Abstract
Previous studies have identified that both CKLF-like MARVEL transmembrane domain-containing member (CMTM6) and Neuropilin-1 (NRP1) played an essential part in regulating tumorigenesis and immune response. However, the potential connection between CMTM6 and NRP1 in oral squamous cell carcinoma (OSCC) remains unknown. In this study, we investigated the clinicopathologic significance of CMTM6 and NRP1 in OSCC. We examined the co-expression of CMTM6 and NRP1 in both OSCC tissues and cell lines. Co-overexpression of CMTM6 and NRP1 was generally highly expressed in cancer tissues and is associated with poor prognosis. Gain- and loss-of-function assays confirmed the oncogenic properties of CMTM6 in OSCC cells. Depletion of NRP1 abrogated tumorigenesis induced by CMTM6. By performing co-immunoprecipitation (co-IP), we discovered a potential interaction between CMTM6 and NRP1. Meanwhile, the stability of CMTM6 was significantly decreased in the NRP1-silencing cells, indicating the involvement of NRP1 in the degradation process of CMTM6. The crosstalk between CMTM6 and NRP1 provided a new insight into the progression of OSCC, which may indicate an alternative strategy for OSCC treatment.
Keywords: OSCC, CMTM6, NRP1, EMT, tumorigenesis
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common cancers in the world with a 5-year survival rate less than 50% and the exact etiology of OSCC is still unclear [1,2]. Although progress has been made in the treatment of OSCC, the overall survival rate remains unsatisfactory [3]. Therefore, an improved understanding of the molecular of its occurrence and progress is of great urgency [4,5].
CKLF-like MARVEL transmembrane domain-containing member (CMTM) is a novel gene family of proteins that link classical chemokines and transmembrane-4 superfamily (TM4SF) and was first reported in 2003 [6]. CMTM6 has been proved to play a key role in secretory proteins and plays multiple roles in physiological and pathological processes [6-10]. Recently, Marian L Burr and colleagues discovered that CMTM6 could bind to PD-L1 to maintain its cell surface expression [11]. Riccardo mezzadra et al. reported that CMTM6 could associate with the PD-L1 protein, reduce its ubiquitination and increase PD-L1 protein half-life [7]. CMTM6 depletion ameliorated PD-L1-mediated T-cell suppression and suggested a potential value of CMTM6 as therapeutic targets in PD-L1-PD-1 blocking therapies. Other related studies have been demonstrated in non-small cell lung cancer, gastric cancer, triple-negative breast cancer and pancreatic cancer [9,10,12,13]. Furthermore, CMTM6 was reported to be overexpressed in OSCC patients and participated in the regulation of cancer stem cells (CSCs) and correlated with Wnt/β-catenin-induced epithelial-mesenchymal transition (EMT) phenotype alteration [14].
Neuropilins (NRPs) were characterized originally for their role in the development of nervous system [15,16]. NRP1 is a 120 kDa type I transmembrane protein involved in a wide range of physiological and pathological processes. NRP1 has been identified as a co-receptor for multiple growth factors, including VEGF-A, FGF, HGF and others, and is expressed on both epithelial and tumor cells [16-18]. In breast cancer, positive regulation of the VEGF/NRP1 axis on the tumorigenesis and metastasis was associated with the enhancing EMT process and the NF-κB and β-catenin signaling [19]. Our group has previously demonstrated that higher NRP1 expression levels were associated with lymph node metastasis and poor prognosis in OSCC patients [20]. NRP1 promoted the EMT process through NF-κB activation. More recently, Dario AA Vignali et al. showed that a high percentage of intratumoral NRP1+ Tregs were correlated with poor prognosis in OSCC [21]. Additionally, NRP1 was expressed on a subset of activated human CD4+ and CD8+ TIL displaying PD-1hi status in NSCLC tumors, indicating the crucial role of NRP1 in PD-1 treatment reactivity. As both CMTM6 and NRP1 played an essential part in regulating tumorigenesis and immune response, we doubted whether CMTM6 could interact with NRP1 thus mediate cell proliferation and invasion.
In this study, we analyzed CMTM6 and NRP1 expression by IHC staining in OSCC tissue microarrays. We examined the co-expression of CMTM6 and NRP1 in both OSCC tissues and cell lines. CMTM6 was generally highly expressed in cancer tissues and is associated with OSCC metastasis and patient prognosis. Gain- and loss-of-function assays confirmed the oncogenic properties of CMTM6 in OSCC cells. Depletion of NRP1 abrogated tumorigenesis induced by CMTM6. By performing co-immunoprecipitation, we discovered a potential interation between CMTM6 and NRP1. The crosstalk between CMTM6 and NRP1 provided a new insight into the progression of OSCC, which may indicate an alternative strategy for OSCC treatment.
Materials and methods
Tissue samples collection
This study was approved by the ethics committee of Nanjing Medical University. In brief, we used 3 sets of OSCC tissue microarrays, including 244 cases of primary OSCC who had undergone surgical resection between 2009 and 2014 in Stomatological Hospital of Jiangsu Province and 32 cases of normal oral mucosae (more details of the pathological characteristics were demonstrated in Table 1). Patients were excluded who had been treated with molecular targeted therapy, chemotherapy or radiotherapy before surgery. The clinicopathologic information was obtained from the patients’ electronic medical records including age, gender, tumor size, histological grade, lymph node and clinical stage (defined according to American Joint Committee on Cancer 7th edition) and follow-up information for overall survival rates. Informed consent was signed by all of the recruited patients.
Table 1.
Correlation between CMTM6 or NRP1 and clinicopathologic characteristics in 244 OSCC cases
| Pathologic characteristics | n | CMTM6 | P value | NRP1 | P value | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Overexpression (number of cases) | Nonoverexpression (number of cases) | Overexpression (number of cases) | Nonoverexpression (number of cases) | ||||
| Age, years | |||||||
| ≥ 60 | 134 | 77 | 57 | 0.6889 | 61 | 73 | 0.1484 |
| < 60 | 110 | 66 | 44 | 40 | 70 | ||
| Sex | |||||||
| Male | 148 | 73 | 75 | 0.2901 | 59 | 89 | 0.5932 |
| Female | 96 | 54 | 42 | 35 | 61 | ||
| Smoking | |||||||
| Yes | 109 | 61 | 48 | 0.3247 | 63 | 46 | 0.8138 |
| No | 135 | 67 | 68 | 76 | 59 | ||
| Drinking | |||||||
| Yes | 90 | 44 | 46 | 0.5371 | 51 | 39 | 0.1625 |
| No | 154 | 69 | 85 | 73 | 81 | ||
| Location | |||||||
| Palate | 23 | 8 | 15 | 0.024 | 11 | 12 | 0.1879 |
| Tongue | 82 | 53 | 29 | 39 | 43 | ||
| Gingiva | 46 | 20 | 26 | 28 | 18 | ||
| Buccal | 68 | 30 | 38 | 42 | 26 | ||
| Mouth floor | 25 | 11 | 14 | 10 | 15 | ||
| Tumor stage | |||||||
| T1 | 109 | 44 | 65 | T1 vs T2 < 0.001 | 40 | 69 | T1 vs T2 = 0.1638 |
| T2 | 95 | 74 | 21 | T1 vs T3-4 = 0.0163 | 44 | 51 | T1 vs T3-4 = 0.0109 |
| T3-T4 | 40 | 25 | 15 | 24 | 16 | ||
| Lymph node status | |||||||
| N0 | 148 | 76 | 72 | N0 vs N1 < 0.001 | 64 | 84 | N0 vs N1 = 0.0045 |
| N1 | 41 | 33 | 8 | N0 vs N2-3 < 0.001 | 28 | 13 | N0 vs N2-3 < 0.001 |
| N2-3 | 55 | 45 | 10 | 42 | 13 | ||
| Pathological grade | |||||||
| I | 135 | 62 | 73 | I vs II = 0.4608 | 76 | 59 | I vs II < 0.001 |
| II | 28 | 15 | 13 | I vs III < 0.001 | 6 | 22 | I vs III < 0.001 |
| III | 81 | 66 | 15 | 69 | 12 | ||
The P values represent probabilities for CMTM6 or NRP1 expression levels between variable subgroups determined by a χ 2 test.
We next collected cancer tissues and matched adjacent normal tissue samples from 50 patients with histologically diagnosed OSCC cancer from Stomatological Hospital of Jiang-su Province between 2018 and 2019. The corresponding clinicopathological data were presented in Table S1. After collection, all samples were immediately frozen in liquid nitrogen and stored at -80°C until further use. Written informed consent was obtained from patients included in the study. Both tumor and matched adjacent normal tissues were collected and histologically confirmed by the Department of Pathology, Stomatological Hospital of Jiangsu Province.
Cell culture
Human OSCC cell lines (HN4, HN6, HN13, CAL27, SCC4 and SCC7) were obtained as previously described [22,23]. Human normal oral keratinocytes (HOK) cells were obtained from the American Type Culture Collection (ATCC). All cells were incubated in the corresponding medium containing 10% fetal calf serum (FBS, HyClone, USA). Cells were cultured in a humidified at mosphere at 37°C with 5% CO2. MG132 and Cycloheximide (CHX) were bought from Selleck (Selleck Chem, Houston). Dimethyl sulfoxide (DMSO) was used for control.
Vector construction and transfection
The full-length coding region of CMTM6 and NRP1 cDNA was inserted into pcDNA 3.1 vectors with myc-tag and were constructed by Generay Biotech (Shanghai, China). The shRNA targeting sequences specific for the human CMTM6 and the siRNA targeting sequences specific for the human NRP1 were synthesized by Shanghai Genepharma (Shanghai, China). The sequences were as follows: shCMTM6-1: 5’-TGGAGAACGGAGCGGTGTACA-3’; shCMTM6-2: 5’-GAGTCTCCTTATACTGATTGT-3’; shCMTM6-3: 5’-GCTGGCCTTCATCTGTGAAGA-3’; shNC: 5’-TTCTCCGAACGTGTCACGT-3’; si-NRP1-1: 5’-GCUCUGGAAUGUUGGGUAUTT-3’; si-NRP1-2: 5’-CCUUACAUCUCCUGGUUAUTT-3’; si-NRP1-3: 5’-CCACAAGUCUCUGAAACUUTT-3’; si-NC: 5’-GGGTATCGACGATTACAAA-3’. The interference efficiency was determined by qRT-PCR and was shown in Figure S1. The sequences of shCMTM6-1 and si-NRP1-2 and the corresponding control were introduced in this study. Cells utilized for transfection (5 × 105 cells/well) were grown to ~60% confluence in recommended growth medium, and cells were starved in serum-free medium and incubated for 16 hours. HN6 and CAL27 cells were transformed with the purified plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For stably CMTM6 and shCMTM6 transfected cells selection, 400 μg/ul puromycin (Gibco) was added into the medium for about 2 wk to generate stable expressing cells.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Cells and tissue samples were collected to extract total RNA using TRIzol (Invitrogen, Carlsbad, CA, USA) reagent and cDNA was generated using Superscript (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Relative expression levels of related genes were measured by the 2-ΔΔCT methods. The primers were listed as follows: CMTM6: F: 5’-TTTCCACACATGACAGGACTTC-3’, R: 5’-GGCTTCAGCCCTAGTGGTAT-3’; NRP1: F: 5’-CAGGTGATGACTTCCAGCTCA-3’, R: 5’-CCCAGTGGCAGAAGGTCTTG-3’; GAPDH: F: 5’-GAAGGTGAAGGTCGGAGTC-3’, R: 5’-GAGATGGTGATGGGATTTC-3’.
Western blot analysis
Western blot analysis was performed as described before [22]. The proteins were incubated with primary antibodies against CMTM6 (HPA026980, Sigma-Aldrich), NRP1 (#81321, Abcam), cyclin E1 (#4129, CST), cyclin D1 (#55506, CST), cyclin D3 (#2936, CST), CDK2 (#2546, CST), CDK4 (#12790, CST), CDK6 (#3136, CST), E-cadherin (#3195, CST), N-cadherin (ab18203, Abcam), snail (#3879, CST), slug (#9585, CST), Vimentin (#5741, CST), β-actin (AP0733, Bioworld, China) at 4°C overnight. The β-actin was regarded as the internal control.
Immunofluorescence staining
Briefly, cells were incubated with primary antibodies against E-cadherin (#3195, CST), N-cadherin (ab18203, Abcam), Vimentin (#5741, CST), CMTM6 (HPA026980, Sigma-Aldrich) or NRP1 (60067-1-Ig, Proteintech) at a dilution of 1:100 overnight. Cells were further incubated with FITC or Cy3-labeled goat anti-rabbit IgG or anti-mouse IgG (Proteintech) at a dilution of 1:500 for 30 minutes and then counterstained with 4’,6-di-amidino-2-phenylindole (DAPI; Sigma Chemicals). Plates were blindly examined and taken by a fluorescence microscope (DM4000B, Leica, Germany). Images were overlayed and analyzed by ImageJ software.
Cell viability CCK-8 assay and colony formation assay
For CCK-8 assay, 1000 OSCC cells were plated in 96-well plates. Cell viabilities were determined at 0, 1, 2, 3 and 4 days after cell attachment. At the end of each timing, 10 μL CCK-8 reagent (Dojindo, Japan) was introduced to each well. Cell growth curves were plotted according to the average absorption values of each experiment. For colony formation assay, 500 stably transformed cells were plated in 6-well plates. After 2 wk of incubation, colonies were fixed in 5% formalin and then stained with crystal violet. Cell colony images were counted under the microscope (DM4000B, Leica, Germany) and analyzed by ImageJ.
Wound healing and cell invasion assays
Artificial wounds were prepared with a 200 μl sterile pipette tip across the cell surface. Images of the same area of the wound were taken at 0H and 12H for calculating the closure of the wound. Cell invasion was measured by 24-well plates in a matrigel-coated 8-μm pore size chamber (BD Biosciences). HN6 and CAL27 cells were respectively seeded on the chambers at densities of 1 × 105 cells per well. After 24 hours, cells attached to the lower layer were fixed with methanol and stained with methylene blue. The results were analyzed by counting the stained cells using microscopy in 3 randomly selected fields.
Flow cytometry cell cycle assay
1 × 106 cells/well HN6 stable transfected cells were plated in 6-well plates. Cells were then harvested and washed twice with PBS, and resuspended in 70% ice-cold ethanol for 2 days. Then cells were washed and centrifuged and resuspended with 0.5 mL propidium iodide (PI) staining buffer for 30 min in the dark at room temperature. The cell cycle profiles were assessed by FACS can cytometry at 488 nm.
Immunoprecipitation
Cells were harvested and lysed in 600 μL of RIPA buffer (Beyotime) with protease inhibitors. Then cells were scraped up on ice and the supernatants were collected by centrifugation. The supernatants of cell lysates were interacted with indicated antibodies, NRP1 (#81321, Abcam) or myc-tag (#ab32, Abcam) and Protein A/G PLUS-Agarose beads (Sigma-Aldrich) at 4°C for 12 hours. After immunoprecipitation, the beads were washed thoroughly with cell lysis buffer. 60 μL of immunoprecipitated proteins and 1 × SDS PAGE was boiled for 10 minutes and then the precipitated proteins were analyzed.
Animal experiments
All animal investigations were approved by the institutional guidelines of Nanjing Medical University. Generally, 20 male nude mice (5 weeks old) were bought from the Model Animal Research Institute of Nanjing University. A total of 4 groups were assigned as followed: CMTM6, CMTM6-Vector, shCMTM6 and shNC. Stable transfected HN6 cells were centrifuged and resuspended in 50% matrigel and were subcutaneously injected into the flank of the nude mice (2 × 107 cells/200 μL). Xenograft tumor size was examined by vernier caliper every three days, and tumor volume was measured according to the formula: volume = (length × width2)/2. After 18 days of injection, all nude mice were executed to assess tumor volume, weigh as well as immunostaining.
Immunohistochemistry
Both tissue microarrays and xenograft tumor tissue sections were stained with primary antibodies against CMTM6 (HPA026980, Sigma-Aldrich) or NRP1 (#81321, Abcam) overnight following secondary antibody incubation for 30 minutes. All of the sections were counterstained using haematoxylin, dehydrated, cleared and mounted before examination using a microscope (DM4000B, Leica, Germany). CMTM6 and NRP1 immunoreactivity in microarray samples was calculated according to staining concentration and proportion semi-quantitatively. The score for the scale of positive cells was demonstrated as follows: 0, negative; 1, < 20%; 2, 20-50%; 3, 51-75%; and 4, > 75% positive cells. For staining strength, grading system was classified as below: 0, no staining; 1, light yellow; 2, brownish yellow; 3, dark brownish yellow. The result was calculated by multiplying the two scores as mentioned above. Scores for > 4 points were regarded as positive.
Statistical analysis
The Chi-square test and t-test were utilized to analyze the correlation between CMTM6 or NRP1 expression levels and clinicopathological parameters. The association between CMTM6 and NRP1 co-overexpression and OSCC overall survival (OS) was estimated using the log-rank test. OS was determined as the outcome variable. All images represent at least three individual experiments. The data were presented as the mean ± SD. Statistical significance was evaluated using Graghpad Prism 7.0. P < 0.05 was considered as statistical significance for all tests (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
Clinicopathologic characteristics of CMTM6 and NRP1 expression by tissue microarrays
To determine the potential role of CMTM6 and NRP1 in OSCC, we examined the expression of CMTM6 and NRP1 in tissue microarrays consisting of human OSCC (n = 244) and normal mucosal tissues (n = 32). Both CMTM6 and NRP1 were highly expressed in tumor tissues compared with normal oral mucosa (Figure 1A, 1B). In normal oral mucosa, CMTM6 was mostly confined to the nucleus with lower expression compared with tumor tissues (Figure 1A). Notably, OSCC samples tended to have stronger membrane and cytoplasmic staining of CMTM6, and the staining was co-expressed with NRP1 in OSCC (Figure 1C). We then analyzed the connection between CMTM6 and NRP1 expression with clinicopathological features (Table 1). Results showed that either increased CMTM6 or NRP1 expression was correlated with lymph node metastasis (Figure 1D) and higher pathological grade (Figure 1E). Elevated CMTM6 or NRP1 staining was detected in higher clinical tumor stages (Figure 1F). By performing the Spearman’s rank correlation coefficient test and linear tendency test, we found that NRP1 expression in OSCC was significantly correlated with CMTM6 (P < 0.001, r = 0.4906) (Figure 1G). The Kaplan-Meier curves indicated that coexpression of increased CMTM6 and NRP1 was related to lower overall survival in OSCC patients, p (CMTM+/NRP1+ vs CMTM-/NRP1-) = 0.0018 (Figure 1H).
Figure 1.
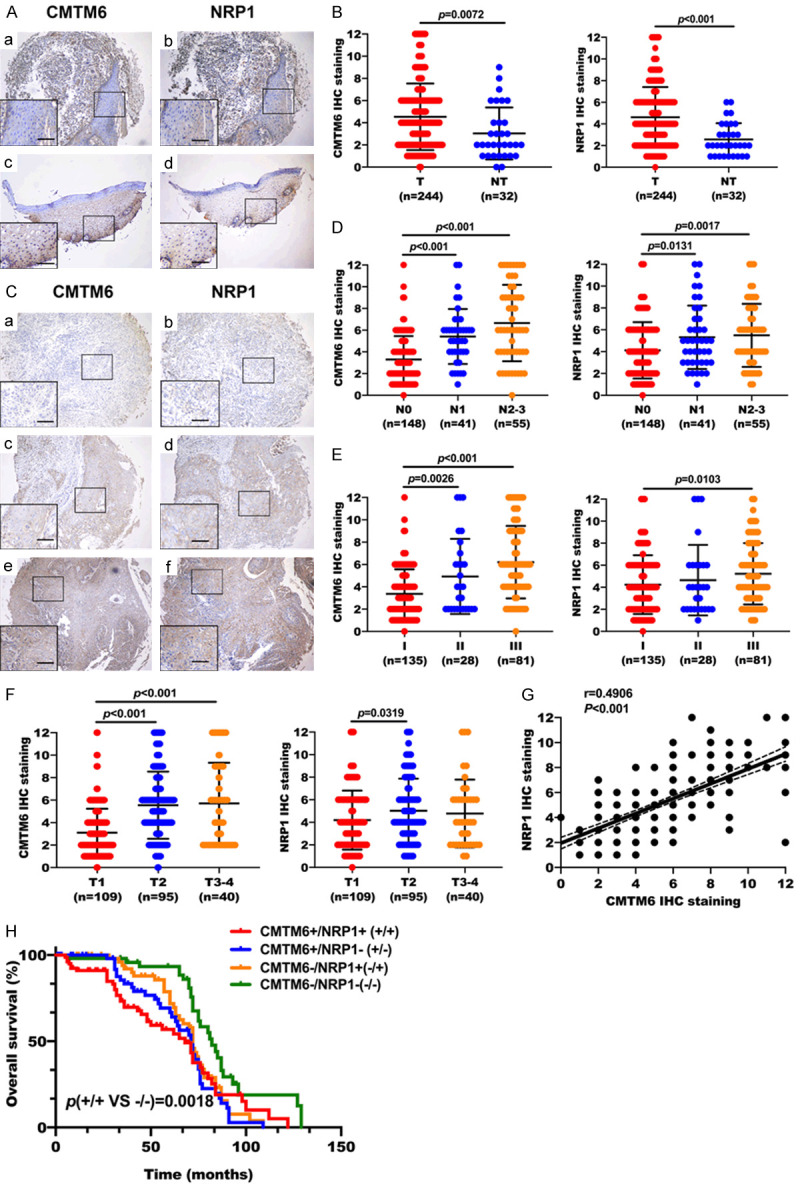
Clinicopathologic characteristics of CMTM6 and NRP1 expression by tissue microarrays. (A) Expression of CMTM6 and NRP1 in normal oral mucosa. (a, b) Both CMTM6 and NRP1 IHC staining was relatively low in normal oral tissues. (c, d) CMTM6 staining was mostly confined to the nucleus with lower expression in normal oral mucosa. NRP1 protein was co-expressed with CMTM6. (B) IHC staining analysis of CMTM6 and NRP1 expression in 244 OSCC tumors (T) and 32 normal oral tissues (NT). (C) The weak (a, b), moderate (c, d) and strong (e, f) levels of CMTM6 and NRP1 staining were demonstrated. OSCC samples tended to have stronger membrane and cytoplasmic staining of CMTM6, and the staining was co-expressed with NRP1. (D) Quantification analysis of CMTM6 and NRP1 staining in OSCC with different lymph node metastasis status. (E) Quantification analysis of CMTM6 and NRP1 staining in OSCC with different pathological grades. (F) Scores for CMTM6 and NRP1 staining in different tumor stages. (G) Scatter plots showed a positive correlation between CMTM6 and NRP1 IHC staining in tissue microarrays. R = 0.4906, P < 0.001. (H) Kaplan-Meier survival analysis showed that co-overexpression of CMTM6 and NRP1 had a shortened overall survival (OS) compared with the nonoverexpression of CMTM6 and NRP1. All error bar values represent the SD. Scale bar, 100 μm.
Correlation between CMTM6 and NRP1 in OSCC samples and cell lines
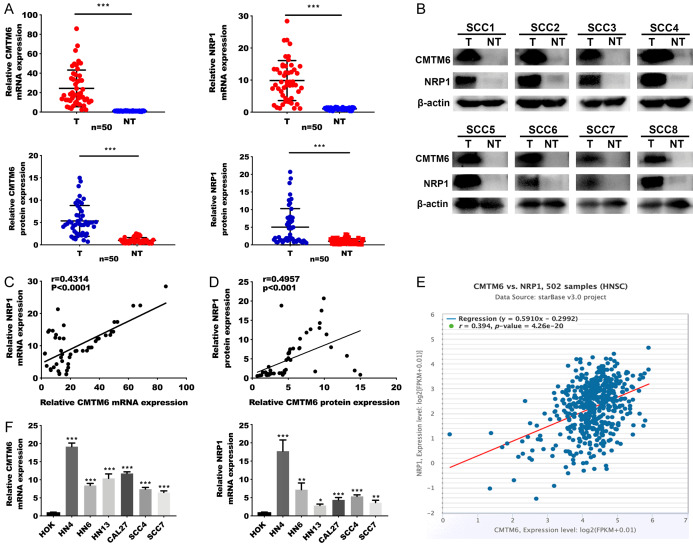
To further clarify the role of CMTM6 and NRP1 in OSCC, we collected tissue biopsies from patients who were clinically diagnosed as OSCC. We first tested the mRNA and protein levels of CMTM6 and NRP1 in 50 OSCC tissue samples and the corresponding adjacent tissues. The qRT-PCR results demonstrated that the CMTM6 and NRP1 mRNA relative fold changes in tumor tissues were markedly higher than that in paired normal tissues (Figure 2A). In addition, both CMTM6 and NRP1 protein expression levels were significantly elevated in OSCC tissues compared to nontumor tissues (Figure 2B). Scatter plots showed that CMTM6 and NRP1 mRNA and protein levels were positively correlated in OSCC samples (Figure 2C, 2D). Consistent with the results in tissue samples, analysis in The Cancer Genome Atlas (TCGA) of multiple cancer types showed that CMTM6 mRNA expression levels were positively related to NRP1 mRNA levels, including head and neck cancer (Figures 2E, S2). We then assessed the CMTM6 and NRP1 mRNA expression levels (Figure 2F) in OSCC cell lines and human normal oral keratinocytes (HOK). The results revealed that both CMTM6 and NRP1 mRNA levels were higher than that in HOK cells.
Figure 2.
Correlation between CMTM6 and NRP1 in OSCC samples and cell lines. (A) qRT-PCR analysis of CMTM6 and NRP1 mRNA expression in 50 OSCC tumors (T) and peritumoral oral tissues (NT). (B) Representative Western blotting analysis of CMTM6 and NRP1 protein expression. (C, D) Scatter plots show a positive correlation between CMTM6 and NRP1 at the mRNA (C) and protein (D) levels in 50 pairs of OSCCs (r = 0.4314, P < 0.0001 and r = 0.4957, P < 0.001). (E) Expression correlation between CMTM6 and NRP1 mRNA expression in the TCGA HNSCC database (starBase v3.0 project). Correlation was analyzed using Spearman’s correlation coefficient test, r = 0.394, P < 0.001, n = 502. (F) Quantification of CMTM6 and NRP1 mRNA expression levels using qRT-PCR analysis in 6 OSCC cell lines and human normal oral keratinocytes (HOK) cells. *P < 0.05, **P < 0.01, ***P < 0.001.
CMTM6 accelerates the proliferation of OSCC cells
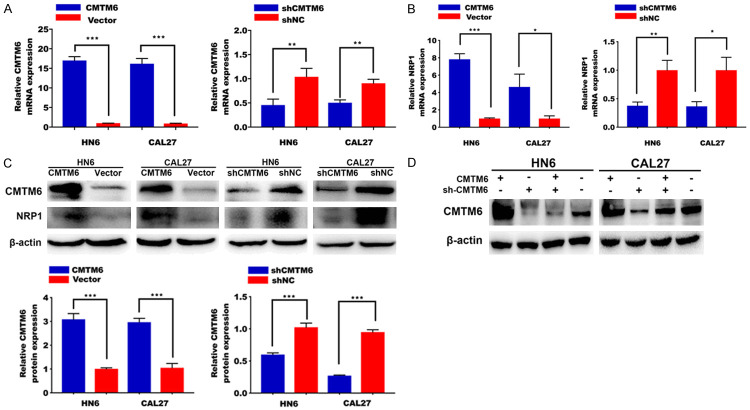
To further explore the effect of the CMTM6 in OSCC cell lines, we transfected HN6 and CAL27 cells with CMTM6 overexpressed plasmid or CMTM6 short hairpin RNA (shRNA). After stable cells selection, the efficiency of transfection was confirmed by qRT-PCR (Figure 3A). Importantly, mRNA levels of NRP1 were elevated after CMTM6 overexpression and reduced by CMTM6 silencing (Figure 3B). Western blotting analysis was consistent with qRT-PCR results (Figure 3C), indicating the plausible interaction between CMTM6 and NRP1. Rescue experiments showed the effective inhibition of shCMTM6 plasmids in both HN6 and CAL27 cells (Figure 3D).
Figure 3.
Efficiency of CMTM6 overexpression and knock-down was assessed by qRT-PCR and Western blotting analysis. A. CMTM6 overexpressed plasmids or CMTM6 short hairpin RNA (shRNA) plasmids were transformed in HN6 and CAL27 cells. After stable cells selection, the efficiency was confirmed by qRT-PCR analysis. B. NRP1 mRNA expression in cells stably transfected with the CMTM6 or shCMTM6 plasmids. C. Western blot analysis of CMTM6 and NRP1 expression in HN6 and CAL27 cells stably transfected with the CMTM6 overexpressed plasmids or the shCMTM6 plasmids. D. Rescue experiments were confirmed by Western blotting analysis.
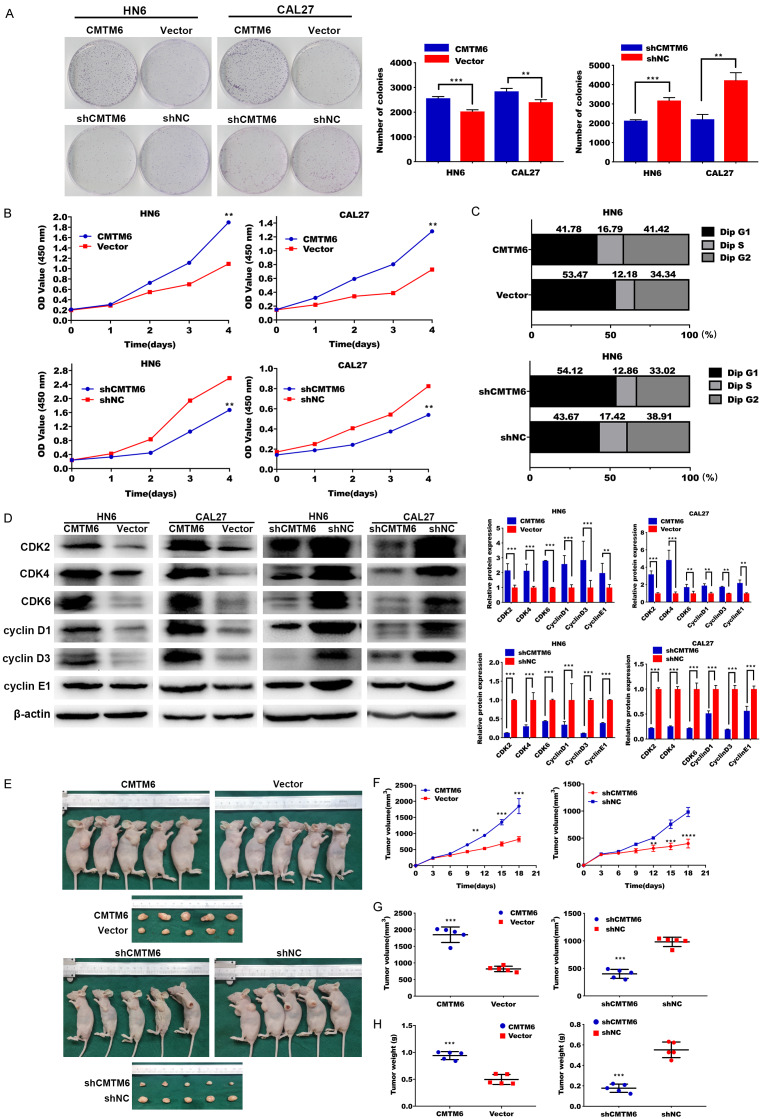
We then tested cell proliferative potential in OSCC cell lines. Much higher cell densities for the CMTM6-overexpressing cells were observed by colony formation assay when compared to the vector controls, and CMTM6 silencing inhibited colony formation compared with negative control (Figure 4A). Consistently, CCK-8 assays demonstrated that CMTM6 overexpression accelerated cell proliferation and cell proliferative rate was restrained after CMTM6 suppression (Figure 4B).
Figure 4.
CMTM6 accelerates the proliferation of OSCC cells. (A) Colony formation assay demonstrated much higher cell densities for the CMTM6-overexpressing cells compared with cells transfected with empty vector. Meanwhile, CMTM6 silencing inhibited colony formation compared with negative control. (B) CCK-8 assay showed that CMTM6 overexpression accelerated cell proliferation and cell proliferative rate was restrained after CMTM6 suppression. (C) CMTM6 transfected cells presented a significantly lower percentage of G1 phase and higher ratio of S phase than the control cells. By contrary, shCMTM6 ceased the cell cycle. (D) Proteins related to cell-cycle progression were detected by Western blot analysis. (E) CMTM6 over-expression promoted tumor growth in vivo. Depletion of CMTM6 inhibited tumor progression in vivo. The tumors dissected from mice were presented below. (F-H) Evaluation on xenografted tumor growth curves (F), tumor volumes (G) and tumor weight (H). *P < 0.05, **P < 0.01, ***P < 0.001.
Moreover, cell cycle analysis was performed to determine whether CMTM6 enhanced cell proliferation via alteration of the cell cycle. As expected, CMTM6 overexpression triggered G1 to S + G2 phases of cell cycle and CMTM6 suppression blocked the cell cycle at G1 phase (Figure 4C). We then analyzed related cell-cycle proteins by western blot [24]. Overexpression or silencing of CMTM6 was related to up-regulated or down-regulated cyclin-dependent kinases levels (Figure 4D).
We then observed the in vivo tumor formation in a mouse xenograft model. CMTM6 overexpression significantly induced tumor growth compared with control. By the contrary, transfection of stale shCMTM6 inhibited tumor progression in vivo (Figure 4E). Tumor proliferative curve, volumes and weight of tumors were presented in Figure 4F-H.
CMTM6 promotes cell migration and invasion in vitro
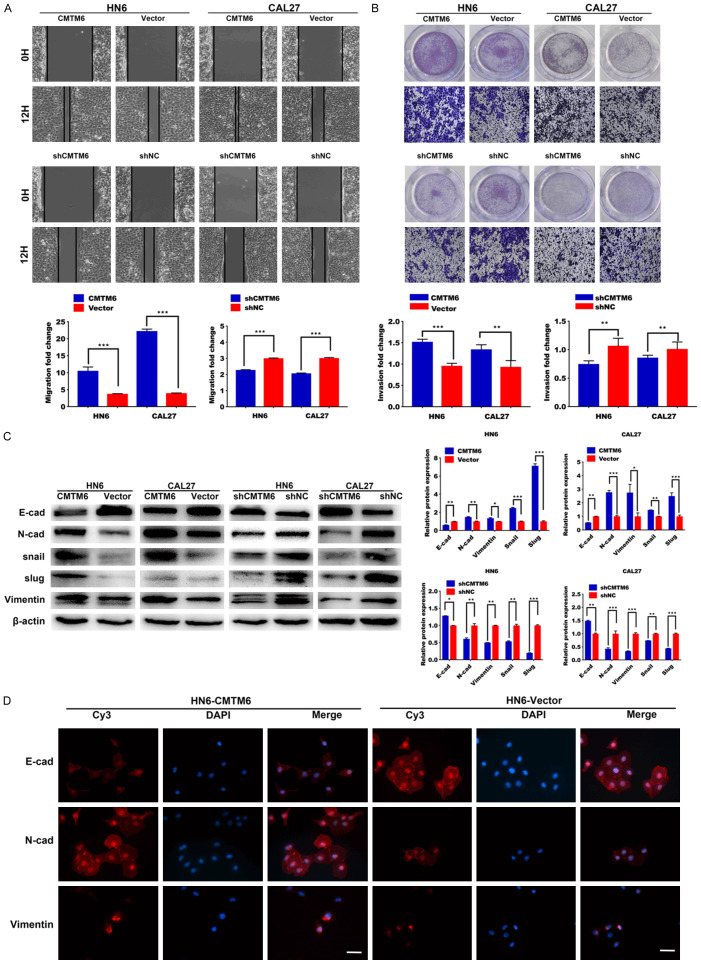
Wound healing and Transwell assays were performed to determine the alteration of cell migrative and invasive ability in OSCC cell lines. In wound healing assay, we found that the cell migration ability was elevated in the CMTM6 overexpression group (Figure 5A). Moreover, we observed that the CMTM6 over-expressed OSCC cells invaded through the matrix quicker than in the control group (Figure 5B). Accordingly, CMTM6 silencing greatly inhibited cell migration and invasion in HN6 and CAL27 cells (Figure 5A, 5B).
Figure 5.
CMTM6 promotes cell migration and invasion in vitro. A. Wound healing assay showed that the cell migration ability was significantly increased in the CMTM6-overexpression group. Meanwhile, the wound closure was delayed in CMTM6 stable knockdown cells compared with shNC control at both the 12-hour time point. B. Transwell assays of HN6 and CAL27 cells with stable CMTM6 overexpression or CMTM6 knockdown. Magnification: × 100. C. EMT-related proteins were introduced in the experiment. CMTM6 promoted the expression of mesenchymal markers (N-cadherin, Vimentin, snail and slug) and reduced the expression of E-cadherin. Downregulation of CMTM6 restrained the EMT process. D. Immunofluorescence staining showed that the epithelial markers E-cadherin were downregulated in CMTM6-overexpressed cells, with an increase in the expression of N-cadherin and Vimentin. Scale bar, 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
There is considerable evidence linking EMT to tumor invasion and metastasis, and EMT is frequently observed in OSCC [25,26]. Generally, EMT is characterized by the down-regulation of epithelial markers (e.g., E-cadherin) and the up-regulation of mesenchymal markers (e.g., N-cadherin and Vimentin) [27]. To verify the alteration of EMT markers regulated by CMTM6, we analyzed E-cadherin, N-cadherin, Vimentin and EMT-inducers in OSCC cells. We noticed a decrease in E-cadherin, accompanied by increased N-cadherin, Vimentin and snail family protein expression in CMTM6-overexpressed cells (Figure 5C). Downregulation of CMTM6 restrained the EMT process, as revealed by increased E-cadherin and decreased mesenchymal markers (Figure 5C). Similarly, Immunofluorescence staining showed that the epithelial markers E-cadherin were downregulated in CMTM6-overexpressed cells, with an increase in the expression of N-cadherin and Vimentin (Figure 5D).
NRP1 is essential for CMTM6-mediated OSCC proliferation and invasion
We have previously noticed that NRP1 expression was positively associated with CMTM6 levels in OSCC. To discover the interaction between CMTM6 and NRP1 in OSCC, we first examined the co-localization of CMTM6 and NRP1 by confocal microscopy. The microscopy detected a similar localization of CMTM6 and NRP1, which suggested a potential interaction between the two putative proteins (Figure 6A).
Figure 6.
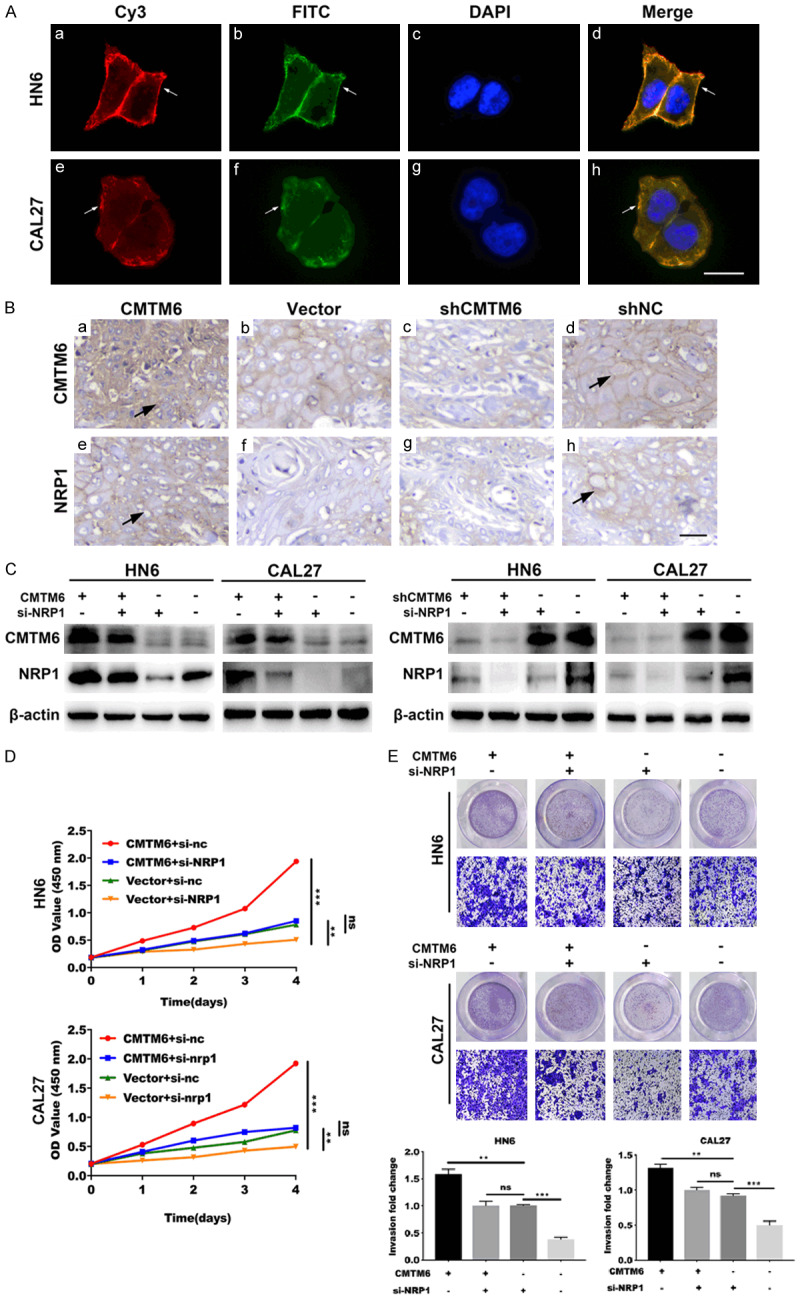
NRP1 is essential for CMTM6-mediated OSCC proliferation and invasion. (A) Confocal microscopy was utilized to capture the subcellular localization of CMTM6 (red) and NRP1 (green) expression in HN6 and CAL27 cells, followed by DAPI nuclear counterstaining (blue). Scale bar, 20 μm. (B) Mice xenograft tumors were analyzed by IHC to detect the co-expression of CMTM6 (a-d) and NRP1 (e-h). Overexpression of CMTM6 (a) resulted in higher NRP1 (e) protein staining. Scale bar, 50 μm. (C) Western blotting analysis was utilized to assess the expression of NRP1 in CMTM6 overexpressed or knockdown cells with the transfection of si-NRP1. (D) CCK-8 assay showed that the depletion of NRP1 significantly reduced the cell proliferation in CMTM6-overexpressed cells. (E) Transwell assay showed an increased invasive ability in CMTM6-transfected cells and the ability was restrained by NRP1 knockdown. Magnification: × 100. *P < 0.05, **P < 0.01, ***P < 0.001.
Mice xenograft tumors were also analyzed by IHC to detect the co-expression of CMTM6 and NRP1. Both CMTM6 and NRP1 proteins were mainly presented on the membrane. Meanwhile, overexpression of CMTM6 resulted in higher NRP1 protein staining (Figure 6B).
To further confirm that CMTM6 promoted OSCC proliferation and invasion by regulating NRP1, we decreased the expression of NRP1 in CMTM6 overexpressed or knockdown cells. We then discovered the protein expression of CMTM6 and NRP1 as well as cell proliferation and invasion abilities by Western blot, CCK-8 and Transwell assays. Immunoblotting results demonstrated that the upregulation of CMTM6 increased the protein levels of NRP1, whereas the NRP1-depletion restrained the upregulation of NRP1 induced by CMTM6 (Figure 6C). Meanwhile, depletion of CMTM6 significantly reduced the expression of NRP1 (Figure 6C). In addition, NRP1 silencing suppressed the elevated cell proliferative and invasive ability promoted by CMTM6 (Figure 6D, 6E). Thus, these results indicated the essential role of NRP1 in CMTM6-mediated OSCC proliferation and invasion.
NRP1 interacts with CMTM6 protein in OSCC cells
To further verify whether there was a relationship between endogenous CMTM6 and NRP1 protein, we performed co-IP and the Yeast two-hybrid assays. The western blotting results confirmed a potential interaction between CMTM6 and NRP1 in OSCC cells (Figure 7A). However, we did not discover a direct activation through the Yeast two-hybrid system (Figure S3). Endogenous CMTM6 and NRP1 accumulation correlated with the concentration and duration of exposure to the proteasome inhibitor MG132 (Figure 7B). Cycloheximide (CHX) experiments were then carried out in HN6 and CAL27 cells. Both CMTM6 and NRP1 proteins were increased in response to CHX (Figure 7C). To explore whether NRP1 played a role in regulating the degradation of CMTM6 protein, we transformed the CMTM6 plasmids into HN6 and CAL27 cells and detected the effects of CMTM6 on NRP1 expression, either with or without CHX (Figure 7D). The degradation dynamics assay showed that the half-life of CMTM6 was increased in the NRP1-overexpressed cells compared to that in the control cells. Finally, we added MG132 to cells transfected with CMTM6 and the lysates were mixed with a myc-tag antibody for immunoprecipitation (Figure 7E). Results showed that MG132 promoted the level of CMTM6 and NRP1, suggesting that NRP1 stabilized CMTM6 protein expression in OSCC cells.
Figure 7.
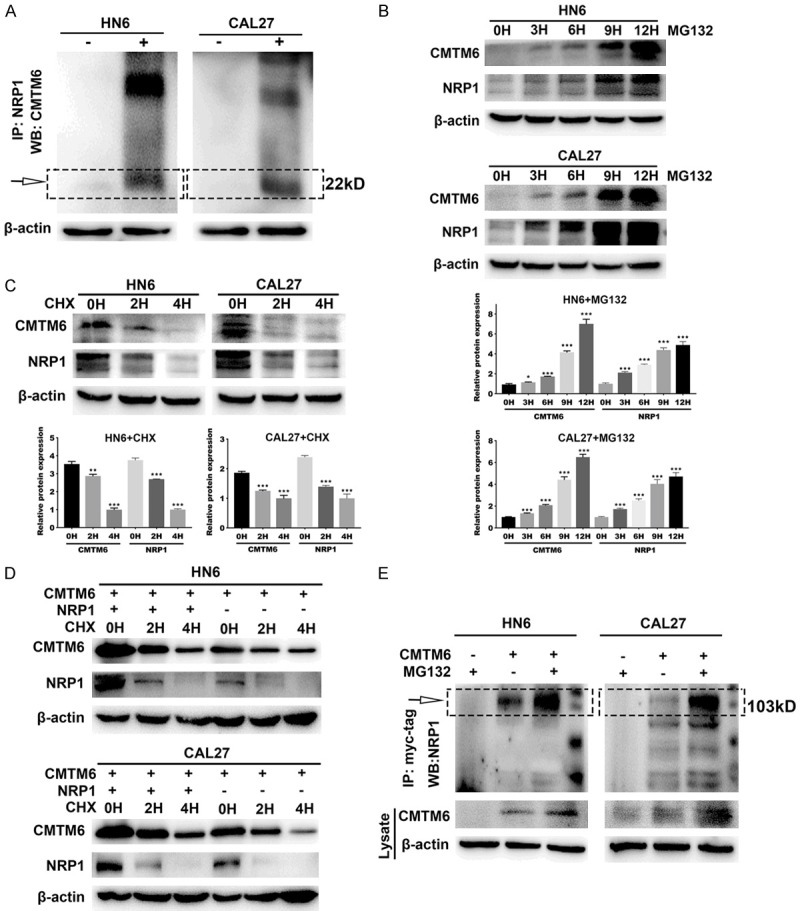
NRP1 interacts with CMTM6 protein in OSCC cells. A. Co-IP between endogenous CMTM6 and NRP1 with NRP1 antibody or igG in HN6 and CAL27 cells. B. HN6 and CAL27 cells were treated with MG132 (10 µM) for the indicated times, and then the levels of CMTM6 and NRP1 were detected by Western blot analysis. C. HN6 and CAL27 cells were subjected to cycloheximide (CHX) (20 µM) exposure at the indicated times and the levels of CMTM6 and NRP1 were determined by Western blot analysis. D. HN6 and CAL27 cells were transfected with the plasmid encoding CMTM6 either in combination with the NRP1 plasmid or without combination. Then, the cells were treated to CHX (20 µM) exposure at the indicated times, and the expression of CMTM6 and NRP1 was detected. E. Cell lysates were prepared and subjected to immunoprecipitation with anti-myc-tag antibody. The level of NRP1 was detected by Western blotting analysis. **P < 0.01, ***P < 0.001.
Discussion
OSCC is one of the most common malignant tumors with high mortality rate in the world [28]. However, because of the complex mechanism, the exact carcinogenesis of OSCC is still unknown. Recently, two papers have indicated that CMTM6 was a critical regulator of PD-L1 [7,11]. CMTM6 stabilized PD-L1 expression in tumor cells against T cells. On the contrary, the depletion of CMTM6 relieved T cell immunosuppression. Although CMTM6 and immune responses have been explored in recent years, the endogenous connection between CMTM6 and other regulating genes is poorly understood. Recently, there has been an increasing interest in NRP1 as a mediator of tumor development and progression [29]. Overexpression of NRP1 was associated with tumor progression and poor clinical outcome. However, the mechanism by which this activity is mediated remains unclear.
To explore the potential connection between CMTM6 and NRP1 in OSCC, we first analyzed the expression of CMTM6 and NRP1 in tissue microarrays. CMTM6 and NRP1 were co-overexpressed in tumor tissues and were significantly correlated with the clinicopathological characteristics and prognosis of patients with OSCC. Our gain- and loss-of-function studies revealed that CMTM6 promoted the cell proliferation, migration and invasion, indicating that CMTM6 led to carcinogenesis in OSCC. To further determine the regulation of NRP1 on CMTM6 in OSCC cells, we performed immunofluorescence staining and rescue experiments. We found that endogenous CMTM6 and NRP1 co-localized on the surface of tumor cells. Western blotting analysis showed that the upregulation of CMTM6 increased NRP1 expression, whereas the downregulation of NRP1 attenuated the upregulation of NRP1 expression in CMTM6 overexpressed OSCC cells. Studies on phenotypic alteration experiments showed that the silencing of NRP1 greatly reduced cell proliferation and invasion caused by CMTM6 overexpression. We further discovered the involvement of NRP1 in the degradation process of CMTM6. Endogenous CMTM6 and NRP1 accumulation correlated with the duration of exposure to the proteasome inhibitor. Meanwhile, the half-life of CMTM6 was significantly increased in the NRP1-overexpressed cells compared to that in the control cells. However, we did not discover an explicit activation between NRP1 and CMTM6 in the Yeast two-hybrid system. The exceeded length of NRP1 full-length protein may lower the possibility of interaction between the two proteins in vitro.
As a co-receptor for multiple growth factors, including VEGF-A, FGF, HGF and others, NRP1 has been identified to be expressed on both endothelial and tumor cells. Specifically, VEGF/NRP1 axis contributed to key aspects of tumorigenesis including the self-renewal and survival of CSCs [16,18,19]. In our current study, we noticed a positive staining on CMTM6 and NRP1 in tumor endothelial cells (data not shown). In addition to the significant roles on neoplastic epithelial cells, CMTM6 may also have positive effect on tumor endothelial cells. The potential mechanism of CMTM6/NRP1 axis on angiogenesis may provide a new insight into cancer therapy.
EMT is a biological process in which nonmotor epithelial cells are transformed into mesenchymal phenotypes and is thought to be the primary mechanism of cancer cell migration and invasion [30,31]. It is characterized by loss of E-cadherin, enhanced cell movement, and acquisition of N-cadherin and vimentin [25]. In the present study, we found that overexpression of CMTM6 significantly promoted cell migration and invasion. In addition, we observed that overexpression of CMTM6 attenuated the expression of E-cadherin and elevated the protein levels of N-cadherin, Vimentin and EMT-inducers in HN6 and CAL27 cells. Except for the above EMT makers, we also tested the level of β-catenin which is proved to be formed a complex with E-cadherin at the cell membrane and played a role in maintaining the adhesion of homotype cells and preventing cell movement [32]. However, we did not notice a visible change on the total level of β-catenin in both CMTM6 overexpressed or silencing tumor cells (data not shown). The abnormity of β-catenin expression may reflect the potential interaction between CMTM6 and Wnt/β-catenin signaling [14].
In summary, our study reported a regulatory network of CMTM6 and NRP1 expression in OSCC. Identification of the clinical and molecular association between CMTM6 and NRP1 expression in OSCC may provide a new therapeutic insight and thus improve the current treatment.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (81402236, 81772887), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2018-87), Jiangsu Provincial Medical Innovation Team (CXTDA2017036), Natural Science Foundation of Jiangsu Province of China (BK20171488) and Jiangsu Provincial Medical Youth Talent (QNRC2016854).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Li CC, Shen Z, Bavarian R, Yang F, Bhattacharya A. Oral cancer: genetics and the role of precision medicine. Surg Oncol Clin N Am. 2020;29:127–144. doi: 10.1016/j.soc.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Chow LQM. Head and neck cancer. N Engl J Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 3.Gau M, Karabajakian A, Reverdy T, Neidhardt EM, Fayette J. Induction chemotherapy in head and neck cancers: results and controversies. Oral Oncol. 2019;95:164–169. doi: 10.1016/j.oraloncology.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26:123–141. doi: 10.1016/j.coms.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Wang Z, Ding X, Dong Y, Zhang W, Zhang W, Zhong Y, Gu W, Wu Y, Song X. Combined Erlotinib and PF-03084014 treatment contributes to synthetic lethality in head and neck squamous cell carcinoma. Cell Prolif. 2018;51:e12424. doi: 10.1111/cpr.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu K, Li X, Gu H, Yang Q, Liu Y, Wang L. Research advances in CKLF-like MARVEL transmembrane domain-containing family in non-small cell lung cancer. Int J Biol Sci. 2019;15:2576–2583. doi: 10.7150/ijbs.33733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, Broeks A, Horlings HM, Wessels LFA, Blank CU, Xiao Y, Heck AJR, Borst J, Brummelkamp TR, Schumacher TNM. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Li J, Su Y, Fan Y, Guo X, Li L, Su X, Rong R, Ying J, Mo X, Liu K, Zhang Z, Yang F, Jiang G, Wang J, Zhang Y, Ma D, Tao Q, Han W. A novel 3p22.3 gene CMTM7 represses oncogenic EGFR signaling and inhibits cancer cell growth. Oncogene. 2014;33:3109–3118. doi: 10.1038/onc.2013.282. [DOI] [PubMed] [Google Scholar]
- 9.Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine. 2018;35:233–243. doi: 10.1016/j.ebiom.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao F, Chen J, Wang J, Li P, Wu S, Wang J, Ji Y. CMTM6, the newly identified PD-L1 regulator, correlates with PD-L1 expression in lung cancers. Biochem Biophys Rep. 2019;20:100690. doi: 10.1016/j.bbrep.2019.100690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, Gilan O, Bloor S, Noori T, Morgens DW, Bassik MC, Neeson PJ, Behren A, Darcy PK, Dawson SJ, Voskoboinik I, Trapani JA, Cebon J, Lehner PJ, Dawson MA. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Qi G, Li C, Bei C, Tan C, Zhang Y, Shi W, Zeng W, Kong J, Fu Y, Tan S. Expression and clinical significance of CMTM6 in hepatocellular carcinoma. DNA Cell Biol. 2019;38:193–197. doi: 10.1089/dna.2018.4513. [DOI] [PubMed] [Google Scholar]
- 13.Jin MH, Nam AR, Park JE, Bang JH, Bang YJ, Oh DY. Therapeutic co-targeting of WEE1 and ATM downregulates PD-L1 expression in pancreatic cancer. Cancer Res Treat. 2020;52:149–166. doi: 10.4143/crt.2019.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, Xiao Y, Bu LL, Zhang WF, Sun ZJ. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol Res. 2020;8:179–191. doi: 10.1158/2326-6066.CIR-19-0394. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhary B, Khaled YS, Ammori BJ, Elkord E. Neuropilin 1: function and therapeutic potential in cancer. Cancer Immunol Immunother. 2014;63:81–99. doi: 10.1007/s00262-013-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercurio AM. VEGF/Neuropilin signaling in cancer stem cells. Int J Mol Sci. 2019;20:490. doi: 10.3390/ijms20030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vivekanandhan S, Mukhopadhyay D. Genetic status of KRAS influences transforming growth factor-beta (TGF-beta) signaling: an insight into neuropilin-1 (NRP1) mediated tumorigenesis. Semin Cancer Biol. 2019;54:72–79. doi: 10.1016/j.semcancer.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plein A, Fantin A, Ruhrberg C. Neuropilin regulation of angiogenesis, arteriogenesis, and vascular permeability. Microcirculation. 2014;21:315–323. doi: 10.1111/micc.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo M, Hou L, Li J, Shao S, Huang S, Meng D, Liu L, Feng L, Xia P, Qin T, Zhao X. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-kappaB and beta-catenin. Cancer Lett. 2016;373:1–11. doi: 10.1016/j.canlet.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Chu W, Song X, Yang X, Ma L, Zhu J, He M, Wang Z, Wu Y. Neuropilin-1 promotes epithelial-to-mesenchymal transition by stimulating nuclear factor-kappa B and is associated with poor prognosis in human oral squamous cell carcinoma. PLoS One. 2014;9:e101931. doi: 10.1371/journal.pone.0101931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overacre-Delgoffe AE, Chikina M, Dadey RE, Yano H, Brunazzi EA, Shayan G, Horne W, Moskovitz JM, Kolls JK, Sander C, Shuai Y, Normolle DP, Kirkwood JM, Ferris RL, Delgoffe GM, Bruno TC, Workman CJ, Vignali DAA. Interferon-gamma drives treg fragility to promote anti-tumor immunity. Cell. 2017;169:1130–1141. e11. doi: 10.1016/j.cell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y, Wang Z, Xiong X, Zhong Y, Zhang W, Dong Y, Li J, Zhu Z, Zhang W, Wu H, Gu W, Wu Y, Wang X, Song X. Membrane-tethered Notch1 exhibits oncogenic property via activation of EGFR-PI3K-AKT pathway in oral squamous cell carcinoma. J Cell Physiol. 2019;234:5940–5952. doi: 10.1002/jcp.27022. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y, Wang Z, Ding X, Zhang W, Li G, Liu L, Wu H, Gu W, Wu Y, Song X. A novel Notch1 missense mutation (C1133Y) in the Abruptex domain exhibits enhanced proliferation and invasion in oral squamous cell carcinoma. Cancer Cell Int. 2018;18:6. doi: 10.1186/s12935-017-0496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Xu D, Li X, Zhang J, Xu W, Hou J, Zhang W, Tang J. Latest overview of the cyclin-dependent kinases 4/6 inhibitors in breast cancer: the past, the present and the future. J Cancer. 2019;10:6608–6617. doi: 10.7150/jca.33079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanlon CS, Van Tubergen EA, Inglehart RC, D’Silva NJ. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92:114–121. doi: 10.1177/0022034512467352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thierauf J, Veit JA, Hess J. Epithelial-to-mesenchymal transition in the pathogenesis and therapy of head and neck cancer. Cancers (Basel) 2017;9:76. doi: 10.3390/cancers9070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Zimmermann M, Tinhofer I, Kaufmann AM, Albers AE. Epithelial-to-mesenchymal transition and cancer stem(-like) cells in head and neck squamous cell carcinoma. Cancer Lett. 2013;338:47–56. doi: 10.1016/j.canlet.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Ding X, Zheng Y, Wang Z, Zhang W, Dong Y, Chen W, Li J, Chu W, Zhang W, Zhong Y, Mao L, Song X, Wu Y. Expression and oncogenic properties of membranous Notch1 in oral leukoplakia and oral squamous cell carcinoma. Oncol Rep. 2018;39:2584–2594. doi: 10.3892/or.2018.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adham SA, Al Harrasi I, Al Haddabi I, Al Rashdi A, Al Sinawi S, Al Maniri A, Ba-Omar T, Coomber BL. Immunohistological insight into the correlation between neuropilin-1 and epithelial-mesenchymal transition markers in epithelial ovarian cancer. J Histochem Cytochem. 2014;62:619–631. doi: 10.1369/0022155414538821. [DOI] [PubMed] [Google Scholar]
- 30.Fazilaty H, Rago L, Kass Youssef K, Ocana OH, Garcia-Asencio F, Arcas A, Galceran J, Nieto MA. A gene regulatory network to control EMT programs in development and disease. Nat Commun. 2019;10:5115. doi: 10.1038/s41467-019-13091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tripathi S, Levine H, Jolly MK. The physics of cellular decision making during epithelial-mesenchymal transition. Annu Rev Biophys. 2020;49:1–18. doi: 10.1146/annurev-biophys-121219-081557. [DOI] [PubMed] [Google Scholar]
- 32.Shiah SG, Shieh YS, Chang JY. The role of wnt signaling in squamous cell carcinoma. J Dent Res. 2016;95:129–134. doi: 10.1177/0022034515613507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.