polito L, Bolognesi A, Ricci F, Salvi S, Gargaglione V, Mantero S, Alberghini M, Ferrara GB and Pistillo MP: CTLA-4 is constitutively expressed on tumor cells and can trigger apoptosis upon ligand interaction. Int J Cancer 117(4): 538–550, 2005. DOI: 10.1002/ijc.21155 [DOI] [PubMed] [Google Scholar]
- 52.De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, Budhu S, Ghosh A, Pink M, Tchaicha J, Douglas M, Tibbitts T, Sharma S, Proctor J, Kosmider N, White K, Stern H, Soglia J, Adams J, Palombella VJ, McGovern K, Kutok JL, Wolchok JD and Merghoub T: Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 539(7629): 443–447, 2016. DOI: 10.1038/nature20554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traina TA, Miller K, Yardley DA, Eakle J, Schwartzberg LS, O’Shaughnessy J, Gradishar W, Schmid P, Winer E, Kelly C, Nanda R, Gucalp A, Awada A, Garcia-Estevez L, Trudeau ME, Steinberg J, Uppal H, Tudor IC, Peterson A and Cortes J: Enzalutamide for the treatment of androgen receptor-expressing triple-negative breast cancer. J Clin Oncol 36(9): 884–890, 2018. DOI: 10.1200/JOT.2016.71.3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collignon J, Lousberg L, Schroeder H and Jerusalem G: Triple-negative breast cancer: treatment challenges and solutions. Breast Cancer (Dove Med Press) 8: 93–107, 2016. DOI: 10.2147/BCTT.S69488 [DOI] [PMC free article] [PubMed] [Google Scholar]


