Abstract
Breast Cancer stem cells (BCSCs) have been extensively studied and have been used directly as a therapeutic target, but how the BCSCs themselves are regulated remain unclear. Here we reported identification of miR-205 that may act as a tumor suppressor and negatively-regulate BCSCs stemness and tumor malignance. By qRT-PCR analysis, we have shown that miR-205 was decreased in CD44+/CD24-/low BCSCs compared with non-BCSCs. We have also shown that miR-205 expression level was very low in MB-231 cells with high BCSC percentage, while relatively high in MCF-7 cells with low BCSC percentage. We then overexpressed miR-205 in MB-231 and SUM-149 cells and knocked it down in MCF-7 and BT-474 cells respectively. Our results showed that overexpression of miR-205 could reduce CD44+/CD24-/low population percentage in MB-231 cells. The mechanism might associate with mesenchymal-epithelial transition (MET). Finally, we found an important transcriptional factor and oncogene, RunX2, was a target gene of miR-205. miR-205 overexpression could inhibit breast cancer malignancy by regulating RunX2 both in vitro and in vivo. A rescue experiment by cotransfection of RunX2 and miR-205 into the MCF-7 cell line attenuate cell proliferation, invasion, migration, CD44+/CD24-/low population, mammosphere formation abilities and xengraft tumor formation. Together, our results support that miR-205 is a tumor suppressor during breast cancer development.
Keywords: microRNA-205, breast cancer, breast cancer stem cell, RunX2
Introduction
Breast cancer is the most common cancer among women. Both incidence and mortality are the highest among all cancers in women [1]. Breast cancer cells are heterogeneous. A small number of cells with strong tumor formation ability that show the characteristics of stem cells with the ability of self-renewal and multipotent differentiation are termed breast cancer stem cells (BCSCs) [2-4]. In 2003, Al-Hajj first discovered and isolated BCSCs with the cell surface markers CD44+/CD24-/low [3]. BCSCs were first found and confirmed in solid tumors. They are believed to play a key role in the initiation, recurrence and chemotherapy resistance of breast cancer [2,5]. However, how BCSCs regulate these processes remains unclear.
MicroRNAs (miRs) are small noncoding RNAs that usually hybridize to the 3’UTR of mRNAs, facilitating their degradation, which results in reduced protein expression [6]. miRs control many cellular functions in eukaryotic organisms, including development, differentiation, proliferation, and apoptosis, among others [7]. Recently, the role of miRNAs in cancer stem cells has been explored and confirmed in many kinds of tumors [8]. miR-205 is expressed in the myoepithelial/basal cell compartment of mammary ducts and lobules and plays an important role in the progression of mammary cell differentiation and tissue development [9,10]. Some experimental data show that miR-205 expression is highly reduced in the basal tumors and in TNBC cell lines, which are more malignant and have a higher ratio of BCSCs [11,12]. However, the regulatory mechanism of miR-205 in breast cancer and breast cancer stem cells is not yet clear. Therefore, in this study, we aimed to investigate how miR-205 regulates breast cancer cell stemness and tumor malignancy and the mechanism involved in it.
Materials and methods
Cells culture and infection
MCF-7-CD44+/CD24-/low, MDA-MB-231-CD44+/CD24-/low, MCF-7-ALDH+, and MDA-MB-231-ALDH+ cells were generous gifts from Prof. Liu’s lab at the University of Science and Technolog of China. MCF-10A cells were cultured in DMEM/F12, which contained 5% horse serum, 100 ng/ml cholera enterot oxin, 10 µg/ml bovine insulin, 20 ng/ml recombinant human EGF, and 0.5 μg/ml hydrocortisone. SUM-149 cells were cultured in Ham’s F12 medium containing 5% FBS, 4 ug/ml gentamicin, 5 ug/ml insulin and 1 ug/ml hydrocortisone. MCF-7, BT-474 and MDA-MB-231 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere with 5% CO2. For cell infection, the lentivirus used in the experiment was obtained from GenePharma (Shanghai, China). The cells (5×104/well) were seeded in 24-well plates and incubated for 24 h. Then, the MDA-MB-231 and SUM-149 cells were infected with Lv-hsa-miR-205 mimics (mimic) or Lv-hsa-miR-205-negative-control (NC). MCF-7 and BT-474 cells were infected with Lv-hsa-miR-205 inhibitor (inhibitor) or Lv-hsa-miR-205-negative-control (NC). Polybrene (Sigma-Aldrich) was added to the culture medium after the cells successfully infected with lentivirus.
miRNA microarray
miRNA microarray analysis was performed to compare the expression profiles of miRNAs among MCF-7-CD44+/CD24-/low, MDA-MB-231-CD44+/CD24-/low, MCF-7-ALDH+, MDA-MB-231-ALDH+, MCF-7 and MDA-MB-231 cells. These cells were collected and total RNA was extracted using TRIzol reagent (CWBio company, China), followed by reverse transcribing and labeling with miRNA complete labeling and hybrid kit in accordance with the manufacturer’s protocol (Agilent Technologies, USA). Quality control was performed before performing the microarray. The signals were detected by a microarray scanner. The data were analyzed using Agilent Feature Extraction software (v10.7). Triplicate samples were analyzed to generate the data.
Quantitative real-time PCR and reverse transcription PCR
Total RNA was extracted using TRIzol reagent (CWBio company, China), according to the manufacturer’s instructions. For quantitative real-time PCR (qRT-PCR), the relative expression of miRNA-205 was quantified using a microRNA and U6 snRNA Normalization RT-PCR Quantitation Kit (GenePharma, Shanghai, China) in accordance with the manufacturer’s instructions. U6 was selected as the internal reference, and the 2-ΔΔCt method was used to calculate the relative expression. For reverse transcription PCR (RT-PCR), RNA was reverse-transcribed according to the manufacturer’s protocol for cDNA synthesis using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA). The primers used in the study were designed as follows: hsa-miR-205, forward primer: 5’-CATACCTCCTTCATTCCACCG-3’, hsa-miR-205, reverse primer: 5’-TATGGTTGTTCACGACTCCTTCAC-3’, U6, forward primer: 5’-CAGCACATATACTAAAATTGGAACG-3’, U6, reverse primer: 5’-ACGAATTTGCGTGTCATCC-3’, RunX2, forward primer: 5’-CCGGAATGCCTCTGCTGTTATGA-3’ and RunX2, reverse primer: 5’-ACTGAGGCGGTCAGAGAACAAACT-3’.
Western blotting
The protein extraction reagent RIPA solubilized the cells and collected the lytic cells. The total protein concentration was determined with the BCA protein assay kit (Keygen, China). Equal amounts of total proteins were separated on 10% SDS polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocking in TBST containing 5% non-fat dried milk for 1 h, then incubated overnight at 4°C with primary antibodies (1:1000, CST, Cambridge, UK), followed by secondary antibody (1:2000, Biosharp, China). Protein bands were visualized by ECL chemiluminescence method.
Luciferase assays
RunX2 WT-3’UTR, RunX2 WT-3’UTR, miR-205 mimic/NC were cotransfected into 293T cells in 24-well plates. After 48 h of transfection, cells were collected and treated with luciferase reporter gene reagent (Beyotime, China), and firefly luciferase activity was normalized to Renilla luciferase activity.
Transwell assays
The transwell chambers (BD Biosciences, Franklin Lakes, US) were loaded into 24-well plates (Corning, NY, USA), which divided the well into two parts with an upper chamber and a lower chamber. Cells (5×104) were seeded in serum-free culture medium in the upper chamber. The lower chamber was filled with 500 µl of culture medium containing 10% FBS. After 24 h incubation for cells, the cells migrated towards the lower chamber membrane surface and were fixed and stained with 10% formalin containing 0.1% crystal violet. Images of five randomly selected microscope fields were taken, and cell numbers were counted for analysis.
MTT assay
The cell proliferation activities with miR-205 inhibitor or miR-205 mimic were determined by MTT assay at 0, 24, 48, 72 and 96 h after the cells were adherent. MTT reagent (KeyGen Biotech) was added into each well at the indicated time points. Then, the cells were incubated in the cell incubator for 4 h at 37°C. After removing the medium, 150 µl of DMSO was added to each well. The optical density (OD) was detected at 490 nm. All experiments were repeated at least 3 times.
Colony formation assay
The cells (5×102) were trypsinized and seeded into six-well plates (Corning, NY, USA). The 6-well plates were placed in a 37°C, 5% CO2 incubator for approximately 2 weeks and then fixed with methanol and stained with 0.1% crystal violet. Visual counting and imaging of the colonies was performed for preservation. The experiment was repeated three times.
Soft agar colony formation assay
1.2% and 0.7% agarose (Biosharp, China) were prepared and melted in a water bath. Then, 1.5 ml of the bottom layer mixture, which contained 0.6% agar and 1×DMEM with 10% FBS, was gently added into each well of a 6-well culture plate without forming any air bubbles, and 1.2 ml of the cell-containing layer mixture, which contained 0.35% agar, 1×DMEM with 10% FBS. The cells (3×103) was gently added onto the bottom layer mixture of the 6-well culture plate. After 2 weeks of cell growth in the soft agar, the number of colonies in each well was counted using a light microscope (Zeiss, Oberkochen, Germany).
Mammospheres assay
The protocol was performed as we previously described (Xu xiaodan et al, 2018). 5000 cells/well were plated as single cell suspension in ultra-low attachment 6-well plates, which coated with agarose gel to avoid cell adhesion. Cells were grown in DMEM/F12 (Gibco, NY, USA) supplemented with 10 ng/ml hr-bFGF (PeproTech, Rocky Hill, NJ) and 20 ng/ml EGF (Thermo Fisher Scientific) and 2% B27 (Gibco, NY, USA). After culturing for 6-7 days, mammospheres with diameters of >50 μm were counted.
Tumor xenografts in nude mice
Female BALB/c nude mice (3-4 weeks) were purchased from the Nanjing Model Animal Center. The mice were equally divided into four groups (n = 6 in each group) and were allowed to acclimatize to the new conditions for 1 week prior to the experiments. The nude mice were injected with 5×106 tumor cells into the right mammary fat pad. The tumor volumes were measured every three days after implantation, and the tumors were removed and weighed approximately 30 days after tumor formation. All animal care and experiments were approved by the Anhui Medical University Institutional Animal Care and Use Committee and Ethics Committee.
Statistical analysis
The results are presented as the mean ± SD. The statistical data for all experiments were analyzed by t-test or analysis of variance. Data were considered significantly different when P<0.05. The number of asterisks indicates the p value range as follows: *p value <0.05, **p value <0.01, ***p value <0.001. All statistical results were calculated using GraphPad 7.0 software (GraphPad Software Inc.).
Results
The miR-205 expression level is negatively related to BCSC numbers and breast cancer cell metastasis level
First, using a microRNA array, we detected miRNA expression in breast cancer stem cells (MCF-7-CD44+/CD24-/low, MDA-MB-231-CD44+/CD24-/low, MCF-7-ALDH+, and MDA-MB-231-ALDH+) and breast cancer cells (MCF-7 and MDA-MB-231). We found several differentially expressed miRNAs between these cell lines. One of them, miR-205, was severely decreased in both the CD44+/CD24-/low and ALDH1+ BCSC populations (Figure 1A). Then, we confirmed the expression level of miR-205 in those cell lines by RT-PCR (Figure 1B). Furthermore, we detected the percentage of CD44+/CD24-/low BCSCs in normal breast epithelial cells MCF-10A, MCF-7 and MDA-MB-231 cells by flow cytometry and found that MDA-MB-231 cells had the highest BCSC ratio and that MCF-7 had a lower BCSC ratio (Figure 1C). Furthermore, we detected the expression of miR-205 in normal breast epithelial cell MCF-10A, and in human breast cancer cell lines MCF-7, BT-474, MB-231, SUM-149 by RT-PCR (Figure 1D) and qRT-PCR (Figure 1E). The results showed that miR-205 was significantly lower in breast cancer cell lines compared with normal MCF-10A cell. These data indicated that miR-205 might be involved in CD44+/CD24-/low BCSC regulation in breast cancer.
Figure 1.
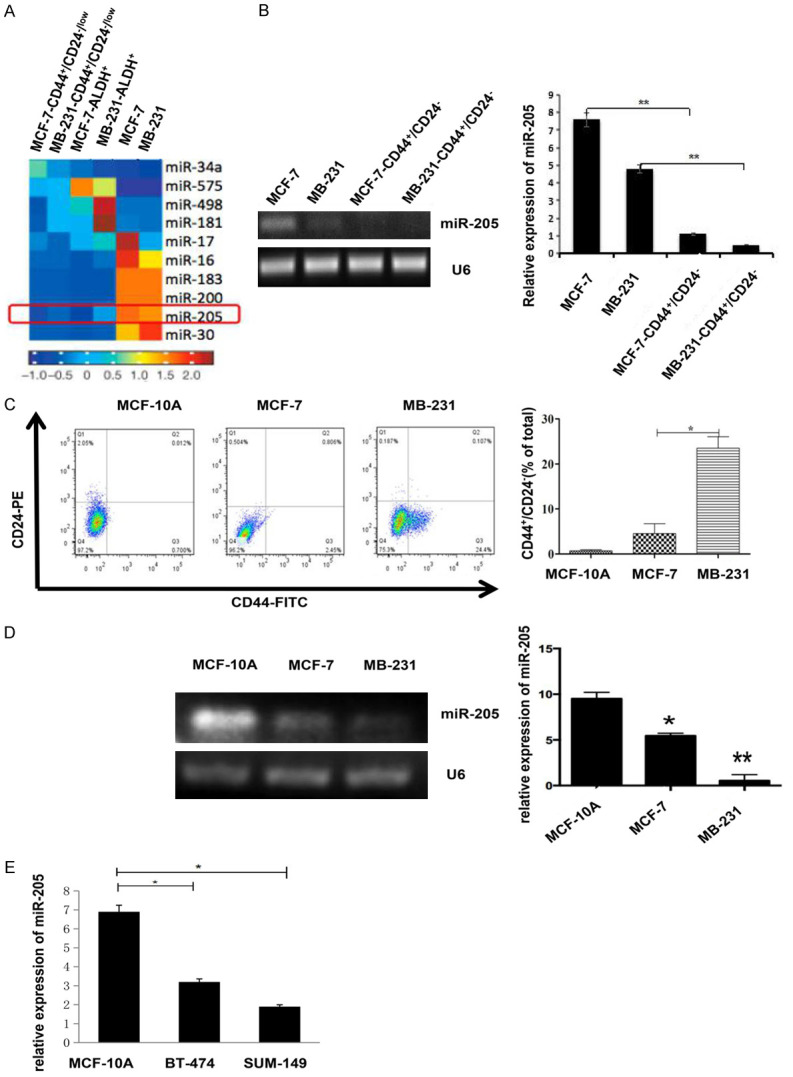
miR-205 is significantly reduced in both breast cancer cells and breast cancer stem cells. A. Heatmap analysis of microarray data of representative miRNA expression in different subsets of breast cancer cells. Red indicates high expression, and blue indicates low expression. B. The expression levels of miR-205 in MCF-7-CD44+/CD24-/low, MCF-7, MDA-MB-231, MDA-MB-231-CD44+/CD24-/low cells were detected by RT-PCR (**P<0.01). C. Flow cytometry was used to detect breast cancer stem cell (BCSC) subpopulations of different breast cancer cell lines (*P<0.05). Each experiment mentioned above was repeated at least three times, and the results are presented as the mean ± SD. D. The expression of miR-205 in normal breast epithelial cells and breast cancer cell lines was detected by RT-PCR (*P<0.05, **P<0.01). E. The expression of miR-205 in normal breast epithelial cell MCF-10A and breast cancer cell lines (SUM-149 and BT-474) were detected by RT-qPCR (*P<0.05).
miR-205 can inhibit the growth and invasion of breast cancer cells as a tumor suppressor
Because BCSCs can substantially promote cancer malignancy, we investigated whether miR-205 could also affect malignant breast cancer cell phenotypes, such as proliferation, migration and invasion ability. We modified miR-205 expression using miR-205-specific Lv-hsa-miR-205 mimics and Lv-hsa-miR-205 inhibitor in MDA-MB-231 and MCF-7 cells, respectively, and established that miR-205 was overexpressed in the MDA-MB-231 cell line (referred to as ‘mimic’ in the figures) and miR-205 was knocked down in the MCF-7 cell line (referred to as ‘inhibitor’ in the figures). The untreated group (control) and the miRNA negative control group (NC) were used as negative controls. Besides, We also choose SUM-149 cells were transfected with miR-205 mimic to up-regulate miR-205 expression, and BT-474 cells were transfected with miR-205 inhibitor to down-regulate miR-205 expression. The transfection efficacies of miR-205 mimic and inhibitor in SUM-149 and BT-474 cells were verified by qRT-PCR (Figure 2A). The MTT assay was used to detect cell proliferation ability. We found that cell proliferation significantly inhibited after overexpression miR-205 expression in MDA-MB-231 and SUM-149 cells (Figure 2B). In contrast, cell growth was significantly increased after reducing miR-205 expression in MCF-7 and BT-474 cells (Figure 2C). Overexpression of miR-205 inhibited colony formation ability, while knockdown of miR-205 had the opposite effect (Figure 2D, 2E). Overexpression of miR-205 in MDA-MB-231 and SUM-149 cells reduced cell invasion, while downregulation of miR-205 in MCF-7 and BT-474 cells significantly increased invasion ability (Figure 2F, 2G). The above data indicated that miR-205 could negatively regulate cell growth and metastasis in breast cancer. Considering the decreased expression of miR-205 in BCSCs, we hypothesized that miR-205 may act as a tumor suppressor in breast cancer by affecting BCSCs.
Figure 2.
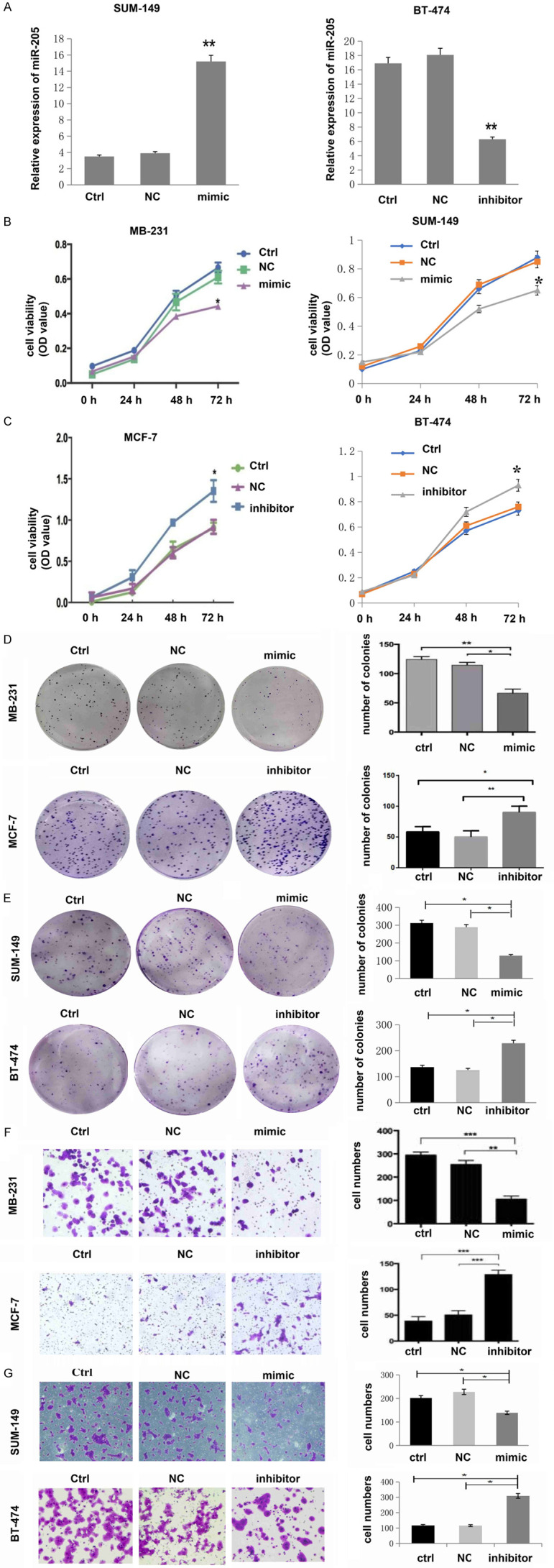
miR-205 is negatively associated with the malignant level of breast cancer cells. A. RT-PCR was used to detect the relative expression of miR-205 in cells transfected with miR-205 mimic or miR-205 inhibitor. **P<0.01, compared to Ctrl and NC. Data analysis was performed by an unpaired Student’s t-test. B. Cell proliferation was decreased after upregulation of miR-205 by infection with miR-205 mimic in MDA-MB-231 and SUM-149 cells (*P<0.05). C. Cell proliferation was increased after downregulation of miR-205 by infection with miR-205 inhibitor in MCF-7 and BT-474 cells (*P<0.05). D, E. Cell colony numbers were decreased after upregulation of miR-205 by infection with miR-205 mimic in MDA-MB-231 and SUM-149 cells, while they increased after downregulation of miR-205 by infection with a miR-205 inhibitor in MCF-7 cells and BT-474 cells (*P<0.05, **P<0.01). F, G. Cell migration ability was decreased after upregulation of miR-205 by infection with miR-205 mimics. However, the expression of miR-205 was increased after downregulation of miR-205 by infection with the miR-205 inhibitor (*P<0.05, **P<0.01, ***P<0.001). Each experiment was repeated three times, and the results are presented as the mean ± SD.
miR-205 negatively regulates stemness directly
To demonstrate that miR-205 could affect BCSC stemness directly, we detected the expression of the BCSC surface markers CD44 and CD24 by western blotting. We found that overexpression of miR-205 in MDA-MB-231 and SUM-149 cells led to decreased CD44 expression and increased CD24 levels (Figure 3A). However, knockdown of miR-205 in MCF-7 and BT-474 cells led to increased CD44 expression and decreased CD24 levels (Figure 3B). The soft agar colony formation experiment showed that overexpression of miR-205 in MDA-MB-231 cells resulted in fewer colony numbers compared to the control group. The results were reversed when miR-205 was knocked down in MCF-7 cells (Figure 3C). Mammosphere formation experiments also showed that overexpression of miR-205 could significantly inhibit BCSC sphere formation, which suggested that the self-renewal ability of BCSCs is decreased, whereas knockdown of miR-205 had the opposite effect (Figure 3D). These data indicate that miR-205 can negatively regulate stemness directly.
Figure 3.
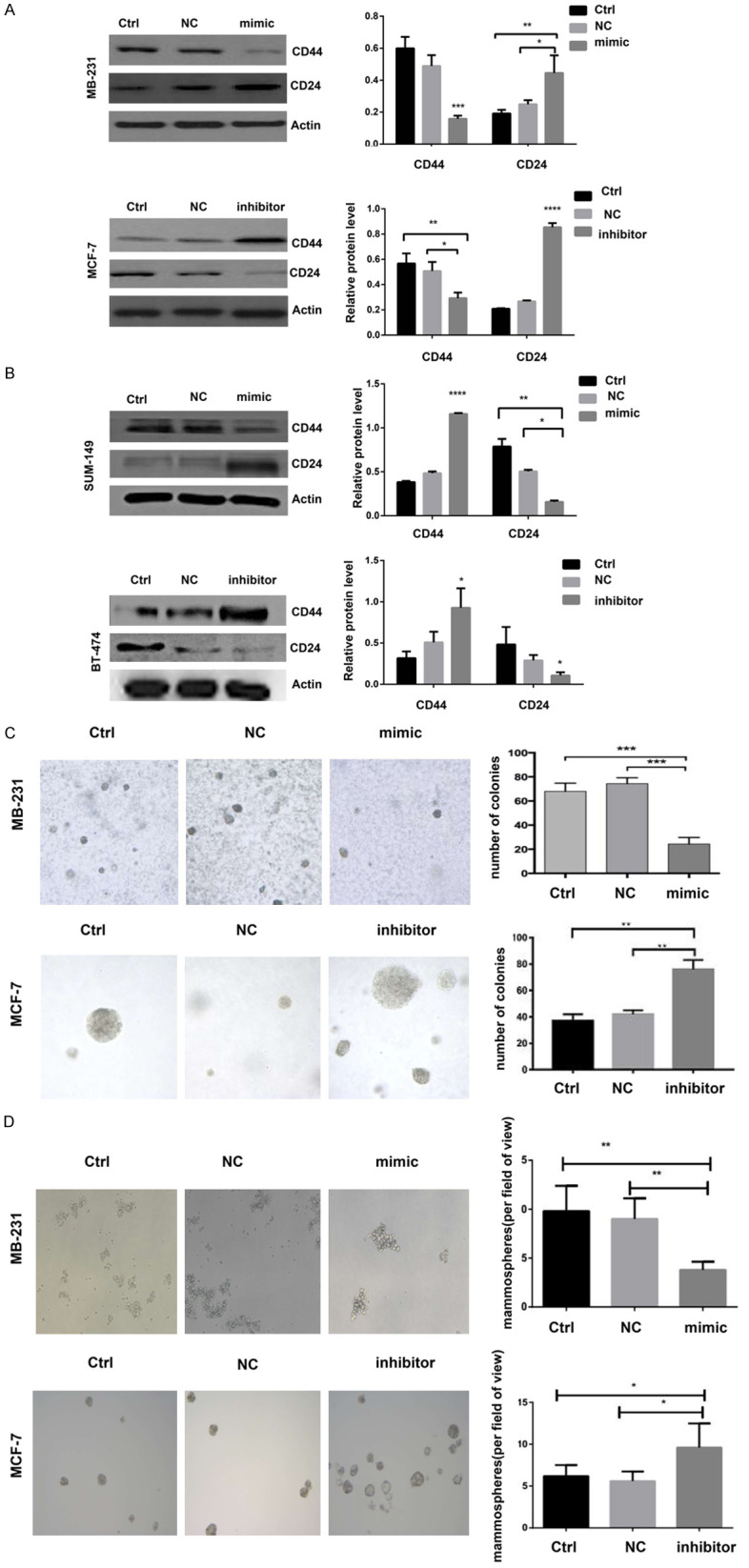
miR-205 could negatively regulate the stemness of breast cancer cells. (A and B) Western blot analysis of the protein expression of CD44 and CD24 in MDA-MB-231, MCF-7, SUM-149 and BT-474 cells after infection with miR-205 mimics or miR-205 inhibitor. Each experiment was repeated three times, and the results are presented as the mean ± SD. *P<0.05, **P<0.01, ****P<0.0001 compared to Ctrl and NC (C) Soft agar colony formation in MDA-MB-231 and MCF-7 cells after infection with miR-205 mimics or miR-205 inhibitor (**P<0.01, ***P<0.001). (D) The effect of miR-205 mimics or miR-205 inhibitor on MB-231 and MCF-7 cells were determined (*P<0.05, **P<0.01).
miR-205 could affect the EMT process of breast cancer cells
During cell culture, we observed morphological changes of MDA-MB-231 cells from mesenchymal-like spindle forms to organized epithelial-like squamous shapes after infection with miR-205 mimics (Figure 4A) and its mechanism might related to the Wnt/β-catenin signaling pathway (Figure 4B). It is known that EMT is a very important process during BCSC formation. Therefore, we detected the EMT markers E-cadherin, vimentin, MMP3, and MMP9 by western blot assay. The results showed that overexpression of miR-205 could increase the expression of the epithelial marker E-cadherin and inhibit the expression of the mesenchymal markers vimentin, MMP3, and MMP9. In contrast, the knockdown of miR-205 had the opposite effect (Figure 4C). These data indicated that miR-205 could negatively regulate breast cancer cell stemness and that the mechanism might be related to EMT regulation.
Figure 4.
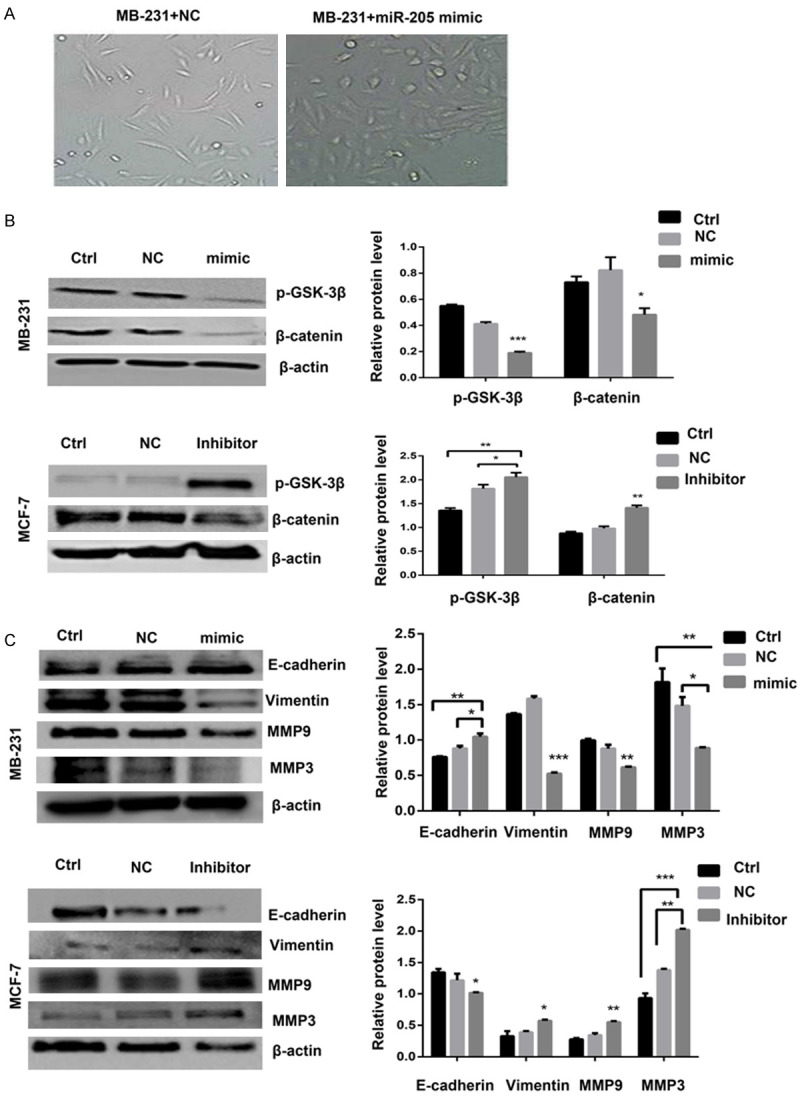
Morphological changes and western blotting results indicated that miR-205 could inhibit the breast cancer stem cell EMT process. A. Images of representative cell morphology after MDA-MB-231 infection with miR-205 mimics or NC vector. B. Western blotting was performed to determine the protein expression levels of the Wnt/β-catenin pathway-related markers p-GSK-3β and β-catenin. *P<0.05, **P<0.01, ***P<0.001 compared to Ctrl and NC. C. Western blot results showed the change in EMT marker expression when the expression of miR-205 was altered in MCF-7 and MDA-MB-231 cells. Each experiment was repeated three times, and the results are presented as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001 compared to Ctrl and NC.
MiR-205 can directly target the transcriptional factor RunX2
To further investigate the molecular mechanism of miR-205 in breast cancer, we predicted potential target genes of miR-205 by TargetScan and found that there was a nucleotide sequence completely complementary to the miR-205 sequence at position 1124-1131 of the 3’UTR of RunX2, which is an important transcriptional factor and oncogene during breast cancer development (Figure 5A). The dual luciferase reporter gene assay showed that the fluorescence activity was significantly decreased upon coinfection of miR-205 and wild-type RunX2 plasmids, while coinfection of miR-205 and the mutant RunX2 plasmid had no significant effect on fluorescence activity, which confirmed that RunX2 is a direct target of miR-205 (Figure 5B). We detected the expression of Runx2 and miR-205 in MCF-10A, MCF-7, and MB-231 cells by RT-PCR. The results showed that compared with normal breast epithelial cells, miR-205 was lowly expressed in breast cancer cells, Runx2 was highly expressed in breast cancer cells (Figure 5C). Western blot also showed that miR-205 mimics could reduce RunX2 protein levels, while miR-205 inhibitor could directly increase RunX2 protein levels (Figure 5D).
Figure 5.

miR-205 directly targeted RunX2 in breast cancer cells. A. In silico analysis of two target prediction databases showed that miR-205 targeted the 3’UTR of RunX2 mRNA, (http://mirdb.org/cgi-bin/target_detail.cgi?targetID=547892), (http://www.targetscan.org/cgi-bin/targetscan/vert_71/targetscan.cgi?species=Human&mir_sc=miR-205-5p). B. Dual luciferase reporter assays verified that RunX2 was the target gene of miR-205 (**P<0.01). C. RT-PCR result of the mRNA expression levels of miR-205 and RunX2 in different breast cell lines. D. Western blot was used to detect RunX2 expression levels after infection with miR-205 mimics in MDA-MB-231 cells and infection with miR-205 inhibitor in MCF-7 cells.
miR-205 could attenuate breast cancer stem cell stemness by inhibiting RunX2
Some results of our study have already proved that RunX2 could act as a breast cancer promoter to increase tumor malignancy and stemness. To investigate whether miR-205 acts through RunX2 on BCSCs, we performed a rescue experiment to observe the changes in breast cancer cell stemness by cotransfecting miR-205 mimics and over expression RunX2 lentiviral vector (LV-Runx2). We used RT-qPCR confirmed that lentivirals were successfully transfected into MCF-7 cells (Figure 6A). RT-PCR analysis and western blotting was performed to measure RunX2 mRNA and protein expression. The results demonstrated that RunX2 was higher at the mRNA and protein levels cell after co-transfected with miR-205+RunX2 mimic compared with miR-205 mimic group (Figure 6B). Then we examined cell stemness. We found that overexpression of RunX2 could increase BCSC marker CD44 levels and reduce CD24 levels in MCF-7 cells, while cotransfection of miR-205 could inhibit the CD44 increase and CD24 decrease induced by RunX2 (Figure 6C). Accordingly, flow cytometry results showed that overexpression of RunX2 increased the CD44+/CD24- BCSC population, while cotransfection of miR-205 inhibited the RunX2-induced increase in BCSCs (Figure 6D). Mammosphere culture also showed that cotransfection of miR-205 could significantly inhibit BCSC sphere formation induced by RunX2 (Figure 6E). Therefore, we believe that miR-205 could reverse breast cancer stem cell stemness by inhibiting RunX2.
Figure 6.

miR-205 attenuated breast cancer stem cell stemness by inhibiting RunX2. A. RT-qPCR was used to assess the expression of miR-205 and RunX2 mRNA after transfection with miR-205 mimic or LV-RunX2 and either miR-205+RunX2 mimic or miR-NC. Data analysis was performed by One-way ANOVA. **P<0.01, ***P<0.001. B. The expression levels of RunX2 mRNA and protein were detected by RT-PCR and western blot assay. C. Western blot was used to examine the expression of RunX2, CD44 and CD24. D. The breast cancer stem cell markers CD44 and CD24 were assessed by flow cytometry using a Beckman Coulter and CytExpert software. E. The Spheres Formation Efficiency of MCF-7 in different groups was determined using a mammosphere formation assay. Only spheres ≥ 50 µM were counted, and images were taken at 10× magnification (*P<0.05 and ***P<0.001).
miR-205 could attenuate breast cancer cell growth and metastasis by inhibiting RunX2 in vitro
To investigate whether miR-205 could inhibit breast cancer malignancy through RunX2, we performed a rescue experiment to observe the changes in breast cancer cell growth and metastasis by cotransfecting miR-205 mimics and RunX2. Then, we examined cell malignant behavior. We found that overexpression of RunX2 could increase MCF-7 cell growth, while cotransfection of miR-205 could inhibit the cell growth increase induced by RunX2 (Figure 7A). Cell migration and invasion assays also showed that cotransfection with miR-205 significantly inhibited the cell migration and invasion enhanced by RunX2 (Figure 7B, 7C). Overexpression of RunX2 increased the colony formation ability of MCF-7 cells, while cotransfection of miR-205 inhibited the effect of RunX2 (Figure 7D). miR-205 reduced breast cancer cell growth and metastasis by inhibiting RunX2.
Figure 7.
miR-205 attenuated the RunX2-mediated proliferation, migration, invasion, and non-anchored growth ability of MCF-7 cells. (A) MTT, (B) migration, (C) invasion and (D) soft agar colony formation assays were performed on cells transfected with miR-negative control (NC) mimic, LV- RunX2 , miR-205 mimic, or cotransfected with miR-205+RunX2 mimic. Nontransfected MCF-7 cells were used as controls. *P<0.05, **P<0.01, vs miR-205 mimic.One-way ANOVA was used for comparisons among multiple groups.
miR-205 can attenuate the tumor growth ability promoted by RunX2 in vivo
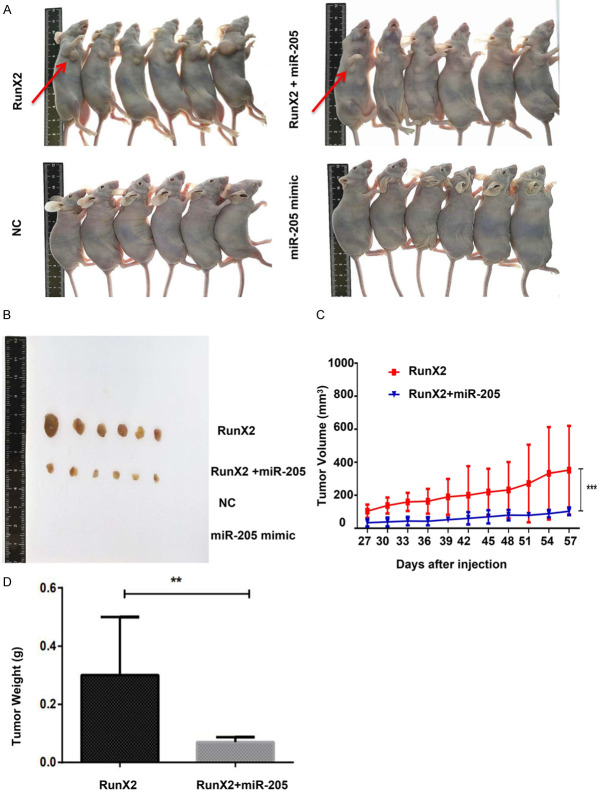
To further explore the role of miR-205 in RunX2-mediated tumor development, we performed tumor formation experiments in nude mice. Nude mice were subcutaneously injected with MCF-7 cells stably transfected with miR-NC, LV- RunX2, miR-205 mimic and RunX2+miR-205 mimic. Tumor growth was observed for 28 days after injection. As shown in Figure 8A and 8B, the miR-NC and miR-205 groups did not exhibit tumor formation. Tumors formed in mice injected with the LV- RunX2, but injection of cells transfected with RunX2+miR-205 led to a slower growth. Tumor diameter was measured every 3 days, and a growth curve was calculated. The tumor volume in xenograft tumors of the RunX2+miR-205 group was lower than that of the RunX2 group (Figure 8C). The mice were sacrificed 30 days after tumor growth. Compared to the RunX2 group, mice injected with miR-205-transfected cells, showed a decrease in tumor weight (Figure 8D). These results indicate that RunX2 overexpression promotes tumor growth and that this effect can be reversed by increasing the expression of miR-205.
Figure 8.
miR-205 suppressed RunX2 expression and negatively regulated tumor growth. A and B. Tumor growth in nude mice. C. Tumor growth curve of nude mice in each group (***P<0.001). D. Tumor weight of nude mice in each group (**P<0.05). The results are presented as the mean ± SD.
Discussion
MicroRNAs (miRNAs) are a group of small RNAs that negatively regulate gene expression by targeting the 3’-untranslated region (UTR) of mRNAs through formation of the RNA-induced silencing complex (RISC) [13]. The mechanism is complicated and includes many aspects, such as regulating cytokines, oncogenes, tumor suppressors, and EMT. The role of miR-205 is still not very clear. Some studies have shown that it acts as a tumor promoter to induce endometrial carcinoma development [14]. Other studies show that it is a tumor suppressor in glioma, renal cell carcinoma, and thyroid cancer [15-17]. For breast cancer, it was reported that the expression of miR-205 was downregulated in radio-resistant subpopulations of breast cancer cells [18]. To further determine its precise function in breast cancer, we detected the expression of miR-205 in different breast cancer cells and breast cancer stem cells and proved that the miR-205 expression level was negatively related to breast cancer metastatic level and stemness. miR-205 could inhibit the proliferation, migration, invasion, EMT and stemness of breast cancer cells. Therefore, miR-205 is a tumor suppressor in breast cancer.
Furthermore, we found that RunX2 is a direct downstream target of miR-205. RunX2 is a member of the RunX (runt-related transcription factor) family and has been demonstrated to play an important role in different biological processes of osteoblasts and chondrocytes [19,20]. In recent research, RunX2 was found to be overexpressed in several tumor tissues, such as prostate cancer, lung cancer, malignant melanoma, and pancreatic cancer, and to be related to tumor malignant behaviors and poor outcomes [21-24]. In particular, RunX2 is overexpressed and promotes breast tumor initiation, progression and invasion, especially during bone metastases of breast cancer [25]. RunX2 can promote CD44+/CD24-/low breast cancer stem cell properties. RunX2 regulates the malignant phenotype of breast cancer by promoting the proportion of CD44+/CD24-/low breast cancer stem cells population. Important question are why RunX2 is overexpressed and how to control it. Here, we hypothesized that the reduced expression of miR-205 in breast cancer leads to RunX2 uncontrolled expression and plays a tumorigenic function during breast cancer development. We proved this hypothesis and found that miR-205 overexpression could inhibit breast cancer malignancy by regulating RunX2 both in vitro and in vivo. The underlying mechanism might be related to the negative regulation of BCSC stemness by miR-205. Knockdown miR-205 expression led to enhanced BCSC activation by increasing the EMT process. Therefore, tumor growth and metastasis were promoted. The signal involved in this process still needs to be investigated. One of our other studies found that the TGF-β-RunX2 axis plays a vital role in triple-negative breast cancer (TNBC) drug resistance by regulating stemness and EMT [26]. Next, we will further explore whether miR-205 is also involved in the TGF-β-RunX2 axis and regulates breast cancer drug resistance.
Our study reveals the beneficial role of miR-205 in preventing breast cancer development. This finding could indicate a potential therapeutic intervention by using miR-205 in the future in breast cancer patients.
Acknowledgements
The work described in this article was funded by grants from the Youth National Natural Science Foundation of China (project number: 81302319, FL), the Major Research Project of Anhui Colleges and Universities Natural Science Foundation (project number: KJ2018ZD018, FL), and the Anhui Medical University Excellent Youth Training Program (project number: GJYQ-1403, FL). Thanks the support from Shenzhen Science and Technology Program (Grant No.: KQTD20170810154011370, QZ).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Geng SQ, Alexandrou AT, Li JJ. Breast cancer stem cells: multiple capacities in tumor metastasis. Cancer Lett. 2014;349:1–7. doi: 10.1016/j.canlet.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wicha MS. Cancer stem cells and metastasis: lethal seeds. Clin Cancer Res. 2006;12:5606–7. doi: 10.1158/1078-0432.CCR-06-1537. [DOI] [PubMed] [Google Scholar]
- 5.Fan X, Chen W, Fu Z, Zeng L, Yin Y, Yuan H. MicroRNAs, a subpopulation of regulators, are involved in breast cancer progression through regulating breast cancer stem cells. Oncol Lett. 2017;14:5069–5076. doi: 10.3892/ol.2017.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 7.Schmittgen TD. Regulation of microRNA processing in development, differentiation and cancer. J Cell Mol Med. 2008;12:1811–9. doi: 10.1111/j.1582-4934.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao J, Zhao S, Zhang Y, Zhao Z, Ye R, Wen J, Li J. Emerging role of microRNAs in cancer and cancer stem cells. J Cell Biochem. 2014;115:605–10. doi: 10.1002/jcb.24702. [DOI] [PubMed] [Google Scholar]
- 9.Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10:507–17. doi: 10.4161/cc.10.3.14754. [DOI] [PubMed] [Google Scholar]
- 10.Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, Tortoreto M, Sasso M, Plantamura I, Triulzi T, Taccioli C, Tagliabue E, Iorio MV, Croce CM. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol. 2012;6:458–72. doi: 10.1016/j.molonc.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huo L, Wang Y, Gong Y, Krishnamurthy S, Wang J, Diao L, Liu CG, Liu X, Lin F, Symmans WF, Wei W, Zhang X, Sun L, Alvarez RH, Ueno NT, Fouad TM, Harano K, Debeb BG, Wu Y, Reuben J, Cristofanilli M, Zuo Z. MicroRNA expression profiling identifies decreased expression of miR-205 in inflammatory breast cancer. Mod Pathol. 2016;29:330–46. doi: 10.1038/modpathol.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piasecka D, Braun M, Kordek R, Sadej R, Romanska H. MicroRNAs in regulation of triple-negative breast cancer progression. J Cancer Res Clin Oncol. 2018;144:1401–1411. doi: 10.1007/s00432-018-2689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–55. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su N, Qiu H, Chen Y, Yang T, Yan Q, Wan X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol Rep. 2013;29:2297–302. doi: 10.3892/or.2013.2400. [DOI] [PubMed] [Google Scholar]
- 15.Ji T, Zhang X, Li W. microRNA-205 acts as a tumor suppressor and directly targets YAP1 in glioma. Mol Med Rep. 2017;16:1431–1438. doi: 10.3892/mmr.2017.6748. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Chen B, Duan B, Zheng J, Wu X. miR-205 suppresses cell proliferation, invasion, and metastasis via regulation of the PTEN/AKT pathway in renal cell carcinoma. Mol Med Rep. 2016;14:3343–9. doi: 10.3892/mmr.2016.5589. [DOI] [PubMed] [Google Scholar]
- 17.Li D, Wang Q, Li N, Zhang S. miR-205 targets YAP1 and inhibits proliferation and invasion in thyroid cancer cells. Mol Med Rep. 2018;18:1674–1681. doi: 10.3892/mmr.2018.9074. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Wang L, Rodriguez-Aguayo C, Yuan Y, Debeb BG, Chen D, Sun Y, You MJ, Liu Y, Dean DC, Woodward WA, Liang H, Yang X, Lopez-Berestein G, Sood AK, Hu Y, Ang KK, Chen J, Ma L. miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat Commun. 2014;5:5671. doi: 10.1038/ncomms6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Mi R, Fan C, Zhang P, Zhu T, Wang Q, Lu Y, Gu J, Zheng Q. Runx2-interacting genes identified by yeast two-hybrid screening of libraries generated from hypertrophic chondrocytes. Am J Transl Res. 2016;8:5465–74. [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Ghori-Javed FY, Rashid H, Adhami MD, Serra R, Gutierrez SE, Javed A. Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. J Bone Miner Res. 2014;29:2653–65. doi: 10.1002/jbmr.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugimoto H, Nakamura M, Yoda H, Hiraoka K, Shinohara K, Sang M, Fujiwara K, Shimozato O, Nagase H, Ozaki T. Silencing of RUNX2 enhances gemcitabine sensitivity of p53-deficient human pancreatic cancer AsPC-1 cells through the stimulation of TAp63-mediated cell death. Cell Death Discov. 2015;1:15010. doi: 10.1038/cddiscovery.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Bai Y, He Y, Zhao Y, Chen J, Ma L, Pan Y, Hinten M, Zhang J, Karnes RJ, Kohli M, Westendorf JJ, Li B, Zhu R, Huang H, Xu W. PTEN loss promotes intratumoral androgen synthesis and tumor microenvironment remodeling via aberrant activation of RUNX2 in castration-resistant prostate cancer. Clin Cancer Res. 2018;24:834–846. doi: 10.1158/1078-0432.CCR-17-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai X, Meng L, Sun H, Li Z, Zhang X, Hua S. MicroRNA-196b inhibits cell growth and metastasis of lung cancer cells by targeting Runx2. Cell Physiol Biochem. 2017;43:757–767. doi: 10.1159/000481559. [DOI] [PubMed] [Google Scholar]
- 24.Li XQ, Lu JT, Tan CC, Wang QS, Feng YM. RUNX2 promotes breast cancer bone metastasis by increasing integrin α5-mediated colonization. Cancer Lett. 2016;380:78–86. doi: 10.1016/j.canlet.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W, Huang JA, Liu Z. Repression of Smad4 by miR-205 moderates TGF-β-induced epithelial-mesenchymal transition in A549 cell lines. Int J Oncol. 2016;49:700–8. doi: 10.3892/ijo.2016.3547. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Zhang L, He X, Zhang P, Sun C, Xu X, Lu Y, Li F. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem Biophys Res Commun. 2018;502:160–165. doi: 10.1016/j.bbrc.2018.05.139. [DOI] [PubMed] [Google Scholar]